10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(2):771-782. doi:10.7150/ijbs.66218 This issue Cite
Review
N6-Methyladenosine Modifications in the Female Reproductive System: Roles in Gonad Development and Diseases
1. Institute of Reproductive Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
2. Center of Reproductive Medicine, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
Received 2021-8-18; Accepted 2021-11-15; Published 2022-1-1
Abstract
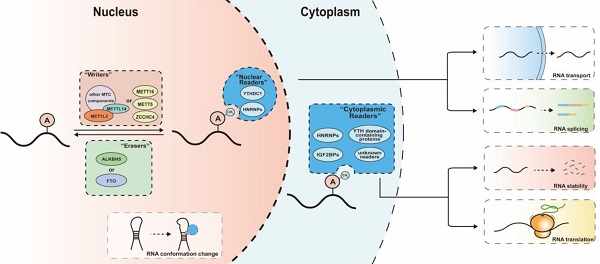
N6-methyladenosine (m6A) is the most prevalent chemical modification in eukaryotic messenger RNAs. By participating in various RNA-related bioprocesses including RNA decay, splicing, transport and translation, m6A serves as a pivotal regulator of RNA fate and plays an irreplaceable role in cellular activities. The m6A modifications of transcripts are coordinately regulated by methyltransferase “writers” and demethylase “erasers”, and produce variable effects via different m6A reading protein “readers”. There is emerging evidence that m6A modifications play a critical role in a variety of physiological and pathological processes in the female reproductive system, subsequently affecting female fertility. Here, we introduce recent advances in research on m6A regulators and their functions, then highlight the role of m6A in gonad development and female reproductive diseases, as well as the underlying mechanisms driving these processes.
Keywords: N6-methyladenosine, epigenetics, oogenesis, fertility, Female reproductive system
Introduction
More than 170 kinds of RNA modifications were found to be widely distributed among various organisms [1], which are collectively known as the 'epitranscriptome'. Substantial studies aimed to uncover the mechanisms and characterizations of these modifications, such as N6-methyladenosine (m6A), N1-methyladenosine, 5-methylcytidine and pseudouridine [2], unveiling a brand-new landscape of epigenetic regulatory mechanisms.
Among all of these modifications, m6A, which has been reported as early as 1974 [3, 4], was considered to be the most prevalent chemical modification in eukaryotic messenger RNAs [2, 5, 6]. The m6A modification is preferentially enriched around stop codons, 5'- and 3'-untranslated regions (UTR), as well as in long internal exons with the consensus motif RRACH (R = A or G, H = A, C or U) [7, 8]. Due to its involvement in various RNA-related bioprocesses, including RNA decay, splicing, transport and translation [9-12], m6A is a pivotal regulator of RNA fate and plays an irreplaceable role in cell differentiation, development, metabolism, stress response and other cellular activities [2, 13-15].
Epigenetic molecules and signaling pathways in the female reproductive system have various effects and are important for female fertility [16-19]. With the deepening of m6A research, there is emerging evidence for the involvement of m6A in various physiological and pathological processes in the female reproductive system, including oogenesis and diseases, subsequently affecting female fertility. Here, we introduce recent advances in research on m6A regulators and their functions, and highlight the role of m6A in oogenesis and female reproductive diseases, as well as the underlying mechanisms driving these processes.
mechanism of m6A modifications. The m6A modification is catalyzed by writers including multicomponent methyltransferase complex (METTL3, METTL14 and other auxiliary components) and several independent methyltransferase (METTL16, METTL5, ZCCHC4), removed by erasers (ALKBH5 or FTO) and recognized by m6A readers (YTH domain-containing proteins, HNRNPs and IGF2BPs). Readers bind to their target RNAs and meditate various downstream effect including RNA transport, splicing, degradation and translation. The m6A modifications can regulate the interaction between RNA and m6A readers by meditating the conformation changes of RNAs.
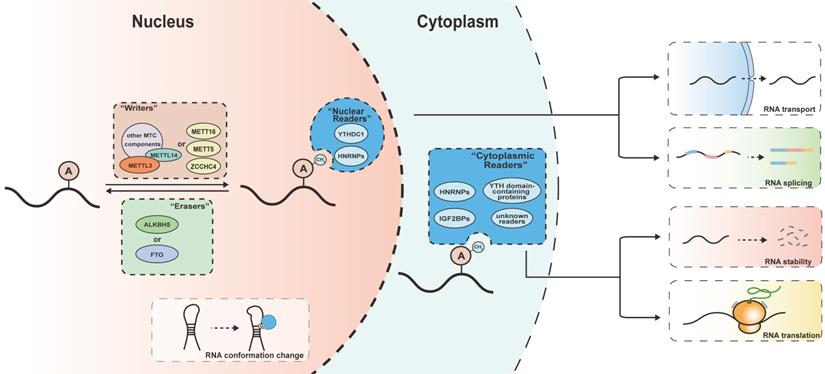
Regulators of m6A
The m6A modification is a reversable process dynamically controlled by m6A writers and erasers, and its variable downstream effects rely on the recognition of m6A modifications by m6A readers (Figure 1).
m6A writers
The introduction of the m6A modification is accomplished by the multicomponent methyltransferase complex (MTC). The MTC consists of the catalytic subunit METLL3 and auxiliary subunits including METTL14, WTAP, KIAA1429, RBM15 and ZC3H13 [20]. METTL3 serves as the core component of the MTC by installing the m6A modification by adding a methyl group to the N6 position of adenine. METTL14 forms a stable heterodimer core complex with METTL3, enabling the latter to recognize its substrates [21-23]. WTAP is indispensable for the correct localization of the METTL3-METTL14 complex in the nuclear speckles, which guarantees the correct continuation of m6A-related processes [24, 25]. KIAA1429 and RBM15 maintain the m6A levels by recruiting the MTC to specific RNA regions [24, 26, 27]. ZC3H13 and its Drosophila homolog Flacc function as a bridge between the MTC and its mRNA targets, and guide RMB15 to WTAP to facilitate m6A modification [28, 29]. Hakai is a newly identified member of the MTC in Drosophila and human cells that helps stabilize other subunits of the MTC through its ubiquitination domain [30]. Given that METTL3 only produces a part of RNA m6A modifications, there must be other writers meditating the other methyl transfer reactions [31]. METTL16 is a newly identified m6A methyltransferase targeting pre-mRNAs and various non-coding RNAs [32, 33]. It consists of two structural domains, whereby the N-terminal methyltransferase domain independently recognizes the 5′-UACAGAGAA motif and the C-terminal vertebrate conserved region binds to internal stem-loops that facilitate the methyltransferase activity of METTL16[34]. In addition, METTL5 and ZCCHC4 were found to be m6A writers that respectively act on the 18S and 28S rRNAs in an MTC-independent manner. METTL5 forms a heterodimer with TRMT112 to maintain its metabolic stability and serves as an exclusive m6A rRNA methyltransferase.[35]
m6A Erasers
Erasers are demethylases that remove m6A modifications in a ferrous iron and α-ketoglutaric acid dependent manner, dynamically regulating the m6A labeling of mRNAs and other types of transcripts in the nucleus. Erasers determine the m6A levels of transcripts together with m6A writers, thus modulating the subsequent effects of m6A readers. There is evidence that FTO controls RNA splicing by modulating he interaction of RNAs with SRSF2 protein [36]. ALKBH5, another known m6A eraser, has been proved to colocalize with mRNA-processing factors in nuclear speckles, and it plays critical roles in RNA transport, synthesis and stability. ALKBH5-dependent demethylation precisely controls the splicing process and length of the 3'-UTR region [37].
m6A Readers
Recognition and binding of m6A readers to their target RNAs is required for m6A modification to execute its important functions in regulating RNA fate. The m6A readers discovered to date include YT521-B homology (YTH) domain-containing proteins, heterogeneous nuclear ribonucleoproteins (HNRNPs), insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) and eukaryotic initiation factor 3 (eIF3)[38, 39].
These m6A readers take part in different RNA metabolic processes and affect different aspects of RNA behavior. YTH domain-containing proteins, the most researched m6A readers, recognize m6A modifications through the YTH domain and subsequently participate in various RNA-related processes [40]. YTHDF1 positively regulates the translation of its targeted RNAs in a m6A-denpend manner [11, 41], YTHDF2 selectively induces the degradation of m6A-modifed mRNAs, decreasing the abundance of its targets [12, 42], while YTHDF3 facilitated decay and translation of RNAs by interacting with YTHDF1 and YTHDF2 [43, 44]. As a nuclear m6A reader, YTHDC1 meditates nuclear RNA export and is required for alternative splicing [45, 46]. YTHDC2 promotes translational activities and also accelerates the degradation of its targets, showing a double-sided effect [47]. HNRNPs and IGF2BPs are also protein families that function as m6A readers. Specifically, the binding affinity of HNRNPC for transcripts can be increased by m6A-mediated RNA structure alteration, leading to the m6A-switch effect [48]. HNRNPA2B1 binds RNA through its RNA recognition motif and recognizes m6A-modified RNA in a non-specific manner, enhancing METTL3-dependent microRNA processing and production [49, 50]. HNRNPG interacts with the phosphorylated carboxy-terminal domain of RNA polymerase II through its RGG motifs and co-transcriptionally regulates alternative splicing [51]. IGF2BPs are a group of RNA binding proteins that selectively bind m6A-modified RNA and share overlapping RNA targets. IGF2BPs enhance mRNA stability and promote mRNA translation, thus upregulating gene expression [52-54].
Biological functions of m6A signals
The introduction of m6A into transcripts mainly occurs in the nucleus, making it a nuclear imprint of RNA fate [55, 56]. The m6A levels determined by the m6A MTC and m6A erasers re closely related to RNA metabolism outcomes in the cytoplasm. Through reversible and dynamic methylation of adenine residues, m6A regulates the stability, splicing, transport and translation of RNAs [21, 22], playing an important role in a variety of physiological and pathological processes.
m6A in mRNA stability
Possible involvement of m6A modification in regulating RNA stability was first explored in 1978. Researchers deduced that rapid elimination of m6A in the cytoplasm may be caused by the shorter half-life of mRNAs with a high m6A content [57]. This hypothesis that mRNAs harboring more m6A were susceptible to degradation has been proved later, and Mettl3 knockout, which significantly decreased the m6A levels of RNAs, remarkably increased their half-life [58].
There is substantial evidence that m6A readers are the main regulators of RNA stability. RNA decay takes place in cytoplasmic processing bodies (P bodies) [59], and YTHDF2 was found to colocalizes with P bodies and its cognate mRNAs were intended to have shorter half-live, demonstrating YTHDF2's role in destabilizing its mRNA targets. The deadenylation-dependent decay pathway was also a part of mechanism driving YTHDF2-mediated RNA decay [12, 42]. YTHDF3 shares targets with its family members and promotes RNA degradation in cooperation with YTHDF2 [44]. Conversely, IGF2BPs enhances mRNA stability by recognizing m6A modifications, increasing the abundance of its mRNA targets. IGF2BPs silencing led to comprehensive downregulation of its target genes [54, 60].
It is worth mentioning that m6A modifications are also deposited in the RNAs of pseudogenes. These RNAs have higher m6A levels than their cognate mRNAs, which facilitates the degradation of processed pseudogene transcripts in the cytoplasm. Given that processed pseudogenes cannot be recognized and cleaned by canonical nonsense-mediated decay process, m6A-meditated cytosolic elimination of these nonfunctional RNAs provides a novel mechanism of RNA surveillance [61].
m6A in mRNA splicing
The m6A modification plays a key role in sex determination in Drosophila by modulating the splicing of Sex lethal transcripts [62-64], revealing a novel posttranscriptional regulatory mechanism of gene expression. However, the effects and functions of m6A modification in RNA splicing in mammals have not been fully elucidated. Some researchers proposed that m6A does not have a major effect on splicing [58]. However, there is accumulating evidence that m6A modification is involved in RNA splicing through multiple mechanisms. Mettl3-depletion partially affected RNA splicing in terms of frequency and type of alternative splicing events [15]. The demethylases FTO and ALKBH5 control this process by regulating m6A levels in alternative exons cassettes and coding sequences, respectively[36, 65]. The nucleus-located m6A reader, YTHDC1, participates in the processing of transcripts together with splicing regulators such as CPSF6, SRSF3, and SRSF7 [10, 66].
Mechanistically, the distribution pattern of m6A modifications may be a key factor driving the impact of m6A on alternative splicing. It was found that m6A is preferentially located in long exons, near the stop codons, and in 3′-UTR [48]. Yet another study found that m6A was enriched near splice sites in both exonic and intronic regions [67]. This differential distribution may lead to region-specific binding of splicing-related proteins, thereby mediating alternative splicing. The indirect effect of m6A on RNA splicing should also be taken into consideration. By controlling the expression of highly m6A-modified splicing regulators such as SON44, HNRNPC38 and HNRNPF [68], m6A may also alter RNA splicing outcomes without direct participation.
m6A in mRNA transport
Correct subcellular localization of transcripts is closely related to their function, making m6A modification an important regulator of RNA fate by affecting nuclear RNA export. A conditional knockout of Mettl14 led to nuclear retention of m6A-modified mRNAs by impairing the preferential binding of FMRP [69]. ALKBH5-deficient cells with increased m6A levels showed accelerated nuclear export and massive cytoplasmic accumulation of mRNA, leading to significant changes in the ratio of nuclear and cytoplasmic mRNA content [9].The m6A reader YTHDC1 was found to enhance the binding of m6A-modified RNAs to nuclear export adaptor protein SRSF3 and mRNA export receptor NXF1, which facilitates the nuclear export of its target mRNAs[46]. These studies proved the involvement of m6A in nuclear RNA export, but its effect on the whole transcriptome needs to be further elucidated.
m6A in mRNA Translation
Some researchers have shown that m6A facilitates mRNA translation by interacting with eukaryotic translation initiation factor eIF3 via two different pathways [11, 70]. As one of the most important translation initiation factors in eukaryotic cells eIF3 is usually bound to the 5′ untranslated regions of mRNAs and recruits the small ribosomal subunit around its binding sites, thereby regulating translation initiation [71]. It has been shown that the effect of the m6A reader YTHDF1 on protein expression is dependent on the eIF3-related translation machinery. YTHDF1-binding mRNAs showed a higher ribosome-bound fraction and translation efficiency [11]. Conversely, YTHDF1 silencing led to significantly downregulated protein production of its target mRNAs. Since YTHDF1 usually binds RNA close to stop codons in mammalian cells, the effect of 3'UTR-enriched YTHDF1 was based on eIF4G-induced spatial proximity to translation initiation sites [11]. Direct eIF3 recruitment around 5'UTR also initiated mRNA translation. Cell-stress associated mRNAs contain m6A modifications around the 5'UTR region and exhibit enhanced cap-independent translation [70]. Additionally, YTHDF1 was found to augment the translation of EIF3C in an m6A-dependent manner and form a YTHDF1-EIF3C axis promoting general protein synthesis [41].
Furthermore, METTL3, which was generally known as a methylase acting in the nucleus, also took part in mRNA regulation by interacting with the translation initiation machinery. Cytoplasmic METTL3 physically and functionally associates with eIF3 subunits, bridging its binding sites to the 5′-end of the mRNA to meditate mRNA circularization [72, 73]. METTL3 depletion remarkably downregulated the expression of its target mRNAs [73].
m6A in non-coding RNA
In addition to mRNAs, the deposition of m6A modifications also affects the fate of modified non-coding RNA (ncRNA) in various ncRNA-related bioprocesses [74]. METTL3 induced m6A hyper-methylation facilitated stabilizing lncRNA, thus enhancing its expression [75, 76]. FTO decreased the m6A level on the LINC00022 transcript and inhibited its degradation via the m6A reader YTHDF2[77].HNRNPA2B1, a nuclear m6A reader, showed a METTL3-like effect in RNA splicing and promoted primary miRNA processing by interacting with the microRNA microprocessor complex protein DGCR8[50]. YTHDC1 was involved in the circRNA transport process. YTHDC1 silencing led to significant nuclear retention of circNSUN2, which was rescued by enforced expression of wild-type YTHDC1 [78]. Moreover, the m6A modifications can initiate circRNA translation through YTHDF3-mediated recruitment of the translation initiation complex [79].
The m6A modification plays a key role in modulating interaction between ncRNAs and their targeted RNAs. For example, m6A modification promoted miR-133a binding to its targets and enhanced the miR-133a repression [80]. LncRNA ILF3-AS1 recruited METTL3 to ILF3 mRNA, which increased its m6A level and strengthened its stability [81]. The m6A-dependent RNA structural changes regulate the accessibility of RNA binding motifs of proteins to affect RNA-protein interactions, which is termed as “m6A-switch”. The m6A modification destabilizes hairpin stem structure of lncRNA MALAT1, thus making it more accessible for the RNA binding proteins [48].
m6A in female gonad development and gametogenesis
Establishment of gonadal structure and function is dependent on the development of both germline cells and somatic cells. The oocyte and its surrounding somatic cells form the basic functional unit of the ovary, the follicle. Normal follicle development and gametogenesis are prerequisites for female fertility.
Follicle development, also known as folliculogenesis, is a long and complex process in which the recruited primordial follicle either develops into a mature Graafian follicle to ovulate or undergoes atresia. Folliculogenesis is synchronized with the growth and maturation of recruited oocytes, providing necessary sex steroids and establishing an appropriate microenvironment for oocytes.
In a broad sense, oogenesis is a highly sophisticated and coordinated process by which primordial germ cells (PGCs) develop into mature oocytes to prepare for fertilization. In humans, PGCs are specified as early as the peri-implantation window of the embryo at day 12 post-fertilization [82]. PGCs exist transiently and soon differentiate into oogonia after undergoing genome-wide epigenetic reprogramming [83]. The primary oocytes derived from diploid oogonia then enter meiosis and remain arrested in prophase I at birth. The prophase I oocytes have an intact germinal vesicle (GV) and remain dormant until gonadotropin stimulation in puberty, which can lead to germinal vesicle breakdown (GVBD). Following GVBD, the oocytes continue to complete meiosis I with formation of metaphase I spindles and separation of paired homologous chromosomes. Afterwards, the oocytes directly enter meiosis II and arrest for the second time in metaphase II (MII) until fertilization triggers the completion of meiosis.
Gene expression is precisely and dynamically controlled during the whole female gonad developmental process. In follicles, proliferation and differentiation of somatic cells relies on the adequate alteration of gene expression patterns, while in oocytes, the changes are even more intricate. Transcription of oocyte genes is active in the early stage of follicle formation and ceases at the GV stage[84]. Maternal mRNAs synthesized in this period dominate the subsequent developmental phases of oocytes and early embryos until zygotic genome activation. As the oocytes emerge from meiotic arrest, maternal mRNAs are constantly degraded and decrease to the lowest level in the two-cell embryo [85]. All these processes largely rely on the post-transcriptional regulation of mRNA metabolism and expression. There is wide-ranging evidence for the importance of epigenetic modification in folliculogenesis and oogenesis [31, 86, 87], and the roles of m6A modification in these processes have begun to be partially elucidated in recent years. The m6A level was found to generally increase along with the development of the mouse ovary, reaching the highest point at the luteal phase. The m6A associated regulators also demonstrated a similar trend in accordance with the m6A levels, whereby m6A writers increased and erasers decreased from 12.5 days post coitum to the mature phase in the ovary, suggesting the potential impact of m6A modification during female gonad development [88]. There is increasing evidence for the involvement of m6A in specific stages and aspects of this process (Figure 2).
Follicle development
Follicle development is a dynamic and continuous process that happens simultaneously with oocyte growth. Before the resumption of meiosis, oocytes remain in prophase I stage and continue accumulating water, ions, carbohydrates and lipids until GVBD. Follicles provide a developmental microenvironment for oocyte growth and help sustain meiotic arrest [89, 90]. MeRIP-seq identified changes in the abundance and distribution of m6A modifications in granulosa cells during follicular development [91].
Mettl3 mutation reduced m6A levels in zebrafish ovaries. These Mettl3-defective females had smaller gonads than wild-type fish, and a large fraction of follicles failed to fully develop [92]. KIAA1429 was found to be expressed in the oocyte nucleus and granulosa cells from follicles in different stages. Increased numbers of primary follicles, decreased secondary, preantral and antral follicles, as well as multiple oocyte follicles were found in Kiaa1429 ZP3-cKO mice, indicating that Kiaa1429 deficiency led to arrest and disorganization of follicle developmental. Moreover, Kiaa1429 deficiency inhibited the proliferation and promoted the apoptosis of granulosa cells. All of these findings suggested that KIAA1429 is required for normal folliculogenesis in mice [93].
Role of m6A modification in gonad development. The m6A modification regulates oogenesis in terms of follicle development, oocyte maturation, maternal-to-zygote maturation and self-renewal of female germline stem cells, which partially determine oocyte competence and female fertility.
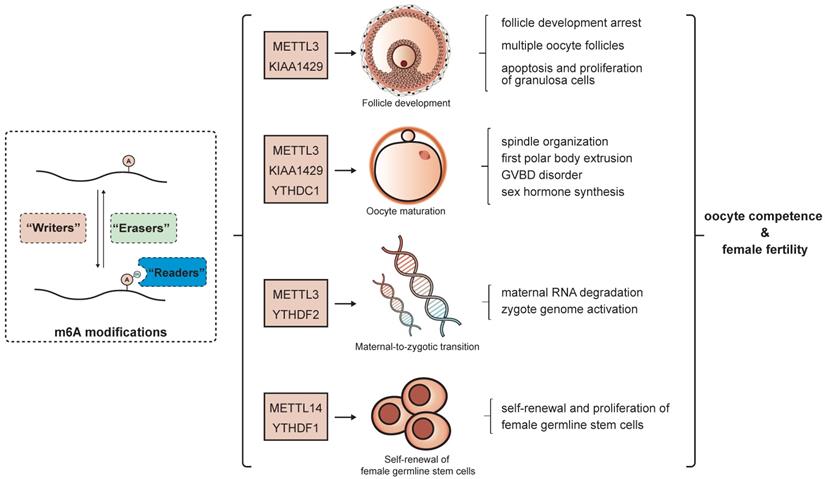
Oocyte maturation
Under the stimulation of LH surge, fully grown oocytes emerge from meiotic arrest and complete subsequent developmental progression up to MII stage prior to ovulation and fertilization. This process, which marks the maturation of oocytes, is largely dependent on precise epigenetic regulation in oocytes [16]. The roles of m6A in oocyte maturation encompass multiple aspects, including RNA splicing, degradation, transport and translation.
METTL3 expression shares a similar pattern with m6A levels in the processes of oogenesis and early embryonic development. They are both abundant in GV and MII oocytes, then dramatically decrease in the zygote and gradually increase during early embryogenesis. Mettl3 knockdown at the GV stage hindered spindle organization and first polar body extrusion in MII oocytes, which could be partially attributed to decreased translational efficiency of several meiosis-related genes [94]. In zebrafish, GVBD disorder and oocyte incompetence were observed in mettl3-mutant females. The mettl3 mutation disturbed sex hormone levels by regulating genes involved in sex hormone synthesis and gonadotropin signaling. A decreased GVBD rate could be partially rescued by hCG or 17α-20β-DHP treatment [92]. Oocyte-specific conditional knockout (cKO) of Kiaa1429 led to a significantly reduced GVBD rate and first polar body extrusion rate. Mechanistically, Kiaa1429 cKO impaired the correct localization of splicing-related factors SRSF3 and YTHDC1, perturbed the alternative splicing of oogenesis-related transcripts, and led to meiotic arrest [93]. The nuclear m6A reader YTHDC1 regulates RNA splicing in oocytes by interacting with pre-mRNA processing factors CPSF6, SRSF3, and SRSF7. Loss of YTHDC1 changed the alternative polyadenylation pattern and led to massive alternative splicing defects in the mouse oocyte nucleus [66]. It is worth mentioning that both Kiaa1429-dificient and Ythdf1-dificient oocytes showed abnormal cytoplasmic RNA granules. Considering that both KIAA1429 and YTHDF1 are closely related to m6A-mediated RNA splicing, this RNA mislocalization may be caused by inappropriate posttranscriptional processing and transport [66, 93]. However, the impact of these RNA granules is still unclear.
Maternal-to-zygotic transition
The maternal-to-zygotic transition (MZT) consists of selective maternal mRNA degradation and zygotic genome activation. The MZT starts at the resumption of oocyte meiosis rather than fertilization [95]. Correct elimination of maternal transcripts lays the foundation for zygotic genome activation and the upcoming embryogenesis [16, 96], and is part of the oocyte competence built up in preceding developmental processes. It has been demonstrated that MZT is partially controlled m6A modification.
Mettl3 knockdown impeded embryonic development from the two-cell stage to four-cell stage in activated and diploidized MII oocytes. The Mettl3-mropholino treated group showed remarkably decreased global transcription. RNA-seq assays detected hundreds of upregulated genes, part of which are enriched in GO terms related to RNA metabolic processes and cell cycle, demonstrating that METTL3 knockdown may affect MZT by impairing mRNA degradation[94]. The clearance of maternal RNAs was disturbed in ythdf2-dificient zebrafish. Upregulated maternal transcripts and downregulated zygotic transcripts were observed in both ythdf2 homozygous mutant oocytes and ythdf2-morpholino treated oocytes. The ythdf2-dificiency-meditated RNA retention occurred in a m6A preferential manner, leading to obvious embryonic developmental delay in zebrafish [97]. In mice, Ythdf2 knockout did not affect follicle development or oocyte meiosis, but caused serious embryonic developmental defects, resulting in complete female infertility. YTHDF2 deficiency mainly affected gene expression in MII oocytes instead of GV oocytes. This suggested that YTHDF2 did not regulate the maternal transcriptome but was required to maintain the appropriate transcript dosage [98].
Self-renewal of female germline stem cells
The discovery of female germline stem cells (FGSCs) provided a theoretical basis to obtain additional oocytes even after birth. Zou et al. established a neonatal mouse FGSC line and produced offspring using FGSCs retrieved from adult mice, providing a novel idea for research on oogenesis [99].
RNA m6A profiling revealed differential m6A patterns between FGSCs and somatic STO cells. Compared to STO cells, FGSCs had higher m6A abundance and manifested dissimilar mRNA expression levels of m6A regulators. Knockdown of the m6A reader YTHDF1 hindered the proliferation of FGSCs, but the specific mechanisms have not yet been explored [100]. METTL14 participates in the self-renewal of FGSCs by facilitating the m6A modification of circGFRα1, which acts as a sponge of miR-449 and promotes the expression of their common target GFRα1 to sustain FGSCs proliferation. METTL14 knockdown resulted in decreased m6A modification of circGFRα1, which did not change its expression level but led to its nuclear retention. Similarly, m6A-motif-mutated circGFRα1 mainly accumulated in the nucleus after its overexpression, suggesting that cytoplasmic transport was dependent on m6A [101]. All these findings support the involvement of m6A in postnatal oocyte production from FGSCs.
m6A in female reproductive diseases
There is evidence that m6A modification takes part in the process of inflammation and immune activity [102, 103]. Considering that inflammatory or immune disorders play a critical role in many female reproductive diseases such as endometriosis and adenomyosis, polycystic ovary syndrome, preeclampsia and spontaneous abortion [104, 105], it stands to reason that m6A modification may also contribute to the pathological mechanisms underlying these diseases.
Endometriosis and adenomyosis
Bioinformatic analyses revealed that most of the m6A regulators in the eutopic endometrium were aberrantly expressed compared to normal or eutopic endometrium. Among all the regulators, HNRNPA2B1 and HNRNPC were negatively correlated with the severity of endometriosis, and might influence infiltrating immune cells [106]. The m6A content was found to be dramatically decreased in eutopic and ectopic samples, which was consistent with decreased METTL3 and increased ALKBH5 levels. METTL3 regulates the migration and invasion of endometrial stromal cells by facilitating DGCR8-mediated maturation of miR126, contributing to endometriosis development [107]. Similarly, the expression pattern of m6A regulators in the myometrium was also found to be different from normal tissues. The cluster of m6A regulators may potentially affect the immune response and cell adhesion according to data mining, playing roles in the myometrium dysfunction [108].
Polycystic ovary syndrome
Polycystic ovary syndrome (PCOS) is often accompanied by insulin resistance. FTO overexpression diminished m6A modification in FLOT2 transcripts and enhanced its expression in granulosa cell line KGN. FLOT2 is associated with insulin resistance and was found to be upregulated in granulosa cells from PCOS patients. FTO was found to regulate cell proliferation, apoptosis and insulin resistance in KGN cells through a FLOT2-dependent manner, indicating that FTO might participate in regulating insulin resistance in PCOS [109]. Luteinized granulosa cells from non-obese PCOS patients had higher m6A levels and exhibited a dissimilar distribution pattern of m6A peaks in comparison to the control group. Mechanistically, the FOXO3 transcript was m6A modified and underwent YTHDF2‐mediated decay in controls, which was in contrast to the samples from PCOS patients. Reduced m6A modification of the FOXO3 mRNA in controls enhanced the expression of FOXO3 after METTL3 or METTL14 knockdown, while having the inverse effect after FTO knockdown. However, this effect was absent from the PCOS granulosa cells[110]. These findings illustrate a potential mechanism driving insulin resistance in PCOS patients.
Premature ovarian insufficiency
In addition to increased m6A modification in naturally aging mouse ovaries [111], the levels of m6A modification ware also found to be obviously upregulated in either ovarian tissues or human granulosa cells from premature ovarian insufficiency (POI) patients. This elevation was associated with a relative reduction of FTO compared to ALKBH5. FTO knockdown impeded cell proliferation, alleviated apoptosis and disturbed marker expression in human granulosa cells[112]. Furthermore, m6A was also increased in a mouse model of cyclophosphamide (CTX)-induced premature ovarian insufficiency in a concentration- and time-dependent manner. CTX treatment upregulated the m6A writers METTL3, METTL14, ZC3H13 and KIAA1429, while downregulating the demethylase FTO and several m6A readers[112, 113].These results indicate that m6A modification may act as a potential biomarker of ovarian dysfunction [112, 113].
Preeclampsia
Dysfunction of the placenta in preeclampsia and eclampsia patients usually leads to abnormal fetal growth and serious complications for the pregnant women. Previous studies have identified the role of epigenetics in preeclampsia and fetal growth restriction [114, 115]. The potential involvement of m6A modifications in the pathological processes of preeclampsia has been proposed based on several lines of evidence. MeRIP-seq analysis demonstrated a correlation between higher m6A levels at the 5′‐UTR in placental mRNAs and small‐for‐date children, whereas decreased m6A levels near stop codons were dominant in heavy‐for‐date placenta samples. Higher SMPD1 protein levels in preeclampsia placenta samples, in particular, are more likely to result from m6A enrichment at 5′‐UTR instead of increased mRNA levels [116]. Enhanced METTL3 expression was found to be responsible for increased m6A methylation in placental trophoblasts from preeclampsia patients. These aberrant m6A changes lead to the upregulation of hnRNPC1/C2 expression, which may induce vitamin D deficiency by inducing VDR and trophoblast dysfunction [117]. Furthermore, METTL3 was found to facilitate the maturation of miR-497-5p/195-5p by mediating DGCR8's recognition of pri-miR-497-5p/195-5p in an m6A-dependent manner. Increased miR-497-5p/195-5p in the preeclampsia-affected placenta was found to suppress WWP1 expression, disturbing trophoblast proliferation, migration, and invasion, eventually aggravating preeclampsia progression [118].
Role of m6A in female reproductive diseases.
| diseases | m6A regulator | aberrant expression | target | effect of m6A on target RNA | effect of m6A in diseases | reference |
|---|---|---|---|---|---|---|
| Endometriosis | METTL3 | ↓ | miR-126 | Enhanced DGCR8-mediated maturation of pri-miR126 | Decreased METTL3 and m6A promoted the migration and invasion of endometrial stromal cells in endometriosis | [107] |
| Polycystic ovary syndrome | FTO | - | FLOT2 | FTO overexpression diminished m6A modification in FLOT2 transcripts and enhanced its expression in granulosa cell line KGN | FTO might regulate insulin resistance process in PCOS by enhancing FLOT2 expression | [109] |
| YTHDF2 | - | FOXO3 | Lower m6A levels inhibited YTHDF2-meditated FOXO3 transcript decay | m6A might regulate function of granulosa cells by controlling FOXO3 expression | [110] | |
| Premature ovarian insufficiency | FTO | ↓ | - | - | FTO knockdown impeded cell proliferation, alleviated cell apoptosis and disturbed expression of cell markers in human granulosa cells | [112] |
| Preeclampsia | - | - | SMPD1 | m6A modification of SMPD1 at the 5′‐UTR promoted protein translation at posttranscriptional level | m6A levels in placenta were related to birth weight | [116] |
| METTL3 | ↑ | hnRNPC1/C2 | METTL3 knockdown significantly reduced hnRNPC1/C2 expression in trophoblast cells | - | [117] | |
| METTL3 | - | miR-497-5p/195-5p | METTL3 facilitated maturation of miR-497-5p/195-5p by mediating recognition of pri-miR-497-5p/195-5p by DGCR8 | METTL3-meditated upregulation of miR-497-5p/195-5p disturbed trophoblast proliferation, migration, and invasion, eventually aggravating preeclampsia progression | [118] | |
| Recurrent miscarriage/ spontaneous abortion | METTL14 | ↑ | lncHZ01 | METTL14 stabilized lncHZ01 and formed a positive feedback loop with LncHZ01 | METTL14 enhanced lncHZ01 expression to inhibit trophoblast cell proliferation and induces miscarriage. | [122] |
| ALKBH5 | ↑ | CYR61 | ALKBH5 diminished m6A modification on CYR61 mRNA, impaired its stability and subsequent protein expression | inhibited proliferation and invasion of trophoblast at the maternal-fetal face in early pregnancy | [121] |
Recurrent miscarriage/spontaneous abortion
The pathogenesis of recurrent miscarriage (RM) has not been fully elucidated yet. In addition to the known factors including genetic, endocrine, anatomical, and immunological disorders [119], epigenetic abnormalities are also part of the underlying mechanisms [120]. So far, the research on the role of m6A in RM has mainly focused on its effects on trophoblast function. Higher ALKBH5 expression and lower m6A levels were found in chorionic villi from RM patients. ALKBH5 diminished m6A modification of CYR61 mRNA, reducing its stability and subsequent protein expression. These changes inhibited the proliferation and invasion of trophoblast cell at the maternal-fetal interface in early pregnancy, suggesting that ALKBH5 may be a potential key factor in the pathogenesis of RM [121]. The m6A modification was also found to be involved in BPDE-related RM. BPDE, a metabolite of environmental benzo(a)pyrene, was found to upregulate lncHZ01 in trophoblast cells, and lncHZ01-meditated MXD1 upregulation promoted METTL14 transcription, which increased m6A modification and enhanced the stability of lncHZ01, forming a positive feedback loop that eventually inhibited trophoblast cell proliferation and induced miscarriage[122]. Higher m6A levels and several aberrantly expressed m6A regulators were found in spontaneous abortion (SA) patients. FTO, in particular, was significantly downregulated in the chorionic villi and trophoblasts of SA patients. Lower FTO changed the m6A patterns of several genes involved in immunotolerance, immune cell infiltration and angiogenesis, demonstrating a possible role of m6A in SA progression [123].
Conclusions
The research on m6A RNA modification has expanded broad horizons for researchers and uncovered several new epigenetic mechanisms underlying a number of pathophysiological processes. This reversible modification is controlled by three kinds of regulators and influences various RNA-related processes such as RNA degradation, splicing, transport and translation, regulating gene expression from multiple aspects at the post-transcriptional level. The involvement of m6A modifications has been observed in various bioprocesses including cell differentiation, development, metabolism, stress response and other cellular activities [2, 13-15], serving as an important epigenetic regulatory mechanism underlying both physiological and pathological processes.
In the female reproductive system, m6A modifications regulate multiple stages of oogenesis, including follicle development, oocyte maturation, and the maternal-to-zygotic transition, as well as the self-renewal of female germline stem cells artificially established in vitro. Furthermore, m6A participates in the pathogenesis of a variety of female reproductive diseases including endometriosis and adenomyosis, polycystic ovary syndrome, premature ovarian insufficiency, preeclampsia, and recurrent miscarriage, all of which underscore the impact of m6A on female fertility.
Recently several researches have revealed the clinical potential of m6A-targeting strategies. METTL3 inhibitors exhibited the promising anti-cancer effect against acute myeloid leukemia [124, 125]. pharmacological inhibition of FTO significantly suppressed leukemia stem/initiating cell self-renewal and sensitized leukemia cells to T cell cytotoxicity[126]. However, m6A was only considered as a prospective therapeutic target of cancer treatment in current evidences [127]. Considering its crucial participation in female gonad development and reproductive diseases, m6A-targeting interventions should also be taken into account as promising approaches in treatment of infertility and reproductive disorders.
Nevertheless, our understanding of the full impact of m6A is far from complete. For example, how different m6A readers recognize m6A-modified transcripts and lead to different transcript outcomes is still unclear. More importantly, translational studies aiming to utilize m6A as a novel target for therapy are in their infancy. All these unsolved questions merit further studies in the future.
Acknowledgements
Funding: This work was supported by the National Natural Science Foundation of China (NSFC 92068120).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A. et al. MODOMICS: a database of RNA modification pathways-2013 update. Nucleic acids research. 2013;41:D262-7
2. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nature reviews Molecular cell biology. 2017;18:31-42
3. Perry RP, Kelley DE. Existence of methylated messenger RNA in mouse L cells. Cell. 1974;1:37-42
4. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:3971-5
5. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nature reviews Molecular cell biology. 2014;15:313-26
6. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nature reviews Genetics. 2014;15:293-306
7. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
8. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-46
9. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
10. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF. et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507-19
11. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-99
12. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-20
13. Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan X. et al. N(6)-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol Cell. 2018;69:636-47.e7
14. Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L. et al. The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron. 2018;99:389-403.e9
15. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science (New York, NY). 2015;347:1002-6
16. He M, Zhang T, Yang Y, Wang C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Frontiers in cell and developmental biology. 2021;9:654028
17. Ramarao-Milne P, Kondrashova O, Barry S, Hooper JD, Lee JS, Waddell N. Histone Modifying Enzymes in Gynaecological Cancers. Cancers. 2021 13
18. Bar-Sadeh B, Rudnizky S, Pnueli L, Bentley GR, Stöger R, Kaplan A. et al. Unravelling the role of epigenetics in reproductive adaptations to early-life environment. Nat Rev Endocrinol. 2020;16:519-33
19. Pacini G, Paolino S, Andreoli L, Tincani A, Gerosa M, Caporali R. et al. Epigenetics, pregnancy and autoimmune rheumatic diseases. Autoimmun Rev. 2020;19:102685
20. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187-200
21. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z. et al. Corrigendum: Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2017;542:260
22. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63:306-17
23. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology. 2014;10:93-5
24. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell reports. 2014;8:284-96
25. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24:177-89
26. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-73
27. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z. et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell discovery. 2018;4:10
28. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes & development. 2018;32:415-29
29. Wen J, Lv R, Ma H, Shen H, He C, Wang J. et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018;69:1028-38.e6
30. Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D. et al. Hakai is required for stabilization of core components of the m(6)A mRNA methylation machinery. Nature communications. 2021;12:3778
31. Christou-Kent M, Dhellemmes M, Lambert E, Ray PF, Arnoult C. Diversity of RNA-Binding Proteins Modulating Post-Transcriptional Regulation of Protein Expression in the Maturing Mammalian Oocyte. Cells. 2020 9
32. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C. et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004-14
33. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP. et al. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824-35.e14
34. Aoyama T, Yamashita S, Tomita K. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic acids research. 2020;48:5157-68
35. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic acids research. 2019;47:7719-33
36. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research. 2014;24:1403-19
37. Zheng G, Dahl John A, Niu Y, Fedorcsak P, Huang C-M, Li Charles J. et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Molecular Cell. 2013;49:18-29
38. Meyer KD, Jaffrey SR. Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319-42
39. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Molecular Cancer. 2019 18
40. Liao S, Sun H, Xu C. YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinformatics. 2018;16:99-107
41. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y. et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic acids research. 2020;48:3816-31
42. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M. et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nature communications. 2016;7:12626
43. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y. et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell research. 2017;27:444-7
44. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell research. 2017;27:315-28
45. Xiang Y, Laurent B, Hsu C-H, Nachtergaele S, Lu Z, Sheng W. et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573-6
46. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y. et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife. 2017 6
47. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y. et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell research. 2017;27:1115-27
48. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
49. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR. et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nature communications. 2018;9:420
50. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299-308
51. Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN. et al. Regulation of Co-transcriptional Pre-mRNA Splicing by m(6)A through the Low-Complexity Protein hnRNPG. Mol Cell. 2019;76:70-81.e9
52. Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z. et al. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clinical and translational medicine. 2021;11:e426
53. Bechara R, Amatya N, Bailey RD, Li Y, Aggor FEY, Li DD. et al. The m(6)A reader IMP2 directs autoimmune inflammation through an IL-17- and TNFα-dependent C/EBP transcription factor axis. Sci Immunol. 2021 6
54. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
55. Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M. et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567:414-9
56. Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L. et al. The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256-9
57. Sommer S, Lavi U, Darnell JE Jr. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. Journal of molecular biology. 1978;124:487-99
58. Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S. et al. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes & development. 2017;31:990-1006
59. Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science (New York, NY). 2003;300:805-8
60. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN. et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112
61. Tan L, Cheng W, Liu F, Wang DO, Wu L, Cao N. et al. Positive natural selection of N6-methyladenosine on the RNAs of processed pseudogenes. Genome Biol. 2021;22:180
62. Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH. et al. m(6)A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242-7
63. Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim KS. et al. The m(6)A pathway facilitates sex determination in Drosophila. Nature communications. 2017;8:15737
64. Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG. et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301-4
65. Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y. et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E325-e33
66. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS genetics. 2018;14:e1007412
67. Louloupi A, Ntini E, Conrad T, Ørom UAV. Transient N-6-Methyladenosine Transcriptome Sequencing Reveals a Regulatory Role of m6A in Splicing Efficiency. Cell reports. 2018;23:3429-37
68. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nature reviews Molecular cell biology. 2019;20:608-24
69. Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H. et al. FMRP Modulates Neural Differentiation through m(6)A-Dependent mRNA Nuclear Export. Cell reports. 2019;28:845-54.e5
70. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O. et al. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999-1010
71. Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111-4
72. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62:335-45
73. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J. et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556-60
74. Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer cell. 2020;37:270-88
75. Chen JQ, Tao YP, Hong YG, Li HF, Huang ZP, Xu XF. et al. M(6)A-mediated up-regulation of LncRNA LIFR-AS1 enhances the progression of pancreatic cancer via miRNA-150-5p/ VEGFA/Akt signaling. Cell cycle (Georgetown, Tex). 2021:1-12
76. Liu HT, Zou YX, Zhu WJ, Sen L, Zhang GH, Ma RR. et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell death and differentiation. 2021
77. Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y. et al. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. Journal of experimental & clinical cancer research: CR. 2021;40:294
78. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nature communications. 2019;10:4695
79. Sinha T, Panigrahi C, Das D, Chandra Panda A. Circular RNA translation, a path to hidden proteome. Wiley interdisciplinary reviews RNA. 2021: e1685.
80. Qian B, Wang P, Zhang D, Wu L. m6A modification promotes miR-133a repression during cardiac development and hypertrophy via IGF2BP2. Cell Death Discov. 2021;7:157
81. Bo C, Li N, He L, Zhang S, An Y. Long non-coding RNA ILF3-AS1 facilitates hepatocellular carcinoma progression by stabilizing ILF3 mRNA in an m(6)A-dependent manner. Hum Cell. 2021;34:1843-54
82. Chen D, Sun N, Hou L, Kim R, Faith J, Aslanyan M. et al. Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell reports. 2019;29:4568-82.e5
83. Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nature reviews Genetics. 2016;17:585-600
84. Eppig JJ, Schroeder AC. Capacity of mouse oocytes from preantral follicles to undergo embryogenesis and development to live young after growth, maturation, and fertilization in vitro. Biol Reprod. 1989;41:268-76
85. Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development (Cambridge, England). 2009;136:3033-42
86. Pan Z, Zhang J, Li Q, Li Y, Shi F, Xie Z. et al. Current advances in epigenetic modification and alteration during mammalian ovarian folliculogenesis. J Genet Genomics. 2012;39:111-23
87. Kageyama S, Liu H, Kaneko N, Ooga M, Nagata M, Aoki F. Alterations in epigenetic modifications during oocyte growth in mice. Reproduction (Cambridge, England). 2007;133:85-94
88. Sun X, Zhang J, Jia Y, Shen W, Cao H. Characterization of m6A in mouse ovary and testis. Clinical and translational medicine. 2020;10:e141
89. Zhang M, Su YQ, Sugiura K, Xia G, Eppig JJ. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science (New York, NY). 2010;330:366-9
90. Tsuji T, Kiyosu C, Akiyama K, Kunieda T. CNP/NPR2 signaling maintains oocyte meiotic arrest in early antral follicles and is suppressed by EGFR-mediated signaling in preovulatory follicles. Mol Reprod Dev. 2012;79:795-802
91. Cao Z, Zhang D, Wang Y, Tong X, Avalos LFC, Khan IM. et al. Identification and functional annotation of m6A methylation modification in granulosa cells during antral follicle development in pigs. Animal reproduction science. 2020;219:106510
92. Xia H, Zhong C, Wu X, Chen J, Tao B, Xia X. et al. Mettl3 Mutation Disrupts Gamete Maturation and Reduces Fertility in Zebrafish. Genetics. 2018;208:729-43
93. Hu Y, Ouyang Z, Sui X, Qi M, Li M, He Y. et al. Oocyte competence is maintained by m(6)A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell death and differentiation. 2020;27:2468-83
94. Sui X, Hu Y, Ren C, Cao Q, Zhou S, Cao Y. et al. METTL3-mediated m(6)A is required for murine oocyte maturation and maternal-to-zygotic transition. Cell cycle (Georgetown, Tex). 2020;19:391-404
95. Sha QQ, Zhang J, Fan HY. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals†. Biol Reprod. 2019;101:579-90
96. Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J. et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104-17
97. Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A. et al. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475-8
98. Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN. et al. The RNA m(6)A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol Cell. 2017;67:1059-67.e4
99. Zou K, Yuan Z, Yang Z, Luo H, Sun K, Zhou L. et al. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat Cell Biol. 2009;11:631-6
100. Zhao X, Tian GG, Fang Q, Pei X, Wang Z, Wu J. Comparison of RNA m(6)A and DNA methylation profiles between mouse female germline stem cells and STO cells. Mol Ther Nucleic Acids. 2021;23:431-9
101. Li X, Tian G, Wu J. Novel circGFRα1 Promotes Self-Renewal of Female Germline Stem Cells Mediated by m(6)A Writer METTL14. Frontiers in cell and developmental biology. 2021;9:640402
102. Shulman Z, Stern-Ginossar N. The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat Immunol. 2020;21:501-12
103. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L. et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell. 2019;76:96-109.e9
104. Vannuccini S, Clifton VL, Fraser IS, Taylor HS, Critchley H, Giudice LC. et al. Infertility and reproductive disorders: impact of hormonal and inflammatory mechanisms on pregnancy outcome. Hum Reprod Update. 2016;22:104-15
105. Zhang X, Wei H. Role of Decidual Natural Killer Cells in Human Pregnancy and Related Pregnancy Complications. Front Immunol. 2021;12:728291
106. Jiang L, Zhang M, Wu J, Wang S, Yang X, Yi M. et al. Exploring diagnostic m6A regulators in endometriosis. Aging. 2020 12
107. Li X, Xiong W, Long X, Dai X, Peng Y, Xu Y. et al. Inhibition of METTL3/m6A/miR126 promotes the migration and invasion of endometrial stromal cells in endometriosis. Biol Reprod. 2021
108. Zhai J, Li S, Sen S, Opoku-Anane J, Du Y, Chen ZJ. et al. m(6)A RNA Methylation Regulators Contribute to Eutopic Endometrium and Myometrium Dysfunction in Adenomyosis. Frontiers in genetics. 2020;11:716
109. Zhou L, Han X, Li W, Wang N, Yao L, Zhao Y. et al. N6-methyladenosine Demethylase FTO Induces the Dysfunctions of Ovarian Granulosa Cells by Upregulating Flotillin 2. Reprod Sci. 2021
110. Zhang S, Deng W, Liu Q, Wang P, Yang W, Ni W. Altered m(6) A modification is involved in up-regulated expression of FOXO3 in luteinized granulosa cells of non-obese polycystic ovary syndrome patients. Journal of cellular and molecular medicine. 2020;24:11874-82
111. Sun X, Zhang Y, Hu Y, An J, Li L, Wang Y. et al. Decreased expression of m(6)A demethylase FTO in ovarian aging. Archives of gynecology and obstetrics. 2021;303:1363-9
112. Ding C, Zou Q, Ding J, Ling M, Wang W, Li H. et al. Increased N6-methyladenosine causes infertility is associated with FTO expression. Journal of cellular physiology. 2018;233:7055-66
113. Huang B, Ding C, Zou Q, Wang W, Li H. Cyclophosphamide Regulates N6-Methyladenosine and m6A RNA Enzyme Levels in Human Granulosa Cells and in Ovaries of a Premature Ovarian Aging Mouse Model. Frontiers in endocrinology. 2019;10:415
114. Suvakov S, Ghamrawi R, Cubro H, Tu H, White WM, Tobah YSB. et al. Epigenetic and senescence markers indicate an accelerated ageing-like state in women with preeclamptic pregnancies. EBioMedicine. 2021;70:103536
115. Sui L, An L, Tan K, Wang Z, Wang S, Miao K. et al. Dynamic proteomic profiles of in vivo- and in vitro-produced mouse postimplantation extraembryonic tissues and placentas. Biol Reprod. 2014;91:155
116. Taniguchi K, Kawai T, Kitawaki J, Tomikawa J, Nakabayashi K, Okamura K. et al. Epitranscriptomic profiling in human placenta: N6-methyladenosine modification at the 5'-untranslated region is related to fetal growth and preeclampsia. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2020;34:494-512
117. Gu Y, Chu X, Morgan JA, Lewis DF, Wang Y. Upregulation of METTL3 expression and m6A RNA methylation in placental trophoblasts in preeclampsia. Placenta. 2021;103:43-9
118. Li R, Qiu X, He M, Qiao J, He J, Zhong M. METTL3-mediated mature miR-497-5p/195-5p inhibits trophoblast migration and invasion by targeting WWP1 in preeclampsia. Cell cycle (Georgetown, Tex). 2021:1-16
119. Garrido-Gimenez C, Alijotas-Reig J. Recurrent miscarriage: causes, evaluation and management. Postgrad Med J. 2015;91:151-62
120. Arias-Sosa LA, Acosta ID, Lucena-Quevedo E, Moreno-Ortiz H, Esteban-Pérez C, Forero-Castro M. Genetic and epigenetic variations associated with idiopathic recurrent pregnancy loss. Journal of assisted reproduction and genetics. 2018;35:355-66
121. Li XC, Jin F, Wang BY, Yin XJ, Hong W, Tian FJ. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9:3853-65
122. Xu Z, Tian P, Guo J, Mi C, Liang T, Xie J. et al. Lnc-HZ01 with m6A RNA methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating BPDE-activated lnc-HZ01/MXD1 positive feedback loop. Sci Total Environ. 2021;776:145950
123. Qiu W, Zhou Y, Wu H, Lv X, Yang L, Ren Z. et al. RNA Demethylase FTO Mediated RNA m(6)A Modification Is Involved in Maintaining Maternal-Fetal Interface in Spontaneous Abortion. Frontiers in cell and developmental biology. 2021;9:617172
124. Moroz-Omori EV, Huang D, Kumar Bedi R, Cheriyamkunnel SJ, Bochenkova E, Dolbois A. et al. METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes. ChemMedChem. 2021;16:3035-43
125. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597-601
126. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M. et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer cell. 2020;38:79-96.e11
127. Huang W, Chen TQ, Fang K, Zeng ZC, Ye H, Chen YQ. N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J Hematol Oncol. 2021;14:117
Author contact
![]() Corresponding author: Wenpei Xiang: Email: wpxiang2010com
Corresponding author: Wenpei Xiang: Email: wpxiang2010com

 Global reach, higher impact
Global reach, higher impact