10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(9):1604-1615. doi:10.7150/ijbs.38930 This issue Cite
Research Paper
The lncRNA Rhno1/miR-6979-5p/BMP2 Axis Modulates Osteoblast Differentiation
1. Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
2. Division of Plastic Surgery, Brigham and Women's Hospital, Harvard Medical School, Boston, 02215, USA.
*These authors contributed equally to this work.
Received 2019-8-1; Accepted 2020-2-26; Published 2020-3-12
Abstract
The roles of long non-coding RNAs (lncRNAs) and micro RNAs (miRNAs) as regulators of mRNA expression in various diseases have recently been reported. Osteoblast differentiation is the vital process which mediates bone formation and fracture healing. In present study, we found microRNA-6979-5p (miR-6979-5p) to be the most differentially expressed miRNA between normal bone and calluses of mice, and overexpression of miR-6979-5p was negatively associated with osteoblast differentiation. Through luciferase assays, we found evidence that bone morphogenetic protein 2 (BMP2) is a miR-6979-5p target gene that positively regulates osteoblast differentiation. We further identified the lncRNA Rhno1 as a competing endogenous RNA (ceRNA) of miR-6979-5p, and we verified that it was able to influence osteoblast differentiation both in vitro and in vivo. In summary, our data indicates that the lncRNA Rhno1/miR-6979-5p/BMP2 axis is a significant regulatory mechanism controlling osteoblast differentiation, and it may thus offer a novel therapeutic strategy for fracture healing.
Keywords: LncRNA, miRNA, mRNA, Fracture, BMP2
Introduction
Fracture healing is a complex physiological process that occurs in individuals after injury, and osteoblast differentiation is a vital component of this process [1,2]. Previous studies have suggested that osteogenic differentiation is triggered via the up- or down-regulation of hormones and growth factors that ultimately modulate osteoblast differentiation and proliferation [3-5]. For example, orexin signals through orexin receptor 2 in the brain to enhance bone formation, and exerts opposing regulatory roles in the context of osteoporosis [6]. Similarly, targeting integrins had been reported to be a potential therapeutic strategy for counteracting bone loss [7]. In recent years, there has been increasing research regarding the field of osteoblast differentiation, and specifically of the regulatory roles of the lncRNA/miRNA/mRNA axis in this process [8-10].
LncRNAs are long RNAs that lack coding ability, but which are transcribed and processed [11,12]. There is now clear evidence that many lncRNAs play a critical role in regulation of gene expression via transcriptional or post-transcriptional mechanisms, and thereby participate in physiological and pathological processes [13,14]. lncRNAs have also been reported to function as competing endogenous RNAs (ceRNAs), binding to specific miRNA response elements to effectively sequester specific miRNAs, thereby regulating processes such as osteogenic-related gene expression [15]. This strategy effectively prevents miRNAs from binding to their target mRNA molecules, which they are normally able to repress the translation of in a highly specific manner via binding to their 3'-untranslated region (3'-UTR) [16]. This lncRNA/miRNA/mRNA axis had been reported to widely exist in various disease contexts, but there is limited research regarding how this axis functions in the context of osteogenic differentiation, making the investigation of these functions and the underlying molecular mechanisms an important research pursuit.
Rhno1 (aka RHINO, RAD9-Hus1-Rad1 Interacting Nuclear Orphan) was reported as a factor to DNA damage checkpoint signaling, which physically couples the 9-1-1 (RAD9-HUS1-RAD1) checkpoint clamp to DNA Topoisomerase II Binding Protein 1 [17]. Ensemble mirror database (http://useast.ensembl.org) was used to search for the details of Rhno1. The results showed that Rhno1 have a lot of transcript ID and many of them have the ability of protein coding. However, a non-coding transcript ID (ENSMUST00000132604.1) named Rhno1-204 can also be found in this database. Thus, in the present research, we used this transcript ID as lncRNA Rhno1 and explore the association between lncRNA Rhno1 and osteoblast differentiation. From literature reviewing, few studies concerning miR-6979-5p, we then search for the miRBase database (www.mirbase.org) to get the more details of miR-6979-5p and the results indicated that the accession number of miR-6979-5p is MI0022827, and the sequence is: GGGGAGGCGCAGAGACUGAGCUGCUCAGUGCGCGCUUGUGUCUGUCUGGCUCCCAG.
BMP2, as a significant member in the transforming growth factor-β family, has demonstrated a regulatory role in the formation of cartilage [18]. Furthermore, previous research reported that BMP2 can accelerate fracture healing by stimulating the bone marrow mesenchymal stem cells differentiate into osteoblasts [19]. Additionally, it is shown that BMP2 can induce the osteoclast activity and the remodeling process of fracture healing [20]. And a recent study reported that the expression of BMP2 was significantly decreased in patients with bone none union compared with fracture healing patients [21]. Therefore, BMP2 is a key target gene for fracture healing and the formation of callus.
We aimed to investigate the ability of the lncRNA Rhno1/miR-6979-5p/BMP2 axis to regulate osteoblast differentiation in vivo and in vitro. We found that miR-6979-5p exerts a negative effect on osteoblast differentiation via directly targeting BMP2, and local injection of miR-6979-5p into fracture sites was able to counteract fracture healing. Furthermore, lncRNA Rhno1 was able to promote osteoblast differentiation in MC3T3-E1 cells via serving as a miR-6979-5p ceRNA, and local injection of lncRNA Rhno1 was able to enhance fracture healing in vivo. This is the first report to our knowledge to demonstrate the role of the lncRNA Rhno1/miR-6979-5p/BMP2 axis in osteoblast differentiation, and to outline the underlying mechanisms.
Materials and Methods
Animal fracture models
The Animal Research Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology approved all study experiments. Male C57BL/6J mice (age: 6 weeks) from the Center of Experimental Animal, Tongji Medical College, Huazhong University of Science and Technology were used for all experiments, with anesthesia conducted using 10% chloral hydrate (300mg/kg body weight). A longitudinal incision was made, and then blunt separation of the underlying muscles was performed to produce a murine femoral fracture model. Subsequently, we used a diamond disk to cut the femur in order to produce a transverse osteotomy in the mid-diaphysis region. All fractures were then stabilized using 23-gauge intramedullary needles. After 14 days, half of the mice were euthanized, and the calluses at the fracture location were harvested for subsequent analysis. Similarly, the calluses of remaining mice were harvested for subsequent analysis on day 21 post injury.
Imaging of small animals in vivo
We performed small animal imaging at the Central laboratory of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan Union Hospital; http://www.whuh.com/). On days 0, 4, 7, 10, and 14; post-local direct 100µl injections of 200 µM Cy3-labled agomiR-6979-5p were made (GenePharma, Shanghai, China). At 15 min post-injection, 200 µL of 1% chloral hydrate solution was used for anesthetization and were imaged using an In-Vivo FX PRO imaging system (BRUKER, Karlsruhe, Germany) with a 1-minute exposure time.
Radiographic images
On days 7, 14, and 21 post-injury, all animals were given X-rays using an In-Vivo FX PRO imaging system (BRUKER, Karlsruhe, Germany) with a 10 second exposure time.
micro-CT analysis
The fracture site was scanned using a BRUKER SkyScan 1176 scanner micro-CT system (BRUKER, Karlsruhe, Germany) to provide images at 2400 views, 5 frames/view, 37 kV, and 121mA, and three-dimensional images were rendered and evaluated using CT-Vox 2.1 (BRUKER Minimal Intensity Projection Software, Karlsruhe, Germany). Soft tissues were cleaned thoroughly. After scanning, calluses were preserved at -80°C for miRNA or protein extraction for qRT-PCR and western blotting, respectively. Parameters measured were measured by CT Analyser, Version: 1.15.4.0 evaluation software, as follows: bone volume (BV), total volume (TV), BV/TV, and bone mineral density (BMD).
Cell cultures and transfection
MC3T3-E1 cells were grown in DMEM/F12 media (Hyclone, UT, USA, #SH30023.01B) with 10% FBS (Gibco, GI, USA, #10099141) and 1% penicillin/streptomycin (Hyclone, UT, USA, #SV30010). Transfection of agomiR-6979-5p, agomiR-NC, antagomiR-6979-5p, antagomiR-NC, lncRNA Rhno1, silncRNA Rhno1, and siRNA-NC (GenePharma, Shanghai, China) at 200uM was conducted using Lipofectamine 3000 (ThermoFisher Scientific, MA, USA, #L3000001) based on provided directions. Lipofectamine 3000 was also used to transfect cells with miRNAs or siRNA oligos. BMP2 siRNA, silncRNA Rhno1, and siRNA-NC (GenePharma, Shanghai, China) were transfected at 50 nM.
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed as in previous studies [42]. Trizol (ThermoFisher Scientific, MA, USA, #L15596026) was used for total RNA extraction, after which a qPCR RT Master Mix (Toyobo, Osaka, Japan) was used for cDNA synthesis. The 2-ΔΔCt method was used for relative gene expression quantification, with GAPDH and U6 used to normalize mRNA and miRNA expression. Primers are listed in Table 1.
Western blotting
NETN buffer (20 mM TrisHCl, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% Nonidet P-40) was used to prepare cell lysates, which were separated via SDS-PAGE and transferred onto PVDF membranes. These were then blocked using 5% skim milk, and probed at 4°C overnight using primary antibodies, after which appropriate HRP-conjugated secondary antibodies (Aspen, Johannesburg, South Africa, #AS1058) were used for antigen detection. A chemiluminescence detection system (Canon, Tokyo, Japan, #LiDE110) was used to visualize protein based on provided directions. Antibodies used were as follows: anti-collagen I (1:500, Abcam, MA, USA, #ab34710), anti-ALP (1:1,000, Abcam, MA, USA, #ab95462), anti-Osteocalcin (1:500, Abcam, MA, USA, #ab93876), anti-RunX2 (1:500, Abcam, MA, USA, #ab23981), anti-BMP2 (1:2,000, CST, MA, USA), and anti-GAPDH (1:10,000, Abcam, MA, USA, #ab37168).
Alkaline Phosphatase (ALP) Staining
A BCIP/NBT alkaline phosphatase color development kit (Beyotime, Shanghai, China, #C3206) used based on provided directions to assess ALP staining. Samples were washed twice in PBS, and then fixed with 10% formalin for 15 minutes. The BCIP/NBT liquid substrate was then applied to cells for 24 h. Samples were prepared in the dark at room temperature. Color change was obtained under a charge-coupled device (CCD) microscope, and a scanner (EPSON V600, Nagano prefecture, Japan) was used for imaging. Samples were analyzed in triplicate.
miRNAs, lncRNAs, and mRNA primer sequence
| microRNAs or gene name | Primer sequence |
|---|---|
| Mmu - miR - 6979 - 5p - Forward | ACACTCCAGCTGGGGGGGAGGCGCAGAG |
| Mmu - miR - 6979 - 5p - Reverse | TGGTGTCGTGGAGTCG |
| Mmu - lncRNA - Rhno1 - Forward | CTATGAGACAGGAGGCAGGGT |
| Mmu - lncRNA - Rhno1 - Reverse | AATGTGAGGCCCTTTTGTCTCA |
| U6-Forward | CTCGCTTCGGCAGCACA |
| U6-Reverse | AACGCTTCACGAATTTGCGT |
| Mmu - BMP2 - Forward | GACATCCTGAGCGAGTTCGA |
| Mmu - BMP2 - Reverse | CACTTGTTTCTGGCAGTTCTTC |
| Mmu - ALP - Forward | TGACTACCACTCGGGTGAACC |
| Mmu - ALP - Reverse | TGATATGCGATGTCCTTGCAG |
| Mmu - COL1A1 - Forward | CTGACTGGAAGAGCGGAGAG |
| Mmu - COL1A1 - Reverse | CGGCTGAGTAGGGAACACAC |
| Mmu - OCN - Forward | TTCTGCTCACTCTGCTGACCC |
| Mmu - OCN - Reverse | CTGATAGCTCGTCACAAGCAGG |
| Mmu - Runx2 - Forward | CGCCACCACTCACTACCACAC |
| Mmu - Runx2 - Reverse | TGGATTTAATAGCGTGCTGCC |
| Mmu - GAPDH - Forward | AGAGTGTTTCCTCGTCCCG |
| Mmu - GAPDH - Reverse | CCGTTGAATTTGCCGTGA |
Alizarin Red Staining
MC3T3-E1 cells were grown in osteogenic whole-media (Cyagen, Guangzhou, China, #HUXMA-90021) in 6-well plates to induce osteoblast mineralization. Cells were fixed with 10% formalin for 15 minutes, after which cells were washed using PBS and stained using 1 mL 0.5% alizarin red staining solution at room temperature for 15 min. Cells were then rinsed with distilled water for 5 minutes while shaking, and red mineralized nodules were assessed via CCD microscope and imaged (EPSON V600, Nagano prefecture, Japan). All experiments were repeated three times.
Luciferase Reporter Assay
MC3T3-E1 cells were plated in 24-well plates and transfected using a dual-luciferase vector (BMP2 WT, BMP2 Mut, lncRNA Rhno1 WT and lncRNA Rhno1 Mut) as well as either miR-6979-5p mimics or negative control (NC mimics). Following a 24 h incubation, the dualluciferase reporter assay system (Promega, Wisconsin, USA) was utilized as a means of measuring luciferase activity.
RNA immunoprecipitation (RIP) experiments
An RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA, #17-701) was used for RIP assays based on provided directions. Briefly, RIP lysis buffer (Solarbio, Beijing, China, #N8031) was used for MC3T3-E1 cell lysis, and lysates were combined with either anti-Argonaute2 (anti-Ago2) or anti-IgG control overnight at 4°C. Proteinase K was then used for sample treatment, and immunoprecipitated RNA was collected and analyzed via qRT-PCR.
RNA-pull down assay
The Biotin RNA Labeling Mix (Roche, Basel, Switzerland, #11685597910) was used to labled isolated RNAs, while the T7 RNA polymerase (Ambion Life, TX, USA) was used for in vitro transcription. An RNeasy Plus Mini Kit (Qiagen, Munich, Germany, #74134) together with DNase I (Qiagen, Munich, Germany, #19101) was used for RNA purification. Cell lysates were then mixed together with these RNAs (positive control, negative control, or biotinylated), and magnetic beads were added to all samples followed by a room temperature incubation. Bead-bound RNA complexed were then eluted and extracted, after which RNA was analyzed via qRT-PCR.
RNA fluorescent in situ hybridization
Cy3-labeled-lncRNA Rhno1 (GenePharma, Shanghai, China) and DAPI-labeled U6 probes (RiboBio, Guangzhou, China) were designed and synthesized. A FISH kit was used for RNA-FISH experiments based on provided protocols (ThermoFisher Scientific, MA, USA, #F32954).
Histological analysis
Following decalcification, 7 µm thick paraffin-embedded tissue samples were prepared and subjected to hematoxylin-eosin (H&E) and Alcian blue staining. Sections were then imaged and measured with an Olympus BX51 microscope and a DP73 CCD Olympus Imaging System (Olympus Corporation, Tokyo).
Statistical analysis
Data are means ± SD, and GraphPad Prism 8.0 (GraphPad Software, Inc, La Jolla, CA) was used for all analyses unless otherwise noted. Three or more samples were compared via a one-way ANOVA with Tukey's post-hoc test, while two-tailed Student's tests were used to compare two groups. P < 0.05 was the threshold of significance.
Results
MiR-6979-5p inhibits fracture healing in vivo
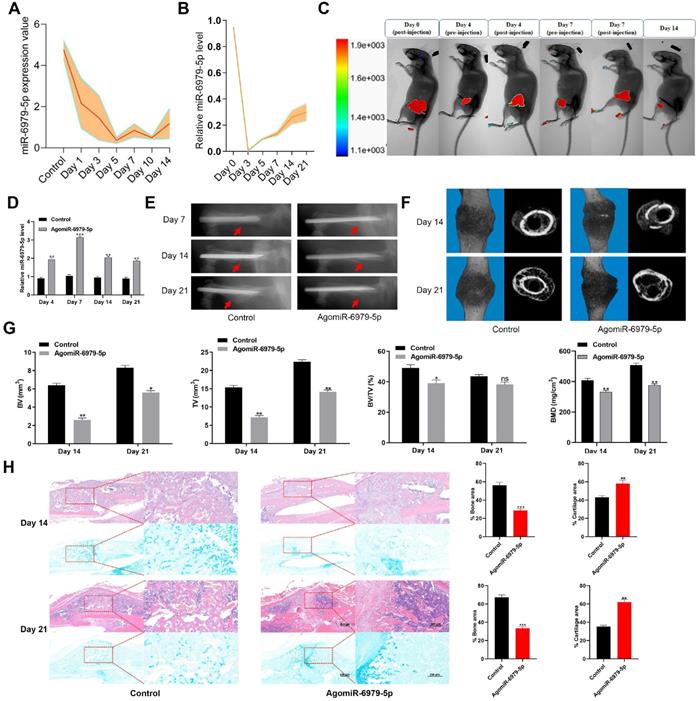
To explore how miR-6979-5p affects fracture healing, we measured the relative levels of miR-6979-5p in fracture gene chips (Figure 1A). In addition, we generated a murine fracture model, and calluses from fracture sites were harvested to measure miR-6979-5p levels during early fracture healing stages (days 1 to 21) (Figure 1B), revealing this levels to be markedly decreased during this early healing period. Next, mice were divided randomly into two groups (control group and agomiR-6979-5p group), with agomiR-6979-5p animals receiving local direct injections of Cy3-labeled miR-6979-5p at the fracture sites on day 0, day 4, and day 7 post-injury, after which miR-6979-5p levels in fracture sites were assessed via in vivo imaging in these animals (Figure 1C). On days 4, 7, 14 and 21 post-injury, 3 mice in each group were euthanatized and calluses were collected from the fracture sites to measure the level of miR-6979-5p and, as shown in Figure 1D, miR-6979-5p levels were higher in mice administered agomiR-6979-5p than in control animals at both of these time points. All animals were also assessed via X-rays and micro-CT examinations during the fracture healing process, and those animals treated with agomiR-6979-5p exhibited a smaller callus volume and a larger fracture gap relative to control animals (Figure 1E-F). Additionally, micro-CT scanning datas indicated the presence of a smaller total and bone callus volume in agomiR-6979-5p-treated animals than in control animals on post-fracture days 14 and 21 (Figure 1G). Moreover, H&E/Alcian-blue staining results revealed a reduced bone area and increased cartilage area at the fracture junction in the agomiR-6979-5p group, suggesting less bone remodeling for these animals (Figure 1H). These results indicated that miR-6979-5p may negatively regulate the fracture healing process.
MiR-6979-5p negatively regulates in vitro osteoblast differentiation
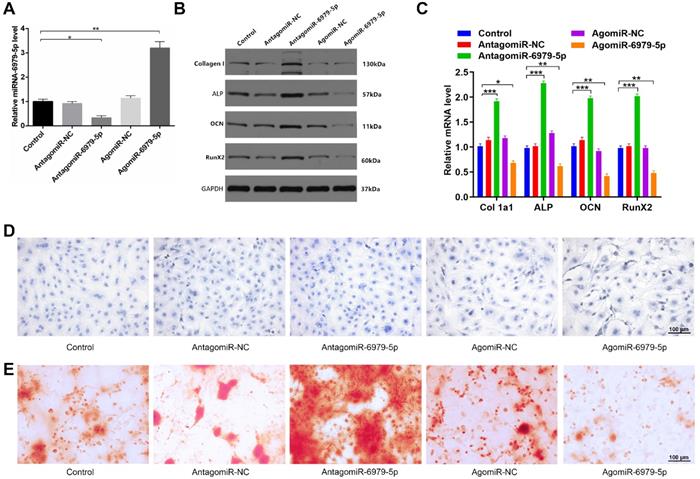
We next explored how miR-6979-5p affects osteogenic differentiation in vitro. MC3T3-E1 cells were transfected with lipofectamine 3000 alone, or together with an antagomiR-negative control (antagomiR-NC), antagomiR-6979-5p, agomiR-negative control (agomiR-NC), or agomiR-6979-5p. Overexpression of miR-6979-5p was evident in the agomiR-6979-5p group by PCR analysis (Figure 2A). Next, expression of collagen I, ALP, OCN, and RunX2, which are osteogenic genes, were measured among the five groups via western blotting. As shown in Figure 2B and 2C, higher mRNA levels of relative osteogenic genes were detectable in the antagomiR-6979-5p group than in the other experimental groups. Furthermore, we found overexpression of miR-6979-5p reduced ALP activity and staining, whereas knockdown of miR-6979-5p via antagomiR-6979-5p transfection enhanced ALP activity and staining (Figure 2D). In addition, to assess whether miR-6979-5p stimulates or suppresses extracellular matrix mineralization, cells were continuously cultured for 21 days, after which we observed reduced mineral deposition in agomiR-6979-5p-transfected cells relative to antagomiR-6979-5p-transfected cells (Figure 2E). These data indicated that miR-6979-5p inhibits the osteoblast differentiation of MC3T3-E1 cells.
MiR-6979-5p regulates osteogenic differentiation via directly targeting BMP2
Local administration of agomiR-6979-5p impaired healing in mice. Levels of miR-6979-5p were generally decreasing in gene chips (A) and fracture animal models (B) at the early stages of fracture healing. (C) In vivo imaging of agomiR-6979-5p metabolism at the fracture sites. (D) High level of miR-6979-5p were found in calluses of agomiR-6979-5p animals on day 4 and day 7 by qRT-PCR analysis. n=3, per group. Mice treated with agomiR-6979-5p exhibited a smaller callus volume and a disrupted fracture gap relative to control animals upon X ray (E) and three dimensional m-CT analyses (F). (G) Smaller total and bone callus volume can be seen in agomiR-6979-5p-treated animals relative to control animals on post-fracture days 14 and 21 by micro-CT data analysis. (H) H&E/Alcian-blue staining results revealed a reduced bone area and increased cartilage area at the fracture junction in the agomiR-6979-5p group. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
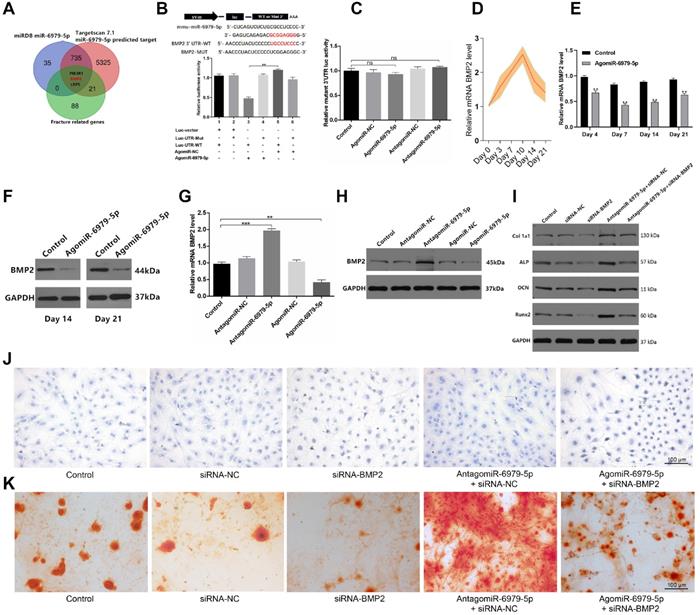
We next sought to explore miR-6979-5p downstream targets via screening miRDB datasets, Targetscan datasets, and GEO datasets for potential targets. we combined the results of the above three databases to enhance the strength of the evidence, and in order to show the process of the method of selecting target gene more clearly, we produced a Venn diagram and as shown in Figure 3A, three candidate genes were identified of interest - PI3KR1, BMP2, and LRP5. Following a literature review, although LRP5 is involved in Wnt/LRP5/β-Catenin signaling pathway, and thereby mediate the osteoblast differentiation process [22], BMP2 is a osteogenic mediator demonstrated widely in much more previous researches [23-25]. Thus, we select BMP2 from the three candidates as a target gene for miR-6979-5p. We next assessed whether miR-6979-5p directly targets BMP2, using either wild-type (WT) BMP2 3′ UTR or mutated (Mut) BMP2 3' UTR constructs fused to luciferase reporters. With these constructs, we found that agomiR-6979-5p substantially attenuated WT BMP2 3′ UTR reporter activity (Figure 3B), whereas it failed to influence the activity of the mutated 3′ UTR BMP2 reporter (Figure 3C). Furthermore, BMP2 mRNA levels in fracture calluses were measured over a 21 day post-injury period, revealing an apparent increase in expression over time (Figure 3D). To assess changes in BMP2 mRNA levels during the early stages of fracture healing in vivo, calluses samples from both animal groups were collected on days 4, 7, 14, and 21 for PCR analysis. Lower BMP2 expression was evidence in agomiR-6979-5p-treated animals relative to control animals at each time point (Figure 3E). Similarly, fracture calluses were collected from both animal groups on days 14 and 21 for western blotting, and the results indicated that BMP2 expression was significant lower in the agomiR-6979-5p group relative to controls (Figure 3F). Next, MC3T3-E1 cells were transfected using either agomiR or antagomiR constructs as above, and BMP2 mRNA expression was assessed via PCR and western blotting. As shown in Figure 3G-H, higher BMP2 mRNA levels were evident in the antagomiR-6979-5p group relative to other experimental groups. Additionally, to explore whether osteoblast differentiation is BMP2-dependent, we next tested whether antagomiR-6979-5p was able to rescue the negative effects of siRNA-BMP2 on osteogenic differentiation. Western blotting indicated that antagomiR-6979-5p was able to restore BMP2-dependent osteogenesis-associated gene expression (collagen I, ALP, OCN, and Runx2) (Figure 3I). Moreover, we found overexpression of BMP induced ALP activity and staining, whereas antagomiR-6979-5p was able to restore the effect of BMP2 on ALP activity and staining (Figure 3J). To observe whether miR-6979-5p stimulates or suppresses extracellular matrix mineralization, cells were next continuously cultured for 21 days, and we observed reduced mineral deposition in siBMP2-transfected cells relative to the antagomiR-6979-5p+siRNA-NC group (Figure 3K). Collectively, these results indicated miR-6979-5p directly targets the BMP2 3' UTR in the context of osteogenic differentiation.
miR-6979-5p inhibits osteoblast differentiation in vitro. MC3T3-E1 cells were transfected with lipofectamine 3000 only, antagomiR-NC, antagomiR-6979-5p, agomiR-NC, or agomiR-6979-5p. MiR-6979-5p levels were up-regulated in cells treated with agomiR-6979-5p as measured via qRT-PCR analysis. (B-C) Western blotting and qRT-PCR analyses of the osteogenic markers among the treated groups. ALP staining (D) and alizarin red-mediated calcium staining in (E) for cells treated as in (A). Scale bar, 100 µm. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
BMP2 is a miR-6979-5p target gene. Three genes (PI3KR1, BMP2, and LRP5) were selected as potential targets of miR-6979-5p. (B) AgomiR-6979-5p substantially attenuated WT BMP2 3' UTR reporter activity. (C) No significant differential activity of the mutated 3' UTR BMP2 reporter was evident between groups. (D) Elevated levels of BMP2 in calluses of fracture model animals were detectable at the early stage of fracture healing (days 1 to 21). (E) Lower BMP2 mRNA levels in calluses samples were detected by qRT-PCR analysis in agomiR-6979-5p-treated animals than in those in the other groups at time day points (days 4, 7, 14, and 21). (F) Western blotting of BMP2 in calluses samples between the two groups on day 14 and day 21. (G) qRT-PCR analysis of BMP2 mRNA levels in the five transfected groups (Control group, agomiR-NC group, agomiR-6979-5p group, antagomiR-NC group, and antagomiR-6979-5p). (H) Western blotting analysis of BMP2 mRNA levels in (G). (I) Western blotting of collagen I, ALP, OCN, and Runx2 mRNA levels in five groups (Control group, siRNA-NC, siRNA-BMP2 group, agomiR-6979-5p+siRNA-BMP2, and agomiR-6979-5p+siRNA-NC group). ALP staining (J) and alizarin red-mediated calcium staining (K) results in (A). Scale bar, 100 µm. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
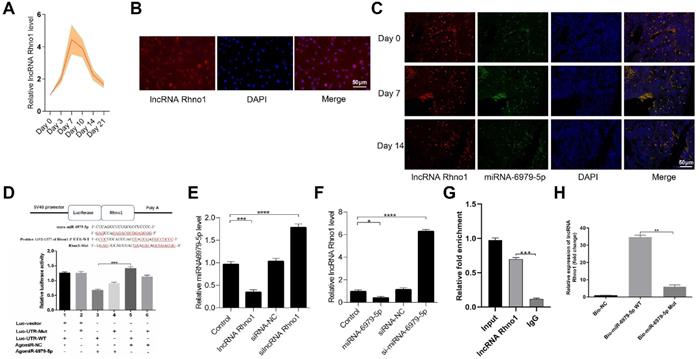
LncRNA Rhno1 serves to sequester miR-6979-5p
Next, to further elucidate the upstream regulatory mechanisms of miR-6979-5p, we searched the LncBase Experimental V.2 Database to identify its putative regulators, and a possible interaction between the lncRNA Rhno1 and miR-6979-5p was found. Consistent with this, in fracture animal models we found the level of lncRNA Rhno1 to trend upwards during the early stages of fracture healing (day 1 to 21 post-injury) (Figure 4A). Next, we assessed lncRNA Rhno1 localization in MC3T3-E1 cells via fluorescence in situ hybridization (FISH), revealing that lncRNA Rhno1 is localized mainly in the cytoplasm of MC3T3-E1 cells (Figure 4B). Furthermore, FISH assays were used to assess lncRNA Rhno1 localization in bone or calluses among animal models on days 0, 7, 14 post-injury, which suggesting lncRNA Rhno1 is localized mainly in the cytoplasm of bone tissues or calluses (Figure 4C). Moreover, using StarBase v2.0 (http://starbase.sysu.edu.cn/starbase2/mirLncRNA.php), we were able to identify the site of interaction between lncRNA Rhno1 and miR-6979-5p as shown in Figure 4D. A luciferase reporter gene assay was next conducted, and results in Figure 4D confirmed that the lncRNA Rhno1 directly bound to miR-6979-5p in MC3T3-E1 cells. To further validate the association between lncRNA Rhno1 and miR-6979-5p, MC3T3-E1 cells were next transfected with lipofectamine 3000 either alone, or together with silncRNA control (silncRNA-NC), or silncRNA Rhno1, and after a 48 hours cell culture period miR-6979-5p levels were measured by qRT-PCR analysis. This led miR-6979-5p levels in the silncRNA Rhno1-treated cells to be significantly higher than in the other experimental groups (Figure 4E). Similarly, the lncRNA Rhno1 levels were much higher in the antagomiR-6979-5p group than other groups, especially relative to the agomiR-6979-5p group (Figure 4F). Furthermore, the interaction between lncRNA Rhno1 and miR-6979-5p was further demonstrated via an RNA RIP assay, which revealed that lncRNA Rhno1 increased Ago2-containing miRNA ribonucleoprotein complex enrichment than when mixed with samples imunoprecipitated using control IgG (Figure 4G). Consistent with this, a pull-down assay provided evidence that lncRNA Rhno1 was readily bound by biomiR-6979-5p (Figure 4H), but not by a mutated isoform of this bio-miR-6979-5p. Collectively, these results suggest that lncRNA Rhno1 may serve as a ceRNA that functions as a molecular sponge to sequester miR-6979-5p.
LncRNA Rhno1 suppressed miR-6979-5p expression by sequestering this miRNA MC3T3-E1 cells. Levels of Rhno1 are increased in fracture animal models at early stages of fracture healing. (B) Localization of lncRNA Rhno1 by RNA-FISH in MC3T3-E1 cells. LncRNA Rhno1 is stained red (DAPI), and nuclei are stained blue (Scale bar = 50 µm). (C) Localization of lncRNA Rhno1 by RNA-FISH in bone tissues and calluses on days 7 and 14 post injury. LncRNA Rhno1 is stained red (DAPI), miR-6979-5p is stained green (DAPI) and nuclei are stained blue (Scale bar = 50 µm). (D) AgomiR-6979-5p substantially attenuated WT lncRNA Rhno1 3' UTR reporter activity. (E) miR-6979-5p levels in silncRNA Rhno1-treated samples were significantly higher than other groups. (F) Similarly, lncRNA Rhno1 levels were much higher in the antagomiR-6979-5p-treated group than in other groups, particularly relative to the agomiR-6979-5p group. (G) The interaction of miR-6979-5p and lncRNA Rhno1 with Ago2 was assessed via RIP assays. RNA levels were presented as fold enrichment in Ago2 relative to IgG immunoprecipitates (lower panel). (H) qRT-PCR was used to detect lncRNA Rhno1 expression in the sample pulled down by biotinylated miR-6979-5p WT and miR-6979-5p Mut probes. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
LncRNA Rhno1 promotes in vitro osteoblast differentiation
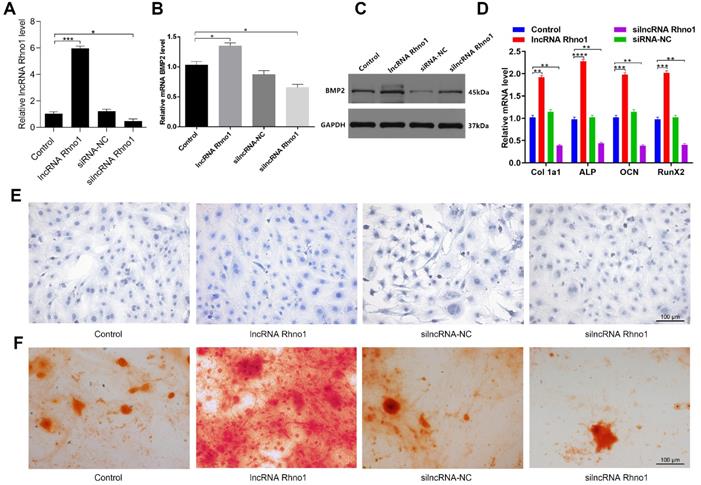
To explore how the lncRNA Rhno1 influences osteogenic differentiation, MC3T3-E1 cells were treated with lipofectamine 3000 alone, or together with lncRNA Rhno1, silncRNA-NC, or silncRNA Rhno1 (Figure 5A). After continuous culture for 48 hours, overexpression of lncRNA Rhno1 and BMP2 was then detectable in lncRNA Rhno1-transfected cells by qRT-PCR analysis (Figure 5B). The results of western blotting analyses of BMP2 levels in these four groups were also consistent with the qRT-PCR results (Figure 5C). Consistent with these changes, we found that the expression of genes involved in osteoblast differentiation, such as collagen I, ALP, OCN, and RunX2, were elevated in the lncRNA Rhno1 group relative to the other groups (Figure 5D, Supplementary Figure 1). In addition, overexpression of lncRNA Rhno1 induced ALP activity and staining, whereas lncRNA Rhno1 silencing attenuated ALP activity and staining (Figure 5E). Moreover, to observe the effect of lncRNA Rhno1 on extracellular matrix mineralization, cells were continuously cultured for 21 days, after which we observed more mineral deposition in lncRNA Rhno1-transfected cells (Figure 5F). This indicates that lncRNA Rhno1 enhances the expression of BMP2 and exerts a positive role during osteogenic differentiation.
LncRNA Rhno1 accelerates fracture healing in vivo
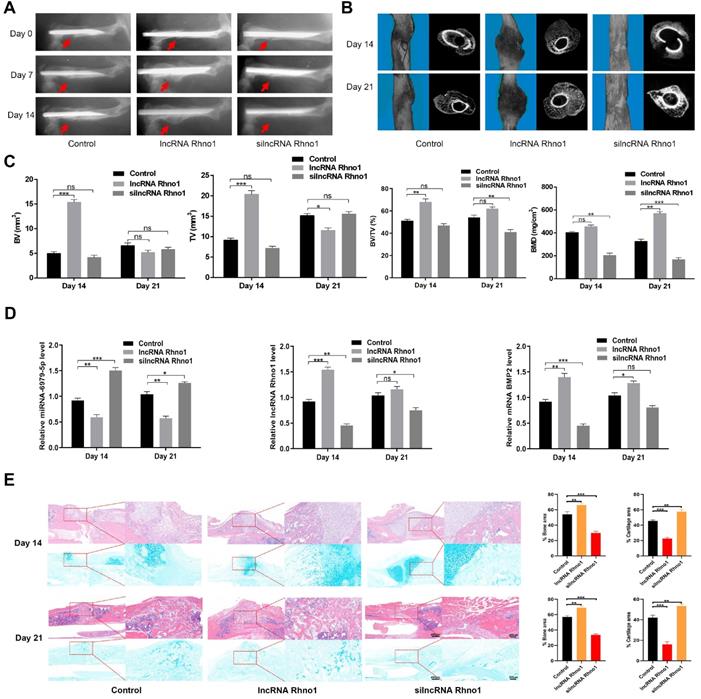
To evaluate the effects of lncRNA Rhno1 on fracture healing, fracture model mice were locally injected with phosphate buffered saline (PBS), lncRNA Rhno1, or an equal volume of silncRNA Rhno1 in the fracture sites on days 0, 4, and 7 after surgery. X rays and CT examinations were performed to assess the process of fracture healing. Mice treated with lncRNA Rhno1 exhibited a larger callus volume and smaller fracture gap relative to control animals (Figure 6A-B). In addition, a larger total and bone callus volume was evident in lncRNA Rhno1-treated animals relative to other groups on post-fracture days 14 and 21, whereas silncRNA Rhno1-treated animals had the smallest total and bone callus volume among the three groups (Figure 6C). Furthermore, on days 14 and 21 post micro-CT scanning, 3 mice from each group were euthanatized and calluses were collected to measure the levels of miR-6979-5p, BMP2, and lncRNA Rhno1, and as shown in Figure 6D, the level of miR-6979-5p decreased in lncRNA Rhno1-treated animals, whereas the levels of BMP2 and lncRNA Rhno1 increased. Moreover, H&E/Alcian-blue staining results revealed a reduced cartilage area and increased bone area at the fracture junction in the lncRNA Rhno1 group, suggesting more bone remodeling for these animals (Figure 6E). In summary, these data suggest that lncRNA Rhno1 downregulates miR-6979-5p and induces the expression of BMP2, thereby promoting fracture healing in vivo.
LncRNA Rhno1 promotes the osteoblastic differentiation of MC3T3-E1 cells. MC3T3-E1 cells were transfected with lipofectamine 3000 only, lncRNA Rhno1, siRNA-NC, or silncRNA Rhno1. Overexpression of lncRNA Rhno1 was detectable in lncRNA Rhno1-transfected cells via qRT-PCR analysis. (B-C) Overexpression of BMP2 were detectable in lncRNA Rhno1-transfected cells in (A). (D) Collagen I, ALP, OCN, and RunX2 were upregulated in (A). ALP staining (E) and alizarin red-mediated calcium staining (F) in MC3T3-E1 cells treated as in (A). Scale bar, 100 µm. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
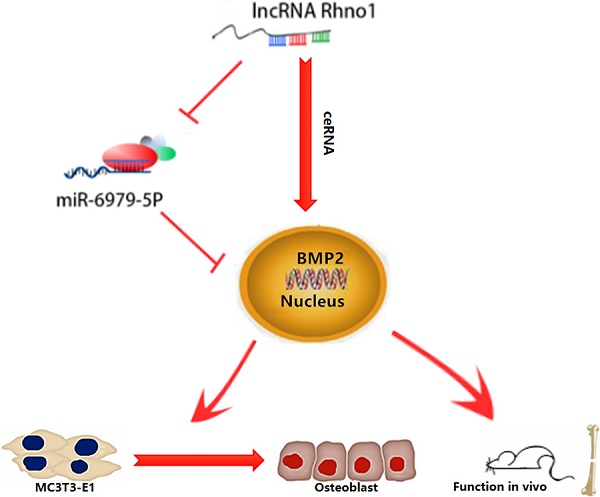
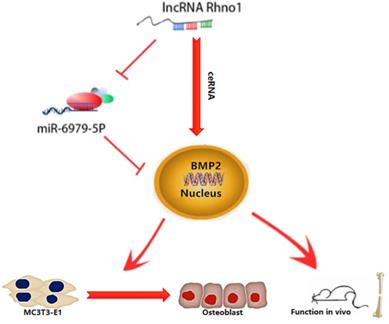
This study provides the first verification that local injection of lncRNA Rhno1 into murine fracture sites induces a prominent regenerative effect on the bone and its micro-architecture, as evidenced by more rapid healing and larger callus volumes in treated animals. We further found in vitro that lncRNA Rhno1 could serve as a ceRNA for miR-6979-5p, thereby counteracting the negative effects of miR-6979-5p on osteoblast differentiation and fracture healing. Moreover, we explored miR-6979-5p downstream targets, revealing BMP2 to be a relevant target gene for this miRNA. Thus, as shown in Figure 7, our results suggest that lncRNA Rhno1 may induce osteoblast differentiation by competitively binding miR-6979-5p to upregulate BMP2 mRNA levels, and thus lncRNA Rhno1/miR-6979-5p/BMP2 axis plays a significant role in the regulation of osteoblast differentiation.
In recent decades, miRNAs have been reported to serve as important regulatory factors in the context of osteogenic differentiation, and the underlying mechanisms have been the focus of substantial research [26-29]. We therefore sought to identify miRNAs with differential expression between intact control bone and calluses of post-fracture mice, leading to our selection of miR-6979-5p from the GEO database. We found no articles regarding the function of miR-6979-5p in the reported literature, and thus it was necessary to investigate the biological functions of this miRNA. In this study, the effect of miR-6979-5p on fracture healing was investigated in vivo, with the results suggesting that miR-6979-5p inhibits fracture healing, and we were able to confirm this negative impact on osteogenic differentiation using MC3T3-E1 cells. As previous studies [30] have reported miRNAs can bind to target mRNAs to exert specific effects, we subsequently explored the downstream targets of miR-6979-5p using luciferase assays.
Local administration of lncRNA Rhno1 promotes fracture healing in mice. X ray comparisons of fracture healing between control, lncRNA Rhno1, and silncRNA Rhno1 treated animals at days 7, 14, and 21 post-operation. (B) micro-CT images in (A) at day 14, and 21 after injury. (C) BV, TV, and BV/TV of the callus in (A) on days 14 and 21 post-operation were established via micro-CT. n=5 mice, per group. (D) The levels of miR-6979-5p were decreased in lncRNA Rhno1-treated animals, whereas the levels of BMP2 and lncRNA Rhno1 increased in these samples, as measured by qRT-PCR analysis. (E) H&E/Alcian-blue staining results revealed a reduced cartilage area and increased bone area at the fracture junction in the lncRNA Rhno1 group. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
BMP2, which we confirmed to be a target for miR-6979-5p, is able to stimulate the differentiation of myoblasts into osteoblasts via the EIF2AK3-EIF2A-ATF4 pathway [31]. The activation of EIF2AK3 can in turn stimulate phosphorylation of EIF2A, leading to increased ATF4 expression, which plays a central role in osteoblast differentiation [32,33]. Indeed, several studies have previously demonstrated that BMP2 plays a regulatory role in various biological processes [34-36]. For example, one group found that a BMP activity gradient is essential for normal dorsoventral patterning during gastrulation [37]. In our study, we demonstrated that BMP2 is a direct target of miR-6979-5p, and downregulation of miR-6979-5p increases the BMP2 mRNA levels, thereby promoting osteogenesis.
In addition, lncRNAs are known to affect disease progression by serving as molecular ceRNA sponges that can sequester miRNAs [38]. For example, one group found that the lncRNA AGAP2-AS1 plays a competitive role in hepatocellular carcinoma via acting as a miR-16-5p sponge [39]. In present study, we searched for potential upstream regulators of miR-6979-5p using the LncBase Experimental V.2 Database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-experimental), leading to our identification of the lncRNA Rhno1. As a growing body of evidence suggests that cytoplasmic lncRNAs can serve as decoys for other noncoding RNAs such as miRNAs [40,41], we determined the localization of this lncRNA Rhno1 via a FISH approach, revealing that lncRNA Rhno1 mainly located in cytoplasm, which is consistent with other published findings. RIP and pull down assays further confirmed the ability of lncRNA Rhno1 and miR-6979-5p to directly interact, thereby providing additional support to our model wherein the lncRNA Rhno1/miR-6979-5p/BMP2 axis modulates osteoblast differentiation.
A model of the lncRNA Rhno1/miR-6979-5p/BMP2 axis regulation of osteoblast differentiation. LncRNA Rhno1 serves as a ceRNA that competitively inhibits the expression of miR-6979-5p, thereby elevating the BMP2 mRNA level, and thus promoting osteoblast differentiation.
There were a number of limitations in the present study. First, it remains to be determined whether more frequent injections of lncRNA Rhno1 can induce similar or much better bone formation effects in fracture mice. Moreover, we were not able to conduct clinical trials regarding the beneficial effects of lncRNA Rhno1 on fracture healing in the present study. In addition, to produce more definitive evidence of the role of the lncRNA Rhno1/miR-6979-5p/BMP2 axis in osteogenesis, transgenic mice that specifically lack relevant genes in osteoblast lineages are necessary.
Taken together, our results indicate that the lncRNA Rhno1/miR-6979-5p/BMP2 axis can effectively modulate the differentiation of osteoblasts; potentially due to the lncRNA Rhno1 serving as a ceRNA that competitively inhibits the expression of miR-6979-5p, thereby elevating BMP2 mRNA levels and promoting osteoblast differentiation. Our findings suggest that the lncRNA Rhno1/miR-6979-5p/BMP2 axis may represent a novel therapeutic target for accelerating fracture healing.
Supplementary Material
Supplementary figure S1.
Acknowledgements
This study was supported by the National Science Foundation of China (No.81772345), Healthy Commission Key Project of Hubei Province (No.WJ2019Z009), Science and Technology Department of Hubei Province (No.2016CFB424), Development Center for Medical Science and Technology National Health and Family Planning Commission of the People's Republic of China (ZX-01-C2016024), and the Funds of Wuhan Union Hospital (02.03.2017-13), Wuhan Science and Technology Bureau (No.2017060201010192).
Author contributions
BM and GL conceived and designed the study; YX, CY and LC supervised the study; YX, YE and YS performed experiments; WZ, HX, YH and LH analyzed the data; QW, DC, FD, CY and LC provided advice and technical assistance; and YX wrote the manuscript. YX was supported by the China Scholarship Council for 18 months study at the Harvard Medical School. All authors approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Debnath S, Yallowitz AR, McCormick J. et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018;562:133-9
2. Farr JN, Xu M, Weivoda MM. et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat Med. 2017;23:1072-9
3. Yu SB, Zhu K, Lai YM. et al. atf4 promotes β-catenin expression and osteoblastic differentiation of bone marrow mesenchymal stem cells. Int. J. Biol. Sci. 2013;9:256-66
4. Wei J, Shimazu J, Makinistoglu MP. et al. Glucose Uptake and Runx2 Synergize to Orchestrate Osteoblast Differentiation and Bone Formation. Cell. 2015;161:1576-91
5. Moverare-Skrtic S, Henning P, Liu X. et al. Osteoblast-derived WNT16 represses osteoclastogenesis and prevents cortical bone fragility fractures. Nat Med. 2014;20:1279-88
6. Wei W, Motoike T, Krzeszinski JY. et al. Orexin regulates bone remodeling via a dominant positive central action and a subordinate negative peripheral action. Cell Metab. 2014;19:927-40
7. Marie PJ. Targeting integrins to promote bone formation and repair. Nat Rev Endocrinol. 2013;9:288-95
8. Cho TJ, Gerstenfeld LC, Einhorn TA. Differential temporal expression of members of the transforming growth factor beta superfamily during murine fracture healing. J Bone Miner Res. 2002;17:513-520
9. Liu DD, Zhang JC, Zhang Q. et al. TGF-beta/BMP signaling pathway is involved in cerium-promoted osteogenic differentiation of mesenchymal stem cells. J Cell Biochem. 2013;114:1105-1114
10. Govender S, Csimma C, Genant HK. et al. Recombinant human bone morphogenetic protein-2 for treatment of open tibial fractures: a prospective, controlled, randomized study of four hundred and fifty patients. J Bone Joint Surg Am. 2002;84(A):2123-2134
11. Peng H, Lu S-L, Bai Y. et al. MiR-133a inhibits fracture healing via targeting RUNX2/BMP2. Eur Rev Med Pharmacol Sci. 2018;22:2519-2526
12. Wang X, Guo B, Li Q. et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19:93-100
13. Kusuyama J, Amir MS, Albertson BG. et al. JNK inactivation suppresses osteogenic differentiation, but robustly induces osteopontin expression in osteoblasts through the induction of inhibitor of DNA binding 4 (Id4). FASEB J. 2019 fj201802465R
14. Xiong Y, Chen L, Yan C. et al. Circulating Exosomal miR-20b-5p Inhibition Restores Wnt9b Signaling and Reverses Diabetes-Associated Impaired Wound Healing. Small. 2020;16:e1904044
15. Kim J, Piao HL, Kim BJ. et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705-15
16. Han P, Li W, Lin CH. et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102-6
17. Day M, Rappas M, Ptasinska K. et al. BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands. Elife. 2018;7:undefined
18. Xiao L, Wu J, Wang JY. et al. Long Noncoding RNA uc.173 Promotes Renewal of the Intestinal Mucosa by Inducing Degradation of MicroRNA 195. Gastroenterology. 2018;154:599-611
19. Zheng J, Huang X, Tan W. et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48:747-57
20. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-83
21. Zeng HC, Bae Y, Dawson BC. et al. MicroRNA miR-23a cluster promotes osteocyte differentiation by regulating TGF-beta signalling in osteoblasts. Nat Commun. 2017;8:15000
22. Kim SP, Da H, Li Z. et al. Lrp4 expression by adipocytes and osteoblasts differentially impacts sclerostin's endocrine effects on body composition and glucose metabolism. J. Biol. Chem. 2019;294:6899-6911
23. Son HE, Kim EJ, Jang WG. Curcumin induces osteoblast differentiation through mild-endoplasmic reticulum stress-mediated such as BMP2 on osteoblast cells. Life Sci. 2018;193:34-39
24. Salazar VS, Ohte S, Capelo LP. et al. Specification of osteoblast cell fate by canonical Wnt signaling requires Bmp2. Development. 2016;143:4352-4367
25. Zappitelli T, Chen F, Aubin JE. Up-regulation of BMP2/4 signaling increases both osteoblast-specific marker expression and bone marrow adipogenesis in Gja1Jrt/+ stromal cell cultures. Mol. Biol. Cell. 2015;26:832-42
26. Hashimoto K, Ochi H, Sunamura S. et al. Cancer-secreted hsa-miR-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc Natl Acad Sci U S A. 2018;115:2204-9
27. Xiong Y, Cao F, Hu L. et al. miRNA-26a-5p Accelerates Healing via Downregulation of PTEN in Fracture Patients with Traumatic Brain Injury. Mol Ther Nucleic Acids. 2019;17:223-234
28. Li Y, Fan L, Liu S. et al. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials. 2013;34:5048-58
29. Sun Y, Xiong Y, Yan C. et al. Downregulation of microRNA-16-5p accelerates fracture healing by promoting proliferation and inhibiting apoptosis of osteoblasts in patients with traumatic brain injury. Am J Transl Res. 2019;11:4746-4760
30. Xiong Y, Yan C, Chen L. et al. IL-10 induces MC3T3-E1 cells differentiation towards osteoblastic fate in murine model. J. Cell. Mol. Med. 2020;24:1076-1086
31. Zhou X, Feng W, Qiu K. et al. BMP-2 Derived Peptide and Dexamethasone Incorporated Mesoporous Silica Nanoparticles for Enhanced Osteogenic Differentiation of Bone Mesenchymal Stem Cells. ACS Appl Mater Interfaces. 2015;7:15777-89
32. Drevelle O, Daviau A, Lauzon MA. et al. Effect of BMP-2 and/or BMP-9 on preosteoblasts attached to polycaprolactone functionalized by adhesive peptides derived from bone sialoprotein. Biomaterials. 2013;34:1051-62
33. Kawasaki T, Niki Y, Miyamoto T. et al. The effect of timing in the administration of hepatocyte growth factor to modulate BMP-2-induced osteoblast differentiation. Biomaterials. 2010;31(6):1191-8
34. Mi B, Xiong Y, Chen L. et al. CircRNA AFF4 promotes osteoblast cells proliferation and inhibits apoptosis via the Mir-7223-5p/PIK3R1 axis. Aging (Albany NY). 2019;11:11988-12001
35. Lee H, Cho Y. An Innovative Strategy for the Fabrication of Functional Cell Sheets Using an Electroactive Conducting Polymer. Theranostics. 2015;5:1021-9
36. Chen L, Xiong Y, Yan C. et al. LncRNA KCNQ1OT1 accelerates fracture healing via modulating miR-701-3p/FGFR3 axis. FASEB J. 2020 undefined
37. Xue Y, Zheng X, Huang L. et al. Organizer-derived Bmp2 is required for the formation of a correct Bmp activity gradient during embryonic development. Nat Commun. 2014;5:3766
38. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113-21
39. Liu Z, Wang Y, Wang L. et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:194
40. Li MJ, Zhang J, Liang Q, Xuan C. et al. Exploring genetic associations with ceRNA regulation in the human genome. Nucleic Acids Res. 2017;45:5653-65
41. Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56:347-59
42. Qi X, Zhang JY, Yuan H. et al. Exosomes Secreted by Human-Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells Repair Critical-Sized Bone Defects through Enhanced Angiogenesis and Osteogenesis in Osteoporotic Rats. Int. J. Biol. Sci. 2016;12:836-49
Author contact
![]() Corresponding authors: Bobin Mi, Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Avenue, Wuhan 430022, China. E-mail: mi19882com; Guohui Liu, Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Avenue, Wuhan 430022, China. E-mail: liuguohuiedu.cn
Corresponding authors: Bobin Mi, Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Avenue, Wuhan 430022, China. E-mail: mi19882com; Guohui Liu, Department of Orthopaedics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Avenue, Wuhan 430022, China. E-mail: liuguohuiedu.cn

 Global reach, higher impact
Global reach, higher impact