10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(15):3018-3027. doi:10.7150/ijbs.49302 This issue Cite
Review
S-adenosylmethionine tRNA modification: unexpected/unsuspected implications of former/new players
Department of Health Science University of Milan via A. di Rudinì 8 20142 Milan.
Received 2020-6-10; Accepted 2020-9-10; Published 2020-9-30
Abstract
S-adenosylmethionine supplies methyl groups to many acceptors, including lipids, proteins, RNA, DNA, and a wide range of small molecules. It acts as the precursor in the biosynthesis of metal ion chelating compounds, such as nicotianamine and phytosiderophores, of the polyamines spermidine and spermine and of some plant hormones. Finally, it is the source of catalytic 5′-deoxyadenosyl radicals.
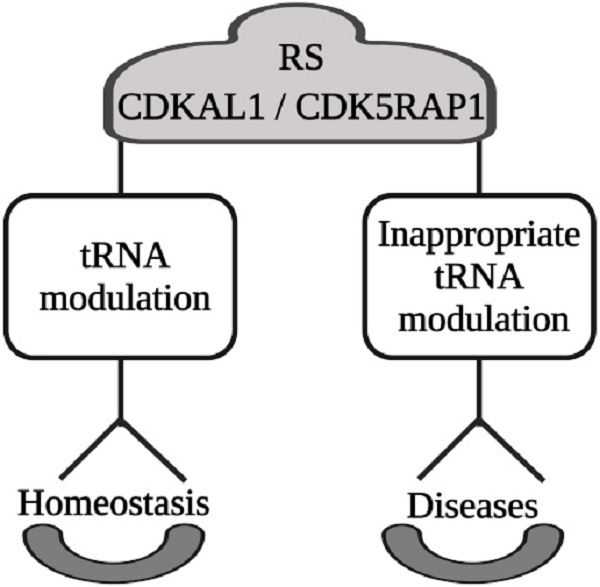
Radical S-adenosylmethionine (SAM) enzymes (RS) represent one of the most abundant groups (more than 100,000) of enzymes, exerting a plethora of biological functions, some of which are still unknown.
In this work, we will focus on two RS: CDK5RAP1 and CDKAL1, both of which are involved in tRNA modifications that result in important tRNA folding and stability and in maintaining high translational fidelity. Based on this crucial role, their impairment can be important in the development of different human diseases.
Keywords: Radical S-adenosylmethionine enzymes, Methylthiotransferases, CDK5RAP1, CDKAL1, tRNA modifications, human diseases
Introduction
S-adenosylmethionine (SAM) enzymes
S-adenosyl-L-methionine (SAM) enzymes were discovered nearly 60 years ago [1]. These enzymes represent the main biological methyl donor synthesized in all mammalian cells, but some tissue is more involved than others; for instance, in the liver, 85% of all transmethylation reactions and 50% of all methionine metabolism occur [2]. These reactions happen because of many different enzymes. More than 114,000 enzymes have been found in all domains of life: Archaea, Bacteria, and Eukarya [3] and were categorized as a superfamily in 2001 [4], being named radical S-adenosylmethionine (RS) enzymes. A [4Fe-4S] cluster of the enzyme and the SAM are involved in the initiation of various radical reactions, generating 5′-deoxyadenosyl radical intermediate [5]. RS superfamily members have a limited sequence homology; a CX3CX2C motif is their common feature, contributing in the coordination of three out of the four irons of the [4Fe-4S] cluster at the active site of the enzyme [5]. Within all the different RS enzymes, eight have been found in humans: CDK5 Regulatory Subunit Associated Protein 1 Like 1 (CDKAL1) for methylthio-N6-threonylcarbamoyladenosine biosynthesis; CDK5 Regulatory Subunit Associated Protein 1 (CDK5RAP1), for 2-methylthio-N6-isopentenyladenosine biosynthesis; tRNA-YW Synthesizing Protein 1 Homolog (TYW1), for wybutosine biosynthesis; Elongator Acetyltransferase Complex Subunit 3 (ELP3), 5-for methoxycarbonylmethyl uridine biosynthesis; Molybdenum Cofactor Synthesis 1 (MOCS1); Lipoic Acid Synthetase (LIAS), for lipoic acid biosynthesis; Radical S-Adenosyl Methionine Domain Containing 1 (RSAD1) and viperin, with the last having currently unknown functions. Biochemical and bioinformatics analyses indicate that in humans, these reactions are involved in several modifications, some of which alter transfer RNAs (tRNAs).
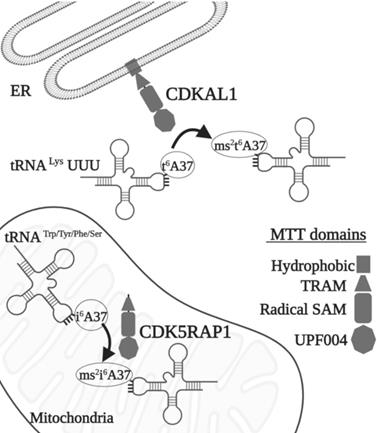
Methylthiotransferases (MTTases) are a subclass of RS. They use two [4Fe-4S] cluster cofactors and are bound to an N-terminal MTT domain and a central radical SAM domain; their function is to add a methylthiol moiety (-SCH3) at specific locations on tRNAs. MTTases have been classified into three families that are represented by the bacterial enzymes Methanol-corrinoid protein co-methyltransferase (MtaB), tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase (MiaB), and Ribosomal protein S12 methylthiotransferase (RimO) [3]. The MiaB family catalyzes the bound of a methylthiol group of C2 of N6-isopentenyladenosine (i6A) and produces 2-methylthio-N6- isopentenyladenosine (ms2i6A), whereas the MtaB family catalyzes the methylthiolation of the carbon in the same position of N6-threonylcarbamoyladenosine (t6A) to synthetize 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A). CDKAL1 and CDK5RAP1 are the human orthologs of MtaB and MiaB, respectively, with which they share 18% and 23% of the protein domain structure (Fig. 1) [3].
Scheme of MTTase action. Effects of CDKAL1 and CDK5RAP1. These two enzymes act in the ER and in the mitochondria, respectively. Different domains are indicated as follows: square: hydrophobic, triangle: TRAM, rectangle: radical SAM; octagon UPF004. For the abbreviations, see the text.
tRNA post-transcriptional modification
Transfer RNAs are best known as a essential class of adapter that are central players in the flow of information between mRNA and protein, with their main role as tranferring the information of the genes to the information of the proteins. tRNAs, however, carry out many other functions in both prokaryotes and eukaryotes, including, but not limited to, amino acid delivery to membrane lipids, the synthesis of peptidoglycan, the synthesis of antibiotics, and, under stress conditions, producing the molecules that act in the signaling pathway as regulators of gene expression. tRNAs exert these functions through of their own cleavage [6, 7]. tRNA biogenesis takes place over many steps: the transcription of a tRNA gene leading to the production of a pre-tRNA transcript, which is then subjected to a number of post-transcriptional modifications that differ based on the organisms and the identity of the tRNA; this can include the removal of the 5'-leader and 3'-trailer sequences, the incorporation of the 3'-CCA amino acid accepting sequence, the removal of intronic sequences, and many post-transcriptional chemical modifications. The final product is a mature molecule with a cloverleaf secondary structure that is composed of five main regions: the T-arm, the variable region, the anticodon-arm, the D-arm, and acceptor stem [8].
tRNA processing is carried out with very complex and intricate mechanisms, most likely because we do not understand all the regulatory steps that take place in this system; for example, post-transcriptional modifications appear across the entire process before or after the processing of the extremities and before or after the removal of the introns [8]. Many of these modifications are introduced to tRNA independently; however, several of them are interconnected with a modification that drives the formation of following one [8].
About 150 post-transcriptional modifications change up to 20% of all nucleotides in tRNAs [7] and introduce a functional diversity that allows the four RNA base residues to acquire new functions, hence directly influencing RNA structure. This is achieved by modifying specific intramolecular interactions that change its flexibility, refining the specificity and decoding the frame for protein translation. These changes also influence the translational speed and affect its interactions with other molecules, such as proteins [9].
The most commonly occurring changes comprise methylation of the ribose or base, such as base isomerization, base reduction, and base thiolation. More complicated changes are also present and include the addition of larger chemical groups or require multiple modification steps [8].
A total of 112 modified nucleosides have been reported in RNA, with the largest number, 93, being found in tRNA [10, 11]; however, there are some differences among Archaea, Bacteria, and Eukarya (which are more heavily modified) in terms of their composition and density [12]. These modifications are mostly located in the anticodon loop (ACL) region, where the limited sequence and structural information can be magnified [13]; for instance, of the 28 distinct modifications in cytoplasmic tRNAs in humans, 17 are in the ACL region, and 11 are in the main body [12].
One question, though, arises: Why do these modifications occur?
To answer this, we have to consider that the primary sequence of the tRNA is quite simple, short, and formed by the same building blocks (nucleotides G, C, A, and T) and permit the formation of a variety of different base pairs that lead to the production of alternative secondary and tertiary structures. For this reason, tRNA will not fold into its biologically active conformation because other alternative structures can have a similar level of free energy [14]. Indeed, these modifications induce major rearrangements of the tRNA structure, allowing for the formation of the canonical cloverleaf shape.
Base modification has local implications, which, however, are highly important for tRNA function. For instance, positions 34 and 37 are often hypermodified (Fig. 1). These ACL modifications achieve two different purposes: the modulation of the interaction possibilities between the codon and anticodon. They fine-tune the tRNA structure; specifically, the base modifications in the first base of the anticodon (34) have a role in the wobble interaction with the third position of the codon.
The second function of these modifications in ACL is a structural one reinforcing the loop structure for efficient translation in the anticodon and in the tRNA core region site, in which occurs interactions between the D- and T-loop [14]. Among the various modifications that are present in the tRNA, sulfur modifications are especially pivotal for tRNA functions. Four different thionucleoside are found in tRNAs, and two of them are found in the ACL region at position 34 (2-thiouridine derivatives xm5s2U), position 37 (2-methylthioadenosine derivatives ms2x6A), and position 37 (where “x” represents several functional groups differing between species and organelles) [15].
Position A37 modifications are the most represented; nonetheless, they are heterogeneous between species (Fig. 1) [16]. N6-isopentenyladenosine derivatives (ms2i6A, io6A, and ms2io6A) from the conversion of N6-isopentenyladenosine were found in this position, but although i6A is present in many species, the modified i6A types were present only in a few species. For instance, ms2i6A modification was found in eukaryotes only as Sus scrofa, either in mitochondria and cytoplasm (of the heart), whereas i6A was detected in almost all eukaryotes [16]. Moreover, N6-threonylcarbamoyladenosine (t6A) is modified into 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A) in tRNA. Also, this modification was found mainly in Sus scrofa but not in other eukaryotes, where t6A was widely present instead [16].
Within the different RS, CDK5RAP1 and CDKAL1, which, respectively, catalyze methylthiolation at i6A37 and t6A37, exert a pivotal action on the A37 nucleotide, which is located adjacent to the third nucleotide (position 36) of the anticodon (Fig. 1). This hypermodified nucleotide, outside the anticodon region, is not essential for mRNA translation, but seems to stabilize the base pairing between tRNAs and mRNA maintaining translational fidelity [17, 18].
CDKAL1
CDKAL1 is a 579 AA protein (in human), an ortholog of MtaB RS, which is ubiquitously expressed. It is also a homolog to CDK5RAP1 (CDK5 (cyclin-dependent kinase 5) regulator subunit-associated protein 1). CDKAL1 is composed of three distinct domains, a radical SAM, a tRNA methyltransferase 2 and MiaB (TRAM), and a hydrophobic domain (Fig. 1). The catalytic radical SAM domain and the TRAM domain (a potential tRNA-binding domain) are conserved among mammals and bacteria, whereas the C-terminus hydrophobic domain, which carries the endoplasmic reticulum (ER)-localization signal, is present only in mammalian CDKAL1 [19-21]. Because CDK5RAP1 binds the activators (p35 and p39 [22]) of CDK5, CDKAL1 was also postulated as interacting with CDK5. Even though CDK5 is involved in many aspects of insulin production—such as the regulation of β-cell functions, and β-cell differentiation, cell survival, and insulin secretion (where CDKAL1 is implicated as well)—there are no indications that CDKAL1 directly or indirectly regulates CDK5 kinase activity [23]. Using a Real-Time PCR analysis of cDNAs from 20 different tissues or cell types, CDKAL1 (full length but not short form) expression was detected at a high level in skeletal muscle, pancreas, testis, and immune cells, such as CD4+, CD8+, CD9+, and monocytes [24].
From an Ensembl genome analysis, it is possible to determine that at least two CDKAL1 transcript isoforms exist: the full-length cDNA and a shorter cDNA lacking exons 4 and 13 [24]. Moreover, a splice variant CDKAL1-v1, a noncoding transcript, can regulate the CDKAL1 level by competitive binding to a CDKAL1-targeting miRNA (miR-494) [25].
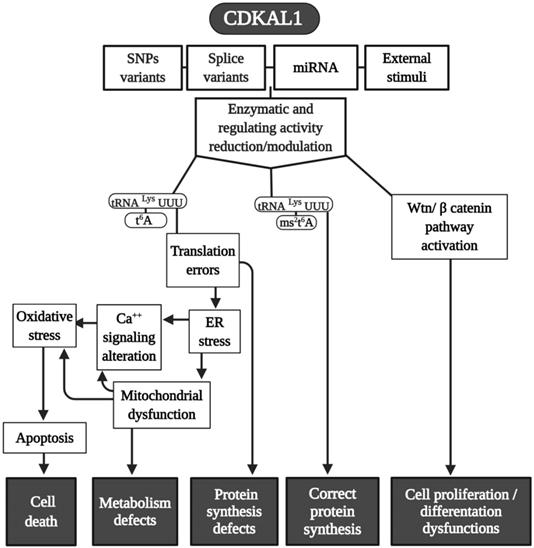
In 2007, four genome-wide association studies (GWAS) conducted in African, Asian, North American, and European populations indicated that single-nucleotide polymorphism (SNPs) (there are at least 13 variants) exist in a previously uncharacterized gene: CDKAL1 was found to be a risk factor for type 2 diabetes (TD2) [26-32]. After this, more than 70 scientific studies on SNPs in CDKAL1 involvement in T2D have been conducted [23]. SPNs in the CDKAL1 gene have been found to be related to a reduction in insulin secretion and subsequent development of T2D [26, 28] (Fig. 2).
In term of tRNA modifications, CDKAL1 catalyzes the ms2t6A change in tRNALys(UUU) in mammalian cells [20], and its functional loss affects the accuracy of protein translation and, consequently, the improper synthesis of proinsulin; as an additional element, the expression of ER stress-related genes can determine an abnormally structured ER [20]. Moreover, because the CDKAL1 activity is tightly regulated by iron levels, which are an integral component of its SAM catalytic domain needed for its methylthiotransferase activity, cellular iron deficiency impairs its function in altering proinsulin synthesis [33].
In addition to its direct effects in T2D, it was recently found that SNPs (rs7756992) of CDKAL1 significantly increased the risk for many forms of cancer, including cancers of female reproductive organs, pancreas, breast, colorectal, liver, and urinary tract (Fig. 2) [34, 35]. Indeed, CDKAL1 is highly expressed in patients with chromosomal 6p22 amplification, with the most frequent changes seen in high-grade muscle-invasive bladder cancer [36, 37].
CDKAL1-mediated ms2 is decreased in growth hormone-producing pituitary adenomas (GHPAs), and a knockdown of CDKAL1 determines an increase of growth hormone (GH) biosynthesis, most likely altering calcium signaling at the ER level (Fig. 2) [38]. The excess of GH modifies insulin sensitivity and can alter the function of pancreatic β-cells [39].
The downregulation of CDKAL1 in insulinoma cells affects the levels of at least three proteins:
- insulin, along with the other two components of the insulin secretory granules;
- islet cell autoantigen 512 (ICA512/IA-2);
- chromogranin [21].
Another metabolic implication of CDKAL1 is related to its involvement in adipose tissue differentiation (Fig. 2). Its knockdown promotes differentiation of adipocytes, most likely activating the Wnt/β-catenin pathway; this is known to be an inhibitory regulator of adipocyte differentiation [40] and might also be related to birth weight (Fig. 2) [41].
Diseases in which MTTases are involved. Dark gray are effects of CDKAL1, and light gray are effects of CDK5RAP1. SAPHO: synovitis, acne, pustulosis, hyperostosis and osteitis. The asterisk present in the glioblastoma and breast cancer boxes indicates that in these two diseases is also implicated CDKAL1.
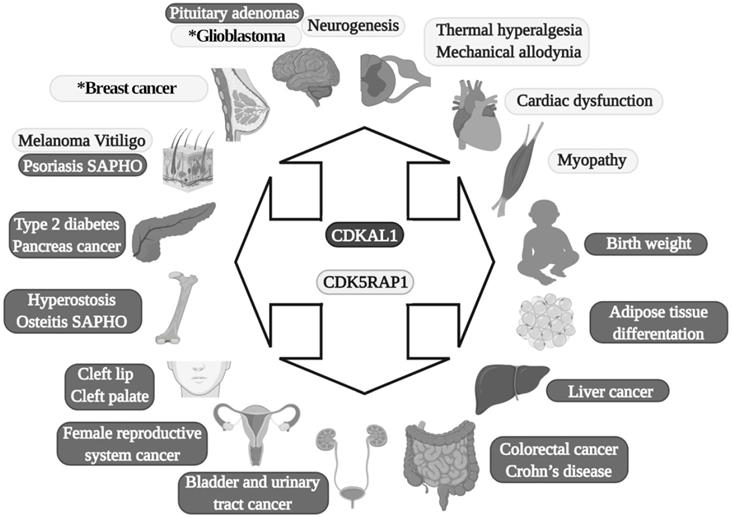
Moreover, CDKAL1 seems to be associated with Crohn's disease (CD) [42] and psoriasis [43] (where low levels of CDKAL1 were present in the colon, small intestine, and skin-keratinocytes (Fig. 2) [24]), but the CDKAL1 SNP associated with T2D does not confer susceptibility to psoriasis or CD [24, 44]. Moreover, syndrome synovitis, acne, pustulosis, hyperostosis, and osteitis (SAPHO) (another autoimmune disease of unknown etiology) were found to be associated with CDKAL1 SNPs [45].
Finally, an association of the CDKAL1 SPN variant that comes with the risk of nonsyndromic cleft lip with or without a cleft palate was detected (Fig. 2) [46].
CDK5RAP1
CDK5RAP1, formerly called C42 [22], was discovered to inhibit active CDK5, a predominantly neural-specific serine/threonine kinase, and is a major player in the organization of the central nervous system (CNS), such as in the regulation of neurotransmitter release, synaptic plasticity, neurite outgrowth, and neuron migration. CDK5 needs the binding of p35, p39, or p25 (a calpain proteolytic fragment of p35 also named CDK5R1, which is detected in the neurons of Alzheimer patients) for its activation [47]. CDK5RAP1 specifically inhibits the activation of CDK5 by P35, most likely forming a complex with the latter [22, 48]. CDK5RAP1 contains a mitochondria-targeting sequence at its N-terminus that directs the enzyme to the inner membrane of mitochondria (Fig. 1) [49]. Indeed, its main RS activity is performed in the mitochondria [50], although in previous work, it was hypothesized that this enzyme could have a role in the modification of cytoplasmic tRNA [51]. The most important contributing element to modification dynamics might be the level of intracellular compartmentalization, which is present in all organisms but is the most represented in Eukarya, where two genome-containing compartments - the nucleus and the mitochondria - are found [52]. Certainly, the nucleus encodes for the majority of tRNA; however, mammal mitochondria synthesize 21 tRNA, which should be sufficient for decoding all the codons necessary for the translation of the organelle [52].
CDK5RAP1 is widely expressed, and its mRNA was found in the human brain, heart, placenta, skeletal muscle, liver, lung, pancreas, and kidney [53]. The protein size is about 66 KDa although some different forms might exist in particular subsets of tissue or cells, due to different post-translational changes (Adami and Bottai unpublished results).
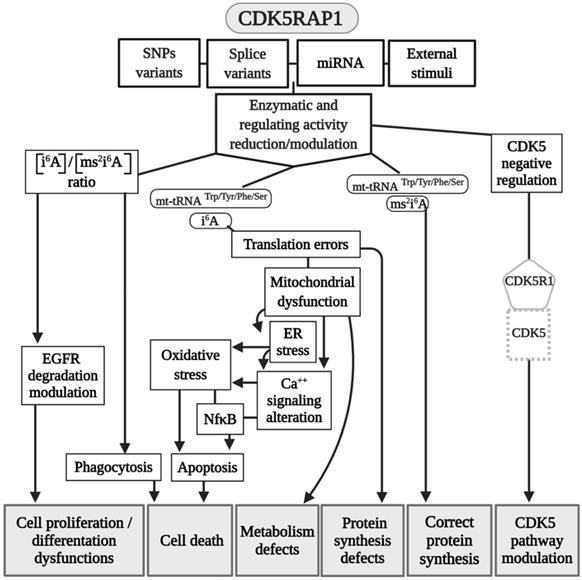
Other than the regulatory effect that CDK5RAP1 has on CDK5, as mentioned earlier, this protein exerts an important role as MTT to modify nucleotide A37 of mt-RNAs, which read codons for Trp, Tyr, Phe, and Ser [49]. CDK5RAP1 is composed by four domains: N-terminal UPF0004 domain (135 residues in length) and a central radical AdoMet domain (235 residues), where there is the cysteine residue, which is crucial for the stability of the [4Fe-4S] clusters. The other two domains are: the mitochondria localization signal (MLS), which allows CDK5RAP1 to localize to the mitochondria, and the TRAM domain, a 60-70-residue-long module which bind tRNA and deliver the RNA-modifying enzymatic domain to their targets (Fig. 1) [54, 55].
CDK5RAP1 is involved in many pathological aspects that can have relevance for further treatments to counteract human diseases.
CDK5RAP1 was hypothesized as having a role in human breast cancer growth. CDK5RAP1 deficiency-induced MCF-7 cells (a cellular model of breast cancer) to the cell cycle arrest in the G2/M phase; apoptosis most likely occurs via the reactive oxygen species (ROS)/JNK signaling pathway, which is known to be heavily involved in the apoptosis process (Fig. 2) [56].
CDK5RAP1 deficiency was also found to induce apoptosis in melanoma A375 cells via the NF‑κB signaling pathway [57], indicating that different tumors might have and share similar pathways related to their development (Figs. 2 and 3).
The relevance of CDK5RAP1 in tumor progression has also been demonstrated by the fact that this enzyme was upregulated in the epithelial human breast cancer cell line MDA-MB-231 and glioblastoma cell line U87VIII [58] after treatment with AC1MMYR2, a small molecule that inhibits CDK5 activity by elevating CDK5RAP1 via miR-21, which then competes with p39 for the activation of CDK5 (Fig. 2) [58].
In other studies, it was demonstrated that CDK5RAP1 deficiency-induced intracellular accumulation of i6A in glioma cells. i6A needs to be converted into ms2i6A to avoid its tumor-suppressive effect; this can be obtained inhibiting the enzymatic machinery which sustains the glioma-initiating cell (GIC) (Fig. 2) [59]. The high level of activity of MTTases is essential for i6A (t6A) modification to ms2i6A (ms2t6A), and as we mentioned earlier, this is pivotal for the correct translation of many proteins. Another intriguing aspect, however, concerns the levels of i6A and ms2i6A (and maybe t6A and ms2t6A). ms2i6A is 9.6 times enriched in the cell culture medium of GIC than inside cells, suggesting that, most likely, CDK5RAP1 decreases [i6A] by promoting the 2-methylthio conversion of i6A in the mitochondria and by its secretion outside the cell [59]. The lower the [i6A] is, the lesser cytotoxic effects on GIC are; indeed, the exogenous application of i6A exerts an antitumor effect in vitro, inducing autophagic cell death and suggesting that i6A is a promising therapeutic molecule to target GICs. Unfortunately, the dosage has not been translated into a therapeutic protocol because it was quite high [59]. The antitumor activity of i6A has been shown in several reports as either acting with antiangiogenic [60] and immunomodulatory activities [61, 62]; later, the same group depicted the role of i6A as an anticancer in glioblastoma multiforme (GMB), the most common brain tumor type [63], revealing that i6A causes epidermal growth factor receptor (EGFR) proteasome degradation and consequent deregulation [64].
Modeling CDK5RAP1 mechanisms. A summary scheme of the various pathways in which CDK5RAP1 is involved. Many of the final effects of the enzyme are shared in common wit CDKAL1. Abbreviations are as in the text.
Epidermal growth factor receptor (EGFR) is particularly abundant in neural stem cells (NSCs) [65, 66]; indeed, the growth of NSCs is driven by the addition of epidermal growth factor (EGF) in the medium [67, 68]. Based on this, we can speculate that a reduction of the proliferative capacity of NSCs, which have been found in an animal model of movement constraints, in which also the level of CDK5RAP1 was drastically reduced [69], could be related to an increase of the i6A intracellular form, which can induce a cytotoxic effect and increase autophagy, hence reducing cell proliferation, as demonstrated in a CIG model [59]. Indeed, the CDK5RAP1 level was found to be reduced in NSCs obtained from mice that underwent movement restraint [69], most likely because of the alteration of the proliferative and differentiative capabilities of these cells (Fig. 2). This could be an important aspect to be considered because many diseases and conditions could reduce movement capabilities [67, 70-72].
The inadequate quality control system in mitochondria, for example, which is induced by CDK5RAP1 KO (animal models), can contribute to the development of myopathy in vivo [49]. Indeed, CDK5RAP1 KO is more likely to determine stress-induced mitochondrial remodeling and exhibit accelerated myopathy and cardiac dysfunction [49]. We can speculate that the high level of oxidative stress present in some different diseases can result in an inhibition of CDK5RAP1 by oxidation of the [4Fe-4S] clusters [19, 49]; indeed, treatment with H2O2 causes a rapid decrease of ms2 modifications, which are reversed by treatment with antioxidants such as pyruvate [49]. The potential role of CDK5RAP1 in mitochondrial metabolism can provide a key to the development of new clinical treatments for cancer.
Interestingly, CDK5RAP1 is involved directly in some neurological effects. For instance, miR-21 is involved in CDK5-axis regulation; moreover, with piRNADQ541777, a Piwi-interacting RNAs (piRNA) present in the spinal cord, mir-21was found to be increased in a mouse model of sciatic nerve injury. Spinal cord piR-DQ541777 ablation relieved thermal hyperalgesia and mechanical allodynia and spinal neuronal sensitization (Fig. 2); its overexpression had the opposite effect [73]. This miR seems to act through the induction of the methylation CpG islands in the cdk5rap1 promoter and results in a consequent reduction of the expression of Cdk5rap1 [73].
Finally, CDK5RAP1 SNPs could have a role in vitiligo patients (Fig. 2) [74].
Conclusions
MTTs have important roles in the regulation of many pivotal cellular functions; they seem to be pivotal epitranscriptomic regulators that can answer and adapt to environmental alterations and stresses by employing several signaling pathways.
One of their main targets is the base A37 of the tRNA. CDKLA1 mostly works in the cytoplasmic site, whereas CDK5RAP1 exerts its main action in the mitochondria. The effects induced by tRNA modifications, mostly ms2t6A and ms2i6A, respectively, represent an important cellular physiological mechanism that can be altered in different diseases.
Because CDK5RAP1 paucity can reduce the proliferative capability of human malignant melanoma, it enhances the formation of ROS [57]. Also, CDKAL1 seems to act throughout ROS modification. For instance, metallothione (MT) gene family expression (which works as free radical scavengers and can relieve ER stress in some cellular dysfunction) is remarkably reduced in CDKAL1-/- cells that are used as models of insulin-expressing pancreatic β-cells [75], and induced expression of MT can improve an impairment of β-cells in vitro and in vivo. Like CDK5RAP1, CDKAL1 could be implicated in some diseases via ROS alteration (Fig. 4), which can increase the level of cellular stress This may be an interesting common mechanism that could be a target of pharmaceutical intervention.
Modeling CDKAL1 mechanisms. A summary scheme of the various pathways in which CDKAL1 is involved. Many of the final effects of the enzymes are shared in common with CDK5RAP1. Abbreviations are as in the text.
Abbreviations
SAM: radical S-adenosylmethionine; RS: SAM enzymes; CDKAL1: CDK5 regulatory subunit associated protein 1 like 1; CDK5RAP1: CDK5 regulatory subunit associated protein 1; TYW1: tRNA-YW synthesizing protein 1 homolog; ELP3: elongator acetyltransferase complex subunit 3; MOCS1: molybdenum cofactor synthesis 1; LIAS: lipoic acid synthetase; RSAD1: radical S-adenosyl methionine domain containing 1; tRNAs: transfer RNAs; MTTases: methylthiotransferases; MtaB: methanol-corrinoid protein co-methyltransferase; MiaB: tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase; RimO: Ribosomal protein S12 methylthiotransferase; i6A: N6-isopentenyladenosine; t6A: N6-threonylcarbamoyladenosine; ACL: anticodon loop; ms2t6A: methylthio-N6-threonylcarbamoyladenosine; TRAM: tRNA methyltransferase 2 and MiaB; ER: endoplasmic reticulum; GWAS: genome-wide association studies; SNPs: single-nucleotide polymorphism; TD2: type 2 diabetes; GHPAs: growth hormone-producing pituitary adenomas; GH: growth hormone; CD: Crohn's disease; SAPHO: syndrome synovitis, acne, pustulosis, hyperostosis, and osteitis; MLS: mitochondria localization signal; GIC: glioma-initiating cell; EGFR: epidermal growth factor receptor; NSCs: neural stem cells; EGF: epidermal growth factor; piRNA: Piwi-interacting RNAs; MY: metallothione.
Acknowledgements
The editing of the manuscript was performed by International Research Promotion, 14 Hanover Street, Mayfair, London W1S 1YH (http://cert.researchpromotion.com/media/documents/IRP-2020-ELES-26588_1.pdf). Graphical abstract and Figures 1, 2, 3 and 4 were created with BioRender.com.
The authors would like to thank Asamsi ONLUS, via Prosciutta, 23-48018 Faenza (BD), Italy and Vertical Foundation, via Carlo Bernari 13, 00139 Roma (IT), Italy (BD).
Author Biography
Daniele Bottai
Dr. Daniele Bottai obtained his Ph. D. in Basic Neuroscience in 1995 at the University of Pisa. Then he moved at McGill University (Montreal, Canada) in Pierre Drapeau's and Rober Dunn's Labs for his first postdoctoral training, and in Peter Seeburg Lab in Max Plank Institute in Heidelberg (Germany) for his second postdoctoral training in 1997. In 2002, then, he came back to Italy at DIBIT San Raffaele in Angelo Vescovi's Lab. He obtained an Assistant Professor position at the University of Milan in 2006 and the tenure in 2009.
His major interest has always been Neuroscience, in learning and memory in the early career using various models such as leech, electric fish, and mouse, later in neurological diseases using various mouse models. In the last 18 years, he has been involved in the study of stem cells of various origin such as neural stem cells, mesenchymal stem cells, and embryonic or induced pluripotent stem cells as tools for the study of neurological diseases and their therapy. Lately, he grew a strong interest in the effects of the motor deprivation in the interaction between muscle and neurogenic regions. Since the patients affected by neuromuscular diseases have a reduction in motor activity, he hypothesizes that the limitation in motor activity could exacerbate the effect of the disease itself. Moreover, other categories of individuals that undergo a reduction in muscular activity are the astronauts which are subject to a reduction of gravitational force. This is particularly important now that long inter-planet travels, on the Moon and March, are scheduled.
Dr. Bottai teaches Pharmacology at the University of Milan in the classes of Medicine (5th year) and Nursing (2nd year) and in Specialty Schools of Otolaryngology, Maxillofacial, Ophthalmology and Child Neuropsychiatry. Moreover, he trained many students for their under graded and Ph. D. studies.
Raffaella Adami
Dr. Adami graduated in Biology at the University of Tuscia and was awarded a Ph.D. in Cellular and molecular biology at the University of Ferrara. She has been working at the University of Milan since 2005 and is involved in microscopy research as a confocal microscopy technician. Dr. Adami collaborates with many groups within the Department, in particular for neuroscience, stem cells, cancers, and rare disease research.
Author Contributions
DB wrote the paper; RA prepared graphical abstract and the figures; RA and DB revised and discussed the text, the graphical abstract and figures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cantoni GL. The nature of the active methyl donor formed enzymatically from L-methionine and adenosinetriphosphate. Journal of the American Chemical Society. 1952;74:2942-3
2. Finkelstein JD. Methionine metabolism in mammals. The Journal of nutritional biochemistry. 1990;1:228-37
3. Landgraf BJ, McCarthy EL, Booker SJ. Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annual review of biochemistry. 2016;85:485-514
4. Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic acids research. 2001;29:1097-106
5. Broderick JB, Duffus BR, Duschene KS, Shepard EM. Radical S-adenosylmethionine enzymes. Chemical reviews. 2014;114:4229-317
6. Raina M, Ibba M. tRNAs as regulators of biological processes. Frontiers in genetics. 2014;5:171
7. Oberbauer V, Schaefer MR. tRNA-Derived Small RNAs: Biogenesis, Modification, Function and Potential Impact on Human Disease Development. Genes (Basel). 2018;9:607
8. Barraud P, Tisne C. To be or not to be modified: Miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB life. 2019;71:1126-40
9. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic acids research. 2018;46:D303-D7
10. Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X. et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic acids research. 2011;39:D195-201
11. Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic acids research. 2014;42:7346-57
12. Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA biology. 2014;11:1619-29
13. Han L, Phizicky EM. A rationale for tRNA modification circuits in the anticodon loop. Rna. 2018;24:1277-84
14. Lorenz C, Lunse CE, Morl M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017;7:35
15. Shigi N. Recent Advances in Our Understanding of the Biosynthesis of Sulfur Modifications in tRNAs. Front Microbiol. 2018;9:2679
16. Globisch D, Pearson D, Hienzsch A, Bruckl T, Wagner M, Thoma I. et al. Systems-based analysis of modified tRNA bases. Angewandte Chemie. 2011;50:9739-42
17. Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. The EMBO journal. 2001;20:4863-73
18. Krutyholowa R, Zakrzewski K, Glatt S. Charging the code - tRNA modification complexes. Current opinion in structural biology. 2019;55:138-46
19. Arragain S, Handelman SK, Forouhar F, Wei FY, Tomizawa K, Hunt JF. et al. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. The Journal of biological chemistry. 2010;285:28425-33
20. Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A. et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. The Journal of clinical investigation. 2011;121:3598-608
21. Brambillasca S, Altkrueger A, Colombo SF, Friederich A, Eickelmann P, Mark M. et al. CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) is a tail-anchored protein in the endoplasmic reticulum (ER) of insulinoma cells. J Biol Chem. 2012;287:41808-19
22. Ching YP, Pang AS, Lam WH, Qi RZ, Wang JH. Identification of a neuronal Cdk5 activator-binding protein as Cdk5 inhibitor. The Journal of biological chemistry. 2002;277:15237-40
23. Wei FY, Tomizawa K. tRNA modifications and islet function. Diabetes Obes Metab. 2018;20(Suppl 2):20-7
24. Quaranta M, Burden AD, Griffiths CE, Worthington J, Barker JN, Trembath RC. et al. Differential contribution of CDKAL1 variants to psoriasis, Crohn's disease and type II diabetes. Genes Immun. 2009;10:654-8
25. Zhou B, Wei FY, Kanai N, Fujimura A, Kaitsuka T, Tomizawa K. Identification of a splicing variant that regulates type 2 diabetes risk factor CDKAL1 level by a coding-independent mechanism in human. Human molecular genetics. 2014;23:4639-50
26. Diabetes Genetics Initiative of Broad Institute of H, Mit LU, Novartis Institutes of BioMedical R, Saxena R, Voight BF, Lyssenko V. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331-6
27. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341-5
28. Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB. et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nature genetics. 2007;39:770-5
29. Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336-41
30. Kang J, Guan RC, Zhao Y, Chen Y. Obesity-related loci in TMEM18, CDKAL1 and FAIM2 are associated with obesity and type 2 diabetes in Chinese Han patients. BMC Med Genet. 2020;21:65
31. Krentz NAJ, Gloyn AL. Insights into pancreatic islet cell dysfunction from type 2 diabetes mellitus genetics. Nat Rev Endocrinol. 2020;16:202-12
32. Vatankhah Yazdi K, Kalantar SM, Houshmand M, Rahmanian M, Manaviat MR, Jahani MR. et al. SLC30A8, CDKAL1, TCF7L2, KCNQ1 and IGF2BP2 are Associated with Type 2 Diabetes Mellitus in Iranian Patients. Diabetes Metab Syndr Obes. 2020;13:897-906
33. Santos M, Anderson CP, Neschen S, Zumbrennen-Bullough KB, Romney SJ, Kahle-Stephan M. et al. Irp2 regulates insulin production through iron-mediated Cdkal1-catalyzed tRNA modification. Nature communications. 2020;11:296
34. Larsson SC, Orsini N, Wolk A. Diabetes mellitus and risk of colorectal cancer: a meta-analysis. Journal of the National Cancer Institute. 2005;97:1679-87
35. Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA. et al. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33:1674-85
36. Shen H, Morrison CD, Zhang J, Underwood W 3rd, Yang N, Frangou C. et al. 6p22.3 amplification as a biomarker and potential therapeutic target of advanced stage bladder cancer. Oncotarget. 2013;4:2124-34
37. Bellmunt J. Stem-Like Signature Predicting Disease Progression in Early Stage Bladder Cancer. The Role of E2F3 and SOX4. Biomedicines. 2018;6:85-98
38. Takesue Y, Wei FY, Fukuda H, Tanoue Y, Yamamoto T, Chujo T. et al. Regulation of growth hormone biosynthesis by Cdk5 regulatory subunit associated protein 1-like 1 (CDKAL1) in pituitary adenomas. Endocr J. 2019;66:807-16
39. Hannon AM, Thompson CJ, Sherlock M. Diabetes in Patients With Acromegaly. Curr Diab Rep. 2017;17:8
40. Take K, Waki H, Sun W, Wada T, Yu J, Nakamura M. et al. CDK5 Regulatory Subunit-Associated Protein 1-like 1 Negatively Regulates Adipocyte Differentiation through Activation of Wnt Signaling Pathway. Scientific reports. 2017;7:7326
41. Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ. et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nature genetics. 2013;45:76-82
42. Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature genetics. 2008;40:955-62
43. Wolf N, Quaranta M, Prescott NJ, Allen M, Smith R, Burden AD. et al. Psoriasis is associated with pleiotropic susceptibility loci identified in type II diabetes and Crohn disease. Journal of medical genetics. 2008;45:114-6
44. Blaschke M, Koepp R, Lenz C, Kruppa J, Jung K, Siggelkow H. Crohn's disease patient serum changes protein expression in a human mesenchymal stem cell model in a linear relationship to patients' disease stage and to bone mineral density. J Clin Transl Endocrinol. 2018;13:26-38
45. Li N, Ma J, Li K, Guo C, Ming L. Different Contributions of CDKAL1, KIF21B, and LRRK2/MUC19 Polymorphisms to SAPHO Syndrome, Rheumatoid Arthritis, Ankylosing Spondylitis, and Seronegative Spondyloarthropathy. Genet Test Mol Biomarkers. 2017;21:122-6
46. Gaczkowska A, Zukowski K, Biedziak B, Hozyasz KK, Wojcicki P, Zadurska M. et al. Association of CDKAL1 nucleotide variants with the risk of non-syndromic cleft lip with or without cleft palate. Journal of human genetics. 2018;63:397-406
47. Roufayel R, Murshid N. CDK5: Key Regulator of Apoptosis and Cell Survival. Biomedicines. 2019;7:88-100
48. Lee KY, Rosales JL, Tang D, Wang JH. Interaction of cyclin-dependent kinase 5 (Cdk5) and neuronal Cdk5 activator in bovine brain. The Journal of biological chemistry. 1996;271:1538-43
49. Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H. et al. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell metabolism. 2015;21:428-42
50. Fakruddin M, Wei FY, Emura S, Matsuda S, Yasukawa T, Kang D. et al. Cdk5rap1-mediated 2-methylthio-N6-isopentenyladenosine modification is absent from nuclear-derived RNA species. Nucleic acids research. 2017;45:11954-61
51. Reiter V, Matschkal DM, Wagner M, Globisch D, Kneuttinger AC, Muller M. et al. The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic acids research. 2012;40:6235-40
52. Paris Z, Alfonzo JD. How the intracellular partitioning of tRNA and tRNA modification enzymes affects mitochondrial function. IUBMB life. 2018;70:1207-13
53. Zou X, Ji C, Jin F, Liu J, Wu M, Zheng H. et al. Cloning, characterization and expression of CDK5RAP1_v3 and CDK5RAP1_v4, two novel splice variants of human CDK5RAP1. Genes Genet Syst. 2004;79:177-82
54. Anantharaman V, Koonin EV, Aravind L. TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS microbiology letters. 2001;197:215-21
55. Holliday GL, Akiva E, Meng EC, Brown SD, Calhoun S, Pieper U. et al. Atlas of the Radical SAM Superfamily: Divergent Evolution of Function Using a "Plug and Play" Domain. Methods in enzymology. 2018;606:1-71
56. Wang H, Wei L, Li C, Zhou J, Li Z. CDK5RAP1 deficiency induces cell cycle arrest and apoptosis in human breast cancer cell line by the ROS/JNK signaling pathway. Oncology reports. 2015;33:1089-96
57. Xiong J, Wang Y, Gu Y, Xue Y, Dang L, Li Y. CDK5RAP1 targeting NF-kappaB signaling pathway in human malignant melanoma A375 cell apoptosis. Oncology letters. 2018;15:4767-74
58. Ren Y, Zhou X, Yang JJ, Liu X, Zhao XH, Wang QX. et al. AC1MMYR2 impairs high dose paclitaxel-induced tumor metastasis by targeting miR-21/CDK5 axis. Cancer letters. 2015;362:174-82
59. Yamamoto T, Fujimura A, Wei FY, Shinojima N, Kuroda JI, Mukasa A. et al. 2-Methylthio Conversion of N6-Isopentenyladenosine in Mitochondrial tRNAs by CDK5RAP1 Promotes the Maintenance of Glioma-Initiating Cells. iScience. 2019;21:42-56
60. Pisanti S, Picardi P, Ciaglia E, Margarucci L, Ronca R, Giacomini A. et al. Antiangiogenic effects of N6-isopentenyladenosine, an endogenous isoprenoid end product, mediated by AMPK activation. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2014;28:1132-44
61. Ciaglia E, Pisanti S, Picardi P, Laezza C, Malfitano AM, D'Alessandro A. et al. N6-isopentenyladenosine, an endogenous isoprenoid end product, directly affects cytotoxic and regulatory functions of human NK cells through FDPS modulation. Journal of leukocyte biology. 2013;94:1207-19
62. Ciaglia E, Pisanti S, Picardi P, Laezza C, Sosa S, Tubaro A. et al. N6-isopentenyladenosine affects cytotoxic activity and cytokines production by IL-2 activated NK cells and exerts topical anti-inflammatory activity in mice. Pharmacol Res. 2014;89:1-10
63. Bottai D, Adami R, Paroni R, Ghidoni R. Brain cancer-activated microglia: A potential role for sphingolipids. Current medicinal chemistry. 2020;27:4039-61
64. Ciaglia E, Abate M, Laezza C, Pisanti S, Vitale M, Seneca V. et al. Antiglioma effects of N6-isopentenyladenosine, an endogenous isoprenoid end product, through the downregulation of epidermal growth factor receptor. International journal of cancer Journal international du cancer. 2017;140:959-72
65. Bottai D, Fiocco R, Gelain F, Defilippis L, Galli R, Gritti A. et al. Neural stem cells in the adult nervous system. Journal of hematotherapy & stem cell research. 2003;12:655-70
66. Bottai D, Adami R, Ghidoni R. The crosstalk between glycosphingolipids and neural stem cells. Journal of neurochemistry. 2019;148:698-711
67. Adami R, Scesa G, Bottai D. Stem cell transplantation in neurological diseases: improving effectiveness in animal models. Frontiers in cell and developmental biology. 2014;2:17
68. Bottai D, Spreafico M, Pistocchi A, Fazio G, Adami R, Grazioli P. et al. Modeling Cornelia de Lange syndrome in vitro and in vivo reveals a role for cohesin complex in neuronal survival and differentiation. Human molecular genetics. 2019;28:64-73
69. Adami R, Pagano J, Colombo M, Platonova N, Recchia D, Chiaramonte R. et al. Reduction of movement in neurological diseases: effects on neural stem cells characteristics. Frontiers in neuroscience. 2018;12:336
70. Bottai D, Adami R. Spinal muscular atrophy: new findings for an old pathology. Brain pathology. 2013;23:613-22
71. Adami R, Bottai D. Movement impairment: Focus on the brain. Journal of neuroscience research. 2016;94:310-7
72. Adami R, Bottai D. Spinal Muscular Atrophy Modeling and Treatment Advances by Induced Pluripotent Stem Cells Studies. Stem Cell Rev Rep. 2019;15:795-813
73. Zhang C, Sha H, Peng Y, Wang Y, Liu C, Zhou X. PiRNA-DQ541777 Contributes to Neuropathic Pain via Targeting Cdk5rap1. J Neurosci. 2019;39:9028-39
74. Shin MK, Uhm YK, Lee JH, Kim SK, Chung JH, Lee MH. Association between CDK5RAP1 polymorphisms and susceptibility to vitiligo in the Korean population. European journal of dermatology: EJD. 2012;22:495-9
75. Guo M, Zhang T, Dong X, Xiang JZ, Lei M, Evans T. et al. Using hESCs to Probe the Interaction of the Diabetes-Associated Genes CDKAL1 and MT1E. Cell reports. 2017;19:1512-21
Author contact
![]() Corresponding author: Daniele Bottai, Ph. D. Assistant Professor in Pharmacology, E-mail: daniele.bottaiit; University of Milan, Department of Health Science, San Paolo Hospital via A di Rudinì 8 20142, Milan Italy. Tel.: 0039-02-50323215; Fax: 0039-02-50323245; orcid.org/0000-0002-7951-9512.
Corresponding author: Daniele Bottai, Ph. D. Assistant Professor in Pharmacology, E-mail: daniele.bottaiit; University of Milan, Department of Health Science, San Paolo Hospital via A di Rudinì 8 20142, Milan Italy. Tel.: 0039-02-50323215; Fax: 0039-02-50323245; orcid.org/0000-0002-7951-9512.

 Global reach, higher impact
Global reach, higher impact