10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(8):2080-2088. doi:10.7150/ijbs.60655 This issue Cite
Review
COVID-19 one year later: a retrospect of CRISPR-Cas system in combating COVID-19
1. Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410078, P. R. China; Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, Changsha 410078, P. R. China.
2. Engineering Research Center of Applied Technology of Pharmacogenomics, Ministry of Education, 110 Xiangya Road, Changsha 410078, P. R. China.
3. National Clinical Research Center for Geriatric Disorders, 87 Xiangya Road, Changsha 410008, Hunan, P.R. China.
4. Hunan Key Laboratory of Precise Diagnosis and Treatment of Gastrointestinal Tumor, Changsha 410078, P. R. China.
5. Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008, P. R. China.
Received 2021-3-20; Accepted 2021-4-28; Published 2021-5-13
Abstract
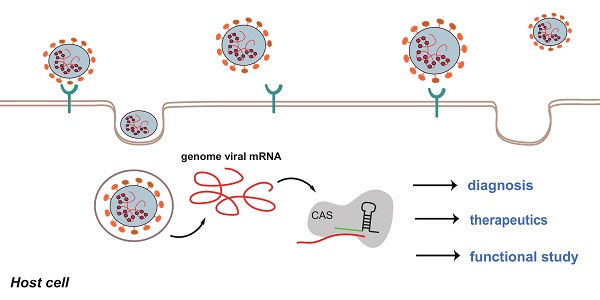
Coronavirus disease 2019 (COVID-19), an infectious disease caused by Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has posed a persistent global threat. The transmission of SARS-CoV-2 is wide and swift. Rapid detection of the viral RNA and effective therapy are imperative to prevent the worldwide spread of the new infectious disease. Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPR)- CRISPR-associated protein (Cas) system is an RNA-directed adaptive immune system, and it has been transformed into a gene editing tool. Applications of CRISPR-Cas system involves in many fields, such as human gene therapy, drug discovery and disease diagnosis. Under the background of COVID-19 pandemic, CRISPR-Cas system shows hidden capacity to fight the emergency in many aspects. This review will focus on the role of gene editing in COVID-19 diagnosis and treatment. We will describe the potential use of CRISPR-Cas-based system in combating COVID-19, from diagnosis to treatment. Furthermore, the limitation and perspectives of this novel technology are also evaluated.
Keywords: COVID-19, CRISPR-Cas, gene editing, SARS-CoV-2, diagnosis, mechanism research, therapeutics
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a coronavirus which caused an infectious disease named coronavirus infectious disease 2019 (COVID-19) [1]. On March 11, 2020, the World Health Organization (WHO) announced that COVID-19 epidemic has constituted a pandemic. COVID-19 has posed a persistent global threat. As of March 2021, more than 100 million people were infected-confirmed and 2 million people died of this disease. Rapid detection of viral RNA and effective therapy are imperative to prevent the global spread of this new infectious diseases.
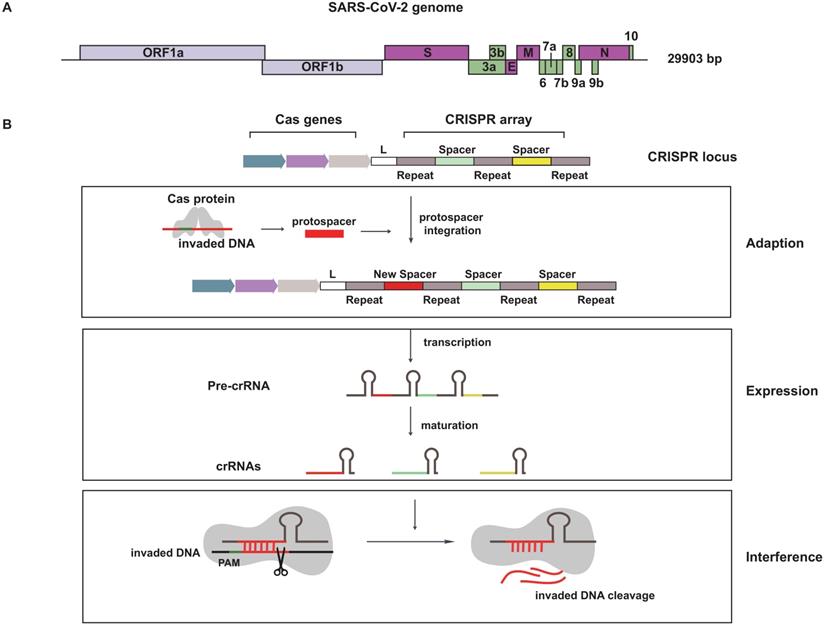
A basic understanding of SARS-CoV-2 is needed for fighting this virus. SARS-CoV-2 is an enveloped positive-sense RNA virus contained a single-stranded RNA genome of 29.9 kb (Figure 1A). The SARS-CoV-2 genome sequence is similar to SARS-CoV (~79%), but is distant from MERS-CoV (~50%). The genome is composed of 14 open reading frames (ORFs), most of them encode non-structural proteins (nsp1~nsp16) that participated in genome replication and RNA synthesis. The remaining parts encode nine accessory proteins (ORF3a~ORF10) and four structural proteins (spike, envelope, membrane and nucleocapsid), that are involved in the process of viral entry, assembly and release [2, 3].
The general detecting methods used for identifying SARS-CoV-2 are Metagenomic next-generation sequencing (mNGS) and Real Time Quantitative PCR (RT-qPCR) [4, 5]. All methods that might work are recommended for treatment [6, 7]. However, it is far from being enough to fight COVID-19 when people are confronted with huge infection burden. Any emerging technology that may be helpful is being valued. CRISPR-Cas system is being assessed. Researchers are making use of this technology in both therapeutic and molecular diagnostics of COVID-19 based on recent advances in this system. As a genome editing tool, CRISPR-Cas system can achieve precise editing, and simplify the experimental steps. It can provide rapid, inexpensive and highly sensitive diagnosis when used in nucleic acid detection. CRISPR is ahead of many new technologies for the features of high quality and cost-effective. We believe that it will play an important role in the fighting against the novel coronavirus.
CRISPR-Cas system
CRISPR is a special repeated DNA sequences family and it is cooperated with Cas to constitute the CRISPR-Cas system [8]. The first appearance of this magic combination is in 1987, but it wasn't until 2005 when CRISPR started catching people's attention. The CRISPR-Cas system is an RNA-directed adaptive immune system, which existed in approximately 48% of bacteria and 95% of archaea. When the invasion of foreign hereditary material is happening, the natural defense mechanism is triggered [9, 10]. The whole defensive process including three stages: adaption, expression and interference (Figure 1B). In the stage of adaption, short direct repeats separated by spacers are short variable DNA sequences to form CRISPR sequence, and foreign DNA called protospacer is cleaved and integrated into CRISPR array. Therefore, these incorporated fragments become new spacers. The second stage is expression, CRISPR array is transcribed as precursor CRISPR-derived RNA (pre-crRNA), then pre-crRNA is matured to yield CRISPR-derived RNA (crRNA). Interference is the third stage, crRNA recruits and guides Cas effectors to specific target for cleaving nucleic acid derived from invading virus [11].
Schematics for SARS-CoV-2 genome and the natural defense mechanism of CRISPR system. (A) The skeleton diagram of SARS-CoV-2 genome. Light purple represents ORF1a and ORF1b, which encode non-structural proteins. Dark purple represents four structural protein-encoding genes. Green represents nine accessory protein-encoding genes. (B) A brief description of CRISPR-Cas system mechanism in defending foreign genetic material.
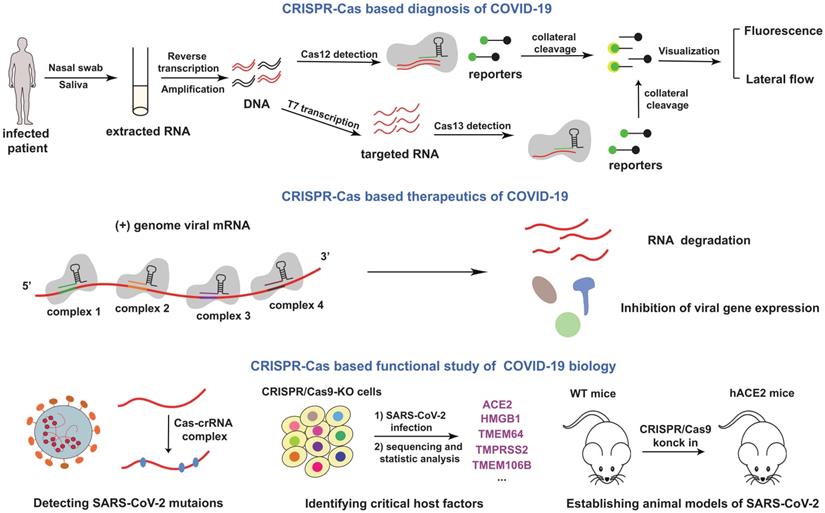
Schematics for CRISPR-Cas based applications in COVID-19. The mentioned contents were summarized in this figure. In the top of the picture, a cardinal principle of CRISPR-based diagnosis was shown. Researchers extracted patient sample RNA from nasopharyngeal or oropharyngeal swabs, and transformed RNA into cDNA with the process of reverse transcription. Then amplified virus cDNA, and use Cas protein targets predefined coronavirus sequences. The collateral cleavage of ssDNA probe confirmed detection of the virus. In the middle of the picture, the mechanism of therapeutics was depicted. Genome viral mRNA was degradation after CRISPR-Cas system cleavage, and the viral gene expression have also been inhibited. In the bottom of the picture, potential utilization of CRISPR in COVID-19 mechanism research was exhibited.
CRISPR-Cas system is divided into two classes based on different organization of effectors. Class 1 CRISPR-Cas system uses a huge effector complex consisting of multiple Cas proteins to deal with foreign nucleic acid, whereas Class 2 CRISPR-Cas system uses a single-protein effector [12, 13]. Based on the effector protein families, class1 system is split into type I, type III, and type IV and class 2 system contains type II, V and VI systems. Cas9, Cas 12 and Cas13 belongs to type II, V, and VI, respectively.
Due to the difficulty of heterologous expression of multiple cascade complexes, class 2 CRISPR-Cas system is more convenient and rapid in dealing with emergencies, such as COVID-19, compared with class 1 system [14]. Class 2 system is used in many fields, for example, (1) genome editing, including gene knock-out or knock-in that can be realized by a type II or type V effector enzyme for determining critical factors and providing potential therapy for diseases [15-17]; (2) Targeting ssDNA/RNA for detection and treatment of viral diseases using type V or type VI effector [18].
At present, the COVID-19 pandemic is still not under control. CRISPR-Cas system has the capacity to fight the disease. Although about 90% of all CRISPR-Cas loci identified in bacteria and archaea belong to the class 1 CRISPR- Cas system, most of the class 1 system are rarely used in eukaryotic genome engineering, because the common Cas proteins in class 1 CRISPR-Cas system have no specific functions. Based on these considerations, in our review, we mainly describe the related applications of class2 CRISPR-Cas system in combating the SARS-CoV-2 infection, which is the topic of general interest (Figure 2).
CRISPR-Cas based diagnosis of COVID-19
COVID-19 is a public health emergency of international concern. Accurate and rapid diagnosis is essential for its early recognition and treatment. mNGS and RT-qPCR assays are the most commonly used technologies in detecting SARS-CoV-2. In the early days of the outbreak, mNGS is used to analyze the bronchoalveolar lavage fluid of patients and identified the virus [19]. The advantages of mNGS are as follows: high throughput, unrestricted culture condition and comprehensive analysis. While its disadvantages are also obvious: high cost, complex operation process and long-time detection. In contrast, RT-qPCR reduces the analysis time, but it has high requirements for experimental operation. All problems mentioned above can be solved by CRISPR. CRISPR-Cas system can speed up the diagnostic process for its convenient operation and low cost. Advances in application of CRISPR-Cas system indicates its significance in fighting against COVID-19.
CRISPR-Cas12 based diagnostic tools of COVID-19
CRISPR-Cas12 system is an extension of genomic editing tool. Cas12a, also called Cpf1, was identified first [20], then Cas12b (C2c1) and other Cas proteins in type V system were confirmed [21]. The team of Jennifer Doudna found that type V CRISPR-associated proteins can stimulate non-specific single-stranded DNase activity to cut nearby dsDNA while Cas12 nucleases target dsDNA upon the mediation of guide RNA (gRNA). Hence, Jennifer et al. reported a method named DETECTER to realize quick and excellent nucleic acid detection for HPV-infected patient in clinics, which achieves attomolar sensitivity [22]. The same principle is applied for the detection of SARS-CoV-2. Broughton et al. develops a CRISPR-Cas12-based assay for detection of SARS-CoV-2, which also called DETECTER. They extract patient sample RNA from nasopharyngeal or oropharyngeal swabs, and RNA is transcribed into cDNA with the process of reverse transcription. Using reverse tranion loop-mediated isothermal amplification (RT-LAMP) for isothermal amplification, the Cas12 targets predefined coronavirus sequences, the E and N genes of SARS-CoV-2, after which cleavage of ssDNA probe confirms detection of the virus. The system can be run in approximately 30-40 min and visualized on a lateral flow strip, which is faster and more specific than traditional RT-qPCR [23]. SARS-CoV-2 DETECTER is a powerful instrument for COVID-19 diagnosis and scientists are contributed to optimizing the diagnostic procedures. Ma et al. described using the MeCas12a system to detect SARS-CoV-2 by targeting the E gene. They screened manganese ion to enhance the signal up to 13‐folds for the tested crRNAs, which enable the limit of detection to reach five copies and improve specificity. It also can distinguish infection of SARS‐CoV‐2 from MERS‐CoV in the simulated clinical samples [24]. Another analogous platform is ENHANCE. Nguyen et al. developed the system with engineered crRNAs and optimized conditions that enabled to detect various clinically relevant nucleic acid targets, including human immunodeficiency virus, hepatitis virus C and SARS-CoV-2, with high sensitivity [25]. Apart from the increasing sensitivity and specificity, researchers setup to simplifying the diagnostic procedures. Ding et al. presented an AIOD-CRISPR assay for simple, ultrasensitive, one-pot, and visual detection of SARS-CoV-2. In the AIOD-CRISPR assay, all components used for nucleic acid amplification and CRISPR-based detection are thoroughly mixed in a single-tank reaction system and incubated at one temperature, eliminating the need for separate pre-amplification and transfer of culture amplification products. Meanwhile, double crRNA without PAM sequence restriction is introduced to launch CRISPR-based SARS-CoV-2 detection [26]. This procedure has fewer steps and dramatically reduces the chance of operation-introduced nucleic acid contamination or cross-contamination among samples. STOPCovid is described as a concise tool targeting N gene for detection, which combines simplified extraction of viral RNA with isothermal amplification and CRISPR-mediated detection. Considering loop-mediated isothermal amplification (LAMP) operates at 55 to 70 °C, researchers choose Cas12b as protein effector which is a thermostable Cas enzyme [27]. This same conditions also apply in CASdetec, a system which acts on RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 [28]. In addition, there have some other optimizations, including introducing microfluidics technology into CRISPR-based diagnostics [29, 30], and adding the quenched fluorescent single-stranded DNA (ssDNA) reporter generating fluorescence signal visible to the naked eye [31, 32]. The summary of Cas12-based SARS-CoV-2 diagnostics in presented in Table 1.
CRISPR-Cas13 based diagnostic tools of COVID-19
Cas13a (C2c2) is an important part of CRISPR-Cas13 system (type VI) specifically targeting RNA [14, 33]. Dr. Zhang and his colleagues confirmed that Cas13a is a single-component programmable RNA-guided RNA-targeting CRISPR effector and they employed Cas13a to develop new RNA-targeting tools [34]. CRISPR-Cas13 based diagnostics can be used to detect invading RNA due to its collateral cleavage of nearby non-targeted RNAs [35]. SHERLOCK was created for this reason. Gootenberg et al. use SHERLOCK to detect Zika and Dengue virus with high sensitivity and specificity [36, 37]. SHERLOCK was also used to identify Ebola virus and Lassa virus [38]. Since the outbreak of CODVID-19, people are eager looking for diagnosis to identify symptomatic, asymptomatic, and pre-symptomatic carriers of the virus. The team of Feng Zhang reformed SHERLOCK to make it feasible for SARS-CoV-2 detection. They redesigned different primers and guide-RNA for targeting open reading frame 1ab (ORF1ab) and spike (S) gene. Clinical validations suggest that test result of SHERLOCK is identical to RT-qPCR [39, 40]. SHERLOCK is not a multi-purpose system. It is time-consuming when dealing with massively multiplexed nucleic acid detection. It also has a lot of hidden dangers. SHINE was created to seek the optimal conditions to realize recombinase polymerase amplification (RPA)-based amplification and Cas13-based detection occurring in one step, which simplifying the whole streamline and available for testing unextracted samples [41]. CARMEN can comprehensive test large SARS-CoV-2 sample sets based on its intrinsic multiplexing and throughput capabilities [42]. DISCoVER is a RNA extraction-free SARS-CoV-2 test which applies saliva-based sample matrix to detect virus [43]. Careful crRNA selection, and proper combinations between crRNA and Cas13 will optimize SARS-CoV-2 detection. Introducing emerging technologies will avoid the need for amplification and reverse transcription. Fozouni et al. designed many crRNAs along the N gene of SARS-CoV-2 to determine the best, and replaced Cas13 with its homolog for highest sensitivity and robust trans-cleavage activity [44]. Tian et al. applied microfluidic technology to CRISPR-Cas13-based system for nucleic acid amplification-free single-molecule RNA detection [45]. There are other platforms show outstanding performance in SARS-CoV-2 detection. All Cas13-based SARS-CoV-2 diagnostics are comprehensively summarized in Table 2.
CRISPR Cas9-based diagnostic tools of COVID-19
CRISPR-Cas9 is well-known for its strong gene-editing capacity [46]. Moreover, Cas9 provides excellent DNA recognition capability without trans-cleavage activity, which is used for development of biosensors. In the COVID-19 diagnosis, scientists utilize dead Cas9 (dCas9), a mutation of Cas9, to realize the function of virus detection. dCas9 was generated by the simultaneous mutation of RuvC1 and HNH regions of cas9 endonuclease. The endonuclease activity of dCas9 disappeared completely, and only the ability of gRNA to guide into the genome was retained. Therefore, dCas9 can achieve multiple functions by binding to other effector proteins. Researchers reported a colorimetric viral detection method based on CRISPR/dCas9 system. The dcas9 gRNA complex was fixed on the well plate, and then viral lysate and biotin-PAMmer were added. After incubation and washing, streptavidin horseradish peroxidase (HRP) and 3,3′,5,5′-tetramethylbenzidine (TMB) reagent were added to the plate. Through the oxidation of TMB, yellow light could be observed for the viral RNA [47].
CRISPR-Cas12 based COVID-19 detection assays
| Platform Name | Cas protein | Time | Sensitivity | Specificity | Visualization | Target genes | Reference |
|---|---|---|---|---|---|---|---|
| contamination-free visual detection | Cas12a | 40 min | 100% | 100% | Lateral flow | ORF1ab, N, E | [69] |
| SENA | Cas12a | N/A | 99% | 99% | Fluorescence | ORF1ab, N | [70] |
| opvCRISPR | Cas12a | 45 min | N/A | N/A | Naked eye | S | [31] |
| iSCAN | Cas12a | 40 min | 86% | 100% | Fluorescence/lateral flow | N, E | [71] |
| ITP-CRISPR | Cas12a | 30-40 min | 93.8% | 100% | Fluorescence | N, E | [29] |
| CRISPR-FDS | Cas12a | 15 min | 100% | 100% | Fluorescence | ORF1ab | [30] |
| CRISPR/Cas12a-NER | Cas12a | 45 min | N/A | N/A | Fluorescence | E | [32] |
| STOPCovid.v1 | Cas12b | 50 min | N/A | N/A | Lateral flow | N | [27] |
| STOPCovid.v2 | Cas12b | 15-45 min | 93.1% | 98.5% | Fluorescence/lateral flow | N | [27] |
| DETECTR | Cas12a | 45 min | 95% | 100% | Lateral flow | E, N | [23] |
| MeCas12a | Cas12a | 45 min | 100% | 100% | Naked eye | E | [24] |
| ENHANCE | Cas12a | 40-60 min | N/A | N/A | Fluorescence/lateral flow | N | [25] |
| AIOD-CRISPR | Cas12a | 20 min | N/A | N/A | Naked eye | N | [26] |
| COVID-19 CRISPR-FDS | Cas12a | 50 min | 100% | 71.4% | Fluorescence | ORF1ab, N | [72] |
| CRISPR-ABC | Cas12a | 30-40 min | 91.2% | 99.2% | Fluorescence | ORF1ab | [73] |
| OR‑DETECTR | Cas12a | 50 min | N/A | N/A | Fluorescence | RdRp, N | [74] |
| CASdetec | Cas12b | 60 min | N/A | N/A | Naked eye | RdRp | [28] |
CRISPR-Cas13/Cas9 based COVID-19 detection assays
| Platform Name | Cas protein | Time | Sensitivity | Specificity | Visualization | Target genes | Reference |
|---|---|---|---|---|---|---|---|
| SHERLOCK | Cas13a | 40 min | 100% | 100% | Fluorescence | ORF1ab, N, S | [39] |
| SHINE | Cas13a | 50 min | 90% | 100% | Smartphone/lateral flow | ORF1ab | [41] |
| DISCoVER | Cas13a | 30 min | N/A | 100% | Fluorescence | N | [43] |
| CARMEN | Cas13a | N/A | N/A | N/A | Fluorescence | N/A | [42] |
| Ultralocalized Cas13a assay | Cas13a | N/A | single-molecule | single-nucleotide | Fluorescence | ORF1a, N | [45] |
| TL-LFA | Cas9 | < 1 h | 100% | 97.1% | Fluorescence | ORF1ab, E | [48] |
Xiong et al. realized the rapid and visual detection of two SARS-CoV-2 genes (ORF1ab and E) in a single test and a single strip at the same time based on CRISPR/Cas9‐Mediated Lateral Flow Assay [48]. There are other detection platforms based on CRISPR-Cas9 [49, 50], such as CASLFA, which has realized accurate diagnosis of Listeria monocytogenes, genetically modified organisms, and African swine fever virus. The application of Cas9-based SARS-CoV-2 diagnostics are presented in Table 2.
CRISPR-Cas based functional study of COVID-19 biology
At present, the functional study of COVID-19 biology mainly considered three aspects: virus, host and their interaction in vivo. We need understand these parts in order to better conquer the virus. CRISPR-Cas system has many contributions in these fields.
Detecting mutations of SARS-CoV-2
Coronavirus is prone to mutation in the process of evolution because of its particular genome structure. Thousands of SARS-CoV-2 genome sequences reveal a number of mutations [51]. D614G is a missense mutation which leads to an amino acid change from aspartate to a glycine residue at position 614 in the S protein [52]. The mutation was first discovered in Europe. After several months of transmission, it has become the main genotype. The ability of SARS-CoV-2 to invade human is enhanced due to D614G mutation. Thus powerful methods for its fast and reliable detection in very important [53]. Wang et al. proposed a method based on a CRISPR-Cas13 amplification principle to profile mutated variants of SARS-CoV-2 [54]. At the same time, Meng et al. used engineered Cas12a gRNA to detect the SARS-CoV-2 D614G mutation [55].
Screening critical host factors for SARS-CoV-2 infection
The first step of the virus attacking human is infection. The spike protein of the virus binds to the receptor located in host cells, and then release a small single stranded RNA into it, which can be used as a messenger RNA to cheat the ribosome and synthesize RNA replicase. RNA replicase will generate RNA negative strand according to the viral RNA, and this RNA negative strand will continue to generate more RNA fragments and RNA positive strand. These different RNA fragments and ribosomes generate more different viral protein structures. Finally, complete viral particles are formed, that can be excluded from cells and infect new cells. Therefore, it is very important to find out the key host factors affecting viral infection. Using genome-wide screening based on CRISPR gene editing technology, scientists identified several potential host factors related to the viral invasion. Daniloski et al. found that knockout of a set of genes (ATP6AP1, ATP6V1A, NPC1, RAB7A, CCDC22, and PIK3C3) induces transcriptional changes in cholesterol biosynthesis pathway and reduces viral entry [56]. Wei et al. revealed that HMGB1 regulates ACE2 expression and is critical for the entry of SARS-CoV-2 [57]. TMEM106, BVMP1, TMEM41, and TMEM64 (VTT) domain-containing protein transmembrane protein 41B (TMEM41B) were significant for infection by SARS-CoV-2 [58, 59]. ACE2 and TMPRSS2 were also screened out and ranked as the top genes [60]. Baggen et al. discovered that lysosomal protein TMEM106B is important for SARS-CoV-2 to infect human cell lines and primary lung cells [61]. Not only were they important in virus infection, but host factors screened by CRSISPR-Cas system also can be used to predict adverse drug response or improve drug efficacy. Akinci et al. found that loss of SLC29A3 mitigates remdesivir toxicity but not change SARS-CoV-2 antiviral potency, and that the mitochondrial adenylate kinase AK2 is a kinase required for the remdesivir efficacy and toxicity [62]. These screened host factors represent a rich resource to develop new therapeutic strategies for combating COVID-19.
Establishing animal models of SARS-CoV-2
Studies in SARS-CoV-2 should not stay at the cellular level, but also need appropriate animal models to seek some clues for combating COVID-19. Human angiotensin-converting enzyme II (ACE2) is a functional receptor for SARS-CoV-2 entry. Researchers use CRISPR-Cas9 knock-in technology to generate a mouse model expressing human ACE2 (hACE2). The transgenic mouse would be a useful tool for studying SARS-CoV-2 transmission and pathogenesis and testing potential vaccines and therapeutics [63-65].
CRISPR-Cas based therapeutics for COVID-19
It is known that CRISPR-Cas13 is an RNA-guided RNA-targeting CRISPR system. The characteristic of this system may be used to destroy the RNA of SARS-CoV-2 and inhibit its replication capacity. Nguyen et al. used CRISPR-Cas13d system to disrupt the virus. CRISPR-Cas13d system consists of two parts, Cas13d protein and gRNAs which contained spacer sequences specifically complement with the virus RNA genome. They designed 10,333 guide-RNA for targeting 10 peptide-coding regions of SARS-CoV-2, and treat adeno-associated virus as a vector to deliver the Cas13d effector [66]. PAC-MAN is created based on the same operating principle. To find a best region for specific targeting viral RNA, Abbott et al. utilized bioinformatic analysis, a group of six crRNAs that can target 91% of sequenced coronaviruses and a group of 22 crRNAs enabled to target all sequenced coronaviruses were revealed finally. PAC-MAN becomes a potential antiviral strategy [67]. The latest study published in February 2021 displayed that CRISPR-Cas13a system can mitigate SARS-CoV-2 infection, and this conclusion is identified in hamsters [68]. In a word, Cas13 can be used in vivo against SARS-CoV-2.
Limitation and future perspective
Diagnostics based on CRISPR-Cas system is a rapid and simple method. At the beginning of using CRISPR system in nucleic acid detection, it requires a large quantity of targeted RNA molecules. In addition, the false positive rate was a concern. With continuous improvement of this technology, CRISPR-Cas-based diagnosis achieves attomolar sensitivity. Compared with RT-qPCR, it avoids aerosol pollution, which reduces false positive rate. mNGS can relieve the disadvantage of RT-qPCR, and improve the positive detection of CRISPR system. However, the analysis time of mNGS is too long, and the results cannot be produced in time. CRISPR system has many advantages.
Mechanistic researches based on CRISPR-Cas system is expanding. Recently, a new viral strain, B1.1.7, was found in UK. The virus strain had 23 mutations, N501Y is a concerning one [69]. It changes the most important part of the spike protein, the receptor binding domain that the virus uses to contact with human respiratory cells. Currently, conventional methods are still used for viral detection. Although there is no relevant literature about CRISPR system applied to N501Y detection, CRISPR will make contributions for its detection.
Therapeutics based on CRISPR-Cas system provide a potential option for treating COVID-19. The risk of off-target cannot be ignored in treatment. There are two solutions for off-target effect, optimize design of sgRNA and improve the specificity of Cas protein. Scientists have come up with a lot of possible measures and the risk of off-target effect has been low thus far. Another limitation is applicability of CRISPR-Cas system. CRISPR is not widely used in human. Most of the current research has been carried out in animals, and this limitation should be overcome one day.
In conclusion, in the context of COVID-19 pandemic, novel technologies will help advance the disease diagnosis and therapeutics.
Abbreviations
SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; COVID-19: coronavirus disease 2019; ORFs: open reading frames; RT-qPCR: Real Time Quantitative PCR; CRISPR: Clustered Regularly-Interspaced Short Palindromic Repeats; Cas: CRISPR-associated protein; pre-crRNA: precursor CRISPR-derived RNA; crRNA: CRISPR-derived RNA; dsDNA: double-stranded DNA; RT-LAMP: reverse tranion loop-mediated isothermal amplification; LAMP: loop-mediated isothermal amplification; PAM: protospacer adjacent motif; RdRp: RNA-dependent RNA polymerase; ssDNA: single-stranded DNA; ORF1ab: open reading frame 1ab; S: spike; N: nucleocapsid; E: envelope; M: membrane; RPA: recombinase polymerase amplification; gRNA: guide RNA; dCas9: dead CRISPR-associated protein 9; PAMmer: adjacent motif -presenting oligonucleotide; HRP: horseradish peroxidase; TMB: 3,3′,5,5′-tetramethylbenzidine; hACE2: human angiotensin-converting enzyme II.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81773823, 82073943) and the Hunan Natural Science Foundation (2019JJ40519).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-33
2. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141-54
3. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497-506
4. Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W. et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9:313-9
5. Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T. et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J Clin Microbiol. 2020 58
6. Asselah T, Durantel D, Pasmant E, Lau G, Schinazi RF. COVID-19: Discovery, diagnostics and drug development. J Hepatol. 2021;74:168-84
7. Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782-93
8. Hanna RE, Doench JG. Design and analysis of CRISPR-Cas experiments. Nat Biotechnol. 2020;38:813-23
9. Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479-91
10. Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475-93
11. Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180-96
12. Makarova KS, Zhang F, Koonin EV. SnapShot: Class 1 CRISPR-Cas Systems. Cell. 2017;168:946-e1
13. Makarova KS, Zhang F, Koonin EV. SnapShot: Class 2 CRISPR-Cas Systems. Cell. 2017;168:328-e1
14. Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E. et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385-97
15. Lek A, Zhang Y, Woodman KG, Huang S, DeSimone AM, Cohen J. et al. Applying genome-wide CRISPR-Cas9 screens for therapeutic discovery in facioscapulohumeral muscular dystrophy. Sci Transl Med. 2020 12
16. Selle K, Fletcher JR, Tuson H, Schmitt DS, McMillan L, Vridhambal GS. et al. In vivo Targeting of Clostridioides difficile Using Phage-Delivered CRISPR-Cas3 Antimicrobials. mBio. 2020 11
17. Fellmann C, Gowen BG, Lin PC, Doudna JA, Corn JE. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. 2017;16:89-100
18. van Dongen JE, Berendsen JTW, Steenbergen RDM, Wolthuis RMF, Eijkel JCT, Segerink LI. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens Bioelectron. 2020;166:112445
19. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG. et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-9
20. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759-71
21. East-Seletsky A, O'Connell MR, Knight SC, Burstein D, Cate JH, Tjian R. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270-3
22. Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436-9
23. Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J. et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870-4
24. Ma P, Meng Q, Sun B, Zhao B, Dang L, Zhong M. et al. MeCas12a, a Highly Sensitive and Specific System for COVID-19 Detection. Adv Sci (Weinh). 2020: 2001300.
25. Nguyen LT, Smith BM, Jain PK. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat Commun. 2020;11:4906
26. Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM. et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11:4711
27. Joung J, Ladha A, Saito M, Kim NG, Woolley AE, Segel M. et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N Engl J Med. 2020;383:1492-4
28. Guo L, Sun X, Wang X, Liang C, Jiang H, Gao Q. et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:34
29. Ramachandran A, Huyke DA, Sharma E, Sahoo MK, Huang C, Banaei N. et al. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:29518-25
30. Ning B, Yu T, Zhang S, Huang Z, Tian D, Lin Z. et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci Adv. 2021 7
31. Wang R, Qian C, Pang Y, Li M, Yang Y, Ma H. et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens Bioelectron. 2021;172:112766
32. Wang X, Zhong M, Liu Y, Ma P, Dang L, Meng Q. et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull (Beijing). 2020;65:1436-9
33. Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ. et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18:67-83
34. Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573
35. East-Seletsky A, O'Connell MR, Burstein D, Knott GJ, Doudna JA. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol Cell. 2017;66:373-83 e3
36. Caliendo AM, Hodinka RL. A CRISPR Way to Diagnose Infectious Diseases. N Engl J Med. 2017;377:1685-7
37. Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438-42
38. Barnes KG, Lachenauer AE, Nitido A, Siddiqui S, Gross R, Beitzel B. et al. Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nat Commun. 2020;11:4131
39. Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P. et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4:1140-9
40. An J, Liao X, Xiao T, Qian S, Yuan J, Ye H. et al. Clinical characteristics of recovered COVID-19 patients with re-detectable positive RNA test. Ann Transl Med. 2020;8:1084
41. Arizti-Sanz J, Freije CA, Stanton AC, Petros BA, Boehm CK, Siddiqui S. et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11:5921
42. Ackerman CM, Myhrvold C, Thakku SG, Freije CA, Metsky HC, Yang DK. et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277-82
43. Agrawal S, Fanton A, Chandrasekaran SS, Prywes N, Lukarska M, Biering SB. et al. Rapid detection of SARS-CoV-2 with Cas13. medRxiv. 2020
44. Fozouni P, Son S, Diaz de Leon Derby M, Knott GJ, Gray CN, D'Ambrosio MV. et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323-33 e9
45. Tian T, Shu B, Jiang Y, Ye M, Liu L, Guo Z. et al. An Ultralocalized Cas13a Assay Enables Universal and Nucleic Acid Amplification-Free Single-Molecule RNA Diagnostics. ACS Nano. 2021;15:1167-78
46. Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230-2
47. Moon J, Kwon HJ, Yong D, Lee IC, Kim H, Kang H. et al. Colorimetric Detection of SARS-CoV-2 and Drug-Resistant pH1N1 Using CRISPR/dCas9. ACS Sens. 2020;5:4017-26
48. Xiong E, Jiang L, Tian T, Hu M, Yue H, Huang M. et al. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay. Angew Chem Int Ed Engl. 2021;60:5307-15
49. Xu X, Luo T, Gao J, Lin N, Li W, Xia X. et al. CRISPR-Assisted DNA Detection: A Novel dCas9-Based DNA Detection Technique. CRISPR J. 2020;3:487-502
50. Wang X, Xiong E, Tian T, Cheng M, Lin W, Wang H. et al. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Mediated Lateral Flow Nucleic Acid Assay. ACS Nano. 2020;14:2497-508
51. Zhang Y, Xi H, Juhas M. Biosensing Detection of the SARS-CoV-2 D614G Mutation. Trends Genet. 2021;37:299-302
52. Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y. et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739-51 e8
53. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W. et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812-27 e19
54. Wang Y, Zhang Y, Chen J, Wang M, Zhang T, Luo W. et al. Detection of SARS-CoV-2 and Its Mutated Variants via CRISPR-Cas13-Based Transcription Amplification. Anal Chem. 2021;93:3393-402
55. Meng Q, Wang X, Wang Y, Dang L, Liu X, Ma X. et al. Detection of the SARS-CoV-2 D614G mutation using engineered Cas12a guide RNA. Biotechnol J. 2021: e2100040.
56. Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M. et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 2021;184:92-105 e16
57. Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL. et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell. 2021;184:76-91 e13
58. Schneider WM, Luna JM, Hoffmann HH, Sanchez-Rivera FJ, Leal AA, Ashbrook AW. et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell. 2021;184:120-32 e14
59. Wang R, Simoneau CR, Kulsuptrakul J, Bouhaddou M, Travisano KA, Hayashi JM. et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell. 2021;184:106-19 e14
60. Zhu Y, Feng F, Hu G, Wang Y, Yu Y, Zhu Y. et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat Commun. 2021;12:961
61. Akinci E, Cha M, Lin L, Yeo G, Hamilton MC, Donahue CJ. et al. Elucidation of remdesivir cytotoxicity pathways through genome-wide CRISPR-Cas9 screening and transcriptomics. bioRxiv. 2020
62. Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY. et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 2020;28:124-33 e4
63. Bao L, Deng W, Huang B, Gao H, Liu J, Ren L. et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830-3
64. Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR. et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell. 2020;182:50-8 e8
65. Nguyen TM, Zhang Y, Pandolfi PP. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189-90
66. Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L. et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell. 2020;181:865-76 e12
67. Blanchard EL, Vanover D, Bawage SS, Tiwari PM, Rotolo L, Beyersdorf J. et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat Biotechnol. 2021
68. Matic N, Lowe CF, Ritchie G, Stefanovic A, Lawson T, Jang W, et al. Rapid Detection of SARS-CoV-2 Variants of Concern, Including B.1.1.28/P.1, in British Columbia, Canada. Emerg Infect Dis. 2021; 27
69. Chen Y, Shi Y, Chen Y, Yang Z, Wu H, Zhou Z. et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens Bioelectron. 2020;169:112642
70. Huang W, Yu L, Wen D, Wei D, Sun Y, Zhao H. et al. A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. EBioMedicine. 2020;61:103036
71. Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T. et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129
72. Huang Z, Tian D, Liu Y, Lin Z, Lyon CJ, Lai W. et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens Bioelectron. 2020;164:112316
73. Huang Z, Ning B, Yang HS, Youngquist BM, Niu A, Lyon CJ. et al. Sensitive tracking of circulating viral RNA through all stages of SARS-CoV-2 infection. J Clin Invest. 2021 131
74. Sun Y, Yu L, Liu C, Ye S, Chen W, Li D. et al. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J Transl Med. 2021;19:74
Author contact
![]() Corresponding authors: Professor Ji-Ye Yin, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008; P. R. China. Tel: +86 731 84805380, Fax: +86 731 82354476, E-mail: yinjiyeedu.cn. ORCID: 0000-0002-1244-5045; Professor Xiang-Ping Li, Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008, P. R. China. Tel: +86 731 84327453. Fax: +86 731 84327453, E‐mail: xylxpingcom. ORCID: 0000-0002-5801-489X.
Corresponding authors: Professor Ji-Ye Yin, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008; P. R. China. Tel: +86 731 84805380, Fax: +86 731 82354476, E-mail: yinjiyeedu.cn. ORCID: 0000-0002-1244-5045; Professor Xiang-Ping Li, Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008, P. R. China. Tel: +86 731 84327453. Fax: +86 731 84327453, E‐mail: xylxpingcom. ORCID: 0000-0002-5801-489X.

 Global reach, higher impact
Global reach, higher impact