10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(8):1324-1334. doi:10.7150/ijbs.40551 This issue Cite
Research Paper
MSTN Mutant Promotes Myogenic Differentiation by Increasing Demethylase TET1 Expression via the SMAD2/SMAD3 Pathway
1. State Key Laboratory of Reproductive Regulation & Breeding of Grassland Livestock, Inner Mongolia University, Hohhot, 010070, China
2. School of Life Science, Inner Mongolia University, Hohhot, 010070, China
Received 2019-9-23; Accepted 2020-1-31; Published 2020-2-21
Abstract
Myostatin (MSTN) is mostly expressed in skeletal muscle and plays crucial roles in the negative regulation of muscle mass development. The methylation and demethylation of myogenesis-specific genes are major regulatory factors in muscle satellite cell differentiation. The present study was designed to investigate the mechanism of myogenic differentiation regulated by MSTN mutation (MT) and the methylation/demethylation state of downstream genes. The results showed that, in the MSTN-/+ satellite cells, a higher myotube fusion index and a larger myotube length were observed compared to the wild type controls; the genes associated with myogenesis were all up-regulated compared to the WT controls. The methylation of the promoters and gene bodies of PAX3, PAX7, MyoD, and MyoG were all down-regulated, while the expression of the key demethylase TET1 was significantly promoted. ChIP-qPCR was used to demonstrate that the SMAD2/SMAD3 complex combined with the promoter of TET1 to inhibit the activity of TET1 promoter, indicating that MSTN may regulate TET1 via SMAD2/SMAD3. The overexpression of TET1 in wild type cells promoted myogenic differentiation, increased the myotube index, and reduced the methylation of the associated genes. On the contrary, the knockdown of TET1 in the MSTN mutant cells resulted in the opposite phenomena as in the overexpressed cells. In conclusion, the myostatin mutant showed an increased transcriptional activity of TET1, inducing higher levels of demethylation and improving the transcriptional activity levels of myogenic differentiation-associated genes. The binding of SMAD2/SMAD3 directly to the TET1 promoter region indicated that the MSTN mutant demethylated the myogenesis-specific genes by up-regulating TET1, which is directly controlled by SMAD2/SMAD3.
Keywords: MSTN mutant, myogenic differentiation, ten-eleven translocation methylcytosine dioxygenase 1 (TET1), SMAD2/SMAD3, DNA methylation
Introduction
Myostatin (MSTN), also known as growth/ differentiation factor-8, is a member of the transforming growth factor β (TGF-β) family; it is mostly expressed in skeletal muscle and plays a crucial role in the negative regulation of muscle mass development [1]. Genetic studies have demonstrated that MSTN deficiency leads to muscle hypertrophy due to a combination of increased myofiber numbers and increased myofiber sizes in multiple species, including humans, sheep, dogs and cattle [1-5], without causing severe adverse consequences. Therefore, extensive efforts have been made to develop effective strategies for blocking the expression of MSTN in order to increase the muscle mass of animals. Luo et al. (2014) reported on the production of MSTN knockout cattle mediated by Zinc finger nuclease [6], Wang et al. (2017) produced MSTN knockout pigs by CRISPR/Cas9 [7], Yu et al. (2016) [8] and Wang et al. (2018) [9] produced MSTN knockout goats by TALEN and CRISPR/Cas9, and Zhang et al. (2018) produced a fat-1 transgenic goat integrated at the MSTN locus. All of these MSTN knockout animals showed improved skeletal muscle development [10].
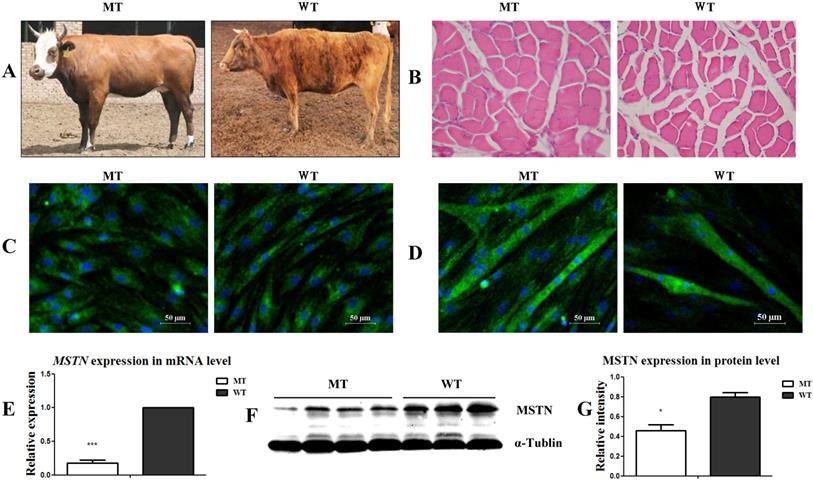
Identification of the muscle satellite cells from MSTN mutant (MT) and wild type (WT) cattle. (A) Samples from MT and WT cattle showing different muscular morphologies. (B) Cross-sectional images of muscle tissues from MT and WT cattle (200×). (C) Identification of satellite cells with MyoD expression. (D) Identification of differentiated myotubes with the expression of MHC. (E) Expression of MSTN mRNA in the satellite cells. (F, G) Expression of MSTN protein in satellite cells. *P<0.05, **P<0.01, ***P<0.001; t-tests were used to calculate the p values.
Epigenetic events regulate the quiescent and proliferation state of muscle satellite cells and their progeny. DNA methylation is a major repressive mechanism of muscle satellite cell differentiation [11, 12], whereas demethylation, along with MyoD and myogenin, are required for the initiation of the differentiation program [11]. Previous studies have proven that the CpG islands of MYF5 undergo hypomethylation during myogenic differentiation [13]. The modulation of methylation via prolonged treatment with 5-azacytidine, an inhibitor of DNA methylation, has been associated with an increased myogenic commitment of fibroblasts, mature adipocyte-derived dedifferentiated fat cells, and cardiac cells [14-17]. Furthermore, in C2C12 cells, treatment with 5-azacytidine resulted in an enhanced expression of muscle-specific genes (including myogenin) and increased myotube maturation, which suggested that the inhibition of methylation was a suitable tool to further boost myogenic differentiation in already committed precursors [18, 19].
Although both MSTN and DNA methylation are related to skeletal muscle development, the mechanism of MSTN and DNA methylation in muscle satellite cell differentiation remains unclear. In the present study, muscle satellite cells derived from MSTN mutant and wild type cattle were isolated and myogenic differentiation was induced. The myotube index including fusion rate and myotube length, expression and methylation levels of genes associated with myogenesis, and the expression of demethylase TETs were investigated. Our findings provide important insights into the roles of MSTN in epigenetic modification during myogenesis.
Results
Isolation and identification of the muscle satellite cells from MSTN mutant and wild type cattle
As shown in Figure 1A, the MSTN mutant (MT) cattle had a visibly stronger muscle morphology than the wild type (WT) cattle. The muscle myofiber cross-sectional areas (CSA) of the longissimus dorsi muscles in the MT cattle were larger than those in the WT cattle (Figure 1B). After culturing the muscle tissues for 4 days, the presumptive satellite cells were obtained. Cell identification showed that the MSCs markers CD90 and CD105 were positively expressed, while the hematopoietic lineage marker CD34 was negatively expressed (Figure S1A) in both the MT- and WT-derived cells. Further identification of the cells by differentiation induction showed that both the MT and WT cells positively expressed the myogenic markers of MyoD (Figure 1C) and MHC (Figure 1D) during myogenic differentiation. The expression of MSTN at both the mRNA (Figure 1E) and protein (Figure 1F, 1G) levels significantly decreased in the MT-derived satellite cells compared to that in the WT cells. The presumptive satellite cells also had the capacity for adipogenic and osteogenic differentiation (Figure S1B, S1C). These results confirm that the isolated cells were MSTN mutant (MT) and wild type (WT) cattle muscle satellite cells.
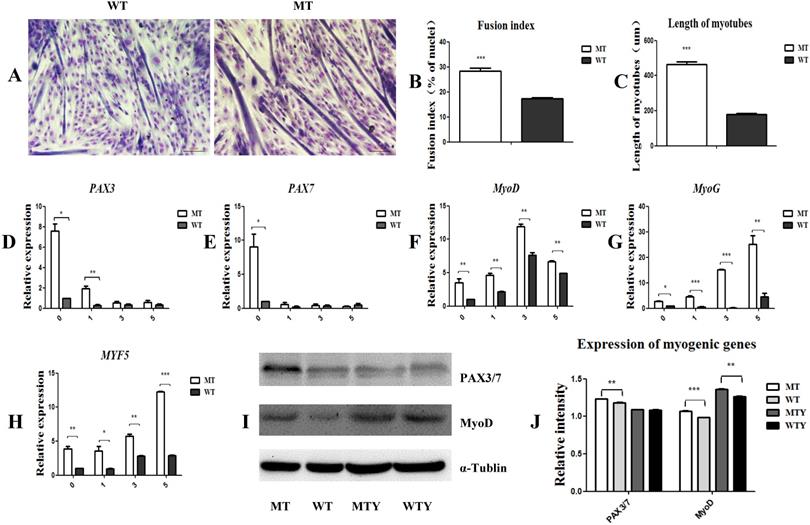
MSTN mutant promotes myogenic differentiation and increases the expression of myogenic differentiation-associated genes. (A) Differentiated myotubes in both MT and WT cells. (B) Fusion index. (C) Length of myotubes. (D-H) mRNA levels of myogenic-associated genes PAX3, PAX7, MyoD, MyoG, and MYF5 in the differentiated MT and WT muscle satellite cells. (I, J) Protein expression of PAX3/7 and MyoD genes in the differentiated MT and WT muscle satellite cells. WTY: Myotubes induced from WT cells; MTY: Myotubes induced from MT cells. *P<0.05, **P<0.01, ***P<0.001; t-tests were used to calculate the p-values.
MSTN mutant promoted myogenic differentiation and increased the expression of myogenic genes
Myogenic differentiation was induced in the satellite cells of both the MT and WT cattle. Three days later, the cells fused to form myotubes with a fusion index of 28.2% in the MT cells, which was significantly higher than that of the WT satellite cells (17.3%). The length of myotubes of the MT cells was higher than that of the WT cells (446.3892 ± 55.5 vs 117.4693 ± 24.1; P<0.05) (Figure 2A, 2B, and 2C). To further detect the expression of genes associated with myogenic differentiation, cells induced for 1, 3, and 5 days were collected, respectively, and the mRNA was extracted. PAX3, PAX7, MyoD, MyoG, and MYF5 were up-regulated in the MT satellite cells compared to those in the WT controls (Figure 2D-2J). These results indicate that the MSTN mutant promoted the myotube fusion index, elongated the myotube length, and increased the expression of the myogenic differentiation-associated genes.
MSTN mutant demethylated myogenesis-specific genes by up-regulation of demethylase TET1
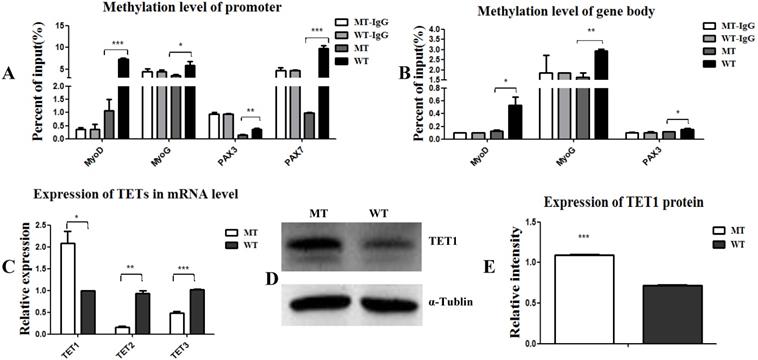
Epigenetic events, such as DNA methylation and demethylation of MyoD, MYF5, and MyoG, are known to play important roles in the regulation and initiation of differentiation [11, 13]. Here, we aimed to examine the methylation state of PAX3, PAX7, MyoD, and MyoG. As shown in Figure 3, the methylation levels of the promoters and gene bodies of PAX3, PAX7, MyoD, and MyoG were all down-regulated (Figure 3A, 3B), suggesting that the MSTN mutant demethylated myogenesis-specific genes, thus promoting the expression of these genes. To confirm this demethylation, we analyzed the activity of the demethylases TET1, TET2, and TET3 and found that TET1 expression was promoted, while TET2 and TET3 expression decreased (Figure 3C, 3D, and 3E). These results demonstrate that the MSTN mutant demethylated the myogenesis-specific genes by up-regulating demethylase TET1, thus promoting myogenic differentiation.
MSTN mutant demethylates myogenesis-specific genes by up-regulating demethylase TET1. (A, B) Methylation levels of myogenesis-specific genes of MyoD, MyoG, PAX3, and PAX7 in promoters (A) and gene bodies (B). (C) Expression of demethylase TET1, TET2, and TET3 at the mRNA levels. (D, E) Expression of demethylase TET1 at the protein level. *P<0.05, **P<0.01, ***P<0.001; t-tests were used to calculate the p-values.
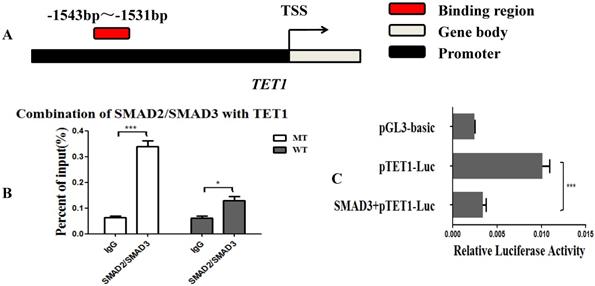
MSTN regulated TET1 expression via SMAD2/SMAD3
Since the MSTN mutant improved the transcriptional activity of TET1, we hypothesized that the SMAD2 and SMAD3 transcription factors, which are found downstream of the MSTN type II receptors, may bind with the promoter of TET1 and regulate the transcriptional activity of TET1. We used the JASPAR database (http://jaspar.binf.ku.dk/cgi-bin/jaspar_db.pl) for prediction analysis and found that SMAD2/SMAD3 bound to the TET1 (NC_037355.1) promoter region from ‒1543 to ‒1531 (Figure 4A, Figure S2A). To confirm this, we performed a ChIP assessment using an anti-SMAD2/SMAD3 monoclonal antibody with an amplified TET1 binding area. The detected regions spanned from ‒1613 to ‒1437 on the TET1 promoter. The ChIP-qPCR results showed that SMAD2/SMAD3 directly binds to the promoter of TET1 (Figure 4B, Figure S2B). To study the effect of SMAD2/SMAD3 on the transcriptional activity of TET1, we detected the transcriptional activity of TET1 using a Luciferase reporter assay after the overexpression of SMAD3. SMAD3 was found to inhibit the transcriptional activity of the TET1 promoter (Figure 4C), and the overexpression of SMAD3 was found to inhibit the expression of TET1 at both the mRNA and protein levels (Figure S3).
Overexpression of TET1 promoted myogenic differentiation
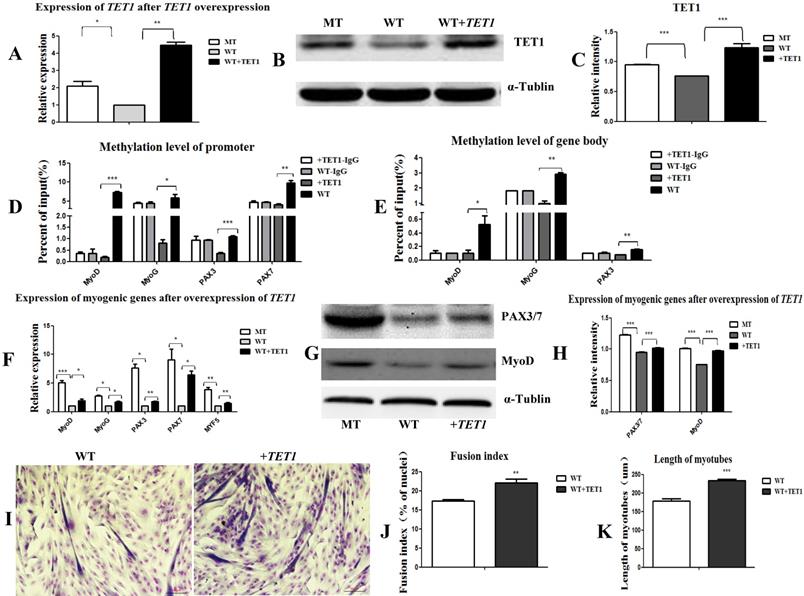
In the following experiment, we transfected the wild type satellite cells with the TET1 overexpression vector (Figure S4B) to further prove the regulatory role of TET1 in myogenic differentiation. As shown in Figure 5A, 5B, and 5C, the mRNA and protein expression levels of TET1 in the transgenic cells increased significantly. The methylation levels in the promoters (Figure 5D) and gene bodies of MyoD, MyoG, PAX3, and PAX7 (Figure 5E) decreased significantly. The mRNA and protein levels of the above genes were significantly promoted (Figure 5F, 5G, and 5H). After the induction of myogenic differentiation, the myotube fusion index (Figure 5I, 5J) was higher and the length of myotubes (Figure 5K) was larger in the TET1 overexpressed cells than in the controls.
Knockdown of TET1 inhibited myogenic differentiation
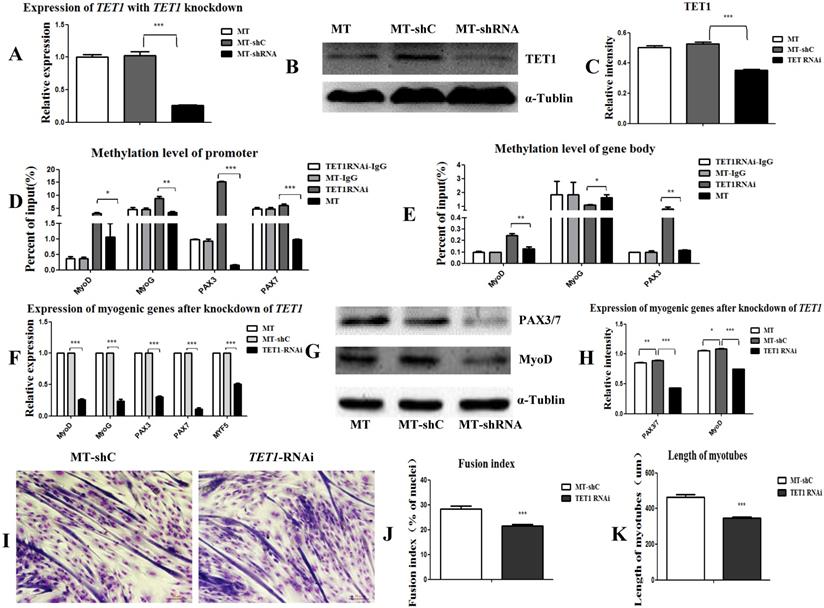
The overexpression of TET1 increased the myogenic differentiation and the expression of the associated genes. For the knockdown of TET1, we used satellite cells derived from the MSTN mutant muscle as target cells for transfection with TET1 shRNA. The results showed that the expression of TET1 at both mRNA and protein level decreased significantly (Figure 6A, 6B, and 6C). The methylation in the promotors (Figure 6D) and gene bodies (Figure 6E) of MyoD, MyoG, PAX3, and PAX7 was significantly promoted after the knockdown of TET1 in the mutation cells. The expression of these genes at both the mRNA and protein level was significantly decreased (Figure 6F, 6G, and 6H). The myotube fusion index was also significantly decreased (Figure 6I, 6J) and the length of the myotubes was shorter after TET1 knockdown (Figure 6K).
MSTN regulates TET1 expression via SMAD2/SMAD3. (A) The binding region of SMAD2/SMAD3 with the promoter of TET1. (B) The combination of TET1 promoter with SMAD2/SMAD3 detected by ChIP-qPCR. The detected binding region was from ‒1613 to ‒1437 on the TET1 promoter. (C) Luciferase reporter assays. HEK293T cells were transfected with luciferase reporter plasmids containing the promoter of TET1 (1800 bp). *P<0.05, **P<0.01, ***P<0.001; t-tests were used to calculate the p-values.
Overexpression of TET1 promotes myogenic differentiation. (A-C) The expression of TET1 at the mRNA and protein level after the overexpression of TET1 in the wild type muscle satellite cells. (D, E) Methylation levels of myogenesis-specific genes in the promoter and gene body. (F) Expression of myogenic genes at the mRNA level. (G, H) Expression of myogenic genes at the protein level. (I) The differentiated myotubes. (J) Myotube fusion index after the overexpression of TET1. (K) Length of myotubes. *P<0.05, **P<0.01, ***P<0.001; t-tests were used to calculate the p-values.
Knockdown of TET1 inhibits myogenic differentiation. (A-C) The expression of TET1 at the mRNA and protein level after knockdown of TET1 in MSTN mutant muscle satellite cells. (D, E) Methylation level of myogenesis-specific genes in promoter and gene body. (F) Expression of myogenic genes at the mRNA level. (G, H) Expression of myogenic genes at the protein level. (I) The differentiated myotubes. (J) Myotube fusion index after TET1 knockdown. (K) Length of myotubes. *P<0.05, **P<0.01, ***P<0.001; t-tests were used to calculate the p-values.
Discussion
MSTN is known to negatively regulate muscle development [1]. As a result, MSTN mutation leads to muscle hypertrophy growth, as reported previously in beef cattle [2], humans [3], goats [4], and dogs [5], resulting from a combination of an increase in the number and size of myofibers [1-5]. Since muscle mass is an important indicator in animal production, extensive efforts have been made to develop effective strategies that block the expression of MSTN for the production of animals with increased muscle masses [20]. Several studies have demonstrated that the overexpression of MSTN inhibits the myogenic process by downregulating the mRNA levels of muscle regulatory factors MyoD and MyoG [21, 22]. In contrast, the shRNA knockdown of endogenous MSTN results in the induction of myogenic differentiation [22]. Previous studies have also shown that the overexpression of MSTN inhibits SMAD3 and MyoD activity, resulting in the failure of myoblasts differentiate into myotubes [23]. In the present study, we isolated muscle satellite cells from MSTN mutant and wild type cattle. The MSTN -/+ satellite cells were found to have a higher myotube fusion index and larger myotube length than the control cells. The expression of genes associated with myogenic differentiation, such as MyoD, MyoG, PAX3, and PAX7, were also significantly higher in the MSTN -/+ satellite cells. These results indicate that the MSTN mutation increases the rate of muscle development by increasing the levels of myogenesis-specific gene expression.
DNA methylation is associated with gene silencing [24]. The levels of methylation are catalyzed by specific methyltransferases (DNMT) and demethylases (TET) [25, 26]. Various studies have demonstrated that DNA methylation is a major repressive mechanism of muscle satellite cell differentiation [11, 13], whereas demethylation, as well as MyoD and MyoG, are required for the initiation of the differentiation program [11]. In C2C12 cells, treatment with 5-azacytidine resulted in an enhanced expression of muscle-specific genes (including myogenin) and increased myotube maturation [18, 19], which suggests that DNA methylation plays an important role in the regulation of the differentiation of muscle satellite cells. In the present study, we found that the MSTN mutant promoted the expression of the demethylase TET1, but reduced the expression of TET2 and TET3. The methylation levels of myogenesis-specific genes were all decreased, especially in the promoter region. These results indicate that the myogenesis-specific genes MyoD, MyoG, PAX3, and PAX7 may be regulated by TET1. Additionally, previous studies have demonstrated that TETs play important roles in the regulation of muscle development [27-29] via the demethylation of myogenesis-specific genes. TET1 and TET2 were found to meditate the conversion of methylated cytosine into 5hmC, which is the first step in the DNA demethylation active pathway [30-32]. However, the overexpression of TET3 was found to not cause an obvious decrease in 5mC staining [33]. In this study, the expression of TET1 was up-regulated and the expression of TET2 and TET3 were down-regulated in the MSTN mutant cells. This may be due to the fact that the gene body region had low levels of methylation, such that the demethylation of myogenic genes mainly occurred in the promoter region. TET1 meditates the conversion of methylated cytosine into 5hmC, particularly in the promoter region [34], such that the demethylation of the MSTN mutant was mainly carried out by TET1. By contrast, TET2 and TET3 did not play an important role in this process. However, the reason for the down-regulation of TET2 and TET3 will require further study.
To further evaluate the role of TET1 during myogenic differentiation, TET1 was overexpressed in the muscle satellite cells of the wild type cattle. The results showed that the methylation levels of myogenesis-specific genes were significantly decreased, while the expression of these genes was visible increased and the myogenic differentiation was promoted. However, upon the knockdown of TET1 in the MSTN mutant muscle satellite cells, the methylation of the myogenic genes increased, while the expression levels of these genes were markedly reduced and the myogenic differentiation inhibited. These results further demonstrate that the MSTN mutant promoted myogenic differentiation via the demethylation of myogenesis-specific genes, particularly in the promoter region, via the up-regulation of TET1.
Since SMAD2 and SMAD3 are known to be downstream of the MSTN type II receptors, the combination of MSTN with its receptors results in the phosphorylation and activation of SMAD2 and SMAD3 [23, 35, 36]. As such, we hypothesized that the SMAD2/SMAD3 transcription factors are likely to interact directly with TET1. We predicted the binding of SMAD2/SMAD3 to the TET1 promoter region using the JASPAR database and performed ChIP assays with an anti-SMAD2/SMAD3 monoclonal antibody. Using ChIP-qPCR, we found that SMAD2/SMAD3 was able to bind with the promoter of TET1. Moreover, a Luciferase reporter assay indicated that the overexpression of SMAD3 inhibited the transcriptional activity of TET1. In the present study, the MSTN mutant was found to result in decreased SMAD2/SMAD3 activity [23, 35], which reduced the inhibition of SMAD2/SMAD3 by the transcriptional activity of TET1, leading to increased levels of TET1 activity. These increased levels of TET1 activity resulted in the demethylation of the myogenesis-specific genes, thereby promoting myogenic differentiation.
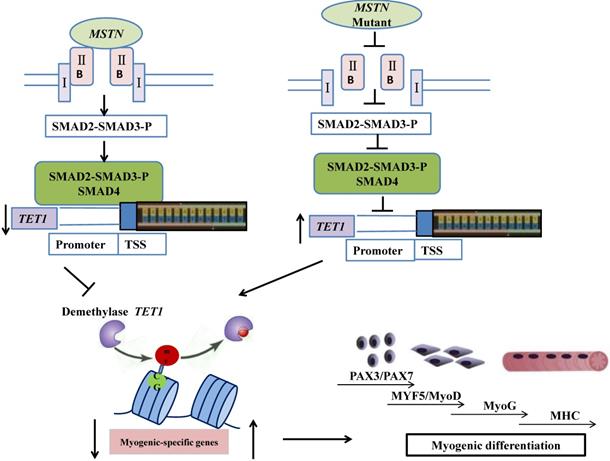
In conclusion, the mutation of myostatin resulted in increased levels of TET1 activity, which induced higher levels of demethylation and myogenic-associated gene activity. SMAD2/SMAD3 bound directly to the TET1 promoter region, which indicated that the MSTN mutant demethylated the myogenesis-specific genes by up-regulating TET1, which is directly controlled by SMAD2/SMAD3. The possible mechanisms by which the MSTN mutation inhibited SMAD2/SMAD3, activated TET1, and increased myogenesis-specific gene expression, subsequently inducing myogenic differentiation, are shown in Figure 7.
Materials and Methods
Ethics statement
All animal procedures were reviewed and approved by the Committee on the Ethics of Animal Experiments at Inner Mongolia University. All methods were performed in accordance with the relevant guidelines and regulations.
Animals
The MSTN -/+ cattle used in this study were a cross of female Mongolian yellow cattle and male MSTN -/- cattle, as our previous report [6]. Five 24-month-old female MSTN -/+ cattle and five wild type female cattle were selected for muscle sampling. The cattle were fed as ordinary beef cattle in a local livestock farm.
Muscle tissue collection
Longissimus dorsi muscles were harvested using a muscle-sampler, and immediately rinsed in 75% ethanol for 3 s, then transferred to phosphate-buffered saline (PBS) (Hyclone, Thermo Scientific, Marietta, OH, USA) containing antibiotics (100 U/ml of penicillin and 100 mg/mL of streptomycin) (Gibco-BRL, Grand Island, NY, USA). The samples were then immediately transferred to a biological safety cabinet (HF-1100, HK, China) for cell isolation. Part of the muscle was frozen in liquid N2 for RNA-Seq, while the rest of the samples were fixed in 4% paraformaldehyde for tissue slice preparation.
Histological preparation
For morphological analysis, the Longissimus dorsi muscle tissues from MSTN mutant (MT) and wild type (WT) cattle were fixed in 4% paraformaldehyde for 24 h, dehydrated using an alcohol-xylene series, and embedded in paraffin wax. The sample blocks were then cut into 5-μm sections using a microtome and routinely stained with hematoxylin and eosin (H&E). Images were captured using a microscope (DS-Ri1, Nikon).
Cell isolation and culture
The muscle samples were rinsed in PBS containing antibiotics three times, then transferred into 75% ethanol for 15 s and quickly transferred into PBS. The samples were rinsed another three times, cut into 1-3 mm slices, added 1 mg/mL collagenase IV, and plated at 38.5°C with 5% CO2 for 2-3 h. After the tissue had dissolved, the mixture was centrifuged at 1500 rpm/min for 5 min to collect the cells. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% fetal bovine serum (FBS) and 10% horse serum (HS) at 38.5℃ with 5% CO2. The cells were passaged after reaching 90% confluence.
Myogenic cells differentiation and immunostaining
When the cultured cells reached 80-90% confluence, myogenic differentiation was induced by low serum medium comprised of DMEM supplemented with 2% horse serum (Hyclone Ltd) for 5 days. Myogenic cells and myotubes were identified by immunostaining, as described previously [37]. Myogenic cells were incubated with primary antibodies against MyoD (1:200; Santa Cruz, USA) and myotubes were incubated with primary antibodies against MHC (1:200; Cell Signaling Technology, USA) at 4℃ overnight, then incubated with fluorescence-tagged secondary antibodies (1:500; Santa Cruz, USA) at 37℃ for 1 h. After incubation and mounting, the samples were stained with 4',6-diamidino-2-phenylindole (DAPI) for 20 min. Samples were observed and photographed using a confocal fluorescence microscope (A1R/A1; Nikon, Japan).
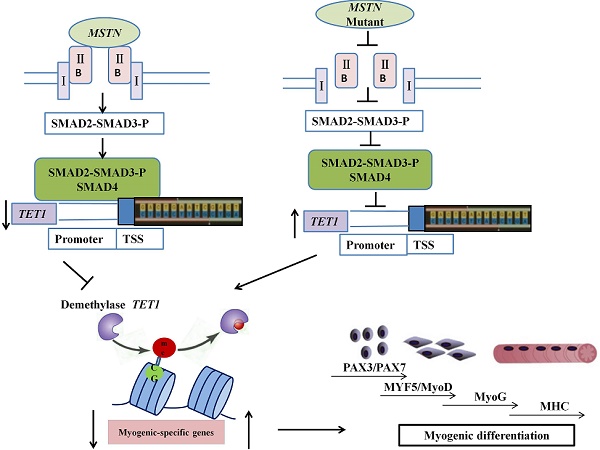
Schematic of mechanisms for the inhibition of SMAD2/SMAD3 and the activation of TET1 signaling by MSTN mutation. In the wild type model (left), MSTN combined with ActRIIB, followed by the combination of the complex with type I receptor, the activation of the GS region (Ser/Thr) of the receptor, and the transmission of the MSTN signal. The activated MSTN signal then combined and phosphorylated SMAD2/SMAD3, whereby pSMAD2/SMAD3 was activated and combined with SMAD4. This complex entered the nucleus to combine with the promoter of demethylase TET1, and inhibited the activity of TET1 promoter. In the MSTN mutant model (the right), the MSTN mutant decreased the binding between pSMAD2/SMAD3 and the promoter of TET1 and enhanced TET1 expression, which increased the demethylation of myogenesis-specific genes, including PAX3, PAX7, MyoD, and MyoG.
Fusion index calculation and myotube length measurement
Myotubes induced for 3 d were fixed with methanol for 10 min, then dyed with Giemsa dye (G1015; Solarbio, Beijing, China). Images were captured using a microscope (DS-Ri1; Nikon). The degree of differentiation was determined by calculating the fusion index (i.e. the ratio of nuclei in multinucleated cells to the total number of nuclei) [38] and measuring the length of myotubes (myotube lengths were carefully measured by drawing a line along the long axis of the myotube between two myotube tips and converting the pixels into the unit of length in ImageJ) [39].
Real-time PCR
The RNA from the myogenic cells and myotubes was isolated using an RNAiso Plus kit (9108; Takara), then reverse-transcribed into cDNA using a PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (RR047A; Takara). We amplified the cDNA using ABI7500 real-time PCR (Applied Biosystems, America) and SYBR Green (RR820A; Takara). The PCR primers are listed in Table S1. The protocol for PCR amplifications was as follows: 95℃ for 30 s, followed by 40 cycles at 95℃ for 5 s, 60℃ for 34 s, and a final melting curve stage. The cycle threshold (Ct) values of the targeted genes were normalized to the housekeeping gene GAPDH using the 2-ΔΔCt method [40].
Western blotting
The cell samples were rinsed with PBS and lysed in 150 μL of ice-cold Radio Immunoprecipitation Assay (RIPA) buffer. The cell lysates were then centrifuged at 4ºC for 30 min at 8000 × g. The supernatant was electrophoresed in a 10% SDS-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane by electroblotting. The membrane was blocked in 5% non-fat milk in Tris-buffered saline with 0.1% Tween-20 (TBST) blocking solution at room temperature for 1 h. The membrane was subsequently incubated with anti-MSTN (sc-134345; Santa Cruz, USA), anti-PAX3/7 (sc-7749; Santa Cruz, USA), anti-MyoD (Santa Cruz, USA), and anti-TET1 (sc-293186; Santa Cruz, USA) monoclonal antibodies at 1:500 in TBST. The membrane was then incubated for 1 h with a horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit secondary antibody at 1:10000 in TBST, followed by detection using the chemiluminescence labeling detection reagent ECL Plus (Thermo Scientific).
Prediction of CpG island
The CpG islands and CpG sites in the promoters and gene bodies of MyoD, MyoG, PAX3, and PAX7 were examined using MethPrimer software online (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi).
Methylated DNA immunoprecipitation (MeDIP)
MeDIP analysis was performed according to the manufacturer's instructions (ab117133; Abcam, USA). Total genomic DNA was sonicated (three pulses for 10-12 s at level 2 with 30-40 s intervals between pulses while resting on ice) to yield DNA fragments between 200 bp and 1000 bp in length (Sonics Vibra, USA). One microgram of fragmented DNA was heat denatured to produce single-stranded DNA. Immunoprecipitation was performed for 2 h at room temperature using 1 μL anti-5mC antibody, with 1 μL of normal mouse IgG as the negative control. The mDNA was released from the DNA/antibody complex using Proteinase K at 65℃ for 1 h. Finally, the mDNA was captured, eluted, and used for real-time PCR detection. The PCR primers are listed in Table S2.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed according to the manufacturer's instructions (26157; Thermo Fisher Scientific) using a previously described protocol [41]. Briefly, approximately 1 × 107 cells were fixed with 1% formaldehyde and quenched by glycine. The cells were washed three times with PBS, then harvested in ice-cold PBS containing 1% Halt Cocktail. The DNA was lysed and digested using MNase. The mixture was then sonicated on ice to break the nuclear membrane. The resulting lysate was incubated with anti-SMAD2+SMAD3 (ab202445; Abcam, USA) and protein G beads at 4℃ overnight. Normal rabbit IgG was used as a negative control. The beads were washed four times before eluting the DNA using ChIP elution buffer. The elution was incubated at 65℃ for 1.5 h. The DNA was then revived using a DNA purification kit. The purified DNA was assayed by quantitative PCR. Assays were repeated at least three times. The data represent the average values ± standard deviation (SD) of the representative experiments. The sequences of the primers used are provided in Table S3.
Luciferase reporter
The SMAD3 cDNA was amplified from the bovine cDNA and inserted into the pCAG-EGFP vector to obtain an overexpression vector of SMAD3. The promoter region of the TET1 gene was amplified from the bovine genomic DNA and inserted into the pGL3-Basic vector (Promega, Madison, WI, USA). For the luciferase reporter assays, HEK293T cells were seeded in 96-well plates and transfected with the indicated plasmids using Lipofectamine LTX Reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). Cells were collected 48 h after transfection. Luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega).
Transfection with TET1 short hairpin RNA (shRNA) and overexpression vector
TET1 shRNA and control shRNA were purchased form Santa Cruz (sc-154204-SH and sc-108060). The TET1 overexpression vector pIRES-hrGFPⅡ-TET1 (#83568) was purchased from Addgene. The knockdown and overexpression vectors were transfected using Lipofectamine LTX Reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. The transfected cells were then induced with low serum medium (2% HS) after 48 h of transfection.
Statistical analysis
The data of the MSTN mutant and wild type myoblasts were statistically compared using t-tests. Results are expressed as the mean ± standard error of the means (SEM). Differences with p-values of less than 0.05 were considered statistically significant.
Abbreviations
TGF-β: transforming growth factor β; MT: mutant; WT: wild type; CSA: cross-sectional area; DMEM: Dulbecco's modified Eagle's medium; RIPA: Radio Immunoprecipitation Assay; MeDIP: Methylated DNA immunoprecipitation; ChIP: Chromatin immunoprecipitation; shRNA: short hairpin RNA.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We would like to thank Dr. Yunping Dai, State Key Laboratory for Agrobiotechnology China Agricultural University, and Xinyu Zhou, Di Wu, Lin Zhu, Xueqiao Wang, Anqi Di, Guanghua Su and Xuefei Liu, School of Life Science Inner Mongolia University for their technical assistances. This work was supported by the National Science and Technology Major Project of China grant 2016ZX08007-002, Inner Mongolia Science and Technology Innovation Guide Project grant KCBJ2018002.
Author Contributions
LG and GL conceived and designed the experiments; LG and MY performed the experiments; MG, CB and ZW analyzed the data; LY and YW helped to collect muscle samples. LG and GL drafted and revised the manuscript. All authors read and approved the final version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mcpherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457-12461
2. Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in Double-Muscled Belgian Blue and Piedmontese Cattle. Genome Research. 1997;7:910-916
3. Schuelke M, Wagner KR, Stolz LE, Hübner C, Riebel T, Kömen W. et al. Myostatin mutation associated with gross muscle hypertrophy in a Child. New England J Med. 2004;350:2682-2688
4. Alex C, Fabienne M, Haruko T, Dimitri P, Xavier T, Bernard B. et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nature Genetics. 2006;38:813-818
5. Mosher DS, Pascale Q, Bustamante CD, Sutter NB, Mellersh CS, Parker HG. et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. Plos Genetics. 2007;3:e79
6. Luo J, Song Z, Yu S, Cui D, Wang B, Ding F. et al. Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PloS One. 2014;9(4):e95225
7. Wang K, Tang X, Xie Z, Zou X, Li M, Yuan H. et al. CRISPR/Cas9-mediated knockout of myostatin in Chinese indigenous Erhualian pigs. Transgenic Res. 2017;26:1-7
8. Yu B, Lu R, Yuan Y, Zhang T, Song S, Qi Z. et al. Efficient TALEN-mediated myostatin gene editing in goats.BMC Develop Biol. 2016;16:26.
9. Wang X, Niu Y, Zhou J, Zhu H, Ma B, Yu H. et al. CRISPR/Cas9-mediated MSTN disruption and heritable mutagenesis in goats causes increased body mass. Animal Genetics. 2018;49:43-51
10. Zhang J, Cui ML, Nie YW, Dai B, Li FR, Liu DJ. et al. CRISPR/Cas9-mediated specific integration of fat-1 at the goat MSTN locus. FEBS J. 2018;285(15):2828-2839
11. Daniela P, Pier LP. The epigenetic network regulating muscle development and regeneration. J Cell Physiol. 2010;207:1-11
12. Bigot A, Duddy WJ, Ouandaogo ZG, Negroni E, Mariot V, Ghimbovschi S. et al. Age-associated methylation suppresses SPRY1, leading to a failure of re-quiescence and loss of the reserve stem cell pool in elderly muscle. Cell Reports. 2015;13:1172-1182
13. Carrio E, Diezvillanueva A, Lois S, Mallona I, Cases I, Forn M. et al. Deconstruction of DNA methylation patterns during myogenesis reveals specific epigenetic events in the establishment of the skeletal muscle lineage. Stem Cells. 2015;33:2025-2036
14. Kazama T, Fujie M, Endo T, Kano K. Mature adipocyte-derived dedifferentiated fat cells can transdifferentiate into skeletal myocytes in vitro. Biochem Biophys Res Commun. 2008;377:780-785
15. Kaur K, Yang J, Eisenberg CA, Eisenberg LM. 5-azacytidine promotes the transdifferentiation of cardiac cells to skeletal myocytes. Cell Reprogram. 2014;16:324-330
16. Taylor SM, Jones PA. Multiple new phenotypes induced in 10T12 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771-779
17. Liu L, Harrington M, Jones PA. Characterization of myogenic cell lines derived by 5-azacytidine treatment. Develop Biol. 1986;117:331-336
18. Senesi P, Luzi L, Montesano A, Terruzzi I. DNA demethylation enhances myoblasts hypertrophy during the late phase of myogenesis activating the IGF-I pathway. Endocrine. 2014;47:244-254
19. Scarpa S, Lucarelli M, Palitti F, Carotti D, Strom R. Simultaneous myogenin expression and overall DNA hypomethylation promote in vitro myoblast differentiation. Cell Growth Differ. 1996;7:1051-1058
20. Fiems LO. Double muscling in cattle: Genes, husbandry, carcasses and meat. Animals. 2012;2(3):472-506
21. Ramón R, Isabel C, Arce VM, Jesús D. Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol. 2002;282:993-999
22. Liu C, Li W, Zhang X, Zhang N, He S, Huang J. et al. The critical role of myostatin in differentiation of sheep myoblasts. Biochem Biophys Res Commun. 2012;422:381-386
23. Langley B, Thomas MA, Sharma M, Sharma M, Gilmour S, Kambadur R. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J Biol Chem. 2002;277:49831-49840
24. Breuls N, Giacomazzi G, Sampaolesi M. (Epi)genetic modifications in myogenic stem cells: From novel insights to therapeutic perspectives. Cell. 2019;8(5):429
25. Smith ZD, Alexander M. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14:204-220
26. Aguirre-Arteta AM, Grunewald I, Cardoso MC, Leonhardt H. Expression of an alternative Dnmt1 isoform during muscle differentiation. Cell Growth Differ. 2000;11:551
27. Zhang X, Nie Y, Cai S, Ding S, Fu B, Wei H. et al. Earlier demethylation of myogenic genes contributes to embryonic precocious terminal differentiation of myoblasts in miniature pigs. FASEB J. 2019;33:9638-9655
28. Zhong X, Wang QQ, Li JW, Zhang YM, An XR, Hou J. Ten-Eleven Translocation-2 (Tet2) is involved in myogenic differentiation of skeletal myoblast cells in vitro. Scientific Reports. 2017;7:43539
29. Fei W, Minakhina S, Tran H, Changela N, Kramer J, Steward R. Tet protein function during Drosophila development. Plos One. 2018;13:e0190367
30. Hao W, Yi Z. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Develop. 2011;25:2436-2452
31. Myunggon K, Bandukwala HS, Jungeun A, Edward DL, Elizabeth CT, Ryan H. et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108:14566-14571
32. Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Develop. 2016;30:733
33. Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES cell self-renewal, and ICM specification. Nature. 2010;466(7310):1129-1133
34. Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J. et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111:1361-1366
35. Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mole Cell Biol. 2003;23:7230-7242
36. Roberta S, Giulia M, Maria P, Cristina M, Bert B, Reimar A. et al. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol. 2009;296:1248-1257
37. Wei ZY, Li DF, Zhu L, Yang L, Chen C, Bai C. et al. Omega 3 polyunsaturated fatty acids inhibit cell proliferation by regulating cell cycle in fad3b transgenic mouse embryonic stem cells. Lipids Health Dis. 2018;17:210
38. Oláh T, Fodor J, Ruzsnavszky O, Vincze J, Berbey C, Allard B. et al. Overexpression of transient receptor potential canonical type 1 (TRPC1) alters both store operated calcium entry and depolarization-evoked calcium signals in C2C12 cells. Cell Calcium. 2011;49:415-425
39. Hayashi S, Yonekura S. Thermal stimulation at 39°C facilitates the fusion and elongation of C2C12 myoblasts. Anim Sci J. 2019;90(8):1008-1017
40. Shen SJ, Zhang YH, Gu XX, Jiang SJ, Xu LJ. Yangfei Kongliu Formula, a compound Chinese herbal medicine, combined with cisplatin, inhibits growth of lung cancer cells through transforming growth factor-β1 signaling pathway. J Integrative Med. 2017;15:242-251
41. Gao L, Yang MM, Wang XQ, Yang L, Bai C, Li G. Mstn knockdown decreases the trans-differentiation from myocytes to adipocytes by reducing Jmjd3 expression via the SMAD2/SMAD3 complex. Biosci Biotech Biochem. 2019;83(11):2090-2096
Author contact
![]() Corresponding author: Guangpeng Li, State Key Laboratory of Reproductive Regulation and Breeding of Grassland Livestock, Inner Mongolia University, Hohhot, 010070, China. Tel: 86-10-471-5298583; E-mail: gpengliedu.cn
Corresponding author: Guangpeng Li, State Key Laboratory of Reproductive Regulation and Breeding of Grassland Livestock, Inner Mongolia University, Hohhot, 010070, China. Tel: 86-10-471-5298583; E-mail: gpengliedu.cn

 Global reach, higher impact
Global reach, higher impact