10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(9):2262-2277. doi:10.7150/ijbs.60131 This issue Cite
Review
The Role of N6-Methyladenosine Modified Circular RNA in Pathophysiological Processes
Mechanobiology and Regenerative Medicine Laboratory, Bioengineering College, Chongqing University, Chongqing, 400044, China.
Received 2021-3-5; Accepted 2021-5-21; Published 2021-6-1
Abstract
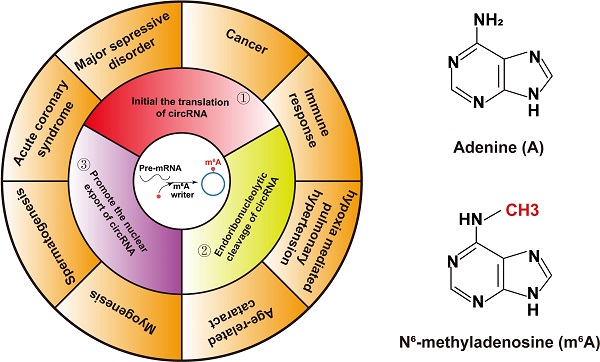
Circular RNA (circRNA) is a type of covalently closed and endogenous non-coding RNA (ncRNA) with tissue- and cell-specific expression patterns generated by a non-canonical splicing event. Previous reports have indicated that circRNAs exert their functions in different ways, thereby participating in various pathophysiological processes. N6-methyladenosine (m6A) methylation occurs in the N6-position, which is the most abundant and conserved internal transcriptional modification in eukaryotes, including mRNA and ncRNAs. Accumulating evidences confirm that m6A modification also exists in the circRNA and greatly affects the biological functions of circRNA. Their dysregulated expression can be a cause of various pathophysiological processes, such as spermatogenesis, myoblast differentiation, cancer, cardiovascular disease, mental illness and so on. Understanding the role of m6A-modified circRNAs in pathophysiological processes may contribute to better understanding the physiological mechanisms and develop new biomarkers. This review summarizes the regulatory mechanism of m6A modification on circRNA metabolism and the role of m6A-modified circRNAs in pathophysiological processes. This article may pave the way for a better understanding of the role of epigenetically modified circRNAs in pathophysiological process.
Keywords: Circular RNA, N6-methyladenosine (m6A), Biogenesis, M6A-modified circRNA, Pathophysiological processes
Introduction
Two percent of human genome transcripts are mRNAs, the rest are noncoding RNAs (ncRNAs), which can regulate genes expression in growth and development of organisms [1,2]. Circular RNAs (circRNAs) are single-stranded and covalently closed endogenous ncRNAs that mainly exist in cytoplasm and have evolutionary conservation and specific expression pattern of tissue and developmental stage [3,4]. Furthermore, the circular structure makes circRNAs more stable and not easily degraded by ribonucleases [3].
Although circRNAs were emerged in 1976 [5,6], they were all considered as the “junk” of abnormal splicing events. Up to 2018, 30,000 circRNAs have been identified [7]. Increasing studies have shown that aberrant circRNAs are related to many human diseases, such as cardiovascular disease (CVD) [8], neurological disorders [9], diabetes mellitus [10], autoimmune disease [11] and cancers [12]. CircRNAs are promising in serving the biological targets for diagnosis and treatment due to their peculiarity. CircRNAs are involved in the regulation of biological processes by sponging disease-related microRNA (miRNA) [13] or proteins [14], or translating into proteins [15]. Currently, it has been discovered that the post-transcriptional modification N6-methyladenosine (m6A), occurring in N6-position of adenosine, also exists in circRNAs and participates in the metabolism and function of circRNAs [16].
Post-transcriptional modification has become a critical regulatory factor in many physiological processes and disease progression [17]. The deposition of chemically modified RNA has emerged as a basic mechanism that regulates the fate of lineages in cell transcriptome and proteome during development [18]. Around 10 million peaks collected from 672 samples have been recorded in the RNA EPItranscriptome Collection database. Even though the high-resolution mapping is available, only a few amounts of modifications are mapped [20]. Among those modifications, the m6A modification of mRNA is the most prevalent regulator in the eukaryotes [21]. In addition, the dynamic and reversible m6A modification makes it possible to regulate the complex and delicate expression of genes. Numerous studies have indicated that m6A modification can respond to different physiological or pathological changes [18, 22].
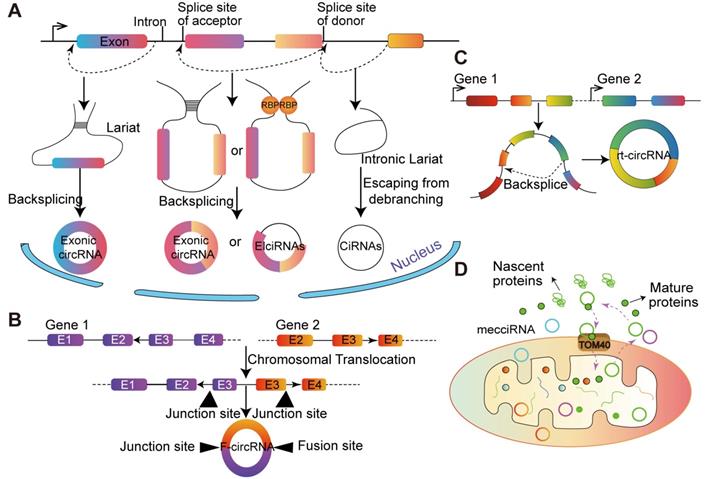
The biogenesis of circRNAs and special types of circRNA. (A) Lariat-driven circularization. Exon skipping events result in lariat structure, then the lariat is removed, and finally circRNA is formed. Intron-pairing-driven circularization. There are complementary sequence motifs between the introns sequences flanking the exon the base complementary forms a circular structure and the introns are removed or retained to form ecircRNAs or EIciRNAs. RBPs-pairing-driven circularization. The RBP interaction on the exon flanking sequence promotes the formation of circRNAs. CiRNAs. The ciRNAs were obtained from intronic lariat escape from debranching. (B) F-circRNA. CircRNA derived from the back-splicing of fusion-gene. (C) Rt-circRNA. Stop-codon read-through from read-through circRNA rt-circRNA. (D) MecciRNAs. CircRNAs encoded by the mitochondrial genome.
Similar to mRNA, m6A-modified circRNAs are also installed by a multicomponent methyltransferase complex and removed by demethylases enzymes [23]. Although relevant studies are still in infancy, growing evidences have shown that the biogenesis, decay, export and translation of circRNAs are affected by m6A modification [24]. Although similar researches are still in a few, people have discovered the role of m6A-modified circRNA in pathophysiological processes. For instance, a groundbreaking research has summarized circRNAs with m6A modification are widely linked with several human diseases [23,24]. Furthermore, researches demonstrated that m6A modification devotes to myogenesis and spermatogenesis [25,26]. This review briefly addresses current studies about circRNAs and summarizes the regulatory effect of m6A modification on circRNA metabolism. Particularly, the role of m6A-modified circRNAs in the formation of some pathophysiological processes is emphasized.
CircRNA
CircRNAs, a class of endogenous ncRNAs, are generated by variable splicing events during transcription and then form a covalently closed loop structure. Specifically, circRNAs are expressed in the different developmental stages and tissues, and play a role in plenty of important biological functions, such as affecting the development process [27], modulating immune response [28], promoting cardiac muscle repair [29], tumorigenesis or anti-tumorigenesis [30,31] and nerve injury [32]. This section will introduce the biogenesis models, types and functions of circRNAs.
Biogenesis of circRNAs
According to gene annotation, circRNA can be divided into three types according to gene annotation information: exonic circRNAs (ecircRNAs) [33], intronic circRNAs (ciRNAs) [34] and exon-intron circRNAs (EIciRNA) [1] (Figure 1). Although the sources of circRNAs are different, back-splicing circularization is a dominant way to produce circRNAs [35]. Generally, cis-regulatory elements and trans-acting factors regulate the biogenesis of cirRNAs by controlling the splices [36].
The majorities of circRNAs are generated by protein-coding genes and contain complete exons, which implies that RNA polymerase (Pol II) participates in their transcription and spliceosome maybe involved in their biogenesis [14]. Typically, circRNAs are generated from non-canonical splicing sites, and dependent on the non-canonical splicing machinery [37]. A study in Drosophila melanogaster demonstrated that depleting one component of the spliceosome dramatically decreased the ratio of linear RNA to circRNA. The above results indicated that the processing of pre-mRNA has slowed down and nascent RNA can be used for back-splicing [14]. Additionally, Kramer et al. [38] showed that depletion of splicing factors increased the level of circRNA, and multiple factors are taken additive effects, each splicing factor is not redundant in the formation of circRNA. The back-splicing hypothesis is catalyzed by canonical spliceosome machinery [39], which means the 5' and 3' ends of transcribed exons and/or introns are covalently linked. During this process, the down-stream splice site of donor exon/intron is joined to the upstream splice site of acceptor exon/intron on a pre-mRNA molecule [40] (Figure 1).
Multiple splice variants of circRNA transcripts can be generated from a single gene, depending on the exons selected during the back-splicing [41]. In the process of biogenesis, part of the RNA is transcribed from pre-mRNA, and exons are skipped as RNA folds. These structural changes generate some areas called lariat structures, in which discontinuous extrapolation zones initially become very close as the interior changes. CircRNA is formed after removing the subsequence from the lariat structure. This splicing process is called as 'lariat-driven circularization'. Due to the internalized feedback sequences of the former mRNA, the complementary pairing of introns on both sides forms circRNA. Another lariat-driven circRNA is ciRNA, whose formation processes depend on consensus RNA motifs near 5' splice site and branchpoint [42]. Studies in Homo sapiens have identified hundreds of ciRNAs [37]. CiRNAs are largely restricted to the nucleus and regulate gene expression in a cis manner.
General speaking, endogenous human circRNAs comprise two or three exons, or even more [42], and back-splicing requires cis-elements in intron flanking circularized exons [34]. Mutagenesis results indicate that the circularization of exons is dependent on the presence of short (~30- to 40- nucleotide) inverted repeats, such as the Alu element [43]. Besides, hundreds of trans-regulators, such as RNA-binding proteins (RBPs) and miRNA-containing ribonucleoprotein complexes, control the gene expression at the posttranscriptional level in eukaryotes [44]. For example, circRNAs can be produced when the synthetic Quaking (QKI)-binding sites insert into introns [45]. RBPs in different systems and organisms can also regulate exon circularization, such as adenosine deaminases acting on RNA, nuclear factors NF90/NF110, fused in sarcoma, DExH-Box helicase 9, serine/arginine-rich proteins and epithelial splicing regulatory protein 1 [46].
Types and characteristics of circRNAs
Generally, circRNA can be divided into three types, and some specific sources of circRNA have recently been discovered (Figure 1). It has been demonstrated that fusion-gene is the driver in many tumors, including leukemia [47] and lung cancer [48,49]. CircRNAs derived from fusion-gene (F-circRNA) (Figure 1B), such as PML/RARα and MLL/AF9 fusion genes, have a carcinogenic effect in vivo model [45]. F- circEA-2a, derived from EML4-ALK-v3b with 'AA' motif at the junction site, promotes the migration and invasion of non-small cell lung cancer cell and acts as a diagnostic marker for lung cancer [48,49]. Additionally, Wu et al. [50] demonstrated that F-circrSR1 and F-circrSR2 originated from the same fusion gene solute carrier family 34 member 2 and reactive oxygen species proto-oncogene 1 (SLC34A2-ROS1), both of which promoted the migration of lung cancer cells, but did not affect cell proliferation.
Commonly, mRNA translation will not terminate until meet the stop codon. However, in some unconventional conditions, the ribosome goes past the stop codon and continues translating into an otherwise untranslated region (UTR) of a transcript, which is known as 'stop-codon read-through' [51]. It is reported that the transcriptional read-through process is related to the formation of circRNA. Genes that interfere with the regulation of transcriptional termination may contribute to the production of transcriptional read-through products and promote the generation of circRNA from downstream gene sources [52]. Moreover, based on more than 2,000 human tumor specimens from different tissue sources, and a novel type of circular RNA transcript was uncovered, termed as read-through circRNA (rt-circRNA) (Figure 1C) [12]. Lately, Liu et al. [53] reported that the mitochondrial genome of humans and mice encode hundreds of circRNAs. These mecciRNAs (Figure 1D) act as a molecular chaperone to facilitate nuclear-encoded proteins entry into mitochondria. This is the first report of circRNA encoded by mammalian mitochondrial genome.
Functions of circRNAs
Modulate miRNA functions
Only small parts of the biological function of circRNAs have been investigated, one of the most typical mechanisms is to act as a miRNA sponge [13]. Many circRNAs have specific binding sites with miRNAs, which decreased miRNA activity and increase miRNA-target gene activity. Piwecka et al. [54] found that miR-7 expression was significantly decreased in CDR1as knockout mice, while some targets of miR-7 increased in CDR1as-knockout mice brain. Recently, it was demonstrated that Cyrano binds and targets miR-7 for degradation in CRISPR-Cas9 engineered mice [55]. This effect of Cyrano on miR-7 indirectly modulated the degradation of CDR1as by miR-671, which reveals a molecular regulatory network composed of non-coding RNAs.
Many studies illustrated that circRNAs participate in pathological conditions by sponging miRNAs. For example, high expression of circRNA-000284 in cervical cancer tissues promotes the migration, proliferation and invasion of cancer cells through the sponge of miR-506 [56]. CircRNAs, hsa_circ_0001564 and hsa_circ_0009910 can facilitate osteosarcoma cell proliferation, migration and invasion by targeting miR-29c-3p and miR-449a, respectively [57,58]. CircSEPT9 activates the LIF/STAT3 pathway by competitively binding to miR-637 in triple-negative breast cancer [59]. Jost et al. [60] successfully designed an artificial circRNA that can sponge miR-122 in hepatitis C virus (HCV) cells, thereby preventing the formation of HCV viral proteins and alleviating hepatitis C. In neonatal mouse ventricular cardiomyocytes, circRNA_000203 aggravated cardiac hypertrophy via specific adsorption miR-26b-5p and miR-140-3p [61]. Notably, most previously identified mammalian circRNAs are expressed at low level and do not have miRNA binding sites. Thus, circRNAs may not only act as miRNA sponges [35]. The latest findings showed that hsa_circ_0008558 (circLONP2) can regulate the intercellular miRNAs maturation and metastasis, accelerating the metastasis of colorectal cancer cell to other organs [62].
Regulate parental gene transcription
Some scholars have pointed out that circRNAs also can participate in gene transcription regulation. ANRIL is an antisense ncRNA in cyclin-dependent kinase 4 inhibitor (INK4) locus and is found in melanoma [63]. Increased expression or mutation of ANRIL is associated with coronary atherosclerotic heart disease and atherosclerosis [64]. ANRIL also inhibits the transcription of the INK4 and its alternative reading frame (ARF) by interacting with the polycomb group complex. Burd et al. [65] speculated that the circular ANRIL (cANRIL, product of ANRIL back-splicing) may regulate the expression of INK4/ARF. Most ciRNAs are abundant in the nucleus and have few miRNA target sites. Importantly, knockdown of ciRNAs hinders the corresponding parent gene transcription. Ci-ankrd52 (ciRNA from the gene ANKRD52) deposits in the transcription initiation region of the gene, associates with the Pol II extension mechanism, and promotes the function of RNA Pol II [34].
In addition, EIciRNAs, such as EIciPAIP2 and EIciEIF3J, promote host gene transcription by interacting with the U1 small nuclear ribonucleoproteins (snRNP) in RNA-RNA conjunction to form the EIciRNAs-U1 snRNP complex [66]. In Arabidopsis, exon 6 of the SEPALLATA3 gene cyclizes and forms an R-loop structure of RNA-DNA hybrid complex by circRNA strongly binding the cognate DNA locus of host gene. R-loop structure inhibits the transcription of this region and allows the exon skipping [67]. Subsequently, this circRNA functions by modulating the alternative back-splicing of parental transcript [68]. All these functions suggest that circRNA can bind to genomic DNA to regulate alternative splicing and cis-modulate diseases genes.
Interact with proteins
Some circRNAs have one or more RBPs sites, which can be used as proteins sponges to isolate and inhibit the biological function of proteins. For example, cADR1as and sex-determining region Y circRNAs can bind to miRNA response factor Argonaute to be degraded [68,69]. MBL circRNA (circMbl) flanking intron has many muscleblind (MBL) binding sites. Studies have shown that MBL is related to the biological synthesis of circMbl. When MBL protein is redundant, it will reduce the level of its own mRNA by promoting the generation of circMbl [14]. CircPABPN1 is derived from the poly (A) binding protein 1 (PABPN1) gene, and the Hu-antigen R (HuR) regulates the abundance of PABPN1 instead of circPABPN1. Interestingly, circPABPN1 and PABPN1 competes for the HuR protein binding sites [70].
Furthermore, circRNAs participate in a variety of physiological processes by interacting with proteins. Du et al [71] found circ-Foxo3 could be retained in the cytoplasm through interactions with anti-aging and anti-stress protein-related factors, such as inhibitors of DNA-binding 1 (ID-1) protein, focal adhesion kinase and hypoxia inducible factor 1α, thereby hindering the progress of the corresponding resistance. By binding to mouse double-minute 2 (MDM2) and p53, circFoxo3 can promote the ubiquitination of p53 induced by MDM2, leading to the overall degradation of p53 [72]. Recently, some studies have found that two or more proteins are assemble into greater protein complexes, such as enzymes and their substrates, in which circRNA can serve as protein scaffolds [73]. For example, circ-Foxo3 forms a ternary complex with p21 and CDK2, which can inhibit the function of CDK2 and block cell-cycle process [74]. Another example is the acute myeloid leukemia (AML)-associated circMYBL2. This circRNA regulates the translation efficiency of oncogene FMS-like tyrosine kinase-3 (FLT3) by recruiting PTBP1, resulting in progression of FLT3-internal tandem duplication AML [75]. Designing an exogenous circRNA to target cancer, diabetes, or other disease-related protein may be useful for medical treatments.
Translate into proteins
Although circRNAs have been well-described as miRNA sponges, many of them do not contain miRNA capture sites. Researches indicated that many circRNAs are generated from exons localized in the cytoplasm, suggesting that they can be translated into peptides by loading into ribosomes [33]. The genome of hepatitis delta (D) virus (HDV) has been identified with a natural translatable circRNA. The core circRNA of HDV encoded the HDV antigen, which plays critical role in the hepatitis disease development [76]. Guo et al. [77] found that circRNAs could be translated in human osteosarcoma U2OS cells, while most circular isoforms are far less efficiently compared to their linear isoforms. Because circRNAs do not have a 5'cap structure and cannot be translated into protein through cap-dependent mechanisms. Another way of RNA translation is to use the internal ribosome entry site (IRES) sequence as an internal ribosome entry sites for translatable circRNAs [78]. Artificial circRNAs with a RES can be translated in vitro or in vivo [79-81]. The covalently closed circular RNA is found in rice yellow mottle virus, which possesses an internal ribosome entry site and utilizes two (or three) open reading frames (ORFs) that directly translate into a 16-kDa highly basic protein [82].
Although many circRNAs have ORFs and with an upstream IRES elements, 46 of these are translated into corresponding proteins according to mass spectrometry [83]. So far, only circular RNA zinc finger protein 609 (circ-ZNF609), circMb1, circ-FBXW7, circPINTTaxon2, circ-SHPRH and circβ-catenin have the coding ability in related studies. Circ-ANF609 is related with heavy polysomes and can be translated in an IRES-dependent mode, even though the translation activity of the circular template is much lower than the linear counterpart template [84]. Furthermore, decreases of circ-ZNF609 expression cause damage to the proliferation of human and mouse myoblasts, and the translation of circ-ZNF609 can be significantly induced by heat shock. Circ-Mb1 shares the same start codon with the host RNA, encoding a peptide in a cap-independent manner in Drosophila, starvation can regulate the circ-Mb1 production and/or stability depending on 4E-BP and FOXO proteins [15].
Additionally, circ-FBXW7 [85], circ-SHPRH [86, 87] and circPINTTaxon2 [88] can produce FBXW7-185aa, PINT87aa and SHPRH-146aa, respectively. All of them can inhibit human glioblastomas. Circ-FBXW7 is extremely abundant in the brain, and IRES drives a 21-kDa protein named FBXW7-185aa. FBXW7-185aa directly interacts with de-ubiquitinating enzyme USP28, which can stabilize the oncoprotein c-Myc by inhibiting FBXW7α (the most abundant isoform of the FBXW7 gene) activity. FBXW7-185aa can inhibit proliferation and cell cycle of U251 and U373 cell lines by this way [85]. Two reports have successively reported that circ-SHPRH (originate from the sucrose non fermenting 2 histone linker PHD RING helicase (SHPRH) gene) employs overlapping genetic codes to encode a 17-kDa peptide named SHPRH-146aa [85,86]. Theoretically, SHPRH-146aa protects full-length SHPRH from degradation by ubiquitin proteasomes. E3 ubiquitin protein ligase SHPRH increases its anti-tumor function by targeting proliferating cell nuclear antigen. Finally, circPINTTaxon2 is generated from long intergenic non-protein-coding RNA p53-induced transcript gene and encodes a peptide named PINT87aa, which can inhibit multiple oncogenes though interacting with polymerase associated factor complexes and act as a tumor suppressor [88]. Polymerase associated factor complex is involved in the recruitment of RNA Pol II and the transcriptional elongation of downstream genes [89]. Circβ-catenin encodes a 370-amino acid β-catenin isoform by IERS, which prevents β-catenin from phosphorylation and degradation by antagonizing glycogen synthase kinase 3β and subsequently promotes tumor growth through activating the Wnt pathway [90]. The information about circRNA-encoded proteins is scattered in many published papers, which is not conducive to further exploration on circRNA translation. Liu et al. [91] presented an ncEP database, which recorded all published articles containing proteins or peptides encoded by ncRNAs. These discoveries imply that the coding potential of circRNAs has been largely disregarded, which may help to deepen our understanding of circRNAs.
Pseudogenes derived from circRNAs
Around 19,000 conserved pseudogenes have been revealed in human by sequencing efforts and their transcription patterns display tissue and pathological condition specificity [92]. Researchers developed computational pipeline to identify circRNAs-originated pseudogene, called CIRCpseudo, to determine whether stable circRNAs can be retrotranscribed and integrated into the host genome. Dozens of circRNAs-derived pseudogenes have been identified [93]. Among them, at least 33 circRNAs at the ring finger and WD repeat domain 2 (RFWD2) locus-derived pseudogenes (circRFWD2) were found in different mouse strains, characterized by the exon 6-exon 2 anchor in a reversed order. The pseudogene derived from circSATB1 can specifically bind to CCCTC-binding factor and/or Rad21-binding sites in several mouse cell lines to regulate gene expression. In addition, the insertion of retrotransposed circRNAs into the genome changes the composition of genomic DNA and regulates the potential for gene expression [93].
CircRNA acts as a ribozyme
Satellite virus, viroid and HDV RNA exist in a circular form, and RNA is part of a replication ring that contains self-dividing sequences (ribozymes). Through the research of the classical type I hammerhead ribozyme (HHR) motif in diverse metazoans genomes, a novel and conserved small ribozyme has been found to efficiently synthesize circular RNA [94]. A systematic analysis of the genomes of cnidaria (a coral), mollusca (a mussel) and chordata (a salamander) has revealed that there are abundant type I HHR motifs in the DNA tandem repeats of their genomes, with a length of about 170-400 nt, mostly in the form of linear and circRNAs. These modifications confirmed the existence of novel natural pathways for circRNA biosynthesis through a conserved autocatalytic RNA in metazoan.
M6A modification
Epigenetic modifications are involved in cell fate and respond to environmental stimuli by regulating gene expression [95]. These modifications are implicated in the development of organism [18,96,97]. While the role of epitranscriptomic modifications in gene regulation is scarcely acquainted. Recently, considerable studies have revealed the role of post-transcriptional RNA modifications in modeling an epitranscriptomic landscape of gene expression [23,98]. RNA modifications tend to deposit in highly abundant RNA species to regulate RNA splicing, transport, translation and turnover [99].
There are more than 170 known RNA modifications, such as m6A, N1-methyladenosine, inosine, 5-methylcytidine, 5-hydroxymethylcytidine, pseudouridine, N6,2′-O-dimethyladenosine, N4-acetylcytidine, N7-methylguanosine, 8-oxoguanosine and 2′-O-methyl [100]. Among them, m6A is the most abundant modifications of mRNAs found in all eukaryotes [22]. Since around 20-40% of mammalian transcripts are m6A methylated, and the methylated mRNAs tend to have multiple m6A modifications [21]. Furthermore, recovering the mechanisms of deposition, removing and recognition RNA modifications (more preferably known as writers, erasers and readers, respectively) have helped to understand the fates of those post-transcriptional modifications in cellular processes, such as cell growth, body development and diseases.
Regulators of m6A
M6A is similar to DNA and histone methylation, a dynamic and reversible event [101], and the methyltransferases and demethylases coordinately regulate the deposition and decay of m6A, also named with m6A writer and m6A eraser respectively (Figure 2). M6A is installed by a multiprotein methyltransferase complex (MTC, also termed m6A 'writer'). The core of MTC is a heterodimer core catalytic subunit composed of methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) [102-104]. There are many other accessory subunits, including wilms tumor 1 associated protein (WTAP) [105], vir-like m6A methyltransferase-associated (also known as Virilizer or KIAA1429) [106], RNA binding motif protein 15 [107], cbl proto oncogene-like protein 1 (also known as Hakai) [108] and zinc finger CCCH domain-containing protein 13 (ZC3H13) [107]. METTL3 is the catalytic subunit and METTL14 acts as the RNA-binding platform. Moreover, the METTL3 homolog methyltransferase-like 16 (METTL16) can catalyze the m6A onto the U6 small nuclear RNA (snRNA) and some structured RNAs [110]. In addition to the above cofactors, other catalytic subunits join the methyltransferase complex, which is designed for precise post-transcriptional regulation [85]. Two demethylases, fat mass and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5), can remove the m6A modification, acting as erasers [101,111].
Mechanism of m6A. The dynamic modification of m6A modification is installed by the methylransferase complex (writers) composed of METTL3, METTL14, WTAP, KIA1429, ZC3H13 and RBM15/RBM15b, and removed by RNA demethylases (erasers) FTO and ALKBH5. Methyl-specific binding proteins (readers) in are the YTH family, which can achieve the biological functions of m6A modification.
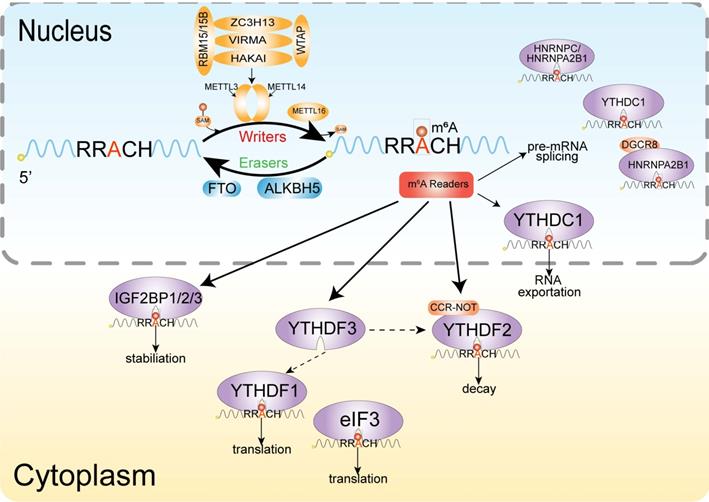
The m6A modification realizes its biological function specific recognition and binding of RNA binding proteins (readers), thereby affecting RNA fate by regulating RNA metabolism. Several proteins or complexes that specifically recognize m6A sites have been identified, including YT521-B homology (YTH) domain-containing protein [112], eukaryotic initiation factor 3 [96], IGF2 mRNA binding proteins (IGF2BP) families [113] and heterogeneous nuclear ribonucleoprotein protein family. The YTH domain can recognize m6A through a conserved aromatic cage and another two proteins FMRA, LRPPRC 'read' this modification [114].
Characteristics and functions of m6A
M6A modification exists in different types of mammalian cell, including blood, liver, neuronal and muscle cells and so on. The deposition of m6A modification affects the process of cell metabolism, transcription, splicing, translation, degradation and localization.
The distribution of m6A modification on mRNA is sequence-specific and often appears on a consensus RNA motif of RRACH (R = G or A; H = A, C or U), coding sequence, 3'UTRs, especially near stop codons [115]. M6A modification can cause changes in RNA secondary structure by affecting RNA pairing, regulate RNA stability and its interaction with proteins [116,117]. M6A is also highly enriched in the exon region near the splicing site, and has significant spatial overlap with the splicing factor SRSF1 and SRSF2 RNA binding sites. Experimental studies have revealed that m6A-modified exons tended to be retained during splicing [118]. Ke et al. [119] illustrated the molecular mechanism of m6A-modified exons being retained during the splicing process: m6A binding protein YTHDC1 promotes its retention by promoting SRSF3 and suppressing SRSF10 in combination with m6A-modified exons. M6A also shows a tendency of enrichment in the last exon of a transcript, which may be related to the regulation of the alternative polyadenylation of mRNA. The unstable pairing of m6A-U causes the melting of double-stranded RNAs and transformation of secondary structure.
M6A methylation on circRNAs metabolism
Chemical covalent modification of nucleotides molecules is a ubiquitous life process and many studies have illustrated chemical modifications in DNA and RNA. There have been identified more than 100 types of chemical modifications of mRNAs, rRNAs, snRNAs and snoRNAs have been identified in organisms. It has been reported that linear mRNA and lncRNA have m6A modification. Currently, m6A modification has been identified and characterized in circRNA, but the potential regulatory mechanism has not been fully elucidated [120]. The researches on circRNAs with m6A-modification, and the function of these m6A-modifications are described in Figure 3.
M6A on circRNA translation
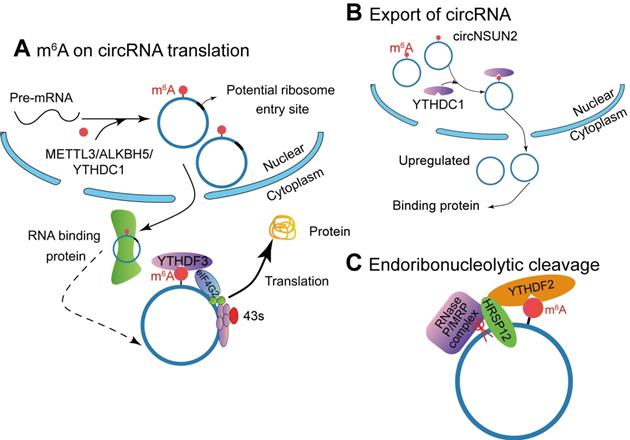
Studies have shown that circrRNAs have coding potential. Yang et al. [16] described a new mechanism that circRNA with m6A residues can be translated in a cap-independent manner (Figure 3A). These translatable circRNAs contain a large number of m6A consensus motifs (13% of total circRNAs), and a single m6A site is enough to initiate circRNA translation. The translation of m6A-modified circRNA depends on IF eIF4G2 (non-canonical eIF4G) protein and m6A reader YTHDF3. The m6A-driven translation is reversible, because methyltransferase METTL3/14 can increase the translation efficiency, while inhibited by the demethyla FTO. Moreover, mRNA with m6A modification in its 5'UTR can be translated under specific cellular stress (such as amino acid starvation) and through 5'cap-independent way to regulate the translation [121]. Here, they proved that m6A modification continuously activates the translation of circRNA with continuation of heat shock stimulation or overexpression of METTL13/14. They also authenticated 250 circRNAs associated with polysome. Puromycin can significantly reduce the amount of circRNAs, which indicates that circRNAs with polysomes may be actively translated [16]. However, how m6A modified circRNA is generated has not been clarified yet.
Tang et al. [26] found that m6A modification could promote the formation of ORF-carrying circRNA by studying the development process of male germ cells in mice. Sequencing analysis during spermatogenesis has found that a large number of circRNAs are generated while the corresponding linear mRNA decreased. Some of the circRNAs often carry high levels of m6A modification on both sides of the reverse junction point, and m6A is often enriched around the ORF start and stop codons of the mRNA (Figure 3A). Interestingly, Tang et al. [26] found that nearly half of these circRNAs carry longer ORFs whose start codons are modified by m6A to bind ribosomes. Hundreds of peptides encoded by these circRNAs were detected by liquid chromatography and mass spectrometry. This discovery indicates that m6A can mediate the generation of circRNA and reports a new mechanism that relies on circRNA to achieve the stable expression of protein products after linear RNA deletion. Di Timoteod et al. [121] further demonstrated this point by taking circ-ZNF609 as a study case. The generation of circ-ZNF609 modified by m6A is regulated by METTL3/YTHDC1, and the recognition of m6A site by YTHDF3 and eIF4G2 promotes circ-ZNF609 translation.
M6A modification affects the function of circRNAs. (A) METTL3, ALKBH5 and YTHDC1 involve in the biogenesis of circRNA, and m6A-modified circRNA can be translated into proteins in a cap-independent manner with YTHDF3 and translation initiation factors eIF4G2 and eIF3A. (B) The m6A reader YTHDC1 promotes circRNA cytoplasmic export. (C) The YTHDF2-HRSP12 mediated RNase P/MRP complex binds and degrades circRNAs.
Endoribonucleolytic cleavage of circRNA
M6A is involved in regulating multiple steps of mRNA modification, including stability, and the degradation of mRNA containing m6A modification is mediated by YTHDF2 (Figure 3B). In detail, it depends on the presence of the adaptor protein heat-responsive protein 12 (HRSP12)-binding site in the messenger ribonucleoprotein. If it exists, deadenylation pathway mediated by CCR4/NOT complex is activated, leading to deadenylation and decay of m6A-containing mRNAs [122] or endoribonucleolytic cleavage pathway by the YTHDF2-HRSP12-RNase P/mitochondrial RNA-processing (MRP) (endoribonuclease) complex causing degradation of YTHDF2-bound RNAs [123]. Comparing to mRNA, circRNAs have a covalently closed loop and do not have a 3' polyadenylated tail, they are naturally more stable than their homologous linear RNAs in both intracellular and extracellular environments and their degradation can be avoided [124]. Therefore, circRNAs can only be degraded by endoribonucleolytic cleavage. Park et al. [123] demonstrated that circRNAs containing m6A can be decayed through YTHDF2-HRSP12-RNase P/MRP-mediated endoribonucleolytic cleavage. The abundance of circRNAs containing m6A increased after a component of RNase P/MRP was down-regulated [123].
Promote the nuclear export of circRNA
Following biogenesis, most circRNAs are efficiently exported to cytoplasm. However, the intron-containing circRNAs accumulate in the nucleus [68,69]. Some nondividing cells (e.g. neurons) highly express circRNAs. Therefore, the transport process regulates the localization of circRNAs, depending on nuclear envelope breakdown during mitosis. Based on RNA interference screening in Drosophila, the interference of Hel25E significantly led to the enrichment of circRNA in the nucleus. Further identification showed that Hel25E was an important regulator of post-transcriptional nuclear export of circRNAs. In human cells, circRNAs are transported from the nucleus in a transcript-length-dependent manner to the cytoplasm via Hel25E homogenous proteins: ATP-dependent RNA helicase DDX39A (also known as nuclear RNA helicase URH49 or URH49) and spliceosome RNA helicase DDX39B (also known as DEAD box protein UAP56 or UAP56) [125].
Knockout of m6A demethylase ALKBH5 accelerates the nuclear export of mRNA (Figure 3C) [126]. Mechanism studies have proved that YTHDC1 interacts with splice factor SRSF3 to recruit NXF1, thereby promoting the nucleation of m6A mRNA [127]. Chen et al. [128] found that m6A-modified circRNA NOP2/Sun RNA methyltransferase family member 2 (NSUN2) locus (circNSUN2) exports from the nucleus to cytoplasm by binding to YTHDC1. Then cytoplasmic circNSUN2 interacts with RBP and insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) to promote the stability of high mobility group AT-hook 2 (HMGA2) transcript [128]. Compared with paracancerous tissues, circNSUN2 is highly expressed in colorectal cancer tissues during the progression of lymph node metastasis and liver metastasis in colorectal cancer, and the expression level of circNUSN2 gradually increases. Further, in vivo nude mouse metastasis and in vitro cell function experiments show that circNSUN2 promotes the metastasis of colorectal cancer tumours. This circRNA binds to the m6A binding protein YTHDC1 in the nucleus, and YTHDF1 regulates the nuclear localization of circNSUN2 in an m6A-dependent manner. The cytoplasm of circNSUN2 can bind to the RNA-binding protein IGF2BP2 and the downstream HMGA2 (a member of the high motility group, HMG) mRNA to form a circNSUN2/IGF2BP2/HMGA2 RNA-protein ternary complex, which promotes the stability of HMGA2 mRNA. Ultimately, m6A modified circNSUN2 promotes liver metastasis of colorectal cancer tumors by accommodating the stability of HMGA2 mRNA [128]. It is suggested that the interacting proteins can help to discover inner mechanism of circRNAs.
m6A-modified circRNA associated with pathophysiological processes
With the gradual understanding of the structure and function of m6A modification and circRNA, researchers have investigated the role of m6A-modified circRNAs in physiological and pathological processes. Additionally, many evidences were showed that m6A-modified circRNA are related with the progress of various cancer [24, 128-132], immune response [133], CVD [134], mental illness [135], myoblast differentiation [25], spermatogenesis [26] and so on. The pathophysiological processes regulated by m6A-modified circRNAs are summarized in here.
m6A-modified circRNA in pathological processes
Although most m6A-modified circRNAs are thought play a role in pathological processes, the function of m6A modification on circRNA is unknown. Table 1 lists the proposed mechanism and functions of m6A-modified circRNAs, and is discussed below.
The mechanism of m6A-modified circRNAs in pathological processes
| Type of disease | circRNA | Mechanisms of action | Putative biological function | Refs |
|---|---|---|---|---|
| Gastric cancer (GC) | circPVRL3 | miRNA sponge or template for translation | Proliferation and migration of GC cells | 129 |
| Poorly differentiated gastric adenocarcinoma (PDGA) | A set of circRNA (has-circRNA-0077837) | - | Progression of PDGA | 130 |
| Colorectal cancer (CC) | circNSUN2 | Protein scaffold (forms a circNUSN2/ IGF2BP2/ HMGA2 RNA-protein ternary complex) | Liver metastasis of CC | 128 |
| Hepatocellular carcinoma (HCC) | circRNA-SORE | miRNA sponge (miR-103a-2-5p and miR-660-3p) | Sorafenib resistance of HCC | 132 |
| Esophageal squamous cell carcinoma | circSLC7A5 | Template for translation | Biomarker for cancer diagnosis and prognosis | 140 |
| Cervical carcinoma | circE7 | Template for translation (E7 oncoprotein) | Link to the transforming properties of some human papillomaviruses | 143 |
| Acute coronary syndrome | Has_circ_029589 | - | Induce macrophage pyrptosis | 146 |
| Hypoxia mediated pulmonary hypertension | A set of circRNAs (circ-Xpo6, circ-Tmtc3 ) | Affect the circRNA-miRNA-mRNA network | Potential therapy targets | 147 |
| Major sepressive disorder | circSTAG1 | Protein partner (Binding with ALKBH5) | Depressive | 135 |
Cancers
Wealth of recent studies have revealed that circRNA and m6A modification exert a regulatory function in different diseases, and many of them highlighted a role in various cancer [17,136]. Cancer is a leading cause of mortality worldwide [137]. Based on the structure and function of m6A-modified circRNA, researchers have found that they play an important role in the progression, proliferation and metastasis of cancers, such as colorectal cancer [128], gastric cancer [129], hepatocellular carcinoma (HCC) [130,131] and poorly differentiated gastric adenocarcinoma [132].
A study on gastric cancer (GC) found that knockdown of circRNA poliovirus receptor-related 3 (circPVRL3) significantly promoted cell proliferation [129]. The internal ribosomal entry sites, open reading frame and m6A modification are present on circPVRL3, indicating that circPVRL3 has coding potential. Overall, this study pointed to the m6A-modified circPVRL) in the carcinogenesis [129]. 30 upregulated circRNAs and 35 downregulated circRNAs were evaluated in poorly differentiated gastric adenocarcinoma (PDGA) by circRNA microarray. The level of m6A was positive relative with circRNA in PDGA, which indicated that m6A-modified circRNA plays a role in GC progression [130].
Liver cancer has a high morbidity and mortality rate worldwide [137]. In HCC, circ_KIAA1429 (originate from KIAA1429) can accelerate the progress of HCC through the m6A-YTHDF3-Zeb1 mechanism, and may be a potential target for the HCC therapy [131]. In addition to modifying m6A modification process, circRNA can also affect the stability of circRNA via changing the methylation states. In HCC, circRNA-SORE acts as a competitive endogenous RNA, which induces sorafenib resistance by competitively activating the Wnt/β-catenin pathway by adsorbing miR-103a-2-5p and miR-660-3p. They found that the increasing m6A level of circRNA-SORE can maintain its stability and promotes the development of drug resistance [132]. This paper expounds the new mechanism of drug resistance in targeted therapy of liver cancer and provides new clues for finding new therapeutic targets for advanced liver cancer patients.
Lung cancer is the leading cause of cancer death in men [138]. Recently, a study indicated that circNDUFB2, derived from NADH: ubiquinone oxidoreductase subunit B2, is frequently downregulated in non-small cell lung cancer (NSCLC), and negatively correlated with the malignant characteristics of NSCLC. CircNDUFB2 acts as a scaffold of tripartite motif containing 25 and IGF2BPs to form a ternary complex, which promotes the degradation of IGF2BPs and inhibits the growth and metastasis of NSCLC cells. Furthermore, m6A-modified circNDUFB2 can enhance this effect [139]. In addition, circNDUFB2 recognized by RIG-I can active RIG-I-MAVS signaling cascades, thereby recruiting immune cells into the tumor microenvironment. Apart from enhancing IGF2BPs stability, the down-regulated circNDUFB2 in NSCLC also leads to immune evasion and promotes tumor progression [139].
In addition, some m6A-modified circRNAs have not yet clarified their unctions. In esophageal squamous cell carcinoma (ESCC), circ-SLC7A5 was significantly up-regulated in the plasma of ESCC patients and high levels of circ-SLC7A5 are correlated with shorter survival time. Bioinformatics predicts that circ-SLC7A5 has m6A modification structure with translation potential [140]. Zhou et al. [32] developed a computational pipeline together with experimental results, which revealed m6A circRNAs are widely spread in circRNAs (they accounts for 41% of the total circRNAs) and are significantly different between human embryonic stem cells (hESCs) and HeLa cells. Moreover, even if circRNAs are originated from the same mRNA, they cannot be detected in different cell types. Up to 65% of m6A-modified circRNA derived from HeLa cells cannot be detected in hESCs. Furthermore, about 41% of HeLa-generated circRNA do not appear in hESCs. Zhou et al. [32] also found some other features of m6A-modified circRNA: 1) the distribution of m6A modification sites on circRNAs and corresponding mRNAs are different; 2) the parental exons of m6A circRNAs are relatively greater than those of non-methylated circRNAs; 3) circRNAs share the same m6A modification enzymes with linear RNA; 4) the host mRNA is unstable. The m6A expression level is negatively associated with circRNA expression. This article found that circRNA has a cell-specific m6A modification state, which can be used to diagnose diseases.
Immune response
CircRNA is automatically spliced and cyclized after in vitro transcription to transfected cells. Unexpectedly, this type of circRNA can induce the activation of innate immune response and inhibition of the RNA virus infection process by retinoic acid-inducible gene-I (RIG-I) [134]. Endogenous circRNA with m6A modification can suppress innate immune responses by inhibiting RIG-I activation [141]. When cells are stimulated by polyinosinic acid-polycytidylic acid or infected by viruses, the endonuclease RNase L activates and degrades circRNA molecules. Protein kinase (PKR) is released and activates the downstream antiviral mechanism systematic lupus erythematosus (SLE). The expression of circRNA in the peripheral blood mononuclear cells (PBMC) of SLE patients decreased and the activity of PKR increased. However, overexpression of circRNA in PBMC derived from SLE patients can reduce the activity of PKR, which may be helpful for the treatment of autoimmune diseases such as SLE [134].
Chen et al. [134] found that the lack of sample purity might be caused by the non-specificity of the substituted linear RNA with 5' phosphate effect. Thus, Chen et al. [134] did some other experiments to show that their research system is reliable and produced a novel mechanism. The m6A reader YTHDF2 may be reason for the RNA degradation [19, 24]. Studies have shown that m6A modification controls immune responses [142]. CircE7 was modified by m6A and translated to produce E7 oncoprotein [143]. CircE7 exists in TCGA RNA-Seq data of human papillomavirus (HPV)-positive cancer cells and in cell lines where only free HPV is present. These results prove that virus-derived that can encode proteins has biological functions and is related to certain HPV transformation characteristics [144].
Wesselhoeft et al. [144] found that exogenously synthesized circRNA transfected into cells did not induce toll-like receptor/RIG-I-mediated innate immune response, and circRNAs can more effectively translate into proteins in mouse tissues. Deposition of m6A modification in circRNA can recruit and bind YTHDF2, inhibit the activation of RIG-I/K63-Ubn/circRNA complex and do not induce the innate immune response [144]. Collectively, these finding proved that m6A modification of circRNAs is a key regulator in circRNA related immune responses.
CVD
CVD causes 17.5 million annual deaths and increases the burden of public health [145]. Cell death is one of the critical pathological mechanism of atherosclerosis (AS). IFN regulatory factor (IRF)-1 has been demonstrated play a critical role in regulating cell death of AS. The relative RNA expression level of hsa_circ_0029589 in macrophages of acute coronary syndrome decreased, while the m6A level of hsa_circ_0029589 and the m6A METL3 were significantly increased [146]. In addition, the overexpression of IRF-1 inhibited the expression of hsa_circ_0029589 in macrophages, and simultaneously induced the expression of m6A and METL3. Overexpression of hsa_circ_0029589 or inhibition of METTL3 can significantly increase the expression of hsa_circ_0029589 and reduce macrophage apoptosis [146]. Increased m6A abundance in hypoxia mediated pulmonary hypertension (HPH) reduces the total circRNAs abundance in hypoxia in vitro [147]. M6A affects the co-expression network of circRNA-siRNA-mRNA during hypoxia. Specifically, the m6A-modified circXpo6 and circTmtc3 may be used as HPH biomarkers due to their different enrichments in specific pathological condition [146]. The difference of m6A circRNAs in cells or tissues suggests that they may be involved in progression of CVD or act as a pathological marker.
Age-related cataract
Age-related cataract (ARC) is the leading cause of world blindness, which causes ~50% blindness worldwide [148]. Few circRNA has been identified in cataract. For instance, circKMT2E sponges miR-204 to regulate the pathogenesis of diabetic cataract [149]. Total circRNA expression level was decreased in cortical of ARC (ARCC). Compared with non-m6A modified circRNAs, the expression of highly abundant m6A circRNAs were mostly reduced. Bioinformatics analysis predicted that ALKBH5 was significantly upregulated in lens epithelium cells (LECs) of ARCC among the five major methyltransferases. Those results indicate that M6A modification of circRNAs may be associated with LEC lesions by regulating genes/pathways related to the onset of ARC [150].
Major depressive disorder
Major depressive disorder (MDD) is a severe mental disorder with high incidence [151]. Currently, several studies have demonstrated circRNAs play a role in MDD. CircDYM is originated from DYM gene exon 4, 5, 6 circulation, and was found to be significantly decreased in plasma of MDD patients [152]. Huang et al. found that circRNA stromal antigen 1 and 2 (STAG1) play a crucial role in attenuating depressive-like behaviors. circSTAG1 can bind ALKBH5 to inhibit its nuclear entry, thereby changing the m6A modification of total RNA and increasing the level of m6A modification of RNA, including fatty acid amide hydrolase (FAAH) mRNA [135]. The expression of circSTAG1 in the hippocampus of CUS depressed mice was significantly reduced, and the m6A modification of FAAH mRNA was changed by ALKBH5, which ultimately led to the depression phenotype. 345-395aa of ALKBH5 may be the region that binds to circSTAG1. This study revealed that circRNA regulates the process of m6A modification by combining to m6A modification enzyme.
m6A-modified circRNA in physical processes
Myogenesis
Increasing evidences have shown that circRNAs are abundant in skeletal muscles during the differentiation myoblasts, and the global expression level of circRNA changes dynamically [153]. Additionally, several circRNAs (such as circ-ZNF609 and circular RNA supervillin have been proved played an important role in the differentiation and development of skeletal muscle [154]. Here, newly study has found that 581 circRNAs are differentially expressed between skeletal muscle C2C12 myoblasts and myotubes. These myogenic-specific genes, 91 miRNA and the top 30 upregulated circRNAs forming a regulating network with 239 edges. Among the 581 circRNAs, 224 circRNAs have been identified as having coding potential. In addition, the number of m6A motifs in 224 cicrRNAs with encoding potential was also determined. Totally, 44 cicrRNAs had an m6A motif, 43 cicrRNAs had two m6A motifs, and 137 cicrRNAs had three or more m6A motifs. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were performed that 75 cicrRNAs based on the linear counterparts were related with actin cytoskeleton and metabolic pathways [25]. These annotations may offer novel insights in the development and differentiation of skeletal muscle and provide new therapeutic strategies foe muscular diseases.
Spermatogenesis
Spermatogenesis is a highly complex and specialized cell differential process during which diploid spermatogonial stem cells produce spermatozoa [155]. Studies have shown that depletion of m6A writers (Mettl3 and Mettl14 or their homologs) can impair gametogenesis in multiple organisms [156]. Recent research demonstrated that m6A is dynamically regulated and plays an important role in shaping gene expression during spermatogenesis [157]. Moreover, a large number of circRNA will be generated from the thick line stage after meiosis to the round cell stage and the corresponding linear mRNA expression decreases during spermatogenesis [26]. Tang et al. [26] studied the developmental process of male germ cells in mice that m6A modification can promote the formation of ORF-carrying circRNA. Sequencing analysis during spermatogenesis has found that a large number of circRNAs were generated while the corresponding linear mRNA was decreased. Some circRNAs often have high levels of m6A modification on both sides of the reverse junction point, and m6A is often enriched around the ORF start and stop codons of the mRNA. Interestingly, Tang et al. [26] found that nearly half of these circRNAs carry longer ORFs whose start codon is modified by m6A to bind ribosomes. Hundreds of peptides encoded by these circRNAs were detected by liquid chromatography and mass spectrometry. This research not only proved that m6A can mediate the generation of circRNA, but also discovered a novel mechanism for stable expression of circRNA-dependent protein product after linear RNA deletion.
Conclusions and future directions
Recent researches on circRNA with m6A modification uncovered that epigenetic modification can also affect the circRNAs involved cellular process. CircRNA interacts with m6A-related proteins can regulate the progression and development of cancer. There are very few studies about circRNAs with m6A modification, so more understanding about the regulation and functions of this modification in circRNAs is still needed. Furthermore, m6A is most common in linear RNA and circRNA is considered to be the shaper of the RNA world [158]. These findings indicate that the m6A modification may be a means for circRNA to achieve this reshape function. Therefore, m6A modifications in circRNA can uncover more information about the function of epigenetic modification in RNA world. However, limitations still exist in current researches. Further exploration can be conducted from the following aspects: 1) Determining the molecular mechanism of circRNA m6A modification (Are the 'writers', 'erasers' and 'readers' completely consistent with linear RNA?); 2) Elucidating the formation of cell types or tissues specific circRNAs (Why do circRNAs have specific modifications? Do they have a special function? Are there circRNAs that are not modified by m6A?); 3) Differentiating between specific exons and classical exons with m6A modification. Further studies are needed to unravel those mysteries.
Furthermore, this review describes the role, mechanism and application of m6A-modified circRNA in the pathophysiological processes. The most important role of circRNAs is in the development of various diseases and life processes. Nevertheless, the exact mechanism for m6A-modified circRNA in life progression is still unclear, because m6A modification can act as a double-edged sword. In spite of epigenetically modification play a significant regulatory role in the biological function of cell, while the terminal aim of medical researches is to be applied in the clinic. Designing targeted therapeutic drugs based on these newly elucidated molecular mechanisms the subject.
Abbreviations
CircRNAs: Circular RNAs; ncRNA: Noncoding RNA; m6A: N6-methyladenosine; miRNA: MicroRNA; EcircRNAs: Exonic circRNAs; CiRNAs: Intronic circRNAs; EIciRNA: Exon-intron circRNAs; Pol II: RNA polymerase II; RBPs: RNA-binding proteins; QKI: Quaking; F-circRNA: CircRNAs derived from fusion-gene; ROS: Reactive oxygen species; SLC34A2-ROS1: Solute carrier family 34 member 2 and ROS proto-oncogene 1; UTR: Untranslated region; rt-circRNA: Read-through circRNA; HCV: Hepatitis C virus; NMVCs: Neonatal mouse ventricular cardiomyocytes; CRC: Colorectal cancer; ANRIL: The antisense ncRNA in cyclin-dependent kinase 4 inhibitor locus; INK4: Cyclin-dependent kinase 4 inhibitor; ARF: Alternative reading frame; SnRNP: Small nuclear ribonucleoproteins; MBL: Muscleblind; circMbl: Muscleblind circRNA; PABPN1: Poly(A) binding protein 1; HuR: Hu-antigen R; MDM2: Mouse double-minute 2; AML: Acute myeloid leukemia; FLT3: FMS-like tyrosine kinase-3; HDV: Hepatitis delta (D) virus; IRESs: Internal ribosome entry sites; ORFs: Open reading frames; SHPRH: Sucrose non fermenting 2 (SNF2) histone linker PHD RING helicase; LINC-PINT: Long intergenic non-protein-coding RNA p53-induced transcript; RFWD2: Ring finger and WD repeat domain 2; CTCF: CCCTC-binding factor; HHR: Hammerhead ribozyme; MTC: Methyltransferase complex; METTL: Methyltransferase-like; WTAP: Wilms Tumor 1 associated protein; ZC3H13: Zinc finger CCCH domain-containing protein 13; SnRNA: Small nuclear RNA; FTO: Obesity-associated protein; ALKBH5: α-ketoglutarate-dependent dioxygenase alkB homolog 5; YTH: YT521-B homology; IGF2BP: IGF2 mRNA binding proteins; mRNP: Messenger ribonucleoprotein; MRP: Mitochondrial RNA-processing; NSUN2: NOP2/Sun RNA methyltransferase family member 2; IGF2BP2: Insulin-like growth factor 2 mRNA-binding protein 2; HMGA2: High mobility group AT-hook 2; GC: Gastric cancer; PDGA: Poorly differentiated gastric adenocarcinoma; CC: Colorectal cancer; HCC: Hepatocellular carcinoma; circPVRL3: circRNA poliovirus receptor-related 3; ESCC: esophageal squamous cell carcinoma; hESCs: Human embryonic stem cells; RIG-I: Retinoic acid-inducible gene-I; PKR: Protein kinase; SLE: Systematic lupus erythematosus; PBMC: Peripheral blood mononuclear cells; HPV: Human papillomavirus; AS: Atherosclerosis; HPH: Hypoxia mediated pulmonary hypertension; ARC: Age-related cataract; ARCC: Cortical of ARC; LECs: Lens epithelium cells; MDD: Major depressive disorder; STAG: Stromal antigen 1 and 2; FAAH: fatty acid amide hydrolase; RNase: Ribonuclease; LM: Liver metastasis; RNPs: Ribonucleoprotein particles; NSCLC: Non-small cell lung cancer.
Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (11872134, 11672051), Natural Science Foundation of Chongqing, China (cstc2020jcyj-msxmX0035) and sharing fund of Chongqing university's large-scale equipment (202103150044).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Salzman J, Chen RE, Olsen MN. et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777
2. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167-79
3. Wang Y, Mo Y, Gong Z. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25-32
4. Rybak-Wolf A, Stottmeister C, Glažar P. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870-85
5. Sanger HL, Klotz G, Riesner D. et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852-56
6. Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547-55
7. Si-Tu J, Cai Y, Feng T. et al. Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. J Biol Macromol. 2019;122:1235-43
8. Holdt LM, Stahringer A, Sass K. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nature Commun. 2016;7:12429-42
9. Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. 2017;14:1028-34
10. Wu H, Wu S, Zhu Y. et al. Hsa_circRNA_0054633 is highly expressed in gestational diabetes mellitus and closely related to glycosylation index. Clin Epigenetics. 2019;11:22-34
11. Zhang C, Wang X, Chen Y. et al. The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4(+)T cells of systemic lupus erythematous. Clin Sci (Lond). 2018;132:2285-98
12. Vo JN, Cieslik M, Zhang Y. et al. The landscape of circular RNA in cancer. Cell. 2019;176:869-81
13. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
14. Ashwal-Fluss R, Meyer M, Pamudurti NR. et al. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
15. Pamudurti NR, Bartok O, Jens M. et al. Translation of circRNAs. Mol Cell. 2017;66:9-21
16. Yang Y, Fan X, Mao M. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-41
17. Kadumuri RV, Janga SC. Epitranscriptomic code and its alterations in human disease. Trends Mol Med. 2018;24:886-903
18. Frye M, Harada BT, Behm M. et al. RNA modifications modulate gene expression during development. Science. 2018;361:1346-49
19. Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21:552-9
20. Yoon KJ, Ringeling FR, Vissers C. et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877-89
21. Livneh I, Moshitch-Moshkovitz S, Amariglio N. et al. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020;21:36-51
22. Yang Y, Hsu PJ, Chen YS. et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616-24
23. Zhang L, Hou C, Chen C. et al. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020;19:105-15
24. Ma S, Chen C, Ji X. et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121-35
25. Chen R, Jiang T, Lei S. et al. Expression of circular RNAs during C2C12 myoblast differentiation and prediction of coding potential based on the number of open reading frames and N6-methyladenosine motifs. Cell Cycle. 2018;17:1832-1845
26. Tang C, Xie Y, Yu T. et al. M6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020;30:211-28
27. Tay ML, Pek JW. Maternally inherited stable intronic sequence RNA triggers a self-reinforcing feedback loop during development. Curr Biol. 2017;27:1062-67
28. Chen YG, Kim MV, Chen X. et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67:228-38
29. Zeng Y, Du WW, Wu Y. et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842-55
30. Yang Q, Du WW, Wu N. et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609-20
31. Yao Z, Luo J, Hu K. et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422-37
32. Zhou J, Xiong Q, Chen H. et al. Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front Mol Neurosci. 2017;10:91-112
33. Salzman J, Gawad C, Wang PL. et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7:e30733
34. Zhang Y, Zhang XO, Chen T. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
35. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428-42
36. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-11
37. Jeck WR, Sorrentino JA, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
38. Kramer MC, Liang D, Tatomer DC. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168-82
39. Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13-4
40. Ragan C, Goodall GJ, Shirokikh NE. et al. Insights into the biogenesis and potential functions of exonic circular RNA. Sci Rep. 2019;9:2048-65
41. Zhang XO, Dong R, Zhang Y. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277-87
42. Zhang XO, Wang HB, Zhang Y. et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
43. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233-47
44. Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129-41
45. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
46. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836
47. Guarnerio J, Bezzi M, Jeong JC. et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289-302
48. Tan S, Gou Q, Pu W. et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018;28:693-5
49. Tan S, Sun D, Pu W. et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. 2018;17:138-42
50. Wu K, Liao X, Gong Y. et al. Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol Cancer. 2019;18:98-103
51. Li C, Zhang J. Stop-codon read-through arises largely from molecular errors and is generally nonadaptive. PLoS Genet. 2019;15:e1008141
52. Liang D, Tatomer DC, Luo Z. et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell. 2017;68:940-54
53. Liu X, Wang X, Li J, Hu S, Deng Y, Yin H. et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci China Life Sci. 2020;63:1429-49
54. Piwecka M, Glažar P, Hernandez-Miranda LR. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526
55. Kleaveland B, Shi CY, Stefano J. et al. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350-62
56. Ma HB, Yao YN, Yu JJ. et al. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. American journal of translational research. 2018;10:592-604
57. Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495:2369-75
58. Deng N, Li L, Gao J. et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495:189-96
59. Zheng X, Huang M, Xing L. et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19:73-92
60. Jost I, Shalamova LA, Gerresheim GK. et al. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018;15:1032-9
61. Li H, Xu JD, Fang XH. et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. 2020;116:1323-34
62. Han K, Wang FW, Cao CH. et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60-77
63. Pasmant E, Laurendeau I, Héron D. et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963-9
64. Kong Y, Hsieh CH, Alonso L C. ANRIL: a lncRNA at the cdkn2a/b locus with roles in cancer and metabolic disease. Front Endocrinol (Lausanne). 2018;9:405-17
65. Burd CE, Jeck WR. et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233
66. Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
67. Conn VM, Hugouvieux V, Nayak A. et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053-7
68. Hansen TB, Wiklund ED, Bramsen JB. et al. MiRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414-22
69. Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
70. Abdelmohsen K, Panda AC, Munk R. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
71. Du WW, Yang W, Chen Y. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European Heart Journal. 2017;38:1402-12
72. Du WW, Fang L, Yang W. et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357-70
73. Hansen TB, Veno MT, Damgaard CK. et al. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58
74. Du WW, Yang W, Liu E. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846-58
75. Sun YM, Wang WT, Zeng ZC. et al. CircMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134:1533-46
76. Kos A, Dijkema R, Arnberg AC. et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558-60
77. Guo JU, Agarwal V, Guo H. et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409-22
78. Abe N, Matsumoto K, Nishihara M. et al. Rolling circle translation of circular RNA in living human cells. Scientific Rep. 2015;5:16435-43
79. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415-7
80. Qu S, Yang X, Li X. et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141-8
81. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172-9
82. AbouHaidar MG, Venkataraman S, Golshani A. et al. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA. 2014;111:14542-7
83. Chen X, Han P, Zhou T. et al. CircRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985
84. Legnini I, Di Timoteo G, Rossi F. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:2-37
85. Yang Y, Gao X, Zhang M. et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304-15
86. Zhang M, Huang N, Yang X. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-14
87. Begum S, Yiu A, Stebbing J. et al. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37:4055-7
88. Zhang M, Zhao K, Xu X. et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nature Commun. 2018;9:4475-91
89. Chen FX, Woodfin AR, Gardini A. et al. PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell. 2015;162:1003-15
90. Liang WC, Wong CW, Liang PP. et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84-95
91. Liu H, Zhou X, Yuan M. et al. NcEP: a manually curated database for experimentally validated ncRNA-encoded proteins or peptides. J Mol Bio. 2020;432:3364-8
92. Pink RC, Wicks K, Caley DP. et al. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792-8
93. Dong R, Zhang XO, Zhang Y. et al. CircRNA-derived pseudogenes. Cell Res. 2016;26:747-50
94. Cervera A, de la Peña M. Small circRNAs with self-cleaving ribozymes are highly expressed in diverse metazoan transcriptomes. Nucleic Acids Res. 2020;48:5054-64
95. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635-8
96. Yao B, Christian KM, He C. et al. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17:537-49
97. Hwang JY, Aromolaran KA, Zukin RS. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat Rev Neurosci. 2017;18:347-61
98. Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017;33:319-42
99. Roundtree IA, Evans ME, Pan T. et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187-200
100. Kadumuri RV, Janga SC. Epitranscriptomic code and its alterations in human disease. Trends Mol Med. 2018;24:886-903
101. Jia G, Fu Y, Zhao X. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
102. Bokar JA, Shambaugh ME, Polayes D. et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233-47
103. Liu J, Yue Y, Han D. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-5
104. Schöller E, Weichmann F, Treiber T. et al. Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA. 2018;24:499-512
105. Agarwala SD, Blitzblau HG, Hochwagen A. et al. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8:e1002732
106. Schwartz S, Mumbach MR, Jovanovic M. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284-96
107. Patil DP, Chen CK, Pickering BF. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-73
108. Knuckles P, Lence T, Haussmann IU. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415-29
109. Lence T, Paolantoni C, Worpenberg L. et al. Mechanistic insights into m6A RNA enzymes. Biochim Biophys Acta Gene Regul Mech. 2019;1862:222-9
110. Pendleton KE, Chen B, Liu K. et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824-35
111. Zheng G, Dahl JA, Niu Y. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
112. Haussmann IU, Bodi Z, Sanchez-Moran E. et al. M6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301-4
113. Müller S, Glaß M, Singh AK. et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019;47:375-90
114. Arguello AE, DeLiberto AN, Kleiner RE. RNA chemical proteomics reveals the N6-methyladenosine (m6A)-regulated protein-rna interactome. J Am Chem Soc. 2017;139:17249-52
115. Dominissini D, Moshitch-Moshkovitz S, Schwartz S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
116. Dominissini D. Genomics and proteomics. Roadmap to the epitranscriptome. Science. 2014;346:1192-4
117. Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2016;14:23-31
118. Zhao X, Yang Y, Sun BF. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403-19
119. Ke S, Alemu EA, Mertens C. et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 2015;29:2037-53
120. Yi YC, Chen XY, Zhang J. et al. Novel insights into the interplay between m6A modification and noncoding RNAs in cancer. Mol Cancer. 2020;19:121-30
121. Di Timoteo G, Dattilo D, Centrón-Broco A. et al. Modulation of circRNA metabolism by m6A modification. Cell Rep. 2020;31:107641-56
122. Du H, Zhao Y, He J. et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Comm. 2016;7:12626 -
123. Park OH, Ha H, Lee Y. et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNAse P/MRP complex. Mol Cell. 2019;74:494-507
124. Liu L, Wang J, Khanabdali R. et al. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 2017;14:1715-21
125. Huang C, Liang D, Tatomer DC. et al. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639-44
126. Fustin JM, Doi M, Yamaguchi Y. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793-806
127. Roundtree IA, Luo GZ, Zhang Z. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311
128. Chen RX, Chen X, Xia LP. et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695-709
129. Sun HD, Xu ZP, Sun ZQ. et al. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8:10111-23
130. Zhang C, Wang J, Geng X. et al. Circular RNA expression profile and m6A modification analysis in poorly differentiated adenocarcinoma of the stomach. Epigenomics. 2020;12:1027-40
131. Wang M, Yang Y, Yang J. et al. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m6A-YTHDF3-Zeb1. Life Sci. 2020;257:118082-8
132. Xu J, Wan Z, Tang M. et al. N6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. 2020;19:163-78
133. Chen YG, Chen R, Ahmad S. et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96-109
134. Jakobi T, Siede D, Eschenbach J. et al. Deep characterization of circular RNAs from human cardiovascular cell models and cardiac tissue. Cells. 2020;9:1616-41
135. Huang R, Zhang Y, Bai Y. et al. N6-Methyladenosine modification of fatty acid amide hydrolase messenger rna in Circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry. 2020;88:392-404
136. Jeyaraman S, Hanif EAM, Ab Mutalib NS, Jamal R, Abu N. Circular RNAs: potential regulators of treatment resistance in human cancers. Front Genet. 2020;10:1369-77
137. Pan XY, Huang C, Li J. The emerging roles of m6A modification in liver carcinogenesis. Int J Biol Sci. 2021;17:271-84
138. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021
139. Li B, Zhu L, Lu C. et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12:295-310
140. Wang Q, Liu H, Liu Z. et al. Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genet. 2020;240:33-39
141. Liu CX, Li X, Nan F. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865-80
142. Zhang C, Fu J, Zhou Y. A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front Immunol. 2019;10:922-30
143. Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, Zhan X, Laimins L, Wang RC. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10:2300-11
144. Wesselhoeft RA, Kowalski PS, Parker-Hale FC. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol cell. 2019;74:508-20
145. Pazoki R, Evangelou E, Mosen-Ansorena D. et al. GWAS for urinary sodium and potassium excretion highlights pathways shared with cardiovascular traits. Nat Commun. 2019Aug13;10(1):3653-63
146. Guo M, Yan R. et al. IFN regulatory factor-1 induced macrophage pyroptosis by modulating m6A modification of circ_0029589 in patients with acute coronary syndrome. Int Immunopharmacol. 2020;86:106800-12
147. Su H, Wang G, Wu L. et al. Transcriptome-wide map of m6A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics. 2020;21:39-53
148. Ling J, Tan K, Lu L. et al. lncRNA MIAT increases cell viability, migration, EMT and ECM production in age-related cataracts by regulating the miR-181a/CTGF/ERK signaling pathway. Exp Ther Med. 2020;20:1053-63
149. Fan C, Liu X, Li W. et al. Circular RNA circ KMT2E is up-regulated in diabetic cataract lenses and is associated with miR-204-5p sponge function. Gene. 2019;710:170-77
150. Li P, Yu H, Zhang G. et al. Identification and characterization of N6-methyladenosine circRNAs and methyltransferases in the lens epithelium cells from age-related cataract. Invest Ophthalmol Vis Sci. 2020;61:13-25
151. LeMoult J, Humphreys KL, Tracy A. et al. Meta-analysis: exposure to early life stress and risk for depression in childhood and adolescence. J Am Acad Child Adolesc Psychiatry. 2020;59:842-55
152. Zhang Y, Du L, Bai Y. et al. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol Psychiatry. 2020;25:1175-90
153. Yue B, Wang J, Ru W. et al. The circular RNA circHUWE1 sponges the miR-29b-AKT3 axis to regulate myoblast development. Mol Ther Nucleic Acids. 2020;19:1086-97
154. Zhang P, Chao Z, Zhang R. et al. Circular RNA regulation of myogenesis. cells. 2019;8:885-97
155. Griswold MD. Spermatogenesis: the commitment to meiosis. Physiol Rev. 2016;96:1-17
156. Schwartz S, Agarwala SD, Mumbach MR. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409-21
157. Lin Z, Hsu PJ, Xing X. et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27:1216-30
158. Kosik KS. Molecular biology: circles reshape the RNA world. Nature. 2013;495:322-4
Author contact
![]() Corresponding author: Dr. Yonggang Lv, Professor, ORCID: 0000-0003-3966-8975; Mechanobiology and Regenerative Medicine Laboratory, Bioengineering College, Chongqing University, 174 Shazheng Street, Shapingba District, Chongqing 400044, China. Tel: 86-23-65102507; Fax: 86-23-65102507; E-mail: yglvedu.cn; yglvbmecom; Web: http://yglvbme.com.
Corresponding author: Dr. Yonggang Lv, Professor, ORCID: 0000-0003-3966-8975; Mechanobiology and Regenerative Medicine Laboratory, Bioengineering College, Chongqing University, 174 Shazheng Street, Shapingba District, Chongqing 400044, China. Tel: 86-23-65102507; Fax: 86-23-65102507; E-mail: yglvedu.cn; yglvbmecom; Web: http://yglvbme.com.

 Global reach, higher impact
Global reach, higher impact