10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(5):1865-1877. doi:10.7150/ijbs.66369 This issue Cite
Review
Immune response and potential therapeutic strategies for the SARS-CoV-2 associated with the COVID-19 pandemic
1. Institute of Biomedical Informatics, School of Basic Medical Sciences, Henan University, Kaifeng 475004, China.
2. Henan International Joint Laboratory of Nuclear Protein Regulation, School of Basic Medical Science, Henan University, Kaifeng, Henan 475004, China.
Received 2021-8-22; Accepted 2022-1-28; Published 2022-2-14
Abstract
Following onset of the first recorded case of Coronavirus disease 2019 (COVID-19) in December 2019, more than 269 million cases and over 5.3 million deaths have been confirmed worldwide. COVID-19 is a highly infectious pneumonia, caused by a novel virus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Currently, it poses a severe threat to human health across the globe, a trend that is likely to persist in the foreseeable future. This paper reviews SARS-CoV-2 immunity, the latest development of anti-SARS-CoV-2 drugs as well as exploring in detail, immune escape induced by SARS-CoV-2. We expect that the findings will provide a basis for COVID-19 prevention and treatment.
Keywords: COVID-19, SARS-CoV-2, immunity, potential drugs
Introduction
In December 2019, China reported a few cases of acute atypical respiratory disease in Wuhan, which rapidly spreads across the country. On February 11, 2020, World Health Organization (WHO) nominated the acute respiratory infectious disease “COVID-19” [1], while the International Committee on Taxonomy of Viruses (ICTV) immediately named this virus isolate “SARS-CoV-2”. Coronavirus, a spherical or polymorphic RNA virus, contains a single-stranded sense RNA genome and is the longest RNA virus so far reported [2]. According to the systematic classification of viruses, coronavirus is part of Coronaviridae genus, the order Nidovirales, and can be further grouped into four genera, α, β, γ and δ based on their genome sequence homology [3]. As a large family of viruses, coronavirus specifically is associated with common flu and severe infection. Previously, seven coronaviruses had been reported to cause human respiratory diseases [4], including NL63, human coronavirus 229E, HKU1, and OC43, which specifically infect the upper respiratory tract with mild symptoms [5]. The other three coronaviruses include the Middle East respiratory syndrome (MERS-CoV), severe acute respiratory syndrome (SARS-CoV), and the recent SARS-CoV-2, which are capable of infecting the lower respiratory tract, as well as may bring about fatal pneumonia. SARS-CoV-2 represents different similarities to SARS-CoV (79%), bat coronavirus RaTG13 (96.2%), and pangolin coronavirus (92.4%) [6, 7]. Herein, we explore immune reaction of SARS-CoV-2 infection together with the latest development of antiviral drugs in view of the recently published literature.
Innate immune response to SARS-CoV-2 infections
SARS-CoV-2 invades the host cell where it undergoes massive proliferation. Consequently, the cell lyses, and then releases virus particles and intracellular components. The pattern recognition receptors (PRRs) in innate immune cells trigger the inducible response of innate immune system as soon as they recognize the virus particles and intracellular components [8]. However, SARS-CoV-2 is also able to avoid or delay the stimulation of type I IFN-related response in vivo.
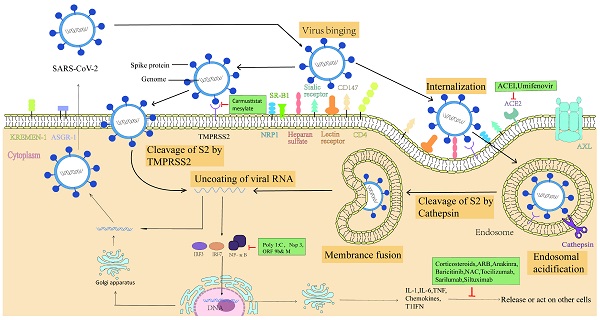
Entry point of SARS-CoV-2
The spike protein of SARS-CoV-2 mediates major entry steps: including receptor binding and membrane fusion. Furin and TMPRSS2 are the two major proteases involved in viral attachment and membrane fusion [9]. Furin, a ubiquitous proprotein convertase in the surface of Golgi apparatus, is capable of cleaving S protein into S1 and S2 subunits for virus maturation. Therefore, the S protein of the mature SARS-CoV-2 is comprised of two non-covalently subunits: the S1 subunit binds the ACE2 receptor and the S2 subunit anchors the S protein to the virion membrane and mediates membrane fusion. ACE2 engagement by SARS-CoV-2 will exposes an additional cleavage site in S2 subunits, termed S2' site. The S2' site can be cleaved by TMPRSS2 on the cell surface or cathepsin L in the endosomal compartment following ACE2-mediated endocytosis to release the fusion peptide and then initiate fusion pore formation, which allows the SARS-CoV-2 genome to reach the cytoplasm.
Although SARS-CoV-2 utilize ACE2 as the main receptor for cell entry, cells in the main target organs including the lungs and bronchi display low levels of ACE2 expression. Moreover, SARS-CoV-2 can also efficiently infect the upper respiratory tract and other tissues/organs, including the pharynx, heart, liver, brain, kidneys, and the gastrointestinal tract. These results suggest that SARS-CoV-2 employs other receptors for colonization and entry [10-12]. Recent studies have demonstrated that a few cell surface molecules consisting of AXL, KREMEN1, CD147, KIM-1, TfR and DPP4 also are potential alternative receptor independent of ACE2. Meanwhile, heparan sulfate, GRP78, lectin and neuropilin-1 are considered as auxiliary attachment receptors dependent on ACE2 (Table 1).
Recognition of SARS-CoV-2 infection
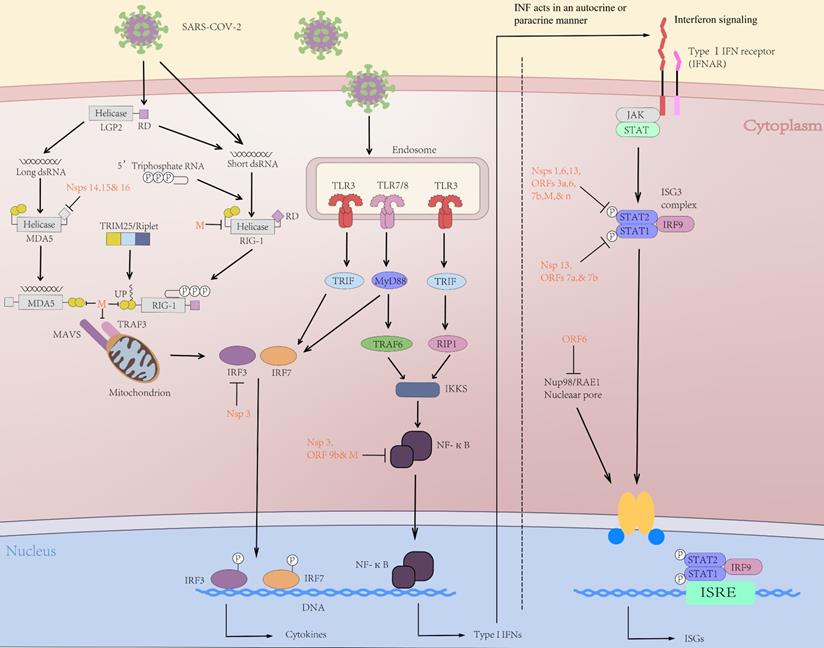
Following the human infected by SARS-CoV-2, virus genomic RNA or the intermediate product of virus replication (dsRNA) is recognized by the immune system via pattern recognition receptors (PRRs) [28]. In PRRs, toll-like receptors (TLRs), RIG-I-like receptors (RLRs), for instance, melanoma differentiation-associated protein 5 (MDA5), and retinoic acid-induced gene protein I (RIG-I) are crucial in viral RNA recognition [29]. Subsequently, the activated receptors cause a cascade of downstream immune signals, including IRF3/7 activation and nuclear translocation of NF-κB (nuclear factor kappa B), which thus can induce the expression of interferon (IFN), proinflammatory cytokines and chemokines, etc. (Figure 1) [30]. The chemokines mentioned above are important mediators for giving rise to the release of innate immune cells from the bone marrow tissue.
Receptors, co-receptors, and cofactors involved in SARS-CoV-2 entry
| No. | Molecule | Function | Virus type | Functional annotation | Refs |
|---|---|---|---|---|---|
| 1 | AXL | Receptor | SARS-CoV-2 | A potential alternative receptor independent of ACE2 | [13] |
| 2 | KREMEN1 | Receptor | SARS-CoV-2 | A potential alternative receptor independent of ACE2 | [14] |
| 3 | CD147 | Receptor | SARS-CoV-2, SARS-CoV | A potential alternative receptor independent of ACE2 | [15] |
| 4 | KIM-1 | Receptor | SARS-CoV-2, SARS-CoV, MERS-CoV | A potential alternative receptor independent of ACE2 | [16] |
| 5 | TfR | Receptor | SARS-CoV-2 | A potential alternative receptor independent of ACE2 | [17] |
| 6 | DPP4 | Receptor | SARS-CoV-2, MERS-CoV | A potential alternative receptor independent of ACE2 | [18] |
| 7 | Heparan sulfate | Coreceptor | SARS-CoV-2, SARS-CoV, MERS-CoV | Auxiliary attachment receptor, dependent on ACE2 | [19] |
| 8 | GRP78 | Coreceptor | SARS-CoV-2, MERS-CoV | Auxiliary attachment receptor, dependent on ACE2 | [20] |
| 9 | Lectin receptors | Coreceptor | SARS-CoV-2, SARS-CoV, MERS-CoV | Auxiliary attachment receptor, dependent on ACE2 | [21] |
| 10 | Neuropilin 1 | Coreceptor | SARS-CoV-2 | Auxiliary attachment receptor, dependent on ACE2 | [22, 23] |
| 11 | Furin | Cofactor | SARS-CoV-2, MERS-CoV | Proteolysis of S protein at the S1/S2 site | [24, 25] |
| 12 | Cathepsins | Cofactor | SARS-CoV-2, SARS-CoV, MERS-CoV | Proteolysis of S protein at the S1/S2 and S2′ sites | [26] |
| 13 | TMPRSS2 | Cofactor | SARS-CoV-2, SARS-CoV, MERS-CoV | Proteolysis of S protein at the S1/S2 and S2′ sites | [27] |
Cellular innate immune response to SARS-CoV-2
SARS-CoV-2 infection of the human lung triggers lung inflammatory innate immune response, which results in the recruitment of innate immune cells including monocytes, neutrophils, NK cells, DCs to the lungs. These innate immune cells are capable of releasing cytokines and prime adaptive T and B cell immune responses.
Phenotypic studies of infiltrating monocytes in the bronchoalveolar lavage (BAL) of COVID-19 patients have revealed that a surge of CD169+ monocytes and depletion of CD16+CD14-monocytes were observed early after symptom onset [31, 32]. CD169+ monocytes have a strong association with proinflammatory cytokines, including IFN-γ, MCP-2, IL-18 and IL-6 [32]. At later stages, CD16+ monocytes were present in patients with severe disease, dominated by high CCL3 and CCL4 abundance [32]. In the same way, the high concentration of neutrophil-released NETs (Neutrophil extracellular traps) are detected in COVID-19 patients' tracheal aspirate, which are considered to be related to the COVID-19 severity [33]. SARS-CoV-2-triggered NETs characterized by extracellular exposure of DNA, myeloperoxidase (MPO) and histone, capacity to induce tissue damage and promote systemic immunothrombosis [33, 34].
Initial immune response to SARS-CoV-2 infection. After the initial immune cell infection, RIG-I and MDA5 can trigger the activation of the adaptor molecule MAVS on the mitochondrial membrane, thus activating TRAF3 upstream of IFN regulatory factors (IRFs) 3/7. In the nucleus, NF-κB up-regulates expression of genes encoding interferon, pro-inflammatory cytokines (IL-1, IL-6, TNF-α, etc.) and chemokines that promote transcription of inflammatory acute phase-related proteins. The tyrosine kinases JAK1 and STAT were activated through the binding of Type I interferon and IFNAR. Activated JAK1/STAT can phosphorylate STAT1 and STAT2 to form heterodimers. ISGF3 is then transferred to the nucleus and initiate interferon-stimulated genes (ISGs) transcription. The ISG-encoded proteins exert antiviral activity by preventing virus invasion, replication, and budding. SARS-CoV-2-encoded proteins (red) inhibit multiple aspects of these pathways, resulting in decreased IFN and altered proinflammatory cytokine expression.
It is widely reported that overall NK cell numbers are substantially reduced in the blood of COVID-19 patients relative to healthy controls [31]. NK cells exert anti-viral activity but are functionally impaired with increased IFN-γ and TNF production in severe COVID-19 [35]. Further, NK cell dysfunction may be relevant to the development of fibrotic lung disease, as NK cells exhibited impaired anti-fibrotic activity [35].
As specialized producers of IFN-I, pDCs can secrete a great quantity of IFN-I isoforms immediately after SARS-CoV-2 infection. Notably, several days after SARS-CoV-2 infection, pDCs tune down their capacity for IFN-I production, which can favor prolonged viral replication and opportunistic secondary infections, termed “pDC exhaustion” [36]. Furthermore, the expression of CD86 and HLA-DR in peripheral mDCs are reduced in severe COVID-19, which are crucial for antigen presentation and the induction of helper T cell response [37]. This result may imply that the decreased DC capacity in response to TLR stimulation is associated to the diminished virus-specific T cell response in severe COVID-19 cases [37].
Cell transcriptomic profiling demonstrated that SARS-CoV-2 infection result in the activation of alveolar macrophages which promote the production of T cell chemoattractant [38]. Then, T cells recruited by chemokines are capable of inducing the production of IFN-γ, which can in turn stimulate macrophages to produce a large number of inflammatory cytokines and further promote T cell activation [38]. In each new area of infection, positive feedback loop between alveolar macrophages containing the virus and activated T cells could promote persistent injury and inflammation.
Adaptive immune response induced by SARS-CoV-2
Adaptive immune response has been shown to play important roles in the control of almost all viral infections and promoting viral clearance, which consist of three major cell types: CD4+ T cells, CD8+ T cells and B cells. During SARS-CoV-2 infection, initiation of adaptive immune responses (6-10 days) was indispensable for naive cells undergoing proliferation and differentiation into effector cells [39]. Meanwhile, the adaptive immune response will be not triggered by the time of the innate immune activation, which will result in high viral burden [40] and excessive lung immunopathology [41].
CD4+ T cells
CD4+ T cell immune responses have close relationships with control of mainly SARS-CoV-2 infection more than CD8+ T cells [42-44]. Among the COVID-19 cases in recovery period, almost all infections caused by SARS-CoV-2 proteins could initiate CD4+ T immune cell responses [42]. And S, M, and N proteins were the most prominent targets of SARS-CoV-2-specific CD4+ T cells. Notably, compared with B and CD8+ T cells, the elevated levels of CD4+ T cells in acute COVID-19 had the strongest association with mild disease and rapid viral clearance [45]. In contrast, rapid reduction of CD4+ T cells was related to severe COVID-19 [46]. T follicular helper cells (Tfh) and Th1 cells are mainly cell subtypes differentiated from CD4+ T cells. Th1 cells exert antiviral activities via IFN-γ production and secretion of cytokines IL-12, IL-3 and IL-2. Further, Th1 cells can induce CTL differentiation and activate B cells to secrete cytokines IL-10, IL-5 and IL-4. Tfh cells are able to assist differentiation of memory B cells and maintain long-lasting humoral immunity which is important for the progress of neutralizing antibody response [47].
CD8+ T cells
CD8+ T cells have an important role in prevention of viral infections, on account of the availability of eliminating infected cells. In virus infections, the presence of virus-specific CD8+ T cells is closely related with the better outcome of patients with COVID-19 [48]. Several viral antigens consisting of S, N, M and Orf3a can be specifically recognized by SARS-CoV-2 specific CD8+ T cells [49, 50]. In acute COVID-19, viral specific CD8+ T cell responses were activated swiftly, with a study of specific CD8+ T cells as early as the first day after COVID symptom onset [51]. In acute SARS-CoV-2 infection, virus-specific CD8+ T cells indicate high expression of molecules related with potent cytotoxic effector functions consisting of CD107a, perforin, granzyme B and IFN-γ [51].
Antibodies and B cells
Most SARS-CoV-2 infected persons seroconversion occurred were within 5-15 days of PSO, especially about 90% within 10 days of PSO [52-54]. The main antigens involved in detection of seroconversion in infected individuals are Spike and nucleocapsid proteins [55]. It is relatively easy to synthesis neutralizing antibodies against SARS-CoV-2 by many B cells with little affinity maturation required [56-59]. There is a positive correlation between neutralizing-antibody titers and COVID-19 disease severity in large-scale cohort studies [60]. Both SARS-CoV and MERS share the similar observations [61]. After infection, the adaptive immune response occurs late, and many host cells are actually infected until an antibody response develops. This may be why the titer of SARS-CoV-2 neutralizing antibody is not associated with the reduction of disease severity in primary COVID-19 [62].
Dysfunctional immune responses to SARS-CoV-2
Severe COVID-19 is usually associated to a dysregulated innate immune response, which is widely characterized by a delayed IFN response relative to peak virus replication and an excessive inflammatory response. After SARS-CoV-2 invasion, timely activation of IFN-I is critical for suppression of viral replication [63]. Notably, SARS-CoV-2 produces a variety of effector molecules, including structural proteins, non-structural proteins and accessory proteins (Table 1), which selectively counteract the IFN response, thereby evading the immune system [31]. Although the specific molecular mechanisms remain to be determined, suppression of the IFN-I is considered to occur at several stages consisting of inhibiting the PRR recognition of SARS-CoV-2 RNA and preventing translation via degradation of host cell mRNA [64]. The resultant suppressed or delayed IFN-I cascade enhances viral replication and causes inflammatory cytokine storm [65]. Recent study found that cytokine profiles from COVID-19 patients with cytokine storm syndrome (CSS) were linked to lung damage, multi-organ failure, and a hostile prognosis in serious COVID- 19 patients [66, 67].
Based on the strong association between the increased cytokine levels and severity of COVID-19, modulation of inflammatory cytokines provides therapy strategies to mitigate severe disease. Cytokine-targeted therapies have become the preferable option as they have fewer potential adverse effects compared to conventional antiviral therapy, for instance JAK/STAT inhibitors, IL-1R inhibitors and IL-6R inhibitors [68, 69].
Therapeutic drugs
No specific drug is available for COVID-19 cure based on the current understanding of the pathophysiology [86]. The drug treatment options mainly focus on broad-spectrum antiviral drugs, membrane fusion inhibitors, protease inhibitors, virus spike protein targeted drugs, and immune modulators, etc. [87, 88]. These drugs can prevent the virus interaction with the host cell receptor, or inhibit virus replication as well as excessive antiviral immune responses. Several potential anti-SARS-CoV-2 drugs are currently in clinical trials with different phases (Figure 2).
Innate immune antagonism by SARS-CoV-2 proteins
| Protein | Function in Virus Life Cycle | Function in Immune Escape | Refs |
|---|---|---|---|
| NSP1 | mRNA degradation | Inhibiting IFN-β production | [70, 71] |
| NSP3 | polyproteins cleavage | Antagonizing IRF3 and NF-κB signaling pathways | [70, 72] |
| NSP6 | Forming membrane vesicles | Inhibiting STAT1 phosphorylation | [72, 73] |
| NSP12 | RNA-dependent RNA polymerase | Inhibiting IFN-β production | [70, 74] |
| NSP13 | RNA helicase | Abolishing the TBK-1 phosphorylation | [70, 73, 75] |
| Nsp14 | 3'-5' exonuclease | RNA capping to evade PRR detection | [70, 75, 76] |
| Nsp15 | Endonuclease | Degrading RNA to evade PRR detection | [75, 77] |
| Nsp16 | Methyltransferase | RNA capping to evade PRR detection | [78, 79] |
| ORF3a | Perforating host cell membranes | Inhibiting STAT1 phosphorylation | [70, 73] |
| ORF3b | Inhibiting IFN-I production | Blocking IRF3 nuclear translocation | [80] |
| ORF6 | Blocking the transmission of signals | Blocking IRF3 nuclear translocation | [70, 73, 75] |
| ORF7a | Inducing suicide of infected cells | Inhibiting STAT2 phosphorylation | [73] |
| ORF7b | Inducing apoptosis | Inhibiting STAT2 phosphorylation | [73] |
| ORF8 | - | Blocking IRF3 nuclear translocation | [70, 81] |
| ORF9b | - | Disrupting MAVS/TRAF3/TRAF6 signalosome | [82] |
| S | Mediating virus entry into host cells | Inhibiting IFNβ production | [75] |
| M | Participating virus assembly | inhibiting STAT1 phosphorylation | [83, 84] |
| N | Packaging the viral genome into a nucleocapsid | Inhibiting ISG production | [85] |
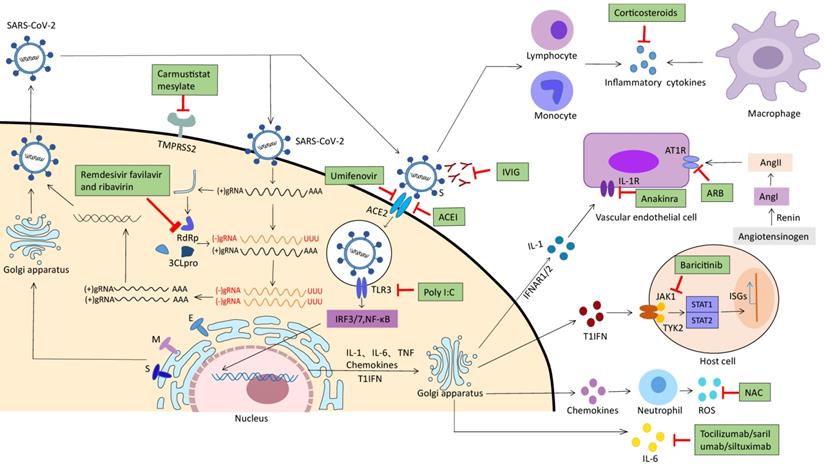
Potential therapeutic strategies for virus receptor binding, replication proliferation and immune response. Potential antivirals target the different steps of SARS- CoV-2 infection, ranging from receptor binding, entry and fusion to replication. Camostat mesylate, inhibits TMPRSS2; Remdesivir,favipiravir and ribavarin, inhibits RdRp; Umifenovir, inhibits S-ACE2 interaction and membrane fusion; Corticosteroids, inhibits inflammatory cytokines and neutrophil infiltration; Tocilizumab,sarilumab and siltuximab, binds IL-6 specifically and inhibits IL-6 signalling; IVIG, neutralizes the virus; Poly I:C, inhibits TLR3; NAC, scavenges ROS; ACEI, inhibits S-ACE2 interaction and membrane fusion; ARB, inhibits angiotensin II type 1 receptor(AT1R); Anakinra, inhibits IL-1 receptor.
RNA polymerase-RNA dependent inhibitors
Nucleoside analogues, such as remdesivir (GS-5734) [89], favipiravir [90] and ribavirin [91] are presently considered as potential drug candidates for the treatment of COVID-19. All these drugs are available for blocking viral RNA-dependent RNA polymerase (RdRp) during RNA synthesis, as a mechanism to block virus replication [92]. Remdesivir, a broad-spectrum anti-virus' drug, was originally used for the treatment of Ebola and Marburg virus infections. And several animal studies have indicated that it is highly effective against the coronaviruses [93]. A clinical study revealed that remdesivir exerts anti-virus activity against SARS-CoV-2 through in vitro and in vivo test [94, 95]. Preliminary results of clinical trials showed that remdesivir could shorten the recovery time of COVID-19 patients by a couple of days compared to placebo, with no significant difference in mortality [96-98]. FDA has authorized remdesivir for emergency use in severe COVID-19 cases [99]. However, some international phase III clinical trials are currently evaluating the safety and efficacy of remdesivir for COVID-19 management. Favipiravir (T-705), an antiviral drug developed in Japan, has been approved in China, Russia, and India for COVID-19 treatment [100]. A clinical study in China suggested that favipiravir significantly decreased the signs on chest imaging and shortened the time interval of virus infection [101]. Moreover, a preliminary report from Japan showed that mild COVID-19 patients exhibited clinical improvement rates on the 7th and 14th day being 73.8% and 87.8%, respectively [102]. Similarly, for severe COVID-19 patients, the clinical improvement rates on the 7th and 14th day were 40.1% and 60.3%, respectively [103]. Also, there are several clinical trials on ribavirin for use in COVID-19 treatment. Although ribavirin has received extensive application in coronavirus treatment, it is associated with adverse reactions; therefore, it should be used with caution [104].
Enzyme inhibitors prevent viral invasion
Camostat mesylate
Human transmembrane protease-serine 2 (TMPRSS2) is an essential host cell protease for SARS-CoV-2 spike protein activation [105]. Camostat mesylate, an effective inhibitor of TMPRSS2, is currently under investigation for its effectiveness in COVID-19 patients. A possible advantage of blocking a critical host component like TMPRSS2 rather than targeting the virus itself is that its effect is not likely to be negated by mutations in the virus genome [106]. Camostat mesylate is available for blocking SARS-CoV invasion and protects mice from lethal infection via inhibiting TMPRSS2 [107]. Recently, Clinical observation demonstrated that Camostat mesylate blocked SARS-CoV-2 entry into human lung cells [108]. In addition, a few animal trials have exhibited that the treatment of SARS-CoV-2-infected mice with a therapeutic dose of Camostat mesylate can reduce the mortality of mice from 100% to 35% [109]. Furthermore, these studies also revealed that Camostat mesylate reduces TGF-β levels which would alleviate COVID-19-induced acute respiratory distress syndrome (ARDS). Obviously, as a potential antiviral drug against SARS-CoV-2 infection, sufficient clinical data are still needed to support its efficacy.
Baricitinib
Baricitinib, often used to treat rheumatoid arthritis previously, was urgently approved by FDA to be used in combination with remdesivir in the treatment of patients with COVID-19 in November19, 2020. Baricitinib is available for inhibiting SARS-CoV-2 entry into target cells via preventing AP2-associated protein kinase 1(AAK1) and cyclin G-related kinase (GAK) activation [110-112]. Moreover, Baricitinib is also available for reducing IL-6 levels and down-regulating CD80/CD86 expression in human monocyte, thereby impeding the release of type I IFN [113, 114]. Collectively, Baricitinib can prevent cytokine storm syndrome via inhibiting the release of these cytokines, demonstrating that Baricitinib could be essential at every viral infection stage. Ultimately, the virus entry into cells in the early stage is reduced and then exerts anti-inflammatory effects in the late period [115].
ACEIs/ARBs
A large number of studies have shown that renin-angiotensin system (RAS) deregulation is responsible for ARDS, which is the main clinical manifestation of severe COVID-19. Although excessive angiotensin II is responsible for the exacerbation of COVID-19, yet the reduction of angiotensin II or blocking RAS pathway is able to limit the severity of acute lung injury caused by SARS-CoV-2 infection. Angiotensin-converting enzyme inhibitors (ACEIs) and angiotensin receptor blockers (ARBs) are considered to exert inhibition effects on the renin-angiotensin system (RAS) [116]. Zhang et al found that the mortality of COVID-19 sufferers treated with ACEIs/ARBs was much lower than those treated without ACEIs/ARBs [117]. Several clinical trials have also demonstrated that the up-regulation ACE2 expression levels induced by ARBs and ACEIs are supposed to reduce lung injury in SARS-CoV-2 infection presumably via decreasing ACE-derived Ang II [118, 119]. While several organizations have issued statements advising the continuation of treatment of COVID-19 with ACEIs/ARBs, more large-scale clinical studies are still warranted to elucidate whether it could be associated with other unknown damages to the human body [120].
Membrane fusion inhibitor
Umifenovir (arbidol), a derivative of indole carboxylic acids used for the treatment of influenza A and B virus infection, has been applied to in the treatment of SARS-CoV-2 infections [121]. Umifenovir was available for inhibiting the interaction of S protein with ACE2 via incorporating into cell membranes and disturbing with the hydrogen bonding network of phospholipids, which blocks the membrane fusion of the virus. A study determined that Umifenovir monotherapy was superior to Liponavir/Ritonavir against COVID-19[122]. COVID-19 patients treated with Umifenovir and Liponavir/ritonavir exhibited better prognosis compared with patients treated with Liponavir/Ritonavir alone [123]. Paradoxically, a clinical study in Wuhan revealed that Umfenovir did not increase the clearance rate of SARS-CoV-2 or accelerate the recovery of patients in any way [124]. Thereby, the effect of Umfenovir in the treatment of SARS-CoV-2 needs further clinical study.
IL-6 receptor-specific antibodies
Abnormally high levels of IL-6 are an indicator of poor outcome in COVID-19 patients with pneumonia and ARDS. Anti-IL-6 biological drugs, such as tocilizumab and sarilumab, have recently been adopted to treat COVID-19 patients with pneumonia [125, 126]. Tocilizumab is a recombinant humanized anti-human IL-6 receptor monoclonal antibody which is capable of blocking the membrane-bound IL-6 receptor (mIL6R), thereby inhibiting signal transduction [127]. COVID-19 patients treated with tocilizumab showed significant fever reduction within several days, and 75% patients were observed a reduced oxygen requirement [128]. Another IL-6 receptor antagonist, sarilumab, may potentially prevent cytokine release syndrome (CRS) and pulmonary symptoms in severe COVID-19 patients [93]. The efficacy and safety of sarilumab in the treatment of hospitalized patients with COVID-19 were evaluated in a randomized, double-blind study, which illustrated that sarilumab was effective in critical COVID-19 patients, but not in severe patients [129].
IL-1 receptor antagonist
Anakinra is a recombinant non-glycosylated human IL-1 receptor antagonist [130, 131]. It is initially approved for the treatment of auto-inflammatory disorders characterized by excess cytokines production which sharing clinical and molecular characteristics with COVID-19 hyper-inflammation [132, 133]. Thus, anakinra was used to test the clinical efficacy of treatment in critical COVID-19 patients. Gilles Hayem et al found that anakinra treatment prominently decreased the mortality of severe COVID-19 patients [130]. Another research has also demonstrated that the early administration of anakinra significantly diminished the requirement for invasive mechanical ventilation in COVID-19 ICU patients [93]. Also, preliminary findings of Randomized Evaluation of COVID-19 Therapy (RECOVERY) trial exhibited that anakinra combined with corticosteroids in COVID-19 treatment is more efficacious than the single use [134].
Reactive oxygen species (ROS) inhibitor
N-acetylcysteine (NAC) is an L-cysteine precursor which directly scavenges free radicals, especially reactive oxygen species (ROS) [135]. In vivo and in vitro, NAC has been successfully adopted to manage numerous diseases, including chronic inflammation, acetaminophen poisoning and asthma [136]. For COVID-19 infection, NAC was demonstrated to decrease inflammatory cytokines serum levels significantly. Furthermore, NAC can elevate the level of glutathione (GSH) in human T cells and prevent the T lymphocytes apoptosis induced by virus infection [132]. In a randomized trial, ventilator associated pneumonia (VAP) patients related to COVID-19 treated with NAC had a shorter duration of hospital stay and a higher incidence of full restoration. NAC may alleviate insulin resistance by vitamin D deficiency, revealing NAC's potential benefit for diabetics with COVID-19 disease [137]. The adverse effects are uncommon including hypersensitivity reactions, pyrexia and low blood pressure [138, 139].
Polyclonal IgG antibody
Intravenous immunoglobulins (IVIGs) are polyclonal IgG antibodies from the plasma of SARS-CoV-2-infected blood donors [140]. They potentially regulate systemic inflammation via capturing the activated complement factors, blocking the Fc γ-receptors and inhibiting lymphocyte differentiation and activation. Several studies showed that the transition from early to late course of COVID-19 is accompanied by the raised immunoglobulin levels, which indicating that IVIG therapy might be conducive to COVID-19 patients' rehabilitation [141, 142]. In another clinical study, treatment with IVIG for COVID-19 pneumonia within 48 h reduced ventilator days and shortened ICU and hospital length of stay, eventually decreased 28-day mortality [143]. An open randomized clinical trial indicated that adjuvant IVIG therapy was associated with a significantly shorter median time to real-time polymerase chain reaction negativity compared to standard of care (7 vs. 18 days) [144]. Collectively, higher titer polyclonal immunoglobulins from the plasma of convalescent patients and neutralizing antibodies from lung lavage fluid of severe COVID-19 patients are potentially new treatment options in the management of COVID-19 [145].
Others
Interferon type I
Interferon type I can induce the expression of various interferon stimulating genes (ISGs) through the JAK-STAT pathway which interferes with every virus replication step [73]. Previous studies have reported that IFN-I could be an appealing option for SARS-CoV treatment [146]. In vitro, study of experimental data indicated that the sensitivity of SARS-CoV-2 to IFN-I is higher than SARS-CoV. Several studies exhibited that the joint application of nebulized IFN-α2b, corticosteroids and lopinavir/ritonavir decrease mortality rate to zero in COVID-19 patients. A study involving about 100 COVID-19 patients exhibited that IFN inhalation was very effective in treating SARS-CoV-2 infection, lowing the risk of developing severe disease by 79% [147]. These observations support the option for the emergency application of IFN-I to treat the early COVID-19 pandemic [148].
Mesenchymal stem cell
Mesenchymal stem cells (MSCs) are pluripotent stem self-renewed cells, also known as adult stem cells. In regenerative medicine, they can improve impaired tissues and organs by differentiating into mature cells to release various cytokines [149, 150]. Lately, Leng et al discovered that MSCs graft improved lung function in COVID-19 patients with pneumonia [151] via decreasing the number of inflammatory cells and elevating the level of lymphocytes [152]. Intravenous transplantation of hUC-MSCs for the treatment of severe COVID-19 exhibited that the 28-day mortality rate in the hUC-MSC treatment group was 0, whereas 10.34% in the control group [153]. Meanwhile, MSC-derived exosomes were tested clinically for treatment of COVID-19 patients and the results showed that 71% of the patients recovered [154, 155]. In addition, MSC-derived exosomes also exhibited prominent amelioration in neutrophil as well as CD4+ and CD8+ lymphocyte numbers, with a decline in C-reactive protein (CRP) and Ferritin [153]. Based on the existed evidence, MSC and its exosomes appear to be an appropriate option for treating patients with COVID-19 pneumonia [156, 157].
Corticosteroids (dexamethasone/methylprednisolone)
Following SARS-CoV-2 invasion, type II alveolar epithelial cells release inflammatory signals to recruit leukocytes, resulting in a "cytokine storm". Corticosteroids can decrease capillary permeability and reduce leukocyte migration to inflammatory sites [158]. Moreover, it can inhibit neutrophil apoptosis and aggregation [159]. Dexamethasone and methylprednisolone are synthetic corticosteroids [160], and both can inhibit the transcription activation of pro-inflammatory cytokines and cell adhesion genes with an anti-inflammatory effect. In moderate to severe COVID-19 patients, methylprednisolone treatment with a short course significantly reduced escalation of care from ward to ICU [161]. Surprisingly, a preliminary report from the United Kingdom showed that dexamethasone treatment is only beneficial to the survival of patients with severe COVID-19 requiring respiratory support, but not mild COVID-19 patients who do not require respiratory support. In two other Brazilian studies, dexamethasone or methylprednisolone treatment conferred survival benefits to hospitalized patients with moderate to severe acute respiratory distress syndrome or over 60 years of age [162]. However, early pulse dose therapy or long-term use of high-dose corticosteroids may be related to delayed virus clearance and high mortality [163]. Therefore, it is imperative to use low doses of corticosteroids in accordance with the severity and clinical indications of COVID-19 patients.
Antithrombotic therapy
Several evidences suggest that hospitalized COVID-19 patients often suffer from an acute infection-related coagulopathy and elevated risks of microvascular thrombosis [164]. Strategies to prevent thrombosis are of critical importance for reducing the high mortality rate in COVID-19[165]. Several antithrombotic drugs have been proposed as potential therapies to prevent COVID-19-associated thrombosis, including heparin, and dipyridamole, both of which also possess pleiotropic anti-inflammatory or antiviral effects [164, 166].
Heparins, including unfractionated heparin (UFH) and low molecular weight heparin (LMWH), were mostly used for anticoagulant therapy related to SARS-CoV-2 infection [164, 167]. Compared to COVID-19 patients treated with high-dose of LMWH, the mortality of hospitalized patients who did not received any anticoagulant treatment or received low-dose LMWH, was 6.2-fold and 2.0-fold increased, respectively [168]. Also, there is general agreement that thromboprophylaxis should be administered to all hospitalized patients with COVID-19 infection with the use of standard dose of UFH or LMWH [169].
Dipyridamole (DIP), an antiplatelet agent, is often used for thromboprophylaxis combined with vitamin K antagonists following mechanical heart valve replacement [170]. Previous study regarding coagulation function exhibited that the higher levels of D-dimer was presented in severe COVID-19 patients, becoming more significant with disease progression [171].In analysis of 12 cases of COVID-19 patients with prophylactic anti-coagulation therapy, it was found that DIP treatment was related to significantly diminished D-dimer levels in comparison to control groups [166]. Half a month after initiation of DIP supplementation, 3 of the 6 severe COVID-19 patients and all 4 of the mild COVID-19 patients were discharged from the hospital [166].
Conclusion
COVID-19 is the third most deadly human coronavirus disease. While its mortality rate is lower than SARS-CoV and the MERS, COVID-19 is highly infectious and poses threat to global health in this century. More than one year after the start of the pandemic, there seems to be cautious hope for its control and vaccination programs. However, there is still an urgent need for effective drug therapy to prevent severe diseases and limit the long-term complications of COVID-19. We believe that effective drug therapy may require a variety of methods, including early antiviral treatment to prevent virus replication, immunomodulatory treatment to deal with the late high inflammatory state and anticoagulation to prevent the sequelae of venous thromboembolism and micro-thrombosis. In this paper, the characteristics of innate and adaptive immunity induced by SARS-CoV-2 are reviewed, and the current treatment strategies are summarized in detail.
Importantly, with the continuous in-depth study, a lot of research reports have displayed that age, sex and heredity were also risk factors for severe COVID-19 [39, 172]. Firstly, there is little possibility for the older to develop a coordinated adaptive immune response to SARS-CoV-2 than the young people [44]. T cell response against virus infection is determined by the repertoire of naive T cells, which will be declined substantially with age [173]. Compared with 20-year-olds, 65-year-olds in the United States have a 90-fold higher risk of COVID-19 death, as well as 75-year-olds have a 200-fold higher risk of death. Secondly, men with severe COVID-19 have slightly higher risk than that in women due to sex differences in immune responses, although no obvious functional differences have been found in the adaptive immune response [174]. However, in some studies, men infected with SARS-CoV-2 produced higher antibody titer [175]. Interestingly, 10% of the severe COVID-19 patients were IFN-I auto-antibodies, while among them more than 90% were male [176]. Therefore, severe gender differences in COVID-19 can be attributed to differences in innate immune response of IFN-I. Lastly, early genomic and transcriptome studies exhibited that a gene cluster on chromosome 3 was considered as a genetic susceptibility locus in patients with respiratory failure [177]. The immune-related genes loci on chromosome 3 consist of CCR1, CCR2, CCR9, XCR1 and CXCR6. Among them, CCR1 is identified as a receptor of several chemokines (CCL3, 5, 7 and CCL23) [178]. In addition, CCR1 deficient mice could be conducive to the protection of this receptor against excessive inflammation and reduce susceptibility to viruses and fungi [179].
Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 81803573), Postdoctoral Science Foundation (No. 2021M690095), Key R&D and Promotion Projects in Henan Province (Nos. 222102310621 and 222102310013), College Students' Innovative Entrepreneurial Training Plan Program (Nos. 202110475070, 202110475090, 202110475006 and 202110475067).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-3
2. Wu A, Peng Y, Huang B, Ding X, Jiang T. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell host & microbe. 2020 27
3. Malik YA. Properties of Coronavirus and SARS-CoV-2. Malays J Pathol. 2020;42:3-11
4. Fung TS, Liu DX. Human Coronavirus: Host-Pathogen Interaction. Annu Rev Microbiol. 2019;73:529-57
5. van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC. et al. Identification of a new human coronavirus. Nat Med. 2004;10:368-73
6. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141-54
7. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-74
8. Li K, Hao Z, Zhao X, Du J, Zhou Y. SARS-CoV-2 infection-induced immune responses: Friends or foes? Scand J Immunol. 2020;92:e12895
9. Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. 2009;106:5871-6
10. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185-92
11. Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16:e9610
12. Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH 3rd. et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429-46.e14
13. Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T. et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126-40
14. Gu Y, Cao J, Zhang X, Gao H, Wang Y, Wang J. et al. Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv. 2020
15. Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P. et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Tar. 2020;5:283
16. Yang C, Zhang Y, Zeng X, Chen H, Chen Y, Yang D. et al. Kidney injury molecule-1 is a potential receptor for SARS-CoV-2. J Mol Cell Biol. 2021;13:185-96
17. Tang X, Yang M, Duan Z, Liao Z, Liu L, Cheng R. et al. Transferrin receptor is another receptor for SARS-CoV-2 entry. BioRxiv. 2020
18. Cameron K, Rozano L, Falasca M, Mancera RL. Does the SARS-CoV-2 Spike Protein Receptor Binding Domain Interact Effectively with the DPP4 (CD26) Receptor? A Molecular Docking Study. Int J Mol Sci. 2021 22
19. Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD. et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043-57.e15
20. Carlos AJ, Ha DP, Yeh DW, Van Krieken R, Tseng CC, Zhang P. et al. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J Biol Chem. 2021;296:100759
21. Thépaut M, Luczkowiak J, Vivès C, Labiod N, Bally I, Lasala F. et al. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2021;17:e1009576
22. Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856-60
23. Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C. et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861-5
24. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L. et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620
25. Tang T, Jaimes JA, Bidon MK, Straus MR, Daniel S, Whittaker GR. Proteolytic Activation of SARS-CoV-2 Spike at the S1/S2 Boundary: Potential Role of Proteases beyond Furin. ACS Infect Dis. 2021;7:264-72
26. Jaimes JA, Millet JK, Whittaker GR. Proteolytic Cleavage of the SARS-CoV-2 Spike Protein and the Role of the Novel S1/S2 Site. iScience. 2020;23:101212
27. Djomkam ALZ, Olwal CO, Sala TB, Paemka L. Commentary: SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Front Oncol. 2020;10:1448
28. Jiang M, Österlund P, Poranen MM, Julkunen I. In vitro production of synthetic viral RNAs and their delivery into mammalian cells and the application of viral RNAs in the study of innate interferon responses. Methods. 2020;183:21-9
29. Szabo A, Rajnavolgyi E. Collaboration of Toll-like and RIG-I-like receptors in human dendritic cells: tRIGgering antiviral innate immune responses. Am J Clin Exp Immunol. 2013;2:195-207
30. Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: A review. Clin Immunol. 2020;215:108427
31. Lowery SA, Sariol A, Perlman S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe. 2021;29:1052-62
32. Chevrier S, Zurbuchen Y, Cervia C, Adamo S, Raeber ME, de Souza N. et al. A distinct innate immune signature marks progression from mild to severe COVID-19. Cell Rep Med. 2021;2:100166
33. Veras FP, Pontelli MC, Silva CM, Toller-Kawahisa JE, de Lima M, Nascimento DC. et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J Exp Med. 2020 217
34. Middleton EA, He XY, Denorme F, Campbell RA, Ng D, Salvatore SP. et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169-79
35. Krämer B, Knoll R, Bonaguro L, ToVinh M, Raabe J, Astaburuaga-García R. et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity. 2021
36. Greene TT, Zuniga EI. Type I Interferon Induction and Exhaustion during Viral Infection: Plasmacytoid Dendritic Cells and Emerging COVID-19 Findings. Viruses. 2021 13
37. Ruenjaiman V, Hirankarn N, Palaga T. Innate immunity in COVID-19: Drivers of pathogenesis and potential therapeutic targets. Asian Pac J Allergy Immunol. 2021;39:69-77
38. Grant RA, Morales-Nebreda L, Markov NS, Swaminathan S, Querrey M, Guzman ER. et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635-41
39. Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861-80
40. Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J. et al. Impact of Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients With Coronavirus Disease 2019. Clin Infect Dis. 2021;73:e4197-e205
41. Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR. et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020 5
42. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR. et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489-501.e15
43. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A. et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158-68.e14
44. Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D. et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996-1012.e19
45. Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K. et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728
46. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F. et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270-4
47. Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50:1132-48
48. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D. et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336-45
49. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457-62
50. Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M. et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22:74-85
51. Schulien I, Kemming J, Oberhardt V, Wild K, Seidel LM, Killmer S. et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med. 2021;27:78-85
52. Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK. et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845-8
53. Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A. et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 2020 5
54. Ripperger TJ, Uhrlaub JL, Watanabe M, Wong R, Castaneda Y, Pizzato HA. et al. Orthogonal SARS-CoV-2 Serological Assays Enable Surveillance of Low-Prevalence Communities and Reveal Durable Humoral Immunity. Immunity. 2020;53:925-33.e4
55. Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M. et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024-42.e21
56. Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S. et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339-43
57. Nguyen-Contant P, Embong AK, Kanagaiah P, Chaves FA, Yang H, Branche AR. et al. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. mBio. 2020 11
58. Shrock E, Fujimura E, Kula T, Timms RT, Lee IH, Leng Y. et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020 370
59. Anderson EM, Goodwin EC, Verma A, Arevalo CP, Bolton MJ, Weirick ME. et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858-64.e10
60. Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A. et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437-42
61. Sariol A, Perlman S. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity. 2020;53:248-63
62. Baumgarth N, Nikolich-Žugich J, Lee FE, Bhattacharya D. Antibody Responses to SARS-CoV-2: Let's Stick to Known Knowns. J Immunol. 2020;205:2342-50
63. Acharya D, Liu G, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. 2020;20:397-8
64. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363-74
65. Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat Med. 2021;27:28-33
66. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506
67. Gao Y, Li T, Han M, Li X, Wu D, Xu Y. et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92:791-6
68. Yang L, Xie X, Tu Z, Fu J, Xu D, Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct Tar. 2021;6:255
69. Cavalli G, De Luca G, Campochiaro C, Della-Torre E, Ripa M, Canetti D. et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325-e31
70. Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z. et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11:3810
71. Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M. et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249-55
72. Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013 4
73. Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H. et al. Evasion of Type I Interferon by SARS-CoV-2. Cell reports. 2020;33:108234
74. Imbert I, Guillemot JC, Bourhis JM, Bussetta C, Coutard B, Egloff MP. et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. Embo j. 2006;25:4933-42
75. Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H. et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418-28
76. Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B. et al. Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A. 2006;103:5108-13
77. Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL. et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991-1004
78. DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM, Regla-Nava JA, Castaño-Rodriguez C, Fernandez-Delgado R. et al. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124-37
79. Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B. et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863
80. Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y. et al. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell reports. 2020;32:108185
81. Rashid F, Dzakah EE, Wang H, Tang S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021;296:198350
82. Wu J, Shi Y, Pan X, Wu S, Hou R, Zhang Y. et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34:108761
83. Fu YZ, Wang SY, Zheng ZQ, Yi H, Li WW, Xu ZS. et al. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol Immunol. 2021;18:613-20
84. Zheng Y, Zhuang MW, Han L, Zhang J, Nan ML, Zhan P. et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct Target Ther. 2020;5:299
85. Mu J, Fang Y, Yang Q, Shu T, Wang A, Huang M. et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:65
86. Ahsan W, Javed S, Bratty MA, Alhazmi HA, Najmi A. Treatment of SARS-CoV-2: How far have we reached? Drug Discov Ther. 2020;14:67-72
87. McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859
88. Wu R, Wang L, Kuo HD, Shannar A, Peter R, Chou PJ. et al. An Update on Current Therapeutic Drugs Treating COVID-19. Curr Pharmacol Rep. 2020:1-15
89. Reina J. [Remdesivir, the antiviral hope against SARS-CoV-2]. Rev Esp Quimioter. 2020;33:176-9
90. Boretti A. Favipiravir use for SARS CoV-2 infection. Pharmacol Rep. 2020;72:1542-52
91. Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253:117592
92. Song Y, Zhang M, Yin L, Wang K, Zhou Y, Zhou M. et al. COVID-19 treatment: close to a cure? A rapid review of pharmacotherapies for the novel coronavirus (SARS-CoV-2). Int J Antimicrob Agents. 2020;56:106080
93. Varghese PM, Tsolaki AG, Yasmin H, Shastri A, Ferluga J, Vatish M. et al. Host-pathogen interaction in COVID-19: Pathogenesis, potential therapeutics and vaccination strategies. Immunobiology. 2020;225:152008
94. Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H. et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499-504
95. Pruijssers AJ, George AS, Schäfer A, Leist SR, Gralinksi LE, Dinnon KH 3rd. et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020;32:107940
96. Durante-Mangoni E, Andini R, Bertolino L, Mele F, Florio LL, Murino P. et al. Early experience with remdesivir in SARS-CoV-2 pneumonia. Infection. 2020;48:779-82
97. Spinner CD, Gottlieb RL, Criner GJ, Arribas López JR, Cattelan AM, Soriano Viladomiu A. et al. Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients With Moderate COVID-19: A Randomized Clinical Trial. Jama. 2020;324:1048-57
98. Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R. et al. Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. N Engl J Med. 2020;383:1827-37
99. Drożdżal S, Rosik J, Lechowicz K, Machaj F, Kotfis K, Ghavami S. et al. FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resist Updat. 2020;53:100719
100. Shannon A, Le NT, Selisko B, Eydoux C, Alvarez K, Guillemot JC. et al. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178:104793
101. Kaptein SJF, Jacobs S, Langendries L, Seldeslachts L, Ter Horst S, Liesenborghs L. et al. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc Natl Acad Sci U S A. 2020;117:26955-65
102. McCullough PA. Favipiravir and the Need for Early Ambulatory Treatment of SARS-CoV-2 Infection (COVID-19). Antimicrob Agents Chemother. 2020 64
103. Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J. et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering (Beijing). 2020;6:1192-8
104. Khalili JS, Zhu H, Mak NSA, Yan Y, Zhu Y. Novel coronavirus treatment with ribavirin: Groundwork for an evaluation concerning COVID-19. J Med Virol. 2020;92:740-6
105. Baughn LB, Sharma N, Elhaik E, Sekulic A, Bryce AH, Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clin Proc. 2020;95:1989-99
106. Prussia A, Thepchatri P, Snyder JP, Plemper RK. Systematic approaches towards the development of host-directed antiviral therapeutics. Int J Mol Sci. 2011;12:4027-52
107. Breining P, Frølund AL, Højen JF, Gunst JD, Staerke NB, Saedder E. et al. Camostat mesylate against SARS-CoV-2 and COVID-19-Rationale, dosing and safety. Basic Clin Pharmacol Toxicol. 2021;128:204-12
108. Hoffmann M, Hofmann-Winkler H, Smith JC, Krüger N, Sørensen LK, Søgaard OS. et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. bioRxiv. 2020
109. Ghareeb DA, Saleh SR, Nofal MS, Kaddah MMY, Hassan SF, Seif IK. et al. Potential therapeutic and pharmacological strategies for SARS-CoV2. J Pharm Investig. 2021:1-16
110. Zhang X, Zhang Y, Qiao W, Zhang J, Qi Z. Baricitinib, a drug with potential effect to prevent SARS-COV-2 from entering target cells and control cytokine storm induced by COVID-19. Int Immunopharmacol. 2020;86:106749
111. Bronte V, Ugel S, Tinazzi E, Vella A, De Sanctis F, Canè S. et al. Baricitinib restrains the immune dysregulation in patients with severe COVID-19. J Clin Invest. 2020;130:6409-16
112. Stebbing J, Krishnan V, de Bono S, Ottaviani S, Casalini G, Richardson PJ. et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol Med. 2020;12:e12697
113. Hoang TN, Pino M, Boddapati AK, Viox EG, Starke CE, Upadhyay AA. et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell. 2021;184:460-75.e21
114. Jorgensen SCJ, Tse CLY, Burry L, Dresser LD. Baricitinib: A Review of Pharmacology, Safety, and Emerging Clinical Experience in COVID-19. Pharmacotherapy. 2020;40:843-56
115. Cantini F, Niccoli L, Nannini C, Matarrese D, Natale MED, Lotti P. et al. Beneficial impact of Baricitinib in COVID-19 moderate pneumonia; multicentre study. J Infect. 2020;81:647-79
116. Ciulla MM. SARS-CoV-2 downregulation of ACE2 and pleiotropic effects of ACEIs/ARBs. Hypertens Res. 2020;43:985-6
117. Zhang J, Wang M, Ding W, Wan J. The interaction of RAAS inhibitors with COVID-19: Current progress, perspective and future. Life Sci. 2020;257:118142
118. Kuster GM, Pfister O, Burkard T, Zhou Q, Twerenbold R, Haaf P. et al. SARS-CoV2: should inhibitors of the renin-angiotensin system be withdrawn in patients with COVID-19? Eur Heart J. 2020;41:1801-3
119. John TJ, John K. Renin-angiotensin-aldosterone system dysregulation and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Eur Heart J. 2020;41:2126-7
120. Aleksova A, Ferro F, Gagno G, Cappelletto C, Santon D, Rossi M. et al. COVID-19 and renin-angiotensin system inhibition: role of angiotensin converting enzyme 2 (ACE2) - Is there any scientific evidence for controversy? J Intern Med. 2020;288:410-21
121. Costanzo M, De Giglio MAR, Roviello GN. SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus. Curr Med Chem. 2020;27:4536-41
122. Vankadari N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int J Antimicrob Agents. 2020;56:105998
123. Deng L, Li C, Zeng Q, Liu X, Li X, Zhang H. et al. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study. J Infect. 2020;81:e1-e5
124. Lian N, Xie H, Lin S, Huang J, Zhao J, Lin Q. Umifenovir treatment is not associated with improved outcomes in patients with coronavirus disease 2019: a retrospective study. Clin Microbiol Infect. 2020;26:917-21
125. Saghazadeh A, Rezaei N. Towards treatment planning of COVID-19: Rationale and hypothesis for the use of multiple immunosuppressive agents: Anti-antibodies, immunoglobulins, and corticosteroids. Int Immunopharmacol. 2020;84:106560
126. Ucciferri C, Vecchiet J, Falasca K. Role of monoclonal antibody drugs in the treatment of COVID-19. World J Clin Cases. 2020;8:4280-5
127. Rizk JG, Kalantar-Zadeh K, Mehra MR, Lavie CJ, Rizk Y, Forthal DN. Pharmaco-Immunomodulatory Therapy in COVID-19. Drugs. 2020;80:1267-92
128. Masiá M, Fernández-González M, Padilla S, Ortega P, García JA, Agulló V. et al. Impact of interleukin-6 blockade with tocilizumab on SARS-CoV-2 viral kinetics and antibody responses in patients with COVID-19: A prospective cohort study. EBioMedicine. 2020;60:102999
129. Lescure FX, Honda H, Fowler RA, Lazar JS, Shi G, Wung P. et al. Sarilumab in patients admitted to hospital with severe or critical COVID-19: a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Respir Med. 2021;9:522-32
130. Huet T, Beaussier H, Voisin O, Jouveshomme S, Dauriat G, Lazareth I. et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393-e400
131. Kooistra EJ, Waalders NJB, Grondman I, Janssen NAF, de Nooijer AH, Netea MG. et al. Anakinra treatment in critically ill COVID-19 patients: a prospective cohort study. Crit Care. 2020;24:688
132. Shi Z, Puyo CA. N-Acetylcysteine to Combat COVID-19: An Evidence Review. Ther Clin Risk Manag. 2020;16:1047-55
133. Zhou N, Yang X, Huang A, Chen Z. The potential mechanism of N-acetylcysteine in treating COVID-19. Curr Pharm Biotechnol. 2020
134. Ma S, Xu C, Liu S, Sun X, Li R, Mao M. et al. Efficacy and safety of systematic corticosteroids among severe COVID-19 patients: a systematic review and meta-analysis of randomized controlled trials. Signal Transduct Target Ther. 2021;6:83
135. Bourgonje AR, Offringa AK, van Eijk LE, Abdulle AE, Hillebrands JL, van der Voort PHJ. et al. N-acetylcysteine (NAC) and Hydrogen Sulfide (H(2)S) in Coronavirus Disease 2019 (COVID-19). Antioxid Redox Signal. 2021
136. Poe FL, Corn J. N-Acetylcysteine: A potential therapeutic agent for SARS-CoV-2. Med Hypotheses. 2020;143:109862
137. Andreou A, Trantza S, Filippou D, Sipsas N, Tsiodras S. COVID-19: The Potential Role of Copper and N-acetylcysteine (NAC) in a Combination of Candidate Antiviral Treatments Against SARS-CoV-2. In vivo. 2020;34:1567-88
138. Jorge-Aarón RM, Rosa-Ester MP. N-acetylcysteine as a potential treatment for COVID-19. Future Microbiol. 2020;15:959-62
139. Nasi A, McArdle S, Gaudernack G, Westman G, Melief C, Rockberg J. et al. Reactive oxygen species as an initiator of toxic innate immune responses in retort to SARS-CoV-2 in an ageing population, consider N-acetylcysteine as early therapeutic intervention. Toxicol Rep. 2020;7:768-71
140. Díez JM, Romero C, Vergara-Alert J, Belló-Perez M, Rodon J, Honrubia JM. et al. Cross-neutralization activity against SARS-CoV-2 is present in currently available intravenous immunoglobulins. Immunotherapy. 2020;12:1247-55
141. Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. Clinical performance of different SARS-CoV-2 IgG antibody tests. J Med Virol. 2020;92:2243-7
142. Liu X, Cao W, Li T. High-Dose Intravenous Immunoglobulins in the Treatment of Severe Acute Viral Pneumonia: The Known Mechanisms and Clinical Effects. Front Immunol. 2020;11:1660
143. Ji YL, Wu Y, Qiu Z, Ming H, Zhang Y, Zhang AN. et al. The Pathogenesis and Treatment of COVID-19: A System Review. Biomed Environ Sci. 2021;34:50-60
144. Barnes CO, West AP Jr, Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR. et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828-42.e16
145. Sharun K, Tiwari R, Iqbal Yatoo M, Patel SK, Natesan S, Dhama J. et al. Antibody-based immunotherapeutics and use of convalescent plasma to counter COVID-19: advances and prospects. Expert Opin Biol Ther. 2020;20:1033-46
146. Sa Ribero M, Jouvenet N, Dreux M, Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737
147. Lin F, Shen K. Type I interferon: From innate response to treatment for COVID-19. Pediatr Investig. 2020;4:275-80
148. Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K. et al. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem. 2020;295:13958-64
149. Tsuchiya A, Takeuchi S, Iwasawa T, Kumagai M, Sato T, Motegi S. et al. Therapeutic potential of mesenchymal stem cells and their exosomes in severe novel coronavirus disease 2019 (COVID-19) cases. Inflamm Regen. 2020;40:14
150. Yu F, Jia R, Tang Y, Liu J, Wei B. SARS-CoV-2 infection and stem cells: Interaction and intervention. Stem Cell Res. 2020;46:101859
151. Leng Z, Zhu R, Hou W, Feng Y, Yang Y, Han Q. et al. Transplantation of ACE2(-) Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020;11:216-28
152. Al-Khawaga S, Abdelalim EM. Potential application of mesenchymal stem cells and their exosomes in lung injury: an emerging therapeutic option for COVID-19 patients. Stem Cell Res Ther. 2020;11:437
153. Rezakhani L, Kelishadrokhi AF, Soleimanizadeh A, Rahmati S. Mesenchymal stem cell (MSC)-derived exosomes as a cell-free therapy for patients Infected with COVID-19: Real opportunities and range of promises. Chem Phys Lipids. 2021;234:105009
154. Sengupta V, Sengupta S, Lazo A, Woods P, Nolan A, Bremer N. Exosomes Derived from Bone Marrow Mesenchymal Stem Cells as Treatment for Severe COVID-19. Stem Cells Dev. 2020;29:747-54
155. Bamba C, Singh SP, Choudhury S. Can mesenchymal stem cell therapy be the interim management of COVID-19? Drug Discov Ther. 2020;14:139-42
156. Bari E, Ferrarotti I, Saracino L, Perteghella S, Torre ML, Corsico AG. Mesenchymal Stromal Cell Secretome for Severe COVID-19 Infections: Premises for the Therapeutic Use. Cells. 2020 9
157. Xiong J, Bao L, Qi H, Feng Z, Shi Y. Mesenchymal Stem Cell-Based Therapy for COVID-19: Possibility and Potential. Curr Stem Cell Res Ther. 2021;16:105-8
158. Ferraù F, Ceccato F, Cannavò S, Scaroni C. What we have to know about corticosteroids use during Sars-Cov-2 infection. J Endocrinol Invest. 2020:1-9
159. Lee KY, Rhim JW, Kang JH. Early preemptive immunomodulators (corticosteroids) for severe pneumonia patients infected with SARS-CoV-2. Clin Exp Pediatr. 2020;63:117-8
160. Fatima SA, Asif M, Khan KA, Siddique N, Khan AZ. Comparison of efficacy of dexamethasone and methylprednisolone in moderate to severe covid 19 disease. Ann Med Surg (Lond). 2020;60:413-6
161. Rana MA, Hashmi M, Qayyum A, Pervaiz R, Saleem M, Munir MF. et al. Comparison of Efficacy of Dexamethasone and Methylprednisolone in Improving PaO2/FiO2 Ratio Among COVID-19 Patients. Cureus. 2020;12:e10918
162. Ye ZW, Yuan S, Chan JF, Zhang AJ, Yu CY, Ong CP. et al. Beneficial effect of combinational methylprednisolone and remdesivir in hamster model of SARS-CoV-2 infection. Emerg Microbes Infect. 2021;10:291-304
163. Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Potential therapeutic use of corticosteroids as SARS CoV-2 main protease inhibitors: a computational study. J Biomol Struct Dyn. 2020:1-14
164. Godino C, Scotti A, Maugeri N, Mancini N, Fominskiy E, Margonato A. et al. Antithrombotic therapy in patients with COVID-19? -Rationale and Evidence. Int J Cardiol. 2021;324:261-6
165. Bonaventura A, Vecchié A, Dagna L, Martinod K, Dixon DL, Van Tassell BW. et al. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat Rev Immunol. 2021;21:319-29
166. Liu X, Li Z, Liu S, Sun J, Chen Z, Jiang M. et al. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm Sin B. 2020;10:1205-15
167. Moschonas IC, Tselepis AD. SARS-CoV-2 infection and thrombotic complications: a narrative review. J Thromb Thrombolysis. 2021;52:111-23
168. Gonzalez-Porras JR, Belhassen-Garcia M, Lopez-Bernus A, Vaquero-Roncero LM, Rodriguez B, Carbonell C. et al. Low molecular weight heparin is useful in adult COVID-19 inpatients. Experience during the first Spanish wave: observational study. Sao Paulo Med J. 2021
169. Spyropoulos AC, Levy JH, Ageno W, Connors JM, Hunt BJ, Iba T. et al. Scientific and Standardization Committee communication: Clinical guidance on the diagnosis, prevention, and treatment of venous thromboembolism in hospitalized patients with COVID-19. J Thromb Haemost. 2020;18:1859-65
170. Kanthi Y, Knight JS, Zuo Y, Pinsky DJ. New (re)purpose for an old drug: purinergic modulation may extinguish the COVID-19 thromboinflammatory firestorm. JCI Insight. 2020 5
171. Lu H, Chen M, Tang S, Yu W. Association of coagulation disturbances with severity of COVID-19: a longitudinal study. Hematology. 2021;26:656-62
172. Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell. 2021;184:1671-92
173. Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE. et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020 369
174. Bunders MJ, Altfeld M. Implications of Sex Differences in Immunity for SARS-CoV-2 Pathogenesis and Design of Therapeutic Interventions. Immunity. 2020;53:487-95
175. Dan JM, Mateus J, Yu K, Hastie K, Yu ED, Faliti CE. et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science (New York, NY). 2021;371:eabf4063
176. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020 370
177. Casanova JL, Su HC. A Global Effort to Define the Human Genetics of Protective Immunity to SARS-CoV-2 Infection. Cell. 2020;181:1194-9
178. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P. et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med. 2020;383:1522-34
179. Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Baillie JK. Genetic mechanisms of critical illness in Covid-19. Nature. 2020
Author contact
![]() Corresponding authors: E-mail: tengtieshanhenu.edu.cn; Tel.: +86-0371-22892865.
Corresponding authors: E-mail: tengtieshanhenu.edu.cn; Tel.: +86-0371-22892865.

 Global reach, higher impact
Global reach, higher impact