Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(13):3268-3280. doi:10.7150/ijbs.63488 This issue Cite
Review
Emerging Roles of LncRNAs in the EZH2-regulated Oncogenic Network
1. Key Laboratory of Saline-alkali Vegetation Ecology Restoration, Ministry of Education, College of Life Science, Northeast Forestry University, Harbin 150040, China
2. Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin, 150081, China
3. College of Arts and Sciences, Winthrop University, Rock Hill, SC 29733, the United States
Received 2021-6-23; Accepted 2021-7-16; Published 2021-7-25
Abstract
Cancer is a life-threatening disease, but cancer therapies based on epigenetic mechanisms have made great progress. Enhancer of zeste homolog 2 (EZH2) is the key catalytic component of Polycomb repressive complex 2 (PRC2) that mediates the tri-methylation of lysine 27 on histone 3 (H3K27me3), a well-recognized marker of transcriptional repression. Mounting evidence indicates that EZH2 is elevated in various cancers and associates with poor prognosis. In addition, many studies revealed that EZH2 is also involved in transcriptional repression dependent or independent of PRC2. Meanwhile, long non-coding RNAs (lncRNAs) have been reported to regulate numerous and diverse signaling pathways in oncogenesis. In this review, we firstly discuss functional interactions between EZH2 and lncRNAs that determine PRC2-dependent and -independent roles of EZH2. Secondly, we summarize the lncRNAs regulating EZH2 expression at transcription, post-transcription and post-translation levels. Thirdly, we review several oncogenic pathways cooperatively regulated by lncRNAs and EZH2, including the Wnt/β-catenin and p53 pathways. In conclusion, lncRNAs play a key role in the EZH2-regulated oncogenic network with many fertile directions to be explored.
Keywords: cancer, lncRNA, EZH2, PRC2, H3K27me3, non-histone methylation, epigenetic regulation
Introduction
Cancers are characterized by dysregulated genetic and epigenetic homeostasis under environmental and endogenous stimuli, leading to uncontrolled cell division, growth and life-threatening metastasis [1]. Malignant transformations are generally caused by accumulated genetic mutations of oncogenes and tumor suppressors involved in various important signaling pathways, but mechanisms underlying cancer development and progression can be extended to the level of epigenetic regulation [2, 3]. Epigenetics is defined as regulatory mechanisms leading to stably inheritable changes resulting from alterations of gene expression without alterations in DNA sequences [4, 5]. In the past decades, cancer epigenetics has gained considerable attention due to our increasing understanding of its regulation in cell fate decisions, reversible and environment-dependent features, and vulnerability as therapeutic targets [6-8]. DNA methylation and histone modification are two important epigenetic mechanisms regulating gene expression. Meanwhile, noncoding RNAs (ncRNAs), a large population of molecules with currently undetermined numbers, are closely involved in different epigenetic processes, either directly or indirectly, to modulate gene expression, RNA splicing and stability, chromatin remodeling, and genomic stability [9, 10].
With the development of high-throughput technologies and computational algorithms, we have learned that only about 2% of the human genome codes proteins, and most transcripts from over 70% of the genome are ncRNAs [11-13]. In the past decade, ncRNAs have been intensively studied in the context of their regulatory roles in gene expression [9, 10]. Long noncoding RNAs (lncRNAs) are defined as a class of ncRNAs longer than 200 nucleotides without evident protein coding ability. LncRNAs could modulate gene expression by serving as molecular signals, decoys, scaffolds and guides through binding both nucleic acids and proteins [14, 15]. Based on these features, emerging evidence demonstrates that lncRNAs are involved in almost all progressive stages of cancers, including cell migration, invasion, metastasis and angiogenesis, as well as autophagy [16-18]. Since the expression of lncRNAs exhibit more tissue- and cell-specific patterns than protein-coding genes, they have been considered as potential biomarkers and therapeutic targets in cancers [19]. For example, a well-characterized lncRNA, HOTAIR (HOX Transcript Antisense RNA), has been recognized as a prognostic marker of several cancer types [20-22]. Mechanistically, HOTAIR acts as a molecular scaffold for PRC2 (Polycomb repressive complex 2) and other histone modification complexes, such as LSD1, to establish gene silencing [23, 24].
Enhancer of zeste homolog 2 (EZH2), a well-known SET domain-containing protein, is the catalytic core of the PRC2 with methyltransferase activity [25]. PRC2 is essential for transcription silencing through specifically catalyzing the tri-methylation of lysine 27 on histone H3 (H3K27me3) [26]. EZH2 expression is commonly elevated in nearly all solid cancers, including gastric, breast and prostate cancers, which is associated with poor prognosis, suggesting an oncogenic role of EZH2 in these cancers [25]. Noteworthily, in lymphoma and myeloid neoplasms, several EZH2 missense mutations and truncations were reported to associate with oncogenic activities [27-29]. In most studies, EZH2 inhibition could reduce tumor formation and promote cell death. Recent reports also demonstrated EZH2 as a promising target to suppress tumor immune escape and block viral infection [25, 30-32]. Thus, EZH2 has been considered a bona fide therapeutic target in cancer therapies, and inhibitors targeting EZH2 have been increasingly developed, such as tazemetostat (EPZ-6438), valemetostat, CPI-0209 and CPI-1205 [33-36]. Tazemetostat was the first drug approved by the FDA in 2020 for the treatment of patients with locally advanced or metastatic epithelioid sarcoma not eligible for complete resection [37], and phase Ⅰ/Ⅱ clinical trials are still ongoing in the treatments of other cancers, such as diffuse large B-cell lymphoma and malignant mesothelioma [33].
In addition, an increasing number of studies indicate that lncRNAs regulate the expression and function of EZH2 at multiple levels. On the other hand, many lncRNAs involved in different oncogenic signaling pathways are also transcriptional targets of EZH2. In the current review, we will discuss functional interplays between EZH2 and different lncRNAs, and the biological consequences of their interactions, mostly in the contexts relevant to oncogenesis.
LncRNAs regulate a dual role of EZH2 in PRC2-dependent transcription
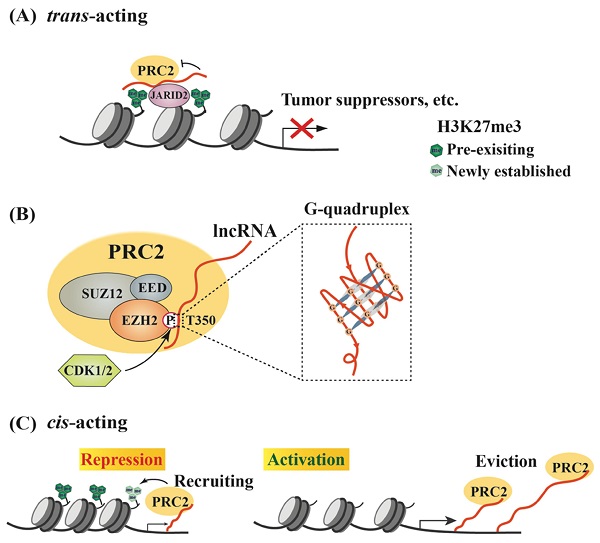
SUZ12, EZH2 and EED comprise the core components of PRC2 that serves as an epigenetic “writer” to switch off genes [26]. Although H3K27 methylation is an important event to determine gene expression status and cell fate, it remains poorly understood how it is catalyzed by PRC2 in a site-specific manner. Early studies revealed that many molecules could act as “readers” or recruiters of PRC2 to promote de novo H3K27me3 [38-42], such as RBBP4/7, JARID2 and YY1 [43-45]. However, a growing number of lncRNAs have been demonstrated to serve as “genomic address codes” for PRC2 [46], including HOTAIR, PVT1 and many other lncRNAs (Table 1). A potential mechanism is that lncRNAs preferentially bind a GA-rich DNA motif in promoters to form a triplex, and then recruit protein complexes, such as PRC2 [47-51]. As an example of these studies, the HITT is an EZH2-binding lncRNA, and its additional region forms an RNA-DNA triplex with the HIF-1α promoter, leading to PRC2 recruitment and reduced HIF-1α expression under normoxic conditions. This suppression is released and HIF-1α expression is activated under hypoxic conditions due to reduced HITT levels [51]. Actually, computational tools, such as Triplexator and Triplex Domain Finder (TDF), were developed to evaluate triplex-forming lncRNAs and their potential target sites in the human genome [49, 52]. In addition, another software, Triplex-Inspector, can also aid the design of sequence-specific ligands and selection of optimal targets to avoid off-target effects in genomic manipulations [53]. The PRC2- or EZH2-associated lncRNAs exert critical regulatory activities in the expression of various genes involved in different oncogenic signaling pathways. Interestingly, many of these lncRNAs inhibit the methyltransferase activity of EZH2 unless PRC2 associates with JARID2, which reduces PRC2-RNA interaction and subsequently promotes EZH2 activity [54-56] (Figure 1A).
To date, a large number of lncRNAs have been identified to interact with PRC2 [95, 96]. Both EZH2 and EED contribute to PRC2 binding to lncRNAs, and EZH2 has much higher affinity to lncRNAs than EED, although no canonical RNA recognition motif (RRM) has been identified in either protein [55, 97]. Thus, the heterodimer EZH2-EED is necessary and sufficient for HOTAIR binding [97]. Phosphorylation at T350 of EZH2 (pT350) mediated by CDK1 and CDK2 is crucial for PRC2 binding to its lncRNA recruiters [98-100] (Figure 1B). Consistently, the phosphorylation of mouse Ezh2 at T345 (corresponding to T350 of human EZH2) was also important for its binding to lncRNAs HOTAIR and Xist, and the T345D mutant also showed increased affinity to the lncRNAs compared to wild type Ezh2 and T345A mutant [101]. In addition, Ezh2-T345 is located in a ncRNA-binding domain (ncRBD1, amino acids 342-370), overlapping with the BRCA1 binding region (amino acids 341-559) on Ezh2 [102]. Therefore, BRCA1 could act as a negative regulator of PRC2 through competing for EZH2 association with lncRNAs [103].
Research efforts have been made to investigate whether any special RNA sequence or motif is favored by PRC2. In HOTAIR, a highly structured domain consisting of 89 nucleotides was identified as an EZH2 binding region [97]. This G-rich region forms alternative G-quadruplex structures depending the presence of potassium ions, which is different from a reported PRC2-binding tandem dual-hairpin motif in Xist RepA [104]. Consistently, several studies demonstrated that PRC2 showed high affinity to G-tract-containing RNA molecules and preferentially bound to G-quadruplex structures, but displayed much lower binding affinity to RNA duplexes [105] (Figure 1B). Importantly, PRC2's preferential binding to RNA G-quadruplex is evolutionarily conserved [106]. Another oncogenic lncRNA, HERES, promotes Wnt signaling pathways through recruiting PRC2 to chromatin in esophageal squamous cell carcinoma and this recruitment is through direct binding of EZH2 to HERES [70]. Interestingly, the GGW (W: A or U) repeats in HERES, which form the G-quadruplex-like motifs, are essential for its interaction with EZH2.
LncRNAs recruiting EZH2 to inhibit gene transcription in cancers.
| LncRNA | Expression | Cancer type | Interacting molecules | Cell process / Clinical feature |
|---|---|---|---|---|
| AGAP2-AS1 | Up | GBM[57] | EZH2, LSD1 | Proliferation, apoptosis, invasion / Shorter OS |
| ANCR | Up | BC*[58], glioma[59] | EZH2 | Proliferation, migration & invasion[58, 59], apoptosis, EMT & stemness[59] / Lymph node metastasis, advanced TNM stages[58] |
| AWPPH | Up | BC#[60], NPC[61] | EZH2[60, 61], LSD1[61] | Proliferation, apoptosis, migration[60, 61], autophagy[60] / - |
| BLACAT1 | Up | PC[62] | EZH2 | Proliferation, migration, AG and MOP / - |
| CASC9 | Up | BC#[63], ESCC[64] | EZH2 | Proliferation[63, 64], migration & invasion[63], apoptosis[64] / Larger tumor size, shorter OS[64] |
| FOXC2-AS1 | Up | Melanoma[65] | EZH2 | Proliferation, apoptosis / Metastasis, shorter OS |
| FOXD2-AS1 | Up | HCC[66] | EZH2 | Proliferation / Larger tumor size, shorter OS and DFS |
| FOXP4-AS1 | Up | NSCLC[67], osteosarcoma[68] | EZH2, LSD1[67, 68] | Proliferation, apoptosis, migration & invasion[67, 68] / Larger tumor size, advanced TNM stage, shorter OS and PFS[67] |
| H19 | Up | BC#[69] | EZH2 | Metastasis / Invasion |
| HERES | Up | ESCC[70] | EZH2 | Cell cycle, apoptosis / Worse survival |
| HOTAIR | Up | BC*[20, 71-73], OSCC[74], PC[75] | EZH2 | Proliferation, apoptosis, migration & invasion[74, 75], EMT & stemness, DNA damage repair, radio-resistance[20, 71-73] / Metastasis and survival[20, 71-73], advanced clinical stage[75], lymph node metastasis and shorter survival[74] |
| HOXD-AS1 | Up | GC[76], osteosarcoma[77] | EZH2[76, 77] | Proliferation[77], cisplatin resistance[76] / Lymph node metastasis, advanced TNM stage[76], shorter OS[76, 77] |
| LINC-PINT | Down | Melanoma[78] | EZH2 | Proliferation, migration / Shorter OS |
| MAGI2-AS3 | Down | Esophageal cancer [79] | EZH2 | Proliferation, apoptosis, radio-resistance / - |
| MALAT1 | Up | RCC[80] | EZH2 | Proliferation, apoptosis, invasion / Advanced clinical stage and shorter OS |
| MEG3 | Down | BC*[47, 81, 82] | EZH2, JARID2 | Proliferation, invasion, angiogenesis / - |
| PVT1 | Up | GC[83], NSCLC[84], thyroid cancer[85] | EZH2 | Proliferation [83-85], apoptosis [84] / Larger tumor size[84], deeper invasion depth[83], lymph node metastasis and advanced TNM stag, shorter survival[83, 84] |
| SNHG6 | Up | Chondrosarcoma[86], GC[87] | EZH2 | Proliferation[86, 87], migration & metastasis / Deeper invasion depth and advanced TNM stage[87], clinical classification[86] |
| SNHG7 | Up | OC[88] | EZH2 | Growth, migration & invasion, metastasis, EMT / - |
| SPRY4-IT1 | Up | CCA[89] | EZH2, LSD1, DNMT1 | Growth, metastasis / Tumor node metastasis, worse OS and PFS |
| TUG1 | Up | SCLC[90] | EZH2 | Proliferation, migration & invasion, apoptosis, chemo-sensitivity / Worse OS |
| Down | NSCLC[91, 92] | EZH2[91, 92], EED[92] | Proliferation[91, 92] / Larger tumor size and advanced pathological stage[91] | |
| Xist | Up | NSCLC[93], NB[94] | EZH2[93, 94] | Growth[93], migration & invasion[94], metastasis[93] / larger tumor size and advanced TNM stage, shorter OS[93] |
BC*: breast cancer; BC#: bladder cancer; CCA: cholangiocarcinoma; ESCC: esophageal squamous cell carcinoma; GBM: glioblastoma; GC: gastric cancer; HCC: hepatocellular carcinoma; NB: neuroblastoma; NPC: nasopharyngeal carcinoma; NSCLC: non-small cell lung cancer; OC: ovarian cancer; OSCC: oral squamous cell carcinoma; PC: pancreatic cancer; RCC: renal cancer Carcinoma; SCLC: small cell lung cancer; AG: aerobic glycolysis; MOP: mitochondrial oxidative phosphorylation; EMT: epithelial-mesenchymal transition; TNM: tumor node metastasis; OS: overall survival; DFS: disease-free survival; PFS: progression-free survival.
Models of lncRNA-mediated EZH2 action to regulate chromatin remodeling. (A) Schematic model of trans lncRNAs recruiting EZH2 to catalyze H3K27me3 and repress gene transcription. LncRNA (red curve, the same below) recruits PRC2 to the promoter of target genes, such as tumor suppressor genes, but inhibits the methyltransferase activity of EZH2, which can be relieved by JARID2 binding. (B) Characteristics of interaction between EZH2 and lncRNAs. EZH2, EED and SUZ12 are the core subunits of PRC2. Phosphorylation of EZH2 at T350 mediated by CDK1/2 is essential for its association with lncRNAs, and preferentially binds to G-quadruplex RNA. (C) Regulation of PRC2 activity by cis-acting RNAs. PRC2 scans nascent RNAs and then binds to G-tract regions. With slow transcription, EZH2 binding to nascent RNAs can promote H3K27me3 on chromatin to repress gene expression; with fast transcription, pre-existing PRC2 can bind nascent RNAs and then be evicted from the promoter, leading to gene activation.
RNA G-quadruplexes are generally present in the 5'-region of the first intron [107], which may explain why nascent RNAs may act as cis-regulatory elements in gene activation through binding to EZH2 and antagonizing the repressive activity of PRC2 [54]. Consistently, a recent report by Beltran et al. verified preferential binding of PRC2 to G-tracts in nascent pre-mRNA and further revealed that RNA G-quadruplex could attract PRC2 to evict it from chromatin, leading to specific gene activation [108]. In addition to nascent RNAs, other lncRNAs, such as LINC-PINT, HOTAIRM1 and PPP1R1B, could also exert cis- or trans-regulatory roles to activate gene expression through sequestering PRC2 [109-111].
Thus, RNAs possess a dual role in regulating PRC2-mediated gene expression. In the cis-acting regulation, nascent RNAs can be scanned by PRC2, which allows it to bind RNA G-tracts or G-quadruplex motifs. The recruited PRC2 can enhance H3K27me3 in the promoter to strengthen transcriptional repression. However, if nascent RNAs are quickly transcribed, the pre-existing PRC2 on chromatin can bind G-rich RNAs and be promptly removed from the promoter, leading to reduced H3K27me3 on a targeted promoter and consequent gene activation. Therefore, the decision between recruitment and eviction of PRC2 on a target gene depends on the status of RNAs, including both nascent RNAs and lncRNAs [26, 47, 105] (Figure 1C). In a study of oncogene-induced senescence, Muniz et al. discovered that an isoform of the circular ANRIL could bind to Polycomb proteins and reduce EZH2 occupation on the p15 and p16 promoters, leading to declined H3K27me3. As a result, the expression of ANRIL or its relative levels versus EZH2 determined the expression of these genes, as well as cell senescence [112]. Noteworthily, many lncRNAs are downstream effectors of EZH2, and over 20% of lncRNAs are regulated by PRC2 (Figure 2A) [113]. Several lncRNAs acting as guide partners of PRC2, such as MEG3 and SPRY4-IT1, were also found to be suppressed by EZH2 [114, 115].
To date, mechanisms of RNA-mediated trans-regulation for EZH2 eviction are still poorly understood, owing to the highly complex and dynamic features of epigenetic regulation. With the applications of RNA-immunoprecipitation followed by high-throughput sequencing (RIP-seq) technology, more PRC2/EZH2-associated lncRNAs have been identified and annotated, and their functional roles will be explored and expectantly revealed in further research [113, 116]. In a report by Ye et al., the authors demonstrated that 2,595 previously annotated lncRNAs were potentially associated with EZH2 in neuroblastoma cells [113] In another study, Wang et al. carried out a global screening for EZH2-binding lncRNAs in different tissues, and identified 1,328 EZH2-associated lncRNAs. Among them, 470 lncRNAs were detected in at least two tissues, while 858 were only present in single tissues. Although a potential EZH2-binding motif was discovered in many lncRNAs, but it was not found in all immunoprecipitated RNAs [116]. Thus, EZH2 is a protein with promiscuous binding affinity to different lncRNAs, and its detailed RNA binding domain(s) needs to be prudently mapped in future studies.
LncRNAs involved in PRC2-independent methylation of non-histone proteins
As a methyltransferase, EZH2 has also been reported to directly promote the methylation of many non-histone proteins. For instance, EZH2-mediated K299 mono-methylation of GATA4 reduced its transcription factor activity through disrupting its binding to p300 and consequently decreasing its acetylation [117]. In another report, Lee et al. discovered methylation-dependent ubiquitination machinery. Through this mechanism, non-histone proteins, such as RORα, were mono-methylated by EZH2, and the methylation status could be recognized by a specific E3 ligase to promote their ubiquitination [118], which is independent of EZH2's activity in mediating H3K27me3.
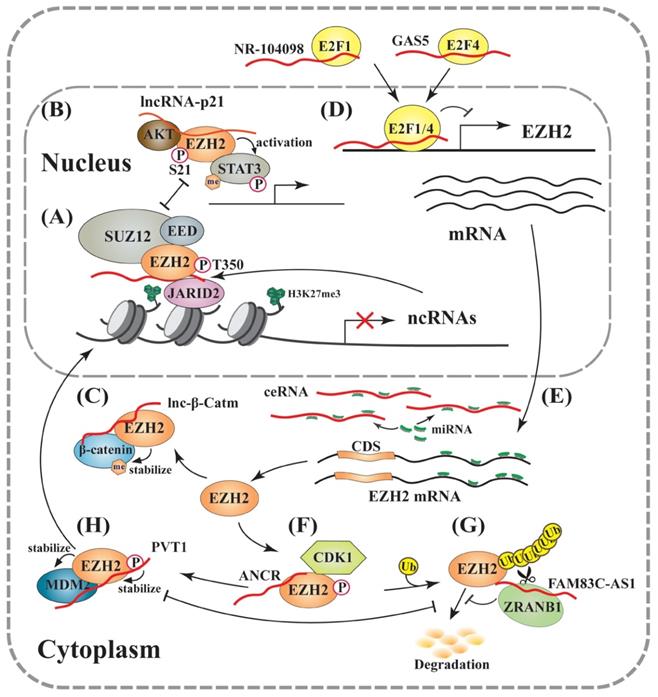
On the other hand, EZH2 could also manifest its oncogenic activity through acting as a coactivator to promote the methylation of transcription factors, such as androgen receptor in prostate cancer and STAT3 in glioblastoma [119, 120]. AKT can mediate EZH2 phosphorylation at S21 (pS21). A number of reports demonstrated that pS21 of EZH2 reduced its activity in promoting H3K27me3 [120-122], or negatively correlated with this gene silencing marker [119, 120]. Consistently, inhibition of the PI3K-AKT pathway could decrease EZH2-pS21 but increase the global levels of H3K27me3 [120], suggesting that pS21 acts as a molecular switch of EZH2 between a repressor and an activator. A recent study revealed that lincRNA-p21 could enhance EZH2-pS21 through inhibiting the EZH2-HOTAIR interaction, leading to PRC2 disruption in prostate cancer cells [123], indicating that lincRNA-p21 determines the outcomes of EZH2-mediated gene expression (Figure 2B). It supports a trans-regulatory mechanism of lncRNAs through competitively binding EZH2 to activate gene expression. In addition, β-catenin, the key effector of the Wnt signaling pathway and a signal transducer to the nucleus, was reported to be methylated at K49 by EZH2, which increased its stability through blocking ubiquitination. In this process, a lncRNA, lnc-β-Catm, could bind to both β-catenin and EZH2 to promote β-catenin methylation and improve its stability [124] (Figure 2C). Importantly, this regulatory activity of EZH2 is independent of PRC2, since depletion of SUZ12 and EED exerted no effect on it.
LncRNAs involved in regulating EZH2 expression and activity
Accumulating evidence indicates that lncRNAs regulate the expression and activity of EZH2 at different levels, including transcription, post-transcription and post-translational modifications.
LncRNA-mediated transcriptional regulation of EZH2
Early studies demonstrated that EZH2 expression was regulated by various transcription factors, including MYC, SOX4 and E2Fs [125-127]. The E2F family is a group of DNA-binding proteins that play a critical role in promoting cell cycle progression [128]. Whether an E2F protein activates or represses gene expression depends on its associated cofactors [129-132]. Two recent studies indicated that both E2F1 and E2F7 could enhance EZH2 transcription through binding to its promoter [133, 134]. Consistently, an E2F binding site was identified in the EZH2 promoter [135-137]. However, Feng et al. reported that E2F1 repressed EZH2 expression when recruited by a lncRNA, NR-104098, to the EZH2 promoter [138] (Figure 2D). Based on this mechanism, treatment of acute myelogenous leukemia cells by ATPR, a derivative of all-trans retinoic acid (ATRA), could markedly stimulate NR-104098 expression and subsequently block EZH2 expression, leading to reduced cell proliferation and enhanced cell differentiation [138].
As a well-characterized lncRNA, GAS5 is downregulated in solid tumors, and its decreased expression correlated with poor prognosis of cancer patients [136, 139-141]. Mechanistically, GAS5 could bind E2F4, recruit it to the EZH2 promoter, and repress EZH2 expression, leading to enhanced apoptosis and reduced viability of bladder cancer cells [136] (Figure 2D). Consistently, Xu et al. also reported that GAS5-mediated EZH2 inhibition could decrease H3K27me3 and consequently upregulate CDKN1C expression, causing accelerated oxidative stress and apoptosis of melanoma cells [142]. All these findings extended our understanding of lncRNAs involved in EZH2 transcription, and suggested new therapeutic targets in cancer therapies.
LncRNAs involved in regulation of EZH2 expression and activity. (A) The PRC2- or EZH2-associated lncRNAs regulate the expression of various genes, including lncRNAs. Many lncRNAs inhibit EZH2 methyltransferase activity, which can be relieved by JARID2 binding leading to reduced PRC2-RNA interaction and subsequently enhanced EZH2 activity. (B) EZH2 directly methylates non-histone proteins independent on PRC2: LincRNA-p21 disrupts PRC2 through competitively binding EZH2, and enhances the EZH2-pS21 by AKT, resulting in the coactivation of methylated STAT3. (C) Lnc-β-Catm stabilizes β-catenin protein through directly promoting its methylation by EZH2. (D) At the transcriptional level: LncRNAs, such as NR-104098 and GAS5, suppress EZH2 transcription through recruiting transcription factors, such as E2F1 and E2F4, respectively. (E) At the post-transcriptional level: ceRNAs scavenge microRNAs, and thus prevent the EZH2 mRNA from degradation and translational inhibition. (F), (G) and (H) At the post-translational level: lncRNAs, such as ANCR, directly bind EZH2 and enhance pT350 through increasing its interaction with CDK1, leading to EZH2 ubiquitination and proteasomal degradation (F). FAM83C-AS1 recruits deubiquitinase ZRANB1 to stabilize EZH2 (G). PVT1 improves EZH2 stability through blocking EZH2-pT350 to antagonize its ubiquitination, and EZH2 can also stabilize MDM2 through direct interaction. PVT1 also recruits EZH2 to repress gene transcription (H).
LncRNAs-mediated post-transcriptional regulation of EZH2
MicroRNAs are small endogenous ncRNAs regulating gene silencing through promoting destabilization and blocking translation of target mRNAs. MicroRNA expression profiles can be used to classify cancers, and the levels of specific microRNAs correlate with diagnosis and prognosis of cancer patients [143, 144]. Many groups, including ours, reported that EZH2 expression was regulated by microRNAs, such as miR-101 and miR-26a [145, 146]. It is also interesting to know that EZH2 can generally inhibit the expression of EZH2-targeting microRNAs through promoting the H3K27me3 of their genomic loci to create a positive feedback loop that maintains high EZH2 expression and malignant status [147]. A microRNA can be specifically blocked by its antisense oligonucleotide, such as a locked nucleic acid (LNA) with its complementary sequence, or an RNA containing multiple copies of its binding site to act as a microRNA sponge, leading to reduced microRNA binding to the target mRNA and its upregulated gene expression [148, 149]. In addition, this regulatory mechanism of microRNAs' action also exists through a concept of competing endogenous RNA (ceRNA) [150, 151] (Figure 2E).
CeRNAs scavenging microRNAs that target EZH2 mRNA.
| CeRNA | Cancer type | MicroRNAs | Cellular activity | Clinical relevance |
|---|---|---|---|---|
| FOXC2-AS1 | PCa[157] | miR-1253 | Proliferation, tumor growth | Shorter survival |
| H19 | NPC[158] | miR-630 | Invasion | - |
| HOTAIR | OC[159] | miR-138-5p | Cisplatin resistance | - |
| MALAT1 | OSCC[160] GC[161] | miR-101[160], miR-124-3p[161] | Proliferation[160, 161], invasion[160], migration, and H2 resistance[161] | - |
| NEAT1 | EC[162] | miR-144-3p | Proliferation, migration & invasion | - |
| PVT1 | NSCLC[163] | mi-526b | Proliferation, migration | Shorter survival |
| SNHG6 | CRC[155], ESCC[154], OCCC[156] | miR-101-3p[154], miR-214[155], miR-26a/b[153, 155] | Apoptosis[154, 156], migration & invasion, metastasis[153, 155, 156], EMT[153] | Deeper invasion depth, lymph node metastasis and advanced TNM stage, shorter survival[155, 156] |
| SPRY4-IT1 | BC#[89, 164] | miR-101-3p[89, 164] | Metastasis[89, 164], proliferation, migration & invasion[164] | Tumor node metastasis, worse OS and PFS[89] |
| TUG1 | PC[165] | miR-382 | Proliferation, migration and EMT | Large tumor size, advanced TNM stage, shorter survival |
| Xist | CRC[166], GC[167] | miR-137[166], miR-101[167] | Proliferation[167], migration & invasion[166, 167] | Large tumor size, lymph node invasion and advanced TNM stage, distant metastasis and worse OS[167] |
| ZNFX1-AS1 | CRC[168] | miR-144-3p | Proliferation, migration & invasion, metastasis | Larger tumor size, deeper invasion depth, lymph node invasion, and advanced TNM stage, shorter survival |
EC: endometrial cancer; OCCC: ovarian clear cell carcinoma; PCa: prostate cancer.
The first reported microRNA targeting EZH2 is miR-26a, which is overexpressed during myogenesis [145, 152]. Recently, the lncRNA, SNHG6, was determined as a ceRNA to trap miR-26a and consequently promote EZH2 expression in colorectal cancer [153]. Importantly, SNHG6 has also been identified as a molecular sponge of miR-101, -214 and -4465, which all target EZH2 in different cancers [154-156]. The term “molecular sponge” was designated as a relatively large ncRNA containing multiple binding sites of one or more microRNA molecules and thus sequestering them. In addition, a number of other lncRNAs could also work as decoys to antagonize different microRNAs and thus promote EZH2 expression, leading to enhanced cancer progression and resistance to chemo- or radiotherapies (Table 2). As we indicated above, many lncRNAs work with the EZH2 protein to mediate H3K27me3 at specific genomic loci. Therefore, lncRNAs can modulate the function of EZH2 at multiple levels.
LncRNA-mediated post-translational modifications of EZH2
EZH2 activity and stability are regulated by different post-translational modifications, including phosphorylation, ubiquitination, methylation and O-GlcNAcylation [98, 121, 169-173], and many lncRNAs contribute to these processes. As discussed above, pT350 is crucial for EZH2 binding to lncRNAs and PRC2 recruitment to chromatin. Activities of the lncRNA ANCR in binding EZH2 and promoting its phosphorylation have been reported by several research groups, but the overall effects of this regulation on cancer cells varied, likely depending on cancer types and genetic backgrounds. Li et al. reported that ANCR could directly bind mouse Ezh2, promote its interaction with CDK1, and consequently increase pT345 and pT487, leading to enhanced Ezh2 ubiquitination and degradation, and reduced breast cancer progression [31] (Figure 2F). Consistently, in another report, ANCR inhibited proliferation, migration and invasion of osteosarcoma cells through binding and attenuating EZH2 [174]. However, a positive regulation of EZH2 expression by ANCR was also reported in colorectal cancer and glioma cells [59, 175], as well as osteoblast cells [176].
Several recent studies revealed the activities of additional lncRNAs in promoting EZH2 ubiquitination and degradation, including MEG3, UCA1 and the circular RNA circ-ADD3 [177-179]. Interestingly, all these lncRNAs exerted their regulation through promoting EZH2's interaction with CDK1 and enhancing its phosphorylation, which subsequently caused EZH2 ubiquitination and degradation. As for the regulatory mechanism, Li et al. proposed that lncRNA binding likely altered EZH2 conformation that improved recognition by CDK1 to facilitate its phosphorylation [25]. Another lncRNA PAR5, downregulated in anaplastic thyroid carcinoma, was also reported to negatively regulate EZH2 activity. PAR5 interacts with EZH2, decreases its protein levels, and reduces its binding to the E-cadherin promoter [180]. Consistently, similar regulation of EZH2 by PAR5 was also observed in human glioma [181].
On the other hand, lncRNA FAM83C-AS1 was shown to stabilize EZH2 protein through promoting its binding to ZRANB1, a deubiquitinase (Figure 2G). Upregulated EZH2 could mediate the H3K27me3 in the promoter of SEMA3F, which promoted colorectal cancer development [182]. In addition, lncRNAs PVT1 and HERES were both reported to promote EZH2 protein stability with undetermined molecular mechanisms [70, 183]. Based on the evidence of available studies, we predict that the trans recruitment of EZH2 by lncRNAs to chromatin can concurrently reduce its ubiquitination through blocking T350 phosphorylation, and consequently stabilizing EZH2.
Several lncRNAs have been reported to negatively regulate EZH2 stability or activity. EDAL is a lncRNA that inhibits the replication of neurotropic viruses in neuronal cells. The mechanism underlying this regulation is EDAL-blocked T309 O-GlcNAcylation of EZH2. Interestingly, although T309 is involved, EDAL did not apparently alter the phosphorylation of EZH2 [32]. As discussed above, lincRNA-p21 can dramatically enhance AKT and EZH2 interaction, which consequently leads to increased pS21 of EZH2, and promotes EZH2-mediated STAT3 methylation [123]. Additionally, pS21 of EZH2 disrupts EZH2 binding to histone H3 and subsequently reduces H3K27me3 of target genes [121].
Actually, a positive feedback regulation exists between lincRNA-p21 and p53; while p53 transactivates lincRNA-p21 gene expression, the lncRNA also antagonizes MDM2-mediated p53 ubiquitination and degradation [184-187]. In addition, EZH2 serves as a signal relay between lncRNA PVT1 and p53; PVT1 improves EZH2 protein stability, while EZH2 also physically interacts with and stabilizes MDM2 (Figure 2H), leading to enhanced p53 degradation [183, 188, 189]. MEG3, which promotes EZH2 degradation [177], was also reported to downregulate MDM2 and subsequently activate the p53 pathway [190]. On other hand, MDM2's physical interaction with EZH2 on chromatin promotes H3K27me3, which suppresses lineage-specific genes and favors efficient generation of induced pluripotent stem cells (iPSCs) [191].
Based on this complex regulatory network, EZH2 is causally relevant to p53 activation. Thus, these lncRNAs regulating EZH2 stability and activity can either directly modulate EZH2-related epigenetic modifications or indirectly alter p53 signaling pathways through a positive EZH2-MDM2 interplay.
Conclusion and prospective
LncRNAs can regulate both the expression and methyltransferase activity of EZH2. In this review, we summarized this complex regulatory network in the following three aspects. First, during transcriptional initiation, lncRNAs and nascent RNAs can either recruit PRC2 to target promoters or evict it from chromatin, which determines the repressive or active expression status of a target gene, respectively (Table 1 and Figure 1C). Second, regarding target selection, lncRNAs regulate EZH2-mediated methylation in either histone or non-histone proteins, which can both determine target proteins' activities and alter global H3K27me3 levels (Figures 2A, 2B and 2C). Third, lncRNAs modulate EZH2 expression through directly regulating its gene transcription and indirectly acting as ceRNAs to scavenge EZH2-targeting microRNAs (Figures 2D, 2E and Table 2). Fourth, at the protein level, lncRNAs are also involved in the regulation of EZH2 post-translational modifications, which impacts its methyltransferase activity and also the activity or stability of its binding partners (Figures 2B, 2C, 2F-2H). Taken together, the complex regulation of EZH2 by lncRNAs at different levels just represents a tip of the iceberg in the whole ncRNA regulatory network. Deep understanding of additional mechanisms underlying lncRNA-mediated EZH2 actions, as well as other key cancer-related genes and proteins, will heavily rely on continuous exploration by scientific researchers and the advance of research technologies. Nevertheless, the current understanding of EZH2 expression and activity mediated by lncRNAs will not only be exemplificative in the studies of other oncogenic networks regulated by lncRNAs, but also provide insights in discovering novel therapeutic targets of cancer treatments.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (2572016AA53) to AH, and the National Natural Science Foundation of China (81872293 and 81672795) to GS.
Author Contributions
A.H., Yu W. and G.S. conceived the review and wrote the manuscript. Yunxuan W. and D.B.S. critically read the manuscript and provided constructive suggestions.
Abbreviations
AGAP2-AS1: AGAP2 antisense RNA 1; AKT: serine/threonine kinase 1; ANCR: differentiation antagonizing non-protein coding RNA; BLACAT1: bladder Cancer associated transcript 1; CASC9: cancer susceptibility 9; CDK1/2: cyclin-dependent kinase ½; CDKN1C: cyclin dependent kinase inhibitor 1C; E2F: E2F transcription factor; EDAL: EZH2 degradation associated lncRNA; EED: Embryonic Ectoderm Development; FAM83C-AS1: FAM83C antisense RNA 1; FOXC2-AS1: FOXC2 antisense RNA 1; FOXD2-AS1: FOXD2 adjacent opposite strand RNA 1; FOXP4-AS1: FOXP4 antisense RNA 1RBBP4/7, RB binding protein 4/7; GAS5: growth arrest specific 5; GATA4: GATA binding protein 4; H19: H19 imprinted maternally expressed transcript; HERES: highly expressed lncRNA in esophageal squamous cell carcinoma; HOTAIRM1: HOXA transcript antisense RNA, myeloid-specific 1; HOXD-AS1: HOXD antisense growth-associated long noncoding RNA; JARID2: jumonji and AT-Rich interaction domain containing 2; lincRNA-p21: tumor protein p53 pathway corepressor 1; LINC-PINT: long intergenic non-protein coding RNA, p53 induced transcript; LSD1: lysine specific demethylase 1; MAGI2-AS3: MAGI2 antisense RNA 3; MALAT1: metastasis associated lung adenocarcinoma transcript 1; MEG3: maternally expressed 3; MYC: proto-oncogene, BHLH transcription factor; NEAT1: nuclear paraspeckle assembly transcript 1; PAR5 : Prader Willi/Angelman region RNA 5; PPP1R1B: protein phosphatase 1 regulatory inhibitor subunit 1B; RBBP4/7: RB binding protein 4/7, chromatin remodeling factor; RORα: RAR related orphan receptor A; SEMA3F: semaphorin 3F; SNHG6: small nucleolar RNA host gene 6; SNHG7: small nucleolar RNA host gene 7; SOX4: SRY-box transcription factor 4; SPRY4-IT1: SPRY4 intronic transcript 1; STAT3: signal transducer and activator of transcription 3; TUG1: taurine up-regulated 1; UCA1: urothelial cancer associated 1; XIST: X inactive specific transcript; YY1: Yin Yang 1; ZNFX1-AS1: ZNFX1 antisense RNA 1; ZRANB1: zinc finger RANBP2-Type containing 1.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Basanta D, Anderson ARA. Homeostasis Back and Forth: An Ecoevolutionary Perspective of Cancer. Cold Spring Harb Perspect Med. 2017;7:a028332
2. Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726-34
3. Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505
4. Waterland RA. Epigenetic mechanisms and gastrointestinal development. J Pediatr. 2006;149:S137-42
5. Haig D. The (dual) origin of epigenetics. Cold Spring Harb Symp Quant Biol. 2004;69:67-70
6. Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630-41
7. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12-27
8. Nebbioso A, Tambaro FP, Dell'Aversana C, Altucci L. Cancer epigenetics: Moving forward. PLoS Genet. 2018;14:e1007362
9. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18
10. Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179:1033-55
11. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
12. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38
13. Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J. et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23:5866-78
14. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-14
15. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-66
16. Bermudez M, Aguilar-Medina M, Lizarraga-Verdugo E, Avendano-Felix M, Silva-Benitez E, Lopez-Camarillo C. et al. LncRNAs as Regulators of Autophagy and Drug Resistance in Colorectal Cancer. Front Oncol. 2019;9:1008
17. Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613-20
18. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661-7
19. Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD. et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529-40
20. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-6
21. Qu X, Alsager S, Zhuo Y, Shan B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019;454:90-7
22. Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151-64
23. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-93
24. Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ. et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942-55
25. Li Z, Li M, Wang D, Hou P, Chen X, Chu S. et al. Post-translational modifications of EZH2 in cancer. Cell Biosci. 2020;10:143
26. Yang Y, Li G. Post-translational modifications of PRC2: signals directing its activity. Epigenetics Chromatin. 2020;13:47
27. Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980-5
28. Ryan RJ, Nitta M, Borger D, Zukerberg LR, Ferry JA, Harris NL. et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One. 2011;6:e28585
29. Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299-313
30. Zhao Y, Wang XX, Wu W, Long H, Huang J, Wang Z. et al. EZH2 regulates PD-L1 expression via HIF-1alpha in non-small cell lung cancer cells. Biochem Biophys Res Commun. 2019;517:201-9
31. Li Z, Hou P, Fan D, Dong M, Ma M, Li H. et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59-71
32. Sui B, Chen D, Liu W, Wu Q, Tian B, Li Y. et al. A novel antiviral lncRNA, EDAL, shields a T309 O-GlcNAcylation site to promote EZH2 lysosomal degradation. Genome Biol. 2020;21:228
33. Eich ML, Athar M, Ferguson JE 3rd, Varambally S. EZH2-Targeted Therapies in Cancer: Hype or a Reality. Cancer Res. 2020;80:5449-58
34. Yan KS, Lin CY, Liao TW, Peng CM, Lee SC, Liu YJ. et al. EZH2 in Cancer Progression and Potential Application in Cancer Therapy: A Friend or Foe? Int J Mol Sci. 2017;18:1172
35. Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13:104
36. Marchesi I, Bagella L. Targeting Enhancer of Zeste Homolog 2 as a promising strategy for cancer treatment. World J Clin Oncol. 2016;7:135-48
37. Hoy SM. Tazemetostat: First Approval. Drugs. 2020;80:513-21
38. Choi J, Bachmann AL, Tauscher K, Benda C, Fierz B, Muller J. DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat Struct Mol Biol. 2017;24:1039-47
39. Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P. et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287-91
40. Hojfeldt JW, Laugesen A, Willumsen BM, Damhofer H, Hedehus L, Tvardovskiy A. et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat Struct Mol Biol. 2018;25:225-32
41. Sharif J, Koseki H. Rewriting the past: de novo activity of PRC2 restores global H3K27 methylation patterns. Nat Struct Mol Biol. 2018;25:197-9
42. Laugesen A, Hojfeldt JW, Helin K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol Cell. 2019;74:8-18
43. Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV. et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077-85
44. Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF. et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618-24
45. Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627-38
46. Nishikawa K, Kinjo AR. Essential role of long non-coding RNAs in de novo chromatin modifications: the genomic address code hypothesis. Biophys Rev. 2017;9:73-7
47. Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B. et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743
48. Kalwa M, Hanzelmann S, Otto S, Kuo CC, Franzen J, Joussen S. et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016;44:10631-43
49. Kuo CC, Hanzelmann S, Senturk Cetin N, Frank S, Zajzon B, Derks JP. et al. Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 2019;47:e32
50. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667-78
51. Wang X, Wang Y, Li L, Xue X, Xie H, Shi H. et al. A lncRNA coordinates with Ezh2 to inhibit HIF-1alpha transcription and suppress cancer cell adaption to hypoxia. Oncogene. 2020;39:1860-74
52. Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372-81
53. Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplex-Inspector: an analysis tool for triplex-mediated targeting of genomic loci. Bioinformatics. 2013;29:1895-7
54. Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258-64
55. Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55:171-85
56. Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW. et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017;24:1028-38
57. Luo W, Li X, Song Z, Zhu X, Zhao S. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging (Albany NY). 2019;11:3811-23
58. Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. 2020;14:309-28
59. Cheng C, Dong Y, Ru X, Xia Y, Ji Y. LncRNA ANCR promotes glioma cells invasion, migration, proliferation and inhibits apoptosis via interacting with EZH2 and repressing PTEN expression. Cancer Gene Ther. 2020 (Online ahead of print)
60. Zhu F, Zhang X, Yu Q, Han G, Diao F, Wu C. et al. LncRNA AWPPH inhibits SMAD4 via EZH2 to regulate bladder cancer progression. J Cell Biochem. 2018;119:4496-505
61. Guo D, Liu F, Zhang L, Bian N, Liu L, Kong L. et al. Long non-coding RNA AWPPH enhances malignant phenotypes in nasopharyngeal carcinoma via silencing PTEN through interacting with LSD1 and EZH2. Biochem Cell Biol. 2020;99:195-202
62. Zhou X, Gao W, Hua H, Ji Z. LncRNA-BLACAT1 Facilitates Proliferation, Migration and Aerobic Glycolysis of Pancreatic Cancer Cells by Repressing CDKN1C via EZH2-Induced H3K27me3. Front Oncol. 2020;10:539805
63. Yuan B, Sun R, Du Y, Jia Z, Yao W, Yang J. STAT3-Induced Upregulation of lncRNA CASC9 Promotes the Progression of Bladder Cancer by Interacting with EZH2 and Affecting the Expression of PTEN. Onco Targets Ther. 2020;13:9147-57
64. Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X. et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150
65. Xu DF, Tao XH, Yu Y, Teng Y, Huang YM, Ma JW. et al. LncRNA FOXC2-AS1 stimulates proliferation of melanoma via silencing p15 by recruiting EZH2. Eur Rev Med Pharmacol Sci. 2020;24:8940-6
66. Xu K, Zhang Z, Qian J, Wang S, Yin S, Xie H. et al. LncRNA FOXD2-AS1 plays an oncogenic role in hepatocellular carcinoma through epigenetically silencing CDKN1B(p27) via EZH2. Exp Cell Res. 2019;380:198-204
67. Chen RY, Ju Q, Feng LM, Yuan Q, Zhang L. The carcinogenic complex lncRNA FOXP4-AS1/EZH2/LSD1 accelerates proliferation, migration and invasion of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:8371-6
68. Yang L, Ge D, Chen X, Qiu J, Yin Z, Zheng S. et al. FOXP4-AS1 participates in the development and progression of osteosarcoma by downregulating LATS1 via binding to LSD1 and EZH2. Biochem Biophys Res Commun. 2018;502:493-500
69. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213-21
70. You BH, Yoon JH, Kang H, Lee EK, Lee SK, Nam JW. HERES, a lncRNA that regulates canonical and noncanonical Wnt signaling pathways via interaction with EZH2. Proc Natl Acad Sci U S A. 2019;116:24620-9
71. Qian L, Fei Q, Zhang H, Qiu M, Zhang B, Wang Q. et al. lncRNA HOTAIR Promotes DNA Repair and Radioresistance of Breast Cancer via EZH2. DNA Cell Biol. 2020 (Online ahead of print)
72. Ren Y, Jia HH, Xu YQ, Zhou X, Zhao XH, Wang YF. et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ss1 secretion. Mol Cancer. 2018;17:5
73. Padua Alves C, Fonseca AS, Muys BR, de Barros ELBR, Burger MC, de Souza JE. et al. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31:2827-32
74. Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X. et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46:2586-94
75. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R. et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616-25
76. Ye Y, Yang S, Han Y, Sun J, Xv L, Wu L. et al. HOXD-AS1 confers cisplatin resistance in gastric cancer through epigenetically silencing PDCD4 via recruiting EZH2. Open Biol. 2019;9:190068
77. Gu W, Zhang E, Song L, Tu L, Wang Z, Tian F. et al. Long noncoding RNA HOXD-AS1 aggravates osteosarcoma carcinogenesis through epigenetically inhibiting p57 via EZH2. Biomed Pharmacother. 2018;106:890-5
78. Xu Y, Wang H, Li F, Heindl LM, He X, Yu J. et al. Long Non-coding RNA LINC-PINT Suppresses Cell Proliferation and Migration of Melanoma via Recruiting EZH2. Front Cell Dev Biol. 2019;7:350
79. Cheng W, Shi X, Lin M, Yao Q, Ma J, Li J. LncRNA MAGI2-AS3 Overexpression Sensitizes Esophageal Cancer Cells to Irradiation Through Down-Regulation of HOXB7 via EZH2. Front Cell Dev Biol. 2020;8:552822
80. Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL. et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322-31
81. Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K. et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290-300
82. Zhang CY, Yu MS, Li X, Zhang Z, Han CR, Yan B. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumour Biol. 2017;39:1010428317701311
83. Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM. et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82
84. Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R. et al. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol Cancer Ther. 2016;15:1082-94
85. Zhou Q, Chen J, Feng J, Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR). Tumour Biol. 2016;37:3105-13
86. Pu FF, Shi DY, Chen T, Liu YX, Zhong BL, Zhang ZC. et al. SP1-induced long non-coding RNA SNHG6 facilitates the carcinogenesis of chondrosarcoma through inhibiting KLF6 by recruiting EZH2. Cell Death Dis. 2021;12:59
87. Li Y, Li D, Zhao M, Huang S, Zhang Q, Lin H. et al. Long noncoding RNA SNHG6 regulates p21 expression via activation of the JNK pathway and regulation of EZH2 in gastric cancer cells. Life Sci. 2018;208:295-304
88. Bai Z, Wu Y, Bai S, Yan Y, Kang H, Ma W. et al. Long non-coding RNA SNGH7 Is activated by SP1 and exerts oncogenic properties by interacting with EZH2 in ovarian cancer. J Cell Mol Med. 2020;24:7479-89
89. Xu Y, Yao Y, Jiang X, Zhong X, Wang Z, Li C. et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37:81
90. Niu Y, Ma F, Huang W, Fang S, Li M, Wei T. et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16:5
91. Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH. et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243
92. Lin PC, Huang HD, Chang CC, Chang YS, Yen JC, Lee CC. et al. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:583
93. Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811-7
94. Zhang J, Li WY, Yang Y, Yan LZ, Zhang SY, He J. et al. LncRNA XIST facilitates cell growth, migration and invasion via modulating H3 histone methylation of DKK1 in neuroblastoma. Cell Cycle. 2019;18:1882-92
95. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667-72
96. Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K. et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939-53
97. Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52:9519-27
98. Zeng X, Chen S, Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10:579-83
99. Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X. et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108-14
100. Wang X, Sehgal L, Jain N, Khashab T, Mathur R, Samaniego F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J Transl Med. 2016;14:346
101. Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA. et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615-20
102. Wang L, Zeng X, Chen S, Ding L, Zhong J, Zhao JC. et al. BRCA1 is a negative modulator of the PRC2 complex. EMBO J. 2013;32:1584-97
103. Li T, Cai J, Ding H, Xu L, Yang Q, Wang Z. EZH2 participates in malignant biological behavior of epithelial ovarian cancer through regulating the expression of BRCA1. Cancer Biol Ther. 2014;15:271-8
104. Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750-6
105. Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD. et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell. 2017;65:1056-67 e5
106. Long Y, Bolanos B, Gong L, Liu W, Goodrich KJ, Yang X. et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife. 2017;6:e31558
107. Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321-33
108. Beltran M, Tavares M, Justin N, Khandelwal G, Ambrose J, Foster BM. et al. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat Struct Mol Biol. 2019;26:899-909
109. Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37:265
110. Kang X, Zhao Y, Van Arsdell G, Nelson SF, Touma M. Ppp1r1b-lncRNA inhibits PRC2 at myogenic regulatory genes to promote cardiac and skeletal muscle development in mouse and human. RNA. 2020;26:481-91
111. Marin-Bejar O, Mas AM, Gonzalez J, Martinez D, Athie A, Morales X. et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017;18:202
112. Muniz L, Lazorthes S, Delmas M, Ouvrard J, Aguirrebengoa M, Trouche D. et al. Circular ANRIL isoforms switch from repressors to activators of p15/CDKN2B expression during RAF1 oncogene-induced senescence. RNA Biol. 2021;18:404-20
113. Ye M, Xie L, Zhang J, Liu B, Liu X, He J. et al. Determination of long non-coding RNAs associated with EZH2 in neuroblastoma by RIP-seq, RNA-seq and ChIP-seq. Oncol Lett. 2020;20:1
114. Qin WX, Shi Y, Zhu D, Li YP, Chen YH, Cui J. et al. EZH2-mediated H3K27me3 enrichment on the lncRNA MEG3 promoter regulates the growth and metastasis of glioma cells by regulating miR-21-3p. Eur Rev Med Pharmacol Sci. 2020;24:3204-14
115. Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R. et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298
116. Wang Y, Xie Y, Li L, He Y, Zheng D, Yu P. et al. EZH2 RIP-seq Identifies Tissue-specific Long Non-coding RNAs. Curr Gene Ther. 2018;18:275-85
117. He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P. et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37-42
118. Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY. et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell. 2012;48:572-86
119. Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465-9
120. Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N. et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839-52
121. Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT. et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306-10
122. Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD. et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70:453-62
123. Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y. et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10:2571
124. Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J. et al. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23:631-9
125. Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669-83
126. Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M. et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768-83
127. Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323-35
128. Slansky JE, Farnham PJ. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1-30
129. Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245-62
130. DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739-48
131. Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804-16
132. Komori H, Goto Y, Kurayoshi K, Ozono E, Iwanaga R, Bradford AP. et al. Differential requirement for dimerization partner DP between E2F-dependent activation of tumor suppressor and growth-related genes. Sci Rep. 2018;8:8438
133. Yang R, Wang M, Zhang G, Bao Y, Wu Y, Li X. et al. E2F7-EZH2 axis regulates PTEN/AKT/mTOR signalling and glioblastoma progression. Br J Cancer. 2020;123:1445-55
134. Du L, Fakih MG, Rosen ST, Chen Y. SUMOylation of E2F1 Regulates Expression of EZH2. Cancer Res. 2020;80:4212-23
135. Rajabi H, Hiraki M, Tagde A, Alam M, Bouillez A, Christensen CL. et al. MUC1-C activates EZH2 expression and function in human cancer cells. Sci Rep. 2017;7:7481
136. Wang M, Guo C, Wang L, Luo G, Huang C, Li Y. et al. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 2018;9:238
137. Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18:1763-77
138. Feng Y, Hu S, Li L, Zhang S, Liu J, Xu X. et al. LncRNA NR-104098 Inhibits AML Proliferation and Induces Differentiation Through Repressing EZH2 Transcription by Interacting With E2F1. Front Cell Dev Biol. 2020;8:142
139. Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP. et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319
140. Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F. et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1-E12
141. Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q. et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13:1541-50
142. Xu W, Yan Z, Hu F, Wei W, Yang C, Sun Z. Long non-coding RNA GAS5 accelerates oxidative stress in melanoma cells by rescuing EZH2-mediated CDKN1C downregulation. Cancer Cell Int. 2020;20:116
143. Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297-308
144. Fabris L, Ceder Y, Chinnaiyan AM, Jenster GW, Sorensen KD, Tomlins S. et al. The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur Urol. 2016;70:312-22
145. Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J. et al. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer. 2010;9:108
146. Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B. et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695-9
147. Liu T, Cai J, Cai J, Wang Z, Cai L. EZH2-miRNA Positive Feedback Promotes Tumor Growth in Ovarian Cancer. Front Oncol. 2020;10:608393
148. Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137-41
149. Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721-6
150. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-8
151. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
152. Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836-43
153. Zhang M, Duan W, Sun W. LncRNA SNHG6 promotes the migration, invasion, and epithelial-mesenchymal transition of colorectal cancer cells by miR-26a/EZH2 axis. Onco Targets Ther. 2019;12:3349-60
154. Wang J, Yang X, Li R, Zhang R, Hu D, Zhang Y. et al. LncRNA SNHG6 Inhibits Apoptosis by Regulating EZH2 Expression via the Sponging of MiR-101-3p in Esophageal Squamous-Cell Carcinoma. Onco Targets Ther. 2020;13:11411-20
155. Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X. et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12:3
156. Wu Y, Deng Y, Guo Q, Zhu J, Cao L, Guo X. et al. Long non-coding RNA SNHG6 promotes cell proliferation and migration through sponging miR-4465 in ovarian clear cell carcinoma. J Cell Mol Med. 2019;23:5025-36
157. Chen Y, Gu M, Liu C, Wan X, Shi Q, Chen Q. et al. Long noncoding RNA FOXC2-AS1 facilitates the proliferation and progression of prostate cancer via targeting miR-1253/EZH2. Gene. 2019;686:37-42
158. Li X, Lin Y, Yang X, Wu X, He X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2016;473:913-9
159. Zhang Y, Ai H, Fan X, Chen S, Wang Y, Liu L. Knockdown of long non-coding RNA HOTAIR reverses cisplatin resistance of ovarian cancer cells through inhibiting miR-138-5p-regulated EZH2 and SIRT1. Biol Res. 2020;53:18
160. Xiao L, Wang W, Zhao J, Xu H, Li S, Yang X. lncRNA MALAT1 promotes cell proliferation and invasion by regulating the miR-101/EZH2 axis in oral squamous cell carcinoma. Oncol Lett. 2020;20:164
161. Zhu B, Cui H, Xu W. Hydrogen inhibits the proliferation and migration of gastric cancer cells by modulating lncRNA MALAT1/miR-124-3p/EZH2 axis. Cancer Cell Int. 2021;21:70
162. Wang W, Ge L, Xu XJ, Yang T, Yuan Y, Ma XL. et al. LncRNA NEAT1 promotes endometrial cancer cell proliferation, migration and invasion by regulating the miR-144-3p/EZH2 axis. Radiol Oncol. 2019;53:434-42
163. Qiu C, Li S, Sun D, Yang S. lncRNA PVT1 accelerates progression of non-small cell lung cancer via targeting miRNA-526b/EZH2 regulatory loop. Oncol Lett. 2020;19:1267-72
164. Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X. et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281-91
165. Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The Lncrna-TUG1/EZH2 Axis Promotes Pancreatic Cancer Cell Proliferation, Migration and EMT Phenotype Formation Through Sponging Mir-382. Cell Physiol Biochem. 2017;42:2145-58
166. Liu X, Cui L, Hua D. Long Noncoding RNA XIST Regulates miR-137-EZH2 Axis to Promote Tumor Metastasis in Colorectal Cancer. Oncol Res. 2018;27:99-106
167. Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS. et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142
168. Shi L, Hong X, Ba L, He X, Xiong Y, Ding Q. et al. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10:150
169. Consalvi S, Brancaccio A, Dall'Agnese A, Puri PL, Palacios D. Praja1 E3 ubiquitin ligase promotes skeletal myogenesis through degradation of EZH2 upon p38alpha activation. Nat Commun. 2017;8:13956
170. Li Z, Wang D, Lu J, Huang B, Wang Y, Dong M. et al. Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ. 2020;27:3226-42
171. Li Z, Wang D, Wang W, Chen X, Tang A, Hou P. et al. Macrophages-stimulated PRMT1-mediated EZH2 methylation promotes breast cancer metastasis. Biochem Biophys Res Commun. 2020;533:679-84
172. Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC. et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A. 2014;111:1355-60
173. Lo PW, Shie JJ, Chen CH, Wu CY, Hsu TL, Wong CH. O-GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2. Proc Natl Acad Sci U S A. 2018;115:7302-7
174. Zhang F, Peng H. LncRNA-ANCR regulates the cell growth of osteosarcoma by interacting with EZH2 and affecting the expression of p21 and p27. J Orthop Surg Res. 2017;12:103
175. Yang ZY, Yang F, Zhang YL, Liu B, Wang M, Hong X. et al. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017;18:95-104
176. Cai N, Li C, Wang F. Silencing of LncRNA-ANCR Promotes the Osteogenesis of Osteoblast Cells in Postmenopausal Osteoporosis via Targeting EZH2 and RUNX2. Yonsei Med J. 2019;60:751-9
177. Jin L, Cai Q, Wang S, Wang S, Mondal T, Wang J. et al. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018;9:1017
178. Dong Z, Gao M, Li C, Xu M, Liu S. LncRNA UCA1 Antagonizes Arsenic-Induced Cell Cycle Arrest through Destabilizing EZH2 and Facilitating NFATc2 Expression. Adv Sci (Weinh). 2020;7:1903630
179. Sun S, Wang W, Luo X, Li Y, Liu B, Li X. et al. Circular RNA circ-ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1-mediated ubiquitination. Am J Cancer Res. 2019;9:1695-707
180. Pellecchia S, Sepe R, Decaussin-Petrucci M, Ivan C, Shimizu M, Coppola C. et al. The Long Non-Coding RNA Prader Willi/Angelman Region RNA5 (PAR5) Is Downregulated in Anaplastic Thyroid Carcinomas Where It Acts as a Tumor Suppressor by Reducing EZH2 Activity. Cancers (Basel). 2020;12:235
181. Wang XP, Shan C, Deng XL, Li LY, Ma W. Long non-coding RNA PAR5 inhibits the proliferation and progression of glioma through interaction with EZH2. Oncol Rep. 2017;38:3177-86
182. Xue W, Wang F, Han P, Liu Y, Zhang B, Gu X. et al. The oncogenic role of LncRNA FAM83C-AS1 in colorectal cancer development by epigenetically inhibits SEMA3F via stabilizing EZH2. Aging (Albany NY). 2020;12:20396-412
183. Guo J, Hao C, Wang C, Li L. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int. 2018;18:98
184. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409-19
185. Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C. et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452-65
186. Wang X, Ruan Y, Wang X, Zhao W, Jiang Q, Jiang C. et al. Long intragenic non-coding RNA lincRNA-p21 suppresses development of human prostate cancer. Cell Prolif. 2017;50:e12318
187. Zhou L, Tian Y, Guo F, Yu B, Li J, Xu H. et al. LincRNA-p21 knockdown reversed tumor-associated macrophages function by promoting MDM2 to antagonize* p53 activation and alleviate breast cancer development. Cancer Immunol Immunother. 2020;69:835-46
188. Yang A, Wang H, Yang X. Long non-coding RNA PVT1 indicates a poor prognosis of glioma and promotes cell proliferation and invasion via target EZH2. Biosci Rep. 2017;37:BSR20170871
189. Huang XM, Shi SS, Jian TM, Tang DR, Wu T, Sun FY. LncRNA PVT1 knockdown affects proliferation and apoptosis of uveal melanoma cells by inhibiting EZH2. Eur Rev Med Pharmacol Sci. 2019;23:2880-7
190. Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R. et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731-42
191. Wienken M, Dickmanns A, Nemajerova A, Kramer D, Najafova Z, Weiss M. et al. MDM2 Associates with Polycomb Repressor Complex 2 and Enhances Stemness-Promoting Chromatin Modifications Independent of p53. Mol Cell. 2016;61:68-83
Author contact
![]() Corresponding authors: wangyuedu.cn; gcsuiedu.cn
Corresponding authors: wangyuedu.cn; gcsuiedu.cn

 Global reach, higher impact
Global reach, higher impact