Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(13):3573-3582. doi:10.7150/ijbs.60551 This issue Cite
Review
A combination therapy of Phages and Antibiotics: Two is better than one
1. Institute of Biomedical Informatics, school of Basic Medical Sciences, Henan University, Kaifeng 475004, China.
2. Henan International Joint Laboratory of Nuclear Protein Regulation, school of Basic Medical Sciences, Henan University, Kaifeng 475004, China.
3. Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, Jouf University, Sakaka 2014, Saudi Arabia.
Received 2021-3-17; Accepted 2021-8-1; Published 2021-8-18
Abstract
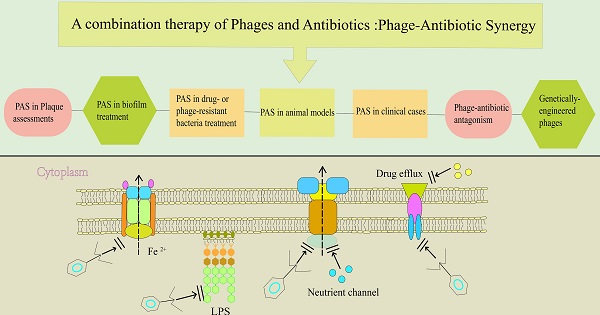
Emergence of antibiotic resistance presents a major setback to global health, and shortage of antibiotic pipelines has created an urgent need for development of alternative therapeutic strategies. Bacteriophage (phage) therapy is considered as a potential approach for treatment of the increasing number of antibiotic-resistant pathogens. Phage-antibiotic synergy (PAS) refers to sublethal concentrations of certain antibiotics that enhance release of progeny phages from bacterial cells. A combination of phages and antibiotics is a promising strategy to reduce the dose of antibiotics and the development of antibiotic resistance during treatment. In this review, we highlight the state-of-the-art advancements of PAS studies, including the analysis of bacterial-killing enhancement, bacterial resistance reduction, and anti-biofilm effect, at both in vitro and in vivo levels. A comprehensive review of the genetic and molecular mechanisms of phage antibiotic synergy is provided, and synthetic biology approaches used to engineer phages, and design novel therapies and diagnostic tools are discussed. In addition, the role of engineered phages in reducing pathogenicity of bacteria is explored.
Keywords: phage-antibiotic synergy, multidrug-resistance, bacterial anti-phage resistance, biofilm, phage therapy
Introduction
Alexander Fleming discovered the first antibiotic, penicillin in 1928 and this marked the beginning of the era of antibiotics [1]. Various antibiotics are extensively used to fight infectious diseases in clinical practice. However, there has been a rapid increase in the levels of bacterial drug-resistance due to lack of effective control system and inappropriate use of antibiotics. World Health Organization (WHO) listed antibiotic resistance as one of the three most important public health threats in the 21st century[2]. According to the estimates of economist Jim O'Neill who was commissioned by the UK Prime Minister, drug-resistant bacteria cause about 700,000 deaths worldwide every year [3]. Furthermore, it is projected that 10 million and 300 million deaths directly and indirectly, associated with infections caused by drug-resistant bacteria will occur by 2050 respectively, exceeding the current number of cancer-related deaths [4]. Therefore, it is likely that the post antibiotic era is slowly approaching and human beings are likely to face a world without effective antimicrobial drugs [5].
To address the looming threat of drug-resistant bacteria, scientists have proposed phage therapy as an alternative to antibiotic therapy. Phages also known as bacterial viruses, are widely distributed in nature and can infect and kill bacteria [6]. Use of phages have been reconsidered as therapeutical tools due to the raised antimicrobial resistance (AMR) [7]. Notably, the ability of phages to counteract multidrug-resistant bacteria has several advantages compared with antibiotics, including high specificity [8, 9], low dosage, low cost of production [10, 11], high safety and antibiofilm activity [12, 13]. Instead of replacing antibiotics with phages, scientists have proposed that a combination of these two types of antibacterial agents may be more effective compared with use of either independently. In addition, the joint approach might confer possible advantages such as enhanced bacterial suppression, stronger effective penetration into biofilms and reduced capacity of bacteria to develop phage and/or antibiotic resistance. In this study, phage-antibiotic synergy refers to an increase in phage production following exposure to sublethal levels of bactericidal antibiotics [14], and this is considered a promising therapeutic strategy.
Interactions between Phage and Antibiotic
Previous studies used various experimental models to determine the synergistic effect of various types of phages and antibiotics. These models include plaque assessments, elimination of drug- or phage-resistant bacteria, reducing the number of bacteria embedded in biofilm and in vivo evaluation[15]. This section focuses on the different evaluation methods. A summary of the types of phages and bacteria, as well as the synergistic effect of phage-antibiotic combinations and the corresponding references is presented in Table 1.
PAS in Plaque assessments
The term “phage-antibiotic synergy” (PAS) was first coined by Comeau AM et al in 2007. In that first report, the researchers observed that stimulation by sublethal concentrations of β-Lactam and quinolone antibiotics resulted in significantly higher diameter and number of plaques, implying that a higher adsorption rate, shorter latent period and larger burst size occurred during plaque formation [14, 16]. In addition, Uchiyama et al [17] screened 21 types of antibiotics with synergistic effect with Pseudomonas aeruginosa phage, most of which were effective in inhibiting bacterial cell wall synthesis or protein synthesis. A similar phenomenon of PAS was reported using species such as Burkholderia cepacia and Staphylococcus aureus by determining the plaque diameter [18, 19]. Besides enlarging plaque size, antibiotics have been shown to significantly affect phage adsorption rates and the latent period during these infections. For example, Ryan et al. [20] treated Escherichia coli with either T4 phage alone or with a combination of cefotaxime and T4. The study findings showed that in absence of the antibiotic, the T4 phage had a latent period of 24 min which was reduced to 18 min upon addition of cefotaxime. Furthermore, the initial concentration of T4 phage increased from 5×106 to 5×107 plaque forming unit (PFU)/ml, indicative of a better replication as well as an increase in the rate of phage adsorption. Another study reported a significant increase in the burst size of E. coli phage ϕMFP following treatment with sublethal concentrations of cefotaxime, which was considered to be the cause for formation of long bacterial filaments [17]. In summary, these findings show that addition of antibiotics causes changes in plaque diameter, latency period and burst size during growth of phages, and can therefore be used to determine the synergistic effect of phages and antibiotics.
PAS in drug- or phage-resistant bacteria treatment
Drug-resistant bacteria pose a major threat to human health during clinical practice. Phage therapy is an alternative to antibiotics and studies report that it is effective in circumventing bacterial resistance. However, an important concern is that bacteria can also develop resistance to phages. Therefore, the recently proposed PAS is a potential approach for management of bacteria resistant to both antibiotics and phages. Several studies report that PAS can significantly reduce bacterial density, especially in phage and drug-resistant bacteria [21-24]. In addition, PAS effectively limits production of bacterial virulence factors [25].
A previous study reported that 8 hours of treatment with a combination of 1/10 minimum inhibitory concentration (MIC) (0.05 mg/L) ciprofloxacin and phage ECA2 caused a significant decrease in the colony forming unit (CFU) of E. coli (decreased by about 7.8 folds). However, there was no significant decrease in bacterial counts following treatment with individual doses of the phage or antibiotic [26]. Furthermore, phage-antibiotic combination treatments suppress pathogen activity and mitigate antibiotic or phage resistance in bacteria. Torres-Barceló et al [27] treated P. aeruginosa strain PA01 with either phage LUZ7 alone or a combination of streptomycin and LUZ7 and observed bacterial regrowth after 24 hours, which can be attributed to mutant phage-resistant subpopulations. However, a combination of streptomycin (100 and 240 mg/mL) with phage LUZ7 (105 PFU/mL) effectively prevented development of phage-resistant PA01 mutants. Moreover, the combination therapy showed a 4-log CFU/ml loss of bacterial viability, compared with either antibiotic (100 and 240 mg/mL) or single-phage (105 PFU/mL) therapy alone. Another PAS analysis was performed on a recombinant P. aeruginosa strain with plasmid pUCP24 that was resistant to gentamicin [28]. The recombinant P. aeruginosa strain was administered with either a monotherapy and combination therapy consisting of gentamicin and filamentous phage Pf1. Analysis of vitality of overnight cultures containing approximately 106 CFU, after exposure to 300 μg/ml gentamicin, showed that the growth of strain PA01 was not significantly affected. However, treatment with 50 μg/ml gentamicin and 108 PFU/ml of Pf1 phage effectively inhibited growth [24].
PAS in biofilm treatment
Bacterial biofilms refer to communities of microbes attached to an abiotic or biotic surface, such as medical implants, including catheters and artificial hip joints. The community of cells is encapsulated in an extracellular matrix comprising extracellular DNA, secreted proteins, lipids and polysaccharides that are collectively referred to as extracellular polymeric substances (EPS). These substances allow adhesion to the surface and provide protection against antimicrobial agents [29, 30]. However, biofilms represent a dangerous reservoir for persistent bacteria, which is a phenotypic variation in bacterial population and confers significant antimicrobial resistance without genetic mutations and drug resistance genes [31-33]. Therefore, biofilms are more difficult to eradicate compared with planktonic bacteria treated with only antibiotic therapy. As a result, biofilms are considered as a potential non-antibiotic therapy. Notably, numerous studies have explored the combined effect of phages and antibiotics for elimination of bacteria in biofilms [16, 20].
For instance, a previous study explored efficacy of phage Sb-1 alone or in combination with different classes of antibiotics for elimination of S. aureus biofilms in a rat model [34]. The findings showed that treatment with phage Sb-1 alone significantly reduced persistent bacteria although it did not eradicate the biofilm. However, simultaneous treatment with phage Sb-1 and rifampicin/daptomycin significantly degraded EPS and eradicated S. aureus biofilm. Moreover, staggered phage Sb-1 and antibiotic treatment is an effective strategy for degradation of biofilms.
After bacterial infection, toxic genes encoded by phage genomes are expressed as specific enzymes, such as peptidoglycan hydrolase and polysaccharide depolymerases, which are involved in cleavage of bacterial peptidoglycans. A combination of hydrolytic enzymes, encoded by phages, with specific antibiotics significantly destroys the structure of bacterial biofilms and releases persistent bacteria embedded in the biofilm into the nutrient environment. This subsequently enhances metabolic activity of persistent bacteria in the nutrient environment, making them more sensitive to antibiotics [5, 35-37]. For instance, a 6-hour treatment with a combination of KPO1K2 and ciprofloxacin significantly eliminated a biofilm formed by Klebsiella pneumoniae, and caused a 4.5-fold reduction in the number of bacteria embedded in the biofilm.
PAS in animal models
Evaluating effects of PAS in vitro has a number of limitations due to lack of an immune effect [38]. Several studies have explored the synergistic effect of phage-antibiotic combinations in vitro, but not in vivo [39-41]. A previous study simulated bacteremia and bladder infection conditions, and reported that serum and urine components completely prevented PAS between phage φHP3 and ceftazidime, by determining the bacterial density which was significantly decreased in the culture medium. This finding indicates that human host conditions suppress PAS on bacterial growth in vivo [42].
Furthermore, animal models can be used to evaluate the effect of phages in weakening inflammatory reaction, in order to ascertain the exact synergistic effect of phages and antibiotics. Results from previous animal models show that phage infection triggers a specific immune response, although other studies report that phages cannot cause significant disease symptoms in vivo[43, 44]. Studies report that liposome-encapsulated phages can significantly alleviate inflammatory responses, thereby providing a basis for phage treatment in vivo. For example, Kaur et al. [45] reported that levels of PCT, IL-1β and TNF-α cytokines in mice by K-wire, surgically implanted into the intra-medullary canal, implanted with a specific phage and linezolid were lower compared with those in a K-wire embedded with phages or antibiotics only. Consequently, the number of intracellular bacteria will be reduced to a certain level, increasing effectiveness of oxidative killing by phagocytes [46]. In this case, the phagocytic function and bactericidal activity of macrophages is significantly enhanced since bacteria are phagocytized by macrophages in vivo. Notably, the pathogen can also rapidly develop resistance to phages in vivo. However, previous studies report that neutrophils are more effective in scavenging phage-resistant, compared with phage-sensitive, bacteria [38]. Moreover, Tiwari et al. [47] reported that phage PA1Ø significantly reduces the lethal rate infected with immune-competent mice compared with neutropenic mice. Notably, use of phages for treatment of bacterial infections in vivo may produce high amounts of residue along with bacterial dissolution, including, lipopolysaccharides, cytoplasmic proteins, membrane particles and large pieces of cell debris [48]. Therefore, it is not clear which components directly elicit an immune response in the body. Although some studies report that the level of inflammatory factors decreases after phage therapy, it is possible that this phenomenon may also be caused by reduction in the number of pathogens in vivo [49].
Phage-antibiotic combinations with confirmed positive interactions
| Pathogens | Strains | Antibiotics | Phage | Synergistic effects | Ref. |
|---|---|---|---|---|---|
| Plaque assessments | |||||
| S. aureus | - | AMX | ϕSZIP1 | ++ | [18] |
| - | TIM | ϕSZIP1 | + | ||
| - | CRO, CHL | ϕSZIP1 | + | ||
| P. aeruginosa | PA5 | CP, CPZ, FOM, etc. | KPP21 | +++ | [17] |
| PAO1, PA4, PA23, etc. | AMK, AZT, CAZ, etc. | KPP22 | +++ | ||
| PA3 | CPZ/SBT, FOM, TOB | KPP23 | +++ | ||
| B. cepacia | K56-2/C6433 | CIP, MEM, TET | KS12/KS14 | +++/++ | [19] |
| E. coli | MFP/ | AZT, CFM, CTX, etc. | ϕMFP/RB32/ RB33/T4 | + | [50] |
| E. coli | ATCC11303 | AML, AMP, CFR, etc. | Φszut/ ϕSZIP1/ϕSZIP2 | + | [20] |
| Drug- or phase-resistant bacteria/biofilm | |||||
| E. coli | ATCC 11303 | TOB | T4 | +b | [22] |
| ATCC 13706 | CIP | ECA2 | +++a | [26] | |
| JE2571 | KAN, RIF | RPD1(T) | +a | [51] | |
| S. aureus | PS80 | GEN | SA5 | +a, +b | [52] |
| MRSA | TEC | Sb-1 | +a, +++b | [53] | |
| MRSA | RFP/AZI | SAP-26 | +a, +b | [23] | |
| MRSA | CIP, TET | PYO | +a | [24] | |
| P. aeruginosa | PAO1 | CAR, GEN, TET, etc. | Pf3 | + | [24] |
| PAK | CAR, GEN, CHL | Pf1 | + | ||
| ATCC 9027 | CRO | σ-1 | ++a | [54] | |
| PA-4U | CRO | δ | +a | ||
| PA-M2 | CRO | 001A | ++a | ||
| PAO1 | STR | LUZ7 | +a | [27] | |
| PA01/PAPS | TET, ERY, CIP, etc. | OMKO1 | ++a | [21] | |
| CHA | CIP, MEN | Cocktail | ++a | [55] | |
| PAO1 | CAZ, CIP | LKD16 | ++a | [56] | |
| PAO1 | CAZ, CIP | LUZ7,14/1 | +a | ||
| PAO1 | CAZ, PIPC | KPP22 | ++a | [57] | |
| PA14 | CAZ, CIP, GEN, etc. | NP1, NP3 | +a | [35] | |
| PAO1 | CST | KTN4 (M) | +a | [56] | |
| PA365707, PA364077 | CIP | PEV20 | ++b | [58] | |
| A. baumannii | AB01, AB04, AB16 | MEM, CIP, MEM | KARL-1 | ++a | [59] |
| K. pneumoniae | KPB5055 | CIP | KPO1K2 | +b | [60] |
| KPB5055 | AMX | Not known | ++a/b | [61] | |
| B. cepacia | K56-2 | CIP, TET, MEM | KS12 | +a | [19] |
| In vivo | |||||
| E. coli | poultry isolate | ENR | SPR02/DAF6 | +++ | [62] |
| S. aureus | ATCC43300 | LZD | MR-10 | + | [63] |
| P. aerginosa | CHA | CIP | cocktail | ++ | [55] |
| B. cepacia | K56-2 | MIN | KS12 | + | [19] |
| K56-2 | MEM | KS12 | ++ | ||
| E. faecalis | V583 | AMP | EFDG1, EFLK1 | +++ | [64] |
| K. pneumoniae | KPB5055 | AMK | SS (P) | + | [65] |
| Clinical cases | |||||
| P. aerginosa | - | CAZ, CIP | OMKO1 | * | [66] |
| K. pneumoniae | ERKp | SMZ-TMP | KP152, KP154, KP155, KP164, KP6377, HD001 | * | [67] |
| K. pneumoniae | ATCC 700603 | MEM | unknown | * | [68] |
“a” represents the effect on planktonic bacteria, “b” represents the effect on biofilms. “-” represents an unknown species of bacteria used, “*” represents clinical trials, “+” shows that the percentage of phage-antibiotic synergistic effect is enhanced compared to phage alone and its number represents the degree of enhancement. “+” indicates 10~50%, “++” indicates 50~80% and “+++” indicates above 80%. The names of antibiotics are abbreviated as follows: Amikacin [AMK], Ampicillin [AMP], Amoxicillin [AMX], Azithromycin [AZI], Aztreonam [AZT], Carbenicillin [CAR], Ceftazidime [CAZ], Cefixime [CFM], Chloramphenicol [CHL], Ciprofloxacin [CIP], Cefoperazone [CPZ], Sulbactam/Cefoperazone [SBT/CPZ], Ceftriaxone [CRO], Colistin [CST], Cefotaxime [CTX], Enrofloxacin [ENR], Erythromycin [ERY], Fosfomycin [FOM], Gentamicin [GEN], Kanamycin [KAN], Linezolid [LZD], Meropenem [MEM], Minocycline [MIN], Piperacillin [PIPC], Rifampicin [RIF], Streptomycin [STR], Teicoplanin [TEC], Tetracycline [TET], Ticarcillin [TIM] and Tobramycin [TOB].
PAS in clinical cases
Although several studies have discussed the effect of combining phages with antibiotics in vitro and in vivo, there were also some successful clinical experiments. Bao et al had reported a case of patient who developed a recurrent urinary tract infection (UTI) with extensively drug-resistant Klebsiella pneumoniae (ERKp) which resisted all tested antibiotics, except tigecycline and polymyxin B [67]. After critical care treatments, including tigecycline administration, the UTI was not cured and became persistent. The patient was enrolled in phage therapy clinical trial after involvement evaluation and informed consent. In vitro, the combination of phage cocktail III (KP152, KP154, KP155, KP164, KP6377 and HD 001) and SMZ-TMP could completely suppress the growth of ERKp for more than 24h. Therefore, after treated with the above therapeutic regimen, including oral administration of trimethoprim-sulfamethoxazole (SMZ-TMP) twice a day, and bladder irrigation with phage cocktails III for five days of continuous treatment, the ERKp couldn't be isolated from the patient's urine, and the symptoms of urinary tract infection disappeared completely. Moreover, there was no sign of recurrence within six months after discharge.
A case of renal transplant patient developed urinary tract infection with an extended-spectrum β-lactamase (ESBL)-positive K. pneumoniae strain in the first month post-transplant was ineffective in the treatment of multiple antibiotics [68]. Although ESBL K. pneumoniae in this case was sensitive to meropenem, the infection recurred eventually evolved into epididymitis after repeated treatment with meropenem. A phage from Georgia exhibited excellent lytic activity against this ESBL-K. pneumoniae isolates. After treatment with meropenem combined with this phage by oral and bladder irrigation, respectively, the urethral symptoms of the patient completely subsided within one day, and urine cultures remained negative for 14 months after treatment.
The phage OMKO1's receptor is the outer membrane protein M of mexAB- and mexXY-multidrug efflux systems of P. aeruginosa, which is essential for antibiotic (eg. ceftazidime and ciprofloxacin) pump-out, thus OMKO1 induced receptor-mutant-resistant strain would be more susceptible to ceftazidime. In a case of therapeutic application of phage OMKO1 to treat a drug-resistant P. aeruginosa infection of an aortic Dacron graft, Benjamin K. Chan et al. conducted an experiment in vitro and found that both phage OMKO1 alone and ceftazidime combined with phage OMKO1 could reduce bacterial density. Especially ceftazidime at 2×MIC + phage OMKO1 treatment could significantly reduce the bacterial density, compared with ceftazidime alone in the treatment of bacterial biofilm infection [66]. In addition, the OMKO1 (107 PFU/ml) and ceftazidime (0.2 g/ml) solution was successfully used in the treatment of patients with aortic perforation infection, and the patient represented stable vital signs and there was no sign of recurrent infection for 18 months.
Phage-antibiotic antagonism
Efficient synergistic bactericidal effects are achieved when these phages are combined with specific antibiotics. Additionally, other phage-antibiotic combinations show no synergistic or antagonistic effects [69] (Table 2). A typical example is rifampicin, which can inhibit the growth of host bacteria by targeting its RNA polymerase. Rifampicin also inhibits the production of phage particle, due to the fact that its replication depends on the host RNA polymerase. This function of rifampicin is shown the antagonistic effect with a number of phages (Table 2). However, when the phage encodes its own RNA polymerase for virion replication rather than the host bacteria's, Rifampicin will not restrain the production of progeny phage, which shows the synergistic bactericidal activity of phages-antibiotics. For instance, P. aeruginosa phage ϕKZ could encode its own RNA polymerase, infect the host bacteria and produce the progeny phage in the presence of 400 µg/mL of rifampin [70]. However, one counter-example, the virion replication of phage LUZ19 depending on the host RNA polymerase was completely inhibited at the same concentration of rifampicin. These results indicate that the interaction effects between phages and antibiotics depended on the type of antibiotics or phages [71].
Genetic mechanism underlying PAS
Initial infection with phages occurs through binding to receptors, such as lipopolysaccharides, teichoic acids, proteins, and flagella, on the surface of bacteria. Although emergence of phage-resistant bacteria is likely inevitable, numerous studies report that phage selective pressure may accelerate bacterial mutations thus promoting them to subvert phage infection, but with a cost to their fitness [90]. Such fitness trade-offs include reduced virulence, limited nutrient uptake, resensitization to antibiotics, and colonization defects. This observation lays a basis for application of phages. In addition to direct killing effect of phages on host bacteria, selective pressure produced by phages is useful in limiting bacterial growth. Surface molecules of bacteria play an important role in disease phenotypes, in a similar mechanism to receptor molecules on phages [91, 92]. These surface components consist of lipopolysaccharides (LPS), outer membrane proteins, teichoic acid, type IV pili, capsules, siderophores receptors and the efflux pumps. Their components are often considered to be virulence factors, antibiotic resistance related factors and normal growth factors, as they can mediate attachment to and damage of hosts and antibiotic efflux, respectively [93-95].
The ferric catecholate receptor, known as FepA on the surface of Salmonella enterica is the key protein for siderophore mediated iron transport in bacteria. However, FepA can also act as a bacterial receptor, playing a role in adsorption of phage H8. Notably, the gene encoding FepA mutates to resist phage infection under high pressure of phage selection. On the other hand, its mutant strain cannot transport iron from the environment, causing its own death due to growth restriction [96]. Previous studies report that phage H8 can force a desired genetic trade-off between phage resistance and growth restriction, a phenomenon that can be beneficial in phage therapy against MDR-S. enterica.
No synergistic or antagonistic effects of the combination of phage and antibiotic
| Pathogens | Antibiotics | Phages | Antagonistic effects | Refs. |
|---|---|---|---|---|
| M. tuberculosis | RIF, INH | TM4, D29 | Inhibiting production of phage particle | [72-76] |
| P. aeruginosa | RIF | LUZ19 | Inhibiting production of phage particle | [70] |
| CIP, MEM | Phage cocktail | No synergistic | [55] | |
| CIP, TOB, GEN | NP1, NP3 | Inhibiting production of phage particle | [35] | |
| B. subtilis | RIF | SPO1 | Reducing bacteriolytic activity | [77] |
| RIF | β22, AR9 | Inhibiting production of phage particle | [78, 79] | |
| Nalidixic Acid | SP50, SP82, etc. | Inhibiting production of phage particle | [80] | |
| E. coli | RIF | λvir, T2, T5, Mu | Inhibiting production of phage particle | [81-84] |
| Nalidixic Acid | ϕR, T2, T7, etc. | Inhibiting production of phage particle | [85, 86] | |
| CHL, TET | ECA2 | Reducing bactericidal activity | [26] | |
| CIP | ELY-1 | Inhibiting production of phage particle | [87] | |
| P. aeruginosa | RIF | PM2 | Inhibiting production of phage particle | [88] |
| R. solanacearum | RIF | ΦRP12, ΦRP31, ΦRSB1, etc. | Inhibiting production of phage particle | [89] |
| S. aureus | GEN, RIF, LZD, etc. | PYO | Inhibiting production of phage particle | [24] |
The outer membrane porin M (OprM) in P. aeruginosa is an indispensable component of drug efflux systems including MexAB-OprM and MexXY-OprM. For example, Chan et al. [97] reported that OprM can be recognized by phage OMKO1 as a key receptor protein. In addition, the gene encoding OprM mutates under selection pressure during phage infection, thus allowing P. aeruginosa to tolerate phage OMKO1 during chronic infection. Moreover, OprM mutation simultaneously causes a deficiency in drug efflux pumps, thus preventing elimination of antibiotics. In summary, these results indicate that phage infection triggers an evolutionary trade-off in P. aeruginosa, where the evolution of bacterial resistance to phages interferes with the function of efflux pumps, thus increasing sensitivity to antibiotics.
Furthermore, ompU gene which encodes the outer membrane porin in Vibrio cholerae and acts as a phage infection receptor in the bacteria, produces evolutionary selection pressure as a result of phage infection [98]. Under this selective pressure, ompU or toxR (regulatory gene of ompU expression) genes in V. cholerae mutate, resulting in resistance to phages. Similarly, V. cholerae displays an evolutionary trade-off between phage resistance and bacterial virulence. Notably, these mutations attenuate virulence by at least 100-fold since the mutant strains are unable to cause cholera and loses the ability of disease transmission. These results indicate that adaptation to phage infection involves trade-offs in evolutionary fitness and provides a molecular basis for understanding the effect of phage infection on transmission of V. cholerae as well as seeding of environmental reservoirs (Figure 1). The findings from these studies show that phages in combination with antibiotics synergistically act against host bacteria and alter expression of bacterial virulence factors, antibiotic resistance and activity of growth factors. These mechanisms, in turn, cause an increase in antibiotic sensitivity or inhibition of bacterial growth (Table 3).
Use of engineered phages in enhancing susceptibility to antibiotics
The current rapid advances in sequencing technology and molecular biology have led to increase in development of genetically-engineered phages. Genetically-engineered phages are effective in eliminating drug-resistant pathogens and provide a key therapy for treatment of patients [5, 106, 107]. The recombinant phage can efficiently restore sensitivity of drug-resistant bacteria, decrease MIC value of antibiotics and target deletion of essential genes in host bacteria. Notably, several studies have reported successful construction of genetically-engineered phages that exhibit significant activity against drug-resistant bacteria (Table 4).
Trade-off in bacteria between phage resistance and bacterial fitness; Profile of proteins found on the surface of bacteria including nutrient channels, lipopolysaccharide (LPS), drug efflux pumps, and siderophore receptors. They are related to bacterial life history traits and initial infection of phage. When the genes encoding these proteins are altered, through events such as mutations, the bacteria exhibit the characteristics of phage resistance. In addition, these changes separately block bacterial intake of nutrients, downregulate virulence factors, and hinder entry of iron ion into bacteria, thus affecting normal growth of bacteria. Moreover, the blocked drug efflux system predisposes bacteria to antibiotics.
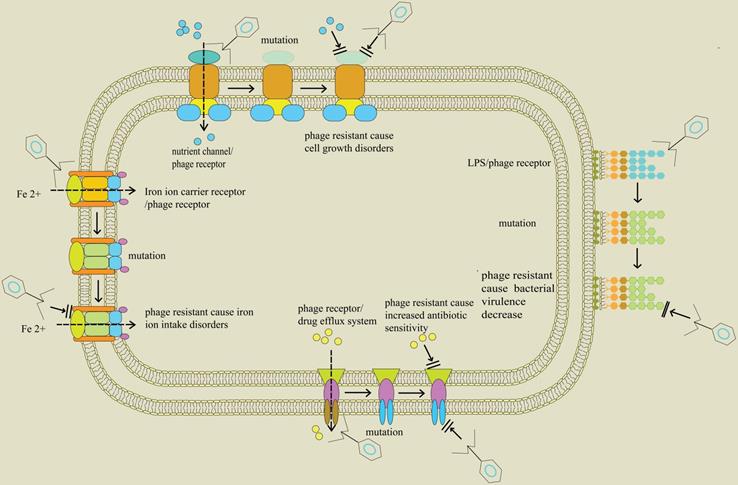
Phage-induced changes in bacterial fitness and antibiotic resistance
| Pathogens | Phage | Target/Effect | Result | Ref. |
|---|---|---|---|---|
| Salmonella | f2αSE, f3αSE, f18αSE | LPS/phage receptor, virulence factor | Attenuating virulence | [99] |
| Salmonella | φ1 | LPS/phage receptor, virulence factor | Attenuating virulence | [100] |
| V. cholerae | ICP2_2013 _A_Haiti | OmpU/phage receptor, virulence factor | Attenuating virulence | [98] |
| E. coli K-1 | NM | OmpA/phage receptor, immune system evasion | Immune system evasion | [101] |
| S. aureus | NM | Teichoic acids/phage receptor, virulence factor | Attenuating virulence | [102] |
| S. aureus | MSa | Teichoic acids/phage receptor, virulence factor | Attenuating virulence | [103] |
| B. cenocepacia | NM | FepA/phage receptor, siderophore receptor | Inhibiting bacterial growth | [104] |
| Salmonella | H8 | Siderophore transporter flagellum/phage | Inhibiting bacterial proliferation | [96] |
| V. anguillarum | Lambda Zap II | receptor, motility, virulence factor | Reducing motility | [105] |
| P. aeruginosa | OMKO1 | OprM/phage receptor, efflux pump | Increasing sensitivity to antibiotics | [97] |
| S. aureus | SA5 | Not known | Reducing antibiotic resistance | [52] |
| P. aeruginosa | LUZ7 | Not known | Increasing sensitivity to antibiotics | [27] |
Previous studies report that the genome of phage M13 can be edited to overexpress the SOS inhibitor, LexA3 [108]. Consequently, the modified M13 inhibited SOS reaction following DNA damage to the bacteria. In addition, the bactericidal effect of the modified M13 combined with antibiotics was significantly augmented. A combination of modified M13 and Ofloxacin showed a 2.7-fold increase in the bactericidal effect and significantly reduced bacterial resistance, compared with single-dose antibiotic treatment [109]. A previous study inserted a streptomycin sensitive gene into the genome of phage lambda (λ). Treatment of a streptomycin-resistant E. coli with this recombinant phage restored their sensitivity to the antibiotic, and the MIC value of the antibiotic against E. coli decreased from 100 to 12.5 mg/ml. The same method was used to restore sensitivity of E. coli to nalidixic acid, achieving a 2-fold reduction in MIC value [110].
Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Cas9 system is an adaptive immune mechanism formed by bacteria during their evolution. Currently, CRISPR/Cas9 nuclease has gained popularity as a major tool for targeted deletion of foreign DNA in pathogens. In a previous study, researchers integrated the genome of phage λ with the CRISPR/Cas9 system to target genes encoding β-lactamases in the E. coli genome, then infected E. coli resistant to β lactams with the modified phage λ. The results showed that the drug-resistant E. coli regained its sensitivity to the antibiotics [111]. In a similar study, Park et al. [112] used Cas9-triggered homologous recombination to integrate a CRISPR/cas9 targeted NUC gene (nuclear gene common to all S. aureus) into the genome of phage ØSaBov. Infecting S. aureus with the recombinant phage resulted in death of all bacteria within eight hours in vitro, whereas the number of pathogens in vivo reduced by 2-fold [112].
Bactericidal effect of genetically engineered phage
| Stain | Phage | Method | Result | Refs. |
|---|---|---|---|---|
| L. monocytogenes | PSA | Removing lysogen module | Improving lytic ability | [113] |
| L. monocytogenes | B025 | Removing lysogen module | Improving lytic ability | [113] |
| S. aureus | ØSaBov | Integrating with CRISPR/Cas | Improving lytic ability | [112] |
| S. aureus | Φ11 | Recombining wit SnCe6 | Improving lytic ability | [114] |
| C. albicans | JM | Recombining with PPA | Improving lytic ability | [115] |
| E. coli | M13 | Recombining with CAP | Improving lytic ability | [116] |
| E. coli | M13 | Overexpressing LexA3 | The synergetic bactericidal efficacy of engineered phage M13 and ofloxacin was increased by 2.7 logs | [109] |
| E. coli | λ | Recombining with streptomycin sensitive genes | The synergetic efficacy of engineered λ and streptomycin reduced MIC value from 100 g/ml to 12.5 g/ml | [110] |
| E. coli | λ | Integrating with CRISPR/Cas | The synergetic efficacy of λCas-CRISPR and streptomycin sensitized and killed antibiotic-resistant bacteria | [111] |
| E. coli | T7 | Recombining with Dsp8 | Improving lytic ability | [117] |
| E. coli | M13 | Recombining with toxin gene | Improving lytic ability | [118] |
| P. aeruginosa | T7 | Recombining with AILA | Improving lytic ability | [119] |
| C. trachomatis | M13 | Recombining with RGD and PmpD | Improving lytic ability | [120] |
| E. coli | T2 | Recombining with tail fiber | Expanding phage host range | [121] |
| E. coli | FD | Rcombining with IKE | Expanding phage host range | [122] |
| E. coli | T3 | Replacing tail fiber gene 17 | Expanding phagehost range | [123] |
| H. pylori | M13 | Rcombining with gene 3 protein | Expanding phage host range | [124] |
| E. faecalis | ΦEf11 | Reorganizing with defective ΦFL1C | Expanding phage host range | [125] |
| E. coli | T2 | Replacing host recognition genes | Expanding phage host range | [126] |
| L. monocytogenes | A511 | Bacteriophages PEGylation | Enhancing the half-life of phage | [127] |
| S. typhi | Felix-O1 | Bacteriophages PEGylation | Enhancing the half-life of phage | [127] |
| E. coli | T7 | Inserting PhoE signal peptide | Enhancing the half-life of phage | [128] |
| S. aureus | P954 | Inserting cat phage genome | Reducing endotoxin production | [129] |
| E. coli | M13 | Recombining with BgIII | Reducing endotoxin production | [130] |
| E. coli | M13 | Recombining with Gef and ChpBK | Reducing endotoxin production | [131] |
| P. aeruginosa | Pf3 | Recombining with endonuclease | Reducing endotoxin production | [132] |
Discussion
A combination of phages and antibiotics has been extensively used to enhance eradication of drug-resistant pathogens, and alleviate the widespread antibiotics resistance worldwide [133]. Numerous experimental models, including plaque analysis, liquid plankton, biofilm tests and animal experiments, have been used to successfully evaluate the synergistic effect of phages and antibiotics. Several studies have explored the underlying mechanism of synergy between phages and antibiotics. Notably, the seesaw effect of host evolution explains the mechanism of this synergistic bactericidal effect [15]. Phage infection can exert selective pressure on bacteria, thus predisposing them to gene mutations [134]. Under this selective pressure, there is loss or down-regulation of some of the host bacteria's important components related to bacterial toxicity, drug sensitivity and growth factors. Studies report that phage-resistant strains exhibit lower toxicity, are more sensitive to antibiotics and have slower growth rate compared with wild strains [12].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929. Bull World Health Organ. 2001;79:780-90
2. World HealthOrganization. (2018). Antibiotic resistance. https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance.
3. Piddock LJV. Reflecting on the final report of the O'Neill Review on Antimicrobial Resistance. Lancet Infect Dis. 2016;16:767-8
4. King A. Antibiotic Resistance Will Kill 300 Million People by 2050. https://www.scientificamerican.com/article/antibiotic-resistance-will-kill-300-million-people-by-2050/. 2014
5. Gordillo Altamirano FL, Barr JJ. Phage Therapy in the Postantibiotic Era. Clin Microbiol Rev. 2019 32
6. Comeau AM, Tétart F, Trojet SN, Prère MF, Krisch HM. [The discovery of a natural phenomenon, "Phage-Antibiotic Synergy". Implications for phage therapy]. Med Sci (Paris). 2008;24:449-51
7. Kutateladze M, Adamia R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends in biotechnology. 2010;28:591-5
8. Abdelkader K, Gerstmans H, Saafan A, Dishisha T, Briers Y. The Preclinical and Clinical Progress of Bacteriophages and Their Lytic Enzymes: The Parts are Easier than the Whole. Viruses. 2019 11
9. D'Accolti M, Soffritti I, Mazzacane S, Caselli E. Bacteriophages as a Potential 360-Degree Pathogen Control Strategy. Microorganisms. 2021 9
10. Samoylova TI, Braden TD, Spencer JA, Bartol FF. Immunocontraception: Filamentous Bacteriophage as a Platform for Vaccine Development. Curr Med Chem. 2017;24:3907-20
11. Samoylova TI, Norris MD, Samoylov AM, Cochran AM, Wolfe KG, Petrenko VA. et al. Infective and inactivated filamentous phage as carriers for immunogenic peptides. J Virol Methods. 2012;183:63-8
12. Międzybrodzki R, Borysowski J, Weber-Dąbrowska B, Fortuna W, Letkiewicz S, Szufnarowski K. et al. Clinical aspects of phage therapy. Advances in virus research. 2012;83:73-121
13. Shlezinger M, Friedman M, Houri-Haddad Y, Hazan R, Beyth N. Phages in a thermoreversible sustained-release formulation targeting E. faecalis in vitro and in vivo. PLoS One. 2019;14:e0219599
14. Kim M, Jo Y, Hwang YJ, Hong HW, Hong SS, Park K. et al. Phage-Antibiotic Synergy via Delayed Lysis. Applied and environmental microbiology. 2018 84
15. Morrisette T, Kebriaei R, Lev KL, Morales S, Rybak MJ. Bacteriophage Therapeutics: A Primer for Clinicians on Phage-Antibiotic Combinations. Pharmacotherapy. 2020;40:153-68
16. Comeau AM, Tétart F, Trojet SN, Prère MF, Krisch HM. Phage-Antibiotic Synergy (PAS): beta-lactam and quinolone antibiotics stimulate virulent phage growth. PloS one. 2007;2:e799
17. Uchiyama J, Shigehisa R, Nasukawa T, Mizukami K, Takemura-Uchiyama I, Ujihara T. et al. Piperacillin and ceftazidime produce the strongest synergistic phage-antibiotic effect in Pseudomonas aeruginosa. Arch Virol. 2018;163:1941-8
18. Iqbal M, Narulita E, Zahra F, Murdiyah S. Effect of Phage-Antibiotic Synergism (PAS) in increasing antibiotic inhibition of bacteria caused of foodborne diseases. J Infect Dev Ctries. 2020;14:488-93
19. Kamal F, Dennis JJ. Burkholderia cepacia complex Phage-Antibiotic Synergy (PAS): antibiotics stimulate lytic phage activity. Appl Environ Microbiol. 2015;81:1132-8
20. Ryan EM, Alkawareek MY, Donnelly RF, Gilmore BF. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS immunology and medical microbiology. 2012;65:395-8
21. Segall AM, Roach DR, Strathdee SA. Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy. Current opinion in microbiology. 2019;51:46-50
22. Coulter LB, McLean RJ, Rohde RE, Aron GM. Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses. 2014;6:3778-86
23. Rahman M, Kim S, Kim SM, Seol SY, Kim J. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling. 2011;27:1087-93
24. Dickey J, Perrot V. Adjunct phage treatment enhances the effectiveness of low antibiotic concentration against Staphylococcus aureus biofilms in vitro. PloS one. 2019;14:e0209390
25. Goerke C, Köller J, Wolz C. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:171-7
26. Valério N, Oliveira C, Jesus V, Branco T, Pereira C, Moreirinha C. et al. Effects of single and combined use of bacteriophages and antibiotics to inactivate Escherichia coli. Virus research. 2017;240:8-17
27. Torres-Barceló C, Arias-Sánchez FI, Vasse M, Ramsayer J, Kaltz O, Hochberg ME. A window of opportunity to control the bacterial pathogen Pseudomonas aeruginosa combining antibiotics and phages. PloS one. 2014;9:e106628
28. Hagens S, Habel A, Bläsi U. Augmentation of the antimicrobial efficacy of antibiotics by filamentous phage. Microb Drug Resist. 2006;12:164-8
29. Pandit S, Ravikumar V, Abdel-Haleem AM, Derouiche A, Mokkapati V, Sihlbom C. et al. Low Concentrations of Vitamin C Reduce the Synthesis of Extracellular Polymers and Destabilize Bacterial Biofilms. Front Microbiol. 2017;8:2599
30. Wu J, Xi C. Enzymatic method for extracting extracellular DNA in biofilm matrix. Cold Spring Harb Protoc. 2010;2010:pdb.prot5456
31. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318-22
32. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623-33
33. Huber B, Eberl L, Feucht W, Polster J. Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z Naturforsch C J Biosci. 2003;58:879-84
34. Yilmaz C, Colak M, Yilmaz BC, Ersoz G, Kutateladze M, Gozlugol M. Bacteriophage Therapy in Implant-Related Infections: An Experimental Study. JBJS. 2013;95:117-25
35. Chaudhry WN, Concepción-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PloS one. 2017;12:e0168615
36. Abedon ST. Ecology of Anti-Biofilm Agents I: Antibiotics versus Bacteriophages. Pharmaceuticals (Basel, Switzerland). 2015;8:525-58
37. Abedon ST. Ecology of Anti-Biofilm Agents II: Bacteriophage Exploitation and Biocontrol of Biofilm Bacteria. Pharmaceuticals (Basel, Switzerland). 2015;8:559-89
38. Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP. et al. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell host & microbe. 2017;22:38-47.e4
39. Hesse S, Malachowa N, Porter AR, Freedman B, Kobayashi SD, Gardner DJ. et al. Bacteriophage Treatment Rescues Mice Infected with Multidrug-Resistant Klebsiella pneumoniae ST258. mBio. 2021 12
40. Esteves NC, Porwollik S, McClelland M, Scharf BE. The multi-drug efflux system AcrABZ-TolC is essential for infection of Salmonella Typhimurium by the flagellum-dependent bacteriophage Chi. Journal of virology. 2021
41. Gordillo Altamirano F, Forsyth JH, Patwa R, Kostoulias X, Trim M, Subedi D. et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nature microbiology. 2021;6:157-61
42. Gu Liu C, Green SI, Min L, Clark JR, Salazar KC, Terwilliger AL. et al. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio. 2020 11
43. Bogovazova GG, Voroshilova NN, Bondarenko VM. [The efficacy of Klebsiella pneumoniae bacteriophage in the therapy of experimental Klebsiella infection]. Zh Mikrobiol Epidemiol Immunobiol. 1991:5-8
44. Bogovazova GG, Voroshilova NN, Bondarenko VM, Gorbatkova GA, Afanas'eva EV, Kazakova TB. et al. [Immunobiological properties and therapeutic effectiveness of preparations from Klebsiella bacteriophages]. Zh Mikrobiol Epidemiol Immunobiol. 1992:30-3
45. Kaur S, Harjai K, Chhibber S. In vivo Assessment of Phage and Linezolid Based Implant Coatings for Treatment of Methicillin Resistant S. aureus (MRSA) Mediated Orthopaedic Device Related Infections. PloS one. 2016;11:e0157626
46. Kaur S, Harjai K, Chhibber S. Bacteriophage-aided intracellular killing of engulfed methicillin-resistant Staphylococcus aureus (MRSA) by murine macrophages. Appl Microbiol Biotechnol. 2014;98:4653-61
47. Tiwari BR, Kim S, Rahman M, Kim J. Antibacterial efficacy of lytic Pseudomonas bacteriophage in normal and neutropenic mice models. The Journal of Microbiology. 2011;49:994-9
48. Sarkisian SA. [Dynamics of phagocytic activity of leukocytes and protein fractions in dysentery patients treated with antibiotics and by a combined method (antibiotics, pentoxyl and bacteriophages)]. Zh Eksp Klin Med. 1968;8:99-103
49. Van Belleghem JD, Dabrowska K, Vaneechoutte M, Barr JJ, Bollyky PL. Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses. 2018 11
50. Comeau AM, Tétart F, Trojet SN, Prère M-F, Krisch HM. Phage-Antibiotic Synergy (PAS): β-Lactam and Quinolone Antibiotics Stimulate Virulent Phage Growth. PLOS ONE. 2007 2:: e799
51. Ojala V, Laitalainen J, Jalasvuori M. Fight evolution with evolution: plasmid-dependent phages with a wide host range prevent the spread of antibiotic resistance. Evolutionary applications. 2013;6:925-32
52. Kirby AE. Synergistic action of gentamicin and bacteriophage in a continuous culture population of Staphylococcus aureus. PloS one. 2012;7:e51017
53. Yilmaz C, Colak M, Yilmaz BC, Ersoz G, Kutateladze M, Gozlugol M. Bacteriophage therapy in implant-related infections: an experimental study. The Journal of bone and joint surgery American volume. 2013;95:117-25
54. Knezevic P, Curcin S, Aleksic V, Petrusic M, Vlaski L. Phage-antibiotic synergism: a possible approach to combatting Pseudomonas aeruginosa. Res Microbiol. 2013;164:55-60
55. Oechslin F, Piccardi P, Mancini S, Gabard J, Moreillon P, Entenza JM. et al. Synergistic Interaction Between Phage Therapy and Antibiotics Clears Pseudomonas Aeruginosa Infection in Endocarditis and Reduces Virulence. The Journal of infectious diseases. 2017;215:703-12
56. Danis-Wlodarczyk K, Vandenheuvel D, Jang HB, Briers Y, Olszak T, Arabski M. et al. A proposed integrated approach for the preclinical evaluation of phage therapy in Pseudomonas infections. Sci Rep. 2016;6:28115
57. Uchiyama J, Shigehisa R, Nasukawa T, Mizukami K, Takemura-Uchiyama I, Ujihara T. et al. Piperacillin and ceftazidime produce the strongest synergistic phage-antibiotic effect in Pseudomonas aeruginosa. Archives of Virology. 2018;163:1941-8
58. Chang RYK, Das T, Manos J, Kutter E, Morales S, Chan H-K. Bacteriophage PEV20 and Ciprofloxacin Combination Treatment Enhances Removal of Pseudomonas aeruginosa Biofilm Isolated from Cystic Fibrosis and Wound Patients. The AAPS Journal. 2019 21
59. Jansen M, Wahida A, Latz S, Krüttgen A, Häfner H, Buhl EM. et al. Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multi-drug resistant Acinetobacter baumannii. Scientific Reports. 2018 8
60. Verma V, Harjai K, Chhibber S. Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling. 2010;26:729-37
61. Bedi MS, Verma V, Chhibber S. Amoxicillin and specific bacteriophage can be used together for eradication of biofilm of Klebsiella pneumoniae B5055. World Journal of Microbiology and Biotechnology. 2009;25:1145
62. Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM. Therapeutic efficacy of bacteriophage and Baytril (enrofloxacin) individually and in combination to treat colibacillosis in broilers. Poult Sci. 2004;83:1944-7
63. Chhibber S, Kaur T, Sandeep K. Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PloS one. 2013;8:e56022
64. Gelman D, Beyth S, Lerer V, Adler K, Poradosu-Cohen R, Coppenhagen-Glazer S. et al. Combined bacteriophages and antibiotics as an efficient therapy against VRE Enterococcus faecalis in a mouse model. Research in microbiology. 2018;169:531-9
65. Chhibber S, Kaur S, Kumari S. Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. Journal of medical microbiology. 2008;57:1508-13
66. Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evolution, medicine, and public health. 2018;2018:60-6
67. Bao J, Wu N, Zeng Y, Chen L, Li L, Yang L. et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerging microbes & infections. 2020;9:771-4
68. Kuipers S, Ruth MM, Mientjes M, de Sévaux RGL, van Ingen J. A Dutch Case Report of Successful Treatment of Chronic Relapsing Urinary Tract Infection with Bacteriophages in a Renal Transplant Patient. Antimicrob Agents Chemother. 2019 64
69. Abedon ST. Phage-Antibiotic Combination Treatments: Antagonistic Impacts of Antibiotics on the Pharmacodynamics of Phage Therapy? Antibiotics (Basel). 2019 8
70. Ceyssens PJ, Minakhin L, Van den Bossche A, Yakunina M, Klimuk E, Blasdel B. et al. Development of giant bacteriophage ϕKZ is independent of the host transcription apparatus. Journal of virology. 2014;88:10501-10
71. Verma V, Harjai K, Chhibber S. Restricting ciprofloxacin-induced resistant variant formation in biofilm of Klebsiella pneumoniae B5055 by complementary bacteriophage treatment. J Antimicrob Chemother. 2009;64:1212-8
72. Jacobs WR Jr, Barletta RG, Udani R, Chan J, Kalkut G, Sosne G. et al. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science (New York, NY). 1993;260:819-22
73. Wilson SM, al-Suwaidi Z, McNerney R, Porter J, Drobniewski F. Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nature medicine. 1997;3:465-8
74. Galí N, Domínguez J, Blanco S, Prat C, Quesada MD, Matas L. et al. Utility of an in-house mycobacteriophage-based assay for rapid detection of rifampin resistance in Mycobacterium tuberculosis clinical isolates. Journal of clinical microbiology. 2003;41:2647-9
75. McNerney R, Mallard K, Urassa HM, Lemma E, Donoghue HD. Colorimetric phage-based assay for detection of rifampin-resistant Mycobacterium tuberculosis. Journal of clinical microbiology. 2007;45:1330-2
76. Pholwat S, Ehdaie B, Foongladda S, Kelly K, Houpt E. Real-time PCR using mycobacteriophage DNA for rapid phenotypic drug susceptibility results for Mycobacterium tuberculosis. Journal of clinical microbiology. 2012;50:754-61
77. Geiduschek EP, Sklar J. Continual requirement for a host RNA polymerase component in a bacteriophage development. Nature. 1969;221:833-6
78. Hemphill HE, Whiteley HR, Brown LR, Doi RH. The effect of rifampin on the production of beta22 phage by Bacillus subtilis. Biochemical and biophysical research communications. 1969;37:559-66
79. Lavysh D, Sokolova M, Slashcheva M, Förstner KU, Severinov K. Transcription Profiling of Bacillus subtilis Cells Infected with AR9, a Giant Phage Encoding Two Multisubunit RNA Polymerases. mBio. 2017 8
80. Price AR, Fogt SM. Effect of nalidixic acid on PBS2 bacteriophage infection of Bacillus subtilis. Journal of virology. 1973;12:405-7
81. Takeda Y, Oyama Y, Nakajima K, Yura T. Role of host RNA polymerase for lambda phage development. Biochemical and biophysical research communications. 1969;36:533-8
82. Beckman LD, Witonsky P, McCorguodale DJ. Effect of rifampin on the growth of bacteriophage T5. Journal of virology. 1972;10:179-86
83. Toussaint A, Lecocq JP. Sensitivity of bacteriophage Mu-1 development to rifampicin and streptolydigin. Molecular & general genetics: MGG. 1974;129:185-8
84. Sokolova EV, Ovadis MI, Gorlenko ZM, Khesin RB. Localization of streptolydigin resistant mutation in E. coli chromosome and effect of streptolydigin on T2 phage development in stl-r and stl-s strains of E. coli. Biochemical and biophysical research communications. 1970;41:870-6
85. Baird JP, Bourguignon GJ, Sternglanz R. Effect of nalidixic acid on the growth of deoxyribonucleic acid bacteriophages. Journal of virology. 1972;9:17-21
86. Taketo A, Watanabe H. Effect of nalidixic acid on the growth of bacterial viruses. Journal of biochemistry. 1967;61:520-2
87. Lopes A, Pereira C, Almeida A. Sequential Combined Effect of Phages and Antibiotics on the Inactivation of Escherichia coli. Microorganisms. 2018 6
88. Riveros-Moreno V. Effect of rifampicin on bacteriophage PM2 biogenesis. FEBS letters. 1975;51:249-52
89. Matsui T, Yoshikawa G, Mihara T, Chatchawankanphanich O, Kawasaki T, Nakano M. et al. Replications of Two Closely Related Groups of Jumbo Phages Show Different Level of Dependence on Host-encoded RNA Polymerase. Frontiers in microbiology. 2017;8:1010
90. Díaz-Muñoz SL, Koskella B. Bacteria-phage interactions in natural environments. Advances in applied microbiology. 2014;89:135-83
91. Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol. 2010;70:217-48
92. Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol. 2010;59:145-55
93. Brown S, Santa Maria JP Jr, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313-36
94. Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224-32
95. León M, Bastías R. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol. 2015;6:343
96. Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, Schuerch DW. et al. FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J Bacteriol. 2007;189:5658-74
97. Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep. 2016;6:26717
98. Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE. et al. Evolutionary consequences of intra-patient phage predation on microbial populations. Elife. 2014;3:e03497
99. Santander J, Robeson J. Phage-resistance of Salmonella enterica serovar Enteritidis and pathogenesis in Caenorhabditis elegans is mediated by the lipopolysaccharide. Electronic Journal of Biotechnology. 2007;100 -
100. Capparelli R, Nocerino N, Iannaccone M, Ercolini D, Parlato M, Chiara M. et al. Bacteriophage therapy of Salmonella enterica: a fresh appraisal of bacteriophage therapy. The Journal of infectious diseases. 2010;201:52-61
101. Smith SG, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett. 2007;273:1-11
102. Weidenmaier C, Peschel A, Xiong YQ, Kristian SA, Dietz K, Yeaman MR. et al. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. The Journal of infectious diseases. 2005;191:1771-7
103. Capparelli R, Nocerino N, Lanzetta R, Silipo A, Amoresano A, Giangrande C. et al. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PloS one. 2010;5:e11720
104. Tyrrell J, Whelan N, Wright C, Sa-Correia I, McClean S, Thomas M. et al. Investigation of the multifaceted iron acquisition strategies of Burkholderia cenocepacia. Biometals. 2015;28:367-80
105. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310-9
106. Monteiro R, Pires DP, Costa AR, Azeredo J. Phage Therapy: Going Temperate? Trends in microbiology. 2019;27:368-78
107. Chen Y, Batra H, Dong J, Chen C, Rao VB, Tao P. Genetic Engineering of Bacteriophages Against Infectious Diseases. Front Microbiol. 2019;10:954
108. Podlesek Z, Žgur Bertok D. The DNA Damage Inducible SOS Response Is a Key Player in the Generation of Bacterial Persister Cells and Population Wide Tolerance. Front Microbiol. 2020;11:1785
109. Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proceedings of the National Academy of Sciences. 2009;106:4629-34
110. Edgar R, Friedman N, Molshanski-Mor S, Qimron U. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Applied and environmental microbiology. 2012;78:744-51
111. Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:7267-72
112. Park JY, Moon BY, Park JW, Thornton JA, Park YH, Seo KS. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Scientific reports. 2017;7:44929
113. Kilcher S, Studer P, Muessner C, Klumpp J, Loessner MJ. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proceedings of the National Academy of Sciences. 2018;115:567-72
114. Hope CK, Packer S, Wilson M, Nair SP. The inability of a bacteriophage to infect Staphylococcus aureus does not prevent it from specifically delivering a photosensitizer to the bacterium enabling its lethal photosensitization. Journal of Antimicrobial Chemotherapy. 2009;64:59-61
115. Dong S, Shi H, Zhang X, Chen X, Cao D, Mao C. et al. Difunctional bacteriophage conjugated with photosensitizers for Candida albicans-targeting photodynamic inactivation. International journal of nanomedicine. 2018;13:2199-216
116. Moradpour Z, Sepehrizadeh Z, Rahbarizadeh F, Ghasemian A, Yazdi MT, Shahverdi AR. Genetically engineered phage harbouring the lethal catabolite gene activator protein gene with an inducer-independent promoter for biocontrol ofEscherichia coli. FEMS Microbiology Letters. 2009;296:67-71
117. Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11197-202
118. Krom RJ, Bhargava P, Lobritz MA, Collins JJ. Engineered Phagemids for Nonlytic, Targeted Antibacterial Therapies. Nano letters. 2015;15:4808-13
119. Pei R, Lamas-Samanamud GR. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Applied and environmental microbiology. 2014;80:5340-8
120. Bhattarai SR, Yoo SY, Lee SW, Dean D. Engineered phage-based therapeutic materials inhibit Chlamydia trachomatis intracellular infection. Biomaterials. 2012;33:5166-74
121. Mahichi F, Synnott AJ, Yamamichi K, Osada T, Tanji Y. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol Lett. 2009;295:211-7
122. Marzari R, Sblattero D, Righi M, Bradbury A. Extending filamentous phage host range by the grafting of a heterologous receptor binding domain. Gene. 1997;185:27-33
123. Lin TY, Lo YH, Tseng PW, Chang SF, Lin YT, Chen TS. A T3 and T7 recombinant phage acquires efficient adsorption and a broader host range. PLoS One. 2012;7:e30954
124. Cao J, Sun Y, Berglindh T, Mellgård B, Li Z, Mårdh B. et al. Helicobacter pylori-antigen-binding fragments expressed on the filamentous M13 phage prevent bacterial growth. Biochimica et biophysica acta. 2000;1474:107-13
125. Zhang H, Fouts DE, DePew J, Stevens RH. Genetic modifications to temperate Enterococcus faecalis phage ϕEf11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology. 2013;159:1023-35
126. Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. Journal of biotechnology. 2005;115:101-7
127. Kim KP, Cha JD, Jang EH, Klumpp J, Hagens S, Hardt WD. et al. PEGylation of bacteriophages increases blood circulation time and reduces T-helper type 1 immune response. Microbial biotechnology. 2008;1:247-57
128. Nobrega FL, Costa AR, Santos JF, Siliakus MF, van Lent JW, Kengen SW. et al. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Scientific reports. 2016;6:39235
129. Paul VD, Sundarrajan S, Rajagopalan SS, Hariharan S, Kempashanaiah N, Padmanabhan S. et al. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC microbiology. 2011;11:195
130. Hagens S, Bläsi U. Genetically modified filamentous phage as bactericidal agents: a pilot study. Letters in applied microbiology. 2003;37:318-23
131. Westwater C, Kasman LM, Schofield DA, Werner PA, Dolan JW, Schmidt MG. et al. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrobial agents and chemotherapy. 2003;47:1301-7
132. Hagens S, Habel A, von Ahsen U, von Gabain A, Bläsi U. Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrobial agents and chemotherapy. 2004;48:3817-22
133. Tagliaferri TL, Jansen M, Horz HP. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front Cell Infect Microbiol. 2019;9:22
134. Alibayov B, Baba-Moussa L, Sina H, Zdeňková K, Demnerová K. Staphylococcus aureus mobile genetic elements. Molecular biology reports. 2014;41:5005-18
Author contact
![]() Corresponding author: E-mail: xiaoshan1220com; Tel.: +86-0371- 22892865.
Corresponding author: E-mail: xiaoshan1220com; Tel.: +86-0371- 22892865.

 Global reach, higher impact
Global reach, higher impact