10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(8):3458-3469. doi:10.7150/ijbs.71046 This issue Cite
Review
The progress of multimodal imaging combination and subregion based radiomics research of cancers
1. Department of Neurosurgery, First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China.
2. Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China.
3. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Changsha, Hunan, China.
#These authors contributed equally to this work.
Received 2022-1-14; Accepted 2022-4-18; Published 2022-5-9
Abstract
In recent years, with the standardization of radiomics methods; development of tools; and popularization of the concept, radiomics has been widely used in all aspects of tumor diagnosis; treatment; and prognosis. As the study of radiomics in cancer has become more advanced, the currently used methods have revealed their shortcomings. The performance of cancer radiomics based on single-modality medical images, which based on their imaging principles, only partially reflects tumor information, has been necessarily compromised. Using the whole tumor as a region of interest to extract radiomic features inevitably leads to the loss of intra-tumoral heterogeneity of, which also affects the performance of radiomics. Radiomics of multimodal images extracts various aspects of information from images of each modality and then integrates them together for model construction; thus, avoiding missing information. Subregional segmentation based on multimodal medical image combinations allows radiomics features acquired from subregions to retain tumor heterogeneity, further improving the performance of radiomics. In this review, we provide a detailed summary of the current research on the radiomics of multimodal images of cancer and tumor subregion-based radiomics, and then raised some of the research problems and also provide a thorough discussion on these issues.
Keywords: cancer, multimodal imaging, subregion, heterogenous, radiomics
Background
Cancer is a leading public health concern globally. Genetic biomarkers are critical for the suitable treatments for cancer patients (1). For example, IDH mutation (2-4), BRCA mutations (5,6), and EGFR amplification (7,8) have been widely reported for accurate diagnosis and precise treatment of cancers. However, the identification of biomarkers is based on the pathological tissues of cancers. Limited by the unique anatomy of some cancers, the method of obtaining tumor tissue usually involves risky surgery or stereotactic biopsy. Therefore, not all patients with cancer undergo genomic analyses. Even when genomic analysis is performed, the sampling is usually a one-time and single-site process. In addition, most cancer types are highly heterogeneous (9). The results of genomic analysis are highly susceptible to sampling errors and observer variability (10).
Radiomics is the process of converting medical images into mineable high-dimensional data (11,12). It has helped medical imaging expand from diagnostic aid to a clinical decision aid in the context of personalized medicine (13). Radiomics involves combining imaging data with patient characteristics, including clinical data (14), genomics (15,16), and drug response (17,18), to improve the functionality of decision models (19). Radiomics is generally considered a good solution to these issues: First, radiomics can help guide patients on the need for further genetic sequencing, as it can derive information from all parts of the tumor images (19); Second, the correlation between radiomics and genomics can guide biopsy site selection to address the high error rate of histopathology due to sampling errors (20,21); Third, radiomics can accurately predict valuable biomarkers in cancers, such as IDH mutations in glioma (22-24), and EGFR amplification in lung cancer (25).
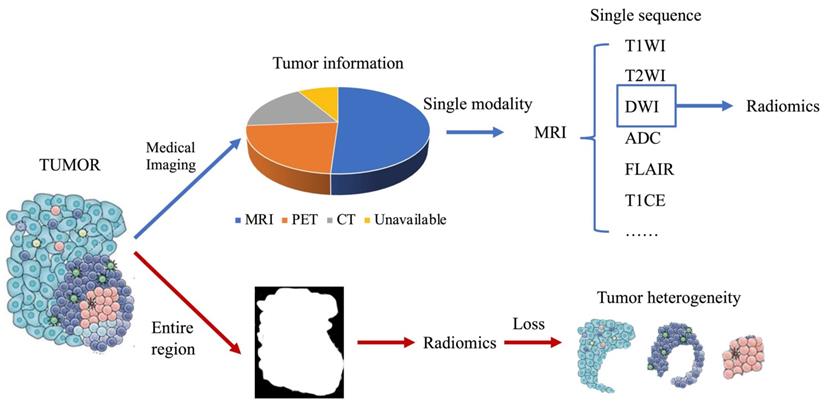
With the increasing popularity of radiomics, its application in cancer research is not only for genetic markers but also for patient survival, drug treatment effects, and other similar classification. However, some problems were also encountered. Previously, single-modality or single-sequence image was usually used for radiomics studies. But one modality or sequence can only reflect certain characteristics of the tumor. Therefore, the radiomics features extracted from single-modality images inevitable miss some tumor information, which will affect subsequent analysis. In addition, almost all cancers highly display intratumoral heterogeneity (9), and their medical images contain such heterogeneous information. However, many current radiomics studies have extracted radiomics features from the entire tumor region, which inevitably loses heterogeneous information in tumor images. These issues have been gradually considered. Multi-modality/sequence images combination have been used for radiomics studies and have yielded better results than single modality or sequence (26). Benefiting from the combination of multi-modality/sequence images, tumors have been segmented into regions reflecting different biological properties, and radiomics studies based on these regions have further improved the capabilities of radiomics (27). To make radiomics an effective diagnostic, prognostic, and predictive tool for cancer, it is important to understand advances in radiomics. This review describes recent advances in multimodality/multisequence studies in radiomics, the challenges that may be involved, and an outlook on the future of radiomics (Fig. 1).
Shortcomings of current radiomics research. (Blue arrow) Different medical images can each obtain partial information about the tumor, and a large number of current radiomics studies tend to use only single modality images, and for MRI even only a few of its sequences are used, resulting in only a small fraction of tumor information being used. (Red arrow) Tumors are heterogeneous, but current radiomics tends to extract radiomics features from the entire tumor region on the image, thus making the information of tumor heterogeneity lost.
Progress of radiomics
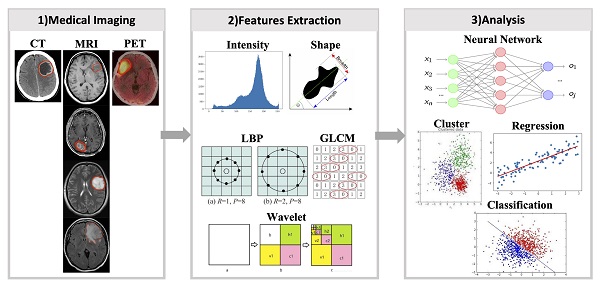
Radiomics is the process of extracting, analyzing, and building predictive models for radiomic features associated with predictive targets, and quantitative mapping between medical images and targets (28). It comprises a series of steps, including image acquisition, registration, segmentation, feature extraction, feature dimensionality reduction, and predictive modelling. 1) Image acquisition is the acquisition of clinical images from conventional imaging tools, such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET). 2) Registration involves establishing spatial correspondences between different image acquisitions (29). 3) Segmentation is the process of using clinical, imaging, and pathological knowledge to delineate the region of interest (ROI) in medical images. 4) Feature extraction is mining high-dimensional features from ROIs. 5) Feature dimensionality reduction involves cleaning redundant, irrelevant, or useless features. 6) Building robust predictive models is the ultimate purpose of radiomics research, which can help make clinical judgments and decisions (Fig. 2).
Diagram of radiomics process. 1) Radiomics first requires obtaining medical images of the patient and segmenting the target area. 2) Next step, various types of radiomics features were extracted from the images using a radiomics approach. 3) Using radiomics features, the models are constructed by machine learning or deep learning methods.
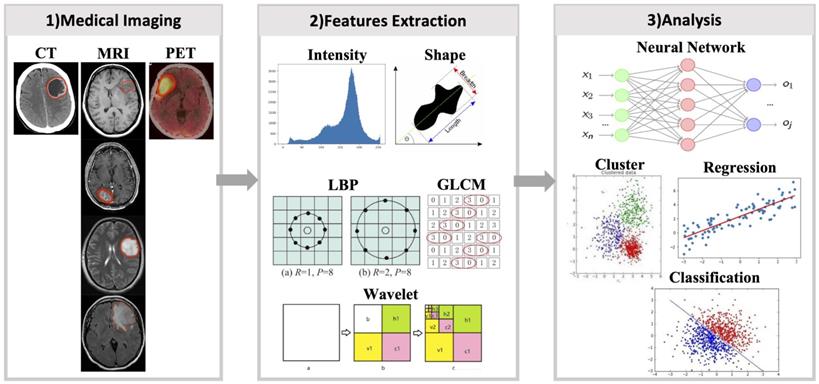
Medical imaging commonly used in clinical includes X-ray, ultrasound, CT, MRI, and PET. Each type of imaging technique is suitable for various diseases. For example, for lung diseases, X-rays are usually selected first and CT is used to obtain more information; MRI are more common for brain and spinal cord diseases. Different MRI sequences can provide the most accurate data on different types of data for patients. Although there are radiomics studies of X-rays (30) and ultrasound (31), the main research of radiomics is focused on CT, MRI, and PET. Therefore, this study mainly focused on radiomics studies of these three imaging types (Table 1).
Information of radiomic studies in human cancer
| Years | Cited reference | Tumor Types | Imaging combination |
|---|---|---|---|
| 2019-2020 | 26, 43-53 | Glioma, Breast cancer, Prostate cancer | Multimodal MRI sequences |
| 2019-2021 | 58-62 | Lung cancer, Nasopharyngeal carcinoma, Oropharyngeal squamous cell carcinoma | PET and CT |
| 2018-2021 | 63-65 | Brain metastasis, Rectal cancers, Breast cancer | PET and MRI |
| 2020-2021 | 78-80 | Rectal cancers | MRI and CT |
| 2019-2021 | 74-77 | Lung cancer, Pancreatic cancer, Colorectal liver metastases | CT and CE-CT |
| 2015-2021 | 54-57, 66-70 | Glioma, Colorectal cancer, Cervical cancer, Nasopharyngeal carcinoma, Lung cancer | Subregion |
MRI
MRI can be classified into structural MRI, functional MRI, diffusion-weighted imaging (DWI), susceptibility-weighted imaging (SWI), and other medical and physiological applications. Different types of MRIs have different pathological and physiological characteristics. Therefore, MRI has received considerable attention in radiomics research, especially for precise diagnosis (32-34), treatment response prediction (35-37), molecular typing (38-40), and prognostic analysis (41,42).
Compared with models built by radiomics features only from structural MRI, it has been found that combining structural MRI with other MRI sequences can build better models. For example, in gliomas, radiomic features can be extracted from T1 contrast enhancement (T1CE), T2-weighting (T2WI), and apparent diffusion coefficient (ADC). The nomogram established by integrating the radiomics features extracted from these three MRIs had the best predictive value, which was higher than that established by any single sequence (26). Combining six types of MRI sequences (T2WI, DWI, ADC, fractional anisotropy, and mean kurtosis) for radiomics can accurately distinguish glioblastoma and low-grade glioma with an accuracy rate as high as 91% (43). A model for determining glioma grade was constructed with radiomics features from seven different sequences (T1-weighted (T1WI), T1CE, T2WI, FLAIR, diffusion tensor imaging (DTI), diffusion perfusion imaging, and 1H-MR spectra) achieving 95.5% accuracy, 95% sensitivity, 96% specificity, and 95.5% AUC (44). Pseudo-progression is a diagnostic challenge for glioblastoma after early treatment, and AUC of predictive model built by 12 radiomics features from T1CE, FLAIR, ADC, and cerebral blood volume (CBV) mapping is 0.90, which is much higher than those built using only features from conventional sequences (AUC=0.76), ADC (AUC=0.78), and CBV (AUC=0.8) (45). It is widely known that IDH mutations play a pivotal role in gliomas. The AUC of the model for predicting IDH mutation status using conventional MRI radiomics features was 0.835, which increased to 0.9 after combining the radiomics features of DTI (46). In another radiomics study to predict IDH mutations, the model achieved an accuracy of 0.823 by using radiomics features from T1CE, T2WI, and ASL images (47). The AUC of the nomogram to predict IDH mutations using T1CE, FLAIR, and ADC radiomics features combined with age was 0.913 (48). In addition, other key signaling pathways can also be assessed using MRI radiomics. Using radiomics features of conventional MRI and DWI, the AUC of the model predicting RTK in glioma was 0.88, TP53 was 0.76, and RB1 was 0.81, which was greater than that of the model using only conventional MRI (49).
In addition to gliomas, the combination approach has been widely used for other tumors. The radiomics features from T2WI, T1WI, diffusion kurtosis imaging (DKI), ADC, and dynamic contrast-enhanced (DCE) pharmacokinetic parameter maps were used to build breast cancer diagnosis models, and the best model (combination of T2WI, DKI, and DCE) AUC was 0.921, accuracy was 0.833, and the AUCs of the model based on single T1WI, T2WI, ADC, DKI, and DCE pharmacokinetic parameter maps were 0.730, 0.791, 0.770, 0.788, and 0.836, respectively (50). To predict whether breast cancer will produce a pathologic complete response to neoadjuvant chemotherapy, a prediction model jointly built using the radiomics features of T2WI, DWI, and T1CE had an AUC of 0.79 in the training set and reached 0.86 in the validation set (51). In prostate cancer, the need for biopsy at PSA levels of 4-10 ng/mL is a clinical question, and a prediction model built using radiomic features extracted from T2WI, DWI, and T1CE achieved an AUC of 0.956 in the training set and 0.933 in the validation set (52). Good performance was obtained for models predicting prostate cancer with a Gleason score of 8 or higher (AUC = 0.72), built from radiomics features extracted from T2WI, ADC, and DKI; better performance was also obtained for models predicting prostate cancer with a Decipher score of 0.6 or higher (AUC 0.84) (53).
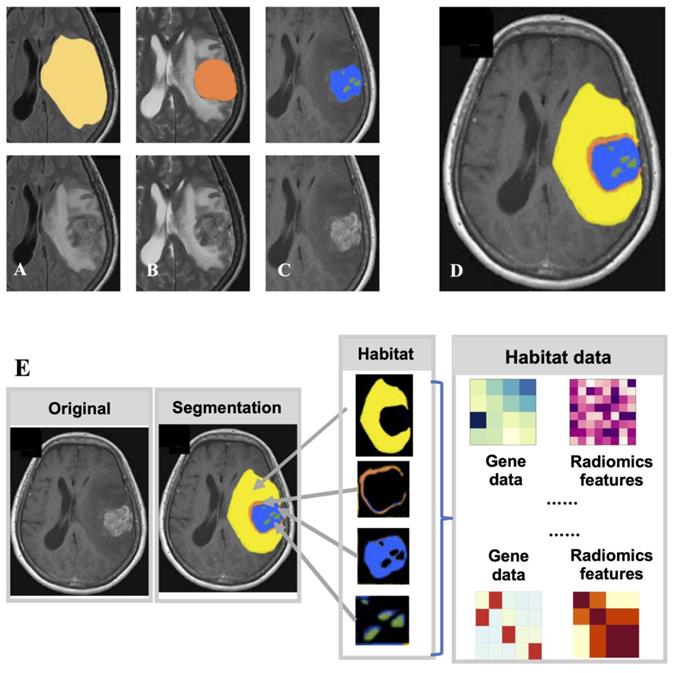
Because of the unique biophysical principles that control signal generation, MRI sequences provide a non-invasive way of physiologically reflecting different local microenvironments. Solid tumors are genetically and physiologically heterogeneous. Different physiological regions exert different selective pressures, resulting in the growth of tumor cell clones with an adapted genome/proteome. These regions are also called "Habitats" (27), and research on radiomics has gradually shifted from the entire tumor area to the habitats (Fig. 3). T1WI, FLAIR, and ADC can divide the ROI into two habitats, the tumor and the edema around the tumor, and then extract radiomics features from the two habitats. The AUC of the glioma MGMT methylation prediction model constructed using the combination of radiomics features from the two habitats reached 0.925 in the training set and 0.902 in the validation set (54). To predict the main pathological growth pattern of liver metastases from colorectal cancer, radiomics features were extracted from multiple MRI sequences (T1WI, T2WI, DWI, ADC, and T1CE) from the tumor-liver interface region, i.e., the region between the tumor margin expanding outward by 2 mm and shrinking inward by 2 mm, and the prediction model incorporating these radiomics features had an AUC of 0.912. superior to the model constructed from radiomics features extracted from tumor regions (AUC=0.879) (55). A total of nine MRI sequences of sagittal T2WI, axial T1WI, axial T2-FS, DWI (b=0 and b=800), ADC, and T1CE (sagittal, axial, and coronal) in advanced cervical cancer were manually outlined by the radiologists, the radiomics features were extracted, and the AUC of the model constructed by combining the radiomics features of multiple sequences reached 0.82 (56). To assess the survival risk of glioblastoma and predict its progression-free survival, T1CE, T1WI, and FLAIR sequences were used to delineate and annotate the necrotic core, enhancement part, and surrounding edema of glioblastoma. Radiomics features were extracted from these features, which can be used to accurately stratify the survival risk of patients with glioblastoma (57).
PET
Cancer cells usually receive more imaging agents, resulting in malignant tumors appearing as bright spots on PET. The amount of imaging agent taken by a malignant tumor depends on the metabolic level of the malignant tumor; therefore, PET radiomics is also called metabolic radiomics.
PET is usually combined with CT, the most common clinical practice is PET-CT. Using the radiomics features from PET and CT in patients with suspected lung cancer, the AUC of the established predictive nomogram was 0.96, which is far higher than the 0.79 of only CT radiomics features, and 0.93 of only PET radiomics features (58). For accurate differentiation between active pulmonary tuberculosis and lung cancer, three radiomics signatures were constructed. The PET radiomics signature yielded AUC of 0.79 (95 % CI: 0.67-0.86]; the CT radiomics signature displayed higher discrimination performance relative to the PET radiomics signature (AUC, 0.86; 95 % CI, 0.79-0.91); the PET/CT radiomics signature incorporating both PET and CT radiomics features further improved the discriminatory ability with the highest AUC (0.91, 95 % CI, 0.84-0.95) (59). To distinguish squamous cell carcinoma from adenocarcinoma of lung, the combined model, comprising 7 PET radiomics and 3 CT radiomic parameters, had the highest predictive efficiency and clinical utility in predicting the non-small cell lung cancer subtypes compared to the use of these parameters alone in both the training and validation sets (AUCs (95% CIs) = 0.932 (0.900-0.964), 0.901 (0.840-0.957), respectively) (p < 0.05) (60). For prediction of the prognosis of patients with NPC, combining PET and CT features with clinical parameters showed equal or higher prognostic performance than models with PET, CT, or clinical parameters alone (C-index 0.71-0.76 vs. 0.67-0.73 and 0.62-0.75 vs. 0.54-0.75 for training and validation cohorts, respectively), while the prognostic performance was significantly improved in the locally advanced regional cohort (C-index 0.67-0.84 vs. 0.64-0.77, p value 0.001-0.059) (61). To predict HPV status in oropharyngeal squamous cell carcinoma, radiomic features were extracted from primary tumor lesions and metastatic cervical lymph nodes on PET and non-contrast CT scans. Single PET or CT models yielded similar classification performance without significant difference in independent validation; however, models combining PET and CT features outperformed single-modality PET- or CT-based models, with AUC of 0.78, and 0.77, respectively, in cross-validation and independent validation, respectively (62).
Radiomics research based on subregion of tumor generated by multimodal medical image. (A) is FLAIR, from which the edematous part (yellow), (B) is T2, from which the overall tumor core can be seen, and (C) is T1CE, from which the enhancing part (blue) and necrotic part (green) of the tumor core can be obtained. (D) After superimposing the labels together, the final habitat segmentation results can be obtained, where the non-enhanced part of the tumor core (red) is the overall tumor core minus the enhanced and necrotic parts. (E) After segmenting the tumor area into different subregions, radiomic studies were performed from each subregion.
Compared to PET/CT combinations, PET/MRI combinations emerged much later and are currently less used; however, some progress has been made in dual-modality radiomics studies of PET and MRI. To investigate the potential of combined textural feature of contrast-enhanced MRI (CE-MRI) and static O-(2-[18F]fluoroethyl)-L-tyrosine (FET) PET for differentiating between local recurrent brain metastasis and radiation injury, radiomic features were extracted from above two imagines individually. CE-MRI texture features had a diagnostic accuracy of 81% (sensitivity, 67%; specificity, 90%). FET PET textural features revealed a slightly higher diagnostic accuracy of 83% (sensitivity, 88%; specificity, 75%). However, the highest diagnostic accuracy was obtained CE-MRI and FET PET features were combined (accuracy, 89%; sensitivity, 85%; specificity, 96%) (63). Approximately 15-30% of locally advanced rectal cancers (LARC) will produce a pathologic complete response (pR) to neoadjuvant therapy. To predict whether LARC responds to neoadjuvant therapy, radiomic features were extracted from PET and MRI. A model containing 6 radiomics features (5 from PET and 1 from MRI) yielded the highest predictability in distinguishing between pR+ and pR- patients (AUC = 0.86; sensitivity = 0.86; sensitivity = 86%, and specificity = 83%) (64). For prediction of hormone receptor status and proliferation rate in breast cancer, a model constructed by combining radiomics features of PET and MRI achieved the best results for AUC (estrogen receptor, 0.87; progesterone receptor, 0.88; Ki-67, 0.997) (65).
As mentioned above, tumors are highly heterogeneous, and this heterogeneity can be reflected in PET images. Their general assessments are made using the intratumoral heterogeneity index (SUVmax/SUVmean) which can help predict more accurately the prognosis of patients with tumors (66, 67). In addition, similar to MRI, PET can be used to segment tumors into subregions (“Habitats”) with different physiological characteristics based on imaging principles. The radiomics features from the subregions retained the heterogeneous information of the region and avoided the loss of this heterogeneous information. To predict PFS in patients with NPC, each tumor in the PET/CT imaging was partitioned into several phenotypically consistent subregions. For each subregion, 202 radiomic features were extracted to construct the models. Three subregions (denoted as S1, S2, and S3) with distinct PET/CT imaging characteristics were identified. The prognostic performance of the model from S3 outperformed the model from the whole tumor (C-index, 0.69 vs. 0.58; log-rank test, p < 0.001 vs. p = 0.552) (68). To preoperatively discriminate between non-small cell lung cancer and benign inflammatory diseases, PET/CT was separated into variant subregions based on the adapted clustering method. The AUC of the subregion-based PET/CT radiomics models was 0.7270 ± 0.0147, which showed a significantly improved discrimination performance compared to conventional methods (p <.001) (69). Radiomic features were extracted from whole PET, CT, and subregions to estimate the prognosis of patients with locally advanced cervical cancer treated with chemoradiotherapy. The radiomics signatures that included habitat features achieved significantly higher C-indexes of 0.78 and 0.76 for PFS estimation, and 0.83 and 0.78 for OS estimation in the training and test cohorts, respectively, compared with radiomics signatures without habitat features with C-indexes of 0.72 and 0.68 for PFS estimation (P = .004, z test). and 0.79 and 0.72 for OS estimation (P = .048, z test) (70).
CT
CT reflects the spatial distribution of tissue strength in the detected area. It is the most commonly used medical radiological examination in clinical practice. Radiomics of CT in cancer has been extensively researched (71-73).
Recently, radiomics studies combining plain CT with contrast-enhanced CT (CE-CT) have achieved better results than plain CT alone. When using CE-CT and CT plain scans to predict EGFR mutation status in patients with non-small cell lung cancer, the general radiomics signature (CT+CE-CT) yielded the highest AUC of 0.756 and 0.739 in the two test sets, and the performance of the general radiomics signature was always similar to or higher than that of models built using only CT or CE-CT features (74). CE-CT can be divided into different phases, which contain different tumor information. To develop and validate a radiomics-based nomogram for preoperatively predicting grade 1 and grade 2/3 tumors in patients with pancreatic NETs, radiomic features were extracted from arterial phase (AP) and portal venous phase (PVP) CT images. The fusion radiomic signature (CT+CE-CT) achieved optimal performance in both the training (AUC 0.970; 95% CI 0.943-0.997) and validation (AUC 0.881; 95% CI 0.760-1) cohorts (75). To predict histopathologic growth patterns (HGPs) in colorectal liver metastases (CRLMs) using a radiomics model, radiomic features were extracted from the AP and PVP CT images. The phase-fused radiomics signature demonstrated the best predictive performance for distinguishing between replacement and desmoplastic HGPs (AUCs of 0.926 and 0.939 in the training and external validation cohorts, respectively) (76). A total of 384 radiomics features were extracted from AP or PVP images to preoperatively discriminate pancreatic ductal adenocarcinoma (PDAC) in stages I-II and III-IV, and predict overall survival. The AP+PVP radiomics signature showed the best performance among the three radiomics signatures (training cohort: AUC = 0.919; validation cohort: AUC = 0.831) (77).
In clinical practice, CT and MRI are the most commonly used imaging modalities. The performance of combined CT and MRI radiomics is usually better than that of either alone. To predict the treatment response to neoadjuvant chemotherapy for LARC, radiomics features were extracted from CT, ADC, T1CE, and high-resolution T2-weighted imaging (HR-T2WI). For an individual sequence, the HR-T2WI model performed better (AUC = 0.859, ACC = 0.896) than CT (AUC = 0.766, ACC = 0.792), T1CE (AUC = 0.812, ACC = 0.854), and ADC (AUC = 0.828, ACC = 0.833) models in the validation set; the combined radiomics model (AUC = 0.908, ACC = 0.812) had a better performance than the individual T1CE, HR-T2WI, and ADC models (78). To build a predictive model of lymphatic vascular infiltration (LVI) in rectal cancer, radiomic features were extracted from T2WI, DWI, and CE-CT. The combined model achieved the highest AUC of all single-modal models (AUC = 0.884 and 0.876, respectively), with high sensitivity and specificity in the training and validation cohorts (sensitivity = 0.938 and 0.929, specificity = 0.727 and 0.800, respectively) (79). To predict pCR in patients with rectal cancer after neoadjuvant treatment, a radiomics model based on pretreatment CT was built first. Even the CT radiomics model yielded the highest AUCs of 0.997 [95% CI 0.990-1.000] in the primary cohort and 0.822 [95% CI, 0.649-0.995] in the validation cohort. When the MRI-based radiomics signature was added to the previous CT radiomics model, the performance of the integrated model (CT-MRI) was significantly better than that of CT (P = 0.005) or MRI (P = 0.003) alone (AIC: 75.49 vs. 81.34 vs. 82.39%) (80).
CT is often combined with PET for radiomic studies of tumor subregions, as summarized in the PET section above. Theoretically, CT and CE-CT, or CT and MRI could also be combined with each other for tumor subregion studies, but to the best of our knowledge, we have not found any literature on this yet.
Discussion
By combining multimodal images, radiomics can obtain more comprehensive image information and better radiomic models, and even segment "subregions" with different biological characteristics and perform more refined radiomic analysis based on these subregions. However, it must be noted that the current tumor genomics data, both in public databases and clinical practice, are derived from single-site sampling of tumors (Fig. 4). The genomic data obtained inherently ignore the heterogeneity of tumors and have the potential for substantial bias (81). The true efficacy of radiomics models trained using biased data is questionable. For example, in a radiomics study to predict the presence of IDH mutations in gliomas, if the labels in a supervised learning model are incorrect because of the genetic heterogeneity of the gliomas, then the model is bound to be unreliable. Tumor heterogeneity will also indirectly affect the study of a range of factors, such as treatment response, prognosis, and tumor recurrence. An ideal way to solve this problem is to obtain heterogeneous genomic data of the tumor. However, it is unrealistic based on technology and costs. Therefore, a method to obtain the genomic heterogeneity of tumors as realistically as possible is an unavoidable challenge in the continued development of future radiomics. Medical images can analyze different subregions to inform tissue biopsy sites (82). The more accurate conclusions can be drawn by analyzing genomic information sampled from different tumor subregions. The reliability of different subregions in radiomics can be further verified using genomic data obtained from different subregions. However, to date, there is no public database with both genomic and imaging data of tumor subregions.
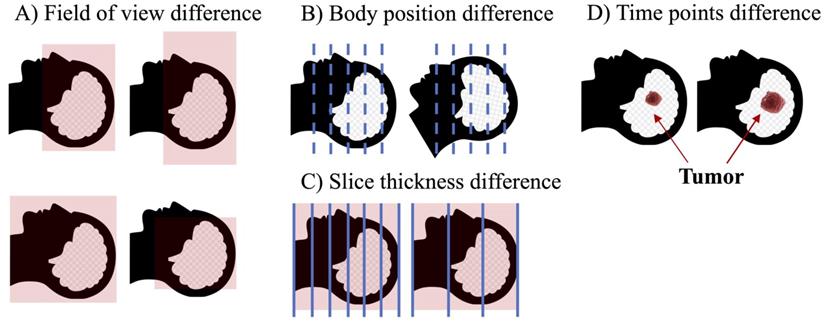
Although the combination of multimodal images has certainly helped the advancement of radiomics, the implementation process still has many problems. For example, for studies that require the combination of CT and MRI, it is likely that medical images of different modalities will have different spatial structure because they usually come from different points in time, and the patient's position, field of view of the device, and number of slices scanned may differ, even if the patient is the same. In this case, it is necessary to align medical images of different modalities (Fig 5). However, although many image alignment tools have been developed, they do not always yield ideal alignment results (83,84), especially for diseases such as tumors, which can cause significant changes in normal anatomy. Cancer-specific tumor alignment tools have been used to address this problem (85), but there are so many different cancer types that the cost of this approach remains exorbitant. Development of a universal and reliable registration tool is a possible solution to this problem.
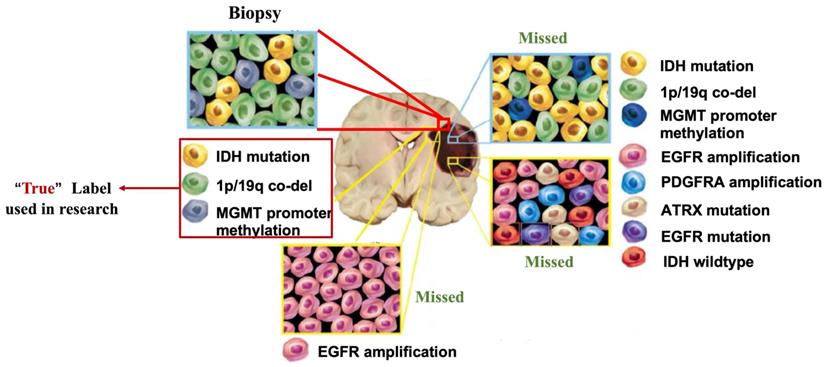
Tumor heterogeneity can lead to bias in genomics data. Tumors are highly heterogeneous, with cells of different genotypes in different subregions. “Biopsy”: one site can only obtain the genotypes of cells in a localized region of the tumor. “Missed”: The genotypes of other parts of the tumor are lost. However, most of the current studies have used this biased data as a "true" label.
Factors leading to differences in the spatial structure of medical images of different modalities. (A) Field of view may be different for different modality images at the time of examination. (B) Images of different modalities are often not completed simultaneously, and the patient's position cannot be guaranteed to be identical during multiple imaging examination. (C) Images of different modalities usually have different slice thicknesses. (D) The same patient may be imaged at different times, which may result in structural changes for some rapidly progressing tumors.
In addition, multimodal images also indicate that the researcher must outline the ROIs for multiple images. For researchers who manually outline ROIs, this workload has undoubtedly increased by several times. Even for researchers who use or develop their own semi-automatic or fully automatic segmentation tools, more effort is needed to develop different segmentation tools, as a single segmentation tool is unlikely to cope with images of different modalities. Radiomics studies of multimodal images require more reliable segmentation. The current segmentation for multimodal images involves multiple iterations of the segmentation of a single-modal image (86). Therefore, each error in multimodal image segmentation magnifies the error in the final segmentation result (87). Such errors have a greater impact on “subregion” based imaging studies, where even small segmentation errors may determine the presence or absence of certain “subregions.” Addressing this issue should be a future focus in radiomics.
To verify the capabilities of radiomics and expand its application range, “big” data with a large number of various data types are required (88). Powerful public databases such as TCIA (89) and TCGA (90) are effective solutions for this. However, the implementation of big data is difficult. The data in public databases come from different institutions, such as TCIA, resulting in variable quality of their data information. While big data can draw reliable conclusions from relatively "dirty" data, in the case of radiomics, the different parameters of the image acquisition or the noise in the image can cause serious interference with the radiomics features (91,92), which will inevitably affect the model's ability for generalization of other databases (93). To solve this problem, it is necessary to develop worldwide database access standards. In addition, it is necessary to utilize this standard to clean the data in existing public databases. However, despite the existence of public databases, the amount of data in these databases is extremely limited. Although many institutions have a large amount of image data, they are still independent of each other. Making use of all data from all sources can greatly expand the number of samples and produce more reliable results. This can be accomplished by facilitating the federation of medical centers, allowing them to request access to each other's data, or by commissioning a trusted third-party organization that can manage the data of all participating institutions. Although it is costly and difficult to unite institutions or find a third party to build a high-quality public database, the benefits are enormous.
The relationship between radiomics and clinical symptoms has been widely documented, and correlations between radiomics and other data types, such as genomics, transcriptomics, proteomics, and metabolomics, are major directions for future development. The correlation between radiomics and genomics has been extensively studied in cancers. Radiomics features can also be used to represent transcriptomics. Research on the combination of radiomics with proteomics and metabolomics is just beginning, and more effort is required. The combined research of radiomics and another kind of omics, the combined research of multiple omics, is an area worthy of future research. However, omics data are independent of each other, that is, samples with genomic data may not have radiomics data, or samples may have radiomics data but lack transcriptomic data. A high-quality multi-omics database is a good way to break this isolation, but the cost is huge.
Conclusions
In this review, we summarized the current advances and hotspots in radiomics research. Multimodality/multisequence medical image fusion radiomics and habitat-based radiomics research fully consider the uniqueness of each medical imaging modality, compensate for the shortcomings of omitted information in previous radiomics studies, and greatly improve the prediction accuracy of radiomics, which is a future development trend. With the improvement in radiomics technology, expansion of public databases, and advancement of deep learning algorithms, radiomics will definitely play an important role in future clinical diagnosis, treatment, and prognosis.
Abbreviations
CT, computed tomography; MRI, magnetic resonance imaging; PET, positron emission tomography; ROI, region of interest; DWI, diffusion-weighted imaging; SWI, susceptibility-weighted imaging; T1CE, T1 contrast enhancement; T2WI, T2-weighting; ADC, apparent diffusion coefficient; CBV, cerebral blood volume; DKI, diffusion kurtosis imaging; DCE, dynamic contrast-enhanced; CE-MRI, contrast-enhanced MRI; FET PET, O-(2-[18F]fluoroethyl)-L-tyrosine (FET) PET; LARC, locally advanced rectal cancers; HGPs, histopathologic growth patterns; CRLMs, colorectal liver metastases; AP, arterial phase; PVP, portal venous phase; PDAC, pancreatic ductal adenocarcinoma; HR-T2WI, high-resolution T2-weighted imaging; RTK, receptor tyrosine kinase.
Acknowledgements
Funding
This Research was funded by the National Natural Science Foundation of China (82071285, 82171118), Key Research & Development Plan of Zhejiang Province (No.2019C030340), The China Postdoctoral Science Foundation funded project (2021M693567, 2021TQ0374), Hunan Postdoctoral Program for Innovative Talent (2021RC2017), Natural Science Foundation of Zhejiang Province (GF22H168636) and Natural Science Foundation of Hunan Province (2021JJ41027).
Availability of data and materials
All relevant data are provided in the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bates SE. It's all about the test: the complexity of companion diagnostic co-development in personalized medicine. Clin Cancer Res. 2014;20:1418
2. Karsy M, Guan J, Cohen AL, Jensen RL, Colman H. New Molecular Considerations for Glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep. 2017;17:19
3. Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV. et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372:2499-2508
4. Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71:1319-1325
5. Godet I, Gilkes DM. BRCA1 and BRCA2 mutations and treatment strategies for breast cancer. Integr Cancer Sci Ther. 2017;4:doi 10.15761/ICST.1000228
6. Kotsopoulos J. BRCA Mutations and Breast Cancer Prevention. Cancers. 2018;10:doi 10.3390/cancers10120524
7. An Z, Aksoy O, Zheng T, Fan Q-W, Weiss WA. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene. 2018;37:1561-1575
8. Saadeh FS, Mahfouz R, Assi HI. EGFR as a clinical marker in glioblastomas and other gliomas. Int J Biol Markers. 2018;33:22-32
9. Reardon DA, Wen PY. Glioma in 2014: unravelling tumour heterogeneity-implications for therapy. Nat Rev Clin Oncol. 2015;12:69-70
10. Ji B, Wang S, Liu Z, Weinberg BD, Yang X, Liu T, Wang L, Mao H. Revealing hemodynamic heterogeneity of gliomas based on signal profile features of dynamic susceptibility contrast-enhanced MRI. Neuroimage Clin. 2019;23:101864
11. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, Zegers CML, Gillies R, Boellard R, Dekker A. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-446
12. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJWL, Dekker A, Fenstermacher D. et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234-1248
13. Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006
14. Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RTH, Hermann G, Lambin P, Haibe-Kains B, Mak RH, Aerts HJWL. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345-350
15. Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, Aldape K, Cha S, Kuo MD. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105:5213-5218
16. Gevaert O, Xu J, Hoang CD, Leung AN, Xu Y, Quon A, Rubin DL, Napel S, Plevritis SK. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data-methods and preliminary results. Radiology. 2012;264:387-396
17. Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Jänne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A. et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442-2449
18. Burstein HJ, Harris LN, Marcom PK, Lambert-Falls R, Havlin K, Overmoyer B, Friedlander RJ Jr, Gargiulo J, Strenger R, Vogel CL. et al. Trastuzumab and vinorelbine as first-line therapy for HER2-overexpressing metastatic breast cancer: multicenter phase II trial with clinical outcomes, analysis of serum tumor markers as predictive factors, and cardiac surveillance algorithm. J Clin Oncol. 2003;21:2889-2895
19. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577
20. Clauson J, Hsieh YC, Acharya S, Rademaker AW, Morrow M. Results of the Lynn Sage Second-Opinion Program for local therapy in patients with breast carcinoma. Changes in management and determinants of where care is delivered. Cancer. 2002;94:889-894
21. Nguyen PL, Schultz D, Renshaw AA, Vollmer RT, Welch WR, Cote K, D'Amico AV. The impact of pathology review on treatment recommendations for patients with adenocarcinoma of the prostate. Urol Oncol. 2004;22:295-299
22. Lohmann P, Lerche C, Bauer EK, Steger J, Stoffels G, Blau T, Dunkl V, Kocher M, Viswanathan S, Filss CP. et al. Predicting IDH genotype in gliomas using FET PET radiomics. Sci Rep. 2018;8:13328
23. Liu X, Li Y, Li S, Fan X, Sun Z, Yang Z, Wang K, Zhang Z, Jiang T, Liu Y. et al. IDH mutation-specific radiomic signature in lower-grade gliomas. Aging. 2019;11:673-696
24. Wu S, Meng J, Yu Q, Li P, Fu S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J Cancer Res Clin Oncol. 2019;145:543-550
25. Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, Liu Y, Gevaert O, Wang K, Zhu Y. et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53:doi 10.1183/13993003.00986-2018
26. Wang Q, Li Q, Mi R, Ye H, Zhang H, Chen B, Li Y, Huang G, Xia J. Radiomics Nomogram Building from Multiparametric MRI to Predict Grade in Patients with Glioma: A Cohort Study. J Magn Reson Imaging. 2019;49:825-833
27. Jardim-Perassi BV, Huang S, Dominguez-Viqueira W, Poleszczuk J, Budzevich MM, Abdalah MA, Pillai SR, Ruiz E, Bui MM, Zuccari DAPC. et al. Multiparametric MRI and Coregistered Histology Identify Tumor Habitats in Breast Cancer Mouse Models. Cancer Res. 2019;79:3952-3964
28. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762
29. Song G, Han J, Zhao Y, Wang Z, Du H. A Review on Medical Image Registration as an Optimization Problem. Curr Med Imaging Rev. 2017;13:274-283
30. Zhang H-X, Sun Z-Q, Cheng Y-G, Mao G-Q. A pilot study of radiomics technology based on X-ray mammography in patients with triple-negative breast cancer. J Xray Sci Technol. 2019;27:485-492
31. Hu H-T, Wang Z, Huang X-W, Chen S-L, Zheng X, Ruan S-M, Xie X-Y, Lu M, Yu J, Tian J. et al. Ultrasound-based radiomics score: a potential biomarker for the prediction of microvascular invasion in hepatocellular carcinoma. Eur Radiol. 2019;29:2890-2901
32. Corino VDA, Montin E, Messina A, Casali PG, Gronchi A, Marchianò A, Mainardi LT. Radiomic analysis of soft tissues sarcomas can distinguish intermediate from high-grade lesions. J Magn Reson Imaging. 2018;47:829-840
33. Ortiz-Ramon R, Larroza A, Arana E, Moratal D. A radiomics evaluation of 2D and 3D MRI texture features to classify brain metastases from lung cancer and melanoma. Conf Proc IEEE Eng Med Biol Soc. 2017;2017:493-496
34. Han Y, Yang Y, Shi Z-S, Zhang A-D, Yan L-F, Hu Y-C, Feng L-L, Ma J, Wang W, Cui G-B. Distinguishing brain inflammation from grade II glioma in population without contrast enhancement: a radiomics analysis based on conventional MRI. Eur J Radiol. 2020;134:109467
35. Lopez CJ, Nagornaya N, Parra NA, Kwon D, Ishkanian F, Markoe AM, Maudsley A, Stoyanova R. Association of Radiomics and Metabolic Tumor Volumes in Radiation Treatment of Glioblastoma Multiforme. Int J Radiat Oncol Biol Phys. 2017;97:586-595
36. Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, Plecha D, Madabhushi A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19:57
37. Liu Z, Zhang X-Y, Shi Y-J, Wang L, Zhu H-T, Tang Z, Wang S, Li X-T, Tian J, Sun Y-S. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin Cancer Res. 2017;23:7253-7262
38. Lu C-F, Hsu F-T, Hsieh KL-C, Kao Y-CJ, Cheng S-J, Hsu JB-K, Tsai P-H, Chen R-J, Huang C-C, Yen Y. et al. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin Cancer Res. 2018;24:4429-4436
39. Yu J, Shi Z, Lian Y, Li Z, Liu T, Gao Y, Wang Y, Chen L, Mao Y. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol. 2017;27:3509-3522
40. Li Y, Liu X, Xu K, Qian Z, Wang K, Fan X, Li S, Wang Y, Jiang T. MRI features can predict EGFR expression in lower grade gliomas: A voxel-based radiomic analysis. Eur Radiol. 2018;28:356-362
41. Zhang B, Tian J, Dong D, Gu D, Dong Y, Zhang L, Lian Z, Liu J, Luo X, Pei S. et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clin Cancer Res. 2017;23:4259-4269
42. Zhang B, He X, Ouyang F, Gu D, Dong Y, Zhang L, Mo X, Huang W, Tian J, Zhang S. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017;403:21-27
43. Takahashi S, Takahashi W, Tanaka S, Haga A, Nakamoto T, Suzuki Y, Mukasa A, Takayanagi S, Kitagawa Y, Hana T. et al. Radiomics Analysis for Glioma Malignancy Evaluation Using Diffusion Kurtosis and Tensor Imaging. Int J Radiat Oncol Biol Phys. 2019;105:784-791
44. Vamvakas A, Williams SC, Theodorou K, Kapsalaki E, Fountas K, Kappas C, Vassiou K, Tsougos I. Imaging biomarker analysis of advanced multiparametric MRI for glioma grading. Phys Med. 2019;60:188-198
45. Kim JY, Park JE, Jo Y, Shim WH, Nam SJ, Kim JH, Yoo R-E, Choi SH, Kim HS. Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol. 2019;21:404-414
46. Park CJ, Choi YS, Park YW, Ahn SS, Kang S-G, Chang J-H, Kim SH, Lee S-K. Diffusion tensor imaging radiomics in lower-grade glioma: improving subtyping of isocitrate dehydrogenase mutation status. Neuroradiology. 2020;62:319-326
47. Peng H, Huo J, Li B, Cui Y, Zhang H, Zhang L, Ma L. Predicting Isocitrate Dehydrogenase. IDH; Mutation Status in Gliomas Using Multiparameter MRI Radiomics Features. J Magn Reson Imaging. 2020 doi:10.1002/jmri.27434
48. Tan Y, Zhang S-T, Wei J-W, Dong D, Wang X-C, Yang G-Q, Tian J, Zhang H. A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur Radiol. 2019;29:3325-3337
49. Park JE, Kim HS, Park SY, Nam SJ, Chun S-M, Jo Y, Kim JH. Prediction of Core Signaling Pathway by Using Diffusion- and Perfusion-based MRI Radiomics and Next-generation Sequencing in Isocitrate Dehydrogenase Wild-type Glioblastoma. Radiology. 2020;294:388-397
50. Zhang Q, Peng Y, Liu W, Bai J, Zheng J, Yang X, Zhou L. Radiomics Based on Multimodal MRI for the Differential Diagnosis of Benign and Malignant Breast Lesions. J Magn Reson Imaging. 2020;52:596-607
51. Liu Z, Li Z, Qu J, Zhang R, Zhou X, Li L, Sun K, Tang Z, Jiang H, Li H. et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin Cancer Res. 2019;25:3538-3547
52. Qi Y, Zhang S, Wei J, Zhang G, Lei J, Yan W, Xiao Y, Yan S, Xue H, Feng F. et al. Multiparametric MRI-Based Radiomics for Prostate Cancer Screening with PSA in 4-10 ng/mL to Reduce Unnecessary Biopsies. J Magn Reson Imaging. 2020;51:1890-1899
53. Hectors SJ, Cherny M, Yadav KK, Beksaç AT, Thulasidass H, Lewis S, Davicioni E, Wang P, Tewari AK, Taouli B. Radiomics Features Measured with Multiparametric Magnetic Resonance Imaging Predict Prostate Cancer Aggressiveness. J Urol. 2019;202:498-505
54. Wei J, Yang G, Hao X, Gu D, Tan Y, Wang X, Dong D, Zhang S, Wang L, Zhang H. et al. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur Radiol. 2019;29:877-888
55. Han Y, Chai F, Wei J, Yue Y, Cheng J, Gu D, Zhang Y, Tong T, Sheng W, Hong N. et al. Identification of Predominant Histopathological Growth Patterns of Colorectal Liver Metastasis by Multi-Habitat and Multi-Sequence Based Radiomics Analysis. Front Oncol. 2020;10:1363
56. Fang M, Kan Y, Dong D, Yu T, Zhao N, Jiang W, Zhong L, Hu C, Luo Y, Tian J. Multi-Habitat Based Radiomics for the Prediction of Treatment Response to Concurrent Chemotherapy and Radiation Therapy in Locally Advanced Cervical Cancer. Front Oncol. 2020;10:563
57. Beig N, Bera K, Prasanna P, Antunes J, Correa R, Singh S, Saeed Bamashmos A, Ismail M, Braman N, Verma R. et al. Radiogenomic-Based Survival Risk Stratification of Tumor Habitat on Gd-T1w MRI Is Associated with Biological Processes in Glioblastoma. Clin Cancer Res. 2020;26:1866-1876
58. Kang F, Mu W, Gong J, Wang S, Li G, Li G, Qin W, Tian J, Wang J. Integrating manual diagnosis into radiomics for reducing the false positive rate of 18F-FDG PET/CT diagnosis in patients with suspected lung cancer. Eur J Nucl Med Mol Imaging. 2019;46:2770-2779
59. Du D, Gu J, Chen X, Lv W, Feng Q, Rahmim A, Wu H, Lu L. Integration of PET/CT Radiomics and Semantic Features for Differentiation between Active Pulmonary Tuberculosis and Lung Cancer. Mol Imaging Biol. 2021;23:287-298
60. Ren C, Zhang J, Qi M, Zhang J, Zhang Y, Song S, Sun Y, Cheng J. Machine learning based on clinico-biological features integrated 18 F-FDG PET/CT radiomics for distinguishing squamous cell carcinoma from adenocarcinoma of lung. Eur J Nucl Med Mol Imaging. 2021;8:1538-1549
61. Lv W, Yuan Q, Wang Q, Ma J, Feng Q, Chen W, Rahmim A, Lu L. Radiomics Analysis of PET and CT Components of PET/CT Imaging Integrated with Clinical Parameters: Application to Prognosis for Nasopharyngeal Carcinoma. Mol Imaging Biol. 2019;21:954-964
62. Haider S, Mahajan A, Zeevi T, Baumeister P, Reichel C, Sharaf K, Forghani R, Kucukkaya A, Kann B, Judson B, Prasad M, Burtness B, Payabvash S. PET/CT radiomics signature of human papilloma virus association in oropharyngeal squamous cell carcinoma. Eur J Nucl Med Mol Imaging. 2020;47:2978-2991
63. Lohmann P, Kocher M, Ceccon G, Bauer E, Stoffels G, Viswanathan S, Ruge M, Neumaier B, Shah N, Fink G, Langen K, Galldiks N. Combined FET PET/MRI radiomics differentiates radiation injury from recurrent brain metastasis. Neuroimage Clin. 2018;20:537-542
64. Giannini V, Mazzetti S, Bertotto I, Chiarenza C, Cauda S, Delmastro E, Bracco C, Dia A, Leone F, Medico E, Pisacane A, Ribero D, Stasi M, Regge D. Predicting locally advanced rectal cancer response to neoadjuvant therapy with 18 F-FDG PET and MRI radiomics features. Eur J Nucl Med Mol Imaging. 2019;46:878-888
65. Umutlu L, Kirchner J, Bruckmann N, Morawitz J, Antoch G, Ingenwerth M, Bittner A, Hoffmann O, Haubold J, Grueneisen J, Quick H, Rischpler C, Herrmann K, Gibbs P, Domenig K. Multiparametric Integrated 18 F-FDG PET/MRI-Based Radiomics for Breast Cancer Phenotyping and Tumor Decoding. Cancers. 2021;13:2928
66. Gong C, Ma G, Hu X, Zhang Y, Wang Z, Zhang J, Zhao Y, Li Y, Xie Y, Yang Z, Wang B. Pretreatment 18 F-FDG Uptake Heterogeneity Predicts Treatment Outcome of First-Line Chemotherapy in Patients with Metastatic Triple-Negative Breast Cancer. Oncologist. 2018;23:1144-1152
67. Yang Z, Shi Q, Zhang Y, Pan H, Yao Z, Hu S, Shi W, Zhu B, Zhang Y, Hu C. Pretreatment. 18)F-FDG uptake heterogeneity can predict survival in patients with locally advanced nasopharyngeal carcinoma-a retrospective study. Radiat Oncol. 2015;10:4
68. Xu H, Lv W, Feng H, Du D, Yuan Q, Wang Q, Dai Z, Yang W, Feng Q, Ma J, Lu L. Subregional Radiomics Analysis of PET/CT Imaging with Intratumor Partitioning: Application to Prognosis for Nasopharyngeal Carcinoma. Mol Imaging Biol. 2020;22:1414-1426
69. Chen L, Liu K, Zhao X, Shen H, Zhao K, Zhu W. Habitat Imaging-Based 18 F-FDG PET/CT Radiomics for the Preoperative Discrimination of Non-small Cell Lung Cancer and Benign Inflammatory Diseases. Front Oncol. 2021;11:759897
70. Mu W, Liang Y, Hall L, Tan Y, Balagurunathan Y, Wenham R, Wu N, Tian J, Gillies R. 18 F-FDG PET/CT Habitat Radiomics Predicts Outcome of Patients with Cervical Cancer Treated with Chemoradiotherapy. Radiol Artif Intell. 2020;2:e190218
71. Liu A, Wang Z, Yang Y, Wang J, Dai X, Wang L, Lu Y, Xue F. Preoperative diagnosis of malignant pulmonary nodules in lung cancer screening with a radiomics nomogram. Cancer Commun. 2020;40:16-24
72. Fan L, Fang M, Li Z, Tu W, Wang S, Chen W, Tian J, Dong D, Liu S. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol. 2019;29:889-897
73. Beig N, Khorrami M, Alilou M, Prasanna P, Braman N, Orooji M, Rakshit S, Bera K, Rajiah P, Ginsberg J. et al. Perinodular and Intranodular Radiomic Features on Lung CT Images Distinguish Adenocarcinomas from Granulomas. Radiology. 2019;290:783-792
74. Yang X, Liu M, Ren Y, Chen H, Yu P, Wang S, Zhang R, Dai H, Wang C. Using contrast-enhanced CT and non-contrast-enhanced CT to predict EGFR mutation status in NSCLC patients-a radiomics nomogram analysis. Eur Radiol. 2021 Online ahead of print
75. Gu D, Hu Y, Ding H, Wei J, Chen K, Liu H, Zeng M, Tian J. CT radiomics may predict the grade of pancreatic neuroendocrine tumors: a multicenter study. Eur Radiol. 2019;29:6880-6890
76. Cheng J, Wei J, Tong T, Sheng W, Zhang Y, Han Y, Gu D, Hong N, Ye Y, Tian J, Wang Y. Prediction of Histopathologic Growth Patterns of Colorectal Liver Metastases with a Noninvasive Imaging Method. Ann Surg Oncol. 2019;26:4587-4598
77. Cen C, Liu L, Li X, Wu A, Liu H, Wang X, Wu H, Wang C, Han P, Wang S. Pancreatic Ductal Adenocarcinoma at CT: A Combined Nomogram Model to Preoperatively Predict Cancer Stage and Survival Outcome. Front Oncol. 2021;11:594510
78. Li Z, Wang X, Li M, Liu X, Ye Z, Song B, Yuan F, Yuan Y, Xia C, Zhang X, Li Q. Multi-modal radiomics model to predict treatment response to neoadjuvant chemotherapy for locally advanced rectal cancer. World J Gastroenterol. 2020;26:2388-2402
79. Zhang Y, He K, Guo Y, Liu X, Yang Q, Zhang C, Xie Y, Mu S, Guo Y, Fu Y, Zhang H. A Novel Multimodal Radiomics Model for Preoperative Prediction of Lymphovascular Invasion in Rectal Cancer. Front Oncol. 2020;10:457
80. Zhuang Z, Liu Z, Li J, Wang X, Xie P, Xiong F, Hu J, Meng X, Huang M, Deng Y, Lan P, Yu H, Luo Y. Radiomic signature of the FOWARC trial predicts pathological response to neoadjuvant treatment in rectal cancer. J Transl Med. 2021;19:256
81. Kros JM, van Run PR, Alers JC, Avezaat CJ, Luider TM, van Dekken H. Spatial variability of genomic aberrations in a large glioblastoma resection specimen. Acta Neuropathol. 2001;102:103-109
82. Klaeser B, Wiskirchen J, Wartenberg J, Weitzel T, Schmid RA, Mueller MD, Krause T. PET/CT-guided biopsies of metabolically active bone lesions: applications and clinical impact. Eur J Nucl Med Mol Imaging. 2010;37:2027-2036
83. Ou Y, Akbari H, Bilello M, Da X, Davatzikos C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE Trans Med Imaging. 2014;33:2039-2065
84. Lin JS, Fuentes DT, Chandler A, Prabhu SS, Weinberg JS, Baladandayuthapani V, Hazle JD, Schellingerhout D. Performance Assessment for Brain MR Imaging Registration Methods. AJNR Am J Neuroradiol. 2017;38:973-980
85. Gooya A, Pohl KM, Bilello M, Cirillo L, Biros G, Melhem ER, Davatzikos C. GLISTR: glioma image segmentation and registration. IEEE Trans Med Imaging. 2012;31:1941-1954
86. Chaddad A, Kucharczyk MJ, Daniel P, Sabri S, Jean-Claude BJ, Niazi T, Abdulkarim B. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front Oncol. 2019;9:374
87. Traverso A, Wee L, Dekker A, Gillies R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int J Radiat Oncol Biol Phys. 2018;102:1143-1158
88. McNutt TR, Benedict SH, Low DA, Moore K, Shpitser I, Jiang W, Lakshminarayanan P, Cheng Z, Han P, Hui X. et al. Using Big Data Analytics to Advance Precision Radiation Oncology. Int J Radiat Oncol Biol Phys. 2018;101:285-291
89. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M. et al. The Cancer Imaging Archive. TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045-1057
90. Lee H, Palm J, Grimes SM, Ji HP. The Cancer Genome Atlas Clinical Explorer: a web and mobile interface for identifying clinical-genomic driver associations. Genome Med. 2015;7:112
91. Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study. Med Phys. 2009;36:1236-1243
92. Lerski RA, Schad LR, Luypaert R, Amorison A, Muller RN, Mascaro L, Ring P, Spisni A, Zhu X, Bruno A. Multicentre magnetic resonance texture analysis trial using reticulated foam test objects. Magn Reson Imaging. 1999;17:1025-1031
93. Mayo CS, Kessler ML, Eisbruch A, Weyburne G, Feng M, Hayman JA, Jolly S, El Naqa I, Moran JM, Matuszak MM. et al. The big data effort in radiation oncology: Data mining or data farming? Adv Radiat Oncol. 2016;1:260-271
Author contact
![]() Corresponding authors: Weihong Jiang, weihongjiangedu.cn, Renya Zhan, 1196057edu.cn & Jian Shen, 1314006edu.cn
Corresponding authors: Weihong Jiang, weihongjiangedu.cn, Renya Zhan, 1196057edu.cn & Jian Shen, 1314006edu.cn

 Global reach, higher impact
Global reach, higher impact