ISSN: 1449-2288
Int J Biol Sci 2020; 16(7):1153-1165. doi:10.7150/ijbs.41587 This issue Cite
Research Paper
Development and Validation of a Prognostic Nomogram for Gastric Cancer Based on DNA Methylation-Driven Differentially Expressed Genes
1. Department of Liver Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College (CAMS & PUMC), Beijing, China.
2. Department of Immunology, Beijing Key Laboratory for Cancer Invasion and Metastasis, Advanced Innovation Center for Human Brain Protection, School of Basic Medical Sciences, Capital Medical University, Beijing, China.
3. Department of Pancreatic and Gastric Surgery, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
4. Department of Hepatobiliary Surgery, First Central Hospital, Tianjin, China.
*These authors contributed equally to this work.
Abstract
Background/Aims: The incidence of gastric cancer (GC) ranks fifth among common tumors and GC is the third leading cause of cancer-related death worldwide. The aim of this study was to develop and validate a nomogram for predicting the overall survival (OS) of patients with GC.
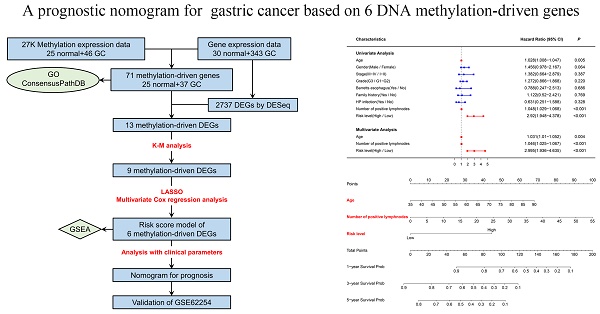
Methods: DNA methylation (DNAm)-driven genes were identified by integrating DNAm and gene expression profiling analyses from The Cancer Genome Atlas (TCGA) GC cohort. Then, a risk score model was built based on Kaplan-Meier (K-M), least absolute shrinkage and selector operation (LASSO), and multivariate Cox regression analyses. After analyzing the clinical parameters, a nomogram was constructed and assessed. Another cohort (GSE62254) was used for external validation.
Results: Thirteen differentially expressed DNAm-driven genes were narrowed down to a six-gene signature (PODN, NPY, MICU3, TUBB6 and RHOJ were hypermethylated, and MYO1A was hypomethylated), which was associated with OS (P < 0.05) after survival and LASSO regression analyses. These differentially expressed genes (DEGs) with altered DNAm statuses were included in the prognostic risk score model. The univariate Cox regression analysis indicated that risk score, age, and number of positive lymph nodes were significantly associated with survival time in GC patients. The multivariate Cox regression analysis also indicated that these variables were significant prognostic factors for GC. A nomogram including these variables was constructed, and its performance in predicting the 1-, 3- and 5-year survival outcomes of GC patients was estimated through time-dependent receiver operating characteristic (ROC) curves. In addition, the clinical benefit of this model was revealed by decision curve analysis (DCA). Pathway enrichment analysis suggested that these DNAm-driven genes might impact tumor progression by affecting signaling pathways such as the “ECM RECEPTOR INTERACTION” and “DNA REPLICATION” pathways.
Conclusions: The altered status of the DNAm-driven gene signature (PODN, MYO1A, NPY, MICU3, TUBB6 and RHOJ) was significantly associated with the OS of GC patients. A nomogram incorporating risk score, age and number of positive lymph nodes can be conveniently used to facilitate the individualized prediction of OS in patients with GC.
Keywords: nomogram, risk score, gastric cancer, DNA methylation, prognosis

 Global reach, higher impact
Global reach, higher impact