10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(6):2583-2596. doi:10.7150/ijbs.71519 This issue Cite
Research Paper
Downregulation of lncRNA APCDD1L-AS1 due to DNA hypermethylation and loss of VHL protein expression promotes the progression of clear cell renal cell carcinoma
1. Department of Urology, Peking University First Hospital, Beijing 100034, P.R. China.
2. Hereditary Kidney Cancer Research Center, Peking University First Hospital, Beijing 100034, P.R. China.
3. Institute of Urology, Peking University, Beijing 100034, P.R. China.
*These authors contributed equally to this work.
Received 2022-1-28; Accepted 2022-3-5; Published 2022-3-21
Abstract
Background: The current studies only indicated that long non-coding RNA (lncRNA) APCDD1L-AS1, as a novel lncRNA, may play a role in oral squamous cell carcinoma and lung cancer. However, its potential role in clear cell renal cell carcinoma (ccRCC) and its possible mechanism of action remain vague.
Methods: TCGA-KIRC and GEO data and qRT-PCR and pyrosequencing results of clinical specimens were used to identify the expression level and DNA methylation status of APCDD1L-AS1. The effects of APCDD1L-AS1 overexpression on ccRCC growth and metastasis were determined by function experiments. Western blot and Tandem mass tags (TMT) were utilized to explore the relationship between APCDD1L-AS1 and VHL expression and its downstream underlying mechanisms.
Results: The expression of APCDD1L-AS1 was downregulated in ccRCC. Decreased APCDD1L-AS1 expression was related to higher tumor stage and histological grade and shorter RFS (Relapse-free survival). Besides, APCDD1L-AS1 overexpression restrained the growth and metastasis of ccRCC cells in vitro and in vivo. Moreover, reduced APCDD1L-AS1 expression could be caused by DNA hypermethylation and loss of von Hippel Lindau (VHL) protein expression. Furthermore, the dysregulation of histones expression caused by APCDD1L-AS1 overexpression may be one of the important mechanisms to suppress the progression of ccRCC.
Conclusion: APCDD1L-AS1 was able to inhibit the progression of ccRCC, and its decreased expression could be caused by DNA hypermethylation and loss of VHL protein expression. Therefore, APCDD1L-AS1 may serve as a new therapeutic target in the treatment of ccRCC.
Keywords: long non-coding RNA, APCDD1L-AS1, clear cell renal cell carcinoma, DNA hypermethylation, von Hippel Lindau
Introduction
Renal cell carcinoma (RCC), one of the three major tumors of the urinary system, accounts for 2-3% of global cancer diagnoses and deaths, and its incidence has been on the rise for over 20 years [1]. Besides, 60% of patients are diagnosed at the metastatic stage or develop metastases during the disease process [2, 3], and the 5-year survival rate for patients with metastatic RCC is as low as 12% [4].
There are several subtypes of RCC, and more than 70% of individuals are diagnosed with clear cell RCC (ccRCC) [5]. Loss or mutation of the von Hippel Lindau (VHL) gene is generally regarded as one of the inevitable initial steps in the development of ccRCC. VHL protein is an E3 ubiquitin ligase, and its best-known function is the ubiquitination of the prolyl hydroxylation transcription factors hypoxia-inducible factors (HIF1α and HIF2α) [6, 7], and leads to their subsequent proteolytic degradation under normoxic conditions [8]. HIF1α and HIF2α can regulate the transcription of many genes involved in angiogenesis, metabolism and chromatin remodeling, which are associated with the development of ccRCC [9]. Based on this biological property of ccRCC, several targeted drugs with antiangiogenic activity have been approved for the treatment of advanced RCC, including mammalian target of rapamycin inhibitors, monoclonal antibody that interferes with vascular endothelial growth factor and tyrosine kinase inhibitors [10]. Unfortunately, current drug treatments are not effective enough, have severe side effects, and are prone to developing drug resistance during the course of treatment. Therefore, there is an urgent need to find more effective and safe therapeutic targets for ccRCC.
In recent years, numerous studies have proved that long non-coding RNA (lncRNA) plays a crucial role in the carcinogenesis and protein coding gene expression disorders of multiple tumors, and they can act as both oncogenes and tumor suppressors [11]. Currently, lncRNAs are known to be involved in various cellular processes including cell differentiation, autophagy, apoptosis, chemoresistance and metastasis [12-14]. Moreover, lncRNAs also play important roles in the development of ccRCC. For example, ZNF582-AS1 restrained the growth and metastasis of ccRCC by regulating the N(6)-methyladenosine modification of MT-RNR1 [15], and PVT1 promoted the tumorigenesis and metastasis of ccRCC by stabilizing HIF2α [16]. APCDD1L-AS1 was a novel lncRNA and located at Chromosome 20: 58,515,379-58,619,888. Previous studies have shown that APCDD1L-AS1 was related to the prognosis of patients with lung squamous cell carcinoma, induced 5-fluorouracil resistance in oral squamous cell carcinoma (OSCC) and icotinib resistance in lung adenocarcinoma (LUAD) [17-19]. Nevertheless, the explicit role of APCDD1L-AS1 in ccRCC is still vague.
In the present study, based on data from TCGA-KIRC and GEO databases and qRT-PCR results of clinical tumor specimens, our results suggested that the expression of lncRNA APCDD1L-AS1 was significantly lower in ccRCC than that in adjacent normal renal (AN) tissue. Overexpression of APCDD1L-AS1 increased cell apoptosis and inhibited cell proliferative, migratory and invasive ability in vitro and in vivo. Moreover, reduced expression of APCDD1L-AS1 could be caused by DNA hypermethylation and loss of VHL protein expression in ccRCC. Furthermore, APCDD1L-AS1 overexpression could lead to alterations in the expression of histones.
Materials and methods
Ethics statement
This study was approved by the ethics committee of Ministry of Science and Technology of the People's Republic of China (Approval no. 2021SLCJ2189, 2021.09.14, Beijing, China), and conducted in accordance with the Declaration of Helsinki (1975). Informed consent signed by each patient has been obtained in this study.
TCGA-KIRC and GEO data acquisition
Transcriptome sequencing data and DNA methylation data of ccRCC patients were gained from TCGA-KIRC (The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma). GSE53757, GSE66272 and GSE105260 data were obtained from Gene Expression Omnibus (GEO) DataSets.
Clinical samples collection
Samples from 54 patients diagnosed with ccRCC were collected in the study. All samples were provided by the Department of Urology, Peking University First Hospital. Clinicopathological and survival information data of these 54 ccRCC cases were also obtained.
Cell culture
The normal human renal tubular epithelial cell line HK2, and six RCC cell lines A498, ACHN, OSRC2, Caki-1, 786-O and RCC4 were used in this study, and these cell lines were cultured according to conditions specified by the provider. APCDD1L-AS1 overexpression plasmid, VHL overexpression and knockdown plasmids, HIF1α and HIF2α knockdown plasmids and HIF1α overexpression plasmid were constructed. Accordingly, the stably transfected cell lines were established by lentivirus infection.
Quantitative real-time PCR (qRT-PCR)
Total RNA of 54 pairs of ccRCC tissue specimens and the transfected cell lines was extracted using the TRIzol reagent (Invitrogen; USA). cDNA was produced using the RevertAid First Strand cDNA Synthesis Kit (Thermo, K1622, USA). qRT-PCR was performed according to the manufacturer's recommended procedure, and normalized to β-actin. All experiments were repeated at least three times. The primer sequences of APCDD1L-AS1 are as follows: Forward primer: GTTCCTGCTCGGTTTCTGGA; Reverse primer: TTTTGCCTGCACAGCATTCC.
Cell proliferation assays
EdU Apollo DNA in vitro kit (RiboBio, Guangzhou, China) and BeyoClick™ EdU Cell Proliferation Kit with DAB (Beyotime, China) were utilized to identify the cell proliferation ability. Besides, the proliferation ability of cells was also examined using the MTT Cell Proliferation and Cytotoxicity Assay Kit (Beyotime, China).
Cell apoptosis assays
One Step TUNEL Apoptosis Assay Kit (Beyotime, China) was utilized to test cell apoptotic status in cell slides. Colorimetric TUNEL Apoptosis Assay Kit (Beyotime, China) was utilized to examine cell apoptotic status in the paraffin sections of mice tumors. In addition, cell apoptosis was also assayed by staining with Annexin V-FITC and PI (Beyotime, China), and a flow cytometer was utilized to detect the fluorescence level of the cells.
Cell cycle assays
Cell cycle was detected using the Cell Cycle and Apoptosis Analysis Kit (Beyotime, China) according to the instructions.
Cell transwell migratory and invasive assays
For cell transwell migration assay, 2×103 OSRC2 and Caki-1 cells were plated into the upper chambers with 150 μL serum-free DMEM. The lower chambers were filled with 650 μL DMEM containing 15% FBS. After 48 h, cells under the surface of the lower chamber were washed with PBS and stained with 0.5% crystal violet for 20 min. For cell invasion assay, 2×103 cells were seeded on upper chambers coated with 150 μL Matrigel (1:6 dilution in PBS). The culture conditions were the same as transwell migration assay. 48 h later, adherent cells on the lower surface were stained with 0.5% crystal violet for 20 min. The number of cells on the lower surface was then counted under a microscope.
Wound healing assay
Cell migration ability was also determined by wound-healing assay. Briefly, approximately 1×106 cells were seeded in 6-well plates at equal densities and grown to 85% ~ 95% confluency. Then, artificial gaps were generated by a 1 ml sterile pipette tip after transfection with the corresponding lentivirus. Wounded areas were marked and photographed under a microscope.
Mouse model experiments
Eighteen 4-week-old male BALB/c nude mice were purchased from Vitalriver, Beijing, China. About 5×106 APCDD1L-AS1 overexpressed OSRC2 cells and its control cells were implanted subcutaneously into the right side of the mice. Tumor size was measured every four days and calculated using the formula: (length×width2)/2. For EdU incorporation assay, ethynyl-2-deoxyuridine (EdU, 50 mg/kg; Beyotime, China) was intraperitoneally injected three hours before mice were euthanized. For the lung metastasis experiment, ten 5-week-old male B-NDG severely immunodeficient mice were purchased from BIOCYTOGEN, Beijing, China. About 50×105 CON-Luc and APCDD1L-AS1-Luc Caki-1 cells were suspended in 100 ul PBS and injected into the lateral tail veins of each mouse. Forty days after injection, mice were anesthetized with tribromoethanol, and bioluminescence imaging was performed as described previously [15]. The procedures of animal experiments were approved by the Institutional Animal Care and Use Committee at Peking University First Hospital following the Guideline for the Care and Use of Laboratory Animals (Approval no. 202056, 2020.09.01, Beijing, China).
Immunohistochemistry
Immunohistochemical staining was utilized to measure the protein expression levels of E-cadherin and N-cadherin in the paraffin sections of mice lung metastases, and the detailed primary antibody information is as follows: anti-E-cadherin (1:500; Abcam, ab40772) and anti-N-cadherin (1:1000; Abcam, ab19348).
Pyrosequencing
Pyrosequencing was performed as described previously [20]. The primer sequences of used for pyrosequencing are as follows: F1: GTTTAATTTGTTAGAAGAGGTGGAGATA; R1: CCAAAACTTAAAAAAAACCTCCTTAC; S1: AAAACCTCCTTACCCA.
Western blot
RIPA lysis buffer was used to extract the total protein form cells and tissues, and the protein level was quantified using the BCA protein assay Kit (APPLYGEN). The detailed information on the primary antibodies used is as follows: anti-Cleaved Caspase-3 (1:1000, 9664S, CST), anti-Bcl-2 (1:2000, Proteintech, China), anti-VHL (1:1000, 68547S, CST); anti-HIF1α (1:1000, 36169S, CST), anti-HIF2α (1:1000, 71565S, CST); anti-Histone H3.1 (1:1000, NBP2-75524, Novus Biologicals), Histone H4 (1:1000, Proteintech, China), Histone H3 (1:1000, Proteintech, China), Histone H1.3 (1:1000, ab183736, Abcam), Histone H1.5 (1:1000, DF13539, Affinity), Histone H1.4 (1:1000, A20550, ABclonal), Histone H1.2 (1:1000, Proteintech, China), and anti-GAPDH (1:8000, Proteintech, China).
TMT (Tandem mass tags) quantitative proteomics test
The TMT test was completed by the Beijing Liuhe BGI Technology Co., LTD. The brief steps were as follows: (1) Pre-experimental stage: sample protein extraction, protein quantification and gel electrophoresis, FASP enzymatic hydrolysis, pre-mass spectrometry analysis, database comparison, and pre-experiment pass. (2) Formal experimental stage: peptide segment TMT labeling, High PH RP classification, mass spectrometry analysis, database comparison, peptide segment mass deviation, bioinformatics analysis and formal experiment report.
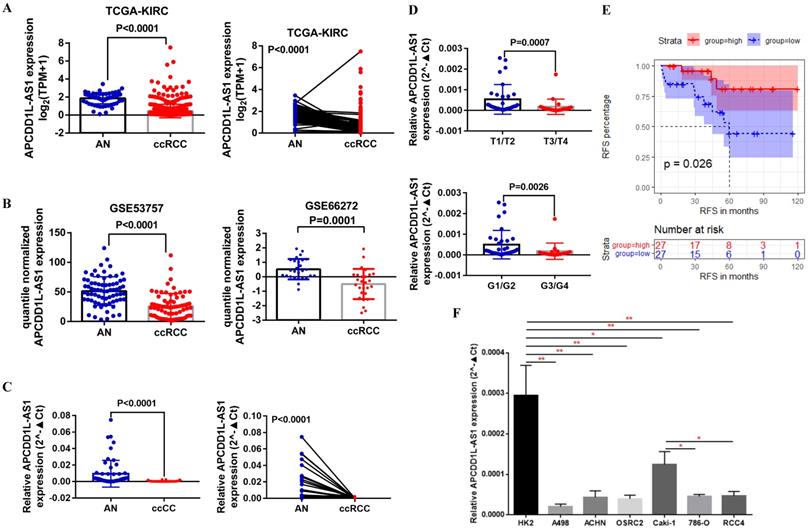
APCDD1L-AS1 expression was decreased in ccRCC. A Comparison of APCDD1L-AS1 expression in ccRCC (n=539) and adjacent normal renal (AN) tissues (n=72) based on TCGA-KIRC data. B Comparison of APCDD1L-AS1 expression in ccRCC and AN tissue based on GSE53757 (n=72) and GSE66272 (n=27) data. C and D Comparison of APCDD1L-AS1 expression in 54 pairs of ccRCC and AN tissue, and its correlation with tumor tumor stage and histological grade. E The association of APCDDL1-AS1 expression with the RFS of these 54 ccRCC patients. F Comparison of APCDD1L-AS1 expression in the control HK2 cell line and six RCC cell lines (A498, ACHN, OSRC2, Caki-1, 786-O and RCC4). *P< 0.05, **P< 0.01.
Statistical analyses
Student's t-test was used to detect differences in continuous variables. Pearson's correlation analysis was utilized to examine the correlation between APCDD1L-AS1 expression and VHL expression, the DNA methylation level of cg23487201 and APCDD1L-AS1 expression. All statistical tests were two-sided, and a P value of < 0.05 was regarded as statistical difference.
Results
The expression of lncRNA APCDD1L-AS1 in ccRCC was significantly downregulated
First, we analyzed APCDD1L-AS1 expression in ccRCC and AN tissue using the transcriptome sequencing data of TCGA-KIRC, and the paired and unpaired t test results indicated that APCDD1L-AS1 expression was significantly reduced in ccRCC compared with AN tissue (Fig. 1A). Besides, we also compared the expression of APCDD1L-AS1 in ccRCC and AN tissue using the transcriptome sequencing data of two GEO DataSets (GSE53757 and GSE66272) and obtained the consistent results (Fig. 1B). Moreover, we used qRT-PCR to examine the expression of APCDD1L-AS1 in 54 pairs of ccRCC and AN tissue, and the analysis results identified the low expression status of APCDD1L-AS1 in ccRCC (Fig. 1C). Combining the clinicopathological and survival information of these 54 patients, our analysis results showed that the expression of APCDD1L-AS1 was lower in ccRCC with higher tumor stage and histological grade (Fig. 1D) and patients in APCDD1L-AS1 low expression group had a shorter RFS (Relapse-free survival) (Fig. 1E). The detailed clinical data of these 54 ccRCC patients are shown in Table 1. Finally, we examined the expression of APCDD1L-AS1 in the control HK2 cell line and six RCC cell lines (A498, ACHN, OSRC2, Caki-1, 786-O and RCC4). Our results identified that APCDD1L-AS1 expression was significantly decreased in these RCC cell lines compared with HK2 cell line, and the expression of APCDD1L-AS1 was also remarkably reduced in 786-O and RCC4 cell lines (with a VHL mutant) compared with Caki-1 cell line (VHL wild-type) (Fig. 1F).
The clinicopathologic characteristics of 54 ccRCC patients
| Clinicopathologic characteristics | n (%) |
|---|---|
| Age | |
| <60 | 29 (53.7) |
| ≥60 | 25 (46.3) |
| Tumor size | |
| < 2cm | 7 (13.0) |
| ≥ 2cm, < 5cm | 29 (53.7) |
| ≥ 5cm | 18 (33.3) |
| Gender | |
| Male | 39 (72.2) |
| Female | 15 (27.8) |
| Tumor stage | |
| T1/T2 | 32 (59.3) |
| T3/T4 | 22 (40.7) |
| Histological grade | |
| G1/G2 | 36 (66.7) |
| G3/G4 | 18 (33.3) |
| Relapse-free survival | |
| Non-replased | 41 (75.9) |
| Replased | 13 (24.1) |
LncRNA APCDD1L-AS1 overexpression promoted cell apoptosis and weakened cell proliferation, migration and invasion in vitro
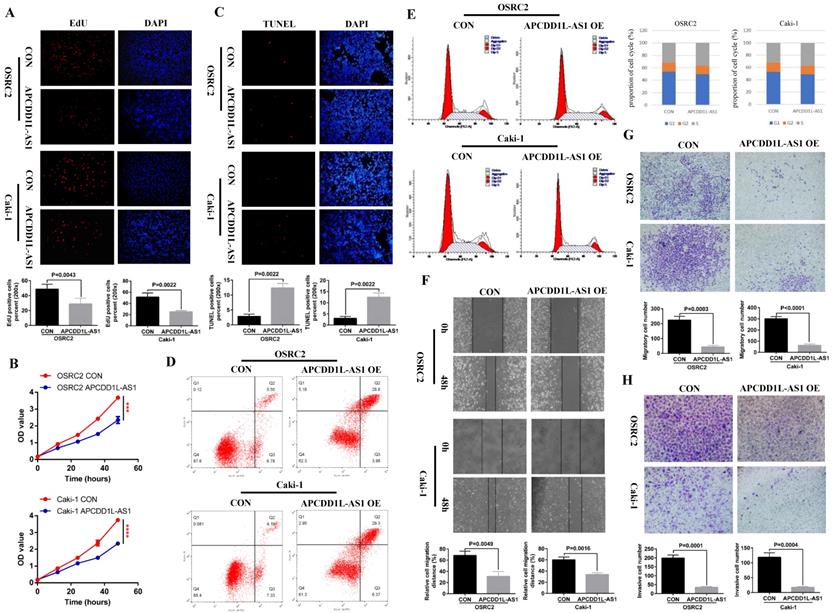
According to the results of the previous section, we have determined the low expression state of APCDD1L-AS1 in ccRCC, and then we needed to know whether overexpression of APCDD1L-AS1 affects the growth state of ccRCC cells? To this end, we constructed OSRC2 and Caki-1 cell lines stably transfected with APCDD1L-AS1. The effect of APCDD1L-AS1 expression increase on the proliferation of ccRCC cells was detected by immunofluorescence EdU method and MTT assay, and the effect of APCDD1L-AS1 expression increase on ccRCC cells apoptosis was detected by immunofluorescence TUNEL method and flow cytometry assay. The cell cycle was also detected. The effect of APCDD1L-AS1 expression increase on the invasion and metastasis ability of ccRCC cells was detected by cell wound healing, migration and invasion assays. Our results indicated that overexpression of APCDD1L-AS1 significantly restrained cell growth and increased cell apoptosis in OSRC2 and Caki-1 cells (Fig. 2A-D). The results of cell cycle assay showed that cells in S-phase were slightly increased in the APCDD1L-AS1 overexpression OSRC2 and Caki-1 cells compared with their control cells (Fig. 2E). Besides, APCDD1L-AS1 overexpression also attenuated the migration and invasion abilities of OSRC2 and Caki-1 cells (Fig. 2F-H).
LncRNA APCDD1L-AS1 overexpression promoted cell apoptosis and attenuated cell proliferation and lung metastasis in vivo
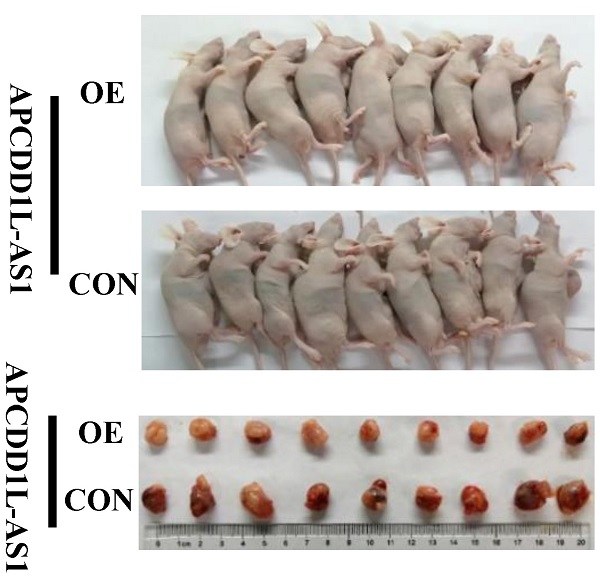
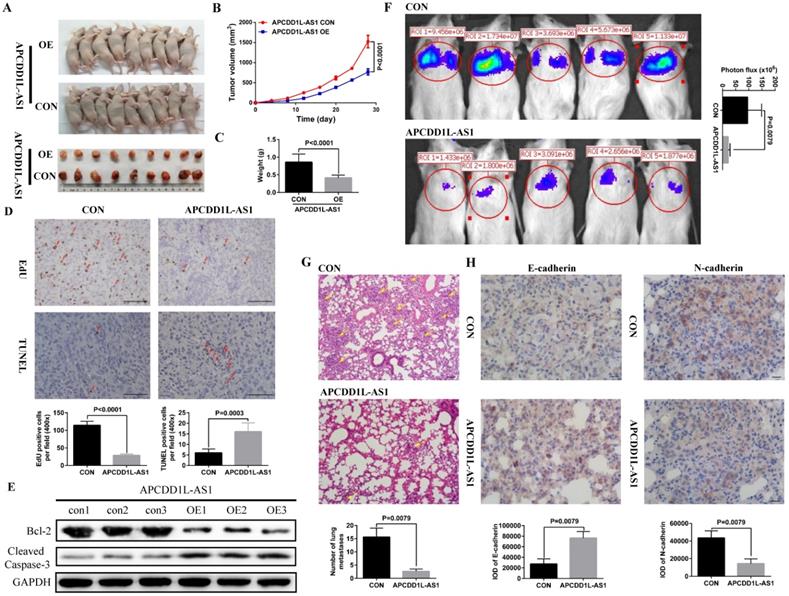
To further verify the effect of increased APCDD1L-AS1 expression on the phenotype of ccRCC cells, we constructed ccRCC cells xenograft and lung metastasis mice models. Our xenograft experiment results showed that the tumor growth in the APCDD1L-AS1 overexpressed group was significantly slower than that in the control group (Fig. 3A-C), and the results of immunohistochemistry also indicated that cell proliferation was decreased and cell apoptosis was increased in APCDD1L-AS1 overexpressed tumors compared with the control tumors (Fig. 3D). Besides, compared with the control group, the protein expression of Cleaved Caspase-3 was increased and Bcl-2 expression was decreased in the APCDD1L-AS1 overexpressed group (Fig. 3E). The mice lung metastasis experiments results determined that the luciferase signal in the mouse lung was significantly attenuated after APCDD1L-AS1 overexpression (Fig. 3F), and the results of hematoxylin-eosin staining of lung tissue also identified that the number of lung metastases in mice was significantly reduced after APCDD1L-AS1 overexpression (Fig. 3G). Epithelial-mesenchymal transition (EMT) is a key factor in tumor progression, promoting tumor cell migration and invasion, thereby supporting metastasis [21]. Thus, we also examined the expression of two EMT markers (E-cadherin and N-cadherin) in the lung metastases, and our immunohistochemistry results found that the expression of E-cadherin was increased and the expression of N-cadherin was decreased in the pulmonary metastases of APCDD1L-AS1 overexpressed group (Fig. 3H). Taken together, our results confirmed the role of APCDD1L-AS1 as a tumor suppressor gene in ccRCC.
DNA methylation regulated the expression of lncRNA APCDD1L-AS1
Now that we have demonstrated the role of reduced APCDD1L-AS1 in ccRCC, what are the mechanisms that lead to the decrease of APCDD1L-AS1? DNA methylation, as an epigenetic modification, can include global hypomethylation and regional hypermethylation, in which regional hypermethylation is usually associated with gene silencing. Previous studies have also pointed out that the decreased expression of lncRNA SNHG3, SNHG15 and ZNF582-AS1 caused by DNA hypermethylation were closely related to the progression of ccRCC [15, 20]. Thus, we obtained the methylation data of 10 CpG sites (cg07402669, cg10354244, cg13778709, cg14546153, cg14724471, cg20600210, cg23418591, cg23487201, cg24115032 and cg25970377) in APCDD1L-AS1 DNA from TCGA-KIRC. All of these CpG sites were located in the promoter region of APCDD1L-AS1, and the specific information of these CpG sites was shown in Table 2. Our analysis results suggested that the DNA methylation levels of all of these CpG sites were significantly higher in ccRCC compared with AN tissue (Fig. 4A and B), and the DNA methylation levels of nine of these 10 CpG sites were significantly negatively correlated with the APCDD1L-AS1 expression level (Fig. 4C).
APCDD1L-AS1 overexpression promoted cell apoptosis and weakened cell proliferation, migration and invasion in vitro. A Comparison of the proportion of EdU positive cells in the APCDD1L-AS1 overexpression and the control OSRC2 and Caki-1 cells by immunofluorescence (200X). B Comparison of the proliferation ability of APCDD1L-AS1 overexpression and the control OSRC2 and Caki-1 cells by MTT assay. C Comparison of the proportion of TUNEL positive cells in the APCDD1L-AS1 overexpression and the control OSRC2 and Caki-1 cells by immunofluorescence (200X). D Comparison of the apoptosis of APCDD1L-AS1 overexpression and the control OSRC2 and Caki-1 cells by flow cytometry assay. E The proportion of cell cycle of APCDD1L-AS1 overexpression and the control OSRC2 and Caki-1 cells. F wound healing assay determined the migratory distances of APCDD1L-AS1 overexpressed OSRC2 and Caki-1 cells and their control cells (100X). G and H Comparison of cell migration and cell invasion ability in the APCDD1L-AS1 overexpression and the control Caki-1 and OSRC2 cells (100X). ***P< 0.001, ****P< 0.0001.
The detailed information of 10 CpG sites in APCDD1L-AS1 DNA based on TCGA-KIRC data
| Composite Element REF | Chromosome | Start | End | CGI_Coordinate | Feature_Type |
|---|---|---|---|---|---|
| cg07402669 | chr20 | 58514959 | 58514960 | CGI:chr20:58514404-58515181 | Island |
| cg10354244 | chr20 | 58515034 | 58515035 | CGI:chr20:58514404-58515181 | Island |
| cg13778709 | chr20 | 58514905 | 58514906 | CGI:chr20:58514404-58515181 | Island |
| cg14546153 | chr20 | 58515259 | 58515260 | CGI:chr20:58514404-58515181 | S_Shore |
| cg14724471 | chr20 | 58514405 | 58514406 | CGI:chr20:58514404-58515181 | Island |
| cg20600210 | chr20 | 58514957 | 58514958 | CGI:chr20:58514404-58515181 | Island |
| cg23418591 | chr20 | 58515261 | 58515262 | CGI:chr20:58514404-58515181 | S_Shore |
| cg23487201 | chr20 | 58515066 | 58515067 | CGI:chr20:58514404-58515181 | Island |
| cg24115032 | chr20 | 58514897 | 58514898 | CGI:chr20:58514404-58515181 | Island |
| cg25970377 | chr20 | 58514877 | 58514878 | CGI:chr20:58514404-58515181 | Island |
APCDD1L-AS1 overexpression promoted cell apoptosis and attenuated cell proliferation and lung metastasis in vivo. A Tumors collected from mice. B and C Tumor volume curves and tumor weights of the APCDD1L-AS1 overexpressed and the control groups were measured and compared. D The cell proliferation and apoptosis of tumors were examined. E The protein expression of Cleaved Caspase-3 was increased and Bcl-2 expression was decreased in the APCDD1L-AS1 overexpressed group. F The luciferase signals in the APCDD1L-AS1 overexpressed group were remarkably lower than those in the control group. G The results of hematoxylin-eosin staining of mice lung tissue. H APCDD1L-AS1 overexpression increased E-cadherin expression and decreased N-cadherin expression in mice pulmonary metastases.
Additionally, the DNA methylation level of cg23487201, whose DNA methylation level was most negatively correlated with APCDD1L-AS1 expression based on TCGA-KIRC data, was also higher in ccRCC compared with AN tissue based on the data from GSE105260 (Fig. 5A). Besides, we examined the methylation level of cg23487201 in 15 pairs of clinical ccRCC and AN tissue through pyrosequencing. The pyrosequencing results also determined that cg23487201 methylation level was significantly upregulated in ccRCC compared with AN tissue, and cg23487201 methylation level was significantly negatively correlated with APCDD1L-AS1 expression level (r=-0.7562, P=0.0011) (Fig. 5B). The representative results of pyrosequencing for cg23487201 in ccRCC and AN tissue were shown in Fig. 5C. In addition, the methylation level of cg23487201 in ccRCC cell lines were also tested, and our results identified that cg23487201 DNA methylation level was generally upregulated in ccRCC cells (Fig. 5D). Furthermore, the expression of APCDD1L-AS1 was remarkably increased after 5-Aza-2′-deoxycytidine (5-aza-dC, 5-Aza, A) and Trichostatin A (TSA, T) induced demethylation of APCDD1L-AS1 promoter in OSRC2 and Caki-1 cells (Fig. 5E). Thus, DNA hypermethylation may be one of the important causes for the downregulation of APCDD1L-AS1 expression in ccRCC.
VHL protein affected the expression of lncRNA APCDD1L-AS1
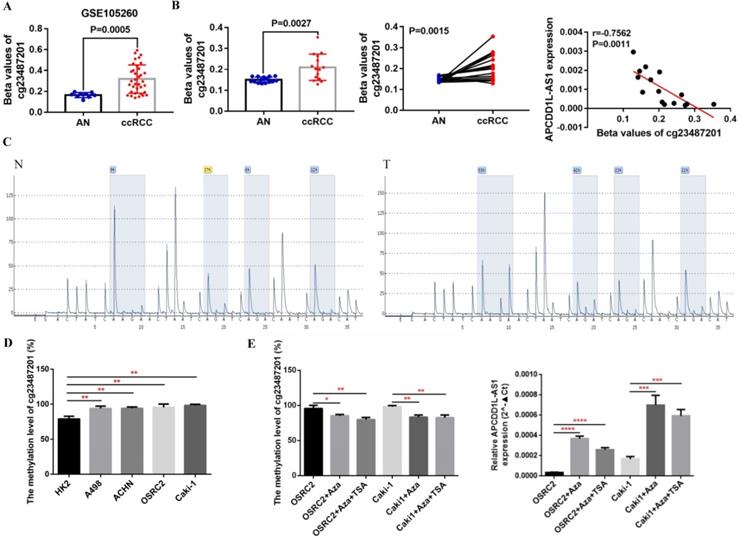
VHL gene inactivation is known to be by far the most common oncogenic driver event in ccRCC [22]. Previous studies also indicated that the expression of many lncRNAs that play important roles in renal cancer were regulated by VHL or HIFs [23]. By analyzing the RNA-seq data from TCGA-KIRC and GSE53757, we found that the expression of APCDD1L-AS1 was significantly positively correlated with the expression of VHL (Fig. 6A). To explore the specific relationship between VHL and APCDD1L-AS1, we overexpressed VHL expression in OSRC2 cells and knocked down VHL expression in Caki-1 cells. The qRT-PCR results showed that APCDD1L-AS1 expression was significantly increased after overexpression of VHL in OSRC2 cells (Fig. 6B), while APCDD1L-AS1 expression was significantly decreased after knockdown of VHL expression in Caki-1 cells (Fig. 6C). To further explore whether VHL protein affects the expression of APCDD1L-AS1 by regulating HIFs, we knocked down the expression of HIF1α and HIF2α in OSRC2 and Caki-1 cells. The qRT-PCR results demonstrated that APCDD1L-AS1 expression was significantly increased after knockdown of HIF1α expression in OSRC2 and Caki-1 cells (Fig. 6D), whereas there was no obvious alteration in APCDD1L-AS1 expression after knockdown of HIF2α expression in OSRC2 and Caki-1 cells (Fig. 6E). Furthermore, after overexpression of HIF1α in 786-O cells, APCDD1L-AS1 expression was significantly downregulated (Fig. 6F). Therefore, the above results determined that APCDD1L-AS1 expression could be regulated by VHL/HIF1α axis in ccRCC.
APCDD1L-AS1 overexpression induced histones expression disorders
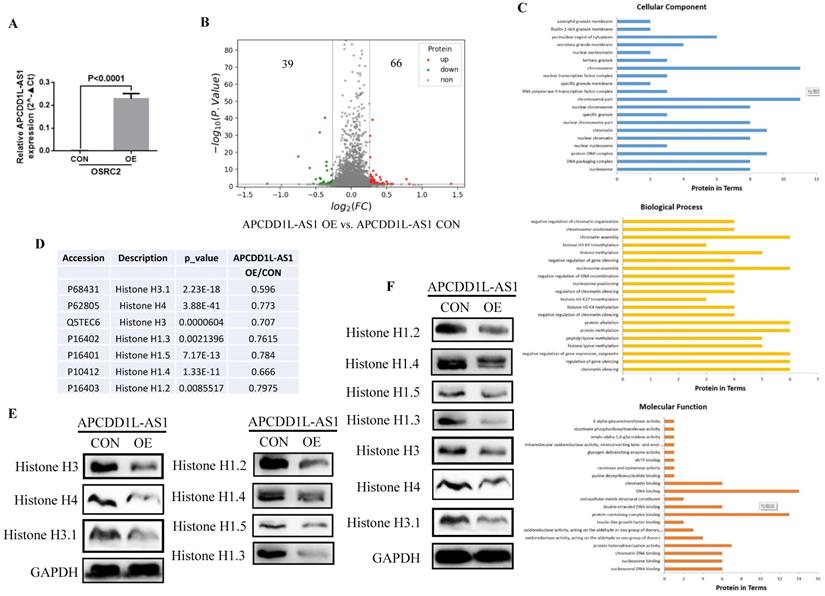
Then, we utilized the TMT method to explore the potential downstream targets of APCDD1L-AS1 and its mechanism of action. ANOVA variance was used to evaluate the significance of differences in the TMT results, and the proteins with p value less than 0.05, ratio≥1.2 or ratio≤0.83 were regarded as differential proteins. The analysis results indicated a total of 105 differential proteins, including 66 upregulated proteins and 39 downregulated proteins in APCDD1L-AS1 overexpressed cells (Fig. 7A and B). Besides, the Biological Process GO term enrichment analysis of these 105 statistically significant proteins revealed that P68431 (Histone H3.1), P62805 (Histone H4), Q5TEC6 (Histone H3), P16402 (Histone H1.3), P16401 (Histone H1.5), P10412 (Histone H1.4), P16403 (Histone H1.2) proteins were included in the most of the top 20 enriched terms of Cellular Component, Biological Process and Molecular Function (Fig. 7C and Table 3). Moreover, we used western blot to examine the expression of these six proteins (Histone H3.1, Histone H4, Histone H3, Histone H1.3, Histone H1.5, Histone H1.4 and Histone H1.2) in the same cell protein samples. Consistent with the results of TMT (Fig. 7D), the western blot results identified that the expression of these six histones was remarkably lower in APCDD1L-AS1 overexpressed cells (Fig. 7E). Furthermore, we also examined the expression of these six histones in mice tumors by western blot and obtained the same results (Fig. 7F).
DNA methylation level of APCDD1L-AS1 DNA was enhanced in ccRCC. A and B Heatmap and statistical comparison of the difference in the DNA methylation levels of 10 CpG sites of APCDD1L-AS1 promoter in ccRCC (n=325) and AN tissue (n=160). C The correlation between the DNA methylation levels of these CpG sites and APCDD1L-AS1 expression.
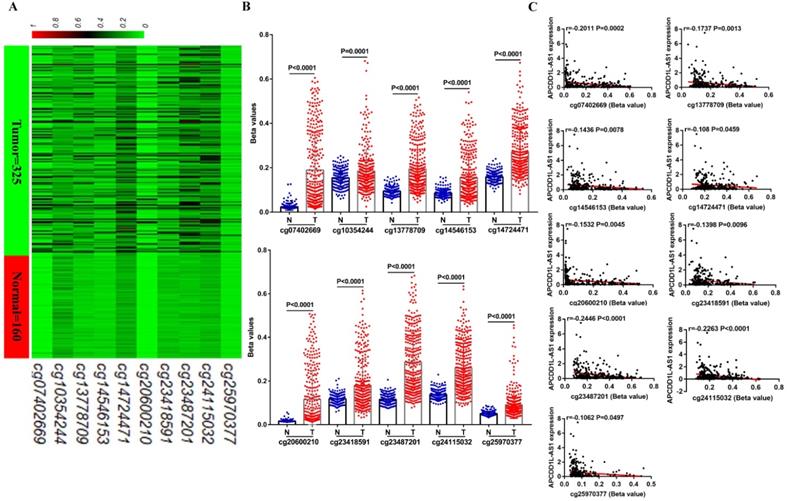
APCDD1L-AS1 expression was negatively regulated by DNA hypermethylation. A Comparison of the DNA methylation levels of cg23487201 in ccRCC (n=35) and AN (n=9) based on GSE105260 data. B Test of the DNA methylation levels of cg23487201 in 15 pairs of ccRCC and AN tissue through pyrosequencing. C Representative images of the pyrosequencing results. D Test of the DNA methylation levels of cg23487201 in HK2 cell lines and RCC cell lines. E Treatment with 5-aza-dC and TSA demethylated APCDD1L-AS1 promoter and increased APCDD1L-AS1 expression in OSRC2 and Caki-1 cells. N: adjacent normal renal tissue, T: ccRCC, *P< 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
GO term enrichment analysis of the complete 105 statistically significant proteins
| Term | Desc | p-value | Protein involve |
|---|---|---|---|
| Cellular Component | |||
| GO:0000786 | nucleosome | 5.42657E-09 | P68431, P62805, Q5TEC6, P16402 |
| P16401, P10412, P16403, Q8IUE6 | |||
| GO:0044815 | DNA packaging complex | 1.94097E-08 | P68431, P62805, Q5TEC6, P16402 |
| P16401, P10412, P16403, Q8IUE6 | |||
| GO:0000788 | nuclear nucleosome | 3.42286E-05 | P68431, P62805, P10412 |
| GO:0000790 | nuclear chromatin | 0.000563299 | P68431, P62805, P16402, P16401 |
| P10412, P16403, O00268, Q8IUE6, | |||
| GO:0000785 | chromatin | 0.0015933 | P68431, P62805, Q5TEC6, P16402, P16401 |
| P10412, P16403, O00268, Q8IUE6 | |||
| GO:0044454 | nuclear chromosome part | 0.002548084 | P68431, P62805, P16402, P16401 |
| P10412, P16403, O00268, Q8IUE6 | |||
| GO:0042581 | specific granule | 0.003470143 | Q9NX76, Q9NQR4, Q5T4S7 |
| GO:0000228 | nuclear chromosome | 0.004498937 | P68431, P62805, P16402, P16401 |
| P10412, P16403, O00268, Q8IUE6 | |||
| GO:0044427 | chromosomal part | 0.006060522 | P68431, P62805, Q5TEC6, P16402 |
| P16401, P10412, B7Z7S9, P16403 | |||
| O00268, Q8IUE6, Q9NVF7 | |||
| GO:0090575 | RNA polymerase II | 0.01167711 | O00268, P25208, A0A0B4J1Z5 |
| transcription factor complex | |||
| GO:0035579 | specific granule membrane | 0.01464028 | Q9NX76, Q5T4S7 |
| GO:0044798 | nuclear transcription factor complex | 0.01664205 | O00268, P25208, A0A0B4J1Z5 |
| GO:0005694 | chromosome | 0.01855285 | P68431, P62805, Q5TEC6, P16402 |
| P16401, P10412, B7Z7S9, P16403 | |||
| O00268, Q8IUE6, Q9NVF7 | |||
| GO:0070820 | tertiary granule | 0.01950291 | Q9NQR4, Q5T9A4, Q5T4S7 |
| GO:0005719 | nuclear euchromatin | 0.02006129 | P16402, P16403 |
| GO:0030667 | secretory granule membrane | 0.02145841 | Q9NX76, O95716, Q5T9A4, Q5T4S7 |
| GO:0048471 | perinuclear region of cytoplasm | 0.02266291 | P04732, P16989, Q96J84, P15531 |
| Q9BR76, P15121 | |||
| GO:0101003 | ficolin-1-rich granule membrane | 0.02618205 | Q5T9A4, Q5T4S7 |
| GO:0035577 | azurophil granule membrane | 0.02618205 | Q9NX76, O95716 |
| GO:0030659 | cytoplasmic vesicle membrane | 0.027415 | Q9NX76, O75396, O95716 |
| Q5T9A4, Q5T4S7, G3V5X8 | |||
| Biological Process | |||
| GO:0006342 | chromatin silencing | 1.53002E-06 | P68431, P62805, P16402 |
| P16401, P10412, P16403 | |||
| GO:0060968 | regulation of gene silencing | 4.22941E-06 | P68431, P62805, P16402 |
| P16401, P10412, P16403 | |||
| GO:0045814 | negative regulation of gene expression, epigenetic | 4.22941E-06 | P68431, P62805, P16402 |
| P16401, P10412, P16403 | |||
| GO:0034968 | histone lysine methylation | 1.35777E-05 | P16402, P16401, P10412 |
| P16403, Q9C005 | |||
| GO:0018022 | peptidyl-lysine methylation | 1.35777E-05 | P16402, P16401, P10412 |
| P16403, Q9C005 | |||
| GO:0006479 | protein methylation | 1.4475E-05 | P16402, P16401, P10412 |
| P16403, O95716, Q9C005 | |||
| GO:0008213 | protein alkylation | 1.4475E-05 | P16402, P16401, P10412 |
| P16403, O95716, Q9C005 | |||
| GO:0031936 | negative regulation of chromatin silencing | 1.5545E-05 | P16402, P16401, P10412, P16403 |
| GO:0051568 | histone H3-K4 methylation | 1.5545E-05 | P16402, P10412, P16403, Q9C005 |
| GO:0098532 | histone H3-K27 trimethylation | 3.42286E-05 | P16402, P10412, P16403 |
| GO:0031935 | regulation of chromatin silencing | 3.53573E-05 | P16402, P16401, P10412, P16403 |
| GO:0016584 | nucleosome positioning | 3.53573E-05 | P16402, P16401, P10412, P16403 |
| GO:0045910 | negative regulation of DNA recombination | 3.53573E-05 | P16402, P16401, P10412, P16403 |
| GO:0006334 | nucleosome assembly | 5.22695E-05 | P68431, P62805, P16402 |
| P16401, P10412, P16403 | |||
| GO:0060969 | negative regulation of gene silencing | 6.89335E-05 | P16402, P16401, P10412, P16403 |
| GO:0016571 | histone methylation | 7.94447E-05 | P16402, P16401, P10412 |
| P16403, Q9C005 | |||
| GO:0080182 | histone H3-K4 trimethylation | 0.000133642 | P16402, P10412, P16403 |
| GO:0031497 | chromatin assembly | 0.000181423 | P68431, P62805, P16402 |
| P16401, P10412, P16403 | |||
| GO:0030261 | chromosome condensation | 0.000196527 | P16402, P16401, P10412, P16403 |
| GO:1905268 | negative regulation of chromatin organization | 0.000196527 | P16402, P16401, P10412, P16403 |
| Molecular Function | |||
| GO:0031492 | nucleosomal DNA binding | 2.60664E-06 | P68431, Q5TEC6, P16402 |
| P16401, P10412, P16403 | |||
| GO:0031491 | nucleosome binding | 2.05966E-05 | P68431, Q5TEC6, P16402 |
| P16401, P10412, P16403 | |||
| GO:0031490 | chromatin DNA binding | 2.05966E-05 | P68431, Q5TEC6, P16402 |
| P16401, P10412, P16403 | |||
| GO:0046982 | protein heterodimerization activity | 0.00029803 | P68431, P62805, Q5TEC6, O00268 |
| P25208, P51116, Q8IUE6 | |||
| GO:0016903 | oxidoreductase activity, acting on the aldehyde or oxo group of donors | 0.000846244 | P08559, A0A1B0GTY9 |
| Q5ZEY3, P15121 | |||
| GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | 0.003470143 | P08559, A0A1B0GTY9, Q5ZEY3 |
| GO:0005520 | insulin-like growth factor binding | 0.006113907 | Q16270, B3KRV6 |
| GO:0044877 | protein-containing complex binding | 0.006324178 | P68431, Q5TEC6, P16402, P16401, P10412 |
| P16403, Q13751, P16989, P25208 | |||
| P15531, P05386, Q9BR76, F8WBH5 | |||
| GO:0003690 | double-stranded DNA binding | 0.007727656 | P16402, P16401, P10412 |
| P16403, P16989, P15531 | |||
| GO:0005201 | extracellular matrix structural constituent | 0.01464028 | Q16270, Q13751 |
| GO:0003677 | DNA binding | 0.01591233 | P68431, P62805, Q5TEC6, P16402 |
| P16401, P10412, P16403, Q6DD87 | |||
| P16989, O00268, P25208, P15531 | |||
| O00287, Q8IUE6 | |||
| GO:0003682 | chromatin binding | 0.01982527 | P68431, Q5TEC6, P16402 |
| P16401, P10412, P16403 | |||
| GO:0032554 | purine deoxyribonucleotide binding | 0.03277154 | P10412 |
| GO:0016854 | racemase and epimerase activity | 0.03277154 | C9J6A7 |
| GO:0032564 | dATP binding | 0.03277154 | P10412 |
| GO:0004133 | glycogen debranching enzyme activity | 0.03277154 | A0A0M4G3H8 |
| GO:0016862 | intramolecular oxidoreductase activity interconverting keto- and enol-groups | 0.03277154 | P14174 |
| GO:0004135 | amylo-alpha-1,6-glucosidase activity | 0.03277154 | A0A0M4G3H8 |
| GO:0004516 | nicotinate phosphoribosyltransferase activity | 0.03277154 | C9J8U2 |
| GO:0004134 | 4-alpha-glucanotransferase activity | 0.03277154 | A0A0M4G3H8 |
Discussion
Several recent studies have shown that lncRNAs can function as tumor suppressor genes in ccRCC. For instance, ADAMTS9-AS2 inhibited cell proliferation and decreased chemoresistance in ccRCC [24]; lnc-DILC stabilized PTEN and inhibited the progression of ccRCC [25]; lncRNA MAGI2-AS3 restrained tumor progression and angiogenesis by regulating ACY1 in ccRCC [26]. In the study, we identified a novel lncRNA, APCDD1L-AS1, which was downregulated in ccRCC, and overexpression of APCDD1L-AS1 restrained the growth and metastasis of ccRCC cells. However, previous studies have pointed out that APCDD1L-AS1 may also function as an oncogene in other tumors. For example, APCDD1L-AS1 induced icotinib resistance in LUAD and resulted in 5-fluorouracil resistance in OSCC [18, 19]. Thus, APCDD1L-AS1 may play different roles in different tumors.
DNA methylation is a key epigenetic regulator of gene expression and well known as impactful makers for cancer detection [27]. It is well known that epigenetic alterations, such as DNA methylation of promoter, can play a crucial role in renal tumorigenesis by silencing tumor suppressor genes [28]. In RCC, promoters of >200 genes have been reported to be methylated, and many of these frequently methylated genes in RCC influence cancer hallmarks, including invasion, cell cycle regulation, apoptosis, and cell metabolism [29]. In the present study, we found the DNA methylation levels of 10 CpG sites in APCDD1L-AS1 promoter were significantly upregulated in ccRCC, and the DNA methylation levels of nine of these 10 CpG sites were significantly negatively correlated with the expression levels of APCDD1L-AS1. Moreover, APCDD1L-AS1 expression in ccRCC cells was significantly increased after demethylation of APCDD1L-AS1 promoter.
Recently, there is increasing evidence that hypoxia promotes tumor progression and resistance to treatment, including ccRCC, pheochromocytoma and paraganglioma [30]. Besides, hypoxia is also one of the major drivers of metabolic changes in ischemic disease and myocardial infarction [31]. It can be seen that hypoxia not only plays an important role in tumors, but also affects the progression of non-tumor diseases. Therefore, it is very important to explore new hypoxia sensing pathway. Loss of VHL protein function leads to the activation of HIFs pathway, thereby activating hundreds of genes involved in many oncogenic pathways, which is one of the important mechanisms to promote the development or progression of ccRCC [32]. Previous studies have pointed out that several lncRNAs that play vital roles in renal cancer are regulated by the VHL/HIFs axis. SARCC could inhibit the progression of hypoxic cell cycle in the VHL-mutant RCC cells while derepress it in the VHL-restored RCC cells [33]. NICI was highly expressed in ccRCC, and its expression could be regulated by VHL [34]. PVT1 interacted with HIF2α protein to enhance its stability by protecting it from ubiquitin-dependent degradation, thereby contributing to the development and metastasis of ccRCC [16]. During this study, our result determined that overexpression of VHL upregulated the expression of APCDDL-AS1 and knockdown of VHL inhibited APCDD1L-AS1 expression. Moreover, APCDD1L-AS1 expression was increased after knockdown of HIF1α and decreased after overexpression of HIF1α. Similarly, previous study has indicated that lncRNA SNHG11 expression was also regulated by DNA methylation in colorectal cancer and could interact with and stabilize HIF1α and upregulate the expression of HIF1α target genes [35]. Hence, our results confirmed that APCDD1L-AS1 was a downstream target regulated by both DNA methylation and the VHL/HIF1α axis in ccRCC.
VHL protein expression affected the expression of APCDD1L-AS1. A The correlation between VHL expression and APCDD1L-AS1 expression based on TCGA-KIRC and GSE53757 data. B Comparison of APCDD1L-AS1 expression in VHL overexpressed OSRC2 cells and its control cells. C Comparison of APCDD1L-AS1 expression in VHL knockdown Caki-1 cells and its control cells. D Comparison of APCDD1L-AS1 expression in HIF1α knockdown OSRC2 and Caki-1 cells and their control cells. E Comparison of APCDD1L-AS1 expression in HIF2α knockdown OSRC2 and Caki-1 cells and their control cells. F Comparison of APCDD1L-AS1 expression in HIF1α overexpression 786-O cells and its control cells.
APCDD1L-AS1 overexpression induced histones expression disorders. A The overexpression efficiency of APCDD1L-AS1 in OSRC2 cells. B The TMT results showed that 39 proteins were downregulated and 66 proteins were upregulated in APCDD1L-AS1 overexpressed OSRC2 cells compared with its control cells. C Biological Process GO term enrichment analysis results of the complete 105 statistically significant proteins. D The TMT results of Histone H3.1, Histone H4, Histone H3, Histone H1.3, Histone H1.5, Histone H1.4 and Histone H1.2. E The protein expression of Histone H3.1, Histone H4, Histone H3, Histone H1.3, Histone H1.5, Histone H1.4 and Histone H1.2 in the same cell protein samples. F The protein expression of Histone H3.1, Histone H4, Histone H3, Histone H1.3, Histone H1.5, Histone H1.4 and Histone H1.2 in the in mice tumors.
Although our results suggested that APCDD1L-AS1 expression was significantly increased after knockdown of HIF1α expression, the mechanism by which HIF1α regulates APCDD1L-AS1 remains unclear. Previous studies have indicated that HIF1α could transcriptionally activate the expression of lncRNA. For instance, HIF1α directly bond to the promoter of KDM4A-AS1 and upregulated it expression in hepatocellular carcinoma [36], and HIF1α upregulated PVT1 transcription by binding to its promoter region in pancreatic cancer [37]. Whether HIF1α can transcriptionally regulate the expression of APCDD1L-AS by binding to its promoter will be addressed in the near future.
Chromatin, a complex of DNA polymers and associated proteins, is the physiological substrate for all genomic processes, including gene expression, damage repair, replication, and the organization and segregation of chromosomes. The major components of chromatin are histones, including four core isoforms, H2A, H2B, H3, and H4, which together with associated DNA make up nucleosomes, and H1-linked histones, which associate with nucleosomes in dyads Combine at or near the shaft to form “chromosome” particle [38, 39]. Previous several studies have also suggested that histone alterations are important causes of specific cancers, including Histone H3 and H3.3 alterations in glioblastoma [40], Histone H3.1 change in a congenital-onset soft tissue neoplasm [41], and histone H1 mutation in lymphoma [42]. Besides, histone modifications seem to play an important role in renal cancer progression and can serve as prognostic biomarkers. Total histone H4 acetylation level was inversely correlated with pathological stage and nuclear grade, and decreased H3 acetylation level was correlated with systemic metastatic spread and tumor progression in RCC [43]; lower total acetylation level of lysine 18 of histone H3 was associated with poorer survival in localized RCC [44]. In addition, hypoxia could induce alterations in histone marks, which were associated with both repression and activation of genes [45]. Previous study has also shown that hypoxia increased global demethylation level of lysine 9 of histone H3, thereby enhancing methyltransferase G9a activity, leading to gene repression in human embryonic renal cell line HEK293 [46]. In this study, the results of TMT and western blot confirmed that the expression of Histone H3.1, Histone H4, Histone H3, Histone H1.3, Histone H1.5, Histone H1.4 and Histone H1.2 were significantly reduced after APCDD1L-AS1 overexpression. Therefore, APCDD1L-AS1 may inhibit the growth and metastasis of ccRCC by causing the dysregulation of histones. Additionally, the mechanism of how APCDD1L-AS1 regulates the expression of histones will be more thoroughly investigated in the near future.
Conclusions
Our study demonstrated that lncRNA APCDD1L-AS1 functioned as a tumor suppressor gene and its expression was downregulated in ccRCC. DNA hypermethylation and loss of VHL expression could lead to the decrease in APCDD1L-AS1 expression. APCDD1L-AS1 overexpression caused dysregulation of the expression of histones and restrained the development of ccRCC. Therefore, APCDD1L-AS1 may be a potential therapeutic target in ccRCC treatment.
Abbreviations
ccRCC: clear cell renal cell carcinoma; lncRNAs: long non-coding RNAs; VHL: von Hippel Lindau; HIFs: hypoxia-inducible factors; OSCC: oral squamous cell carcinoma; LUAD: lung adenocarcinoma; AN: adjacent normal renal; TCGA-KIRC: The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma; GEO: Gene Expression Omnibus; qRT-PCR: Quantitative real-time PCR; TMT: Tandem mass tags; RFS: Relapse-free survival; EMT: Epithelial-mesenchymal transition.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82141103; 82172617; 82172665; 81872081), the Basic Research Fund of Peking University First Hospital (No. 2021BR02), the Scientific Research Seed Fund of Peking University First Hospital (No. 2021SF01), the 2021 “Outstanding Doctoral Student Innovation Fund” of Peking University School of Medicine, Special Health Development Research Project of Capital, Beijing Key Laboratory of Urogenital diseases (male) molecular diagnosis and treatment center, and Sino-Russian Mathematics Center.
Author Contributions
WY and JZ: Design and performance of experiments, analysis and explanation of data, prepared the figures and tables, and drafting of manuscript. ZZ, KZ, YX, LL and LC: Acquisition of data and statistical analysis. YG and KG: Administrative support, gain of funding, supervision, wrote and edited the manuscript. WY and JZ contributed equally to this work. All authors read and approved the final manuscript.
Data sharing statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bui TO, Dao VT, Nguyen VT, Feugeas J-P, Pamoukdjian F, Bousquet G. Genomics of Clear-cell Renal Cell Carcinoma: A Systematic Review and Meta-analysis. European urology. 2022
2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer. 2010;127:2893-917
3. Volpe A, Patard JJ. Prognostic factors in renal cell carcinoma. World journal of urology. 2010;28:319-27
4. Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A, Vakiti A. et al. Epidemiology of Renal Cell Carcinoma. World journal of oncology. 2020;11:79-87
5. Jonasch E, Walker CL, Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nature reviews Nephrology. 2021;17:245-61
6. Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ. et al. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8104-9
7. Flamme I, Krieg M, Plate KH. Up-regulation of vascular endothelial growth factor in stromal cells of hemangioblastomas is correlated with up-regulation of the transcription factor HRF/HIF-2alpha. The American journal of pathology. 1998;153:25-9
8. Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME. et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271-5
9. Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nature reviews Cancer. 2011;12:9-22
10. Comandone A, Vana F, Comandone T, Tucci M. Antiangiogenic Therapy in Clear Cell Renal Carcinoma (CCRC): Pharmacological Basis and Clinical Results. Cancers. 2021 13
11. Braga EA, Fridman MV, Filippova EA, Loginov VI, Pronina IV, Burdennyy AM. et al. LncRNAs in the Regulation of Genes and Signaling Pathways through miRNA-Mediated and Other Mechanisms in Clear Cell Renal Cell Carcinoma. International journal of molecular sciences. 2021 22
12. Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics, proteomics & bioinformatics. 2017;15:177-86
13. Chen X, Xie R, Gu P, Huang M, Han J, Dong W. et al. Long Noncoding RNA LBCS Inhibits Self-Renewal and Chemoresistance of Bladder Cancer Stem Cells through Epigenetic Silencing of SOX2. Clinical cancer research: an official journal of the American Association for Cancer Research. 2019;25:1389-403
14. Chen Z, Chen X, Xie R, Huang M, Dong W, Han J. et al. DANCR Promotes Metastasis and Proliferation in Bladder Cancer Cells by Enhancing IL-11-STAT3 Signaling and CCND1 Expression. Molecular therapy: the journal of the American Society of Gene Therapy. 2019;27:326-41
15. Yang W, Zhang K, Li L, Xu Y, Ma K, Xie H. et al. Downregulation of lncRNA ZNF582-AS1 due to DNA hypermethylation promotes clear cell renal cell carcinoma growth and metastasis by regulating the N(6)-methyladenosine modification of MT-RNR1. Journal of experimental & clinical cancer research: CR. 2021;40:92
16. Zhang MX, Zhang LZ, Fu LM, Yao HH, Tan L, Feng ZH. et al. Positive feedback regulation of lncRNA PVT1 and HIF2α contributes to clear cell renal cell carcinoma tumorigenesis and metastasis. Oncogene. 2021;40:5639-50
17. Luo Y, Xuan Z, Zhu X, Zhan P, Wang Z. Long non-coding RNAs RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9 were associated with the prognosis of lung squamous cell carcinoma. Molecular medicine reports. 2018;17:7238-48
18. Li S, Shi Z, Fu S, Li Q, Li B, Sang L. et al. Exosomal-mediated transfer of APCDD1L-AS1 induces 5-fluorouracil resistance in oral squamous cell carcinoma via miR-1224-5p/nuclear receptor binding SET domain protein 2 (NSD2) axis. Bioengineered. 2021;12:7188-204
19. Wu J, Zheng C, Wang Y, Yang Z, Li C, Fang W. et al. LncRNA APCDD1L-AS1 induces icotinib resistance by inhibition of EGFR autophagic degradation via the miR-1322/miR-1972/miR-324-3p-SIRT5 axis in lung adenocarcinoma. Biomarker research. 2021;9:9
20. Yang W, Zhang K, Li L, Ma K, Hong B, Gong Y. et al. Discovery and validation of the prognostic value of the lncRNAs encoding snoRNAs in patients with clear cell renal cell carcinoma. Aging. 2020;12:4424-44
21. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature reviews Molecular cell biology. 2006;7:131-42
22. Carril-Ajuria L, Santos M, Roldán-Romero JM, Rodriguez-Antona C, de Velasco G. Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma. Cancers. 2019 12
23. Yang W, Zhou J, Zhang K, Li L, Xu Y, Ma K. et al. Identification and validation of the clinical roles of the VHL-related LncRNAs in clear cell renal cell carcinoma. Journal of Cancer. 2021;12:2702-14
24. Song EL, Xing L, Wang L, Song WT, Li DB, Wang Y. et al. LncRNA ADAMTS9-AS2 inhibits cell proliferation and decreases chemoresistance in clear cell renal cell carcinoma via the miR-27a-3p/FOXO1 axis. Aging. 2019;11:5705-25
25. Zhang H, Wei P, Lv W, Han X, Yang J, Qin S. Long noncoding RNA lnc-DILC stabilizes PTEN and suppresses clear cell renal cell carcinoma progression. Cell & bioscience. 2019;9:81
26. Wang G, Li H, Hou Y. LncRNA MAGI2-AS3 inhibits tumor progression and angiogenesis by regulating ACY1 via interacting with transcription factor HEY1 in clear cell renal cell carcinoma. Cancer gene therapy. 2021
27. Chen X, Zhang J, Ruan W, Huang M, Wang C, Wang H. et al. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. The Journal of clinical investigation. 2020;130:6278-89
28. Joosten SC, Deckers IA, Aarts MJ, Hoeben A, van Roermund JG, Smits KM. et al. Prognostic DNA methylation markers for renal cell carcinoma: a systematic review. Epigenomics. 2017;9:1243-57
29. Joosten SC, Smits KM, Aarts MJ, Melotte V, Koch A, Tjan-Heijnen VC. et al. Epigenetics in renal cell cancer: mechanisms and clinical applications. Nature reviews Urology. 2018;15:430-51
30. Jochmanová I, Yang C, Zhuang Z, Pacak K. Hypoxia-inducible factor signaling in pheochromocytoma: turning the rudder in the right direction. Journal of the National Cancer Institute. 2013;105:1270-83
31. Sousa Fialho MDL, Abd Jamil AH, Stannard GA, Heather LC. Hypoxia-inducible factor 1 signalling, metabolism and its therapeutic potential in cardiovascular disease. Biochimica et biophysica acta Molecular basis of disease. 2019;1865:831-43
32. Mehdi A, Riazalhosseini Y. Epigenome Aberrations: Emerging Driving Factors of the Clear Cell Renal Cell Carcinoma. International journal of molecular sciences. 2017 18
33. Zhai W, Sun Y, Jiang M, Wang M, Gasiewicz TA, Zheng J. et al. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2α/C-MYC axis under hypoxia. Oncogene. 2016;35:4866-80
34. Lauer V, Grampp S, Platt J, Lafleur V, Lombardi O, Choudhry H. et al. Hypoxia drives glucose transporter 3 expression through hypoxia-inducible transcription factor (HIF)-mediated induction of the long noncoding RNA NICI. The Journal of biological chemistry. 2020;295:4065-78
35. Xu L, Huan L, Guo T, Wu Y, Liu Y, Wang Q. et al. LncRNA SNHG11 facilitates tumor metastasis by interacting with and stabilizing HIF-1α. Oncogene. 2020;39:7005-18
36. Chen T, Liu R, Niu Y, Mo H, Wang H, Lu Y. et al. HIF-1α-activated long non-coding RNA KDM4A-AS1 promotes hepatocellular carcinoma progression via the miR-411-5p/KPNA2/AKT pathway. Cell death & disease. 2021;12:1152
37. Zhu Y, Wu F, Gui W, Zhang N, Matro E, Zhu L. et al. A positive feedback regulatory loop involving the lncRNA PVT1 and HIF-1α in pancreatic cancer. Journal of molecular cell biology. 2021;13:676-89
38. McGinty RK, Tan S. Nucleosome structure and function. Chemical reviews. 2015;115:2255-73
39. Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nature reviews Molecular cell biology. 2018;19:192-206
40. Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226-31
41. Kernohan KD, Grynspan D, Ramphal R, Bareke E, Wang YC, Nizalik E, et al. H3.1 K36M mutation in a congenital-onset soft tissue neoplasm. Pediatric blood & cancer. 2017; 64
42. Soshnev AA, Allis CD, Cesarman E, Melnick AM. Histone H1 Mutations in Lymphoma: A Link(er) between Chromatin Organization, Developmental Reprogramming, and Cancer. Cancer research. 2021;81:6061-70
43. Mosashvilli D, Kahl P, Mertens C, Holzapfel S, Rogenhofer S, Hauser S. et al. Global histone acetylation levels: prognostic relevance in patients with renal cell carcinoma. Cancer science. 2010;101:2664-9
44. Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S. et al. Global levels of histone modifications predict prognosis in different cancers. The American journal of pathology. 2009;174:1619-28
45. Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutation research. 2008;640:174-9
46. Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer research. 2006;66:9009-16
Author contact
![]() Corresponding authors: Dr. Yanqing Gong, E-mail: yqgongedu.cn; Mailing address: No. 8, Xishiku Street, Xicheng District, Beijing 100034, China. Dr. Kan Gong, E-mail: gongkan_pkucom; Mailing address: No. 8, Xishiku Street, Xicheng District, Beijing 100034, China; Tel: +86-010-83575101; Fax: +86-010-66551122.
Corresponding authors: Dr. Yanqing Gong, E-mail: yqgongedu.cn; Mailing address: No. 8, Xishiku Street, Xicheng District, Beijing 100034, China. Dr. Kan Gong, E-mail: gongkan_pkucom; Mailing address: No. 8, Xishiku Street, Xicheng District, Beijing 100034, China; Tel: +86-010-83575101; Fax: +86-010-66551122.

 Global reach, higher impact
Global reach, higher impact