10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(15):3039-3049. doi:10.7150/ijbs.47510 This issue Cite
Research Paper
Multiple Developmental Defects in sox11a Mutant Zebrafish with Features of Coffin-Siris Syndrome
1. State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China.
2. Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, China.
3. University of Chinese Academy of Sciences, Beijing 100049, China.
#These authors contributed equally to this work.
Received 2020-4-27; Accepted 2020-9-16; Published 2020-10-3
Abstract
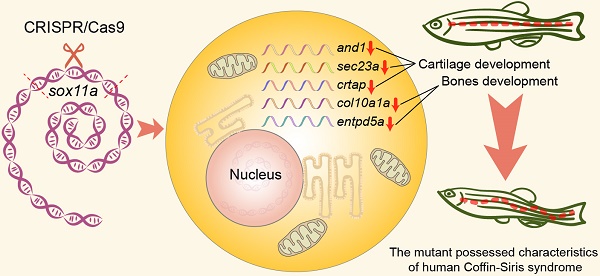
A previous study suggested that human Coffin-Siris syndrome is related to the mutation of SOX11. Since the homozygous SOX11 mutant mice died soon after birth, no suitable model was available for the study of the pathogenic mechanism of Coffin-Siris syndrome. To solve this problem, we generated two viable homozygous zebrafish mutants, sox11am/m and sox11bm/m. We found that the sox11am/m mutant possessed Coffin-Siris syndrome features. The sox11am/m mutants exhibited growth deficiency from 3.3 hpf embryos to adulthood. Furthermore, the sox11am/m mutant also displayed microcephaly, narrow pupillary distance, achondroplasia, and bone deformity in adults. Growth deficiency could be rescued by the injection of sox11a mRNA at the one-cell stage. In addition, the expression levels of genes related to cartilage and bone were downregulated in the sox11am/m mutant, indicating that sox11a mainly affected the growth and development of zebrafish by regulating the expression of genes related to skeletal development. Our results indicate that sox11am/m mutant zebrafish offered a potential model system to help with the search for pathogenic mechanisms of human Coffin-Siris syndrome.
Keywords: sox11, zebrafish, Coffin-Siris, CRISPR/Cas9
Introduction
There are seven members in the sox family, namely soxA, soxB1, soxB2, soxC, soxD, soxE, soxF, and soxG [1]. The functional domain of the SOX protein is an HMG domain consisting of three α helices that can bind to the ATTGTT sequence or other related sequences [2,3]. Therefore, all sox family members are transcription factors. sox11 comes from the soxC sub-family, which also includes sox4 and sox12 [4]. Studies have illustrated that sox11 is involved in multiple biological processes, including neurogenesis, organogenesis, craniofacial, and skeletal development [5,6].
Coffin-Siris syndrome is a disease that has affected humans for a long time, and most of the patients show retardation of motor and language expression abilities and bone development, besides small body size and low intelligence level [7-9]. Clinical data has showed that the disease is related to SOX11 mutation. Hempel et al. reported that the cause of Coffin-Siris syndrome in 10 patients was associated with SOX11 deletion or mutation [7]. However, the specific pathogenic mechanism of Coffin-Siris syndrome is not clear. The function of Sox11 is relevant to tissue remodeling, and Sox11 mutant mice have been shown to develop symptoms of congenital cyanosis, congenital heart disease, cranial hypoplasia, and non-cranial dysplasia, besides embryo death immediately after birth [10]. However, the difficulty of obtaining viable homozygote mutants hinders the study of the function and underlying mechanisms of Sox11 in mammals.
Zebrafish are widely used as model organisms in vertebrate development research and disease model. There are two sox11 genes in zebrafish including sox11a and sox11b. However, the role and potential mechanisms of sox11a/b in zebrafish remain largely unknown, and there is no sox11a/b mutation model in zebrafish. Therefore, our study aimed to further elucidate the mechanism of Coffin-Siris syndrome diseases and establish appropriate disease models by revealing and comparing sox11a/b functions in zebrafish development. For this, we obtained viable sox11am/m and sox11bm/m mutants for the first time using the CRISPR/Cas9 system. We found that sox11am/m mutant zebrafish exhibited reduced size and abnormal cartilages and bones, with smaller heads and lower pupillary distances compared to non-mutant zebrafish. However, there were no obvious differences between the sox11bm/m mutants and WT individuals. Therefore, only sox11am/m mutant zebrafish can be used to explore the pathogenic mechanism of Coffin-Siris syndrome.
Materials and Methods
Zebrafish maintenance
In this study, AB strain zebrafish were used and maintained according to the zebrafish book [11], raised in recirculation systems at 28 °C and under a 14 h: 10 h light: dark cycle. All zebrafish experiments were performed according to the Guiding Principles for the Care and Use of Laboratory Animals approved by the Institute of Hydrobiology, Chinese Academy of Sciences.
Generation of sox11a and sox11b mutant zebrafish lines and double mutant lines
Disruptions of sox11a and sox11b in zebrafish were accomplished via CRISPR/Cas9 technology. Zebrafish sox11a and sox11b sgRNA were designed using tools provided on a website (http://www.crisprscan.org/?page=sequence), and the target sites were GGTCGCTTTATGTGTC NGG and GGGTCGTTTGATGTGGC NGG, respectively. The primers were designed according to the pT7gRNA and target sites. pT7gRNA was used as a template to synthesize the gRNA in vitro transcription template using PCR. The forward and reverse primers of the two target sites were shown in Figure S1. The transcription kit (Invitrogen, USA) was used for in vitro gRNA transcript and the gRNA was purified using the gRNA purification kit (QIAGEN, Germany). Cas9 mRNA was obtained using an in vitro transcription kit (Invitrogen, USA) with the linearizing Cas9 plasmid as template and purified by ammonium acetate precipitation. The gRNA and Cas9 mRNA were diluted to 50 ng/µL and 500 ng/µL, respectively, and then mixed according to a 1:1 ratio with a small amount of phenol red as an indicator. The prepared sample was injected into the animal pole of the embryos at the one-cell stage and then cultured in 0.3 × Danieau' s buffer for P0 generations. Then, P0 generation adult fish were crossed with WT to produce the F1 generation. The screened heterozygote F1 generation was self-crossed to produce the F2 generation, from which the homozygote mutation could be identified. The homozygote F2 generation mutation was then self-crossed to produce the F3 generation, which was used in the following experiments.
The sox11am/+sox11bm/+ double heterozygous genotype mutants were produced by crossing the sox11am/m mutants with the sox11bm/m mutants. We expected to obtain the sox11am/msox11bm/m double mutants by genotyping offspring which was produced by sox11am/+sox11bm/+ double heterozygous self-crossing. In theory, the following genotype mutants could be produced as shown in Figure S7. However, the number of the sox11am/msox11bm/m double mutants was zero after genotyping 46 offspring. Then we picked up three groups including sox11am/msox11bm/+ self-crossing, sox11am/+sox11bm/m self-crossing and crossing the sox11am/+sox11bm/m mutant with the sox11am/+sox11bm/+ mutant to produce the double mutants.
Real-time quantitative PCR (qPCR)
Total RNA from zebrafish tissues and embryos was extracted with TRIzol (Invitrogen, USA) according to the manufacturer's instructions. The obtained RNA was analyzed by agarose gel electrophoresis, and its concentration was measured using a spectrophotometer (Thermo, USA). Then, 1 µg of RNA was removed for DNA removal, and a kit (Toyobo, Japan) was used for reverse transcription. cDNA was obtained using the ReverTra Ace reverse transcriptase kit (Toyobo, Japan) and random primers. The fluorescence quantitative PCR assay was performed using the SYBR green mix (Toyobo, Japan) according to the manufacturer's instructions: 2×SYBR Green real-time master mix, 10 µL; cDNA, 2 µL; F, 0.4 µL; R, 0.4 µL; H2O, 7.2 µL. The PCR amplification conditions were: 95 °C for 3 min; 45 cycles × (95 °C for 15 s; 55 °C for 20 s; 72 °C for 30 s); 65 °C for 0.06 s; 95 ℃, 0.5 s. The primers were listed in Figure S1.
Assessment of growth parameters
The body length of the zebrafish was measured based on photographs using the ImageJ software. Body weight was determined using an electronic analytical balance (Shimadzu, Japan) after the individual's body was dried using filter paper.
Histological analysis
We observed adult zebrafish muscle development and embryo brain growth using paraffin sections. The samples were fixed in Bouin's fixative overnight. After dehydration, penetration, and wax immersion, the samples were embedded in paraffin wax. The samples were then sectioned into 6-μm slices and stained using the hematoxylin and eosin method. The images were taken using a microscope (OLYMPUS, Japan).
Immunofluorescence staining
Whole mount muscles of zebrafish larvae were analyzed by immunofluorescent staining. For this, the samples were immersed in 4% PFA overnight at 4 °C. The fixed samples were washed with PBS every 5 min three times and permeabilized for 30 min in 0.1% Triton PBS. The samples were then stained with phalloidin (1:500) overnight at 4 °C. The nuclei were stained with DAPI (1:1000). After washing three times with PBS, the samples were sealed with mounting medium (Vectorlabs, USA) and the images were taken using a confocal microscope (Leica, Germany).
Alcian blue staining
To observe the cartilage development of sox11am/m mutant zebrafish, the samples were stained with Alcian blue. The 5-day post fertilization (dpf) zebrafish larvae were fixed in 4% PFA overnight at 4 °C. The samples were then transferred into 70% methanol. The samples were stained in an Alcian blue buffer (35 mL 100% MeOH, 4 mg MgCl2, 10 mg Alcian blue, 50 mL final volume) for 4 h after incubating the samples in 50% ethanol for 10 min on a decolorizing rocker. The samples were washed in PBST every 5 min four times before rinsing in rinsing buffer (30% H2O2 5 mL, 1% KOH 25 mL, and 20 mL ddH2O, 50 mL final volume). Then, the fish body was digested with 1 mg/mL trypsin (100 mg trypsin, 100 mL 60% saturated sodium borate) near transparent after washing the samples in PBST every 5 min four times. Images were taken after washing the samples in PBST every 5 min twice.
Micro-CT
For this procedure, a sox11am/m mutant and a WT zebrafish of the same age were used. The body surfaces were dried using filter paper after anesthesia with MS222. Then, the bodies were wrapped with the bubble film matching the instrument and placed on the appropriate sample table before the scanning was carried out. When the scanning was completed, image reconstruction and processing was performed.
Whole-mount in situ hybridization
Digoxigenin-labeled probes against sox11a, sox11b, and1, crtap, sec23a and col10a1a were generated by PCR using the primer pairs shown in Figure S1 with an added T7 polymerase binding site and synthesized using a T7 in vitro transcription kit (Invitrogen, USA). Whole-mount in situ hybridization was performed according to the method of Thisse et al. [12], and the images were taken with a Nikon digital camera.
Rescue experiment
The sox11a and gfp cDNA sequences were cloned into pcsII vectors. The plasmids were linearized using NotI (NEB, USA). Then, the linearized vector was transcribed using the Sp6 mMessage mMachine kit (Invitrogen, USA) and purified with the RNA purification kit (Sigma, USA) according to the manufacturer' s instructions. The primers are listed in Figure S1. The samples were injected into the embryos at the one-cell stage at a concentration of 150 ng/µL.
Results
Conservation and expression patterns of sox11a and sox11b in zebrafish
Two sox11 genes, sox11a and sox11b, occur in zebrafish. The amino acid sequence homology analysis results are shown in Figure S2. The BLASTP results showed that the similarity between zebrafish sox11a and its homologous gene in medaka, mouse, rat, and human was 74.25%, 58.44%, 58.29%, and 58.5%, respectively. The similarity between zebrafish sox11b and its homologous gene in medaka, mouse, rat, and human were 67.46%, 58.6%, 58.6%, and 81.43%, respectively. This indicates that both genes are evolutionarily conserved.
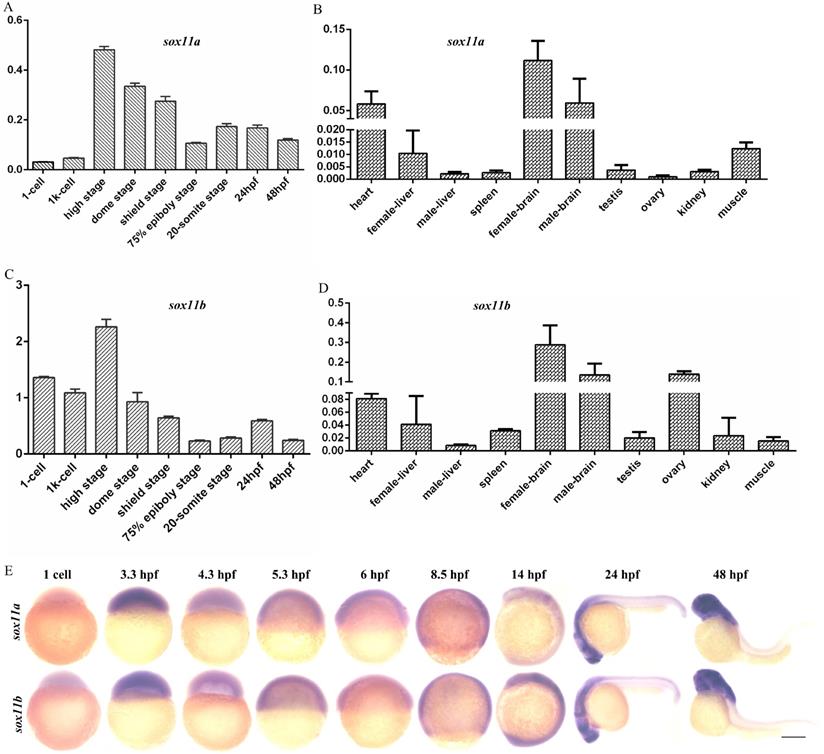
The expression levels of sox11a and sox11b in embryos of different stages and in different tissues of WT zebrafish were analyzed. The results indicated that sox11a and sox11b were expressed during various periods of embryonic development, with the highest expression in the high stage and the expression level gradually decreasing as the embryo developed (Fig. 1A, C). The mRNA expression pattern in adult tissues was that sox11a and sox11b mRNAs were mainly detected in the heart, brain, liver, muscle, and other tissues, with the highest levels in the brain (Fig. 1B, D). The expression of sox11a and sox11b was analyzed by whole-mount in situ hybridization (Fig. 1E).
Generation of sox11a and sox11b mutants by CRISPR/Cas9
To further elucidate the function of sox11 in zebrafish in vivo, sox11a and sox11b mutants were generated via CRISPR/Cas9. Two sox11a mutant lines were obtained, including a 7-bp deletion and a 11-bp deletion, which had a consistent phenotype (Fig. 2A, S3A-C). The F3 generation 7-bp deletion homozygote mutant line was used for subsequent experiments marked as sox11am/m. The sox11b mutant line included a 1- bp deletion and a 7-bp deletion in two lines, which also had a consistent phenotype (Fig. 2B, S3D-F). The F3 generation 7-bp deletion homozygote mutant line was used for subsequent experiments marked as sox11bm/m.
The sox11am/m homozygote mutant was confirmed by sequencing and screened using the high-resolution melting curve method, and the results are listed in Figure 2E, G. The amino acid sequence analysis of the sox11am/m mutant line showed that it would produce a frameshift mutation and the translated amino acid sequence would terminate in front of the SOX-TCF_HMG-box domain (Fig. 2C). The whole-mount in situ hybridization results showed an obvious decrease in sox11a expression in the sox11am/m mutant (Fig. 2I). And the qPCR results also indicated the expression level of sox11a in sox11am/m mutant was lower than WT (Fig. 2J).
The sox11bm/m homozygote mutant was also checked using the high-resolution melting curve method (Fig. 2F, H). The translated amino acid sequence of the sox11bm/m mutant would produce a frameshift mutation and terminate ahead of the SOX-TCF_HMG-box domain (Fig. 2D). An apparent lower sox11b level in the sox11bm/m mutant could be found by whole-mount in situ hybridization (Fig. 2K). And the qPCR data also indicated the expression level of sox11b in sox11bm/m mutant was lower than WT (Fig. 2L).
The expression patterns of sox11a and sox11b in zebrafish. A: qPCR analysis of zebrafish sox11a expression during early embryonic development. B: qPCR analysis of zebrafish sox11a expression in different tissues. C: qPCR analysis of zebrafish sox11b expression during early embryonic development. D: qPCR analysis of zebrafish sox11b expression in different tissues. The qPCR data were analyzed by 2-∆CT method and ꞵ-actin was chosen as reference gene. E: The expression levels of sox11a and sox11b during early embryonic development by whole-mount in situ hybridization.
Establishment of sox11am/m and sox11bm/m mutant line via CRISPR/Cas9 technology in zebrafish. A: The location of the sgRNA targeting sites on zebrafish sox11a. B: The location of sgRNA targeting sites on zebrafish sox11b. C: Schematic representation of the putative peptides of WT and the mutated sox11a peptides from the targeted allele. D: Schematic representation of the putative peptides of WT and the mutated sox11b peptides from the targeted allele. E: Confirmation of sox11a homozygote mutant by sequencing. F: Confirmation of sox11b homozygote mutant by sequencing. G: Screen of homozygote mutant by high resolution melting curve method. (red line, homozygote; green line, heterozygote; blue line, WT.) H: Screen of homozygote mutant by high resolution melting curve method. red line, homozygote; green line, heterozygote; blue line, WT. I: Whole-mount in situ hybridization of WT and sox11am/m mutant. Antisense probes against sox11a were visualized at 4 dpf. Scale bar, 200 µm. J: The expression level of sox11a in WT and sox11am/m mutant by qPCR. K: Whole-mount in situ hybridization of WT and sox11bm/m mutant. Antisense probes against sox11bm/m were visualized at 24 hpf. Scale bar, 200 µm. L: The expression level of sox11b in WT and sox11bm/m mutant by qPCR. The qPCR data were analyzed by 2-∆∆CT method and ꞵ-actin was chosen as reference gene (*P<0.05). M: The generation of sox11am/msox11bm/m double mutant.
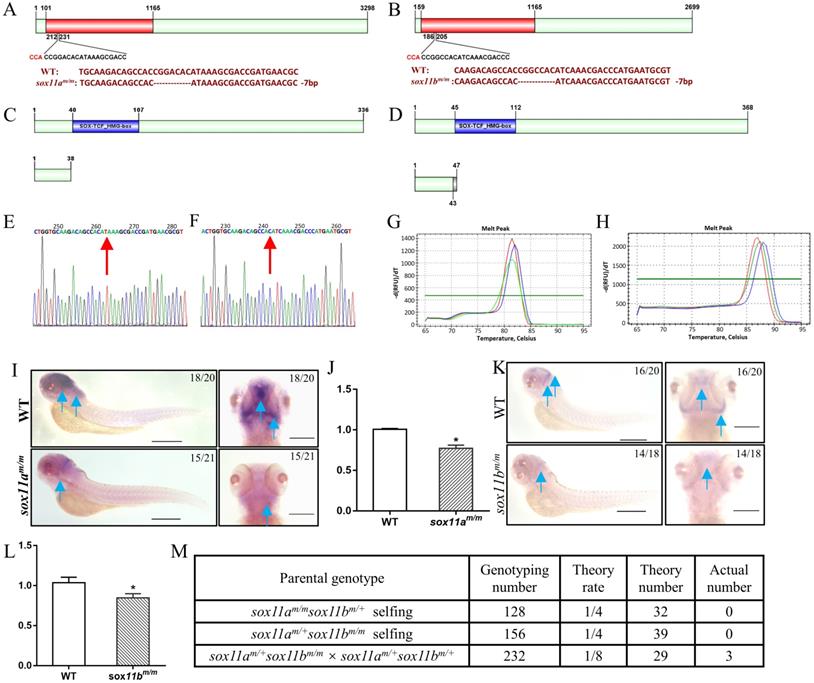
In addition, we have tried to generate sox11am/msox11bm/m double mutant by three groups including: sox11am/msox11bm/+ self-crossing, sox11am/+sox11bm/m self-crossing and crossing the sox11am/+sox11bm/m mutant with the sox11am/+sox11bm/+ mutant. However, only three adult sox11am/msox11bm/m double mutants were confirmed after genotyping 516 offspring from the three groups which deviated from the law of independent assortment. The detailed information of this result could be found in Figure 2M. In conclusion we failed to generate a double mutant family since the three sox11am/msox11bm/m double mutants couldn't produce offspring.
sox11am/m mutant zebrafish displayed growth and developmental delay while sox11bm/m mutant development normally
First, the early embryo development process was observed by comparing sox11am/m mutants with WT embryos. We found that sox11am/m mutant embryos began to present some delay compared with WT embryos from 3.3 h post fertilization (hpf). This phenomenon was maintained throughout early embryo development. At 10 hpf, the WT embryos had developed to 100% epiboly stage, while the sox11am/m mutant embryos fell behind. At 21 hpf, the sox11am/m mutant embryo tail was clearly shorter than that of WT embryos (Fig. 3A).
The sox11am/m mutant exhibit growth retardation while the sox11bm/m mutant developed normally. A: The morphology of WT and sox11am/m mutant larvae from 2 hpf to 21 hpf. Scale bar, 200 µm. B: The body length of sox11am/m mutants was significantly shorter than that of WT from 5 dpf to 30 dpf (****P<0.0001). C: The 70 dpf female body length and body weight of sox11am/m mutants were significantly smaller than that of WT. And the 70 dpf male body length and body weight of sox11am/m mutants were also significantly smaller than that of WT (*P<0.05, **P<0.01). D: The morphology of WT and sox11am/m mutant larvae from 3.3 hpf to 24 hpf. Scale bar, 200 µm. E: There were no significant difference between the 70 dpf female or male sox11bm/m mutants and their respective WT counterpart regarding body length and body weight.
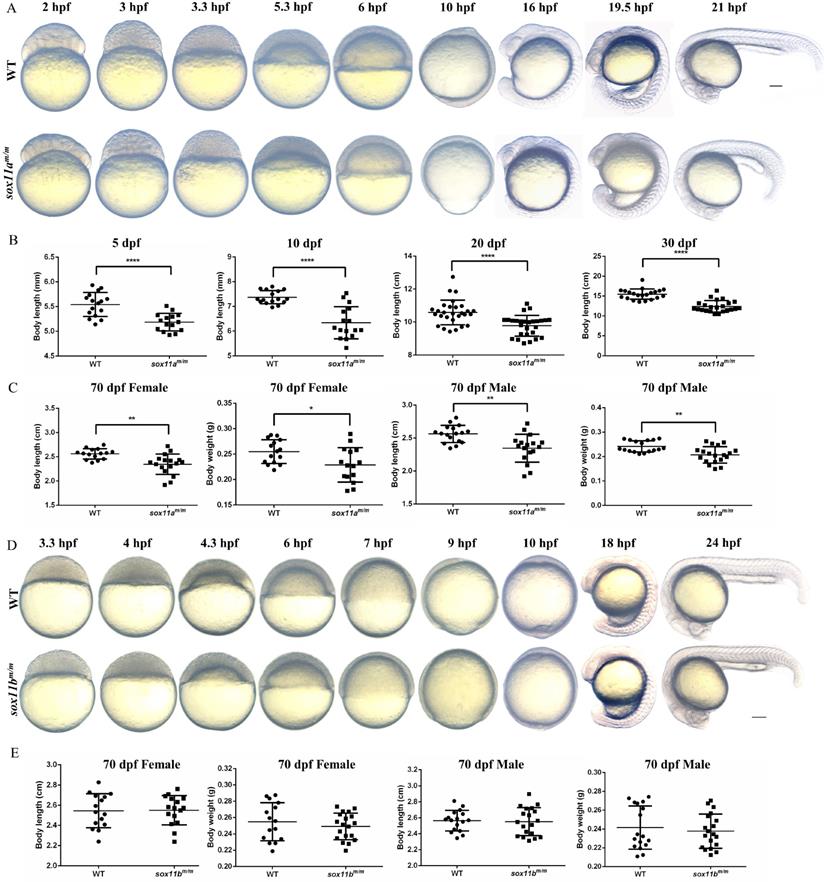
Then, juvenile fish development was analyzed by measuring the body length of sox11am/m mutants and WTs bred under the same conditions. Four points were chosen for statistical analysis including 5 dpf, 10 dpf, 20 dpf, and 30 dpf (Fig. 3B). The results indicated that the body length of sox11am/m mutants was significantly smaller than that of WTs (****P<0.0001).
The 70 dpf adult zebrafish were separated into males and females before analysis. Then, the body length and body weight were measured and counted for each group separately (Fig. 3C). The results suggested that the body lengths of male and female sox11am/m mutants were significantly lower than those of male and female WTs, respectively (**P<0.01). The body weight of female sox11am/m mutants was lower than that of WT females (*P<0.05), as was that of male sox11am/m mutants compared to WT males (**P<0.01).
In addition, the sox11bm/m mutant zebrafish was also observed and analyzed. The developmental process of the sox11bm/m mutant embryo was in accordance with that of WT during the early embryo development stage (Fig. 3D). Subsequently, we measured the body length and body weight of the adult zebrafish, including males and females (Fig. 3E). The results showed that there was no significant difference in body length between males and females. The body weight of the sox11bm/m mutant and WT showed no difference either (P>0.05).
sox11am/m mutant zebrafish showed microcephaly, normal muscle and curved spine
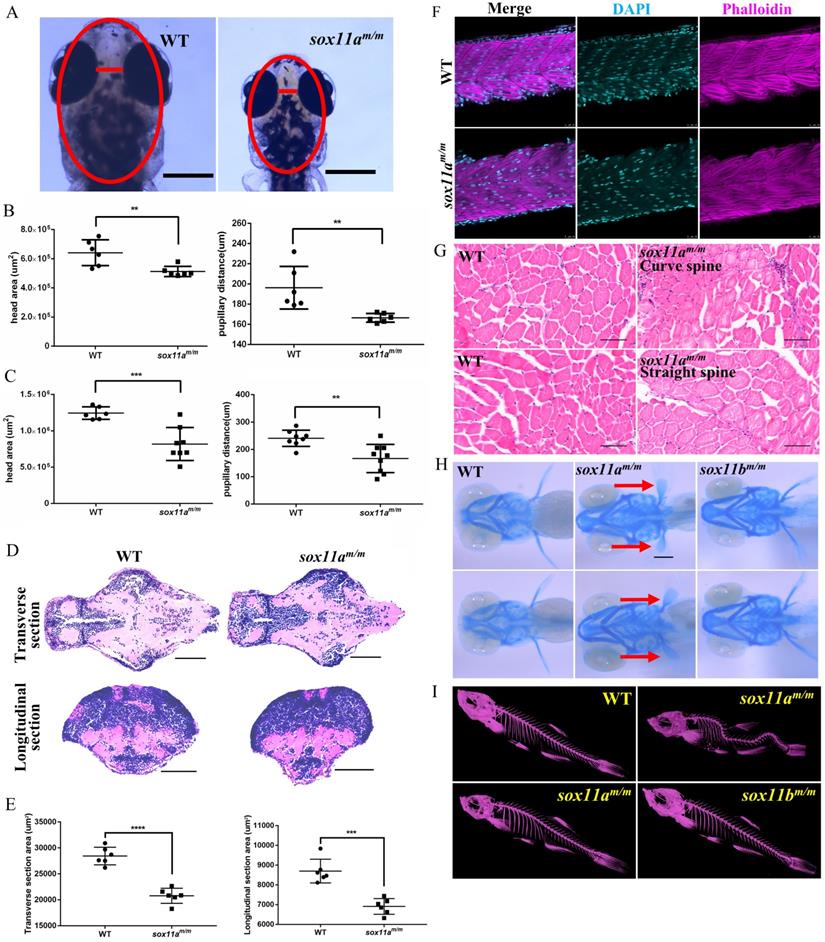
sox11a is essential not only for growth but also for head development. The head area and pupillary distance of the embryos were measured using Image J, based on the scale bars in the photographs (Fig. 4A). The results showed that both the head area and pupillary distance of the sox11am/m mutants at 5 dpf were significantly smaller than those of WTs (Fig. 4B, **P<0.01). At 10 dpf, the head area of the sox11am/m mutants was much smaller than that of the WTs (***P<0.001), as was the pupillary distance (Fig. 4C, **P<0.01). The paraffin section indicated the brain of sox11am/m mutant was smaller than WT including the transverse and longitudinal sections (Fig. 4D, E ***P<0.001, ****P<0.0001). These data conformably indicated that the sox11am/m mutants had microcephaly defects.
To investigate why sox11am/m mutants display developmental delay, the myoblast fusion of the larvae was first examined. Larvae at 48 hpf were stained with phalloidin, and the results showed no significant difference between sox11am/m mutants and WT larvae regarding the myoblast fusion (Fig. 4F). Furthermore, the muscles of adult zebrafish were studied using paraffin sections. The results showed that there was no obvious difference in muscle development among sox11am/m mutant with curve body, sox11am/m mutant with straight body and WTs (Fig. 4G).
To further determine whether the sox11am/m mutant growth defect was caused by the skeleton, Alcian blue staining was conducted to observe cartilage development (Fig. 4H). We noted that the pectoral fin development of sox11am/m mutant zebrafish was clearly slower than that of WTs and sox11bm/m mutants. Then, the bone development of adult zebrafish was checked using micro-CT. Figure 4I shows that some sox11am/m mutants presented curved spines, whereas the bones of WTs and sox11bm/m mutants were normal. A total of 33% (±2%) of sox11am/m mutants presented severely curved spines (Fig. S5, ****P<0.0001).
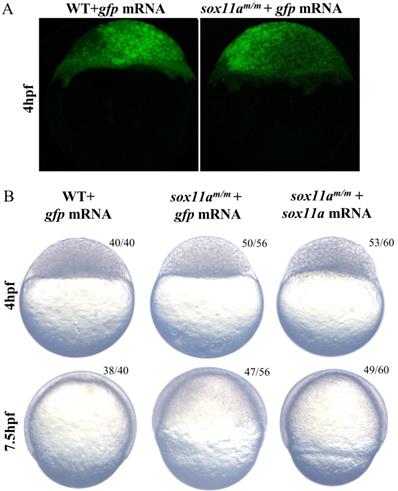
The sox11am/m development delay could be rescued by sox11a mRNA
To confirm whether the phenotype was caused by the disruption of sox11a, the sox11am/m mutants were rescued by the injection of sox11a mRNA, while the two control groups were set by injection of exogenous gfp mRNA (Fig. 5A). The results showed that sox11am/m mutants injected with zebrafish sox11a mRNA developed significantly faster than the mutant group injected with gfp mRNA, but its process was still a little slower than that of the WT group injected with gfp mRNA. At 7.5 h after injection, we observed that the sox11am/m mutants injected with sox11a mRNA developed into 65% epiboly stage, while the sox11am/m mutants and WTs injected with gfp mRNA developed into 50% and 75% epiboly, respectively (Fig. 5B).
Targeted disruption of sox11a in zebrafish affects the expression of genes related to cartilage and bone development
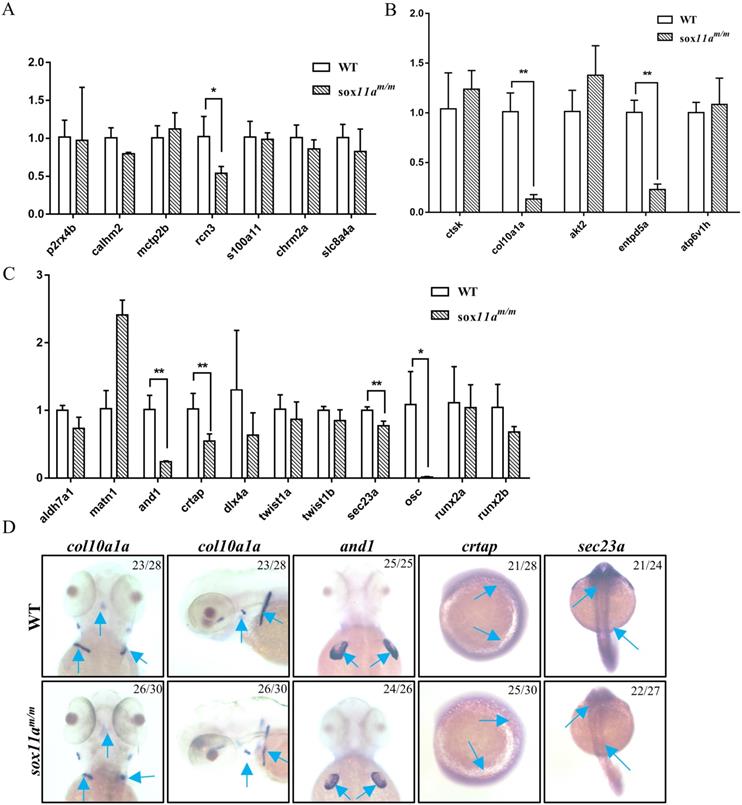
To further elucidate the mechanism of disruption of sox11a affecting skeleton development, the expression level of genes related to calcium signaling pathway, cartilage development, and bone development in adult zebrafish were examined by qPCR. We found no significant changes in the calcium signaling pathway, except for a slight decrease in rcn3 expression levels in sox11am/m mutant zebrafish (Fig. 6A). In the bone development-related pathway, we detected significant downregulation in the expression levels of col10a1a and entpd5a (Fig. 6B). Moreover, there were also significantly lower expression levels of and1, crtap, and sec23a in the sox11am/m mutant, which is involved in cartilage development (Fig. 6C). Furthermore, the expression levels of col10a1a, and1, crtap, and sec23a were checked in embryos by whole-mount in situ hybridization. The results indicated that the expression levels of these genes were all downregulated, which is consistent with the qPCR experimental results (Fig. 6D). These data suggested that skeletal dysplasia in sox11am/m mutant zebrafish was mainly a result of the abnormal expression levels of skeletal-related signaling pathways.
Discussion
In the present study, sox11am/m and sox11bm/m mutant zebrafish were generated via CRISPR/Cas9 for the first time. The results herein obtained suggested that the sox11am/m mutant zebrafish could survive and develop into adults but display growth and developmental delay, besides microcephaly with typical Coffin-Siris syndrome characteristics. However, no differences were found between the sox11bm/m mutant zebrafish and WT zebrafish.
The sox11am/m mutant zebrafish exhibit microcephaly, normal muscle and curved spine. A: The head area and pupillary distance of sox11am/m mutant were significantly smaller than WT. Scale bar, 500 µm. B: The head area and pupillary distance of sox11am/m mutants were significantly smaller than that of WT in 5 dpf (**P<0.01). C: The head area and pupillary distance of sox11am/m mutants were significantly smaller than that of WT in 10 dpf (**P<0.01, ***P<0.001). D: The brain section of sox11am/m mutant at 5 dpf was obviously smaller than WT including the transverse and longitudinal sections. Scale bar, 100 µm. E: Statics of the area of the brain section. F: The myoblast fusion in 48 hpf larvae. (Cyan: DAPI; Magenta: Phalloidin.) Scale bar, 25 µm. G: Arrangement of muscle fibers of muscles in adult zebrafish. sox11am/m Curve spine: the mutant with curved spine; sox11am/m Straight spine: the mutant without curved spine. Scare bar, 50 µm. H: The pectoral fin development of sox11am/m mutant zebrafish obviously lagged behind that of WT and sox11bm/m mutant. I: Micro-CT of adult zebrafish. Some sox11am/m mutant had curved spine compared with WT while all the sox11bm/m mutants were normal.
Partial rescue of development delay by sox11a mRNA injection. A: The sox11am/m mutant and WT could produce green fluorescence after injection of gfp mRNA, which indicated the microinjection was effective. B: The sox11am/m mutant was rescued by sox11a mRNA while the injection of gfp mRNA as controls. The sox11am/m mutant injected with zebrafish sox11a mRNA could be partially rescued at 4 hpf and 7.5 hpf.
The sox11am/m mutant zebrafish possessed characteristics of human Coffin-Siris syndrome
In clinical statistics, human Coffin-Siris syndrome is related to the mutation of SOX11 [7]. However, Sox11 homozygous mutant mice normally die soon after birth [10], which hinders the study of the pathogenic mechanism of Coffin-Siris syndrome using mice as models. Several researchers have attempted to build disease models using the morpholino technique to knock down Sox11 for further research. Hempel et al. used morpholino knockdown Sox11 in Xenopus laevis, which resulted in a significant reduction in head area and interpupillary distance compared with controls [7]. Tsurusaki et al. injected sox11a or sox11b morpholino into zebrafish embryos, and the injected embryos had smaller heads and significantly higher mortality rates compared with the control embryos [8]. Nevertheless, it is known that morpholino itself is somewhat toxic and can therefore affect the embryo development [13]. In addition, the morpholino technique is only suitable for knockdown of target genes during embryonic development. Therefore, neither MO Xenopus laevis nor MO zebrafish are ideal models for Coffin-Siris syndrome.
We herein generated heritable sox11am/m and sox11bm/m mutants using CRISPR/Cas9. The homozygous mutant was obtained by self-crossing, and this method was able to eliminate the maternal effect and produce maternal and zygotic deficiency mutant lines. The sox11am/m mutant zebrafish obtained had growth deficiency, pectoral fin development delay, and microcephaly; these features are characteristic of human Coffin-Siris syndrome. Some sox11am/m mutants even had a deformed skeleton, which was clearly observed in the Micro-CT scans; however, this effect was not obvious in juvenile fish under 20 dpf. Unlike the lethal effect of Sox11 knockout in mice, the sox11am/m mutant and sox11bm/m mutant zebrafish herein obtained were viable. This is the first time that viable sox11 mutants have been obtained in all animals. Thus, the sox11am/m mutant zebrafish may be helpful for finding the target gene of sox11 and it also is an adequate model for further studying the pathogenic mechanisms of Coffin-Siris syndrome.
The genes related to cartilage and bone development were downregulated, which affected the development of the sox11am/m mutant
Skeletons and muscles are of vital importance in body shape development. The length of bones and the number of muscles play a decisive role in human height and weight. Many growth-related diseases in humans are caused by abnormal skeletal and muscular development. Zebrafish skeletal development patterns conform to classical skeletal development patterns [14]. The process of skeletal development is easier to observe in zebrafish than in mammals. Therefore, many human skeletal disease models have been established based on studies on zebrafish [15]. For example, the sec23a mutation in humans causes growth delay, and its mutation in zebrafish causes a shorter body length, which has been used as a disease model for humans [16-18].
The sox11am/m mutant zebrafish presented growth and developmental delay, and this phenomenon could last to adulthood. Muscle development in the sox11am/m mutants presented no obvious abnormalities such as in the number of myoblast nuclei and the arrangement of muscle fibers (Fig. 4F, G). Since the skeleton is another important factor affecting growth in development, sox11am/m mutant skeletal development was subsequently examined (Fig. 4H, I). Alcian blue staining indicated that cartilage development in 5 dpf sox11am/m mutant was slower than in WTs. Moreover, the analysis of the Micro-CT scans suggested that some 20 dpf mutants developed bone malformations. Hence, we believe that the defects in the sox11am/m mutants were mainly related to skeleton development. We further found that the expression levels of genes related to cartilage development were significantly downregulated (Fig. 6). A previous study showed that and1 was mainly related to zebrafish fin regeneration [19]. Other studies showed that disruption of sec23a in zebrafish leads to a shorter body length [20] and that crtap plays an important role in bone formation [21]. In addition, bone development-related genes, including col10a1a and entpd5a, were significantly downregulated. As previously reported, col10a1a was considered to be related to osteoblast development [22], whereas entpd5a, which is expressed in zebrafish osteoblasts, was considered as being related to skeletal mineralization [23]. These data suggest that sox11a mainly affects the growth and development of zebrafish by regulating the expression of genes related to skeleton development.
Analysis of the expression of genes related to skeleton development by qPCR and whole-mount in situ hybridization. A: Expression profile of genes involved in calcium signaling pathway (*P<0.05). B: Expression profile of genes involved in cartilage development (**P<0.01). C: Expression profile of genes involved in bone development (*P<0.05, **P<0.01). The qPCR data analysis use by 2-∆∆CT and ꞵ-actin was chosen as reference gene. D: Analysis of the expression of genes involved in skeleton development by whole-mount in situ hybridization staining for zebrafish col10a1a, and1, crtap and sec23a during embryogenesis.
The sox11bm/m mutant zebrafish didn't show any phenotype including growth or development defects. Nevertheless, this did not mean that we failed to disrupt sox11b or the sox11b was unfunctional. we had detected the down-regulated of sox11b in sox11bm/m mutant by qPCR and whole-mount in situ hybridization, which may be mediated by nonsense-mediated mRNA decay [24,25]. The expression of sox11a in sox11bm/m mutant was lower than WT (Fig. S6), and the expression of sox11b in sox11am/m mutant was also lower than WT (Fig. S7). Based on our data, we inferred the sox11a and sox11b may function together. Moreover, the process of generating sox11am/msox11bm/m double mutant further indicated the double mutant was associated with a more severe phenotype than either of the single sox11am/m or sox11bm/m mutants. There was only one copy of the gene in mice, human and Xenopus laevis [7,9,10]. In particular, the homozygote Sox11 in mice lead to embryo lethality [10], which was resemble to the double mutant of sox11a and sox11b in zebrafish.
In conclusion, the disruption of sox11a affects skeleton development, which leads to the sox11am/m mutant possessing a series of Coffin-Siris syndrome features. To the best of our knowledge, this is the first study to analyze the function of sox11 by establishing a sox11am/m mutant model in vertebrates. Moreover, the sox11am/m mutant zebrafish is expected to be a disease model of Coffin-Siris syndrome, which may be expedient to study the pathogenesis mechanism and identify target drugs for this syndrome.
Abbreviations
dpf: days post fertilization; hpf: hours post fertilization; WT: wild type; MO: morpholino.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was financially supported by National Key R&D Program of China (2018YFA0801000), the National Natural Science Foundation of China (31721005), and the Chinese Academy of Sciences (2019FBZ05). The authors acknowledge with great appreciation the help of Ms. Ming Li for care of animals. We would like to thank Ms. Fang Zhou and Mr. Xin Wang at The Analysis and Testing Center of Institute of Hydrobiology, Chinese Academy of Sciences for their assistance with providing confocal services and Micro-CT analysis.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129-44
2. Badis G, Berger MF, Philippakis AA. et al. Diversity and complexity in DNA recognition by transcription factors. Science (New York, NY). 2009;324:1720-3
3. Kondoh H, Kamachi Y. SOX-partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. Int J Biochem Cell Biol. 2010;42:391-9
4. Dy P, Penzo-Mendez A, Wang H. et al. The three SoxC proteins-Sox4, Sox11 and Sox12-exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101-17
5. Bhattaram P, Penzo-Mendez A, Sock E. et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 2010;1:9
6. Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42:425-8
7. Hempel A, Pagnamenta AT, Blyth M. et al. Deletions and de novo mutations of SOX11 are associated with a neurodevelopmental disorder with features of Coffin-Siris syndrome. J Med Genet. 2016;53:152-62
8. Tsurusaki Y, Koshimizu E, Ohashi H. et al. De novo SOX11 mutations cause Coffin-Siris syndrome. Nat Commun. 2014;5:4011
9. Turan S, Boerstler T, Kavyanifar A. et al. A novel human stem cell model for Coffin-Siris Syndrome like syndrome reveals the importance of SOX11 dosage for neuronal differentiation and survival. Hum Mol Genet. 2019;28:2589-99
10. Sock E, Rettig SD, Enderich J. et al. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24:6635-44
11. Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). Eugene, USA: University of Oregon Press. 1995
12. Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3:59-69
13. Czopka T, Lyons DA. Dissecting mechanisms of myelinated axon formation using zebrafish. Methods Cell Biol. 2011;105:25-62
14. Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191-220
15. Luderman LN, Unlu G, Knapik EW. Zebrafish Developmental Models of Skeletal Diseases. Curr Top Dev Biol. 2017;124:81-124
16. Boyadjiev SA, Fromme JC, Ben J. et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38:1192-7
17. Fromme JC, Ravazzola M, Hamamoto S. et al. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623-34
18. Lang MR, Lapierre LA, Frotscher M. et al. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet. 2006;38:1198-203
19. Konig D, Page L, Chassot B. et al. Dynamics of actinotrichia regeneration in the adult zebrafish fin. Dev Biol. 2018;433:416-32
20. Khoriaty R, Hesketh GG, Bernard A. et al. Functions of the COPII gene paralogs SEC23A and SEC23B are interchangeable in vivo. Proc Natl Acad Sci USA. 2018;115:E7748-E57
21. Trancozo M, Moraes MVD, Silva DA. et al. Osteogenesis imperfecta in Brazilian patients. Genet Mol Biol. 2019;42:344-50
22. Azetsu Y, Inohaya K, Takano Y. et al. The sp7 gene is required for maturation of osteoblast-lineage cells in medaka (Oryzias latipes) vertebral column development. Dev Biol. 2017;431:252-62
23. Huitema LF, Apschner A, Logister I. et al. Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish. Proc Natl Acad Sci U S A. 2012;109:21372-7
24. Shum EY, Jones SH, Shao A. et al. The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay. Cell. 2016;165:382-95
25. Ma Z, Zhu P, Shi H. et al. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019;568:259-63
Author contact
![]() Corresponding author: Dr. Wei Hu, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China. Tel: 86-27-68780051; Fax: 86-27-68780051; E-mail: huweiac.cn.
Corresponding author: Dr. Wei Hu, State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China. Tel: 86-27-68780051; Fax: 86-27-68780051; E-mail: huweiac.cn.

 Global reach, higher impact
Global reach, higher impact