10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(15):2964-2973. doi:10.7150/ijbs.47959 This issue Cite
Research Paper
RAB37 multiple alleles, transcription activation and evolution in mammals
Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Received 2020-5-9; Accepted 2020-8-23; Published 2020-9-19
Abstract
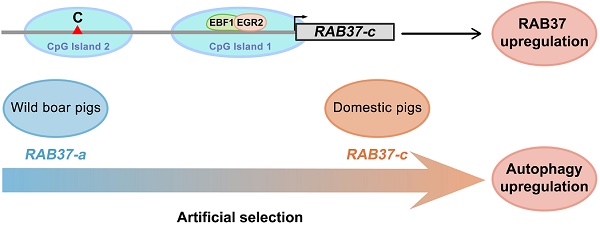
Detecting selection signatures in genomes that relates to transcription regulation has been challenges in genetic analysis. Here, we report a set of transcription factors EBF1, E2F1 and EGR2 for transcription activation of RAB37 promoter by a comparative analysis of promoter activities of RAB37 in humans, mice, and pigs. Two of the transcription factors bound to and co-regulated RAB37 promoter in each species. SNPs were further screened in pig RAB37 gene by population genomics in pig populations from both China and Europe. Three SNPs were identified in second CpG island upstream of core promoter of RAB37. These SNP variations led to at least 5 haplotypes, representing 5 multiple alleles of RAB37 in pig population. Distribution of these alleles in different genetic background of breeds showed a role of artificial selection for the variations of these multiple alleles. Of them, RAB37-c acquired the highest ability to activate gene expression in comparison with the other promoters, thus enhanced autophagy efficiently. These findings provide better understanding of transcription activation of RAB37 and artificial selection via RAB37 for autophagy regulation.
Keywords: transcription factors, genomic variation, RAB37, SNPs, mammals
Introduction
Macroautophagy (hereafter autophagy) is an evolutionarily conserved process of catabolism, in which intracellular unwanted components and organelles are sequestered by double-membrane vesicle and delivered to the lysosome for degradation and recycling. Autophagy pathways usually degrade cytoplasmic components, and yet selective nuclear autophagy (nucleophagy) has been observed in eukaryotic cells [1]. Thus, autophagy controls both cytoplasmic and nuclear degradation, which are essential for cell survival, tissue homeostasis and organ development. Accordingly, dysregulation of autophagy is associated with a large variety of diseases, including metabolic disorders, neurodegeneration, and cancer [2].
Autophagy occurs when cells encounter different forms of stress in various physiological and pathological conditions, including nutrient deprivation and hypoxia. A range of signaling activated phagophore induction process, which were involved in a set of ATGs proteins [3]. Upon starvation induction, the ULK1/ATG1 kinase complex was first activated, which consisted of ULK1, ULK2, ATG13, RBCC1/FIP200, and ATG101. The PtdIns3K kinase complex was then activated, including PIK3C3/VPS34, PIK3R4/VPS15, BECN1, ATG14, and UVRAG) [4]. For phagophore expansion, two ubiquitin-like conjugation complexes had essential roles [5]. ATG5 conjugated to ATG12 and interacted with ATG16L1 to form the ATG5-ATG12-ATG16L1 complex via the action of ATG7 and ATG10. The LC3B/ATG8 conjugated with ATG3 via the action of ATG4 and ATG7 sequentially. Finally, conjugation of LC3B to PE (phosphatidylethanolamine) formed a lipidated LC3B-PE and promoted autophagosome expansion. The ATG5-ATG12-ATG16L1 complex plays a key role in the process of LC3B lipidation [6]. Autophagosome was then fused with lysosome to form autolysosome, and sequestered substances were degraded by lysosomal enzymes and recycled for cell reuse. Although, extensive studies detail molecular mechanisms in regulation of autophagosome formation under various physiological and pathological conditions, in particular, remains to be further explored.
RAB37, as a small GTPase, plays important roles in several cellular processes through intracellular membrane traffic. RAB37 can regulate insulin secretion in β-cells [7] and TNF-α secretion from activated macrophages [8]. In allergic responses, RAB37 regulated mast cell degranulation [9]. RAB37 has been identified to be a tumor suppressor and it regulated exocytosis of several proteins including TIMP metallopeptidase inhibitor 1 (TIMP1) [10, 11], secreted frizzled-related protein-1 (SFRP1) [12], thrombospondin-1 (TSP1) [13], and soluble ST2 [14]. In addition, promoter hypermethylation of RAB37 was associated with metastasis [15]. Thus, RAB37 had multiple functions in development, metabolism, immunity, and cancer.
As the core machinery of autophagy, the complex ATG5-ATG12-ATG16L1 was essential for LC3B lipidation and thus promoted autophagosome membrane biogenesis [6]. Accordingly, upstream regulations of the complex ATG5-ATG12-ATG16L1 will be key steps in autophagy. Accumulated evidence showed that RAB37-mediated autophagy regulation was an important cellular event. Recently, our group found that RAB37 functioned as an organizer for autophagosome biogenesis [16, 17]. RAB37 regulated autophagosome formation by modulating ATG5-ATG12-ATG16L1 complex assembly. GTP-bound RAB37 can interact directly with ATG5 and promoted interaction of the ATG5-ATG12 with ATG16L1. Multimeric RAB37-ATG5-ATG12-ATG16L1 complex was recruited and then lipidated LC3B to form active LC3B-PE, which accelerated autophagosome formation [16]. However, transcription regulations of RAB37 remain elusive. To further explore transcription activation of RAB37, in this study, we comparatively analyzed promoter activities of RAB37 in humans, mice, and pigs, investigated transcription factors and their binding abilities to the promoters of RAB37 genes in these species, screened polymorphism in pig RAB37 gene, and identified RAB37 alleles in pig populations worldwide. Further roles of these RAB37 alleles in autophagy were analyzed. These results highlight the importance of transcription activation of RAB37 in autophagy.
Methods
Antibodies and reagents
Anti-LC3B (Cat# 3868) was purchased from Cell Signalling Technology, Danvers, MA, USA. Anti-p62 (SQSTM1) (Cat# 18420-1-AP) was from Proteintech Group, Rosemont, IL, USA. Anti-FLAG antibody (Cat# F3165) was from Sigma-Aldrich, St Louis, MO, USA. Goat anti-rabbit IgG (H+L), horseradish peroxidase (HRP) conjugated antibody (Cat# 31460), goat anti-mouse IgG (H+L), HRP conjugated antibody (Cat# 31430), and TRITC-conjugated ImmunoPure goat anti-rabbit IgG (H+L) (A16071) were obtained from Invitrogen, Carlsbad, USA. DAPI (Cat# C1002) was purchased from Beyotime Institute of Biotechnology (Jiangsu, China). EBSS (Cat# SH30029.02) was from HyClone, Logan, USA.
Plasmid constructs
Eight deletion fragments of the pig RAB37 (ENSSSCT00025032574.1) promoter were amplified from pig genomic DNA, and cloned into pGL3-basic (E1751, Promega, Madison, WI, USA) vector digested by XhoΙ and HindⅢ. Full-length EBF1 (ENSSSCT00000040359.2) and EGR2 (ENSSSCT00025106300.1) of pig were cloned into pcDNA3.0 using HindⅢ/XhoΙ and HindⅢ/EcoRΙ double digestion to generate pCMV-pig-EBF1 and pCMV-pig-EGR2. Site-directed mutagenesis for the EBF1 and EGR2 binding sites were performed using the primers described in Table S1. PS-EBF1-MUT was used as a template for constructing PS-EBF1+EGR2-MUT.
Four different promoter fragments of pig RAB37 containing relevant alleles were cloned into pGL3-basic vector (digested by XhoΙ and HindⅢ) to generate pGL3-RAB37 (pGL3-RAB37-a, pGL3-RAB37-b, pGL3-RAB37-c, and pGL3-RAB37-d). These promoter fragments were cloned into RAB37-FLAG and RAB37-GFP [16], and replaced the transcription elements of the vector to construct RAB37-FLAG (RAB37-a-FLAG, RAB37-b-FLAG, RAB37-c-FLAG, and RAB37-d-FLAG) and RAB37-EGFP (RAB37-a-EGFP, RAB37-b-EGFP, RAB37-c-EGFP, and RAB37-d-EGFP). The primers and PCR conditions were listed in Table S1. All constructs were sequenced.
In silico sequence and SNP analysis
Genomatix database (www.genomatix.de) was used to predict transcription factors in RAB37 promoters. MethPrimer software (www.urogene.org) was used to predict the CpG islands in promoter with island size > 100, observed/expected ratios > 0.6, and GC percent > 50.0. JASPAR database (jaspar.genereg.net/) was used to search sequences and logos of transcription factors.
Genomic DNA samples from Meishan, Xihuzhou large white and Tongcheng pigs were used to amplify the RAB37 promoter regions by polymerase chain reaction and primers described in Table S1. The sequences were aligned with available genomic data in Sus scrofa 11.1 using Ensembl and corresponding SNP position of the sequences was determined in the region of chr12: 6493414-6494172. Positions on the chromosome of SNP variations were at nt 6493746 (SNP1), nt 6493872 (SNP2), and nt 6493991 (SNP3). SNP sites were blasted against available SNP data in the Ensembl databases. SNP frequencies were calculated based on these data.
Cell culture, transfection, and dual-luciferase reporter assays
COS-7, 293T and CHO cells were obtained from China Center for Type Culture Collection. COS-7 and 293T cells were cultured in DMEM (Cat# SH30022.01B, HyClone, Logan, USA) with 10% FBS (Cat# P30-330250, PAN-Biotech, Aidenbach, Germany). CHO cells were cultured in MEM (Cat# SH30023.01, HyClone, Logan, USA) with 10% FBS.
Dual-luciferase reporter assays were performed according to our previous protocols [18]. Cells per 48-well were transfected with 0.5 μg recombinant construct and 10 ng pRL-TK (Cat# E2241, Promega) using LipofectamineTM 2000 (Cat# 11668027, Invitrogen). Luciferase activities were tested by a dual-luciferase reporter assay system (Promega, Madison, USA) and recorded by a Modulus Single Tube Multimode Reader (Turner Biosystems, Sunnyvale, USA) according to the manufacturer's protocol. The experiments were repeated at least 3 times, and analyzed as the means ± SD.
Western blot analysis
Western blots were performed following our previous protocols [19]. 293T cells were transfected with RAB37-FLAG or vector control. At 48 h after transfection, the whole protein extracts were extracted from the cells and used for SDS-PAGE. The gels were transferred onto a 0.45 μm PVDF membrane (Cat# NK0414, Roche Diagnostics, Indianapolis, IN, USA). The membranes were blocked with 5% skim milk powder (Cat# 1172GR100, BioFroxx Gmbh) in TBST and incubated with the primary antibody overnight at 4 °C followed by the horseradish peroxidase-conjugated secondary antibody at room temperature for at least 1 h. ECL Plus detecting reagents (Millipore, Billerica, MA, USA) were used to visualize the protein bands. ImageJ (version J2, NIH, Maryland, US) was used to analyze band intensity values.
Immunofluorescence analysis
Immunofluorescence analysis was performed according to our previous study [16]. CHO cells were cultured on glass coverslips. At 48 h after transfection using LipofectamineTM 2000, the cells were starved in EBSS medium for 1 h. After fixed and permeabilized, the samples were incubated with primary antibody overnight at 4 °C and subjected to indirect immunofluorescence secondary antibody. The DAPI was used to stain the nuclei. Confocal fluorescence microscope (Olympus, FV1000, Tokyo, Japan) was used to capture images.
Statistics analysis
The data were presented as the means ± standard deviation from at least three independent experiments. One-way analysis of variance was tested for comparisons among more than two groups. Statistics analysis was performed using GraphPad Prism 7 software package (GraphPad Software, La Jolla, USA). For all analyses, P value < 0.05 was considered to be statistically significant.
Results
EBF1 and EGR2 activate the pig RAB37 promoter
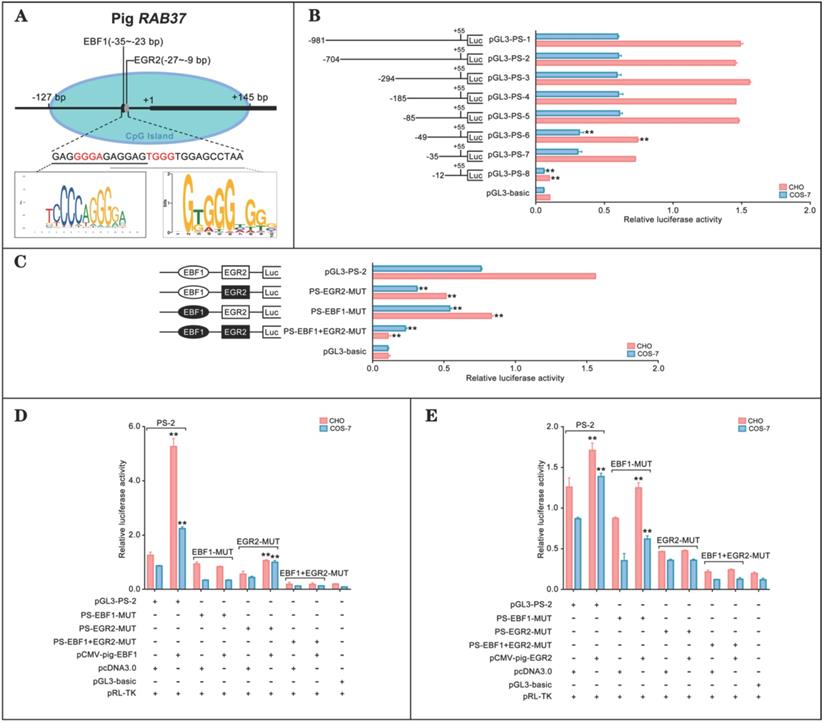
To explore transcription regulation of RAB37, we analyzed potential transcription factors to activate RAB37 transcription in pig. CpG island and transcription factors were first predicted. An EBF1 binding site (-35 bp to -23 bp) and an E2F1 binding site (-27 bp to -9 bp) were detected in a CpG island in the potential promoter in pig (Figure 1A). Deletion analysis and luciferase analysis showed that sequence from -85 bp to -12 bp in the 5' flanking region was essential for the promoter activity (Fig. 1B). To determine roles of these two sites for transcription factors EBF1 and EGR2, site-directed mutants were constructed using the wild-type pGL3-PS-2 as a template. Compared with the pGL3-PS-2 construct, activities of single mutants PS-EBF1-MUT and PS-EGR2-MUT, and double mutant PS-EBF1+EGR2-MUT were significantly decreased, while the activities were obviously low in double mutant PS-EBF1+EGR2-MUT (Figure 1C). These results suggested that the binding sites for the transcription factors EBF1 and EGR2 were important for the promoter activity of pig RAB37.
The binding abilities of EBF1 and EGR2 to the corresponding sites were further tested by luciferase assays, which showed that EBF1 could increase the luciferase activities of wild-type pGL3-PS-2 and mutant PS-EGR2-MUT, but not its single mutant PS-EBF1-MUT and double mutant PS-EBF1+EGR2-MUT (Figure 1D). On the other hand, EGR2 could also activate the activities of wild-type pGL3-PS-2 and single mutant PS-EBF1-MUT, but had no contribution to the activities of single mutant PS-EGR2-MUT and double mutant PS-EBF1+EGR2-MUT (Figure 1E). Thus, EBF1 and EGR2 were the transcription factors of the pig RAB37 promoter.
Promoter activity and transcription activation of pig RAB37. (A) Schematic diagram of the pig RAB37 promoter. An EGR2 binding site and an EBF1 binding site are located in the CpG island (blue oval). Sequence logos of EBF1 and EGR2 binding sites are shown in the lower panel, and logos are based on JASPAR database. (B) Luciferase assays showing activities of a series of deleted constructs in both CHO and COS-7 cells. Left panel indicates each deleted mutant linked with the luciferase gene in the pGL3-basic vector. Right panel shows relative activities of these deleted constructs, as determined by luciferase assays. One-way ANOVA was performed. **P < 0.01. (C) Luciferase assays of point mutations in core promoter. The pGL3-PS-2 construct of 704 bp was used as a basic construct for the analysis. Luciferase assays were used to determine the relative activities. The intact binding sites of EBF1 and EGR2 are indicated by open ovals and boxes respectively. The filled ovals and boxes show the corresponding mutations. The pGL3-basic vector was used as a control. One-way ANOVA was performed. **P < 0.01. (D) Overexpression of EBF1 activates the pig RAB37 promoter. In total, 0.4 mg pGL3-PS-2 or its site mutants (PS-EBF1-MUT, PS-EGR2-MUT, or PS-EBF1+EGR2-MUT) were cotransfected with 0.1 mg EBF1 expressing plasmid (pCMV-pig-EBF1). EBF1 overexpression can increase the promoter activity, except PS-EBF1-MUT and PS-EBF1+EGR2-MUT. One-way ANOVA was performed. **P < 0.01. (E) Overexpression of EGR2 activates the pig RAB37 promoter. In each transfection, 0.4 mg pGL3-PS-2 or its site mutants (PS-EBF1-MUT, PS-EGR2-MUT, or PS-EBF1+EGR2-MUT) were cotransfected with 0.1mg EGR2 expression plasmid (pCMV-pig-EGR2). EGR2 overexpression increases the promoter activity, except the PS-EGR2-MUT and PS-EBF1+EGR2-MUT. One-way ANOVA was performed. **P < 0.01.
Transcription factor-associated evolution in RAB37 promoter
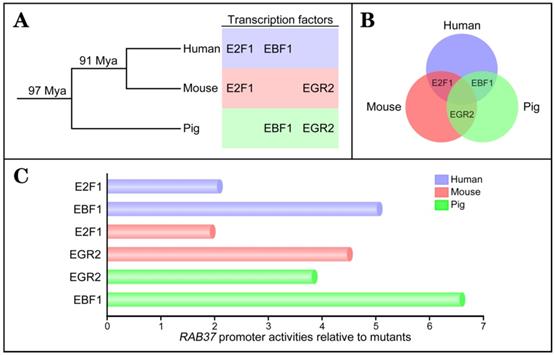
To investigate conservation of transcription factors in regulation of RAB37 gene, the same approaches were used to identify transcription factors to activate the RAB37 promoters in human and mouse. The analysis results showed that binding sites for the transcription factors EBF1 and E2F1 were important for the promoter activity of human RAB37 (Supplementary Figure S1), while EGR2 and E2F1 were the transcription factors of the mouse Rab37 promoter (Supplementary Figure S2). To realize phylogenetic relationship of RAB37 genes in human, mouse, and pig, phylogenetic analysis of binding sites of the transcription factors in the promoters were performed among these species. Based on phylogeny time of mammals [20], the evolution analysis showed that EBF1 and EGR2 were evolved in pig ~97 Mya, and EBF1 was replaced by E2F1 in mouse ~91 Mya. However, EGR2 site was degenerated and both EBF1 and E2F1 sites were evolved in human (Figure 2A). Venn diagram showed alternative usage of two transcription factors from EGR2, E2F1, and EBF1 in RAB37 promoters among human, mouse, and pig (Figure 2B).
Comparisons of transcriptional activation abilities of transcription factors EGR2, E2F1, and EBF1 in the RAB37 promoters among human, mouse, and pig revealed that each species had a dominant transcription factor (EBF1 in human and pig; EGR2 in mouse), while another transcription factor was also essential for transcription activation of the RAB37 gene (Figure 2C). These results suggested earlier evolved transcription factors EBF1 and EGR2 had a dominant contribution to RAB37 transcription, although divergent evolution of binding sites of transcription factors in the RAB37 promoters existed in different lineages.
Artificial selection generates RAB37 alleles in pig populations
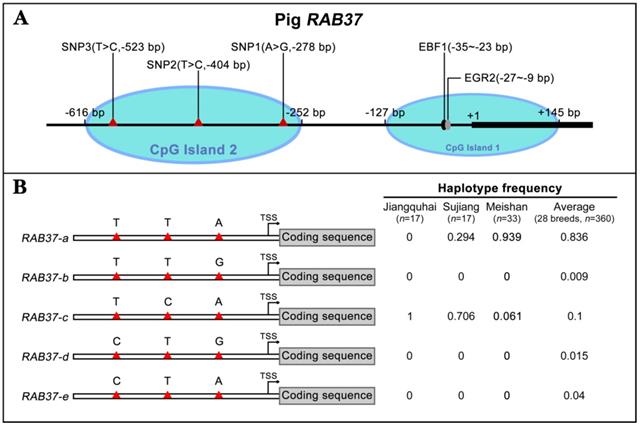
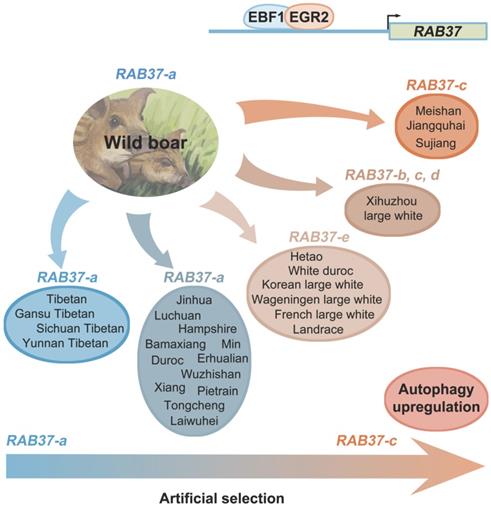
To further explore polymorphism in pig RAB37, genomic sequences of the pig RAB37 promoter were PCR amplified from genomic DNA samples of Meishan, Tongcheng, and Xihuzhou large white pigs. Sequencing analysis revealed 3 SNP sites in the CpG island 2 (Figure 3A). SNP1 (A>G) was located at 278 bp upstream of transcriptional start site (TSS), SNP2 (T>C) ws at 404 bp upstream of the TSS, SNP3 (T>C) at 523 bp upstream of the TSS (Figure 3A). Frequency of these SNPs in 28 different breeds, including both Asian and European wild pigs, 17 Chinese breeds, and 4 breeds with large white descent, and 5 Western breeds, were further analyzed (Table 1), which showed that biased frequency distributions of these SNPs in these breeds.
Haplotype analysis of these SNPs showed that 5 haplotypes existed in these pig breeds (Figure 3B). RAB37-a was a major haplotype. Compared with RAB37-a, RAB37-b had a variation A>G in SNP1 site, RAB37-c with T>C in SNP2 site, and RAB37-e with T>C in SNP3 site, while RAB37-d had A>G in SNP1 site as well T>C in SNP3 site. In pig populations, these haplotypes represented 5 alleles of RAB37 gene.
SNP haplotyping revealed both Asian and European wild pigs, Tibetan pigs, most of local breeds in China, and Duroc, and Hampshire and Pietrain pigs of Western breeds, had only RAB37-a, indicating that RAB37-a was a wild-type allele. Landrace, White duroc, Hetao pigs, and French-, Korean-, and Wageningen-large white pigs had a small proportion of RAB37-e, in addition to wild-type RAB37-a. Xihuzhou large white pigs from China had RAB37-b, RAB37-c, RAB37-d, and the wild-type RAB37-a, suggesting a mixed population. Interestingly, Chinese local breeds, Jiangquhai, Sujiang, and Meishan pigs had RAB37-c, in addition to wild-type RAB37-a, and Jiangquhai pigs had only RAB37-c in particular (Table 1; Figure 3B). Thus, wild boars from both Chinese and European, and Tibetan pigs had only wild-type RAB37-a, new alleles RAB37-b,-e were mainly detected in domestic pig breeds, with 100% allele RAB37-c in Jiangquhai pigs (Figure 4). These data suggested that RAB37 multiple alleles were generated by artificial selection in pig populations.
Evolution of transcription factors in RAB37 promoter. (A) Phylogenetic relationship of transcription factors EGR2, E2F1, and EBF1 in the promoter of RAB37 gene. Mya, million years ago. (B) Venn diagram showing conservation of transcription factors EGR2, E2F1, and EBF1 for RAB37 among human (blue circle), mouse (red circle), and pig (green circle). E2F1 is a common transcription factor between human and mouse, EBF1 is a common transcription factor between human and pig, EGR2 is a common transcription factor between mouse and pig. (C) Comparisons of transcriptional activation abilities of transcription factors EGR2, E2F1, and EBF1 in the RAB37 promoter among human (blue), mouse (red) and pig (green). The RAB37 promoter activities were determined by luciferase assays in COS-7 cells and calculated by relative values to their corresponding point mutants.
Identification of RAB37 SNPs and haplotypes in pig population. (A) Schematic diagram of the SNPs in the pig RAB37 promoter. Three SNPs (A>G, -278 bp; T>C, -404 bp; T>C, -523 bp) are in CpG island 2, whereas EGR2 and EBF1 binding sites are located in CpG island 1. The blue ovals indicate the CpG islands. (B) Identification of RAB37 haplotypes in pig population. In total, 5 haplotypes of RAB37 were detected from 28 pig breeds. Haplotypes frequencies were calculated based on all available data of 28 pig breeds. RAB37-a is a major haplotype, RAB37-b has a variation A>G in SNP1 site, RAB37-c has T>C in SNP2 site, RAB37-e has T>C in SNP1 site, while RAB37-d has A>G in SNP1 site as well as T>C in SNP3 site. Haplotype frequencies in Jiangquhai, Sujiang, Meishan pigs and average of all 28 breeds are shown in the right panel.
Haplotype frequency of RAB37 in pig breeds
| Breed | n | RAB37-a (TTA) | RAB37-b (TTG) | RAB37-c (TCA) | RAB37-d (CTG) | RAB37-e (CTA) |
|---|---|---|---|---|---|---|
| Wild boar | ||||||
| Asian wild boar | 10 | 1 | 0 | 0 | 0 | 0 |
| European wild boar | 17 | 1 | 0 | 0 | 0 | 0 |
| Local breed in China | ||||||
| Tibetan | 12 | 1 | 0 | 0 | 0 | 0 |
| Gansu Tibetan | 10 | 1 | 0 | 0 | 0 | 0 |
| Sichuan Tibetan | 12 | 1 | 0 | 0 | 0 | 0 |
| Yunnan Tibetan | 12 | 1 | 0 | 0 | 0 | 0 |
| Bamaxiang | 6 | 1 | 0 | 0 | 0 | 0 |
| Jinhua | 6 | 1 | 0 | 0 | 0 | 0 |
| Luchuan | 6 | 1 | 0 | 0 | 0 | 0 |
| Laiwuhei | 6 | 1 | 0 | 0 | 0 | 0 |
| Min | 6 | 1 | 0 | 0 | 0 | 0 |
| Wuzhishan | 6 | 1 | 0 | 0 | 0 | 0 |
| Xiang | 2 | 1 | 0 | 0 | 0 | 0 |
| Tongcheng | 18 | 1 | 0 | 0 | 0 | 0 |
| Hetao | 6 | 0.917 | 0 | 0 | 0 | 0.083 |
| Erhualian | 21 | 1 | 0 | 0 | 0 | 0 |
| Meishan | 33 | 0.939 | 0 | 0.061 | 0 | 0 |
| Sujiang | 17 | 0.294 | 0 | 0.706 | 0 | 0 |
| Jiangquhai | 17 | 0 | 0 | 1 | 0 | 0 |
| Large white descent | ||||||
| Xihuzhou large white | 26 | 0.481 | 0.115 | 0.192 | 0.212 | 0 |
| French large white | 36 | 0.931 | 0 | 0 | 0 | 0.069 |
| Korean large white | 14 | 0.786 | 0 | 0 | 0 | 0.214 |
| Wageningen large white | 14 | 0.5 | 0 | 0 | 0 | 0.5 |
| Europe-breed | ||||||
| Duroc | 22 | 1 | 0 | 0 | 0 | 0 |
| White duroc | 2 | 0.75 | 0 | 0 | 0 | 0.25 |
| Hampshire | 2 | 1 | 0 | 0 | 0 | 0 |
| Pietrain | 5 | 1 | 0 | 0 | 0 | 0 |
| Landrace | 16 | 0.938 | 0 | 0 | 0 | 0.062 |
| Average | 0.836 | 0.009 | 0.1 | 0.015 | 0.04 | |
Promoter activities and roles of RAB37 alleles in autophagy
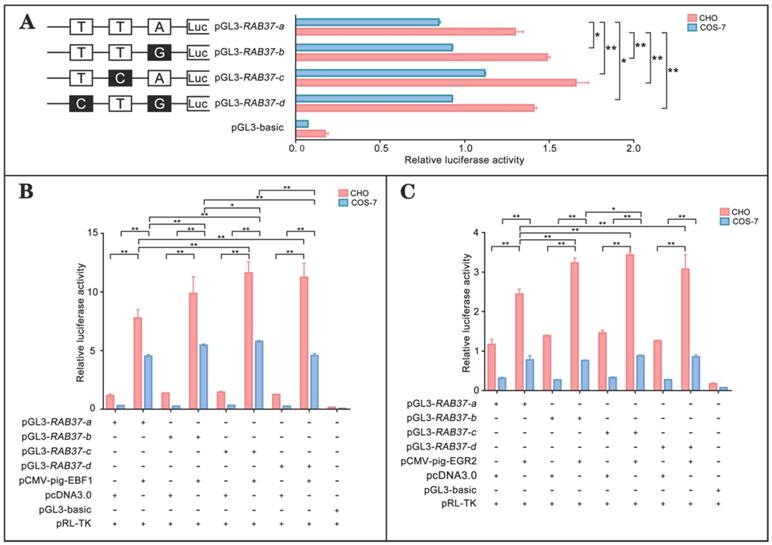
To realize roles of these alleles, promoter regions of the alleles were linked with luciferase gene in the pGL3-basic vector and their activities were tested. The analysis showed that luciferase activities of the constructs with SNP mutants (pGL3-RAB37-b, pGL3-RAB37-c, pGL3-RAB37-d) were higher than that of wild-type pGL3-RAB37-a in CHO cells (Figure 5A). To further explore influence of SNP mutants on transcription activation of the transcription factors, these constructs were cotransfected with EBF1 or EGR2 in CHO and COS-7 cells, and luciferase activities were tested. The luciferase reporter analysis showed that both EBF1 and EGR2 could increase promoter activities of all these constructions with the highest in pGL3-RAB37-c (Figure 5B and 5C). These data suggested that these SNP variations in the promoter could upregulate activity of the pig RAB37 promoter.
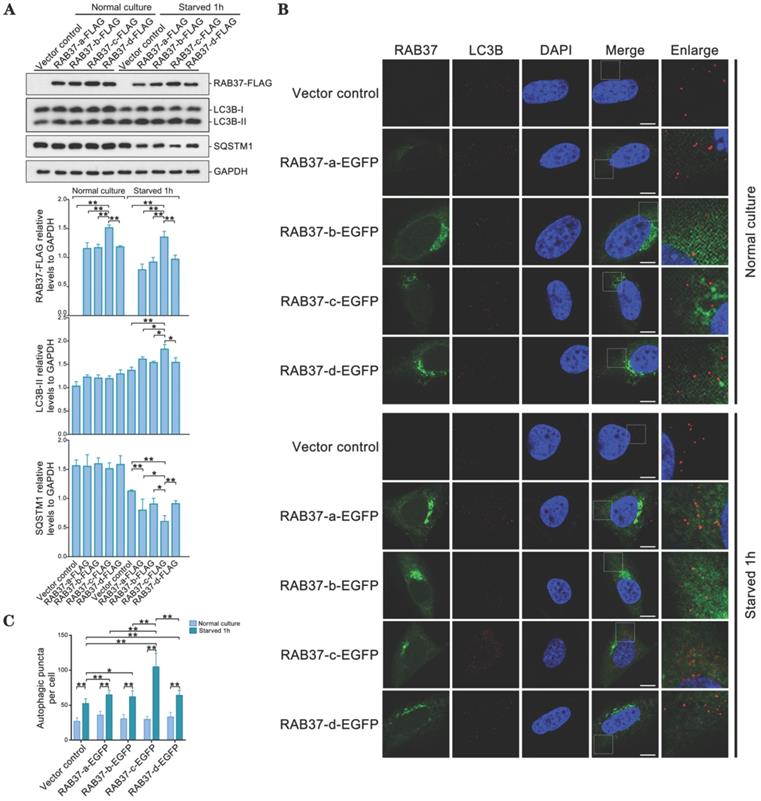
As RAB37 can promote autophagy [16], we further compared roles of these alleles in enhancing autophagy. RAB37 alleles with various SNP variations were transfected into 293T cells and starved for 1 h. Western blots showed that LC3B-II was upregulated, while downstream substrate SQSTM1 was downregulated upon starvation (Figure 6A). RAB37 alleles with various SNPs were overexpressed in CHO cells and endogenous LC3B puncta were further analyzed under starvation. In comparison with normal culture, autophagic puncta in RAB37-c were significantly higher than those in RAB37-a, RAB37-b, and RAB37-d, while these alleles could enhance autophagy upon starvation (Figure 6B and 6C). These results suggested these SNP variations in RAB37 promoter promoted autophagy.
Discussion
Considering the importance of RAB37 as an organizer for autophagosome biogenesis, mechanistic insights into transcription regulation of RAB37 will provide better understanding of roles and mechanisms of autophagy in cellular homeostasis and development. In this study, we characterized RAB37 promoters in humans, mice, and pigs. Transcription factors and their binding abilities to the promoters were determined. We further screened SNPs in pig RAB37 gene and identified RAB37 alleles in pig populations from both China and Europe. In addition, roles of RAB37 alleles in autophagy were confirmed. These findings provide better understanding of transcription activation of RAB37 and suggest RAB37 as a potential gene for artificial selection of autophagy regulation in pig breeding practice.
By comparative analysis of promoter activities of RAB37 genes among humans, mice and pigs, a set of related transcription factors were identified. Transcription factors EBF1 and E2F1 were essential for transcription activation of RAB37 gene in human, while EBF1 was a dominant regulator in RAB37 in both human and pig. EBF1 belongs to a transcription factors family containing an atypical zinc finger DNA binding domain and a nonbasic helix-loop-helix dimerization domain, which mainly regulated adipocyte morphology and lipolysis [21] and B cell differentiation [22]. EBF1 can interact with the methylcytosine dioxygenase enzyme TET2 in several cancer types, suggesting a role of EBF1 in regulating DNA methylation [23]. Indeed, EBF1 binding site in RAB37 promoter is located in a CpG island in both human and pig, hinting a role of epigenetic regulation of EBF1 in RAB37 transcription. Considering a key role of RAB37 in regulation of autophagosome formation via modulating ATG5-ATG12-ATG16L1 complex assembly [16, 17], EBF1-mediated RAB37 pathway is probably involved in cellular homeostasis. EBF1 as a transcription factor of RAB37 suggests their synergistic role in autophagy regulation. EBF1 knockdown affected lipid and glucose levels, thus energy metabolism [21], which supports importance of EBF1-mediated RAB37 pathway in autophagy regulation and cellular homeostasis.
Artificial selection generates RAB37 multiple alleles in pig population. Wild boars have only wild-type RAB37-a. Tibetan pigs experienced a weak pressure of artificial selection and retain only RAB37-a. In domestic pigs, artificial selection generates new alleles RAB37-b, RAB37-c, RAB37-d, and RAB37-e in some breeds, while RAB37-a is still only allele in many breeds. In addition, transcription factors EBF1 and EGR2 have be selected to activate RAB37 transcription in the pig lineage.
Promoter activities and transcription activation of RAB37 alleles in pig. (A) Luciferase assays showing promoter activities of RAB37 alleles in both CHO and COS-7 cells. SNPs are indicated by boxes, while the filled boxes show corresponding SNPs with lower frequency relative to those in allele RAB37-a. Right panel shows relative activities of these promoters containing SNPs, as determined by luciferase assays. One-way ANOVA was performed. *P < 0.05; **P < 0.01. (B) Overexpression of EBF1 activates these allele promoters. In total, 0.4 mg pGL3-RAB37-a or its SNP mutants (pGL3-RAB37-b, pGL3-RAB37-c, or pGL3-RAB37-d) were cotransfected with 0.1 mg EBF1 expressing plasmid (pCMV-pig-EBF1). EBF1 overexpression can increase the activities of all these promoters containing SNPs with the highest in pGL3-RAB37-c. One-way ANOVA was performed. *P < 0.05; **P < 0.01. (C) Overexpression of EGR2 activates these allele promoters. In total, 0.4 mg pGL3-RAB37-a or its site mutants (pGL3-RAB37-b, pGL3-RAB37-c, or pGL3-RAB37-d) were cotransfected with 0.1 mg EGR2 expression plasmid (pCMV-pig-EGR2). EGR2 overexpression increases activities of these allele promoters containing SNPs with the highest in pGL3-RAB37-c. One-way ANOVA was performed. *P < 0.05; **P < 0.01.
In addition to EBF1, co-regulation of transcription factors EGR2 and E2F1 differentially evolved for RAB37 genes in different lineages of human, mouse, and pig. Further, a dominant transcription factor (EBF1 in human and pig; EGR2 in mouse) for RAB37 genes was acquired in these species. These data suggest that these transcription factors regulate RAB37 via transcription factor-associated evolution in RAB37 promoter for maintaining cellular homeostasis. These transcription factors can also directly regulate transcription of autophagy genes. For example, E2F1 can bind to the promoter regions of autophagy genes LC3B, ATG1, and DRAM to activate transcription of these genes, thus enhanced autophagy [24]. In human adipocytes, EBF1 chromatin immunoprecipitation showed potential EBF1 target genes including autophagy related genes ATG12, ATG16L2, ATG9B, ULK3, SQSTM1 and RAB37 [21]. Thus, transcription factors EBF1 and E2F1 maintain cellular homeostasis by either directly regulating some autophagy genes or indirectly modulating autophagy via activating RAB37. These two modes of regulation probably exert roles synergistically, which is important for maintaining cellular homeostasis upon different forms of stress. Further studies are needed to clarify this mechanism.
Long-term artificial selection of female fecundity and meat production has changed gene expression pattern in pig breeding [25]. Here we further showed that the artificial selection generated SNP variations in the pig RAB37 promoter, in addition to transcription factors EBF1 and EGR2 selected to activate RAB37 promoter in the pig lineage. Three SNPs are located in second CpG island upstream of core promoter of RAB37, which could affect binding of EBF1 and EGR2 to RAB37 promoter by mediating spatial structure of transcription complex. These SNP variations resulted in at least 5 haplotypes, representing 5 alleles of RAB37. Of them, RAB37-c acquired the highest ability to activate expression in comparison with the other promoters, thus enhanced autophagy efficiently. A large scale of population genome analysis from both China and Europe demonstrated that artificial selection led to RAB37 multiple alleles in pig population (Figure 4). Both Chinese and European wild boars have only wild-type RAB37-a. Tibetan pigs also have only RAB37-a, as experienced a weak pressure of artificial selection in the Tibetan Plateau. However, new alleles RAB37-b, RAB37-c, RAB37-d, and RAB37-e appeared in domestic pig breeds. In particular, Jiangquhai pig, a Chinese local breed with a high litter size (n=13.5 at third birth), acquired 100% frequency of the allele RAB37-c. Considering a high female fecundity in Jiangquhai pigs and Meishan pigs, the allele RAB37-c could be artificially selected for autophagy regulation. Further analysis to illuminate the molecular mechanism will help in genetics and breeding practice in pigs.
Alleles of pig RAB37 promote autophagosome formation. (A) Western blot analysis of RAB37-FLAG, LC3B, and SQSTM1 in 293T cells. The cells were transfected with RAB37-FLAG constructs (RAB37-a-FLAG, RAB37-b-FLAG, RAB37-c-FLAG, RAB37-d-FLAG, and negative control vector) and cultured in normal (control) or EBSS medium for 1 h, respectively. The cell lysates were analyzed by immunoblotting with antibodies as indicated. GAPDH was used as an endogenous control. Bottom panels: western blots quantified for RAB37-FLAG/GAPDH ratio, LC3BII/GAPDH ratio and the SQSTM1/GAPDH ratio. Data are presented as means ± S.D. *P < 0.05, **P < 0.01 (n = 3 independent experiments). (B) Immunofluorescence analysis of RAB37-EGFP and LC3B in CHO cells. Representative images of the cells transfected with these RAB37-EGFP constructs (RAB37-a-EGFP, RAB37-b-EGFP, RAB37-c-EGFP, and RAB37-d-EGFP). The cells were cultured in normal (control) or EBSS medium for 1 h, respectively. Endogenous LC3B (red) was detected first by anti-LC3B and then TRITC-conjugated ImmunoPure goat anti-rabbit IgG (H + L). The nuclei were stained by DAPI (blue). Vector control is a negative control without promoter. Single channel (red, green or blue) and merged images were taken by confocal microscopy. The enlarged images originated from the white squares in merge panels. Scale bar, 10 µm. (C) Statistics of autophagic puncta per cell from A. The dots were counted from 20 cells. *P < 0.05; **P < 0.01.
Conclusions
In this study, we performed a comparative analysis of promoter activities and related transcription factors of RAB37 in humans, mice and pigs. Transcription factors and their binding abilities to the promoters were determined. SNPs in the pig RAB37 gene were screened and multiple alleles of RAB37 in pig populations from both China and Europe were identified. Population genomics showed a role of artificial selection in variations of these alleles. Furthermore, roles of RAB37 alleles in autophagy regulation were confirmed. These findings provided a better understanding of transcription activation of RAB37 and suggested RAB37 as a potential gene for artificial selection of autophagy regulation in pig breeding practice.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the Key Transgenic New Organism Project of China (2018ZX08009-27B) and the National Natural Science Foundation of China (31771487, 31771370 and 31970539).
Author contributions
R. Z. and H. C. conceived and designed research. X. X. and R. Z. analysed and interpreted the data and wrote the paper. X. X., M. H., R. Y., J. Z., L.L. carried out the experimental work. All authors read and approved the final manuscript.
Ethics statement
All animal experiments and methods were performed in accordance with the relevant approved guidelines and regulations, as well as under the approval of the Ethics Committee of Wuhan University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Luo M, Zhao X, Song Y, Cheng H, Zhou R. Nuclear autophagy: an evolutionarily conserved mechanism of nuclear degradation in the cytoplasm. Autophagy. 2016;12:1973-83
2. Yang Y, Klionsky DJ. Autophagy and disease: unanswered questions. Cell Death Differ. 2020;27:858-71
3. Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem. 2017;86:225-44
4. Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207-15
5. Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2019;63:1-10
6. Lystad AH, Carlsson SR, Simonsen A. Toward the function of mammalian ATG12-ATG5-ATG16L1 complex in autophagy and related processes. Autophagy. 2019;15:1485-6
7. Ljubicic S, Bezzi P, Brajkovic S, Nesca V, Guay C, Ohbayashi N. et al. The GTPase Rab37 participates in the control of insulin exocytosis. PLoS One. 2013;8:e68255-e
8. Mori R, Ikematsu K, Kitaguchi T, Kim SE, Okamoto M, Chiba T. et al. Release of TNF-alpha from macrophages is mediated by small GTPase Rab37. Eur J Immunol. 2011;41:3230-9
9. Higashio H, Satoh Y-i, Saino T. Mast cell degranulation is negatively regulated by the Munc13-4-binding small-guanosine triphosphatase Rab37. Sci Rep. 2016;6:22539 -
10. Tsai CH, Cheng HC, Wang YS, Lin P, Jen J, Kuo IY. et al. Small GTPase Rab37 targets tissue inhibitor of metalloproteinase 1 for exocytosis and thus suppresses tumour metastasis. Nat Commun. 2014;5:4804
11. Wang YS, Tzeng HT, Tsai CH, Cheng HC, Lai WW, Liu HS. et al. VAMP8, a vesicle-SNARE required for RAB37-mediated exocytosis, possesses a tumor metastasis suppressor function. Cancer Lett. 2018;437:79-88
12. Cho S-H, Kuo IY, Lu P-JF, Tzeng H-T, Lai W-W, Su W-C. et al. Rab37 mediates exocytosis of secreted frizzled-related protein 1 to inhibit Wnt signaling and thus suppress lung cancer stemness. Cell Death Dis. 2018;9:868 -
13. Tzeng HT, Tsai CH, Yen YT, Cheng HC, Chen YC, Pu SW. et al. Dysregulation of Rab37-mediated cross-talk between cancer cells and endothelial cells via thrombospondin-1 promotes tumor neovasculature and metastasis. Clin Cancer Res. 2017;23:2335-45
14. Tzeng HT, Su CC, Chang CP, Lai WW, Su WC, Wang YC. Rab37 in lung cancer mediates exocytosis of soluble ST2 and thus skews macrophages toward tumor-suppressing phenotype. Int J Cancer. 2018;143:1753-63
15. Li Y, Yang X, Du X, Lei Y, He Q, Hong X. et al. RAB37 Hypermethylation regulates metastasis and resistance to docetaxel-Based induction chemotherapy in nasopharyngeal carcinoma. Clin Cancer Res. 2018;24:6495-508
16. Sheng Y, Song Y, Li Z, Wang Y, Lin H, Cheng H. et al. RAB37 interacts directly with ATG5 and promotes autophagosome formation via regulating ATG5-12-16 complex assembly. Cell Death Differ. 2018;25:918-34
17. Song Y, Shang D, Cheng H, Zhou R. The small GTPase RAB37 functions as an organizer for autophagosome biogenesis. Autophagy. 2018;14:727-9
18. Hong Q, Li C, Ying R, Lin H, Li J, Zhao Y. et al. Loss-of-function of sox3 causes follicle development retardation and reduces fecundity in zebrafish. Protein & Cell. 2019;10:347-64
19. Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H, Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11:1081-98
20. Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF. et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393-8
21. Gao H, Mejhert N, Fretz JA, Arner E, Lorente-Cebrian S, Ehrlund A. et al. Early B cell factor 1 regulates adipocyte morphology and lipolysis in white adipose tissue. Cell Metab. 2014;19:981-92
22. Boller S, Li R, Grosschedl R. Defining B cell chromatin: lessons from EBF1. Trends Genet. 2018;34:257-69
23. Guilhamon P, Eskandarpour M, Halai D, Wilson GA, Feber A, Teschendorff AE. et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun. 2013;4:2166
24. Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860-4
25. Chen K, Huang C, Yuan J, Cheng H, Zhou R. Long-term artificial selection reveals a role of TCTP in autophagy in mammalian cells. Mol Biol Evol. 2014;31:2194-211
Author contact
![]() Corresponding authors: Professors Rongjia Zhou and Hanhua Cheng, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China, Fax: 0086-27-68756253, E-mail: hhchengedu.cn; rjzhouedu.cn.
Corresponding authors: Professors Rongjia Zhou and Hanhua Cheng, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China, Fax: 0086-27-68756253, E-mail: hhchengedu.cn; rjzhouedu.cn.

 Global reach, higher impact
Global reach, higher impact