10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(1):38-48. doi:10.7150/ijbs.34422 This issue Cite
Review
The Function of Pre-mRNA Alternative Splicing in Mammal Spermatogenesis
1. Key Lab of Swine Genetics and Breeding of Ministry of Agriculture and Rural Affairs & Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education, Huazhong Agricultural University, Wuhan 430070, PR China
2. The Cooperative Innovation Center for Sustainable Pig Production, Wuhan 430070, PR China
Received 2019-2-26; Accepted 2019-9-20; Published 2020-1-1
Abstract
Alternative pre-mRNA splicing plays important roles in co-transcriptional and post-transcriptional regulation of gene expression functioned during many developmental processes, such as spermatogenesis. The studies focusing on alternative splicing on spermatogenesis supported the notion that the development of testis is regulated by a higher level of alternative splicing than other tissues. Here, we aim to review the mechanisms underlying alternative splicing, particularly the splicing variants functioned in the process of spermatogenesis and the male infertility. There are five points regarding the alternative splicing including ⅰ) a brief introduction of alternative pre-mRNA splicing; ⅱ) the alternative splicing events in spermatogenesis-associated genes enriched in different stages of spermatogenesis; ⅲ) the mechanisms of alternative splicing regulation, such as splicing factors and m6A demethylation; ⅳ) the splice site recognition and alternative splicing, including the production and degradation of abnormal transcripts caused by gene variations and nonsense-mediated mRNA decay, respectively; ⅴ) abnormal alternative splicing correlated with male infertility. Taking together, this review highlights the impacts of alternative splicing and splicing variants in mammal spermatogenesis and provides new insights of the potential application of the alternative splicing into the therapy of male infertility.
Keywords: Alternative splicing, Mammal spermatogenesis, Splicing factors, Male infertility
Introduction
Alternative splicing is one of the most popular co-transcriptional and post-transcriptional regulatory mechanisms that result in a large number of mRNA and protein isoforms from a single gene, and the protein isoforms always show different or mutually antagonistic functional and structural characteristics [1, 2]. Recent high-throughput analyses have shown the abundance of alternative splicing events reaching > 95-100% in human genes and 63% in mouse genes [3, 4], and identified several modes of alternative transcript events, including exon-skipping (ES), intron-retention (IR), alternative 5' splice site (A5SS), alternative 3' splice site (A3SS), alternative first exon (AFE), alternative last exon (ALE) and mutually exclusive exon (MXE) [5]. The exon skipping events are accumulated in the brain and testis, suggesting a tissue-specific nature of alternative splicing [6, 7].
The process of spermatogenesis occurs in the seminiferous tubules of testis. Asingle spermatogonia self-renew and form spermatogonial stem cells to ensure the maintenance of the stem cell pool, while Apaired spermatogonia usually differentiate into two B spermatogonia [8, 9]. B spermatogonia come into the process of mitosis and proliferate into primary spermatocytes. Subsequently, primary spermatocytes divide twice during meiosis and form haploid round spermatids. In this phase, genetic material recombines mutually and the cells constantly split twice [10]. Finally, round spermatids go through the process of spermiogenesis to form spermatozoa. In this phase, the morphological changes have taken place in spermatids, including the condensation of nucleus, the formation of acrosome and tail, the elimination of residual body and the transient appearance of manchette [11]. The complicated physiological process requires specific genes to perform certain regulatory functions. Recently, high-throughput sequencing has revealed the importance of alternative splicing in the testis proteome diversification and spermatogenesis [5, 12-15]. In this review, we will provide the functions of alternative splicing in spermatogenesis, clarify the mechanisms of alternative splicing in spermatogenesis, and explain the potential correlations between alternative splicing and male infertility.
Alternative splicing of key genes involved in spermatogenesis
Spermatogenesis is a highly complex process that initiates shortly after birth and continues until old age [16]. The alternative splicing events are particularly prevalent in the testis, just less popular than in the brain. Spermatogenesis is an extremely complex and coordinated process, and the various genes are dynamically expressed in each type of spermatogenic cells. For better understanding the precision of gene regulation in spermatogenesis, we offer a series of alternative splicing events in the process of spermatogenesis.
GO analysis had identified 242 differently expressed genes enriched in spermatogenesis (GO: 0007283) in the testes from Large White pig at the age of 60 days (60-d) and 180 days (180-d) by RNA-seq [5]. Several genes was identified with alternative splicing to generate different transcripts encoding numerous protein isoforms, such as Spata3 [17], Spata19 [18], Crem [19, 20], Dazl [21], Hsf1 [22], Acrbp [23], Ybx3 [24]. Two members of SPATA family, Spata3, Spata19 can generate two splicing variants by exon skipping or alternative 3' splice site, respectively. Crem, a transcription factor regulated by cAMP controls the developmental progression of germ cells, and has ten splicing variants via several modes of alternative splicing. In postnatal testis developmental stages, the Dazl-Δ8 isoform is constantly expressed, along with Dazl-FL isoform. The information about spermatogenesis-enriched genes and their alternative splicing events are summarized in Table 1.
The information of genes enriched in spermatogenesis and their alternative splicing modes.
| Gene name | Species | Ensembl ID | Chromosome location | Variants number | Transcript ID | Modes of alternative splicing | Accession number | Conventional transcript or not | References |
|---|---|---|---|---|---|---|---|---|---|
| Spata3 | Mouse | ENSMUSG00000026226 | 1 | 2 | ENSMUST00000052854 | _ | NM_027300 | Yes | [17] |
| ENSMUST00000152501 | Exon skipping | NM_027029 | No | ||||||
| Spata19 | Mouse | ENSMUSG00000031991 | 9 | 2 | ENSMUST00000034473 | _ | NM_029299 | Yes | [18] |
| ENSMUST00000214287 | Alternative 3' splicing site | NM_001305058 | No | ||||||
| Crem | Rat [19] | ENSMUSG00000063889 | 18 | 10 | ENSMUST00000025069 | _ | NM_001271506 | Yes | [19,20] |
| Human [20] | ENSMUST00000049942 | Alternative first exon | NM_001311067 | No | |||||
| ENSMUST00000082141 | Exon skipping | NM_001271505 | No | ||||||
| ENSMUST00000122958 | Alternative first exon | NM_001110853 | No | ||||||
| ENSMUST00000124747 | Alternative first exon | NM_001311066 | No | ||||||
| ENSMUST00000130599 | Alternative first exon, Exon skipping | NM_001110857 | No | ||||||
| ENSMUST00000137568 | Alternative first exon, Exon skipping | NM_001110850 | No | ||||||
| ENSMUST00000142690 | Alternative last exon | NM_001110851 | No | ||||||
| ENSMUST00000146265 | Exon skipping | NM_001271503 | No | ||||||
| ENSMUST00000149803 | Alternative 3' splicing site | NM_001110852 | No | ||||||
| Dazl | Mouse | ENSMUSG00000010592 | 17 | 2 | _ | _ | NM_010021 | Yes | [21] |
| ENSMUST00000010736 | Exon skipping | NM_001277863 | No | ||||||
| Hsf1 | Mouse | ENSMUSG00000022556 | 15 | 4 | ENSMUST00000072838 | _ | NM_008296 | Yes | [22] |
| ENSMUST00000226872 | Alternative 3' splicing site | NM_001331214 | No | ||||||
| ENSMUST00000227478 | Alternative 3' splicing site | NM_001331154 | No | ||||||
| ENSMUST00000228371 | Exon skipping | NM_001331153 | No | ||||||
| Acrbp | Mouse | ENSMUSG00000072770 | 6 | 2 | ENSMUST00000088294 | _ | NM_016845 | Yes | [23] |
| ENSMUST00000112414 | Alternative 3' splicing site | NM_001127340 | No | ||||||
| Ybx3 | Mouse | ENSMUSG00000030189 | 6 | 2 | ENSMUST00000032309 | _ | NM_139117 | Yes | [24] |
| ENSMUST00000087865 | Exon skipping | NM_011733 | No |
Note:'_'means that there is no information.
The critical genes and their alternative splicing transcripts in the first wave of spermatogenesis. C-kit is the marker of differentiated spermatogonial cells. Two transcripts, full length transcript and the exon skipped transcript were detected in mouse testis [26, 27]. Sycp3 and Dmc1 are the markers of spermatocytes. The A3SS transcript and retained intron transcript of Sycp3 were identified in the testes from 60-d and 180-d Large White pigs [5]. The exon skipped Dmc1-d transcript was expressed in both male and female germ cells [31]. Akap4 is the marker of spermatids. Two Akap4 alternative splicing variants - Akap82 and Fsc1 were different in their 5' UTR [33].
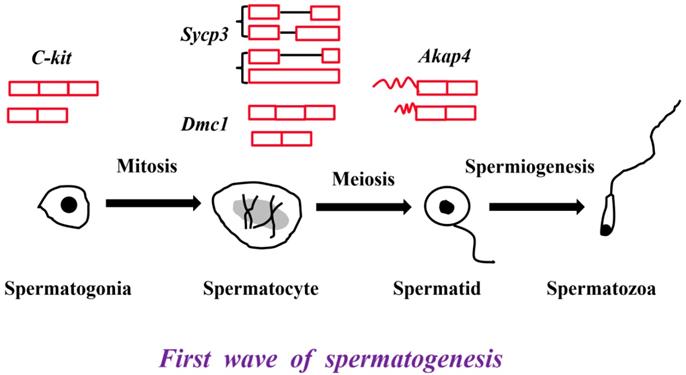
Many germ cell-specific transcripts are developmentally regulated and stage specific [25]. C-kit, proto-oncogene receptor tyrosine kinase, plays an indispensable role in the differentiation of spermatogonial cells [26]. A truncated form tr-kit is specifically expressed in spermatids and spermatozoa, and acts as a putative sperm factor required for triggering activation of mouse eggs at fertilization [26, 27]. Synaptonemal complex protein 3 (Sycp3), specifically localized in spermatocytes, is necessary for male meiosis and spermatogenesis [28, 29]. In our previous study, an A3SS transcript and a retained intron transcript of Sycp3 gene were identified in the testes from 60-d and 180-d Large White pigs [5]. Disrupted meiotic cDNA1 (Dmc1) is required for double-strand break repair and plays a pivotal role during meiotic homologous recombination [30]. An exon skipped transcript Dmc1-d is expressed in both male and female germ cells, which indicates a novel role in meiosis [31]. A-kinase anchoring protein 4 (Akap4) is transcribed only in the post-meiotic phase of spermatogenesis and Akap4 knockout mice exhibit defects in sperm flagellum and motility [32]. Akap82 and Fsc1 belonging to Akap4 alternative splicing variants are different in 5' UTR, and therefore encode the identical proteins, but Akap82 transcript is more abundantly expressed in spermatids than Fsc1 transcript (Figure 1) [33].
Sperm-associated antigen families are the proteins that highly expressed in the testis and are essential for motile cilia and flagella [34]. Previous studies have indicated that alternative splicing of sperm-associated antigen genes play important roles in spermatogenesis [5, 35-40]. Three isoforms of human Spag11b including Spag11b-a, Spag11b-d and Spag11b-g were different in their 3D fold structure [35]. Spag11b-d isoform could interact with tryptase alpha/beta 1 (Tpsab1), tetraspanin 7 (Tspan7), and attractin (Atrn), and then played major roles in immunity and fertility [36]. Two Spag11 transcripts including Spag11c and the exon-skipped transcript Spag11t were detected in rat [37]. Spag11c was expressed in epididymis and testis, while Spag11t was only confined to the caput region in the epididymis and was absent from testis and seminal vesicle [37]. Spag16 is the homologous to Chlamydomonas reinhardtii PF20 and is associated with the axonemal central apparatus [38]. In human testis, the long Spag16 isoform was localized to the central microtubule of the sperm, whereas the short isoform was located around the nucleus of spermatogenic cells in the late stage of meiosis [39, 40]. RNA-sequencing of immature and mature porcine testes identified several alternative splicing events of Spag6 gene [5]. Overall, the alternative splicing events of sperm-associated antigen family are spatio-temporal expressed, which illustrate the crucial role in spermatogenesis.
The mechanisms of alternative splicing regulation
Eukaryotic genes are composed of short exons and long introns. A majority of pre-mRNAs exist constitutive splicing and alternative splicing. Both types of splicing follow the “GT-AG” rule and are regulated by cis-acting splicing regulatory elements (SRE) and trans-acting splicing factors. The SREs consist of exonic splicing enhancers and silencers (ESE and ESS), intronic splicing enhancers and silencers (ISE and ISS) [41, 42]. The trans-acting factors are some RNA binding proteins such as the serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoprotein (hnRNP) family. SR proteins usually promote pre-mRNA splicing by binding ESE or ISE sequences via the N-terminal RNA recognition motifs (RRMs) or by interacting with other proteins via the C-terminal arginine-and serine-rich domain (RS), while hnRNP family usually inhibits pre-mRNA splicing by binding ESS or ISS sequences via their RRMs [43]. Generally, SR and hnRNP proteins have the opposite function in RNA splicing. However, the recent study in Drosophila indicates that SR and hnRNP proteins tend to act coordinately but not antagonistically [44].
RNA binding proteins can act as splicing enhancers or repressors, depending on which region of a skipped exon they bind to [45, 46]. Recent researches have revealed the critical role of RNA binding proteins in mammal spermatogenesis [47-50]. The polypyrimidine tract binding protein 2 (Ptbp2) is a strong regulator of alternative splicing [48, 52, 53]. In mammal spermatogenesis, Ptbp2 binds 3'SS of the cassette exons in critical genes, inhibits the exon skipping and then regulates alternative splicing in a stage-specific manner [48, 51]. Furthermore, numerous mis-spliced isoforms in those genes that are essential for Sertoli cell cytoskeleton and germ cell-Sertoli cell crosstalk appeared in Ptbp2-deficient mouse testis [48]. Dazap1 is a ubiquitous hnRNP protein that is expressed most abundantly in the testis and knock down of Dazap1 in 293T cells exhibits alternative splicing changes in those genes involved in cell cycle, DNA replication, transcriptional control and metabolism [49]. Another study revealed that a missense mutation (R263P) in the second RRM of RNA binding motif protein 5 (Rbm5) affected pre-mRNA splicing, produced aberrantly spliced transcripts and displayed spermatid differentiation arrest, azoospermia and male sterility [50].
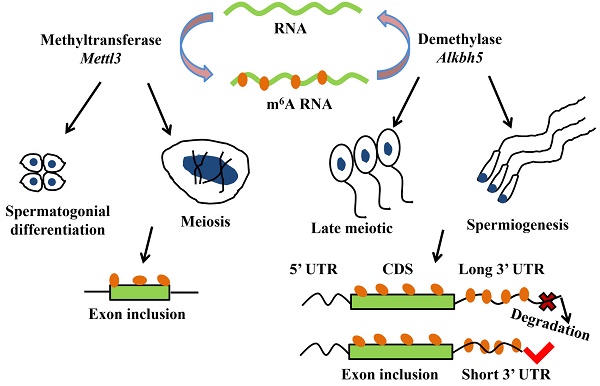
N6-methyladenosine (m6A) is a kind of the most abundant modifications in messenger RNAs and plays a pivotal role in regulating alternative splicing and RNA degradation [54]. RNA m6A methylation is a dynamic and reversible modification that mediated by m6A “writers”, “erasers” and “readers” [55-61]. m6A “writers” mainly consist of methyltransferases like 3 and 14 (Mettl3 and Mettl14), WT1 associated protein (Wtap) and vir like m6A methyltransferase associated (Virma) [55-57]. While alpha-ketoglutarate dependent dioxygenase (Fto) and alkB homolog 5 (Alkbh5) as demethylases (m6A “erasers”) can reverse the m6A methylation [58, 59]. And currently, m6A “readers” have been found including the binding proteins family such as heterogeneous nuclear ribonucleoprotein family and YTH N6-methyladenosine RNA binding proteins [60, 61].
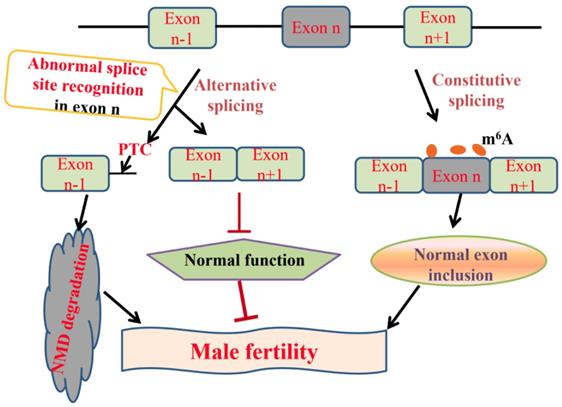
More and more researches show m6A modification plays a critical role in mRNA alternative splicing and stability in mammal spermatogenesis [59, 62]. m6A methyltransferase Mettl3 knockout mouse shows the defects in spermatogonial differentiation and meiosis, with a lower exon inclusion level in those transcripts containing m6A and alternative splicing in several critical genes involved in spermatogenesis including Dazl, Sohlh1, Nasp and Cdk11b [63]. During late spermiogenesis, the increased production of transcripts with shorter 3'-UTRs allows for an efficient translation and a quick mRNA/protein turnover [64]. m6A demethylase Alkbh5 is required for the late meiotic and haploid phases of spermatogenesis [65]. In spermatocytes and round spermatids, Alkbh5 tends to mark the coding sequences and 3'-UTRs of longer mRNAs that are destined to be degraded and controls correct splicing of long 3'-UTR transcripts (Figure 2) [66, 67].
The splice site recognition and alternative splicing
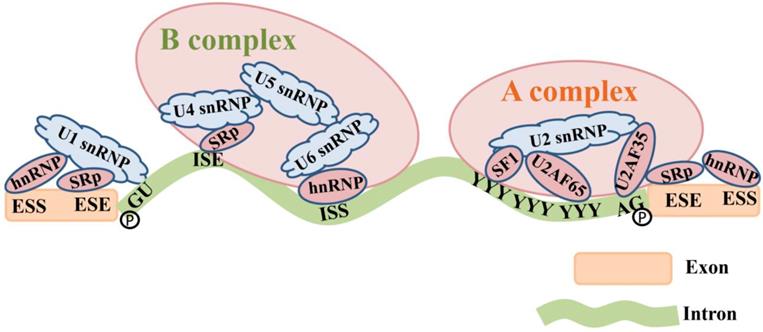
The pre-mRNA splicing is originated by spliceosomes that bind to sequences located at the 5' and 3' ends of introns [68]. Spliceosome assembly comprises five types of small nuclear ribonucleoproteins (snRNPs). U1 snRNP binds to the 5' GU. U2 snRNP and splicing factor 1 (SF1) bind to the branch site under the assistance of the mRNA splicing factor U2 associated factor (U2AF) and form the splicing complex precursor (A complex) [69]. U4-U5-U6 snRNP trimer forms the splicing complex (B complex) through interactions between RNA-RNA (SR protein) and RNA-protein (hnRNP) [1]. Subsequently, 5' end of the intron is cleaved from the upstream exon and joined to the branch site by a 2', 5'-phosphodiester linkage. The 3' end of the intron is cleaved from the downstream exon, and the two exons are joined by a phosphodiester bond [41]. The intron is then released in lariat form and degraded [70] (Figure 3).
m6A modification plays a critical role in mRNA alternative splicing and stability in mammal spermatogenesis. m6A methyltransferase Mettl3 is essential for spermatogonial differentiation and meiosis during mouse spermatogenesis. m6A modification that mediated by Mettl3 ensures exons containing m6A sites have the correct exon inclusion levels [63]. m6A demethylase Alkbh5 is required for the late meiotic and spermiogenesis during spermatogenesis [65]. m6A tends to mark the 3'-UTRs of longer mRNAs that are destined to be degraded and Alkbh5 controls correct splicing of exon and 3'-UTR which have m6A sites [66, 67].
The composition of the spliceosome complex and the pre-mRNA splicing process. U1 snRNP binds to the 5' GU. U2 snRNP and SF1 bind to the branch site under the assistance of the mRNA splicing factor U2 associated factor (U2AF65 and U2AF35) and form the splicing complex precursor (A complex) [69]. U4-U5-U6 snRNP trimer forms the splicing complex (B complex) through interactions between RNA-RNA (SR proteins) that bind to ISE and RNA-proteins (hnRNP) that bind to ISS. The 5' end and 3' end of the intron are cleaved from the upstream and downstream exons by a 2', 5'-phosphodiester linkage, and therefore exons are connected together.
Alternative splicing always comes with the emergence of the improper splice site recognition, which is always influenced by genetic variants or abnormal expression of splicing factors [41]. For example, an exonic SNP (c.2851G>T) in sperm flagellar 2 (Spef2) is associated with semen deformity rate and post-thaw cryopreserved sperm motility in Holstein bulls, potentially leads to the production of Spef2-SV3 transcript which is only detected in testis and epididymis [71]. Inner centromere protein (Incenp) is involved in cell division and sister chromatid separation as the main member of chromosomal passenger protein complex and a mutation (g.19970A>G) in its intron 11 results in exon 12 skipping and creates several novel binding sites for the splicing factors SRSF1, SRSF5, and SRSF6 [72]. The SR proteins, such as SRp38 and 9G8, are highly expressed in germ cells [73, 74]. The SNPs in these SR proteins are significantly associated with the risk of non-obstructive azoospermia in Chinese men, which provides more evidence for the role of splicing activity in human spermatogenesis [75]. A SNP in exonuclease 1 (Exo1) intron 8 potentially results in the production of truncated forms-tr1-Exo1 and tr2-Exo1, which partly explains metaphase-specific apoptosis in MRL/MpJ mice [76]. Zrsr1 is a U2AF35-like splicing factor which recognizes the 3' splice site during spliceosome assembly, and the Zrsr1 mutant mice containing truncating mutations within its RRM exhibits the abundant intron retention events in genes associated with spermatogenesis, and germ cell apoptosis, azoospermia and male sterility [77]. The conditional deficiency of another pre-mRNA splicing factor breast cancer amplified sequence 2 (Bcas2) in male germ cells results in male infertility and aberrant splicing of Dazl, Ehmt2 and Hmga1 genes [78].
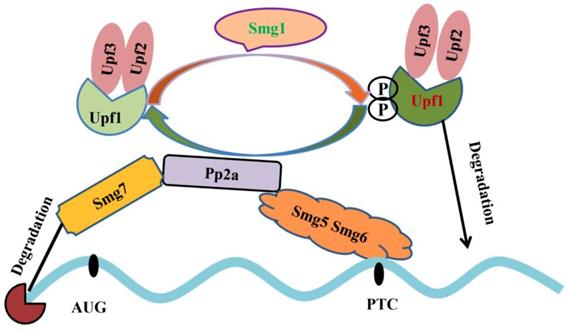
Alternative splicing coupled to nonsense-mediated mRNA decay (NMD) is an efficient strategy to regulate gene expression. The main feature of NMD is that the last exon generates a premature termination codon (PTC) producing a transcript with a C-terminally truncated polypeptide. NMD pathway is a post-transcriptional RNA surveillance mechanism that rapidly degrades the toxic truncated protein, preventing it from damaging cells [79, 80]. Up-frameshift mutant (UPF) and suppressor with morphological effect on genitalia (SMG) family are important factors in NMD pathway. Upf1 recognizes abnormal translation termination and then interacts with Upf2 and Upf3 [81]. Upf1 phosphorylation is mediated by Smg1. Protein phosphatase 2A, Pp2a interacts with Smg5, Smg6 and Smg7, and then promotes Upf1 de-phosphorylation. Smg5 and Smg6 identify PTCs by their special domains and degrade mRNAs (Figure 4) [82-84]. The mRNAs that contain PTCs or long 3' UTRs could be the potential substrates of NMD [85]. Emerging evidence provide an indispensable role of NMD in spermatogenesis. Inactivation of Upf2, a component of the chromatoid body, causes azoospermia and male sterility. In Upf2-null mouse spermatocytes and round spermatids, the long 3' UTR transcripts from ubiquitously expressed genes are abundantly accumulated and trigger nonsense-mediated mRNA decay [86]. Tudor domain containing 6 (Tdrd6) is essential for Upf1-Upf2 interaction. The long 3' UTR-stimulated NMD is impaired in Tdrd6-/- spermatids by interfering with Upf1-mRNA binding, thus perturbing mRNA processing [87].
Alternative splicing and male infertility
Spermatogenesis is a continuous hormone-dependent cell proliferation and differentiation process. Follicle-stimulating hormone (FSH) and luteotropic hormone (LH) can induce the secretion of testosterone that is important for spermatogenesis [88]. In addition, FSH and LH are also essential for the initiation and maintenance of normal spermatogenesis [89]. Multiple studies have found that alternative splicing of hormone receptor genes influences male infertility [93-96]. For example, a homozygous G>A mutation in intron 10-exon 11 boundary of LHR gene results in skipping of partial exon 11. The male patient appears delayed puberty, micropenis and oligospermia, and two of his sisters are infertile [90]. Several splicing variants in FSHR gene are identified in infertile patients, including the exon 9 skipped variant which leads to the removal of two highly conserved cysteine residues at positions 275 and 276 [91, 92]. Another study reveals several FSHR splicing variants lack of exon 2, exon 2 and 5, exon 5 and 6, exon 2, 5 and 6 in mouse testis [93]. The exon 5 and 6 skipped transcript produces a mutant receptor that can't bind to FSH and then leads to male infertility.
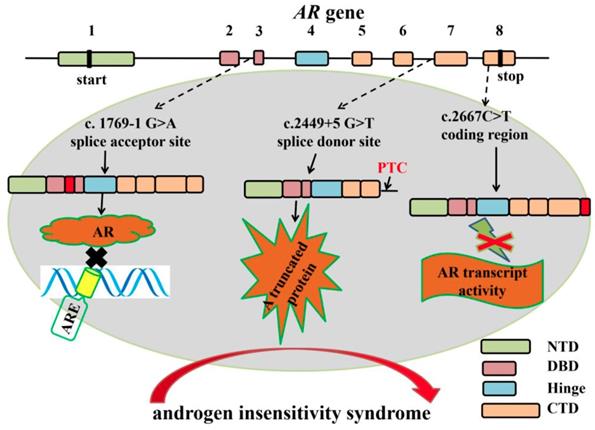
Previous studies showed that Sertoli-cell specific androgen receptor (AR) knockout mouse exhibited spermatogenic arrest and male infertile, suggesting the indispensable role of AR in spermatogenesis [94-97]. Human AR protein consists of four functional domains: N-terminal transactivation domain (NTD) encoded by exon 1; DNA-binding domain (DBD) encoded by exon 2 and 3; Hinge domain encoded by exon 4 and ligand-binding domain (LBD) encoded by exon 5-8 [98]. Several mutations in AR gene splice sites are closely associated with androgen insensitivity syndrome [99-102]. The c.1769-1G>A mutation in intron 2 splice acceptor site of AR gene results in an insertion of 69 nucleotides, which creates an insertion of 23 amino acids [99]. The insertion that located between the two zinc fingers of the AR DBD domain impairs the specific contact from AR and its hormone response element [100]. A c.2449+5 G>T mutation in intron 6 boundary splice donor site prevents the normal splicing of intron 6 and gives rise to a truncated protein that contains a PTC in retained intron 6. The truncated protein lacks part of C-terminal ligand-binding domain, and the truncated transcript is highly expressed in an 11-year-old girl, which probably explains the partial androgen insensitivity syndrome [101]. A synonymous mutation (C>T) in exon 8 is identified in a patient with partial androgen insensitivity syndrome [102]. The mutation produces an aberrant splicing variant that leads to partial skipping of exon 8 and a shortened 3'-untranslated region and the androgen-induced transcriptional activity is inhibited (Figure 5). Overall, aberrant alternative splicing of hormone receptor genes is closely associated with spermatogenesis and male infertility.
Summary and prospects
We have summarized the current knowledge regarding genes that regulate alternative splicing in spermatogenesis and also investigated the relationships between alternative splicing and male infertility (Figure 6). Spermatogenesis is a strictly regulated process, at both the transcriptional and the post-transcriptional level. Understanding the alternative splicing in spermatogenesis will be helpful to improve the sperm quality and cure male infertility.
Mechanistic model underlying nonsense-mediated mRNA decay. Upf1 interacts with Upf2, Upf3 and then undergoes Smg1-mediated phosphorylation. Pp2a interacts with Smg5, Smg6 and Smg7 and promotes Upf1 de-phosphorylation. Smg5 and Smg6 identify PTC; Smg7 degrade mRNA 5'cap structure and phosphorylated-Upf1 degrades mRNAs [81-84].
The mutations in AR gene splice sites result in aberrant splicing which are closely associated with human androgen insensitivity syndrome. Boxes represent exons of human AR gene. Exon 1 encodes N-terminal transactivation domain (NTD); Exon 2 and 3 encode the first and second zinc fingers of DNA-binding domain (DBD); Exon 4 encodes the hinge region; Exon 5-8 encode the COOH-terminal domain (CTD). Several mutations in AR gene splice sites result in aberrant splicing in androgen insensitivity syndrome patients. For example, the c.1769-1G>A mutation in intron 2 splice acceptor site results in an insertion of 69 nucleotides. The insertion between the two zinc fingers of the AR DNA binding domain (DBD domain) impairs the specific binding of AR response elements to AR [99, 100]. A c.2449+5G>T mutation in intron 6 boundary splice donor site gives rise to a truncated protein that lacks part of C-terminal ligand-binding domain [101]. A c2667C>T mutation in exon 8 produces an aberrant splicing variant that leads to partial skipping of exon 8 and a shortened 3'-untranslated region and the androgen-induced transcriptional activity is inhibited [102].
Pre-mRNA splicing in mammal spermatogenesis and male infertility. In normal condition, pre-mRNA undergoes constitutive splicing, removes introns and joins adjacent exons. m6A modification maintains correct exon inclusion levels and plays a critical role in mammal spermatogenesis [62-67]. Alternative splicing often comes into being along with the emergence of the abnormal splice site recognition. On the one hand, alternative splicing generates abnormal transcripts with a premature termination codon (PTC), which are degraded by nonsense-mediated mRNA decay (NMD) [65, 86, 87]. On the other hand, the mutations in splice sites result in the production of exon-skipped transcripts which are closely associated with male infertility [75, 76, 99-102].
It's well known that germ cells differentiate into spermatozoa, and transmit genetic and epigenetic information across generations [103]. This review provides a large amount of information about alternative splicing events in germ cells of the genes critical for spermatogenesis. However, testis is a highly complex tissue which contains Leydig cells, Sertoli cells, germ cells and peritubular myoid cells. These cells cooperate with each other and ensure normal spermatogenesis. The Sertoli cells are essential for creating a microenvironment that enables to produce the functional spermatozoa. The communication between Sertoli-Sertoli cell and Sertoli-germ cell constitutes ectoplasmic specialization and blood-testis barrier (BTB) which protect germ cells from immunological attack and provide nutrients for germ cells [104]. In addition, Leydig cells that between seminiferous tubules produce growth factors and secrete testosterone. A previous study has shown that LHR plays a key role in testosterone production and eight splicing variants of LHR gene are detected in bovine Leydig cells [105]. The androgenic stimulation of peritubular myoid cells around seminiferous tubules is also essential for normal germ cell development [106]. However, the functions of alternative splicing patterns of critical genes in Sertoli cells, Leydig cells and peritubular myoid cells are worth being explored.
Recently, there have been many researches on non-coding RNAs in spermatogenesis. Circular RNAs (circRNAs), unlike miRNAs and long non-coding RNAs (lncRNAs), are a novel type of non-coding RNAs originated from introns, intergenic regions, untranslated regions and exhibit distinct patterns of alternative back-splicing and alternative splicing [107, 108]. The roles of circRNAs in self-renewal and differentiation of spermatogonial stem cells and sperm motility are gradually being studied [109-112]. However, the relationships between alternative splicing that involved by circRNAs and spermatogenesis was seldom reported and needed further study.
High-throughput sequencing of short cDNA fragments (RNA-seq) generates tens of thousands alternative splicing events in numerous tissues. Meanwhile, the identification of important alternative splicing transcripts has become an urgent issue. There are two commonly used methods to estimate alternative splicing events at present. One is percent spliced in (PSI), proposed by Wang et al [113]. ΔPSI is used to detect differential alternative splicing events in two samples of RNA-seq data. When ΔPSI ≥ 10%, the changes of splicing events in different samples are considered important [114]. The other popular method to recognize alternative splicing is alternative splicing detector (ASD), which detects differential alternative splicing exons in different samples of RNA-seq data [115]. This software considers the altered junction reads and altered coverage of AS-exons while calculating a P value. When P < 0.05, the AS event is statistically significant. Currently, both methods have been widely applied in numerous tissues [116-118], but scarcely in the testis. Therefore, the important alternative splicing events in the development of spermatogenesis deserve further excavation and investigation. In general, this study reviews some progresses in alternative splicing of spermatogenesis and also provides new ideas in the therapy of male infertility.
Abbreviations
ES: exon-skipping; IR: intron-retention; A5SS: alternative 5' splice site; A3SS: alternative 3' splice site; AFE: alternative first exon; ALE: alternative last exon; MXE: mutually exclusive exon; SRE: splicing regulatory elements; ESE: exonic splicing enhancers; ESS: exonic splicing silencers; ISE: intronic splicing enhancers; ISS: intronic splicing silencers; SR: serine/arginine-rich; hnRNP: heterogeneous nuclear ribonucleoprotein; m6A: N6-methyladenosine; snRNPs: small nuclear ribonucleoproteins; NMD: nonsense-mediated mRNA decay; PTC: premature termination codon; UPF: Up-frameshift; FSH: follicle-stimulating hormone; LH: luteotropic hormone; AR: androgen receptor; NTD: N-terminal transactivation domain; DBD: DNA-binding domain; LBD: ligand-binding domain; BTB: blood-testis barrier; circRNAs: circular RNAs; PSI: percent spliced in; ASD: alternative splicing detector.
Acknowledgements
We thank Prof. Chunyan Mou (Huazhong Agricultural University) for her efforts in revising this manuscript. This work was supported financially by the National Natural Science Foundation of China (31772561, 31572362), The Trans-gene Key Project of China (2018ZX08009-27B), The Natural Science Foundation of Hubei Province (2018CFA015) and the Fundamental Research Funds for the Central Universities (2662018PY037, 2662019PY017).
Author contributions
H.S. and F.L. planned and wrote the manuscript. L.W. and D.C. helped with the planning and writing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291-323
2. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457-63
3. Merkin J, Russell C, Chen P. et al. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593-9
4. Pan Q, Shai O, Lee LJ. et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413-5
5. Song H, Zhu L, Li Y. et al. Exploiting RNA-sequencing data from the porcine testes to identify the key genes involved in spermatogenesis in Large White pigs. Gene. 2015;573:303-9
6. Schmid R, Grellscheid SN, Ehrmann I. et al. The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res. 2013;41:10170-84
7. Yeo G, Holste D, Kreiman G. et al. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74
8. Jan SZ, Hamer G, Repping S. et al. Molecular control of rodent spermatogenesis. Biochim Biophys Acta. 2012;1822:1838-50
9. Aponte PM, van Bragt MP, de Rooij DG. et al. Spermatogonial stem cells: characteristics and experimental possibilities. APMIS. 2005;113:727-42
10. Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124-36
11. O'Donnell L, Nicholls PK, O'Bryan MK. et al. Spermiation: The process of sperm release. Spermatogenesis. 2011;1:14-35
12. Margolin G, Khil PP, Kim J. et al. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics. 2014;18:39
13. Zuo H, Zhang J, Zhang L. et al. Transcriptomic variation during spermiogenesis in mouse germ cells. PLoS One. 2016;11:e0164874
14. Yang J, Zhang Z, Zhang Y. et al. CLOCK interacts with RANBP9 and is involved in alternative splicing in spermatogenesis. Gene. 2018;5:199-204
15. Ding H, Luo Y, Liu M. et al. Histological and transcriptome analyses of testes from Duroc and Meishan boars. Sci Rep. 2016;11:20758
16. Kopera IA, Bilinska B, Cheng CY. et al. Sertoli-germ cell junctions in the testis: a review of recent data. Philos Trans R Soc Lond B Biol Sci. 2010;365:1593-605
17. Li L, Liu G, Fu JJ. et al. Molecular cloning and characterization of a novel transcript variant of Mtsarg1 gene. Mol Biol Rep. 2009;36:1023-32
18. Nourashrafeddin S, Ebrahimzadeh-Vesal R, Modarressi MH. et al. Identification of Spata-19 new variant with expression beyond meiotic phase of mouse testis development. Rep Biochem Mol Biol. 2014;2:89-93
19. Walker WH, Sanborn BM, Habener JF. An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription. Proc Natl Acad Sci U S A. 1994;91:12423-7
20. Guo Q. Cyclic AMP-responsive element modulator α polymorphisms are potential genetic risks for systemic lupus erythematosus. J Immunol Res. 2015;2015:906086
21. Xu X, Tan X, Lin Q. et al. Mouse Dazl and its novel splice variant functions in translational repression of target mRNAs in embryonic stem cells. Biochim Biophys Acta. 2013;1829:425-35
22. Neueder A, Achilli F, Moussaoui S. et al. Novel isoforms of heat shock transcription factor 1, HSF1γα and HSF1γβ, regulate chaperone protein gene transcription. J Biol Chem. 2014;289:19894-906
23. Kanemori Y, Ryu JH, Sudo M. et al. Two functional forms of ACRBP/sp32 are produced by pre-mRNA alternative splicing in the mouse. Biol Reprod. 2013;88:105
24. Uchibe K, Shimizu H, Yokoyama S. et al. Identification of novel transcription-regulating genes expressed during murine molar development. Dev Dyn. 2012;241:1217-26
25. Eddy EM. Male germ cell gene expression. Recent Prog Horm Res. 2002;57:103-28
26. Sette C, Dolci S, Geremia R. et al. The role of stem cell factor and of alternative c-kit gene products in the establishment, maintenance and function of germ cells. Int J Dev Biol. 2000;44:599-608
27. Sette C, Bevilacqua A, Bianchini A. et al. Parthenogenetic activation of mouse eggs by microinjection of a truncated c-kit tyrosine kinase present in spermatozoa. Development. 1997;124:2267-74
28. Wang S, Pan Z, Zhang Q. et al. Differential mRNA expression and promoter methylation status of SYCP3 gene in testes of yaks and cattle-yaks. Reprod Domest Anim. 2012;47:455-62
29. Ozaki Y, Saito K, Shinya M. et al. Evaluation of Sycp3, Plzf and Cyclin B3 expression and suitability as spermatogonia and spermatocyte markers in zebrafish. Gene Expr Patterns. 2011;11:309-15
30. Ahlawat S, Chopra M, Jaiswal L. et al. Exon skipping creates novel splice variants of DMC1 gene in ruminants. Mol Cell Probes. 2016;30:66-73
31. Habu T, Taki T, West A. et al. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 1996;24:470-7
32. Miki K, Willis WD, Brown PR. et al. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol. 2002;248:331-42
33. Nipper RW, Chennothukuzhi V, Tutuncu L. et al. Differential RNA expression and polyribosome loading of alternative transcripts of the Akap4 gene in murine spermatids. Mol Reprod Dev. 2005;70:397-405
34. Nagarkatti-Gude DR, Jaimez R, Henderson SC. et al. Spag16, an axonemal central apparatus gene, encodes a male germ cell nuclear speckle protein that regulates SPAG16 mRNA expression. PLoS One. 2011;6:e20625
35. Narmadha G, Yenugu S. Molecular modeling of the human sperm associated antigen 11 B (SPAG11B) proteins. Syst Biol Reprod Med. 2015;61:78-88
36. Radhakrishnan Y, Hamil KG, Tan JA. et al. Novel partners of SPAG11B isoform D in the human male reproductive tract. Biol Reprod. 2009;81:647-56
37. Yenugu S, Hamil KG, Grossman G. et al. Identification, cloning and functional characterization of novel sperm associated antigen 11 (SPAG11) isoforms in the rat. Reprod Biol Endocrinol. 2006;28:23
38. Alciaturi J, Anesetti G, Irigoin F. et al. Distribution of sperm antigen 6 (SPAG6) and 16 (SPAG16) in mouse ciliated and non-ciliated tissues. J Mol Histol. 2019;50:189-202
39. Zhang Z, Shen X, Gude DR. et al. MEIG1 is essential for spermiogenesis in mice. Proc Natl Acad Sci U S A. 2009;106:17055-60
40. Zhang Z, Kostetskii I, Tang W. et al. Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol Reprod. 2006;74:751-9
41. Li Y, Yuan Y. Alternative RNA splicing and gastric cancer. Mutat Res. 2017;773:263-73
42. Kelemen O, Convertini P, Zhang Z. et al. Function of alternative splicing. Gene. 2013;514:1-30
43. Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479-90
44. Brooks AN, Duff MO, May G. et al. Regulation of alternative splicing in Drosophila by 56 RNA binding proteins. Genome Res. 2015;25:1771-80
45. König J, Zarnack K, Luscombe NM. et al. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13:77-83
46. Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689-701
47. Paronetto MP, Sette C. Role of RNA-binding proteins in mammalian spermatogenesis. Int J Androl. 2010;33:2-12
48. Hannigan MM, Zagore LL, Licatalosi DD. Ptbp2 controls an alternative splicing network required for cell communication during spermatogenesis. Cell Rep. 2017;19:2598-612
49. Choudhury R, Roy SG, Tsai YS. et al. The splicing activator DAZAP1 integrates splicing control into MEK/Erk-regulated cell proliferation and migration. Nat Commun. 2014;5:3078
50. O'Bryan MK, Clark BJ, McLaughlin EA. et al. RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet. 2013;9:e1003628
51. Boutz PL, Stoilov P, Li Q. et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636-52
52. Licatalosi DD, Yano M, Fak JJ. et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626-42
53. Xu M, Hecht NB. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3'UTR. Biol Reprod. 2007;76:1025-33
54. Cao G, Li HB, Yin Z. et al. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003
55. Ping XL, Sun BF, Wang L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177-89
56. Wang Y, Li Y, Toth JI. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191-8
57. Schwartz S, Mumbach MR, Jovanovic M. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284-96
58. Jia G, Fu Y, Zhao X. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
59. Zheng G, Dahl JA, Niu Y. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
60. Liu N, Dai Q, Zheng G. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
61. Wang X, Lu Z, Gomez A. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-20
62. Lin Z, Hsu PJ, Xing X. et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27:1216-30
63. Xu K, Yang Y, Feng GH. et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100-14
64. Zhang Y, Tang C, Yu T. et al. MicroRNAs control mRNA fate by compartmentalization based on 3' UTR length in male germ cells. Genome Biol. 2017;18:105
65. Zheng G, Dahl JA, Niu Y. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
66. Bao J, Vitting-Seerup K, Waage J. et al. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3'UTR transcripts. PLoS Genet. 2016;12:e1005863
67. Tang C, Klukovich R, Peng H. et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2018;115:E325-33
68. Hoskins AA, Friedman LJ, Gallagher SS. et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289-95
69. Fox-Walsh KL, Dou Y, Lam BJ. et al. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci U S A. 2005;102:16176-81
70. Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386-98
71. Guo F, Yang B, Ju ZH. et al. Alternative splicing, promoter methylation, and functional SNPs of sperm flagella 2 gene in testis and mature spermatozoa of Holstein bulls. Reproduction. 2013;147:241-52
72. Liu J, Sun Y, Yang C. et al. Functional SNPs of INCENP affect semen quality by alternative splicing mode and binding affinity with the target Bta-miR-378 in Chinese Holstein bulls. PLoS One. 2016;11:e0162730
73. Xiao PJ, Hu L, Li J. et al. NSSR1 is regulated in testes development and cryptorchidism and promotes the exon 5-included splicing of CREB transcripts. Mol Reprod Dev. 2007;74:1363-72
74. Dreumont N, Bourgeois CF, Lejeune F. et al. Human RBMY regulates germline-specific splicing events by modulating the function of the serine/arginine-rich proteins 9G8 and Tra2-b. J Cell Sci. 2010;123:40-50
75. Ni B, Ma H, Lin Y. et al. Genetic variants in Ser-Arg protein-coding genes are associated with the risk of nonobstructive azoospermia in Chinese men. Fertil Steril. 2014;101:1711-7
76. Namiki Y, Endoh D, Kon Y. Genetic mutation associated with meiotic metaphase-specific apoptosis in MRL/MpJ mice. Mol Reprod Dev. 2003;64:179-88
77. Horiuchi K, Perez-Cerezales S, Papasaikas P. et al. Impaired spermatogenesis, muscle, and erythrocyte function in U12 intron splicing-defective Zrsr1 mutant mice. Cell Rep. 2018;23:143-55
78. Liu W, Wang F, Xu Q. et al. BCAS2 is involved in alternative mRNA splicing in spermatogonia and the transition to meiosis. Nat Commun. 2017;8:14182
79. Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189-92
80. Tabrez SS, Sharma RD, Jain V. et al. Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity. Nat Commun. 2017;8:306
81. Hilleren P, Parker R. mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA. 1999;5:711-9
82. Chakrabarti S, Bonneau F, Schüssler S. et al. Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res. 2014;42:9447-60
83. Kurosaki T, Li W, Hoque M. et al. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28:1900-16
84. Ohnishi T, Yamashita A, Kashima I. et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187-200
85. Maquat LE. Nonsense-mediated mRNA decay. Curr Biol. 2002;12:R196-7
86. Mühlemann O. Spermatogenesis studies reveal a distinct nonsense-mediated mRNA decay (NMD) mechanism for mRNAs with long 3'UTRs. PLoS Genet. 2016;12:e1005979
87. Fanourgakis G, Lesche M, Akpinar M. et al. Chromatoid body protein TDRD6 supports long 3' UTR triggered nonsense mediated mRNA decay. PLoS Genet. 2016;12:e1005857
88. Wang C, Swerdloff RS. Male contraception. Best Pract Res Clin Obstet Gynaecol. 2002;16:193-203
89. O'Shaughnessy PJ. Hormonal control of germ cell development and spermatogenesis. Semin Cell Dev Biol. 2014;29:55-65
90. Bruysters M, Christin-Maitre S, Verhoef-Post M. et al. A new LH receptor splice mutation responsible for male hypogonadism with subnormal sperm production in the propositus, and infertility with regular cycles in an affected sister. Hum Reprod. 2008;23:1917-23
91. Song GJ, Park YS, Lee YS. et al. Alternatively spliced variants of the follicle-stimulating hormone receptor gene in the testis of infertile men. Fertil Steril. 2002;77:499-504
92. Gromoll J, Gudermann T, Nieschlag E. Molecular cloning of a truncated isoform of the human follicle stimulating hormone receptor. Biochem Biophys Res Commun. 1992;188:1077-83
93. Tena-Sempere M, Manna PR, Huhtaniemi I. Molecular cloning of the mouse follicle-stimulating hormone receptor complementary deoxyribonucleic acid: functional expression of alternatively spliced variants and receptor inactivation by a C566T transition in exon 7 of the coding sequence. Biol Reprod. 1999;60:1515-27
94. O'Hara L, Smith LB. Androgen receptor roles in spermatogenesis and infertility. Best Pract Res Clin Endocrinol Metab. 2015;29:595-605
95. Chang C, Chen YT, Yeh SD. et al. Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc Natl Acad Sci U S A. 2004;101:6876-81
96. Denolet E, De Gendt K, Allemeersch J. et al. The effect of a sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Mol Endocrinol. 2006;20:321-34
97. Lim P, Robson M, Spaliviero J. et al. Sertoli cell androgen receptor DNA binding domain is essential for the completion of spermatogenesis. Endocrinology. 2009;150:4755-65
98. Cutress ML, Whitaker HC, Mills IG. et al. Structural basis for the nuclear import of the human androgen receptor. J Cell Sci. 2008;121:957-68
99. Jääskeläinen J, Mongan NP, Harland S. et al. Five novel androgen receptor gene mutations associated with complete androgen insensitivity syndrome. Hum Mutat. 2006;27:291
100. Wickert L, Selbig J. Structural analysis of the DNA-binding domain of alternatively spliced steroid receptors. J Endocrinol. 2002;173:429-36
101. Sammarco I, Grimaldi P, Rossi P. et al. Novel point mutation in the splice donor site of exon-intron junction 6 of the androgen receptor gene in a patient with partial androgen insensitivity syndrome. J Clin Endocrinol Metab. 2000;85:3256-61
102. Hellwinkel OJ, Holterhus PM, Struve D. et al. A unique exonic splicing mutation in the human androgen receptor gene indicates a physiologic relevance of regular androgen receptor transcript variants. J Clin Endocrinol Metab. 2001;86:2569-75
103. Neto FT, Bach PV, Najari BB. et al. Spermatogenesis in humans and its affecting factors. Semin Cell Dev Biol. 2016;59:10-26
104. Cheng CY, Wong EW, Yan HH. et al. Regulation of spermatogenesis in the microenvironment of the seminiferous epithelium: new insights and advances. Mol Cell Endocrinol. 2010;315:49-56
105. Ma TH, Xiong QH, Yuan B. et al. Luteinizing hormone receptor splicing variants in bovine Leydig cells. Genet Mol Res. 2012;11:1721-30
106. Welsh M, Saunders PT, Atanassova N. et al. Androgen action via testicular peritubular myoid cells is essential for male fertility. FASEB Journal. 2009;23:4218-30
107. Liu Q, Zhang X, Hu X. et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'Sponge' in human cartilage degradation. Sci Rep. 2016;6:22572
108. Zhang XO, Dong R, Zhang Y. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277-87
109. Zhou F, Chen W, Jiang Y. et al. Regulation of long non-coding RNAs and circular RNAs in spermatogonial stem cells. Reproduction. 2019 Apr 1. pii: REP-18-0517. R2
110. Gao Y, Wu M, Fan Y. et al. Identification and characterization of circular RNAs in Qinchuan cattle testis. R Soc Open Sci. 2018;5:180413
111. Zhou T, Xie X, Li M. et al. Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages. RNA. 2018;24:1443-56
112. Dong WW, Li HM, Qing XR. et al. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci Rep. 2016;6:39080
113. Wang ET, Sandberg R, Luo S. et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-6
114. Bondy-Chorney E, Crawford Parks TE, Ravel-Chapuis A. et al. Staufen1 regulates multiple alternative splicing events either positively or negatively in DM1 indicating its role as a disease modifier. PLoS Genet. 2016;12:e1005827
115. Zhou X, Wu W, Li H. et al. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res. 2014;42:4019-30
116. Ryan M, Wong WC, Brown R. et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016;44:D1018-22
117. Cheng TL, Chen J, Wan H. et al. Regulation of mRNA splicing by MeCP2 via epigenetic modifications in the brain. Sci Rep. 2017;17:42790
118. Yan Q, Xia X, Sun Z. et al. Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet. 2017;13:e1006663
Author contact
![]() Corresponding author: Dr. Fenge Li. Affiliation: College of Animal Science, Huazhong Agricultural University, Wuhan, 430070, P.R. China; Tel: 0086-27-87282091; Fax: 0086-27-87280408; E-mail: lifenerhzau.edu.cn
Corresponding author: Dr. Fenge Li. Affiliation: College of Animal Science, Huazhong Agricultural University, Wuhan, 430070, P.R. China; Tel: 0086-27-87282091; Fax: 0086-27-87280408; E-mail: lifenerhzau.edu.cn

 Global reach, higher impact
Global reach, higher impact