Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(7):1230-1237. doi:10.7150/ijbs.39161 This issue Cite
Research Paper
Development and validation of a novel nomogram to predict overall survival in gastric cancer with lymph node metastasis
1. Department of Laboratory Medicine, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, China; Sun Yat-sen University Cancer Center, Guangzhou, China
2. State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, China
3. Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin's Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin, China
4. Guangzhou Medical University, Guangzhou, China
5. Department of Clinical Laboratory, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong, China
* Minjie Mao, Ao Zhang, Yi He and Lin Zhang contributed equally to this work.
Received 2019-8-9; Accepted 2019-12-26; Published 2020-2-10
Abstract
Gastric cancer (GC) with lymph node metastasis (LNM) at diagnosis is associated with a unstable prognosis and indefinite survival times. The aim of the present study was to construct and validate a model for the Overall survival (OS) estimation for patients with LNM. The nomogram was constructed to predict the OS for LNM-positive GC using the primary group of 836 patients and validated using an independent cohort of 411 patients. Factors in the nomogram were identified by multivariate Cox hazard analysis. The predictive capability of nomogram was evaluated by calibration analysis and decision curve analysis. Multivariate analysis suggested that eight pre-treatment characteristics were used for developing the nomogram. In the primary cohort, the C-index for OS prediction was 0.788 (95% CI: 0.753-0.823), while in validation cohort, the C-index for OS prediction was 0.769 (95% CI: 0. 720-0.818). The calibration plot for the probability of OS and decision curve analyses showed an optimal agreement. Based on the nomogram, we could divided patients into three groups: low-risk group, middle-risk group and a high-risk group(p <0.001).Taken together, we have provided an easy-to-used and accurate tool for predicting OS, furthermore could be used for risk stratification of OS of LNM-positive GC patients.
Keywords: gastric cancer, prognosis, nomogram, lymph node metastasis
Introduction
Gastric cancer (GC) is one of the leading causes of cancer mortality worldwide with about one million new cases reported every year, posing a burden to patients across the globe [1-3]. Lymph node metastasis (LNM) is the most common metastasis form of GC and the high mortality rate in GC is mostly influenced by formation of LNM. Therefore, LNM-positive GC patients are often nonresectable in advanced stage at the time of diagnosis, and is generally associated with a poor prognosis [4]. Lymph nodes are invaded by tumor cells to influence the immune system, which in turn is closely related to inflammation, and inflammation promotes tumor metastasis [5]. Recently, a novel systemic immune-inflammation index (SII) was developed and has proved to be a powerful prognostic indicator of poor outcome for hepatocellular carcinoma patients and colorectal cancer [6]. Besides, coagulation pathway is essential for the establishment of metastasis in experimental model systems. High serum LDH has been shown reported in previous studies to confer prognostic information in lymphoma [7], multiple myeloma [8], and small cell lung cancer [9]. Therefore, LNM-positive GC comprises a heterogeneous group of patients, and variation in inflammation, metabolism of tumor, blood routine examination and coagulation, we should pay more attention to accurate assessment.
Reliable prognostic information is desired for monitor of individual patients with LNM-positive GC. The most extensive staging system for GC is the Union for International Cancer Control (UICC)/American Joint Committee on Cancer (AJCC) tumor, lymph node and metastases (TNM) staging system[3,10]. According to the depth of primary tumor invasion (T stage), regional lymph node metastasis (N stage) and distant metastasis (M stage), the TNM staging system divides patients with GC into different stages [11]. TNM staging system ignores the biological heterogeneity of LNM-positive GC patients and it is inadequate for predicting recurrence, which cause large variations in the clinical practice even patients with similar treatment strategies [12,13]. Thus, some other factors should be considered for as a new prognostic nomogram for LNM-positive GC, such as SII based on lymphocyte counts, platelet counts, and neutrophil counts, tumor metabolism including LDH, and biomarker of GC including CA199, CEA.
The aim of this study was to generate and internally validate a nomogram based on widely available pretreatment clinical and laboratory data to improve our ability in predicting survival in LNM-positive GC patients.
Methods
Patient selection
A total of 1247 GC patients with Lymph node metastasis admitted to Sun Yat-sen University Cancer Center (SYSUCC, Guangdong, China) between December 2010 and July 2017 were enrolled. We randomly allocated the patients into two cohorts: primary cohort and validation cohort. All the patients were classified as the first record of hospitalizations and the clinical information and serum biomarkers were extracted from Electronic Medical Record (EMR) system and Laboratory Information System (LIS). The inclusion criteria were as follows: (1) patients with a confirmed histologically diagnosed of GC; (2) patients with Lymph node metastasis positive; (3) patients without second tumor, or indefinite diagnoses; (4) patients with complete clinical data; (5) patients without concomitant diseases associated with influenced plasma coagulation levels (i.e., VTE, pulmonary embolism, or disseminated intravascular coagulation (DIC) within 1 month of study onset or during the subsequent treatment); (6) patients who not regularly took procoagulant or anticoagulant therapy or took blood transfusions within 1 month of study onset. All patients provided written informed consent. The Institute Research Ethics Committee of the Sun Yat-Sen University Cancer Center, Guangzhou, China approved this study. It was conducted in accordance with the ethical standards of the World Medical Association Declaration of Helsinki. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www.researchdata.org.cn), with the approval RDD number as RDDA2020001396.
Predictor variables
The following variables of interest were collected for each patient: age, sex, TNM stage, Tumor stage, Metastasis, Lauren, platelet-lymphocyte ratio (PLR), neutrophil-lymphocyte ratio (NLR), (lymphocyte-mononuclyte)LMR, lactate dehydrogenase (LDH), prothrombin time (PT), activated partial thromboplastin time (APTT), thrombin time (TT), fibrinogen (Fbg), carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA199). Statistical analyses were performed using SPSS 16.0 (IBM, Chicago, IL, USA). The optimal cut-off points in our study were evaluated by minimum P value from log-rank ×2 statistics using the X-tile program [14] and continuous variables were transformed to categorical variables, while the categorical variables were classified based on clinical findings. Univariate and multivariate regression analysis was used to analyze the risk factors in the primary cohort.
Outcomes
The outcome of our study was overall survival (OS). OS was defined as the time from the diagnosis of HCC to the date of the last follow-up or death. All GC patients were advised to receive regular follow-ups after completion of the primary therapy according to clinical guidelines. Patients who did not visit our hospital as scheduled were telephoned for follow-ups to obtain the treatment information and living status (performed by The Medical Information Unit in our Cancer Center). The last follow-up occurred in September 2018.
Statistical analysis
Statistical analyses were performed using R for Windows (version 3.4.2, http://www.r-project.org/). A nomogram was formulated based on the results of multivariate analysis by the package of rms. We tested the accuracy of the nomograms by discrimination and calibration both in primary and externa validation cohort. The discrimination of the nomogram was measured by Harrell's C-index (C-index). The value of the C-index ranges from 0.5 to 1.0, with 0.5 indicating random chance and 1.0 indicating a perfect ability to correctly discriminate the outcome with the model. Then, the calibration curve of the nomogram model for the OS and decision curve analyses were performed. The total points of each patient were calculated according to the established Cox regression model, 3 groups of patients with different risk of prognosis (based on the total points) were delineated using the X-tile program [14] Survival curves were depicted by the Kaplan-Meier method, and using the dichotomized risk group as a factor, finally, compared using the log-rank test. All statistical tests were two-sided, and P values of less than 0.05 were considered to be statistically significant.
Results
Basic characteristics
In the development cohort, we included 836 consecutive LNM-positive GC patients, with 190(22.73%) patients died. While in the validation cohort, 411 patients were screened with 97(263.60%) patients died respectively. There are no significant differences between the two cohorts and basic characteristics are given in Table 1.
Biomarker selection
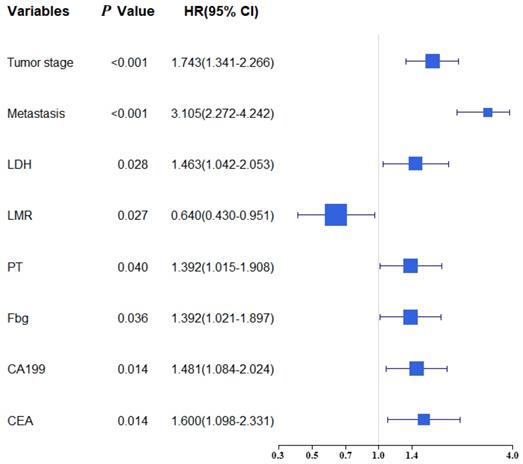
Univariate and multivariate analysis were selected for evaluating the clinicopathologic characteristics and blood biomarkers (Table 2). The univariate analyse indicated age, TNM stage, Tumor stage, Metastasis, PLR, NLR, LMR, LDH, PT, APTT, TT, Fbg, CA199 and CEA were related to OS. In the multivariate analysis, eight of fourteen biomarkers were contained in final model (Tumor stage, Metastasis, LDH, LMR, Fbg, PT, CA199 and CEA). A forest plot shows the hazard ratios and 95% confidence intervals for OS according to the Cox proportional hazards regression analysis (Figure 1).
Development and Validation of the Prediction Model
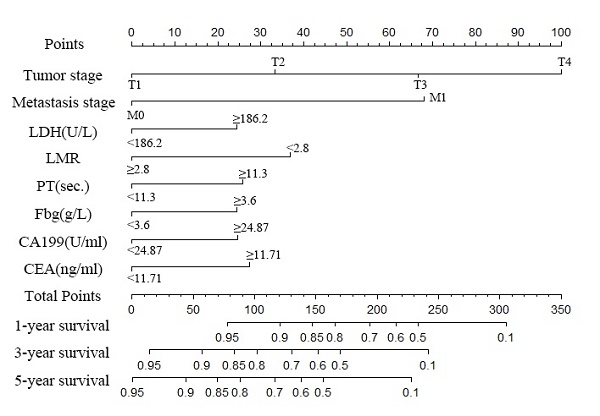
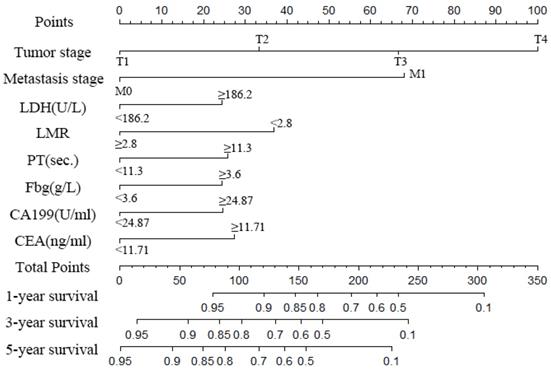
A nomogram was created to predict the probability of a particular outcome. Figure 2 shows the nomogram predicting 1-, 3- and 5- year OS that was constructed that incorporated the above independent predictors. In the primary cohort, the C-index for OS prediction was 0.788 (95% CI: 0.753-0.823). Figure 3 showed that the calibration plot for the probability of OS at 1, 3 or 5 year after therapy demonstrated good agreement between the prediction by nomogram and actual observation. Furthermore, the discrimination of the nomogram have been compared with AJCC TNM Stage, the C-index of nomogram was 0.788 (95% CI: 0.753-0.823), which was superior to the C-index of AJCC TNM Stage 0.719 (95% CI: 0.687-0.751).
Baseline clinical features
| Development cohort (n=836) | Validation Cohort(n=411) | |
|---|---|---|
| Characteristics | Mean±SD/ No(%) | Mean±SD/ No(%) |
| Age, year | 57.10±11.94 | 57.89±11.58 |
| Sex | ||
| Male | 527(63.04%) | 272(66.18%) |
| Female | 309(36.96%) | 139(33.82%) |
| TNM stage | ||
| Ⅰ | 17(2.03%) | 5 (1.22%) |
| Ⅱ | 99(11.84%) | 45(10.95%) |
| Ⅲ | 453(54.19%) | 218(53.04%) |
| Ⅳ | 267(31.94%) | 143(34.79%) |
| Tumor stage | ||
| T1 | 37(4.43%) | 13 (3.16%) |
| T2 | 49(5.86%) | 20(4.87%) |
| T3 | 308(36.84%) | 151(36.74%) |
| T4 | 442(52.87%) | 227(55.23%) |
| Metastasis | ||
| No | 569(68.06%) | 268(65.21%) |
| Yes | 267(31.94%) | 143(34.79%) |
| Lauren | ||
| 1 | 270(32.30%) | 134(32.60%) |
| 2 | 345(41.27%) | 155(37.71%) |
| 3 | 221(26.44%) | 122(29.68%) |
| PLR | ||
| ≤290.8 | 729(87.20%) | 362(88.08%) |
| >290.8 | 107(12.80%) | 49(11.92%) |
| NLR | ||
| ≤2.8 | 513(61.36%) | 256(62.29%) |
| >2.8 | 323(38.63%) | 155(37.71%) |
| LMR | ||
| ≤2.8 | 220(26.31%) | 116(28.22%) |
| >2.8 | 616(73.68%) | 295(71.78%) |
| LDH | ||
| ≤186.2 | 145.12±21.22 | 148.01±21.16 |
| >186.2 | 255.51±241.22 | 245.99±90.26 |
| PT, S | ||
| ≤11.3 | 10.69±0.39 | 10.65±0.40 |
| >11.3 | 12.11±1.00 | 11.97±0.60 |
| APTT | ||
| ≤31.2 | 25.45±2.94 | 25.47±2.84 |
| >31.2 | 34.16±3.20 | 33.25±1.68 |
| TT | ||
| ≤18.0 | 16.98±0.71 | 16.94±0.79 |
| >18.0 | 19.20±1.34 | 19.13±1.20 |
| Fbg,g/L | ||
| ≤3.6 | 2.79±0.52 | 2.74±0.53 |
| >3.6 | 4.39±0.65 | 4.36±0.63 |
| CEA | ||
| ≤11.71 | 2.73±2.11 | 2.84±2.14 |
| >11.71 | 116.45±259.56 | 256.67±929.68 |
| CA199 | ||
| ≤24.87 | 9.56±5.82 | 10.01±6.27 |
| >24.87 | 523.52±1775.40 | 467.01±1347.50 |
Data are presented as mean (SD) or N (%).
Univariate and multivariate cox hazards analysis between clinical features and OS
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Characteristics | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, year | 1.437(1.076-1.919) | 0.014 | 1.339(0.988-1.815) | 0.060 |
| Sex | ||||
| Male/Female | 0.892(0.762-1.367) | 0.892 | ||
| TNM stage | ||||
| Ⅰ/Ⅱ/Ⅲ/Ⅳ | 3.537(2.751-4.547) | <0.001 | ||
| Tumor stage | ||||
| T1/ T2/ T3/ T4 | 2.040(1.601-2.599) | <0.001 | 1.743(1.341-2.266) | <0.001 |
| Metastasis | ||||
| No/ Yes | 4.271(3.199-5.702 | <0.001 | 3.105(2.272-4.242) | <0.001 |
| Lauren | ||||
| 1/2/3 | 1.006(0.836-1.211) | 0.952 | ||
| PLR | ||||
| ≤290.8/>290.8 | 2.118(1.494-3.003) | <0.001 | 1.432(0.935-2.194) | 0.099 |
| NLR | ||||
| ≤2.8/>2.8 | 2.092(1.573-2.783) | <0.001 | 1.007(0.699-1.449) | 0.972 |
| LMR | ||||
| ≤2.8/>2.8 | 0.402(0.300-0.538) | <0.001 | 0.640(0.430-0.951) | 0.027 |
| LDH | ||||
| ≤26.9/>26.9 | 1.885(1.382-2.571) | <0.001 | 1.463(1.042-2.053) | 0.028 |
| PT, S | ||||
| ≤11.9/>11.9 | 1.770(1.321-2.373) | <0.001 | 1.392(1.015-1.908) | 0.040 |
| APTT | ||||
| ≤11.9/>11.9 | 1.570(1.036-2.379) | 0.033 | 1.297(0.830-2.027) | 0.253 |
| TT | ||||
| ≤11.9/>11.9 | 0.535(0.398-0.720) | <0.001 | 0.836(0.609-1.148) | 0.269 |
| Fbg,g/L | ||||
| ≤3.6/>3.6 | 2.050(1.525-2.757) | <0.001 | 1.392(1.021-1.897) | 0.036 |
| CEA | ||||
| ≤11.71/>11.71 | 2.688(1.926-3.752) | <0.001 | 1.600(1.098-2.331) | 0.014 |
| CA199 | ||||
| ≤24.87/>24.87 | 2.416(1.798-3.246) | <0.001 | 1.481(1.084-2.024) | 0.014 |
Validation the Predictive Accuracy of Nomograms for OS
Validation was performed by using the other cohort of 411 LNM-positive GC patients. In the validation cohort, independent risk factors included in the nomogram were examined. Also, the C-index for OS prediction was up to 0.769 (95% CI: 0. 720-0.818). Figure 3 showed that the calibration plot for the probability of OS at 1, 3 or 5 year after therapy demonstrated good agreement between the prediction by nomogram and actual observation.
Decision curve analysis
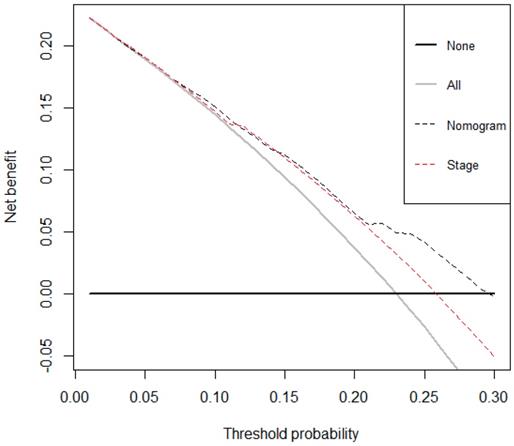
The decision curve analysis for the nomogram and TNM staging systems is presented in Figure 4. The decision curve demonstrated that if the threshold probability of a patient is > 10%, the developed nomogram and TNM staging system in predicting OS is more benefit than all patients dead scheme or none patients dead scheme. Furthermore, the net benefit was comparable, the nomogram in predicting OS is more benefit than that of TNM staging system in this range.
Forest plot showed the hazard ratio and 95% confidence interval for OS according to the Cox regression analysis.
Nomogram to predict the probability of one-, three- and five-year overall survival (OS), including tumour stage, metastases stage, and LDH, LMR, PT, Fbg, CEA and CA199 levels in GC patients with LNM. The nomogram can be used to obtain the probability of one-, three- and five-year survival by adding up the points identified on the point scale for each variable.
Calibration curve of the nomogram both in the primary and validation cohort. A. One-year OS in the primary cohort; B. Three-year survival OS in the primary cohort; C. Five-year survival OS in the primary cohort; D. One-year OS in the validation cohort; E. Three-year survival OS in the validation cohort; F. Five-year survival OS in the validation cohort.

Risk stratification of OS by the nomogram model
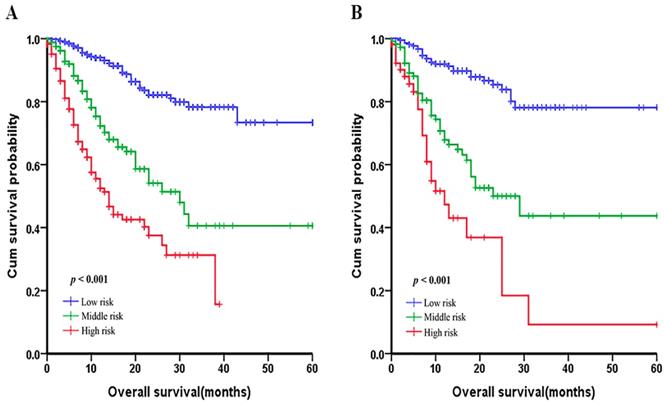
In order to evaluate the subgroups of patients that were positively influenced by nomogram, we divided patients into three groups: low-risk group, middle-risk group and a high-risk group both in development cohort and validation cohort, which showed good prognostic classification for LNM-positive GC patients. In the primary cohort, the OS between the three risk groups were (17.35±11.90) months, (13.21±11.06) months, (7.25±6.54) months (p <0.001). Also, in the validation cohort, the mean OS between the three risk groups were (17.47±11.93) months, (14.30±11.23) months and (8.62±9.85) months (p <0.001) (Figure 5).
Decision curve analysis for overall survival. Black line: All patients died. Grey line: None of the patients died. Dashed black line: Nomogram model. Dashed red line: TNM staging system model.
Graphs showing the Kaplan-Meier curves for all three groups based on the predictors from the nomogram model in the primary cohort (A) and those in the validation cohort (B).
Discussion
The present prognostic nomogram derived from prospectively collected data on 836 patients was shown to provide improved ability for individualized survival prediction in patients with LNM. By using this nomogram, individualization of patient counselling and decision-making regarding management can be improved.
As one of the most common malignant neoplasms in the digestive system, gastric cancer results in the death of thousands every year [15]. Recurrence and metastasis is the major cause of GC treatment failure and death. Lymph node metastasis and organ invasion at diagnosis are the most important poor prognostic factors [16]. In LNM-positive GC patients, the possibility to extend survival has been a topic of exploration for investigators globally, as improving life is always a consistent goal. Compared with LNM-negative patients, the overall recurrence rate in LNM-positive GC patients is obviously higher and the overall survival of LNM-positive GC patients is significantly shorter [17,18]. Thus, the accurate tumor prognosis after definitive treatment is indispensable for LNM-positive GC patients.
Some studies have demonstrated that Systemic immune-inflammation index (SII) was developed and has proved to be a powerful prognostic indicator in hepatocellular carcinoma [19], and renal cell carcinoma patients [20], which based lymphocyte counts, neutrophil counts, and platelet counts. It is a close and complicated in peripheral blood where inflammation-based cells interact with tumor distant metastasis [5]. The abnormal counts and ratio of neutrophil, lymphocyte, monocytes in the peripheral blood of cancer patients may be associated with tumor development [21]. Platelet lymphocyte ratio (PLR) has been reported as an independent risk factor in gastric cancer [6,22]. And it is reported that the coagulation pathway is essential for the establishment of metastasis in experimental model systems [23,24]. Therefore, inflammatory markers in combination with other clinical characteristics may provide more reliable value for the LNM-positive GC patients.
At present, the TNM staging system as a tool to evaluate prognosis of patients who had gastric cancer, is current and accepted by commons, but it exists some limitations [25]. The system only considers the anatomical extent of GC, ignoring the systemic immune-inflammation and coagulation heterogeneity of LNM-positive GC patients [26]. Hence, we aimed to generate and validate a nomogram to overcome the above mentioned shorts of the TNM staging system as a means for survival prediction and treatment strategies guidance in patients with LNM. In our study, we found that T stage, M stage, LDH, LMR, PT, Fbg, CA199 and CEA were the factors that influenced prognosis of patients according to the multivariate analysis. Patients with an earlier stage of T stage, M stage, and a lower LDH, LMR, PT, Fbg, CA199 and CEA level have improved survival rates. Not only should we consider the impact of anatomical extent, but comprehensive systemic inflammatory biomarkers, metabolism of tumor, coagulation and traditional biomarkers, thus we developed more accurate prediction of the patient's prognosis.
Our nomogram is also a useful tool that utilizes conveniently available clinical information to provide simple prognostic information for oncologists and patients from complex statistical analysis. However, a major problem is to provide an accurate estimate of prognosis, especially, in patients with incurable cancers. Compared with the traditional TNM staging system, our method is more accurate and has a higher coincidence rate for patients with LNM. Our method combines other biomarkers including the systemic inflammatory biomarkers, coagulation, metabolism of tumor and traditional tumor biomarkers with the TNM stage, taking into account anatomy and LNM conditions, and more accurately predicts patients 1-year OS, 3- year OS, and 5-year OS. Simultaneously, the decision curve showed that the nomogram in predicting OS is better than that of TNM staging system in all range.
There are also some limitations in our study. First, lacking multi-center research data, the nomogram was created based on single data source and tested with only one external cohort in China. Second, there are no molecular or genetic biomarkers included in nomogram that have been reported to have independent prognostic value. Finally, in the validation cohort, the follow-up time was shorter, and patients in the validation cohort still needed close monitoring and five-year follow-up data. In addition, future research can incorporate the LNM-positive GC patient's quality of life into the research system, and the nutritional status and quality of life of LNM-positive GC patients during the survival period have the same important status as the prolonged survival time. Despite these limitations, this nomogram represents a prognostic effect on patients with LNM-positive GC. We anticipate that this nomogram will stimulate ongoing research and multiple-center clinical research with wider geographic recruitment will further improve and validate it.
Conclusions
The present nomogram can predict the prognosis of LNM-positive GC patients, potentially facilitating highly tailored patient management. This nomogram represents an improvement in prognostication over the current TNM stage.
Abbreviations
GC: gastric cancer; LNM: lymph node metastasis; OS: Overall Survival; SII: systemic immune-inflammation index; TNM: tumor, lymph node and metastases; DIC: disseminated intravascular coagulation; PLR: platelet-lymphocyte ratio; NLR: rationeutrophil-lymphocyte ratio; LMR: lymphocyte-mononuclyte ratio; LDH: lactate dehydrogenas; PT: Prothrombin time; APTT: activated partial thromboplastin time; TT: thrombin time; Fbg: fibrinogen; CEA: carcinoembryonic antigen; CA199: carbohydrate antigen 19-9
Acknowledgements
We thank the staff of the biochemical laboratory of Sun Yat-sen University Cancer Center who provided various biochemical markers, and all of the staff who supported our study.
Author Contributions
Xueping Wang and Guanmin Jiang contributed to the conception and design of the study and drafted the manuscript; Minjie Mao, Yi He and Lin Zhang contributed to data analysis and interpretation; Ao Zhang contributed to manuscipt modification; Wen Liu, Yiling Song and Shuqi Chen participated in data collection and literature research. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Duarte HO, Gomes J, Machado JC, Reis CA. Gastric cancer: Basic aspects. HELICOBACTER. 2018;23(Suppl 1):e12523
2. He Y, Mao M, Shi W, He Z, Zhang L, Wang X. Development and validation of a prognostic nomogram in gastric cancer with hepatitis B virus infection. J TRANSL MED. 2019;17:98
3. Chae S, Lee A, Lee JH. The effectiveness of the new (7th) UICC N classification in the prognosis evaluation of gastric cancer patients: A comparative study between the 5th/6th and 7th UICC N classification. GASTRIC CANCER. 2011;14:166-71
4. Yang B, Jing C, Wang J, Guo X, Chen Y, Xu R, Peng L, Liu J, Li L. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. CLIN TRANSL ONCOL. 2014;16:374-9
5. Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. MOL CANCER RES. 2006;4:221-33
6. Zheng L, Zou K, Yang C, Chen F, Guo T, Xiong B. Inflammation-based indexes and clinicopathologic features are strong predictive values of preoperative circulating tumor cell detection in gastric cancer patients. CLIN TRANSL ONCOL. 2017;19:1125-1132
7. Zhong H, Chen J, Cheng S, Chen S, Shen R, Shi Q, Xu P, Huang H, Zhang M, Wang L, Wu D, Zhao W. Prognostic nomogram incorporating inflammatory cytokines for overall survival in patients with aggressive non-Hodgkin's lymphoma. EBIOMEDICINE. 2019;41:167-174
8. Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, Richardson P, Caltagirone S, Lahuerta JJ, Facon T, Bringhen S, Gay F, Attal M, Passera R, Spencer A, Offidani M, Kumar S, Musto P, Lonial S, Petrucci MT, Orlowski RZ, Zamagni E, Morgan G, Dimopoulos MA, Durie BG, Anderson KC, Sonneveld P, San MJ, Cavo M, Rajkumar SV, Moreau P. Revised international staging system for multiple myeloma: A report from international myeloma working group. J CLIN ONCOL. 2015;33:2863-9
9. Hsieh AH, Tahkar H, Koczwara B, Kichenadasse G, Beckmann K, Karapetis C, Sukumaran S. Pre-treatment serum lactate dehydrogenase as a biomarker in small cell lung cancer. Asia Pac J Clin Oncol. 2018;14:e64-e70
10. Marano L, Boccardi V, Braccio B, Esposito G, Grassia M, Petrillo M, Pezzella M, Porfidia R, Reda G, Romano A, Schettino M, Cosenza A, Izzo G, Di Martino N. Comparison of the 6th and 7th editions of the AJCC/UICC TNM staging system for gastric cancer focusing on the "N" parameter-related survival: The monoinstitutional NodUs Italian study. WORLD J SURG ONCOL. 2015;13:215
11. Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93-99
12. Jiao XG, Deng JY, Zhang RP, Wu LL, Wang L, Liu HG, Hao XS, Liang H. Prognostic value of number of examined lymph nodes in patients with node-negative gastric cancer. World J Gastroenterol. 2014;20:3640-8
13. Mita MT, Marchesi F, Cecchini S, Tartamella F, Ricco' M, Abongwa HK, Roncoroni L. Prognostic assessment of gastric cancer: Retrospective analysis of two decades. Acta Biomed. 2016;87:205-11
14. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. CLIN CANCER RES. 2004;10:7252-9
15. Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. The incidences and mortalities of major cancers in China, 2009. CHIN J CANCER. 2013;32:106-12
16. Ma M, Chen S, Zhu BY, Zhao BW, Wang HS, Xiang J, Wu XB, Lin YJ, Zhou ZW, Peng JS, Chen YB. The clinical significance and risk factors of solitary lymph node metastasis in gastric cancer. PLOS ONE. 2015;10:e0114939
17. Suzuki H, Oda I, Abe S, Sekiguchi M, Mori G, Nonaka S, Yoshinaga S, Saito Y. High rate of 5-year survival among patients with early gastric cancer undergoing curative endoscopic submucosal dissection. GASTRIC CANCER. 2016;19:198-205
18. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
19. Hu B, Yang XR, Xu Y, Sun YF, Sun C, Guo W, Zhang X, Wang WM, Qiu SJ, Zhou J, Fan J. Systemic immune-inflammation index predicts prognosis of patients after curative resection for hepatocellular carcinoma. CLIN CANCER RES. 2014;20:6212-22
20. Chrom P, Zolnierek J, Bodnar L, Stec R, Szczylik C. External validation of the systemic immune-inflammation index as a prognostic factor in metastatic renal cell carcinoma and its implementation within the international metastatic renal cell carcinoma database consortium model. INT J CLIN ONCOL. 2019;24:526-532
21. Botta C, Barbieri V, Ciliberto D, Rossi A, Rocco D, Addeo R, Staropoli N, Pastina P, Marvaso G, Martellucci I, Guglielmo A, Pirtoli L, Sperlongano P, Gridelli C, Caraglia M, Tassone P, Tagliaferri P, Correale P. Systemic inflammatory status at baseline predicts bevacizumab benefit in advanced non-small cell lung cancer patients. CANCER BIOL THER. 2013;14:469-75
22. Hirahara T, Arigami T, Yanagita S, Matsushita D, Uchikado Y, Kita Y, Mori S, Sasaki K, Omoto I, Kurahara H, Maemura K, Okubo K, Uenosono Y, Ishigami S, Natsugoe S. Combined neutrophil-lymphocyte ratio and platelet-lymphocyte ratio predicts chemotherapy response and prognosis in patients with advanced gastric cancer. BMC CANCER. 2019;19:672
23. Gil-Bernabe AM, Lucotti S, Muschel RJ. Coagulation and metastasis: What does the experimental literature tell us? Br J Haematol. 2013;162:433-41
24. Falanga A, Marchetti M, Vignoli A. Coagulation and cancer: Biological and clinical aspects. J THROMB HAEMOST. 2013;11:223-33
25. Mahar AL, Zagorski B, Kagedan D, Dixon M, El-Sedfy A, Vasilevska-Ristovska J, Cortinovis D, Rowsell C, Law C, Helyer L, Paszat L, Coburn N. Evaluating TNM stage prognostic ability in a population-based cohort of gastric adenocarcinoma patients in a low-incidence country. CAN J PUBLIC HEALTH. 2018;109:480-488
26. Barchi LC, Yagi OK, Jacob CE, Mucerino DR, Ribeiro UJ, Marrelli D, Roviello F, Cecconello I, Zilberstein B. Predicting recurrence after curative resection for gastric cancer: External validation of the Italian Research Group for Gastric Cancer (GIRCG) prognostic scoring system. Eur J Surg Oncol. 2016;42:123-31
Author contact
![]() Corresponding authors: Xueping Wang, Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, 651 Dong Feng Road East, Guangzhou 510060, Guangdong Province, China. Tel: +86-20-87343804, Email: wangxueporg.cn. Guanmin Jiang, Department of Clinical Laboratory, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai 519000, Guangdong Province, China. Email: jianggm3sysu.edu.cn.
Corresponding authors: Xueping Wang, Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, 651 Dong Feng Road East, Guangzhou 510060, Guangdong Province, China. Tel: +86-20-87343804, Email: wangxueporg.cn. Guanmin Jiang, Department of Clinical Laboratory, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai 519000, Guangdong Province, China. Email: jianggm3sysu.edu.cn.

 Global reach, higher impact
Global reach, higher impact