10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(11):2737-2755. doi:10.7150/ijbs.61410 This issue Cite
Review
The Multifaceted Role of Long Non-Coding RNA in Gastric Cancer: Current Status and Future Perspectives
1. Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China.
2. South Sichuan Institute of Translational Medicine, Luzhou 646000, Sichuan, China.
3. Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province,Sichuan Industrial Institute of Antibiotics, School of Pharmacy, Chengdu University, Chengdu 610106, Sichuan, China.
4. Department of Pharmacy, the Affiliated Hospital of Southwest Medical University, Luzhou 646000, Sichuan, China.
5. Department of Gastroenterology, Shenzhen Hospital, Southern Medical University, Shenzhen 518000, Guangzhou, China.
#These authors contributed equally to the work.
Received 2021-4-9; Accepted 2021-6-8; Published 2021-6-26
Abstract
Gastric cancer (GC) is one of the major public health concerns. Long non-coding RNAs (lncRNAs) have been increasingly demonstrated to possess a strong correlation with GC and play a critical role in GC occurrence, progression, metastasis and drug resistance. Many studies have shed light on the understanding of the underlying mechanisms of lncRNAs in GC. In this review, we summarized the updated research about lncRNAs in GC, focusing on their roles in Helicobacter pylori infection, GC metastasis, tumor microenvironment regulation, drug resistance and associated signaling pathways. LncRNAs may serve as novel biomarkers for diagnosis and prognosis of GC and potential therapeutic targets. The research gaps and future directions were also discussed.
Keywords: long non-coding RNA, gastric cancer, drug resistance, helicobacter pylori, tumor microenvironment
Introduction
Gastric cancer (GC) is among the most common human malignancies and is the third leading cause of cancer death worldwide [1]. Approximately one million new cases are diagnosed each year. Although great advances have been gained in early diagnosis and management of GC [2], it still remains a major public health problem. GC is caused by the interaction of host genetic factors and complex environmental factors and manifested by complex cross talks within tumor microenvironment. Helicobacter pylori (H. pylori) infection has been suggested as one of the most important drivers in GC tumorigenesis [3]. Most GC patients can prolong their overall survival and improve life quality through the combinational use of chemotherapeutics and targeted therapy. However, the multi-drug resistance (MDR) of GC is a major restrain in clinical practice.
It is revealed that protein-coding RNAs account for less than 2% of the total transcripts, while the rest of them are non-coding RNAs (ncRNAs) that lack the ability to code proteins. However, the increasing evidence shows that ncRNAs play a crucial role in different cellular processes from normal development to disease process [4]. The ncRNAs are separated into two major types based on their size: (1) small ncRNAs with a molecular length of no longer than 200 base pairs (bps), represented by microRNAs and transfer RNAs; (2) long ncRNAs (lncRNAs) with more than 200 bps in length. In the past decades, for small ncRNAs, especially the microRNAs (miRNAs), their biological functions have been extensively explored. On the other hand, the underlying knowledge about the functional role of lncRNAs is rather limited, as they were often considered to be transcription noises. Owing to the advantages of next-generation sequencing technologies, lncRNAs have received extensive attention in different areas. Emerging evidence indicates that lncRNAs play a vital role in regulating various cellular processes through several ways including sponging miRNAs, interaction with proteins and modulation of target gene expressions [5]. Specifically, given the multifaceted roles of lncRNAs in GC tumorigenesis and progression, in this work, we will comprehensively review the function of lncRNAs in GC development and discuss their interactions with GC tumor microenvironment and the impact on GC drug resistance.
LncRNAs and H. pylori Infection
H. pylori infection exhibits a pivotal role in gastric tumorigenesis, which is infected by more than 70% of all patients [3]. During the past years, numerous studies have demonstrated that the crucial role of protein-coding genes and genomic variations in the pathogenesis of GC. It is increasingly realized that lncRNAs, which fail to code protein, may become a big part of the occurrence and progression of various diseases. It has become apparent that lncRNAs are associated with GC induced by H. pylori infection. In this work, we summarized the interactions between lncRNAs and H. pylori-associated GC.
Association of the Expression Level of LncRNAs with H. pylori-Related GC
Some lncRNAs play an oncogenic role in GC. A recent study showed that lncRNA H19 was significantly upregulated in tumors of GC patients with H. pylori infection [6, 7]. Overexpression of lncRNA H19 was correlated with all inflammatory indexes and promoted proliferation, migration and invasion in GC cells infected by H. pylori. Notably, Receiver operating characteristic (ROC) analysis aiming at exploring the potential markers for distinguishing stages III to Ⅳ of GC showed that lncRNA H19 preformed high diagnostic accuracy (95.5%), specificity (100%) and sensitivity (90.9%) [6, 7]. This study suggested that lncRNA H19 could be a diagnostic marker for H. pylori-associated GC. Jin et al. found a significant increase of lncRNA HULC in the serum of GC patients, and overexpressed lncRNA HULC was significantly correlated with tumor growth and metastasis, as well as H. pylori infection [8]. In Jing et al.'s study [9], they found that the expression of lncRNA PVT1 was remarkably elevated in normal gastric epithelial cells GES-1 which were infected by H. pylori. Intriguingly, the inflammatory markers caused by H. pylori infection were inhibited upon knockdown of lncRNA PVT1. The result suggested that lncRNA PVT1 may function as a pro-inflammatory factor and led to the tumorigenesis of H. pylori-associated GC [9]. Moreover, it was found that the aberrant expression of GC-associated lncRNA 1 (GClnc1) contributed to pathological differentiation, vascular invasion, tumor size and poor prognosis in GC [10]. GClnc1 was increasingly expressed from normal stomach tissue to intestinal metaplasia (IM), to dysplasia, and to GC, suggesting that overexpressed GClnc1 may be a diagnostic index for early stage of GC. Mechanistically, GClnc1 could interact with the complex of WDR5 (a key component of histone methyltransferase complex) and KAT2A histone acetyltransferase, leading to an alteration of the target gene histone modification pattern and thus enhancing the progression of GC. Similarly, overexpression of GClnc1 was also observed in GC tissues with H. pylori infection. However, the accurate role of GClnc1 on H. pylori-induced GC development was indistinct [10].
On the contrary, evidence also suggests that lncRNAs function as tumor suppressor, which had been implicated in the development of GC following H. pylori infection. For example, lncRNA AF147447 was able to suppress the expression of MUC2 through interacting with miRNA-34c, and the lncRNA AF147447 promoter could recruit the transcription factor E2F1 [11]. This suggested that lncRNA AF147447 was involved in GC development acting as a tumor suppressor, which was inhibited by H. pylori infection. In addition, lncRNA NR_026827 was found downregulated in GES-1 cells infected by H. pylori, and had a lower expression in GC tissues, comparing with normal tissues. However, the exact role of lncRNA NR_026827 in H. pylori-associated GC remains inconclusive [12].
Many studies have investigated the profiles of lncRNAs in H. pylori-infected cells and tumors. By using microarray analysis, Yang et al. identified some lncRNAs which were aberrantly expressed in H. pylori-infected gastric epithelial cells [13]. They found that expressions of 23 lncRNAs were upregulated, while 21 lncRNAs were downregulated. Further analysis revealed that, among the aforementioned 44 lncRNAs, only the expression level of XLOC-004122 and XLOC-014388 was decreased in the gastric mucosal samples of patients infected by H. pylori [13]. Another study by Yang et al. also recognized that some lncRNAs were abnormally regulated in H. pylori-associated GC, which may have prognostic values [14]. Notably, RP11-169F17.1 and RP11-669N7.2 were found significantly correlated with poor overall survival and were remarkably related with H. pylori infection [14]. Moreover, in Yang et al.'s earlier investigations, 765 H. pylori infection-associated genes were obtained based on miRNA-mRNA interaction network [15]. Using Venn diagram, they indicated that 41% of RP11-169F17.1-related targets and 40.9% of RP11-669N7.2-related targets were overlapped with genes associated with H. pylori infection, respectively. There were an obvious positive association between the expression levels of two mentioned lncRNAs and their target genes. Additionally, these target genes were closely associated with GC [15]. Taken together, the results suggest that RP11-169F17.1 and RP11-669N7.2 lncRNAs actively participate in the process of H. pylori-promoted gastrointestinal diseases including GC. It would be more convincing if the correlation between RP11-169F17/RP11-669N7.2 lncRNAs and H. pylori-associated gastric diseases is further clarified in animal models.
Additionally, it was found that 303 lncRNAs as well as 565 mRNAs were recognized as aberrantly expressed RNAs (p < 0.05) in the cells infected by H. pylori comparing with control [16]. Quantitative real time polymerase chain reaction (qRT-PCR) analysis verified the expression profile of 8 lncRNAs in H. pylori-infected cells, including n345630, XLOC_004787, LINC00152, n378726, XLOC_005517, LINC00473, XLOC_13370 and n408024. Four lncRNAs were found down-regulated in the cells infected by H. pylori, which may be significantly correlated with the pathological effects of H. pylori [16]. Nonetheless, the precise mechanisms and downstream targets are required to be explored in future studies.
In conclusion, many lncRNAs are found abnormally expressed in H. pylori-associated GC, some of which may have diagnostic or prognostic values. Nevertheless, in most cases, in vitro and in vivo experiments should be conducted in future investigations to confirm the exact roles and mechanisms of these lncRNAs in H. pylori-infected GC.
Regulatory Networks of LncRNAs in H. pylori-Associated GC
It has been suggested that lncRNAs participated in regulation of H. pylori-associated GC by targeting specific genes via different signaling pathways. Glucocorticoid-inducible kinase 1 (SGK1) is an effective stimulator of multiple ion channels, which is capable of regulating cell proliferation, migration, cell volume, exocytosis, excitation and epithelial transport, and thus promotes tumor growth [17, 18]. Interestingly, Yao et al. found that in intertumoral or peripheral T lymphocytes an increase in expression levels of SGK1 and its upstream lncRNA named lnc-SGK1 were observed in H. pylori-associated gastric carcinoma, which had a strong association with H. pylori infection and a high-salt diet (HSD). Moreover, lnc-SGK1 increased the transcription of SGK1 in a cis-regulatory way, and lnc-SGK1 may increase T helper 2 (Th2) and Th17 while decrease the differentiation of Th1 through SGK1/JUNB pathway. An elevated expression of lnc-SGK1 in serum upon H. pylori infection and/or HSD and in T lymphocytes was related to poor prognosis of GC patients, thus indicated that it may be an ideal diagnostic marker in human GC [18].
In another study, the lnc-GNAT1-1 was dramatically downregulated in GC cells infected by H. pylori, comparing to normal cells. H. pylori infection could suppress the expression level of lnc-GNAT1-1 in tumor xenografts. Overexpression of lnc-GNAT1-1 inhibited cell proliferation, migration and invasion in SGC-7901 and MKN45 cells via suppression of Wnt/β‑catenin signaling [19]. These data revealed that lnc-GNAT1-1 played a role in regulating Wnt/β‑catenin pathway in H. pylori-associated GC.
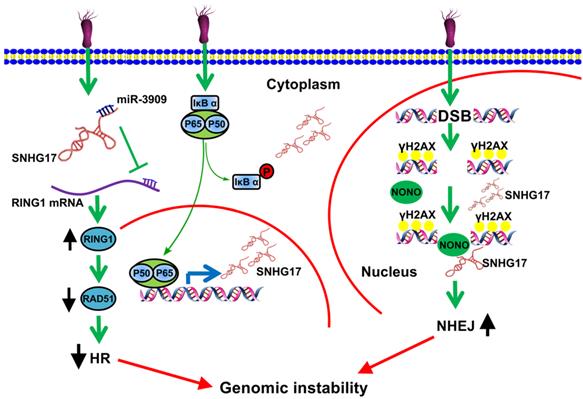
Numerous studies have revealed that H. pylori infection causes DNA damage and influences DNA repair [20-23]. DNA damage can lead to DNA double-strand breaks (DSBs) [24]. To deal with these physical damages, there are two major repairing patterns in eukaryotic cells: one is through homologous recombination (HR), which utilizes homologous DNA sequences as templates and guarantees error-free repairing; another is through nonhomologous end joining (NHEJ), which recruits DNA-dependent protein kinase catalytic submit (DNA-Pkcs) to DNA break ends upon lacking the homologous recombination. Of great interest, Han et al. found that lncRNA SNHG17 was elevated due to H. pylori infection and highly-expressed lncRNA SNHG17 was correlated with low survival rate of GC patients [25]. Notably, up-regulated nuclear lncRNA SNHG17 caused the recruitment of NONO, which together functioned as an abduction of miRNA-3909, modulating Rad51 expression level and thus resulting in an alteration of DSBs repairing pathways from HR to NHEJ. These findings indicated that lncRNA SNHG17/miRNA-3909/Rad51 pathway may contributed to progression of GC induced by H. pylori through affecting the DNA repair pathway (Fig. 1) [25].
Based on current findings, although some studies identified some novel lncRNAs which were aberrantly expressed in H. pylori-associated GC and paid attention to the regulatory network of lncRNAs, most of the downstream targets and action mechanisms were still mysterious. There still remain questions. For example, cytotoxin associated gene A (cagA) and vacuolating cytotoxin A (vacA) are two of the most important pathogenic factors in H. pylori, few studies had addressed the possible interactions between lncRNAs and these pathogenic markers. Thus, the potential contribution of dysregulated lncRNAs in multiple pathogenic factors induced by H. pylori needs more investigations.
LncRNA SNHG17 participated in deregulation of NONO and miRNA-3909/RING1/Rad51 pathway during H. pylori infection and promoted the cells more prone to genetic rearrangements, therefore enhancing tumorigenesis in GC [25]. Reprinted with permission [25].
In summary, the current findings extend our understanding in the crucial roles of dysregulation of lncRNAs and open novel investigation lines toward the potential lncRNA-related target for treatment or diagnosis of H. pylori-associated GC.
LncRNAs and Metastatic GC
Cancer metastasis is one of the main features of GC due to late diagnosis, leading to an acceleration of malignant progression. Many lncRNAs, as one of key drivers, have been demonstrated to promote GC metastasis. A recent study showed that the expression of lncRNA AC093818.1 was positively correlated with invasion, lymphatic metastasis, distal metastasis, and tumor-node-metastasis stage of GC, which promoted GC metastasis through epigenetically inducing PDK1 expression both in vitro and in vivo [26]. On the other hand, Zhang et al. found that lncRNA DRAIC could suppress GC metastasis of GC cells via through influencing NFRKB de-ubiquitination induced by UCHL5 [27].
EMT is considered to be a core factor of tumor metastasis, and it is clear that a variety of lncRNAs participated in GC development by regulating this cell program. For instance, Li et al. observed that lncRNA MAG12-AS3 was overexpressed in GC tissues based on bioinformatics analysis, and high-expressed MAG12-AS3 was positively correlated with poor prognosis [28]. Mechanically, further studies indicated that MAG12-AS3 was identified to an EMT-related lncRNA as it could significantly regulate ZEB1 (Zincfinger ebox binding homeobox 1) expression, while it sponged miRNA-141/200a and suppressed their expressions in GC [28]. Moreover, it was confirmed that the expression level of lncRNA DLX6-AS1 was remarkably elevated in GC cells, and high-expressed DLX6-AS1 could enhance cell proliferation, migration and EMT process of GC cells [29]. MAP4K1 (Mitogen-activated protein kinase 1) was positively regulated by lncRNA DLX6-AS1, and this biological process depended on the mediation of Fused sarcoma translocated in liposarcoma (FUS) [29].
Evidence has shown that lncRNAs have played an essential role in GC metastasis. However, till now it remains largely elusive that to what extent lncRNAs participate in promoting or inhibiting GC metastasis as well as whether and how they cooperate with other factors.
LncRNAs and Tumor Microenvironment
The hypothesis of “seed and soil” is one of the most influential theories in cancer biology. This theory insists that the initiation and progression of cancer are not only the alterations in tumor cytogenetics and epigenetics, but also influenced by surrounding tumor microenvironment (TME), as the “fertile soil” for the growth of cancerous cells, interacting with each other and coevolving to promote the generations of tumors [30]. TME composed of multiple immune cells, stromal cells, extracellular matrix and active mediators in addition to tumor cells, which can be broadly divided into immune microenvironment dominated by immune cells and non-immune microenvironment dominated by fibroblasts and numerous inflammatory factors (Fig. 2) [31]. Recently, the advent of tumor immunotherapy, especially targeting immune checkpoints, leads to promising results in the treatments of GC. However, emerging evidence has been reported that heterogeneity of TME is a major hurdle in tumor immunotherapy, which can mediate immunosuppression, leave cancer patients insensitive to immunotherapy, eventually lead to a poor prognosis in the patients with GC. Lifting the immunosuppression of TME is beneficial to the recovery and reconstruction of autoimmunity. Therefore, the therapeutic approaches to TME are promising. Notably, it has become increasingly apparent that lncRNAs have a significant correlation with the TME, which participate in tumorigenicity and progression of GC. Understanding the complex interaction between lncRNAs and TME will eventually excavate prognostic immune markers, which is beneficial to explore both new mechanisms and pathways for blocking tumor immunosuppression, subsequently leading to the stimulation of the host immune response through activating different subsets of immune cells.
Regulation of lncRNAs in the immune tumor microenvironment. The tumor immune microenvironment consists of not only numerous cancer cells and cancer stem cells, but also a great variety of immune cells, such as regulatory T cells and tumor-associated macrophages. Tumor non-immune microenvironment contains chemokines and cytokines. LncRNAs such as linc-POU3F3 have been shown to interact with intracellular targets and induce T-reg cell differentiation, creating immunosuppressive microenvironment to promote GC cell proliferation [43]. Some lncRNAs including lncRNA H19 induce M1 to M2 macrophage polarization that influences release of IL-10, VEGF and MMP-9 and mediates T cell suppression, tumor blood vessel and ECM formation [49].
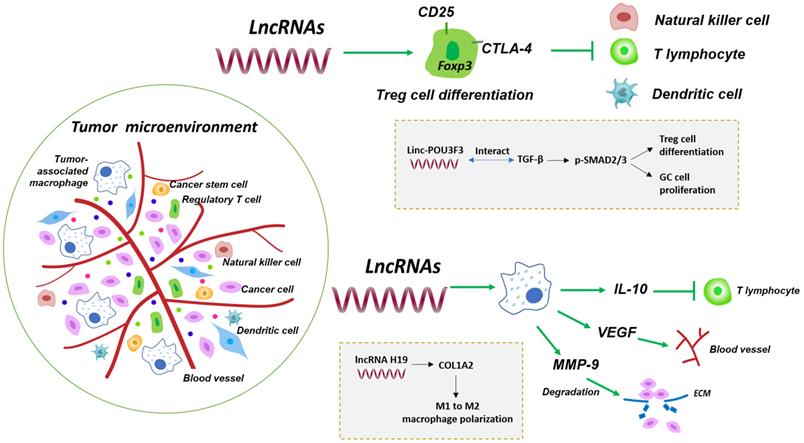
The host's own immune response consists of innate immunity, which is represented by macrophages, natural killer (NK) cells and dendritic cells (DC), and specific adaptive immunity, which is dominated by T and B lymphocytes. It seems that immune cells exhibit a bidirectional role in the progression of cancers. Immune cells are responsible for eliminating cancerous cells in the initial stages of tumor invasion, while tumor infiltration lymphocytes (TILs) suppress the effector T lymphocytes activation to help cancer cells immune escape and proliferation during the development of cancer, acting as the accomplice of cancer cells. Among lymphocytes infiltrating in tumor, regulatory T cells (T-regs) and tumor-associated macrophages (TAMs) are the most effective components of the tumor immunosuppressive microenvironment [32].
T-regs are a subset of CD4+ T lymphocytes which constitutively expressed interleukin (IL)-2 receptor α-chain (CD25), cytotoxic T lymphocyte antigen-4 (CTLA-4) and the most important lineage differentiation specific factor Foxp3, showing distinct functions of immune impotence and suppression [33]. For immune suppression, an elevated expression of IL-2 receptor on T-reg cells could deprive the IL-2 of effectors T cells and inhibit their proliferation [34]. On the other hand, the aforementioned another T-reg cell-specific surface molecule, CTLA-4, besides its central role in impeding the activated T cells responses, is involved in T-reg cells-mediated immune suppression [35]. Given the indispensable role of T-reg cells on immune suppression, more and more evidence indicated that T-reg cells were implicated in the development of cancers [36-40]. Recently, it is recognized that lncRNAs could influence the progression of cancers through regulating the differentiation and functions of T-reg cells (Fig. 2) [41, 42]. Xiong et al. found that the number of CD4+CD25+Foxp3+ T-reg cells were increased in peripheral blood of GC patients, comparing to the normal samples [43]. Further studies indicated that linc-POU3F3 activated the distribution of T-reg and promote the proliferation of GC cells. Mechanically, they observed that linc-POU3F3 could interact with TGF-β which improved the SMAD2/3 phosphorylation level [43]. This study showed that the regulation of lncRNAs was capable of promoting the differentiation of T-regs in GC.
Cancer-related inflammation is one of the biological characteristics of tumors [44]. Infiltrating macrophages in tissue are derived from bone marrow monocyte precursors, which are mainly divided into M1- and M2-type. Tumor-associated macrophages (TAMs), as the central component of infiltrating leukocytes in tumors, are more similar to the functional phenotype of M2-type macrophages, playing an extremely important role in mediating the occurrence of tumor inflammation and tumor progression [45-48]. It is recently found that lncRNA H19 was overexpressed in GC, and was correlated with poor outcomes in patients [49]. Meanwhile, lncRNA H19 could regulate the COL1A2 expression level via miRNA-29a-3p. COL1A2 was significantly associated with immune infiltrating cells and lymphatic metastasis, indicating that lncRNA H19 could alter COL1A2 level to enhance the polarization process of macrophages from M1 to M2 in GC, thus promoted the development of GC [49].
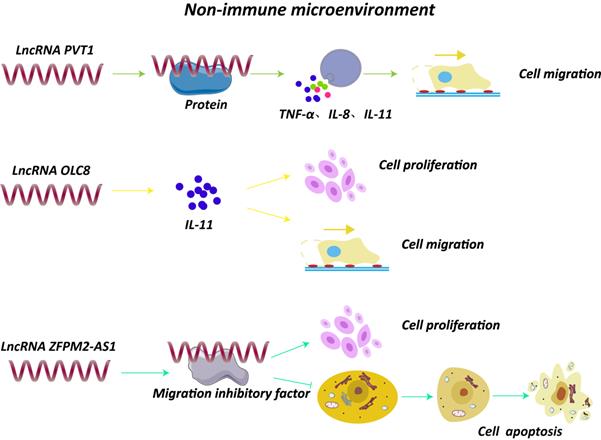
Besides the immune cells, non-immune TME is composed of multiple extracellular molecules including cytokines and chemokines. Notably, lncRNAs could affect the secretion and synthesis of these small molecule compounds, which may exert a vital role in GC progression (Fig. 3). For example, knockdown of lncRNA PVT1 remarkably decreased the expression of inflammatory factors, such as tumor necrosis factor-α (TNF-α), IL-1β, IL-8, leading to a reduced ability of cell migration [9]. A study found that lncRNA H19 induced by H. pylori infection was overexpressed in GC cells and tissues, and could activate NF-κB signaling pathway, resulting in an upregulation of pro-inflammatory cytokines and enhancing the progression of GC [7]. Moreover, lncRNA OLC8, which was observed highly expressed in GC cell lines and samples, could bind to IL-11, and the complex significantly suppressed the degradation of IL-11 mRNAs, thus elevated IL-11 expression and promoted cell proliferation and migration [50]. In addition, a novel lncRNA ZFPM2-AS1 was found overexpressed in GC tissues and was able to interact with macrophage migration inhibitory factor (MIF), protecting it from degradation [51]. Of note, down-regulation of MIF could attenuate the role of lncRNA ZFPM2-AS1 on p53, revealing that lncRNA ZFPM2-AS1 may participate in GC development through ZFPM2-AS1/MIF/p53 pathway [51].
Interaction of lncRNAs with components of tumor non-immune microenvironment. LncRNA PVT1 increased expression of TNF-α, IL-1β and IL-8 and promoted cell migration [9]. LncRNA OLC8 bound to IL-11, and the complex significantly suppressed the degradation of IL-11 mRNAs, thus elevating IL-11 expression and promoting cell proliferation and migration [50]. LncRNA ZFPM2-AS1 interacted with MIF and prevented degradation of MIF [51].
LncRNAs affects TGF-β1 signaling and epithelial-mesenchymal transition. For instance, lncRNA ELIT-1, functioning as Smad cofactor, could be activated by TGF-β1 through the TGF-β1/Smad axis [52]. miRNA-185 could inhibit lncRNA XIST and TGF-β1, thus suppressed GC development [53].
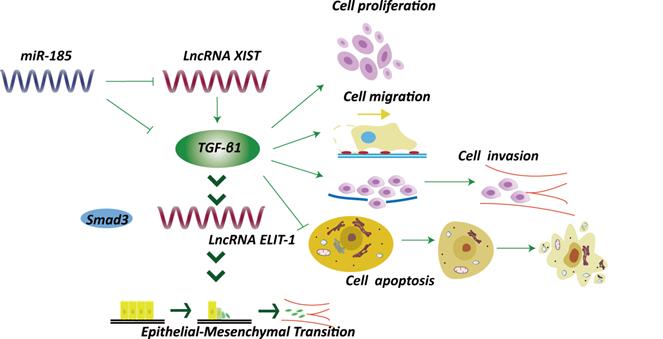
TGF-β1 has long been correlated with epithelial-mesenchymal transition (EMT) and tumor progression (Fig. 4). Sakai et al. identified a novel lncRNA ELIT-1, which acts as Smad cofactor and could be activated by TGF-β1 through the TGF-β1/Smad axis [52]. Knockdown of ELIT-1 inhibited EMT progression mediated by TGF-β1, as well as the expression of downstream target of TGF-β1, including some transcription molecules essential for EMT such as the Snail. Further investigations indicated that lncRNA ELIT-1 directly bound with Smad3, thus positively modulated the target genes activities through recruiting Smad3 to their promoters. The study thus illustrated the roles of lncRNA ELIT-1 in EMT [52]. Another lncRNA XIST was overexpressed in GC cells [53]. Intriguingly, a negative correlation between lncRNA XIST and miRNA-185 was observed based on bioinformatics analysis, and TGF-β1 was identified as a downstream target of miRNA-185, which could counter-regulate TGF-β1. Knockdown of lncRNA XIST suppressed GC development through elevating miRNA-185 to inhibit TGF-β1. The results revealed that lncRNA XIST/miRNA-185/TGF-β1 pathway was involved in GC development (Fig. 4) [53].
However, the studies on the regulatory role of lncRNAs within GC TME were considered insufficient. The facts that GC is a molecularly and phenotypically highly-heterogeneous disease and has different subtypes with striking distinct clinical features may obstruct the exploration of the accurate and specific lncRNAs which could regulate the T-regs or other components to confer the suppressive activity on tumor immunity. Therefore, to explore the interaction between lncRNA and TME and to find out the exact mechanisms underlying lncRNAs regulate components of TME are among the most urgent and fundamental challenges.
LncRNAs and Drug Resistance
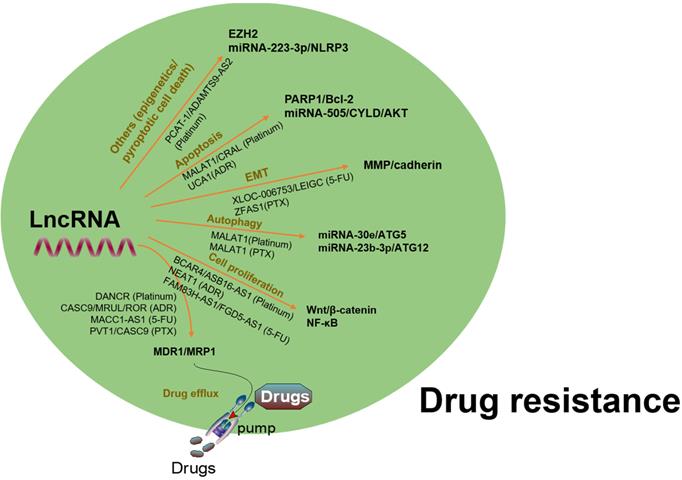
To date, chemotherapy and targeted therapy are commonly used in the treatment of GC. However, the emergence of MDR in GC cells is a major challenge in clinical practice. A variety of lncRNAs have been found to be abnormally expressed in GC and contributed to MDR via regulation of varied target genes and pathways (Fig. 5). Several oncogenic lncRNAs such as PCAT-1, SNHG5, BCAR4, GHET1, HOTAIR, PVT1, MALAT1, UCA1 and NEAT1, as well as certain tumor suppressive lncRNAs, have been demonstrated to involve in MDR of GC (Table 1).
LncRNAs and Platinum Resistance
Numerous lncRNAs are involved in platinum drugs resistance. For example, Guo et al. found that the expression level of lncRNA PCAT-1 was significantly increased in both GC tissues and cells, which contributed to cisplatin drug (DDP) resistance to GC cells [54]. Subsequent experiments indicated that lncRNA PCAT-1 could enhance DDP resistance of GC cells via regulating the miRNA-128/ZEB1 axis [54]. Another study also demonstrated that knockdown of lncRNA PCAT-1 reversed DDP resistance in GC cells [55]. Mechanically, resistance of GC cells to DDP modulated by lncRNA PCAT-1 was attributed to silenced phosphatase and tensin homolog (PTEN) via interacting with enhancer of zeste homolog 2 (EZH2) [55]. A recently study demonstrated that lncRNA UCA1 had been correlated with DDP resistance in GC [56]. It was found that high expression of lncRNA UCA1 enhanced cell proliferation while suppressed apoptosis induced by DDP, which was related with poor prognosis in GC patients based on TCGA and GEO database. Moreover, lncRNA UCA1 could interact with EZH2 and activate PI3K/AKT signaling pathway, thereby promote the GC cells resistance to DDP [56]. Homoplastically, lncRNA DNACR was identified to be overexpressed in GC cells resistant to cisplatin drugs. Knockdown of lncRNA DANCR in BGC823 and SGC7901 GC cells could enhance cell apoptosis as well as suppress cell growth. While overexpressing lncRNA DANCR led to an increase of MDR1, MRP1 expression level which contributed to DDP-resistance in GC cells [57]. Yan et al. demonstrated that lncRNA HOTAIR, which was remarkably upregulated in DDP-resistant GC cells and tissues, resulting in an enhancement of cancer cell proliferation but an inhibition of cancer cells apoptosis, and accelerated transition of cell cycle G1/S [58]. Upregulation of HOTAIR contributed to DDP resistance of GC through interacting with miRNA-126 and decreasing miRNA-126 expression to activate PI3K/AKT/MRP1 signaling pathways [58]. Of great interest, overexpression of lncRNA BCAR4 promoted DDP resistance of GC via stimulating the Wnt pathway, while reducing the expression of BCAR4 increased the sensitivity of GC cell to cisplatin [59]. It had been reported that lncRNA MALAT1 was emerging player in DDP-resistance of various types of cancers [60-62]. A recent study found that GC cells showing cisplatin resistance were often accompanied by an upregulation of lncRNA MALAT1, while using propofol could suppress MALAT1 expression and enhance GC cell apoptosis [63]. Mechanically, MALAT1 inhibited autophagy through binding with miRNA-30e, thus regulated ATG5 expression. Collectively, propofol may promote autophagy, thereby enhance the sensitivity of cisplatin drugs in GC through downregulating MALAT1 [63]. Moreover, lncRNAs HOXD-AS1 [64], FAM84B-AS [65], TP73-AS1 [66], ASB16-AS1 [67], PVT1 [68] and HOTTIP [69] also conferred the resistance of GC cells in response to DDP via regulating expression levels of MDR-associated genes.
LncRNAs associated with MDR in GC
| LncRNAs | Role | Expression | Target genes and/or pathways | Reference |
|---|---|---|---|---|
| Platinum resistance | ||||
| PCAT-1 | Oncogene | Upregulated | miRNA-128/EZH2 | [54] |
| UCA1 | Oncogene | Upregulated | PI3K/AKT | [56] |
| DANCR | Oncogene | Upregulated | MDR1, MRP1 | [57] |
| HOTAIR | Oncogene | Upregulated | miRNA-126; PI3K/AKT | [58] |
| BCAR4 | Oncogene | Upregulated | Wnt/β-catenin | [59] |
| HOXD-AS1 | Oncogene | Upregulated | EZH2 | [64] |
| FAM84B-AS | Oncogene | Upregulated | Unknown | [65] |
| TP73-AS1 | Oncogene | Upregulated | HMGB1/RAGE | [66] |
| ASB16-AS1 | Oncogene | Upregulated | NF-κB | [67] |
| HOTTIP | Oncogene | Upregulated | HMGA1/miRNA-218 | [69] |
| MALAT1 | Oncogene | Upregulated | miRNA-30e/ATG5 | [60-63] |
| PVT1 | Oncogene | Upregulated | miRNA-3619-5p/TBL1XR1 | [68] |
| ADAMTS9-AS2 | Tumor suppressor | Downregulated | miRNA-223-3p/NLRP3 | [70] |
| CRAL | Tumor suppressor | Downregulated | miRNA-505/CYLD | [71] |
| DOX resistance | ||||
| GAS5 | Tumor suppressor | Downregulated | Unknown | [72] |
| HOTAIR | Oncogene | Upregulated | miRNA-17-5p | [74] |
| CASC9 | Oncogene | Upregulated | MDR1 | [73] |
| MRUL | Oncogene | Upregulated | ABCB1 | [75] |
| UCA1 | Oncogene | Upregulated | PARP1, Bcl-2, miRNA-27b | [76, 134] |
| D63785 | Oncogene | Upregulated | miRNA-422a | [78] |
| NEAT1 | Oncogene | Upregulated | Unknown | [79] |
| ROR | Oncogene | Upregulated | MRP1 | [81] |
| 5-FU resistance | ||||
| FAM83H-AS1 | Oncogene | Upregulated | miRNA-145-5p | [82] |
| MACC1-AS1 | Oncogene | Upregulated | MDR1, MRP1 | [83] |
| ANRIL | Oncogene | Upregulated | PI3K/AKT/mTOR | [84] |
| XLOC_006753 | Oncogene | Upregulated | miRNA-153-5p/CITED2 | [85] |
| FGD5-AS1 | Oncogene | Upregulated | Unknown | [86] |
| LEIGC | Tumor suppressor | Downregulated | miRNA-145-5p | [87] |
| Paclitaxel resistance | ||||
| ZFAS1 | Oncogene | Upregulated | Wnt/β-catenin | [89] |
| MALAT1 | Oncogene | Upregulated | miRNA-23b-3p | [90] |
| PVT1 | Oncogene | Upregulated | MDR1, MRP, mTOR and HIF-1α | [91] |
| HOTAIR | Oncogene | Upregulated | miRNA-217 | [92] |
| CASC9 | Oncogene | Upregulated | MDR1 | [73] |
A summary of regulation of lncRNAs in chemoresistance via interaction with different targets and pathways. LncRNAs have been found to be involved in drug resistance in GC by influencing drug efflux, apoptosis, DNA repair, cell cycle, proliferation, autophagy, epithelial-mesenchymal transition, and cancer stem cell through regulating expression of potential target genes and related signaling pathways.
Despite numerous lncRNAs had been corroborated to be the contributors of DDP-resistance in GC, some lncRNAs function as tumor suppressor and are able to sensitize DDP-resistant GC cells. For example, lncRNA ADAMTS9-AS2 was under-expressed in GC tissues and cells, which could inhibit GC progression by sponging miRNA-223-3p to suppress miRNA-223-3p expression in GC cells [70]. In addition, NLRP3 was a downstream target of lncRNA ADAMTS9-AS2 binding with miRNA-223-3p, transposed DDP-resistant GC cells to cisplatin via activating NLRP3 induced pyroptotic cell death [70]. Another lncRNA CRAL was identified to be downregulated in cisplatin-resistant GC cells, thereby attenuated cisplatin-induced DNA damage and apoptosis [71]. The results indicated that lncRNA CRAL could sponge miRNA-505 to upregulate the cylindromatosis gene (CYLD) expression, which subsequently inhibited AKT activation and resulted in an enhancement in the sensitivity of GC cells to DDP [71].
LncRNAs and Doxorubicin Resistance
Doxorubicin (DOX) is an antibiotic reagent that is widely used in various cancers, which can inhibit DNA topoisomerase II in tumor cells and induce DNA damage and cell apoptosis. Here, we summarized some lncRNAs that were involved in DOX resistance in GC, including the lncRNAs GAS5, HOTAIR, CASC9, MRUL, UCA1, D63785, HULC, NEAT1 and ROR. Zhang et al. found that lncRNA GAS5 expression was downregulated in both GC tissues and cells [72]. GAS5 had a lower level in GC cells that were resistant to DOX. Upon restoring GAS5 expression, an inhibition of cell growth while an enhancement in cell apoptosis were observed, suggested that GAS5 functioned as a tumor suppressor and could reverse DOX resistance in GC cells [72]. LncRNA CASC9, which was involved in poor differentiation, invasion and metastasis, could promote DOX resistance in GC cells [73]. Shenqi-fuzheng formula (SQFZ), a Chinese medicine, was able to remarkably boost the sensitivity of DOX to GC cells, and this therapeutic effect was counteracted by the regulation of lncRNA HOTAIR in miRNA-17-5p/PTEN axis. This may indicate that lncRNA HOTAIR was associated with DOX resistance in GC [74].
Moreover, the expression of lncRNA MRUL was remarkably elevated in two chemoresistance GC cells, which could positively affect ABCB1 expression, causing the resistance to DOX in GC [75]. Of great interest, lncRNA UCA1 had been found to confer DOX resistance to GC cells via affecting apoptosis-related genes of PARP1 and Bcl-2 [76], or through sponging miR-27b [77]. Another research found that aberrant expression of lncRNA D63785 could enhance cell growth, migration and invasion of GC cells, and was inversely correlated with microR-422a expression. Further study showed that lncRNA D63785 caused DOX resistance in GC via suppressing miRNA-422-dependent inhibition of the myocyte enhancer factor-2D (MEF2D) [78]. In addition, lncRNA NEAT1, which was verified to be overexpressed in GC, could promote cancer cells proliferation and invasion capability [79]. Silence of lncRNA NEAT1 in SGC7901 cells reversed the DOX resistance of GC cells [79]. LncRNA HULC and ROR were also associated with increased GC cells resistant to DOX [80, 81].
LncRNAs and 5-Fluoruoracil Resistance
5-Fluoruoracil (5-FU) is a thymidylate synthase inhibitor that affects the synthesis of DNA and proteins. However, cancer cells with 5-FU resistance become a major obstacle to successful chemotherapy for GC patients. There are several oncogenic or tumor suppressive lncRNAs that have been associated with 5-FU resistance, such as FAM83H-AS1, MACC-AS1, HULC, ANRIL, XLOC-006753, FGD5-AS1 and LE1GC. For instance, it was found that the regulation of FAM83H-AS1 in 5-FU resistance was associated with Wnt/β-catenin pathway [82]. Multiple lines of evidence indicated that mesenchymal stem cells (MSCs) were related with chemotherapy resistance, but with inconclusive molecular mechanisms. Recently, He et al. found that lncRNA MACC-AS1 could be regulated by MSCs, which was able to enhance stemness and 5-FU as well as oxaliplatin resistance through fatty acid oxidation in GC [83]. Moreover, overexpression of lncRNAs ANRIL and XLOC-006753 were found to promote 5-FU resistance in GC cells [84, 85]. LncRNA XLOC-006753 affected G1/S phase transition, increasing some markers of MDR and EMT expression levels in GC cells [85]. In addition, knockdown of lncRNA FGD5-AS1 could inhibit cell growth and sensitivity of GC cells to 5-FU. Further investigations demonstrated that miRNA-153-3p/CITED2 pathway was the downstream target of lncRNA FGD5-AS1 in GC [86]. In contrast, tumor suppressor lncRNAs can sensitize the resistance to 5-FU in GC. For instance, overexpression of lncRNA LEIGC was observed to increase the sensitivity of GC cells to 5-FU through suppressing the EMT in GC [87].
LncRNAs and Paclitaxel Resistance
Paclitaxel (PTX) is one of the first-line chemotherapeutics to handle GC, which results in G2/M cell cycle arrest [88]. Five oncogenic lncRNAs have been identified to involve in PTX resistance, including the lncRNAs ZFAS1, MALAT1, PVT-1, HOTAIR and CASC9. For instance, lncRNA ZFAS1 contributed to PTX resistance in GC by changing the expression levels of EMT makers, including E-cadherin, N-cadherin, vimentin, matrix metalloproteinase (MMP)-2 and MMP-14, as well as cell cycle related markers (cyclin D1, cyclin E and cyclin B1) [89]. Furthermore, it was demonstrated that lncRNA ZFAS1 may stimulate Wnt/β-catenin pathway to enhance PTX resistance in GC [89]. LncRNA MALAT1 functioned as a competing endogenous RNA for miRNA-23b-3p and attenuated the suppressive effect of miRNA-23b-3p on ATG12, resulting in PTX chemoresistance in GC cells [90]. In addition, lncRNA PVT1was found to overexpress in human GC samples and cells which were resistant to PTX, suggesting that aberrant expression of lncRNA PVT1 may be correlated with chemotherapy efficacy in GC [91]. In another study, upregulation of lncRNA HOTAIR contributed to the resistance of PTX in GC. Intriguingly, lncRNA HOTAIR expression had negative correlation with miRNA-217 expression in GC, which indicated that elevated lncRNA HOTAIR may boost PTX resistance in GC cells via suppressing miRNA-217 [92]. In addition, lncRNA CASC9 was also correlated with PTX resistance of GC through regulating MDR1 protein expression [73].
In conclusion, multiple lncRNAs have been found to be correlated with drug resistance of GC. DDP, DOX, PTX and VCR are all first-line chemotherapeutic reagents to treat GC. However, MDR becomes the primary obstacle of clinical application, which leads to recurrence and poor prognosis. Fortunately, an increasing number of researches have exposed the underlying mechanisms of MDR-related lncRNAs in GC. Treatments towards these abnormally expressed lncRNAs are a promising method to overcome MDR. Furthermore, combination of lncRNAs-based treatment interventions with conventional chemotherapeutics may be a considerable strategy for GC.
Influences of LncRNAs on Regulating Cellular Signaling Pathways
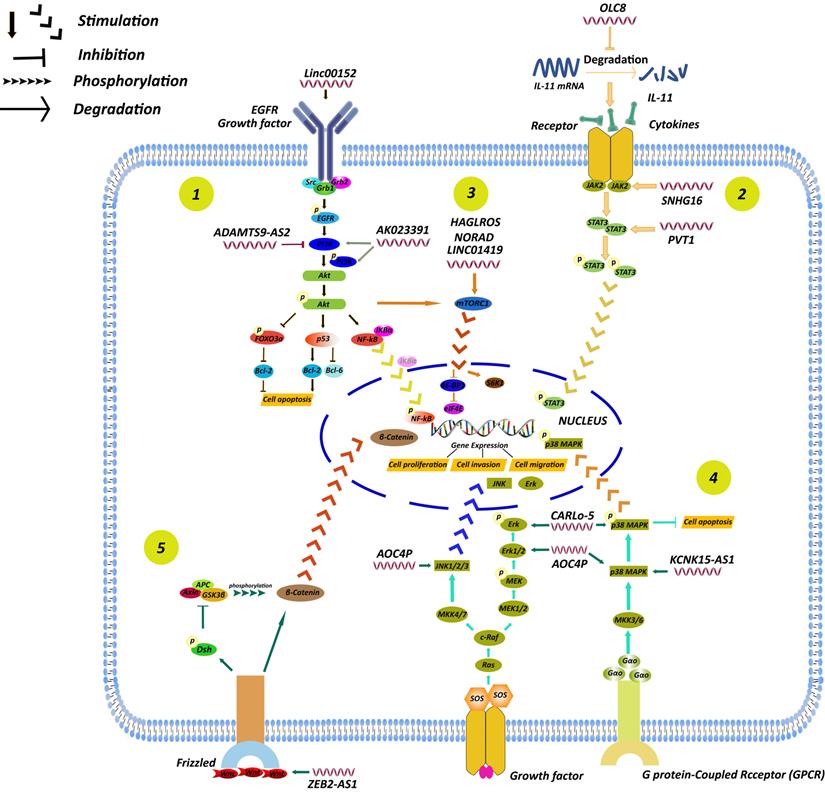
Accumulating evidence has indicated that a series of cancer phenotypes attributed to abnormal function of cellular regulator networks. As a given cell signaling pathway may consists of multiple transcription factors, these signaling molecules play a central role in cellular physiological functions and processes. Of great interest, it has become increasingly apparent that lncRNAs are able to participate in the regulation of signaling pathways to affect the development of GC (Fig. 6).
A summary of regulation of lncRNAs in GC through different signaling pathways, including the PI3K/AKT, STAT3, mTOR, MAPK and Wnt/β-catenin signaling pathways. (1) LncRNA AK023391 [96] and lncRNA ADAMTS9-AS2 [97] participated in GC development by affecting PI3K/AKT signaling pathway. (2) LncRNA PVT1 [99], SNHG16 [100], and OLC8 [50] participated in GC development by affecting STAT3 signaling pathway. (3) LncRNA LINC01419 [105], HAGLROS [106], NORAD [107] participated in GC development by affecting mTOR signaling pathway. (4) LncRNA AOC4P [109], CARlo-5 [110], KCNK15-AS1 [111] participated in GC development by affecting MAPK signaling pathway. (5) Some lncRNAs enhanced GC progression by activating other signaling pathways, including Wnt/β-catenin signaling pathways [112].
AKT Signaling Pathway
AKT signaling pathway is a crucial pathway in tumor development, including cell proliferation, migration and invasion, and other biological processes [93]. Intriguingly, lncRNAs can influence tumor development by regulating key genes in the AKT pathway. For example, it was demonstrated that linc00152 was closely correlated with cell migration, invasion, apoptosis, cell cycle arrest, and EMT in GC [94]. Zhou et al. found that down-regulated expression of linc00152 was able to inhibit tumor growth in vivo and in vitro, and RNA immunoprecipitation (RIP) experiment showed that linc00152 could directly bind to epidermal growth factor receptor (EGFR). Downregulation of linc00152 led to suppressed p-EGFR, p-AKT or p-PI3K, revealing that linc00152 may enhance tumor growth via activation of PI3K-AKT signaling pathway [95].
Recently, a novel lncRNA AK023391 was observed highly expressed in GC tissues comparing with the paired adjust normal tissues, and was closely correlated with lower survival in GC patients [96]. Further studies indicated that an elevation of lncRNA AK023391 could promote proliferation, migration and invasion of GC cells [96]. Bioinformatics analysis showed that PI3K/AKT and FOXO signaling pathways had the most significant enrichment among the pathways regulated by lncRNA AK023391. It was further confirmed that expression levels of the genes associated with these two mentioned pathways had changed to varying degrees. Downregulation of lncRNA AK023391 repressed tumor growth through PI3K/AKT pathway [96]. Therefore, lncRNA AK023391 activated the PI3K/AKT pathway and further affected transcription factors associated with cell proliferation, apoptosis and invasion, resulting in promoting GC tumorigenesis and progression.
On the contrary, lncRNA ADAMTS9-AS2, functioned as a tumor suppressor, was identified to be significantly down-regulated both in GC tissues and cells [97]. Cell proliferation, migration and invasion was suppressed in SGC-7901 cells that were transfected with pc-ADAMTS9-AS2 [97]. Mechanically, p-PI3K and p-AKT expression level was significantly down-regulated in GC cells overexpressed with lncRNA ADAMTS9-AS2. Furthermore, treatment with LY294002 (a PI3K inhibitor) on SGC-7901 cells could reverse the enhanced ability of cell proliferation, cell migration and invasion but a suppression of cell apoptosis when lncRNA ADAMTS9-AS2 expression was down-regulated. These results indicated that the activation of P13K/AKT pathway may participate in the modulation of lncRNA ADAMTS9-AS2 in GC [97].
STAT3 Signaling Pathway
It is generally known that the signal transducer and activator of transcription 3 (STAT3) pathway participate in a series of biological processes in human cancers, including cell proliferation, survival and apoptosis [98]. Vasculogenic mimicry (VM), a vessel-like network which generated by cancer cells with strong invasion ability, exerts a central role in tumor progression. According to Zhao et al.'s research, lncRNA PVT1 contributed to VM formation both in vitro and in vivo [99]. Moreover, they observed that lncRNA PVT1 could activate STAT3 and assist STAT3 to recruit Slug promoter, thereby increasing Slug expression and leading to EMT and VM formation [99]. Hence, their findings showed that the lncRNA PVT1/STAT3 axis may be a possible therapeutic target in GC [99]. Furthermore, lncRNA SNHG16 could act as an oncogene in GC which was attributed to the activation of JAK2/STAT3 signaling pathway via blocking the function of miRNA-135 [100]. Zhou et al. found that lncRNA OLC8 could interfere with IL-11 mRNA degradation so that the expression level of IL-11 was significantly increased. With no doubt, an elevated expression of IL-11 could enhance STAT3 activation to favor GC development [50].
mTOR Signaling Pathway
The mammalian target of rapamycin (mTOR) pathway exhibits a pivotal role on cell growth and migration in human cancers, including GC [101, 102]. Moreover, mTOR exerts its role through two complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which are completely different both in function and structure [103, 104].
Wang et al. found that lncRNA LINC01419 was significantly upregulated in GC tissues [105]. Downregulation of lncRNA LINC01419 or inhibition of the mTOR pathway, or both, could lead to a suppressive role in migration and invasion of GC cells while promoted cell autophagy [105]. AKT1 and mTOR phosphorylation level was decreased upon the inhibition of LINC01419 expression, which may reveal that lncRNA LINC01419 participated in the progression of GC through activating the AKT/mTOR pathway [105].
Autophagy is one of the most essential biological processes with the capable of maintaining cellular homeostasis and protecting against invading pathogens. A study showed that overexpression of lncRNA HAGLROS promoted GC developments and represented poor prognosis via regulating mTOR signaling pathway to interact with mTORC1, which resulted in autophagy suppression [106]. Another investigation demonstrated that lncRNA NORAD was overexpressed in GC tissues and cell lines, while miRNA-214 was remarkably down-regulated. LncRNA NORAD accelerated GC cell growth both in vivo and in vitro. However, an elevated miRNA-214 expression could reserve this effect. Further investigation suggested that lncRNA NORAD may contribute the progression of GC through regulating mTOR signaling pathways [107].
MAPK Signaling Pathway
Evidences demonstrate that the mitogen-activated protein kinase (MAPK) pathway plays a crucial role in cell growth and consists of key cascades of the ERK1/2, JNK1/2/3, and p38-MAPK [108].
Qu et al. found that lncRNA AOC4P was overexpressed in GC tissues, comparing with the paired adjacent tissue, and its high expression was closely related with poor survival in GC patients [109]. Knockdown of lncRNA ACO4P led to the inhibition of cell proliferation, migration and invasion, while enhanced the apoptosis in GC cells [109]. Expression level of the related proteins, including ERK1, p38 and JNK, were decreased upon the inhibition of lncRNA ACO4P. These data suggested that lncRNA ACO4P participated in GC development via activating the MAPK signaling pathway [109]. Similarly, lncRNA CARlo-5 was overexpressed in GC tissues which contributed to tumor growth [110]. Knockdown of lncRNA CARlo-5 could inactivate the ERK/MAPK signaling pathway, thereby induced G0/G1 cycle arrest and apoptosis and dramatically inhibited GC cell proliferation [110]. By contrast, overexpression of lncRNA KCNK15-AS1 could inhibit cell growth, while increase cell apoptosis in GC. Subsequent experiment revealed that both DNA methyltransferase (cytosine-5) 1 (DNMT1) and histone deacetylase 1 (HDAC1) were involved in the process of KCNK15-AS1 methylation in GC, and this interaction led to the activation of the MAPK and AKT signaling pathway [111].
Other Signaling Pathways
Besides the mentioned above signaling pathways, there are other signaling pathways involved in regulation of lncRNAs in GC. For instance, the expression level of lncRNA ZEB2-AS1 was up-regulated in GC cells and clinical samples [112]. Knockdown of lncRNA ZEB2-AS1 via infecting ZEB2-AS1-shRNA was capable of inhibiting cell proliferation, metastasis and EMT, while promoting the apoptosis of GC cells [112]. Furthermore, western blot experiment indicated that ZEB2-AS1 may contribute to development of GC via modulating the Wnt/β-catenin signaling pathway [112]. There was a report revealed that the well-known lncRNA, HOTAIR, was highly expressed in GC cells and tissues via the activation of CXCR4 and RhoA signaling pathways [113]. Additionally, lncRNA BANCR and ANRIL were both involved in GC progression via regulating NF-κB [114, 115].
LncRNAs as Biomarkers and Therapeutic Targets in GC
Diagnostic and Prognostic Role of LncRNAs
Owing to the lack of conspicuous symptoms in early stage, most GC patients are at advanced stage in diagnosis. Hence, it is urgent to identify effective biomarkers for diagnosis. Some established tumor-specific biomarkers, including carcinoembryonic antigen (CEA), carbohydrate antigen (CA) 242, CA724, and serum pepsinogen (SPG), are known to have limitations in the diagnosis of GC [116]. LncRNAs can be detected in a highly stable form in plasma and are therefore considered to be promising diagnostic indices. Recently, studies reported that lncRNAs were enriched and stabilized in exosomes that were secreted by cancer cells [117, 118]. It was reported that lncRNAs such as ZFAS1, HOTTIP and UUEGC1 could be detected in plasma exosomes and there were distinct differences between GC patients and the controls [119-121]. Thus, abundant lncRNAs could be the potential biomarkers in GC diagnosis (Table 2).
Emerging evidence indicated that the levels of lncRNAs in plasma could be satisfactory tumor biomarkers in GC diagnosis. For instance, the expressions of lncRNA H19 in plasma of GC patients were remarkably higher than that in controls, and subsequent ROC analysis showed that the area under the curve (AUC) was 0.838. Furthermore, the expression of H19 distinguished early-stage GC from controls with an AUC of 0.877. These results demonstrated that plasma H19 may be an available detecting biomarker for GC [122].
Beyond that the use of a single lncRNA, the multiple plasma lncRNA combinations could become more powerful for GC diagnosis. Studies suggested that the levels of plasma lncRNAs of FAM49B-AS1, GUSBP11 and CTDHUT in GC patients were significantly higher than that in healthy controls [123]. Moreover, the combinational use of these three lncRNAs with CA242 or CA724 was superior to the CA242 or CA724 alone for GC diagnosis [123]. Consequently, plasma FAM49B-AS1, GUSBP11 and CTDHUT had great potential as promising tumor biomarkers for early GC diagnosis.
Evaluation of prognosis is essential for assessing treatment status and adjusting treatment strategy. More strikingly, lncRNAs are able to act as prognostic biomarkers in GC (Table 3). For instance, the expression of lncRNA MIR100HG was observed to be overexpressed in clinical samples from GC patients and GC cells, compared with controls or the adjacent normal gastric mucosa tissues. Further study indicated that an elevated level of lncRNA MIR100HG was positively correlated with clinical stage, tumor invasion, lymph node metastasis and distant metastasis. Moreover, survival analysis revealed that MIR100HG expression was negatively related with clinical outcome in patients with GC [124]. Therefore, MIR100HG, an oncogenic lncRNA, could be a valuable biomarker in GC prognosis. Similarly, lncRNA Sox2 overlapping transcript (Sox2ot) was highly expressed in GC cell lines and tissues based on Zhang et al.'s study [125]. The relationship between lncRNA Sox2ot and clinicopathological features was analyzed subsequently. The results showed that the level of Sox2ot was correlated with clinical stage, tumor depth, lymph node metastasis and distant metastasis [125], suggesting that Sox2ot was a potential prognostic marker for GC patients.
LncRNAs as diagnostic biomarkers in GC
| LncRNAs | Expression | Diagnostic accuracy | Reference |
|---|---|---|---|
| H19 | Upregulated | AUC, 0.838; sensitivity, 82.9%; specificity, 72.9% | [122] |
| FAM49B-AS, GUSBP11, CTDHUT | Upregulated | AUC, 0.818 | [123] |
| GNAQ-6:1 | Downregulated | AUC, 0.732 | [137] |
| B3GALT5-AS1 | Upregulated | AUC, 0.816 | [138] |
| UCA1 | Upregulated | AUC, 0.759; sensitivity, 66.67%; specificity, 87.04% | [139] |
| UEGC1 | Upregulated | AUC, 0.8406 | [121] |
| PANDAR, FOXD2-AS1, SMARCC2 | Upregulated | AUC, 0.839 | [140] |
| TINCR, CCAT2, AOC4P, BANCR, LINC00857 | Upregulated | AUC, 0.91 | [141] |
Note: AUC, area under the curve of ROC analysis.
LncRNAs as prognostic biomarkers in GC
| LncRNAs | Expression | Prognostic implications | Reference |
|---|---|---|---|
| SNHG6 | Upregulated | Correlated with invasion depth, lymph node metastasis, and distant metastasis | [142] |
| SNHG8 | Upregulated | Predicted poor prognosis and shorter survival time | [143] |
| Sox2ot | Upregulated | Correlated with TNM stage, tumor depth, lymph node metastasis, and distant metastasis | [125] |
| MALAT1 | Upregulated | Correlated with distant metastasis | [144] |
| NEAT1 | Upregulated | Correlated with clinical stage, histological type, lymph node metastasis, and distant metastasis | [145] |
| MIR100HG | Upregulated | Correlated with TNM stage, tumor invasion, lymph node metastasis, and distant metastasis | [124] |
| CTD-2510F5.4 | - | Correlated with pathological grade, vascular or nerve invasion, TNM stage and OS, shorter MST | [146] |
Note: TNM, Tumor node metastasis; OS, Overall survival; MST, Median survival time.
LncRNAs as Therapeutic Targets for GC
Given the fact that lncRNAs are involved in many biological and pathological functions, exerting a crucial role in GC development, progression and metastasis, it is possible to consider exploiting lncRNAs as therapeutic targets in GC. One of advantages of targeting lncRNAs is that they may have pleiotropic effects via influencing multiple tumor-related signaling pathways. As a result, targeting lncRNAs may yield pomissing treatment outcome based on small interference RNAs (siRNAs), antisense oligonucleotides (ASOs), or Crispr-Cas9 techniques.
Some drugs may also target lncRNAs to influence GC growth. Accumulating vigorous evidence indicated that lncRNA PVT1 was overexpressed in a number of cancers including GC [126], and was significantly associated with tumor disease processes [127-130]. Cardamonin (CARD, 2′,4′-dihydroxy-6′-methoxychalcone), which is a kind of chalcone, is an active ingredient isolated from several plants such as the Alpinia [131]. Wang et al. found that CARD was able to inhibit cell proliferation and migration, as well as induce apoptosis in GC cell lines. Furthermore, they observed that CARD remarkably decreased lncRNA PVT1 expression, leading to an inhibition of STAT3 pathway [132]. The findings firstly indicated that some drugs may regulate lncRNAs to inhibit GC, and lncRNAs serve as potential targets. Despite that there are few studies on drugs targeting lncRNAs in GC at present, it is clear that there is great potential for developing drugs targeting lncRNAs for the clinical treatment of GC.
Conclusions and Future Perspectives
GC is one of the most common malignant tumors with high morbidity and mortality worldwide. Emerging evidence indicates that a large number of lncRNAs play a vital role in the maintenance of human homeostasis. Alteration of lncRNAs expression may affect a series of disease process, including cell growth, metastasis, apoptosis, EMT and MDR. Hence, it is essential to clarify the detail mechanisms between lncRNAs and gastric tumorigenesis and developments.
H. pylori infection exhibits a pivotal role in GC tumorigenesis, and differentially expressed lncRNAs are associated with H. pylori-associated GC. In this review, we summarized the current findings regarding the role of lncRNAs in H. pylori-associated GC. Some lncRNAs may be potential diagnostic marker and prognostic factor. Despite the fact that most studies revealed the potential association of lncRNAs in H. pylori-induced GC, the relationships between multiple virulence factors considering H. pylori infection and the alterations of lncRNAs in gastric diseases, especially GC, are required to illuminate in further investigations.
In decades, it is realized that finding out the key molecules and/or signaling pathways in the TME is essential for future cancer therapy. In this regard, lncRNAs has been found to implicated in regulation of multiple immune cells and cytokines, such as TGF-β superfamily [9, 43, 52, 53, 133], IFN family [134], IL family [9, 50], and related signaling pathways [7, 51], thus regulating the TME, changing the biological functions of tumor, and gradually promoting or inhibiting the progression of GC. However, the studies on the regulatory role of lncRNAs within GC TME were considered insufficient, which may be due to the facts that GC is a molecularly and phenotypically highly-heterogeneous disease and has different subtypes with striking distinct clinical features. Therefore, to explore the interaction between lncRNA and TME and to find out the exact mechanisms underlying the regulation of lncRNAs in TME are among the most urgent and fundamental challenges.
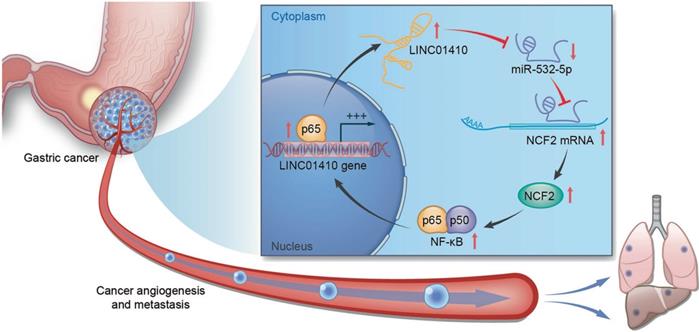
The feedback loop between LINC01410 and NCF2. LINC01410 upregulated NCF2 and NF-κB signal via binding to and inhibiting miRNA-532-5p. NCF2 in turn increased LINC01410 via NF-κB [135]. Reprinted with permission [135].
Alteration of lncRNAs were found closely correlated with drug resistance. Nevertheless, the detailed mechanisms may be complicate. The results suggested that knockdown of lncRNAs through small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) may benefit to extend understanding of molecular mechanisms underlying MDR of GC as well as the GC treatment. Furthermore, combination of lncRNAs-based therapeutic interventions with traditional chemotherapy seems to be an approach to overcome MDR in GC.
It has become increasingly apparent that lncRNAs are capable of regulating cellular physiological processes and other biological functions by targeting the key genes or regulator through different signaling pathways in GC. Here, we summarized the abnormally expressed of lncRNAs in GC progression as well as the signaling pathway correlated with the tumorigenesis and development of GC. Studies reveal that lncRNAs could become important indexes for GC diagnose and prognosis, which might provide novel therapeutic targets in future GC treatment. Moreover, lncRNAs can also interact with miRNAs that affect gene expression, thereby promoting or suppressing GC progression. For example, miRNA-532-5p was able to suppress GC angiogenesis and metastasis, while linc01410 could bind with miRNA-532-5p which repressed its function. Furthermore, there had been a positive feedback loop between linc01410 and neutrophil cytosolic factor 2 (NCF2) through the NF-κB pathway that promoted GC development (Fig. 7) [135]. Thus, this opens novel investigation lines towards the relationship between the dysregulated lncRNAs and GC progression, thereby providing brand new treatment concept.
Many studies have revealed the potential role of lncRNAs as biomarkers in GC. LncRNAs can be detected in a highly stable form in plasma, and testing lncRNAs in plasma is simple, non-invasive and easy to be accepted for GC diagnosis. Either a single lncRNA or the combinations of plasm lncRNAs has been reported to be employed as biomarkers for GC diagnosis. Moreover, lncRNAs can be a credible complement to classic cancer detecting indices. In addition to the use of diagnosis, more and more studies have shown that lncRNAs may act as prognostic factors in GC owing to a closely correlation between the expression levels of lncRNAs and clinical features of GC patients. Furthermore, it is distinct to notice an obvious alteration of levels of lncRNAs after certain clinic treatments. Thus, novel lncRNAs that can be stably used in the diagnosis and prognosis of GC is a promising filed which is needed for further evaluation.
It is as well convincing that lncRNAs can be therapeutic targets in GC, since lncRNAs control tumor micro-regulatory networks, and targeting lncRNAs may affect different tumor-associated pathways [136]. Targeting lncRNAs may yield reliable therapeutic results based on small interference RNAs (siRNAs), antisense oligonucleotides (ASOs), Crispr-Cas9 techniques, lncRNA-targeting drugs and so on. However, there are still limitations for translating knowledge on lncRNAs to the clinical practice. First, the current status of mechanism study of lncRNAs in GC is considered inadequate. Second, the vast majority of studies associated with lncRNAs in GC focused on their expression profiles, but the role of lncRNA mutants in GC progression is still a mystery. Nonetheless, there is no doubt about the significance of lncRNAs applications in clinic treatment in future.
Collectively, despite of numerous gaps, recent researches have indicated that a large number of lncRNAs play a crucial role in biological process in GC, including cell growth, metastasis, apoptosis, drug resistance and so on. Studies dedicating to decode the underlying mechanisms of lncRNA-associated GC may provide a rationale in the fight against GC, regarding both biomarkers and therapeutic targets.
Abbreviations
DOX: Doxorubicin; CagA: Cytotoxin associated gene A; DDP: Cisplatin drug; DSBs: Double-strand breaks; EMT: Epithelial-mesenchymal transition; ERK: Extracellular signal-related kinases; GC: Gastric cancer; H. pylori: Helicobacter pylori; HR: Homologous recombination; IL: Interleukin; JNK: Jun amino terminal kinases; MAPK: Mitogen-activated protein kinase; MDR: Multi-drug resistance; MIF: Macrophage migration inhibitory factor; miRNA: MicroRNA; MMP: Matrix metalloproteinase; MSCs: Mesenchymal stem cells; mTOR: Mammalian target of rapamycin; ncRNA: Non-coding RNA; NHEJ: Nonhomologous end joining; PTX: Paclitaxel; STAT3: Signal transducer and activator of transcription 3; TAMs: Tumor-associated macrophages; TILs: Tumor infiltration lymphocytes; TME: Tumor microenvironment; T-reg: Regulatory T cell; VacA: Vacuolating cytotoxin A; 5-FU: 5-Fluoruoracil; TNM: Tumor node metastasis; OS: Overall survival; MST: Median survival time.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 81803237, 81803812 and 81770562), the Joint Funds of the Southwest Medical University & Luzhou, China (grant no. 2018LZXNYD-ZK34), Science and Technology Planning Project of Luzhou, China (grant no. 2018-RCM-64) and National Key R&D Project of China (grant no. 2018YFC0115301).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Figueiredo C, Costa S, Karameris A, Machado JC. Pathogenesis of Gastric Cancer. Helicobacter. 2015;20(Suppl 1):30-5
2. Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S. et al. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029-44
3. Dastmalchi N, Khojasteh SMB, Nargesi MM, Safaralizadeh R. The correlation between lncRNAs and Helicobacter pylori in gastric cancer. Pathog Dis. 2019 77
4. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18
5. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661-7
6. Schaalan M, Mohammad W, Fathy S. MiRNA-200c, MiRNA-139 and ln RNA H19; new predictors of treatment response in H-pylori- induced gastric ulcer or progression to gastric cancer. Microb Pathog. 2020: 104442.
7. Zhang Y, Yan J, Li C, Wang X, Dong Y, Shen X. et al. LncRNA H19 induced by helicobacter pylori infection promotes gastric cancer cell growth via enhancing NF-kappaB-induced inflammation. J Inflamm (Lond). 2019;16:23
8. Jin C, Shi W, Wang F, Shen X, Qi J, Cong H. et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget. 2016;7:51763-72
9. Jing XH, Li LX, Han TT, Shi J. Effects of Long Non-coding RNA Plasmacytoma Variant Translocation 1 Gene on Inflammatory Response and Cell Migration in Helicobacter Pylori Infected Gastric Epithelial Cell Line. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2020;42:228-35
10. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC. et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784-801
11. Zhou X, Chen H, Zhu L, Hao B, Zhang W, Hua J. et al. Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget. 2016;7:82770-82
12. Zhong F, Zhu M, Gao K, Xu P, Yang H, Hu D. et al. Low expression of the long non-coding RNA NR_026827 in gastric cancer. Am J Transl Res. 2018;10:2706-11
13. Yang L, Long Y, Li C, Cao L, Gan H, Huang K. et al. Genome-wide analysis of long noncoding RNA profile in human gastric epithelial cell response to Helicobacter pylori. Jpn J Infect Dis. 2015;68:63-6
14. Yang J, Song H. Identification of long noncoding RNA RP11-169F17.1 and RP11-669N7.2 as novel prognostic biomarkers of stomach adenocarcinoma based on integrated bioinformatics analysis. Epigenomics. 2019;11:1307-21
15. Yang J, Song H, Cao K, Song J, Zhou J. Comprehensive analysis of Helicobacter pylori infection-associated diseases based on miRNA-mRNA interaction network. Brief Bioinform. 2019;20:1492-501
16. Zhu H, Wang Q, Yao Y, Fang J, Sun F, Ni Y. et al. Microarray analysis of Long non-coding RNA expression profiles in human gastric cells and tissues with Helicobacter pylori Infection. BMC Med Genomics. 2015;8:84
17. Lang F, Shumilina E. Regulation of ion channels by the serum- and glucocorticoid-inducible kinase SGK1. FASEB J. 2013;27:3-12
18. Yao Y, Jiang Q, Jiang L, Wu J, Zhang Q, Wang J. et al. Lnc-SGK1 induced by Helicobacter pylori infection and highsalt diet promote Th2 and Th17 differentiation in human gastric cancer by SGK1/Jun B signaling. Oncotarget. 2016;7:20549-60
19. Liu L, Shuai T, Li B, Zhu L, Li X. Long noncoding RNA lncGNAT11 inhibits gastric cancer cell proliferation and invasion through the Wnt/betacatenin pathway in Helicobacter pylori infection. Mol Med Rep. 2018;18:4009-15
20. Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181-211
21. Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M. et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci U S A. 2011;108:14944-9
22. Koeppel M, Garcia-Alcalde F, Glowinski F, Schlaermann P, Meyer TF. Helicobacter pylori Infection Causes Characteristic DNA Damage Patterns in Human Cells. Cell Rep. 2015;11:1703-13
23. Dorer MS, Fero J, Salama NR. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 2010;6:e1001026
24. Song J, Bent AF. Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLoS Pathog. 2014;10:e1004030
25. Han T, Jing X, Bao J, Zhao L, Zhang A, Miao R. et al. H. pylori infection alters repair of DNA double-strand breaks via SNHG17. J Clin Invest. 2020;130:3901-18
26. Ba MC, Ba Z, Long H, Cui SZ, Gong YF, Yan ZF. et al. LncRNA AC093818.1 accelerates gastric cancer metastasis by epigenetically promoting PDK1 expression. Cell Death Dis. 2020;11:64
27. Zhang Z, Hu X, Kuang J, Liao J, Yuan Q. LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell Mol Biol Lett. 2020;25:29
28. Li D, Wang J, Zhang M, Hu X, She J, Qiu X. et al. LncRNA MAGI2-AS3 Is Regulated by BRD4 and Promotes Gastric Cancer Progression via Maintaining ZEB1 Overexpression by Sponging miR-141/200a. Mol Ther Nucleic Acids. 2020;19:109-23
29. Wu Q, Ma J, Meng W, Hui P. DLX6-AS1 promotes cell proliferation, migration and EMT of gastric cancer through FUS-regulated MAP4K1. Cancer Biol Ther. 2020;21:17-25
30. Langley RR, Fidler IJ. The seed and soil hypothesis revisited-the role of tumor-stroma interactions in metastasis to different organs. Int J Cancer. 2011;128:2527-35
31. Wu X, Hu W, Lu L, Zhao Y, Zhou Y, Xiao Z. et al. Repurposing vitamin D for treatment of human malignancies via targeting tumor microenvironment. Acta Pharm Sin B. 2019;9:203-19
32. Hou CM, Qu XM, Zhang J, Ding TT, Han W, Ji GC. et al. Fibroblast activation proteins-alpha suppress tumor immunity by regulating T cells and tumor-associated macrophages. Exp Mol Pathol. 2018;104:29-37
33. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531-64
34. Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat Immunol. 2007;8:1353-62
35. Bachmann MF, Kohler G, Ecabert B, Mak TW, Kopf M. Cutting edge: lymphoproliferative disease in the absence of CTLA-4 is not T cell autonomous. J Immunol. 1999;163:1128-31
36. Wang X, Lang M, Zhao T, Feng X, Zheng C, Huang C. et al. Cancer-FOXP3 directly activated CCL5 to recruit FOXP3(+)Treg cells in pancreatic ductal adenocarcinoma. Oncogene. 2017;36:3048-58
37. Jang JE, Hajdu CH, Liot C, Miller G, Dustin ML, Bar-Sagi D. Crosstalk between Regulatory T Cells and Tumor-Associated Dendritic Cells Negates Anti-tumor Immunity in Pancreatic Cancer. Cell Rep. 2017;20:558-71
38. Martinez LM, Robila V, Clark NM, Du W, Idowu MO, Rutkowski MR. et al. Regulatory T Cells Control the Switch From in situ to Invasive Breast Cancer. Front Immunol. 2019;10:1942
39. Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K. et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679-84
40. Marshall EA, Ng KW, Kung SH, Conway EM, Martinez VD, Halvorsen EC. et al. Emerging roles of T helper 17 and regulatory T cells in lung cancer progression and metastasis. Mol Cancer. 2016;15:67
41. Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol. 2018;118:24-30
42. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y. et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129
43. Xiong G, Yang L, Chen Y, Fan Z. Linc-POU3F3 promotes cell proliferation in gastric cancer via increasing T-reg distribution. Am J Transl Res. 2015;7:2262-9
44. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
45. Su S, Liu Q, Chen J, Chen J, Chen F, He C. et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605-20
46. Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X. et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18:64
47. Georgoudaki AM, Prokopec KE, Boura VF, Hellqvist E, Sohn S, Ostling J. et al. Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep. 2016;15:2000-11
48. Weichand B, Popp R, Dziumbla S, Mora J, Strack E, Elwakeel E. et al. S1PR1 on tumor-associated macrophages promotes lymphangiogenesis and metastasis via NLRP3/IL-1beta. J Exp Med. 2017;214:2695-713
49. Nie K, Zheng Z, Wen Y, Pan J, Liu Y, Jiang X. et al. A novel ceRNA axis involves in regulating immune infiltrates and macrophage polarization in gastric cancer. Int Immunopharmacol. 2020;87:106845
50. Zhou R, Wu Z, Deng X, Chen H. The long non-coding RNA OLC8 enhances gastric cancer by interaction with IL-11. J Clin Lab Anal. 2019;33:e22962
51. Kong F, Deng X, Kong X, Du Y, Li L, Zhu H. et al. ZFPM2-AS1, a novel lncRNA, attenuates the p53 pathway and promotes gastric carcinogenesis by stabilizing MIF. Oncogene. 2018;37:5982-96
52. Sakai S, Ohhata T, Kitagawa K, Uchida C, Aoshima T, Niida H. et al. Long Noncoding RNA ELIT-1 Acts as a Smad3 Cofactor to Facilitate TGFbeta/Smad Signaling and Promote Epithelial-Mesenchymal Transition. Cancer Res. 2019;79:2821-38
53. Zhang Q, Chen B, Liu P, Yang J. XIST promotes gastric cancer (GC) progression through TGF-beta1 via targeting miR-185. J Cell Biochem. 2018;119:2787-96
54. Guo Y, Yue P, Wang Y, Chen G, Li Y. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother. 2019;118:109255
55. Li H, Ma X, Yang D, Suo Z, Dai R, Liu C. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem. 2020;121:1353-61
56. Dai Q, Zhang T, Pan J, Li C. LncRNA UCA1 promotes cisplatin resistance in gastric cancer via recruiting EZH2 and activating PI3K/AKT pathway. J Cancer. 2020;11:3882-92
57. Xu YD, Shang J, Li M, Zhang YY. LncRNA DANCR accelerates the development of multidrug resistance of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:2794-802
58. Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol. 2016
59. Wang L, Chunyan Q, Zhou Y, He Q, Ma Y, Ga Y. et al. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:4064-70
60. Wang R, Lu X, Yu R. lncRNA MALAT1 Promotes EMT Process and Cisplatin Resistance of Oral Squamous Cell Carcinoma via PI3K/AKT/m-TOR Signal Pathway. Onco Targets Ther. 2020;13:4049-61
61. Kangboonruang K, Wongtrakoongate P, Lertsuwan K, Khachonkham S, Changkaew P, Tangboonduangjit P. et al. MALAT1 Decreases the Sensitivity of Head and Neck Squamous Cell Carcinoma Cells to Radiation and Cisplatin. Anticancer Res. 2020;40:2645-55
62. Wang N, Hou MS, Zhan Y, Shen XB, Xue HY. MALAT1 promotes cisplatin resistance in cervical cancer by activating the PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:7653-9
63. Zhang YF, Li CS, Zhou Y, Lu XH. Propofol facilitates cisplatin sensitivity via lncRNA MALAT1/miR-30e/ATG5 axis through suppressing autophagy in gastric cancer. Life Sci. 2020;244:117280
64. Ye Y, Yang S, Han Y, Sun J, Xv L, Wu L. et al. HOXD-AS1 confers cisplatin resistance in gastric cancer through epigenetically silencing PDCD4 via recruiting EZH2. Open Biol. 2019;9:190068
65. Zhang Y, Li Q, Yu S, Zhu C, Zhang Z, Cao H. et al. Long non-coding RNA FAM84B-AS promotes resistance of gastric cancer to platinum drugs through inhibition of FAM84B expression. Biochem Biophys Res Commun. 2019;509:753-62
66. Peng J. si-TP73-AS1 suppressed proliferation and increased the chemotherapeutic response of GC cells to cisplatin. Oncol Lett. 2018;16:3706-14
67. Fu T, Ji K, Jin L, Zhang J, Wu X, Ji X. et al. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-kappaB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer. 2020
68. Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer. 2020;19:96
69. Wang J, Lv B, Su Y, Wang X, Bu J, Yao L. Exosome-Mediated Transfer of lncRNA HOTTIP Promotes Cisplatin Resistance in Gastric Cancer Cells by Regulating HMGA1/miR-218 Axis. Onco Targets Ther. 2019;12:11325-38
70. Ren N, Jiang T, Wang C, Xie S, Xing Y, Piao D. et al. LncRNA ADAMTS9-AS2 inhibits gastric cancer (GC) development and sensitizes chemoresistant GC cells to cisplatin by regulating miR-223-3p/NLRP3 axis. Aging (Albany NY). 2020;12:11025-41
71. Wang Z, Wang Q, Xu G, Meng N, Huang X, Jiang Z. et al. The long noncoding RNA CRAL reverses cisplatin resistance via the miR-505/CYLD/AKT axis in human gastric cancer cells. RNA Biol. 2020:1-14
72. Zhang N, Wang AY, Wang XK, Sun XM, Xue HZ. GAS5 is downregulated in gastric cancer cells by promoter hypermethylation and regulates adriamycin sensitivity. Eur Rev Med Pharmacol Sci. 2016;20:3199-205
73. Shang C, Sun L, Zhang J, Zhao B, Chen X, Xu H. et al. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393-8
74. Jia J, Zhan D, Li J, Li Z, Li H, Qian J. The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and Shenqifuzheng injection on chemosensitivity of gastric cancer cells. J Cell Mol Med. 2019;23:656-69
75. Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182-93
76. Shang C, Guo Y, Zhang J, Huang B. Silence of long noncoding RNA UCA1 inhibits malignant proliferation and chemotherapy resistance to adriamycin in gastric cancer. Cancer Chemother Pharmacol. 2016;77:1061-7
77. Fang Q, Chen X, Zhi X. Long Non-Coding RNA (LncRNA) Urothelial Carcinoma Associated 1 (UCA1) Increases Multi-Drug Resistance of Gastric Cancer via Downregulating miR-27b. Med Sci Monit. 2016;22:3506-13
78. Zhou Z, Lin Z, He Y, Pang X, Wang Y, Ponnusamy M. et al. The Long Noncoding RNA D63785 Regulates Chemotherapy Sensitivity in Human Gastric Cancer by Targeting miR-422a. Mol Ther Nucleic Acids. 2018;12:405-19
79. Zhang J, Zhao B, Chen X, Wang Z, Xu H, Huang B. Silence of Long Noncoding RNA NEAT1 Inhibits Malignant Biological Behaviors and Chemotherapy Resistance in Gastric Cancer. Pathol Oncol Res. 2018;24:109-13
80. Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC Enhances Chemotherapy Induced Apoptosis in Human Gastric Cancer. J Med Biochem. 2016;35:137-43
81. Wang S, Chen W, Yu H, Song Z, Li Q, Shen X. et al. lncRNA ROR Promotes Gastric Cancer Drug Resistance. Cancer Control. 2020;27:1073274820904694
82. Wang B, Guan G, Zhao D. Silence of FAM83H-AS1 promotes chemosensitivity of gastric cancer through Wnt/beta-catenin signaling pathway. Biomed Pharmacother. 2020;125:109961
83. He W, Liang B, Wang C, Li S, Zhao Y, Huang Q. et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38:4637-54
84. Lan WG, Xu DH, Xu C, Ding CL, Ning FL, Zhou YL. et al. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol Rep. 2016;36:263-70
85. Zeng L, Liao Q, Zou Z, Wen Y, Wang J, Liu C. et al. Long Non-Coding RNA XLOC_006753 Promotes the Development of Multidrug Resistance in Gastric Cancer Cells Through the PI3K/AKT/mTOR Signaling Pathway. Cell Physiol Biochem. 2018;51:1221-36
86. Gao Y, Xie M, Guo Y, Yang Q, Hu S, Li Z. Long Non-coding RNA FGD5-AS1 Regulates Cancer Cell Proliferation and Chemoresistance in Gastric Cancer Through miR-153-3p/CITED2 Axis. Front Genet. 2020;11:715
87. Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J. et al. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer. 2014;14:932
88. Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L. et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62
89. Xu W, He L, Li Y, Tan Y, Zhang F, Xu H. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/beta-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem. 2018;82:456-65
90. YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C. et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174
91. Zhang XW, Bu P, Liu L, Zhang XZ, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227-32
92. Wang H, Qin R, Guan A, Yao Y, Huang Y, Jia H. et al. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J Cell Biochem. 2018;119:7226-34
93. Murugan AK. Special issue: PI3K/Akt signaling in human cancer. Semin Cancer Biol. 2019;59:1-2
94. Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P. et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112-23
95. Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J. et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135
96. Huang Y, Zhang J, Hou L, Wang G, Liu H, Zhang R. et al. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36:194
97. Cao B, Liu C, Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed Pharmacother. 2018;107:185-93
98. Furtek SL, Backos DS, Matheson CJ, Reigan P. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS Chem Biol. 2016;11:308-18
99. Zhao J, Wu J, Qin Y, Zhang W, Huang G, Qin L. LncRNA PVT1 induces aggressive vasculogenic mimicry formation through activating the STAT3/Slug axis and epithelial-to-mesenchymal transition in gastric cancer. Cell Oncol (Dordr). 2020;43:863-76
100. Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 Functions as an Oncogene by Sponging MiR-135a and Promotes JAK2/STAT3 Signal Pathway in Gastric Cancer. J Cancer. 2019;10:1013-22
101. Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12:71
102. Rong L, Li Z, Leng X, Li H, Ma Y, Chen Y. et al. Salidroside induces apoptosis and protective autophagy in human gastric cancer AGS cells through the PI3K/Akt/mTOR pathway. Biomed Pharmacother. 2020;122:109726
103. Conciatori F, Ciuffreda L, Bazzichetto C, Falcone I, Pilotto S, Bria E. et al. mTOR Cross-Talk in Cancer and Potential for Combination Therapy. Cancers (Basel). 2018 10
104. Holroyd AK, Michie AM. The role of mTOR-mediated signaling in the regulation of cellular migration. Immunol Lett. 2018;196:74-9
105. Wang LL, Zhang L, Cui XF. Downregulation of long noncoding RNA LINC01419 inhibits cell migration, invasion, and tumor growth and promotes autophagy via inactivation of the PI3K/Akt1/mTOR pathway in gastric cancer. Ther Adv Med Oncol. 2019;11:1758835919874651
106. Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY. et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6
107. Tao W, Li Y, Zhu M, Li C, Li P. LncRNA NORAD Promotes Proliferation And Inhibits Apoptosis Of Gastric Cancer By Regulating miR-214/Akt/mTOR Axis. Onco Targets Ther. 2019;12:8841-51
108. Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600-4
109. Qu CX, Shi XC, Bi H, Zhai LQ, Yang Q. LncRNA AOC4P affects biological behavior of gastric cancer cells through MAPK signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:8852-60
110. Zhang Y, Ma M, Liu W, Ding W, Yu H. Enhanced expression of long noncoding RNA CARLo-5 is associated with the development of gastric cancer. Int J Clin Exp Pathol. 2014;7:8471-9
111. Zhang H, Zhang Z, Wang D. Epigenetic regulation of IncRNA KCNKI5-ASI in gastric cancer. Cancer Manag Res. 2019;11:8589-602
112. Wang F, Zhu W, Yang R, Xie W, Wang D. LncRNA ZEB2-AS1 contributes to the tumorigenesis of gastric cancer via activating the Wnt/beta-catenin pathway. Mol Cell Biochem. 2019;456:73-83
113. Xiao J, Lai H, Wei SH, Ye ZS, Gong FS, Chen LC. lncRNA HOTAIR promotes gastric cancer proliferation and metastasis via targeting miR-126 to active CXCR4 and RhoA signaling pathway. Cancer Med. 2019;8:6768-79
114. Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF, Yuan CT. et al. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-kappaB1. Biochem Biophys Res Commun. 2015;465:225-31
115. Deng W, Zhang Y, Cai J, Zhang J, Liu X, Yin J. et al. LncRNA-ANRIL promotes gastric cancer progression by enhancing NF-kB signaling. Exp Biol Med (Maywood). 2019;244:953-9
116. Tian S-B, Yu J-C, Kang W-M, Ma Z-Q, Ye X, Cao Z-J. et al. Combined detection of CEA, CA 19-9, CA 242 and CA 50 in the diagnosis and prognosis of resectable gastric cancer. Asian Pac J Cancer Prev. 2014;15:6295-300
117. Bae S, Brumbaugh J, Bonavida B. Exosomes derived from cancerous and non-cancerous cells regulate the anti-tumor response in the tumor microenvironment. Genes Cancer. 2018;9:87-100
118. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83
119. Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W. et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143:991-1004
120. Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X. et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68
121. Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH. et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84
122. Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516
123. Zheng R, Liang J, Lu J, Li S, Zhang G, Wang X. et al. Genome-wide long non-coding RNAs identified a panel of novel plasma biomarkers for gastric cancer diagnosis. Gastric Cancer. 2019;22:731-41
124. Li J, Xu Q, Wang W, Sun S. MIR100HG: a credible prognostic biomarker and an oncogenic lncRNA in gastric cancer. Biosci Rep. 2019 39
125. Zhang Y, Yang R, Lian J, Xu H. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016;8:5035-43
126. Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J. et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094-109
127. Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer biology & medicine. 2018;15:238-50
128. Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X. et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 2020;19:118
129. Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W. et al. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37:4723-34
130. Jiang B, Yang B, Wang Q, Zheng X, Guo Y, Lu W. lncRNA PVT1 promotes hepatitis B virus-positive liver cancer progression by disturbing histone methylation on the c-Myc promoter. Oncol Rep. 2020;43:718-26
131. Lee JH, Jung HS, Giang PM, Jin X, Lee S, Son PT. et al. Blockade of nuclear factor-kappaB signaling pathway and anti-inflammatory activity of cardamomin, a chalcone analog from Alpinia conchigera. The Journal of pharmacology and experimental therapeutics. 2006;316:271-8
132. Wang Z, Tang X, Wu X, Yang M, Wang W, Wang L. et al. Cardamonin exerts anti-gastric cancer activity via inhibiting LncRNA-PVT1-STAT3 axis. Biosci Rep. 2019 39
133. Lei K, Liang X, Gao Y, Xu B, Xu Y, Li Y. et al. Lnc-ATB contributes to gastric cancer growth through a MiR-141-3p/TGFbeta2 feedback loop. Biochem Biophys Res Commun. 2017;484:514-21
134. Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q. et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18:115
135. Zhang JX, Chen ZH, Chen DL, Tian XP, Wang CY, Zhou ZW. et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660-75
136. Tan H, Zhang S, Zhang J, Zhu L, Chen Y, Yang H. et al. Long non-coding RNAs in gastric cancer: New emerging biological functions and therapeutic implications. Theranostics. 2020;10:8880-902
137. Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J. et al. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta. 2020;501:252-7
138. Feng W, Zong W, Li Y, Shen X, Cui X, Ju S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. J Cell Biochem. 2020;121:557-65
139. Shan L, Liu C, Ma C. High Expression of Serum UCA1 may be a Potential Biomarker for Clinical Diagnosis of Gastric Cancer. Clin Lab. 2019 65
140. Yang Z, Sun Y, Liu R, Shi Y, Ding S. Plasma long noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 as potential novel diagnostic biomarkers for gastric cancer. Cancer Manag Res. 2019;11:6175-84
141. Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y. et al. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics. 2017;7:213-27
142. Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999-1012
143. Lin Y, Hu D, Zhou Q, Lin X, Lin J, Peng F. Correction: The fasting blood glucose and long non-coding RNA SNHG8 predict poor prognosis in patients with gastric carcinoma after radical gastrectomy. Aging (Albany NY). 2018;10:4294
144. Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q. et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209-18
145. Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571-9
146. Wang Z, Qin B. Prognostic and clinicopathological significance of long noncoding RNA CTD-2510F5.4 in gastric cancer. Gastric Cancer. 2019;22:692-704
Author contact
![]() Corresponding authors: Xu Wu, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-13882770623. E-mail: wuxulzcom. Mingxing Li, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-13882773663. E-mail: star.leecom.
Corresponding authors: Xu Wu, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-13882770623. E-mail: wuxulzcom. Mingxing Li, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-13882773663. E-mail: star.leecom.

 Global reach, higher impact
Global reach, higher impact