10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(14):2628-2647. doi:10.7150/ijbs.47203 This issue Cite
Review
miRNA-based biomarkers, therapies, and resistance in Cancer
1. Department of Thoracic Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China.
2. Hunan Key Laboratory of Early Diagnosis and Precision Therapy, Department of Thoracic Surgery, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China.
3. Key Laboratory of Carcinogenesis and Cancer Invasion, Ministry of Education, Department of Pathology, Xiangya Hospital, Central South University, Hunan, 410078 China.
4. NHC Key Laboratory of Carcinogenesis (Central South University), Cancer Research Institute and School of Basic Medicine, Central South University, Changsha, Hunan, 410078 China.
Received 2020-4-19; Accepted 2020-7-4; Published 2020-7-19
Abstract
MicroRNAs (miRNAs), small non-coding RNAs (ncRNAs) of about 22 nucleotides in size, play important roles in gene regulation, and their dysregulation is implicated in human diseases including cancer. A variety of miRNAs could take roles in the cancer progression, participate in the process of tumor immune, and function with miRNA sponges. During the last two decades, the connection between miRNAs and various cancers has been widely researched. Based on evidence about miRNA, numerous potential cancer biomarkers for the diagnosis and prognosis have been put forward, providing a new perspective on cancer screening. Besides, there are several miRNA-based therapies among different cancers being conducted, advanced treatments such as the combination of synergistic strategies and the use of complementary miRNAs provide significant clinical benefits to cancer patients potentially. Furthermore, it is demonstrated that many miRNAs are engaged in the resistance of cancer therapies with their complex underlying regulatory mechanisms, whose comprehensive cognition can help clinicians and improve patient prognosis. With the belief that studies about miRNAs in human cancer would have great clinical implications, we attempt to summarize the current situation and potential development prospects in this review.
Keywords: MicroRNAs, Cancer, miRNA-based biomarkers, miRNA-based therapies, Cancer therapy resistance
Introduction
MicroRNAs (miRNAs) are small, evolutionarily conserved, non-coding RNAs (ncRNAs) of about 22 nucleotides in size that take significant roles in gene regulation [1]. It was first described in 1993 that such ncRNAs were made in the proper timing of Caenorhabditis elegans [2]. Later, many miRNAs were discovered in species such as animals and plants. There are about 40 thousand entries of hairpin forerunner forms in the 2018 version (miRBase 22), demonstrating about 50 thousand mature miRNAs in 271 species (http:www.mirbase.org) [3]. It has been estimated that miRNAs can target more than 60% of human protein-coding genes [4]. And nowadays, more than 2,000 miRNAs have been illustrated to have the ability to regulate the expression situation of about one-third genes in the human genome [5].
To be mentioned, the online miRNA resource miRBase provides systematic nomenclature for miRNAs dedicatedly [3]. Take “hsa-miR-136-5p” for instance, “hsa” indicates the organism, “miR” signifies mature miRNA while precursors would be shown as “mir”, and “5p” there means this miRNA is from the 5' arm whereas “3p” would refer to those from the 3' arm when two miRNAs from a single predicted precursor present similar relative abundancies. Besides, a suffixed letter was used to distinguish close-related miRNAs, which we can see in miR-34a, miR-29b, miR-519c, among others. In keeping with miRBase, the HUGO Gene Nomenclature Committee (HGNC) also makes special gene symbols for miRNA genes [6, 7], which we have not introduced in detail here.
The control of gene expression with miRNAs is owned to virtually all known cancer cells. In the course of tumor development, miRNAs have shown its biology characteristics, indicating the possibilities in cancer classification and prognosis improvement. In this Review, we represent the biogenesis and regulation of miRNAs, as well as describe how their functioning mode works, with the links to cancer development. We also summarize the findings in which miRNAs act as biomarkers for cancer diagnosis and cancer prognosis prediction, and highlight the situation of miRNA-based cancer therapies.
Biogenesis, regulation, and functioning mode of miRNA
Biogenesis of miRNA
Canonical biogenesis
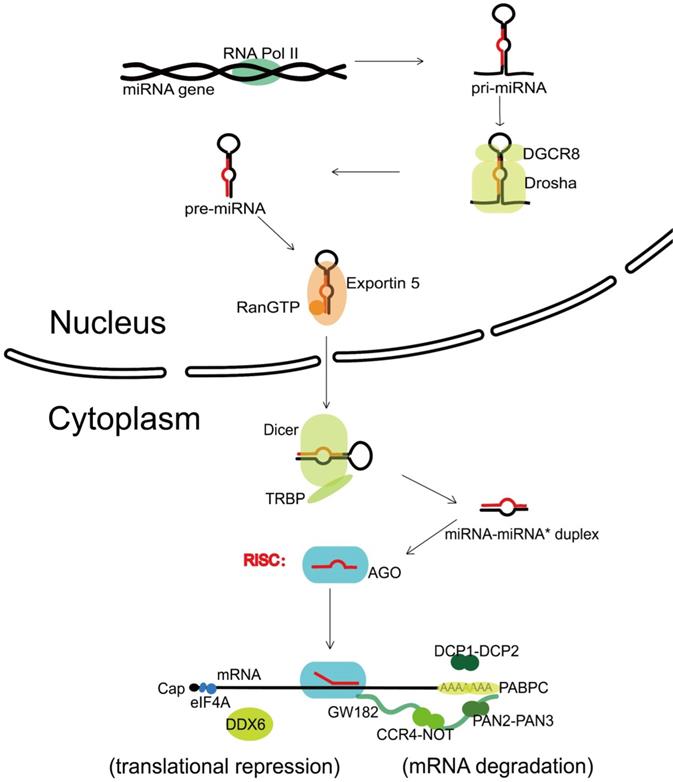
The canonical biogenesis of miRNA is a complex pathway with both nuclear and cytoplasmic steps. Transcribed by the RNA polymerase II (Pol II) [8], one nuclear miRNA gene produces a hairpin intermediate called “pri-miRNA” [9], which is then recognized by a microprocessor basically made up by one molecule of Drosha (a kind of RNase III enzyme with two RNase III domains) and two molecules of its cofactor, DiGeorge syndrome critical region gene 8 (DGCR8) [10]. Next, Drosha cleaves two strands of the stem in the pri-miRNA hairpin, and form a stem-loop named “pre-miRNA” [11]. Following that, the pre-miRNA formed in the nucleus is transported to the cytoplasm via Exportin 5 and Ran guanosine triphosphate (RanGTP) [12]. In the cytoplasm, the pre-miRNA is further recognized by Dicer, an endonuclease with two RNase III domains too, and associates with a partner protein called TRBP (HIV-1 TAR RNA binding protein, also named as TARBP2) [13-15]. Dicer cuts off the terminal loop from the hairpin and generates the miRNA-miRNA* duplex, in which the “miRNA*” is a passenger strand paired to the miRNA. The miRNA-miRNA* duplex is then loaded into an Argonaute (Ago) family protein and is unwound driven by the N-terminal domain of Ago [16]. Notably, in the process of assorting among Ago complexes, the nucleotide composition of miRNA-miRNA* duplex also impacts on the strand selection [17]. Finally, the miRNA* strand is commonly degraded, while the other strand, also known as guide strand (to become the mature form of miRNA) is retained and form the functional RNA-induced silencing complex (RISC) eventually (Figure 1).
Non-canonical biogenesis
The re-evaluation about the roles of key factors in the canonical biogenesis shows that there are alternative mechanisms for miRNA biogenesis [18]. In fact, there are more and more miRNAs with non-canonical biogenesis reported as more high-throughput sequencing datasets are used, and those miRNAs characterized as Drosha-independent or Dicer-independent.
Drosha-independent miRNAs were classified into three classes. The first is “mirtrons”, which refer to certain debranched introns that feature as pre-miRNAs and can be exported out of the nucleus immediately for Dicer-mediated processing [19]. Evidence showed mammalian mirtrons have many features different from Drosha-generated pre-miRNAs, such as substantially longer mirtron hairpins, frequently 3'-uridylation, and atypically heterogeneous 5' termini [20]. The mirtron pathway is a permanent class of non-canonical biogenesis and is thought to provide an option for the miRNA emergence prior to the advent of Drosha [19]. A second class is originated from other ncRNAs like small nucleolar RNAs (snoRNAs) and transfer RNAs (tRNAs) [21, 22], whose gene transcription often relies on the transcriptional and processing key factors of various RNAs to help produce the hairpins serving as Dicer substrates [1]. However, although some snoRNA and tRNA fragments can originate potential miRNAs, and even be loaded into RISC, most Ago-associated snoRNA and tRNA fragments may remain other functions unidentified now [23]. For example, studies suggested the DNA damage response (DDR) could be mediated by such ncRNAs [24, 25]. Besides, a kind of tRNA-derived fragments was demonstrated to enhance mRNA translation by regulating ribosomal proteins such as RPS28 [26, 27]. The third class involves 7-methylguanosine (m7G)-capped pre-miRNAs, which are synthesized from the 5' end of genes by Pol II directly, characterized by small RNA Cap-Seq, and exported by Exportin 1 [28].
Dicer-independent miRNAs are much fewer compared to Drosha-independent ones. Such miRNA biogenesis method needs Ago2 slicer catalytic activity [29]. A typical small RNA taking a great role in erythropoiesis, miR-451, is processed by Drosha but does not require Dicer for maturation. Its pre-miRNA gets cleaved by the catalytic center of the Ago loaded, generating an intermediate 3' end which is further trimmed by poly(A)-specific ribonuclease (PARN) [30, 31].
Canonical miRNA biogenesis and miRNA-mediated gene silencing. Transcribed by RNA Pol II, one nuclear miRNA gene produces a pri-miRNA, which is then recognized and cleaved into pre-miRNA by a microprocessor basically consisting of Drosha and DGCR8. Next, the pre-miRNA is exported to the cytoplasm with the help of Exportin 5 and RanGTP, further recognized and cut by Dicer and TRBP, and generates the miRNA-miRNA* duplex. After loaded into an Ago protein, the miRNA* strand is commonly degraded, while the miRNA strand forms the functional RISC. Upon loaded into RISC, the mature miRNA guides RISC to complementary target sequences located generally in the 3' untranslated region (3'-UTR) of mRNAs, then can function with the mode of mRNA degradation or translational repression. In the mRNA degradation mode, by the interactions with Ago protein, the GW182 recruits PABPC and binds the cytoplasmic deadenylase complexes PAN2-PAN3 and CCR4-NOT, rapid 5′-3′ mRNA degradation can also be in progress with the participation of DCP1-DCP2 complex. In the translational repression mode, the interaction of GW182 and CCR4-NOT can recruit DDX6 (a known translational inhibitor), and the release of eIF4AI or eIF4AII can inhibit the translation initiation. RNA Pol II, RNA polymerase II; DGCR8, DiGeorge syndrome critical region gene 8; RanGTP, Ran guanosine triphosphate; TRBP, HIV-1 TAR RNA binding protein; Ago, Argonaute; RISC, RNA-induced silencing complex; PABPC, polyadenylatebinding protein; PAN2-PAN3, poly(A)-nuclease deadenylation complex subunit 2 (PAN2)-PAN3; CCR4-NOT, carbon catabolite repressor protein 4 (CCR4)-NOT; DCP1-DCP2, mRNA-decapping enzyme subunit 1 (DCP1)-DCP2; DDX6, DEAD-box protein 6; eIF4A, eukaryotic translation initiation factor 4 A.
Regulation of microRNA biogenesis
As described in the canonical pathway, efficient miRNAs biogenesis relies on a series of elements and proper local RNA structures. In particular, the RISC slicer Ago2 can cut the pre-miRNA to additional processing intermediate termed ac-pre-miRNA, which can work as a Dicer substrate to mature into the active miRNA, which may facilitate nicked passenger strand removal [32]. Regulatory post-translational modifications (PTMs), like the phosphorylation, ubiquitylation, and sumoylation of miRNA biogenesis factors, can also affect miRNA processing by connecting miRNA expression with cell signaling pathways [23]. For example, a newly published study indicated that through the PTM of DDX17 (a cofactor of the Drosha-DGCR8 complex), miRNA biogenesis and histone modifications can get concerted regulation, which underlies many cancer stem-like features [33]. The target messenger RNA (mRNA) can also regulate the biogenesis of cognate miRNAs [34]. Besides, several recent studies suggested that RNA-binding proteins (RBPs) and long non-coding RNAs (lncRNAs) can control miRNAs biogenesis [35, 36]. Even, a lncRNA of hyper-conservative regional origin was illustrated to block pri-miRNA cleavage by Drosha [37]. In brief, the biogenesis process of miRNAs is in tight temporal and spatial regulation, and faulty biogenesis in human body would be associated with a great number of diseases, particularly cancer.
Effects of miRNA
RISC is an effector ribonucleoprotein complex, in which the miRNA works as a target-recognition element, an Ago family protein serves as a vital component, and a range of accessory components are along with them [38]. RISC assembly is a key process for miRNAs exerting functions, in which four Ago proteins named Ago1-4 encoded by the mammalian genome are available. Ago2 particularly, with additional special features introduced ahead, is the most highly expressed. Human miRNAs are thought to associate with all Ago proteins, even though some of them preferentially loaded into specific Ago proteins [39].
Generally, miRNAs are guided to specific target sites which are located in the 3'-untranslated region (3'-UTR) of mRNAs, then direct their post-transcriptional repression effects with the mode of mRNA degradation or translational repression. Nucleotide 1 of miRNA counted from 5' end is permanently unavailable for pairing, while complement miRNA nucleotides 2-8 form the seed which is pivotal for the recognition of target mRNA [40]. In the target mRNA, an adenine (A) opposite to miRNA nucleotide 1 (known as “t1A”) is recognized by an Ago binding pocket and confers enhanced target repression, supplementing the seed [41]. Although target sites that complement miRNA nucleotides 2-8 with t1A are the strongest, those with complementarity to miRNA nucleotides 2-7 or 3-8 are considered canonical too, as non-canonical sites (which lack a contiguous 6 nucleotides pairing to the seed region) are thought inefficacy [42]. By the way, in a subset of sites, the 3′ end pairing can make help for target recognition, particularly nucleotides 13-16 (the supplementary region) [43]. Recent findings have also shown that 3′ end pairing on uridylated miRNAs can also contribute to novel target recognition [44].
The mRNA decay requires the consecutive action of a 182 kDa glycine-tryptophan protein (GW182) and the mRNA-decapping enzyme subunit 1 (DCP1)-DCP2 complex [45]. Following the multivalent protein actions with Ago proteins [46], GW182 recruits cytosolic polyadenylate-binding protein (PABPC), which promotes mRNA deadenylation through binding the poly(A)-nuclease deadenylation complex subunit 2 (PAN2)-PAN3 complex and carbon catabolite repressor protein 4 (CCR4)-NOT complex [47]. Deadenylation is followed by decapping via the DCP1-DCP2 complex, leading to rapid mRNA degradation with 5′-3′ exoribonuclease 1 (XRN1) [48]. Apart from the degradation of miRNA targets, GW182-mediated interaction with CCR4-NOT can also promote translational repression by the recruitment of a known translational inhibitor, the DEAD-box ATPase DEAD-box protein 6 (DDX6) [49]. And the translation initiation can be inhibited by releasing eukaryotic translation initiation factor 4 A-I (eIF4AI, the founding member of DEAD-box helicases) or its paralogue eIF4AII from target mRNAs [50], which is proved to be GW182 independent in Drosophila even [51]. However, in 2018, a study reported novel miRNA-recognition elements (MREs) that only work in protein coding sequences (CDS) regions, which is in requirement of extensive 3' side base-pairing rather than the 5' seed, and is Ago-dependent but GW182-independent [52]; In 2016, another study reported that Ago Tryptophan-binding pockets mediating GW182 binding are required for translation inhibition in human cells as well as D. melanogaster cells [53], both showing the complete mechanism of translational repression still remains unresolved (Figure 1).
It is revealed that two modes of miRNA-mediated gene silencing are connected with each other [54], and global steady-state measurements verified that the effect of mRNA decay dominates for generally explaining 66% - >90% of such silencing [55]. Besides, there are complex interaction networks formed by miRNAs, as each miRNA can influence widespread genes with the advantage of their small size, and multiple miRNAs can silence the same gene, although the effect of each miRNA on each gene is relatively mild [43]. Typically, the miRNA abundance in any cell is less than the effective target abundance (the count of sites that have to be combined when achieving half-maximal de-inhibition on the targets), which illustrates why lowly expressed miRNAs function negligibly and suggests that even the most repressed genes can be silenced by increased miRNAs [1]. By reinforcing normal transcripts and attenuating abnormal transcriptional programs, miRNAs can buffer against random fluctuations in the post-transcriptional level [56]. What needs to be emphasized is, cytoplasmic miRNAs can localize in the endoplasmic reticulum, mitochondria, and other compartments with some differences in their pathways [57].
Intriguingly, different from cytoplasmic functions mentioned above, miRNA can affect regulation both post-transcriptionally and transcriptionally in nuclear by working with promoters in three models including RNA-RNA hybrid, RNA-DNA hybrid, and RNA-DNA triplex [58]. Besides, nucleus miRNAs can serve as enhancer triggers to activate gene transcription epigenetically [59]. All of those are related to Ago proteins. Recently, a study illustrated that overexpressed nuclear Ago in embryonic stem cells has an important place at the beginning of differentiation [60]. Furthermore, nucleoplasmic protein Sfpq can directly modulate nucleoplasmic and cytoplasmic miRNA functions [61]. These findings present a complicated miRNA-related modulation network, and it will be more perfect as new discoveries continuing to appear.
Cell-free miRNAs found in various biological fluids indicate that miRNAs could contribute to intercellular signaling, and are potential biomarkers available for a variety of diseases [62]. Exosomes, the naturally equipped biological vehicles, have been thought to contain most circulating miRNAs but with some controversy [39], and adipose tissue is proved to constitutes a primary source for circulating exosomal miRNAs [63]. Besides, although exosomal Ago2-loaded miRNAs and single-stranded miRNAs have been detected, evidence of cell-cell communication via exosomal miRNAs in a physiological context remains elusive [39]. Recently, a study found miR-494 was up-expressed in exosomes of melanoma samples, which can be reduced by Rab27a depletion [64], showing the feasibility of transferred exosome-shuttled miRNAs as therapeutic methods. Thus, we require more studies to fully understand their mechanism and function.
Biological functions of miRNA in cancer
miRNAs in cancer progression
In 2002, the deletion and low-expression of miR-15 and miR-16 cluster in chronic lymphocytic leukemia were demonstrated, firstly suggesting the role of miRNAs in the progression of cancers [65]. During the past period, miRNAs have been linked to virtually all known cancer processes. According to the target gene, some miRNAs often negatively influence protein-coding oncogenes, while some other miRNAs can inhibit known tumor suppressors, therefore miRNAs could act as onco-miRNAs or tumor-suppressor miRNAs [66].
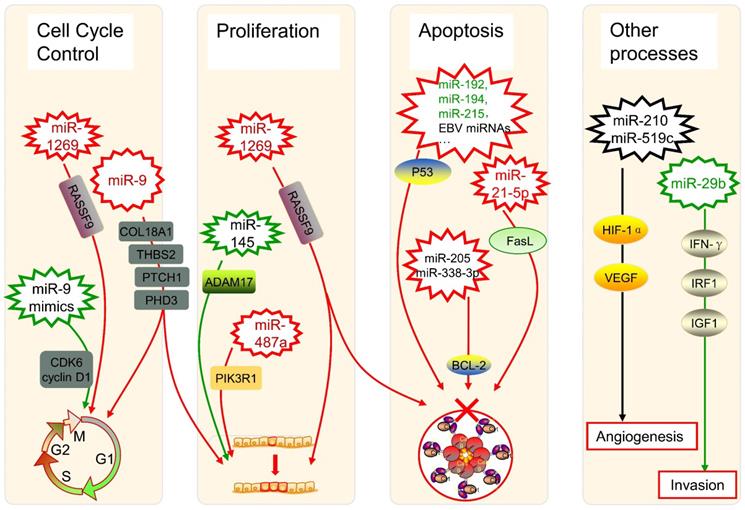
The dysregulation of miRNAs can also link to alterations in genes that govern cancer progression. During the formation and progression of gastric cancer, the expression of miR-1269 is upregulated, which promotes cancer cell proliferation as well as cell cycle G1-S transition and suppresses cell apoptosis by regulation of the AKT signaling pathway and the Bax/Bcl-2 signaling pathway, with the target of RASSF9 [67]. In oral squamous cell carcinoma (OSCC) patients, miR-9 is downregulated, whose expression induces G0/G1 cell cycle arrest, while the use of miR-9 mimics significantly halts cell proliferation by repressing cyclin-dependent kinase 6 (CDK6) and cyclin D1 [68]. Besides, miR-9 has an expanded scope of target RNAs because of its pri-miRNA paralogs [69]. As one of the most abundant miRNAs in the brain, miR-9 was verified to target COL18A1, THBS2, PTCH1, and PHD3 directly in glioma cells, whose overexpression takes positive roles in processes such as proliferation and cell cycle progression [70]. Therefore, the regulatory characterization of miRNAs in the cell cycle regulation is complex, maybe with some critical pathways still to be found.
As for cell proliferation, it was found that the miR-145 targeting A disintegrin and metalloproteinase 17 (ADAM17) could inhibit such a process in liver cancer cells [71]. Another study about hepatocellular carcinoma (HCC) showed overexpressed miR-487a happened in poor prognostic patients, which increases cell proliferation by phosphoinositide-3-Kinase regulatory subunit 1 (PIK3R1) induced AKT signaling [72]. Those findings can provide novel promising treatment strategies in the therapy of HCC, as well as other cancers.
Several miRNAs can take roles in programmed cell death, especially in the apoptosis of cancer cells, too. The dysregulation of p53 has been widely reported to enable cancer cells against apoptosis, which could be triggered by the low-expression of miR-192, miR-194, and miR-215 in multiple myeloma [73]. It was also shown the p53 diversification happened because of several miRNAs in a research of soft tissue sarcoma as well as breast cancer [74], while some Epstein-Barr virus (EBV) miRNAs were associated with p53 regulation in nasopharyngeal cancer as well as EBV-associated gastric cancer [75]. Besides, miR-205 and miR-338-3p can restrain the apoptosis of prostate cancer cells by targeting one of its inhibition genes, B-cell lymphoma 2 (BCL-2) [76]. In the extrinsic apoptotic pathway, miRNAs can inhibit pivotal elements like Fas ligand (FasL). It was characterized that FasL is directly targeted by miR-21-5p, and the expression of its mRNA and protein both can be negatively influenced by the up-regulation of miR-21-5p in HCC cells [77].
The angiogenesis of tumors can be controlled by miRNAs such as miR-210 and miR-519c under hypoxic conditions via modulating Hypoxia-inducible factor 1α (HIF-1α) and vascular endothelial growth factor (VEGF) [78]. Regulatory networks of miRNAs are considered to be a characteristic of epithelial-mesenchymal transition (EMT) in the physiopathology of OSCC [79]. An aggressive phenotype of colorectal cancer (CRC) is significantly associated with depression of miR-29b, whose existence restricts the development and incursion of cancer cells, with the potential interaction of interferon-γ (IFN-γ), IRF1, and IGF1 [80]. All those highlighted showed that miRNAs are implicated in tumor progression including cell cycle, cell proliferation, cell apoptosis, angiogenesis, EMT, and tumor invasion, acting important and complex roles in the regulatory network of cancers (Figure 2).
miRNAs affect tumor immunity
The dysregulation of patient immune systems is significant in cancer development, and a variety of miRNAs implicate in the process of tumor immune surveillance as well as tumor immune escape. As discussed ahead, miRNAs can be loaded by exosomes and then involved in intercellular communication. In cancer, exosomes can affect many aspects including the progression, metastasis, multidrug resistance, and EMT [81]. Some kinds of immune cells can be directly interacted with, such as the function of natural killer (NK) cells limited by miR-23a, the function of dendritic cells diminished by miR-212-3p and miR-203, and the inhibition of CD8+ cells while the activation of CD4+ regulatory T cells effected by tumor-derived miR-24-3p [81]. Especially, talking about NK cells, emerging evidence shows that some miRNAs can, directly and indirectly, modulate their anti-tumor potential for instance by controlling the surface expression of immune checkpoints on NK cells or that of their ligands on tumor cells [82-85].
Apart from the oncogenic and tumor-suppressive function, miRNAs were reported to impress on tumor immunogenicity and antitumor immune responses. For example, a study confirmed that miR-124-3p is the target of LINC00240, and could promote cervical cancer development with the action of STAT3 and MHC class I-related proteins A (MICA), mediating natural killer T (NKT) cell tolerance [86]. While, under the HLA class I antigen processing machinery (APM) components, regulation of the B7 family members, and expression of IFN-γ signaling molecules, other miRNAs including miR-346, miR-451, and miR-148a are summarized to be implicated [81]. In fact, miRNAs are interacted with many immune checkpoint proteins (ICPs) other than the B7 family [87], presenting significant advantages for the personalized treatment and prognostic prediction of cancer patients.
What is more, miRNAs have become the critical regulators for antitumor immune cells. Here we present several related studies. For tumor-infiltrating lymphocytes (TILs), scientists characterized the tight connection among miR-574-3p, PD-L1 expression, and TIL levels in chordoma [88], while mechanically another group has presented the function of the p53/miR-34/PD-L1 axis in immune evasion of lung cancer [89]. For myeloid-derived suppressor cells (MDSCs), miR-34a can also regulate oncoprotein MUC1 and then results in the MDSC increase in acute myeloid leukemia (AML) [90]. For tumor-associated macrophages (TAMs), miR-98 directly targets the gene of interleukin 10 (IL-10) in TAMs, suppressing the progress of HCC [91]. For cancer-associated fibroblasts (CAFs), miR-21 acts a role in the metabolically altering process of CAFs thereby affects pancreatic cancer cells [92].
Involved miRNA sponges in cancer
A pool of different transcripts that compete synergistically to attract miRNAs for interactions is known as competing endogenous RNAs (ceRNAs), mainly including some experimentally confirmed ncRNAs, even some pseudogenes and protein-coding RNAs. Through the titration mechanism, the ceRNAs can regulate each other indirectly, while ceRNA-ceRNA interactions are also called miRNA sponge interactions [93].
In addition to miRNAs, ncRNAs contain several types including lncRNAs, circular RNA (circRNAs), small nuclear RNAs (snRNAs), and so on, making up 98% in the human genome [94]. Among these, lncRNA is the one first found as miRNA sponge [93], and many recent studies illustrated the role lncRNAs take in miRNA-related tumor regulation. In a study about lung adenocarcinoma stem cells, miR-146 is referred to as the post-transcriptional regulation of NUMB, while one of its sponge TUSC-7, a strong suppressive lncRNA, could abolish the degradation toward to NUMB by inactivating NOTCH signaling. This research proved the decrease of tumor-suppressive miR-146 was essential in TUSC-7 function, showing complex non-coding genes employed in renewal repression regulation [95]. In nasopharyngeal carcinoma (NPC), the lncRNA FAM225A was over-expressed as a sponge of miR-590-3p and miR-1275, promoting NPC development by up-regulating integrin β3, the target of the two miRNAs [96]. Besides, a novel lncRNA, prostate cancer-associated transcript 7 (PCAT7), was considered to promote the tumor progression of NPC as a miR-134-5p sponge [97]. In addition to these, the miRNA sponge-lncRNAs have also been studied in numerous kinds of cancers such as a miR-34a sponge MALAT1 in melanoma [98], a miR-15a/16 sponge LINC00461 in multiple myeloma [99], a miR-330-5p sponge LINC00958 in pancreatic cancer [100], a miR-7-5p sponge RP4 in colorectal cancer [101], a miR-324-5p sponge TPT1-AS1 [102] and a miR-186 sponge antisense noncoding RNA of the INK4 locus (ANRIL) [103] in cervical cancer, a miR-96 sponge TP53TG1 in pancreatic ductal adenocarcinoma [104], and a miR-300 sponge TUG1 in gallbladder carcinoma (GBC) [105].
circRNAs are some endogenous ncRNAs with a covalently closed successive circle, working as miRNA sponges is one of its important roles [106]. The circRNAs expressions were abnormal in tumor cell lines, tissues, and even plasma samples of cancer patients with the advantage of RNA sequencing technology [107]. In one study, circMMP9 was found to act as a sponge directly targeting miR-124, quickening up the growth and migration of glioblastoma multiforme (GBM) cells. Furthermore, they found that the cyclization and increased expression of circMMP9 were induced by eukaryotic initiation factor 4A3 (eIF4A3), whereas eIF4A3 tied up with the mRNA transcript of MMP9 in GBM [108]. Besides, circRNAs were verified as miRNA sponges in OSCC [109], HCC [110], lung cancer [111], breast cancer [112], gastric cancer [113], and so on.
Pseudogenes are genomic elements that lose protein-coding abilities because of various endogenous inactivating mutations [114] and some of them have also been found to be miRNA sponges such as PTENP1 and OCT4P4 [115]. Some types of protein-coding RNAs can sponge to miRNAs, too [93]. The tumor suppressor gene PTEN has been demonstrated to be a miRNA sponge [116], while TNRC6B was found to be a PTEN ceRNA then, showing hints at complex interactions among different pathways in cancer [117]. Apart from them, an antisense RNA of the TP73 gene, TP73-AS1, was verified to sponge to human-specific miRNA miR-941 by Ago protein immunoprecipitation experiments, wound-healing assay, and massive transcriptome data analyses. Moreover, the duo represents of TP73-AS1 and miR-941 showed a fast alteration of noncoding regulators that influences cell migration, proliferation, and tumorigenesis [118].
Several miRNAs in cancer progression. The up-expression or down-expression of miRNAs (in red or green, if mentioned) could influence specific cancer progression process (red frames and lines mean promote cancer progression, while green ones mean inhibit). RASSF9: Ras-association domain family 9; CDK6: cyclin-dependent kinase 6; EBV: Epstein-Barr virus; BCL-2: B-cell lymphoma 2; FasL: Fas ligand; ADAM17: A disintegrin and metalloproteinase 17; PIK3R1: phosphoinositide-3-Kinase regulatory subunit 1; HIF-1α: Hypoxia-inducible factor 1α; VEGF: vascular endothelial growth factor; IFN-γ: interferon-γ; IRF1: interferon regulatory factor 1; IGF1: insulin-like growth factor 1.
However, some questions had been raised about the hypothesis of ceRNAs. In 2014, a stoichiometry on miR-122 and its targets shown little influence exerted by ceRNAs [119]. Another study on global properties also doubted the sponge function of the vast majority of circRNAs [120]. Furthermore, researchers found the phenomenon of target mRNAs or ncRNAs triggering miRNA tailing, trimming, and decay, which was termed as target RNA-directed miRNA degradation (TDMD) [121]. A recent study showed this is a process triggered by target RNAs with an enzymatic attack activity from an Ago2 PAZ domain around miRNA 3' end [122]. Compare to ceRNAs, TDMD needs much fewer target RNAs to make obvious miRNA reduction [123]. Based on the TDMD study, a regulatory network containing a lncRNA (Cyrano), a circRNA (Cdr1as), and two miRNAs (miR-7, miR-671) was even established for neuronal activity [124]. In short, we think further investigations are needed to reach a consensus and theorem about those ncRNAs participating in the biological action of miRNAs in cancer.
Clinical utilization of miRNA
Potential miRNA-based biomarkers
As so many functions miRNAs have in cancer development, the dysregulation of one specific miRNA or a group of miRNAs may be closely associated with human cancer progression. There are thousands of papers published by scientists all over the world in PubMed about potential miRNAs that are available for cancer diagnosis and prognosis. We searched recent journal articles reported since Jan 1st, 2017, and presented some of them here (Table 1). What should be noted is, a lot of them were not verified by Northern Blot—the gold standard for analyzing RNA, so the utilization of those potential biomarkers in clinical practice at once is not suitable. However, these studies might provide novel insights and promising tools for us.
miRNA-based biomarkers for diagnosis
The miRNAs from different sample sources might have idiographic meanings. The urinary cell-free miRNAs are appropriate potential markers for some urologic neoplasms, for example, researchers performed a multicenter study by 543 urine samples and proposed a noninvasive diagnostic tool with a > 90% (p < 0.001) sensitivity to discriminate bladder cancer from benign hematuria, which could reduce unnecessary cystoscopies at a certain point [125]. Another study even reported the relationship between urinary levels of miR-126 and children proliferating hemangiomas [126]. Besides, a research of head and neck squamous cell carcinoma (HNSCC) put forward a combination of seven methylations of genomic loci encoding microRNA (mgmiRs) with sensitivity and specificity of 76.7% and 86.1% respectively in saliva, even though lower than that in tissues as 92.6% and 92.4%, still a novel and promising screening tool [127]. What is interesting, the measurement of several miRNAs on cerebrospinal fluid (CSF) samples suggests high sensitivity as well as specificity for the distinction of primary central nervous system lymphoma (PCNSL) [128]. And, some doctors came up with a feasible idea of detecting significant miRNAs on fine-needle aspiration (FNA) samples with the method of endoscopic ultrasonography for pancreatic cancer early detection [129]. These sources like urine, saliva, CSF, FNA samples, as well as common serum or plasma used for miRNA detection showed us excellent potential in their clinical applications.
A variety of detection methods were used to explore the potential miRNA biomarkers. What impressed us most is the electrochemical nano-genosensor. Such nano-genosensors had a remarkable signal amplification effect, resulting from the unique structure of nitrogen-doped functionalized graphene, nano-sized silver particles, and polyaniline [130]. By applying differential pulse voltammetry (DPV), researchers performed a detection after the functionalizing of that nanocomposite and immobilizing of the circumstantial miR-21 aminated complementary oligonucleotide sequence, as miR-21 is a foregone biomarker in breast cancer. By virtue of highly sensitive and label-free, that nano-biosensor is utilizable for breast cancer early discovery via detecting the miR-21 on human samples directly without the need for the pretreatment, RNA extraction, or amplification [130]. Equally exciting is, some researchers exploited a digital amplification-free quantification manner by the use of NanoString Technologies, the nCounter technology, which can identify dysregulated exosomal serum miRNAs (ex-miRNAs) for progressive prostate cancer [131]. The above studies provide new ideas, and we look forward to more advanced technologies applied to the detection of miRNAs.
Those diagnosis methods put out based on miRNAs, though with some differences, have great significance. Unlike those who use sole miRNA as biomarkers, some scientists identified a panel of miRNAs for a kind of cancer, like the six miRNAs significantly overexpressed in gastric cancer [132]. Three exosomal miRNAs together also presented higher accuracy for the diagnosis of metastatic lung cancer than a single one [133]. After the verification of Northern Blot and TaqMan miRNA qRT-PCR assays, scientists noticed the considerably higher ratio of miR-25/92a group to the miR-22/29a group in cervical specimens with cervical intraepithelial neoplasia, then proposed an early diagnostic method for cervical cancers [134]. Another group identified the DNA methylation of genomic loci encoding miRNAs (mgmiRs) in vitro cell lines, and successfully developed a seven-mgmiR diagnostic panel for HNSCC [127]. A more instance, four miRNAs (miR-498, miR-183, miR-205, and miR-31), which are suitable indicators in urine for the presurgical identification of representative benign renal oncocytoma, can well conduct preoperative plans and avoid some unnecessary stress for patients [135]. These miRNA panels may be more effective than sole miRNA used, but, both with outstanding significance for cancer patients.
However, there still exist challenges unresolved for the application of miRNAs as cancer diagnosis biomarkers. There are three plasma miRNAs (miR-222-3p, miR-423-5p, and miR-150-5p) for the detection of OSCC identified, former two of which are considered inversely associated with cancer progression related to T stage, lymph node metastasis, and clinical stage, while the latter one in plasma is up-regulated in OSCC. Nevertheless, what the Cancer Genome Atlas (TCGA) analysis reveals is not matched to their findings, as OSCC miR-222-3p increased in tissue whereas not in plasma, and over-expressed miR-150-5p just happened in plasma [136]. Talking about the inconsistency of miRNA levels between plasma and tissue, the explain maybe the selective release of miRNA [137], or maybe various parameters (such as drugs) affecting circulating miRNAs [138], or maybe something we haven't discovered yet. Furthermore, as we introduced, there are many miRNA sponges involved in cancer so some of them may be used as biomarkers such as lncRNA MIR31HG (a miR-575 sponge) for HCC [139] and circRNA cTFRC (a ceRNA for miR-107) for bladder cancer [140], providing a new perspective on cancer diagnosis.
miRNA-based biomarkers for prognosis
Many articles concerning miRNA biomarkers have revealed the mechanisms behind. Publishing in an article about ESCC in April 2019, researchers first determined the miRNA and mRNA expression situations in ESCC tissues by the application of microarray; following this, they constructed a miRNA-mRNA regulatory network by multistep bioinformatics methods; then they measured miR-1 expression using in situ hybridization (ISH) and explored its connection with clinicopathological characteristics and outcome of ESCC patients. Moreover, they studied the functional role of miR-1 with several experiments and verified a direct target gene—Fibronectin 1 (FN1)—by applying technologies like qRT-PCR, Western blot, and luciferase reporter assays [141]. In short, these results provide a valuable therapeutic target more than only a potential prognostic predictor.
As we mentioned earlier, miRNA sponges might be significant components for the post-transcriptional gene regulation, and some miRNA sponges have been indicated to be prospective biomarkers of cancer. For example, markedly up-regulated lncRNAs CCAT1—a sponge of let-7 miRNA family—is linked with AFP, tumor size, and microvascular invasion in HCC, which presented as a poor prognostic biomarker for HCC patients [142]. With the consideration of different molecular profiles in breast cancer subtypes, researchers found the subtype-specific sponge interactions are quite distinct [143], which show high prognostic potential for breast cancer subtypes and may expand research perspectives for other tumors. By the way, another group analyzed the interactions of the lncRNA-mRNA ceRNA network based on the four subtypes of breast cancer systematically [144], providing new insights for other scientists.
Some researchers identified prognostic factors by using expressed databases such as TCGA and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC). For instance, the METABRIC cohort data was utilized to further explore the expression and survival data of miR-196a in breast cancer, following with genomic data analysis and chromatin interaction analysis [145]. Not only can these databases save a lot of time and effort, but they also remove the need for patient consent or ethics approval.
Currently, computer-aided biomarker discovery is full of attractiveness. Take a novel network vulnerability analysis model reported for example, scientists identified five miRNAs as potential biological markers for prostate cancer metastasis based upon their bioinformatics model, and get convinced by the prediction action, reported articles, and functional enrichment analysis [146]. As experimental methods are difficult to find the driver or pivotal molecules in the viewpoint of systems, such a concentrated network in the model can serve as a biological system for biomarker prediction, which focuses on special regulatory patterns of stability altering [146].
Taken together, as more and more miRNA biomarker candidates in cancers have been come up with, studies with larger cohorts are needed, and complete mechanisms of miRNA dysregulation are warranted to be understood.
Potential miRNA biomarkers for diagnosis and prognosis in cancers
| Cancer type | miRNA | Dysregulation | Sample type | Detection method | Ref. |
|---|---|---|---|---|---|
| for diagnosis | |||||
| Pheochromocytomas or paragangliomas (PPGLs) | miR-210 | down | serum | qRT-PCR | [196] |
| Colorectal cancer (CRC) | miR-103a-3p, miR-127-3p, miR-151a-5p, miR-17-5p, miR-181a-5p, miR-18a-5p and miR-18b-5p | up | plasma | qRT-PCR | [197] |
| Cervical cancer | the ratio between miR-25/92a and miR-22/29a groups | up | cervical specimens | Northern Blot and TaqMan miR qRT-PCR assays | [134] |
| Gastric cancer | miR-491-5p | down | Tissue and serum | qRT-PCR | [198] |
| Invasive breast cancer | exosomal miR-223-3p | up | plasma exosomes | TaqMan miR assays | [199] |
| Glioma | miR-21, miR-222 and miR-124-3p | up | serum exosomes | qRT-PCR | [200] |
| breast cancer | miRNA-21 | up | blood | electrochemical nano-genosensor | [130] |
| Lung cancer | miR-33a-5p and miR-128-3p | down | tissue and blood | qRT-PCR | [201] |
| Prostate cancer | miR-214 | down | cell lines | qRT-PCR | [202] |
| Hepatocellular carcinoma (HCC) | miR-221 | up | cell lines, tissues, and serum | qRT-PCR | [203] |
| Oral squamous cell carcinoma (OSCC) | miR-222-3p, and miR-423-5p / miR-150-5p | down/up | plasma | qRT-PCR | [136] |
| Head and neck squamous cell carcinoma (HNSCC) | mgmiR9-1, mgmiR124-1, mgmiR124-2, mgmiR124-3, mgmiR129-2, mgmiR137, and mgmiR148a | up | tissues, cell lines, and saliva | qMS-PCR | [127] |
| Primary central nervous system lymphoma (PCNSL) | miR-21, miR-19b, and miR-92a | up | cerebrospinal fluid (CSF) | RT-qPCR | [128] |
| Germ cell tumours (GCTs) | miR-371a-3p | up | serum | RT-qPCR | [204] |
| Synovial sarcoma (SS) | miR-92b-3p | up | serum and cell lines | miRNA microarray and RT-qPCR | [205] |
| for poor prognosis | |||||
| Esophageal squamous cell carcinoma (ESCC) | miR-1 | down | tissues | ISH | [141] |
| Advanced cervical cancer | miR-944 | up | cell lines and FFPE tissues | RT-qPCR | [206] |
| Oral squamous cell carcinoma (OSCC) | miR-1290 | down | plasma | qRT-PCR | [207] |
| Colorectal cancer (CRC) | Circ_0026344 (a miRNA sponge for miR-21 and miR-31) | down | tissues and cell lines | qRT-PCR | [208] |
| Pancreatic ductal adenocarcinoma (PDAC) | exosomal miR-451a | up | plasma | TaqMan MiR assays | [209] |
| Small intestinal neuroendocrine tumors | miR-375 | down | biopsies | ISH | [210] |
| TNM stage II and III colon cancer | a 16-miRNA signature including miR-143-5p, miR-27a-3p, miR-31-5p, miR-181a-5p, miR-30b-5p, miR-30d-5p, miR-146a-5p, miR-23a-3p, miR-150-5p, miR-210-3p, miR-25-3p, miR-196a-5p, miR-148a-3p, miR-222-3p, miR-30c-5p and miR-223-3p | up | fresh frozen biopsies | RT-qPCR | [211] |
| Gastric cancer | miR-155 | up | tissues | qRT-PCR | [212] |
| Pancreatic ductal adenocarcinoma (PDAC) | miR-296-5p | up | tissues and cell lines | miRNA array and RT-PCR | [213] |
| Ovarian cancer | miR-135a-3p | down | serum | miRNA array and qRT-PCR | [214] |
| Chronic lymphocytic leukemia (CLL) | miR-155-5p | up | peripheral blood mononuclear cells (PBMCs) | an in-house-developed qRT-PCR | [215] |
| Prostate cancer | miR-424-3p | up | tissues | ISH | [216] |
| Breast cancer | miR-362-3p | up | data from TCGA | - | [217] |
| Clear cell renal cell carcinoma (ccRCC) | miR-130b, miR-18a, and miR-223 | up | data from TCGA | - | [218] |
| Breast cancer | miR-196a | up | data from METABRIC | - | [145] |
qRT-PCR: quantitative real-time polymerase chain reaction; RT-qPCR: Reverse transcription-quantitative PCR; mgmiR: methylation of genomic loci encoding microRNA; qMS-PCR: quantitative methylation-specific PCR; ISH: in situ hybridization; FFPE: formalin-fixed paraffin-embedded; TCGA: the Cancer Genome Atlas; METABRIC: the Molecular Taxonomy of Breast Cancer International Consortium.
miRNA-based therapies
As we discussed earlier, miRNAs play a non-negligible role in the regulation of cancers and can, therefore, be used as alternative therapeutic targets. Many miRNAs have been entered the clinical stage of cancer treatment, along with many related experiments in vitro and in vivo, as introduced later (Table 2). Here, we would like to picture some advanced treatment methods including the following four forms (Figure 3).
Combination of miRNA therapeutics and chemotherapy
Recently, numerous researches have been conducted on chemo with miRNA therapeutics. In completed pharmacological experiments, miR-126 inhibits CDK4/6 and PIK3CA to prevent cell cycle progression along with the mitosis. In related experiments, it has been confirmed that transfection of miR-126 mimic into breast cancer cells can enhance its sensitivity to fourteen chemotherapy drugs such as trimetinib and alpelisib, and prevent the emergence of drug resistance [147]. Moreover, temozolomide-resistant glioblastoma cells sensitized by miR-151a was restored with the moderation of XRCC4-mediated DNA repair after the transfection of miR-151a mimic into the glioblastoma cell line [148]. Investigators studied the tumor-suppressive function of miR-205 in gemcitabine-resistant pancreatic ductal adenocarcinoma, finding that lentiviral vectors can inhibit cancer stem cell proliferation and resensitize those gemcitabine-resistant cells when miR-205 is overexpressed. Thus, it supplied a platform for the combined treatment of miR-205 and gemcitabine as a feasible method to treat advanced pancreatic cancer [149]. Another science group used pancreatic cancer miR-1291 prodrug with gemcitabine and nab-paclitaxel in cancer model mice, noticing that miR-1291 up-regulated ARID3B and further induced programmed cell death, DNA damage, and mitotic block, which significantly inhibited the growth of tumor cells [150]. Correspondingly, it was confirmed that forced expression of miR-634 can induce programmed cell death in a variety of cell lines containing pancreatic cancer cells, and intravenous injection of lipid nanoparticles containing miR-634 can greatly reduce xenograft tumor progress in mouse BxPC-3 cells [151]. In another study, raised miR-149 level restrained cell proliferation and colony formation in neuroblastoma cells, nevertheless promoted apoptosis and chemical sensitivity to doxorubicin (Dox) [152]. Dox was also found to induce autophagy and interact with miR-137, while miR-137 can make it easier for pancreatic cancer cells to react with chemotherapy [153]. The combinatorial strategy of anti-tumor miRNAs with chemo drugs can synergistically enhance the therapeutic efficacy, thus showing a hopeful research direction for cancer therapies.
Combination of miRNA therapeutics and radiotherapy
The prime mechanism of radiotherapy is inducing DNA double-strand break (DSB). It is known that miR-34a acts as a regulator in various human neoplasms [154]. In an animal experiment, scientists studied the cooperative relationship between miR-34a and radiation therapy, finding that overexpression of miR-34a (which bound to the 3'-UTR site of RAD51) enhanced γ-H2AX foci and inhibited the formation of HR and RAD51 foci, thereby inhibiting tumor cell DNA repair. They transfected MRX34 (a liposomal nanoparticle bond to miR-34a mimics) into a mouse model of non-small cell lung cancer (NSCLC), leading to significantly enhanced radiotherapy effect and suppressed tumor cell growth [155].
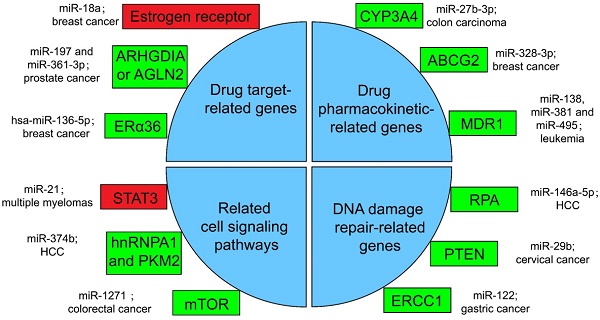
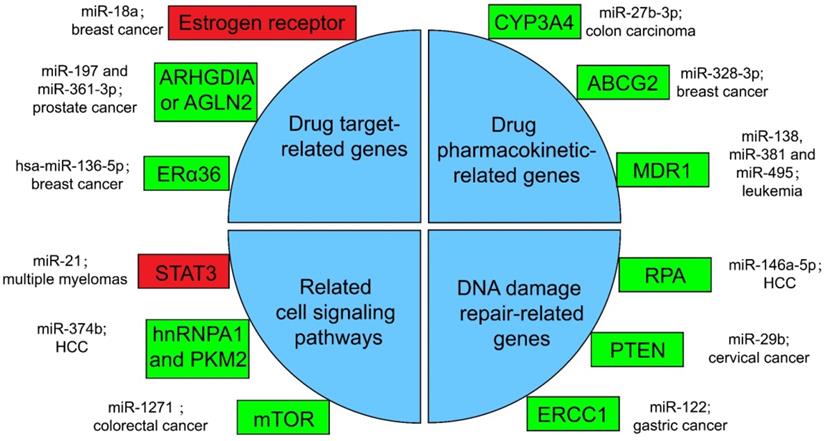
Several miRNA-related cancer therapy resistance. There are four categories along with some regulators presented in the text, where the rectangle in green or in red shows whether the function of miRNAs in mentioned cancer type is positive or negative for the corresponding therapy. ERα36: estrogen receptor alpha 36; CYP: cytochrome P450; ABCG2: ATP-binding cassette sub-family G member 2; MDR1: multidrug resistance 1; STAT3: signal transducer and transcriptional activating factor 3; MYC: myelocytomatosis oncogene; HnRNPA1: heterogeneous nuclear ribonucleoprotein A1; PKM2: the Pyruvate kinase muscle 2; mTOR: mammalian target of rapamycin; RPA: replication Protein A; PTEN: phosphatase and tensin homologous protein; ERCC1: excision repair cross-complementing 1.
Clinical trials with miRNA-based therapeutics
| Targeted miRNA | Developmental drug or format of miRNA therapeutics | Indication | Phase |
|---|---|---|---|
| miR-16 | Mesomir | Malignant pleural mesothelioma | Clinical Trial Phase I [219] |
| miR-34a | MRX34 | melanoma | Clinical Trial Phase II [220] |
| miR-122 | Miravirsen | Hepatitis C | Clinical Trial Phase II [221] |
| miR-34a | MRX34 | NSCLC | Clinical Trial Phase I [155] |
| miR-634 | miR-634-LNP | Pancreatic Cancer | in vitro [151] |
| miR-429 | Pre-miR-429 | Pancreatic Cancer | in vitro [222] |
| miR-126 | miRNA mimic | Breast cancer | in vitro [147] |
| miR-151a | miRNA mimic | Glioblastoma | in vitro and in vivo [148] |
| miR-155 | Cobomarsen (MRG-106) | chronic lymphocytic leukemia | Clinical Trial Phase II [223] |
| miR-1294 | miRNA mimic | Glioma | in vitro and in vivo [224] |
| miR-383-5p | miRNA mimic | Ovarian cancer | in vivo [225] |
| miR145 | miRNA145 mimics | colorectal cancer | in vivo [226] |
| miR-1258 | miR-1258 mimic | myeloma | in vitro and in vivo [160] |
| miR-497 | miR-497 mimic | NSCLC | in vitro [227] |
The sharp phosphorylation of histone H2AX to γ-H2AX presents a biological marker for DNA repair after radiotherapy. miR-328-3p can directly interact with the 3'-UTR of H2AX transcript and reduce its protein expression level, whose overexpression promotes radiation-induced cellular DNA damage. Investigators then introduced miR-328-3p mimic into animal models of NSCLC, and the tumor cells were significantly reduced in mice after radiotherapy, proving that overexpressed miR-328-3p can improve cell radiosensitivity with the change of DNA damage/repair signaling pathways [156]. In conclusion, miRNA targeting approaches are able to promote the sensitivity and effect of radiation therapy, and employing them together would potentially help many carcinoma patients improve their prognosis.
Combination of miRNA therapeutics and immunotherapy
Immunotherapy is one of the main treatments for current cancer treatments, one of its principles is to inject genetically modified tumor-specific T cells for malignant tumor patients to identify, aim, and kill tumor cells. However, the persistence and effectiveness of T cell therapy are not satisfactory. In addition, miRNAs are key controllers for T cell stimulation, proliferation, living, differentiation, and effector function, all of which are dominant causes for judgment of adoptive immunotherapy treatment. With the in-depth understanding of miRNA and the exploration of its application in treatment, the application of miRNA will solve many problems in immunotherapy [157]. For example, miR-143 overexpression enhances specific killing of HER2-CAR (human epidermal growth factor receptor 2-chimeric antigen receptor) T cells which target esophageal carcinoma cell line TE-7, because miR-143 enhances the tumor-treat effect of T cells by supporting memory T cell differentiation and metabolic reprogramming via targeting glucose transporter 1 (Glut-1) gene [158]. A novel lentiviral vector of one miRNA cluster and epidermal growth factor receptor (EGFR)-variant III was introduced into CAR T cells to enhance the durability of its therapeutic response in glioblastoma, which effectively improved the efficacy of adoptive T cells [159]. Besides, it was illustrated that miR-1258 targeted the 3′-UTR of programmed death ligand 1 (PDL1), and thus affected the treatment of myeloma by inhibiting PDL1 [160]. With the introduction of miRNAs, we may deal with those difficulties in immunotherapies, and promote their application in human cancers.
Replacement therapy or miRNA mimics
Through research on miRNA-based cancer gene therapy, it has been found that single miRNA has limited therapeutic effects on cancer. This is because many miRNAs participate in the appearance and development of malignant neoplasms by regulating broad target genes or various signal transduction processes, and cancer cells can get tolerance to miRNA involvement within a special period and restore proliferative capability via other routes readily [161]. Therefore, interventions targeting no less than two miRNAs with complementary relations may restrain multiform signaling pathways and improve the efficacy of miRNA mimic therapy.
Up-regulated miR-34a in cell line HCT116 remarkably restrained cell growth, migration, aggression, and metastasis, along with evoked apoptosis, G1 phase arrest, and p53 transcriptional state [162]. Scientists have also effectively co-expressed miR-126 and miR-34a through oncolytic adenovirus vectors in a mouse model of pancreatic adenocarcinoma (PAC) and noticed that their co-expression can lead to enhanced or synergistic treatment functions in PAC [161].
Besides, up-regulated miR-34a can inhibit NSCLC regeneration and metastasis [163]. Simultaneous addition of miR-34 and let-7 in a mouse model of NSCLC leads to extensive suppression of key oncogenes and brings survival advantages [164]. MRX34, a miRNA mimic of miR-34a, is in phase I and phase II clinical trials for melanoma at present [165], as well as a phase I clinical trial for NSCLC [155]. In another study, overexpression of E2F1 in high-grade tumors resulted in decreased outcome and chemoresistance of patients. Compared with the effect of transfection with miR-205-5p or miR-342-3p into tumor cell lines alone, the co-transfection of these two miRNAs into tumor cell lines inhibited E2F1 significantly more, thereby reducing the chemically resistant better [166, 167].
In summary, miRNA-based cancer therapies have shown promising results. Methods by combining with different strategies synergistically, applying complementary miRNAs together, or developing novel forms of miRNA provide significant clinical benefits to cancer patients potentially. Further investigations are needed in this field to achieve the ultimate goal of improving patient OS and DFS.
miRNA-related cancer therapy resistance
It is well established that drug-resistance takes major responsibility for the failure of advanced cancer patients treated with chemotherapy. At present, the main drug resistance mechanism of cancer cells is the miRNA-targeted regulation of therapy-related mRNA [168]. Specifically, it can work through the following four methods.
Resistance with drug target-related genes
Drugs against cancers are connected with target proteins, and abnormal regulation of these proteins often leads to drug resistance [168]. It was confirmed that up-regulated miR-18a serves as a potential marker for bad overall survival (OS) and disease-free survival (DFS) after neoadjuvant chemotherapy (nCT), as miR-18a increased significantly in the remaining breast cancer cells after nCT. While researches proved that the remaining breast cancer cells are resistant to tamoxifen, which may associate the function of miR-18a to inhibit the translation of the estrogen receptor, but does not affect its transcription process. Moreover, miR-18a has been verified to link with the mitotic hallmarks and cell cycle checkpoints, and does not affect tumor invasion and migration [169]. In addition, in the study of castration treatment of prostate cancer, investigators justified the increase of miR-197 and miR-361-3p could silence tumor suppressors ARHGDIA or TAGLN2. ARHGDIA or TAGLN2 can stabilize the activity of the androgen receptor, the level of mRNA and protein, and cause the effect of castrate-resistant. It was revealed that miR-197 and miR-361-3p can work as biological markers for castrate-resistant [170]. Low level of hsa-miR-136-5p in tamoxifen treatment of breast cancer often indicates poor prognosis and metastasis of tumor cells. The expression of estrogen receptor alpha 36 (ERα36) was negatively related to the content of hsa-miR-136-5p. It has been found that the up-regulation of ERα36 is in connection with the inhibition of methylation in the promoter region, which may suggest that hypermethylation may inhibit miRNA biogenesis and processing, but the specific mechanism has not been clearly studied [171, 172].
Drug resistance according to the drug target-related genes regulated by miRNAs is the most direct reason for the failure of chemo agents. Only by excavating these regulators, can we then take actions such as choosing undisturbed drug targets or applying combined therapies.
Resistance with drug pharmacokinetic-related genes
Pharmacokinetics is the study facing the absorption, distribution, metabolism, or excretion (ADME) of drugs in the body [173]. CYP3A4 is an important drugmetabolizing enzyme in the liver and intestine, which belongs to a group of well-known drug-metabolizing enzymes, cytochrome P450 (CYP) [174]. CYP3A4 is often highly expressed in drug-resistant tumor samples, and the inhibition of CYP3A4 activity is a considerable part for solving the drug resistance of tumor cells [175]. By the way, the content of miR-27b-3p was proved to be limited in drug-resistant tumor cells, while it has been proved that the up-regulation of miR-27b-3p can inhibit the expression of CYP3A4 and change the drug resistance of LS-180 cells—a kind of colon carcinoma cell line [176]. Similarly, the expression level of miR-328-3p was low in breast cancer, but the ATP-binding cassette sub-family G member 2 (ABCG2) level was high. ABCG2 is a very important protein related to ADME. When it is highly expressed, it will accelerate the metabolism of mitoxantrone and produce drug resistance in tumor cells [177]. When cancer cells overexpress miR-328-3p, it can obviously limit the expression of ABCG2 and decrease the drug resistance of breast cancer cells [176]. Also, miR-27b can directly act on CYP1B1 to reduce its expression [178]. miR-148a-3p, miR-148b-3p, or miR-152-3p can act on SPIN1 and inhibit its expression, then affect the drug metabolism enzyme CYP2C8, UGT2B4, UGT2B17, and ABCB4, and decrease the drug resistance in cancer cells [179].
As for the regulation of drug transport, overexpressed products of drug efflux pump genes take a vital part in drug resistance. Proteins such as ABCC1, ABCG2, ABCB1, and MRP1 can exclude drugs from tumor cells and keep tumor cells alive [180]. miRNA can also directly or indirectly inhibit the overexpression of drug efflux pump genes, and decrease drug resistance. For example, for the treatment of leukemia patients, researchers found that miR-138 can restrain the expression of NF-kB/p65 and then inhibit the production of Multidrug resistance 1 (MDR1). MDR1 is a drug efflux pump P-glycoprotein (P-gp) that encodes proteins that lead to chemotherapy resistance in patients with leukemia. Therefore, overexpression of miR-138 can inhibit chemotherapy resistance in patients with leukemia [181]. Likewise, miR-381 and miR-495 are low-expressed in drug-resistant cells of leukemia patients, too. Up-regulation of miR-381 and miR-495 can effectively inhibit the production of MDR1 and P-gp, and decrease the drug resistance of tumor cells in patients with leukemia [182].
The study of pharmacokinetics can help deal with the contradiction between the safety and efficacy of anti-cancer drugs because we must consider the appropriate dose for cancer patients. So it is significant to take miRNA regulations into consideration of ADME.
Resistance with related cell signaling pathways
A great number of anticancer drugs play a role by the inhibition of cell proliferation and the guidance of apoptosis. These mechanisms are inhibited to a certain extent in drug-resistant cells, resulting in drug resistance [168]. Some miRNAs can influence drug resistance by controlling significant genes in cancer cell-related signaling pathways. For example, the signal transducer and transcriptional activating factor 3 (STAT3) functions importantly in multiple myelomas. The regulation of miR-21 is closely related to STAT3, which can specifically link with the enhancer part of miR-21 and enhance the expression of miR-21, whereas such miRNA is an anti-apoptotic factor and eventually leads to drug resistance in tumor cells [183]. In the treatment of HCC, miR-374b can inhibit the expression of PKM2 by inhibiting the antagonistic glycolysis pathway, thus making resistant cells re-sensitive to sorafenib [184]. HnRNPA1 and PKM2 were both highly expressed in sorafenib resistant cells [185]. Among them, hnRNPA1 can promote the high expression of PKM2. Related studies have demonstrated the expression of miR-374b in drug-resistant HCC cells with a negative correlation with hnRNPA1 and PKM2. Down-regulation of hnRNPA1 in vitro can inhibit the expression of PKM2, but the mechanism has not been evidently studied [186]. In a CRC study, the expression of mammalian target of rapamycin (mTOR) was increased in the remaining tumor cells after cisplatin (a non-specific cell cycle drug) treatment, while mTOR has been shown to participate in the progress and metastatic process of several cancers, behaving like a carcinogenic gene through a variety of mechanisms [187]. In contrast, miR-1271 in tumor cells is low in CRC. And the overexpression of miR-1271 in vitro cell experiments will significantly inhibit the expression of mTOR, then decrease the drug resistance of tumor cells [188].
There are rather complicated regulation networks in multiple cancer cell development processes such as proliferation, metastasis, and apoptosis. We must realize that cancer treatment is an organic whole, and interventions that target only one aspect of the signaling pathway without considering related regulatory mechanisms may not achieve satisfactory results.
Resistance with DNA damage repair-related genes
In many drug-resistant cells, drugs achieve therapeutic effects by destroying tumor cell genes. However, some enzymes can repair these damaged DNAs, and produce drug resistance in tumor cells [175]. In HCC cells, miR-146a-5p was usually expressed low, while Replication Protein A (RPA) was highly expressed [189]. RPA can inhibit DNA repair pathway [190], and then promote tumor cell growth and resist apoptosis in radiotherapy of liver cancer. After radiotherapy, miR-146a-5p expression was raised in HCC cells, while RPA expression was suppressed, and the DNA repair pathway was activated to promote the sensitivity to radiotherapy [191]. Similarly, the decrease of miR-29b level and the increase of phosphatase and tensin homologous protein (PTEN) expression in residual tumor cells happened after radiotherapy for cervical cancer. High expression of PTEN will inhibit the AKT signaling pathway, which will lead to DNA DSB repair, therefore produce tumor cells radioresistance [192]. If miR-29b was overexpressed in the remaining cervical cancer cells after radiotherapy, it will significantly inhibit the expression of PTEN, activate the AKT signal pathway and inhibit DSB, and enhance the sensitivity to radiotherapy [193, 194]. Besides, the miR-122 level was decreased in drug-resistant gastric cancer cells treated with cisplatin. On the other hand, miR-122 directly acted on excision repair cross-complementing 1 (ERCC1) 3′-UTR and inhibited its expression. Therefore, in drug-resistant tumor cells, the expression of ERCC1 will be increased, which can repair the genes of tumor cells destroyed during chemotherapy and produce drug resistance of tumor cells [195].
A comprehensive cognition of the roles miRNAs take in tumor resistance will help us improve existing therapies and explore new drugs, so that related researches can help clinicians and truly improve patient prognosis.
Conclusion
The biogenesis of miRNA is an intricate activity classified into canonical and non-canonical pathway, many factors could influence the expression of miRNAs and their functions. Dysregulation of miRNAs can link to cancer development processes such as interfering with the cell cycle and evading immune destruction. Because of the ability of modulating tens to hundreds of specific genes, miRNAs as vital biomarkers in cancer diagnosis or prognosis prediction are appearing. Some kinds of miRNA-related cancer therapy resistance exist according to the control of drug target-related genes, drug pharmacokinetic-related genes, DNA damage repair-related genes, and related cell signaling pathways. Besides, miRNA therapeutics can be combined with chemotherapy, radiation therapy, and immunotherapy, providing new methods for miRNA-based therapy, as well as the use of replacement therapy and miRNA mimics. It is obvious that miRNAs, along with other ncRNAs such as lncRNAs and circRNAs, would have great clinical implications in the pathogenesis, diagnosis, and treatment of human cancers. More studies related should be followed to understand the mechanism completely and improve targeted therapies in cancer.
Abbreviations
miRNAs: MicroRNAs; ncRNAs: non-coding RNAs; HUGO: Human Genome Organisation; HGNC: the HUGO Gene Nomenclature Committee; Pol II: RNA polymerase II; DGCR8: DiGeorge syndrome critical region gene 8; RanGTP: Ran guanosine triphosphate; TRBP: HIV-1 TAR RNA binding protein; Ago: Argonaute; N-domain: N-terminal domain; RISC: RNA-induced silencing complex; snoRNAs: small nucleolar RNAs; tRNAs: transfer RNAs; m7G: 7-methylguanosine; (PARN): poly(A)-specific ribonuclease; PTMs: post-translational modifications; mRNA: messenger RNA; RBPs: RNA-binding proteins; lncRNAs: long non-coding RNAs; 3'-UTR: 3'-untranslated region; A: adenine; GW182: a 182 kDa glycine-tryptophan protein; DCP1-DCP2: mRNA-decapping enzyme subunit 1 (DCP1)-DCP2; PABPC: cytosolic polyadenylate-binding protein; PAN2-PAN3: poly(A)-nuclease deadenylation complex subunit 2 (PAN2)-PAN3; CCR4-NOT: carbon catabolite repressor protein 4 (CCR4)-NOT; XRN1: 5′-3′ exoribonuclease 1; DDX6: DEAD-box protein 6; eIF4A: eukaryotic translation initiation factor 4 A; MREs: miRNA-recognition elements; CDS: coding sequences; OSCC: oral squamous cell carcinoma; CDK6: cyclin-dependent kinase 6; ADAM17: A disintegrin and metalloproteinase 17; HCC: hepatocellular carcinoma; PIK3R1: phosphoinositide-3-Kinase regulatory subunit 1; CRC: colorectal cancer; EBV: Epstein-Barr virus; NSCLC: non-small cell lung cancer; BCL-2: B-cell lymphoma 2; FasL: Fas ligand; HIF-1α: Hypoxia-inducible factor 1α; VEGF: vascular endothelial growth factor; EMT: epithelial-mesenchymal transition; IFN-γ: interferon-γ; NK: natural killer; MHC: major histocompatibility complex; MICA: major histocompatibility complex class I-related proteins A; NKT: natural killer T; HLA: human leukocyte antigens; APM: class I antigen processing machinery; ICPs: immune checkpoint proteins; TILs: tumor-infiltrating lymphocytes; Th1 cells: T helper 1 cells; CTLs: Cytotoxic T-cell lymphocytes; MDSCs: myeloid-derived suppressor cells; AML: acute myeloid leukemia; TAMs: tumor-associated macrophages; IL-10: interleukin 10; CAFs: cancer-associated fibroblasts; ceRNAs: endogenous RNAs; circRNAs: circular RNAs; snRNAs: small nuclear RNAs; NPC: nasopharyngeal carcinoma; PCAT7: prostate cancer associated transcript 7; ESCC: esophageal squamous cell carcinoma; ANRIL: antisense noncoding RNA in the INK4 locus; GBC: gallbladder carcinoma; GBM: glioblastoma multiforme; eIF4A3: eukaryotic initiation factor 4A3; PPIAP43: peptidyl-prolyl cis-trans isomerase A pseudogene 43; PPIA: peptidyl-prolyl cis-trans isomerase A; SCLC: small cell lung cancer; TDMD: target RNA-directed miRNA degradation; qRT-PCR: quantitative real-time polymerase chain reaction; RT-qPCR: Reverse transcription-quantitative PCR; mgmiR: methylation of genomic loci encoding microRNA; qMS-PCR: quantitative methylation-specific PCR; FFPE: formalin-fixed paraffin-embedded; CSF: cerebrospinal fluid; PBMCs: peripheral blood mononuclear cells; TCGA: the Cancer Genome Atlas; METABRIC: the Molecular Taxonomy of Breast Cancer International Consortium; PPGLs: pheochromocytomas or paragangliomas; HNSCC: head and neck squamous cell carcinoma; FNA: fine needle aspiration; CNS: central nervous system; PCNSL: primary central nervous system lymphoma; GCTs: Germ cell tumours; SS: Synovial sarcoma; TNBC: triple-negative breast cancer; ESCC: esophageal squamous cell carcinoma; PDAC: pancreatic ductal adenocarcinoma; CLL: chronic lymphocytic leukemia; EC: endometrial cancer; ccRCC: clear cell renal cell carcinoma; DPV: differential pulse voltammetry; ex-miRNAs: exosomal serum miRNAs; ISH: in situ hybridization; FN1: Fibronectin 1; SNHG1: small nucleolar RNA host gene 1; Dox: doxorubicin; DSB: double-strand break; HER2: human epidermal growth factor receptor 2; CAR: chimeric antigen receptor; EGFR: epidermal growth factor receptor; Glut-1: glucose transporter 1; EGFR: epidermal growth factor receptor; PDL1: programmed death ligand 1; PAC: pancreatic adenocarcinoma; OS: overall survival; DFS: disease-free survival; nCT: neoadjuvant chemotherapy; ERα36: estrogen receptor alpha 36; ADME: absorption distribution metabolism and excretion; CYP: cytochrome P450; ABCG2: ATP-binding cassette sub-family G member 2; MDR1: multidrug resistance 1; P-gp: P-glycoprotein; STAT3: signal transducer and transcriptional activating factor 3; mTOR: mammalian target of rapamycin; RPA: replication Protein A; PTEN: phosphatase and tensin homologous protein; DSB: double-strand break; ERCC1: excision repair cross-complementing 1.
Acknowledgements
The authors thank Prof. Deyun Feng, Dr. Hongjing Zang, Dr. Jianlun Peng, and Dr. Chengyu Huang for their comments and suggestions throughout the writing process. It is with regret that not all relevant studies could be cited due to space limitations.
Ethics approval and consent to participate
This study was conducted at the Second Xiangya Hospital, Central South University, Hunan, China. All of the protocols were reviewed and approved by the Joint Ethics Committee of the Central South University Health Authority and performed in accordance with national guidelines.
Funding
This work was supported by the National Natural Science Foundation of China (81672308, X. Wang; 81672787, Y. Tao), the National Basic Research Program of China (2015CB553903, Y. Tao), and the Hunan Provincial Key Area R&D Programmes (2019SK2253, X. Wang).
Authors' contributions
BH, ZZ, YZ, PZ, SS, HX, XP, WY designed the study and wrote the paper. BH designed and drew the figures. BH and ZZ drew tables. QC helped the paper revision. YT, XW were the originators of the concept of this report. All of the authors read and approved this manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51
2. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855-62
3. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic acids research. 2019;47:D155-d62
4. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92-105
5. Norouzi M, Yasamineh S, Montazeri M, Dadashpour M, Sheervalilou R, Abasi M. et al. Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection. Materials science & engineering C, Materials for biological applications. 2019;104:110007
6. Braschi B, Denny P, Gray K, Jones T, Seal R, Tweedie S. et al. Genenames.org: the HGNC and VGNC resources in 2019. Nucleic acids research. 2019;47:D786-d92
7. Seal RL, Chen L-L, Griffiths-Jones S, Lowe TM, Mathews MB, O'Reilly D. et al. A guide to naming human non-coding RNA genes. EMBO J. 2020;39:e103777-e
8. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-60
9. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA (New York, NY). 2004;10:1957-66
10. Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S. et al. Functional Anatomy of the Human Microprocessor. Cell. 2015;161:1374-87
11. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-9
12. Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA (New York, NY). 2004;10:185-91
13. Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D. et al. Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature. 2011;475:201-5
14. Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57-68
15. Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell. 2015;57:397-407
16. Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol. 2012;19:145-51
17. Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ. et al. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9208-13
18. Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E1881-9
19. Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83-6
20. Wen J, Ladewig E, Shenker S, Mohammed J, Lai EC. Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates. PLoS Comput Biol. 2015;11:e1004441
21. Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W. et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519-28
22. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA (New York, NY). 2010;16:673-95
23. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5-20
24. Burger K, Schlackow M, Potts M, Hester S, Mohammed S, Gullerova M. Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. The Journal of cell biology. 2017;216:2373-89
25. Burger K, Schlackow M, Gullerova M. Tyrosine kinase c-Abl couples RNA polymerase II transcription to DNA double-strand breaks. Nucleic acids research. 2019;47:3467-84
26. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57-62
27. Kim HK, Xu J, Chu K, Park H, Jang H, Li P. et al. A tRNA-Derived Small RNA Regulates Ribosomal Protein S28 Protein Levels after Translation Initiation in Humans and Mice. Cell reports. 2019;29:3816-24.e4
28. Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V. et al. Mammalian 5'-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568-80
29. Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S. et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694-8
30. Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584-9
31. Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ. et al. Poly(A)-specific ribonuclease mediates 3'-end trimming of Argonaute2-cleaved precursor microRNAs. Cell reports. 2013;5:715-26
32. Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097-108
33. Kao SH, Cheng WC, Wang YT, Wu HT, Yeh HY, Chen YJ. et al. Regulation of miRNA Biogenesis and Histone Modification by K63-Polyubiquitinated DDX17 Controls Cancer Stem-like Features. Cancer Res. 2019;79:2549-63
34. Bose M, Bhattacharyya SN. Target-dependent biogenesis of cognate microRNAs in human cells. Nat Commun. 2016;7:12200
35. Treiber T, Treiber N, Plessmann U, Harlander S, Daiss JL, Eichner N. et al. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol Cell. 2017;66:270-84.e13
36. Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B. et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. 2017;24:816-24
37. Liz J, Portela A, Soler M, Gómez A, Ling H, Michlewski G. et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55:138-47
38. Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368-76
39. Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21-37
40. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
41. Schirle NT, Sheu-Gruttadauria J, Chandradoss SD, Joo C, MacRae IJ. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife. 2015;4:e07646
42. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005
43. Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Cell. 2016;64:320-33
44. Yang A, Bofill-De Ros X, Shao TJ, Jiang M, Li K, Villanueva P. et al. 3' Uridylation Confers miRNAs with Non-canonical Target Repertoires. Mol Cell. 2019;75:511-22.e4
45. Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA (New York, NY). 2005;11:1640-7
46. Elkayam E, Faehnle CR, Morales M, Sun J, Li H, Joshua-Tor L. Multivalent Recruitment of Human Argonaute by GW182. Mol Cell. 2017;67:646-58 e3
47. Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T. et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211-7
48. Braun JE, Truffault V, Boland A, Huntzinger E, Chang CT, Haas G. et al. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5' exonucleolytic degradation. Nat Struct Mol Biol. 2012;19:1324-31
49. Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A. et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014;54:751-65
50. Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J. et al. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell. 2014;56:79-89
51. Fukaya T, Iwakawa HO, Tomari Y. MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Mol Cell. 2014;56:67-78
52. Zhang K, Zhang X, Cai Z, Zhou J, Cao R, Zhao Y. et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat Struct Mol Biol. 2018;25:1019-27
53. Kuzuoglu-Ozturk D, Bhandari D, Huntzinger E, Fauser M, Helms S, Izaurralde E. miRISC and the CCR4-NOT complex silence mRNA targets independently of 43S ribosomal scanning. EMBO J. 2016;35:1186-203
54. Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421-33
55. Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D. et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56:104-15
56. Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515-24
57. Trabucchi M, Mategot R. Subcellular Heterogeneity of the microRNA Machinery. Trends in genetics: TIG. 2019;35:15-28
58. Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer. 2018;17:64
59. Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y. et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017;14:1326-34
60. Sarshad AA, Juan AH, Muler AIC, Anastasakis DG, Wang X, Genzor P. et al. Argonaute-miRNA Complexes Silence Target mRNAs in the Nucleus of Mammalian Stem Cells. Mol Cell. 2018;71:1040-50 e8
61. Bottini S, Hamouda-Tekaya N, Mategot R, Zaragosi LE, Audebert S, Pisano S. et al. Post-transcriptional gene silencing mediated by microRNAs is controlled by nucleoplasmic Sfpq. Nat Commun. 2017;8:1189
62. Makarova JA, Shkurnikov MU, Wicklein D, Lange T, Samatov TR, Turchinovich AA. et al. Intracellular and extracellular microRNA: An update on localization and biological role. Prog Histochem Cytochem. 2016;51:33-49
63. Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C. et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450-5
64. Li J, Chen J, Wang S, Li P, Zheng C, Zhou X. et al. Blockage of transferred exosome-shuttled miR-494 inhibits melanoma growth and metastasis. Journal of cellular physiology. 2019 [Epub ahead of print]
65. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15524-9
66. Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347-55
67. Liu WL, Wang HX, Shi CX, Shi FY, Zhao LY, Zhao W. et al. MicroRNA-1269 promotes cell proliferation via the AKT signaling pathway by targeting RASSF9 in human gastric cancer. Cancer Cell Int. 2019;19:308
68. Shang A, Lu WY, Yang M, Zhou C, Zhang H, Cai ZX. et al. miR-9 induces cell arrest and apoptosis of oral squamous cell carcinoma via CDK 4/6 pathway. Artif Cells Nanomed Biotechnol. 2018;46:1754-62
69. Bofill-De Ros X, Kasprzak WK, Bhandari Y, Fan L, Cavanaugh Q, Jiang M. et al. Structural Differences between Pri-miRNA Paralogs Promote Alternative Drosha Cleavage and Expand Target Repertoires. Cell reports. 2019;26:447-59.e4
70. Chen X, Yang F, Zhang T, Wang W, Xi W, Li Y. et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. Journal of experimental & clinical cancer research: CR. 2019;38:99
71. Liu Y, Wu C, Wang Y, Wen S, Wang J, Chen Z. et al. MicroRNA-145 inhibits cell proliferation by directly targeting ADAM17 in hepatocellular carcinoma. Oncology reports. 2014;32:1923-30
72. Chang RM, Xiao S, Lei X, Yang H, Fang F, Yang LY. miRNA-487a Promotes Proliferation and Metastasis in Hepatocellular Carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:2593-604
73. Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R. et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 Impairs the p53/MDM2 Autoregulatory Loop in Multiple Myeloma Development. Cancer Cell. 2016;30:349-51
74. Deng Q, Hu H, Yu X, Liu S, Wang L, Chen W. et al. Tissue-specific microRNA expression alters cancer susceptibility conferred by a TP53 noncoding variant. Nat Commun. 2019;10:5061
75. Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y. et al. Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression. Journal of virology. 2018;92(23):e01022-18
76. Zhang X, Pan Y, Fu H, Zhang J. microRNA-205 and microRNA-338-3p Reduces Cell Apoptosis in Prostate Carcinoma Tissue and LNCaP Prostate Carcinoma Cells by Directly Targeting the B-Cell Lymphoma 2 (Bcl-2) Gene. Medical science monitor: international medical journal of experimental and clinical research. 2019;25:1122-32
77. Chen S, Yang C, Sun C, Sun Y, Yang Z, Cheng S. et al. miR-21-5p Suppressed the Sensitivity of Hepatocellular Carcinoma Cells to Cisplatin by Targeting FASLG. DNA Cell Biol. 2019;38:865-73
78. Omar HA, El-Serafi AT, Hersi F, Arafa EA, Zaher DM, Madkour M. et al. Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019;286:3540-57
79. Domingues C, Serambeque BP, Laranjo Candido MS, Marto CMM, Veiga FJB, Sarmento Antunes Cruz Ribeiro AB. et al. Epithelial-mesenchymal transition and microRNAs: Challenges and future perspectives in oral cancer. Head Neck. 2018;40:2304-13
80. Yuan L, Zhou C, Lu Y, Hong M, Zhang Z, Zhang Z. et al. IFN-gamma-mediated IRF1/miR-29b feedback loop suppresses colorectal cancer cell growth and metastasis by repressing IGF1. Cancer Lett. 2015;359:136-47
81. Eichmuller SB, Osen W, Mandelboim O, Seliger B. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. Journal of the National Cancer Institute. 2017;109(10):10
82. Wang H, Zhang Y, Wu X, Wang Y, Cui H, Li X. et al. Regulation of Human Natural Killer Cell IFN-γ Production by MicroRNA-146a via Targeting the NF-κB Signaling Pathway. Frontiers in immunology. 2018;9:293
83. Xu D, Han Q, Hou Z, Zhang C, Zhang J. miR-146a negatively regulates NK cell functions via STAT1 signaling. Cellular & molecular immunology. 2017;14:712-20
84. Abdelrahman MM, Fawzy IO, Bassiouni AA, Gomaa AI, Esmat G, Waked I. et al. Enhancing NK cell cytotoxicity by miR-182 in hepatocellular carcinoma. Human immunology. 2016;77:667-73
85. Pesce S, Squillario M, Greppi M, Loiacono F, Moretta L, Moretta A. et al. New miRNA Signature Heralds Human NK Cell Subsets at Different Maturation Steps: Involvement of miR-146a-5p in the Regulation of KIR Expression. Frontiers in immunology. 2018;9:2360
86. Zhang Y, Li X, Zhang J, Liang H. Natural killer T cell cytotoxic activity in cervical cancer is facilitated by the LINC00240/microRNA-124-3p/STAT3/MICA axis. Cancer Lett. 2020;474:63-73
87. Yang Q, Cao W, Wang Z, Zhang B, Liu J. Regulation of cancer immune escape: The roles of miRNAs in immune checkpoint proteins. Cancer Lett. 2018;431:73-84
88. Zou MX, Guo KM, Lv GH, Huang W, Li J, Wang XB. et al. Clinicopathologic implications of CD8(+)/Foxp3(+) ratio and miR-574-3p/PD-L1 axis in spinal chordoma patients. Cancer immunology, immunotherapy: CII. 2018;67:209-24
89. Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y. et al. PDL1 Regulation by p53 via miR-34. Journal of the National Cancer Institute. 2015;108(1):djv303
90. Pyzer AR, Stroopinsky D, Rajabi H, Washington A, Tagde A, Coll M. et al. MUC1-mediated induction of myeloid-derived suppressor cells in patients with acute myeloid leukemia. Blood. 2017;129:1791-801
91. Li L, Sun P, Zhang C, Li Z, Zhou W. MiR-98 suppresses the effects of tumor-associated macrophages on promoting migration and invasion of hepatocellular carcinoma cells by regulating IL-10. Biochimie. 2018;150:23-30
92. Chen S, Chen X, Shan T, Ma J, Lin W, Li W. et al. MiR-21-mediated Metabolic Alteration of Cancer-associated Fibroblasts and Its Effect on Pancreatic Cancer Cell Behavior. International journal of biological sciences. 2018;14:100-10
93. Le TD, Zhang J, Liu L, Li J. Computational methods for identifying miRNA sponge interactions. Briefings in bioinformatics. 2017;18:577-90
94. Zhang X, Xie K, Zhou H, Wu Y, Li C, Liu Y. et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol Cancer. 2020;19:47
95. Huang G, Wang M, Li X, Wu J, Chen S, Du N. et al. TUSC7 suppression of Notch activation through sponging MiR-146 recapitulated the asymmetric cell division in lung adenocarcinoma stem cells. Life sciences. 2019;232:116630
96. Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW. et al. Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 2019;79:4612-26
97. Liu Y, Tao Z, Qu J, Zhou X, Zhang C. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochemical and biophysical research communications. 2017;491:374-81
98. Li F, Li X, Qiao L, Liu W, Xu C, Wang X. MALAT1 regulates miR-34a expression in melanoma cells. Cell death & disease. 2019;10:389
99. Deng M, Yuan H, Liu S, Hu Z, Xiao H. Exosome-transmitted LINC00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microRNA/BCL-2 expression. Cytotherapy. 2019;21:96-106
100. Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML. et al. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019;446:49-61
101. Liu ML, Zhang Q, Yuan X, Jin L, Wang LL, Fang TT. et al. Long noncoding RNA RP4 functions as a competing endogenous RNA through miR-7-5p sponge activity in colorectal cancer. World journal of gastroenterology. 2018;24:1004-12
102. Jiang H, Huang G, Zhao N, Zhang T, Jiang M, He Y. et al. Long non-coding RNA TPT1-AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR-324-5p. Journal of experimental & clinical cancer research: CR. 2018;37:169
103. Zhang JJ, Wang DD, Du CX, Wang Y. Long Noncoding RNA ANRIL Promotes Cervical Cancer Development by Acting as a Sponge of miR-186. Oncology research. 2018;26:345-52
104. Zhang Y, Yang H, Du Y, Liu P, Zhang J, Li Y. et al. Long noncoding RNA TP53TG1 promotes pancreatic ductal adenocarcinoma development by acting as a molecular sponge of microRNA-96. Cancer science. 2019;110:2760-72
105. Ma F, Wang SH, Cai Q, Jin LY, Zhou D, Ding J. et al. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;88:863-9
106. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-91
107. Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8-18
108. Wang R, Zhang S, Chen X, Li N, Li J, Jia R. et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166
109. Wang L, Wei Y, Yan Y, Wang H, Yang J, Zheng Z. et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in OSCC. Oncology reports. 2018;39:951-66
110. Qiu L, Huang Y, Li Z, Dong X, Chen G, Xu H. et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Molecular oncology. 2019;13:441-55
111. Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif Cells Nanomed Biotechnol. 2019;47:3410-6
112. Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong SL, Zhu LP. et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10:1229-42
113. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z. et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20
114. Chen X, Wan L, Wang W, Xi WJ, Yang AG, Wang T. Re-recognition of pseudogenes: From molecular to clinical applications. Theranostics. 2020;10:1479-99
115. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nature Reviews Genetics. 2016;17:272
116. Phelps M, Coss C, Wang H, Cook M. Registered report: Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Elife. 2016;5:e12470
117. Zarringhalam K, Tay Y, Kulkarni P, Bester AC, Pandolfi PP, Kulkarni RV. Identification of competing endogenous RNAs of the tumor suppressor gene PTEN: A probabilistic approach. Scientific reports. 2017;7:7755
118. Hu H, Liu JM, Hu Z, Jiang X, Yang X, Li J. et al. Recently Evolved Tumor Suppressor Transcript TP73-AS1 Functions as Sponge of Human-Specific miR-941. Molecular biology and evolution. 2018;35:1063-77
119. Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766-76
120. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome biology. 2014;15:409
121. de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P. et al. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO reports. 2015;16:500-11
122. Sheu-Gruttadauria J, Pawlica P, Klum SM, Wang S, Yario TA, Schirle Oakdale NT. et al. Structural Basis for Target-Directed MicroRNA Degradation. Mol Cell. 2019;75:1243-55.e7
123. Fuchs Wightman F, Giono LE, Fededa JP, de la Mata M. Target RNAs Strike Back on MicroRNAs. Frontiers in genetics. 2018;9:435
124. Kleaveland B, Shi CY, Stefano J, Bartel DP. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell. 2018;174:350-62.e17
125. Piao XM, Jeong P, Kim YH, Byun YJ, Xu Y, Kang HW. et al. Urinary cell-free microRNA biomarker could discriminate bladder cancer from benign hematuria. Int J Cancer. 2019;144:380-8
126. Pavlidis L, Spyropoulou GA, Papas A, Demiri E. Urinary Excretion of MicroRNA-126 Is a Biomarker for Hemangioma Proliferation. Plast Reconstr Surg. 2018;141:319e-20e
127. Cao Y, Green K, Quattlebaum S, Milam B, Lu L, Gao D. et al. Methylated genomic loci encoding microRNA as a biomarker panel in tissue and saliva for head and neck squamous cell carcinoma. Clin Epigenetics. 2018;10:43
128. Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R. et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140-6
129. Vila-Navarro E, Vila-Casadesus M, Moreira L, Duran-Sanchon S, Sinha R, Gines A. et al. MicroRNAs for Detection of Pancreatic Neoplasia: Biomarker Discovery by Next-generation Sequencing and Validation in 2 Independent Cohorts. Ann Surg. 2017;265:1226-34
130. Salahandish R, Ghaffarinejad A, Omidinia E, Zargartalebi H, Majidzadeh AK, Naghib SM. et al. Label-free ultrasensitive detection of breast cancer miRNA-21 biomarker employing electrochemical nano-genosensor based on sandwiched AgNPs in PANI and N-doped graphene. Biosens Bioelectron. 2018;120:129-36
131. Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V. et al. microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res. 2018;78:1833-44
132. Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L. et al. Six Serum-Based miRNAs as Potential Diagnostic Biomarkers for Gastric Cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2017;26:188-96
133. Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L. et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. 2019;18:40
134. Wang X, Wang HK, Li Y, Hafner M, Banerjee NS, Tang S. et al. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4262-7
135. von Brandenstein M, Schlosser M, Herden J, Heidenreich A, Storkel S, Fries JWU. MicroRNAs as Urinary Biomarker for Oncocytoma. Dis Markers. 2018;2018:6979073
136. Chang YA, Weng SL, Yang SF, Chou CH, Huang WC, Tu SJ. et al. A Three-MicroRNA Signature as a Potential Biomarker for the Early Detection of Oral Cancer. Int J Mol Sci. 2018;19(3):758
137. Gon Y, Maruoka S, Inoue T, Kuroda K, Yamagishi K, Kozu Y. et al. Selective release of miRNAs via extracellular vesicles is associated with house-dust mite allergen-induced airway inflammation. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2017;47:1586-98
138. Russo F, Di Bella S, Bonnici V, Lagana A, Rainaldi G, Pellegrini M. et al. A knowledge base for the discovery of function, diagnostic potential and drug effects on cellular and extracellular miRNAs. BMC genomics. 2014;15(Suppl 3):S4
139. Yan S, Tang Z, Chen K, Liu Y, Yu G, Chen Q. et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. Journal of experimental & clinical cancer research: CR. 2018;37:214
140. Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D. et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27
141. Wei Q, Li X, Yu W, Zhao K, Qin G, Chen H. et al. microRNA-messenger RNA regulatory network of esophageal squamous cell carcinoma and the identification of miR-1 as a biomarker of patient survival. J Cell Biochem. 2019;120(8):12259-12272
142. Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. Journal of experimental & clinical cancer research: CR. 2015;34:18
143. Olgun G, Sahin O, Tastan O. Discovering lncRNA mediated sponge interactions in breast cancer molecular subtypes. BMC genomics. 2018;19:650
144. Zhou S, Wang L, Yang Q, Liu H, Meng Q, Jiang L. et al. Systematical analysis of lncRNA-mRNA competing endogenous RNA network in breast cancer subtypes. Breast cancer research and treatment. 2018;169:267-75
145. Milevskiy MJG, Gujral U, Del Lama Marques C, Stone A, Northwood K, Burke LJ. et al. MicroRNA-196a is regulated by ER and is a prognostic biomarker in ER+ breast cancer. Br J Cancer. 2019;120:621-32
146. Lin Y, Chen F, Shen L, Tang X, Du C, Sun Z. et al. Biomarker microRNAs for prostate cancer metastasis: screened with a network vulnerability analysis model. J Transl Med. 2018;16:134
147. Baldassari F, Zerbinati C, Galasso M, Corra F, Minotti L, Agnoletto C. et al. Screen for MicroRNA and Drug Interactions in Breast Cancer Cell Lines Points to miR-126 as a Modulator of CDK4/6 and PIK3CA Inhibitors. Frontiers in genetics. 2018;9:174
148. Zeng A, Wei Z, Yan W, Yin J, Huang X, Zhou X. et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018;436:10-21
149. Chaudhary AK, Mondal G, Kumar V, Kattel K, Mahato RI. Chemosensitization and inhibition of pancreatic cancer stem cell proliferation by overexpression of microRNA-205. Cancer Lett. 2017;402:1-8
150. Tu MJ, Ho PY, Zhang QY, Jian C, Qiu JX, Kim EJ. et al. Bioengineered miRNA-1291 prodrug therapy in pancreatic cancer cells and patient-derived xenograft mouse models. Cancer Lett. 2019;442:82-90
151. Gokita K, Inoue J, Ishihara H, Kojima K, Inazawa J. Therapeutic Potential of LNP-Mediated Delivery of miR-634 for Cancer Therapy. Mol Ther Nucleic Acids. 2020;19:330-8
152. Mao F, Zhang J, Cheng X, Xu Q. miR-149 inhibits cell proliferation and enhances chemosensitivity by targeting CDC42 and BCL2 in neuroblastoma. Cancer Cell Int. 2019;19:357
153. Wang ZC, Huang FZ, Xu HB, Sun JC, Wang CF. MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic cancer cells by targeting ATG5. The international journal of biochemistry & cell biology. 2019;111:63-71
154. Nie D, Fu J, Chen H, Cheng J, Fu J. Roles of MicroRNA-34a in Epithelial to Mesenchymal Transition, Competing Endogenous RNA Sponging and Its Therapeutic Potential. Int J Mol Sci. 2019;20(4):861
155. Cortez MA, Valdecanas D, Niknam S, Peltier HJ, Diao L, Giri U. et al. In vivo Delivery of miR-34a Sensitizes Lung Tumors to Radiation Through RAD51 Regulation. Mol Ther Nucleic Acids. 2015;4:e270
156. Ma W, Ma CN, Zhou NN, Li XD, Zhang YJ. Up- regulation of miR-328-3p sensitizes non-small cell lung cancer to radiotherapy. Scientific reports. 2016;6:31651
157. Ji Y, Hocker JD, Gattinoni L. Enhancing adoptive T cell immunotherapy with microRNA therapeutics. Semin Immunol. 2016;28:45-53
158. Zhang T, Zhang Z, Li F, Ping Y, Qin G, Zhang C. et al. miR-143 Regulates Memory T Cell Differentiation by Reprogramming T Cell Metabolism. J Immunol. 2018;201:2165-75
159. Ohno M, Ohkuri T, Kosaka A, Tanahashi K, June CH, Natsume A. et al. Expression of miR-17-92 enhances anti-tumor activity of T-cells transduced with the anti-EGFRvIII chimeric antigen receptor in mice bearing human GBM xenografts. J Immunother Cancer. 2013;1:21
160. Wang LQ, Kumar S, Calin GA, Li Z, Chim CS. Frequent methylation of the tumour suppressor miR-1258 targeting PDL1: implication in multiple myeloma-specific cytotoxicity and prognostification. Br J Haematol. 2020 [Epub ahead of print]
161. Feng SD, Mao Z, Liu C, Nie YS, Sun B, Guo M. et al. Simultaneous overexpression of miR-126 and miR-34a induces a superior antitumor efficacy in pancreatic adenocarcinoma. Onco Targets Ther. 2017;10:5591-604
162. Li Y, Lu H. Noncoding RNAs: 'our turn' to join the p53 network. J Mol Cell Biol. 2014;6:179-80
163. Sherman EJ, Mitchell DC, Garner AL. The RNA-binding protein SART3 promotes miR-34a biogenesis and G1 cell cycle arrest in lung cancer cells. J Biol Chem. 2019;294:17188-96
164. Kasinski AL, Kelnar K, Stahlhut C, Orellana E, Zhao J, Shimer E. et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34:3547-55
165. Huang CK, Kafert-Kasting S, Thum T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ Res. 2020;126:663-78
166. Lai X, Gupta SK, Schmitz U, Marquardt S, Knoll S, Spitschak A. et al. MiR-205-5p and miR-342-3p cooperate in the repression of the E2F1 transcription factor in the context of anticancer chemotherapy resistance. Theranostics. 2018;8:1106-20
167. Orellana EA, Li C, Lisevick A, Kasinski AL. Identification and validation of microRNAs that synergize with miR-34a - a basis for combinatorial microRNA therapeutics. Cell Cycle. 2019;18:1798-811
168. Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. 2019;11:25
169. Luengo-Gil G, Garcia-Martinez E, Chaves-Benito A, Conesa-Zamora P, Navarro-Manzano E, Gonzalez-Billalabeitia E. et al. Clinical and biological impact of miR-18a expression in breast cancer after neoadjuvant chemotherapy. Cellular oncology (Dordrecht). 2019;42(5):627-644
170. Fletcher CE, Sulpice E, Combe S, Shibakawa A, Leach DA, Hamilton MP. et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene. 2019;38(28):5700-5724
171. Thiebaut C, Chesnel A, Merlin JL, Chesnel M, Leroux A, Harle A. et al. Dual Epigenetic Regulation of ERalpha36 Expression in Breast Cancer Cells. Int J Mol Sci. 2019;20(11):2637
172. Ward A, Balwierz A, Zhang JD, Kublbeck M, Pawitan Y, Hielscher T. et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173-82
173. Yang C, Tavassolian N, Haddad WM, Bailey JM, Gholami B. A Fast Parameter Identification Framework for Personalized Pharmacokinetics. Scientific reports. 2019;9:14143
174. Hu DG, Marri S, McKinnon RA, Mackenzie PI, Meech R. Deregulation of the Genes that Are Involved in Drug Absorption, Distribution, Metabolism, and Excretion in Hepatocellular Carcinoma. The Journal of pharmacology and experimental therapeutics. 2019;368:363-81
175. Nakano M, Nakajima M. Current knowledge of microRNA-mediated regulation of drug metabolism in humans. Expert opinion on drug metabolism & toxicology. 2018;14:493-504
176. Li X, Tian Y, Tu MJ, Ho PY, Batra N, Yu AM. Bioengineered miR-27b-3p and miR-328-3p modulate drug metabolism and disposition via the regulation of target ADME gene expression. Acta pharmaceutica Sinica B. 2019;9:639-47
177. Sjostedt N, Salminen TA, Kidron H. Endogenous, cholesterol-activated ATP-dependent transport in membrane vesicles from Spodoptera frugiperda cells. Eur J Pharm Sci. 2019;137:104963
178. Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090-8
179. Chen X, Wang YW, Gao P. SPIN1, negatively regulated by miR-148/152, enhances Adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. Journal of experimental & clinical cancer research: CR. 2018;37:100
180. Baguley BC. Multiple drug resistance mechanisms in cancer. Molecular biotechnology. 2010;46:308-16
181. Requenez-Contreras JL, Lopez-Castillejos ES, Hernandez-Flores R, Moreno-Eutimio MA, Granados-Riveron JT, Martinez-Ruiz GU. et al. MiR-138 indirectly regulates the MDR1 promoter by NF-kappaB/p65 silencing. Biochemical and biophysical research communications. 2017;484:648-55
182. Xu Y, Ohms SJ, Li Z, Wang Q, Gong G, Hu Y. et al. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PloS one. 2013;8:e82062
183. Chong PSY, Chng WJ, de Mel S. STAT3: A Promising Therapeutic Target in Multiple Myeloma. Cancers (Basel). 2019;11(5):731
184. Cun J, Yang Q. Bioinformatics-based interaction analysis of miR-92a-3p and key genes in tamoxifen-resistant breast cancer cells. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;107:117-28
185. Kuranaga Y, Sugito N, Shinohara H, Tsujino T, Taniguchi K, Komura K. et al. SRSF3, a Splicer of the PKM Gene, Regulates Cell Growth and Maintenance of Cancer-Specific Energy Metabolism in Colon Cancer Cells. Int J Mol Sci. 2018;19(10):3012
186. Zhang M, Zhang H, Hong H, Zhang Z. MiR-374b re-sensitizes hepatocellular carcinoma cells to sorafenib therapy by antagonizing PKM2-mediated glycolysis pathway. American journal of cancer research. 2019;9:765-78
187. Fujishita T, Kojima Y, Kajino-Sakamoto R, Taketo MM, Aoki M. Tumor microenvironment confers mTOR inhibitor resistance in invasive intestinal adenocarcinoma. Oncogene. 2017;36:6480-9
188. Yao H, Sun Q, Zhu J. miR-1271 enhances the sensitivity of colorectal cancer cells to cisplatin. Experimental and therapeutic medicine. 2019;17:4363-70
189. Domingo-Prim J, Endara-Coll M, Bonath F, Jimeno S, Prados-Carvajal R, Friedlander MR. et al. EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nat Commun. 2019;10:2135
190. Wang J, Yang T, Chen H, Li H, Zheng S. Oncogene RPA1 promotes proliferation of hepatocellular carcinoma via CDK4/Cyclin-D pathway. Biochemical and biophysical research communications. 2018;498:424-30
191. Luo J, Si ZZ, Li T, Li JQ, Zhang ZQ, Chen GS. et al. MicroRNA-146a-5p enhances radiosensitivity in hepatocellular carcinoma through replication protein A3-induced activation of the DNA repair pathway. American journal of physiology Cell physiology. 2019;316:C299-C311
192. Liu Q, Turner KM, Alfred Yung WK, Chen K, Zhang W. Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro Oncol. 2014;16:1313-23
193. Zheng T, Shi Y, Zhang J, Peng J, Zhang X, Chen K. et al. MiR-130a exerts neuroprotective effects against ischemic stroke through PTEN/PI3K/AKT pathway. Biomed Pharmacother. 2019;117:109117
194. Zhang T, Xue X, Peng H. Therapeutic Delivery of miR-29b Enhances Radiosensitivity in Cervical Cancer. Molecular therapy: the journal of the American Society of Gene Therapy. 2019;27:1183-94
195. Song AL, Zhao L, Wang YW, He DQ, Li YM. Chemoresistance in gastric cancer is attributed to the overexpression of excision repair cross-complementing 1 (ERCC1) caused by microRNA-122 dysregulation. Journal of cellular physiology. 2019;234(12):22485-22492
196. Ruff SM, Ayabe RI, Malekzadeh P, Good ML, Wach MM, Gonzales MK. et al. MicroRNA-210 May Be a Preoperative Biomarker of Malignant Pheochromocytomas and Paragangliomas. J Surg Res. 2019;243:1-7
197. Zhang H, Zhu M, Shan X, Zhou X, Wang T, Zhang J. et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene. 2019;687:246-54
198. Zhang J, Ren J, Hao S, Ma F, Xin Y, Jia W. et al. MiRNA-491-5p inhibits cell proliferation, invasion and migration via targeting JMJD2B and serves as a potential biomarker in gastric cancer. Am J Transl Res. 2018;10:525-34
199. Yoshikawa M, Iinuma H, Umemoto Y, Yanagisawa T, Matsumoto A, Jinno H. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett. 2018;15:9584-92
200. Santangelo A, Imbruce P, Gardenghi B, Belli L, Agushi R, Tamanini A. et al. A microRNA signature from serum exosomes of patients with glioma as complementary diagnostic biomarker. J Neurooncol. 2018;136:51-62
201. Pan J, Zhou C, Zhao X, He J, Tian H, Shen W. et al. A two-miRNA signature (miR-33a-5p and miR-128-3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Scientific reports. 2018;8:16699
202. Cagle P, Niture S, Srivastava A, Ramalinga M, Aqeel R, Rios-Colon L. et al. MicroRNA-214 targets PTK6 to inhibit tumorigenic potential and increase drug sensitivity of prostate cancer cells. Scientific reports. 2019;9:9776
203. Li F, Wang F, Zhu C, Wei Q, Zhang T, Zhou YL. miR-221 suppression through nanoparticle-based miRNA delivery system for hepatocellular carcinoma therapy and its diagnosis as a potential biomarker. Int J Nanomedicine. 2018;13:2295-307
204. Dieckmann KP, Radtke A, Spiekermann M, Balks T, Matthies C, Becker P. et al. Serum Levels of MicroRNA miR-371a-3p: A Sensitive and Specific New Biomarker for Germ Cell Tumours. Eur Urol. 2017;71:213-20
205. Uotani K, Fujiwara T, Yoshida A, Iwata S, Morita T, Kiyono M. et al. Circulating MicroRNA-92b-3p as a Novel Biomarker for Monitoring of Synovial Sarcoma. Scientific reports. 2017;7:14634
206. Park S, Kim J, Eom K, Oh S, Kim S, Kim G. et al. microRNA-944 overexpression is a biomarker for poor prognosis of advanced cervical cancer. BMC Cancer. 2019;19:419
207. Nakashima H, Yoshida R, Hirosue A, Kawahara K, Sakata J, Arita H. et al. Circulating miRNA-1290 as a potential biomarker for response to chemoradiotherapy and prognosis of patients with advanced oral squamous cell carcinoma: A single-center retrospective study. Tumour Biol. 2019;41:1010428319826853
208. Yuan Y, Liu W, Zhang Y, Zhang Y, Sun S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503:870-5
209. Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M. et al. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25:155-61
210. Arvidsson Y, Rehammar A, Bergstrom A, Andersson E, Altiparmak G, Sward C. et al. miRNA profiling of small intestinal neuroendocrine tumors defines novel molecular subtypes and identifies miR-375 as a biomarker of patient survival. Mod Pathol. 2018;31:1302-17
211. Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, Myklebust MP. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8:87837-47
212. Zhou Z, Zhang H, Deng T, Ning T, Liu R, Liu D. et al. Exosomes Carrying MicroRNA-155 Target Forkhead Box O3 of Endothelial Cells and Promote Angiogenesis in Gastric Cancer. Mol Ther Oncolytics. 2019;15:223-33
213. Okazaki J, Tanahashi T, Sato Y, Miyoshi J, Nakagawa T, Kimura T. et al. MicroRNA-296-5p Promotes Cell Invasion and Drug Resistance by Targeting Bcl2-Related Ovarian Killer, Leading to a Poor Prognosis in Pancreatic Cancer. Digestion. 2019:1-13
214. Fukagawa S, Miyata K, Yotsumoto F, Kiyoshima C, Nam SO, Anan H. et al. MicroRNA-135a-3p as a promising biomarker and nucleic acid therapeutic agent for ovarian cancer. Cancer science. 2017;108:886-96
215. Papageorgiou SG, Kontos CK, Diamantopoulos MA, Bouchla A, Glezou E, Bazani E. et al. MicroRNA-155-5p Overexpression in Peripheral Blood Mononuclear Cells of Chronic Lymphocytic Leukemia Patients Is a Novel, Independent Molecular Biomarker of Poor Prognosis. Dis Markers. 2017;2017:2046545
216. Richardsen E, Andersen S, Al-Saad S, Rakaee M, Nordby Y, Pedersen MI. et al. Low Expression of miR-424-3p is Highly Correlated with Clinical Failure in Prostate Cancer. Scientific reports. 2019;9:10662
217. Assiri AA, Mourad N, Shao M, Kiel P, Liu W, Skaar TC. et al. MicroRNA 362-3p Reduces hERG-related Current and Inhibits Breast Cancer Cells Proliferation. Cancer genomics & proteomics. 2019;16:433-42
218. Luo Y, Chen L, Wang G, Xiao Y, Ju L, Wang X. Identification of a three-miRNA signature as a novel potential prognostic biomarker in patients with clear cell renal cell carcinoma. J Cell Biochem. 2019;120(8):13751-13764
219. Reid G, Johnson TG, van Zandwijk N. Manipulating microRNAs for the Treatment of Malignant Pleural Mesothelioma: Past, Present and Future. Front Oncol. 2020;10:105
220. Ling H, Girnita L, Buda O, Calin GA. Non-coding RNAs: the cancer genome dark matter that matters!. Clin Chem Lab Med. 2017;55:705-14
221. Wong RR, Abd-Aziz N, Affendi S, Poh CL. Role of microRNAs in antiviral responses to dengue infection. J Biomed Sci. 2020;27:4
222. Diaz-Riascos ZV, Ginesta MM, Fabregat J, Serrano T, Busquets J, Buscail L. et al. Expression and Role of MicroRNAs from the miR-200 Family in the Tumor Formation and Metastatic Propensity of Pancreatic Cancer. Mol Ther Nucleic Acids. 2019;17:491-503
223. Raeisi F, Mahmoudi E, Dehghani-Samani M, Hosseini SSE, Ghahfarrokhi AM, Arshi A. et al. Differential Expression Profile of miR-27b, miR-29a, and miR-155 in Chronic Lymphocytic Leukemia and Breast Cancer Patients. Mol Ther Oncolytics. 2020;16:230-7
224. Chen H, Liu L, Li X, Shi Y, Liu N. MicroRNA-1294 inhibits the proliferation and enhances the chemosensitivity of glioma to temozolomide via the direct targeting of TPX2. American journal of cancer research. 2018;8:291-301
225. Jiang J, Xie C, Liu Y, Shi Q, Chen Y. Up-regulation of miR-383-5p suppresses proliferation and enhances chemosensitivity in ovarian cancer cells by targeting TRIM27. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;109:595-601
226. Nagachinta S, Bouzo BL, Vazquez-Rios AJ, Lopez R, Fuente M. Sphingomyelin-Based Nanosystems (SNs) for the Development of Anticancer miRNA Therapeutics. Pharmaceutics. 2020;12(2):189
227. Jeong K, Yu YJ, You JY, Rhee WJ, Kim JA. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip. 2020;20:548-57
Author contact
![]() Corresponding author: Xiang Wang (E-mail: wangxiangedu.cn) and Yongguang Tao (E-mail: taoyongedu.cn).
Corresponding author: Xiang Wang (E-mail: wangxiangedu.cn) and Yongguang Tao (E-mail: taoyongedu.cn).

 Global reach, higher impact
Global reach, higher impact