10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(2):552-571. doi:10.7150/ijbs.64373 This issue Cite
Review
Advanced high resolution three-dimensional imaging to visualize the cerebral neurovascular network in stroke
1. Department of Neurology, Xiangya Hospital of Central South University, Changsha, Hunan, China, 410008
2. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China, 410008
Received 2021-7-1; Accepted 2021-10-28; Published 2022-1-1
Abstract
As an important method to accurately and timely diagnose stroke and study physiological characteristics and pathological mechanism in it, imaging technology has gone through more than a century of iteration. The interaction of cells densely packed in the brain is three-dimensional (3D), but the flat images brought by traditional visualization methods show only a few cells and ignore connections outside the slices. The increased resolution allows for a more microscopic and underlying view. Today's intuitive 3D imagings of micron or even nanometer scale are showing its essentiality in stroke. In recent years, 3D imaging technology has gained rapid development. With the overhaul of imaging mediums and the innovation of imaging mode, the resolution has been significantly improved, endowing researchers with the capability of holistic observation of a large volume, real-time monitoring of tiny voxels, and quantitative measurement of spatial parameters. In this review, we will summarize the current methods of high-resolution 3D imaging applied in stroke.
Keywords: High resolution 3D imaging, Stroke, Neurovascular network, Synchrotron radiation, Two-photon microscopy, Photoacoustic imaging, Magnetic resonance imaging, Light-sheet microscopy
1. Introduction
Stroke is an ischemic or hemorrhagic disease mainly occurring in the cerebral microvascular system, leading to injury of cerebral vasculature and irreversible damage to the neurological function and even to death [1]. The importance of better understanding of its pathogenesis to help find more effective treatments cannot be overlooked. However, neurons that stretch in multiple directions and blood vessels make up a complex spatial network. The research of stroke is no longer satisfied with the information only on two-dimensional planes. Their nature can only be demonstrated in three-dimensional (3D) imaging. Thus, an advanced 3D brain atlas is released as an assist for the study of neural 3D structures [2]. Meanwhile, keeping the tight and orchestrated neurovascular relationship between nerves and blood vessels is the basis for maintaining normal physiological activities of the brain. In stroke, a specific association will emerge. For example, angiogenesis after vascular injury depends on repair-associated VEGF-expressing microglia [3], and endothelial cells of cerebral vessels secrete trophic factors that can be directly neuroprotective [4].
Therefore, the neural networks, cerebrovascular networks, and interaction between them have become an intriguing subject of study for stroke. As noted above, the temporal and spatial studies using microscopic observation methods with multiple resolutions (from the nanoscale to the microscale) provides new insights beyond perspectives from biochemical analysis and genetic testing. Biochemical analyses such as gel electrophoresis and chromatography, and genetic tests such as polymerase chain reaction can quantitatively or qualitatively understand regional effector molecules to explain pathological changes, but lack visualization of spatial distribution and relationships. Thus, the intricate spatial interaction of neurons and vessels is unobtained. For decades, microscopes designed for visualization naturally shift their focus from flat to spatial relationships, and even to spatial dynamics, leading to the development of the imaging from 2D to 3D and even 4D (3D + time). Wang K et al. simultaneously visualized the flow of blood cells through thousands of blood vessels in the cortex of awake mice [5].
Take the study of blood vessels as an example. Dysfunction of small blood vessels is an important process of stroke, which often includes changes in cerebral blood flow and vascular structure [6, 7]. Histological studies are commonly employed for standard analysis of biological specimens. However, histological analysis as a two-dimensional technique has some inherent limitations: the 3D structural integrity of the brain is destructed by the sectioning, bias or error may also occur in slice selection which is based on the assumption that the sample is relatively homogeneous, and sectional observation is discontinuous and lacks description of spatial relations [8, 9]. And the view of the electron microscope is too small. These disadvantages have led researchers to call for the utilization of high-resolution 3D imaging and stimulated the development of the higher resolution of magnetic resonance imaging (MRI), the two-photon microscope, the computer imaging model, and suchlike [10].
The development of the microscope closely followed the improvement of resolution. Microscopy and ultrahigh-resolution microscopy techniques do help explain the brain at the nanoscale micro level, which is used to discover new intracellular structures, analyze biomolecular interactions, and quantify molecular counts [11]. Representative work was that Zhuang et al. analyzed the membrane-associated periodic skeleton in nerve cells and observed that the ring structure of actin formed a periodic structure with an interval of 180-190 nm [12]. However, at the micron level (1-10 μm), a 3D imaging platform is still needed to solve the spatial relationship between hard and soft tissue and provide 3D quantitative characterization [13]. The use of new imaging mediums, like synchrotron radiation, or new modes, like light-sheet microscopy has greatly improved the resolution of 3D imaging. Based on different methods, including light, acoustics, and nuclear magnetic resonance, high-resolution 3D imaging technology is gradually being applied to stroke to investigate cerebral refined structure, function, and metabolic status, etc.
2. Light
Synchrotron Radiation
SR's wide spectrum ranges from infrared to hard x-rays. X-rays, which are the most commonly used SR source in biomedicine, make better use of imaging with smaller wavelengths and greater penetration. This advantage is enhanced by synchrotron radiation (SR) X-ray which is the electromagnetic wave emitted along the tangent direction of the orbit when the magnetic field of the storage ring changes the motion direction of electrons at sub-light speeds. It has the characteristics of high intensity, monochromaticity, collimating coherence, and high spatial resolution around 1,000 times higher than that of conventional X-Ray absorption imaging [14]. There are currently more than 20 synchrotrons worldwide [15]. The third generation of high-performance light sources consists of two types: one with high electron energy (6 GeV), such as American Light Source (APS) [16], and the European Synchrotron Radiation Facility (ESRF) [17], another with medium electronic energy (2-3 GeV) such as Swiss Light Source (SLS) [18], and Shanghai Synchrotron Radiation Facility (SSRF) [19]. Based on those, synchrotron radiation light source was developed to the fourth generation of free-electron laser light source, like Swedish Max-Ⅳ [20]. But third-generation sources are more commonly used, such as SSRF which produces X-rays from the electronic storage ring with an acceleration energy of 3.5 GeV and an average beam of 200 mA. With a tunable energy range of about 8-70kev, the resolution eventually reaches micron-scale or even sub-micron scale [21, 22].
As the importance of brain microvasculature in stroke, SR has provided a mighty impetus in the study of stroke [14]. Conventional SR uses absorption angiography, including K‐edge subtraction angiography (KESA) for large structures and single‐energy temporal subtraction (SETS) for higher resolution [21, 23, 24]. Phase contrast imaging (PCI) can detect micron or submicron microvessels without angiography, which can be divided into inline X-ray phase contrast imaging (ILPCI), grating interferometry (GI), and crystal interferometry (CI), etc [25, 26].
3D imaging using absorption angiography is mainly used in vivo. Immunohistochemical staining can only show the number of cerebral vessels rather than the number of perfusion vessels, but SR can realize dynamic observation of brain microvessels [27]. Yang G et al. continuously achieved the dynamic detection and assessment of thrombolysis and recanalization at a 9μm resolution in rats [28].
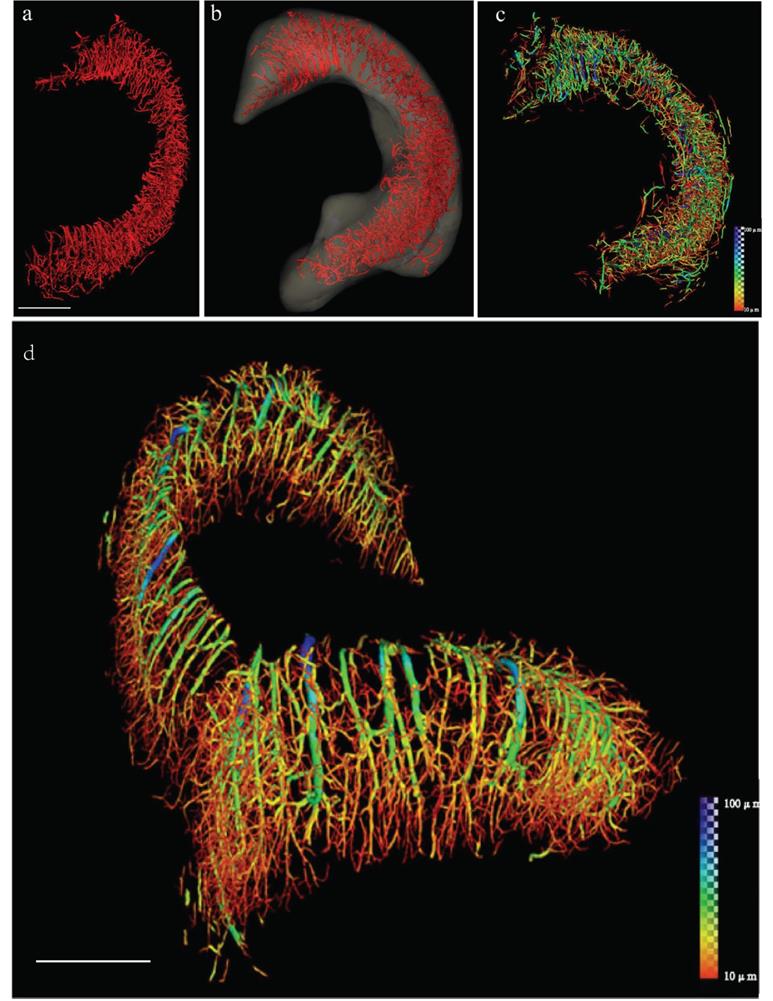
Phase contrast 3D imaging is mostly used in vitro, which is more suitable for neurovascular networks in stroke, and can provide more multi-dimensional and multi-scale information without contrast agents compared with absorption imaging [29]. Unlike absorption contrast imaging (ACI) where incomplete filling or leakage of the contrast agent will inevitably affect the quality of the image obtained [30], PCI shows good applicability for vascular structure [31, 32]. In addition to vascular structure and hemodynamics, PCI can also be used for semiquantitative studies on angiogenesis. It was found that the perfusion vascular density in the area around the ischemic infarction of middle cerebral artery occlusion (MCAO) rats treated with Dl-3-N-butylphthalide was higher than that of the silicone oil group, indicating that butylphthalide treatment promoted angiogenesis in the area around MCAO infarction [33]. In MCAO mice, ILPCI was used to reconstruct the 3D vascular structure, showing that the vascular trees are arranged in a tangled pattern to form a network, identifying vessels with diameters of about 11.8 microns [14]. Epilepsy also presents with microvascular changes like stroke, SR 3D imaging was used to measure microvessel parameters, including vessel surface area, number of vascular segments, and number of vascular bifurcations, with a pixel size of 5.2 μm (Figure 1) [34]. Combined with microcomputed tomography (μCT), SR was used to detect the three-dimensional structural changes of brain microvessels in rats 4 hours, 6 hours, 3 days, and 18 days after ischemia [22].
For nerves, Golgi-cox staining is a basic method [35]. However, based on changes in electron density, Mareike Topperwien et al. successfully visualized and automatically located about 106 nerves in different layers of the cerebellum without any additional staining [36]. Fonseca MC et al. combined μCT SR with Golgi-Cox mercury-based impregnation protocol in the brain of mice, achieving an adjustable pixel size of 0.82 μm for high resolution/small field of view (FOV) imaging without tissue slicing or clearing [37].
At present, SR 3D imaging is still mostly used in microvascular studies in vitro, but the interaction between microvascular network and nervous system in stroke is inevitable in imaging. The imaging of soft tissues by using no X-ray contrast agent can be easily redirected and resegmented virtually in a computer with a resolution of 5-10 μm [38]. The update of μCT allows scientists to identify neurons, glial cells, and vascular systems in the 3D imaging of SR, and effectively measures the cell spacing and distance between cells and blood vessels [39]. Anna Khimchenko et al. used hard X-ray in ESRF to make the effective pixel size up to 130nm, which can distinguish subcellular structures such as nuclear membrane and nuclear pore of Purkinje cells [40]. However, improving the resolution to observe subcellular structures necessarily confines the sample to the micron scale and takes time to match the sample's anatomical markers [9].
In addition to the 3D reconstruction of microvascular network and nervous system, multiple other techniques based on SR can be combined to capture various types of data in a high-resolution manner. For instance, SR-infrared spectrum (SR-IR) or SR micro-X-ray fluorescence (SR-μXRF) can observe the biochemical properties of neurons and vessels. Metabolic changes occurring in neurovascular network are markers of stroke [41]. SR-IR is 100-1000 times brighter than the traditional light source, enabling fourier transform infrared spectro-microtomography (FTIR-μCT) to reconstruct the image in the micron scale. By SR X-ray correction [42, 43], IR was used to construct a 3D mapping of in-situ content of glucose, glycogen, and lactate in the brain of mice [44]. Transition metal homeostasis, such as copper, iron, and zinc, is an important property of brain and plays an important role in stroke and other brain diseases [45-47]. As studies on ion distribution in stroke increase gradually like post-stroke neuroplasticity or ischemic stroke, SR-μXRF has become a powerful technique for ion distribution in biological samples by utilizing the secondary characteristic radiation induced by X-ray [48, 49]. Although not yet thoroughly used in the brain 3D imaging, SR-μXRF will show great potential in the field of stroke.
Two-photon/Multi-photon microscopy
Based on the nonlinear optical microscope driven by the high peak power that is possible with ultrashort pulses, the two-photon microscopy (TPM) first appeared in 1990, invented by Webb, etc [50]. TPM uses near-infrared (NIR) light instead of the visible or ultraviolet light used by conventional laser confocal microscopy. Under the high photonic flux required by the nonlinear optical process, the fluorescent molecule absorbs two long-wavelength photons at the same time, and then emits a photon with a shorter wavelength after reaching the excited state, achieving imaging. Because two-photon or multi-photon imaging requires high photon density of exciting light, only fluorescent molecules in very small volumes in and around the focal point can be excited, which enables TPM and multi-photon microscopy (MPM) to have very high three-dimensional resolution [51]. Currently, a fast high-resolution two-photon microscope invented by Cheng H's team can achieve a horizontal resolution of 0.64 μm, an axial resolution of 3.35 μm, and an imaging frame frequency of 40 Hz [52]. At the same time, short pulses with longer wavelengths are used to allow two or more atoms to be absorbed to reduce scattering and achieve deeper imaging [53]. Compared with conventional laser confocal microscopy, the n-photon absorption probability decreases rapidly, thus photobleaching and phototoxicity occur only at the focus [54]. Other variations were also developed like third-harmonic generation [55].
Reconstruction of the 3D angioarchitecture of the hippocampus in normal rat brain by SR. (a, b) By SR-ILPCI reconstruction, the spatial structure distribution of the right hippocampal three-dimensional network was extracted from the whole brain (a) and accurately outlined (b) (scale bar, 600 μm). (c) Hippocampal vessel diameter rendering identifies the distribution of vessels with different diameters. (d) The original distribution of intracerebral vessels within the hippocampus with continuous pseudocolor changes depicting the distribution spectrum of vessel diameters, ranging from 10 μm to 100 μm (dark blue) (scale bar, 700 μm). Reproduced with permission from Reference 34.
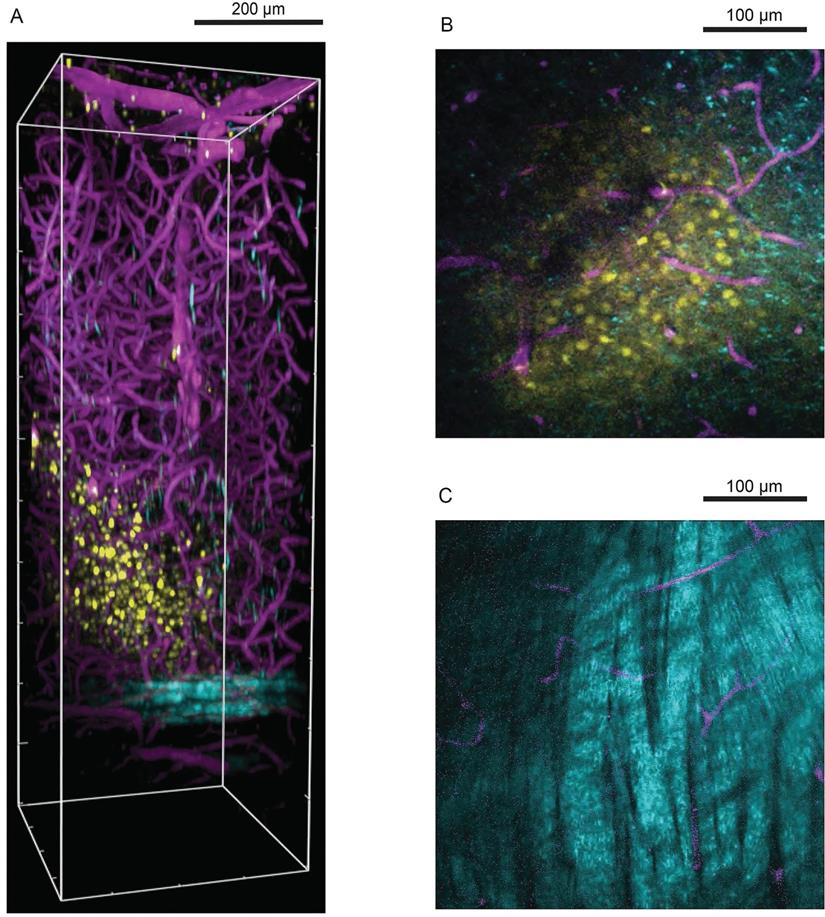
The application of TPM in stroke is primarily to explore cerebral information in live behaving mammals, The confocal laser scanning microscope allows to scan structures about 100 mm below the surface, but the imaging of structures such as the neuron group requires deeper penetration, about 300-500 micrometers below the surface. TPM/MPM can alleviate this problem by providing high-resolution, non-invasive, deep tissue observations [56]. But to scan the brain, the covered opaque skull must be thinned down to a thickness of 15 µm or partially removed to form a cranial window [57]. Subcortical imaging of the neocortex can now be achieved by improving coverslip to minimize the effect of spherical aberration [58]. The methods of fluorochromes administration include intravenous delivery, electroporation, and virus transduction; and for cerebrovascular labeling, the intravenous delivery of a fluorescent dye can efficiently stain an entire vascular tree from the organ [59]. The detection of neuronal activity is necessary for the studies of injured brain, so the TPM/MPM applied to neurons in stroke is mostly functional. Calcium imaging is an essential tool for neural imaging [60]. TPM calcium imaging was used to study how microglia influence neuronal activity after stroke [61]. Lamiae Abdeladim et al. designed a chromatic multiphoton serial (ChroMS) microscope to provide high-contrast multicolor imaging and tracked 261 chromophores formed by 1055 astrocytes in the 1.2×2×1 mm3 area, with the voxel size of only 0.54×0.54×1.5 µm3 [62]. In addition to calcium ions, TPM can also visualize the distribution of magnesium, zinc, and nickel ions [63, 64]. For deeper parts, with a two-photon absorption fluorescent probe, microvessels more than 1mm deep beneath the skin can be imaged in three dimensions [65]. Gradient-index (GRIN) fiber probe implants can be used to go much deeper, distinguishing synapses in the hypothalamus that are 5mm deep[66]. MPM enabled high-resolution imaging in deeper brain areas (Figure 2) [67]. Chris Xu et al. successfully used a three-photon microscope to image the vascular structure in the hippocampus of mice and neurons labeled with red fluorescent protein, with a resolution of about 4.4μm at a depth of about 900 μm [68].
Another characteristic application of TPM is the miniaturization of free-moving mice. It makes up for the absence of a benchtop microscope in the study of long duration behavior or social behavior. Baris N. Ozbay et al. used head-mounted miniature TPM to observe about 200 oligodendrocytes labeled with a green fluorescent protein in the FOV with a diameter of 240μm, with a transverse resolution of 1.8 μm and an axial resolution of 10 μm [69]. However, the miniaturization of TPM still requires effective fluorescence collection, compact and fast scanning mechanisms, and reduction of motion artifacts [70]. Recently, Angus Silver's team achieved real-time movement-corrected 3D two-photon imaging with submicrometer precision [71].
It is worth mentioning that wavefront distortions constrict the achievable resolution with penetration depth, but adaptive optics systems (AOS) enable wavefront correction. Meanwhile, it can reduce the power requirement for two-photon imaging of brain activity in awake mice by more than 30 times [72]. AOS was applied to high-speed 3D dynamic imaging of calcium, blood vasculature, microglia, and structure of the neuronal network in vivo mammalian brain [73]. TPM/MPM is still limited by the speed at which scanning can be realized. Multi-focal excitation generated by adaptive optics is an improved method [74].
Light-sheet microscopy
Due to the photobleaching and phototoxic effects of excitation light, most optical imaging modes cannot achieve the long-term observation of living cells and the recording of intracellular life activities [75]. The light-sheet microscopy (LSM) mode explained by Ernst Stelzer has become an important turning point in the development of imaging technology. It uses excitation and detection objectives arranged at right angles to form layered light and separate the excitation and emission light paths to ensure selective excitation of specific planes; thus it reduces ineffective exposure, photobleaching, and phototoxicity [76]. In 2004, a light sheet microscope named Selective Plane Illumination Microscope (SPIM) was formally born. It is widely used for 3D imaging of large samples due to its huge advantage of shortening imaging time and reducing phototoxicity [77]. Later, scientists are seeking a resolution that meets both the large FOV and the diffraction limit close to the Rayleigh Abbe rate [78, 79]. The thinner a Gaussian beam is, the higher the axial resolution is. The Gaussian beam whose cross-section radius track is hyperbolic makes the area with a relatively thin thickness limited, which means that if the light sheet is thin, the light sheet is short[80]. It means that it is difficult to have both the FOV and the resolution. In contrast to the Gaussian beam, Bessel beam has no diffraction and low phototoxicity, enabling faster acquisition speeds and higher signal-to-noise ratios. Betzig Lab first applied them to light-sheet microscopes, resulting in an isotropic resolution of 300 nm [81]. Later, the lattice light-sheet microscope (LLSM) used a spatial light modulator to adjust the shape of the beam entering the illumination objective to multi-beam Bessel interference, creating an optical lattice, and finally forming an ultra-thin light sheet. It makes it possible to achieve high spatiotemporal resolution, large FOV, and low phototoxicity [82]. Multi-view imaging is implemented in diSPIM and isoView, and becomes a potential candidate for high-volume high-resolution imaging [83, 84]. For example, isoView microscopy can achieve an isotropic spatial resolution of 400 nm, and the volume throughput per second is greater than 108 µm3 [84]. Two-photon excitation allows them to reach greater depths [85].
Three-photon microscopy in visualizing cells deep in mouse visual cortex relative to blood vessels and white matter. (A) Side view perspective of a three-dimensional reconstruction of the three-photon image volume depicting the field of cells with respect to blood vessels and the white matter. Oregon Green labelled cells in layers 5-6 are shown in yellow, blood vessels labelled with Texas Red Dextran are shown in magenta and cortical white matter imaged with THG is shown in cyan. x-y-z volume dimensions: 375 µm × 375 µm × 1100 µm. (B) Single z plane three-photon image at 750 µm below brain surface showing labelled cells and blood vessels. (C) Single z plane three-photon image in the white matter at 900 µm below brain surface. Reproduced with permission from Reference 67.
LSM is limited by artifacts when used in large tissue of stroke brains. For example, if the sample contains high-density structures such as pigments that block excitation light, or lipids that cause light scattering and uneven illumination, it will result in some dark lines. Tissue removal is currently a common solution. Its effect of improving penetrating power and reducing light scattering is well adapted to the three-dimensional imaging of brain tissues, especially the whole brain so that people can obtain volume images of the entire brain without physical slicing. It should be noted that the tissue removal technique is not limited to LSM, but is used in a variety of microscopy techniques such as MRI [86]. The earliest attempt was to use a mixture of benzyl alcohol and benzyl benzoate (BABB/Murray's clear) to process the specimen to observe the brain neural network, and at present, the commonly used tissue removal techniques can be divided into three methods: hydrophobic, hydrophilic and hydrogel-based [87]. For example, CLARITY [88] and DISCO [89] can obtain sub-cell-level resolution. Samples with or without tissue removal are most commonly embedded in agarose or placed in a liquid-filled chamber, but not limited to this [90].
The LSM played a great role in imaging the structure and function of nerves in stroke. It enables researchers to perform whole-brain functional imaging and optogenetics research at a single-cell resolution [91, 92]. Observation objects changed from rodents to humans [93]. The use of ultra-microscopy beyond the diffraction limit will increase the resolution by nanometers. The joint use of expansion microscopy (ExM) and LLSM can visualize and quantify nerve groups, subcellular structures, and synaptic connections with a resolution of 60 × 60 × 90 nm3 [94]. And the lateral average accuracy obtained by combining single-molecule positioning technology can reach 7.2 nm [95]. Non-invasive research has always been the goal. Swept confocally-aligned planar excitation (SCAPE) microscopy through the cranial window can monitor neuronal network activity in the brain of awake and freely moving mice, and distinguish single capillaries of 5-10 μm in depth at 140 μm [96]. And SCAPE2.0 penetration depth can reach 450 μm [97]. Near-Infrared II Window LSM has further realized the live imaging of the head in depth at 750 μm [98].
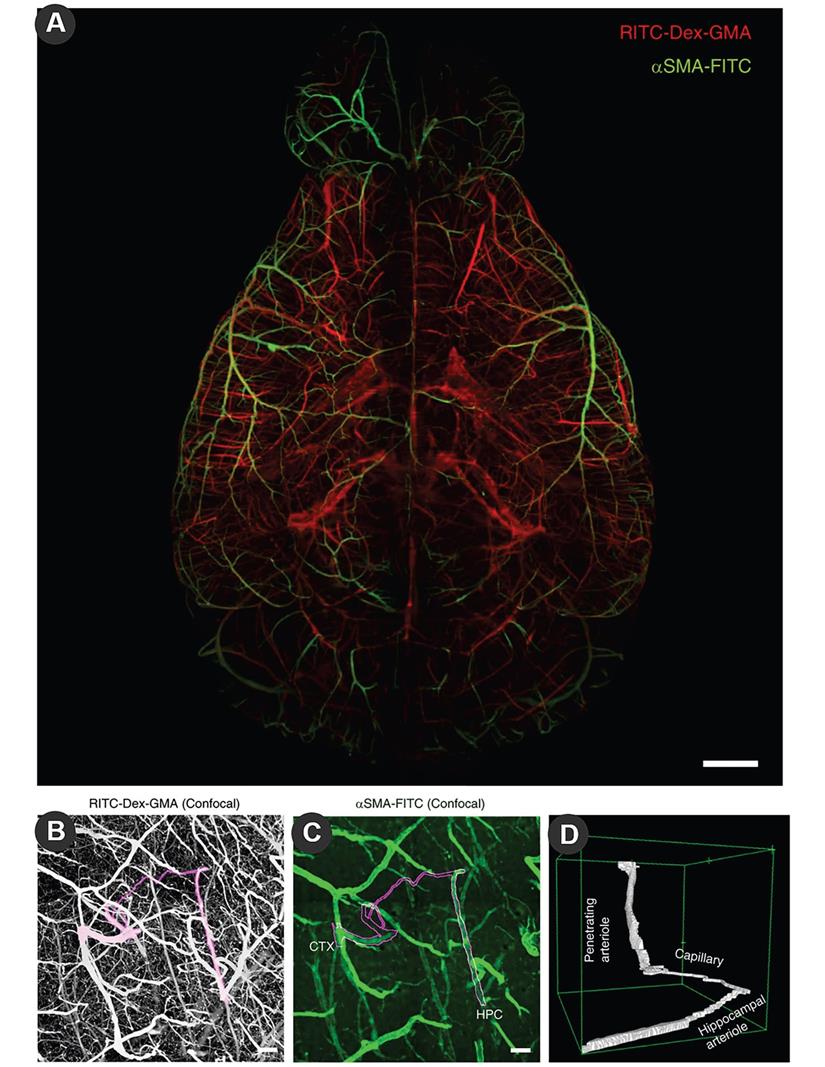
The vasculature of the brain can also take full advantage of LSM through dye filling strategies, such as the special shape of cortical microvessels [99], and changes in blood vessels after cerebral ischemia [100]. The introduction of tissue removal provides three-dimensional imaging of the vasculature. TubeMap designed by Nicolas Renier uses iDISCO to construct, analyze and visualize the blood vessel map with micron resolution containing 100 million blood vessel segments [101]. Fluorescein isothiocyanate (FITC)-conjugated albumin was injected with light film microscopy to assess large-scale microvessels in the ischemic brain [102]. Jonas Gregorius et al. determined microvascular network characteristics after focal cerebral ischemia with a resolution of 2 µm, including microvascular length density, branching point density and microvascular tortuosity [103]. In order to reduce the false negatives of vascular imaging of conventional molecular characterization, the newly developed SeeNet method improves the dye to enable three-dimensional imaging of almost whole brain microvessels (Figure 3) [104].
Simultaneous three-dimensional imaging of neurons and vessels is an important development direction for stroke. LSM can simultaneously study qualitatively and quantitatively neurovascular networks [105]. So far it has been used in neurodegeneration and neurodevelopment [106, 107].
In the existing high-resolution large-sample imaging methods, it is a common solution, like IsoView LSM, to use a tiling strategy to reconstruct 3D samples of the entire nervous system, but it also means complex graphics processing algorithm [108]. Better strategies are being studied, such as the axially swept light-sheet microscopy, which maintains an isotropic resolution of 390 nm over a larger FOV [109]. In addition, a multiangle-resolved subvoxel selective plane illumination microscope (Mars-SPIM) can collect an atlas of the entire mouse brain in 30 minutes with an isotropic resolution of 2μm [110]. A new algorithm for LSM devised by Shroff H's team realized imaging processing time ten-fold to several thousand-fold faster compared to previous methods [111]. Of course, LSM still needs technology that can image large samples with a co-directional resolution of less than 1μm and a faster and more accurate algorithm for processing data sets over tens of megabytes [87].
Imaging System Combined Serial sectioning
Accurate observations of deep nerves and vessels take an increasingly integral role in stroke, and then a new idea boldly alternates imaging with slicing, overcoming depth to produce high-resolution 3D images of the whole brain. Micro-optical sectioning tomography (MOST) devised by Luo Q et al. is a new type of neural optical imaging system, which can simultaneously scan ultra-thin slices processed by diamond cutter for optical imaging and obtain sub-micron brain data with high throughput [112, 113]. When the thickness of the tissue slice is less than the optical diffraction limit, the axial resolution is the slice thickness, and the transverse resolution is determined by the optical imaging configuration. MOST allows 3D imaging of neurons and neuronal connections to reconstruct a digital mouse brain with a voxel size of 0.33×0.33×1.0 μm3 [112]. 3D representation of cerebrovascular network also shows great advantages in quantitative analysis of microvascular distribution at the resolution of micron [114]. The improved Nissl staining method can simultaneously image brain cells and angiography at a resolution of 1 micron voxel [115]. MOST can obtain a voxel resolution of 0.35 × 0.4 × 1 µm in MCAO mice to reconstruct microvasculature [116]. The combination of rabies virus markers with fluorescent MOST (fMOST) can show the input of single neurons in the whole brain, providing a reliable option for optogenetics. The imaging speed was also improved, and the entire mouse brain could be imaged in an hour at a voxel size of 1.30×1.30×0.92 μm3 with a section thickness of 40 μm, without optical clearance [117]. The drawback is that the data set is too large and thus makes data processing more difficult, but this is the inevitable result of this kind of idea.
In fact, slicing and imaging combined with new strategy still show vast potential, and multiple techniques appeared based on this concept, such as serial two-photon tomography (STP), block-face serial microscopy (FAST). The resolution of FAST can reach 0.7×0.7×5 μm3, and the images are collected at z-sampling intervals of 50-100μm; its rapid imaging ability reduces the time of imaging the mouse brain to 2.4 hours[118, 119]. Mereuta OM et al. analyzed ultrastructure of clots in acute ischemic stroke at a resolution of 50 nm with FAST [120]. STP can also conduct 3D imaging of the brain at the resolution of micron and submicron, and avoid embedding the tissue to weaken the intensity of fluorescence signal [121]. It facilitates the construction of 3D neural connections [13]. Later studies were more in-depth. For example, Stowe AM et al. used STP to track CD8+T cells to visualize the whole brain of post-stroke neuroinflammation (Figure 4) [122], and monitor the migration of B cells to visualize neurogenesis and functional recovery after stroke [123]. Deep learning can help to reconstruct the 3D blood vessel and white matter network at the same time [124]. However, STP also has manifest disadvantages. It needs to express the components of fluorescence and imaging is too time-consuming [125].
Optical coherence tomography
Optical coherence tomograph (OCT) is an intravascular imaging modality that utilizes near-infrared light to generate images by interferometrically measuring the amplitude and delay of reflected or backscattered light. It stands out because it provides 3D imaging of deep brain tissue at a micron-scale resolution, a reasonable field of view of several millimeters, and feasible temporal resolution, enabling real-time measurements [126]. Traditional 3D OCT was invented for ophthalmology with resolution limited to ~10 μm [127]. And it has been gradually introduced into cerebrovascular imaging, relying on tracking of blood cells [128]. Recent OCT systems can get uniform spatial and axial resolution of 2.48 μm in trophoblast organoids [129]. Moreover, OCT endoscopy enables 3D microscopy of the intracranial arterial wall at a resolution approaching 10 µm and a penetration depth of ~ 1 mm [130]. With specific microprobe, endoscopic OCT is probably capable of < 3 µm resolution and potentially better imaging contrast [131].
In stroke, the most promising area of OCT is hemodynamics. Optical Doppler tomography (ODT) combined Laser Doppler Flowmeter to OCT, which permits 3D visualization of cerebrovascular networks and quantitative measure for cerebral blood flow (CBF) over a large FOV (e.g., a volume of 2×3 mm2 with >1 mm of depth) [132]. Pan Y et al. used contrast-enhanced ODT with intralipid to achieve an axial solution of 1.8 μm in visualization of microvascular trees and CBF [133]. Optical coherence tomography-based angiography (OCTA) estimates the scattering from moving red blood cells, visualizing functional micro-vessel networks in vivo in a rapid, non-invasive fashion [134]. Approximately 10 μm resolution can be obtained in visualizing capillaries on the surface of the pial artery [135]. It goes beyond Doppler OCT in that spatial and temporal changes in the inner diameter of lumen can be simultaneously monitored by OCTA [135]. However, OCTA does not distinguish descending vessels from ascending vessels like ODT. Normalized field autocorrelation function was utilized to resolve it.
Dynamic light scattering method
Laser speckle imaging
Laser speckle imaging (LSI) is a full-field imaging technology for mapping blood flow with high spatio-temporal resolution using simple apparatus, in which areas of higher flow are indicated by lower speckle contrast values [136]. LSI can resolve individual blood vessels and provides relative measures of CBF and can be used in cases such as penumbra of cerebral ischemia [137]. It has a resolution of 100 μm per se and is rarely used alone for 3D imaging [138]. Another limitation is the shallow imaging depth, A normal solution, using microendoscopy, can make a time-lapse blood flow detection in subcortical regions of the brain [139].
Large-scale molecular phenotyping-compatible 3D imaging of cerebral vasculature. (A) Maximum projection of light-sheet fluorescence microscopic images of the entire vasculature in a SeeNet-treated brain (1.6× zoom, ×0.63 objective lens, numerical aperture: 0.15, working distance: 87 mm) into a single stacked photo. Casted vessels are shown in red. Arterioles immunolabeled with anti-α smooth muscle actin (anti-αSMA) are shown in green. Scale bar = 1 mm. (B) Maximum projection of confocal images of casted vasculatures from the pia to the hippocampus. A cortico-hippocampal vascular path is shown in magenta. Scale bar = 100 μm. (C) The anti-αSMA immunosignal in the same field is shown on a green scale. Scale bar = 100 μm. (D) A 3D-rendered view of the cortico-hippocampal vascular path. Reproduced with permission from Reference 104.
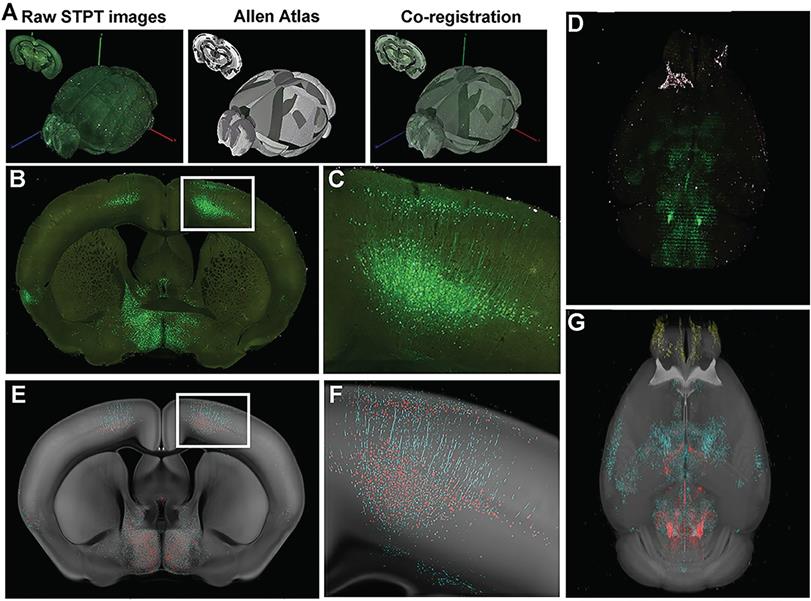
Serial two-photon tomography of the whole mouse brain. (A) Overview of STP data acquisition and registration. (B) Representative coronal section raw fluorescence image, with (C) expansion of areas identified in white box in B showing green fluorescent protein (GFP)+ neuronal cell bodies and processes. (D) Coronal images are stacked into a 3D representation and co-registered to the Common Coordinate Framework (CCF 3.0; Allen Institute for Brain Science) Atlas. (E) Probability map output of the same section shown in (B) depicting pixels classified via machine learning (ML) as either GFP+ cell bodies (red) or GFP+ neuronal processes (cyan) overlaid onto the corresponding section from the atlas (CCF 3.0 average template) shown in gray. (F) Enlarged image of boxed area in (E) showing fine detail and accurate classification of neuronal cell bodies and processes corresponding to the raw fluorescence image shown in (C). (G) 3-D rendering of the probability maps corresponding to the raw fluorescence images of the brain shown in D. GFP+ cell bodies, red, GFP+ neuronal processes, cyan, atlas images, gray. Reproduced with permission from Reference 122.
It measures changes in blood flow, blood volume, and hemoglobin oxygenation, thus it can be combined with OCT to 3D visualize hemodynamics and oxygenation of vessels at 0.7-0.9mm under the cortical surface at 10μm resolution simultaneously [137]. Furthermore, multimodal imaging that combines LSI, OCT, and calcium fluorescence imaging can simultaneously detect cortical hemodynamics, cerebral metabolism, and neuronal activities of the animal brain [140].
Diffused optical imaging
Diffuse optical imaging uses NIR to measure physiology millimeters to centimeters deep in the tissue by measuring spatial variations in tissue refractive index and bulk scattering properties. It is used for structural, functional, and metabolic 3D visualization in a low resolution, including neural activity [141], neuronal death, and brain inflammation [142]. Diffuse optical tomography (DOT) images changes in cerebral hemoglobin concentration without providing anatomical information. It's powerful in imaging deep, but the resolution is currently stopping at the millimeter level [143, 144]. Similarly, diffuse optical correlation tomography (DCT) monitors fluctuation of speckle patterns caused by moving red blood cells to take hemodynamic measurements [145]. DCT obtains 3D images of CBF in tissues (~4mm) deep below the cortex in a resolution of ~0.5mm [146]. Detection of neuronal cell motion will help DCT achieve sub-second latency [147]. DCT combined with LSI allows for probing depths up to ∼10 mm [148].
3. Acoustics
Photoacoustic imaging
Photoacoustic imaging (PAI) combines high contrast based on spectral specificity with the high spatial resolution of ultrasound imaging. PAI can be viewed as ultrasound images whose contrast depends on optical absorption rather than the mechanical properties of tissue [149]. In PAI, the most used light is visible light and NIR light with a wavelength between 550 nm and 900 nm. The near-infrared spectrum ranges from 600nm to 900 nm and has a maximum penetration depth of several centimeters [150]. Particularly, with the help of contrast agents, the imaging depth can reach 5-6cm [151-153]. The theoretical resolution of NIR-PAI is 200 nm, that is, Rayleigh/Abbe diffraction limit [154]. However, due to the attenuation effect of photoacoustic, the resolution decreases with the increase of the penetration depth. PAI can be divided into optical-resolution (OR) and acoustic-resolution (AR) PAI. OR-PAI can reach a horizontal resolution of 0.32 mm and a penetration depth of 1mm, while AR-PAI can reach a penetration depth of 3 mm but the resolution is reduced to 45 mm [155]. The resolution and penetration depth are like contradictory properties in PAI, while the resolution can be less than 10 nm at depths of several hundred microns, but reduces to submillimeter at depths of centimeters [156]. In recent years, the wavelengths of NIR-Ⅱ (wavelength 1000-1700 nm), which allow higher pulse energy, have also been increasingly valued in deep tissue imaging [157]. PAI doesn't require cranial windows, and even the use of tiny optical fibers for deep information is minimally invasive [158]. Additionally, endogenous tissue chromophores like oxyhemoglobin and melanin provide abundant absorption contrast, for different tissues absorbing light in varying wavelength ranges [159]. PAI methods include Photoacoustic tomography (PAT), Photoacoustic microscopy (PAM), Photoacoustic endoscopy, etc., in which PAT is the most common and least restrictive method.
PAI has been adopted to clinical monitoring of stroke [160]. In animal models, PAI can be used for structural and functional imaging, but mainly for the latter. Nie L et al. used PAM to observe the microvascular changes in ischemic stroke in the infarcted area at the microscopic level [161]. In addition to hemodynamic parameters, oxygen saturation and angiogenesis can be measured by PAI as well [162]. The measurement of blood oxygenation is its most important function [163]. Song Hu et al. reconstructed the microvascular network in the ears of nude mice with PAM and quantified local oxygen consumption, with an axial resolution of 15 μm [164]. Huabei Jiang et al. monitored intracranial hemorrhage in vivo noninvasively by photoacoustic tomography, and the spatial resolution of the system was 180 μm, possibly due to the limitation of attenuation effect [165]. The algorithm update can solve this problem. Héctor Estrada et al. corrected the image distortion caused by the PA microscopy due to the skull through the virtual craniotomy deconvolution algorithm and then depicted the 3D skull and vascular anatomical structure [166]. The excitation light wavelength was 578 nm, and the resolution could reach 15 μm. The hybrid dual-wavelength PA also improved the axial resolution to 15μm and provided a three-dimensional view of the entire cortical blood vessels of the mice [167]. Lihong V Wang's team improved the resolution by using section-assisted photoacoustic microscopy to perform endogenous and natural staining of the mouse brain and cerebellum, showing the three-dimensional distribution of the nucleus with a transverse resolution of 0.91 μm and an axial resolution of about 20 μm [168]. But the technology is still in its early stages, and future applications will require it to detect specific endogenous molecules or cells, such as neurons in the brain. Chulhong Kim et al. reported a new positioning PA system with a temporal resolution of 2 ms and a horizontal and vertical resolution of 0.7 and 2.5 μm, respectively (Figure 5) [169]. The new PAI technology can already display multiple hemodynamic photoacoustic data noninvasively in real time, but the spatial resolution is only 200μm, which misses microvascular system [170]. Nevertheless, it shows great potential in the field of stroke, especially neuroimaging [171]. Neuroimaging of PA can also be based on hemoglobin gradient and oxygen saturation changes, but the resolution is limited to several hundred microns [172]. GCaMP is primarily used, while it cannot be used to image neurons deep in the brains of living mammals [173, 174].
4. Magnetic resonance imaging
Magnetic resonance imaging (MRI) is best known for its extensive use in clinical practice. MRI is an indispensable imaging method for the evaluation of cerebral small-vessel diseases, such as microstructural changes in white matter and gray matter, microhemorrhage near small arteries, and subcortical infarction of the cerebellum [175]. MRI is mostly done by measuring the radio frequency signals emitted by hydrogen atoms after applying particular radio frequency (RF) waves from RF coils, and positioning the signals using spatially varying magnetic gradients for noninvasive imaging, while also avoiding radiation damage by providing 3D information of soft tissues from water content. The contrast of each voxel depends on the proton density in the voxel and the properties of the local tissue microenvironment [176]. Conventional MRI is primarily used for anatomical and functional imaging of cm-scale samples with a spatial resolution of about 1 mm because its resolution is constrained by the inherent low signal-to-noise ratio (SNR) of MRI [177]. Higher magnetic field intensity leads to higher spatial resolution, but the related increase in spectral line width is a necessary factor to be considered [178]. Therefore, the high SNR and strong gradient required for accurate MRI datasets determine that it is more suitable for ultra-high-field imaging, which avoids deviation from near field compared with low field [179]. Professor Budinger is advocating raising the magnetic field from 14 T to 20 T, highlighting the benefits of the technique for studying the human brain [180]. The high and ultra-high fields do bring high resolution, with an amazing 100 nm resolution of 3D imaging on a large sample such as the whole brain [181]. However, it cannot cater to the smaller micron scale structure. In fact, micron-scale 3D MRI images have been available for more than ten years. L.Ciobanu used a spatial resolution of 3.7×3.3×3.3 μm3 to image a single cell and observed the helical arrangement of chloroplast [182]. Several methods have emerged in recent years to improve the resolution of MRI—a previously considered "macro-imaging" technique- for studies of micron neurovascular structures in brain in stroke.
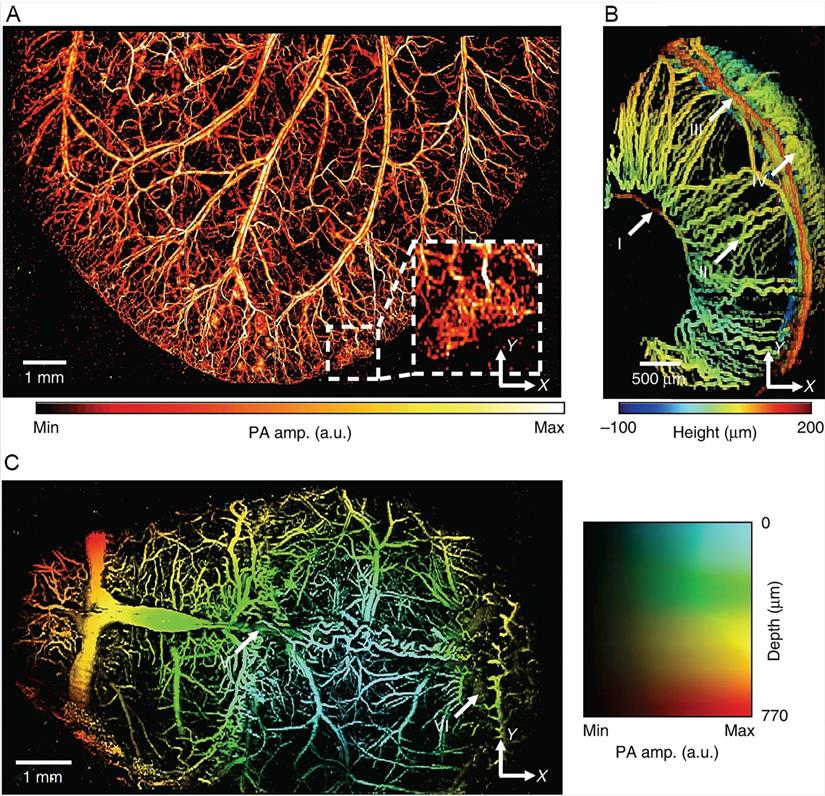
PA images of microvasculatures in small animals in vivo. (A) Wide-FOV PA image of a mouse ear. The region including a capillary bed is outlined by the white dashed box. (B) Depth-encoded PA image of a mouse eye. Circulus arteriosus major (I), iris (II), circulus arteriosus minor (III) and choroid (IV) blood vessels are highlighted by the white arrows. (C) Wide FOV PA MAP image of a mouse brain with color-encoded depths and amplitudes. Superior sagittal sinus (V) and transverse sinus (VI) are highlighted by the white arrows. FOV, field of view; MAP, maximum amplitude projection. Reproduced with permission from Reference 169.
MRI contrast agent is feasible to improve the resolution. The contrast agent can be distributed to extracellular fluid, blood vessels, and even specific organs. By shortening T1 or T2 relaxation time, the signal intensity on T1-weighted images increases or the signal intensity on the T2-weighted image decreases [183]. T1-weighted contrast agents for brighter signals are more suitable for higher resolution Paramagnetic contrast agents except dysprosium compounds are positive contrast agents; and the superparamagnetic or ferromagnetic materials that form negative contrast agents can also change particle size or coating to become positive contrast agents [184, 185]. Gadobutrol, a kind of contrast agent, was used to enhance the resolution of MRI in observing post-stroke brain edema and achieved an isotropic spatial resolution of 100 μm [186]. The application of the contrast agent Magnevist (a kind of gadolinium chelate) can obtain 3D imaging of structures such as hippocampus at 40μm resolution [187]. Manganese enhanced magnetic resonance imaging (MEMRI) can be used for both functional and structural imaging. Paramagnetic manganese ions mimic Ca2+ and accumulate in neurons through Ca2+ channels and are therefore used to study neuronal pathways or pathophysiological processes in stroke [188]. Chika Sato et al. improved MEMRI technology in vitro and obtained isotropic 3D neuroimaging with 25 μm resolution using low temperature RF coils [189].
Low temperature RF technology is equally vital in improving resolution. At low temperatures of tens of degrees Kelvin, the strength of the nuclear magnetic resonance signal can be enhanced by a large spin polarization at thermal equilibrium, thus reducing thermal noise. Meanwhile, dynamic nuclear polarization (DNP) can transfer polarization from electron spin to nuclear spin, and finally, obtain a resolution better than 1 μm [190, 191]. The whole brain MRI scan was reduced to 11 minutes, showing both nerve and vascular lesions at the same time [191]. Robert Tycko et al. obtained the 1H MRI image with an isotropic resolution of 2.8 µm in glycerin/water at 28 K [192]. The low temperature also fulfilled high quality 13C and 39K MRI images for metabolic studies [193, 194].
In addition to the relaxation of T1 and T2, MRI can be based on another contrast mechanism—Brownian motion of water molecules, including diffusion weighted imaging (DWI) and more advanced diffusion tensor imaging (DTI), and their grayscale pixel values depend on the basic diffusivity of these voxels [195]. DWI and DTI have been widely used in the clinical diagnosis and research of brain diseases, including cerebral small vascular disease [196]. DWI can detect the differential distribution of apparent dispersion coefficient (ADC) [197]. The higher the ADC values, the stronger the dispersibility of water molecules is. Choong H. Lee et al. achieved an isotropic spatial resolution of 10μm of drosophila brain using DWI with RF microsurface coils [198]. The research conducted by Chen et al. utilized DWI in human and porcine adjacent α-neurons also obtained 3D reconstructed images with an isotropic resolution of 6.25 μm [199]. The DTI estimates the degree of diffusion anisotropy along the direction of the main fibers, producing features such as mean diffusivity and fractional anisotropy, thus can characterize the neural structures of gray and white matter, but with relatively low resolution [200, 201]. DWI is very sensitive to motion, and it is inevitable to prolong scanning time while avoiding ghosting, but for DTI, the large amount of diffusion directions required often means to sacrifice spatial resolution [195].
5. Conclusions and perspectives
The physiological process of stroke is based on the tight and orchestrated relationship between nerves and vasculature. Nerves are closely related to the surrounding blood vessels. The concept “neurovascular unit” (NVU) focused on the coupling between neural activity and blood flow to explain how neuronal signals modulate nearby microvessels to support cerebral function [202]. Meanwhile, the molecular pathways shared by the neurons and vessels play a role in vascular influence on the homeostasis of the nervous system [203]. Therefore, neurovascular structural and functional imaging is important to explore the physiological or pathological processes of stroke. Over decades of years, many 3D imaging technologies have been developed or improved in resolution. But the need for further research still raises questions about these techniques.
In order to conform to the physiological state, it is necessary to study the nerve and blood activity in the waking state. Fluorescence-labeled TPM and LSM require preparation of cranial windows and are therefore invasive. PAI and MRI are noninvasive. Updates to traditional methods, such as μCT with blood-pool liposomal-iodinated contrast agent [204], fluorescence imaging through a near-infrared window [205], and ultrasound microscopy with special contrast agents [206], also yielded noninvasive, high-resolution three-dimensional images, but mainly for cerebral blood vessels. Observation of living bodies is always limited to shallow structures, while deeper structures or observations of large animals such as humans require deeper penetration. Cumulative scattering limits the penetration depth of optical microscopy. The depth of TPM is stronger than that of LSM, which can reach 1mm. Through the near-infrared window, many optical microscopy techniques have improved the penetration depth and the maximum penetration depth is about 3mm [205]. However, it can still only observe the superficial nerves and vessels. PAI can penetrate several centimeters deep, but it severely sacrifices resolution. MRI and CT that can obtain the structure of the whole brain are difficult to obtain micron resolution. Optical fiber is a viable attempt to achieve multiple penetration depths in both TPM and LSM. Further, we are pursuing the imaging of free-moving animals. The wearable PAT can only achieve a spatial resolution of 200µm, while high-resolution hemodynamic observations lack 3D imaging results [207, 208]. At a micron-resolution scale, tiny motions can also greatly affect image quality. TPM is a more feasible approach that can simultaneously image multiple functional areas of the brain, but it tends to study signals in neural circuits [209-211]. Thus, it is necessary to make a comparison of these imaging techniques (Table 1).
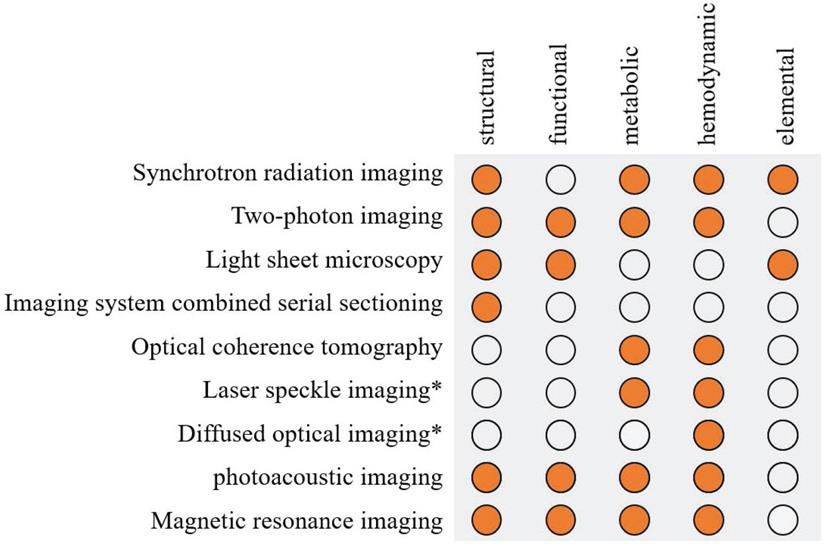
With the diversification of detection indexes and the refinement of detection, multi-mode imaging technology combining complementary microscopy techniques has been developed. PAI, which stands for multimodal, is a combination of photoexcitation and acoustic detection, maintaining high resolution at deeper imaging depths. Combining with CT is a common multi-mode method to improve resolution, such as SR-CT and photoacoustic computed tomography (PACT) [212]. Multi-modality also provides richer detection results, for example, combining the functional information of DOT with a high resolution structural imaging will achieve a high resolution functional imaging [213]. LSM combined with nonlinear two-photon excitation can improve the imaging depth and speed [85]. Fluorescence imaging has been applied in a great assortment of modalities (e.g., PAT and MRI) to study molecular events, thus makes a difference in structural, functional or molecular imaging [214]. Figure 6 summaries the application of high resolution imaging method.
It is worth noting that there are currently several techniques that play an important role in functional, hemodynamic, and metabolic imaging, but fail to achieve micron resolution, such as functional near-infrared spectroscopy (fNIRS) [215], intrinsic signal optical imaging (ISOI) [216], and single-photon emission computed tomography (SPECT) [217]. However, diffused optical imaging is also a low resolution technique, but it achieves high resolution through multimodality.
Optical microscopy is obviously the most important of the above techniques based on different principles, but its resolution is always limited to the scattering limit. In recent years, the development of ultramicroscopy with super-resolution facilitates the resolution beyond the scattering limit and can reveal the landscape of the cellular organelle interactome or observe small intact animals [218, 219]. It has provided innovation from the micron level to the nanoscale level, which is introduced briefly in the part of LSM. Of course, it is used in PAI, TPM, and other technologies as well [220, 221]. It can focus our attention on more subtle structures, but as is discussed earlier, we have both nanoscale and macroscopic means of observation, while ignoring spatial relationships on the micron level; we have a wealth of techniques for monitoring nerves and vessels separately, but we still cannot observe and interpret neurovascular coupling well. At the same time, we are increasingly aware of the close relationship between nerves and vasculature in stroke, thus high-resolution 3D imaging may inevitably become the key to further research.
Advanced 3D imaging technology and characterizations in imaging of small mammals.
| Advanced 3D imaging technology | Focus point | ||||
|---|---|---|---|---|---|
| Whole brain scale | Imaging depth | Living animals | Behaving animals | ||
| Light | Synchrotron radiation imaging | √ | Absorption imaging | ||
| Two-photon imaging | up to mm-level | √ | √ | ||
| Light sheet microscopy | up to mm-level | some methods | some methods like SCAPE | ||
| Imaging system combined serial sectioning | √ | ||||
| Optical coherence tomography | up to mm-level | √ | √ | ||
| Dynamic light scattering method | up to mm-level | √ | √ | ||
| Acoustics | Photoacoustic imaging | up to cm-level | √ | √ | |
| Magnetics | MRI | √ | √ | √ | |
Multimodal imaging: combined with the different techniques in the table above, or super-resolution microscopy, etc.
On the right side of the table are some separate focus points for imaging the brains of animal models, particularly rodents, including whether they are used for whole-brain imaging, the depth of imaging, and whether they are used in living or behaving animals. SCAPE: swept, confocally-aligned planar excitation. μCT: microcomputed tomography.
Summary of the application of high resolution imaging method in stroke. Solid orange circles indicate the use of this technology. *Laser speckle imaging and diffused optical imaging are low-resolution imaging methods but can be combined with other structural imaging to obtain high resolution.
Abbreviations
3D: three-dimensional; MRI: magnetic resonance imaging; SR: synchrotron radiation; APS: American Light Source; ESRF: European Synchrotron Radiation Facility; SLS: Swiss Light Source; SSRF: Shanghai Synchrotron Radiation Facility (SSRF); KESA: K‐edge subtraction angiography; SETS: single‐energy temporal subtraction; PCI: Phase contrast imaging; ILPCI: inline X-ray phase contrast imaging; GI: grating interferometry; CI: crystal interferometry; ACI: absorption contrast imaging; MCAO: middle cerebral artery occlusion; µCT: microcomputed tomography; FOV: fieldof view; SR-IR: SR-infrared spectrumor; SR-μXRF: SR micro-X-ray fluorescence; FTIR-μCT: fourier transform infrared spectro-microtomography; TPM: two-photon microscopy; NIR: near-infrared; MPM: multi-photon microscopy; ChroMS: chromatic multiphoton serial; GRIN: gradient-index; AOS: adaptiveoptics systems; LSM: light-sheet microscopy; SPIM: Selective Plane Illumination Microscope; LLSM: lattice light-sheet microscope; SCAPE: swept confocally-aligned planar excitation; FITC: Fluorescein isothiocyanate; Mars-SPIM: multiangle-resolved subvoxel selective plane illumination microscope; anti-αSMA: anti-αsmooth muscle actin; MOST: micro-optical sectioning tomography; fMOST: fluorescent micro-optical sectioning tomography; STP: serial two-photon tomography; FAST: block-face serial microscopy; GFP: green fluorescent protein; OCT: optical coherence tomograph; ODT: optical Doppler tomography; CBF: cerebral blood flow; OCTA: optical coherence tomography-based angiography; LSI: laser speckle imaging; DOT: diffuse optical tomography; DCT: diffuse optical correlation tomography; PAI: photoacoustic imaging; OR: optical-resolution; AR: acoustic-resolution; PAT: photoacoustic tomography; PAM: photoacoustic microscopy; MAP: maximum amplitude projection; RF: radio frequency; SNR: signal-to-noise ratio; MEMRI: Manganese enhanced magnetic resonance imaging; DNP: dynamic nuclear polarization; DWI: diffusion weighted imaging; DTI: diffusion tensor imaging; ADC: apparent dispersion coefficient; NVU: neurovascular unit; PACT: photoacoustic computed tomography; fNIRS: functional near-infrared spectroscopy; ISOI: intrinsic signal optical imaging; SPECT: single-photon emissioncomputed tomography.
Acknowledgements
This research was funded by the Natural Science Foundations for Excellent Young Scholars of Hunan Province (No. 2021JJ20095), the Key Research and Development Program of Hunan Province (No. 2020SK2063), Research Project on Education and Teaching Innovation of Central South University (2021jy145), the Natural Science Foundations of Hunan Province (No. 2020JJ4134), and the National Natural Science Foundation of China (No. 81501025).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kleindorfer DO, Towfighi A, Chaturvedi S, Cockroft KM, Gutierrez J, Lombardi-Hill D. et al. 2021 Guideline for the Prevention of Stroke in Patients With Stroke and Transient Ischemic Attack: A Guideline From the American Heart Association/American Stroke Association. Stroke. 2021;52:e364-e467
2. Wang Q, Ding S-L, Li Y, Royall J, Feng D, Lesnar P. et al. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell. 2020;181:936-53.e20
3. Mastorakos P, Mihelson N, Luby M, Burks SR, Johnson K, Hsia AW. et al. Temporally distinct myeloid cell responses mediate damage and repair after cerebrovascular injury. Nat Neurosci. 2021;24:245-58
4. Guo S, Kim WJ, Lok J, Lee S-R, Besancon E, Luo B-H. et al. Neuroprotection via matrix-trophic coupling between cerebral endothelial cells and neurons. Proceedings of the National Academy of Sciences. 2008;105:7582
5. Zhang Z, Bai L, Cong L, Yu P, Zhang T, Shi W. et al. Imaging volumetric dynamics at high speed in mouse and zebrafish brain with confocal light field microscopy. Nat Biotechnol. 2020
6. Shi Y, Thrippleton MJ, Makin SD, Marshall I, Geerlings MI, de Craen AJM. et al. Cerebral blood flow in small vessel disease: A systematic review and meta-analysis. J Cereb Blood Flow Metab. 2016;36:1653-67
7. Held F, Morris AWJ, Pirici D, Niklass S, Sharp MMG, Garz C. et al. Vascular basement membrane alterations and β-amyloid accumulations in an animal model of cerebral small vessel disease. Clin Sci. 2017;131:1001
8. Pichat J, Iglesias JE, Yousry T, Ourselin S, Modat M. A Survey of Methods for 3D Histology Reconstruction. Med Image Anal. 2018 46
9. Khimchenko A, Deyhle H, Schulz G, Schweighauser G, Hench J, Chicherova N. et al. Extending two-dimensional histology into the third dimension through conventional micro computed tomography. Neuroimage. 2016;139:26-36
10. Erdener ŞE, Dalkara T. Small Vessels Are a Big Problem in Neurodegeneration and Neuroprotection. Frontiers in Neurology. 2019;10:889
11. Sigal YM, Zhou R, Zhuang X. Visualizing and discovering cellular structures with super-resolution microscopy. Science. 2018;361:880-7
12. Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452-6
13. Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S. et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207-14
14. Zhang M, Peng G, Sun D, Xie Y, Xia J, Long H. et al. Synchrotron radiation imaging is a powerful tool to image brain microvasculature. Med Phys. 2014;41:031907
15. Owen RL, Juanhuix J, Fuchs M. Current advances in synchrotron radiation instrumentation for MX experiments. Arch Biochem Biophys. 2016;602:21-31
16. Moncton DE. The advanced photon source: performance and results from early operation. J Synchrotron Radiat. 1998;5:155-7
17. Martinez-Criado G, Tucoulou R, Cloetens P, Bleuet P, Bohic S, Cauzid J. et al. Status of the hard X-ray microprobe beamline ID22 of the European Synchrotron Radiation Facility. 2012; 19: 10-8.
18. Böge M, Boksberger HU, Busse-Gravitz M, Dehler M, Dohan D, George D. et al. The Swiss Light Source Accelerator Complex: An Overview. Proc 1998 6th European Particle Acc Conf(EPAC-98), Stockholm, Pub: IoP, UK. 1998 p. 623
19. Zhao ZT, Xu HJ, Ding H. Status of the Shanghai Synchrotron Radiation Facility. Particle Accelerator Conference. 2005
20. Tavares PF, Al-Dmour E, Andersson A, Cullinan F, Jensen BN, Olsson D. et al. Commissioning and first-year operational results of the MAX IV 3 GeV ring. J Synchrotron Radiat. 2018;25:1291-316
21. Gu S, Xue J, Xi Y, Tang R, Jin W, Chen J-J. et al. Evaluating the effect of Avastin on breast cancer angiogenesis using synchrotron radiation. Quant Imaging Med Surg. 2019;9:418-26
22. Luo Y, Yin X, Shi S, Ren X, Zhang H, Wang Z. et al. Non-destructive 3D microtomography of cerebral angioarchitecture changes following ischemic stroke in rats using synchrotron radiation. Front Neuroanat. 2019;13:5 -
23. Lee Y, Lee AC, Kim H-J. A Monte Carlo simulation study of an improved K-edge log-subtraction X-ray imaging using a photon counting CdTe detector. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment. 2016 p. 381-90
24. Li HL, Ding H, Yin XZ, Chen ZH, Tang B, Sun JY. et al. Comparison of high-resolution synchrotron-radiation-based phase-contrast imaging and absorption-contrast imaging for evaluating microstructure of vascular networks in rat brain: from 2D to 3D views. J Synchrotron Radiat. 2019;26:2024-32
25. Bravin A, Coan P, Suortti P. X-ray phase-contrast imaging: from pre-clinical applications towards clinics. Phys Med Biol. 2012;58:R1-R35
26. Xi Y, Kou B, Sun H, Qi J, Sun J, Mohr J. et al. X-ray grating interferometer for biomedical imaging applications at Shanghai Synchrotron Radiation Facility. J Synchrotron Radiat. 2012;19:821-6
27. Mu Z-H, Jiang Z, Lin X-J, Wang L-P, Xi Y, Zhang Z-J. et al. Vessel Dilation Attenuates Endothelial Dysfunction Following Middle Cerebral Artery Occlusion in Hyperglycemic Rats. CNS Neurosci Ther. 2016;22:316-24
28. Wang L, Zhou P, Mu Z, Lin X, Jiang L, Cheng Z. et al. Dynamic Detection of Thrombolysis in Embolic Stroke Rats by Synchrotron Radiation Angiography. Transl Stroke Res. 2019;10:695-704
29. Fukuyama N, Tsukamoto Y, Takizawa S, Ikeya Y, Fujii T, Shinozaki Y. et al. Altered blood flow in cerebral perforating arteries of rat models of diabetes: A synchrotron radiation microangiographic study toward clinical evaluation of white matter hyperintensities. Geriatr Gerontol Int. 2015;15:74-80
30. Zhang M-Q, Zhou L, Deng Q-F, Xie Y-Y, Xiao T-Q, Cao Y-Z. et al. Ultra-high-resolution 3D digitalized imaging of the cerebral angioarchitecture in rats using synchrotron radiation. Sci Rep. 2015;5:14982 -
31. Shi S, Zhang H, Yin X, Wang Z, Tang B, Luo Y. et al. 3D digital anatomic angioarchitecture of the mouse brain using synchrotron-radiation-based propagation phase-contrast imaging. J Synchrotron Radiat. 2019;26:1742-50
32. Cao Y, Zhang M, Ding H, Chen Z, Tang B, Wu T. et al. Synchrotron radiation micro-tomography for high-resolution neurovascular network morphology investigation. J Synchrotron Radiat. 2019;26:607-18
33. Zhou P-T, Wang L-P, Qu M-J, Shen H, Zheng H-R, Deng L-D. et al. Dl-3-N-butylphthalide promotes angiogenesis and upregulates sonic hedgehog expression after cerebral ischemia in rats. CNS Neurosci Ther. 2019;25:748-58
34. Gu P, Xu Z-H, Cao Y-Z, Liao S-H, Deng Q-F, Yin X-Z. et al. Synchrotron Radiation-Based Three-Dimensional Visualization of Angioarchitectural Remodeling in Hippocampus of Epileptic Rats. Neurosci Bull. 2019
35. Shuntong KANG TW, Zhuohui CHEN, Mengqi ZHANG. 3D imaging of rat brain neural network using synchrotron radiation. Nuclear Techniques. 2020;43:70101-070101
36. Töpperwien M, van der Meer F, Stadelmann C, Salditt T. Three-dimensional virtual histology of human cerebellum by X-ray phase-contrast tomography. Proc Natl Acad Sci U S A. 2018;115:6940-5
37. Fonseca MDC, Araujo BHS, Dias CSB, Archilha NL, Neto DPA, Cavalheiro E. et al. High-resolution synchrotron-based X-ray microtomography as a tool to unveil the three-dimensional neuronal architecture of the brain. Sci Rep. 2018;8:12074 -
38. Katsamenis OL, Olding M, Warner JA, Chatelet DS, Jones MG, Sgalla G. et al. X-ray Micro-Computed Tomography for Nondestructive Three-Dimensional (3D) X-ray Histology. Am J Pathol. 2019;189:1608-20
39. Dyer EL, Gray Roncal W, Prasad JA, Fernandes HL, Gürsoy D, De Andrade V. et al. Quantifying Mesoscale Neuroanatomy Using X-Ray Microtomography. eNeuro. 2017;4:ENEURO.0195-17.2017
40. Khimchenko A, Bikis C, Pacureanu A, Hieber SE, Thalmann P, Deyhle H. et al. Hard X-Ray Nanoholotomography: Large-Scale, Label-Free, 3D Neuroimaging beyond Optical Limit. Adv Sci. 2018;5:1700694 -
41. Au A. Metabolomics and Lipidomics of Ischemic Stroke. Adv Clin Chem. 2018;85:31-69
42. Miller LM, Dumas P. Chemical imaging of biological tissue with synchrotron infrared light. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2006;1758:846-57
43. Martin MC, Dabat-Blondeau C, Unger M, Sedlmair J, Parkinson DY, Bechtel HA. et al. 3D spectral imaging with synchrotron Fourier transform infrared spectro-microtomography. Nat Meth. 2013;10:861
44. Ogunleke A, Recur B, Balacey H, Chen H-H, Delugin M, Hwu Y. et al. 3D chemical imaging of the brain using quantitative IR spectro-microscopy. Chem Sci. 2017;9:189-98
45. Ackerman CM, Chang CJ. Copper signaling in the brain and beyond. J Biol Chem. 2018;293:4628-35
46. Portbury SD, Adlard PA. Zinc Signal in Brain Diseases. Int J Mol Sci. 2017;18:2506
47. Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014;13:1045-60
48. Alaverdashvili M, Paterson PG. Mapping the dynamics of cortical neuroplasticity of skilled motor learning using micro X-ray fluorescence and histofluorescence imaging of zinc in the rat. Behav Brain Res. 2017;318:52-60
49. Shi S-P, Wang H, Chen Z-H, Li X-H, Liu S-X, Zhang M-Q. Characterization of metal element distributions in the rat brain following ischemic stroke by synchrotron radiation micro-fluorescence analysis. Nuclear Science and Techniques. 2020;31:96
50. Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73
51. Williams RM, Piston DW, Webb WW. Two-photon molecular excitation provides intrinsic 3-dimensional resolution for laser-based microscopy and microphotochemistry. FASEB J. 1994;8:804-13
52. Zong W, Wu R, Li M, Hu Y, Li Y, Li J. et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat Meth. 2017;14:713
53. Centonze VE, White JG. Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging. Biophys J. 1998;75:2015-24
54. Klaiber M, Briggs JS. Crossover from tunneling to multiphoton ionization of atoms. Phys Rev A. 2016;94:053405
55. Zheng Z, Li D, Liu Z, Peng H-Q, Sung HHY, Kwok RTK. et al. Aggregation-Induced Nonlinear Optical Effects of AIEgen Nanocrystals for Ultradeep In Vivo Bioimaging. Adv Mater. 2019;31:1904799
56. Santisakultarm TP, Kersbergen CJ, Bandy DK, Ide DC, Choi S-H, Silva AC. Two-photon imaging of cerebral hemodynamics and neural activity in awake and anesthetized marmosets. J Neurosci Methods. 2016;271:55-64
57. Isshiki M, Okabe S. Evaluation of cranial window types for in vivo two-photon imaging of brain microstructures. Microscopy. 2013;63:53-63
58. Estrada G, Beetle C, Schummers J. Simple method to improve spatial resolution for in vivo two-photon fluorescence imaging. Appl Opt. 2015;54:10044-50
59. Ricard C, Arroyo ED, He CX, Portera-Cailliau C, Lepousez G, Canepari M. et al. Two-photon probes for in vivo multicolor microscopy of the structure and signals of brain cells. Brain Struct Funct. 2018;223:3011-43
60. Weisenburger S, Tejera F, Demas J, Chen B, Manley J, Sparks FT. et al. Volumetric Ca(2+) Imaging in the Mouse Brain Using Hybrid Multiplexed Sculpted Light Microscopy. Cell. 2019;177:1050-66.e14
61. Szalay G, Martinecz B, Lénárt N, Környei Z, Orsolits B, Judák L. et al. Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat Commun. 2016;7:11499
62. Abdeladim L, Matho KS, Clavreul S, Mahou P, Sintes J-M, Solinas X. et al. Multicolor multiscale brain imaging with chromatic multiphoton serial microscopy. Nat Commun. 2019;10:1662
63. Sarkar AR, Kang DE, Kim HM, Cho BR. Two-photon fluorescent probes for metal ions in live tissues. Inorg Chem. 2014;53:1794-803
64. Kang MY, Lim CS, Kim HS, Seo EW, Kim HM, Kwon O. et al. Detection of nickel in fish organs with a two-photon fluorescent probe. Chemistry. 2012;18:1953-60
65. Yanez CO, Morales AR, Yue X, Urakami T, Komatsu M, Järvinen TAH. et al. Deep vascular imaging in wounds by two-photon fluorescence microscopy. PLoS ONE. 2013;8:e67559-e
66. Meng G, Liang Y, Sarsfield S, Jiang W-C, Lu R, Dudman JT. et al. High-throughput synapse-resolving two-photon fluorescence microendoscopy for deep-brain volumetric imaging in vivo. eLife. 2019;8:e40805
67. Liu CJ, Roy A, Simons AA, Farinella DM, Kara P. Three-photon imaging of synthetic dyes in deep layers of the neocortex. Sci Rep. 2020;10:16351
68. Horton NG, Wang K, Kobat D, Clark CG, Wise FW, Schaffer CB. et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics. 2013;7:205
69. Ozbay BN, Futia GL, Ma M, Bright VM, Gopinath JT, Hughes EG. et al. Three dimensional two-photon brain imaging in freely moving mice using a miniature fiber coupled microscope with active axial-scanning. Sci Rep. 2018;8:8108 -
70. Chen S, Wang Z, Zhang D, Wang A, Chen L, Cheng H. et al. Miniature Fluorescence Microscopy for Imaging Brain Activity in Freely-Behaving Animals. Neurosci Bull. 2020;36:1182-90
71. Griffiths VA, Valera AM, Lau JYN, Roš H, Younts TJ, Marin B. et al. Real-time 3D movement correction for two-photon imaging in behaving animals. Nat Meth. 2020;17:741-8
72. Li B, Wu C, Wang M, Charan K, Xu C. An adaptive excitation source for high-speed multiphoton microscopy. Nat Meth. 2020;17:163-6
73. Park J-H, Kong L, Zhou Y, Cui M. Large-field-of-view imaging by multi-pupil adaptive optics. Nat Meth. 2017;14:581-3
74. Qu J, Liu L, Shao Y, Niu H, Gao BZ. Recent progress in multifocal multiphoton microscopy. J Innovative Opt Health Sci. 2012;5:10.1142 /S1793545812500186
75. Strobl F, Schmitz A, Stelzer EHK. Improving your four-dimensional image: traveling through a decade of light-sheet-based fluorescence microscopy research. Nat Protoc. 2017;12:1103-9
76. Power RM, Huisken J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Meth. 2017;14:360-73
77. Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK. Optical Sectioning Deep Inside Live Embryos by Selective Plane Illumination Microscopy. Science. 2004;305:1007
78. Keller PJ, Ahrens MB. Visualizing Whole-Brain Activity and Development at the Single-Cell Level Using Light-Sheet Microscopy. Neuron. 2015;85:462-83
79. Elisa Z, Toon B, De Smedt SC, Katrien R, Kristiaan N, Kevin B. Technical implementations of light sheet microscopy. Microsc Res Tech. 2018;81:941-58
80. Whitehead LW, McArthur K, Geoghegan ND, Rogers KL. The reinvention of twentieth century microscopy for three-dimensional imaging. Immunol Cell Biol. 2017;95:520-4
81. Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Meth. 2011;8:417-23
82. Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998
83. Wu Y, Wawrzusin P, Senseney J, Fischer RS, Christensen R, Santella A. et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol. 2013;31:1032-8
84. Chhetri RK, Amat F, Wan Y, Höckendorf B, Lemon WC, Keller PJ. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Meth. 2015;12:1171-8
85. Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Meth. 2011;8:757-60
86. Morawski M, Kirilina E, Scherf N, Jäger C, Reimann K, Trampel R. et al. Developing 3D microscopy with CLARITY on human brain tissue: Towards a tool for informing and validating MRI-based histology. Neuroimage. 2018;182:417-28
87. Ueda HR, Ertürk A, Chung K, Gradinaru V, Chédotal A, Tomancak P. et al. Tissue clearing and its applications in neuroscience. Nat Rev Neurosci. 2020;21:61-79
88. Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ. et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332-7
89. Cai R, Pan C, Ghasemigharagoz A, Todorov MI, Förstera B, Zhao S. et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat Neurosci. 2019;22:317-27
90. Corsetti S, Gunn-Moore F, Dholakia K. Light sheet fluorescence microscopy for neuroscience. J Neurosci Methods. 2019;319:16-27
91. Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Meth. 2013;10:413-20
92. Vladimirov N, Wang C, Höckendorf B, Pujala A, Tanimoto M, Mu Y. et al. Brain-wide circuit interrogation at the cellular level guided by online analysis of neuronal function. Nat Meth. 2018;15:1117-25
93. Lai HM, Liu AKL, Ng HHM, Goldfinger MH, Chau TW, DeFelice J. et al. Next generation histology methods for three-dimensional imaging of fresh and archival human brain tissues. Nat Commun. 2018;9:1066 -
94. Gao R, Asano SM, Upadhyayula S, Pisarev I, Milkie DE, Liu T-L. et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science. 2019;363:eaau8302
95. Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS. et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science. 2016;354:aaf3928
96. Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat Photonics. 2015;9:113-9
97. Voleti V, Patel KB, Li W, Perez Campos C, Bharadwaj S, Yu H. et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat Meth. 2019;16:1054-62
98. Wang F, Wan H, Ma Z, Zhong Y, Sun Q, Tian Y. et al. Light-sheet microscopy in the near-infrared II window. Nat Meth. 2019;16:545-52
99. Harrison CH, Buckland GR, Brooks SE, Johnston DA, Chatelet DS, Liu AKL. et al. A novel method to visualise the three-dimensional organisation of the human cerebral cortical vasculature. J Anat. 2018;232:1025-30
100. Zhang L-Y, Lin P, Pan J, Ma Y, Wei Z, Jiang L. et al. CLARITY for High-resolution Imaging and Quantification of Vasculature in the Whole Mouse Brain. Aging Dis. 2018;9:262-72
101. Kirst C, Skriabine S, Vieites-Prado A, Topilko T, Bertin P, Gerschenfeld G. et al. Mapping the Fine-Scale Organization and Plasticity of the Brain Vasculature. Cell. 2020;180:780-95.e25
102. Lugo-Hernandez E, Squire A, Hagemann N, Brenzel A, Sardari M, Schlechter J. et al. 3D visualization and quantification of microvessels in the whole ischemic mouse brain using solvent-based clearing and light sheet microscopy. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2017;37:3355-67
103. Gregorius J, Wang C, Stambouli O, Hussner T, Qi Y, Tertel T. et al. Small extracellular vesicles obtained from hypoxic mesenchymal stromal cells have unique characteristics that promote cerebral angiogenesis, brain remodeling and neurological recovery after focal cerebral ischemia in mice. Basic Res Cardiol. 2021;116:40 -
104. Miyawaki T, Morikawa S, Susaki EA, Nakashima A, Takeuchi H, Yamaguchi S. et al. Visualization and molecular characterization of whole-brain vascular networks with capillary resolution. Nat Commun. 2020;11:1104 -
105. França CM, Riggers R, Muschler JL, Widbiller M, Lococo PM, Diogenes A. et al. 3D-Imaging of Whole Neuronal and Vascular Networks of the Human Dental Pulp via CLARITY and Light Sheet Microscopy. Sci Rep. 2019;9:10860 -
106. Phillips J, Laude A, Lightowlers R, Morris CM, Turnbull DM, Lax NZ. Development of passive CLARITY and immunofluorescent labelling of multiple proteins in human cerebellum: understanding mechanisms of neurodegeneration in mitochondrial disease. Sci Rep. 2016;6:26013 -
107. Park OK, Kwak J, Jung YJ, Kim YH, Hong H-S, Hwang BJ. et al. 3D Light-Sheet Fluorescence Microscopy of Cranial Neurons and Vasculature during Zebrafish Embryogenesis. Mol Cells. 2015;38:975-81
108. Hörl D, Rojas Rusak F, Preusser F, Tillberg P, Randel N, Chhetri RK. et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat Meth. 2019;16:870-4
109. Chakraborty T, Chen B, Daetwyler S, Chang B-J, Vanderpoorten O, Sapoznik E. et al. Converting lateral scanning into axial focusing to speed up three-dimensional microscopy. Light-Sci Appl. 2020;9:165
110. Nie J, Liu S, Yu T, Li Y, Ping J, Wan P. et al. Fast, 3D Isotropic Imaging of Whole Mouse Brain Using Multiangle-Resolved Subvoxel SPIM. Adv Sci. 2019;7:1901891 -
111. Guo M, Li Y, Su Y, Lambert T, Nogare DD, Moyle MW. et al. Rapid image deconvolution and multiview fusion for optical microscopy. Nat Biotechnol. 2020
112. Li A, Gong H, Zhang B, Wang Q, Yan C, Wu J. et al. Micro-Optical Sectioning Tomography to Obtain a High-Resolution Atlas of the Mouse Brain. Science. 2010;330:1404
113. Zheng T, Feng Z, Wang X, Jiang T, Jin R, Zhao P. et al. Review of micro-optical sectioning tomography (MOST): technology and applications for whole-brain optical imaging. Biomed Opt Express. 2019;10:4075-96
114. Xiong B, Li A, Lou Y, Chen S, Long B, Peng J. et al. Precise cerebral vascular atlas in stereotaxic coordinates of whole mouse brain. Front Neuroanat. 2017;11:128
115. Wu J, He Y, Yang Z, Guo C, Luo Q, Zhou W. et al. 3D BrainCV: Simultaneous visualization and analysis of cells and capillaries in a whole mouse brain with one-micron voxel resolution. Neuroimage. 2014;87:199-208
116. Zhou Y-F, Chen A-Q, Wu J-H, Mao L, Xia Y-P, Jin H-J. et al. Sema3E/PlexinD1 signaling inhibits postischemic angiogenesis by regulating endothelial DLL4 and filopodia formation in a rat model of ischemic stroke. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33:4947-61
117. Zhou C, Zheng T, Luo T, Yan C, Sun Q, Ren M. et al. Continuous imaging of large-volume tissues with a machinable optical clearing method at subcellular resolution. Biomed Opt Express. 2020;11:7132-49
118. Seiriki K, Kasai A, Hashimoto T, Schulze W, Niu M, Yamaguchi S. et al. High-Speed and Scalable Whole-Brain Imaging in Rodents and Primates. Neuron. 2017;94:1085-100.e6
119. Seiriki K, Kasai A, Nakazawa T, Niu M, Naka Y, Tanuma M. et al. Whole-brain block-face serial microscopy tomography at subcellular resolution using FAST. Nat Protoc. 2019;14:1509-29
120. Mereuta OM, Fitzgerald S, Christensen TA, Jaspersen AL, Dai D, Abbasi M. et al. High-resolution scanning electron microscopy for the analysis of three-dimensional ultrastructure of clots in acute ischemic stroke. J Neurointerv Surg. 2021;13:906-11
121. Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J. et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Meth. 2012;9:255-8
122. Poinsatte K, Betz D, Torres VO, Ajay AD, Mirza S, Selvaraj UM. et al. Visualization and Quantification of Post-stroke Neural Connectivity and Neuroinflammation Using Serial Two-Photon Tomography in the Whole Mouse Brain. Front Neurosci. 2019;13:1055
123. Ortega SB, Torres VO, Latchney SE, Whoolery CW, Noorbhai IZ, Poinsatte K. et al. B cells migrate into remote brain areas and support neurogenesis and functional recovery after focal stroke in mice. Proc Natl Acad Sci U S A. 2020;117:4983-93
124. Li T, Liu C, Akkin T. Contrast-enhanced serial optical coherence scanner with deep learning network reveals vasculature and white matter organization of mouse brain. Neurophotonics. 2019;6:035004
125. Amato SP, Pan F, Schwartz J, Ragan TM. Whole Brain Imaging with Serial Two-Photon Tomography. Front Neuroanat. 2016;10:31 -
126. Dolezyczek H, Tamborski S, Majka P, Sampson D, Wojtkowski M, Wilczyński G. et al. In vivo brain imaging with multimodal optical coherence microscopy in a mouse model of thromboembolic photochemical stroke. Neurophotonics. 2020;7:015002 -
127. Gabriele ML, Wollstein G, Ishikawa H, Xu J, Kim J, Kagemann L. et al. Three dimensional optical coherence tomography imaging: advantages and advances. Prog Retin Eye Res. 2010;29:556-79
128. Chen C-J, Kumar JS, Chen SH, Ding D, Buell TJ, Sur S. et al. Optical Coherence Tomography: Future Applications in Cerebrovascular Imaging. Stroke. 2018;49:1044-50
129. Deloria AJ, Haider S, Dietrich B, Kunihs V, Oberhofer S, Knofler M. et al. Ultra-High-Resolution 3D Optical Coherence Tomography Reveals Inner Structures of Human Placenta-Derived Trophoblast Organoids. IEEE Trans Biomed Eng. 2021;68:2368-76
130. Ughi GJ, Marosfoi MG, King RM, Caroff J, Peterson LM, Duncan BH. et al. A neurovascular high-frequency optical coherence tomography system enables in situ cerebrovascular volumetric microscopy. Nat Commun. 2020;11:3851 -
131. Yuan W, Brown R, Mitzner W, Yarmus L, Li X. Super-achromatic monolithic microprobe for ultrahigh-resolution endoscopic optical coherence tomography at 800 nm. Nat Commun. 2017;8:1531
132. Ren H, Du C, Yuan Z, Park K, Volkow ND, Pan Y. Cocaine-induced cortical microischemia in the rodent brain: clinical implications. Mol Psychiatry. 2012;17:1017-25
133. Pan Y, You J, Volkow ND, Park K, Du C. Ultrasensitive detection of 3D cerebral microvascular network dynamics in vivo. Neuroimage. 2014;103:492-501
134. Spaide RF, Fujimoto JG, Waheed NK, Sadda SR, Staurenghi G. Optical coherence tomography angiography. Prog Retin Eye Res. 2018 64
135. Shin P, Choi W, Joo J, Oh W-Y. Quantitative hemodynamic analysis of cerebral blood flow and neurovascular coupling using optical coherence tomography angiography. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2019;39:1983-94
136. Kazmi SMS, Richards LM, Schrandt CJ, Davis MA, Dunn AK. Expanding applications, accuracy, and interpretation of laser speckle contrast imaging of cerebral blood flow. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2015;35:1076-84
137. Yuan Z, Luo Z, Volkow ND, Pan Y, Du C. Imaging separation of neuronal from vascular effects of cocaine on rat cortical brain in vivo. Neuroimage. 2011;54:1130-9
138. Sullender CT, Mark AE, Clark TA, Esipova TV, Vinogradov SA, Jones TA. et al. Imaging of cortical oxygen tension and blood flow following targeted photothrombotic stroke. Neurophotonics. 2018;5:035003 -
139. Chen M, Wen D, Huang S, Gui S, Zhang Z, Lu J. et al. Laser speckle contrast imaging of blood flow in the deep brain using microendoscopy. Opt Lett. 2018;43:5627-30
140. Du C, Pan Y. Optical detection of brain function: simultaneous imaging of cerebral vascular response, tissue metabolism, and cellular activity in vivo. Rev Neurosci. 2011;22:695-709
141. Zhou W, Kholiqov O, Zhu J, Zhao M, Zimmermann LL, Martin RM. et al. Functional interferometric diffusing wave spectroscopy of the human brain. Sci Adv. 2021 7
142. Lin AJ, Castello NA, Lee G, Green KN, Durkin AJ, Choi B. et al. In vivo optical signatures of neuronal death in a mouse model of Alzheimer's disease. Lasers Surg Med. 2014;46:27-33
143. Zhao Y, Raghuram A, Kim HK, Hielscher AH, Robinson JT, Veeraraghavan A. High Resolution, Deep Imaging Using Confocal Time-of-Flight Diffuse Optical Tomography. IEEE Trans Pattern Anal Mach Intell. 2021;43:2206-19
144. Lin Z-J, Ren M, Li L, Liu Y, Su J, Yang S-H. et al. Interleaved imaging of cerebral hemodynamics and blood flow index to monitor ischemic stroke and treatment in rat by volumetric diffuse optical tomography. Neuroimage. 2014 85 Pt 1: 566-82
145. Sutin J, Zimmerman B, Tyulmankov D, Tamborini D, Wu KC, Selb J. et al. Time-domain diffuse correlation spectroscopy. Optica. 2016;3:1006-13
146. Zhou C, Yu G, Furuya D, Greenberg J, Yodh A, Durduran T. Diffuse optical correlation tomography of cerebral blood flow during cortical spreading depression in rat brain. Opt Express. 2006;14:1125-44
147. Cheng X, Sie EJ, Naufel S, Boas DA, Marsili F. Measuring neuronal activity with diffuse correlation spectroscopy: a theoretical investigation. Neurophotonics. 2021;8:035004
148. Huang C, Mazdeyasna S, Chen L, Abu Jawdeh EG, Bada HS, Saatman KE. et al. Noninvasive noncontact speckle contrast diffuse correlation tomography of cerebral blood flow in rats. Neuroimage. 2019;198:160-9
149. Zhang HF, Maslov K, Stoica G, Wang LV. Functional photoacoustic microscopy for high-resolution and noninvasive in vivo imaging. Nat Biotechnol. 2006;24:848-51
150. Sathiyamoorthy K, Strohm EM, Kolios MC. Low-power noncontact photoacoustic microscope for bioimaging applications. J Biomed Opt. 2017;22:046001
151. Ku G, Wang LV. Deeply penetrating photoacoustic tomography in biological tissues enhanced with an optical contrast agent. Opt Lett. 2005;30:507-9
152. Nie L, Chen X. Structural and functional photoacoustic molecular tomography aided by emerging contrast agents. Chem Soc Rev. 2014;43:7132-70
153. Wang S, Lin J, Wang T, Chen X, Huang P. Recent advances in photoacoustic imaging for deep-tissue biomedical applications. Theranostics. 2016;6:2394-413
154. Wang LV, Hu S. Photoacoustic Tomography: In Vivo Imaging from Organelles to Organs. Science. 2012;335:1458-62
155. Moothanchery M, Bi R, Kim JY, Balasundaram G, Kim C, Olivo M. High-speed simultaneous multiscale photoacoustic microscopy. J Biomed Opt. 2019;24:1-7
156. Beard P. Biomedical photoacoustic imaging. Interface focus. 2011;1:602-31
157. Upputuri PK, Pramanik M. Photoacoustic imaging in the second near-infrared window: a review. J Biomed Opt. 2019;24:1-20
158. Zhao T, Desjardins AE, Ourselin S, Vercauteren T, Xia W. Minimally invasive photoacoustic imaging: Current status and future perspectives. Photoacoustics. 2019;16:100146 -
159. Upputuri PK, Pramanik M. Recent advances in photoacoustic contrast agents for in vivo imaging. Wiley Interdiscip Rev-Nanomed Nanobiotechnol. 2020 n/a: e1618
160. Yang X, Chen Y-H, Xia F, Sawan M. Photoacoustic imaging for monitoring of stroke diseases: A review. Photoacoustics. 2021;23:100287
161. Lv J, Li S, Zhang J, Duan F, Wu Z, Chen R. et al. In vivo photoacoustic imaging dynamically monitors the structural and functional changes of ischemic stroke at a very early stage. Theranostics. 2020;10:816-28
162. Haedicke K, Agemy L, Omar M, Berezhnoi A, Roberts S, Longo-Machado C. et al. High-resolution optoacoustic imaging of tissue responses to vascular-targeted therapies. Nat Biomed Eng. 2020;4:286-97
163. Li M, Tang Y, Yao J. Photoacoustic tomography of blood oxygenation: A mini review. Photoacoustics. 2018;10:65-73
164. Hu S, Maslov K, Wang LV. Noninvasive label-free imaging of microhemodynamics by optical-resolution photoacoustic microscopy. Opt Express. 2009;17:7688-93
165. Wu D, Yang J, Zhang G, Jiang H. Noninvasive in vivo monitoring of collagenase induced intracerebral hemorrhage by photoacoustic tomography. Biomed Opt Express. 2017;8:2276-86
166. Estrada H, Huang X, Rebling J, Zwack M, Gottschalk S, Razansky D. Virtual craniotomy for high-resolution optoacoustic brain microscopy. Sci Rep. 2018;8:1459 -
167. Rebling J, Estrada H, Gottschalk S, Sela G, Zwack M, Wissmeyer G. et al. Dual-wavelength hybrid optoacoustic-ultrasound biomicroscopy for functional imaging of large-scale cerebral vascular networks. J Biophotonics. 2018;11:e201800057
168. Wong TTW, Zhang R, Zhang C, Hsu H-C, Maslov KI, Wang L. et al. Label-free automated three-dimensional imaging of whole organs by microtomy-assisted photoacoustic microscopy. Nat Commun. 2017;8:1386 -
169. Kim J, Kim JY, Jeon S, Baik JW, Cho SH, Kim C. Super-resolution localization photoacoustic microscopy using intrinsic red blood cells as contrast absorbers. Light-Sci Appl. 2019;8:103
170. Gottschalk S, Fehm TF, Deán-Ben XL, Razansky D. Noninvasive real-time visualization of multiple cerebral hemodynamic parameters in whole mouse brains using five-dimensional optoacoustic tomography. J Cereb Blood Flow Metab. 2015;35:531-5
171. Ovsepian SV, Olefir I, Ntziachristos V. Advances in Optoacoustic Neurotomography of Animal Models. Trends Biotechnol. 2019;37:1315-26
172. Olefir I, Ghazaryan A, Yang H, Malekzadeh-Najafabadi J, Glasl S, Symvoulidis P. et al. Spatial and Spectral Mapping and Decomposition of Neural Dynamics and Organization of the Mouse Brain with Multispectral Optoacoustic Tomography. Cell Rep. 2019;26:2833-46.e3
173. Gottschalk S, Degtyaruk O, Mc Larney B, Rebling J, Deán-Ben XL, Shoham S. et al. Isolated murine brain model for large-scale optoacoustic calcium imaging. Front Neurosci. 2019;13:290 -
174. Gottschalk S, Degtyaruk O, Mc Larney B, Rebling J, Hutter MA, Deán-Ben XL. et al. Rapid volumetric optoacoustic imaging of neural dynamics across the mouse brain. Nat Biomed Eng. 2019;3:392-401
175. de Roos A, van der Grond J, Mitchell G, Westenberg J. Magnetic Resonance Imaging of Cardiovascular Function and the Brain: Is Dementia a Cardiovascular-Driven Disease? Circulation. 2017;135:2178-95
176. Lerch JP, van der Kouwe AJW, Raznahan A, Paus T, Johansen-Berg H, Miller KL. et al. Studying neuroanatomy using MRI. Nat Neurosci. 2017;20:314-26
177. Glover P, Mansfield SP. Limits to magnetic resonance microscopy. Rep Prog Phys. 2002;65:1489-511
178. Ciobanu L, Webb A, Pennington C. Magnetic resonance imaging of biological cells. Prog Nucl Magn Reson Spectrosc. 2003;42:69-93
179. Uğurbil K. Imaging at ultrahigh magnetic fields: History, challenges, and solutions. Neuroimage. 2018;168:7-32
180. Budinger TF, Bird MD. MRI and MRS of the human brain at magnetic fields of 14T to 20T: Technical feasibility, safety, and neuroscience horizons. Neuroimage. 2018;168:509-31
181. Edlow BL, Mareyam A, Horn A, Polimeni JR, Witzel T, Tisdall MD. et al. 7 Tesla MRI of the ex vivo human brain at 100 micron resolution. Sci Data. 2019;6:244 -
182. Ciobanu L, Pennington CH. 3D micron-scale MRI of single biological cells. Solid State Nucl Magn Reson. 2004;25:138-41
183. Xiao Y-D, Paudel R, Liu J, Ma C, Zhang Z-S, Zhou S-K. MRI contrast agents: Classification and application (Review). Int J Mol Med. 2016;38:1319-26
184. Zeng L, Wu D, Zou R, Chen T, Zhang J, Wu A. Paramagnetic and Superparamagnetic Inorganic Nanoparticles for T1-Weighted Magnetic Resonance Imaging. Curr Med Chem. 2018;25:2970-86
185. Shen Z, Chen T, Ma X, Ren W, Zhou Z, Zhu G. et al. Multifunctional Theranostic Nanoparticles Based on Exceedingly Small Magnetic Iron Oxide Nanoparticles for T1-Weighted Magnetic Resonance Imaging and Chemotherapy. ACS Nano. 2017;11:10992-1004
186. Mestre H, Du T, Sweeney AM, Liu G, Samson AJ, Peng W. et al. Cerebrospinal fluid influx drives acute ischemic tissue swelling. Science. 2020 367
187. Cleary JO, Wiseman FK, Norris FC, Price AN, Choy M, Tybulewicz VLJ. et al. Structural correlates of active-staining following magnetic resonance microscopy in the mouse brain. Neuroimage. 2011;56:974-83
188. Aoki I, Naruse S, Tanaka C. Manganese-enhanced magnetic resonance imaging (MEMRI) of brain activity and applications to early detection of brain ischemia. NMR Biomed. 2004;17:569-80
189. Sato C, Sawada K, Wright D, Higashi T, Aoki I. Isotropic 25-Micron 3D Neuroimaging Using ex vivo Microstructural Manganese-Enhanced MRI (MEMRI). Front Neural Circuits. 2018;12:110 -
190. Thurber KR, Yau W-M, Tycko R. Low-temperature dynamic nuclear polarization at 9.4 T with a 30 mW microwave source. J Magn Reson Imaging. 2010;204:303-13
191. Chen H-Y, Tycko R. Temperature-Dependent Nuclear Spin Relaxation Due to Paramagnetic Dopants Below 30 K: Relevance to DNP-Enhanced Magnetic Resonance Imaging. J Phys Chem B. 2018;122:11731-42
192. Chen H-Y, Tycko R. Low-temperature magnetic resonance imaging with 2.8 μm isotropic resolution. J Magn Reson Imaging. 2018;287:47-55
193. Sánchez-Heredia JD, Baron R, Hansen ESS, Laustsen C, Zhurbenko V, Ardenkjær-Larsen JH. Autonomous cryogenic RF receive coil for 13C imaging of rodents at 3 T. Magn Reson Med. 2019 n/a
194. Elabyad IA, Kalayciyan R, Shanbhag NC, Schad LR. First In Vivo Potassium-39 (K-39) MRI at 9.4 T Using Conventional Copper Radio Frequency Surface Coil Cooled to 77 K. IEEE Trans Biomed Eng. 2014;61:334-45
195. Holdsworth SJ, Bammer R. Magnetic resonance imaging techniques: fMRI, DWI, and PWI. Semin Neurol. 2008;28:395-406
196. ter Telgte A, Wiegertjes K, Gesierich B, Baskaran BS, Marques JP, Kuijf HJ. et al. Temporal Dynamics of Cortical Microinfarcts in Cerebral Small Vessel Disease. JAMA Neurol. 2020
197. Holdsworth SJ, O'Halloran R, Setsompop K. The quest for high spatial resolution diffusion-weighted imaging of the human brain in vivo. NMR Biomed. 2019;32:e4056
198. Lee CH, Blackband SJ, Fernandez-Funez P. Visualization of synaptic domains in the Drosophila brain by magnetic resonance microscopy at 10 micron isotropic resolution. Sci Rep. 2015;5:8920 -
199. Flint JJ, Hansen B, Portnoy S, Lee C-H, King MA, Fey M. et al. Magnetic resonance microscopy of human and porcine neurons and cellular processes. Neuroimage. 2012;60:1404-11
200. Feng L, Jeon T, Yu Q, Ouyang M, Peng Q, Mishra V. et al. Population-averaged macaque brain atlas with high-resolution ex vivo DTI integrated into in vivo space. Brain Struct Funct. 2017;222:4131-47
201. Horgusluoglu-Moloch E, Xiao G, Wang M, Wang Q, Zhou X, Nho K. et al. Systems modeling of white matter microstructural abnormalities in Alzheimer's disease. NeuroImage-Clin. 2020;26:102203
202. Schaeffer S, Iadecola C. Revisiting the neurovascular unit. Nat Neurosci. 2021
203. Segarra M, Aburto MR, Hefendehl J, Acker-Palmer A. Neurovascular Interactions in the Nervous System. Annu Rev Cell Dev Biol. 2019;35:615-35
204. Starosolski Z, Villamizar CA, Rendon D, Paldino MJ, Milewicz DM, Ghaghada KB. et al. Ultra High-Resolution In vivo Computed Tomography Imaging of Mouse Cerebrovasculature Using a Long Circulating Blood Pool Contrast Agent. Sci Rep. 2015;5:10178 -
205. Hong G, Diao S, Chang J, Antaris AL, Chen C, Zhang B. et al. Through-skull fluorescence imaging of the brain in a new near-infrared window. Nat Photonics. 2014;8:723-30
206. Errico C, Pierre J, Pezet S, Desailly Y, Lenkei Z, Couture O. et al. Ultrafast ultrasound localization microscopy for deep super-resolution vascular imaging. Nature. 2015;527:499-502
207. Xi L, Jin T, Zhou J, Carney P, Jiang H. Hybrid photoacoustic and electrophysiological recording of neurovascular communications in freely-moving rats. Neuroimage. 2017;161:232-40
208. Tang J, Coleman JE, Dai X, Jiang H. Wearable 3-D Photoacoustic Tomography for Functional Brain Imaging in Behaving Rats. Sci Rep. 2016;6:25470 -
209. Yang M, Zhou Z, Zhang J, Jia S, Li T, Guan J. et al. MATRIEX imaging: multiarea two-photon real-time in vivo explorer. Light-Sci Appl. 2019;8:109 -
210. Nadella KMNS, Roš H, Baragli C, Griffiths VA, Konstantinou G, Koimtzis T. et al. Random-access scanning microscopy for 3D imaging in awake behaving animals. Nat Meth. 2016;13:1001-4
211. Inoue M, Takeuchi A, Manita S, Horigane S-i, Sakamoto M, Kawakami R. et al. Rational engineering of XCaMPs, a multicolor GECI suite for in vivo imaging of complex brain circuit dynamics. Cell. 2019;177:1346-60.e24
212. Zhang P, Li L, Lin L, Hu P, Shi J, He Y. et al. High-resolution deep functional imaging of the whole mouse brain by photoacoustic computed tomography in vivo. J Biophotonics. 2018;11:10.1002 /jbio.201700024
213. Kircher MF, de la Zerda A, Jokerst JV, Zavaleta CL, Kempen PJ, Mittra E. et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat Med. 2012;18:829-34
214. Zhao J, Chen J, Ma S, Liu Q, Huang L, Chen X. et al. Recent developments in multimodality fluorescence imaging probes. Acta Pharm Sin B. 2018;8:320-38
215. Pinti P, Tachtsidis I, Hamilton A, Hirsch J, Aichelburg C, Gilbert S. et al. The present and future use of functional near-infrared spectroscopy (fNIRS) for cognitive neuroscience. Ann N Y Acad Sci. 2020 1464
216. Lu HD, Chen G, Cai J, Roe AW. Intrinsic signal optical imaging of visual brain activity: Tracking of fast cortical dynamics. Neuroimage. 2017;148:160-8
217. Nguyen MP, Goorden MC, Kamphuis C, Beekman FJ. Evaluation of pinhole collimator materials for micron-resolution ex vivo SPECT. Phys Med Biol. 2019;64:105017
218. Zwettler FU, Reinhard S, Gambarotto D, Bell TDM, Hamel V, Guichard P. et al. Molecular resolution imaging by post-labeling expansion single-molecule localization microscopy (Ex-SMLM). Nat Commun. 2020;11:3388
219. Kim J, Wojcik M, Wang Y, Moon S, Zin EA, Marnani N. et al. Oblique-plane single-molecule localization microscopy for tissues and small intact animals. Nat Meth. 2019;16:853-7
220. Shnaiderman R, Wissmeyer G, Ülgen O, Mustafa Q, Chmyrov A, Ntziachristos V. A submicrometre silicon-on-insulator resonator for ultrasound detection. Nature. 2020;585:372-8
221. Liu C, Liu W, Wang S, Li H, Lv Z, Zhang F. et al. Super-resolution nanoscopy by coherent control on nanoparticle emission. Sci Adv. 2020;6:eaaw6579
Author contact
![]() Corresponding author: Mengqi Zhang (Email: zhangmengqi8912edu.cn)
Corresponding author: Mengqi Zhang (Email: zhangmengqi8912edu.cn)

 Global reach, higher impact
Global reach, higher impact