10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(3):897-910. doi:10.7150/ijbs.54055 This issue Cite
Research Paper
Evaluation and characterization of HSPA5 (GRP78) expression profiles in normal individuals and cancer patients with COVID-19
1. Key Laboratory of Epigenetics and Oncology, the Research Center for Preclinical Medicine, Southwest Medical University, Luzhou 646000, Sichuan, China.
2. Department of Pathology, the Affiliated Huaian No. 1 People's Hospital of Nanjing Medical University, Huai'an 223300, Jiangsu, China.
3. PG Department of Biotechnology, Teresian College, University of Mysore, Mysore 570011, Karnataka, India.
4. Department of Thoracic Surgery, The Third Affiliated Hospital of Southern Medical University, Guangzhou 510000, China.
5. Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, Houston 77030, Texas, USA.
*Equal contributions
Received 2020-10-3; Accepted 2021-2-5; Published 2021-2-18
Abstract
HSPA5 (BiP, GRP78) has been reported as a potential host-cell receptor for SARS-Cov-2, but its expression profiles on different tissues including tumors, its susceptibility to SARS-Cov-2 virus and severity of its adverse effects on malignant patients are unclear. In the current study, HSPA5 has been found to be expressed ubiquitously in normal tissues and significantly increased in 14 of 31 types of cancer tissues. In lung cancer, mRNA levels of HSPA5 were 253-fold increase than that of ACE2. Meanwhile, in both malignant tumors and matched normal samples across almost all cancer types, mRNA levels of HSPA5 were much higher than those of ACE2. Higher expression of HSPA5 significantly decreased patient overall survival (OS) in 7 types of cancers. Moreover, systematic analyses found that 7.15% of 5,068 COVID-19 cases have malignant cancer coincidental situations, and the rate of severe events of COVID-19 patients with cancers present a higher trend than that for all COVID-19 patients, showing a significant difference (33.33% vs 16.09%, p<0.01). Collectively, these data imply that the tissues with high HSPA5 expression, not low ACE2 expression, are susceptible to be invaded by SARS-CoV-2. Taken together, this study not only indicates the clinical significance of HSPA5 in COVID-19 disease and cancers, but also provides potential clues for further medical treatments and managements of COVID-19 patients.
Keywords: cancer, HSPA5, prognostics, SARS-CoV-2, transcriptomics.
1. Introduction
Heat shock protein family A (Hsp70) member 5 (HSPA5) (OMIM: 138120), also called binding immunoglobulin protein (BiP) or glucose regulating protein 78 (GRP78), is a protein, that in humans, is encoded by the HSPA5 gene. HSPA5 is commonly positioned in the endoplasmic reticulum (ER). When the ER is stressed, HSPA5 can translocate to the nucleus, the mitochondria and cell surface complexing with other proteins. On the cell surface, HSPA5 plays a multi-functional role in cell proliferation, cell viability, apoptosis, and regulation of innate and adaptive immunity [1]. HSPA5 is the master chaperone protein for unfolded protein response for ER function when unfolded or misfolded proteins accumulated [2]. HSPA5 involves in the correct folding and degradation of misfolded proteins through interacting with DNAJC10/ERdj5, facilitating DNAJC10/ERdj5 release from substrates. Dysregulations of these stress proteins including HSPA5 are associated with many human diseases including cancers, immunological diseases, cardiovascular diseases, neurodegenerative diseases, obesity, stroke and infectious diseases [3-6]. Targeting HSPA5 may be potential in therapy for human diseases [7, 8] as well as COVID-19 (Coronavirus Disease 2019) [9-11].
SARS-Cov-2 (severe acute respiratory syndrome coronavirus 2), which causes the disease of COVID-19, is a member of beta coronaviruses like the previous coronaviruses SARS (severe acute respiratory syndrome) and MERS (middle east respiratory syndrome). COVID-19 virus causes global pandemic events since the first outbreak in Wuhan of Hubei, China in late December 2019. As of the January 11, 2021, the total confirmed cases are 90,833,894, and deaths are 1,942,974 worldwide based on the report from the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (https://coronavirus.jhu.edu/). The host cell entry of coronavirus was regulated by the viral spike protein ( ̴1300 amino acids), originate in homotrimeric state over the virion particle and characterize coronaviruses [12, 13]. Various host cell receptors or entry related-proteins are identified for different coronaviruses such as heparan sulfate proteoglycans, angiotensin-converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), aminopeptidase N, HSPA5, furin, and O-acetylated sialic acid [14-17]. Molecular chaperones are involved in multiple pathophysiological processes including viral infection by spike protein attacks [18]. The cell-surface receptor HSPA5 is susceptible to viral recognition through the substrate-binding domain (SBD), thereby mediating the virus entry into the cells [10, 19, 20]. The spike binding site to HSPA5 is predicted by molecular model docking and structural bioinformatics, and revealed that the binding is more favorable at the regions III (C391-C525) and IV (C480-C488) in the spike protein [15], and region IV is the major driving force for HSPA5 binding which may be useful for developing therapeutics specific against COVID-19. Indeed, recently virtual screening studies revealed that known HSPA5 inhibitors interfere with SARS-Cov-2 infection [21]. Thus HSPA5 may be a receptor for SARS-CoV-2 attachment and entry [10, 15, 22, 23]. The expression levels of HSPA5 were found to be higher in the SARS-COV-2-positive group compared to the other groups [24]. Pep42, a cyclic peptide, binds to HSPA5 at the surface of cancer cells [25, 26].
The expression levels of HSPA5 in different tissues might closely related to the susceptibility and severity of the viral infection. Organ dysfunctions, such as shock, acute cardiac injury, acute respiratory distress syndrome (ARDS), acute kidney injury (AKI), and death can occur in severe events of COVID-19 disease [27, 28]. Older people with comorbidities, such as high blood pressure, diabetes, cardiovascular disease and cerebrovascular disease have been reported to affect the COVID-19 severity [29, 30].
Patients with malignant cancers affected survival status and gene expression in tumor tissues. The incidence of malignant cancers is getting higher and higher, and recently was found to be the common comorbidity of COVID-19. Although one study showed that there was no significant difference in the severity of COVID-19 in cancer patients, as a receptor of SARS-Cov-2 [31], dysregulation of HSPA5 expression in cancer patients' tissues, particularly in the lungs, should affect the susceptibility and severity of this virus infection. Targeting HSPA5 may help to develop and design novel therapeutic strategies against virus infections [32] including SARS-Cov-2, which might also associate with human carcinoma between endoplasmic reticulum stress and anti-viral activities [23, 33]. Hence understanding of the HSPA5 expression profiles on different normal tissues and malignant tumors is important. But the tumor patients with its receptor expression files for HSPA5 in this outbreak have not been reported. In this study, the differences in HSPA5 expression in various types of normal and cancer tissues were evaluated. The influences of these differences on the impacts of SARS-CoV-2 infections were dissected. The cancer patients with COVID-19 were also estimated to assess the susceptibility and severity.
2. Materials and methods
2.1. Sources for data analysis and ethical concerns
The mRNA and protein expressions for HSPA5 from different normal tissues were obtained in the database of the Human Protein Atlas (HPA) (https://www.proteinatlas.org/ENSG00000044574-HSPA5) [34, 35]. The immunohistochemistry (IHC) or immunofluorescence (IF) images of HSPA5 (Ensembl ID: ENSG00000044574.7) were also gained from the HPA database (https://www.proteinatlas.org/ENSG00000044574-HSPA5/cell), (https://www.proteinatlas.org/ENSG00000044574-HSPA5/pathology), or (https://www.proteinatlas.org/ENSG00000044574-HSPA5/cell#human), respectively [36, 37]. The expressions of HSPA5 were verified using Genotype Tissue Expression (GTEx) projects. The Gene Expression Profiling Interactive Analysis (GEPIA) dataset (http://gepia.cancer-pku.cn) or GEPIA 2 (http://gepia2.cancer-pku.cn/#index), an updated and enhanced version of GEPIA, which were developed recently [38], and ONCOMINE (https://www.oncomine.org), were used to compare the expressions between tumors and normal tissues. FANTOM5 database come from https://fantom.gsc.riken.jp/5/. The NCBI database (https://www.nih.gov/) was used. All datasets and clinical data for COVID-19 patients infected with SARS-CoV-2 were retrieved from the published literatures with statements of written informed consent. Thus no local ethics committee was required to approve this study.
2.2. Homology analysis
Homologs for HSPA5 were conducted by the NCBI program (https://www.ncbi.nlm.nih.gov/homologene/?term=Homo+sapiens+HSPA5) [39].
2.3. HPA analysis for HSPA5
The HSPA5 expressions in mRNA and protein were analyzed differentially in human normal and tumor tissues from the HPA database, which includes IHC-based expression for approximately 20 different types of common cancers in 216 cancer patients (maximum 12 patients in a group) [37]. The mRNA levels for HSPA5 in different normal tissues were obtained from the consensus datasets of three sources (HPA, GTEx and FANTOM5). Consensus normalized expression levels for 54 tissue types and 7 blood cell types were created from the above three datasets with the normalization pipeline (https://www.proteinatlas.org/about/assays+annotation#normalization_rna). Protein expression data were shown for each of the 44 normal tissues. Two antibodies for HSPA5 (cat #: CAB005221, sc-1050, Santa Cruz Biotechnology; or cat #: HPA038845, Sigma-Aldrich) were used for IHC staining in these data [40].
2.4. GEPIA analysis for HSPA5 and verification
The mRNA expressions of HSPA5 in tumors and normal tissues were analyzed in the GEPIA dataset, for analyzing the RNA sequencing (RNA-seq) expression data of 9,736 tumors and 8,587 normal samples from the Cancer Genome Atlas and GTEx projects, using a standard processing pipeline [38]. The gene expressions of HSPA5 in cancers and those in normal samples were verified by using ONCOMINE databases. HSPA5 expressions for samples in overall survival (OS) analysis were divided into high and low of two groups using a median expression, and analyzed by a Kaplan-Meier survival plot using the log-rank test. Logrank p < 0.01 was considered as significant differences.
2.5. Systematic reviews of malignant tumors in COVID-19 patients infected with SARS-CoV-2
We searched PubMed, Medline, and Google Scholar on November 25, 2020 from published studies describing the clinical characteristics of COVID-19 due to SARS-CoV-2 and cancers. The search terms “cancer” and “2019-nCoV” or “COVID-19” with no time restrictions were performed. The related works of literature were screened and analyzed, clinical signs and symptoms caused by COVID-19 and cancers were collected, and studies describing patients' malignant cancer status were conducted. The number of patients and the rate of severity combined with malignant cancers were calculated. Studies of incomplete symptom descriptions were excluded. The patient severe events were defined as the admission to ICU, requiring mechanical ventilation, or death of COVID-19. All the selected articles were analyzed by two independent investigators.
3. Results
3.1. HSPA5 is highly conserved
Homologs of the HSPA5 protein showed that it is highly conserved in different species, including H.sapiens, chimpanzee, Rhesus monkey, mouse, dog, cow, rat, chicken, zebrafish, fruit fly, mosquito, C.elegans, S.cerevisiae, K.lactis, E.gossypii, S.pombe, M.oryzae, N.crassa, A.thaliana, rice, and frog (Supplementary figure 1A&B). This implied that HSPA5, similar to ACE2 in animals of different species [39, 41], have the potentials to bind to the receptor binding domain (RBD) of the spike glycoprotein, making it a probable natural host of SARS-CoV-2.
3.2. Expression of HSPA5 in normal tissues
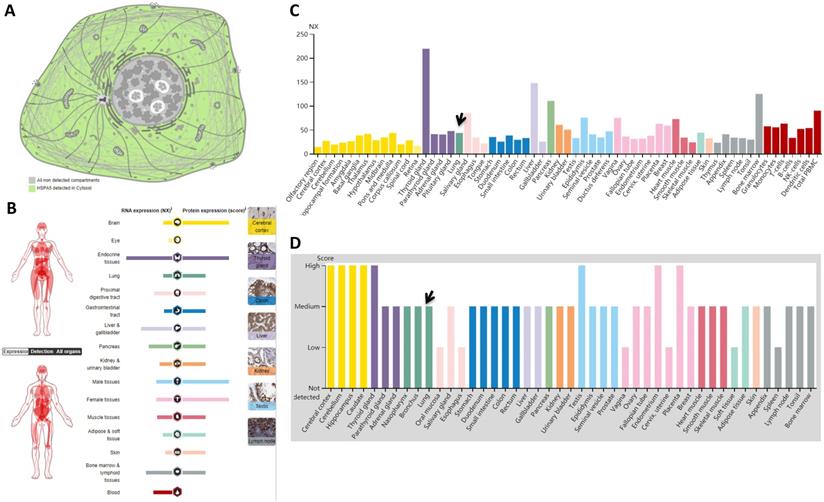
Subcellular locations from HPA data revealed that HSPA5 is localized to the cytoplasm (Fig. 1A). Expression of HSPA5 in RNA level showed low tissue specificity, with highest in the thyroid gland (NX: 219.3) and lowest in the olfactory region (NX: 14.3) (Fig. 1B&C), and in protein level showed cytoplasmic expression ubiquitously, highly abundant in immune, neuronal cells and thyroid follicular cells, specifically from 8 tissues of the cerebral cortex, cerebellum, hippocampus, caudate, thyroid gland, testis, endometrium, and placenta (Fig. 1B&D). The other 31 tissues including the lungs showed medium levels of protein, and only 6 tissues showed low levels (Fig. 1D).
Localizations and expressions of HSPA5 in normal tissues. A. Cellular localization of the HSPA5 protein. Green in color indicates HSPA5 detected in cytoplasm, whereas gray in color indicates the absence. B. The summary of mRNA and protein expressions of HSPA5. Color-coding columns are based on tissue groups, each consisting of tissues with functional features in common. The respective images for normal tissues with staining of HSPA5 protein in the HPA (scale bar 200 µm). C. The mRNA expressions of HSPA5 in normal tissues. Consensus dataset of mRNA level are derived from HPA dataset, GTEx dataset, and FANTOM5 dataset. A NX value of 1.0 is defined as a threshold for HSPA5 mRNA expression. D. The HSPA5 protein expressions in normal tissues from the HPA. Protein expression data are shown for each of the 44 tissues. Arrows indicate the lung tissue. HPA, Human Protein Atlas.
HSPA5 RNA expression in different datasets
| Dataset | Value | Unit |
|---|---|---|
| HPA | 247.6 | pTPM |
| GTEx | 275.1 | pTPM |
| FANTOM5 | 449.0 | Scaled Tags Per Million |
| Consensus | 43.6 | NX |
Note: pTPM: protein-transcripts per million; NX: normalized expression.
3.3. Expression of HSPA5 in normal lungs of humans
The main route of transmission of SARS-CoV-2 is by droplets from the respiratory tract, thus causing the severe acute respiratory syndrome. The expression levels of receptors in the lungs are important. However, from our previous study and other studies, ACE2 expression was very low in human lungs, showing moderate expression in the alveolar macrophages, and a few in the type I alveolar epithelial cells and type Ⅱ alveolar epithelial cells, but most of the typeⅡalveolar epithelial cells were ACE2 negative [39, 42]. Thus we then investigated HSPA5 mRNA expression on normal human lungs and found that, in the lung tissues, the mRNA level shows the NX value 43.6 (Fig. 1C, arrow) and the protein level is medium (Fig. 1D, arrow). This consensus 43.6 NX (Table 1) was derived from databases of LUNG- HPA RNA-seq, LUNG - GTEx RNA-seq (Supplementary figure 2A), and LUNG - FANTOM5 CAGE (Supplementary figure 2B).
The comparison of mRNA expressions of ACE2 and HSPA5 were conducted through analyzing datasets from HPA, GTEx, and FANTOM5 in human normal lungs, and found that mRNA levels of HSPA5 is 54.4-fold higher than that of ACE2 mRNA levels (Supplementary figure 2C), demonstrating that, in addition to ACE2, HSPA5 might play very important roles for SARS-CoV-2 entry.
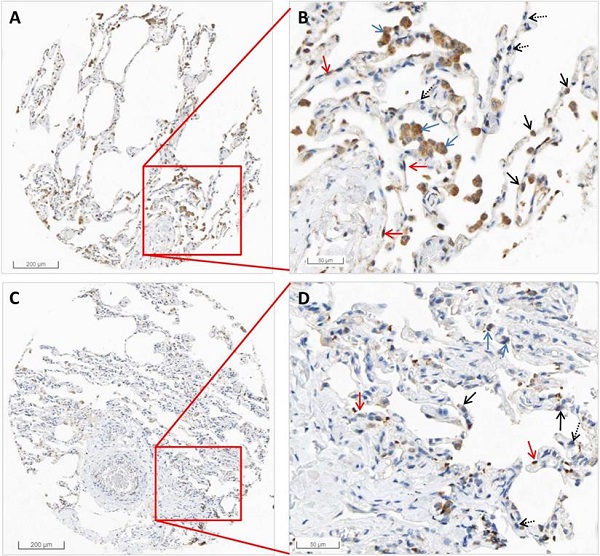
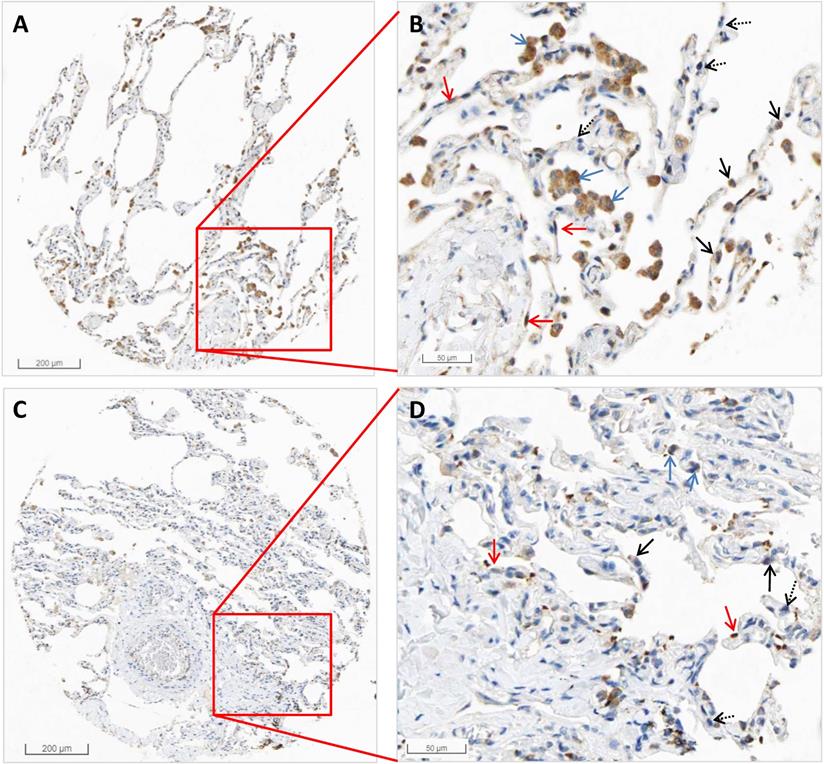
From the HPA RNA-seq data, we found that the mRNA expression of HSPA5 in pneumocytes accounts for 31.67%, endothelial cells 28.33%, macrophages 10%, bronchial epithelium 5%, and other cell types 25%. The results are shown in Table 2. Protein expression of HSPA5 by IHC in both macrophages and pneumocytes of lungs were medium. The representative IHC images of normal lung tissue are shown in Figure 2. From these results, we found that HSPA5 expressions using the HSPA5 antibody (cat# CAB005221) in alveolar macrophages are high and are mainly located in the cytoplasm and a few in the nucleus (Fig. 2B&D, blue arrows). Intensive positive staining of HSPA5 was localized at cytoplasm of type I alveolar epithelial cells (Fig. 2B&D, red arrows) and type Ⅱ alveolar epithelial cells (Fig. 2B&D, black arrow); but some of the typeⅡ alveolar epithelial cells were HSPA5 negative stained (Fig. 2B&D, dashed black arrows). Interestingly, different antibodies from HPA project showed slightly different results and found that the HSPA5 antibody (cat #: HPA038845) showed high expression in both macrophages and pneumocytes (Data not shown). Collectively, these IHC results for protein levels are consistent with those by RNA-seq for mRNA levels of HSPA5, showing much higher than that of ACE2 [39, 43, 44].
HSPA5 RNA expression in different cells from lung tissue
| Cell types | Percentages (%) |
|---|---|
| Pneumocytes | 31.67 |
| Bronchial epithelium | 5.00 |
| Endothelial cells | 28.33 |
| Macrophages | 10.00 |
| Other cell types | 25.00 |
Note: Data was normalized to nine samples by HPA RNA-sequencing.
The representative images by IHC in normal tissues of the lungs of HSPA5. The protein expressions for HSPA5 from normal tissues were obtained in the database of the Human Protein Atlas (HPA) (https://www.proteinatlas.org/ENSG00000044574-HSPA5). A. IHC images from the tissue of normal lungs from a female of age 49 (Patient id: 2268). B. Enlarged picture from A. C. IHC images from the tissue of normal lungs from a male of age 21 (Patient id: 2101). D. Enlarged picture from C. Arrows in blue indicate the representative positive results for macrophages, arrows in red indicate the representative positive results for type I alveolar epithelial cells, arrows in black indicate the representative positive results for type Ⅱ alveolar epithelial cells (Fig.2C, black arrow), and arrows in dashed black indicate the representative negative staining for type Ⅱ alveolar epithelial cells. The scale bars for 200 µm and 50 µm are indicated.
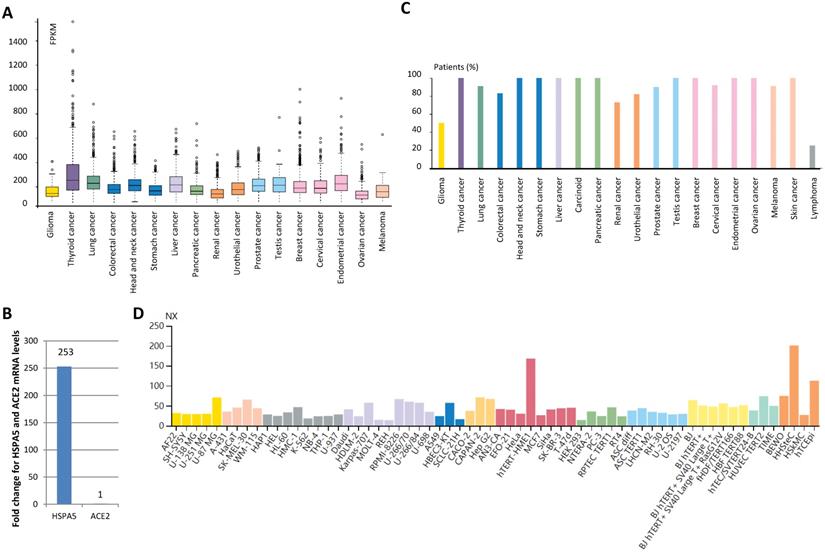
Expression value of HSPA5 in malignant tumor tissues and cancer cells. A. HSPA5 RNA expression in malignant tumor tissues of different types of cancers. B. Comparison for HSPA5 and ACE2 mRNA levels in lung cancer by analysis of TCGA dataset. C. The protein expression of HSPA5. D. HSPA5 RNA expression in the cancer cell lines. For each cancer type, color-coded bars indicate the percentage of patients (≤12 patients) with high and medium expressed of protein level. The cancer types are color-coded according to which type of normal organ the cancer originates from. The cell lines we analyzed are divided into 12 color-coded groups according to the organ from.
3.4. Expression values of HSPA5 in malignant tumors and cancer cell lines
By analyzing patient malignant tissues of different types of cancers, we found that low tissue specificity for the HSPA5 mRNA level is detected in all by RNA-seq (Fig. 3A). In order to test which genes changes more in lung cancer, we analyzed HSPA5 mRNA and ACE2 mRNA in TCGA dataset from 994 samples and found that HSPA5 mRNA levels was 253-fold than that of ACE2 (Fig. 3B), indicating that HSPA5 might play important roles for SARS-Cov-2 entry in cancer patients through lungs. This was supported partially by a systematic review of malignant cancers in COVID-19 patients that men and lung cancer patients were more likely to have COVID-19 when studied for ACE2 expression [31].
The protein expression of HSPA5 was found to be ubiquitously cytoplasmic high expressed, highly abundant in immune, neuronal cells, and thyroid follicular cells (Fig. 3C, Supplementary figure 3A~E). Malignant cells showed moderate to strong cytoplasmic staining (Supplementary figure 3F~I). Membranous staining was observed in a few cases of ovarian and pancreatic cancers (Data not shown). In the cancer cell lines, HSPA5 RNA expressions were enhanced, compared to the matched normal tissues (Fig. 3D, data not shown).
3.5. The expression of HSPA5 is higher in malignant tumors than that in matched normal samples
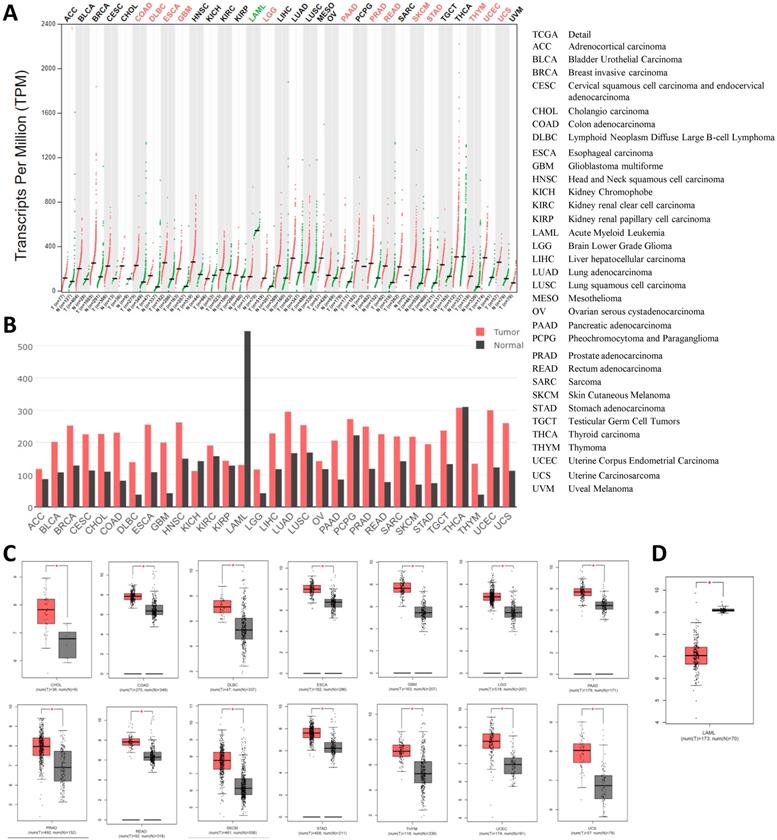
Then, we compared to the HSPA5 mRNA expression profile across all tumor samples and their paired normal tissues in 31 types of cancers using the GEPIA dataset. The results showed that all cancer tissues can express HSPA5, and the highest expression levels were noticed in thyroid carcinoma (Fig. 4A&B). The expressions of HSPA5 were significantly increased in 14 types of cancers, including cholangio carcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), brain lower grade glioma (LGG), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma(PRAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thymoma, uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS) (Fig. 4C, p<0.01). The expressions of HSPA5 were significantly decreased only in acute myeloid leukemia (LAML) (Fig. 4D, p<0.01). But the levels of HSPA5 in LAML of paired normal tissue are much higher than that of the highest one in the thyroid carcinoma tissue. The gene expressions of HSPA5 in tumors and those in normal samples were verified using the database of ONCOMINE (Data not shown). Other cancer types of tissues, except kidney chromophobe (KICH) and thyroid carcinoma (THCA), the expressions of HSPA5 were increased but not significantly (Fig. 4, A&B). In addition, HSPA5 expressions in lung cancers were upregulated compared to normal tissues from TCGA dataset (Data not shown). Altogether, those results indicated that the HSPA5 might play more important roles for SARS-Cov-2 entry in most of the cancer patients through different malignant tissues, or be prone to attack in most of the different types of cancer patients.
Expression values of HSPA5 in malignant tumors and paired normal samples. A. The HSPA5 expression profiles across all cancer samples and paired normal tissues by dot plots. B. The HSPA5 expression profiles across all cancer samples and paired normal tissues by bar plots. The height of bar represents the median expression of certain cancer type or matched normal tissue. C. HSPA5 was overexpressed in fourteen cancer types by box plots. D. HSPA5 was decreased in one cancer of LAML by box plots. HSPA5 mRNA expressions in caners and matched normal tissues were gained from the dataset of GEPIA. GEPIA, Gene Expression Profiling Interactive Analysis. *, p<0.01.
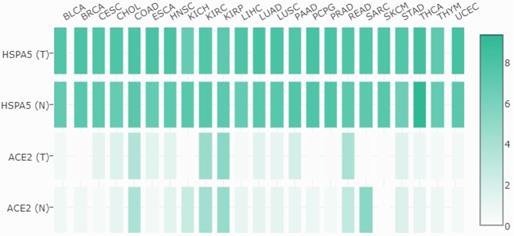
Expression comparisons between HSPA5 and ACE2 in both malignant tumors and matched normal samples in TCGA normal datasets. The cancer types are indicated on the top, and full names are shown in the figure 4. The gene for HSPA5 with/without tumor are indicated as HSPA5 (T)/HSPA5(N), and for ACE2 with/without tumor are indicated as ACE2 (T)/ACE2(N) on the left, respectively. The density of color in each block represents the median expression value of a gene in a given tissue, normalized by the maximum median expression value across all blocks. Different genes in same tumors or normal tissues can be compared in one plot, and the values can be obtained through online (http://gepia.cancer-pku.cn/detail.php?gene=HSPA5###).
3.6. The expressions of HSPA5 are much higher than those of ACE2 in both malignant tumors and matched normal samples
Then, we compared the mRNA expressions between HSPA5 and ACE2 in both malignant tumors and matched normal tissues, and the results are shown in Figure 5. From these results, we found that, unlike HSPA5, the mRNA expressions of ACE2 were significantly overexpressed in some types of cancers, including colon adenocarcinoma (COAD), kidney renal papillary cell carcinoma (KIRC), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ) and stomach adenocarcinoma (STAD), but significantly lower expressions in other types of cancers including kidney chromophobe (KICH), sarcoma, testicular germ cell tumors (TGCT) and thyroid carcinoma (THCA), than those in normal tissues (Figure 5, and data not shown), which has been reported recently for ACE2 [31]. Surprisingly, the mRNA expressions of HSPA5 are much higher than those of ACE2 in both malignant tumors and normal samples across almost kinds of cancer types (Figure 5).
3.7. Prognostic values of HSPA5 in malignant tumors
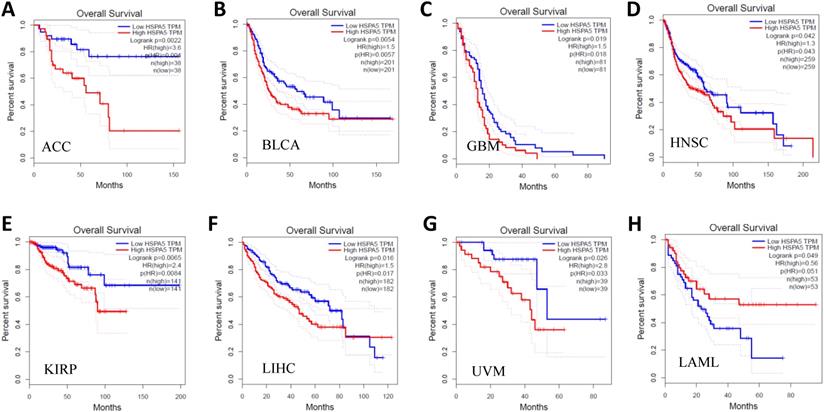
After that, we further investigated the prognostic values of HSPA5 in a pan-cancer. Cancer patients in survival analysis were divided into high expressed and low expressed groups using median HSPA5 expression and analyzed by overall survival (OS) Kaplan-Meier plots. The results are shown in Figure 6, and we found that the over-expressions of HSPA5 significantly decreased patient OS in the indicated seven types of cancers, including adrenocortical carcinoma (ACC), breast invasive carcinoma (BLCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), and uveal melanoma (UVM) (Fig. 6, A~G); whereas low expressions of HSPA5 significantly decreased patient OS only in acute myeloid leukemia (LAML) (Fig. 6, H). These results indicate that overall survivals are reduced significantly in most types of malignant tumor patients when HSPA5 is overexpressed.
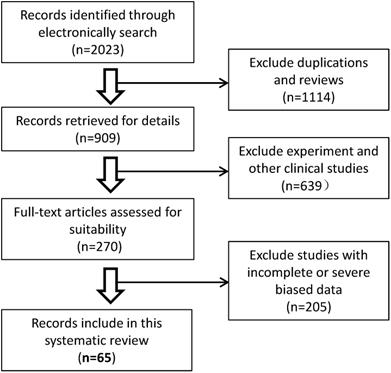
3.8. Characteristics of malignant cancer patients infected with SARS-CoV-2
The recently published studies describing the clinical characteristics of COVID-19 and malignant cancers were screened and analyzed by systematic review. Overall, sixty-five published studies that evaluated patients' malignant cancer statuses were included from 2023 potentially relevant studies in our systematic review. A schematic flow diagram for the selection of the included studies with eligible trials and exclusion criteria is shown in Fig. 7. A total of 70,874 COVID-19 cases in China, USA, Belgium, Spain, Italy, España, Korea, Iran, Poland, Turkey, Germany, France, Turkey, UK, Switzerland, and Israel, or international multicenter were included. Among them, 5,068 COVID-19 cases (7.15%, 5068/708744) had comorbidities of malignant cancer (Table 3). Among these cancer patients, men and lung cancers were more likely to have COVID-19 (Data not shown). The rate of severe events for COVID-19 with malignant cancer patients was 33.33% (1689/5068), while the rate of severe events for all patients of COVID-19 was 16.09% (11404/70874) (Table 3), which is significantly higher for malignant patients with COVID-19 disease (33.33% vs 16.09%, p<0.01), suggesting overexpression of HSPA5 might contribute to the severity of COVID-19 patients. These data are consistent with higher expression of HSPA5 in tumor tissues from different types of cancer patients, but likely through different mechanisms.
Summary of number and severity of malignant cancer patients with COVID-19
| Total cases | Malignant cancers (%) | Severe events of cancer patients | Total severe events (%) | Resources | References | |
|---|---|---|---|---|---|---|
| Yes (%) | No (%) | |||||
| 138 | 10 (7.25) | 4 (40) | 6 (60) | 36 (26.09) | China | PMID: 32031570[29] |
| 41 | 1 (2.44) | 0 | 1 | 13 (31.71) | China | PMID: 31986264[27] |
| 1590 | 18 (1.13) | 9 (50) | 9 (50) | 131 (8.24) | China | PMID: 32066541[47] |
| 641 | 105 (16.38) | 20 (19.05) | 85 (80.95) | 43 (6.71) | China | PMID: 32345594[48] |
| 1276 | 28 (2.19) | 15 (53.57) | 13 (46.43) | 56 (4.39) | China | PMID: 32224151[49] |
| 1090 | 218 (20) | 61 (27.98) | 157 (72.02) | 149 (13.67) | USA | PMID: 32357994[50] |
| 751 | 232 (30.89) | 148 (63.79) | 84 (36.21) | 314 (41.81) | China | PMID: 32479790[51] |
| 5688 | 334 (5.87) | 37 (11.08) | 297 (88.92) | 555 (9.76) | USA | PMID: 32330541[52] |
| 10486 | 892 (8.51) | 327 (36.66) | 565 (63.34) | 3094 (29.51) | Belgium | PMID: 32978251[53] |
| 1878 | 45 (2.40) | 29 (64.44) | 16 (35.56) | 192 (10.22) | Spain | PMID: 32449128[54] |
| 56 | 25 (44.64) | 9 (36) | 16 (64) | 14 (25) | Italy | PMID: 32403946[55] |
| 1069 | 36 (3.37) | 15 (41.67) | 21 (58.33) | 132 (12.35) | España | PMID: 32507536[56] |
| 334 | 167 (50) | 56 (33.53) | 111 (66.47) | 94 (28.14) | Spain | PMID: 33077708[57] |
| 188 | 53 (28.19) | 32 (60.38) | 21 (39.62 ) | 81 (43.09 ) | USA | PMID: 33043705[58] |
| 352 | 15 (4.26) | 4 (26.67) | 11 (73.33) | 24 (6.82) | Korea | PMID: 32924343[59] |
| 459 | 52 (11.33) | 14 (26.92) | 38 (73.08) | 63 (13.73) | Iran | PMID: 32908083[60] |
| 1476 | 29 (1.96) | 20 (68.97) | 9 (31.03) | 757 (51.29) | China | PMID: 32857662[61] |
| 135 | 4 (2.96) | 3 (75) | 1 (25) | 40 (29.63) | China | PMID: 32198776[62] |
| 336 | 14 (4.17) | 6 (42.86) | 8 (57.14) | 133 (39.58) | China | PMID: 32883943[63] |
| 2665 | 109 (4.09) | 32 (29.36) | 77 (70.64) | 293 (10.99) | China | PMID: 32522278[64] |
| 15 | 10 (66.67) | 7 (70) | 3 (30) | 7 (46.67) | Poland | PMID: 32769026[65] |
| 240 | 17 (7.08) | 11 (64.71) | 6 (35.29) | 120 (50) | China | PMID: 33120785[66] |
| 64 | 16 (25) | 1 (6.25) | 15 (93.75) | 12 (18.75) | Turkey | PMID: 32854573[67] |
| 210 | 5 (2.38) | 5 (100) | 0 | 87 (41.43 ) | China | PMID: 32641174[68] |
| 276 | 3 (1.09 ) | 1 (33.33) | 2 (66.67) | 14 (5.07) | China | PMID: 32727456[69] |
| 410 | 22 (5.37) | 11 (50) | 11 (50) | 95 (23.17) | Italy | PMID: 32535188[70] |
| 167 | 39 (23.35) | 16 (41.03) | 23 (58.97) | 52 (31.14) | Germany | PMID: 32931637[71] |
| 901 | 22 (2.44 ) | 1 (4.55) | 21 (95.45) | 124 (13.76) | China | PMID: 33121497[72] |
| 64 | 1 (1.56) | 1 (100) | 0 | 21 (32.81) | China | PMID: 32741931[73] |
| 1603 | 122 (7.61) | 95 (77.87) | 27 (22.13) | 192 (11.98) | Italy | PMID: 32579597[74] |
| 204 | 95 (46.57) | 38 (40) | 57 (60) | 64 (31.37) | Italy | PMID: 32910456[75] |
| 171 | 116 (67.84) | 62 (53.45) | 54 (46.55) | 73 (42.69) | China | PMID: 33194660[76] |
| 1590 | 18 (1.13) | 7 (38.89) | 11 (61.11) | 131 (8.24 ) | China | PMID: 32396163[77] |
| 217 | 112 (51.61) | 40 (35.71) | 72 (64.29) | 56 (25.81) | China | PMID: 33192519[78] |
| 2964 | 17 (0.57) | 1 (5.88) | 16 (94.12) | 239 (8.06) | Iran | PMID: 32353762[79] |
| 1385 | 98 (7.04) | 48 (48.98) | 50 (51.02) | 293 (21.16) | Spain | PMID: 33172949[80] |
| 26 | 3 (11.54) | 3 (100) | 0 | 19 (73.08) | France | PMID: 32941618[81] |
| 238 | 2 (0.84) | 2 (100) | 0 | 48 (20.17) | China | PMID: 32554861[82] |
| 154 | 5 (3.25) | 2 (40) | 3 (60) | 2 (1.3) | Turkey | PMID: 33104780[83] |
| 55 | 52 (94.55 ) | 24 (46.15) | 28 (53.85) | 25 (45.45) | UK | PMID: 32678948[84] |
| 716 | 12 (1.68) | 4 (33.33) | 8 (66.67) | 58 (8.1) | China | PMID: 33033507[85] |
| 67 | 1 (1.49) | 0 | 1 (100) | 13 (19.40) | USA | PMID: 32407719[86] |
| 69 | 4 (5.80) | 1 (25) | 3 (75) | 14 (20.29) | China | PMID: 32176772[87] |
| 170 | 23 (13.53) | 11 (47.83) | 12 (52.17) | 90 (52.94) | USA | PMID: 32946859[88] |
| 599 | 17 (2.84) | 1 (5.88) | 16 (94.12) | 83 (13.86) | China | PMID: 32716901[89] |
| 68 | 3 (4.41) | 3 (100) | 0 | 48 (70.59) | USA | PMID: 32619411[90] |
| 676 | 33 (4.88) | 16 (48.48) | 17 (51.52) | 140 (20.71) | China | PMID: 33166991[91] |
| 61 | 3 (4.92) | 1 (33.33) | 2 (66.67) | 22 (36.07) | Iran | PMID:32910458[92] |
| 1226 | 138 (11.26) | 47 (34.06 ) | 91 (65.94) | 330 (26.92) | Italy | PMID: 32707770[93] |
| 299 | 16 (5.35) | 4 (25 ) | 12 (75) | 71 (23.75 ) | Intl mctr | PMID: 32616597[94] |
| 5154 | 312 (6.05) | 66 (21.15) | 246 (78.85) | 484 (9.39) | USA | PMID: 33112411[95] |
| 323 | 5 (1.55) | 1 (20) | 4 (80) | 26 (8.05) | China | PMID: 32361738[96] |
| 2476 | 195 (7.88) | 63 (32.31) | 132 (67.69) | 320 (12.92) | USA | PMID: 33142266[97] |
| 120 | 30 (25) | 11 (36.67 ) | 19 (63.33) | 47 (39.17) | UK | PMID: 32968429[98] |
| 105 | 16 (15.24) | 11 (68.75) | 5 (31.25) | 51 (48.57) | USA | PMID: 32444880[99] |
| 57 | 24 (42.11) | 5 (20.83) | 19 (79.17) | 7 (12.28) | Switzerland | PMID: 32571972[100] |
| 1480 | 740 (50) | 140 (18.92) | 600 (81.08) | 225 (15.2) | Turkey | PMID: 32776581[101] |
| 585 | 117 (20) | 29 (24.79) | 88 (75.21) | 129 (22.05) | USA | PMID: 32986528[102] |
| 3014 | 100 (3.32) | 30 (30) | 70 (70) | 755 (25.05) | USA | PMID: 32997958[103] |
| 120 | 7 (5.83 ) | 5 (71.43) | 2 (28.57) | 30 (25) | China | PMID: 32279115[104] |
| 483 | 5 (1.04) | 0 | 5 (100) | 62 (12.84) | China | PMID: 32683596[105] |
| 961 | 21 (2.19) | 10 (47.62) | 11 (52.38) | 242 (25.18) | China | PMID: 32716553[106] |
| 49 | 6 (12.24) | 4 (66.67) | 2 (33.33) | 38 (77.55) | Israel | PMID: 32923991[107] |
| 10237 | 76 (0.74 ) | 9 (11.84) | 67 (88.16) | 228 (2.23) | Korea | PMID: 33127965[108] |
| 156 | 2 (1.28) | 0 | 2 (100) | 3 (1.92 ) | Spain | PMID: 33220760[109] |
| 70874 | 5068 (7.15)a | 1689 (33.33)b | 3379 (66.67)c | 11404 (16.09)d | Total | |
Note: a: 5068/70874; b: 1689/5068; c: 3379/5068; d: 11404/70874; Severe events include ICU and died cases; Intl mctr: International multicentre.
Prognostic values of HSPA5 in pan-cancer. The prognostic value of HSPA5 in eight cancer types from the GEPIA dataset. A~H. ACC, BLCA, GBM, HNSC, KIRP, LAML, LIHC, UVM, respectively. *, P < 0.01. The cancer types of full names are shown in Figure 4. GEPIA, Gene Expression Profiling Interactive Analysis. HR, Hazards Ratio.
Schematic flow diagram for the selection of the included studies in the systematic review.
4. Discussions
The COVID-19 pandemic became a global public health issue. Understanding the expression levels and localizations of candidate SARS-CoV-2 receptors in host tissues may provide insights into therapeutics that reduce disease spread, viral replication, disease severity or disease pathology. ACE2 has been implicated in SARS-CoV-2 viral infection [42, 45, 46]. Additional host molecules including HSPA5 may also function as receptors for SARS-CoV-2 recognition [15, 43]. Thus, the same as ACE2, HSPA5 protein would be closely related to this COVID-19 virus entry, and the distributions and expression levels of this receptor might reflect the susceptibility to the virus and viral replication. However, the impacts of HSPA5 on SARS-CoV-2 susceptibility and the characterization of malignant cancer patients in the COVID-19 outbreaks are unknown. Understanding of the HSPA5 expressions in different normal tissues and malignant tumors is important. In the current study, HSPA5 has been found to be highly expressed in almost all the normal tissues and increased in most tumor tissues, indicating that all the organs will be potentially infected, higher susceptible to SARS-CoV-2 in those with tumors. More importantly, HSPA5 mRNA levels increase 54.4 fold than that of ACE2 in normal lung, and 253 fold in lung cancer, indicating that HSPA5 should play important roles for SARS-Cov-2 entry in cancer patients through the lungs. This was supported partially by a systematic review of malignant cancers in COVID-19 patients that men and lung cancer were more likely to have the risk of COVID-19, when studied the ACE2 expression [31]. Surprisingly, the mRNA expressions of HSPA5 are much higher than those of ACE2 in both malignant tumors and normal individuals across almost all kinds of cancer types. Thus, these data implied that, comparing the SARS-CoV-2 that invaded tissues with low expressed ACE2, this virus may more likely invade the highly HSPA5 expressed tissues.
Moreover, patients with malignant tumors are usually weaker and may be more severely affected by SARS-Cov-2. Higher expression of HSPA5 significantly decreased patient survival in OS in 7 types of cancers, including ACC, BLCA, GBM, HNSC, KIRP, LIHC, UVM. Furthermore, our systematic review results indicate that 7.15% of 5,068 COVID-19 cases have malignant cancer coincidental situations, and the rate of more severe events of COVID-19 patients with malignant cancers (33.33%) presented a higher trend than that for all COVID-19 patients (16.09%) with a significant difference (33.33% vs. 16.09%, p<0.01). Since almost all cancer tissues had highly expressed HSPA5, this indicated that all tumor patients are susceptible to the SARS-CoV-2 infection, implying the clinical significance of the role of HSPA5 expression. Hence, the susceptibility of malignant cancer patients and the differences of intensity degree could be estimated by exploring HSAP5 expression. Of course, we should point out, in these studies, the number of tumor patients and the sources from different countries were not sufficient and further studies are needed to confirm our findings. The relationship between HSPA5 expression levels of different specific tumor types and the disease severity should further be explored.
Changes in HSPA5 expression levels may affect the susceptibility for virus infection and the severity of COVID-19 disease. For example, decreasing HSPA5 expression would be the potentials to prevent COVID-19, especially those with malignant tumors. Indeed, HSPA5 has been recently implied as an anticancer drug target [8, 110, 111]. We might consider the treatment potentials such as using HSPA5 inhibitors [112]. Recently virtual screening studies revealed that known HSPA5 inhibitors interferes with the infection by SARS-Cov-2[21]. Two of these drugs, Bosutinib and Ponatinib, are inhibitors of SRC and were patented as also being capable of blocking cell surface HSPA5 expression (http://www.freepatentsonline.com/y2019/0076431.html). Natural products may also interfere with SARS-CoV-2 attachment to stressed cells, which is worth of further investigation [23, 113].
5. Conclusions
In summary, our analyses showed that HSPA5 is expressed in almost all the normal tissues and elevated expression in tumor tissues. HSPA5 mRNA levels increase 253-fold than that of ACE2 in lung cancer, indicating that HSPA5 migh play more important roles for SARS-Cov-2 entry in cancer patients through the lungs. The rate of more severe events for COVID-19 patients with malignant cancers (33.33%) presented a higher trend than that for all COVID-19 patients (16.09%) with a significant difference. Malignant cancer patients are usually weaker and might be more severely affected by COVID-19. Thus, this virus seems more likely to invade tissues with highly expressed HSPA5. Decreasing HSPA5 expression will provide a strategy potentially to prevent COVID-19, especially those with malignant tumors. Collectively, this study may not only imply the clinical significance of the role of HSPA5 in COVID-19 disease and cancers, but also provide a potential clue for further medical treatments and managements of COVID-19 patients.
HSPA5: heat shock protein family A (Hsp70) member 5; BiP: binding immunoglobulin protein; GRP78: glucose regulating protein 78; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; COVID-19: coronavirus disease 2019; SARS: severe acute respiratory syndrome; MERS: middle east respiratory syndrome; ARDS: acute respiratory distress syndrome; ACE2: angiotensin-converting enzyme 2; IHC: immunohistochemistry; HPA: Human Protein Atlas; GEPIA: The Gene Expression Profiling Interactive Analysis; GTEx: Genotype Tissue Expression; OS: overall survival.
Supplementary Material
Supplementary figures.
Acknowledgements
We thank Ms. J. Cheng, S Fu for reading the manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China (81672887 and 30371493), the Joint Research Foundation of Luzhou City and Southwest Medical University (2018LZXNYD-YL01), and The Translational Medicine Foundation of Southwest Medical University (grant nos. 00031476 and 00031477) to JF.
Author contributions
JF conceived and coordinated the study. JiF, LZ, CW, JH, JZ, SS, JP, JF analyzed, and interpreted data. LZ provided pathology expertise. JF wrote the manuscript. JH, and JiF did systematic review. JF, AS and KB edited the manuscript. All of the authors approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tsai YL, Ha DP, Zhao H, Carlos AJ, Wei S, Pun TK. et al. Endoplasmic reticulum stress activates SRC, relocating chaperones to the cell surface where GRP78/CD109 blocks TGF-beta signaling. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E4245-E54
2. Pobre KFR, Poet GJ, Hendershot LM. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. The Journal of biological chemistry. 2019;294:2098-108
3. Enogieru AB, Omoruyi SI, Hiss DC, Ekpo OE. GRP78/BIP/HSPA5 as a Therapeutic Target in Models of Parkinson's Disease: A Mini Review. Advances in pharmacological sciences. 2019;2019:2706783
4. Wang J, Lee J, Liem D, Ping P. HSPA5 Gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene. 2017;618:14-23
5. Araujo N, Hebbar N, Rangnekar VM. GRP78 Is a Targetable Receptor on Cancer and Stromal Cells. EBioMedicine. 2018;33:2-3
6. Ritter A, Kreis NN, Louwen F, Yuan J. Obesity and COVID-19: Molecular Mechanisms Linking Both Pandemics. International journal of molecular sciences. 2020 21
7. Booth L, Roberts JL, Cash DR, Tavallai S, Jean S, Fidanza A. et al. GRP78/BiP/HSPA5/Dna K is a universal therapeutic target for human disease. Journal of cellular physiology. 2015;230:1661-76
8. Lu G, Luo H, Zhu X. Targeting the GRP78 Pathway for Cancer Therapy. Frontiers in medicine. 2020;7:351
9. Elfiky AA, Baghdady AM, Ali SA, Ahmed MI. GRP78 targeting: Hitting two birds with a stone. Life sciences. 2020;260:118317
10. Ha DP, Van Krieken R, Carlos AJ, Lee AS. The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. The Journal of infection. 2020;81:452-82
11. Aoe T. Pathological Aspects of COVID-19 as a Conformational Disease and the Use of Pharmacological Chaperones as a Potential Therapeutic Strategy. Frontiers in pharmacology. 2020;11:1095
12. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011-33
13. Khan RJ, Jha RK, Amera GM, Jain M, Singh E, Pathak A. et al. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2'-O-ribose methyltransferase. Journal of biomolecular structure & dynamics. 2020 p: 1-14
14. Hasan A, Paray BA, Hussain A, Qadir FA, Attar F, Aziz FM. et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of biomolecular structure & dynamics. 2020 p: 1-9
15. Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. The Journal of infection. 2020;80:554-62
16. Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. Journal of virology. 2012;86:6537-45
17. Elfiky AA. SARS-CoV-2 Spike-Heat Shock Protein A5 (GRP78) Recognition may be Related to the Immersed Human Coronaviruses. Frontiers in pharmacology. 2020;11:577467
18. Shu W, Guo Z, Li L, Xiong Z, Wang Z, Yang Y. et al. Regulation of Molecular Chaperone GRP78 by Hepatitis B Virus: Control of Viral Replication and Cell Survival. Molecular and cellular biology. 2020 40
19. Ibrahim IM, Abdelmalek DH, Elfiky AA. GRP78: A cell's response to stress. Life sciences. 2019;226:156-63
20. Chu H, Chan CM, Zhang X, Wang Y, Yuan S, Zhou J. et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. The Journal of biological chemistry. 2018;293:11709-26
21. Palmeira A, Sousa E, Koseler A, Sabirli R, Goren T, Turkcuer I. et al. Preliminary Virtual Screening Studies to Identify GRP78 Inhibitors Which May Interfere with SARS-CoV-2 Infection. Pharmaceuticals. 2020 13
22. Saghazadeh A, Rezaei N. Towards treatment planning of COVID-19: Rationale and hypothesis for the use of multiple immunosuppressive agents: Anti-antibodies, immunoglobulins, and corticosteroids. International immunopharmacology. 2020;84:106560
23. Elfiky AA. Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of biomolecular structure & dynamics. 2020 p: 1-10
24. Koseler A, Sabirli R, Goren T, Turkcuer I, Kurt O. Endoplasmic Reticulum Stress Markers in SARS-COV-2 Infection and Pneumonia: Case-Control Study. In vivo. 2020;34:1645-50
25. Kim Y, Lillo AM, Steiniger SC, Liu Y, Ballatore C, Anichini A. et al. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45:9434-44
26. Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R. et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251-4
27. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506
28. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-13
29. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J. et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama. 2020;323:1061-9
30. Mueller AL, McNamara MS, Sinclair DA. Why does COVID-19 disproportionately affect older people? Aging. 2020;12:9959-81
31. Ren P, Gong C, Ma S. Evaluation of COVID-19 based on ACE2 expression in normal and cancer patients. Open Medicine. 2020;15:613-22
32. Quimque MTJ, Notarte KIR, Fernandez RAT, Mendoza MAO, Liman RAD, Lim JAK. et al. Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. Journal of biomolecular structure & dynamics. 2020 p: 1-18
33. Banerjee A, Czinn SJ, Reiter RJ, Blanchard TG. Crosstalk between endoplasmic reticulum stress and anti-viral activities: A novel therapeutic target for COVID-19. Life sciences. 2020;255:117842
34. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419
35. Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M. et al. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28:1248-50
36. Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H. et al. A subcellular map of the human proteome. Science. 2017 356
37. Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G. et al. A pathology atlas of the human cancer transcriptome. Science. 2017 357
38. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017;45:W98-W102
39. Fu J, Zhou B, Zhang L, Balaji KS, Wei C, Liu X. et al. Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Molecular biology reports. 2020;47:4383-92
40. Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA. et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Molecular & cellular proteomics: MCP. 2008;7:2019-27
41. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444-8
42. Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Molecular systems biology. 2020;16:e9610
43. Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A. et al. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. The European respiratory journal. 2020 56
44. Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infectious diseases of poverty. 2020;9:45
45. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215-20
46. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H. et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221-4
47. Liang W, Guan W, Chen R, Wang W, Li J, Xu K. et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. The Lancet Oncology. 2020;21:335-7
48. Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z. et al. Patients with Cancer Appear More Vulnerable to SARS-CoV-2: A Multicenter Study during the COVID-19 Outbreak. Cancer discovery. 2020;10:783-91
49. Zhang L, Zhu F, Xie L, Wang C, Wang J, Chen R. et al. Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Annals of oncology: official journal of the European Society for Medical Oncology. 2020;31:894-901
50. Mehta V, Goel S, Kabarriti R, Cole D, Goldfinger M, Acuna-Villaorduna A. et al. Case Fatality Rate of Cancer Patients with COVID-19 in a New York Hospital System. Cancer discovery. 2020;10:935-41
51. Tian J, Yuan X, Xiao J, Zhong Q, Yang C, Liu B. et al. Clinical characteristics and risk factors associated with COVID-19 disease severity in patients with cancer in Wuhan, China: a multicentre, retrospective, cohort study. The Lancet Oncology. 2020;21:893-903
52. Miyashita H, Mikami T, Chopra N, Yamada T, Chernyavsky S, Rizk D. et al. Do patients with cancer have a poorer prognosis of COVID-19? An experience in New York City. Annals of oncology: official journal of the European Society for Medical Oncology. 2020;31:1088-9
53. de Azambuja E, Brandao M, Wildiers H, Laenen A, Aspeslagh S, Fontaine C. et al. Impact of solid cancer on in-hospital mortality overall and among different subgroups of patients with COVID-19: a nationwide, population-based analysis. ESMO open. 2020 5
54. Rogado J, Obispo B, Pangua C, Serrano-Montero G, Martin Marino A, Perez-Perez M. et al. Covid-19 transmission, outcome and associated risk factors in cancer patients at the first month of the pandemic in a Spanish hospital in Madrid. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2020;22:2364-8
55. Stroppa EM, Toscani I, Citterio C, Anselmi E, Zaffignani E, Codeluppi M. et al. Coronavirus disease-2019 in cancer patients. A report of the first 25 cancer patients in a western country (Italy). Future oncology. 2020;16:1425-32
56. Lara Alvarez MA, Rogado Revuelta J, Obispo Portero B, Pangua Mendez C, Serrano Montero G, Lopez Alfonso A. [COVID-19 mortality in cancer patients in a Madrid hospital during the first 3 weeks of the epidemic]. Medicina clinica. 2020;155:202-4
57. Martinez-Lopez J, Mateos MV, Encinas C, Sureda A, Hernandez-Rivas JA, Lopez de la Guia A. et al. Multiple myeloma and SARS-CoV-2 infection: clinical characteristics and prognostic factors of inpatient mortality. Blood cancer journal. 2020;10:103
58. Ramachandran P, Kathirvelu B, Chakraborti A, Gajendran M, Zhahid U, Ghanta S. et al. COVID-19 in Cancer Patients From New York City: A Comparative Single Center Retrospective Analysis. Cancer control: journal of the Moffitt Cancer Center. 2020;27:1073274820960457
59. Moon SS, Lee K, Park J, Yun S, Lee YS, Lee DS. Clinical Characteristics and Mortality Predictors of COVID-19 Patients Hospitalized at Nationally-Designated Treatment Hospitals. Journal of Korean medical science. 2020;35:e328
60. Alamdari NM, Afaghi S, Rahimi FS, Tarki FE, Tavana S, Zali A. et al. Mortality Risk Factors among Hospitalized COVID-19 Patients in a Major Referral Center in Iran. The Tohoku journal of experimental medicine. 2020;252:73-84
61. Cheng WT, Ke YH, Yang GY, Sun H, Chen Y, Ying RY. et al. Analysis of clinical features of COVID-19 in cancer patients. Acta oncologica. 2020;59:1393-6
62. Wan S, Xiang Y, Fang W, Zheng Y, Li B, Hu Y. et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. Journal of medical virology. 2020;92:797-806
63. He Y, Xie M, Zhao J, Liu X. Clinical Characteristics and Outcomes of Patients with Severe COVID-19 and Chronic Obstructive Pulmonary Disease (COPD). Medical science monitor: international medical journal of experimental and clinical research. 2020;26:e927212
64. Meng Y, Lu W, Guo E, Liu J, Yang B, Wu P. et al. Cancer history is an independent risk factor for mortality in hospitalized COVID-19 patients: a propensity score-matched analysis. Journal of hematology & oncology. 2020;13:75
65. Biernat MM, Zinczuk A, Biernat P, Bogucka-Fedorczuk A, Kwiatkowski J, Kalicinska E. et al. Nosocomial outbreak of SARS-CoV-2 infection in a haematological unit - High mortality rate in infected patients with haematologic malignancies. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2020;130:104574
66. Rao X, Wu C, Wang S, Tong S, Wang G, Wu G. et al. The importance of overweight in COVID-19: A retrospective analysis in a single center of Wuhan, China. Medicine. 2020;99:e22766
67. Basci S, Ata N, Altuntas F, Yigenoglu TN, Dal MS, Korkmaz S. et al. Outcome of COVID-19 in patients with chronic myeloid leukemia receiving tyrosine kinase inhibitors. Journal of oncology pharmacy practice: official publication of the International Society of Oncology Pharmacy Practitioners. 2020;26:1676-82
68. Kong M, Zhang H, Cao X, Mao X, Lu Z. Higher level of neutrophil-to-lymphocyte is associated with severe COVID-19. Epidemiology and infection. 2020;148:e139
69. Wei Y, Zeng W, Huang X, Li J, Qiu X, Li H. et al. Clinical characteristics of 276 hospitalized patients with coronavirus disease 2019 in Zengdu District, Hubei Province: a single-center descriptive study. BMC infectious diseases. 2020;20:549
70. Ciceri F, Castagna A, Rovere-Querini P, De Cobelli F, Ruggeri A, Galli L. et al. Early predictors of clinical outcomes of COVID-19 outbreak in Milan, Italy. Clinical immunology. 2020;217:108509
71. Shoumariyeh K, Biavasco F, Ihorst G, Rieg S, Nieters A, Kern WV. et al. Covid-19 in patients with hematological and solid cancers at a Comprehensive Cancer Center in Germany. Cancer medicine. 2020;9:8412-22
72. Zhang J, Hao Y, Ou W, Ming F, Liang G, Qian Y. et al. Serum interleukin-6 is an indicator for severity in 901 patients with SARS-CoV-2 infection: a cohort study. Journal of translational medicine. 2020;18:406
73. Huang H, Song B, Xu Z, Jiao Y, Huang L, Zhao P. et al. Predictors of coronavirus disease 2019 severity: A retrospective study of 64 cases. Japanese journal of infectious diseases. 2020
74. Bravi F, Flacco ME, Carradori T, Volta CA, Cosenza G, De Togni A. et al. Predictors of severe or lethal COVID-19, including Angiotensin Converting Enzyme inhibitors and Angiotensin II Receptor Blockers, in a sample of infected Italian citizens. PloS one. 2020;15:e0235248
75. Cattaneo C, Daffini R, Pagani C, Salvetti M, Mancini V, Borlenghi E. et al. Clinical characteristics and risk factors for mortality in hematologic patients affected by COVID-19. Cancer. 2020;126:5069-76
76. Zhang B, Xie R, Hubert SM, Yu Y, Zhang Y, Lei X. et al. Characteristics and Outcomes of 35 Breast Cancer Patients Infected With COVID-19. Frontiers in oncology. 2020;10:570130
77. Liang W, Liang H, Ou L, Chen B, Chen A, Li C. et al. Development and Validation of a Clinical Risk Score to Predict the Occurrence of Critical Illness in Hospitalized Patients With COVID-19. JAMA internal medicine. 2020;180:1081-9
78. Zhang B, Yu Y, Hubert SM, Zhang Y, Lu J, Liu S. et al. Prognostic Value of Pro-Inflammatory Neutrophils and C-Reactive Protein in Cancer Patient With Coronavirus Disease 2019: A Multi-Center, Retrospective Study. Frontiers in pharmacology. 2020;11:576994
79. Nikpouraghdam M, Jalali Farahani A, Alishiri G, Heydari S, Ebrahimnia M, Samadinia H. et al. Epidemiological characteristics of coronavirus disease 2019 (COVID-19) patients in IRAN: A single center study. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2020;127:104378
80. Jimenez E, Fontan-Vela M, Valencia J, Fernandez-Jimenez I, Alvaro-Alonso EA, Izquierdo-Garcia E. et al. Characteristics, complications and outcomes among 1549 patients hospitalised with COVID-19 in a secondary hospital in Madrid, Spain: a retrospective case series study. BMJ open. 2020;10:e042398
81. Vitte J, Diallo AB, Boumaza A, Lopez A, Michel M, Allardet-Servent J. et al. A Granulocytic Signature Identifies COVID-19 and Its Severity. The Journal of infectious diseases. 2020;222:1985-96
82. Huang Y, Cai C, Zang J, Xie J, Xu D, Zheng F. et al. Treatment strategies of hospitalized patients with coronavirus disease-19. Aging. 2020;12:11224-37
83. Uyaroglu OA, Basaran NC, Ozisik L, Dizman GT, Eroglu I, Sahin TK. et al. 30-Day Readmission Rate Of Covid-19 Patients Discharged From A Tertiary Care University Hospital In Turkey; An Observational, Single-Center Study. International journal for quality in health care: journal of the International Society for Quality in Health Care. 2020
84. Fox TA, Troy-Barnes E, Kirkwood AA, Chan WY, Day JW, Chavda SJ. et al. Clinical outcomes and risk factors for severe COVID-19 in patients with haematological disorders receiving chemo- or immunotherapy. British journal of haematology. 2020;191:194-206
85. Wang J, Zhang J, Tu Y, Zhou X, Huang H, Shao L. et al. Cancer patients in SARS-CoV-2 infection: a single-center experience from Wuhan. Journal of Cancer. 2020;11:6243-7
86. Chao JY, Derespina KR, Herold BC, Goldman DL, Aldrich M, Weingarten J. et al. Clinical Characteristics and Outcomes of Hospitalized and Critically Ill Children and Adolescents with Coronavirus Disease 2019 at a Tertiary Care Medical Center in New York City. The Journal of pediatrics. 2020;223:14-9 e2
87. Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical Features of 69 Cases With Coronavirus Disease 2019 in Wuhan, China. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020;71:769-77
88. Daniels LB, Sitapati AM, Zhang J, Zou J, Bui QM, Ren J. et al. Relation of Statin Use Prior to Admission to Severity and Recovery Among COVID-19 Inpatients. The American journal of cardiology. 2020;136:149-55
89. Liu D, Wang Y, Wang J, Liu J, Yue Y, Liu W. et al. Characteristics and Outcomes of a Sample of Patients With COVID-19 Identified Through Social Media in Wuhan, China: Observational Study. Journal of medical Internet research. 2020;22:e20108
90. Goshua G, Pine AB, Meizlish ML, Chang CH, Zhang H, Bahel P. et al. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. The Lancet Haematology. 2020;7:e575-e82
91. Huang Y, Lyu X, Li D, Wang L, Wang Y, Zou W. et al. A cohort study of 676 patients indicates D-dimer is a critical risk factor for the mortality of COVID-19. PloS one. 2020;15:e0242045
92. Taghiloo S, Aliyali M, Abedi S, Mehravaran H, Sharifpour A, Zaboli E. et al. Apoptosis and immunophenotyping of peripheral blood lymphocytes in Iranian COVID-19 patients: Clinical and laboratory characteristics. Journal of medical virology. 2020
93. Pinto C, Berselli A, Mangone L, Damato A, Iachetta F, Foracchia M. et al. SARS-CoV-2 Positive Hospitalized Cancer Patients during the Italian Outbreak: The Cohort Study in Reggio Emilia. Biology. 2020 9
94. Wu G, Yang P, Xie Y, Woodruff HC, Rao X, Guiot J. et al. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: an international multicentre study. The European respiratory journal. 2020 56
95. Lunski MJ, Burton J, Tawagi K, Maslov D, Simenson V, Barr D. et al. Multivariate mortality analyses in COVID-19: Comparing patients with cancer and patients without cancer in Louisiana. Cancer. 2020
96. Hu L, Chen S, Fu Y, Gao Z, Long H, Ren HW. et al. Risk Factors Associated With Clinical Outcomes in 323 Coronavirus Disease 2019 (COVID-19) Hospitalized Patients in Wuhan, China. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020;71:2089-98
97. Ganatra S, Dani SS, Redd R, Rieger-Christ K, Patel R, Parikh R. et al. Outcomes of COVID-19 in Patients With a History of Cancer and Comorbid Cardiovascular Disease. Journal of the National Comprehensive Cancer Network: JNCCN. 2020:1-10
98. Joharatnam-Hogan N, Hochhauser D, Shiu KK, Rush H, Crolley V, Wilson W. et al. Outcomes of the 2019 novel coronavirus in patients with or without a history of cancer: a multi-centre North London experience. Therapeutic advances in medical oncology. 2020;12:1758835920956803
99. Buckner FS, McCulloch DJ, Atluri V, Blain M, McGuffin SA, Nalla AK. et al. Clinical Features and Outcomes of 105 Hospitalized Patients With COVID-19 in Seattle, Washington. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020;71:2167-73
100. Becchetti C, Zambelli MF, Pasulo L, Donato MF, Invernizzi F, Detry O. et al. COVID-19 in an international European liver transplant recipient cohort. Gut. 2020;69:1832-40
101. Yigenoglu TN, Ata N, Altuntas F, Basci S, Dal MS, Korkmaz S. et al. The outcome of COVID-19 in patients with hematological malignancy. Journal of medical virology. 2021;93:1099-104
102. Brar G, Pinheiro LC, Shusterman M, Swed B, Reshetnyak E, Soroka O. et al. COVID-19 Severity and Outcomes in Patients With Cancer: A Matched Cohort Study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2020;38:3914-24
103. Westblade LF, Brar G, Pinheiro LC, Paidoussis D, Rajan M, Martin P. et al. SARS-CoV-2 Viral Load Predicts Mortality in Patients with and without Cancer Who Are Hospitalized with COVID-19. Cancer cell. 2020;38:661-71 e2
104. Zhang R, Ouyang H, Fu L, Wang S, Han J, Huang K. et al. CT features of SARS-CoV-2 pneumonia according to clinical presentation: a retrospective analysis of 120 consecutive patients from Wuhan city. European radiology. 2020;30:4417-26
105. Wang B, Wang Z, Zhao J, Zeng X, Wu M, Wang S. et al. Epidemiological and clinical course of 483 patients with COVID-19 in Wuhan, China: a single-center, retrospective study from the mobile cabin hospital. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2020;39:2309-15
106. Song J, Zeng M, Wang H, Qin C, Hou HY, Sun ZY. et al. Distinct effects of asthma and COPD comorbidity on disease expression and outcome in patients with COVID-19. Allergy. 2020
107. Maor Y, Cohen D, Paran N, Israely T, Ezra V, Axelrod O. et al. Compassionate use of convalescent plasma for treatment of moderate and severe pneumonia in COVID-19 patients and association with IgG antibody levels in donated plasma. EClinicalMedicine. 2020;26:100525
108. An C, Lim H, Kim DW, Chang JH, Choi YJ, Kim SW. Machine learning prediction for mortality of patients diagnosed with COVID-19: a nationwide Korean cohort study. Scientific reports. 2020;10:18716
109. Algado-Selles N, Gras-Valenti P, Chico-Sanchez P, Mora-Muriel JG, Soler-Molina VM, Hernandez-Maldonado M. et al. Frequency, Associated Risk Factors, and Characteristics of COVID-19 Among Healthcare Personnel in a Spanish Health Department. American journal of preventive medicine. 2020;59:e221-e9
110. Ge R, Kao C. Cell Surface GRP78 as a Death Receptor and an Anticancer Drug Target. Cancers. 2019 11
111. Kao C, Chandna R, Ghode A, Dsouza C, Chen M, Larsson A. et al. Proapoptotic Cyclic Peptide BC71 Targets Cell-Surface GRP78 and Functions as an Anticancer Therapeutic in Mice. EBioMedicine. 2018;33:22-32
112. Xiao X, Li S, Zhang X, Lu J, Wang W, Zhou S. et al. HHQ-4, a quinoline derivate, preferentially inhibits proliferation of glucose-deprived breast cancer cells as a GRP78 down-regulator. Toxicology and applied pharmacology. 2019;373:10-25
113. Sharma A, Tiwari V, Sowdhamini R. Computational search for potential COVID-19 drugs from FDAapproved drugs and small molecules of natural origin identifies several anti-virals and plant products. Journal of biosciences. 2020 45
Author contact
![]() Corresponding authors: Dr. Junjiang Fu, Key Laboratory of Epigenetics and Oncology, the Research Center for Preclinical Medicine, Southwest Medical University, Luzhou, Sichuan 646000, P R China. Tel/Fax: +86-830-3160283; E-mail: fujunjiangcom; fujunjiangedu.cn; Dr. Jiangzhou Peng, E-mail: jiangzhou83com; Dr. Amrish Sharma, E-mail: ASharma1org
Corresponding authors: Dr. Junjiang Fu, Key Laboratory of Epigenetics and Oncology, the Research Center for Preclinical Medicine, Southwest Medical University, Luzhou, Sichuan 646000, P R China. Tel/Fax: +86-830-3160283; E-mail: fujunjiangcom; fujunjiangedu.cn; Dr. Jiangzhou Peng, E-mail: jiangzhou83com; Dr. Amrish Sharma, E-mail: ASharma1org

 Global reach, higher impact
Global reach, higher impact