10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(8):3421-3434. doi:10.7150/ijbs.73504 This issue Cite
Review
Targeting HMGB1: An available Therapeutic Strategy for Breast Cancer Therapy
Fudan University Shanghai Cancer Center & Institutes of Biomedical Sciences; Cancer Institutes; Key Laboratory of Breast Cancer in Shanghai; The Shanghai Key Laboratory of Medical Epigenetics; Shanghai Key Laboratory of Radiation Oncology; The International Co-laboratory of Medical Epigenetics and Metabolism, Ministry of Science and Technology; Shanghai Medical College; Fudan University, Shanghai 200032, China.
Received 2022-4-1; Accepted 2022-4-28; Published 2022-5-9
Abstract
HMGB1 is a member of highly conserved high mobility group protein superfamily with intracellular and extracellular distribution. Abnormal HMGB1 levels are frequently manifested in various malignant diseases, including breast cancer. Numerous studies have revealed the clinical value of HMGB1 in the diagnosis and therapy of breast cancer. However, the dual function of pro- and anti-tumor makes HMGB1 in cancer progression requires more profound understanding. This review summarizes the functions and mechanisms of HMGB1 on regulating breast cancer, including autophagy, immunogenic cell death, and interaction with the tumor microenvironment. These functions determine the strategies for the development of chemotherapy, radiotherapy, immunotherapy and combination therapies by targeting HMGB1 in breast cancer. Defining the mechanisms of HMGB1 on regulating breast cancer development and progression will facilitate the application of HMGB1 as a therapeutic target for breast cancer.
Keywords: HMGB1, breast cancer, autophagy, immunogenic cell death, therapy
Introduction
Breast cancer remains a high incidence worldwide and represents a significant threat to female health [1]. Although advances in early diagnosis and therapy, to some degree, have contributed to alleviate the crisis of breast cancer, a large proportion of patients still succumb to a variety of complex malignant phenotypes. The alarming situation urges researchers to find innovative and effective therapeutic targets for breast cancer.
High mobility group protein box 1 (HMGB1) is a member of the high mobility group protein superfamily with secretory and intracellular activity [2, 3]. The subcellular localization of HMGB1 determines the manner in which it regulates normal physiological or pathological processes. In the nucleus, HMGB1 acts as a DNA-binding protein, maintaining the stability of chromosomes or interacting with transcription factors to exert co-activator or co-repressor activity through its DNA-binding domains: A-box and B-box [4-6]. HMGB1 translocated to the cytoplasm is mainly engaged in Beclin1-mediated autophagy [7]. Extracellular HMGB1 is identified as an important damage-associated molecular pattern (DAMP) molecule that interacts with other cells by binding to several receptors, such as RAGE and TLR family members [8-11]. Downstream signals activated by HMGB1 are responsible for a variety of biological functions, including autophagy, immunogenic cell death (ICD), release of cytokines and chemokines, angiogenesis and migration [12, 13]. Aberrant expression of HMGB1 has been demonstrated and is closely associated with proliferation and metastasis in breast cancer cells [14, 15]. More importantly, HMGB1 participates in the regulation of chemo- and radiotherapy resistance in breast cancer [9, 16, 17]. Due to the complexity of HMGB1 functions in breast cancer progression, new therapeutic strategies targeting HMGB1 are constantly being developed, such as the identification of potential ICD inducers and the combination therapies with immune checkpoint blockade [18-20].
The structure of HMGB1.

This review focuses on the function of HMGB1 in breast cancer based on its biological characteristics and summarizes the valuable preclinical and clinical evidence in breast cancer treatment, with the aim of providing insights for the innovation of therapeutic strategies targeting HMGB1.
The biology of HMGB1
The distribution of HMGB1
HMGB1 is highly expressed in a range of tumor tissues, and its subcellular localization determines the manner in which HMGB1 exerts its regulatory function on cancer. HMGB1 is a 215 amino acid long protein that consists of two DNA-binding domains (HMG A box and HMG B box) and a C-terminal acidic tail (Figure 1). In the nucleus, HMGB1 performs its binding and bending functions with DNA mainly through its A and B box domains respectively [21]. The C-terminal acidic tail is composed of continuous aspartate and glutamate residues, and has a protective effect on HMG A and B box during the transport of HMGB1 from the nucleus to the cytoplasm [22, 23]. Such strong non-specific DNA affinity of HMGB1 allows its implication in fundamental biological processes such as DNA replication, transcription and modification. Moreover, several transcription factors, such as p53, retinoblastoma (RB) proteins, and NF-κB family members, can enhance their oncogenic activities through direct interaction with HMGB1 [6, 15, 24]. Furthermore, it has been indicated that loss of HMGB1 activity can increase the frequency of DNA damage. HMGB1 can bind to DNA fragments damaged by chemotherapy or radiotherapy and be engaged in the DNA repair pathway [25]. Canonical DNA repair pathways, such as mismatch repair, have been shown to be regulated by nuclear HMGB1 [26, 27]. Moreover, nuclear HMGB1 alters nucleosome stability in a non-enzymatic, ATP-independent manner [28]. There is a partial overlap in the chromatin binding site of HMGB1 and histone H1, which allows HMGB1 to destabilize nucleosome structure by replacing H1 [29]. Consequently, HMGB1 is critical in maintaining genomic stability. However, more in-depth exploration is required regarding how HMGB1 affects cancer progression through its participation in DNA damage repair. Nuclear HMGB1 is also engaged in the development of serval immune cells. For example, in V(D)J recombination, HMGB1 enhances the activity of RAG1/RAG2 by assembling the RAG-RSS-HMG complex, thereby promotes the rearrangement process of antigen receptor genes, resulting in the diversity of T cell receptors [30, 31]. Jiang et al. found that HMGB1 silencing could prevent functional skewing of macrophages and downregulate inflammatory factors, suggesting nuclear HMGB1 may serve as a checkpoint molecule of macrophages [32].
In the cytoplasm, HMGB1 is mainly associated with the regulation of autophagy in cancer cells [33]. It is necessary to emphasize that acetylation and phosphorylation modifications can prevent HMGB1 from re-localizing in the nucleus, thus facilitating the accumulation of HMGB1 in the cytoplasm. Acetylation of HMGB1 is mediated by the activation of JAK/STAT1 pathway. Specifically, LPS and interferon in the inflammatory microenvironment activate the JAK/STAT1 pathway by binding to TLR4 and initiate HMGB1 acetylation modification [34, 35]. Intriguingly, histone deacetylase (HDAC) plays an important role in the deacetylation of HMGB1. For example, SIRT1, a member of class III HDAC, inhibits HMGB1 cytoplasmic translocation through lysine deacetylation [36]. Consistent, the treatment of the valproic acid derivative OH-VPA, a histone deacetylase inhibitor, induces the acetylation of HMGB1 and its translocation from the nucleus to the cytoplasm, thus affecting the proliferation of Hela cells [37]. Phosphorylation modification promotes HMGB1 localization mainly due to the decreased binding ability of HMGB1 to nuclear import protein KAP-aα1 [38]. However, the specific molecular mechanism regarding how HMGB1 translocates from the nucleus to the cytoplasm needs to be elucidated in more details.
HMGB1 secretes extracellularly as an immune signal that stimulates the innate immune system to recruit monocytes to inflammatory sites [39]. Long-term experimental evidence indicated that extracellular HMGB1 possessed abundant biological activity, such as proliferation, migration, tissue regeneration and anti-bacteria [40]. Among them, the most concerned function of extracellular HMGB1 is immune response. HMGB1 induces the release of cytokines and chemokines from immune cells, depending mainly on ligand-receptor interaction. HMGB1 acts as a biomarker of ICD, maturing antigen-presenting dendritic cells and enhancing their antigen-presenting capacity [12]. In monocytes and neutrophils, HMGB1 induces the secretion of several pro-inflammatory cytokines, such as IL1-β, TNF-α, and IL-6 [41]. In addition, extracellular HMGB1 can exert chemotactic effects and support the up-regulation of vascular adhesion molecules, thereby impairing the barrier function of the epithelium [42]. Hence, extracellular HMGB1 has a powerful ability to coordinate different immune responses in the tumor microenvironment. Moreover, the level of extracellular HMGB1 has also been proven to correlate with the differentiation of various cell types, such as T cells, stem cells and cancer cells. For example, Li et al. have shown that HMGB1 can directly or indirectly promote the differentiation of Th0 cells to Th2 and Th17 cells through RAGE or TLR2 signaling in asthma mouse models [43]. In the inflammatory microenvironment, the interaction between HMGB1 and RAGE or TLRs was also implicated in the differentiation of mesenchymal stem cells (MSCs) into vascular endothelial cells and osteoblasts [44, 45]. Conversely, the release of HMGB1 may result in the dedifferentiation of cancer cells. For instance, Zhang et al. found that radiation-induced release of HMGB1 promoted the dedifferentiation of CD133- pancreatic cancer cells into cancer stem cell (CSC) phenotype via the TLR2/Hippo/YAP pathway [46]. Although targeting HMGB1 may contribute to overcoming metastasis and recurrence of cancer, there are few reports about the association between HMGB1 and CSCs.
The receptors of HMGB1
The receptor for advanced glycosylation end products (RAGE) is an immunoglobulin superfamily transmembrane receptor that derives its name from the ability to bind to advanced glycosylation end products (AGEs). Due to its inflammatory function in innate immunity, RAGE is commonly considered to be a pattern recognition receptor (PRR) [47]. Multiple cellular processes, including proliferation and migration, inflammation and chemotaxis, can be activated via the HMGB1/RAGE signaling axis [48-50]. Todorova et al. found that HMGB1 interacts with its receptor RAGE in tumor cells but not in normal cells [51]. The HMGB1/RAGE signaling axis is also widely involved in breast cancer progression, such as epithelial-mesenchymal transition (EMT), metastasis, and interaction with the tumor microenvironment [52, 53]. Interestingly, in addition to the typical characteristics, HMGB1 can also regulate the complications associated with breast cancer. For example, Okui et al. found that HMGB1 secreted by breast cancer cells induces bone pain by binding to RAGE of sensory neurons in breast cancer patients with bone metastases [54].
Toll-like receptors (TLRs) are an important group of PRRs participated in non-innate immunity and are also considered to be a bridge between innate and adaptive immunity [55]. The function of TLRs in cancer progression has been generally demonstrated [56]. Currently, among the TLR family members, TLR2/4/9 were identified as common receptors for HMGB1 [57].
As an inflammation-associated receptor, TLR2 is mainly expressed on megakaryocytes and platelets which is related to inflammation-induced vascular disease [58, 59]. Notably, Lin et al. revealed a non-inflammatory role of the HMGB1/TLR2 axis in breast cancer. They found that in human and mouse breast cancer stem cells, the HMGB1/TLR2 axis promotes NF-κB signaling, production of IL-6 and TGF-β, and activation of STAT3, thereby facilitating self-renewal of CSCs [60].
There are more reports about HMGB1/TLR4 interactions in breast cancer progression. For example, Lv et al. reported that macrophage migration inhibitory factors (MIFs) enhance the phosphorylation of ERK and caveolin-1, both of which promote the release of HMGB1 from the nucleus to the cytoplasm and extracellular compartment respectively. Extracellular HMGB1 activates TLR4 signaling and NF-κB phosphorylation, which is responsible for Snail and Twist transcription and MMP2 activation. The HMGB1/TLR4 axis ultimately modifies the intrinsic tumor properties and host immune response in the tumor microenvironment, leading to breast cancer metastasis [61].
Compared to TLR2/4, clarified reports about the interaction of TLR9 with HMGB1 in breast cancer is still absent. CpG DNA or synthetic CpGoligodeoxynucleotide (ODN) analogs can activate TLR9 redistribution from the endoplasmic reticulum (ER) to early endosomes [62]. Moreover, CpG ODN also possesses the capacity to stimulate macrophages and dendritic cells (DCs) to secrete HMGB1, which enhances the immunostimulatory potential of CpG ODN in a TLR9-dependent manner [10]. In hepatocellular carcinoma (HCC) tumors, HMGB1 mediates tumor growth through interaction with intracellular TLR9 under hypoxic conditions and attracts macrophages to the tumor site, leading to enhanced metastasis [63, 64]. However, whether HMGB1 functions as a CpG ODN binding protein in breast cancer is not well known.
The regulation of HMGB1
As important regulators of post-transcriptional modifications, several miRNAs targeting HMGB1 have been identified, such as miR-107, miR-129-5p, miR-200c and miR-205 [52, 65-68]. These miRNAs targeting HMGB1 are commonly involved in the regulation of proliferation, migration and therapeutic resistance of breast cancer cells. For instance, the miR-129-5p/HMGB1 axis is implicated in regulating autophagy of breast cancer cells and enhancing their sensitivity to radiotherapy and chemotherapy [66, 67]. Particularly, Chen et al. found that circRNA CHIPK3 expression was upregulated and predicted a poor prognosis in breast cancer. Mechanistically, cirCHIPK3 serves as a sponge for miR-193a, thereby reactivating HMGB1/PI3K/AKT signaling to promote proliferation, migration and invasion of breast cancer cells [69]. Another circRNA, Hsa_circ_0003645 have also been proved that can regulate miR-139-3p/HMGB1 axis to promote breast cancer progression [70]. However, there is a paucity of reports on how RNA sponges, or even competing endogenous RNA (ceRNA) regulatory networks, impact HMGB1-mediated breast cancer progression.
Hypoxia is one of the pivotal factors that induce elevated HMGB1 levels in tumor tissues. For example, Huber et al. have shown that hypoxia-induced high expression of HMGB1 promotes melanoma growth and metastasis, as well as infiltration of M2 macrophages. In breast cancer, the association between hypoxia and HMGB1 has also been confirmed. For example, HMGB1 can regulate HIF-1α expression in breast cancer cells through the PI3K/AKT signaling pathway [71]. Of interest, Sun et al. have observed that HMGB1 induces HIF-1α expression while hypoxia may downregulate miR-134-3p, a hypoxia-responsive miRNA, which is involved in the blockade of HMGB1/HIF-1α signaling to inhibit breast cancer progression [72]. This finding provides new evidence for the search of potential hypoxia-related targets.
The expression of HMGB1 is regulated by a variety of transcription factors, and the interactions between HMGB1 and transcription factors are complicated. Canonical transcriptional factors, p53 and its homolog p73 exert different effects on HMGB1 promoter activity through binding to another transcriptional factor CTF2: p53/CTF2 downregulates HMGB1 promoter activity, while p73/CTF2 upregulates [73]. p53 can not only regulate the expression of HMGB1, but also mediate the release of HMGB1 into the extracellular[74]. Of note, the researchers have demonstrated that HMGB1 can interact with p53 and promote the binding of p53 to DNA through the HMG A box and B box respectively, while the oxidation of the C-terminal acidic tail will impair the binding ability of HMGB1 to p53 [75, 76]. c-Myc is another transcription factor widely exhibited in cancer cells, and its target genes are involved in many biological processes such as cell cycle, cell adhesion and metabolism. Okuyama et al. found that the silence of c-Myc down-regulated the intracellular HMGB1 levels, thus promoting the survival of cancer cells under hypoxia and glucose deficiency [77]. The interaction between HMGB1 and transcription factor STAT3 has also been reported in detail. There is one STAT3 binding site on the human HMGB1 promoter [78], and STAT3-mediated upregulation of HMGB1 can also be inhibited by other miRNAs, such as miR-125b and miR-193-3p [79, 80]. On the other hand, HMGB1-mediated the activation of STAT3 signal participates widely in tumor progression [81, 82].
Intriguingly, Xu et al. demonstrated that HMGB1 can enhance the methylation of STAT3 promoter by recruiting TET2/AID/TDG to the STAT3 promoter region, resulting in the high expression of STAT3 in CD4+ T cells, suggesting that HMGB1 may be involved in epigenetic regulation [83].
The functions of HMGB1 in breast cancer
Autophagy regulation
Autophagy is an important mechanism for maintaining cellular homeostasis by removing unnecessary or dysregulated cellular components and recycling metabolic substrates. Various autophagic pathways may be regulated to inhibit or promote tumor progression. Multiple stress signals have the capacity to induce autophagy in tumor cells, including metabolic stress and hypoxic stress [84].
HMGB1, as an immune signal, is widely recognized as an autophagy inducer. It has been reported that the mechanism of HMGB1-induced autophagy depends on its distribution (Figure 2). In the nucleus, HMGB1 acts as a transcriptional factor based on its DNA binding domains. Tang et al. observed that nuclear HMGB1 upregulates the transcriptional level of HSP27 gene [85]. The HMGB1/HSP27 signaling is implicated in the translocation of Parkin and the ubiquitination of VACD1, in which the Parkin/VACD1 pathway plays a critical role in mitophagy [86]. These findings suggest that nuclear HMGB1 is largely dependent on the HSP27 pathway to regulate autophagy. In the cytoplasm, HMGB1 promotes Bcl-2 phosphorylation via the ERK/AMPK pathway and induces dissociation of the Beclin1-Bcl-2 complex [87]. Critically, dissociation of the Beclin1-Bcl-2 complex promotes the formation of the Beclin1-PI3K-III complex, which is essential in the initiation of cytoplasmic autophagy [7, 88]. Especially, extracellular HMGB1 induces the activity of autophagy mainly through receptor-ligand interactions. For example, HMGB1/RAGE signaling can activate Beclin-1-dependent autophagy. Interestingly, Tang et al. found that reducible HMGB1 triggers autophagy, while oxidized HMGB1 promotes apoptosis in cancer cells [89]. Moreover, ERK1/2 and AMPK/mTOR pathways are also downstream targets of HMGB1/RAGE signaling, and these pathways have been shown to participate in autophagy [90-92].
The mechanisms of HMGB1-mediated autophagy in breast cancer.
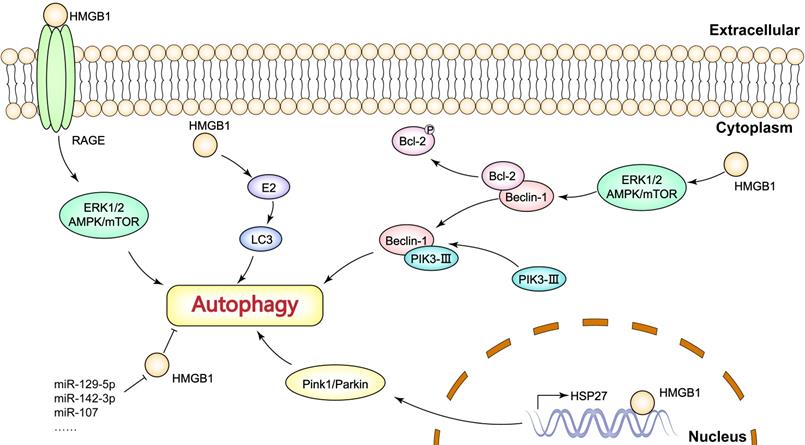
There are several molecules functioning as key regulators of HMGB1-medaited autophagy in breast cancer. For example, Wang et al. validated the role of caveolin-1 and HMGB1 in estradiol (E2)-regulated autophagy [93]. Liu et al. reported that Med19 inhibition suppresses autophagy by downregulating HMGB1 signaling in breast cancer cells, thus increasing adriamycin chemosensitivity [16]. Previous studies have identified several microRNAs that can target HMGB1 to inhibit autophagy in breast cancer cells, such as miR-129-5p, miR-142-3p, miR-107. Particularly, these miRNAs are generally shown to reverse HMGB1-induced resistance to chemotherapy or radiotherapy [65-67, 94], which provide new strategies for breast cancer therapy targeting HMGB1. In addition, HMGB1 is engaged in estradiol (E2)-mediated autophagosome formation in breast cancer. Wang et al. found that HMGB1 knockdown down-regulated E2-mediated LC3-II (microtubule-associated protein 1 light chain 3II, an autophagy-associated protein) and Beclin-1 expression as well as inhibited autophagosome formation in BT474 breast cancer cells [93]. More importantly, Ladoire et al. have reported that nuclear HMGB1 with LC3B (an isoform of LC3) as an independent prognostic marker was significantly associated with prolonged metastasis-free survival [95]. In other words, the combined analysis of HMGB1 and autophagy levels is valuable for predicting breast cancer prognosis.
Immunogenic cell death (ICD) mediation
Immunogenic cell death (ICD) refers to the phenomenon in which tumor cells are transformed from non-immunogenic to immunogenic and mediate anti-tumor immune responses when cancer cells die. ICD involves changes in cell surface components, as well as the release of soluble mediators. These signals, called damage-associated molecular patterns (DAMPs), act on a series of receptors expressed by DCs to stimulate the presentation of tumor antigens to T cells [96]. There are various stressors that can induce ICD, such as traditional chemotherapeutic drugs, targeted anti-cancer drugs, and radiotherapy [97].
HMGB1 is one of the DAMPs of ICD, and its release contains two steps involving crossing the nuclear and plasma membrane. HMGB1 that is secreted into extracellular can bind to cell surface receptors on other immune cells, activating downstream signaling pathways and thus enhancing immunity [98]. HMGB1, serving as a key biomarker of ICD, has been demonstrated in breast cancer. For instance, HMGB1 can induce ICD through interaction with TLR4, which determines the chemo- or radio-resistance of breast cancer [9]. Keita et al. have proved that several chemotherapeutic agents significantly triggered HMGB1-involved ICD by immunohistochemistry assessment in breast cancer [99]. Additionally, PERK/eIF2α/ATF4/CHOP pathway activates the ER- associated ICD, which induces HMGB1 release [18]. These findings suggest that HMGB1-mediated ICD can be a potential target for novel chemotherapy or combination therapy strategies.
Remodeling the tumor microenvironment
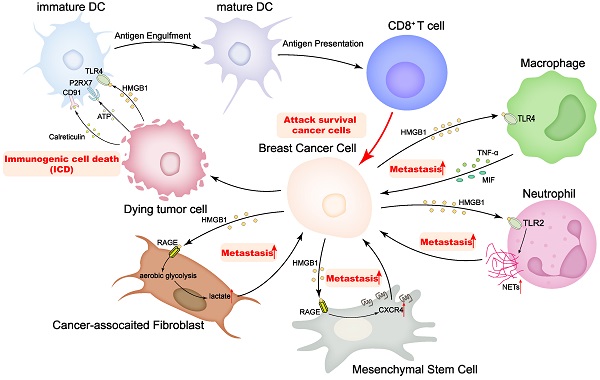
Tumor cells, together with various other cells, including immune cells, cancer-associated fibroblasts (CAFs), and MSCs, compose an intricate tumor microenvironment (TME), and the multiplex interactions between different cells in the TME are critical to tumor progression [100]. There are numerous experiments showing that HMGB1 executes the function that is widely engaged in dynamic interactions with other cells to remodel the TME, influencing cancer progression (Figure 3). Most impressively, HMGB1 operates as a critical marker of the ICD, contributing to the maturation of DCs and the presentation of antigen to cytotoxic T cells (CTLs), thereby activating CTLs to clear neighboring cells. Aoto et al. have found that in breast cancer, HMGB1 interacts with TLR4 on DCs, which is selectively implicated in the initiation of anti-tumor T lymphocytes in vivo [9]. Release of HMGB1 from dying tumor cells may be required to empower DCs to process and present tumor antigens.
Apart from canonical ICD, interactions of HMGB1 with other components of TME have been reported in recent years. It has been suggested that CD62Ldim neutrophils in the pre-metastatic niche are strongly associated with lung metastasis of breast cancer [101]. Wang et al. have found that, in the mouse TNBC model, the ability of CD62Ldim neutrophils to produce neutrophil extracellular traps (NETs) was significantly increased in peripheral blood and lung tissues, which was regulated by tumor-derived HMGB1/TLR2 signaling. Furthermore, the intervention targeting the HMGB1/CD62Ldim neutrophil-NETs axis inhibited lung metastasis [102]. In addition, HMGB1 promotes TNF-α secretion by macrophages via the TLR-4 pathway [103]. Furthermore, MIFs release impacts host immune responses in the TME by regulating the HMGB1/TLR4/NF-κB axis [61], and all these interactions contribute to breast cancer metastasis. Moreover, Hubert et al. have observed that HMGB1 inhibition remodels the breast cancer microenvironment, which involves a dramatic reduction in monocyte/granulocyte myeloid-derived suppressor cells and regulatory T lymphocytes, an elevated M1/M2 ratio of tumor-associated macrophages, and an enhancement of DCs and plasmacytoid DCs activation [104]. The profound impact of HMGB1 on the TME allows the combination of HMGB1 and other immune cells to be a new target for breast cancer prevention and treatment.
HMGB1 plays important roles on regulating the interactions of breast cancer cells and tumor microenvironment.

CAFs are the most prominent cell type in the breast cancer microenvironment, and their abundance correlates with high malignancy and poor prognosis. CAFs promote tumor progression by secreting growth factors, enhancing angiogenesis and remodeling the extracellular matrix. Due to their ability to recruit immune cells and influence tumor-immune system interactions, CAFs have been shown to be key players in mediating tumor-promoting inflammation [105]. Currently, a bidirectional interaction of HMGB1 between breast cancer cells and CAFs has been partially reported. Amornsupak et al. have found that CAFs can induce the production of HMGB1 in breast cancer cells and thus strengthen the chemoresistance to doxorubicin [106]. On the other hand, Chen et al. have reported that HMGB1 secreted by breast cancer cells can promote aerobic glycolysis and subsequently the activation of fibroblasts through the interaction with RAGE, and then the activated fibroblasts contribute to breast cancer cell metastasis by increasing lactate [53]. In addition, tamoxifen elevated HMGB1 expression in CAFs via GPR30/PI3K/AKT signaling, and secreted HMGB1 induced autophagy in an ERK-dependent manner to enhance tamoxifen resistance in MCF-7 cells [107]. According to these findings, HMGB1 plays a pivotal role in CAFs-mediated breast cancer progression and chemoresistance, and targeting HMGB1 may be an effective strategy to diminish chemoresistance in breast cancer.
MSCs are early cells of mesodermal development and are widely observed as undifferentiated cells in various tumor tissues [108]. A consensus has emerged that MSCs residing in the TME can promote cancer development and progression [109]. Ananthula et al. have revealed that extracellular acetylated HMGB1 (Ac-HMGB1) can trigger the expression and activation of RAGE in MSCs through in vivo and in vitro experiments. The activation of RAGE induced CXCR4 expression in MSCs and directed migration to geminin overexpressed (Gem-OE) breast cancer cells. The crosstalk between Gem-OE breast cancer cells and MSCs enhanced the aggressiveness of triple negative breast cancer [110]. The notion that geminin overexpression is closely correlated with the proliferation and invasiveness of breast cancer cells, especially triple-negative breast cancer cells, has been widely demonstrated [111-113]. Therefore, blocking the geminin-HMGB1/RAGE signaling axis has also been attempted as a strategy to inhibit breast cancer cell proliferation and metastasis [114].
HMGB1 regulates breast cancer progression
HMGB1 is engaged in many stages of breast cancer progression. However, the dual anti-tumor and pro-tumor biological functions of HMGB1 make its precise role in breast cancer progression quite elusive. The complex role of HMGB1 in cancer cells may depend on the pattern in which HMGB1 is induced. Chemotherapy- and radiation-induced ICDs lead to the release of HMGB1 from dying tumor cells, which usually functions as an immunogenic enhancer [11, 115, 116]. While the tumor grows, it may permanently express HMGB1 and undergo special post-transcriptional modifications to promote tumor proliferation, invasion and immune tolerance [117, 118].
The direct effect of HMGB1 on breast cancer cell proliferation is accomplished predominantly through the interaction with retinoblastoma (RB) protein. RB, an oncoprotein, acts a bridge for HMGB1-E2F1 contact by binding to the LXCXE motif of HMGB1. HMGB1 enhances the ability of RB to suppress E2F and cyclin A transcription. This suggests that HMGB1-RB interaction is essential for HMGB1-mediated transcriptional repression, cell growth inhibition, G1 cell cycle arrest, apoptosis induction and tumor growth inhibition [17]. The potential association of the LXCXE motif of HMGB1 with RB has been further confirmed. Wang et al. constructed point mutants of the LXCXE motif of HMGB1 and found that the interaction of RB with the HMGB1 mutants was disrupted and that HMGB1 significantly inhibited breast cancer cell proliferation through an LXCXE-dependent mechanism [15].
The ability to invade local tissues and subsequently metastasize to distant organs is an important reason for the high mortality rate of breast cancer. Metastasis is a multistep process that includes cell separation from the primary tumor, entry into circulation, extravasation, and colonization in distant organs. The function of HMGB1 in the metastatic process of breast cancer has been more frequently reported. For example, Ni et al. have found that HMGB1 silencing inhibited the invasion and migration of human breast cells [14]. Mechanistically, HMGB1 can promote angiogenesis and migration of breast cancer through the PI3K/AKT/HIF-1α pathway [71]. Another report revealed that the positive function of HMGB1/RAGE signaling pathway in EMT and invasion in triple-negative breast cancer, which the effect can be reversed by miR-205 [52]. Moreover, hematological and neurological expressed 1-like (HN1L) protein can interact with HSPA9 to upregulate HMGB1 expression and play a key role in promoting breast cancer cell invasion and metastasis [119]. Specially, the function of HN1L to promote the properties of CSCs was revealed in breast cancer and prostate cancer [120, 121]. Notably, as mentioned above, the interaction of HMGB1 with the TME components, such as neutrophils, macrophages, CAFs and MSCs, also contributes to the metastasis of breast cancer cells.
HMGB1 serves a potential target breast cancer therapy
The clinical value of HMGB1 in breast cancer
The clinical value of HMGB1 in breast cancer has been generally established. By immune-histochemical analysis of breast cancer samples, Kostova et al. found that HMGB1 in malignant tissues exhibited stronger immuno-reactivity than HMGB1 in normal tissues [8]. Sun et al. used ELISA to measure serum HMGB1 in the normal population, patients with breast cancer and patients with benign breast disease. They found that the levels of HMGB1 in breast tissues and serum were significantly higher in breast cancer patients than in patients with benign breast disease or the normal population, suggesting that serum HMGB1 may be a useful serological biomarker for the diagnosis of breast cancer [2].
In response to neoadjuvant chemotherapy, Stoetzer et al. suggested that HMGB1 and its receptor RAGE could be used to predict and assess breast cancer treatment efficacy [122]. Arnold et al. found a significant release of HMGB-1 from HCC1143 breast cancer cells treated with epirubicin and docetaxel in vitro assay. More importantly, in vivo experiments showed that 22 of 41 patients treated with the drug combination who exhibited pathologically complete remission or partial remission responders were accompanied by a significant increase in HMGB1 levels, while the remaining 19 patients did not show any significant changes in breast cancer pathological features with plasma HMGB1 levels [123].
Tumor-infiltrating immune cells are strongly associated with clinical diagnosis and treatment, as well as ideal drug targets to improve cancer patient survival. In breast cancer, Lee et al. found that cytoplasmic HMGB1 expression correlated with levels of tumor-infiltrating lymphocytes (TILs) [124]. Additionally, the role of extracellular HMGB1 in remodeling the tumor immune microenvironment was also mentioned above. These findings have enriched the understanding of the profound effect of HMGB1 on tumor immune infiltration.
Altogether, the expression pattern of HMGB1 can be applied to the assessment of prognosis, chemotherapy efficacy and immune infiltration of breast cancer, which provides valuable clinical evidence for breast cancer treatment strategies targeting HMGB1.
HMGB1 and chemotherapy
From different perspectives, HMGB1 enforces different functions in breast cancer chemotherapy. As the most frequently employed approach in breast cancer treatment, the mechanisms in which chemotherapy kills cancer cells depend not only on apoptosis, but also on other non-apoptotic deaths, such as necrosis and autophagy [125]. As mentioned above, HMGB1-mediated autophagy occupies an important part in breast cancer. The ability of HMGB1-mediated autophagy to confer chemoresistance of breast cancer cells has been confirmed in various conventional chemotherapeutic drug trials.
There are a variety of miRNAs that can enhance the chemosensitivity of breast cancer cells by targeting HMGB1-mediated autophagy. Shi et al. reported that miR-129-5p increased the chemosensitivity of MCF-7 cells to paclitaxel by inhibiting HMGB1-mediated autophagy [67]. Liang et al. found that miR-142-3p overexpression inhibited autophagy and promoted drug sensitivity to doxorubicin in breast cancer cells through negative regulation of HMGB1 [94]. The interaction of miRNA with HMGB1 provides new insights into attenuating chemoresistance associated with HMGB1-mediated autophagy.
Moreover, researchers have identified several novel agents that can reverse HMGB1-mediated autophagy-associated chemoresistance. For example, XIAOPI formula was proved to improve breast cancer chemotherapy sensitivity by inhibiting CXCL1/HMGB1-mediated autophagy [126]. Interestingly, Zhang et al. reported that CDK4/6 inhibitors were found to downregulate HMGB1 expression, inhibit the TLR4/NF-κB pathway, and thereby reverse tamoxifen resistance [127]. This suggests the feasibility of HMGB1 as a potential biomarker for screening sensitive patients receiving CDK4/6 inhibitors. Chloroquine, a potent autophagy blocker, has been shown to enhance the chemotherapy sensitivity of cancer [128]. In a clinical trial of chloroquine, HMGB1 was applied to assess the level of autophagy of breast ductal carcinoma in situ (clinical trial NCT01023477) [129]. Furthermore, synergistic therapeutic strategies of different drugs targeting HMGB1 have also been reported. Berberine, a clinically approved alkaloid, exhibits stronger antitumor effects in combination with other chemotherapeutic agents [130]. More importantly, berberine has been identified as a modulator of HMGB1/TLR4 signaling axis [131, 132]. In breast cancer, the synergy of berberine with theophylline induces HMGB1/Bcl-2-mediated apoptosis [133]. Zheng et al. designed biomimetic co-assembled nanodrug of doxorubicin and berberine, and found that it inhibits tumor growth and pulmonary metastasis by blocking HMGB1/TLR4 signaling, which provides novel evidence to develop safe and efficient nanodrugs for breast cancer [134].
HMGB1 and radiotherapy
Radiation therapy is a traditional treatment for many malignancies, including breast cancer, and can significantly improve patient survival, despite the damage to surrounding normal tissues. However, there are conflicting reports on the resistance of HMGB1 to breast cancer radiotherapy.
The ability of HMGB1 to enhance the radiosensitivity of breast cancer cells is generally considered to be largely correlated to its immunogenic effects. Apetoh et al. found that ionizing radiation promotes the release of HMGB1 from dying tumor cells. HMGB1 activates the TLR4/MyD88 signaling pathway in DCs and thus efficiently processes antigens associated with the presentation of dying tumor cells [9]. In addition, the radiation-induced abscopal effect (RIAE) is a miraculous phenomenon that refers to the shrinkage of non-irradiated lesions in contrast to the irradiated tumor lesions. It is now well acknowledged that RIAE relates to the systemic antitumor immune response that is induced by two simultaneous changes in the treatment of tumors: the release of tumor antigens and the exposure of DAMPs [135]. As mentioned above, the reaction that produce these changes are associated with ICD. Zhu et al. reported that HMGB1 released from radiated breast cancer cells could promote TNF-α secretion by surrounding macrophages through the TLR4 pathway, thus contributing to promote the radiation-induced anti-tumor abscopal effect triggered by breast cancer radiotherapy [103]. Although, the exact mechanism of HMGB1 in RIAE has not been adequately elucidated, the role of HMGB1 in bridging the gap between ICD and RIAE remains the importance for the development of immunotherapy in combination with radiotherapy.
Nevertheless, several experiments have shown a positive correlation between HMGB1 and radiotherapy resistance in breast cancer. For example, Luo et al. found that HMGB1-mediated autophagy enhanced radiotherapy resistance in breast cancer cells, and this effect was reversible for miR-129-5p [66]. Additionally, HMGB1 has also been proven to function in replenishing telomeric DNA. Ke et al. demonstrated that HMGB1 knockdown decreased the levels of telomere-binding proteins such as TPP1, TRF1 and TRF2 as well as inhibited DNA damage repair pathways after shRNA-mediated downregulation of HMGB1 [136]. In short, downregulation of HMGB1 disrupts telomere homeostasis, inhibits DNA damage repair and enhances radiosensitivity in human breast cancer cells. However, there are only a few studies on the HMGB1-related DNA damage repair pathway as a therapeutic target, so more investigation is required on HMGB1 knockdown as a potential therapeutic option for radiation therapy.
HMGB1, immunotherapy and combination therapy
Immunotherapy is a therapeutic method to specifically remove small residual tumor lesions, inhibit tumor growth and break immune tolerance by activating immune cells and enhancing the anti-tumor immune response in the body. Based on the powerful immune-modulatory function of HMGB1, immune or combination therapies developed for HMGB1 are of great significance to improve the prognosis and prolong the survival of cancer patients.
Among the currently available HMGB1-related combination therapy strategies, the most frequent is the employment of ICD inducers to activate adaptive anti-tumor immune responses in patients [97]. Conventional antitumor chemotherapeutic agents usually are highly cytotoxic and less targetable. The cell death induced by these chemotherapeutic agents tends to be non-immunogenic, and the dying tumor cells can be cleared without activating the immune response, a tolerogenic process that fails to produce a long-term antitumor effect [137]. In clinical trials, chemotherapeutic agents such as anthracyclines and oxaliplatin have been observed to induce tumor cell death and confer immunogenicity to the dead tumor cells, activating the immune system to fight these dying tumor cell antigens [138, 139]. In breast cancer, researchers have also been identifying new ICD inducers. For instance, oleandrin, a cardiac glycoside, can induce ER stress in breast cancer cells and trigger ICD-mediated immune responses via the PERK/elF2α/ATF4/CHOP pathway [18]. In addition, Somma et al. found that G-quadruplex binders induced ICD in MDA-MB-231 breast cancer cells, accompanied by T-cell activation [140]. Furthermore, Huang et al. indicated that the antioxidant N-acetylcysteine (NAC) enhanced not only the antitumor activity of polyglycolylated arginine deiminase (ADI-PEG20), but also the features of ICD, including HMGB1 and ATP secretion [19]. Nevertheless, the ICD inducers in clinical trials for breast cancer are limited so far. Researchers showed that P2Et, an extract of Caesalpinia Spinosa, induces the release of HMGB1 and has synergistic effects with anthracycline-type chemotherapeutic agents in animal models of breast cancer. Importantly, the results of a phase Ⅰ clinical study in healthy volunteers suggest that the extract is safe, while other factors, such as the optimal dose of P2Et, are still being explored (clinical trial NCT05007444) [141].
Similar to chemotherapeutic agents, oncolytic virus (OV), an in situ tumor vaccine, can generate a pro-inflammatory response by producing pathogen-associated molecular patterns (PAMPs) and releasing tumor-associated antigens (TAAs) [142].Gebremeskel et al. found that lysovirus-induced tumor cell death exhibited significant features of the ICD including elevated HMGB1 levels. More meaningfully, lysovirus and NKT cell immunotherapy can be effectively combined to reduce tumor load and improve survival in mice with metastatic breast cancer [143]. However, while OVs can be significantly improved as powerful cancer treatment candidates, alleviating the limitations of OVs with a combination strategy remains a challenge.
Immune checkpoints refer to the inhibitory signaling pathways that exist in the immune system. Under normal circumstances, the balance of co-stimulatory and co-inhibitory signaling permits the body to maintain immune tolerance. When the body is threatened by tumor burden, the immune checkpoint signaling pathway is generally obstructed, thereby dampening autoimmunity and providing an opportunity for tumor growth and escape [144]. Immune checkpoint blockade disrupts the co-inhibitory signaling pathway and activates the anti-tumor function of T cells, while ICD can enhance the T-cell response to tumors. Therefore, establishing a link between ICD and immune checkpoint blockade is a feasible strategy that promises to break through the inherent restrictions of immunotherapy. In recent years, the dual coordination of ICD and immune checkpoint function of several agents has been gradually revealed [145]. Li et al. discovered that breast cancer cells treated with SR-4835, a CDK12/13 specific inhibitor, exhibited features of ICD, including the release of HMGB1. Critically, enhanced infiltrative activation of DCs and T cells was observed in tumors treated with the combination of SR-4835 and anti-PD-1 antibodies, suggesting that this combination therapy promoted an improved immune response [20]. Shan et al. indicated that pingyangmycin, an ICD inducer, may enhance the therapeutic efficacy of anti-PD-1 antibodies associated with enhanced tumor infiltrating CD8+ T cells, in a similar fashion to SR-4835[146]. However, Hubert et al. showed that blocking extracellular HMGB1 could improve the efficacy of anti-PD-1 immunotherapy [104]. Besides canonical PD-1/PD-L1 blockade, the potential role of HMGB1 involvement in other immune checkpoint blockade has also been reported. For example, the inhibitory receptor TIM-3 limits antitumor immunity in breast cancer by inhibiting CXCL9 production by XCR1+ classical dendritic cells. Pulido et al. found that type I interferon and extracellular DNA are required for enhanced CXCL9 expression by splenic cDC1s after TIM-3 blockade, in which DNA uptake is dependent on the DNA-binding ability of HMGB1. TIM-3 limits the activation of the cGAS-STING pathway in intra-tumoral DCs by inhibiting extracellular DNA uptake [147]. Undoubtedly, this finding affords a novel perspective for understanding the mechanisms of TIM-3-related immunotherapy.
Tumor vaccines carry tumor-specific antigens (TSAs) or TAAs, which attack tumor cells by stimulating adaptive immunity, overcome the immunosuppressive state caused by tumor products, and improve autoimmunity to destroy tumors [148]. Yan et al. infected 4T1 cells with recombinant adenovirus AdVEGFR2 carrying the VEGFR2 gene and found that tumor growth and lung metastasis were inhibited in the vaccinated mice compared to the non-vaccinated mice. In vitro, the expression of HMGB1 and HSP70 was increased in 4T1 cells infected with AdVEGFR2, which was responsible for the activation of tumor antigen-specific T cell immunity [149]. Table 1 summarizes breast cancer therapy strategies targeting HMGB1 and their effects.
Breast cancer therapeutic strategies targeting HMGB1
| Therapeutic Strategies | Mechanism | Effect on breast cancer | Ref. |
|---|---|---|---|
| Chemicals | |||
| XIAOPI formula | Inhibit CXCL1/HMGB1 signal | Enhance taxol chemosensitivity | [126] |
| CDK4/6 inhibitor abemaciclib | Inhibit HMGB1/TLR4/NF-κB pathway | Enahnce tamoxifen chemosensitivity | [127] |
| Triptolide | Inhibit HMGB1/TLR4/NF-κB pathway | Suppress breast cancer growth | [150] |
| Irinotecan | Increase HMGB1 expression | Induce apoptosis; Arrest cell cycle in S phase | [151] |
| ICD inducers | |||
| oleandrin | Induce HMGB1 release | Promote DCs maturation and activation; Enhance CD8+ T cells cytotoxicity | [18] |
| CDK12/13 inhibitor SR-4835 | Induce HMGB1 release | Enhance anti-PD-1 anti-cancer activity | [20] |
| pingyangmycin | Induce HMGB1 release | Enhance anti-PD-1 anti-cancer activity; Augment tumor-infiltrating CD8+ T cells | [146] |
| Synergetic strategy | |||
| Doxorubicin and berberine | Inhibit HMGB1/TLR4 pathway | Inhibit tumor growth and pulmonary metastasis | [134] |
| Theophylline and berberine | Decrease HMGB1 expression | Enhance apoptotic cell death | [133] |
| LiCl and MMC | Decrease HMGB1 expression | Enhance MMC-induced apoptosis | [152] |
| ADI-PEG20 and NAC | Induce HMGB1 release | Enhance NAC-induced apoptosis | [19] |
| Apicidin and docetaxel | Induce HMGB1 release | Enhance apoptotic cell death | [153] |
| ALA and Radiotherapy | Increase HMGB1 expression | Enhance RT-induced cellular senescence | [154] |
| Other | |||
| Recombinant oncolytic VSV | Induce HMGB1 release | Decrease metastatic breast tumor burden | [143] |
| Recombinant adenovirus AdVEGFR2 | Increase HMGB1 expression | Active tumor antigen-specific T cell immunity; Inhibit angiogenesis, tumor growth and pulmonary metastasis | [149] |
LiCl: lithium chloride; MMC: mitomycin C; ADI-PEG20: pegylated arginine deiminase; NAC: N-acetylcysteine; ALA: alpha-lipoic acid; RT: radiotherapy; VSV: vesicular stomatitis virus.
Conclusions
The pro- and anti-tumor activity of HMGB1 have been reported on its carcinogenicity in breast cancer. The different patterns of HMGB1 induction determine its subcellular distribution, influence HMGB1-related biological processes such as autophagy, immunogenic cell death, and ultimately the efficacy of chemotherapy and radiotherapy for breast cancer. Furthermore, HMGB1-mediated immune activity indicates the new potential about immunotherapy for breast cancer, liberating the possibilities for innovation of combination therapeutic strategies. Although HMGB1 is a high potential target for early detection or treatment of cancers, its dual function in breast cancer remains to be elucidated in more detail.
Acknowledgements
This work was supported by The National Key Research and Development Program of China (Stem Cell and Translational Research 2020YFA0112300); National Natural Science Foundation of China (81930075, 81772799); Program for Outstanding Medical Academic Leader in Shanghai (2019LJ04); Program of Shanghai Academic/Technology Research Leader 20XD1400700.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians. 2022;72:7-33
2. Sun S, Zhang W, Cui Z, Chen Q, Xie P, Zhou C. et al. High mobility group box-1 and its clinical value in breast cancer. OncoTargets and therapy. 2015;8:413-9
3. Wang CQ, Huang BF, Wang Y, Hu GR, Wang Q, Shao JK. [Expression of HMGB1 protein in breast cancer and its clinicopathological significance]. Zhonghua bing li xue za zhi = Chinese journal of pathology. 2020;49:57-61
4. Yu Y, Tang D, Kang R. Oxidative stress-mediated HMGB1 biology. Frontiers in physiology. 2015;6:93
5. Stros M, Polanská E, Struncová S, Pospísilová S. HMGB1 and HMGB2 proteins up-regulate cellular expression of human topoisomerase IIalpha. Nucleic acids research. 2009;37:2070-86
6. Shi Q, Wang Y, Dong W, Song E, Song Y. Polychlorinated biphenyl quinone-induced signaling transition from autophagy to apoptosis is regulated by HMGB1 and p53 in human hepatoma HepG2 cells. Toxicology letters. 2019;306:25-34
7. Kang R, Livesey KM, Zeh HJ, Loze MT, Tang D. HMGB1: a novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6:1209-11
8. Kostova N, Zlateva S, Ugrinova I, Pasheva E. The expression of HMGB1 protein and its receptor RAGE in human malignant tumors. Molecular and cellular biochemistry. 2010;337:251-8
9. Apetoh L, Ghiringhelli F, Tesniere A, Criollo A, Ortiz C, Lidereau R. et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunological reviews. 2007;220:47-59
10. Ivanov S, Dragoi AM, Wang X, Dallacosta C, Louten J, Musco G. et al. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007;110:1970-81
11. Curtin JF, Liu N, Candolfi M, Xiong W, Assi H, Yagiz K. et al. HMGB1 mediates endogenous TLR2 activation and brain tumor regression. PLoS medicine. 2009;6:e10
12. Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nature reviews Cancer. 2012;12:860-75
13. Lee SA, Kwak MS, Kim S, Shin JS. The role of high mobility group box 1 in innate immunity. Yonsei medical journal. 2014;55:1165-76
14. Ni P, Zhang Y, Liu Y, Lin X, Su X, Lu H. et al. HMGB1 silence could promote MCF-7 cell apoptosis and inhibit invasion and metastasis. International journal of clinical and experimental pathology. 2015;8:15940-6
15. Wang LL, Meng QH, Jiao Y, Xu JY, Ge CM, Zhou JY. et al. High-mobility group boxes mediate cell proliferation and radiosensitivity via retinoblastoma-interaction-dependent and -independent mechanisms. Cancer biotherapy & radiopharmaceuticals. 2012;27:329-35
16. Liu B, Qi X, Zhang X, Gao D, Fang K, Guo Z. et al. Med19 is involved in chemoresistance by mediating autophagy through HMGB1 in breast cancer. Journal of cellular biochemistry. 2019;120:507-18
17. Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta pharmacologica Sinica. 2007;28:1957-67
18. Li X, Zheng J, Chen S, Meng FD, Ning J, Sun SL. Oleandrin, a cardiac glycoside, induces immunogenic cell death via the PERK/elF2α/ATF4/CHOP pathway in breast cancer. Cell death & disease. 2021;12:314
19. Huang Z, Hu H. Arginine Deiminase Induces Immunogenic Cell Death and Is Enhanced by N-acetylcysteine in Murine MC38 Colorectal Cancer Cells and MDA-MB-231 Human Breast Cancer Cells In vitro. Molecules (Basel, Switzerland). 2021 26
20. Li Y, Zhang H, Li Q, Zou P, Huang X, Wu C. et al. CDK12/13 inhibition induces immunogenic cell death and enhances anti-PD-1 anticancer activity in breast cancer. Cancer letters. 2020;495:12-21
21. Malarkey CS, Churchill ME. The high mobility group box: the ultimate utility player of a cell. Trends in biochemical sciences. 2012;37:553-62
22. Wang Q, Zeng M, Wang W, Tang J. The HMGB1 acidic tail regulates HMGB1 DNA binding specificity by a unique mechanism. Biochemical and biophysical research communications. 2007;360:14-9
23. Blair RH, Horn AE, Pazhani Y, Grado L, Goodrich JA, Kugel JF. The HMGB1 C-Terminal Tail Regulates DNA Bending. Journal of molecular biology. 2016;428:4060-72
24. Liang WJ, Yang HW, Liu HN, Qian W, Chen XL. HMGB1 upregulates NF-kB by inhibiting IKB-α and associates with diabetic retinopathy. Life sciences. 2020;241:117146
25. Stros M. HMGB proteins: interactions with DNA and chromatin. Biochimica et biophysica acta. 2010;1799:101-13
26. Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme. Annual review of biophysics. 2011;40:99-117
27. Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. The Journal of biological chemistry. 2004;279:20935-40
28. Joshi SR, Sarpong YC, Peterson RC, Scovell WM. Nucleosome dynamics: HMGB1 relaxes canonical nucleosome structure to facilitate estrogen receptor binding. Nucleic acids research. 2012;40:10161-71
29. Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochemical Society transactions. 2012;40:341-6
30. Bergeron S, Madathiparambil T, Swanson PC. Both high mobility group (HMG)-boxes and the acidic tail of HMGB1 regulate recombination-activating gene (RAG)-mediated recombination signal synapsis and cleavage in vitro. The Journal of biological chemistry. 2005;280:31314-24
31. Little AJ, Corbett E, Ortega F, Schatz DG. Cooperative recruitment of HMGB1 during V(D)J recombination through interactions with RAG1 and DNA. Nucleic acids research. 2013;41:3289-301
32. Jiang Y, Chen R, Shao X, Ji X, Lu H, Zhou S. et al. HMGB1 silencing in macrophages prevented their functional skewing and ameliorated EAM development: Nuclear HMGB1 may be a checkpoint molecule of macrophage reprogramming. International immunopharmacology. 2018;56:277-84
33. Xu T, Jiang L, Wang Z. The progression of HMGB1-induced autophagy in cancer biology. OncoTargets and therapy. 2019;12:365-77
34. Lu B, Antoine DJ, Kwan K, Lundbäck P, Wähämaa H, Schierbeck H. et al. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3068-73
35. Herzig D, Fang G, Toliver-Kinsky TE, Guo Y, Bohannon J, Sherwood ER. STAT1-deficient mice are resistant to cecal ligation and puncture-induced septic shock. Shock (Augusta, Ga). 2012;38:395-402
36. Rabadi MM, Xavier S, Vasko R, Kaur K, Goligorksy MS, Ratliff BB. High-mobility group box 1 is a novel deacetylation target of Sirtuin1. Kidney international. 2015;87:95-108
37. Contis-Montes de Oca A, Rodarte-Valle E, Rosales-Hernández MC, Abarca-Rojano E, Rojas-Hernández S, Fragoso-Vázquez MJ. et al. N-(2'-Hydroxyphenyl)-2-propylpentanamide (OH-VPA), a histone deacetylase inhibitor, induces the release of nuclear HMGB1 and modifies ROS levels in HeLa cells. Oncotarget. 2018;9:33368-81
38. Youn JH, Shin JS. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. Journal of immunology (Baltimore, Md: 1950). 2006;177:7889-97
39. Dumitriu IE, Baruah P, Manfredi AA, Bianchi ME, Rovere-Querini P. HMGB1: guiding immunity from within. Trends in immunology. 2005;26:381-7
40. Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L. et al. HMGB1 in health and disease. Molecular aspects of medicine. 2014;40:1-116
41. Klune JR, Dhupar R, Cardinal J, Billiar TR, Tsung A. HMGB1: endogenous danger signaling. Molecular medicine (Cambridge, Mass). 2008;14:476-84
42. Liu G, Wang J, Park YJ, Tsuruta Y, Lorne EF, Zhao X. et al. High mobility group protein-1 inhibits phagocytosis of apoptotic neutrophils through binding to phosphatidylserine. Journal of immunology (Baltimore, Md: 1950). 2008;181:4240-6
43. Li R, Wang J, Zhu F, Li R, Liu B, Xu W. et al. HMGB1 regulates T helper 2 and T helper17 cell differentiation both directly and indirectly in asthmatic mice. Molecular immunology. 2018;97:45-55
44. Meng X, Chen M, Su W, Tao X, Sun M, Zou X. et al. The differentiation of mesenchymal stem cells to vascular cells regulated by the HMGB1/RAGE axis: its application in cell therapy for transplant arteriosclerosis. Stem cell research & therapy. 2018;9:85
45. Lin F, Zhang W, Xue D, Zhu T, Li J, Chen E. et al. Signaling pathways involved in the effects of HMGB1 on mesenchymal stem cell migration and osteoblastic differentiation. International journal of molecular medicine. 2016;37:789-97
46. Zhang L, Shi H, Chen H, Gong A, Liu Y, Song L. et al. Dedifferentiation process driven by radiotherapy-induced HMGB1/TLR2/YAP/HIF-1α signaling enhances pancreatic cancer stemness. Cell death & disease. 2019;10:724
47. Teissier T, Boulanger É. The receptor for advanced glycation end-products (RAGE) is an important pattern recognition receptor (PRR) for inflammaging. Biogerontology. 2019;20:279-301
48. Li W, Xu Q, Deng Y, Yang Z, Xing S, Zhao X. et al. High-mobility group box 1 accelerates lipopolysaccharide-induced lung fibroblast proliferation in vitro: involvement of the NF-κB signaling pathway. Laboratory investigation; a journal of technical methods and pathology. 2015;95:635-47
49. El Gazzar M. HMGB1 modulates inflammatory responses in LPS-activated macrophages. Inflammation research: official journal of the European Histamine Research Society [et al]. 2007;56:162-7
50. Degryse B, Bonaldi T, Scaffidi P, Müller S, Resnati M, Sanvito F. et al. The high mobility group (HMG) boxes of the nuclear protein HMG1 induce chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. The Journal of cell biology. 2001;152:1197-206
51. Todorova J, Pasheva E. High mobility group B1 protein interacts with its receptor RAGE in tumor cells but not in normal tissues. Oncology letters. 2012;3:214-8
52. Wang L, Kang FB, Wang J, Yang C, He DW. Downregulation of miR-205 contributes to epithelial-mesenchymal transition and invasion in triple-negative breast cancer by targeting HMGB1-RAGE signaling pathway. Anti-cancer drugs. 2019;30:225-32
53. Chen Y, Cai L, Guo X, Li Z, Liao X, Zhang X. et al. HMGB1-activated fibroblasts promote breast cancer cells metastasis via RAGE/aerobic glycolysis. Neoplasma. 2021;68:71-8
54. Okui T, Hiasa M, Ryumon S, Ono K, Kunisada Y, Ibaragi S. et al. The HMGB1/RAGE axis induces bone pain associated with colonization of 4T1 mouse breast cancer in bone. Journal of bone oncology. 2021;26:100330
55. Fitzgerald KA, Kagan JC. Toll-like Receptors and the Control of Immunity. Cell. 2020;180:1044-66
56. Dajon M, Iribarren K, Cremer I. Toll-like receptor stimulation in cancer: A pro- and anti-tumor double-edged sword. Immunobiology. 2017;222:89-100
57. Venereau E, De Leo F, Mezzapelle R, Careccia G, Musco G, Bianchi ME. HMGB1 as biomarker and drug target. Pharmacological research. 2016;111:534-44
58. Beaulieu LM, Lin E, Morin KM, Tanriverdi K, Freedman JE. Regulatory effects of TLR2 on megakaryocytic cell function. Blood. 2011;117:5963-74
59. Lin E, Freedman JE, Beaulieu LM. Innate immunity and toll-like receptor antagonists: a potential role in the treatment of cardiovascular diseases. Cardiovascular therapeutics. 2009;27:117-23
60. Conti L, Lanzardo S, Arigoni M, Antonazzo R, Radaelli E, Cantarella D. et al. The noninflammatory role of high mobility group box 1/Toll-like receptor 2 axis in the self-renewal of mammary cancer stem cells. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27:4731-44
61. Lv W, Chen N, Lin Y, Ma H, Ruan Y, Li Z. et al. Macrophage migration inhibitory factor promotes breast cancer metastasis via activation of HMGB1/TLR4/NF kappa B axis. Cancer letters. 2016;375:245-55
62. Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF. et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nature immunology. 2004;5:190-8
63. Liu Y, Yan W, Tohme S, Chen M, Fu Y, Tian D. et al. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular carcinoma through Toll-like receptor 9. Journal of hepatology. 2015;63:114-21
64. Jiang J, Wang GZ, Wang Y, Huang HZ, Li WT, Qu XD. Hypoxia-induced HMGB1 expression of HCC promotes tumor invasiveness and metastasis via regulating macrophage-derived IL-6. Experimental cell research. 2018;367:81-8
65. Ai H, Zhou W, Wang Z, Qiong G, Chen Z, Deng S. microRNAs-107 inhibited autophagy, proliferation, and migration of breast cancer cells by targeting HMGB1. Journal of cellular biochemistry. 2018
66. Luo J, Chen J, He L. mir-129-5p Attenuates Irradiation-Induced Autophagy and Decreases Radioresistance of Breast Cancer Cells by Targeting HMGB1. Medical science monitor: international medical journal of experimental and clinical research. 2015;21:4122-9
67. Shi Y, Gong W, Lu L, Wang Y, Ren J. Upregulation of miR-129-5p increases the sensitivity to Taxol through inhibiting HMGB1-mediated cell autophagy in breast cancer MCF-7 cells. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas. 2019;52:e8657
68. Chang BP, Wang DS, Xing JW, Yang SH, Chu Q, Yu SY. miR-200c inhibits metastasis of breast cancer cells by targeting HMGB1. Journal of Huazhong University of Science and Technology Medical sciences = Hua zhong ke ji da xue xue bao Yi xue Ying De wen ban = Huazhong keji daxue xuebao Yixue Yingdewen ban. 2014;34:201-6
69. Chen ZG, Zhao HJ, Lin L, Liu JB, Bai JZ, Wang GS. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thoracic cancer. 2020;11:2660-71
70. Zhang J, Ke S, Zheng W, Zhu Z, Wu Y. Hsa_circ_0003645 Promotes Breast Cancer Progression by Regulating miR-139-3p/HMGB1 Axis. OncoTargets and therapy. 2020;13:10361-72
71. He H, Wang X, Chen J, Sun L, Sun H, Xie K. High-Mobility Group Box 1 (HMGB1) Promotes Angiogenesis and Tumor Migration by Regulating Hypoxia-Inducible Factor 1 (HIF-1α) Expression via the Phosphatidylinositol 3-Kinase (PI3K)/AKT Signaling Pathway in Breast Cancer Cells. Medical science monitor: international medical journal of experimental and clinical research. 2019;25:2352-60
72. Sun S, Ma J, Xie P, Wu Z, Tian X. Hypoxia-responsive miR-141-3p is involved in the progression of breast cancer via mediating the HMGB1/HIF-1α signaling pathway. The journal of gene medicine. 2020;22:e3230
73. Uramoto H, Izumi H, Nagatani G, Ohmori H, Nagasue N, Ise T. et al. Physical interaction of tumour suppressor p53/p73 with CCAAT-binding transcription factor 2 (CTF2) and differential regulation of human high-mobility group 1 (HMG1) gene expression. The Biochemical journal. 2003;371:301-10
74. Davalos AR, Kawahara M, Malhotra GK, Schaum N, Huang J, Ved U. et al. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. The Journal of cell biology. 2013;201:613-29
75. Rowell JP, Simpson KL, Stott K, Watson M, Thomas JO. HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure (London, England: 1993). 2012;20:2014-24
76. Štros M, Kučírek M, Sani SA, Polanská E. HMGB1-mediated DNA bending: Distinct roles in increasing p53 binding to DNA and the transactivation of p53-responsive gene promoters. Biochimica et biophysica acta Gene regulatory mechanisms. 2018;1861:200-10
77. Okuyama H, Endo H, Akashika T, Kato K, Inoue M. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer research. 2010;70:10213-23
78. Wolfson RK, Mapes B, Garcia JGN. Excessive mechanical stress increases HMGB1 expression in human lung microvascular endothelial cells via STAT3. Microvascular research. 2014;92:50-5
79. Yu Y, Ou-Yang WX, Zhang H, Jiang T, Tang L, Tan YF. et al. MiR-125b enhances autophagic flux to improve septic cardiomyopathy via targeting STAT3/HMGB1. Experimental cell research. 2021;409:112842
80. Pan J, Alexan B, Dennis D, Bettina C, Christoph LIM, Tang Y. microRNA-193-3p attenuates myocardial injury of mice with sepsis via STAT3/HMGB1 axis. Journal of translational medicine. 2021;19:386
81. Chen X, Liu X, He B, Pan Y, Sun H, Xu T. et al. MiR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling way in colorectal cancer. American journal of cancer research. 2017;7:2051-69
82. Zhang Y, Ren H, Li J, Xue R, Liu H, Zhu Z. et al. Elevated HMGB1 expression induced by hepatitis B virus X protein promotes epithelial-mesenchymal transition and angiogenesis through STAT3/miR-34a/NF-κB in primary liver cancer. American journal of cancer research. 2021;11:479-94
83. Xu X, Chen Y, Liu E, Fu B, Hua J, Chen X. et al. HMGB1 Recruits TET2/AID/TDG to Induce DNA Demethylation in STAT3 Promoter in CD4(+) T Cells from aGVHD Patients. Journal of immunology research. 2020;2020:7165230
84. Xia H, Green DR, Zou W. Autophagy in tumour immunity and therapy. Nature reviews Cancer. 2021;21:281-97
85. Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B. et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell metabolism. 2011;13:701-11
86. Kang R, Livesey KM, Zeh HJ 3rd, Lotze MT, Tang D. Metabolic regulation by HMGB1-mediated autophagy and mitophagy. Autophagy. 2011;7:1256-8
87. Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P. et al. Endogenous HMGB1 regulates autophagy. The Journal of cell biology. 2010;190:881-92
88. Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell death and differentiation. 2007;14:1576-82
89. Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE. et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299-310
90. Su Z, Wang T, Zhu H, Zhang P, Han R, Liu Y. et al. HMGB1 modulates Lewis cell autophagy and promotes cell survival via RAGE-HMGB1-Erk1/2 positive feedback during nutrient depletion. Immunobiology. 2015;220:539-44
91. Zhang QY, Wu LQ, Zhang T, Han YF, Lin X. Autophagy-mediated HMGB1 release promotes gastric cancer cell survival via RAGE activation of extracellular signal-regulated kinases 1/2. Oncology reports. 2015;33:1630-8
92. Li J, Wu PW, Zhou Y, Dai B, Zhang PF, Zhang YH. et al. Rage induces hepatocellular carcinoma proliferation and sorafenib resistance by modulating autophagy. Cell death & disease. 2018;9:225
93. Wang R, He W, Li Z, Chang W, Xin Y, Huang T. Caveolin-1 functions as a key regulator of 17β-estradiol-mediated autophagy and apoptosis in BT474 breast cancer cells. International journal of molecular medicine. 2014;34:822-7
94. Liang L, Fu J, Wang S, Cen H, Zhang L, Mandukhail SR. et al. MiR-142-3p enhances chemosensitivity of breast cancer cells and inhibits autophagy by targeting HMGB1. Acta pharmaceutica Sinica B. 2020;10:1036-46
95. Ladoire S, Penault-Llorca F, Senovilla L, Dalban C, Enot D, Locher C. et al. Combined evaluation of LC3B puncta and HMGB1 expression predicts residual risk of relapse after adjuvant chemotherapy in breast cancer. Autophagy. 2015;11:1878-90
96. Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annual review of immunology. 2013;31:51-72
97. Fucikova J, Kepp O, Kasikova L, Petroni G, Yamazaki T, Liu P. et al. Detection of immunogenic cell death and its relevance for cancer therapy. Cell death & disease. 2020;11:1013
98. Ahmed A, Tait SWG. Targeting immunogenic cell death in cancer. Molecular oncology. 2020;14:2994-3006
99. Aoto K, Mimura K, Okayama H, Saito M, Chida S, Noda M. et al. Immunogenic tumor cell death induced by chemotherapy in patients with breast cancer and esophageal squamous cell carcinoma. Oncology reports. 2018;39:151-9
100. Fico F, Santamaria-Martinez A. The Tumor Microenvironment as a Driving Force of Breast Cancer Stem Cell Plasticity. Cancers (Basel). 2020 12
101. Wang Z, Yang C, Li L, Zhang Z, Pan J, Su K. et al. CD62L(dim) Neutrophils Specifically Migrate to the Lung and Participate in the Formation of the Pre-Metastatic Niche of Breast Cancer. Front Oncol. 2020;10:540484
102. Wang Z, Yang C, Li L, Jin X, Zhang Z, Zheng H. et al. Tumor-derived HMGB1 induces CD62L(dim) neutrophil polarization and promotes lung metastasis in triple-negative breast cancer. Oncogenesis. 2020;9:82
103. Zhu L, Hu S, Chen Q, Zhang H, Fu J, Zhou Y. et al. Macrophage contributes to radiation-induced anti-tumor abscopal effect on transplanted breast cancer by HMGB1/TNF-α signaling factors. International journal of biological sciences. 2021;17:926-41
104. Hubert P, Roncarati P, Demoulin S, Pilard C, Ancion M, Reynders C. et al. Extracellular HMGB1 blockade inhibits tumor growth through profoundly remodeling immune microenvironment and enhances checkpoint inhibitor-based immunotherapy. Journal for immunotherapy of cancer. 2021 9
105. Houthuijzen JM, Jonkers J. Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer metastasis reviews. 2018;37:577-97
106. Amornsupak K, Insawang T, Thuwajit P, P OC, Eccles SA, Thuwajit C. Cancer-associated fibroblasts induce high mobility group box 1 and contribute to resistance to doxorubicin in breast cancer cells. BMC cancer. 2014;14:955
107. Liu L, Liu S, Luo H, Chen C, Zhang X, He L. et al. GPR30-mediated HMGB1 upregulation in CAFs induces autophagy and tamoxifen resistance in ERα-positive breast cancer cells. Aging. 2021;13:16178-97
108. Li P, Gong Z, Shultz LD, Ren G. Mesenchymal stem cells: From regeneration to cancer. Pharmacology & therapeutics. 2019;200:42-54
109. Kamat P, Schweizer R, Kaenel P, Salemi S, Calcagni M, Giovanoli P. et al. Human Adipose-Derived Mesenchymal Stromal Cells May Promote Breast Cancer Progression and Metastatic Spread. Plastic and reconstructive surgery. 2015;136:76-84
110. Ananthula S, Sinha A, El Gassim M, Batth S, Marshall GD Jr, Gardner LH. et al. Geminin overexpression-dependent recruitment and crosstalk with mesenchymal stem cells enhance aggressiveness in triple negative breast cancers. Oncotarget. 2016;7:20869-89
111. Sundara Rajan S, Hanby AM, Horgan K, Thygesen HH, Speirs V. The potential utility of geminin as a predictive biomarker in breast cancer. Breast cancer research and treatment. 2014;143:91-8
112. Blanchard Z, Mullins N, Ellipeddi P, Lage JM, McKinney S, El-Etriby R. et al. Geminin overexpression promotes imatinib sensitive breast cancer: a novel treatment approach for aggressive breast cancers, including a subset of triple negative. PloS one. 2014;9:e95663
113. Sami E, Bogan D, Molinolo A, Koziol J, ElShamy WM. The molecular underpinning of geminin-overexpressing triple-negative breast cancer cells homing specifically to lungs. Cancer gene therapy. 2021
114. Ryan D, Koziol J, ElShamy WM. Targeting AXL and RAGE to prevent geminin overexpression-induced triple-negative breast cancer metastasis. Scientific reports. 2019;9:19150
115. Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A. et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nature medicine. 2007;13:1050-9
116. Guerriero JL, Ditsworth D, Catanzaro JM, Sabino G, Furie MB, Kew RR. et al. DNA alkylating therapy induces tumor regression through an HMGB1-mediated activation of innate immunity. Journal of immunology (Baltimore, Md: 1950). 2011;186:3517-26
117. Campana L, Bosurgi L, Rovere-Querini P. HMGB1: a two-headed signal regulating tumor progression and immunity. Current opinion in immunology. 2008;20:518-23
118. Srikrishna G, Freeze HH. Endogenous damage-associated molecular pattern molecules at the crossroads of inflammation and cancer. Neoplasia (New York, NY). 2009;11:615-28
119. Jiao D, Zhang J, Chen P, Guo X, Qiao J, Zhu J. et al. HN1L promotes migration and invasion of breast cancer by up-regulating the expression of HMGB1. Journal of cellular and molecular medicine. 2021;25:397-410
120. Liu Y, Choi DS, Sheng J, Ensor JE, Liang DH, Rodriguez-Aguayo C. et al. HN1L Promotes Triple-Negative Breast Cancer Stem Cells through LEPR-STAT3 Pathway. Stem cell reports. 2018;10:212-27
121. Nong S, Wang Z, Wei Z, Ma L, Guan Y, Ni J. HN1L promotes stem cell-like properties by regulating TGF-β signaling pathway through targeting FOXP2 in prostate cancer. Cell biology international. 2022;46:83-95
122. Stoetzer OJ, Fersching DM, Salat C, Steinkohl O, Gabka CJ, Hamann U. et al. Circulating immunogenic cell death biomarkers HMGB1 and RAGE in breast cancer patients during neoadjuvant chemotherapy. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:81-90
123. Arnold T, Michlmayr A, Baumann S, Burghuber C, Pluschnig U, Bartsch R. et al. Plasma HMGB-1 after the initial dose of epirubicin/docetaxel in cancer. European journal of clinical investigation. 2013;43:286-91
124. Lee HJ, Kim A, Song IH, Park IA, Yu JH, Ahn JH. et al. Cytoplasmic expression of high mobility group B1 (HMGB1) is associated with tumor-infiltrating lymphocytes (TILs) in breast cancer. Pathology international. 2016;66:202-9
125. Ricci MS, Zong WX. Chemotherapeutic approaches for targeting cell death pathways. The oncologist. 2006;11:342-57
126. Wang N, Yang B, Muhetaer G, Wang S, Zheng Y, Lu J. et al. XIAOPI formula promotes breast cancer chemosensitivity via inhibiting CXCL1/HMGB1-mediated autophagy. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;120:109519
127. Zhang H, Wang J, Li J, Zhou X, Yin L, Wang Y. et al. HMGB1 is a key factor for tamoxifen resistance and has the potential to predict the efficacy of CDK4/6 inhibitors in breast cancer. Cancer science. 2021;112:1603-13
128. Ferreira PMP, Sousa RWR, Ferreira JRO, Militão GCG, Bezerra DP. Chloroquine and hydroxychloroquine in antitumor therapies based on autophagy-related mechanisms. Pharmacological research. 2021;168:105582
129. Study of the Efficacy of Chloroquine in the Treatment of Ductal Carcinoma in situ (The PINC Trial). https://ClinicalTrials.gov/show/NCT01023477
130. Rauf A, Abu-Izneid T, Khalil AA, Imran M, Shah ZA, Emran TB. et al. Berberine as a Potential Anticancer Agent: A Comprehensive Review. Molecules (Basel, Switzerland). 2021 26
131. Zhang T, Yang S, Du J. Protective Effects of Berberine on Isoproterenol-Induced Acute Myocardial Ischemia in Rats through Regulating HMGB1-TLR4 Axis. Evidence-based complementary and alternative medicine: eCAM. 2014;2014:849783
132. Gendy AM, Elnagar MR, Allam MM, Mousa MR, Khodir AE, El-Haddad AE. et al. Berberine-loaded nanostructured lipid carriers mitigate warm hepatic ischemia/reperfusion-induced lesion through modulation of HMGB1/TLR4/NF-κB signaling and autophagy. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2022;145:112122
133. Hashemi-Niasari F, Rabbani-Chadegani A, Razmi M, Fallah S. Synergy of theophylline reduces necrotic effect of berberine, induces cell cycle arrest and PARP, HMGB1, Bcl-2 family mediated apoptosis in MDA-MB-231 breast cancer cells. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;106:858-67
134. Zheng X, Zhao Y, Jia Y, Shao D, Zhang F, Sun M. et al. Biomimetic co-assembled nanodrug of doxorubicin and berberine suppresses chemotherapy-exacerbated breast cancer metastasis. Biomaterials. 2021;271:120716
135. Rodríguez-Ruiz ME, Vanpouille-Box C, Melero I, Formenti SC, Demaria S. Immunological Mechanisms Responsible for Radiation-Induced Abscopal Effect. Trends in immunology. 2018;39:644-55
136. Ke S, Zhou F, Yang H, Wei Y, Gong J, Mei Z. et al. Downregulation of high mobility group box 1 modulates telomere homeostasis and increases the radiosensitivity of human breast cancer cells. International journal of oncology. 2015;46:1051-8
137. Nagata S, Tanaka M. Programmed cell death and the immune system. Nature reviews Immunology. 2017;17:333-40
138. Ladoire S, Mignot G, Dabakuyo S, Arnould L, Apetoh L, Rébé C. et al. In situ immune response after neoadjuvant chemotherapy for breast cancer predicts survival. The Journal of pathology. 2011;224:389-400
139. Correale P, Rotundo MS, Del Vecchio MT, Remondo C, Migali C, Ginanneschi C. et al. Regulatory (FoxP3+) T-cell tumor infiltration is a favorable prognostic factor in advanced colon cancer patients undergoing chemo or chemoimmunotherapy. Journal of immunotherapy (Hagerstown, Md: 1997). 2010;33:435-41
140. Di Somma S, Amato J, Iaccarino N, Pagano B, Randazzo A, Portella G. et al. G-Quadruplex Binders Induce Immunogenic Cell Death Markers in Aggressive Breast Cancer Cells. Cancers (Basel). 2019 11
141. Safety and Efficacy Evaluation of the P2Et Extract in Patients With Breast Cancer. https://ClinicalTrials.gov/show/NCT05007444
142. Guo ZS, Liu Z, Bartlett DL. Oncolytic Immunotherapy: Dying the Right Way is a Key to Eliciting Potent Antitumor Immunity. Front Oncol. 2014;4:74
143. Gebremeskel S, Nelson A, Walker B, Oliphant T, Lobert L, Mahoney D. et al. Natural killer T cell immunotherapy combined with oncolytic vesicular stomatitis virus or reovirus treatments differentially increases survival in mouse models of ovarian and breast cancer metastasis. Journal for immunotherapy of cancer. 2021 9
144. Kubli SP, Berger T, Araujo DV, Siu LL, Mak TW. Beyond immune checkpoint blockade: emerging immunological strategies. Nature reviews Drug discovery. 2021;20:899-919
145. Wilson AL, Plebanski M, Stephens AN. New Trends in Anti-Cancer Therapy: Combining Conventional Chemotherapeutics with Novel Immunomodulators. Current medicinal chemistry. 2018;25:4758-84
146. Shan CK, Du YB, Zhai XT, Wang YX, Li Y, Gong JH. et al. Pingyangmycin enhances the antitumor efficacy of anti-PD-1 therapy associated with tumor-infiltrating CD8(+) T cell augmentation. Cancer chemotherapy and pharmacology. 2021;87:425-36
147. de Mingo Pulido Á, Hänggi K, Celias DP, Gardner A, Li J, Batista-Bittencourt B. et al. The inhibitory receptor TIM-3 limits activation of the cGAS-STING pathway in intra-tumoral dendritic cells by suppressing extracellular DNA uptake. Immunity. 2021;54:1154-67.e7
148. Guo C, Manjili MH, Subjeck JR, Sarkar D, Fisher PB, Wang XY. Therapeutic cancer vaccines: past, present, and future. Advances in cancer research. 2013;119:421-75
149. Yan HX, Cheng P, Wei HY, Shen GB, Fu LX, Ni J. et al. Active immunotherapy for mouse breast cancer with irradiated whole-cell vaccine expressing VEGFR2. Oncology reports. 2013;29:1510-6
150. Jiang W, Chen M, Xiao C, Yang W, Qin Q, Tan Q. et al. Triptolide Suppresses Growth of Breast Cancer by Targeting HMGB1 in vitro and in vivo. Biological & pharmaceutical bulletin. 2019;42:892-9
151. Keyvani-Ghamsari S, Rabbani-Chadegani A, Sargolzaei J, Shahhoseini M. Effect of irinotecan on HMGB1, MMP9 expression, cell cycle, and cell growth in breast cancer (MCF-7) cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39:1010428317698354
152. Razmi M, Rabbani-Chadegani A, Hashemi-Niasari F, Ghadam P. Lithium chloride attenuates mitomycin C induced necrotic cell death in MDA-MB-231 breast cancer cells via HMGB1 and Bax signaling. Journal of trace elements in medicine and biology: organ of the Society for Minerals and Trace Elements (GMS). 2018;48:87-96
153. Buoncervello M, Borghi P, Romagnoli G, Spadaro F, Belardelli F, Toschi E. et al. Apicidin and docetaxel combination treatment drives CTCFL expression and HMGB1 release acting as potential antitumor immune response inducers in metastatic breast cancer cells. Neoplasia (New York, NY). 2012;14:855-67
154. Choi HS, Kim JH, Jang SJ, Yun JW, Kang KM, Jeong H. et al. Synergistic Tumoricidal Effects of Alpha-Lipoic Acid and Radiotherapy on Human Breast Cancer Cells via HMGB1. Cancer research and treatment. 2021;53:685-94
Author contact
![]() Corresponding authors: Suling Liu, Fudan University Shanghai Cancer Center, Key Laboratory of Breast Cancer in Shanghai, Cancer Institutes, Shanghai 200032, China; Tel/Fax: 086-21-34774409. E-mail: sulingedu.cn; Lu Zhang, Fudan University Shanghai Cancer Center, Key Laboratory of Breast Cancer in Shanghai, Cancer Institutes, Shanghai 200032, China; Tel/Fax: 086-21- 34771071. E-mail: zhanglujennycom.
Corresponding authors: Suling Liu, Fudan University Shanghai Cancer Center, Key Laboratory of Breast Cancer in Shanghai, Cancer Institutes, Shanghai 200032, China; Tel/Fax: 086-21-34774409. E-mail: sulingedu.cn; Lu Zhang, Fudan University Shanghai Cancer Center, Key Laboratory of Breast Cancer in Shanghai, Cancer Institutes, Shanghai 200032, China; Tel/Fax: 086-21- 34771071. E-mail: zhanglujennycom.

 Global reach, higher impact
Global reach, higher impact