10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(4):568-582. doi:10.7150/ijbs.39769 This issue Cite
Research Paper
Comprehensive understanding of B7 family in gastric cancer: expression profile, association with clinicopathological parameters and downstream targets
1. Department of Oncology, The Affiliated Hospital of Southwest Medical University, Luzhou 646000, China.
2. Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, 646000, Sichuan, PR China.
3. South Sichuan Institute of Translational Medicine, Luzhou, 646000, Sichuan, PR China.
4. Shenzhen Aier Aye Hospital, Shenzhen, 518032, Guangdong, PR China
*These authors contributed equally to this work
Received 2019-8-29; Accepted 2019-12-10; Published 2020-1-1
Abstract
Objectives: B7 family members were identified as co-stimulators or co-inhibitors of the immune response and played important roles in cancer immunotherapy; however, their dysregulation in gastric cancer is still unclear.
Methods: Data were obtained from TCGA and GTEX database. B7 mutations, association with DNA methylation and affected proteins were analyzed in cBioportal. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and Gene Ontology (GO) project was studied by DAVID to find the downstream signaling pathway and important metabolic process, respectively. Protein-protein interaction network was analyzed in STRING and Cytoscape. A total of 160 paired specimens in tissue microarray from patients with gastric cancer were used to detect the expression levels of seven B7 family members via immunohistochemical analysis.
Results: Bioinformatics studies revealed dysregulation of B7 members in gastric cancer. Gene and protein alteration were found in B7 family members. Furthermore, DNA methylation and gene alteration may be both involved in B7 member dysregulation in gastric cancer. Importantly, the high expression of B7-H6 is associated with good overall patient survival. B7 family members primarily affect the EGFR tyrosine kinase inhibitor resistance signaling pathway in gastric cancer and TP53 may be an important target of the family. The low expression of B7-1 and high expression of B7-H3 and B7-H7 were validated by IHC staining.
Conclusions: Our results provide insight into B7 family member expression in gastric cancer and stress their importance in stomach tumorigenesis, which may be beneficial for designing future cancer treatments.
Keywords: B7 family members, gastric cancer, bioinformatics, PI3K-AKT signaling pathway
Introduction
Gastrointestinal (GI) cancer is one of the most well-known cancers due to its high lethality and poor prognosis, which are affected by environment, dietary habits, and gender [1]. The complex carcinogenic mechanisms of GI cancer remain unclear; however, variables, such as genetic background, smoking, dietary habits, and geographical environment, are important factors in the progression of GI cancer [2]. Gastric cancer, one type of GI cancer, accounts for approximately 8.6% of newly diagnosed cancer cases worldwide, making it the second highest cause of cancer deaths [3]. Since gastric cancer is generally diagnosed in the advanced stages, treatments are often ineffective [4]. Common treatments include surgery, chemotherapy, and radiotherapy; however, these methods can cause significant physiological distress, economic burden, and poor quality of life [5]. Recently, immunotherapy has been used for cancer treatment, and has potential as a successful tumor therapy and for the prevention of tumor recurrence [6].
B7 family is an important family of costimulatory molecules that promotes or inhibits T cell proliferation and cytokine production, and also plays an important regulatory role in B cell activation and antibody production. Over the past decade, ten members of the B7 gene family have been identified, including B7-1, B7-2, B7-DC, B7-H1, B7-H2, B7-H3, B7-H4, B7-H5, B7-H6 and B7-H7. Each member contains at least 15% conjunct amino acid sequences, which are expressed by antigen-presenting cells (APCs) or tumor cells [7]. B7 family members can be divided into three groups: group I, which includes B7-1/B7-2/CD28/CTLA4 and B7-H2; group II, which contains PD-L1/PD-L2/PD-1, and group III, which consists of B7-H3, B7-H4, and B7-H5. B7 family ligands and receptors play important roles in T-cell regulation of the immune system [8]. Several studies have indicated that B7 family members are overexpressed in tumors and are associated with pathogenesis and malignancy due to their immunological functions in tumor-bearing hosts, as in lung and hematologic cancers [9-11]. Moreover, soluble B7 family ligands have also been detected in the sera of tumor patients; soluble B7-H3 and soluble B7-H4 have proven to be prognostic biomarkers in ovarian and renal cancer [12]. In addition, B7-H1 and B7-H4 were shown to be positively correlated with the depth of tumor invasion, lymph node metastasis, and tumor stage, and are considered to be negative prognostic factors in gastric cancer [13]. B7-H4 is also positively associated with poor prognosis in gastric cancer [14]. B7-H2 was shown to be significantly related to the occurrence of gastric cancer through the SNP rs4819388, which disrupts the regulatory role of miR-24 on B7-H2 expression and promotes the occurrence of gastric cancer [15]. Another study demonstrated that the aberrant expression of B7-H3 promoted cell migration and invasion in gastric cancer patients compared to the controls [16]. Although B7 family members were found to be related to the progression of GI cancer, the expression patterns of each member in GI tumors, especially in gastric cancer, are still not clear. Meanwhile, there is little research progress in the biological function and mechanism of B7 family members in gastric cancer. In this study, we explored the relationship between the expression levels of B7 family members in gastric cancer and show mutations and survival analysis of B7 family members, and association of B7 family with clinicopathological parameters in gastric cancer. Finally, we conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and Gene ontology (GO) analysis of B7 family, and finally we obtained one key protein for further analysis. We hope that these studies will help early detection and treatment strategies for patients with gastric cancer.
Results
The dysregulation of B7 family members in gastric cancer
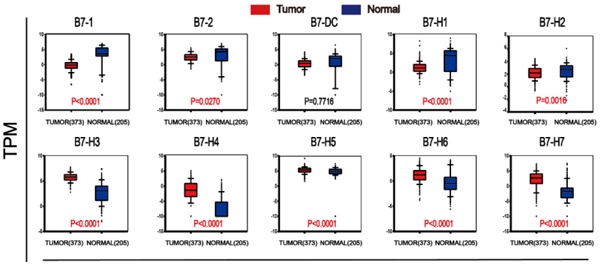
The ten members of the B7 family were shown in Fig. 1A. We compared the expression levels of B7 members in normal and tumor samples (Fig. 1B-C). Results showed that B7-H3, B7-H4, B7-H5, B7-H6 and B7-H7 were significantly upregulated, while B7-1, B7-2, B7-H1 and B7-H2 were significantly downregulated in gastric cancer patients. (Fig. 1C). At the same time, we also compared the expression levels between normal, non-metastatic and metastatic gastric cancer (Fig. 1D). Although there was significant difference between normal and cancer samples, there is no significance between non-metastatic cancer and metastatic gastric cancer. Then, we examined the mRNA expression levels of B7 family members and their relationship to DNA methylation, copy number alterations to obtain an overview of the dysregulation mechanism of B7 family members in gastric cancer (Fig. 1E-F). Gene alterations, including diploid, copy number gain, and gene amplification, were associated with the increased expression of B7 members (Fig. 1E). Moreover, there was a negative correlation between mRNA expression and promotor methylation for B7-1, B7-H3, B7-H4, B7-H5, B7-H6, and B7-H7 (Fig. 1F), suggesting promotor methylation might also be involved in B7 members' dysregulation.
Gene and protein alteration of the B7 family members
In order to analyze mutations of B7 family member in gastric cancer, cBioPortal for Cancer Genomic was used. Overall, about 16% samples (63/393) had genetic changes is analyzed in gastric cancer, as shown in Fig. 2A. The mutation ratio of B7-DC and B7-H1 were relatively higher, 6% and 7% respectively. Meanwhile, the mutation frequency of B7 family member in different types of gastric cancer was shown (Fig. 2B). mRNA overexpression was frequently seen in B7 family members. The frequency of mRNA overexpression was high in B7-1, B7-2, B7-H3, B7-H6 and B7-H7 and B7-H5 showed relatively higher copy number amplification. Changes in protein structure of B7 family member were shown in Fig. 2C. There were more mutation sites in B7-2 and B7-H3 than in other members. The protein domains of B7 family members were shown in Supplementary Fig. 1.
Expression level of B7 family members in gastric cancer. A. Nomenclature of each B7 family member. B. Heat map showing expression levels of B7 family genes between cancer and normal samples based on RNA sequencing data from TCGA database. Data were collected from tumor (n = 373) and normal tissues (n = 32). C. RNA sequencing data from TCGA and GTEX database of 373 cancer patients and 203 healthy people were used to analyze the expression levels of B7 family genes in gastric cancer. D. RNA sequencing data from TCGA and GTEX database were used to evaluate the expression levels of B7 family members in 203 normal people, 291 non-metastasis patients and 82 metastasis patients. E. Association of B7 family member mRNA expression with gene alteration. F. Association of B7 family member mRNA expression with promotor methylation. (* P < 0.05,** P < 0.01, and *** P < 0.001).
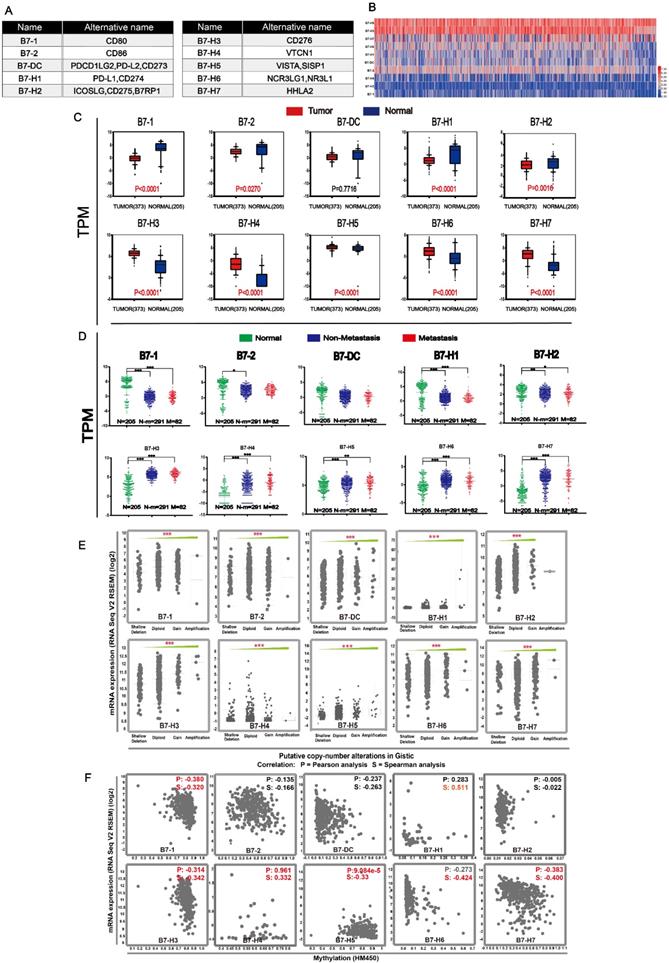
Gene and protein alteration of B7 family members in gastric cancer. A. The mutation rate of B7 family members in gastric cancer. B. Alteration frequency of B7 family genes in gastric cancer, including mutations, amplification, mRNA overexpression and multiple changes were analyzed from the TCGA database and studied in cBioPortal. 478 gastric cancer samples were used for this analysis, and about 14% samples (66/478) showed B7 family alteration. C. Changes in protein structure of B7 family members. SRCSTAD: Signet ring cell carcinoma of the stomach; Tubular stomach adenocarcinoma: TSTAD; Stomach adenocarcinoma: STAD; Diffuse type stomach adenocarcinoma: DTSTAD; Mucinous stomach adenocarcinoma: MSTAD
The association of B7 family with clinicopathological parameters and patient survival in gastric cancer
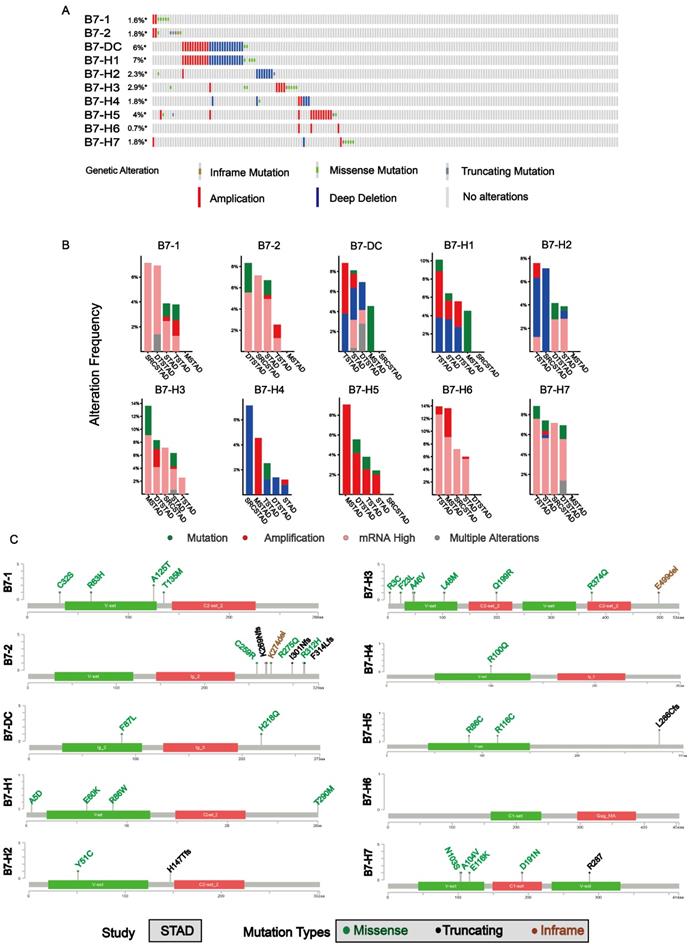
We analyzed the association of B7 family expression with overall survival status (Dead; Alive), gender (Male; Female), cancer status (Tumor; Normal), race (Asian; White; Others), tumor differentiation grade (G1; G2; G3), pathological stage (Stage I; Stage II; Stage III; Stage IV), tumor topography (T1; T2; T3; T4) and lymph node(N1; N2; N3; N4) through chi-square test as shown in Table 1. The results showed that the expression of some B7 family member was significantly associated with tumor differentiation grade, pathological stage, race and tumor topography. The relationship between the four clinical parameters and expression levels of B7 family members was shown in Fig. 3A. For tumor differentiation grade, B7-1, B7-DC and B7-H1 gradually increased from G1 to G3. Conversely, B7-H6 and B7-H7 gradually decreased. For pathological stage, the expression of B7-1, B7-2 and B7-DC were elevated from pathological stage I to IV. Moreover, there are significant differences between B7-DC and B7-H2 in race. For tumor topography, the relationship between the expression of B7-2, B7-DC, B7-H1, B7-H4, B7-H6 and the extent of the primary tumor had significant differences. Next, we combined the data of gene expression and clinical information in the TCGA database to evaluate the influence of B7 family member expression on survival rate in patients with gastric cancer. Kaplan-Meier analysis showed high expression of B7-H6 was significantly associated with longer OS in gastric patients (Fig. 3B).
B7 family's mRNA expression and clinicopathological Characteristics of gastric cancer patients in the TCGA database.
| Characteristics | B7 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | χ2 | P | High | Low | χ2 | P | High | Low | χ2 | P | ||
| OS status | B7-1 | B7-2 | B7-DC | ||||||||||
| Dead | 112 | 117 | 0.3565 | 0.5505 | 111 | 118 | 0.6557 | 0.4181 | 106 | 123 | 3.509 | 0.0610 | |
| Alive | 75 | 69 | 76 | 68 | 81 | 63 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| Dead | 114 | 115 | 0.02946 | 0.8637 | 117 | 112 | 0.2176 | 0.6409 | 106 | 123 | 3.509 | 0.0610 | |
| Alive | 73 | 71 | 70 | 74 | 81 | 63 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| Dead | 121 | 108 | 1.735 | 0.1877 | 115 | 114 | 0.001686 | 0.9672 | 117 | 112 | 0.2176 | 0.6409 | |
| Alive | 66 | 78 | 72 | 72 | 70 | 74 | |||||||
| B7-H7 | |||||||||||||
| Dead | 112 | 117 | 0.3565 | 0.5505 | |||||||||
| Alive | 75 | 69 | |||||||||||
| Gender | B7-1 | B7-2 | B7-DC | ||||||||||
| Male | 72 | 61 | 1.324 | 0.2499 | 67 | 66 | 0.0215 | 0.8834 | 70 | 63 | 0.5157 | 0.4727 | |
| Female | 115 | 125 | 119 | 121 | 117 | 123 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| Male | 73 | 60 | 1.868 | 0.1717 | 70 | 63 | 0.5157 | 0.4727 | 65 | 68 | 0.1317 | 0.7167 | |
| Female | 114 | 126 | 117 | 123 | 122 | 118 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| Male | 60 | 73 | 30.81 | <0.0001 | 66 | 67 | 0.0215 | 0.8834 | 71 | 62 | 0.873 | 0.3501 | |
| Female | 127 | 113 | 121 | 119 | 116 | 124 | |||||||
| B7-H7 | |||||||||||||
| Male | 63 | 70 | 0.6324 | 0.4265 | |||||||||
| Female | 124 | 116 | |||||||||||
| Cancer status | B7-1 | B7-2 | B7-DC | ||||||||||
| Normal | 59 | 61 | 0.07221 | 0.7881 | 61 | 59 | 0.035 | 0.8516 | 63 | 57 | 0.4133 | 0.5203 | |
| Tumor | 109 | 106 | 107 | 108 | 105 | 110 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| Normal | 57 | 63 | 0.5249 | 0.4687 | 59 | 61 | 0.07221 | 0.7881 | 65 | 55 | 1.207 | 0.2719 | |
| With Tumor | 111 | 104 | 109 | 106 | 103 | 112 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| Normal | 63 | 57 | 0.4133 | 0.5203 | 66 | 54 | 1.76 | 0.1846 | 53 | 67 | 2.677 | 0.1018 | |
| Tumor | 105 | 110 | 102 | 113 | 115 | 100 | |||||||
| B7-H7 | |||||||||||||
| Normal | 58 | 62 | 0.2466 | 0.6195 | |||||||||
| Tumor | 110 | 105 | |||||||||||
| Grade | B7-1 | B7-2 | B7-DC | ||||||||||
| G1 | 4 | 6 | 5.088 | 0.0786 | 5 | 5 | 24.25 | <0.0001 | 4 | 6 | 23.34 | <0.0001 | |
| G2 | 58 | 77 | 45 | 90 | 46 | 89 | |||||||
| G3 | 120 | 99 | 132 | 87 | 132 | 87 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| G1 | 4 | 6 | 6.082 | 0.0478 | 6 | 4 | 0.6263 | 0.7311 | 5 | 5 | 0.9699 | 0.6157 | |
| G2 | 57 | 78 | 65 | 70 | 63 | 72 | |||||||
| G3 | 121 | 98 | 111 | 108 | 114 | 105 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| G1 | 6 | 4 | 11.02 | 0.0041 | 4 | 6 | 4.189 | 0.1231 | 7 | 3 | 11.39 | 0.0034 | |
| G2 | 82 | 53 | 59 | 76 | 81 | 54 | |||||||
| G3 | 94 | 125 | 119 | 100 | 94 | 125 | |||||||
| B7-H7 | |||||||||||||
| G1 | 7 | 3 | 7.721 | 0.0211 | |||||||||
| G2 | 78 | 57 | |||||||||||
| G3 | 97 | 122 | |||||||||||
| Race | B7-1 | B7-2 | B7-DC | ||||||||||
| Asian | 34 | 40 | 3.366 | 0.1858 | 36 | 38 | 4.929 | 0.0850 | 33 | 41 | 9.598 | 0.0082 | |
| White | 124 | 112 | 123 | 113 | 127 | 109 | |||||||
| Others | 3 | 8 | 2 | 9 | 1 | 10 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| Asian | 31 | 43 | 3.846 | 0.1462 | 53 | 21 | 17.74 | 0.0001 | 29 | 45 | 5.105 | 0.0779 | |
| White | 126 | 110 | 103 | 133 | 125 | 111 | |||||||
| Others | 4 | 7 | 5 | 6 | 7 | 4 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| Asian | 36 | 38 | 0.8691 | 0.6475 | 40 | 34 | 4.955 | 0.0840 | 35 | 39 | 2.486 | 0.2885 | |
| White | 118 | 118 | 119 | 117 | 118 | 118 | |||||||
| Others | 7 | 4 | 2 | 9 | 8 | .3 | |||||||
| B7-H7 | |||||||||||||
| Asian | 34 | 40 | 2.773 | 0.2499 | |||||||||
| White | 119 | 117 | |||||||||||
| Others | 8 | 3 | |||||||||||
| Pathologic stage | B7-1 | B7-2 | B7-DC | ||||||||||
| Stage I | 21 | 32 | 3.886 | 0.2741 | 18 | 35 | 6.466 | 0.0910 | 18 | 35 | 7.912 | 0.0479 | |
| Stage II | 53 | 58 | 59 | 52 | 55 | 56 | |||||||
| Stage III | 81 | 68 | 78 | 71 | 84 | 65 | |||||||
| Stage IV | 20 | 17 | 20 | 17 | 18 | 19 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| Stage I | 27 | 26 | 1.104 | 0.7760 | 17 | 36 | 8.69 | 0.0337 | 26 | 27 | 2.38 | 0.4973 | |
| Stage II | 51 | 60 | 62 | 49 | 58 | 53 | |||||||
| Stage III | 78 | 71 | 78 | 71 | 69 | 80 | |||||||
| Stage IV | 19 | 18 | 18 | 19 | 22 | 15 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| Stage I | 32 | 21 | 6.208 | 0.1019 | 23 | 30 | 1.237 | 0.7441 | 26 | 27 | 7.085 | 0.0692 | |
| Stage II | 61 | 50 | 58 | 53 | 46 | 65 | |||||||
| Stage III | 67 | 82 | 76 | 73 | 79 | 70 | |||||||
| Stage IV | 15 | 22 | 18 | 19 | 24 | 13 | |||||||
| B7-H7 | |||||||||||||
| Stage I | 27 | 26 | 0.1337 | 0.9875 | |||||||||
| Stage II | 54 | 57 | |||||||||||
| Stage III | 75 | 74 | |||||||||||
| Stage IV | 19 | 18 | |||||||||||
| Tumor topography | B7-1 | B7-2 | B7-DC | ||||||||||
| T1 | 5 | 14 | 7.035 | 0.0708 | 2 | 17 | 14.69 | 0.0021 | 4 | 15 | 12.11 | 0.0070 | |
| T2 | 40 | 40 | 42 | 38 | 41 | 39 | |||||||
| T3 | 81 | 87 | 82 | 86 | 78 | 90 | |||||||
| T4 | 58 | 42 | 58 | 42 | 61 | 39 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| T1 | 5 | 14 | 8.817 | 0.0310 | 10 | 9 | 9.414 | 0.0243 | 5 | 14 | 4.774 | 0.1891 | |
| T2 | 47 | 33 | 28 | 52 | 43 | 37 | |||||||
| T3 | 77 | 91 | 92 | 76 | 85 | 83 | |||||||
| T4 | 55 | 45 | 54 | 46 | 51 | 49 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| T1 | 14 | 5 | 10.4 | 0.0154 | 10 | 9 | 0.3051 | 0.9591 | 11 | 8 | 9.005 | 0.0292 | |
| T2 | 48 | 32 | 40 | 40 | 35 | 45 | |||||||
| T3 | 80 | 88 | 86 | 82 | 76 | 92 | |||||||
| T4 | 42 | 58 | 48 | 52 | 62 | 38 | |||||||
| B7-H7 | |||||||||||||
| T1 | 11 | 8 | 1.092 | 0.7790 | |||||||||
| T2 | 42 | 38 | |||||||||||
| T3 | 80 | 88 | |||||||||||
| T4 | 51 | 49 | |||||||||||
| Lymph Node | B7-1 | B7-2 | B7-DC | ||||||||||
| N1 | 51 | 62 | 3 | 0.3916 | 53 | 60 | 2.471 | 0.4805 | 49 | 64 | 3.196 | 0.3624 | |
| N2 | 54 | 42 | 54 | 42 | 52 | 44 | |||||||
| N3 | 35 | 39 | 34 | 40 | 38 | 36 | |||||||
| N4 | 39 | 35 | 38 | 36 | 40 | 34 | |||||||
| B7-H1 | B7-H2 | B7-H3 | |||||||||||
| N1 | 55 | 58 | 1.47 | 0.6892 | 56 | 57 | 2.264 | 0.5195 | 62 | 51 | 1.984 | 0.5758 | |
| N2 | 49 | 47 | 47 | 49 | 45 | 51 | |||||||
| N3 | 41 | 33 | 42 | 32 | 38 | 36 | |||||||
| N4 | 34 | 40 | 33 | 41 | 34 | 40 | |||||||
| B7-H4 | B7-H5 | B7-H6 | |||||||||||
| N1 | 57 | 56 | 5.578 | 0.1340 | 49 | 64 | 3.087 | 0.3783 | 55 | 58 | 1.825 | 0.6096 | |
| N2 | 46 | 50 | 52 | 44 | 44 | 52 | |||||||
| N3 | 45 | 29 | 39 | 35 | 39 | 35 | |||||||
| N4 | 31 | 43 | 39 | 35 | 41 | 33 | |||||||
| B7-H7 | |||||||||||||
| N1 | 57 | 56 | 1.213 | 0.7498 | |||||||||
| N2 | 44 | 52 | |||||||||||
| N3 | 40 | 34 | |||||||||||
| N4 | 38 | 36 | |||||||||||
Pathway and GO analysis of B7 family members
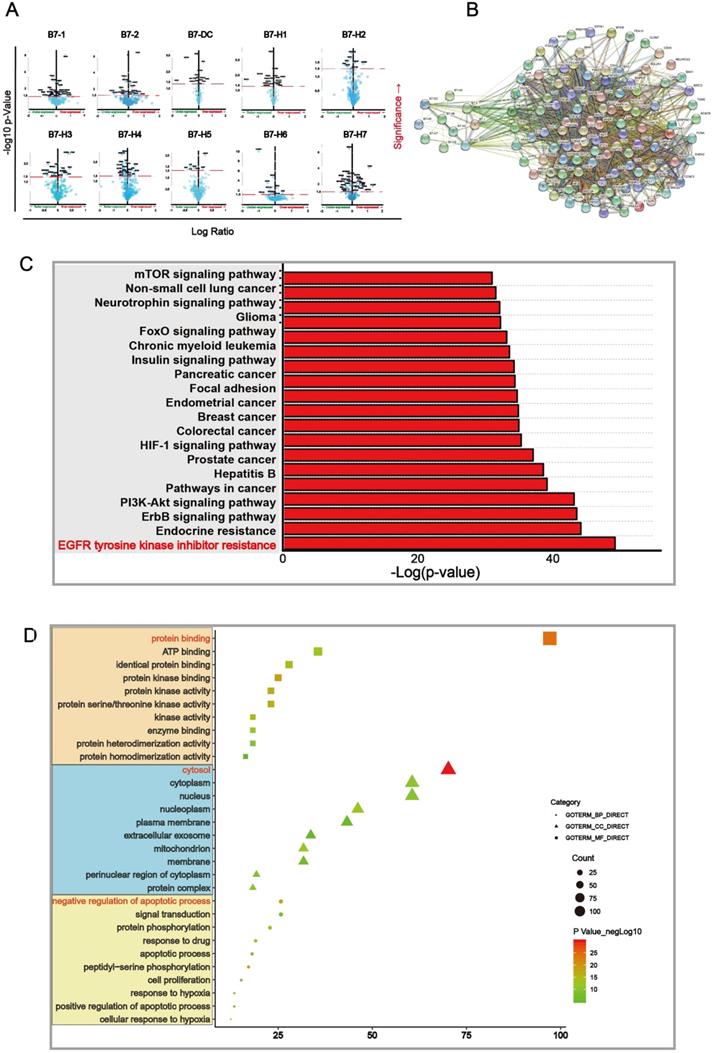
We further found out differential proteins that were altered upon B7 family member mutation. Proteins which have significant changes (p < 0.05) were shown in the volcano map (Fig. 4A) and were used for further analysis. We mapped Protein-Protein Interaction Networks for further understanding the interaction between these differential proteins and B7 family member. (Fig. 4B). In order to identify which pathway the differential protein is mainly enriched in, we analyzed KEGG proteins enrichment by DAVID. According to the p value, EGFR tyrosine kinase inhibitor resistance is considered to be the most important downstream signaling pathway for members of the B7 family in gastric cancer (Fig. 4C). Finally, we analyzed the 105 differential proteins by GO terms biological process enriched via DAVID. The result showed that these differential proteins mainly play an important role in the regulation of protein binding, cytosol, negative regulation of apoptotic process in biological process, cellular component and molecular function respectively (Fig. 4D).
TP53 is closely related to B7 family members in gastric cancer
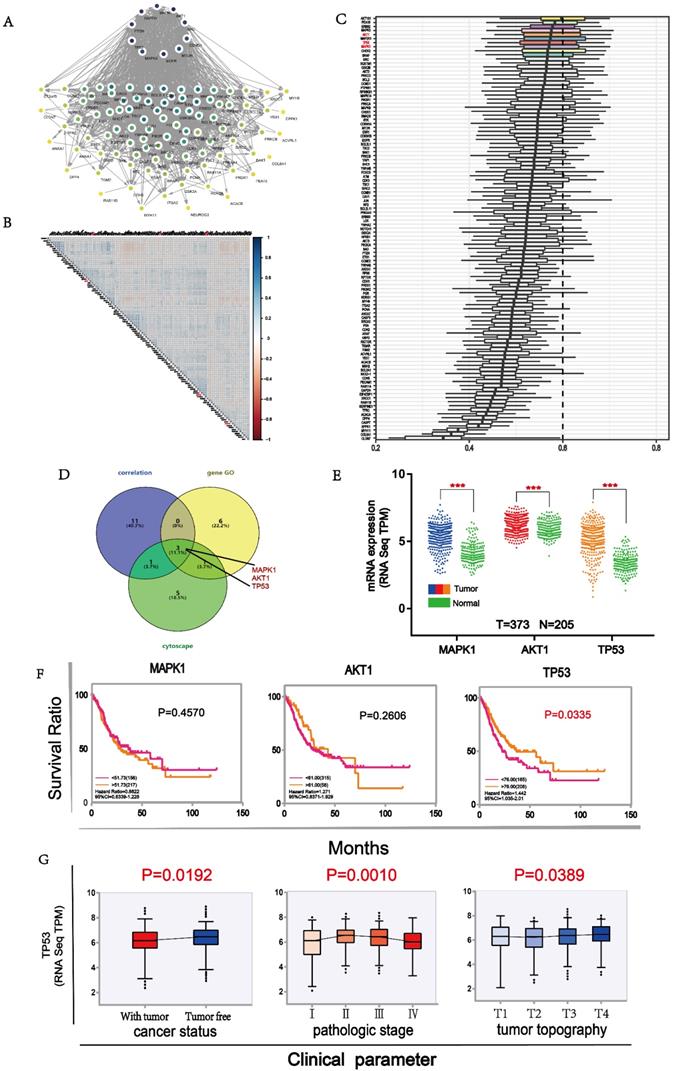
To identify the most important proteins of the 105 differential proteins, we conducted further analysis. First, we mapped Protein-Protein Interaction Networks through cytoscape for understanding the interaction between these differential proteins. Ten proteins demonstrated stronger interaction among all proteins (Fig. 5A). Then, we used the expression level of all differential proteins to conduct a correlation analysis and selected 15 proteins with good correlation with other proteins (Fig. 5B). Next, we compared the importance of functional annotations of different proteins. The results showed that the first ten proteins were strongly involved in the three major categories of GO (Gene Ontology), including biological processes, cell components, and molecular functions. (Fig. 5C). Finally, three proteins, namely MAPK1, AKT1 and TP53, were predicted to be the most important downstream targets by the above 3 approaches (Fig. 5D). Their expressions were significantly correlated with some B7 members (Supplementary Fig. 2). In order to understand the expression of these proteins in gastric cancer, we compared the expression levels of them between tumor and normal samples. The results showed that the expression levels of these proteins in cancer patients were significantly higher than in normal patients (Fig. 5E). Moreover, the high expression of TP53 was significantly associated with longer OS in gastric patients (Fig. 5F). Finally, we conducted clinical parameters analysis of TP53 in gastric cancer. Results showed that the expression level of TP53 was significantly different in cancer status, pathologic stage and tumor topography (Fig. 5G), with its expression lower in patients with tumor than tumor free. In conclusion, we believe that TP53 may be an important downstream target of B7 family members in gastric cancer.
Verification of B7 family expression level in patient samples
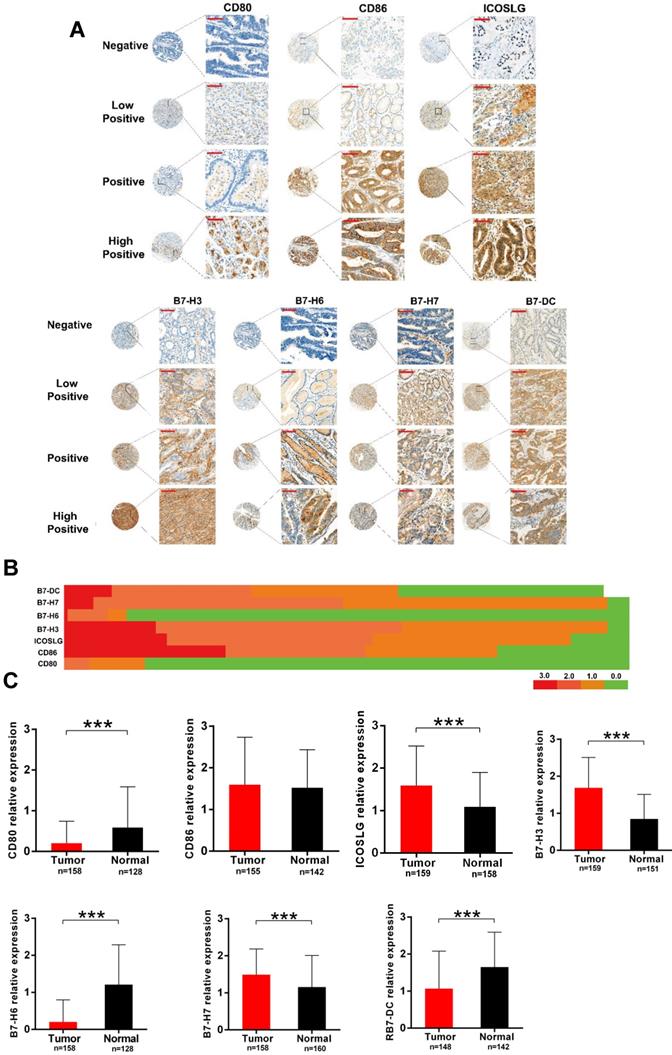
To verify the results of bioinformatics analysis, we performed immunohistochemical (IHC) staining on tissue microarray slides containing 160 gastric cancer tissues. Fig. 6A represents the staining patterns of B7 family members in gastric cancers. In all of the tissues, the staining of B7 family members was observed in the cytoplasm or cell membrane but not the nucleus. Brown, yellow-brown, or buff presented positive staining cells as strong, moderate, and weak, respectively. We then performed heatmap clustering analysis for seven of the B7 family members in gastric cancer samples (Fig. 6B). Results showed higher staining of CD86 (118/157, 75%), ICOSLG (122/157, 77%), B7-H3 (138/157, 88%), B7-H7 (138/157, 88%), and B7-DC (104/157, 66%) and overall lower staining of CD80 and B7-H6 in gastric cancer tissues. The expression levels of B7 family members between tumor and normal tissues were shown in Fig. 6C. Seven B7 family molecules were unequivocally expressed in all stomach tumor samples and paired non-tumor tissues from the same patient. Moreover, ICOSLG (89.6%, 138/154; P < 0.001), B7-H3 (96.1%, 148/154; P < 0.001) and B7-H7 (96.1%, 148/154; P < 0.001) were significantly upregulated in tumors. In contrast, CD80 (14.2%, 22/154; P < 0.001), B7-H6 (11%, 17/154; P <0.001) and B7-DC (59%, 91/154; P < 0.001) were significantly downregulated in tumors, while CD86 (76.6%, 118/154; P = 0.54) was upregulated but not significantly.
The association analysis of B7 family with clinical parameters and Kaplan-Meier survival analysis in gastric cancer. A. Clinical parameter analysis includes race (Asian, White, Others), stage of tumor differentiation (G1, G2 and G3), pathologic stage (Stage I, Stage II, Stage III, Stage IV) and tumor topography (T1, T2, T3, T4) by one-way ANOVA. B. Kaplan-Meier survival curves of B7 family genes in gastric cancer based on the expression level. (* P < 0.05**, P < 0.01, and *** P < 0.001).
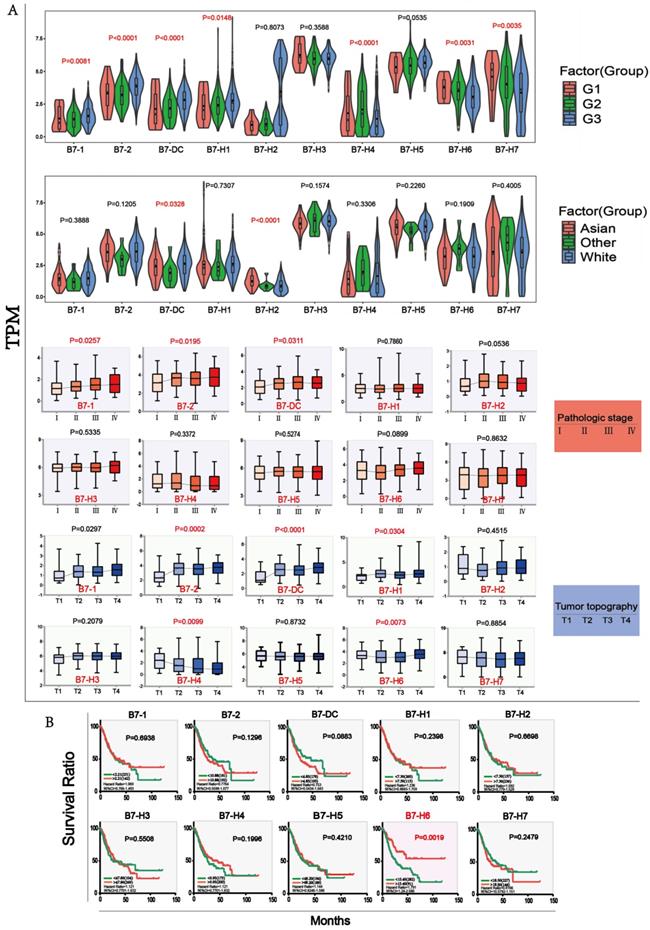
Main signaling pathway affected by B7 family members. A. Volcano plot was drawn to identify differential proteins affected by B7 members analyzed using Reverse Phase Protein Arrays (RPPA) in cBioPortal. The Y axis is the value of fold change of expression level is based on logarithmic ratio (mean of changed expression/mean of unchanged expression). -log10 (p-value)>1.30 is considered to be a significant difference. B. Multicenter protein-protein interaction (PPI) network analysis between B7 family member and differential proteins in STRING database. The stronger the interaction between the two proteins, the thicker the lines. C. Prediction of downstream pathways related to B7 family genes alterations was analyzed by KEGG pathway analysis via DAVID. D. Bubble chart of the GO (Gene Ontology) enrichment analysis of the B7 family was analyzed via DAVID. Gene count and P values were considered to obtain important metabolic process.
Main targets of B7 members. A. The protein interaction network diagram of differential proteins by cytoscape. Ten proteins in black color were strongly connected to other proteins. B. Co-expression analysis of B7 differential proteins. The color intensity reflects the reliability of co-expression. C. GO analysis of differential proteins. Ten proteins were shown to be important in biological processes, cell components, and molecular functions. D. Venn diagram was used to screen important downstream proteins by 3 methods. MAPK1, AKT1, TP53 were predicted to be major downstream targets. E. The comparison of expression level of MAPK1, AKT1 and TP53 between tumor and normal samples. F. Survival analysis of MAPK1, AKT1 and TP53 in gastric cancer. G. Clinical parameters analysis of TP53 in gastric cancer.
Verification of B7 family expression in gastric cancer samples. A. Representative immunohistochemical (IHC) staining showing B7 family expression in tumor tissues. Negative (blue, Score = 0; cases: CD80 135/158, CD86 36/155, ICOSLG 20/159, B7-H3 10/159, B7-H6 140/158, B7-H7 9/158, B7-DC 56/148), Weak (buff, Score = 1; cases: CD80 15/158, CD86 36/155, ICOSLG 54/159, B7-H3 56/159, B7-H6 5/158, B7-H7 72/158, B7-DC 40/148), Moderate (yellow-brown, Score = 2; cases: CD80 7/158, CD86 38/155, ICOSLG 56/159, B7-H3 67/159, B7-H6 12/158, B7-H7 68/158, B7-DC 38/148), and Strong (brown, Score = 3; cases: CD80 1/158, CD86 45/155, ICOSLG 29/159, B7-H3 26/159, B7-H6 1/158, B7-H7 9/158, B7-DC 14/148); (magnification: x400, Scar bar = 50 μm). B. Heatmap clustering showing the expression level of B7 family molecules in gastric cancer samples. C. Quantification of B7 family member levels in paired gastric cancer and adjacent normal tissues by IHC staining. (***p < 0.001).
Discussion
The functions of individual immune system components in different physiological and pathological states are regulated by the functions of opposing factors. The dysregulation of the immune system influences tumor T cell immune activity in the tumor microenvironment, and may accelerate tumor progression, metastasis, and malignancy [17]. The innate and adaptive immune systems play important roles in inhibiting tumor progression through T cell-mediated anti-tumor immune responses [18]. It is well known that B7 family members are involved in immune checkpoints and tumor angiogenesis [19]. The suppression of anti-tumor immune responses is a distinguishing feature of tumorigenesis. B7 co-stimulatory and co-inhibitory family members are involved in this process and have crucial functions in the progress of malignancies, thus they are studied as potential targets of immunotherapeutic strategies for human cancer treatment [9]. B7 family members and their receptors, CD28 family members, play key roles in the regulation of the T cell response [20] and are mainly regarded as secondary signals, in cooperation with the first signals in modulating T cell response. Several B7-CD28 family members have been proven to participate in T cell activation and tolerance in peripheral tissues, including the inhibition of the immune response through the suppression of T cell functions, the regulation of cytokine production, and the stimulation of CD4+ T cell proliferation in synergy with other proteins [21]. To date, only a few studies have mentioned the roles of B7 family members, such as B7-1, B7-2, B7-H3, B7-H4, and B7-H6, in gastric cancer [13,22-24]. The expression patterns and downstream signaling pathway of B7 family members in gastric cancer are not well illustrated. Thus, we performed bioinformatics analysis to determine the regulation and expression patterns of B7 family members in gastric cancer, and verified the results by experiments.
In this study, first, we obtained data from the TCGA database to compare the expression levels of B7 family members with heatmap (Fig. 1B). At the same time, we compare the expression levels in normal and tumor samples using TCGA and GTEX database in box plot. Results demonstrated that B7-H3, B7-H4, B7-H5, B7-H6 and B7-H7 were significantly upregulated in gastric cancer, on the contrary, B7-1, B7-2, B7-H1 and B7-H2 was significantly downregulated (Fig. 1C). Moreover, we performed IHC staining on tissue microarray slides containing 160 gastric cancer tissues. Results showed higher staining of B7-H2, B7-H3, B7-H7 and lower staining of B7-1, B7-H6 and B7-DC in gastric cancer tissues, which is partly consistent with the bioinformatics result. However, the result for the B7-1 and B7-H6 is the opposite (Fig. 6B). Overexpression of B7-H3 in gastric cancer has been reported in different studies and predicts poor patient survival [16,25]. Previous reports suggest that CD80 exhibits anti-tumor functions and is a co-stimulatory factor in the induction of cytotoxic T lymphocytes (CTL) in the suppression of stomach tumor metastasis [26-28]. Decreased expression of CD80 was found in gastric cancer, which is consistent with our experimental results, and gene therapy with CD80 inhibits metastasis [24,27,29]. B7-H6 has been demonstrated to be overexpressed in gastric cancer [30]. And B7-DC was reported to be highly expressed in EBV-associated gastric cancer [31]. Moreover, previous studies revealed that CD86 is highly expressed in metastatic gastric carcinoma [24] and the high expression of B7-H3 is commonly found in different cancers and predicts poor patient survival [32-34]. Therefore, enlargement of the patient sample size and comprehensive study are required to further elucidate the expression and functions of B7 family members in gastric cancer. Next, we analyzed the difference in the expression levels of normal, non-metastatic and metastatic gastric cancers (Fig. 1D). The results showed that there is no significant difference between non-metastatic cancer and metastatic gastric cancer, suggesting B7 family was not strongly involved in gastric tumor metastasis. Further analysis revealed that DNA methylation and gene alteration may both participated in the dysregulation of B7 members (Fig. 1E-F).
Then, we analyzed the mutation ratio (Fig. 2A), mutation frequency (Fig. 2B) and changes in protein structure (Fig. 2C) of B7 family members in gastric cancer, and found that some genes showed higher mutations, such as B7-DC and B7-H1. Meanwhile, The relationship between B7 family member expression and patient clinicopathological features has been investigated in HCC, in which B7-H1/H4 were associated with serosa invasion, lymph node metastasis, and tumor stage, while B7-H3 was associated with clinical stage and distant metastasis [13,35], and B7-H6 was significantly associated with a higher differentiation [22]. However, the relationship between B7 family member expression levels and the gastric cancer clinicopathological parameters has rarely been studied. Thus, we also examined the expression levels of these B7 members with clinicopathological parameters (Fig. 3A). Results showed that except for B7-H2, B7-H3, B7-H5, other genes were significantly associated with tumor differentiation grades, while the expression levels of B7-1, B7-2, and B7-DC were significantly different for the pathological stage. Besides, there are significant differences between B7-DC and B7-H2 for race, and tumor topography was also significantly different for B7-2, B7-DC, B7-H1, B7-H4, and B7-H6. These results indicate that members of the B7 family have important regulatory roles in gastric cancer.
Furthermore, to explore whether the expression of B7 family members is an independent prognostic factor for gastric cancer, Kaplan-Meier analysis was performed and result showed that only B7-H6 was significant related to overall survival (OS). Of note, the higher expression of B7-H6 predicted good patient survival (Fig. 3B), which is on the contrary to the report for other cancers. Previously there was a study indicating that B7-H6 was not predictive of patient survival in gastric cancer [22]. Thus, the importance of B7-H6 as prognosis indicator in gastric cancer awaits further investigation. Several studies have reported that B7 family members were associated with poor survival rates in other cancers, such as B7-H3 in esophageal cancer, B7-H6 in lung cancer, and B7-H1 and B7-H4 in ovarian cancer [11,36,37].
To identify major modulators of B7 family function, we determined proteins affected by B7 expression in gastric cancer (Fig. 4A). Based on those proteins that are significantly different, we conducted a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the pathways in B7 family via DAVID. EGFR tyrosine kinase inhibitor resistance signaling pathway is a key downstream signaling pathways of B7 family in gastric cancer (Fig. 4C). Meanwhile, we also performed gene ontology (GO) and Protein-Protein interaction network analysis (Fig. 4B) on the differential proteins of B7 family members, and found that those proteins are mainly involved in the three regulation of protein binding, cytosol, negative regulation of apoptotic process. (Fig. 4D). Finally, in order to screen the most important proteins downstream of B7 family, we used 3 different approaches (Fig. 5A-C). Then, 3 common proteins from the 3 methods were selected for further analysis, namely MAPK1, ATK1 and TP53 (Fig. 5D). Next, we compared the expression levels and survival analysis of these three proteins respectively (Fig. 5E), and the results showed that high expression of TP53 had a good prognosis in gastric cancer (Fig. 5F). Therefore, we conducted clinical parameter analysis of TP53, further verifying the importance of TP53 in gastric cancer (Fig. 5G).
To our knowledge, this is the first time the expression patterns, mutations and downstream signaling pathway of B7 family members in gastric cancer have been studied. Our results confirmed the overexpression of B7-H3 and B7-H7 by both bioinformatics and experimental analysis. Collectively, our findings provide new insight into the roles of B7 family members in gastric cancer and likely have important implications for future immunotherapy in the treatment of gastric cancer.
Materials and Methods
Data Processing
The expression level and clinical data of B7 family members in gastric cancer were extracted from TCGA database (http://cancergenome.nih.gov) and GTEX database (https://www.gtexportal.org/home/). GISTIC 2.0 database from the cBioportal website (https://www.cbioportal.org/) was used to visualize data such as somatic mutations, copy number changes, DNA methylation and reverse phase protein arrays for further signal pathway analysis. cBioPortal data comes from multiple websites, such as the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/), the ICGC data portal (http://dcc.icgc.org/), the Broad Institute's Genome Data Analysis Center (GDAC) Firehose (http://gdac.broadinstitute.org), the IGV, the University of California, Santa Cruz (UCSC) Cancer Genomics Browser and so on. The association of mRNA expression levels of B7 members with DNA methylation, copy number alterations were analyzed in cBioportal for Cancer Genomics database in gastric cancer samples (N = 408).
Protein Structure Alteration
Lollipop of each protein structure change of gastric cancer were linked to COSMIC. The detailed mutation annotation from OncoKB, CIViC and Hotspot in different genes were displayed in different regions of the protein structure.
R project analysis
R/Bioconductor GOsemsim and org.Hs.eg.db package was used for gene importance analysis of three types of GO, and correlation analysis of differential proteins by R package corrplot.
Pathway Analysis
Proteomic data were collected by Reverse Phase Protein Array (RPPA) based on TCGA data from cBioportal (http://www.cbioportal.org/). For the enriched proteins, significant change in expression was determined by the standard of Log2 based ratio (μ mean altered/ μ mean unaltered) (log>0 for over-expression and log<0 for under-expression) and queried event results in p value<0.05. The -log10 p-value >1.30 proteins were selected for further downstream pathway analysis. Finally, the differential proteins were used to predict the pathway in the DAVID function annotation tool (https://david.ncifcrf.gov).
Gene Ontology analysis
The GO (Gene Ontology) enrichment analysis of B7 family members was analyzed via DAVID function annotation tool. Gene ontology includes three parts: molecular function, biological process and cellular component. The count and P values were considered together to obtain important metabolic process.
Protein-protein interaction network analysis
We conducted the Protein-protein interaction network analysis by using the STRING and Cytoscape software. The database of STRING is a meta resource, including both physical and functional interactions [38].STRING can be reached at http://string-db.org/. Cytoscape can be for visualizing biomedical networks consisting of proteins, genes and other types of interactions, and is one of the most popular open source software tools [39].
Patients and tissue samples
Tumor and adjacent non-tumor tissue (160 cases) collection, informed consent and protocols are from the First Affiliated Hospital, Wannan Medicine College (Wuhu, China). Archived samples were collected from patients during 2011-2012 and the study was conducted in 2017. Each tumor sample was paired with non-tumor tissues (> 5cm from the tumor edge). The tissue samples were fixed in 10% buffered formalin and embedded in paraffin. Authors do not have access to information that could identify individual participants during or after data collection.
Immunohistochemical staining
Pathological sections were used to mark the locations of cancer tissues and adjacent normal tissues on the corresponding wax blocks, and immune chips were constructed according to the cancer tissues and adjacent tissues. IHC was performed on the immune chip using ChemMateTM Envision/HRP technology. Briefly, after the sections were first dewaxed, endogenous peroxidase activity was blocked using H2O2. The primary antibodies for B7-1, B7-2, B7-H2, B7-H3, B7-H6, B7-H7 and B7-DC were incubated with a diluted solution, and then a secondary antibody was added and developed with diaminobenzoquinone (DAB). Finally, the slides were counterstained with hematoxylin.
B7-1, B7-2, B7-H2, B7-H3, B7-H6, B7-H7, and B7-DC positivity were assessed based on overall staining intensity and area. The staining intensity scores were as follows: Negative: 0, Weak positive: 1, Moderate positive: 2, Strong positive: 3. According to the semiquantitative counting method, antibody expression was calculated by measuring both intensity and region of staining: =0%:0; 1%< to <10%:1; 11% to <49%: 2; ≥ 50%: 3. The final score was determined by the intensity of the case, which was calculated by multiplying the area fraction. A final score of 0 was considered as a negative expression, scores of 1 to 3 were considered low to mild positive expression, and scores greater than 3 were considered to be high positive expression. All of the slides were independently evaluated by two researchers blinded to patient identities and clinical conditions.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6 and SPSS 16.0 software. Student's t test was used to compare the difference between two groups, and one-way ANOVA was used to compare multiple groups. Overall survival was shown as a Kaplan-Meier curve, which was calculated using the log-rank test. p<0.05 was considered statistically significant.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant nos. 81602166, 81672444 and 81972643) and Sichuan Science and Technology Plan Project (2018JY0079).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Parkin DM, Bray F, Ferlay J & Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108
2. Compare D, Rocco A & Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010;14:302-308
3. Crew KD & Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354-362
4. Ma YM, Ba CF & Wang YB. Analysis of factors affecting the life quality of the patients with late stomach cancer. J Clin Nurs. 2014;23:1257-1262
5. Menges M. Diagnosis, staging and therapy of gastric cancer. Z Gastroenterol. 2004;42:767-773
6. Wang M, Yin B, Wang HY & Wang RF. Current advances in T-cell-based cancer immunotherapy. Immunotherapy. 2014;6:1265-1278
7. Xu Z, Shen J, Wang MH, Yi T, Yu Y, Zhu Y. et al. Comprehensive molecular profiling of the B7 family of immune-regulatory ligands in breast cancer. Oncoimmunology. 2016;5:e1207841
8. Janakiram M, Shah UA, Liu W, Zhao A, Schoenberg MP & Zang X. The third group of the B7-CD28 immune checkpoint family: HHLA2, TMIGD2, B7x, and B7-H3. Immunol Rev. 2017;276:26-39
9. Seliger B & Quandt D. The expression, function, and clinical relevance of B7 family members in cancer. Cancer Immunol Immunother. 2012;61:1327-1341
10. Greaves P & Gribben JG. The role of B7 family molecules in hematologic malignancy. Blood. 2013;121:734-744
11. Zhang X, Zhang G, Qin Y, Bai R & Huang J. B7-H6 expression in non-small cell lung cancers. Int J Clin Exp Pathol. 2014;7:6936-6942
12. Thompson RH, Zang X, Lohse CM, Leibovich BC, Slovin SF, Reuter VE. et al. Serum-soluble B7x is elevated in renal cell carcinoma patients and is associated with advanced stage. Cancer Res. 2008;68:6054-6058
13. Geng Y, Wang H, Lu C, Li Q, Xu B, Jiang J. et al. Expression of costimulatory molecules B7-H1, B7-H4 and Foxp3+ Tregs in gastric cancer and its clinical significance. Int J Clin Oncol. 2015;20:273-281
14. Cui Y & Li Z. B7-H4 is Predictive of Poor Prognosis in Patients with Gastric Cancer. Med Sci Monit. 2016;22:4233-4237
15. Yang P, Tang R, Zhu J, Zou L, Wu R, Zhou H. et al. A functional variant at miR-24 binding site in B7-H2 alters susceptibility to gastric cancer in a Chinese Han population. Mol Immunol. 2013;56:98-103
16. Dai W, Shen G, Qiu J, Zhao X & Gao Q. Aberrant expression of B7-H3 in gastric adenocarcinoma promotes cancer cell metastasis. Oncol Rep. 2014;32:2086-2092
17. Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263-274
18. Liyanage UK, Moore TT, Joo HG, Tanaka Y, Herrmann V, Doherty G. et al. Prevalence of regulatory T cells is increased in peripheral blood and tumor microenvironment of patients with pancreas or breast adenocarcinoma. J Immunol. 2002;169:2756-2761
19. Ni L & Dong C. New B7 Family Checkpoints in Human Cancers. Mol Cancer Ther. 2017;16:1203-1211
20. Zhao R, Chinai JM, Buhl S, Scandiuzzi L, Ray A, Jeon H. et al. HHLA2 is a member of the B7 family and inhibits human CD4 and CD8 T-cell function. Proc Natl Acad Sci.U S A. 2013;110:9879-9884
21. Greenwald RJ, Freeman GJ & Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515-548
22. Chen XJ, Shen J, Zhang GB & Chen WC. B7-H6 protein expression has no prognostic significance in human gastric carcinoma. Pathol Oncol Res. 2014;20:203-207
23. Wu CP, Jiang JT, Tan M, Zhu YB, Ji M, Xu KF. et al. Relationship between co-stimulatory molecule B7-H3 expression and gastric carcinoma histology and prognosis. World J Gastroenterol. 2006;12:457-459
24. Koyama S, Maruyama T, Adachi S & Nozue M. Expression of costimulatory molecules, B7-1 and B7-2 on human gastric carcinoma. J Cancer Res Clin Oncol. 1998;124:383-388
25. Arigami T, Uenosono Y, Hirata M, Yanagita S, Ishigami S & Natsugoe S. B7-H3 expression in gastric cancer: a novel molecular blood marker for detecting circulating tumor cells. Cancer Sci. 2011;102:1019-1024
26. Sakate Y, Yashiro M, Tanaka H, Sunami T, Kosaka K & Hirakawa K. Usefulness of inhibiting the lymph node metastasis in human gastric carcinoma by B7-1 gene transfection. J Surg Res. 2004;122:89-95
27. Sakate Y, Yashiro M, Tanaka H, Kosaka K, Ohira M & Hirakawa K. Suppression of peritoneal metastasis in human gastric carcinoma by enhanced immunogenicity of B7-1 transfection. Oncol Rep. 2004;12:53-57
28. Zheng SY, Li DC, Zhang ZD, Zhao J & Ge JF. Anti-gastric cancer active immunity induced by FasL/B7-1 gene-modified tumor cells. World J Gastroenterol. 2005;11:3204-3211
29. Kosaka K, Yashiro M, Sakate Y, Tanaka H, Sunami T, Ohira M. et al. CD80 gene therapy for lymph node involvement by gastric carcinoma. Int J Oncol. 2004;25:1319-1325
30. Chen Y, Mo J, Jia X & He Y. The B7 Family Member B7-H6: a New Bane of Tumor. Pathol Oncol Res. 2017
31. Dong M, Wang HY, Zhao XX, Chen JN, Zhang YW, Huang Y. et al. Expression and prognostic roles of PIK3CA, JAK2, PD-L1, and PD-L2 in Epstein-Barr virus-associated gastric carcinoma. Hum Pathol. 2016;53:25-34
32. Ingebrigtsen VA, Boye K, Nesland JM, Nesbakken A, Flatmark K & Fodstad O. B7-H3 expression in colorectal cancer: associations with clinicopathological parameters and patient outcome. BMC Cancer. 2014;14:602
33. Chen JT, Chen CH, Ku KL, Hsiao M, Chiang CP, Hsu TL. et al. Glycoprotein B7-H3 overexpression and aberrant glycosylation in oral cancer and immune response. Proc Natl Acad Sci.U S A. 2015;112:13057-13062
34. Mao Y, Li W, Chen K, Xie Y, Liu Q, Yao M. et al. B7-H1 and B7-H3 are independent predictors of poor prognosis in patients with non-small cell lung cancer. Oncotarget. 2015;6:3452-3461
35. Liu T, Huo Y, Li G, Yu G, Luan X. A negative correlation between B7-H3 expression and the number of CD8+ T cell infiltration in primary hepatocellular carcinoma tissues. 2014;30:1291-1294.
36. Chen L, Chen J, Xu B, Wang Q, Zhou W, Zhang G. et al. B7-H3 expression associates with tumor invasion and patient's poor survival in human esophageal cancer. Am J Transl Res. 2015;7:2646-2660
37. Xu M, Zhang B, Zhang M, Liu Y, Yin FL, Liu X. et al. Clinical relevance of expression of B7-H1 and B7-H4 in ovarian cancer. Oncol Lett. 2016;11:2815-2819
38. Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J. et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412-416
39. Su G, Morris JH, Demchak B & Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47(8 13):11-24
Author contact
![]() Corresponding authors: Zhangang Xiao, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, 646000, Sichuan, PR China.; E-mail: xzg555898com. Bo Qin, Shenzhen Aier Aye Hospital, Shenzhen, 518032, Guangdong, PR China; E-mail: qinbozfcom. Qinglian Wen, Department of Oncology, The Affiliated Hospital of Luzhou Medical College, Luzhou, Sichuan, P.R. China; E-mail: wql73115com
Corresponding authors: Zhangang Xiao, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, 646000, Sichuan, PR China.; E-mail: xzg555898com. Bo Qin, Shenzhen Aier Aye Hospital, Shenzhen, 518032, Guangdong, PR China; E-mail: qinbozfcom. Qinglian Wen, Department of Oncology, The Affiliated Hospital of Luzhou Medical College, Luzhou, Sichuan, P.R. China; E-mail: wql73115com

 Global reach, higher impact
Global reach, higher impact