10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(10):4043-4052. doi:10.7150/ijbs.73616 This issue Cite
Review
Human leukocyte antigens: the unique expression in trophoblasts and their crosstalk with local immune cells
1. Institute of Reproductive Health, Center for Reproductive Medicine, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, P.R. China.
2. Department of Obstetrics, Maternity and Child health care hospital Hubei, Wuhan, PR China.
3. C.S. Mott Center for Human Growth and Development, School of Medicine, Wayne State University, Detroit, MI, USA.
Received 2022-4-4; Accepted 2022-6-1; Published 2022-6-13
Abstract
Trophoblasts differentiate and form the placenta during pregnancy in a complex and finely orchestrated process, which is dependent on the establishment of maternal-fetal immune tolerance and the proper function of trophoblasts. Trophoblasts express HLA-C and non-classical HLA-Ib molecules (HLA-E, HLA-F, and HLA-G). Numerous studies have shown that the unique expression pattern of the HLA molecules is closely linked to the successful acceptance of allogeneic fetus by the mother during pregnancy. However, some controversies still exist concerning the exact expression and recognition patterns of HLA molecules in different trophoblast subpopulations and cell lines. Thus, we summarize three types of trophoblast subpopulations as well as the common trophoblast lineages. Then, the classification and structural characteristics of HLA molecules were elucidated. Finally, the presence of HLA-C and non-classical HLA-Ib molecules (HLA-E, HLA-F, and HLA-G) in various trophoblasts and cell lines, as well as their potential role in establishing and maintaining normal pregnancy were also discussed. Together, this review will help people comprehensively understand the complex immune interactions between maternal and fetal crosstalk during pregnancy and ultimately better understand the physiological and pathological etiologies of pregnancy.
Keywords: pregnancy, cytotrophoblast, syncytiotrophoblast, extravillous trophoblast, HLA-C, HLA-E, HLA-F, HLA-G
Introduction
In 1953, Sir Peter Medawar [1] defined the immunological paradox in pregnancy, whereby the mother would not reject a fetus carrying the father's antigen. The placenta is the interface between the mother and the fetus that separates them anatomically. Nevertheless, trophoblasts, special fetal epithelial cells, have close contact with the maternal immune system. Although it is still a mystery why the maternal immune system does not reject the allogeneic antigens, a preferred immunologic theory has been well recognized, that is the establishment of immune tolerance at the maternal-fetal interface accounts for not attacking the fetus by the maternal immune system [2].
Recent studies have shown that the immunological tolerance state maintained by the maternal immune system during pregnancy is mainly due to the specific human leukocyte antigen (HLA) expression pattern in trophoblasts [3, 4]. These molecules mediate the contact between trophoblasts and maternal immune cells via their receptors. It is well-known that trophoblasts do not express HLA-II class molecules, which are surface markers of strongly immunogenic cells in allograft transplantation [5]. Consequently, the absence of expression of the relevant HLA molecules in trophoblasts that cause allograft rejection may play a key role in long-term fetal survival [6]. Abnormal HLA expression patterns in trophoblasts will lead to pregnancy-related diseases including recurrent miscarriage, preterm delivery, fetal growth restriction, pre-eclampsia, etc. [7].
In this review, we mainly aim to discuss the exact expression pattern of HLA molecules in trophoblasts, their interaction with local immune cells as well as the roles in establishing immune tolerance during normal pregnancy.
Trophoblast types in human placenta
Trophoblasts
A zygote is generated when sperm and egg interact. On the fifth day after fertilization, a preimplantation embryo is created after recurrent cleavage. The inner cell mass eventually develops into the fetus, while the outer trophectoderm proliferates and develops into the placenta. On days 7-9 after fertilization, the outer trophectoderm differentiates into cytotrophoblasts (CTBs), and a deeper, non-proliferative, invasive, multinucleated, thickened mass of primitive syncytium [8]. The primitive syncytium appears to secrete enzymes that digest and loosen the decidua surrounding it, and is present for approximately 7-9 days after initially attaches to the endometrium [8-10]. CTBs are highly proliferative and served as the "stem cells" of the placenta, which can differentiate into the other two types of trophoblasts, namely syncytiotrophoblasts (STBs) and extravillous trophoblast (EVTs) [10-12]. STBs are distinguished by cell fusion to be terminally differentiated and multinucleated, while EVTs are distinguished from CTBs that detach from placental villi and invade into decidua [13].
STBs are located in the outer layer of the placental villi and are in direct contact with maternal blood, forming a barrier between the mother and the fetus [14]. Besides, STBs have endocrine functions, secreting human chorionic gonadotropin, placental prolactin, and pregnancy-specific glycoproteins, and can transport nutrients and oxygen to the developing fetus [15, 16]. EVTs can be divided into endovascular trophoblasts (eEVTs) and interstitial trophoblast (iEVTs) [12, 13]. eEVTs invade the spiral arteries and replace vascular endothelial cells, whereas iEVTs invade the decidual interstitium and eventually form the placental bed in the myometrium [13, 17]. EVTs can interact with the mucosa's local immune cells, reshape spiral arteries, widen previously restricted blood vessels, and boost low-pressure blood flow to the developing placenta, thereby supplying oxygen and nutrients for the growth of the fetus [18].
Trophoblast cell lines
To clarify the role of trophoblasts in establishing maternal-fetal immune tolerance, several human trophoblast cell lines have been set up and widely used. In 1993, Graham et al. [19] established the long-living HTR8/SVneo cell line by introducing the gene-encoding simian virus 40 large T antigen into first-trimester human trophoblasts. In 2005, Feng et al. [20] established a TEV-1 cell line, which stably expressed the human papillomavirus type 16 E6/E7 gene in primary first-trimester trophoblasts via a retroviral vector pLXSN-E6/E7. In 2009, Straszewski-Chavez et al. [21] constructed an early gestational trophoblast cell line Swan 71 by infecting early gestational trophoblasts with human telomerase reverse transcriptase. In addition, several choriocarcinoma cell lines have been established, including the BeWo, JEG3, and JAR cell lines, which are also commonly used to study the properties and functions of trophoblasts [22, 23]. Nonetheless, whether these trophoblast cell lines reflect the properties of primary trophoblasts and the HLA molecule expression patterns are not fully clear and need further investigation. In recent years, a three-dimensional (3D) trophoblast organoid (TO) model [15, 24, 25] and a trophoblast stem cell (TSC) model [26] have also been established, which allows us deeply understand the secrets behind the placental formation.
Classification and structural characteristics of HLA family
The classification of HLA family
The major histocompatibility complex, also known as HLA, is a highly polymorphic gene complex comprising over 200 genes, whose locus is located in the 3Mbp region of the short arm of chromosome 6 [27, 28]. They encode cell surface glycoproteins that can be used to present and recognize self- and non-self-peptides [29]. HLA molecules have been widely studied in transplantation, autoimmune, bacterial, viral infections, and tumor immunotherapy [30]. In addition, the unique expression pattern of non-classical HLA in placenta appears to be the most relevant mechanism for fetus to escape the recognition of maternal immune cells [31].
Usually, HLA is classified into three types based on the function and structure, including HLA-Class I, -Class II, and -Class III molecules [32]. HLA-Class I molecules can be categorized into classical HLA-Ia molecules (HLA-A, HLA-B, and HLA-C) and non-classical HLA-Ib molecules (HLA-E, HLA-F, and HLA-G) [33]. HLA-Class I genes encode commonly expressed proteins participating in antigen presentation, and are ubiquitously expressed on all nucleated cells [34]. HLA-Class II molecules include HLA-DR, HLA-DQ, HLA-DP, HLA-DN, HLA-DM, and HLA-DO [35, 36]. HLA-class II genes encode proteins involved in antigen presentation by the so-called professional antigen-presenting cells, including dendritic cells, macrophages, B cells, and thymic epithelial cells [34, 35]. HLA-Class III molecules cannot be easily classified based on known or assumed functions. HLA-class III genes encode proteins involved in complement activation, hormone synthesis, inflammation, cellular stress, extracellular matrix organization, and immunoglobulin superfamily members [34, 37].
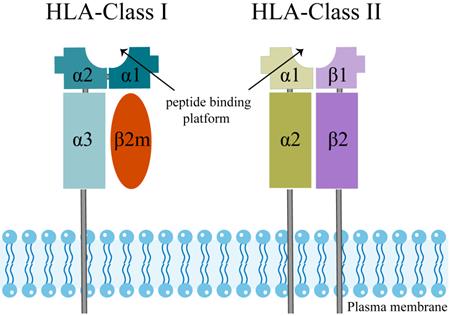
The molecular structure of human leukocyte antigen (HLA) class I and II molecules.
The structural characteristics of the HLA family
The extracellular structural domains of HLA-Class I molecules include heavy chain α1, α2, and α3, where α3 is non-covalently bound to light chain β2-microglobulin (β2M) [38] (Figure 1). The peptide binding platform consists of the α1 and α2 structural domains, while the α3 structural domain serves as a binding site for co-receptors [39, 40]. Besides, all HLA-Class I alpha chains carry a conserved N-glycosylation site, and these glycosylation features directly correlate with the cellular localization of HLA I-like molecules [30].
HLA-Class II molecules have a similar structure to HLA-Class I (Figure 1). The two structural domains, β1 and β2 chains, evolve to form a slightly curved β-fold as a base and two α-helices at the top, which are far enough apart to accommodate a peptide chain [41]. In HLA-Class II molecules, two proximal membrane immunoglobulin (Ig) structural domains support the peptide binding unit, which consists of two structural domains, being the α1 chain and the β1 chain. An Ig domain is present in each chain of HLA-Class II molecules, and a transmembrane helix anchors both chains in the membrane [39, 41]. Trophoblasts do not express the HLA-Class II molecules, which are surface markers of strongly immunogenic cells in allograft transplantation [31]. Therefore, we will not discuss the HLA-Class II molecules below.
Specific expression and function of classical HLA-Ia in trophoblasts
Unlike most nucleated cells, trophoblasts do not express HLA-A and HLA-B molecules. However, trophoblasts can express polymorphic HLA-C [31, 42]. Therefore, in this section, we will focus on summarizing the studies related to the expression and function of HLA-C molecules.
HLA-C in different trophoblasts and cell lines
HLA-C was first discovered in 1970, and it is a highly polymorphic molecule [43]. Although HLA-C is a classical HLA-Ia molecule, it is the only HLA-Ia class molecule expressed on trophoblasts in pregnancy. Most studies did not observe the expression of polymorphic HLA-C in CTBs and STBs by using flow cytometry [44, 45]. However, other study observed that HLA-C can be weakly expressed in the cytoplasm of STBs at 5 gestational weeks and in the nucleus of CTBs at 12 gestational weeks by using immunohistochemistry method [5]. Moreover, the mRNA and protein expression levels of HLA-C in placenta were significantly higher in spontaneous labor than those in non-labored C-section [5]. HLA-C is expressed in all EVTs' population, including eEVTs, iEVTs, and placental bed giant cells [46]. Also, it is expressed in such early pregnancy trophoblasts-derived cell lines as HTR8/SVneo, Swan 71, and TEV-1 [44]. While in the other cell lines, HLA-C was found to express in JEG3 and BeWo, but not in JAR [47-49]. When TO differentiates into EVTs, it can express HLA-C [50].
Function of HLA-C in pregnancy
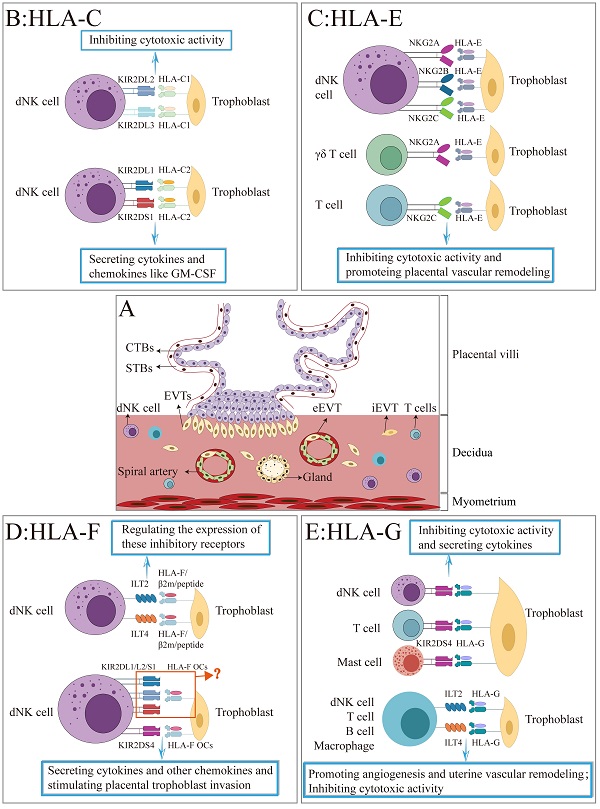
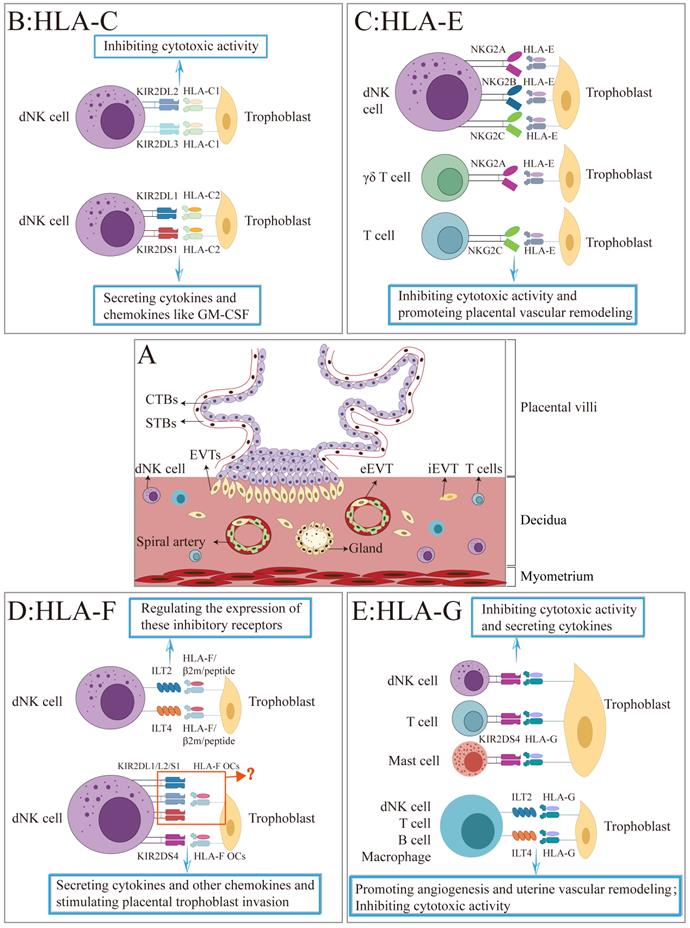
HLA-C is the only classical HLA-Ia molecule identified on trophoblasts. HLA-C can be classified into two types based on C1 or C2 epitopes: HLA-C1 and HLA-C2 [51, 52]; its receptor is the Killer Immunoglobulin-like Receptor (KIR) [53]. KIR is mainly expressed in NK cells, with a small proportion of metaphase T cells expressing KIR [53, 54]. HLA-C1 is a ligand for inhibitory KIR2DL2 and KIR2DL3 and activating KIR2DS2, while HLA-C2 serves as a ligand for activating KIR2DS1 and KIR2DS5 as well as inhibitory KIR2DL1 [55, 56]. It has been found that the inhibitory role of KIR2DL1 with HLA-C2 is more critical, while the weaker inhibitory interaction between HLA-C1 and KIR2DL3 does not seem to have a significant effect on pregnancy outcome [57].
The interaction between HLA-C and inhibitory KIR in NK cells inhibits cytotoxic activity and modulates the secretion of cytokines and growth factors by NK cells, promoting EVTs' invasion and placental vascular remodeling [58]. Sharkey et al [59] found that the percentage of CD56+ cells that expressed KIR in decidual tissues increased, and the mRNA expression levels of KIR also increased in early pregnancy. KIR2DL3, which had the highest level of KIR, led to increased binding of HLA-C1 tetramers to decidual NK (dNK) cells and would increase the production of IFN-γ, which is necessary for normal vascular remodeling and endometrial decidualization in early pregnancy [59]. When the activating receptor KIR2DS1 in dNK cells binds to HLA-C on trophoblasts, it can induce dNK cells to secrete cytokines and chemokines like GM-CSF [60]. The cytokines and chemokines secreted by dNK cells can induce trophoblasts to invade the decidua more effectively, promoting spiral artery remodeling and improving blood supply to the fetus [61]. In addition to its involvement in immune tolerance, HLA-C can directly promote placental growth without interacting with immune cells [62]. In conclusion, HLA-C molecules are critical in regulating the tolerogenic activity of NK cells in the decidua toward the fetus and trophoblast invasion.
Specific expression and function of non-classical HLA-Ib in trophoblasts
Trophoblasts uniquely express non-classical HLA-Ib molecules, including HLA-E, HLA-F, and HLA-G [63]. Therefore, this part will summarize the studies related to the expression and function of non-classical HLA-Ib molecules.
HLA-E
HLA-E in different trophoblasts and cell lines
In 1988, HLA-E was identified in resting T lymphocytes [64]. Moreover, in 1990, it was discovered that HLA-E was expressed in placental and extravillous tissues at all stages of pregnancy [65]. The expression of HLA-E on the cell surface is regulated by the acquisition of peptides derived from the leader sequences of other HLA-I molecules, including the HLA-G and HLA-C molecules [66, 67]. Hackmon et al. [5] found that HLA-E was weakly expressed in CTBs and STBs at 5 weeks of gestation and failed to detect its expression at the other gestational weeks. HLA-E was found to be expressed on the surface of EVTs [68]. HLA-E was localized to iEVT in the decidual stroma by immunohistochemistry, and a small proportion of the cytoplasm of STBs also showed intense HLA-E staining [68]. HTR8/SVneo, Swan 71, and TEV-1 were all found to express HLA-E [44]. HLA-E was expressed on the surface of JEG3 and BeWo and was co-expressed with HLA-G [69]. When TO differentiates into EVTs, it can express HLA-E [24].
Function of HLA-E in pregnancy
Like the classical HLA-Ia molecules, HLA-E is expressed in nearly all nucleated cells [70]. HLA-E was found in 1988, and it only binds to NK cell receptors like CD94/NKG2A, CD94/NKG2B, and CD94/NKG2C, not the KIR receptor family [71-74]. The binding of HLA-E to CD94/NKG2C, a receptor in CD8+ T cell, promotes the expansion of CD8+ T cell subsets and the activation of effector functions [73].
HLA-E is the major ligand of the NK cells inhibitory receptor CD94/NKG2A. Their binding can suppress NK cell-mediated cell lysis and support the fetus in evading maternal immune surveillance [72, 75]. Compared with wild-type mice, NKG2A knockout mice had poor placental angiogenesis remodeling, resulting in low fetal weight and aberrant fetal brain development; both of which are the main characteristics of preeclampsia, indicating that the HLA-E/NKG2A pathway might be related to its pathogenesis [76]. In addition, CD94/NKG2A is expressed in TCRγδ+ T cells [77]. The mutual recognition between HLA-E in trophoblasts and CD94/NKG2 receptors in immune cells may be crucial in maternal-fetal immunological interactions. Further studies are required to determine whether the immune tolerance induced by the interplay of HLA-E in trophoblasts and CD94/NKG2A in decidual immune cells is the root cause of successful pregnancy.
HLA-F
HLA-F in different trophoblasts and cell lines
Since less attention was paid to HLA-F, its expression and potential roles during pregnancy remain largely unknown. Based on limited literature, it is still controversial whether HLA-F is expressed intracellularly or on the cell surface. One study showed that HLA-F was expressed intracellularly in inactivated immune cells and on the surface of activated immune cells [78], while another study showed that HLA-F was expressed on the surface of EVTs [5]. A study by Ishitani et al. [66] showed that low levels of HLA-F staining were observed in CTBs and STBs, but significantly higher levels of HLA-F could be detected in EVTs. Nagamatsu et al. [79] found that HLA-F was only expressed intracellularly in CTBs, STBs, and EVTs, but not on the surface of these cells. In contrary, Shobu et al. [80] discovered that HLA-F was expressed at a low level in the cytoplasm of EVTs during the first trimester but increased in the second trimester, notably in the third trimester, and also expressed on the surface of EVTs. Moreover, Ishitani et al. [66] detected the expression of HLA-F protein in several cell lines and discovered that HLA-F was not expressed on the surface of both JEG3 and Bewo cells. Other research found that HLA-F was expressed in JEG3 [81]. To date, no studies have been conducted to determine whether HLA-F is expressed in the trophoblast cell lines as HTR8/SVneo, Swan 71, and TEV-1.
Function of HLA-F in pregnancy
Although HLA-F was discovered in 1990, the related studies are still lacking [82]. The heavy chain of HLA-F can form a stable complex on the cell surface by binding to β2m and peptide 27 or form HLA open conformers (OCs) if not bound to β2m and peptide 27 [83]. The HLA-F/β2m/peptide 27 complex can bind to the inhibitory immunoglobulin-like transcript (ILT) 2 receptors (LILRB1) and ILT4 (LILRB2) and may act intracellularly to regulate the expression of these inhibitory receptors [84]. HLA-F OCs, a different type of HLA-F expression, can physically and functionally interact with inhibitory receptors KIR3DL1 and KIR3DL2, as well as activating receptors KIR3DS1 and KIR2DS4 [83, 85, 86].
In dNK cells, 45% express the activating receptor KIR2DS4 [87]. HLA-F OCs can bind to KIR2DS4, so HLA-F may be involved in receptor-ligand interactions between trophoblasts and dNK cells [85]. The activation of KIR2DS4 on dNK cells can secrete GM-CSF and other chemokines, which promotes trophoblast invasion [87]. As a result, high levels of HLA-F expression in trophoblasts may be critical for dNK cells to promote blastocyst implantation [88]. HLA-F OCs can also interact with HLA-E, indicating that HLA-F can affect the recognition of HLA-E by CD94/NKG2 heterodimers, and in turn HLA-E may alter interaction of HLA-F with KIR3DL2 [85]. Since HLA-F interacts with MHC class I molecules, KIR3DL2 and KIR2DS4, it is possible that HLA-F is engaged in receptor-ligand interactions between dNK cells and EVTs during pregnancy, and thus contributes to the maternal-fetal immune regulation.
HLA-G
HLA-G in different trophoblasts and cell lines
HLA-G is the first HLA class I molecule found in trophoblasts. It is a non-classical HLA-Ib molecule with low polymorphism. HLA-G is composed of four membrane-bound isoforms (HLA-G1, -G2, -G3, and -G4) as well as three soluble (sHLA-G) isoforms (HLA-G5, -G6, and -G7). The roles of HLA-G in pregnancy have been extensively elucidated. HLA-G was found to localize in EVTs by using immunohistochemical staining with HLA-G monoclonal antibody (mAb) of MEM-G/9 [68]. During the entire pregnancy, HLA-G was found extensively expressed on the surface of EVTs by immunohistochemistry [5]. Although most studies have shown that CTBs and STBs do not express membrane HLA-G, all trophoblast populations, including CTBs and STBs, can secrete sHLA-G [33, 66]. For the detection of HLA-G, anti-HLA-G mAbs including 87G, 16G1, and olG were usually used [66]. 87G detects both soluble and membrane-bound forms of HLA-G; 16G1 only detects the soluble form, and o1G only detects the membrane-bound form of HLA-G [33]. In addition, 16G1 binding was reported to be non-specific [89].
With regards to trophoblast cell lines, Swan 71 [21] and TEV-1 [20] were found to express HLA-G by western blotting assay. However, other study did not detect the HLA-G expression in Swan 71 and TEV-1 by single-color flow cytometry using two different mAbs, G233 and MEMG/11 [44]. Therefore, different experimental methods and/or antibodies could affect the determination of the HLA-G expression on the cell surface. Besides, HTR-8/SVneo, Swan 71 and TEV-1 can express HLA-A and HLA-B molecules and express HLA-class II molecules upon IFN-γ induction [44]. As early as 1990, Kovats et al. [49] discovered that HLA-G was expressed in JEG3 but not in JAR or BeWo when using W6/32 (as a mAb against pan-HLA class I molecules). On the contrary, Liu et al. [90] reported that BeWo expressed HLA-G and may be used to mimic the function of placental trophoblasts in vitro. However, their study did not mention which clone was used to detect HLA-G.
In addition, HLA-G gene was found to be up-regulated in the differentiation of human embryonic stem cells to the trophoblast lineage, and their expression level persisted throughout the differentiation process [91]. Regarding TSCs, Sheridan et al. found they expressed HLA-A, -B, and -C but not HLA-G when using both W6/32 and MEMG-9 mAb [50]. However, when TSCs were differentiated into EVTs, HLA-G expression can be detected; when TSCs were differentiated into STBs, HLA expression was downregulated [50]. In contrast, TO does not express any HLA, but expresses HLA-G when differentiated to EVTs [25, 50].
Function of HLA-G in pregnancy
HLA-G is one of the most well-studied HLA-Ib class molecules, and its role in pregnancy has been extensively reviewed. HLA-G is the first HLA-Ib molecule identified in trophoblasts, and has a unique selective splicing pattern, limited polymorphism, and limited tissue distribution [92]. HLA-G is thought to establish maternal tolerance to allogeneic fetus and could be employed as an inhibitory ligand to enhance tolerance during pregnancy [49, 93]. HLA-G can promote spiral artery remodeling, immune tolerance, and fetal growth through binding to immune cells during pregnancy [94]. Thus, HLA-G, also known as one of the immune checkpoint molecules, has multiple functional effects [95].
HLA expression in trophoblast and their receptors in local immune cells at the maternal-fetal interface.
HLA-G molecules can bind to the inhibitory receptor KIR2DL4, which is expressed in dNK cells [3], mast cells [96], and some T cells [97]. KIR2DL4 could inhibit NK cell-mediated cytotoxicity and induce mast cells to secrete the serine protease MMP-9 and leukemia growth factor, which promotes the invasion and differentiation of trophoblasts, and is conducive to embryo implantation [96, 98, 99]. HLA-G can also inhibit cell lysis of decidual and peripheral NK cells and CD8+ T cells [100-102]. By knocking out HLA-G on TEV-1 through RNA interference, Chen et al. [103] found that NK cells exhibited more robust killing activity against HLA-G knocked out TEV-1 when compared with the control group. The interaction between HLA-G and KIR2DL4 on the surface of dNK cells is significant for immunological tolerance.
HLA-G can also bind to the inhibitory ILT2/LILRB1 and ILT4/LILRB2 [104-105], which are expressed on the surface of NK cells, CD4+ and CD8+ T cells, B cells, macrophages, and monocytes [105]. ILT2/HLA-G can suppress dNK cell's cytotoxicity and increase the secretion of IFN-γ, which attributes to angiogenesis and vascular remodeling during early pregnancy [106]. Furthermore, the HLA-G receptor ILT2/LILRB1 may be transferred from monocytes to activated CD4+ T cells and perform all functions, mediating the inhibitory effect of HLA-G on ILT2+ T cells [42]. ILT2 and ILT4 in CD8+ T cells competitively bind to HLA-G and then prevent the activation of cytotoxic CD8+ T cells, thereby inducing the establishment of tolerance [107]. Besides, when ILT2 binds to HLA-G in decidual dendritic cells (DCs), the expression of IL-10 and IL-6 by DCs increases, and the proliferation of allogeneic lymphocytes is inhibited, finally inducing the formation of tolerant DCs [108]. IL-10-producing DCs drive the differentiation of initial CD4+ T cells into type 1 regulatory T cells via the ILT4/HLA-G signaling pathway, which is essential for maintaining tolerance to self and non-self antigens [109]. ILT2 is also expressed in the decidual stroma, primarily composed of fibroblasts and macrophages, whereas ILT4 is found in the placental vascular smooth muscle [110]. The receptor expression pattern suggests that HLA-G has a unique immunological function during pregnancy. The HLA-G1 homodimer connected by disulfide bonds has a higher affinity for ILT2, and the HLA-G5 free heavy chain has a more vital binding ability to ILT4 [111]. Differences in affinity may regulate the invasion of trophoblasts and the function of the local maternal immune response in the uterus. Recently, a unique subset of decidual memory NK cells was identified, and HLA-G induced its formation, leading to increased secretion of VEGFα and IFN-γ, thereby better supporting the critical process of placental angiogenesis [106].
Conclusion
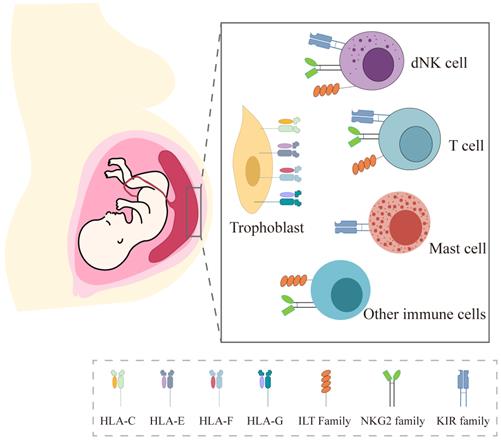
Since Sir Peter Medawar first raised the immunological paradox in pregnancy, a great deal of evidence has subsequently confirmed the importance of immune tolerance underlying the mechanism of allogeneic fetus escaping the attack of maternal immune system. Among the multiple contributing factors, the unique expression of HLA molecules in trophoblasts and the interaction with their receptors in local immune cells are recognized as the key factors (Figure 2).
In this review, we systemically described the expression pattern of HLA-class Ia molecules (HLA-C) and non-classical HLA-Ib class I molecules (HLA-E, HLA-F, and HLA-G) in three types of trophoblasts and the commonly used trophoblast cell lineages, including HTR8/SVneo, Swan 71, TEV-1, BeWo, JAR and JEG3 (Table 1). Also, we summarized how HLA-Class I in trophoblasts interact with their receptors in decidual immune cells and their roles in normal pregnancy (Figure 3). The current evidence will enhance comprehensive understanding on the complex immune mechanisms at the maternal-fetal interface.
A schematic illustration of the interaction between HLA-Class I molecules and their receptors at the maternal-fetal interface. (A) The figure depicts the placental villi consisting of cytotrophoblasts (CTBs), syncytiotrophoblasts (STBs), and extravillous trophoblasts (EVTs) that invade the maternal decidua. (B-F) Summary of the interaction of HLA-C, -E, -F, and -G with their corresponding receptors on the surface of decidual NK cell, T cell, B cell, mast cell, other immune cells, and the roles during pregnancy.
HLA expression in primary trophoblasts and cell lines
| CTBs | STBs | EVTs | HTR8/SVneo | TEV-1 | Swan 71 | BeWo | JEG3 | JAR | |
|---|---|---|---|---|---|---|---|---|---|
| HLA-C | +/- | +/- | + | + | + | + | + | + | - |
| HLA-E | +/- | +/- | + | + | + | + | + | + | - |
| HLA-F | +/- | +/- | + | ? | ? | ? | - | +/- | ? |
| HLA-G | sHLA-G/- | sHLA-G/- | + | +/- | +/- | +/- | +/- | + | - |
| Summary | HLA-C, -E, -F, -G | HLA-C, -E, -F, -G | HLA-C, -E, -F, -G | HLA-C -E -G | HLA-C -E, -G | HLA-C -E, -G | HLA-C, -E, -G | HLA-C, -E, -F, -G | - |
Note: +: express the HLA molecules; -: not express the HLA molecules; +/-: exist contradictory study results; ?: lack of research; CTBs: cytotrophoblasts; STBs: syncytiotrophoblasts; EVTs: extravillous trophoblasts.
Abbreviations
CTBs: cytotrophoblasts; DC: dendritic cells; dNK: decidual NK; eEVT: endovascular trophoblasts; EVTs: extravillous trophoblasts; HLA: human leukocyte antigen; iEVT: interstitial trophoblasts; Ig: immunoglobulin; ILT: immunoglobulin-like transcript; KIR: killer immunoglobulin-like receptor; OCs: open conformers; sHLA-G: soluble HLA-G; STBs: syncytiotrophoblasts; β2m: β2-microglobulin.
Acknowledgements
Funding
This work was supported by the Fundamental Research Funds for the Central Universities, HUST: 2020JYCXJJ022.
Author contributions
Xx. L conceived and prepared the manuscript, Ym. X, Sj. Z and Cy. L helped write the paper and draw figures, G. M revised the manuscript, and Ah. L conceived, revised, and finalized the manuscript. All the authors approved it.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Medawar P. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. In Symp Soc Exp Biol. 1953;7:320-37
2. Mor G, Aldo P, Alvero AB. The unique immunological and microbial aspects of pregnancy. Nat Rev Immunol. 2017;17:469-82
3. Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. HLA-G: At the Interface of Maternal-Fetal Tolerance. Trends Immunol. 2017;38:272-86
4. Persson G, Jorgensen N, Nilsson LL, Andersen LHJ, Hviid TVF. A role for both HLA-F and HLA-G in reproduction and during pregnancy? Hum Immunol. 2020;81:127-33
5. Hackmon R, Pinnaduwage L, Zhang J, Lye SJ, Geraghty DE, Dunk CE. Definitive class I human leukocyte antigen expression in gestational placentation: HLA-F, HLA-E, HLA-C, and HLA-G in extravillous trophoblast invasion on placentation, pregnancy, and parturition. Am J Reprod Immunol. 2017 77
6. Burton GJ, Jauniaux E. What is the placenta? Am J Obstet Gynecol. 2015;213:S6 e1, S6-8
7. Colucci F. The role of KIR and HLA interactions in pregnancy complications. Immunogenetics. 2017;69:557-65
8. Roberts RM, Ezashi T, Schulz LC, Sugimoto J, Schust DJ, Khan T. et al. Syncytins expressed in human placental trophoblast. Placenta. 2021;113:8-14
9. Knofler M, Haider S, Saleh L, Pollheimer J, Gamage T, James J. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol Life Sci. 2019;76:3479-96
10. Hemberger M, Hanna CW, Dean W. Mechanisms of early placental development in mouse and humans. Nat Rev Genet. 2020;21:27-43
11. Gamage TK, Chamley LW, James JL. Stem cell insights into human trophoblast lineage differentiation. Hum Reprod Update. 2016;23:77-103
12. Chang CW, Wakeland AK, Parast MM. Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J Endocrinol. 2018;236:R43-R56
13. Staud F, Karahoda R. Trophoblast: The central unit of fetal growth, protection and programming. Int J Biochem Cell Biol. 2018;105:35-40
14. Carrasco-Wong I, Aguilera-Olguin M, Escalona-Rivano R, Chiarello DI, Barragan-Zuniga LJ, Sosa-Macias M. et al. Syncytiotrophoblast stress in early onset preeclampsia: The issues perpetuating the syndrome. Placenta. 2021;113:57-66
15. Turco MY, Gardner L, Kay RG, Hamilton RS, Prater M, Hollinshead MS. et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature. 2018;564:263-7
16. Horii M, Touma O, Bui T, Parast MM. Modeling human trophoblast, the placental epithelium at the maternal fetal interface. Reproduction. 2020;160:R1-R11
17. Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta. 2006;27:939-58
18. Ji L, Brkic J, Liu M, Fu G, Peng C, Wang YL. Placental trophoblast cell differentiation: physiological regulation and pathological relevance to preeclampsia. Mol Aspects Med. 2013;34:981-1023
19. Graham CH, Hawley TS, Hawley RG, MacDougall JR, Kerbel RS, Khoo N. et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res. 1993;206:204-11
20. Feng HC, Choy MY, Deng W, Wong HL, Lau WM, Cheung AN. et al. Establishment and characterization of a human first-trimester extravillous trophoblast cell line (TEV-1). J Soc Gynecol Investig. 2005;12:e21-32
21. Straszewski-Chavez SL, Abrahams VM, Alvero AB, Aldo PB, Ma Y, Guller S. et al. The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta. 2009;30:939-48
22. Cerneus DP, van der Ende A. Apical and basolateral transferrin receptors in polarized BeWo cells recycle through separate endosomes. J Cell Biol. 1991;114:1149-58
23. Abbas Y, Turco MY, Burton GJ, Moffett A. Investigation of human trophoblast invasion in vitro. Hum Reprod Update. 2020;26:501-13
24. Sheridan MA, Fernando RC, Gardner L, Hollinshead MS, Burton GJ, Moffett A. et al. Establishment and differentiation of long-term trophoblast organoid cultures from the human placenta. Nat Protoc. 2020;15:3441-63
25. Haider S, Meinhardt G, Saleh L, Kunihs V, Gamperl M, Kaindl U. et al. Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta. Stem Cell Reports. 2018;11:537-51
26. Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K. et al. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell. 2018;22:50-63 e6
27. Heath WR, Carbone FR. Cross-presentation in viral immunity and self-tolerance. Nat Rev Immunol. 2001;1:126-34
28. Pujadas E, Cordon-Cardo C. The human leukocyte antigen as a candidate tumor suppressor. Cancer Cell. 2021;39:586-9
29. Sabbatino F, Liguori L, Polcaro G, Salvato I, Caramori G, Salzano FA. et al. Role of Human Leukocyte Antigen System as A Predictive Biomarker for Checkpoint-Based Immunotherapy in Cancer Patients. Int J Mol Sci. 2020;21:7295
30. Hoek M, Demmers LC, Wu W, Heck AJR. Allotype-Specific Glycosylation and Cellular Localization of Human Leukocyte Antigen Class I Proteins. J Proteome Res. 2021;20:4518-28
31. Tersigni C, Meli F, Neri C, Iacoangeli A, Franco R, Lanzone A. et al. Role of Human Leukocyte Antigens at the Feto-Maternal Interface in Normal and Pathological Pregnancy: An Update. Int J Mol Sci. 2020;21:4756
32. The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921-3
33. Dahl M, Hviid TV. Human leucocyte antigen class Ib molecules in pregnancy success and early pregnancy loss. Hum Reprod Update. 2012;18:92-109
34. Schott G, Garcia-Blanco MA. MHC Class III RNA Binding Proteins and Immunity. RNA Biol. 2021;18:640-6
35. Unanue ER, Turk V, Neefjes J. Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu Rev Immunol. 2016;34:265-97
36. Dilthey AT. State-of-the-art genome inference in the human MHC. Int J Biochem Cell Biol. 2021;131:105882
37. Wyatt RC, Lanzoni G, Russell MA, Gerling I, Richardson SJ. What the HLA-I!-Classical and Non-classical HLA Class I and Their Potential Roles in Type 1 Diabetes. Curr Diab Rep. 2019;19:159
38. Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S. et al. Structure of the human MHC-I peptide-loading complex. Nature. 2017;551:525-8
39. Wieczorek M, Abualrous ET, Sticht J, Alvaro-Benito M, Stolzenberg S, Noe F. et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol. 2017;8:292
40. Bouvier M, Wiley DC. Structural characterization of a soluble and partially folded class I major histocompatibility heavy chain/beta 2m heterodimer. Nat Struct Biol. 1998;5:377-84
41. Christophersen A. Peptide-MHC class I and class II tetramers: From flow to mass cytometry. HLA. 2020;95:169-78
42. Alecsandru D, Garcia-Velasco JA. Is there a role for human leukocyte antigen-G typing in infertility treatment? Fertil Steril. 2020;114:515-6
43. Thorsby E, Sandberg L, Lindholm A, Kissmeyer-Nielsen F. The HL-A system: evidence of a third sub-locus. Scand J Haematol. 1970;7:195-200
44. Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 2009;127:26-39
45. Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584-94
46. King A, Burrows TD, Hiby SE, Bowen JM, Joseph S, Verma S. et al. Surface expression of HLA-C antigen by human extravillous trophoblast. Placenta. 2000;21:376-87
47. Persson G, Bork JBS, Isgaard C, Larsen TG, Bordoy AM, Bengtsson MS. et al. Cytokine stimulation of the choriocarcinoma cell line JEG-3 leads to alterations in the HLA-G expression profile. Cell Immunol. 2020;352:104110
48. King A, Boocock C, Sharkey AM, Gardner L, Beretta A, Siccardi AG. et al. Evidence for the expression of HLAA-C class I mRNA and protein by human first trimester trophoblast. J Immunol. 1996;156:2068-76
49. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220-3
50. Sheridan MA, Zhao X, Fernando RC, Gardner L, Perez-Garcia V, Li Q. et al. Characterization of primary models of human trophoblast. Development. 2021;148:dev199749
51. Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201-14
52. Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277-300
53. Apps R, Gardner L, Hiby SE, Sharkey AM, Moffett A. Conformation of human leucocyte antigen-C molecules at the surface of human trophoblast cells. Immunology. 2008;124:322-8
54. Tilburgs T, van der Mast BJ, Nagtzaam NM, Roelen DL, Scherjon SA, Claas FH. Expression of NK cell receptors on decidual T cells in human pregnancy. J Reprod Immunol. 2009;80:22-32
55. Larsen TG, Hackmon R, Geraghty DE, Hviid TVF. Fetal human leukocyte antigen-C and maternal killer-cell immunoglobulin-like receptors in cases of severe preeclampsia. Placenta. 2019;75:27-33
56. Hilton HG, Parham P. Missing or altered self: human NK cell receptors that recognize HLA-C. Immunogenetics. 2017;69:567-79
57. Sharkey AM, Xiong S, Kennedy PR, Gardner L, Farrell LE, Chazara O. et al. Tissue-Specific Education of Decidual NK Cells. J Immunol. 2015;195:3026-32
58. Hiby SE, Walker JJ, O'Shaughnessy K M, Redman CW, Carrington M, Trowsdale J. et al. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957-65
59. Sharkey AM, Gardner L, Hiby S, Farrell L, Apps R, Masters L. et al. Killer Ig-like receptor expression in uterine NK cells is biased toward recognition of HLA-C and alters with gestational age. J Immunol. 2008;181:39-46
60. Xiong S, Sharkey AM, Kennedy PR, Gardner L, Farrell LE, Chazara O. et al. Maternal uterine NK cell-activating receptor KIR2DS1 enhances placentation. J Clin Invest. 2013;123:4264-72
61. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13:133-44
62. Lv H, Zhou Q, Li L, Wang S. HLA-C promotes proliferation and cell cycle progression in trophoblast cells. J Matern Fetal Neonatal Med. 2021;34:512-8
63. Nilsson LL, Hviid TVF. HLA Class Ib-receptor interactions during embryo implantation and early pregnancy. Hum Reprod Update. 2022;28:435-54
64. Koller BH, Geraghty DE, Shimizu Y, DeMars R, Orr HT. HLA-E. A novel HLA class I gene expressed in resting T lymphocytes. J Immunol. 1988;141:897-904
65. Wei XH, Orr HT. Differential expression of HLA-E, HLA-F, and HLA-G transcripts in human tissue. Hum Immunol. 1990;29:131-42
66. Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H. et al. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J Immunol. 2003;171:1376-84
67. King A, Allan DS, Bowen M, Powis SJ, Joseph S, Verma S. et al. HLA-E is expressed on trophoblast and interacts with CD94/NKG2 receptors on decidual NK cells. Eur J Immunol. 2000;30:1623-31
68. Bhalla A, Stone PR, Liddell HS, Zanderigo A, Chamley LW. Comparison of the expression of human leukocyte antigen (HLA)-G and HLA-E in women with normal pregnancy and those with recurrent miscarriage. Reproduction. 2006;131:583-9
69. Shaikly V, Shakhawat A, Withey A, Morrison I, Taranissi M, Dealtry GB. et al. Cell bio-imaging reveals co-expression of HLA-G and HLA-E in human preimplantation embryos. Reprod Biomed Online. 2010;20:223-33
70. Wieten L, Mahaweni NM, Voorter CE, Bos GM, Tilanus MG. Clinical and immunological significance of HLA-E in stem cell transplantation and cancer. Tissue Antigens. 2014;84:523-35
71. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS. et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795-9
72. Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M. et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci U S A. 1998;95:5199-204
73. Guma M, Busch LK, Salazar-Fontana LI, Bellosillo B, Morte C, Garcia P. et al. The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur J Immunol. 2005;35:2071-80
74. Li J, Goldstein I, Glickman-Nir E, Jiang H, Chess L. Induction of TCR Vbeta-specific CD8+ CTLs by TCR Vbeta-derived peptides bound to HLA-E. J Immunol. 2001;167:3800-8
75. Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol. 1997;27:1164-9
76. Shreeve N, Depierreux D, Hawkes D, Traherne JA, Sovio U, Huhn O. et al. The CD94/NKG2A inhibitory receptor educates uterine NK cells to optimize pregnancy outcomes in humans and mice. Immunity. 2021;54:1231-44 e4
77. Barakonyi A, Kovacs KT, Miko E, Szereday L, Varga P, Szekeres-Bartho J. Recognition of nonclassical HLA class I antigens by gamma delta T cells during pregnancy. J Immunol. 2002;168:2683-8
78. Lee N, Ishitani A, Geraghty DE. HLA-F is a surface marker on activated lymphocytes. Eur J Immunol. 2010;40:2308-18
79. Nagamatsu T, Fujii T, Matsumoto J, Yamashita T, Kozuma S, Taketani Y. Human leukocyte antigen F protein is expressed in the extra-villous trophoblasts but not on the cell surface of them. Am J Reprod Immunol. 2006;56:172-7
80. Shobu T, Sageshima N, Tokui H, Omura M, Saito K, Nagatsuka Y. et al. The surface expression of HLA-F on decidual trophoblasts increases from mid to term gestation. J Reprod Immunol. 2006;72:18-32
81. Hakam MS, Miranda-Sayago JM, Hayrabedyan S, Todorova K, Spencer PS, Jabeen A. et al. Preimplantation Factor (PIF) Promotes HLA-G, -E, -F, -C Expression in JEG-3 Choriocarcinoma Cells and Endogenous Progesterone Activity. Cell Physiol Biochem. 2017;43:2277-96
82. Geraghty DE, Wei XH, Orr HT, Koller BH. Human leukocyte antigen F (HLA-F). An expressed HLA gene composed of a class I coding sequence linked to a novel transcribed repetitive element. J Exp Med. 1990;171:1-18
83. Garcia-Beltran WF, Holzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR. et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat Immunol. 2016;17:1067-74
84. Lepin EJ, Bastin JM, Allan DS, Roncador G, Braud VM, Mason DY. et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur J Immunol. 2000;30:3552-61
85. Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol. 2013;191:3553-62
86. Burian A, Wang KL, Finton KA, Lee N, Ishitani A, Strong RK. et al. HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor KIR3DS1. PLoS One. 2016;11:e0163297
87. Kennedy PR, Chazara O, Gardner L, Ivarsson MA, Farrell LE, Xiong S. et al. Activating KIR2DS4 Is Expressed by Uterine NK Cells and Contributes to Successful Pregnancy. J Immunol. 2016;197:4292-300
88. Langkilde CH, Nilsson LL, Jorgensen N, Funck T, Perin TL, Hornstrup MB. et al. Variation in the HLA-F gene locus with functional impact is associated with pregnancy success and time-to-pregnancy after fertility treatment. Hum Reprod. 2020;35:705-17
89. Paul P, Rouas-Freiss N, Moreau P, Cabestre FA, Menier C, Khalil-Daher I. et al. HLA-G, -E, -F preworkshop: tools and protocols for analysis of non-classical class I genes transcription and protein expression. Hum Immunol. 2000;61:1177-95
90. Liu Y, Zhang L, Gao M, Zhang F, Xu X, Liu X. et al. Changes of inhibitory receptors on NK-92 cells and HLA-G on BeWo cells with Toxoplasma gondii infection. Inflammation. 2013;36:1440-7
91. Marchand M, Horcajadas JA, Esteban FJ, McElroy SL, Fisher SJ, Giudice LC. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol Reprod. 2011;84:1258-71
92. Ellis SA, Sargent IL, Redman CW, McMichael AJ. Evidence for a novel HLA antigen found on human extravillous trophoblast and a choriocarcinoma cell line. Immunology. 1986;59:595-601
93. Moffett A, Colucci F. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol Rev. 2015;267:283-97
94. Xu X, Zhou Y, Wei H. Roles of HLA-G in the Maternal-Fetal Immune Microenvironment. Front Immunol. 2020;11:592010
95. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: An Immune Checkpoint Molecule. Adv Immunol. 2015;127:33-144
96. Ueshima C, Kataoka TR, Hirata M, Sugimoto A, Iemura Y, Minamiguchi S. et al. Possible Involvement of Human Mast Cells in the Establishment of Pregnancy via Killer Cell Ig-Like Receptor 2DL4. Am J Pathol. 2018;188:1497-508
97. Nowak I, Wilczynska K, Wilczynski JR, Malinowski A, Radwan P, Radwan M. et al. KIR, LILRB and their Ligands' Genes as Potential Biomarkers in Recurrent Implantation Failure. Arch Immunol Ther Exp (Warsz). 2017;65:391-9
98. Yan WH, Lin A, Chen BG, Zhou MY, Dai MZ, Chen XJ. et al. Possible roles of KIR2DL4 expression on uNK cells in human pregnancy. Am J Reprod Immunol. 2007;57:233-42
99. Nicola NA, Babon JJ. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 2015;26:533-44
100. Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093-100
101. Ponte M, Cantoni C, Biassoni R, Tradori-Cappai A, Bentivoglio G, Vitale C. et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci U S A. 1999;96:5674-9
102. Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci U S A. 1997;94:11520-5
103. Chen LJ, Han ZQ, Zhou H, Zou L, Zou P. Inhibition of HLA-G expression via RNAi abolishes resistance of extravillous trophoblast cell line TEV-1 to NK lysis. Placenta. 2010;31:519-27
104. Biassoni R, Bottino C, Millo R, Moretta L, Moretta A. Natural killer cell-mediated recognition of human trophoblast. Semin Cancer Biol. 1999;9:13-8
105. Colonna M, Samaridis J, Cella M, Angman L, Allen RL, O'Callaghan CA. et al. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J Immunol. 1998;160:3096-100
106. Gamliel M, Goldman-Wohl D, Isaacson B, Gur C, Stein N, Yamin R. et al. Trained Memory of Human Uterine NK Cells Enhances Their Function in Subsequent Pregnancies. Immunity. 2018;48:951-62 e5
107. Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM. et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A. 2003;100:8856-61
108. Apps R, Gardner L, Sharkey AM, Holmes N, Moffett A. A homodimeric complex of HLA-G on normal trophoblast cells modulates antigen-presenting cells via LILRB1. Eur J Immunol. 2007;37:1924-37
109. Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF. et al. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood. 2010;116:935-44
110. McIntire RH, Sifers T, Platt JS, Ganacias KG, Langat DK, Hunt JS. Novel HLA-G-binding leukocyte immunoglobulin-like receptor (LILR) expression patterns in human placentas and umbilical cords. Placenta. 2008;29:631-8
111. Morales PJ, Pace JL, Platt JS, Langat DK, Hunt JS. Synthesis of beta(2)-microglobulin-free, disulphide-linked HLA-G5 homodimers in human placental villous cytotrophoblast cells. Immunology. 2007;122:179-88
Author contact
![]() Corresponding author: Ai-Hua Liao, Ph.D. Address: Institute of Reproductive Health, Center for Reproductive Medicine, Tongji Medical College, Huazhong University of Science and Technology, No. 13 Hangkong Rd, Wuhan, 430030, PR China. E-mail address: aihua_liaoedu.cn (A. Liao). ORCID iD: https://orcid.org/0000-0001-8533-8315.
Corresponding author: Ai-Hua Liao, Ph.D. Address: Institute of Reproductive Health, Center for Reproductive Medicine, Tongji Medical College, Huazhong University of Science and Technology, No. 13 Hangkong Rd, Wuhan, 430030, PR China. E-mail address: aihua_liaoedu.cn (A. Liao). ORCID iD: https://orcid.org/0000-0001-8533-8315.

 Global reach, higher impact
Global reach, higher impact