10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(10):1686-1697. doi:10.7150/ijbs.45472 This issue Cite
Review
Zoonotic origins of human coronaviruses
1. Department of Microbiology, The University of Hong Kong, Pokfulam, Hong Kong.
2. School of Biomedical Sciences, The University of Hong Kong, Pokfulam, Hong Kong.
Abstract
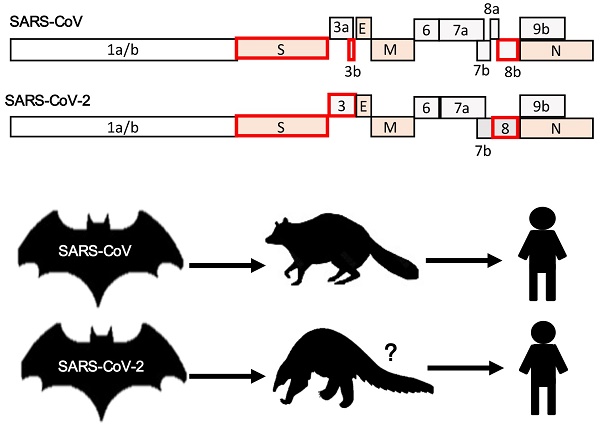
Mutation and adaptation have driven the co-evolution of coronaviruses (CoVs) and their hosts, including human beings, for thousands of years. Before 2003, two human CoVs (HCoVs) were known to cause mild illness, such as common cold. The outbreaks of severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) have flipped the coin to reveal how devastating and life-threatening an HCoV infection could be. The emergence of SARS-CoV-2 in central China at the end of 2019 has thrusted CoVs into the spotlight again and surprised us with its high transmissibility but reduced pathogenicity compared to its sister SARS-CoV. HCoV infection is a zoonosis and understanding the zoonotic origins of HCoVs would serve us well. Most HCoVs originated from bats where they are non-pathogenic. The intermediate reservoir hosts of some HCoVs are also known. Identifying the animal hosts has direct implications in the prevention of human diseases. Investigating CoV-host interactions in animals might also derive important insight on CoV pathogenesis in humans. In this review, we present an overview of the existing knowledge about the seven HCoVs, with a focus on the history of their discovery as well as their zoonotic origins and interspecies transmission. Importantly, we compare and contrast the different HCoVs from a perspective of virus evolution and genome recombination. The current CoV disease 2019 (COVID-19) epidemic is discussed in this context. In addition, the requirements for successful host switches and the implications of virus evolution on disease severity are also highlighted.
Keywords: coronavirus, SARS-CoV, SARS-CoV-2, MERS-CoV, COVID-19, animal reservoir, bats

 Global reach, higher impact
Global reach, higher impact