10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(15):2906-2923. doi:10.7150/ijbs.48812 This issue Cite
Review
Update of the current knowledge on genetics, evolution, immunopathogenesis, and transmission for coronavirus disease 19 (COVID-19)
1. Laboratory Microorganismes and Active Biomolecules, Sciences Faculty of Tunis, University Tunis El Manar, Tunis, Tunisia.
2. Department of Pediatrics, Yonsei University College of Medicine, Seoul, Republic of Korea.
3. Division of Urology, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
4. Department of Global Health and Population, Harvard T.H. Chan School of Public Health, Boston MA, USA.
5. Yonsei University College of Medicine, Seoul, Republic of Korea.
6. University of Florida College of Medicine, Gainesville, FL 32610, USA.
7. The Cambridge Centre for Sport and Exercise Science, Anglia Ruskin University, Cambridge, CB1 1PT, UK.
8. Research and Development Unit, Parc Sanitari Sant Joan de Déu, CIBERSAM, 08830 Barcelona, Spain.
9. ICREA, Pg. Lluis Companys 23, 08010 Barcelona, Spain.
10. Faculty of Medicine, University of Versailles Saint-Quentin-en-Yvelines, 78000 Versailles, France.
11. Department of Internal Medicine IV (Nephrology and Hypertension), Medical University Innsbruck, Innsbruck 6020, Austria.
Received 2020-6-19; Accepted 2020-8-23; Published 2020-9-12
Abstract
In December 2019, an acute respiratory disease caused by novel species of coronavirus (SARS-CoV-2), emerged in China and has spread throughout the world. On 11th March 2020, the World Health Organization (WHO) officially declared coronavirus disease 19 (COVID-19) a pandemic, severe coronavirus-mediated human disease. Based on genomic and phylogenetic studies, SARS-CoV-2 might originate from bat coronaviruses and infects humans directly or through intermediate zoonotic hosts. However, the exact origin or the host intermediate remains unknown. Genetically, SARS-CoV-2 is similar to several existing coronaviruses, particularly SARS-CoV, but differs by silent and non-silent mutations. The virus uses different transmission routes and targets cells and tissues with angiotensin-converting enzyme 2 (ACE2) protein, which makes it contagious. COVID-19 shares both the main clinical features and excessive/dysregulated cell responses with the two previous Middle East respiratory syndrome coronavirus (MERS) and severe acute respiratory syndrome coronavirus (SARS) epidemics. In this review, we provide an update of the current knowledge on the COVID-19 pandemic. Gaining a deeper understanding of SARS-CoV-2 structure, transmission routes, and molecular responses, will assist in the prevention and control of COVID-19 outbreaks in the future.
Keywords: COVID-19, SARS-CoV-2, genetic variation, SARS-CoV-2 genomics, evolution, Immunity, transmission
Introduction
The human community has experienced outbreaks and epidemics of many infectious diseases which are considered as major causes of human morbidity and mortality. Viral pathogens are known to cause outbreaks that have epidemic and pandemic potential. Several factors underlie the emergence of such diseases, including increasing population, increased domestic and global connectivity, social practices, prevalence of immunosuppressive diseases, change in agricultural practices such as mixed farming, and many other related environmental factors [1]. Climate changes and reduction in biodiversity may play a role by forcing species to change geographical range and to survive in seminatural habitats that may bring wild animals closer to humans [2]. Disturbance of natural ecosystems is reported to increase the transfer of disease from wild species to humans and is suggested to increase occurrences of neglected, forgotten and new human diseases [3]. It is well known that pathogen genomics vary considerably, thus, occurring genetic alterations have also been responsible for such outbreaks [4]. Although humanity has made meaningful progress in the battle against pathogens, the re-emergence of viral agents remains a great threat and challenge for the global health community [5]. About 60% of infectious diseases and 70% of emerging infections of humans are zoonotic in origin, with two-thirds originating in wildlife [6]. Over the last decades, several viral diseases had emerged in China, namely, those caused by the severe acute respiratory syndrome- (SARS-) coronavirus (CoV) and the H5N1, H1N1, and H7N9 viruses [7]. Recently, near the end of 2019, an outbreak of an ongoing viral pneumonia with unknown etiology emerged in the city of Wuhan, China. The infectious agent of this viral pneumonia was identified as the 7th member of human coronaviruses (2019-nCoV) [8]. On 11th February 2020, the World Health Organization (WHO) named the novel viral pneumonia “Coronavirus Disease 2019” (COVID-19), while the International Committee on Taxonomy of Viruses (ICTV) suggested this novel coronavirus will be named “SARS-CoV-2” due to the phylogenetic and taxonomic analysis of this novel coronavirus [9]. The SARS-CoV-2 emerged as a highly pathogenic and large-scale epidemic coronavirus compared to the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012. COVID-19 is characterized by a high infectivity during incubation and a time delay between really infected cases and daily observed number of confirmed cases [10]. The novel coronavirus is believed to be infectious during its incubation period, which is reported to be 3-7 days, at most 14 days, when no symptoms are shown in patients [11]. Environmental factors, particularly weather conditions, are suspected to favor emergence and spread of the outbreak. Both drought and cold, which characterized the 2019 winter in Wuhan [12], provided conducive environmental conditions for virus survival [13,14]. During the cold winter, air-dried virus particles are a dangerous form of virus, which survive for a long period of time in airflow [15]. In addition, cold conditions damper humans' innate immunity by reducing blood supply and thus decreasing the provision of immune cells to the nasal mucosa. Low humidity can reduce the capacity of cilia cells in the airway to remove virus particles and secrete mucus as well as repair airway cells [16].
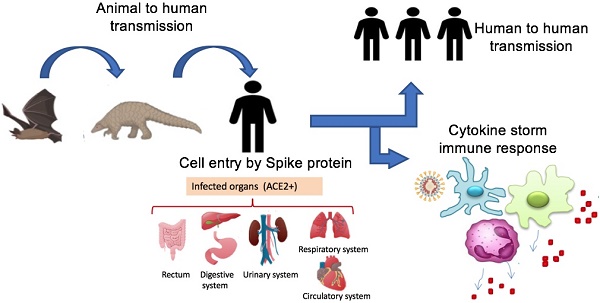
Many COVID-19 patients were potentially exposed to wildlife animals at the Huanan seafood whole sale market, Wuhan, China, where poultry, snake, bats, and other farm animals were also sold [17]. In such wildlife trade markets, animals known to transmit coronaviruses are crammed together under fetid conditions. Thus, humans might become unfortunate hosts for SARS-CoV-2 as a result of some inappropriate interactions with wildlife. Accumulated evidence suggests bats as the origin of the majority of emerging coronaviruses [18, 19,20,21]. Evolution and recombination of these different strains of bat viruses might lead to the creation of various SARS-CoVs capable of cross-species transmission and ultimate infection of human beings [22, 23, 24, 25]. Considering that the earliest COVID-19 patient reported no exposure at the seafood market, the suspected source of first virus infections [17], it becomes vital to identify the intermediate SARS-CoV-2 host to block interspecies transmission. Currently, the research on SARS-CoV-2 is in its primary stages. Based on current published evidence, this review summarizes SARS-CoV-2 origin, genetics, genomics, transmission routes, immune system responses and immunopathogenesis of COVID-19.
Origin of COVID- 19
Recently (at the time of writing this paper), the worldwide scientific community released full genomic sequences and several independent research groups have identified SARS-CoV-2 as a β-coronavirus. It is an enveloped virus containing a non-segmented, positive-sense RNA molecule [8,26,27]. In assigning the SARS-COV-2 to a taxonomic group, phylogenetic analysis of the complete viral genome revealed that the virus was most closely related (89.1% nucleotide similarity) to a group of SARS-like β-coronaviruses (β-CoVs) previously found in bats in China [28]. Sequencing and evolutionary analyses show that the bat was suspected as a natural reservoir host of the virus [19-21]. Indeed, the SARS-CoV-2 genome sequence is identical to those of bat CoVs, particularly to the RaTG13, with similarities from 96.2% to 99.9% [22,26,29-31]. SARS-CoV-2 might be transmitted from bats via unknown intermediate hosts to humans. Bats are the natural reservoir of a wide variety of CoVs, including SARS-CoV-like and MERS-CoV-like viruses [32-34]. The bat SARS-like coronavirus sequence is genetically more similar to the 2019-nCoV than other bat SARS-like coronavirus sequences, but is also more distant from sequences isolated in SARS and MERS coronaviruses [35]. A possible explanation is a past history of recombination in the β-coronavirus group [18]. Coronaviruses are characterized by a high rate of recombination, which may play a role in viral evolution and interspecies infections [36,23].
Origin of SARS-CoV-2 and potential transmission routes to humans. SARS-CoV-2 transmitted to humans from bats directly or by intermediate hosts such as rats, pangolins, snakes, and rats. The virus is spread among humans by different routes including droplets, aerosols, direct contact, and other potential routes such as urine, stool, and fecal swabs. Human organs expressingthe ACE2 receptor are targets for viral infection. Abbreviation: ACE2+, organs having ACE2 receptor.
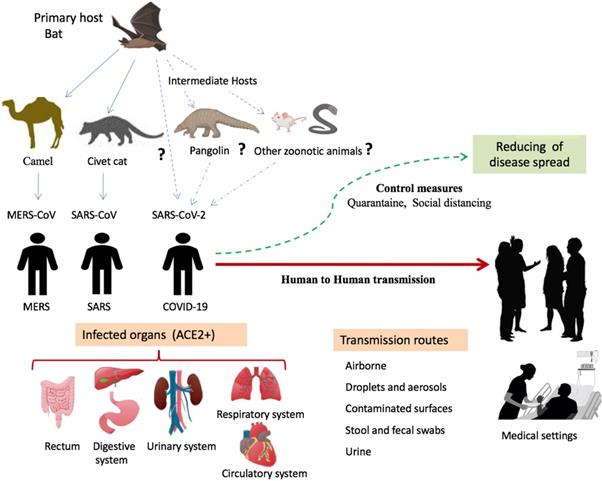
Given that bats were not available for sale in the seafood market in Wuhan and similar residues of virus receptor were observed in many species [24], alternative intermediate hosts, such as turtles, pangolin and snakes were proposed [23,24]. Usually, SARS-CoV and MERS-CoV infect intermediate hosts, such as civets or camels, before leaping to humans [37]. This fact indicates that SARS-CoV-2 was probably transmitted to humans from animals other than bats. Some studies suggested pangolin-CoV is the closest relative of SARS-CoV-2 [38,39]. On 24th October 2019, Liu and colleagues detected the existence of a SARS-CoV-like CoV in lung samples collected from two dead Malayan pangolins [38]. This discovery was made just prior to the COVID-19 outbreak. Interestingly, Pangolin-CoV was composed of a 91.02 % identical genome to SARS-CoV-2 [39]. Notably, Paraskevis et al. [20] reported that the new coronavirus provides a new lineage for almost half of its genome, with no close genetic relationships to other viruses within the subgenus of sarbecovirus, rejecting the hypothesis of SARS-CoV-2 emergence as a result of a recent recombination event. Taken together, genomic and phylogenetic results indicate that the SARS-CoV-2 basal origin is still under debate. Figure 1 summarizes recent findings on the origin of SARS-CoV-2 and potential transmission routes.
Immunopathogenesis
Fever, cough, myalgia, or fatigue with abnormal lung scan findings are features of SARS-CoV-2 infection. The less common symptoms are sputum production, headache, hemoptysis and diarrhea [17,40,41]. Few patients develop acute respiratory distress syndrome (ARDS), respiratory failure, multiple organ failure and even death [17]. The elderly and those with underlying disorders (i.e. hypertension, chronic obstructive pulmonary disease, diabetes, cardiovascular disease), are more likely to develop ARDS, cytokine storm, septic shock, metabolic acidosis, and coagulation dysfunction [11,17,42,43]. Several abnormalities have been observed including cellular immune deficiency, coagulation activation, myocardial injury, hepatic and kidney injury, and secondary bacterial infection [43].Of note, scientists reported that SARS-COV-2 causes an inflammatory response in the lower airway, which may lead to lung injury [44,45]. Viral particles invade the respiratory mucosa, triggering immune responses and a “cytokine storm” closely related to the critical condition of COVID-19 patients [46]. COVID-19 symptoms showed differences in viral tropism compared to SARS-CoV [11,42,45], MERS-CoV [46] and influenza virus [47]. Innate immunity serves to slow viral infection before the adaptive immune response. IFN-γ secretion constitutes the first anti-viral defense barrier. In the case of SARS-CoV-2, IFN-γ secretion might be dampened by viral replication and subsequent inhibition of the adaptive immune response. Moreover, viral replication triggers hyper inflammatory conditions, leading to a high production of pro-inflammatory cytokines and chemokines, particularly by neutrophils and monocytes/macrophages [48-50]. Several plasma cytokines and chemokines were increased in COVID-19 patients, including IL-1, IL-2, IL-4, IL-7, IL-6, IL-10, IL-12, IL-13, IL-17, G-CSF, M-CSF, IP-10, MCP-1, MIP-1α, HGF, IFN-γ and TNF-α [17,24]. Normally, produced cytokines and chemokines play an anti-viral defense role through the recruitment of immune cells. However, in critical cases of COVID-19 patients, a “cytokine storm” is reported and exacerbates immune reactions, initiating viral sepsis and ARDS. These exacerbations lead to death in some cases of elderly and those with underlying disorders [17].
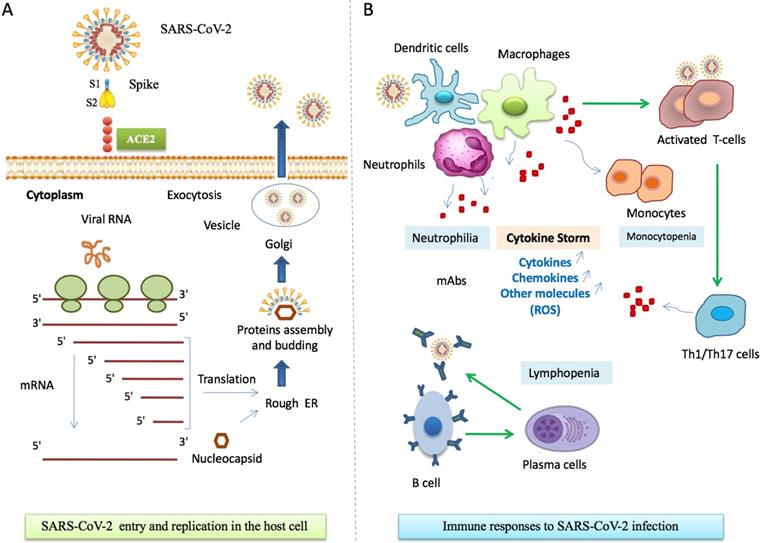
Most patients have normal or decreased white blood cell counts, and lymphocytopenia [51]. However, in severe patients, the neutrophil count, D-dimer, blood urea and creatinine levels are significantly enhanced, while the lymphocyte counts are clearly reduced. The innate immune system recognizes viral 'molecular patterns' (such as double-stranded RNA) [52] and the adaptive immune systems kills virus-infected cells by means of T and B cells that produce pathogen-specific antibodies [53]. Regulated immune response inhibits virus replication, promotes virus clearance, induces tissue repair, and triggers a prolonged adaptive immune response against the viruses [54]. Figure 2 describes the mechanisms of host cell infection by SARS-CoV-2 and gives a simplified schema of the immunopathogenesis of COVID-19.
Immune responses during SARS-CoV-2 infection. A. SARS-CoV-2 entry to host cell. Spike proteins on the surface of the coronavirus bind to angiotensin-converting enzyme 2 (ACE2) receptors on the surface of the target cell. Coronavirus genome replication and transcription takes place in the cytoplasmand involves coordinated processes of both continuous and discontinuous RNA synthesis. Assembly of viral proteins occurred in rough RE. Finally, the virus is released through exocytosis by Golgi. Abbreviations: S1, Spike 1; S2, Spike 2; ACE2, angiotensin-converting enzyme 2, RE, reticulum endoplasmic. B. COVID-19 immuno-pathogenesis. Infected cells present CoV antigens to T cells. This process leads to T cell activation and differentiation, including the production of cytokines associated with the different T cell subsets (Th1/Th17). Cytokines recruit lymphocytes and leukocytes to the site of infection. The adaptive immunity is involved through a subset of differentiated Tcells and activation of B and plasma cells that produce monoclonal antibodies. Activation of immune cells results in excessive productionof chemokines and other cytokines that induce a pro-inflammatory response and attract cells, such as neutrophils and macrophages, to sites of infection. In turn, these cells release injury molecules, such as matrix metalloproteinases and ROS. The “cytokine storm” represents the secretion of large quantities of immune mediators leading to more severe conditions. Regulated immune responses are crucial to clear the infection. Abbreviations: mAbs, Monoclonal Antibodies; Th, T helper; ROS, Reactive Oxygen Species.
SARS-COV-2 genomics and variation
Coronaviruses are single-stranded RNA viruses that belong to the order Nidovirales, family Coronaviridae, and subfamily Coronavirinae [54]. The family Coronavirinae is divided into four genera: α, β, γ, and δ. There are seven human coronaviruses: 229E (α-CoV), NL63 (α-CoV), OC43 (β- CoV), HKU1 (β-CoV), MERS-CoV (β-CoV), SARS-CoV (β-CoV), and SARS-CoV-2 (β-CoV) [55]. The viral genome of SARS-COV-2 is composed of a positive-stranded RNA, and its structures vary considerably [8]. The complete genome of one strain of SARS-CoV-2, isolated from a COVID-19 pneumonia patient working in the Wuhan seafood market, is 29.9 kilo-bases (kb) in size with 29891 nucleotides [56], encoding 9860 amino acids [57], while SARS and MERS-CoVs have RNA genomes of 27.9 kb and 30.1 kb, respectively [58]. Similar to other β-CoVs, the SARS-CoV-2 genome contains two flanking untranslated regions (UTRs) and a variable number (8-13) of open reading frames (ORFs) [59]. Two-thirds of viral RNA, mainly located in the first ORF (ORF1a/b) translates two polyproteins, pp1a and pp1ab, and encodes 16 non-structural proteins (NSP). The remaining ORFs encode accessory and structural proteins [37]. The 2019-nCoV genome is arranged in the order of 5′-replicase (ORF1 /ab)-structural proteins [Spike (S)-Envelope (E)- Membrane (M)-Nucleocapsid (N)]-3′. The 5′- and 3′ -UTR sequences of 2019-nCoV, 265 and 358 nucleotides respectively in length, are similar to those of other β-CoVs, with 83.6% shared nucleotide identity. There are no remarkable differences between the ORFs and NSPS of 2019-nCoV with those of SARS-CoV [57]. The major distinction between 2019-nCoV and SARS-CoV is in spike S1 and ORF8, which were previously shown to be recombination hot spots [57]. In correlation with previous knowledge of CoVs genomics [60], the AT% was higher than GC% in SARS-CoV-2 [49]. In all of its structural genes, SARS-CoV-2 prefers pyrimidine rich codons to purines, and most high-frequency codons ended with A or T. The low-frequency codons ended with G or C [61]. This is in agreement with previous studies of CoVs [62]. SARS-CoV-2 structural proteins showed 5-20 lower ENc values compared to SARS, bat SARS and MERS CoVs. This implies higher codon bias and higher gene expression efficiency of SARS-CoV-2 structural proteins [61]. By meta-transcriptomic sequencing, another study showed that SARS-CoV-2 exhibits some genomic and phylogenetic similarity to SARS-CoV, particularly in the S-glycoprotein gene and receptor-binding domain (RBD) [56]. Most genome-encoded proteins of SARS-CoV-2 are similar to SARS-CoVs [56]. Mutations in NSP2 and NSP3 play a role in the infectious capability and differentiation mechanism of SARS-CoV-2 [63].
Phylogenetic and genomic analyses from distinct countries suggest the newly-emerged SARS-CoV-2 strains are closely related but are distinguished by both synonymous and non-synonymous mutations in different genomic locations [18,19,21,22,29,36,64-70]. Genome sequences of 2019-nCoV sampled from early cases were almost genetically identical [17,21], suggesting a very recent emergence of this virus in humans. The estimated genetic diversity of five newly sampled 2019-nCoV genomes was 0.000094 substitutions per site with an estimated evolutionary rate of 0.0038 substitutions per site per year [65]. Accordingly, the newly identified 2019-nCoV sequences originated from the same isolate about 2 years ago [64]. Evolutionary selection in the human hosts acts on SARS-COV-2 genomes, sometimes with parallel evolution events. Tang et al. [66] conducted a population genetic analysis of 103 SARS-CoV-2 genomes and classified two prevalent evolving types of SARS-CoV-2: L type (~ 70%) and S type (~ 30%). Likewise, the complete genome analysis for the first cases of COVID-19 in Chile detected at least two different viral variants [67]. The new coronavirus could face selective pressures such as diversity in hosts, countries, weather, and other conditions. The phylogeographic patterns are potentially affected by distinctive migratory histories, founder events, and sample sizes [64]. This information contributes to monitoring the spread of the infection and the surveillance for eventual recombination or genome mutations. Population genetics-phylogenetics approach indicated that most sites in the viral ORFs evolved under strong to moderate purifying selection. Particularly, a non-negligible proportion of ORF8 codons had evolved under very weak purifying selection or close to selective neutrality [30]. Positive selection was also detected in the receptor-binding motif (RBM) of the spike protein but most likely resulted from a recombination event that involved the BatCoV RaTG13 sequence [30]. The divergence of SARS-CoV-2 from BatCoV RaTG13 was accompanied by limited episodes of positive selection, suggesting the common ancestor of the two viruses was poised for human infection [30].
Homologous recombination contributes to the 2019‐nCoV cross‐species transmission [23]. Homologous recombination is an important evolutionary force that occurs in many viruses, including Dengue virus [71], human immunodeficiency virus [72], hepatitis B virus [73], and hepatitis C virus [74]. A previous study suggested that recombination of SARS-CoV in the spike glycoprotein genes might have mediated the initial cross-species transmission event from bats to other mammals [25]. It is critical to determine the animal reservoir of the 2019‐nCoV to understand the molecular mechanism of its cross‐species spread. Findings shed a cautiously optimistic light on the possibility of finding effective treatment for this novel coronavirus, starting from already existing anti‐β-coronaviridae compounds [29].
The process of infection in human
In the past two decades, the 2019-nCoV is the third coronavirus to emerge in the human population after the SARS-CoV outbreak in 2002 [75] and the MERS-CoV outbreak in 2012 [76]. Full-genome sequence analysis of 2019-nCoV is different from both MERS-CoV and SARS-CoV that infect humans [8], but all are highly pathogenic zoonotic pathogens. Based on their phylogenetic relationships and genomic structures, human β-coronaviruses (SARS-CoV-2, SARS-CoV, and MERS-CoV) have many similarities but also have differences in their genomic and phenotypic structures that influence their pathogenesis. To date, no therapeutics or vaccines are approved against any human-infecting coronaviruses. Epidemiological investigations suggest SARS-CoV-2 is highly transmissible in humans [77], especially in the elderly and people with underlying diseases [78]. The rapidly increasing number of cases and evidence of human-to-human transmission suggest the virus was more contagious than SARS-CoV and MERS-CoV [79,80]. WHO estimates, the basic reproduction number (R0), as 1.4 to 2.5 less than SARS (2 to 5); but, this number can grow if the pandemic is not controlled by applying quarantine and isolation strategies [21]. The high affinity between ACE2 and 2019-nCoV spike protein also suggests the population with a higher expression of ACE2 might be more susceptible to 2019-nCoV [81]. The SARS-CoV first emerged in China in 2002 and then spread to 37 countries/territories in 2003 and caused a travel-related global outbreak with a 9.6% mortality rate [82]. There is evidence that SARS-CoV originated in bats in China and reached humans after jumping from an intermediate host, the civet (Pagumalarvata) [83]. MERS-CoV, discovered in 2012 in the Middle East [84], is endemic in dromedary camels, from which it can be transmitted to humans [20]. The MERS epidemic is ongoing, and as of December 2019, 2468 cases have been reported [85]. Less virulent coronavirus species cause common colds in humans with relatively mild symptoms [86]. Some coronaviruses are strictly host-specific, while others can be found in a range of hosts [87].
Genetically, SARS-CoV-2 was less similar to SARS-CoV (about 79%) and MERS-CoV (about 50%) [88,89]. The arrangement of nucleocapsid protein (N), envelope protein (E), and membrane protein (M) among β-coronaviruses are different. Both SARS-CoV-2 and SARS-CoV bind to human ACE2 [90] and use transmembrane protease serine 2 (TMPRSS2) to complete cell entry and infection [89,91]. The SARS-COV-2 spike protein has a furin cleavage site in the S1/S2 junction, different from SARS-CoV and other closely related bat viruses [92]. This has implications for viral entry routes. The TMPRSS2 also contributes to the S-protein priming of 2019-nCoV, indicating that the TMPRSS2 inhibitor might constitute a treatment option [89]. There are many similarities between SARS-CoV-2 and the original SARS-CoV [93], and their S proteins share 76.47% identity [93,94]. A total of 53 unique S proteins were selected and their structures modeled according to different subtypes of CoVs, including 2019-nCoV (WH-Human_1), 3 SARS strains, 2 β-CoV strains, and 47 strains from other CoVs. S proteins of 2019-nCoV and SARS strains share high structural similarity with a root-mean-square deviation of 1.21 Å [94]. Wan et al. [94] reported that residue 394 (glutamine) in the SARS-CoV-2 RBD, corresponding to residue 479 in SARS-CoV [95], can be recognized by the critical lysine 31 on the human ACE2 receptor. A highly similar epitope was identified computationally between the 2019-nCoV and SARS-CoV on the binding site of the S proteins to the human ACE2 receptor [55]. Further analysis even suggested SARS-CoV-2 recognizes human ACE2 more efficiently than SARS-CoV, increasing the ability of SARS-CoV-2 to transmit from person to person [94].
Potential transmission routes of SARS-CoV-2
As an emerging acute respiratory infectious disease, COVID-19 primarily spreads through the respiratory tract by droplets, respiratory secretions, saliva, and direct contact for a low infective dose [42,96]. The common transmission routes of novel coronavirus include direct transmission (cough, sneeze, droplet inhalation transmission) and contact transmission with oral, nasal, and eye mucous membranes [97]. Since 2019-nCoV can be passed directly from person to person by respiratory droplets, emerging evidence suggests it may also be transmitted through contact and fomites [98]. The fact that SARS-CoV-2 can infect the human gut epithelium has important implications for fecal-oral transmission and containment of viral spread [99]. The ACE2 receptor is abundantly present throughout the respiratory tract, as well as in the epithelial cells of salivary gland ducts that have been demonstrated to be early targets of SARS-CoV [100]. Both SARS-CoV and SARS-Cov-2 may have much faster replication rates than other coronaviruses infect humans [94] . The 2019-nCoV can produce aerosols, droplets, or particulate matter with high viral loads that increase the viability time of the virus in various environments [100]. After reaching the recipients, the fast replication viruses have a higher chance of successful infection [100]. Of note, a report of one case of 2019-nCoV infection in Germany indicates that transmission of the virus may also occur through contact with asymptomatic patients [97]. In fact, the asymptomatic incubation period for individuals infected with 2019-nCoV has been reported to be ~1-14 days. Thus, those without symptoms can spread the virus [101].
On 10th February 2020, SARS-CoV-2 was isolated from fecal swabs from a severe pneumonia patient in China [102], indicating the possibility of multiple routes of transmission. In addition, a recent pilot experiment showed that 4 out of 62 stool specimens tested positive for 2019-nCoV, and another four patients in a separate cohort who tested positive from rectal swabs also had 2019-nCoV detected in the gastrointestinal tract, saliva, or urine [30]. Thus, in addition to the respiratory droplets and direct contact, fecal-oral transmission might also be a route of transmission for 2019-nCoV [40]. Remarkably, the ACE2 protein presents abundantly on enterocytes in the small intestine [103], which may contribute to this route of infection and disease manifestations. However, the aerosol transmission route and the fecal-oral transmission routes still need to be further studied.
Analysis of conjunctival samples from confirmed and suspected cases of 2019-nCoV suggests eye exposure may provide an effective way for the virus to enter the body [104], and that ocular surfaces may be a potential target for SARS-CoV-2 invasion [98]. However, other studies do not fully support this assumption [105]. ACE2 is mainly expressed in posterior tissues of the eye, such as the retina and the retinal pigment epithelium, not in the human conjunctival and corneal epithelium [106]. Furthermore, tears are constantly renewed by the lacrimal drainage system. Therefore, the virus enters the tears through droplets, which may pass through the naso-lacrimal ducts and then into the respiratory tract [88].
Asymptomatic patients can spread SARS-CoV-2 with high efficiency in noninfectious disease settings such as otolaryngology, which is a high-risk specialty as it closely contacts the upper respiratory tract mucous, secretions, droplets and aerosols during procedures and surgery [107, 108]. Otolaryngologists have been infected with COVID-19 at higher rates than other specialties [107]. Of note, pathogens can be transmitted in dental settings through inhalation of airborne microorganisms that can remain suspended in the air for long periods [109]. Direct contact with blood, oral fluids, contact of conjunctival, nasal, or oral mucosa with droplets and aerosols containing microorganisms generated from an infected individual [110], and indirect contact with contaminated instruments and/or environmental surfaces [111], can rapidly spread viral pathogens. Dental studies show that many dental procedures and contaminated dental instruments or environmental surfaces provide possible routes to the spread of viruses [112,113]. Human coronaviruses such as SARS-CoV, MERS-CoV, or endemic human coronaviruses (HCoV) can persist on surfaces like metal, glass, or plastic for up to a couple of days [114], making contaminated surfaces in healthcare settings a potential source of coronavirus transmission.
Key viral factors
The coronaviral genome encodes four structural proteins, namely, S protein, N protein, M2 protein, and E protein [54,115]. The N protein interacts with the viral RNA to form the ribo-nucleoprotein [116]. The E protein conducts ion channel actions and contributes to virions assembly [117]. Coronavirus entry to host cells is a multi-step process involving several distinct domains in the surface glycoprotein spike (S). The S protein, a trimeric class I fusion protein, exists in a metastable prefusion conformation that undergoes a substantial structural rearrangement to fuse the viral membrane with the host cell membrane [118,119]. The S protein contains the RBD and mediates virus attachment to the cell surface, receptor engagement, protease processing, and membrane fusion, facilitating viral entry into host cells [120-124]. The virion S-glycoprotein on the coronavirus surface can attach to the ACE2 receptor on the surface of human cells [125]. The S proteins mutate and gain capability to recognize host receptors across species [118,126]. For many CoVs, spike is cleaved at the boundary between the S1 and S2 subunits [121]. For SARS-CoV, the cleavage of the trimer S protein is triggered by the cell surface-associated TMPRSS2 [127] and cathepsin [128]. To engage a host cell receptor, the RBD of S1 undergoes hinge-like conformational movements that transiently hide or expose the determinants of receptor binding [129]. The RBD for SARS-CoV-2 has residues and motifs found in all three clades but forms a distinct clade [94]. Receptor binding destabilizes the pre-fusion trimer, resulting in shedding of the S1 subunit and transition of the S2 subunit to a stable post-fusion conformation [130].
After membrane fusion, the RNA viral genome is released into the cytoplasm, and the uncoated RNA translates two polyproteins, pp1a and pp1ab [130], which encode non-structural proteins and form a replication-transcription complex (RTC) in double-membrane vesicles. RTCs replicate and synthesize a nested set of subgenomic RNAs [131], which encode accessory proteins and structural proteins. Mediated by the endoplasmic reticulum (ER) and the Golgi, newly formed genomic RNA, nucleocapsid proteins, and envelope glycoproteins assemble and form viral particle buds [131].
SARS-CoV-2 and ACE2
The ACE2 protein is enriched in the enterocytes in the small intestine and the renal tubules, as well as in the lung alveolar epithelial cells, the heart cells, the arterial smooth muscle cells, and the gastrointestinal system [133,134]. This protein is best known for cleaving several peptides within the renin-angiotensin system and other substrates [135]. ACE2 protein is rare in circulation, but widely expressed in organs, such as kidneys, the gastrointestinal tract, and at relatively lower levels, the lungs [136]. Recently, based on scRNA-seq datasets, Zou et al. [105] constructed a 2019-nCoV infection-related risk map of different organs including nasal mucosa, respiratory tract, bronchus, and lung. They found that pulmonary AT2 cells and respiratory epithelial cells exhibited high ACE2 expression. Similarly, Zhao et al. [81] demonstrated that 83% of ACE2-expressing cells were alveolar epithelial type II cells (AECII), suggesting these cells can serve as a reservoir for viral invasion. In addition, gene ontology enrichment analysis showed ACE2-expressing AECII have high levels of multiple viral process-related genes, including regulatory genes for viral processes, viral life cycle, viral assembly, and viral genome replication [81], this implies that the ACE2-expressing AECII facilitates coronaviral replication in the lung. Taken together, these data suggest that the respiratory tract should be considered as a vulnerable target to SARS-CoV-2 infection.
For the human heart, more than 7.5% of myocardial cells demonstrate positive ACE2 expression, implying that the heart could be at high risk of 2019-nCoV infection, especially in the presence of viremia [105]. Moreover, myocardial infarction may increase ACE2 expression in heart, thereby suggesting that ACE2 plays an important role in cardiac injury [137]. Interestingly, the digestive system, including the esophagus, stomach, ileum, and liver, showed extremely high ACE2 expression in epithelial cells [81,105,138,139]. In addition, proximal tubule cells in the kidney and bladder urothelial cells express ACE2, making kidney and bladder a sat risk for infection [103,105,140]. Taken together, ACE2 tissue distribution in different organs could explain the multi-organ dysfunction and the non-respiratory symptoms observed in some 2019-nCoV pneumonia patients and may help to explain the increased human-to-human transmissibility of this virus [94]. The ACE2 extracellular domain has been demonstrated as a receptor for the S protein of SARS-CoV [141], and recently, for the SARS-CoV-2 [26]. Therefore, accumulated evidence suggests that, similar to SARS-CoV, SARS-CoV-2 uses ACE2 as its host receptor [79,94,105,142] but with higher affinity than does SARS-CoV [79,94,105,143]. The similarity with SARS-CoV is critical because ACE2 is a functional SARS-CoV receptor in vitro [144] and in vivo [145], and this may have implications in therapeutics. Of note, the SARS-CoV-2 does not use other coronavirus receptors such as aminopeptidaseN and dipeptidylpeptidase 4 [31].
During infection, the cleavage of the spike protein required for cell-to-cell fusion depends on both cell type and virus strain [146]. Spike protein is proteolytically cleaved by the cellular enzyme furin into S1 and S2 subunits during intracellular processing [123,147]. The second cleavage S29 exposes the fusion peptide and is thus necessary for viral entry [108], whereas S1 contains the RBD, which directly binds to the peptidase domain (PD) of ACE2 [34]. The ectodomain of the SARS-CoV-2 S protein binds to the PD of ACE2 with a dissociation constant (Kd) of~15nM [126]. The RBD is recognized by the extracellular peptidase domain of ACE2 mainly through polar residues [148]. SARS-CoV-2 and SARS-CoV RBDs show high similarities [94]. However, a number of sequence variations and conformational deviations were found [148]. A single N501T mutation (corresponding to the S487T mutation in SARS-CoV) may significantly enhance the binding affinity between the 2019-nCoV RBD and human ACE2 [94]. Of note, for SARS-CoV pathogenesis, ACE2 is not only the entry receptor of the virus but also has a protective role during acute lung injury in a mouse model [145]. By binding ACE2, SARS-CoV leads to the downregulation of ACE2 expression and might therefore negate the protective effect of ACE2 [145,149]. Human and non-human primates share the identity sequences in the regions and residues, implying that ACE2 from non-human primates may recognize 2019-nCoV and mediate its infection [42]. As a result, non-human primates may be susceptible to 2019-nCoV and serve as animal models for antiviral research or intermediate hosts for cross-species transmission.
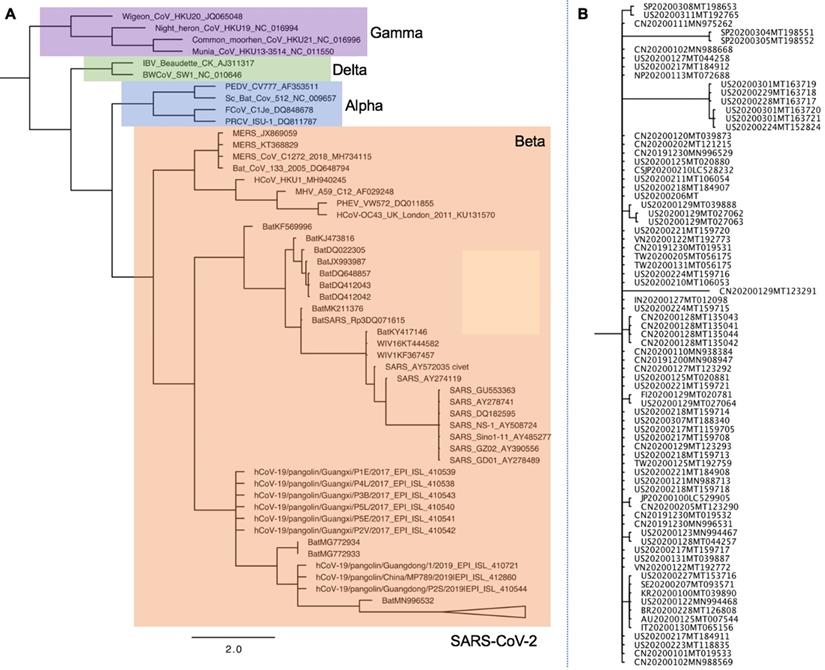
Phylogenetic analysis of full-length genomes of SARS-CoV-2 and representative sequences of genera Alpha, Beta, Delta-, and Gammacoronaviruses. A. Phylogenetic analysis of SARS-CoV-2 with Alpha-, Beta-, Delta- and Gammacoronaviruses full-genomes. Colored boxes denote genera. Purple: Gammacoronavirus. Green: Deltacoronavirus. Blue: Alphacoronavirus. Orange: Betacoronavirus. SARS-CoV-2 strains are collapsed in one branch under the Betacoronaviruses. Tree data were collated from updated sources published until June 4th, 2020 [26,151,152]. Abbreviations: CoV, coronavirus; MHV murine hepatitis virus; PHEV porcine hemagglutinating encephalomyelitis virus; IBV infectious bronchitis virus; MERS Middle East respiratory syndrome; SARS severe acute respiratory syndrome. B. Phylogenetic analysis of SARS-CoV-2 full-genomes. The collapsed SARS-CoV-2 branch from A is shown in full.
Analysis of coding-region variants using the GTEx database in ACE2 and of the expression quantitative trait loci (eQTL) variants, which may affect the expression of ACE2, support the existence of ACE2 mutants resistant to S-protein binding of coronavirus in different populations [150]. East Asian populations have much higher AFs in eQTLvariants associated with higher ACE2 expression in tissues, suggesting different susceptibility or response to SARS-CoV-2 compared to other populations under similar conditions [150]. A recent single-cell RNA-sequencing (RNA-seq) analysis indicated that Asian males may have higher expression of ACE2 [78]. The genetic basis of ACE2 expression and function in different populations is still largely unknown. Increasing understanding of potential functional variants in ACE2 among populations requires further epidemiological investigations of SARS-CoV-2.
Comparison of SARS-CoV-2 sequences with other coronaviruses
Animals have a critical role in COVID-19 outbreak onset and evolution and may act as the virus reservoir. The exact origin of SARS-CoV-2 remains unknown; therefore, phylogenetic analyses are established to find the animal virus reservoir. A phylogenetic analysis comparing the genomes of SARS-CoV-2 with other coronaviruses is shown in Figure 3 [26,151,152]. Nucleocapsid (NC) protein, which is highly conserved and immunogenic, is commonly targeted in studies aimed at developing alternative diagnostic tools [153]. Multiple alignments of the whole protein sequences highlight a very high homology between the NC sequence of SARS-CoV-2 and bat RaTG13 CoV [154]. Tilocca et al showed that some epitopes are shared among a wider range of coronaviruses, while other epitope sequences are more conserved among the most related specimens [154]. Based on the high-homology between the Spike protein epitopes of taxonomically-related coronaviruses, Tilocca et al. hypothesized that past contact with infected dogs shield humans against the circulating SARS-CoV-2 [155]. By using the Immune Epitope Database and Analysis Resource (IEDB), Grifoni et al. found that SARS-CoV has high sequence similarity to SARS-CoV-2, and is the best characterized coronavirus in terms of epitope responses. Multiple specific regions with high homology to the SARS-CoV in addition to a priori potential B and T cell epitopes for SARS-CoV-2 have been [156]. Authors suggest these regions are promising targets for immune recognition of SARS-CoV-2 and can facilitate effective vaccine design against this virus [156].
Compared with other CoV species, bovine CoV is the genetically closest counterpart to human coronaviruses [157,158]. A high similarity was observed between bovine CoV, canine respiratory coronavirus (CRCoV) and human coronavirus OC43 (HCoV-OC43) [159]. More investigations to assess the transmission rate of the bovine and canine respiratory coronaviruses to humans are needed. Homology modelling showed that 2019-nCoV had a similar RBD structure to that of SARS-CoV, despite amino acid variation at some key residues [26]. Lu et al. showed that 2019-nCoV was related (with 88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21 and more distant from SARS-CoV (about 79%) and MERS-CoV (about 50%). However, they also revealed that S gene of 2019-nCoV had the lowest sequence identity with bat-SL-CoVZC45 and bat-SLCoVZXC21, at only around 75% [26]. Zhou et al. demonstrated that the novel virus has 96.2% similarity to a bat SARS-related Coronavirus (SARSr-CoV; RaTG13 (MN996532.1) [31]. Proteins from SARS and SARS-CoV-2 were treated as homologous: identity value > 65%, query coverage, >95% [160]. SARS-CoV-2 has been found to be more distant in relation to SARS-CoV (79%) and MERS-CoV (50%) [161]. Anand et al. found that all sequences showed ~99.98% similarity in the nucleotide sequences, implying a relationship between the currently circulating viruses and suggesting a recent shift to humans [161].
Comparison of the novel coronavirus Wuhan-Hu-1 sequence with that of the closely related human SARS-CoV S strain Tor2 sequence, revealed 76% homology [162]. The S1 RBD was less conserved (64% identity) than the S2 fusion domain (90% identity). The identity between SARS-CoV-2 and SARS-CoV at the S protein amino acid level was 76%, and phylogenetic analyses grouped SARS-CoV-2 in the lineage B of the Betacoronavirus genus, closely related to SARS-CoV, as well as to other CoVs originating in bats [162]. The relatively high degree of sequence identity for the RBD is consistent with the view that SARS-CoV-2, like SARS-CoV, may use ACE2 as its host cell receptor [162]. The amino acid homology of the modeled S proteins in comparison to the template SARS-CoV S was ~ 71% for all the Bat-CoV S, with the exception of the LYRa3 S, which shares a homology of 84.7% with the template S. Overall, all the modeled S proteins shared a similar folding pattern in comparison to SARS-CoV S and both S1 and S2 domains showed a uniform organization [162]. The S protein amino acid identity among the Bat-CoV ranged between 75.3% and 96.7%, with LYRa3 and RaTG13 S proteins having the lowest and highest identity to SARS-CoV-2, respectively [162]. The tertiary structure of the polyprotein isolate SARS-CoV-2_HKU-SZ-001_2020 had 98.94 % identity with SARS-Coronavirus NSP12 bound to NSP7 and NSP8 co-factors [163].
The analysis of fifteen sequences of SARS-CoV-2 S sequences obtained from NCBI and GISAID from China and various export locations worldwide along with representative members of lineages A-D betacoronaviruses showed that all SARS-CoV-2 S sequences clustered very closely with bat SARS-like sequences, with the closest matching sequence corresponding to Bat-SL-RaTG13 [162]. Pairwise comparison between SARS-CoV-2 S protein and that of BatCoV-RaTG13 and representative sequences from Guangxi pangolin (2017, abbreviated GX here) and Guangdong pangolin (2019, abbreviated GD) confirm that overall BatCoV-RaTG13 had the highest identity: 97% overall, 96% and 100% for S1 and S2, respectively [162]. The analysis revealed that pangolin S protein sequences are more divergent overall (92% identity for GX and 89% identity for GD), with most of the divergence concentrating in the S1 domain. the RBD domain of the GD domain was confirmed to be remarkably well conserved compared to SARS-CoV-2 (97% identity compared to 87% identity for GX pangolin and 89% for BatCoV-RaTG13) [162]. Another study showed that he homology of SARS-CoV-2 with the Bat coronavirus isolate RaTG13 strain (MN996532) was 96%, but has no more than 80% homology with other isolates of bat SARS‐like coronavirus [164].
Zhang et al. showed that some pangolin CoV genes show higher amino acid sequence identity to SARS-CoV-2 than to RaTG13 genes [39]. The RBD region within the S1 which is conserved between Pangolin CoV and SARS-CoV2, is phylogenetically closer to pangolin-CoV than RaTG13 pointing potential similarity in their pathogenic properties [39]. At the whole genome sequence level, pangolin CoV and SARSr-CoV RaTG13 show 91.02% and 96.2% similarity with SARS-CoV-2 but the S1 subunit of spike protein of pangolin CoV is more closely related to SARS-CoV-2 than SARSr-CoV RaTG13. Other studies have shown that the homology with a coronavirus strain isolated from pangolin was 99%, suggesting that SARS‐CoV‐2 might have originated from bat and pangolin might have served as the intermediate host between bat and human [165]. The genetic analysis of the currently circulating strains of the pandemic have shown 99.98-100% similarity in their genomes, implying a recent shift to humans [161]. Description of the epitopes distribution over the viral population might provide valuable information driving future researches aimed at setting efficient prophylactic strategies and/or the design of tool capable of differential diagnosis on the basis of serological tests.
Sequence alignment analysis provides evidence of high sequence homology for some of the investigated proteins. In addition, homology modelling of structural epitope mapping revealed a potential immunogenic value for specific sequences scoring a lower identity with SARS-CoV-2 nucleocapsid proteins [154]. Accumulated evidence provide a molecular structural rationale for a potential role in conferring protection from SARS-CoV-2 infection and identifying potential candidates for the development of diagnostic tools. Further experimental studies are desired for a confidential evaluation of the postulated hypotheses. Finally, further studies employing purified forms of the spike proteins and/or its epitopes are needed and should be evaluated carefully.
Current diagnostic tools for COVID-19
Currently, severeral diagnostic tests for coronavirus include RT-PCR, real-time reverse transcription PCR (rRT-PCR), reverse transcription loop-mediated isothermal amplification, as well as real-time RT-LAMP are used [166, 167]. RT-LAMP was used to detect MERS-CoV, it is highly specific and has similar sensitivity to rRT-PCR [168,169].
The China National Health Commission, laboratory examinations established that nasopharyngeal and oropharyngeal swab tests, have become a standard assessment for diagnosis of COVID-19 infection [170]. Two one-step quantitative RT-PCR (qRT-PCR) assays were developed to detect two different regions (ORF1b and N) of the SARS-CoV-2 genome, allowing for earlier identification of patients [170]. Later, three novel RT-PCR assays targeting the RNA-dependent RNA polymerase (RdRp)/helicase (Hel), spike (S), and nucleocapsid (N) genes of SARS-CoV-2 were developed. Among them, the COVID-19-RdRp/Hel assay had the lowest limit of detection in vitro [171]. The SARS-CoV E gene assay was more sensitive than the RdRp gene assay combined with the one-step RT-PCR system [172]. The E gene PCR was sufficient to diagnose a SARS-CoV-2 infection but the RdRp protocol was recommended to confirm a positive result [173,174]. The overall positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan was 38% [175]. In a series of 51 patients with confirmed COVID-19 infection, 71% patients were RT-PCR positive at the first time of testing of throat swab or sputum samples [176]. The RT-PCR results usually become positive after several days (2-8 days) [177]. Chest CT scans can be used to assess the severity of COVID- 19. Despite negative RT-PCR results, COVID-19 infection should be diagnosed with typical chest computerized tomography (CT) characteristics for patients suffering from fever, sore throat, fatigue, coughing or dyspnea [178]. Of 1014 patients, 59% had positive RT-PCR results, and 88% had positive chest CT scans [179]. Assessment of imaging features combined with clinical and laboratory findings could facilitate early diagnosis of COVID-19 pneumonia [180,181,182]. The detection of nucleic acid in the nasal and throat swab sampling or other respiratory tract samplings by real-time PCR and further confirmed by next-generation sequencing is qualified as the most qualified method to diagnosis of COVID-19. Some published sequences comparisons of SARS-CoV-2 with other coronaviruses are presented in Table 1.
Potential therapeutics for COVID-19
Until the writing of this paper, there is no current evidence from randomized controlled trials (RCTs) to recommend any specific anti-SARS-CoV-2 treatment for COVID-19 infection. Antiviral drugs and systemic corticosteroid treatment commonly used for influenza virus including neuraminidase inhibitors, ganciclovir, acyclovir, and ribavirin, as well as methylprednisolone [183] are invalid for COVID-19. However, other viral drugs are found to treat cases of COVID-19 such as remdesivir which has been reported to successfully treat the first case of COVID-19 in the US [184]. Based on the experience accumulated from the SARS and MERS outbreaks, lopinavir (LPV) is a potential treatment option for COVID- 19, LPV inhibits the protease activity of coronavirus in vitro and in animal studies [185]. Ribavirin, a guanosine analogue used to treat several virus infections, showed promising results in a MERS-CoV rhesus macaque model [186] and in SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) model [187]. These features increase its potential as an antiviral against SARS-CoV-2 [188]. Nelfinavir was predicted to be a potential inhibitor of SARS-CoV-2 main protease [188]. The other promising antiviral drugs include nitazoxanide, favipiravir, nafamostat, and so on [189].
Chloroquine is a repurposed drug with great potential to treat COVID-19. It is a widely-used antimalarial and autoimmune disease drug that has been reported to be a potential broad-spectrum antiviral drug [190, 191, 192], and it has been used to treat malaria for many years [193]. The chloroquine antiviral proprieties are investigated, it can inhibit pH-dependent steps of the replication of several viruses [194, 195], it has immunomodulatory effects, suppressing the production/release of TNF-α and IL-6, works as a novel class of autophagy inhibitor [196], which may interfere with viral infection and replication. Several studies have found that chloroquine interfered with the glycosylation of cellular receptors of SARS-CoV [195] and functioned at both entry and at post-entry stages of the COVID-19 infection in Vero E6 cells [197]. Chloroquine was effective in more than 100 COVID-19 patients in terms of reduction of exacerbation of pneumonia, duration of symptoms and delay of viral clearance, all in the absence of severe side effects [198]. Therefore, it has been included in the recommendations for the prevention and treatment of COVID-19 pneumonia [198]. Hydroxychloroquine was found to be more potent than chloroquine Using physiologically-based pharmacokinetic (PBPK) models in SARS-CoV-2-infected Vero cells [199]. Taken together, both chloroquine and hydroxychloroquine have immunomodulatory effects and can suppress the immune response [200], and may be used in prophylaxis as well as curative treatment for individuals exposed to SARS-CoV-2 [201]. The optimal dosage of chloroquine for SARS-CoV-2 will need to be assessed in future trials [201], in addition to the age of the patient and the clinical presentation or stage of the disease [202]. In a study of 41 COVID-19 patients, 21% received corticosteroids, which could suppress lung inflammation [203], in respect to administered dose depending on disease severity [203]. However, the clinical outcomes of coronavirus and similar outbreaks do not support the use of corticosteroids. In a retrospective observational study, 309 MERS patients given corticosteroids were more likely to require mechanical ventilation, vasopressors, and renal replacement therapy [204]. Overall, there are many reasons to prevent treatment of patients with COVID-19 infection with corticosteroids, and such treatment may be harmful [205]. Alternatively, Chinese clinicians combined Chinese and Western medicine treatment including lopinavir/ritonavir, arbidol, and Shufeng Jiedu Capsule (SFJDC, a traditional Chinese medicine) and gained significant improvement in pneumonia associated symptoms in Shanghai Public Health Clinical Center, China [206].
Sequences similarities expressed as % identity, between SARS-CoV-2 and other coronaviruses
| Other Coronaviruses | Bat | SARS | Pangolin | Camel | MERS- | Dromedarius | Bovine | H-Enteric | Canine | Avian |
|---|---|---|---|---|---|---|---|---|---|---|
| Epitopes of SARS-CoV-2 nucleocapsid | ||||||||||
| KHWPQIAQ; FAPSASAFF | 100 | 100 | 100 | 78.571 | 78.571 | 78.571 | 52.941 | 52.941 | 52.941 | - |
| AQFAPSA; SAFFGMSR | 100 | 100 | 71.429 | 78.571 | 78.571 | 78.571 | 52.941 | 52.941 | 52.941 | - |
| AQFAPSA; SAFFGMSR | 100 | 100 | 100 | 71.429 | 71.429 | 71.429 | 63.636 | 63.636 | 63.636 | 58.333 |
| PKGFYAEG; SRGGSQASSR | 100 | 100 | 100 | 61.111 | 61.111 | 61.111 | 61.111 | 48 | 48 | 100 |
| QFAPSASAF; FGMSRIGM | 100 | 100 | 100 | 81.818 | 81.818 | 81.818 | 50 | 50 | 50 | 53.846 |
| QLPQGTTLPKGF; YAEGSRGGSQ | 100 | 100 | 100 | 61.111 | 61.111 | 61.111 | 66.667 | 66.667 | 66.667 | 100 |
| YNVTQAFGR; RGPEQTQGNF | 100 | 100 | 100 | 63.158 | 63.158 | 63.158 | 58.824 | 58.824 | 58.824 | - |
| SARS CoV-2 spike protein | ||||||||||
| 424-437 | - | - | - | - | - | - | 80 | 80 | 80 | - |
| 447-458 | - | - | - | - | - | - | 75 | - | 75 | - |
| 754-764 | - | - | - | - | - | - | 83.33 | 83.33 | 83.33 | - |
| 789-799 | - | - | - | - | - | - | 57.14 | 57.14 | 57.14 | - |
| 1139-1152 | - | - | - | - | - | - | 70 | 100 | 70 | - |
| SARS CoV-2 Spike protein (GI QHR63290) | ||||||||||
| AGO98871 | - | - | - | - | - | - | 38.42 | - | - | - |
| QAY30020 | - | - | - | - | - | - | - | - | 37.68 | - |
| ACJ35486 | - | - | - | - | - | - | - | 37.68 | - | - |
| ACT10865 | - | - | - | - | - | - | 31.23 | - | - | - |
| SARS-CoV-2_HKU-SZ-001_2020 | - | 98.94 | - | - | - | - | - | - | - | - |
| SARS-CoV-2 strain | - | - | 99 | - | - | - | - | - | - | - |
| SARS-CoV-2 (Wuhan-Hu-1) | ||||||||||
| ORF1ab | 95 | 86 | - | - | 50 | - | - | - | - | - |
| S | 80 | 76 | - | - | 30 | - | - | - | - | - |
| ORF3a | 91 | 72 | - | - | - | - | - | - | - | - |
| E | 100 | 94 | - | - | 36 | - | - | - | - | - |
| M | 98 | 90 | - | - | 42 | - | - | - | - | - |
| ORF6 | 93 | 68 | - | - | - | - | - | - | - | - |
| ORF7a | 88 | 85 | - | - | - | - | - | - | - | - |
| ORF8 | 94 | 40 | - | - | - | - | - | - | - | - |
| N | 94 | 90 | - | - | 48 | - | - | - | - | - |
The development of vaccines and therapeutic antibodies against COVID-19 has important implications. The majority of the vaccines being developed for coronaviruses target the spike glycoprotein. The cross-reactivity of anti-SARS-CoV antibodies with the COVID-19 spike protein was assessed because SARS-CoV- 2 and SARS-CoV show a relatively high identity of the receptor-binding domain (RBD). The SARS-CoV-specific human monoclonal antibody CR3022 binds potently with the COVID-19 RBD [207]. However, other SARS-CoV RBD-directed antibodies 230, m396 and 80R cannot bind to the COVID-19 RBD. Thus, Wrapp et al. suggest that CR3022 may be a potential therapeutic candidate, alone or in combination with other neutralizing antibodies, for the prevention and treatment of COVID-19 infections [208]. Monoclonal antibodies can only recognize a single antigen epitope, which limits their use in the treatment of COVID-19. Applying monoclonal antibodies for new pathogens to clinical practice requires time. Alternatively, convalescent plasma has been suggested to treat COVID-19, the method was used early after symptom onset in the treatment of SARS, and the pooled odds of mortality following treatment was reduced compared with placebo or no therapy (odds ratio, 0.25) [209]. Now, the structure of SARS-CoV-2 is revealed allowing to the development of medical countermeasures and optimization vaccination strategies.
Future directions
The emerging pneumonia, COVID-19, caused by SARS-CoV-2, is a contagious pandemic. At the time of writing, the pandemic is still ongoing, with no available treatment. To battle this pandemic and be prepared for future outbreaks, it is crucial to understand its pathogenesis. SARS-CoV-2 exhibits strong infectivity but reduced virulence compared to SARS and MERS-CoVs. Fortunately, the scientific community has made progress in the characterization of the novel coronavirus and is working extensively on therapies and vaccines. Phylogenetic analyses suggest a bat origin of 2019-nCoV, but, considering the widespread distribution of SARS-CoVs in natural reservoirs, such as bats, camels, and pangolins, further research is needed to find novel intermediate hosts and block interspecies transmission. The genomic structure of the newly emerged coronavirus is evolutionary and may be under selective pressure with several mutations and homologous recombination. 2019-nCoV also potentially recognizes ACE2 from a diversity of animal species, implicating these animal species as possible intermediate hosts or animal models for 2019-nCoV infections. These results provide insights into the receptor usage, cell entry, host cell infectivity, and animal origin of 2019-nCoV. Furthermore, epidemic surveillance and preventive measures against SARS-CoV-2 will be enhanced. Structural studies on ACE2 receptor complexes with 2019-nCoV spike protein will contribute to understanding cross-species receptor usage of this novel coronavirus. In vitro studies will be useful for investigating how SARS-CoV-2 modifies gene expression in primary target cells, such as macrophages, dendritic cells, lymphocytes, and pulmonary epithelial cells. Animal models will allow investigators to determine the relationship between viral load and disease outcome, as well as to evaluate the role of infection and immune dysfunction in the disease process. Gained knowledge will facilitate the development of specific therapies designed to minimize pulmonary disease and optimize the anti-SARS-CoV-2 immune response. The striking structural similarity and sequence conservation among the SARS-CoV-2 S and SARS-CoV S glycoproteins emphasize the close relationship between these two viruses, which both recognize human ACE2 to enter target cells. This resemblance is further strengthened by the finding that SARS-CoV spike elicited polyclonal antibodies responses, potently neutralizing SARS-CoV-2 spike-mediated entry into cells [80]. The SARS-CoV-2 RBD binds to ACE2 with a higher affinity than does the SARS-CoV RBD. Thus, potent SARS-CoV-specific neutralizing antibodies that target the receptor binding site of SARS-CoV failed to bind 2019-nCoV S protein, indicating that it is necessary to develop novel monoclonal antibodies that could bind specifically to the 2019-nCoV RBD [210]. Other scientists indicate that the soluble ACE2 may act as a competitive interceptor of SARS-CoV and other coronaviruses by preventing binding of the viral particle to ACE2 [211]. Indeed, in vitro studies showed SARS-CoV replication was blocked by a soluble form of ACE2 in a monkey kidney cell line [212,213]. Moreover, ACE2 fused to the Fc portion of immunoglobulin has just been reported to neutralize SARS-CoV-2 in vitro [214]. In this context, soluble recombinant human ACE2 protein could actually be beneficial as a novel biologic therapeutic to combat or limit infection progression caused by coronaviruses that utilize ACE2 as a receptor.
Acknowledgements
Authorship
All authors made substantial contributions to all of the following: (1) conception and design of the study, data acquisition, or analysis and interpretation of data; (2) drafting or critical revision of the article for intellectual content; and (3) final approval of version to be submitted.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mani RS, Ravi V, Desai A, Madhusudana SN. Emerging viral infections in India. Proc Natl Acad Sci India Sect B Biol Sci. 2012;82:5-21 doi: 10.1007/s40011-011-0001-1
2. Lorentzen HF, Benfield T, Stisen S, Rahbek C. COVID-19 is possibly a consequence of the anthropogenic biodiversity crisis and climate changes. Dan Med J. 2020;67(5):A205025
3. Epstein PR, Chivian E, Frith K. Emerging diseases threaten conservation. Environ Health Perspect. 2003;111(10):A506-7
4. Sarma N. Emerging and re-emerging infectious diseases in South East Asia. Indian J Dermatol. 2017;62:451-5
5. Gao GF. From “A”IV to “Z”IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157-1159
6. World Health Organization Regional Office for the Western Pacific. Asia Pacific strategy for emerging diseases and public health emergencies (APSED III): advancing implementation of the International Health Regulations (2005). 2017. https://iris.wpro.who.int/handle/10665.1/13654
7. Wei P, Cai Z, Hua J, Yu W, Chen J, Kang K. et al. Pains and gains from China's experiences with emerging epidemics: From SARS to H7N9. BioMed Res Int. 2016;2016:1-6 doi: 10.1155/2016/5717108
8. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727-33 doi: 10.1056/NEJMoa2001017
9. Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA. et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and namingit SARS-CoV-2. Nat Microbiol. 2020;5:536-44 doi: 10.1038/s41564-020-0695-z
10. Zhao S, Chen H. Modeling the epidemic dynamics and control of COVID-19 outbreak in China. Quant Biol. 2020;11:1-9 doi: 10.1007/s40484-020-0199-0. [Epub ahead of print]
11. Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J. et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514-23 doi: 10.1016/S0140-6736(20)30154-9
12. Sun Z, Thilakavathy K, Kumar SS, He G, Liu SV. Potential factors influencing repeated SARS outbreaks in China. Int J Environ Res Public Health. 2020;17:1633
13. Casanova LM, Jeon S, Rutala WA, Weber DJ, Sobsey MD. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl Environ Microbiol. 2010;76:2712-7 doi: 10.1128/AEM.02291-09
14. Chan KH, Peiris JS, Lam SY, Poon LL, Yuen KY, Seto WH. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv Virol. 2011;2011:734690. doi: 10.1155/2011/734690
15. Jaakkola K, Saukkoriipi A, Jokelainen J, Juvonen R, Kauppila J, Vainio O. et al. Decline in temperature and humidity increases the occurrence of influenza in cold climate. Environ Health. 2014;13:22. doi: 10.1186/1476-069X-13-22
16. Kudo E, Song E, Yockey LJ, Rakib T, Wong PW, Homer RJ. et al. Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc Natl Acad Sci USA. 2019;116:10905-10 doi: 10.1073/pnas.1902840116
17. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506 doi: 10.1016/S0140-6736(20)30183-5
18. Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA. 2020;117:9241-3 doi: 10.1073/pnas.2004999117
19. Giovanetti M, Benvenuto D, Angeletti S, Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J. Med. Virol. 2020:1-4 doi: 10.1002/jmv.25699
20. Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel coronavirus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212
21. Benvenuto D, Giovanett M, Salemi M, Prosperic M, De FloraC, Alcantara L C J. et al. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathogens and global health. 2020 doi:10.1080/20477724.2020.1725339
22. Hu B, Ge X, Wang L-F, Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12:221
23. Zhang L, Shen FM, Chen F, Lin Z. Origin and evolution of the 2019 novel coronavirus. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa112
24. Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H. et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020 doi: 10. 1002/jmv.25726
25. Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134-3146
26. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574 doi: 10.1016/S0140-6736(20)30251-8
27. Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G. et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265
28. Hu D. et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bat. Emerg Microbes Infect. 2018;7:1-10
29. Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus J Med Virol. 2020; 92: 522-528.
30. Cagliani R, Forni D, Clerici M, Sironi M. Computational inference of selection underlying the evolution of the novel coronavirus SARS-CoV-2. J Virol. 2020 doi: 10.1128/JVI.00411-20
31. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;270:579. doi.org: 10.1038/s41586-020-2012-7
32. Banerjee S, Narayanan K, Mizutani T, Makino S. Murine coronavirus replication-induced p38 mitogen-activatedprotein kinase activation promotes interleukin-6 production and virus replication in cultured cells. J Virol. 2002;76:5937-48
33. Hampton T. Bats may be SARS reservoir. JAMA. 2005;294(18):2291
34. Li F, Li WH, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864-1868 doi: 10.1126/science.1116480
35. Benvenuto D, Giovannetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M. The 2019-new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020 doi: 10.1002/jmv.25688
36. Ji W, Wang W, Zhao X, Zai J, Li X. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J Med Virol. 2020 92(4). doi: 10.1002/jmv.25682
37. Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev. Microbiol. 2019;17:181-192
38. Liu P, Chen W, Chen JP. Viral metagenomics revealed sendai virus and coronavirus infection of Malayan pangolins (Manis javanica). Viruses. 2019;11:979. doi.org: 10. 3390/v11110979
39. Zhang T, Qunfu W, Zhigang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 Outbreak. Curr Biol. 2020;30:1-6 doi: 10.1016/j.cub.2020.03.022
40. Guan W, Ni Z, Hu Y, Liang W, Ou C, He H, Liu L, Shan H, Lei C. et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020;382:1708-1720 doi: 10.1056/NEJMoa2002032
41. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061-9 doi: 10.1001/jama.2020.1585. [Epub ahead of print]
42. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y. et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199-207 doi: 10.1056/NEJMoa2001316
43. Chen N. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513
44. Liu Y, Zhang C, Huang F, Yang Y, Wang F, Yuan J, Yuan J, et al. 2019-novel coronavirus (2019-nCoV) infections trigger an exaggerated cytokine response aggravating lung injury. 2020. http://www.chinaxiv.org/abs/202
45. Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM. et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348(20):1986-94
46. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A. et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13(9):752-61
47. Wang H, Xiao X, Lu J, Chen Z, Li K, Liu H. et al. Factors associated with clinical outcome in 25 patients with avian influenza A (H7N9) infection in Guangzhou, China. BMC Infect Dis. 2016;16(1):534
48. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1-9 doi: 10.12932/ap-200220-0772
49. Cheung CY. et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819-7826
50. Lee CH, Chen RF, Liu JW, Yeh WT, Chang JC, Liu PM. et al. Altered p38 mitogen-activated protein kinase expression in different leukocytes with increment of immunosuppressive mediators in patients with severe acute respiratory syndrome. J Immunol. 2004;172:7841-7
51. Kui L, Fang YY, Deng Y, Liu W, Wang MF, Ma JP. et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl). 2020;133:1025-31
52. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783-801
53. Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P. et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424-432
54. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69
55. Qiu T, Mao T, Wang Y, Zhou M, Qiu J, Wang J. et al. Identification of potential cross-protective epitope between a new type of coronavirus (2019-nCoV) and severe acute respiratory syndrome virus. J Genet Genomics. 2020;47:115-7 doi: 10.1016/j.jgg.2020.01.003
56. Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P. et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325-8 doi: 10.1016/j.chom.2020.02.001
57. Wassenaar T.M. and Zou Y. 2019_nCoV/SARS-CoV-2: rapid classification of betacoronaviruses and identification of traditional Chinese medicine as potential origin of zoonotic coronaviruses. Lett Appl Microbiol. 2020;70:342-348 doi:10.1111/lam.13285
58. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: Recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523-534 https://doi.org/10.1038/ nrmicro.2016.81
59. Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y. et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):E59. doi: 10.3390/v11010059
60. Zhou T, Gu W, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81(1):77-86
61. Kandeel M, Ibrahim A, Fayez M, Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020;92:660-6 doi: 10.1002/jmv.25754. [Epub ahead of print]
62. Kandeel M, Altaher A. Synonymous and biased codon usage by MERS CoV papain-like and 3CL-proteases. Biol Pharm Bull. 2017;40(7):1086-1091
63. Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020 doi. https://doi.org/10.1002/jmv.25719
64. Sah R, Rodriguez-Morales AJ, Jha R, Chu DKW, Gu H, Peiris M. et al. Complete genome sequence of a 2019 novelcoronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol Resour Announc. 2020;9:e00169-20
65. Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433-440
66. Tang X, Wu C, Li X, Song Y, Yao X, Wu X. et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10. 1093/nsr/nwaa036
67. Castillo A, Parra B, Tapia P, Acevedo A, Lagos J, Andrade W. et al. Phylogenetic analysis of the first four SARS-CoV-2 cases in Chile. J Med Virol. 2020 doi: 10.1002/jmv.25797. [Epub ahead of print]
68. Issa E, Merhi G, Panossian B, Salloum T, Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5(3):e00266-20
69. Li X, Giorgi EE, MH Marichannegowda, Foley B, Xiao C, Kong X-P, Chen Y, Gnanakaran S. et al. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci Adv. 2020 10.1126/sciadv.abb9153
70. Tiwari M, Mishra D. Investigating the genomic landscape of novel coronavirus (2019-nCoV) to identify non-synonymous mutations for use in diagnosis and drug design. J Clin Virol. 2020;128(2020):104441
71. Tolou HJ. et al. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J Gen Virol. 2001;82:1283-1290
72. Clavel F, Hoggan MD, Willey RL, Strebel K, Martin MA, Repaske R. Genetic recombination of human immunodeficiency virus. J Virol. 1989;63:1455-1459
73. Bollyky PL, Rambaut A, Harvey PH, Holmes EC. Recombination between sequences of hepatitis B virus from different genotypes. J Mol Evol. 1996;42:97-102
74. Colina R, Casane D, Vasquez S. et al. Evidence of intratypic recombination in natural populations of hepatitis C virus. J Gen Virol. 2004;85:31-37
75. Drosten C, Günther S, Preiser W. et al. Identification of a novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967-1976
76. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814-20 doi: 10.1056/NEJMoa1211721
77. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470-3 doi: 10.1016/S0140-6736(20)30185-9
78. Nikolich-Zugich J, Knox KS, Rios CT, Natt B, Bhattacharya D, Fain MJ. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience. 2020;42:505-14 doi: 10.1007/s11357-020-00186-0
79. Bai Y, Nie X, Wen C. Epidemic prediction of 2019-nCoV in Hubei Province and comparisonwith SARS in Guangdong Province. 2020. https://ssrn.com/abstract=3531427
80. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260-3 doi: 10.1126/science.abb2507
81. Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.01.26.919985
82. Smith RD. Responding to global infectious disease outbreaks: lessons from SARS on the role of risk perception, communication and management. Soc Sci Med. 2006;63:3113-3123
83. Wang L.F. and Eaton, B.T. Bats, civets and the emergence of SARS. Curr Top Microbiol Immunol. 2007;315:325-44
84. Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C. et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290
85. Baharoon S, Memish ZA. MERS-CoV as an emerging respiratory illness: a review of prevention methods. Travel Med Infect Dis. 2019;32:101520. doi: 10.1016/j.tmaid.2019.101520
86. Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J. et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490-502 doi: 10.1016/j.tim.2016.03.003
87. Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45-56 doi: 10.1016/j.antiviral.2013.10.013
88. Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 2020 doi: 10.1101/2020.01.31.929042
89. Guo Y-R, Cao Q-D, Hong Z-S, Tan Y-Y, Chen S-D, Jin H-J. et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreake an update on the status. Mil Med Res. 2020;7(1):1e10
90. Qing H, Li Z, Yang Z, Shi M, Huang Z, Song J. et al. The possibility of COVID-19 transmission from eye to nose. Acta Ophthalmol. 2020;98:e388. doi: 10.1111/aos.14412
91. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271-280.e8 doi: 10.1016/j.cell.2020.02.052
92. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-lik ecleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742
93. Xu X, Chen P, Wang J, Feng J, Zhou H, Li X. et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457-60 doi: 10.1007/s11427-020-1637-5
94. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127-20
95. Wu K, Peng G, Wilken M, Geraghty RJ, Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J Biol Chem. 2012;287:8904-11.doi 10.1074/jbc.M111.325803
96. Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C. et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970-1 doi: 10.1056/NEJMc2001468
97. Lu CW, Liu XF, Jia ZF. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39. doi: 10.1016/S0140-6736(20)30313-5
98. Rodríguez-Morales AJ, MacGregor K, Kanagarajah S, Patel D, Schlagenhauf P. Going global -travel and the 2019 novel coronavirus. Travel Med Infect Dis. 2020;33:101578. doi: 10.1016/j.tmaid.2020.101578
99. Grassia R, Testa S, Pan A, Conti CB. SARS-CoV-2 and gastrointestinal tract: the dark side of the pandemic. Dig Liver Dis. 2020; doi: 10.1016/j.dld. 2020 04.028. [Epub ahead of print]
100. Yang CW, Chen MF. Composition of human-specific slow codons and slow di-codons in SARS-CoV and 2019-nCoV are lower than other coronaviruses suggesting a faster protein synthesis rate of SARS-CoV and 2019-nCoV. J Microbiol Immunol Infect. 2020; doi: 10.1016/j.jmii. 2020 03.002. [Epub ahead of print]
101. Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill. 2020;25(5):2000062
102. Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B. et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386-9 doi: 10.1080/22221751.2020.1729071
103. Hamming I, Timens W, Bulthuis M, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631-7
104. To KK, Tsang OT, Chik-Yan Yip C, Chan KH, Wu TC, Chan JMC. et al. Consistent detection of 2019 novelcoronavirus in saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa149. [Epub ahead of print]
105. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185-92 doi: 10.1007/s11684-020-0754-0
106. Choudhary R, Kapoor MS, Singh A, Bodakhe SH. Therapeutic targets of renin-angiotensin system in ocular disorders. J Curr Ophthalmol. 2016;29:7-16 doi: 10.1016/j.joco.2016.09.009
107. Cheng X, Liu J, Li N, Nisenbaum E, Sun Q, Chen B. et al. Otolaryngology providers must be alert for patients with mild and asymptomatic COVID-19. Otolaryngol Head Neck Surg. 2020 doi: 10.1177/0194599820920649
108. Zhao C, Viana A, Wang Y, Wei H-Q, Yan A-H, Capasso R. Otolaryngology during COVID-19: preventive care and precautionary measures. Am J Otolaryngol. 2020; 22;102508. doi: 10.1016/j.amjoto. 2020 102508
109. Kampf G, Todt D, Pfaender S. Steinmann E. Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. J Hosp Infect. 2020 doi: 10.1016/j.jhin.2020.01.022
110. Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69-71 doi: 10.1016/j.micinf.2020.01.004
111. Liu L, Wei Q, Alvarez X, Wang H, Du Y, Zhu H. et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011;85:4025-30 doi: 10.1128/JVI.02292-10
112. Cleveland JL, Gray SK, Harte JA, Robison VA, Moorman AC, Gooch BF. Transmission of blood-borne pathogens in US dental health care settings: 2016 update. J Am Dent Assoc. 2016;147:729-38 doi: 10.1016/j.adaj.2016.03.020
113. Wei J, Li Y. Airborne spread of infectious agents in the indoor environment. Am J Infect Control. 2016;44(Suppl 9):S102-8
114. Otter JA, Donskey C, Yezli S, Douthwaite S, Goldenberg SD, Weber DJ. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. 2016;92:235-50 doi: 10.1016/j.jhin.2015.08.027
115. Siddell SG, Ziebuhr J, Snijder EJ. Coronaviruses, toroviruses, and arteriviruses. In: Mahy BW, Meulen V, Borriello SP, Murray PR, Funke G, Kaufmann SH, et al. Ed. Topley & Wilson's Microbiology and Microbial Infections. Chichester, UK: John Wiley & Sons, Ltd. 2010
116. Risco C, Antón IM, Enjuanes L, Carrascosa JL. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70(7):4773-4777
117. Ruch T, Machamer C. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363-382
118. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237-261
119. Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spikeprotein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801-11
120. Tortorici MA, Walls AC, Lang Y, Wang C, Li Z, Koerhuis D, Boons GJ, Bosch BJ, Rey FA, de Groot RJ, Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol. 2019;26:481-489
121. Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118e121
122. Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 2013;100:246-254 doi: 10.1016/j.antiviral.2013.08.014
123. Simmons G, Zmora P, Gierer S, Heurich A, Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605-614
124. Matsuyama S. et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658-12664
125. Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93-116
126. Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29-57 doi: 10.1016/bs.aivir.2016.08.004
127. Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120-134
128. Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102(33):11876-81
129. Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch BJ, Rey FA. et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA. 2017;114:11157-62 doi: 10.1073/pnas.1708727114
130. de Wilde AH, Snijder EJ, Kikkert M, van Hemert MJ. Host factors in coronavirus replication. Curr Top Microbiol Immunol. 2018;419:1-42
131. Hussain S, Pan J, Chen Y, Yang Y, Xu J, Peng Y. et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79(9):5288-95
132. Perrier A, Bonnin A, Desmarets L, Danneels A, Goffard A, Rouille Y. et al. The C-terminal domain of the MERS coronavirus M protein contains a trans- Golgi network localization signal. J Biol Chem. 2019;294(39):14406-21
133. Kowalczuk S, Bröer A, Tietze N, Vanslambrouck JM, Rasko JEJ, Bröer S. A protein complex in the brush-border membrane explains a Hartnup disorder allele. FASEB J. 2008;22(8):2880-2887
134. Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532(1-2):107-110
135. Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S. A novel angiotensin-converting enzyme related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87(5):E1-E9
136. Serfozo P, Wysocki J, Gulua G, Schulze A, Ye M, Liu P. et al. Ang II (angiotensin II) conversion to angiotensin-(1-7) in the circulation is POP (prolyloligopeptidase)-dependent and ACE2 (angiotensin-convertingenzyme 2) -independent. Hypertension. 2020;75:173-82 doi: 10.1161/HYPERTENSIONAHA.119.14071
137. Burrell LM, Risvanis J, Kubota E, Dean RG, MacDonald PS, Lu S, Tikellis C, Grant SL, Lew RA, Smith AI, Cooper ME, Johnston CI. Myocardial infarction increases ACE2 expression in rat and humans. Eur Heart J. 2005;26(4):369-375
138. Zhang H, Kang Z, Gong H, Xu D, Wang J, Li Z. et al. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. BioRxiv. 2020 doi: 10.1101/2020.01.30.927806
139. Ren X, Glende J, Al-Falah M, de Vries V, Schwegmann-Wessels C, Qu X. et al. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J Gen Virol. 2006;87:1691-5
140. Ding Y, He L, Zhang Q, Huang Z, Che X, Hou J, Wang H, Shen H, Qiu L. et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203:622-630
141. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV— a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226-36
142. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B beta coronaviruses. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0688-y
143. Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418e23
144. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450-454
145. Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B. et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875-9 doi: 10.1038/nm1267
146. Frana MF, Behnke JN, Sturman LS, Holmes KV. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J Virol. 1985;56:912-20
147. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cellentry mediated by the viral spike protein. Viruses. 2012;4:1011-33 doi: 10.3390/v4061011
148. Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864-1868
149. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444-1448
150. Imai Y, Kuba K, Ohto-Nakanishi T, Penninger JM. Angiotensin-converting enzyme 2 (ACE2) in disease pathogenesis. Circ J. 2010;74:405-410 doi: 10.1253/circj.CJ-10-0045
151. Li T, Liu D, Yang Y. et al. Phylogenetic supertree reveals detailed evolution of SARS-CoV-2. Research Square. 2020 in press. doi:10.21203/rs.3.rs-33194/v1
152. Corman VM, Ithete NL, Richards LR. et al. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88:11297-303 doi: 10.1128/JVI.01498-14
153. Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2565
154. Tilocca B, Soggiu A, Sanguinetti M, Musella V, Britti D, Bonizzi L, Urbani A, Roncada P. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microbes Infect. 2020;22(4):188-194
155. Tilocca B, Soggiu A, Musella V, Britti D, Bonizzi L, Sanguinetti M, Urbani A, Roncada P. Molecular basis of COVID-19 relationships in different species: a one health perspective. Microbes Infect. 2020;22(4):218-220
156. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671-680
157. Vijgen L, Keyaerts E, Lemey P. et al. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. 2006;80:7270e7274
158. Bidokhti MRM, Traven M, Krishna NK. et al. Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains. J Gen Virol. 2013;94:2036e2349
159. Oong XY, Ng KT, Takebe Y. et al. Identification and evolutionary dynamics of two novel human coronavirus OC43 genotypes associated with acute respiratory infections: phylogenetic, spatiotemporal and transmission network analyses. Emerg Microb Infect. 2017;6(1):e3. doi.org: 10.1038/emi.2016.132
160. Xu J, Zhao S, Teng T, Elgaili Abdalla A, Zhu W, Xie L, Wang Y, Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi:10.3390/v12020244
161. Anand KB, Karade S, Sen S, Gupta RM. SARS-CoV-2: Camazotz's curse. Med J Armed Forces India. 2020;76(2):136-141
162. Jaimes JA, André NM, Chappie JS, Millet JK, Whittaker GR. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol. 2020;432(10):3309-3325
163. Shanker A, Bhanu D, Alluri A, Gupta S. Whole genome sequence analysis and homology modelling of a 3C like peptidase and a non-structural protein 3 of the SARS-CoV-2 shows protein ligand interaction with an aza-peptide and a noncovalent lead inhibitor with possible antiviral properties. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11846943.v9
164. Wang N, Luo C, Liu H, Yang X-L, Hu B, Zhang W, Li B. et al. Characterization of a new member of alphacoronavirus with unique genomic features in Rhinolophus bats. Viruses. 2019;11:379
165. Lam T, Shum M, Zhu HZ. et al. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. 2020 945485. doi: 10.1101/2020.02.13.945485
166. Chan JF-W, Choi GK-Y, Tsang AK-L, Tee K-M. et al. Development and evaluation of novel real-time reverse transcription-PCR assays with locked nucleic acid probes targeting leader sequences of human-pathogenic coronaviruses. J Clin Microbiol. 2015;53:2722-2726
167. Wu P, Hao X, Lau EHY, Wong JY, Leung KSM, Wu JT, Cowling BJ, Leung GM. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro. Surveill. 2020;25:2000044
168. Lee SH, Baek YH, Kim YH, Choi YK, Song MS, Ahn JY. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front Microbiol. 2016;7:2166
169. Huang P, Wang H, Cao Z, Jin H, Chi H, Zhao J. et al. A rapid and specific assay for the detection of MERS-CoV. Front Microbiol. 2018;9:1101
170. Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y. et al. Molecular diagnosis of a novel Coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66(4):549-555
171. Chan JF, Yip CC, To KK, Tang TH, Wong SC, Leung KH. et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58(5):e00310-20
172. Konrad R, Eberle U, Dangel A, Treis B, Berger A, Bengs K. et al. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill. 2020;25:2000173
173. Cordes AK, Heim A. Rapid random access detection of the novel SARS-coronavirus- 2 (SARS-CoV-2, previously 2019-nCoV) using an open access protocol for the Panther Fusion. J Clin Virol. 2020;125:104305
174. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045
175. Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y. et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172-5
176. Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P. et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115-E117
177. Huang P, Liu T, Huang L, Liu H, Lei M, Xu W. et al. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295(1):22-33
178. Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296(2):E41-E45
179. Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W. et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32-E40
180. Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J. et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425-34
181. Xu X, Yu C, Qu J, Zhang L, Jiang S, Huang D. et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47:1275-80
182. Wang Y, Kang H, Liu X, Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J Med Virol. 2020;92(6):538-539
183. Li H, Wang YM, Xu JY, Cao B. Potential antiviral therapeutics for 2019 novel coronavirus. Chin J Tuberc Respir Dis. 2020;43(0):E002
184. Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H. et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929-936 doi: 10.1056/NEJMoa2001191
185. Yao T-T, Qian J-D, Zhu W-Y, Wang Y, Wang G-Q. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-a possible reference for coronavirus disease-19 treatment option. J Med Virol. 2020;92(6):556-563
186. Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP. et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313-17
187. Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477
188. Xu Z, Peng C, Shi Y, Zhu Z, Mu K, Wang X, Zhu W. Nelfinavir was predicted to be a potential inhibitor of 2019 nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. bioRxiv. 2020 doi: 10.1101/2020.01.27.921627
189. Guo Y-R, Cao Q-D, Hong Z-S, Tan Y-Y, Chen S-D. et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11
190. Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67-9
191. Rolain JM, Colson P, Raoult D. Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infections in the 21st century. Int J Antimicrob Agents. 2007;30:297-308
192. Yan Y, Zou Z, Sun Y, Li X, Xu KF, Wei Y. et al. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300-2
193. Aguiar ACC, Murce E, Cortopassi WA, Pimentel AS, Almeida M, Barros DCS. et al. Chloroquine analogs as antimalarial candidates with potent in vitro and in vivo activity. Int J Parasitol Drugs Drug Resist. 2018;8(3):459-64
194. Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect Dis. 2003;3(11):722-7
195. Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, Ksiazek TG. et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69
196. Golden EB, Cho HY, Hofman FM, Louie SG, Schonthal AH, Chen TC. Quinoline-based antimalarial drugs: a novel class of autophagy inhibitors. Neurosurg Focus. 2015;38(3):E12
197. Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 doi: 10.1038/s41422-020- 0282-0
198. Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72-3
199. Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P. et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020;71(15):732-739
200. Chen L, Liu HG, Liu W, Liu J, Liu K, Shang J. et al. [Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia]. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:203-8
201. Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020;55(4):105932
202. Touret F, de Lamballerie X. Of chloroquine and COVID-19. Antiviral Res. 2020;177:104762
203. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506
204. Arabi YM, Mandourah Y, Al-Hameed F, Sindi AA, Almekhlafi GA, Hussein MA. et al. Corticosteroid therapy for critically ill patients with Middle East Respiratory Syndrome. Am J Respir Crit Care Med. 2018;197:757-67
205. Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473-5
206. Wang Z, Chen X, Lu Y, Chen F, Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci Trends. 2020;14(1):64-68 doi: 10.5582/bst.2020.01030
207. Tian X, Li C, Huang A, Xia S, Lu S, Shi Z. et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382-5
208. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260-3
209. Mair-Jenkins J, Saavedra-Campos M, Baillie JK, Cleary P, Khaw FM, Lim WS. et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211:80-90
210. Cao Y, Lin Li, Zhimin Feng, Shengqing Wan, Peide Huang, Xiaohui Sun. et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1
211. Tian X, Lia C, Huang A, Xia S, Lua S, Shi Z. et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382-385 doi: 10.1080/22221751.2020.1729069
212. Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci. 2020;134:543-545
213. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S. et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953-1966 doi: 10.1056/NEJMoa030781
214. Lei C, Fu W, Qian K, Li T, Zhang S, Ding M. et al. Potent neutralization of 2019 novel coronavirus by recombinant ace2-ig. BioRxiv. 2020 doi: 10.1101/2020.02.01.929976
Author contact
![]() Corresponding author: Jae Il Shin, M.D., Ph.D. Address: 50 Yonsei-ro, Seodaemun-gu, C.P.O. Box 8044, Department of Pediatrics, Yonsei University College of Medicine, Seoul 03722, Republic of Korea. Tel: +82-2-2228-2050; Fax: +82-2-393-9118; E-mail: shinjiac.
Corresponding author: Jae Il Shin, M.D., Ph.D. Address: 50 Yonsei-ro, Seodaemun-gu, C.P.O. Box 8044, Department of Pediatrics, Yonsei University College of Medicine, Seoul 03722, Republic of Korea. Tel: +82-2-2228-2050; Fax: +82-2-393-9118; E-mail: shinjiac.

 Global reach, higher impact
Global reach, higher impact