10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(8):3358-3373. doi:10.7150/ijbs.68221 This issue Cite
Review
The regulatory function of piRNA/PIWI complex in cancer and other human diseases: The role of DNA methylation
1. Institute for Translational Medicine, The Affiliated Hospital of Qingdao University, College of Medicine, Qingdao University, Qingdao, 266021, China.
2. Department of Vascular Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China.
3. Department of Radiation Oncology, Sun Yat - Sen University Cancer Center, State Key Laboratory of Oncology in Southern China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China.
Received 2021-10-19; Accepted 2022-4-21; Published 2022-5-9
Abstract
Piwi-interacting RNAs (piRNAs) are a class of short chain noncoding RNAs that are constituted by 26-30 nucleotides (nt) and can couple with PIWI protein family. piRNAs were initially described in germline cells and are believed to be critical regulators of the maintenance of reproductive line. Increasing evidence has extended our perspectives on the biological significance of piRNAs and indicated that they could still affect somatic gene expression through DNA methylation, chromatin modification and transposon silencing, etc. Many studies have revealed that the dysregulation of piRNAs might contribute to diverse diseases through epigenetic changes represented by DNA methylation and chromatin modification. In this review, we summarized piRNA/PIWI protein-mediated DNA methylation regulation mechanisms and methylation changes caused by piRNA/PIWI proteins in different diseases, especially cancers. Since DNA methylation and inhibitory chromatin marks represented by histone H3 lysine 9 (H3K9) methylation frequently cooperate to silence genomic regions, we also included methylation in chromatin modification within this discussion. Furthermore, we discussed the potential clinical applications of piRNAs as a new type promising biomarkers for cancer diagnosis, as well as the significance of piRNA/PIWI protein-associated methylation changes in treatment, providing disparate insights into the potential applications of them.
Keywords: piRNA, PIWI, DNA methylation, cancer, disease, diagnosis, therapy
Introduction
Piwi-interacting RNAs (piRNAs) are a class of short chain noncoding RNAs, that are constituted by 26-30 nucleotides (nt) and have a 5'-terminal uridine or tenth position adenosine bias, lacking explicit secondary structure motifs [1]. These short chain RNAs were firstly found in germline cells and are considered critical regulators of germline maintenance. A growing number of findings has now expanded our understanding of piRNAs biological significance, which suggests they could also somatically regulate gene expression via epigenetic alternations such as DNA methylation, transposon silencing and chromatin modification and cause diverse diseases including cancers [2-4]. As a type of chemical modification, DNA methylation is the first described and well-characterized in humans and plays a significant role in long-term gene silencing, especially in the promoter regions [5, 6].
piRNA/PIWI protein
Structure and biosynthesis of piRNAs
Mature piRNAs are 26-30 nt in size with a 2'-O-methylation at the 3' end, which are distinct and unique features of all piRNAs [7]. The difference with miRNAs and siRNAs is that piRNAs precursors are single-stranded and their secondary hairpin structures are not obvious. These precursors are generally produced from specific genomic regions that contain repeating elements, and a dicer-independent pathway typically coordinates this process. Furthermore, additional posttranscriptional modifications are required before nascent piRNAs maturing. Two major pathways are involved in the biogenesis of piRNAs: the primary and secondary amplification cycle - the latter is usually also referred to as “ping-pong cycle” [8] (Fig. 1).
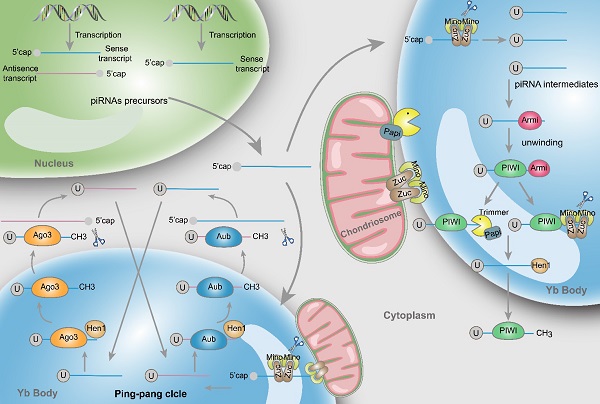
Structure and biosynthesis of piRNAs. Initially, two types piRNA precursors are generated from piRNA clusters within the nucleus and transcribed to the cytoplasmic Yb body to produce the primary piRNA intermediates with a 5' uracil through the incision of Zuc and its co-factors Mino. In primary amplification, after being unwound by Armi, piRNA intermediates couple with PIWI and are cleaved by Zuc or Papi-dependent trimmer to form 3'end. Following methylation of Hen1, the mature piRNA/PIWI complex is produced in cytoplasm. In secondary amplification, two type piRNA intermediates couple with Ago3 or Aub proteins to form piRNA/Ago3 or piRNA/Aub complexes, incise and generate new piRNAs. Newly generated piRNAs synthesize other piRNAs in a similar way.
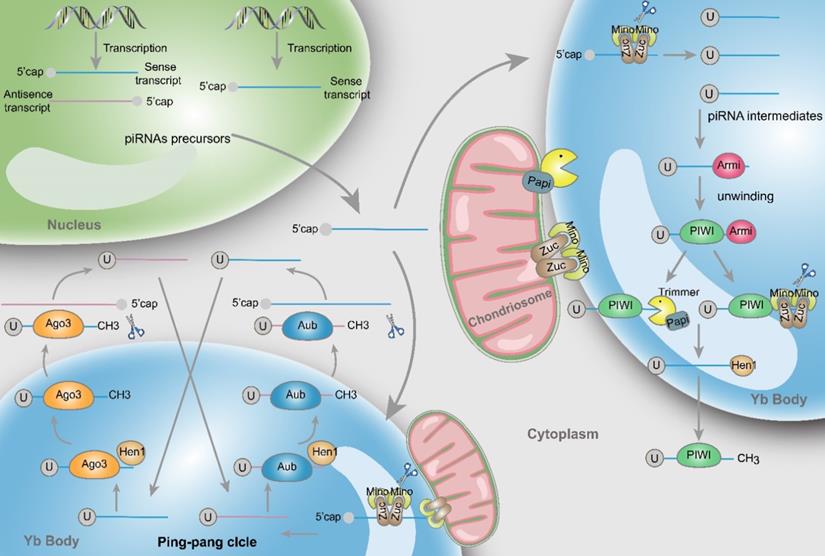
Primary amplification
piRNAs are generated from piRNA clusters, a relatively few and characteristic genomic regions [9]. Then, primary piRNAs are transported to the cytoplasm and Zucchini (Zuc) and its cofactor Minotaur (Mino) cleave them to produce piRNA intermediates with characteristic of 5' uracil [10, 11]. Later, PIWI binds piRNA intermediates through the interaction of the PAZ domain and 5' uracil, and then the maturity of piRNAs is experienced the Zuc endonuclease cleavage at 3'-end, or the Papi-dependent trimmer [12]. Subsequently mature piRNA/PIWI complex is yielded via methylation by Hen1 [13, 14]. Finally, these complexes return to the nucleus where they can activate the transcription silencing machinery and prevent transcription of their target genes.
Secondary amplification (Ping-pong cycle)
In the cytoplasm, after the generation of primary piRNAs, piRNAs are amplified with the ping-pong mechanism [15]. This process is featured by the formation of piRNA/Ago3 or piRNA/Aub complexes, which is different from the primary synthesis associated with PIWI proteins. Specifically, Aub incises sense piRNA precursors by coupling with antisense-strand piRNAs, generating sense piRNAs that load onto Ago3. Inversely, antisense piRNA precursors are incised and generate antisense piRNAs in combination with Aub through Ago3 coupling with sense-strand piRNAs [16]. Ultimately, these piRNAs combine with PIWI proteins and migrate back to the nucleus, silencing the expression of target genes. Actually, through the above mentioned amplifying mechanism, the formation of substrates for other functional piRNAs are produced accompanied by the production of piRNAs [17].
PIWI protein
Across different species, the PIWI protein family is highly conservative [18]. Four PIWI proteins are known in humans, namely PIWIL1, PIWIL2, PIWIL3 and PIWIL4, which are also respectively called HIWI, HILI, HIWI3, HIWI2. In mice, there are three PIWI homologs, PIWIL1 (also termed as MIWI), PIWIL2 (termed as MILI) and PIWIL4 (termed as MIWI2). Three PIWI proteins have also been described in Drosophila, which are named PIWI, Aub and Ago3 [19-21]. PIWI proteins are the members of Argonaute family, PIWI subfamily [18]. As with all Argonaute proteins, PIWI proteins incorporate N, PAZ, MID, and PIWI domains, among which PAZ and MID recognize the 3′ and 5′ ends of piRNA intermediates, respectively [22, 23]. The PIWI domain has endonuclease activity that allows it to incise RNA. PIWI and cancer together with other diseases currently remain an area of active research, because it has been discovered the aberrant expression of PIWI proteins is closely related to adverse clinical outcomes in cancerous patients [24]. In addition, several studies have also revealed that PIWI proteins have a strong correlation with DNA methylation in cancers. For instance, MILI and MIWI2 were initially found to play a crucial part in the occurrence of de novo DNA methylation of retrotransposons [25]. Further studies indicated that MILI and MIWI2 have differential functions on DNA methylation. MILI is responsible for the DNA methylation of a wider range of TEs while MIWI2 is implicated in DNA methylation and transcriptional gene silencing [26, 27].
Function of piRNAs. A. DNA methylation. At transcriptional gene silencing (TGS) level, the piRNA/PIWI complex methylates CpG sites of target gene by recruiting DNMT. B. Histone methylation. At TGS level, the piRNA/PIWI complex recruits repressive H3K9me3 and H3K27me3 marks to target gene and removes active H3K4me2 marks from promoter region. C. mRNA degradation and m6A methylation. At post-transcriptional gene silencing (PTGS) level, piRNA/PIWI complex inhibits the function of targeted RNAs by sequence complementary or block the m6A methylation of mRNA transcripts by interacting with methytransferase-like proteins (METTL) and/or Wilms' tumor 1-associating protein (WTAP). D. The piRNA/PIWI complex binds to transcriptional factor (TF) to regulate posttranslational modifications (PTMs).
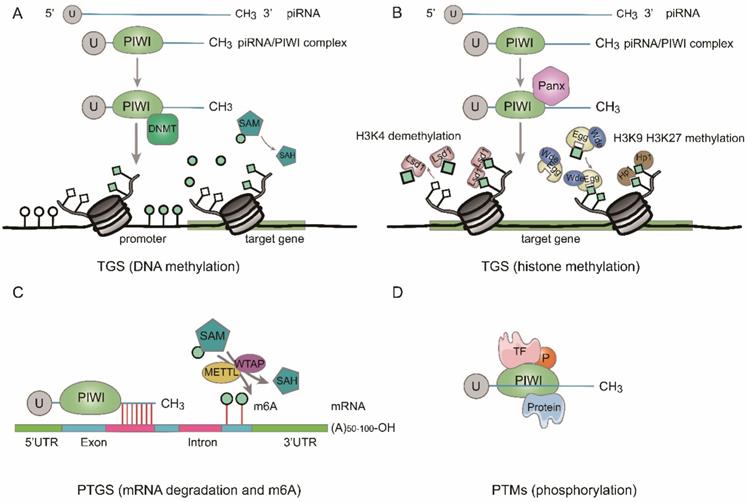
Function of piRNAs
According to previous studies, piRNAs function at the transcriptional, posttranscriptional and posttranslational modifications levels [28-31] (Fig. 2).
Transcriptional gene silencing (TGS)
By binding to the coding regions of gene promoters, piRNAs are involved in the gene regulation process [32]. It was discovered that the piRNA/PIWI pathway is implicated in the transcriptional gene silencing (TGS) through DNA methylation and histone modifications [20, 33]. When formed in the cytoplasm, the piRNA/PIWI complex comes into the nucleus and couples with the genomic target to form a fresh complex. After the new complex combining with Panoramix (Panx), it recruits silencing constituents, and TGS begins. Firstly, activated histone 3 lysine 4 dimethylation (H3K4me2) markers are removed from promoter regions by lysine-specific demethylase 1 (Lsd1), thus the transcription of RNA Pol II is inhibited [34]. Then, repressive histone 3 lysine 9 trimethylation (H3K9me3) markers are added to the target DNA by Eggless (Egg) and its cofactor Windei (Wde). Subsequently, heterochromatin protein 1 (Hp1) is recruited, leading to heterochromatin formation (Fig. 2b). Besides, the piRNA/PIWI complex also methylates genic CpG sites (nontransposon loci) by recruiting DNA methyltransferase (DNMT) and alters transcriptional activity [25] (Fig. 2a).
Posttranscriptional gene silencing (PTGS)
After discovering TGS, researchers found that piRNAs could suppress the function of target genes by regulating posttranscriptional networks, which is similar to what miRNAs form, namely, piRNA-RNA interactions. lncRNAs [35], mRNAs [36], etc. are included in RNAs that can interact with piRNAs. Base pairing at the 5'-end of piRNAs are required in the process of piRNA interaction, with 2-11 nt for strict base pairing and 12-21 nt for less stringent base pairing [37] (Fig. 2c). Moreover, by recruiting CCR4 NOT (carbon catabolite-repressed 4-negative on TATA-less) and Smaug, piRNA/PIWI complex can facilitate RNA repression to construct a specific piRNA-induced silencing complex (pi-RISC), and the latter can cause incomplete base-pairing with RNA [38]. For example, Peng L, et al. discovered that piR-55490 could bind to the 3'-UTR of mammalian target of rapamycin (mTOR) resulting in mRNA degradation and inhibiting the occurrence and development of lung cancer (LC) [36]. Additionally, several studies revealed that piRNA could function in the process of posttranscriptional gene regulation by another way, such as m6A RNA methylation [39, 40] (Fig. 2c). For example, Gao et al. demonstrated that a cardiac-hypertrophy-associated piRNA (CHAPIR) combined with PIWIL4 to form CHAPIR-PIWIL4 complex. Afterward, the CHAPIR-PIWIL4 complex interacted directly with METTL3 and blocked Parp10 mRNA transcripts m6A methylation, resulting in upregulation of Parp10 expression and ultimately leading to the progression of pathological hypertrophy and cardiac remodeling [39]. Additionally, piRNA-30473 was involved in tumorigenesis of diffuse large B-cell lymphoma by manipulating m6A RNA methylation [40].
Posttranslational modifications (PTMs)
The piRNA/PIWI complex can directly interact with certain transcriptional factors (TFs) and regulates their posttranslational modifications such as phosphorylation (Fig. 2d). For example, in colorectal cancer (CRC), researchers found that piR-823 increased the activity of heat shock factor 1 (HSF1) by binding to it and facilitated Ser326 phosphorylation, leading to over-expression of heat shock proteins and proliferation of CRC cells. Moreover, piR-54265/PIWIL2 complex interacted with signal transducer and activator of transcription 3 (STAT3) and phosphorylated-SRC (p-SRC) to form complex and activated STAT3 phosphorylation, in which both of them promoted CRC tumorigenesis [41, 42].
piRNA/PIWI complex affects DNA methylation
Based on the existing research evidence, at the transcriptional level, piRNAs undergo epigenetic changes mainly via sequence-specific aberrant DNA methylation and histone modification in somatic cells [43]. The crosstalk between piRNAs and DNA methylation has profound implications for genomic stability and gene expression, which may lead to aberrations in cell signaling transduction pathways, ultimately disease occurrence [44, 45]. DNA methylation plays an important role in epigenetic mechanisms by silencing transposons and other repetitive elements, and stably repressing specific transgenes and endogenous genes [46]. The methylation site is mostly the 5'-cytosine-phosphate-guanine-3' (CpG) cytosine dinucleotide, and rarely a non-CpG sequence [47]. CpG dinucleotides are localized and clustered to form CpG islands, detected at the majority of mammalian promoter regions [48]. In the human genome, the methylation status of 28 million CpG dinucleotides determines the gene expression level through the grading status of genes and regulatory sections [49]. Four types of DNA methyltransferases (DNMTs) are responsible for DNA methylation and methylation patterns: DNMT1, DNMT3A, DNMT3B and DNMT3L [50]. DNMT3A and DNMT3B contribute to de novo DNA methylation and DNMT3L facilitate the methylation of retrotransposons by interacting with DNMT3A and 3B, whereas DNMT1 recruits methyl groups to hemi-methylated CpG sites to maintain cytosine methylation modification in the new synthetic chain [51].
DNA methylation at transposon loci: the piRNA/PIWI complex mediates TGS by causing transposon DNA methylation
Ago family proteins assemble with small RNAs into RISC, such as miRNAs, siRNAs or piRNAs, mediating sequence-specific target gene silencing [52]. RISC assembly of piRNAs begins by loading single-stranded piRNA intermediates into PIWI subfamily proteins and directing the PIWI proteins to transposon element (TE) targets [52, 53]. Depending on the replication mode, TEs can be divided into two types: 1) retrotransposons, transcribed into RNA intermediates, and 2) DNA transposons, mobilized without transcription [54].
TEs are thought to be kept silent by DNA methylation and packaging into heterochromatin [55]. Previous evidence indicated that piRNAs, together with their PIWI proteins, could promote de novo DNA methylation of retrotransposons by generating complexes, thereby inhibiting transposons and maintaining gene stability [27, 28]. The loss of DNA methylation may stimulate gene expression at TEs including LINE-1 [56]. Notably, by integrating into the genome, active retrotransposons can cause genomic variations, change chromatin structures and alter the proximal gene expression [57]. Uncontrolled transposons threaten the integrity of the genome, and these mutations can be passed on to the next generation [58, 59]. Indeed, activation of LINE-1 may be involved in carcinogenesis and a large proportion of cancer patients have somatic retrotranspositions of LINE-1 elements [56]. In addition to DNA methylation, activated TEs can also be epigenetically silenced through the way of piRNA-dependent heterochromatin formation [60]. Consequently, the piRNA/PIWI complex plays crucial roles in maintaining minimum levels of transposons and protecting genomic stability [61].
DNA methylation at nontransposon loci: piRNA increased the expression of DNMTs and silenced tumor suppressor genes by methylation
Moreover, piRNAs can induce and affect DNA methylation not only at transposons, but also at nontransposon loci [29]. When transfecting piRNA mimics into human somatic cells, methylation changes at multiple genome sites were found, signifying that piRNAs are essential not only for methylation of transposons but also specific genes. According to epigenetic analysis of piRNAs in gene-specific DNA methylation, it is suggested that single-copy piRNAs contribute to DNA methylation at specific genome loci by incompletely combination with genomic DNA at non-TE sites [29]. For example, it was reported that piRNA-823 recruited the DNMT3A and DNMT3B, increased tumor suppressor p16INK4A DNA methylation and inhibited its expression in multiple myeloma cells [44].
The piRNA/PIWI complex affects the occurrence and development of diverse diseases through DNA methylation
Recently, second-generation sequencing technology has provided an extremely effective way to research disease-related genes and noncoding RNAs including piRNAs [62]. Since piRNAs were found to be involved in gene regulation, there has been increasing interest in what roles they play in human diseases. Many studies have shown that the dysregulation of piRNAs could facilitate or inhibit the occurrence and development of multiple diseases, especially cancer. Epigenetic alterations in cancer comprise global DNA hypomethylation, gene-specific DNA hypermethylation, histone modification, etc., causing activation of oncogenes such as Ras and cyclin D2 [63, 64] and silencing antioncogenes such as Rb1 and p16 [65]. Global DNA hypomethylation may be related to genomic instability; for instance, demethylated transposons can damage the genome when transcribed or transposed to other genomic regions [66]. Nonetheless, gene-specific hypermethylation tends to occur in the promoter regions of antioncogenes which may induce or promote cancer by silencing genes involved in DNA repair, apoptosis and tumor-specific signaling pathways [67]. Previous studies have discovered that ncRNAs, including miRNA, circRNA and lncRNA, could regulate gene expression through DNMT mediated DNA methylation [68-70], but the role of piRNAs in the DNA methylation of diseases such as cancer has not been systematically described. Here we summarized the regulation and function of piRNAs/PIWI proteins in DNA methylation in cancers and other human diseases (Tables 1 & 2; Figures 3 & 4). At the same time, we also mentioned chromatin methylation modification, which plays a significant role in gene expression regulation along with DNA methylation.
The piRNA /PIWI complex influences various cancers by regulating DNA methylation
Breast cancer
In women, breast cancer (BC) is the most commonly diagnosed cancer and the leading cause of cancer-related death [71]. Currently, BC affects more than 10% of women worldwide, especially those before the age of 45 [72, 73]. BC consists of mainly four molecular subtypes, namely basal, luminal A, luminal B, HER2-overexpressing, and they have diverse clinical courses, prognoses and therapies [74, 75]. Nevertheless, their underlying molecular mechanisms remain poorly understood. Despite medical advances in early screening and diagnosis, BC mortality rate has continued to rise worldwide over the past 25 years, which could be attributed to the increased incidence and prevalence of this cancer [76]. Therefore, finding new strategies and approaches to prevent BC, improve survival rates, and reduce cancer mortality is crucial and urgent. Recent studies have found that piRNAs could influence BC tumorigenesis, growth, invasiveness, and metastasis by methylating a specific gene site, implying that piRNAs could be used as a class of biomarkers for the early diagnosis of BC and as an alternative treatment [77-80].
First, it was revealed that the changes in DNA methylation caused by piRNAs in BC could promote tumor development. For example, piR-823 was reported to increase the expression of DNMTs including DNMT1, DNMT3A, and DNMT3B, and promote DNA methylation level of the adenomatous polyposis coli (APC) gene, a tumor suppressor, thereby motivating WNT signaling pathway and inducing the formation of cancer cell stemness (CSCs) in the luminal subtype of BC cells, ultimately contributing to BC tumorigenesis [77]. Similarly, piR-932 was also proven to induce BC metastasis through gene-specific hypermethylation [78]. piR-932 expression was significantly higher in BC cells that were induced by EMT, and it could form complexes with PIWIL2. Then, the piR-932/PIWIL2 complex reduced Latexin expression through improving the methylation of CpG island in promoter region. Latexin, also a tumor suppressor, could reduce the transformation of old stem cell into CSCs, decrease the cell replication, and enhance the apoptosis [81, 82]. In contrast, there are some piRNAs that can inhibit BC growth through methylation [79, 80]. The variant piR-021285-mimic transfected into BC cell lines could weaken methylation of pro-invasive gene ARHGAP11A at the region of the 5' UTR/first exon CpG island, leading to increased ARHGAP11A expression and tumor cell invasiveness [79]. Conversely, wild type piR-021285 was proven to inhibit BC cell proliferation and invasion by improving ARHGAP11A methylation. Similarly, a GAS5-derived small RNA (pi-sno75) also played an inhibitory role in tumor growth by inducing methylation/demethylation [80]. Interestingly, pi-sno75 constituted the piRNA/PIWI complex by binding to the PIWIL1/4 proteins and affected gene expression through a different mechanism. The piRNA/PIWI complex coupled with WDR5 and subsequently recruited the COMPASS-like complex containing MLL3/UTX to the promoter region of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL). TRAIL, a proapoptotic protein, was significantly upregulated by increasing H3K4 methylation/H3K27 demethylation, consequently leading to the inhibition of tumor growth [80].
Dysregulated piRNAs in cancers and other diseases
| piRNA | Disease | Dysregulation | Gene | Mechanism | Role | Reference |
|---|---|---|---|---|---|---|
| piR-823 | Breast cancer | up | APC | Increases the expression of DNMTs, promotes DNA methylation of APC gene, thereby activates WNT signaling, induces CSCs and contributes to tumorigenesis | promotor | [77] |
| piR-932 | Breast cancer | up | Latexin | Reduces the expression of Latexin through methylation its promoter region CpG island, induces CSCs and affects metastasis of breast cancer | promotor | [78] |
| piR-021285 | Breast cancer | down | ARHGAP11A | Inhibits cell invasion and proliferation methylating by ARHGAP11A | suppressor | [79] |
| piR-sno75 | Breast cancer | up | TRAIL | Upregulates TRAIL by inducing H3K4 methylation/H3K27 demethylation and inhibits tumor growth | suppressor | [80] |
| piR-34871, piR-52200, piR-35127, piR-46545 | Lung cancer | up | GMIP | Regulates DNA methylation of oncogenes and anti-oncogenes, initiates local tumor growth and inhibits cell apoptosis | promotor | [92] |
| down | A4GALT | |||||
| piR-823 | Multiple myeloma | up | p16INK4A | Increases DNMT3A and 3B, promotes DNA methylation and increases the tumorigenesis | promotor | [44, 102] |
| Has_piR_011186 | Leukemia | up | CDKN2B | Upregulates the methylation of DNA and histone H3 including H3K9 and H3K27 in the region of CDKN2B promoter | promotor | [103] |
| piR-823 | Esophageal cancer | up | - | Induces aberrant DNA methylation through DNMT3B and plays tumor oncogenic role | promotor | [45] |
| piR-823 | Gastric cancer | up | - | Causes cells aberrant “stem-like” state by reducing tumor supporter genes methylation and inhibits proliferation of cancer cells | suppressor | [110, 111] |
| piR-31470 | Prostate cancer | up | GSTP1 | Recruits DNMT1, DNMT3A and methylate CpG island of GSTP1 | promotor | [117] |
| DQ589977, DQ591415, DQ598918, DQ601291, DQ601609 | Testicular cancer | down | PIWIL1, PIWIL2, PIWIL4, TDRD1 | Promoter CpG island hypermethylation-associated silencing | promotor | [118] |
| Aplysia piRNAs | Long-term synaptic facilitation | up | CREB2 | Facilitates CpG island methylation in the CREB2 promoter | promotor | [132] |
| DQ541777 | Neuropathic pain | up | CDK5rap1 | Increases the methylation level of CpG islands by recruiting DNMT3A to CDK5rap1 promoter | promotor | [134] |
| piR-007899, piR-019162 | Gulf War Illness | up | - | Alteration in DNA methylation and piRNAs could be detected | promotor | [135] |
| piR-63076 | Pulmonary hypertension | up | Acadm | Attributes to Acadm DNA methylation and finally increases hypoxia-induced PASMC proliferation | promotor | [139] |
| td-piR(Glu) | Immune | up | CD1a | Recruits SETDB1, SUV39H1, and HP1b to the the region of CD1a promoter and facilitates H3K9 methylation | - | [140] |
piRNAs and DNA methylation involved in diseases in this article. The dysregulation of piRNAs contributes to abnormal expression of genes through DNA methylation or histone methylation and finally promotes or restrains the development of cancers or other diseases in different systems.
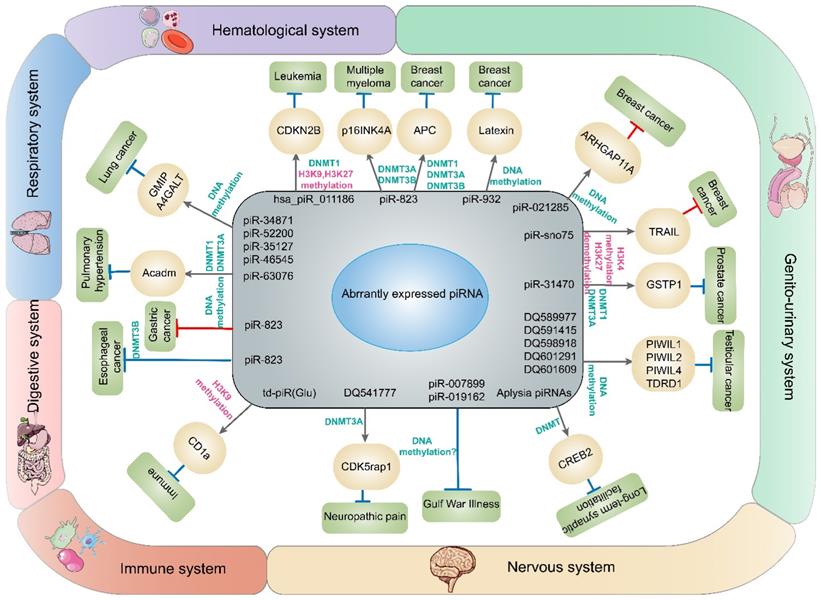
PIWI and DNA methylation involved in diseases in this article. The dysregulation of PIWI protein contributes to abnormal expression of genes through DNA methylation and finally promotes or restrains the development of diseases.
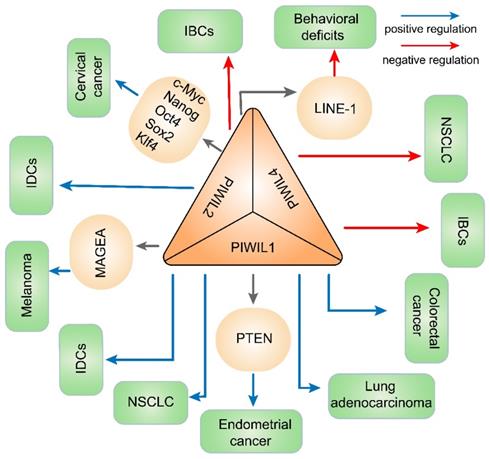
Dysregulated PIWI in cancers and other diseases
| PIWI | Disease | Expression | Gene | Function | Reference |
|---|---|---|---|---|---|
| PIWIL1/HIWI | IDCs | up | - | Promotes cancer development by aberrant DNA methylation | [85] |
| Lung adenocarcinoma | up | - | PIWIL1 expresses aberrantly due to promoter DNA hypo-methylation and promotes proliferation, invasion and migration | [95] | |
| Colorectal cancer | up | - | PIWIL1 gene ectopically activates due to promoter demethylation and correlates with tumor differentiation, lymphnode invasion and metastasis; cancer-specific immunotherapies | [108] | |
| Endometrial cancer | up | PTEN | Causes PTEN underexpression through DNMT1-mediated hypermethylation | [120] | |
| NSCLC | up | Expression could be partly upregulated by methylation | [96] | ||
| PIWIL2/HILI | IBCs | down | - | Probably associated with down-regulation of DNMT1, histone H1, HP1 and SUV39H1 | [83] |
| IDCs | up | - | Aberrant DNA methylation | [85] | |
| Cervical cancer | up | c-Myc, Nanog, Oct4, Sox2, Klf4 | Induces H3K9 acetylation but reduces H3K9 trimethylation | [119] | |
| Melanoma | up | MAGEA | Methylates LINE-1, to regulate MAGEA expression and inhibits melanoma cell migration | [44] | |
| Behavioral deficits | down | LINE-1 | Hypomethylates intergenic areas and LINE-1 promoter areas | [133] | |
| PIWIL4/HIWI2 | IBCs | down | - | Probably solely associated with down-regulation of DNMT1, | [83] |
| NSCLC | down | - | Positively correlated with overall methylation and poor outcomes | [96] |
In addition to piRNAs, PIWI was also significantly associated with DNA methylation in BC [83]. First, epigenetic inactivation of the PIWIL2 gene was caused by hypermethylation of the promoter CpG island and delocalization of the PIWI protein into cytoplasmic stress bodies [84]. Furthermore, DNMT1, histone H1, HP1 and SUV39H1 were downregulated in invasive breast cancers (IBCs) with PIWIL2 underexpression, while DNMT1 was downregulated only in IBCs with PIWIL4 underexpression. Later, molecules involved in genome methylation, chromatin accessibility and increased inflammatory/cytotoxic immune reactions were potentially reduced, and IBCs were thereby promoted [83]. In addition, research indicated that PIWIL1 and PIWIL2 expression were significantly increased in invasive ductal carcinomas (IDCs), inducing a stem-like state of cancer cells through abnormal DNA methylation, generating genomic silencing and ultimately promoting cancer development [85].
Lung cancer
Lung cancer (LC) has the highest cancer-related mortality among men and the second-highest cancer-related mortality (after breast cancer) among women worldwide [86]. Non-small cell lung cancer (NSCLC) is a predominant type of LC, accounting for 85% of all lung cancers, while the incidence of small cell lung cancer is low, accounting for only 15% of all lung cancers [87]. Patients with LC have a poor prognosis and are usually diagnosed late in the disease, leading to a high mortality rate [88]. Epigenetic alterations in LC, including DNA methylation, play critical roles in cancer progression, and epigenetic recognition of LC pathogenesis is increasingly playing a significant role in the discovery of novel diagnostic biomarkers and the development of therapeutic strategies [89].
Firstly, studies have shown that the DLK1-DIO3 locus is methylated aberrantly in current and former smoker patients with NSCLC and aberrant expression of DLK1-DIO3 might contribute to tumorigenesis in LC [90]. Previous studies indicated that the DLK1-DIO3 locus encodes piRNAs in somatic cells and might enhance the prognostic potential of piRNAs to specifically predict patient outcome. In addition, they could have great potential as novel biomarkers and therapeutic targets [91]. However, how abnormal methylation of the DLK1-DIO3 locus affects LC through piRNAs remains to be further studied. In addition, researchers have recently demonstrated a new RASSF1C-PIWIL1-piRNA pathway that facilitates LC cells progression and migration by regulating the methylation of oncogenes and anti-oncogenes [92]. In LC cells, Ras Association Domain Family Member 1 isoform C (RASSF1C) enhanced the PIWIL1 gene expression by the MEK-ERK1/2 pathway [93]. Afterward, PIWIL1 and its interacting piRNAs (piR-34871, piR-52200, piR-35127 and piR-46545) regulated DNA methylation of oncogenes and tumor suppressor genes and inhibited cell apoptosis [2, 94]. In this way, the Gem Interacting Protein (GMIP) gene, which has anticancer properties, was hypermethylated and downregulated. Conversely, the A4GALT gene which serves as an oncogene was upregulated by demethylation. Consequently, cancer cells would initiate local tumor growth at the site of metastasis by transforming from mesenchymal cells to epithelial cells [92]. In terms of PIWI protein, PIWIL1 expressed aberrantly in lung adenocarcinoma and NSCLC due to promoter DNA hypo-methylation and promotes proliferation, invasion and migration of cancer cells [95]. For another PIWI protein, underexpression of PIWIL4 was positively correlated with overall methylation in NSCLC [96]. Moreover, in tumor tissues, PIWIL4 expression was downregulated and patients with lower PIWIL4 expression had a shorter time to relapse (TTR) and overall survival (OS) than others [96].
Hematological neoplasm
Hematological malignancies include leukemia, multiple myeloma (MM) and lymphoma [97]. MM is a B cell-derived malignancy featured with the clonal replication of plasma cells in the bone marrow. Although remarkable progress has been made in treating the disease, drug-resistant relapse usually occurs and MM remains largely incurable [98]. Diffuse large B-cell lymphoma (DLBCL), the most common subtype of malignant lymphoma, is heterogeneous in its clinical presentation, morphology and biological behavior [99]. Although standard therapy cures two-thirds of patients, approximately 30~40% are still not cured [100]. Therefore, further studies on molecular carcinogenesis mechanisms are necessary to provide new therapeutic strategies and improve prognosis.
In MM, CSCs and myeloid-derived suppressor cells (MDSCs) are two important cellular components in the tumor microenvironment, that may change the cancer phenotype and affect patient survival [101]. It was reported that piR-823 expression was increased by granulocytic-MDSCs (G-MDSCs), which in turn promoted DNA methylation and increased the tumorigenic potential of MM cells [102]. In contrast, silencing piR-823 in MM cells resulted in markable decrease in DNMT3A and DNMT 3B at both the mRNA and protein expression levels, which reversely resulted in a reduction in overall DNA methylation and re-expression of the methylation-silenced cancer suppressor gene p16INK4A [44, 102]. Similarly, in the leukemia cell line U937, the CDKN2B-related has_piR_011186 constituted a complex with DNMT1, SUV39H1 and EZH2 proteins, upregulating the methylation level of DNA and histone H3 (including histone H3K9 and H3K27) in the CDKN2B promoter site. Thus, the expression of the CDKN2B gene was inhibited, cell cycle progression was promoted and apoptosis was reduced [103].
Digestive system neoplasm
Digestive malignancies are a group of cancers that occur in the gastrointestinal tract and its related organs. Cancers of the colon and rectum, stomach, liver, esophagus, and pancreas are five of the ten most commonly diagnosed cancers and causes of cancer-related death [104]. Among these digestive cancers, esophageal cancer (EC) is a highly aggressive malignancy that is usually detected in locally advanced stages with distant metastases, limited treatment options, and a five-year survival rate of less than 20% [105]. Colorectal cancer is one of the most commonly diagnosed human cancers and the third reason of cancer-related death [106]. Consequently, identifying fresh and robust diagnostic and therapeutic targets that can facilitate early detection of digestive malignancies or as a specific treatment to reduce mortality remains a priority in cancer research.
Recently, a study revealed that piR-823 induced aberrant DNA methylation through the epigenetic pathway DNMT3B and exerted an oncogenic role in esophageal squamous cell carcinoma (ESCC). Additionally, the expression of piR-823 has high specificity in ESCC, and a higher level of piR-823 signified a higher risk of lymph node metastasis, suggesting its potential as a biomarker for diagnosis and prognosis [45]. Intriguingly, over the past few years, PIWI have also emerged as promising biomarkers for CRC diagnosis and prognosis [107]. The ectopic expression of PIWIL1 has recently been implicated in tumor differentiation and depth of invasion, as well as lymph node invasion and metastasis [108]. In addition, in a significant fraction of CRC, the PIWIL1 gene was ectopically activated accompanied by demethylation of gene promoter, together with production of germline factors required for piRNA, which is a promising application of cancer-specific immunotherapies [109]. For gastric cancer, although studies have shown that piR-823 demonstrated tumor suppressor in gastric cancer cells in vitro and in vivo [110], the roles of piRNA and its associated DNA methylation in the tumorigenesis and development of gastric cancer warrant further study. However, several literatures have revealed that piR-823 inhibits cancer cells proliferation and causes an abnormal “stem-like” state in cells by reducing tumor supporter gene methylation [111].
Genitourinary neoplasms
Genitourinary cancers typically refer to neoplasms of the kidneys, bladder, prostate, testicular, uterus, ovary and so on. Prostate cancer is the most common tumor in men worldwide [112]. Current statistics show that the diagnosis rate and mortality of prostate cancer are as high as 14.3% and 2.6% respectively, or even higher. [113]. In males between 14 and 44 years of age, testicular cancer is the most common solid malignancy and its incidence has risen worldwide during the past two decades [114]. Although the piRNA/PIWI pathway plays a critical role in male germline development [115, 116], the role of piRNAs in genitourinary tumors is scarce and deserves further attention.
It was demonstrated that piRNAs were associated with prostate cancer via specific hypermethylation, which led to their transcriptional inactivation in prostate cancer. First, piR-31470 could form a piR-31470/PIWIL4 complex by binding to PIWIL4. Afterward, this complex coupled with the glutathione S-transferase pi 1 (GSTP1) RNA transcripts and recruited DNMT 1 and DNMT 3A and methylated the CpG island of GSTP1 to activate and sustain the hypermethylation and inactivation of GSTP1. In the process of prostate carcinogenesis, silence of GSTP1 via hypermethylation is an early and common event and functional inactivation of GSTP1 enhanced the susceptibility to oxidative stress and the risk of prostatic cancer progression [117]. Otherwise, under the circumstance of downregulation of piRNA and low DNA methylation of the LINE-1 TEs, it has been observed that the repression of PIWIL1, PIWIL2, PIWIL4, and Tudor domain-containing protein 1 (TDRD1) was associated with hypermethylation of the promoter CpG island in testicular tumors [118].
Regarding to the function of PIWI protein and DNA methylation in genitourinary neoplasms, there are two main types of tumors that are common in women: cervical cancer and endometrial cancer. First, HPV oncoproteins E6 and E7 have been reported to reactivate PIWIL2 during cervical cancer tumorigenesis. Then PIWIL2 induced H3K9 acetylation but reduced H3K9 trimethylation, resulting in epigenetic reprogramming and the maintenance of embryonic stem cells (ESCs) by affecting the expression of genes in c-Myc, Nanog, Oct4, Sox2 and Klf4 [119]. In another female tumor, endometrial cancer, PIWIL1 caused the silence of PTEN expression through DNMT1-mediated hypermethylation [120]. PTEN is a tumor suppressor with growth and survival regulatory functions [121]. Therefore, the PIWIL1/DNMT1/PTEN signaling pathway may play a significant role in the progression of type I endometrial cancer [120].
Melanoma
Melanoma is still a potentially lethal skin malignancy and the occurrence of malignant melanoma has continued to increase for many years [122]. Although most patients have localized disease when diagnosed and are treated by surgical excision of the primary tumor, many patients develop metastases [123]. Nevertheless, piRNA/PIWI protein might become a potential target in melanoma therapy.
MILI, also named PIWIL2 in humans, inhibited melanoma cell metastasis, probably providing a novel perspective on melanoma treatment. Specifically, MILI could methylate the LINE-1 repetitive sequence to regulate MAGEA expression and restrain melanoma cell migration [44]. In addition, it has been demonstrated that aberrant MAGEA expression has a very strong correlation with high-grade lesions and terminal cancer [124].
The piRNA/PIWI complex influences other human diseases through regulating DNA methylation
Reproductive System Diseases
Infertility remains a global problem what is estimated to affect almost 186 million people worldwide, and human infertility contributes to more than 50% of all cases of global childlessness [125]. Infertility patients bear enormous psychological and social pressure, and infertility leads to low fertility and population aging, which increases the social burden [106]. piRNAs were originally discovered in mouse testis cells and further studies confirmed that they play a key role in germ cell generation and maintenance in Drosophila and mouse models. Since piRNAs act as general effectors of gene expression and epigenetic alterations during spermatogenesis, they may be useful and effective tools for diagnosing infertility. In addition, piRNA/PIWI protein can affect spermatogenesis by regulating DNA methylation and thus could also be a promising target of novel drugs for treatment.
Transposons pose a serious challenge to the germline, and mechanisms that inhibit their activity are crucial for transgenerational genomic integrity; thereinto LINE-1 is the most important retrotransposon and is epigenetically repressed by CpG island DNA methylation [126]. There are three important epigenetic mechanisms that inhibit LINE-1 transposons and enhance genomic stability in spermatogenesis [25, 126, 127]. Initially researchers discovered that two PIWI proteins MILI and MIWI2 played critical roles in the construction de novo DNA methylation of retrotransposons containing LINE-1 and intracisternal A particles (IAPs) in germ cells [25]. Further study indicated that MIWI2 as a regulator was indispensable in germ cell's DNA methylation and gene silence [27]. Later five piRNAs in spermatogenic failure samples, DQ589977, DQ591415, DQ598918, DQ601291, and DQ601609 were found to be downregulated and they targeted PIWI2 and TDRD1 gene promoters. In addition, their expression had inverse correlation with the methylation level of PIWI2 and TDRD1 gene promoters [127]. Likewise, a recent study also showed that DNA methylation impacted the piRNA/PIWI complex pathway and was involved in the production of pachytene piRNA and gene expression during the bovine hybrid male sterile spermatogenesis process [128]. DNA hypermethylation and disrupted piRNA production resulted in failure of germ cell development which might drive male sterility in cattle crosses. In addition to these two mechanisms, the euchromatic repressive H3K9 modification co-repressed LINE-1 expression [126]. Subsequent research found that during the DNA hypomethylation stage, protein arginine methyltransferase 5 (PRMT5), together with ubiquitin like with plant homeodomain and ring finger domains 1 (UHRF1), was necessary for the repressive H2A/H4R3me2s chromatin modification on LINE-1 and IAP transposons in primordial germ cells (PGCs) and participated in transposon silencing, while conditional loss of these proteins in early PGCs caused complete male and female sterility [129, 130]. Thereafter, PRMT5 translocated back to the cytoplasm and participated in the piRNA pathway, which promoted transposon silencing through de-novo DNA remethylation [129].
Nervous System Diseases
Many publications have revealed that piRNAs exist extensively in the central and peripheral nervous systems and participate in regulating the physiological and pathological processes of various neurological diseases including anxiety disorders and Parkinson's disease [131]. The study of the role of piRNAs and DNA methylation in neurological diseases may help to further raise our cognitive level of the pathogenesis of neurological diseases and develop novel diagnostic markers and therapeutic targets for these diseases.
Researchers demonstrated for the first time that Aplysia piRNA/PIWI complex facilitated serotonin-dependent methylation at the promoter CpG island of CREB2, a major inhibitory limitation of memory in Aplysia, enhancing long-term synaptic facilitation [132]. Later, a growing body of research showed has shown that piRNAs have epigenetic roles in the mammalian nerve and brain that we have not explored before [133, 134]. The piRNA pathway might play a role in locomotory, exploratory and normal anxiety behavior. It has been demonstrated that intergenic and LINE-1 promoter regions of brain genomic DNA was preferentially hypomethylated across the whole genome of MILI/piRNA-deficient mice, and ultimately, MILI mutant mice presented behavioral deficits such as hyperactivity and decreased anxiety [133]. Furthermore, piR-DQ541777 increased the methylation degree of CpG islands by recruiting DNMT3A to the CDK5rap1 promoter and consequently reduced the expression of CDK5rap1 and regulated neuropathic pain [134]. Otherwise, in Gulf War Illness (GWI), a chronic disorder distinguished by memory dysfunction and depression, changes in methylation throughout the brain and in miRNA and piRNA (piR-007899, piR-019162) expression were detected [135]. However, their connection remains to be further elucidated. Another study indicated that PIWIL1 regulated polarization and radial migration of neurons in part by modulating the expression level of microtubule-associated proteins (MAPs) and mutations of PIWI might be associated with developmental cerebral disorders like autism. However, some literatures pointed out that PIWI might not regulate MAP expression through methylating target gene promoters in cortical neurons [136].
Cardiovascular diseases
Cardiovascular diseases (CVDs) are the main causes of human mortality worldwide [137]. With the advancement of next-generation sequencing technology, piRNAs are differentially expressed in CVDs, indicating that they are involved in the occurrence and progression of cardiac and vascular diseases [138].
Recent research has indicated that the piRNA/PIWI complex might be a new class of molecules that are potentially useful as biomarkers for the classification of pulmonary hypertension as well as therapeutic targets. The expression of piR-63076 was upregulated in pulmonary vessels under hypoxia and partly attributed to DNA methylation of the acyl-CoA dehydrogenase (Acadm) promoter region. Acadm deficiency ultimately increased hypoxia-induced pulmonary-artery smooth muscle cells (PASMCs) proliferation [139].
Others
Although little is known about how piRNAs function in the human immune system, studies have suggested that piRNAs could act as signal transduction mediators in well differentiated immune cells [140].
First, a previous study showed that piR-30840 interacted with PIWIL4 and Ago4 that targeted the intron of IL-4, which inversely caused the degradation of IL-4 pre-mRNA [141]. Subsequently, the reaction restrained the production of IL-4 in CD4 lymphocytes and the differentiation of Th2 CD4-T lymphocytes. The above aberrant expression has been reported in pathological conditions of certain diseases, such as asthma. This finding possibly provides a novel therapeutic target for human allergic diseases. Later, subsequent research indicated that a tRNA-Glu-derived piRNA [td-piR (Glu)] combined with PIWIL4 to form the td-piR (Glu)/PIWIL4 complex and subsequently recruited SETDB1, SUV39H1, and Hp1b to the CD1a promoter region, promoting H3K9 methylation. As a consequence, the transcription of CD1a was notably suppressed. This mechanism may provide a viable and specific method for controlling CD1a expression in the treatment of certain autoimmune diseases, and at the same time, it can also avoid transplant immune rejection and tumor immune evasion caused by abnormal expression of CD1a. Interestingly, IL-4 significantly reduced the production of tRNA-Glu, thereby reducing the generation of td-piR (Glu). However, in another immune disease, rheumatoid arthritis, the piRNA/PIWI system might not play such a significant role [142]. In rheumatoid arthritis synovial fibroblasts (RASFs) and osteoarthritis synovial fibroblasts (OASFs), PIWIL2/4 mRNA expression was potently increased via TNFα+IL1β/TLR- ligands and piR-16659 was increased 4-fold upon poly (I:C) stimulation. However, silencing PIWIL2/4 affected neither LINE-1 methylation nor RASF proliferation. Instead, another system that controls LINE-1 expression, TREX-1, has been detected to be down-regulated in RASF [142].
piRNAs and piRNA-associated methylation as potential biomarkers for cancers
In terms of mortality, globally, cancer is the second cause leading to death (8.97 million deaths) after ischemic heart disease but is expected to become the most common cause of death in 2060. Several cancers, such as esophageal, liver, and pancreatic cancers, have the worst prognosis, usually less than 20% in 5 years [86]. Meanwhile, certain chronic diseases such as heart failure can force a tremendous clinical burden, disrupt social standards, and erode a huge number of economic resources [143]. Accordingly, early diagnosis coupled with effective and accurate treatment is crucial.
More and more evidence point to the fact that piRNAs could represent attractive diagnostic and prognostic biomarker candidates, as some piRNAs have been involved in the development of many diseases [138, 144]. First, piRNAs can be used to distinguish cancer patients from healthy individuals based on the expressive discrepancy in cancer samples and normal controls and the detection sensitivity is higher than CEA and CA19-9 [107, 145]. Similarly, a previous study on piRNAs in gastric cancer indicated that piRNAs provided higher sensitivity and specificity than existing biomarker detection systems based on miRNAs [145]. Second, differences in piRNA expression are useful for predicting the prognosis of TTR and OS in cancer patients. piRNAs in peripheral blood could be used as disease progression indicators and postoperative long-term monitoring due to their remarkable distinct expression among preoperative and postoperative patients [145]. Third, due to the protection of PIWI proteins, circulating piRNAs in peripheral blood or extracellular vesicles are highly stable and abundant, which makes them ideal biomarkers for liquid biopsy [145, 146]. As an example, Lipps et al. was the first to illuminate the function of piRNAs in chronic thromboembolic pulmonary hypertension, demonstrating that piRNAs in the extracellular vesicles can be applied as biomarkers to reflect disease severity [147]. In addition, in recent years, piRNA-associated DNA methylation was detected in the early stages of tumorigenesis and cancer progression and has been proposed as a biomarker for cancer detection, prognosis and prediction of treatment response [148]. For example, in related breast cancer studies, it was found that early breast cancer diagnosis and the prediction of breast cancer risk could be achieved by detecting aberrant methylation of several genes in serum DNA and genome-wide epigenetic changes. Similarly, numerous studies related to colorectal cancers have been conducted to identify specific methylation markers that are important for CRC detection. In reality, clinical assays evaluating the SEPT19 gene and vimentin methylation state became commercially available [148].
piRNA/PIWI complex-associated methylation as a potential therapeutic strategy for cancers
Beyond the role of piRNAs and piRNA-associated methylation as biomarkers, several studies have put forward their potential role as therapeutic tools [44, 78, 109, 149] just like other non-coding RNAs [150-152]. On the basis of the functional characteristics of piRNAs, we offer some thoughts on piRNA as a possible target for cancer therapy as follows. Firstly, at the pre-transcriptional level, specific piRNAs could be engineered to bind PIWI proteins to silence PIWI genes and block harmful outcomes. Secondly, at the transcriptional level, the PIWI/piRNA system might serve as a potential tool for future research to improve human health by manipulating DNA methylation of certain genes. We might be able to hypermethylate the promoter CpG island of PIWI to reduce its expression, thereby blocking piRNA and PIWI binding and its downstream steps. Unlike genetic alterations, DNA methylation is reversible, which makes it extremely interesting for therapeutic approaches. Last, at the posttranscriptional level, piRNAs could block the synthesis of cancer-related proteins through a mechanism that binds to mRNAs, similar to the mechanism by which miRNAs do not require Dicer. Although studies into piRNAs-based therapeutic methods are still in its early stages, more therapeutic outcomes could be achieved in the future as the mechanisms and functions of piRNAs in cancer are increasingly understood.
Conclusion and perspective
Due to the advent of next-generation sequencing technologies, the expression of piRNAs can be easily observed. In fact, the study of piRNAs has deepened our understanding of the complex regulatory network of non-coding RNAs. Numerous studies have reported dysregulated expression of piRNAs in different diseases samples and proposed the underlying mechanisms. In this review, we summarized that piRNAs could guide PIWI proteins to regulate gene expression in a transcriptional way including DNA methylation and histone methylation (Fig. 2a, b) and/or posttranscriptional way including mRNA degradation and m6A methylation and/or posttranslational modifications (Fig. 2c, d). Under physiological conditions, the piRNA/PIWI complex formed by inherent piRNAs can establish an inhibitory chromatin state by histone modification to silence TEs and maintain genomic stability. When certain disease occurs, piRNAs expressions aberrantly increase but, in the others, piRNAs have been seen to be downregulated or they may be absent totally. Newly generated or intrinsic piRNA/PIWI complex affects the expression of different genes through global DNA hypomethylation, gene-specific DNA hypermethylation or posttranscriptional way and they play various roles in different pathophysiological processes, signaling pathways, the cell cycle, DNA repair, cell adhesion, invasion, and metastasis etc. Nevertheless, it is of note that translation and post-translational modification are equally important for the occurrence and development of tumors, so we need to conduct more in-depth research on the level of translation and post-translational modification of piRNAs in the future. piRNA/PIWI complex have a close influence on the occurrence of a variety of diseases including cancers. Owing to the heterogeneity of cancer, piRNAs can be both repressors and promoters in diverse cancers. Actually, the exact molecular mechanisms by which piRNAs are disregulated and impact on human health are still in its fancy and require further investigation. In addition to environmental and genetic factors, epigenetic changes also play a pivotal role in the occurrence of diseases, among which DNA methylation occupies a special position. We also summarized the role of piRNA/PIWI in the occurrence and development of cancer and other human diseases from the perspective of DNA methylation. In addition, there is growing evidence that the expression of piRNAs and methylation levels in some cancer-related genes are related to pathological variables or clinical outcomes; for that reason, they may have significant clinical value as diagnostic biomarkers. Moreover, piRNAs' regulation of disease through DNA methylation appears to offer new insights into epigenetics and great potential for future interventions in disease processes, including cancer.
However, the potential applications of piRNAs for clinical medicine still face plenty of challenges. First of all, it remains unclear that whether the abnormal expression of piRNAs really participate in the development of these diseases or just are a byproduct of other molecular activities. Secondly, to a large extent, the molecular mechanisms of physiological and pathological action of piRNAs are largely unknown, while some studies have proposed potential molecular mechanisms. Thirdly, in clinical practice, the potential application of piRNAs as a diagnostic and therapeutic target is currently at a speculative and theoretical stage, such as how to distinguish between healthy individuals and patients and how to resolve potential side effects, drug resistance, therapeutic effects, and administration mode. Accordingly, in the future, further studies and multi-center clinical trials should be conducted to fully aware of the regulatory mechanism and clinical application of piRNAs.
Abbreviations
Ago3: Argonaute 3; Aub: Aubergine; CDK5rap1: CDK5 regulatory subunit-associated protein 1; CpG: 5'-cytosine-phosphate-guanine-3' ; CSCs: Cancer stem cells; CVDs: Cardiovascular diseases; DNMT: DNA methyltransferase; EMT: Epithelial-mesenchymal transition; H3K9me3: Histone 3 trimethylated on lysine 9; LINE-1: Long interspersed nuclear elements-1; miRNAs: MicroRNAs; Mino: Minotaur; OS: Overall survival; Panx: Panoramix; piRNAs: Piwi-interacting RNAs; PTEN: Phosphatase and tensin homolog deleted on chromosome ten; PTGS: Post-transcriptional gene silencing; PTMs: Posttranslational modifications; RISC: RNA-induced silence compounds; ncRNAs: Non-coding RNAs; siRNAs: Small interfering RNA; TEs: Transposable elements; TGS: Transcriptional gene silencing; Zuc: Zucchini.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (22006084, 81800215); the Qingdao Applied Basic Research Project (19-6-2-49-cg); and Hubei Key Laboratory of Environmental and Health Effects of Persistent Toxic Substances (PTS2019-05).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N. et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203-7
2. Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. Journal of cellular biochemistry. 2012;113:373-80
3. Collins LJ, Penny D. The RNA infrastructure: dark matter of the eukaryotic cell? Trends in genetics: TIG. 2009;25:120-8
4. Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656-68
5. Luo C, Hajkova P, Ecker JR. Dynamic DNA methylation: In the right place at the right time. Science (New York, NY). 2018;361:1336-40
6. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33(Suppl):245-54
7. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199-202
8. Huang Y, Bai JY, Ren HT. PiRNAs biogenesis and its functions. Bioorganicheskaia khimiia. 2014;40:320-6
9. Han BW, Zamore PD. piRNAs. Current biology: CB. 2014;24:R730-3
10. Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. The EMBO journal. 2010;29:3301-17
11. Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L. et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284-7
12. Han BW, Wang W, Li C, Weng Z, Zamore PD. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science (New York, NY). 2015;348:817-21
13. Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annual review of biochemistry. 2015;84:405-33
14. Czech B, Hannon GJ. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends in biochemical sciences. 2016;41:324-37
15. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089-103
16. Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E. et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157:1698-711
17. Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N. et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193-7
18. Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & development. 2002;16:2733-42
19. Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82:323-30
20. Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental cell. 2007;12:503-14
21. Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T. et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science (New York, NY). 2007;315:1587-90
22. Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends in biochemical sciences. 2000;25:481-2
23. Yamaguchi S, Oe A, Nishida KM, Yamashita K, Kajiya A, Hirano S. et al. Crystal structure of Drosophila Piwi. Nature communications. 2020;11:858
24. Weng W, Li H, Goel A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochimica et biophysica acta Reviews on cancer. 2019;1871:160-9
25. Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M. et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & development. 2008;22:908-17
26. Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. MIWI2 and MILI Have Differential Effects on piRNA Biogenesis and DNA Methylation. Cell reports. 2015;12:1234-43
27. Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H. et al. MIWI2 as an Effector of DNA Methylation and Gene Silencing in Embryonic Male Germ Cells. Cell reports. 2016;16:2819-28
28. Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science (New York, NY). 2007;316:744-7
29. Fu A, Jacobs DI, Zhu Y. Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA biology. 2014;11:1301-12
30. Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964-80
31. Zhang P, Kang JY, Gou LT, Wang J, Xue Y, Skogerboe G. et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell research. 2015;25:193-207
32. Shrey K, Suchit A, Nishant M, Vibha R. RNA interference: emerging diagnostics and therapeutics tool. Biochem Biophys Res Commun. 2009;386:273-7
33. Klenov MS, Lavrov SA, Korbut AP, Stolyarenko AD, Yakushev EY, Reuter M. et al. Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries. Nucleic acids research. 2014;42:6208-18
34. Post C, Clark JP, Sytnikova YA, Chirn GW, Lau NC. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA (New York, NY). 2014;20:1977-86
35. Liu X, Zheng J, Xue Y, Yu H, Gong W, Wang P. et al. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics. 2018;8:1084-105
36. Peng L, Song L, Liu C, Lv X, Li X, Jie J. et al. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:2749-56
37. Goh WS, Falciatori I, Tam OH, Burgess R, Meikar O, Kotaja N. et al. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes & development. 2015;29:1032-44
38. Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou Y. et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell research. 2015;25:266
39. Gao XQ, Zhang YH, Liu F, Ponnusamy M, Zhao XM, Zhou LY. et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N(6)-methyladenosine methylation of Parp10 mRNA. Nature cell biology. 2020;22:1319-31
40. Han H, Fan G, Song S, Jiang Y, Qian C, Zhang W. et al. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood. 2020
41. Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R. et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8:5213-30
42. Yin J, Jiang XY, Qi W, Ji CG, Xie XL, Zhang DX. et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer science. 2017;108:1746-56
43. Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353-9
44. Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L. et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196-206
45. Su JF, Zhao F, Gao ZW, Hou YJ, Li YY, Duan LJ. et al. piR-823 demonstrates tumor oncogenic activity in esophageal squamous cell carcinoma through DNA methylation induction via DNA methyltransferase 3B. Pathology, research and practice. 2020;216:152848
46. Zhang H, Zhu JK. RNA-directed DNA methylation. Current opinion in plant biology. 2011;14:142-7
47. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:23-38
48. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484-92
49. Field AE, Robertson NA, Wang T, Havas A, Ideker T, Adams PD. DNA Methylation Clocks in Aging: Categories, Causes, and Consequences. Molecular cell. 2018;71:882-95
50. Michalak EM, Burr ML, Bannister AJ, Dawson MA. The roles of DNA, RNA and histone methylation in ageing and cancer. Nature reviews Molecular cell biology. 2019;20:573-89
51. Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nature reviews Molecular cell biology. 2019;20:590-607
52. Kobayashi H, Tomari Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochimica et biophysica acta. 2016;1859:71-81
53. Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular cell. 2008;31:785-99
54. Hancks DC, Kazazian HH Jr. Active human retrotransposons: variation and disease. Current opinion in genetics & development. 2012;22:191-203
55. Russell SJ, LaMarre J. Transposons and the PIWI pathway: genome defense in gametes and embryos. Reproduction (Cambridge, England). 2018;156:R111-r24
56. Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M. et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science (New York, NY). 2014;345:1251343
57. Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annual review of genetics. 2007;41:331-68
58. Kazazian HH Jr. Mobile elements: drivers of genome evolution. Science (New York, NY). 2004;303:1626-32
59. Chénais B. Transposable elements and human cancer: a causal relationship? Biochimica et biophysica acta. 2013;1835:28-35
60. Lee YC. The Role of piRNA-Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in Drosophila melanogaster. PLoS genetics. 2015;11:e1005269
61. Weick EM, Miska EA. piRNAs: from biogenesis to function. Development (Cambridge, England). 2014;141:3458-71
62. Ling Y, Zhang W, Wang P, Xie W, Yang W, Wang DA. et al. Three-dimensional (3D) hydrogel serves as a platform to identify potential markers of chondrocyte dedifferentiation by combining RNA sequencing. Bioactive materials. 2021;6:2914-26
63. Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y. et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nature cell biology. 2006;8:188-94
64. Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochimica et biophysica acta. 2007;1775:138-62
65. Baylin SB. DNA methylation and gene silencing in cancer. Nature clinical practice Oncology. 2005;2(Suppl 1):S4-11
66. Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nature reviews Genetics. 2007;8:272-85
67. Pfeifer GP. Defining Driver DNA Methylation Changes in Human Cancer. International journal of molecular sciences. 2018 19
68. Xu Y, Chao L, Wang J, Sun Y. miRNA-148a regulates the expression of the estrogen receptor through DNMT1-mediated DNA methylation in breast cancer cells. Oncology letters. 2017;14:4736-40
69. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S. et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome biology. 2018;19:218
70. Tang B. Inference of Crosstalk Effects between DNA Methylation and lncRNA Regulation in NSCLC. BioMed research international. 2018;2018:7602794
71. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
72. Yedjou CG, Sims JN, Miele L, Noubissi F, Lowe L, Fonseca DD. et al. Health and Racial Disparity in Breast Cancer. Advances in experimental medicine and biology. 2019;1152:31-49
73. Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates in surgery. 2017;69:313-7
74. Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT. et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Annals of oncology: official journal of the European Society for Medical Oncology. 2019;30:1194-220
75. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA. et al. Molecular portraits of human breast tumours. Nature. 2000;406:747-52
76. Azamjah N, Soltan-Zadeh Y, Zayeri F. Global Trend of Breast Cancer Mortality Rate: A 25-Year Study. Asian Pacific journal of cancer prevention: APJCP. 2019;20:2015-20
77. Ding X, Li Y, Lü J, Zhao Q, Guo Y, Lu Z. et al. piRNA-823 Is Involved in Cancer Stem Cell Regulation Through Altering DNA Methylation in Association With Luminal Breast Cancer. Frontiers in cell and developmental biology. 2021;9:641052
78. Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surgical oncology. 2013;22:217-23
79. Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36:1094-102
80. He X, Chen X, Zhang X, Duan X, Pan T, Hu Q. et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic acids research. 2015;43:3712-25
81. Liang Y, Jansen M, Aronow B, Geiger H, Van Zant G. The quantitative trait gene latexin influences the size of the hematopoietic stem cell population in mice. Nature genetics. 2007;39:178-88
82. Liang Y, Van Zant G. Aging stem cells, latexin, and longevity. Experimental cell research. 2008;314:1962-72
83. Meseure D, Vacher S, Boudjemaa S, Laé M, Nicolas A, Leclere R. et al. Biopathological Significance of PIWI-piRNA Pathway Deregulation in Invasive Breast Carcinomas. Cancers. 2020 12
84. Gebert M, Bartoszewska S, Janaszak-Jasiecka A, Moszyńska A, Cabaj A, Króliczewski J. et al. PIWI proteins contribute to apoptosis during the UPR in human airway epithelial cells. Scientific reports. 2018;8:16431
85. Litwin M, Szczepańska-Buda A, Michałowska D, Grzegrzółka J, Piotrowska A, Gomułkiewicz A. et al. Aberrant Expression of PIWIL1 and PIWIL2 and Their Clinical Significance in Ductal Breast Carcinoma. Anticancer research. 2018;38:2021-30
86. Mattiuzzi C, Lippi G. Current Cancer Epidemiology. Journal of epidemiology and global health. 2019;9:217-22
87. Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer research. 2004;64:3365-70
88. Li L, Zhu T, Gao YF, Zheng W, Wang CJ, Xiao L. et al. Targeting DNA Damage Response in the Radio(Chemo)therapy of Non-Small Cell Lung Cancer. International journal of molecular sciences. 2016 17
89. Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Current opinion in genetics & development. 2012;22:50-5
90. Molina-Pinelo S, Salinas A, Moreno-Mata N, Ferrer I, Suarez R, Andrés-León E. et al. Impact of DLK1-DIO3 imprinted cluster hypomethylation in smoker patients with lung cancer. Oncotarget. 2018;9:4395-410
91. Enfield KS, Martinez VD, Marshall EA, Stewart GL, Kung SH, Enterina JR. et al. Deregulation of small non-coding RNAs at the DLK1-DIO3 imprinted locus predicts lung cancer patient outcome. Oncotarget. 2016;7:80957-66
92. Amaar YG, Reeves ME. The impact of the RASSF1C and PIWIL1 on DNA methylation: the identification of GMIP as a tumor suppressor. Oncotarget. 2020;11:4082-92
93. Reeves ME, Baldwin ML, Aragon R, Baldwin S, Chen ST, Li X. et al. RASSF1C modulates the expression of a stem cell renewal gene, PIWIL1. BMC research notes. 2012;5:239
94. Liu J, Zhang S, Cheng B. Epigenetic roles of PIWI-interacting RNAs (piRNAs) in cancer metastasis (Review). Oncology reports. 2018;40:2423-34
95. Xie K, Zhang K, Kong J, Wang C, Gu Y, Liang C. et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer medicine. 2018;7:157-66
96. Navarro A, Tejero R, Viñolas N, Cordeiro A, Marrades RM, Fuster D. et al. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget. 2015;6:31544-56
97. Taylor J, Xiao W, Abdel-Wahab O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood. 2017;130:410-23
98. Robak P, Drozdz I, Szemraj J, Robak T. Drug resistance in multiple myeloma. Cancer treatment reviews. 2018;70:199-208
99. Pasqualucci L, Dalla-Favera R. Genetics of diffuse large B-cell lymphoma. Blood. 2018;131:2307-19
100. Coiffier B. Rituximab therapy in malignant lymphoma. Oncogene. 2007;26:3603-13
101. Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A. et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2011;117:553-62
102. Ai L, Mu S, Sun C, Fan F, Yan H, Qin Y. et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol Cancer. 2019;18:88
103. Wu D, Fu H, Zhou H, Su J, Zhang F, Shen J. Effects of Novel ncRNA Molecules, p15-piRNAs, on the Methylation of DNA and Histone H3 of the CDKN2B Promoter Region in U937 Cells. Journal of cellular biochemistry. 2015;116:2744-54
104. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
105. Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P. et al. Oesophageal cancer. Nature reviews Disease primers. 2017;3:17048
106. Sun H, Gong TT, Jiang YT, Zhang S, Zhao YH, Wu QJ. Global, regional, and national prevalence and disability-adjusted life-years for infertility in 195 countries and territories, 1990-2017: results from a global burden of disease study, 2017. Aging. 2019;11:10952-91
107. Vychytilova-Faltejskova P, Stitkovcova K, Radova L, Sachlova M, Kosarova Z, Slaba K. et al. Circulating PIWI-Interacting RNAs piR-5937 and piR-28876 Are Promising Diagnostic Biomarkers of Colon Cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2018;27:1019-28
108. Sun R, Gao CL, Li DH, Li BJ, Ding YH. Expression Status of PIWIL1 as a Prognostic Marker of Colorectal Cancer. Disease markers. 2017;2017:1204937
109. Sellitto A, Geles K, D'Agostino Y, Conte M, Alexandrova E, Rocco D. et al. Molecular and Functional Characterization of the Somatic PIWIL1/piRNA Pathway in Colorectal Cancer Cells. Cells. 2019 8
110. Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z. et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12-7
111. Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H. et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18:123
112. Castillejos-Molina RA, Gabilondo-Navarro FB. Prostate cancer. Salud publica de Mexico. 2016;58:279-84
113. Schatten H. Brief Overview of Prostate Cancer Statistics, Grading, Diagnosis and Treatment Strategies. Advances in experimental medicine and biology. 2018;1095:1-14
114. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
115. Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & development. 2012;26:2361-73
116. Meister G. Argonaute proteins: functional insights and emerging roles. Nature reviews Genetics. 2013;14:447-59
117. Zhang L, Meng X, Pan C, Qu F, Gan W, Xiang Z. et al. piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in prostate cancer via DNA methylation. Cellular signalling. 2020;67:109501
118. Ferreira HJ, Heyn H, Garcia del Muro X, Vidal A, Larriba S, Muñoz C. et al. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics. 2014;9:113-8
119. Feng D, Yan K, Zhou Y, Liang H, Liang J, Zhao W. et al. Piwil2 is reactivated by HPV oncoproteins and initiates cell reprogramming via epigenetic regulation during cervical cancer tumorigenesis. Oncotarget. 2016;7:64575-88
120. Chen Z, Che Q, Jiang FZ, Wang HH, Wang FY, Liao Y. et al. Piwil1 causes epigenetic alteration of PTEN gene via upregulation of DNA methyltransferase in type I endometrial cancer. Biochemical and biophysical research communications. 2015;463:876-80
121. Chen CY, Chen J, He L, Stiles BL. PTEN: Tumor Suppressor and Metabolic Regulator. Frontiers in endocrinology. 2018;9:338
122. MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Annals of oncology: official journal of the European Society for Medical Oncology. 2009;20(Suppl 6):vi1-7
123. Duncan LM. The classification of cutaneous melanoma. Hematology/oncology clinics of North America. 2009;23:501-13 ix
124. Bhan S, Chuang A, Negi SS, Glazer CA, Califano JA. MAGEA4 induces growth in normal oral keratinocytes by inhibiting growth arrest and apoptosis. Oncology reports. 2012;28:1498-502
125. Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Human reproduction update. 2015;21:411-26
126. Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M. et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Molecular cell. 2013;50:601-8
127. Heyn H, Ferreira HJ, Bassas L, Bonache S, Sayols S, Sandoval J. et al. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PloS one. 2012;7:e47892
128. Zhang GW, Wang L, Chen H, Guan J, Wu Y, Zhao J. et al. Promoter hypermethylation of PIWI/piRNA pathway genes associated with diminished pachytene piRNA production in bovine hybrid male sterility. Epigenetics. 2020;15:914-31
129. Kim S, Günesdogan U, Zylicz JJ, Hackett JA, Cougot D, Bao S. et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Molecular cell. 2014;56:564-79
130. Dong J, Wang X, Cao C, Wen Y, Sakashita A, Chen S. et al. UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nature communications. 2019;10:4705
131. Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS. et al. Identification of piRNAs in the central nervous system. RNA (New York, NY). 2011;17:1090-9
132. Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T. et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693-707
133. Nandi S, Chandramohan D, Fioriti L, Melnick AM, Hébert JM, Mason CE. et al. Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:12697-702
134. Zhang C, Sha H, Peng Y, Wang Y, Liu C, Zhou X. PiRNA-DQ541777 Contributes to Neuropathic Pain via Targeting Cdk5rap1. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2019;39:9028-39
135. Pierce LM, Kurata WE, Matsumoto KW, Clark ME, Farmer DM. Long-term epigenetic alterations in a rat model of Gulf War Illness. Neurotoxicology. 2016;55:20-32
136. Zhao PP, Yao MJ, Chang SY, Gou LT, Liu MF, Qiu ZL. et al. Novel function of PIWIL1 in neuronal polarization and migration via regulation of microtubule-associated proteins. Molecular brain. 2015;8:39
137. Gaziano TA, Bitton A, Anand S, Abrahams-Gessel S, Murphy A. Growing epidemic of coronary heart disease in low- and middle-income countries. Current problems in cardiology. 2010;35:72-115
138. Li M, Yang Y, Wang Z, Zong T, Fu X, Aung LHH. et al. Piwi-interacting RNAs (piRNAs) as potential biomarkers and therapeutic targets for cardiovascular diseases. Angiogenesis. 2021;24:19-34
139. Ma C, Zhang L, Wang X, He S, Bai J, Li Q. et al. piRNA-63076 contributes to pulmonary arterial smooth muscle cell proliferation through acyl-CoA dehydrogenase. Journal of cellular and molecular medicine. 2020;24:5260-73
140. Zhang X, He X, Liu C, Liu J, Hu Q, Pan T. et al. IL-4 Inhibits the Biogenesis of an Epigenetically Suppressive PIWI-Interacting RNA To Upregulate CD1a Molecules on Monocytes/Dendritic Cells. Journal of immunology (Baltimore, Md: 1950). 2016;196:1591-603
141. Zhong F, Zhou N, Wu K, Guo Y, Tan W, Zhang H. et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic acids research. 2015;43:10474-91
142. Pleštilová L, Neidhart M, Russo G, Frank-Bertoncelj M, Ospelt C, Ciurea A. et al. Expression and Regulation of PIWIL-Proteins and PIWI-Interacting RNAs in Rheumatoid Arthritis. PloS one. 2016;11:e0166920
143. Chapel JM, Ritchey MD, Zhang D, Wang G. Prevalence and Medical Costs of Chronic Diseases Among Adult Medicaid Beneficiaries. American journal of preventive medicine. 2017;53:S143-s54
144. Kim KW. PIWI Proteins and piRNAs in the Nervous System. Molecules and cells. 2019;42:828-35
145. Cui L, Lou Y, Zhang X, Zhou H, Deng H, Song H. et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clinical biochemistry. 2011;44:1050-7
146. Li B, Hong J, Hong M, Wang Y, Yu T, Zang S. et al. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene. 2019;38:5227-38
147. Lipps C, Northe P, Figueiredo R, Rohde M, Brahmer A, Krämer-Albers EM. et al. Non-Invasive Approach for Evaluation of Pulmonary Hypertension Using Extracellular Vesicle-Associated Small Non-Coding RNA. Biomolecules. 2019 9
148. Dumitrescu RG. Early Epigenetic Markers for Precision Medicine. Methods in molecular biology (Clifton, NJ). 2018;1856:3-17
149. Itou D, Shiromoto Y, Yukiho SY, Ishii C, Nishimura T, Ogonuki N. et al. Induction of DNA methylation by artificial piRNA production in male germ cells. Current biology: CB. 2015;25:901-6
150. Liu Y, Ao X, Wang Y, Li X, Wang J. Long Non-Coding RNA in Gastric Cancer: Mechanisms and Clinical Implications for Drug Resistance. Frontiers in oncology. 2022;12:841411
151. Vrba L, Muñoz-Rodríguez JL, Stampfer MR, Futscher BW. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PloS one. 2013;8:e54398
152. Zhang Y, Jia DD, Zhang YF, Cheng MD, Zhu WX, Li PF. et al. The emerging function and clinical significance of circRNAs in Thyroid Cancer and Autoimmune Thyroid Diseases. International journal of biological sciences. 2021;17:1731-41
Author contact
![]() Corresponding author: Yin-Feng Zhang, ORCID: https://orcid.org/0000-0003-2457-7712; Email address: zhangyinfengedu.cn.
Corresponding author: Yin-Feng Zhang, ORCID: https://orcid.org/0000-0003-2457-7712; Email address: zhangyinfengedu.cn.

 Global reach, higher impact
Global reach, higher impact