10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(11):4414-4431. doi:10.7150/ijbs.72952 This issue Cite
Review
High Mobility Group A1 (HMGA1): Structure, Biological Function, and Therapeutic Potential
1. Institute of Clinical Medicine, The First Affiliated Hospital, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, PR China.
2. Cancer Research Institute, The First Affiliated Hospital, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, PR China.
3. Department of Clinical Laboratory, Shenzhen Traditional Chinese Medicine Hospital, Shenzhen 518033, Guangdong, China.
4. Department of Endocrinology and Metabolism, The First People's Hospital of Chenzhou, First School of Clinical Medicine, University of Southern Medical, Guangzhou 510515, Guangdong, China.
*These authors contributed equally to this work.
Received 2022-3-17; Accepted 2022-6-24; Published 2022-7-4
Abstract
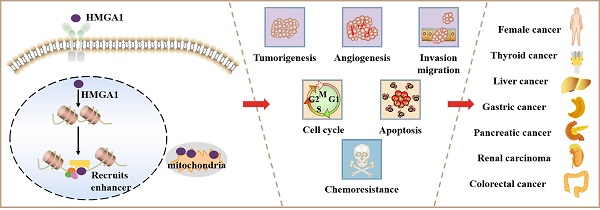
High mobility group A1 (HMGA1) is a nonhistone chromatin structural protein characterized by no transcriptional activity. It mainly plays a regulatory role by modifying the structure of DNA. A large number of studies have confirmed that HMGA1 regulates genes related to tumours in the reproductive system, digestive system, urinary system and haematopoietic system. HMGA1 is rare in adult cells and increases in highly proliferative cells such as embryos. After being stimulated by external factors, it will produce effects through the Wnt/β-catenin, PI3K/Akt, Hippo and MEK/ERK pathways. In addition, HMGA1 also affects the ageing, apoptosis, autophagy and chemotherapy resistance of cancer cells, which are linked to tumorigenesis. In this review, we summarize the mechanisms of HMGA1 in cancer progression and discuss the potential clinical application of targeted HMGA1 therapy, indicating that targeted HMGA1 is of great significance in the diagnosis and treatment of malignancy.
Keywords: High mobility group A1, Malignancies, Mechanisms, Chemoresistance, Targeting treatment
Introduction
High mobility family A (HMGA) consists of three members: HMGA1, HMGA2 and HMGA3. HMGA1 is a nonhistone chromatin structural protein encoded by oncogenes and located in the nucleus. It can change the structure of DNA by binding to the rills of the A+T rich region in dsDNA and interact with transcription factors to regulate transcription. In addition to the expression of HMGA1 during normal development, HMGA1 is overexpressed in most malignancies, Furthermore, the malignant degree of tumours, chemotherapy resistance and drug sensitivity are related to the expression level of HMGA1.
Overview of HMGA1
Biological structure of HMGA1
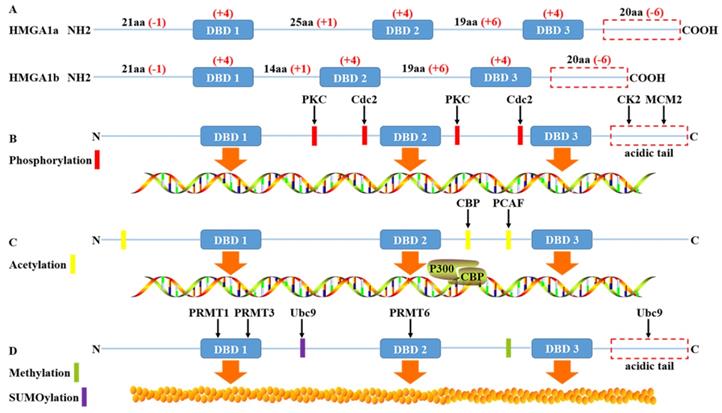
High mobility group protein A1 (HMGA1) is called this because of its rapid migration in polyacrylamide gels. Due to the selective splicing of precursor mRNA, it can be divided into two subtypes: HMGA1a and HMGA1b, also known as HMGI/Y in previous literature [1]. Human HMGA1a and HMGA1b are translated and encoded by the short arm of chromosome 6. The peptide chain length is approximately 100 amino acids (10.7-11.7kDa), belonging to low molecular weight histones. Structurally, it contains three independent conserved AT hook domains (DBDs) and an acidic carboxyl tail. The three positively charged AT hook domains are connected by linker peptides and can bind to AT-rich sequences in double helix DNA. Meanwhile, the P-R-G-R-P palindrome sequence of the AT hook ensures the specificity of the AT hook binding to DNA slots [2]. The acid carboxyl terminal, including 30 consecutive Asp and Glu residues, negatively charged and regulates protein function mainly through phosphorylation.
The charge difference between the AT hook domain and an acidic carboxyl terminal has an effect on the conformation of DNA after binding to DNA, which can lead to DNA bending, straightening, unwinding, looping or topological structure. Then, through conformational changes, DNA can influence transcription by recruiting transcription factors into specific transcriptional regulatory regions through either enhancers or competition with histones or chaperones. In addition, compared with HMGA1a, HMGA1b lacks only 11 amino acid residues between the first and second AT hooks [3], and the rest of the sequences are roughly the same. Therefore, HMGA1a and HMGA1b are highly homologous in function, whether in the process of ageing, embryonic stem cells, type 2 diabetes mellitus or tumour development and progression will be involved in regulation. This regulation also relies on HMGA1 posttranslational modification such as phosphorylation, methylation, acetylation, ubiquitinylation and SUMOylation.
Phosphorylation of HMGA1
HMGA1 is a structural building protein that regulates transcription by changing the conformation of DNA after binding with DNA. Moreover, posttranslational modification of the HMGA1 protein can change its binding affinity with DNA and regulate the function of the HMGA1 protein. Many studies have shown that HMGA1 is the substrate of most protein kinases and can be phosphorylated by tyrosine protein kinase (CK2), cdc2 kinase, protein kinase C(PKC), M-phase specific kinase (H1 kinase) and homeodomain-interacting protein kinase-2 (HIPK2) [4-8]. For instance, the serine and threonine residues of HMGA1 are extensively modified, and the three serines (S98, S101 and S102), mainly located at the C-terminal acid carboxyl end, are phosphorylated by CK2 in a cell cycle-dependent manner [4], without affecting the acetylation and methylation of HMGA1 in vitro [9, 10]. Moreover, the phosphorylation of HMGA1 at the S102 site can be a drug-resistant target in non-small cell lung cancer (NSCLC) resistant to tyrosine kinase inhibitor (TKI), and knockout of HMGA1 can transform TKI resistant NSCLC cells into TKI sensitive cells, which will be helpful for clinical treatment [11]. The phosphorylation of HMGA1 protein by cdc2 kinase weakens the affinity of the N-terminal AT hook with promoter IFNbeta and the inherent bending state of IFNbeta [5]. The phosphorylation of HMGA1 protein in purified maize by cdc2 kinase could also reduce the transcriptional inhibition mediated by histone H1, thus promoting the large-scale synthesis of zeins [12]. HIPK2 can phosphorylate the ser-35, thr-52 and thr-77 of HMGA1a, thr-41 and thr-66 of HMGA1b and reduce the affinity with delta promoter; the degree of decrease is lower than for cdc2 as well. In addition, although the phosphorylation mediated by HIPK2 decreases the affinity of HMGA1a to DNA, the affinity increases in the absence of ATP [8]. PKC alpha, beta, gamma and delta can phosphorylate HMGA1 and reduce its binding capacity with DNA fragments, thus changing its biological function [6]. For example, in invasive and metastatic breast cancer, the phosphorylation level of HMGA1 is greater than that of low invasive primary cancer [13]. Recent studies have shown that minichromosome maintenance protein 2 (MCM2), a factor related to the formation of a replication fork, can promote the proliferation of lung cancer cells by regulating the phosphorylation of ser-99 of HMGA1 [14]. At the same time, HMGA1, as a nuclear target downstream of the insulin receptor (IR) signaling pathway, can be phosphorylated to reduce transcriptional activation. When HMGA1 is phosphorylated, the expression of the insulin receptor gene is decreased, and it is more likely to develop type 2 diabetes [15]. Diana et al. also reported for the first time that the phosphorylation of HMGA1a protein is related to apoptosis, where hyperphosphorylation occurs in the early stage and dephosphorylation occurs in the late stage with the formation of apoptotic bodies [16].
Acetylation of HMGA1
Acetylation is also a common posttranslational modification of HMGA1, and acetylation does not affect CK2-catalysed phosphorylation [17]. Using bottom-up proteomics, HMGA1 was hydrolysed into short peptides before mass spectrometry. The N-terminal lys-14 is acetylated [18]. In addition, histone acetyltransferase P300 and P300/CBP associated factor (PCAF) could induce the acetylation of lys-14, lys-64, lys-66, lys-70 and lys-73 of HMGA1a, and the acetylation mode of HMGA1b was found to be similar [9]. Acetylated HMGA1 is also a crucial structure for enhancer assembly. Enhancers can recruit histone acetyltransferase to activate transcription. HMGA1 can make the enhancer unstable or decompose through the acetylation of CBP at lys-65. The acetylation of PCAF in lys-71 can stabilize the enhancer and prevent the acetylation of CBP to enhance transcription. Two factors coordinate with each other to cause acetylation in HMGA1 to become a transcription switch [19]. In addition, acetylation is detected in all AT hooks of the HMGA1a protein, but the peptide region between the second and third AT hooks is more easily acetylated than other regions. Moreover, HMGA1a isolated from metastatic breast cancer cells had a higher degree of acetylation than that isolated from nonmetastatic breast cancer cells [20]. This also suggests that the modified HMGA1 protein is involved in the regulation of tumour progression [21].
Methylation of HMGA1
HMGA1 is strongly methylated and upregulated in almost all cancer types. Epigenetic changes through GPG methylation of HMGA1 may regulate its gene expression in various cancers [22]. It has been reported that arginine methylation of the HMGA1a protein may be linked to apoptosis, heterochromatin formation and tumour progression and be parallel to dephosphorylation in the process of apoptosis [18, 23, 24]. PRMT1 and PRMT3 of the arginine methyltransferase (PRMT) family mainly methylates the arginine residue of the first AT hook of HMGA1a, and PRMT6 is a good substitute for HMGA1a methylation endogenous enzyme, mainly in the second AT hook methylation [10, 25]. In addition, studies found that HMGA1a was monomethylated, symmetrically demethylated and asymmetric dimethylated in arg-25 [26], and there were also monomethylation modifications independent of acetylation at lys64 and lys70 sites where acetylation can occur, while no methylation was found in HMGA1b [18]. More importantly, methylation of HMGA1 arg-25 does not alter the total charge of residues, so it may not affect protein DNA binding considerably. In contrast, the introduction of hydrophobic methyl groups will affect hydrogen bonds and regulate the interaction between proteins [26].
Ubiquitinylation and SUMOylation of HMGA1
Ubiquitinylation leads to proteasome-mediated degradation of the target protein, while SUMOylation more influences subcellular trafficking or the activity of target protein [27]. GRP75 directly binds to HMGA1 to inhibit the ubiquitinylation and degradation of HMGA1, then upregulates HMGA1 to promote the proliferation and metastasis of lung adenocarcinoma cells by activating JNK/c-JUN signal [28]. The proline-rich segment between AT hooks 1 and 2 and the C-terminal acidic domain of HMGA1b interacts with SUMO E2 Conjugase Ubc9 without being SUMOylated. Ubc9 binding only to the proline-rich segment of HMGA1b enhances the colony forming ability of rat1a cell, whereas the acidic domain negatively impacts on these properties [29]. This also supports the view that HMGA1 has a dual role in some cases. In addition, HMGA1 also undergo ADP-ribosylation, often accompanied by DNA damage [30] (Fig. 1).
The structure and post-translational modification of the HMGA1 protein. (A) The human HMGA1 protein is divided into four functional domains: the DBD 1, DBD 2, DBD 3 and acidic C-terminal tail. The net charge distribution is shown in red. (B) HMGA1 protein is widely phosphorylated by the protein kinases PKC, Cdc2, CK2 and MCM2, affecting the affinity between HMGA1 and DNA to regulate HMGA1 function. The phosphorylation of HMGA1 by CK2 and MCM2 mainly occurs in the acidic C-terminal tail. (C) HMGA1 recruits the assembly of enhancer p300/CRP under the action of histone acetyltransferase CBP and PCAF to play the role of transcriptional regulation. (D) HMGA1 is methylated by PRMT family members and SUMO E2 Conjugase Ubc9 without being SUMOylated to regulate protein interactions.
Interaction between HMGA1 and histone
Histone H1 and HMGA1 are both nuclear proteins and contain specific DNA binding domains, where histones bind to DNA mainly through their own spherical domains [31], and HMGA1 regulates gene transcription by forming multiprotein complexes in promoter/enhancer regions or regulating chromatin structures [32]. However, there is a certain antagonism between the two; HMGA1 increases the activity of DNA ligase IV by forming a complex with DNA-dependent protein kinase (DNA-PK) and counteracts the inhibitory effect of histone H1 on DNA ends, and overexpressed HMGA1 also causes rapid repair of broken DNA double strands [33]. In addition, if cancer cells themselves wish to invade other tissues, they must be plastic. The nucleus is a hard organelle, and cancer cells will invade only when the structure changes. The nuclear building protein HMGA1 can phosphorylate the H1 histone, weaken the affinity between histones and DNA, relax chromatin, and finally achieve the purpose of changing the nuclear hardness and promote the invasion of cancer cells [34].
The role of HMGA1 in cancer
Breast cancer and reproductive system
Breast cancer
Breast cancer is the most common malignant tumour in women worldwide. Compared to other subtypes of breast cancer, triple negative breast cancer (TNBC) is the most aggressive and heterogeneous and often ends with organ-specific metastasis. It has certain resistance to conventional therapy, so prognostic markers and biotherapy are more helpful to the treatment of diseases [35]. TNBC is negative for estrogen (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) [36]. There are likewise four subtypes in clinical practice: (1) luminal androgen receptor (LAR), (2) immunomodulatory, (3) basal-like immune-suppressed, and (4) mesenchymal-like [37]. In breast cancer subtypes, HMGA1 has the highest expression in triple negative breast cancer, implying reduced overall survival [38], and its overexpression is positively correlated with HER-2/neu amplification and PR and negatively correlated with ER [39]. In TNBC, Extracellular HMGA1 can be used as the ligand of receptor for advanced glycation end products (RAGE) to form an HMGA1-RAGE autocrine loop and induce pERK signalling to increase the migration and invasion ability of tumour cells [40]. Cytoplasmic HMGA1 also inhibits p53-mitochondrial apoptosis and correlates with higher invasive tumor histotype [41]. Moreover, when extracellular HMGA1 is blocked, cell adhesion will increase and migration ability will be weakened [42], which indicates that extracellular HMGA1 can be a drug target for TNBC and a biomarker for predicting distant metastasis. In addition, HMGA1a can regulate RAS-extractable signal-related kinase (RAS/ERK) signaling pathway related-genes, including KIT ligand (KL) and caveolins 1 (CAV1) and 2, and increase the sensitivity of MCF-7 cells to the RAS/ERK pathway to promote metastasis [43].
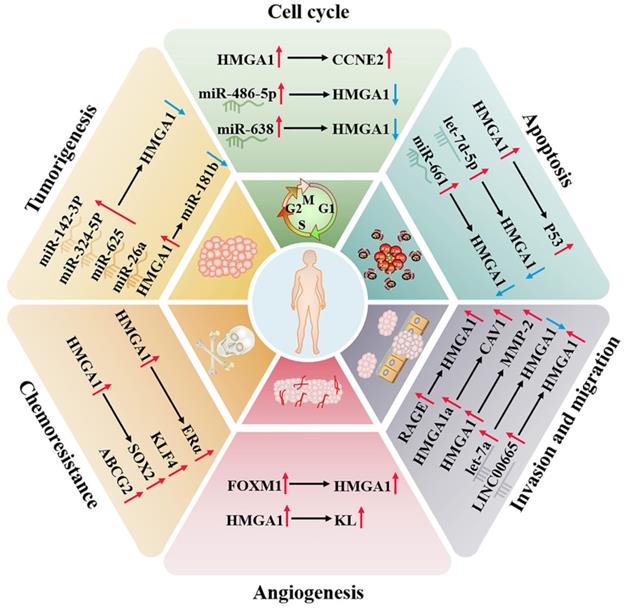
Studies have found that the HMGA1 gene does not rearrange and mutate in breast cancer. This indicates that overexpression is mediated by transcription activation or posttranscriptional regulation [44]. Fra-1 can bind to the enhancer located in the last two introns of HMGA1 and upregulate the expression of HMGA1 mRNA at the transcriptional level, thereby enhancing the malignant metastasis of TNBC [45]. TGF-β1 induces HMGA1 expression by activating HMGA1 promoter activity in MCF-7 and MDA-MB-231 cells, and specificity protein 1 (SP1) also participates in this process [46]. FOXM1 stabilizes in the nucleus by forming a complex with HMGA1, which increases transcriptional activity to shared target genes. At the same time, they can cooperate to drive breast cancer cells to promote tumour angiogenesis to support invasiveness [47].
In addition, the imbalance of noncoding RNAs is a cause of the higher incidence rate and mortality rate of breast cancer, which can directly target HMGA1 to the progression of cancer [48]. For example, let-7 is one of the earliest discovered microRNAs. Let-7a, a member of its family, can significantly downregulate the target gene HMGA1 to affect growth [49]; miR-142-3p and miR-26a can be used as tumour suppressors to inhibit the expression of oncogene HMGA1 [50, 51]; miR-661 induces apoptosis after downregulating HMGA1 [52]; and miR-486-5p leads to G2/M cell cycle arrest and profound cell death of breast cancer cells by downregulating HMGA1 [53]. LINC00963 competes with miR-625 for endogenous RNA and attenuated the inhibitory effect of miR-625 on HMGA1 [54]. HMGA1 can also negatively regulate CBX7, and CBX7 negatively regulates the expression of miR-181b to affect the progression of breast cancer [55].
In breast cancer, the binding activity of HMGA1a and RNA can regulate the alternative splicing of estrogen receptor alpha (ERα) and reduce the sensitivity of tamoxifen-resistant cell lines to tamoxifen [56]. HMGA1P6 and HMGA1P7, two pseudogenes of HMGA1 competing for endogenous RNA, can also lead to cancer when they are dysregulated, and the expression of HMGA1P7 is related to the levels of H19 and IGF2 in breast cancer, which is linked to the high double strand breaks in breast cancer cells [57]. In addition, HMGA1 upregulates the expression of cyclin E2 (CCNE2) to alter Yap nuclear localization and downstream activity of the Hippo pathway and finally regulates the movement of basal-like breast cancer cells [58].
Ovarian cancer
Surgical resection and cisplatin chemotherapy are the preferred choices for the treatment of ovarian cancer, but because early screening factors are not obvious, symptoms only appear in the late stage, and the 5-year survival rate can be as low as 30% [59, 60]. At present, studies have found that HMGA1 can regulate the expression of stem-related genes in ovarian cancer, such as KLF4, SOX2 and ABCG2, a drug-resistant protein, increasing the resistance of ovarian cancer to paclitaxel [61]. HMGA1 can also regulate the expression of the KL promoter in ovarian cancer to maintain angiogenesis, which also indicates that serum KL levels can be used to help the diagnosis of HMGA1-positive carcinomas [62]. HMGA1P6, as a competitive endogenous RNA of HMGA1, interferes with the effect of inhibiting microRNA in HMGA1 synthesis to enhance the malignancy of ovarian cancer cells, and MYC can participate in the transcription of HMGA1P6 [63, 64]. In contrast, let-7d-5p can negatively regulate the expression of HMGA1 and activate the p53 pathway, thereby inhibiting cell proliferation and promoting apoptosis and chemosensitivity [65]. miR-638 negatively regulates HMGA1 to facilitate the cell cycle and suppress apoptosis [66]. In addition, HMGA1 expression was not detected in epithelial cells, which are the common site of ovarian cancer, and compared with low malignant potential tumours, HMGA1 has a higher level in primary ovarian cancer, suggesting that HMGA1 has the potential to distinguish tumour biological behaviour [67].
Endometrial carcinoma
Endometrial cancer is a common disease of the female reproductive system, which mostly occurs in the uterine epithelium of postmenopausal women. Cytology and transvaginal ultrasound are commonly used to detect endometrial cancer, but there is a lack of specificity [68, 69]. The study confirmed that the protein and mRNA levels of HMGA1 are overexpressed in 46 cases of endometrial carcinoma (stages IA to IV), which is positively correlated with tumour stage, grade and size. The results were also verified in TCGA containing 381 endometrial carcinoma tumours (stages IA to IV), which implies that the overexpression of HMGA1 is a potential prognostic factor of endometrial carcinoma [70]. What's more, the expression of HMGA1 pseudogenes increases with tumor stage progression, which is consistent with the expression pattern of HMGA1. It may be the result of the upregulation of HMGA1 pseudogenes suppressing the inhibitory effect of ceRNA on HMGA1 [63, 70]. HMGA1 is involved in nuclear translocation of β-catenin protein and is combined with MMP-2 promoter to participate in the invasion of endometrial cancer [71, 72]. Both LINC00665 and circ 0067835 also upregulate HMGA1 to promote tumour progression. LINC00665 directly binds to the HMGA1 protein to promote cancer cell growth and invasion. Circ0067835 mainly induces the expression of HMGA1 by sponge miR-324-5p [73, 74]. In addition, HMGA1 was truncated by the recurrent breakpoint of chromosome 12q15 translocation and then formed a fusion gene with ectopic DNA sequence to induce the occurrence of uterine leiomyoma [75] (Fig. 2).
Testicular germ cell tumors
Testicular germ cell tumors (TGCTs) are common malignant solid tumors in men aged 20 - 40 years and are one of the most common causes of death in solid tumors at this age [76]. TGCTs are mainly divided into seminomas and nonseminomas germ-cell tumors (NSGCTs) [77]. Previous studies have confirmed that HMGA1 has definite advantages in distinguishing the histologic groups of TGCTs. HMGA1 is only detected in seminomas and embryonic carcinoma belonging to NSGCTs, and HMGA1 is overexpressed in most cases, which is helpful in differential diagnosis [78]. In addition, the overexpression of HMGA1 is partially regulated by the downregulation of Let-7a and miR-26a promoting proliferation, migration and invasion of seminoma cell line [79]. ERβ is an important mediator of germ cell growth and development. The interaction between HMGA1 and androgen receptor co-regulator PATZ1 down regulates ERβ that puts TCam-2 cell line at a higher risk of malignant transformation [80]. Moreover, after treatment with estrogens or estrogen-mimics, ERβ downregulation was also associated with increased cytoplasmic localization of HMGA1 [81]. In general, HMGA1 inhibitor will become a promising molecule to participate in the treatment of TGCTs.
Digestive system
Liver cancer
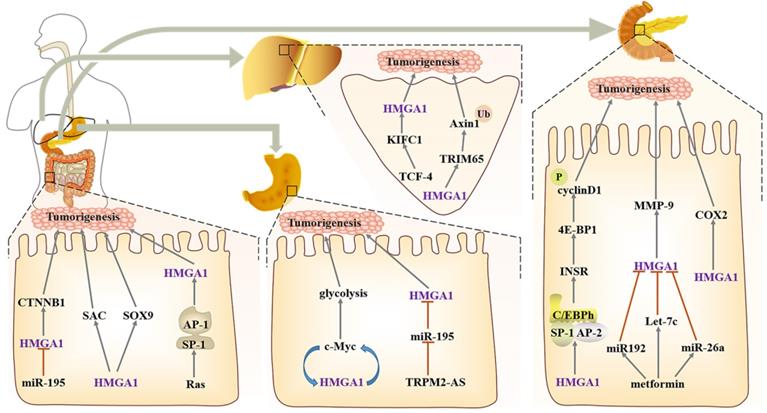
HMGA1 as an oncogene, is often expressed in digestive tract tumours, such as liver cancer, colorectal cancer, pancreatic cancer and gastric cancer [82-85]. In liver cancer, KIFC is located in the nucleus, and after being knocked down, the formation of internal foot and epithelial mesenchymal transformation can be reduced. TCF-4 is an important transcription factor in Wnt/β-catenin pathways, which enhances HMGA1 transcriptional activity by activating KIFC1, and then it promotes the pathogenesis of liver cancer and inhibits the sensitivity of liver cancer cells to paclitaxel [86]. Other studies also confirmed HMGA1 increases monotonously from normal liver to liver cirrhosis and then to liver cancer, both in mRNA and protein levels [87]. Overexpressed HMGA1 blocks the cell cycle and accelerates the proliferation and migration of cancer cells [88]. Hepatocellular carcinoma with high expression of HMGA1 showed a significant increase in the amount of Macrophages M0, while M2 with repair activity decreased, indicating that HMGA1 can affect tumor immune infiltration and help tumor cells escape [89]. Moreover, TRIM65, as an E3 ubiquitin ligase, can bind to Axin1 directly, ubiquitinate it and activate the β-catenin signalling pathway, while HMGA1, as an upstream factor of TRIM65, participates in the regulation [90].
Gastric cancer
In gastric cancer, HMGA1 and the c-Myc promoter contribute to induce c-Myc expression, thereby promoting aerobic glycolysis. After HMGA1 knockdown, glucose uptake and lactate production suffer as a result, and this change is only significant during glycolysis; there is no effect on oxidative phosphorylation in mitochondria, which suggests that HMGA1 regulation of gastric cancer metabolism has specificity [91]. In addition, HMGA1 serves as a downstream target of Wnt/β-catenin pathways; after Wnt/β-catenin activates c-Myc, HMGA1 expression can be induced to maintain the activity of gastric cancer cells, which forms a positive feedback loop between HMGA1 and c-Myc [92]. The downregulation of miR-195 combines with the HMGA1 3'- untranslated region (3' UTR) and induces its expression; this change in HMGA1 enhances the resistance of gastric cancer to 5-FU [93]. In addition, lncRNA TRPM2 antisense RNA (TRPM2-AS) as a miR-195 molecular sponge also indirectly participates in the modulation of HMGA1 [94].
Regulatory network of HMGA1 in the breast cancer and reproductive system. HMGA1 can function as an oncogene, regulating the cell cycle, apoptosis, invasion, migration, angiogenesis, tumorigenesis and chemoresistance by interacting with ncRNAs and targeting genes and ultimately maintaining the malignancy of the reproductive system, making it a promising tumour biomarker.
Colorectal cancer
For colorectal cancer with a rising incidence rate, PD-L1 can evade immune surveillance by inhibiting the function of T cells. PD-L1 is also very expressed in colorectal cancer cells and interacts with HMGA1 to activate HMGA1-dependent pathways, mainly including the PI3K/Akt and MEK/ERK pathways, and it ultimately maintain the self-renewal of colorectal cancer cells [95]. HMGA1 can also be induced by Ras, an oncogene in the distal regulatory region, which promotes the high expression of HMGA1 in tumour cells by activating the Ras GTPase signal and the cooperation between SP1 family members and AP1 factors [96]. In addition, miR-214 is defined as a tumour suppressor due to its regulation of several abnormal signalling pathways. After poor expression in colorectal cancer, it can upregulate the activity of HMGA1 and its binding CTNNB1 by weakening the binding of the 3' untranslated region of wild-type HMGA1 to activate the Wnt pathway [97, 98]. Sox9 is directly activated or indirectly upregulated by HMGA1 changing the chromatin structure to help construct the niche of Paneth cells, thereby amplifying the Wnt signal and maintaining self-renewal. Moreover, based on the overexpression of HMGA1, let-7 may be required for the development of Paneth cells [99].
In the study of faecal metabolomics of polyposis transgenic mice with abnormal expression of HMGA1, it was found that bile acid-related fatty acid metabolites in HMGA1 mice were significantly changed, and the synthesis of carnitine and Fas (myristic acid, palmitic acid, eicosenoic acid) increased, which helped the formation and maintenance of the cell membrane. Moreover, in some cancer samples, citric acid decreased, which reflects the possible existence of enhanced aerobic glycolysis [100]. Similarly, due to the hypomethylation of CpG sites in some abnormal promoters, CAV1 overexpression in colorectal cancer is often associated with the enhancement of aerobic glycolysis. It can stimulate the transcription of glucose transporter SLC2A3/GLUT3 through the HMGA1 binding site in the promoter, increase glucose uptake and ATP production, and protect tumour cells from metabolic stress. At the same time, after consumption of CAV1, autophagy can be induced by activating AMPK-TP53/p53 [101]. These indicate the important role of HMGA1 in carcinogenesis by regulating metabolic pathways.
In addition, dysregulation of the cell cycle is a mechanism of tumorigenesis. The mitotic spindle assembly checkpoint (SAC) is an essential cell cycle control system that can help maintain genomic homeostasis in eukaryotic cells. HMGA1 activates the expression of the SAC gene, which also illustrates a new mechanism of HMGA1 overexpression leading to cancer [102].
Pancreatic cancer
Compared with normal acinar tissue or fibroinflammatory stroma, HMGA1 was significantly increased in pancreatic tissue with intraepithelial neoplasia [103]. Periampullary carcinoma and pancreatic head carcinoma are two types of pancreatic-related cancers, but the expression of HMGA1 is higher in pancreatic head carcinoma and has no relationship with the later survival of patients [104]. Meanwhile, HMGA1 is used as an independent predictor in predicting the survival of patients with pancreatic cancer; it induces tumorigenesis through the PI3-K/Akt mechanism and ensures resistance to gemcitabine and anoikis [83, 105, 106]. When HMGA1 is silenced, there is a certain relationship between the activation of caspase 3 and the apoptosis induced by gemcitabine [105], and after HMGA1 overexpression, it can enhance the tumour invasion ability through Akt-dependent regulation of MMP-9 activity [107]. COX-2 is able to convert arachidonic acid into prostaglandin, and HMGA1 combines with its promoter junction to upregulate its expression. It is worth noting that sulindac and celecoxib, which are COX-2 inhibitors, can significantly block the formation of pancreatic tumours [108]. Metformin is a drug that can be used clinically to treat numerous diseases, such as type 2 diabetes [109], non-small cell lung cancer [110], and ovarian cancer [111]. In the xenotransplantation model of nude mice, metformin can promote the expression of miR-26a, miR-192 and Let-7c in a dose-dependent manner, and upregulated miR-26a can inhibit the expression of HMGA1 and tumour growth, which also suggests why part of the biological effects of metformin play a role through HMGA1 [112]. In Ras signals that can regulate HMGA1, the downstream pathways of Ras and HMGA1 are different, which also explains that there is a synergistic effect between Ras and HMGA1 in transforming normal pancreatic epithelium. Among them, K-Ras mutation is mainly in the early stage of PanIN lesions, and HMGA1 occurs in the late stage, meaning that HMGA1 overexpression can promote the transformation of the premise lesion of K-Ras mutation into a tumour. Interestingly, neither HMGA1 nor K-Ras alone can transform pancreatic epithelial cells [108].
One of the most important characteristics of pancreatic ductal carcinoma cells is the functional inactivation of the Rb-dependent G1 checkpoint. HMGA1 can accelerate the progression of the G1 phase and indirectly hasten the cell cycle of pancreatic ductal adenocarcinoma (PDAC). More importantly, HMGA1, SP1, AP2 and CAAT/enhancer binding protein h (C/EBPh) can form nuclear protein complexes to participate in the regulation of IR gene transcription factors, while IR can regulate the phosphorylation of 4E-BP1, thus controlling the translation of cyclin D1. Finally, the synergistic effect of cyclin D with CDK4/6 can also promote RB inactivation [113] (Fig. 3).
Haemopoietic system
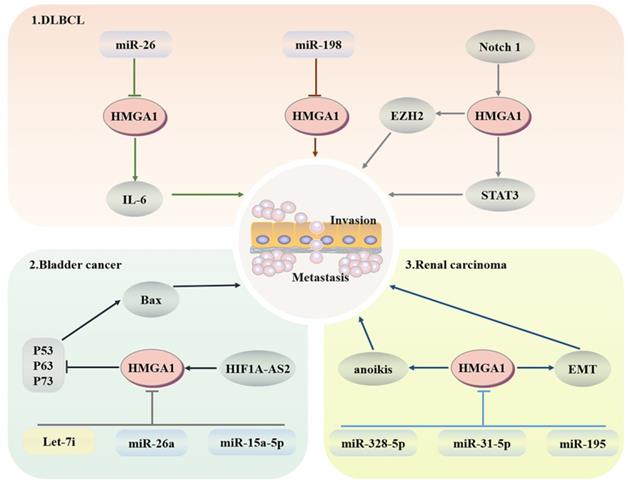
Diffuse large B-cell lymphoma (DLBCL) is a common subtype of non-Hodgkin lymphoma that is invasive and is usually treated with the proteasome inhibitor botizomib. The potential mechanism is that bortezomib upregulates miR-198 in DLBCL, and then miR-198 combines with the 3'-UTR of AT-hook 1 to inhibit the expression of HMGA1, thus inhibiting the proliferation and migration of DLBCL cells [114]. In addition, the imbalance of interleukin expression may lead to damage to T cell differentiation and expansion of B cell clones, resulting in B cell lymphoma. Enhancer of zeste-homolog 2 (EZH2), as an enzymatic subunit of PRC2, plays a cancer promoting role by catalyzing the methylation of histone H3 lysine 27 within PRC2 and regulating the G2/M transition of the cell cycle. HMGA1 combines with the promoter region of EZH2 to promote its transcription to enhance the proliferation and migration of B-cell lymphomas [115, 116]. MiR-26 has been proven to be a multiple tumour suppressor; it does not control IL-6-related inflammation and tumour promoting activity through transcription inhibition but downregulates the production of IL-6 through HMGA1 and MALT1 silencing-induced NF-κB inhibition [117]. HMGA1a targets STAT3 directly, whose activation can redisplay the transformation activity of HMGA1a to fibroblasts and promote the development of invasive lymphoid malignancy [118]. After HMGA1 knockout, the transcriptional activity of the Rag2 protein of V-to-DJ involved in the IgH locus is increased, resulting in the formation of BCR complexes in mature B cells and an increase in the tumour susceptibility of lymphoid tissue [119]. More importantly, the promoter region of HMGA1 can bind to the Notch1 signalling pathway related to leukaemia, and activated HMGA1 regulates cell proliferation through cell cycle regulation [120] (Fig. 4).
Regulatory network of HMGA1 in the digestive system. Liver cancer: TCF-4 enhances the transcriptional activity of HMGA1 and causes resistance to paclitaxel. HMGA1 acts on E3 ubiquitin ligase TRIM65 to activate the β-catenin pathway. Gastric cancer: HMGA1 and c-Myc form positive feedback in the regulation of glycolysis, and miR-195 negatively regulates HMGA1 to cause resistance to 5-FU. Colorectal cancer: HMGA1 upregulates the expression of CTNNB1, SAC and Sox9 to maintain the self-renewal of colon cancer cells. Pancreatic cancer, HMGA1 participates in the regulation of the cell cycle by interacting with the nucleoprotein complexes formed by SP1, AP2 and C/EBPh. Metformin can also indirectly participate in the regulation of HMGA1 to inhibit the progression of colorectal cancer. In addition, HMGA1 can increase the expression of COX-2 and promote pancreatic tumorigenesis.
Urinary system
Bladder cancer
LINC00649, which is located in the cytoplasm of bladder cancer, promotes the progression of bladder cancer by binding to miR-15a-5p to increase the expression of HMGA1 at the posttranscriptional level [121]. HIF1A-AS2 upregulates the expression of HMGA1, thereby inhibiting BAX transcription through P63-, P73- and p53-dependent apoptosis [122]. After HMGA1 silencing, downregulated p53 inducible nuclear protein 1 (TP53INP1) induces the activation of the ERK pathway in a p73-dependent DUSP10 manner. Moreover, TP53INP1 can also interact with LC3 to promote autophagic death, which is not linked to apoptosis [123]. MiR-26a directly targets HMGA1 to block bladder cancer cells in the G1 phase, leading to cell cycle arrest and dyskinesia [124, 125]. In addition, let-7i targets HMGA1 to significantly inhibit the proliferation and migration of T24 and 5637 bladder cancer cells, although let-7i is less expressed in bladder cancer [126].
Renal carcinoma
Studies have confirmed that HMGA1 is mainly expressed in the nucleus of renal carcinoma cells and induces anoikis. After HMGA1 overexpression, the colony-forming ability, invasion and migration ability of ACHN and Caki-1 renal carcinoma cells are enhanced [127]. At the same time, HMGA1 can enhance the expression of miR-671-5p and induce epithelial mesenchymal transformation (EMT) by activating Wnt signalling [128]. Furthermore, miR-328-5p, miR-31-5p and miR-195 have the capacity to prevent the progression of renal carcinoma by downgrading the expression levels of HMGA1 in the form of cell cycle stasis and EMT reversal [129-131]. All the above studies indicate that HMGA1 may be a potential therapeutic target for renal carcinoma (Fig. 4).
Regulatory network of HMGA1 in the haematopoietic system and urinary system. Haematopoietic system: HMGA1 mediates the imbalance of interleukin expression, resulting in B cell lymphoma. The Notch 1 signalling pathway associated with leukaemia activates HMGA1 to regulate cell proliferation. Bladder cancer: HIF1A-AS2 upregulates HMGA1 expression to inhibit apoptosis. MiR-26a targeting HMGA1 leads to cell cycle arrest. After HMAGA1 is inhibited by Let-7i and miR-15a-5p, the proliferation and migration of bladder cancer cells are also weakened. Renal cell carcinoma: HMGA1 promotes migration and metastasis by inducing anoikis and EMT.
Head and neck cancer
Thyroid cancer
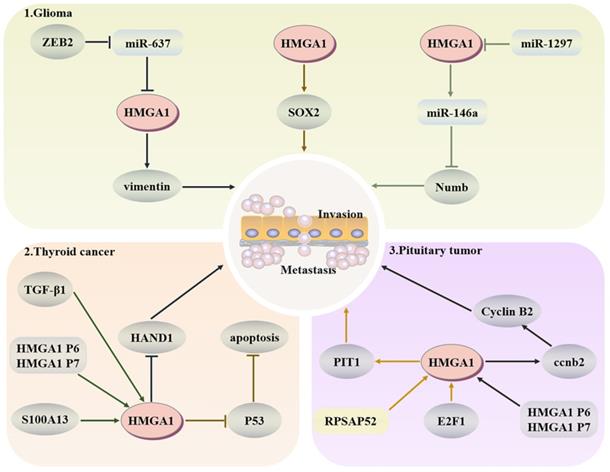
Thyroid cancer is a common malignant endocrine tumour that is often diagnosed by biopsy in the clinic. However, studies have demonstrated that the molecular diagnosis of tumour-specific markers has higher accuracy and sensitivity [132]. TGF-β1 can enhance the activity of the HMGA1 promoter and induce the expression of HMGA1 through extracellular signal-related kinase and phosphoinositide 3-kinase, so HMGA1 may exist as a tumour-specific marker for thyroid cancer [133]. HMGA1 pseudogenes are difficult to detect in well differentiated thyroid cancer, and are only highly expressed in anaplastic thyroid carcinomas, and upregulate the level of HMGA1 protein to exert carcinogenic activity [134]. S100A13, a calcium binding protein in the S100 family, regulates the proliferation and invasion of thyroid cancer by upregulating the expression of HMGA1 [135]. E2F1 interacts functionally with SP1 to promote HMGA1 expression. However, when SP1 binds to HMGA1 structurally, the balance between E2F family members is associated with the regulation of HMGA1 expression during quiescence and proliferation [136]. In addition, HMGA proteins can also induce thyroid cancer transformation but have no impact on its phenotype. By speeding up the cells into S phase and delaying G(2)-M conversion, apoptosis is induced. Moreover, the HMGA1b acetylation site K60 and the third AT-hook are necessary regions to induce apoptosis [137]. HAND1 in the Twist subfamily of Class B bHLH transcription factors is a key transcription factor that encodes trophoblast giant cell differentiation and heart tissue. Its promoter is methylated after binding with HMGA1, increasing the number of anaplastic cancer cell lines and promoting the progression of thyroid cancer [138]. More importantly, the tumour suppressor p53 family is expressed in thyroid cancer, but the inhibitory activity in malignant tumours remains latent. HMGA1 is stably located in mitochondria and can resist the release of cytochrome c by replacing the binding of p53 and Bcl-2 and inhibit p53-mediated apoptosis in a transcription-independent manner [41]. HMGA1 can be involved in the regulation of nuclear factor activity, depending on the interaction of the COOH-terminal oligomerization domain with all P53 family members, weakening the affinity between P53 and DNA, which also indicates that the tumour-promoting function of HMGA1 is partly dependent on the inhibition of tumour suppressor p53 activity [139].
Glioma
Glioma is a common malignant tumour of the brain. It originates from pluripotent neural stem cells and often develops into fatal brain cancer due to a large number of stem cells produced by symmetrical division. Numb has an asymmetric division of stem cells, and HMGA1 can negatively regulate the expression of Numb and promote the development of tumours by binding to the promoter of Numb or affecting miR-146a at the posttranscriptional level [140]. Because HMGA1 is a structural transcription factor, it can change the expression of Sox2 through changes in the chromatin structure to drive the stemness of cells [141]. Moreover, the stemness and temozolamide resistance of glioma cells also decreased after HMGA1 silencing [142]. Zeng et al. indicated that the E-box element in the miR-637 promoter could target HMGA1 and vimentin to promote glioma after binding with zinc finger E-box-binding homeobox-2 (ZEB2) [143]. As an inhibitor of glioma, miR-1297 can also target and negatively regulate HMGA1 to reduce cell viability [144].
Pituitary tumours
In pituitary tumours, HMGA1 can participate in the coding of CCNB2, produce more cyclin B2, and then regulate the cell cycle [145]. RPSAP52 lncRNA can increase the protein level of HMGA1, accelerate the G1-S phase transition and promote tumorigenesis [146]. PIT1, as a key transcription factor in pituitary development, after E2F1 binds to the HMGA1 promoter to increase the expression of HMGA1, and PIT1 tends to show a higher level in tumours [136]. Esposito et al. confirmed that the HMGA1 pseudogene also upregulates the expression of HMGA1 in pituitary tumours to promote tumorigenesis [147]. These results indicate that upregulation of HMGA1 will have a certain cancer-promoting effect in the pituitary (Fig. 5).
HMGA1 and therapeutic strategies of HMGA1 involvement
HMGA1 and Chemoresistance
Chemoresistance is a challenging issue in cancer therapy. Numerous studies have shown that HMGA1 can be involved in drug resistance to cisplatin, gefitinib, 5-FU, gemcitabine, and trabectedin in a TGF-β-, pathway-, microRNA-, and lncRNA-regulated manner in a variety of tumours, such as NSCLC, gastric cancer, ovarian cancer, glioblastoma, cholangiocarcinoma, colorectal cancer, thyroid cancer, and liposarcoma [65, 93, 142, 148-153]. More importantly, HMGA1 silencing can also reduce self-renewal, dryness and spheroid formation in glioblastoma and colorectal cancer, increasing drug sensitivity [95, 142]. As neo-adjuvant chemotherapy, trabectedin reduces HMGA1 expression in the treatment of myxoid LPS to play a potent anti-tumor activity. However, myxoid LPS resistant-cells upregulated the expression of HMGA1 and increased NFκB activity after treatment with trabectedin, resulting in tumor progression, but the cells will re-sensitive when drug-resistant cells treat with NF-κB, a NF-κB inhibitor [153]. HMGA1 in pancreatic cancer is resistant to gemcitabine by an Akt-dependent mechanism, but stable short hairpin RNA targeting HMGA1 can resensitize drug-resistant cells [105]. Therefore, blocking the expression of HMGA1 may be an effective target for chemotherapy resistance (Table 1).
HMGA1 and Autophagy
Autophagy is the degradation of damaged organelles and misfolded proteins in lysosomes under pressure conditions to recycle the available components in cells to maintain cell homeostasis. Autophagy disorder is often shown in cancer and can inhibit or promote tumours [154]. In most cases, cancer can be treated by cytotoxic autophagy, such as radiotherapy and chemotherapy, both of which enhance autophagy [155]. Similarly, HMGA1 depletion increases autophagy and finally influences cancer cell viability by inhibiting the mTOR pathway and upregulating the transcription level of ULK1. However, there is no correlation between the number and maturity of autophagosomes, which leads to a decrease in autophagy flow and affects cell proliferation and survival [156]. The ULk1 complex mainly induces autophagy by phosphorylating ATG13 and FIP200 [157]. In addition, the inhibition of HMGA1 can lead to the death of nerve cells partly through the decrease in autophagic flux induced by MPP+ [158]. Downregulation of HMGA1 can also increase autophagy to inhibit proliferation and migration of bladder cancer by downregulating the level of miR-221 [123]. On the other hand, HMGA1 can directly increase the promoter activity of miR-222 to inhibit autophagy induced by the p27/mTOR pathway and finally aggravate the cardiomyopathy induced by high glucose [157].
HMGA1 and Apoptosis Resistance
P53, a tumour suppressor gene, is mutated in most cancers. It mainly interacts with BCL-xL and Bcl-2 to change the permeability of the mitochondrial membrane to mediate apoptosis [159]. HMGA1, as a nuclear structural factor, can transfer p53 proapoptotic activator homeodomain-interacting protein kinase 2 (HIPK2) from the nucleus to the cytoplasm, modulate the transcription of p53 target genes p21, Bax and Bcl-2, inhibit p53-mediated apoptosis, help cells escape apoptosis and produce cisplatin resistance [122, 160, 161]. Moreover, the inhibitory effect of HMGA1 on p53-mediated mitochondrial apoptosis is mainly related to the inhibition of cytochrome C release and caspase activation. In other words, the cytoplasmic localization of HMGA1 in malignant tumours may be a new mechanism of p53 apoptosis inhibition [41]. For example, HMGA1 knockdown by siRNA can control the growth and invasion of medulloblastoma by regulating apoptosis and forming pseudopodia and stress fibres [162]. ShRNA-mediated HMGA1 silencing in BxPC3 activates caspase-3 and restores the chemosensitivity to gemcitabine [105].
Regulatory network of HMGA1 in the head and neck cancer. Glioma: HMGA1 negatively regulates the expression of Numb to inhibit the asymmetric division of stem cells and positively regulates Sox2 to drive the dryness of cells. ZEB2 also targets HMGA1 and vimentin to promote glioma. Thyroid cancer: TGF-β1 and S100A13 upregulate the expression of HMGA1 to promote invasion and metastasis, and HMGA1 can inhibit p53-mediated apoptosis. Pituitary tumours: HMGA1 participates in the coding of CCNB2 to regulate the cell cycle. After E2F1 binds to HMGA1, PIT1, a key factor of pituitary development, shows a higher level in malignant tumours.
Function of HMGA1 in different tumours
| Disease | Interactors | HMGA1 | Location | Pathway | Function | Reference |
|---|---|---|---|---|---|---|
| Lung adenocarcinoma Non-small cell lung cancer | GRP75 | Up | nucleus | JNK/c-JUN | Proliferation and metastasis | [28] |
| TGFβ1 | Up | nucleus | - | Cisplatin resistance | [148] | |
| LINC01748 | Up | nucleus | - | Increase aggressiveness | [149] | |
| Gastric cancer | miR-195 | Down | nucleus | - | Sensitive to 5-FU | [93] |
| c-Myc | Up | nucleus | Wnt/β-catenin | Warburg effect | [92] | |
| TRPM2-AS | Up | nucleus | MiR-195 molecular sponge | [94] | ||
| Ovarian cancer | Let-7d-5p | Down | nucleus | p53 | Sensitive to Cisplatin and apoptosis | [65] |
| miR-638 | Down | nucleus | - | Suppress apoptosis | [66] | |
| Glioma | miR-637, miR-1297 | Down | nucleus | - | Vimentin inhibition, reduce cell viability, sensitive to temozolomide | [142-144] |
| Pituitary tumours | RPSAP52 | Up | nucleus | - | Accelerate the G1-S phase transition | [146] |
| E2F1 | Up | nucleus | RB/E2F1 | Rb/E2F1 path imbalance | [136] | |
| HMGA1-pseudogene | Up | nucleus | - | Promote tumorigenesis | [147] | |
| Cholangiocarcinoma | INOS, ERBB2 | Up | nucleus | - | Gemcitabine resistance | [150] |
| TMPO-AS1 | Up | nucleus | - | Apoptosis resistance | [151] | |
| Colorectal cancer | PD-L1 | Up | nucleus | PI3K/Akt, MEK/ERK | Enhance dryness | [95] |
| Ras | Up | nucleus | Ras GTPase | Activate Ras GTPase signal | [96] | |
| miR-214 | Down | nucleus | - | Activate Wnt pathway | [97, 98] | |
| CAV1 | Up | nucleus | Wnt | Warburg effect | [101] | |
| Thyroid cancer | TGFβ1, S100A13 | Up | nucleus | ERK | Proliferation and invasion | [133, 135] |
| E2F1 | Up | nucleus | RB/E2F1 | Proliferation | [136] | |
| Breast cancer | RAGE | extracellular | ERK | Migration and invasion | [40] | |
| Bcl-2 | cytoplasm | - | Counteract apoptosis | [41] | ||
| Let-7a, miR-142-3p, miR-625 | Down | nucleus | - | Inhibition of proliferation and invasion | [49, 51, 54] | |
| miR-661 | Down | nucleus | - | Apoptosis | [52] | |
| miR-486-5p | Down | nucleus | - | G2/M cell cycle arrest | [53] | |
| Testicular germ cell tumors | Let-7a, miR-26a | Down | nucleus | - | Proliferation, migration and invasion | [79] |
| ERβ | Up | cytoplasm | - | Malignant transformation | [81] | |
| Liver cancer | TCF-4 | Up | nucleus | - | Paclitaxel resistance, EMT | [86] |
| TRIM65 | Up | nucleus | β-catenin | Activate β-catenin pathway | [90] | |
| Pancreatic cancer | miR-26a, miR-192, Let-7c | Down | nucleus | - | Inhibition of tumour growth | [108] |
| Endometrial carcinoma | LINC00665, circ 0067835 | Up | nucleus | - | Cell proliferation, invasion and migration | [73, 74] |
| Diffuse large B-cell lymphoma | miR-198 | Down | nucleus | - | Proliferation and migration | [114] |
| miR-26 | Down | nucleus | TNF-α/NF-κB | Control inflammation | [117] | |
| Leukaemia | EZH2 | Up | nucleus | - | Proliferation and migration | [116] |
| Notch1 | Up | nucleus | Notch1 | Cell cycle | [120] | |
| Bladder cancer | LINC00649 | Up | nucleus | - | Accelerate progression | [121] |
| miR-26a | Down | nucleus | - | Cell dyskinesia | [125] | |
| HIF1A-AS2 | Up | nucleus | - | Suppress apoptosis | [122] | |
| Let-7i | Down | nucleus | - | Proliferation and migration | [126] | |
| Renal carcinoma | miR-328-5p, miR-31-5p, miR-195 | Down | nucleus | P-Akt, Wnt | Cell cycle stasis and EMT reversal | [129-131] |
Moreover, cell death caused by HMGA1 expression is mainly related to DNA repair damage. Therefore, Baldassarre and others also expressed that HMGA1 is highly expressed in malignant tumours, which helps malignant transformation from the perspective of DNA repair [163].
HMGA1 and Senescence
In response to external pressure or cancer stimulation, cells stop replicating through the mechanism of ageing and inhibit tumour growth independently. Often, the activation of the tumour inhibitory pathway P53 or Rb, cell morphological changes, SA-β-galactosidase activity induction, enhanced protein secretion, and the formation of highly tight heterochromatin lesions are shown [164]. HMGA proteins can change the chromatin structure through the AT hook domain and form senescence-associated heterochromatic foci (SAHFs) to maintain the stability of ageing [165, 166]. Moreover, in mouse embryos overexpressing the HMGA1 P6 pseudogene, the mice grew faster and aged later, which was consistent with the results of pseudogene RNA crosstalk [167]. Some studies have further shown that NOTCH can inhibit the formation of tight chromatin structures caused by HMGA1 and the accessible chromatin region driven by RIS autonomously or nonautonomously in ageing cells induced by Ras. At the same time, the chromatin structure of adjacent cells changes through the connection between cells [168].
HMGA1 and Regulation of Stemness
Normal stem cells have the potential for multidirectional differentiation and proliferation. When their activity is hijacked by tumour cells, tumour cells can invade many parts of the body to escape treatment. Therefore, eliminating the stemness of cancer cells is another strategy for cancer treatment [169]. HMGA1 protein is expressed at low levels or not expressed in adult tissues but is highly expressed in embryonic tissues with strong proliferation ability, which indicates that HMGA1 mainly plays a role in cells with strong proliferation ability [163]. A large number of studies have shown that HMGA1 participates in the regulation of cancer diseases. For example, Wei et al. showed that PD-L1 upregulates the expression of HMGA1 and that the PI3K/Akt and MEK/ERK pathways related to HMGA1 were activated to favour the proliferation of colorectal cancer stem cells [95]; Kim et al. confirmed that HMGA1 overexpression in ovarian cancer could increase the stemness of cancer cells, that cell proliferation, spheroid formation and gene expression related to stemness are enhanced, and that it also has higher resistance to doxorubicin and paclitaxel [61]. Xian et al. indicated that HMGA1 can promote the expression of genes encoding Wnt receptors and Wnt downstream effectors and induce Paneth cells to differentiate into Sox9 to jointly establish an intestinal stem cell (ISC) niche [170]. Wang et al. showed that the extracellular vesicles released by glioblastoma can cause neural stem cells (NSCs) to participate in the recurrence of glioblastoma again, and the imbalance of HMGA1 is involved in this process. All these enable us to find new methods for the treatment of this kind of disease [171]. In addition, stem cells derived from human menstrual blood can regulate the abundance of 5-hydroxymethylcytosine (5-hmC) and 5-methylcytosine (5-mC) in enhancers and inhibit HMGA1 and other genes related to chemotherapy resistance [172]. HMGA1 silencing reduces the stemness of glioblastoma and restores temozolomide sensitivity [142]. HMGA1 itself can also promote the reprogramming of somatic cells to multifunctional stem cells, which will be useful for regenerative medicine and the treatment of poorly differentiated cancer [173].
Targeting HMGA1 and RNA Crosstalk
Destruction of DNA is a basis for cancer treatment. However, due to the tolerance of cancer cells, most tumours will also relapse. HMGA1 plays a major role in tumours recurrence and is often associated with the prediction of poor prognosis [174]. Clinically, doxorubicin, antiestrogen and other drugs target HMGA1 to achieve anticancer therapy [175, 176]. Similarly, HMGA1 itself does not possess transcriptional activity, functioning primarily by changing the structure of the chromosome by binding the DNA region rich in the A-T hook. Dispatycin, a groove-binding drug, affects the binding of DNA rich in the AT region by changing the conformation and sequence specifically, thus alleviating the symptoms of inflammation during endotoxaemia [177]. Adenovirus-mediated block of HMGA1 protein synthesis inhibited tumor growth, but had no effect on normal cells [178]. In addition, many long noncoding RNAs, microRNAs and the two pseudogenes P6 and P7 of HMGA1 can interfere with the expression of HMGA1 by competing with the endogenous RNA of HMGA1. We can use them as target regulators to inhibit cancer development. For example, studies have confirmed that bortezomib upregulates the level of miR-198 to enhance the inhibition of HMGA1 by miR-198 to exert anticancer effects [114]. Metformin upregulates miR-26a, which inhibits HMGA1 expression to affect pancreatic cancer progression [112]. Epidrug mediates the re-expression of HMGA-targeting miRNA to intervene in the progression of pituitary adenomas [179]. This further supports the strategy of targeted HMGA1 therapy.
Small molecule inhibitor
Anti-gene therapy is another breakthrough in clinical drug resistance therapy. Polyetheylenimine polyplexes of spiegelmer NOX-A50 and minor-groove binder (MGB) netropsin compete for the binding of HMGA1 to AT-hook double-stranded DNA, thus affecting the regulation of HMGA [180, 181]. Triplex-forming oligonucleotides (TFO1) and TFO-nitrogen mustard conjugates (TFO2) are two triplet oligonucleotides acting on the HMGA1 promoter. Although they are not stable with deoxynucleotides, they have a great regulatory effect on the transcription level of HMGA1. To improve the curative effect, it can be used together with adriamycin and other anticancer drugs, mainly because TFO is combined with the DNA groove, and adriamycin is embedded in the structure of DNA [182]. The FR900482 class of antitumour drugs cross-links to the minor groove-binding HMGA1 protein to exert antitumour activity [183]. However, these drugs partially affect the activity of HMGA1, so more specific HMGA1 inhibitors and targeted antibodies still need to be explored.
In addition, compared with the silencing of HMGA1 by shRNA and siRNA, the delivery of shRNA and siRNA by nanoparticles to block the transcription of HMGA1 does not interfere with AT binding protein in vivo [184].
Conclusion and Prospects
HMGA1 has been confirmed to affect malignant tumours and has the potential to be a useful biomarker for the diagnosis and treatment of malignant tumours. In addition, HMGA1 is related to the chemoresistance of tumours. However, the function of HMGA1 still divided. For example, individual studies have confirmed that in MCF-7 cells with high HMGA1 expression, the sensitivity to chemotherapeutic drugs will be enhanced due to the decreased expression of BRCA1 with DNA repair activity, so the authors suggested that increasing the expression of HMGA1 can help the malignant transformation of BRCA1-expressing tumours [163]. This may be based on the different cellular environment to affect the function of HMGA1 protein [185]. In B-cell lymphoma caused by an imbalance in interleukin expression, knocking off the HMGA1 gene causes mature B cells to form the BCR complex, increasing the susceptibility of lymphoma [119]. In fact, HMGA1 itself does not have transcriptional activity, which may be because its protein chaperones or regulated genes varies according to cell microenvironment and tumor heterogeneity. Therefore, the detailed role of HMGA1 needs further study.
HMGA1 is an important part of the progression of most tumours. HMGA1 can resist apoptosis and enhance senescence, which stops replication. HMGA1 may play a dual role in the process of disease development. In summary, because of the inconsistency of these functions, the mechanism by which HMGA1 causes these functions has yet to be explored to provide effective guidance for tumour diagnosis and targeted therapy.
Abbreviations
HMGA1: High mobility group A1; PI3K/Akt: Phosphatidylinositol 3 kinase/protein kinase B; MEK/ERK pathways: MAP kinase-ERK kinase/extracellular-signal-regulated kinase; ASP: Aspartic Acid; Glu: Glutamic acid; CK2: Tyrosine protein kinase; PKC: Protein kinase C; H1 kinase: M-phase specific kinase; HIPK2: Homeodomain-interacting protein kinase-2; NSCLC: Non-small cell lung cancer; TKI: Tyrosine kinase inhibitor; Ser: Serine; Thr: Threonine; MCM2: Minichromosome maintenance protein 2; IR: Insulin receptor; Lys: Lysine; PCAF: P300/CBP associated factor; CBP: Histone acetyltransferase binding protein; PRMT1: Arginine methyltransferase 1; PRMT2: Arginine methyltransferase 2; Arg: Arginine; DNA-PK: DNA-dependent protein kinase; TNBC: Triple negative breast cancer; ER: Estrogen receptor; PR: Progesterone receptor; HER2: Human epidermal growth factor receptor 2; LAR: Luminal androgen receptor; RAGE: Rceptor for advanced glycation end products; CAV1: Caveolin-1; RAS/ERK: RAS-extractable signal-related kinase; SP1: Specificity protein 1; ERα: Estrogen receptor alpha; IGF2: Insulin Like Growth Factor-2; CCNE2: Cyclin E2; Yap: Yes-associated protein; KLF4: Kruppel-like factor-4; SOX2: SRY-related high-mobility-group(HMG)-box protein-2; ABCG2: ATP binding cassette subfamily G member 2; KL: KIT ligand; MMP-2: Matrix metalloproteinase 2; TGCTs: Testicular germ cell tumors; NSGCTs: nonseminomas germ-cell tumors; KIFC1: Kinesin family member C1; TCF-4: T cell Factor 4; TRIM65: Tripartite motif‑containing 65; 5-FU: 5-fluorouracil; TRPM2-AS: TRPM2 antisense RNA; PD-L1: Programmed death ligand 1; AP1: Activator protein 1; CTNNB1: Catenin beta 1; SOX9: SRY-Box Transcription Factor 9; SAC: Mitotic spindle assembly checkpoint; MMP-9: Matrix metalloproteinase 9; COX-2: Cyclooxygenase-2; PDAC: Pancreatic ductal adenocarcinoma; C/EBPh: CAAT/enhancer binding protein h; 4E-BP1: 4E-binding protein 1; DLBCL: Diffuse large B-cell lymphoma; EZH2: Enhancer of zeste-homolog 2; PRC2: polycomb repressive complex 2; MALT1: Mucosa-associated lymphoid tissue lymphoma translocation protein 1; TP53INP1: p53 inducible nuclear protein 1; EMT: epithelial mesenchymal transformation; TGF-β1: transforming growth factor β1; E2F1: E2F transcription Factor 1; HAND1: Heart And Neural crest Derivatives expressed 1; ZEB2: Zinc finger E-box-binding homeobox-2; PIT1: Pituitary transcription Factor 1; ULK1: Unc51-like kinase-1; ATG13: Autophagy-related gene 13; MPP+: 1-Methyl-4-phenylpyridinium; HIPK2: Homeodomain-interacting protein kinase 2; SAHF: Senescence-associated heterochromatic foci; ISC: Intestinal stem cell; NSC: neural stem cell; 5-hmC: 5-hydroxymethylcytosine; 5-mC: 5-methylcytosine; MGB: minor-groove binder; TFO1: Triplex-forming oligonucleotide; TFO2: TFO-nitrogen mustard conjugate; BCR: B cell receptor.
Acknowledgements
This work was supported by grants from the Natural Science Foundation of Hunan Province (No. 2019JJ20014), the National Natural Science Foundation of China (No. 81773294, 82170138), the Program of National Health and Family Planning Commission of Hunan (20201929, B2019005), and the Chuanshan Talent Project of the University of South China (No. CS2018-3-35).
Author Contributions
LW wrote the first draft, summarized the tables and made the figures. MX, CL and JZ revised the manuscript and provided suggestions on the figures and tables. JZ and XYZ revised the manuscript and suggested supplementary material. All authors approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta. 2001;1519:13-29
2. Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35-100
3. Watanabe M, Ni S, Lindenberger AL, Cho J, Tinch SL, Kennedy MA. Characterization of the Stoichiometry of HMGA1/DNA Complexes. Open Biochem J. 2013;7:73-81
4. Young NL, Plazas-Mayorca MD, DiMaggio PA, Flaniken IZ, Beltran AJ, Mishra N. et al. Collective mass spectrometry approaches reveal broad and combinatorial modification of high mobility group protein A1a. J Am Soc Mass Spectrom. 2010;21:960-70
5. Piekielko A, Drung A, Rogalla P, Schwanbeck R, Heyduk T, Gerharz M. et al. Distinct organization of DNA complexes of various HMGI/Y family proteins and their modulation upon mitotic phosphorylation. J Biol Chem. 2001;276:1984-92
6. Xiao D, Huang K. [Effect of phosphorylation by protein kinase C on the DNA binding activity of high mobility group protein I]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 1996;18:79-83
7. Meijer L, Ostvold AC, Walass SI, Lund T, Laland SG. High-mobility-group proteins P1, I and Y as substrates of the M-phase-specific p34cdc2/cyclincdc13 kinase. Eur J Biochem. 1991;196:557-67
8. Zhang Q, Wang Y. Homeodomain-interacting protein kinase-2 (HIPK2) phosphorylates HMGA1a at Ser-35, Thr-52, and Thr-77 and modulates its DNA binding affinity. J Proteome Res. 2007;6:4711-9
9. Zhang Q, Zhang K, Zou Y, Perna A, Wang Y. A quantitative study on the in vitro and in vivo acetylation of high mobility group A1 proteins. J Am Soc Mass Spectrom. 2007;18:1569-78
10. Zou Y, Webb K, Perna AD, Zhang Q, Clarke S, Wang Y. A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA, and the effect of C-terminal phosphorylation. Biochemistry. 2007;46:7896-906
11. Wang YT, Pan SH, Tsai CF, Kuo TC, Hsu YL, Yen HY. et al. Phosphoproteomics Reveals HMGA1, a CK2 Substrate, as a Drug-Resistant Target in Non-Small Cell Lung Cancer. Sci Rep. 2017;7:44021
12. Zhao J, Grafi G. The high mobility group I/Y protein is hypophosphorylated in endoreduplicating maize endosperm cells and is involved in alleviating histone H1-mediated transcriptional repression. J Biol Chem. 2000;275:27494-9
13. Zou Y, Wang Y. Mass spectrometric analysis of high-mobility group proteins and their post-translational modifications in normal and cancerous human breast tissues. J Proteome Res. 2007;6:2304-14
14. Cheung CHY, Hsu CL, Chen KP, Chong ST, Wu CH, Huang HC. et al. MCM2-regulated functional networks in lung cancer by multi-dimensional proteomic approach. Sci Rep. 2017;7:13302
15. Chiefari E, Nevolo MT, Arcidiacono B, Maurizio E, Nocera A, Iiritano S. et al. HMGA1 is a novel downstream nuclear target of the insulin receptor signaling pathway. Sci Rep. 2012;2:251
16. Diana F, Sgarra R, Manfioletti G, Rustighi A, Poletto D, Sciortino MT. et al. A link between apoptosis and degree of phosphorylation of high mobility group A1a protein in leukemic cells. J Biol Chem. 2001;276:11354-61
17. Liu H, Liu H, Li X. Use of Serine/Threonine Ligation for the Total Chemical Synthesis of HMGA1a Protein with Site-Specific Lysine Acetylations. Chempluschem. 2019;84:779-785
18. Young NL, Plazas-Mayorca MD, DiMaggio PA, Flaniken IZ, Beltran AJ, Mishra N. et al. Collective mass spectrometry approaches reveal broad and combinatorial modification of high mobility group protein A1a. J Am Soc Mass Spectrom. 2010;21:960-70
19. Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133-6
20. Edberg DD, Bruce JE, Siems WF, Reeves R. In vivo posttranslational modifications of the high mobility group A1a proteins in breast cancer cells of differing metastatic potential. Biochemistry. 2004;43:11500-15
21. Jiang X, Wang Y. Acetylation and phosphorylation of high-mobility group A1 proteins in PC-3 human tumor cells. Biochemistry. 2006;45:7194-201
22. Klett H, Balavarca Y, Toth R, Gigic B, Habermann N, Scherer D. et al. Robust prediction of gene regulation in colorectal cancer tissues from DNA methylation profiles. Epigenetics. 2018;13:386-397
23. Sgarra R, Diana F, Rustighi A, Manfioletti G, Giancotti V. Increase of HMGA1a protein methylation is a distinctive characteristic of leukaemic cells induced to undergo apoptosis. Cell Death Differ. 2003;10:386-9
24. Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V. et al. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem. 2006;281:3764-72
25. Miranda TB, Webb KJ, Edberg DD, Reeves R, Clarke S. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem Biophys Res Commun. 2005;336:831-5
26. Zou Y, Wang Y. Tandem mass spectrometry for the examination of the posttranslational modifications of high-mobility group A1 proteins: symmetric and asymmetric dimethylation of Arg25 in HMGA1a protein. Biochemistry. 2005;44:6293-301
27. Gomarasca M, Lombardi G, Maroni P. SUMOylation and NEDDylation in Primary and Metastatic Cancers to Bone. Front Cell Dev Biol. 2022;10:889002
28. Qiao GB, Wang RT, Wang SN, Tao SL, Tan QY, Jin H. GRP75-mediated upregulation of HMGA1 stimulates stage I lung adenocarcinoma progression by activating JNK/c-JUN signaling. Thorac Cancer. 2021;12:1558-1569
29. Li Y, Lu J, Prochownik EV. Dual role for SUMO E2 conjugase Ubc9 in modulating the transforming and growth-promoting properties of the HMGA1b architectural transcription factor. J Biol Chem. 2007;282:13363-71
30. Giancotti V, Bandiera A, Sindici C, Perissin L, Crane-Robinson C. Calcium-dependent ADP-ribosylation of high-mobility-group I (HMGI) proteins. Biochem J. 1996;317:865-70
31. Hill DA, Pedulla ML, Reeves R. Directional binding of HMG-I(Y) on four-way junction DNA and the molecular basis for competitive binding with HMG-1 and histone H1. Nucleic Acids Res. 1999;27:2135-44
32. Brocher J, Vogel B, Hock R. HMGA1 down-regulation is crucial for chromatin composition and a gene expression profile permitting myogenic differentiation. BMC Cell Biol. 2010;11:64
33. Pellarin I, Arnoldo L, Costantini S, Pegoraro S, Ros G, Penzo C. et al. The Architectural Chromatin Factor High Mobility Group A1 Enhances DNA Ligase IV Activity Influencing DNA Repair. PLoS One. 2016;11:e0164258
34. Senigagliesi B, Penzo C, Severino LU, Maraspini R, Petrosino S, Morales-Navarrete H. et al. The High Mobility Group A1 (HMGA1) Chromatin Architectural Factor Modulates Nuclear Stiffness in Breast Cancer Cells. Int J Mol Sci. 2019;20:2733
35. Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L. et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97
36. Santuario-Facio SK, Cardona-Huerta S, Perez-Paramo YX, Trevino V, Hernandez-Cabrera F, Rojas-Martinez A. et al. A New Gene Expression Signature for Triple Negative Breast Cancer Using Frozen Fresh Tissue before Neoadjuvant Chemotherapy. Mol Med. 2017;23:101-111
37. Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X. et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell. 2019;35:428-440.e5
38. Gorbounov M, Carleton NM, Asch-Kendrick RJ, Xian L, Rooper L, Chia L. et al. High mobility group A1 (HMGA1) protein and gene expression correlate with ER-negativity and poor outcomes in breast cancer. Breast Cancer Res Treat. 2020;179:25-35
39. Sepe R, Piscuoglio S, Quintavalle C, Perrina V, Quagliata L, Formisano U. et al. HMGA1 overexpression is associated with a particular subset of human breast carcinomas. J Clin Pathol. 2016;69:117-21
40. Méndez O, Peg V, Salvans C, Pujals M, Fernández Y, Abasolo I. et al. Extracellular HMGA1 Promotes Tumor Invasion and Metastasis in Triple-Negative Breast Cancer. Clin Cancer Res. 2018;24:6367-6382
41. Esposito F, Tornincasa M, Federico A, Chiappetta G, Pierantoni GM, Fusco A. High-mobility group A1 protein inhibits p53-mediated intrinsic apoptosis by interacting with Bcl-2 at mitochondria. Cell Death Dis. 2012;3:e383
42. Méndez O, Pérez J, Soberino J, Racca F, Cortés J, Villanueva JJIjoms. Clinical Implications of Extracellular HMGA1 in Breast Cancer. Int J Mol Sci. 2019;20:5950
43. Treff NR, Pouchnik D, Dement GA, Britt RL, Reeves R. High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human breast cancer cells. Oncogene. 2004;23:777-85
44. Sgarra R, Pegoraro S, Ros G, Penzo C, Chiefari E, Foti D. et al. High Mobility Group A (HMGA) proteins: Molecular instigators of breast cancer onset and progression. Biochim Biophys Acta Rev Cancer. 2018;1869:216-229
45. Tolza C, Bejjani F, Evanno E, Mahfoud S, Moquet-Torcy G, Gostan T. et al. AP-1 Signaling by Fra-1 Directly Regulates HMGA1 Oncogene Transcription in Triple-Negative Breast Cancers. Mol Cancer Res. 2019;17:1999-2014
46. Zu X, Zhong J, Tan J, Tan L, Yang D, Zhang Q. et al. TGF-β1 induces HMGA1 expression in human breast cancer cells: implications of the involvement of HMGA1 in TGF-β signaling. Int J Mol Med. 2015;35:693-701
47. Zanin R, Pegoraro S, Ros G, Ciani Y, Piazza S, Bossi F. et al. HMGA1 promotes breast cancer angiogenesis supporting the stability, nuclear localization and transcriptional activity of FOXM1. J Exp Clin Cancer Res. 2019;38:313
48. Zhou WB, Zhong CN, Luo XP, Zhang YY, Zhang GY, Zhou DX. et al. miR-625 suppresses cell proliferation and migration by targeting HMGA1 in breast cancer. Biochem Biophys Res Commun. 2016;470:838-44
49. Liu K, Zhang C, Li T, Ding Y, Tu T, Zhou F. et al. Let-7a inhibits growth and migration of breast cancer cells by targeting HMGA1. Int J Oncol. 2015;46:2526-34
50. Jia XP, Meng LL, Fang JC, Wang HW, Chen J, Zhou J. et al. Aberrant Expression of miR-142-3p and its Target Gene HMGA1 and FZD7 in Breast Cancer and its Clinical Significance. Clin Lab. 2018;64:915-921
51. Zhao XX, Yuan QZ, Mu DP, Sun DW, Bo QA, Pan GZ. et al. MicroRNA-26a inhibits proliferation by targeting high mobility group AT-hook 1 in breast cancer. Int J Clin Exp Pathol. 2015;8:368-73
52. Sui C, Qu W, Lian Y, Feng C, Zhan Y. Hsa_circ_0069094 knockdown inhibits cell proliferation, migration, invasion and glycolysis, while induces cell apoptosis by miR-661/HMGA1 axis in breast cancer. Anticancer Drugs. 2021;32:829-841
53. Mansoori B, Najafi S, Mohammadi A, AsadollahSeraj H, Savadi P, Mansoori B, Nazari A, Mokhtarzadeh A, Roshani E, Duijf PH, Cho WC, Baradaran B. The synergy between miR-486-5p and tamoxifen causes profound cell death of tamoxifen-resistant breast cancer cells. Biomed Pharmacother. 2021;141:111925
54. Wu Z, Wang W, Wang Y, Wang X, Sun S, Yao Y. et al. Long noncoding RNA LINC00963 promotes breast cancer progression by functioning as a molecular sponge for microRNA-625 and thereby upregulating HMGA1. Cell Cycle. 2020;19:610-624
55. Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, Esposito F, Iaccarino A, Troncone G, Fusco A. Identification of a New Pathway for Tumor Progression: MicroRNA-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes Cancer. 2010;1:210-24
56. Ohe K, Miyajima S, Abe I, Tanaka T, Hamaguchi Y, Harada Y, Horita Y, Beppu Y, Ito F, Yamasaki T, Terai H, Mori M, Murata Y, Tanabe M, Ashida K, Kobayashi K, Enjoji M, Yanase T, Harada N, Utsumi T, Mayeda A. HMGA1a induces alternative splicing of estrogen receptor alpha in MCF-7 human breast cancer cells. J Steroid Biochem Mol Biol. 2018;182:21-26
57. De Martino M, Forzati F, Marfella M, Pellecchia S, Arra C, Terracciano L, Fusco A, Esposito F. HMGA1P7-pseudogene regulates H19 and Igf2 expression by a competitive endogenous RNA mechanism. Sci Rep. 2016;6:37622
58. Pegoraro S, Ros G, Ciani Y, Sgarra R, Piazza S, Manfioletti G. A novel HMGA1-CCNE2-YAP axis regulates breast cancer aggressiveness. Oncotarget. 2015;6:19087-101
59. Bi F, An Y, Sun T, You Y, Yang Q. PHGDH Is Upregulated at Translational Level and Implicated in Platin-Resistant in Ovarian Cancer Cells. Front Oncol. 2021;11:643129
60. Radu MR, Prădatu A, Duică F, Micu R, Creţoiu SM, Suciu N. et al. Ovarian Cancer: Biomarkers and Targeted Therapy. Biomedicines. 2021;9:693
61. Kim DK, Seo EJ, Choi EJ, Lee SI, Kwon YW, Jang IH. et al. Crucial role of HMGA1 in the self-renewal and drug resistance of ovarian cancer stem cells. Exp Mol Med. 2016;48:e255
62. Treff NR, Dement GA, Adair JE, Britt RL, Nie R, Shima JE. et al. Human KIT ligand promoter is positively regulated by HMGA1 in breast and ovarian cancer cells. Oncogene. 2004;23:8557-62
63. Esposito F, De Martino M, Petti MG, Forzati F, Tornincasa M, Federico A, Arra C, Pierantoni GM, Fusco A. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget. 2014;5:8341-54
64. Tian X, Song J, Zhang X, Yan M, Wang S, Wang Y. et al. MYC-regulated pseudogene HMGA1P6 promotes ovarian cancer malignancy via augmenting the oncogenic HMGA1/2. Cell Death Dis. 2020;11:167
65. Chen YN, Ren CC, Yang L, Nai MM, Xu YM, Zhang F. et al. MicroRNA let-7d-5p rescues ovarian cancer cell apoptosis and restores chemosensitivity by regulating the p53 signaling pathway via HMGA1. Int J Oncol. 2019;54:1771-1784
66. Ma L, Zhang W, Jin Y, Bai X, Yu Q. miR-638 suppresses proliferation by negatively regulating high mobility group A1 in ovarian cancer cells. Exp Ther Med. 2021;22:1319
67. Masciullo V, Baldassarre G, Pentimalli F, Berlingieri MT, Boccia A, Chiappetta G. et al. HMGA1 protein over-expression is a frequent feature of epithelial ovarian carcinomas. Carcinogenesis. 2003;24:1191-8
68. Trojano G, Olivieri C, Tinelli R, Damiani GR, Pellegrino A, Cicinelli E. Conservative treatment in early stage endometrial cancer: a review. Acta Biomed. 2019;90:405-410
69. Urick ME, Bell DW. Clinical actionability of molecular targets in endometrial cancer. Nat Rev Cancer. 2019;19:510-521
70. Palumbo Júnior A, de Sousa VPL, Esposito F, De Martino M, Forzati F, Moreira FCB. et al. Overexpression of HMGA1 Figures as a Potential Prognostic Factor in Endometrioid Endometrial Carcinoma (EEC). Genes (Basel). 2019;10:372
71. Han X, Cao Y, Wang K, Zhu G. HMGA1 facilitates tumor progression through regulating Wnt/β-catenin pathway in endometrial cancer. Biomed Pharmacother. 2016;82:312-8
72. Hillion J, Roy S, Heydarian M, Cope L, Xian L, Koo M. et al. The High Mobility Group A1 (HMGA1) gene is highly overexpressed in human uterine serous carcinomas and carcinosarcomas and drives Matrix Metalloproteinase-2 (MMP-2) in a subset of tumors. Gynecol Oncol. 2016;141:580-587
73. Cai Y, Hao M, Chang Y, Liu Y. LINC00665 enhances tumorigenicity of endometrial carcinoma by interacting with high mobility group AT-hook 1. Cancer Cell Int. 2021;21:8
74. Liu Y, Chang Y, Cai Y. Circ_0067835 sponges miR-324-5p to induce HMGA1 expression in endometrial carcinoma cells. J Cell Mol Med. 2020;24:13927-13937
75. Hennig Y, Wanschura S, Deichert U, Bartnitzke S, Bullerdiek J. Rearrangements of the high mobility group protein family genes and the molecular genetic origin of uterine leiomyomas and endometrial polyps. Mol Hum Reprod. 1996;2:277-83
76. De Martino M, Esposito F, Chieffi P. An update on microRNAs as potential novel therapeutic targets in testicular germ cell tumors. Intractable Rare Dis Res. 2020;9:184-186
77. Chieffi P. Recent advances in molecular and cell biology of testicular germ-cell tumors. Int Rev Cell Mol Biol. 2014;312:79-100
78. Franco R, Esposito F, Fedele M, Liguori G, Pierantoni GM, Botti G. et al. Detection of high-mobility group proteins A1 and A2 represents a valid diagnostic marker in post-pubertal testicular germ cell tumours. J Pathol. 2008;214:58-64
79. De Martino M, Esposito F, Pellecchia S, Cortez Cardoso Penha R, Botti G, Fusco A. et al. HMGA1-Regulating microRNAs Let-7a and miR-26a are Downregulated in Human Seminomas. Int J Mol Sci. 2020;21:3014
80. Chieffi P. New prognostic markers and potential therapeutic targets in human testicular germ cell tumors. Curr Med Chem. 2011;18:5033-40
81. Esposito F, Boscia F, Gigantino V, Tornincasa M, Fusco A, Franco R. et al. The high-mobility group A1-estrogen receptor β nuclear interaction is impaired in human testicular seminomas. J Cell Physiol. 2012;227:3749-55
82. Chang ZG, Yang LY, Wang W, Peng JX, Huang GW, Tao YM. et al. Determination of high mobility group A1 (HMGA1) expression in hepatocellular carcinoma: a potential prognostic marker. Dig Dis Sci. 2005;50:1764-70
83. Liau SS, Rocha F, Matros E, Redston M, Whang E. High mobility group AT-hook 1 (HMGA1) is an independent prognostic factor and novel therapeutic target in pancreatic adenocarcinoma. Cancer. 2008;113:302-14
84. Cui X, Zhang H, Chen T, Yu W, Shen K. Long Noncoding RNA SNHG22 Induces Cell Migration, Invasion, and Angiogenesis of Gastric Cancer Cells via microRNA-361-3p/HMGA1/Wnt/β-Catenin Axis. Cancer Manag Res. 2020;12:12867-12883
85. Yang Q, Wang X, Tang C, Chen X, He J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50:1801-1809
86. Teng K, Wei S, Zhang C, Chen J, Chen J, Xiao K. et al. KIFC1 is activated by TCF-4 and promotes hepatocellular carcinoma pathogenesis by regulating HMGA1 transcriptional activity. J Exp Clin Cancer Res. 2019;38:329
87. Andreozzi M, Quintavalle C, Benz D, Quagliata L, Matter M, Calabrese D. et al. HMGA1 Expression in Human Hepatocellular Carcinoma Correlates with Poor Prognosis and Promotes Tumor Growth and Migration in in vitro Models. Neoplasia. 2016;18:724-731
88. Shi M, Lv X, Zhu M, Dong Y, Hu L, Qian Y, Fan C, Tian N. HMGA1 promotes hepatocellular carcinoma proliferation, migration, and regulates cell cycle via miR-195-5p. Anticancer Drugs. 2022;33:e273-e285
89. Wei YG, Yang CK, Wei ZL, Liao XW, He YF, Zhou X, Huang HS, Lan CL, Han CY, Peng T. High-Mobility Group AT-Hook 1 Served as a Prognosis Biomarker and Associated with Immune Infiltrate in Hepatocellular Carcinoma. Int J Gen Med. 2022;15:609-621
90. Yang YF, Zhang MF, Tian QH, Zhang CZ. TRIM65 triggers β-catenin signaling via ubiquitylation of Axin1 to promote hepatocellular carcinoma. J Cell Sci. 2017;130:3108-3115
91. Cao XP, Cao Y, Zhao H, Yin J, Hou P. HMGA1 promoting gastric cancer oncogenic and glycolytic phenotypes by regulating c-myc expression. Biochem Biophys Res Commun. 2019;516:457-465
92. Akaboshi S, Watanabe S, Hino Y, Sekita Y, Xi Y, Araki K. et al. HMGA1 is induced by Wnt/beta-catenin pathway and maintains cell proliferation in gastric cancer. Am J Pathol. 2009;175:1675-85
93. Wang CQ. MiR-195 reverses 5-FU resistance through targeting HMGA1 in gastric cancer cells. Eur Rev Med Pharmacol Sci. 2019;23:3771-3778
94. Huang B, Chang C, Wang BL, Li H. ELK1-induced upregulation of lncRNA TRPM2-AS promotes tumor progression in gastric cancer by regulating miR-195/ HMGA1 axis. J Cell Biochem. 2019;120:16921-16933
95. Wei F, Zhang T, Deng SC, Wei JC, Yang P, Wang Q. et al. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 2019;450:1-13
96. Cleynen I, Huysmans C, Sasazuki T, Shirasawa S, Van de Ven W, Peeters K. Transcriptional control of the human high mobility group A1 gene: basal and oncogenic Ras-regulated expression. Cancer Res. 2007;67:4620-9
97. Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. miR-214 activates TP53 but suppresses the expression of RELA, CTNNB1, and STAT3 in human cervical and colorectal cancer cells. Cell Biochem Funct. 2017;35:464-471
98. Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. MicroRNA-214 suppresses growth, migration and invasion through a novel target, high mobility group AT-hook 1, in human cervical and colorectal cancer cells. Br J Cancer. 2016;115:741-51
99. Resar L, Chia L, Xian L. Lessons from the Crypt: HMGA1-Amping up Wnt for Stem Cells and Tumor Progression. Cancer Res. 2018;78:1890-1897
100. Williams MD, Xian L, Huso T, Park JJ, Huso D, Cope LM. et al. Fecal Metabolome in Hmga1 Transgenic Mice with Polyposis: Evidence for Potential Screen for Early Detection of Precursor Lesions in Colorectal Cancer. J Proteome Res. 2016;15:4176-4187
101. Ha TK, Chi SG. CAV1/caveolin 1 enhances aerobic glycolysis in colon cancer cells via activation of SLC2A3/GLUT3 transcription. Autophagy. 2012;8:1684-5
102. Pierantoni GM, Conte A, Rinaldo C, Tornincasa M, Gerlini R, Federico A. et al. Deregulation of HMGA1 expression induces chromosome instability through regulation of spindle assembly checkpoint genes. Oncotarget. 2015;6:17342-53
103. Veite-Schmahl MJ, Joesten WC, Kennedy MA. HMGA1 expression levels are elevated in pancreatic intraepithelial neoplasia cells in the Ptf1a-Cre; LSL-KrasG12D transgenic mouse model of pancreatic cancer. Br J Cancer. 2017;117:639-647
104. van der Zee JA, ten Hagen TL, Hop WC, van Dekken H, Dicheva BM, Seynhaeve AL. et al. Differential expression and prognostic value of HMGA1 in pancreatic head and periampullary cancer. Eur J Cancer. 2010;46:3393-9
105. Liau SS, Whang E. HMGA1 is a molecular determinant of chemoresistance to gemcitabine in pancreatic adenocarcinoma. Clin Cancer Res. 2008;14:1470-7
106. Liau SS, Jazag A, Ito K, Whang EE. Overexpression of HMGA1 promotes anoikis resistance and constitutive Akt activation in pancreatic adenocarcinoma cells. Br J Cancer. 2007;96:993-1000
107. Liau S, Jazag A, Whang E. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006;66:11613-22
108. Hillion J, Smail SS, Di Cello F, Belton A, Shah SN, Huso T. et al. The HMGA1-COX-2 axis: a key molecular pathway and potential target in pancreatic adenocarcinoma. Pancreatology. 2012;12:372-9
109. Huang H, Lorenz BR, Zelmanovitz PH, Chan CB. Metformin Preserves β-Cell Compensation in Insulin Secretion and Mass Expansion in Prediabetic Nile Rats. Int J Mol Sci. 2021;22:421
110. Huang S, He T, Yang S, Sheng H, Tang X, Bao F. et al. Metformin reverses chemoresistance in non-small cell lung cancer via accelerating ubiquitination-mediated degradation of Nrf2. Transl Lung Cancer Res. 2020;9:2337-2355
111. Urpilainen E, Puistola U, Boussios S, Karihtala P. Metformin and ovarian cancer: the evidence. Ann Transl Med. 2020;8:1711
112. Li W, Yuan Y, Huang L, Qiao M, Zhang Y. Metformin alters the expression profiles of microRNAs in human pancreatic cancer cells. Diabetes Res Clin Pract. 2012;96:187-95
113. Kolb S, Fritsch R, Saur D, Reichert M, Schmid RM, Schneider G. HMGA1 controls transcription of insulin receptor to regulate cyclin D1 translation in pancreatic cancer cells. Cancer Res. 2007;67:4679-86
114. Wang R, Shen J, Wang Q, Zhang M. Bortezomib inhibited the progression of diffuse large B-cell lymphoma via targeting miR-198. Biomed Pharmacother. 2018;108:43-49
115. De Martino M, Nicolau-Neto P, Ribeiro Pinto LF, Traverse-Glehen A, Bachy E, Gigantino V. et al. HMGA1 induces EZH2 overexpression in human B-cell lymphomas. Am J Cancer Res. 2021;11:2174-2187
116. De Martino M, Esposito F, Fusco A. Critical role of the high mobility group A proteins in hematological malignancies. Hematol Oncol. 2022;40:2-10
117. Chen CY, Chang JT, Ho YF, Shyu AB. MiR-26 down-regulates TNF-α/NF-κB signalling and IL-6 expression by silencing HMGA1 and MALT1. Nucleic Acids Res. 2016;44:3772-87
118. Hillion J, Dhara S, Sumter TF, Mukherjee M, Di Cello F, Belton A. et al. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an achilles heel for hematopoietic malignancies? Cancer Res. 2008;68:10121-7
119. Fedele M, Fidanza V, Battista S, Pentimalli F, Klein-Szanto AJ, Visone R. et al. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 2006;66:2536-43
120. Xi Y, Li YS, Tang HB. High mobility group A1 protein acts as a new target of Notch1 signaling and regulates cell proliferation in T leukemia cells. Mol Cell Biochem. 2013;374:173-80
121. Chen X, Chen S. LINC00649 promotes bladder cancer malignant progression by regulating the miR-15a-5p/HMGA1 axis. Oncol Rep. 2021;45:8
122. Chen X, Liu M, Meng F, Sun B, Jin X, Jia C. The long noncoding RNA HIF1A-AS2 facilitates cisplatin resistance in bladder cancer. J Cell Biochem. 2019;120:243-252
123. Liu X, Zhou Z, Wang Y, Zhu K, Deng W, Li Y. et al. Downregulation of HMGA1 Mediates Autophagy and Inhibits Migration and Invasion in Bladder Cancer via miRNA-221/TP53INP1/p-ERK Axis. Front Oncol. 2020;10:589
124. Lin R, Shen W, Zhi Y, Zhou Z. Prognostic value of miR-26a and HMGA1 in urothelial bladder cancer. Biomed Pharmacother. 2014;68:929-34
125. Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y. et al. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett. 2013;587:2467-73
126. Qin MM, Chai X, Huang HB, Feng G, Li XN, Zhang J. et al. let-7i inhibits proliferation and migration of bladder cancer cells by targeting HMGA1. BMC Urol. 2019;19:53
127. Takaha N, Sowa Y, Takeuchi I, Hongo F, Kawauchi A, Miki T. Expression and role of HMGA1 in renal cell carcinoma. J Urol. 2012;187:2215-22
128. Chi XG, Meng XX, Ding DL, Xuan XH, Chen YZ, Cai Q. et al. HMGA1-mediated miR-671-5p targets APC to promote metastasis of clear cell renal cell carcinoma through Wnt signaling. Neoplasma. 2020;67:46-53
129. Zhang W, Lu Y, Shi H, Li X, Zhang Z, Deng X. et al. LncRNA ITGB2-AS1 promotes the progression of clear cell renal cell carcinoma by modulating miR-328-5p/HMGA1 axis. Hum Cell. 2021;34:1545-1557
130. Liu Z, Wang R, Zhu G. Circ_0035483 Functions as a Tumor Promoter in Renal Cell Carcinoma via the miR-31-5p-Mediated HMGA1 Upregulation. Cancer Manag Res. 2021;13:693-706
131. Dong H, Sun S, Yan T, Liang C, Zhu J, Miao C. et al. MicroRNA-195 inhibits proliferation and metastasis in renal cell carcinoma via regulating HMGA1. Am J Transl Res. 2020;12:2781-2792
132. Rodrigo JP, Rinaldo A, Devaney KO, Shaha AR, Ferlito A. Molecular diagnostic methods in the diagnosis and follow-up of well-differentiated thyroid carcinoma. Head Neck. 2006;28:1032-9
133. Zhong J, Liu C, Zhang QH, Chen L, Shen YY, Chen YJ. et al. TGF-β1 induces HMGA1 expression: The role of HMGA1 in thyroid cancer proliferation and invasion. Int J Oncol. 2017;50:1567-1578
134. Esposito F, De Martino M, Forzati F, Fusco A. HMGA1-pseudogene overexpression contributes to cancer progression. Cell Cycle. 2014;13:3636-9
135. Zhong J, Liu C, Chen YJ, Zhang QH, Yang J, Kang X. et al. The association between S100A13 and HMGA1 in the modulation of thyroid cancer proliferation and invasion. J Transl Med. 2016;14:80
136. Massimi I, Guerrieri F, Petroni M, Veschi V, Truffa S, Screpanti I. et al. The HMGA1 protoncogene frequently deregulated in cancer is a transcriptional target of E2F1. Mol Carcinog. 2013;52:526-34
137. Fedele M, Pierantoni GM, Berlingieri MT, Battista S, Baldassarre G, Munshi N. et al. Overexpression of proteins HMGA1 induces cell cycle deregulation and apoptosis in normal rat thyroid cells. Cancer Res. 2001;61:4583-90
138. Martinez Hoyos J, Ferraro A, Sacchetti S, Keller S, De Martino I, Borbone E. et al. HAND1 gene expression is negatively regulated by the High Mobility Group A1 proteins and is drastically reduced in human thyroid carcinomas. Oncogene. 2009;28:876-85
139. Frasca F, Rustighi A, Malaguarnera R, Altamura S, Vigneri P, Del Sal G. et al. HMGA1 inhibits the function of p53 family members in thyroid cancer cells. Cancer Res. 2006;66:2980-9
140. Puca F, Tosti N, Federico A, Kuzay Y, Pepe A, Morlando S. et al. HMGA1 negatively regulates NUMB expression at transcriptional and post transcriptional level in glioblastoma stem cells. Cell Cycle. 2019;18:1446-1457
141. Lopez-Bertoni H, Lal B, Michelson N, Guerrero-Cázares H, Quiñones-Hinojosa A, Li Y. et al. Epigenetic modulation of a miR-296-5p: HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene. 2016;35:4903-13
142. Colamaio M, Tosti N, Puca F, Mari A, Gattordo R, Kuzay Y. et al. HMGA1 silencing reduces stemness and temozolomide resistance in glioblastoma stem cells. Expert Opin Ther Targets. 2016;20:1169-79
143. Zeng Y, Que T, Lin J, Zhan Z, Xu A, Wu Z. et al. Oncogenic ZEB2/miR-637/HMGA1 signaling axis targeting vimentin promotes the malignant phenotype of glioma. Mol Ther Nucleic Acids. 2021;23:769-782
144. Wang J, Xu X, Mo S, Tian Y, Wu J, Zhang J. et al. Involvement of microRNA-1297, a new regulator of HMGA1, in the regulation of glioma cell growth in vivo and in vitro. Am J Transl Res. 2016;8:2149-58
145. De Martino I, Visone R, Wierinckx A, Palmieri D, Ferraro A, Cappabianca P. et al. HMGA proteins up-regulate CCNB2 gene in mouse and human pituitary adenomas. Cancer Res. 2009;69:1844-50
146. D'Angelo D, Arra C, Fusco A. RPSAP52 lncRNA Inhibits p21Waf1/CIP Expression by Interacting With the RNA Binding Protein HuR. Oncol Res. 2020;28:191-201
147. Esposito F, De Martino M, D'Angelo D, Mussnich P, Raverot G, Jaffrain-Rea ML. et al. HMGA1-pseudogene expression is induced in human pituitary tumors. Cell Cycle. 2015;14:1471-5
148. Zhang R, Tao F, Ruan S, Hu M, Hu Y, Fang Z. et al. The TGFβ1-FOXM1-HMGA1-TGFβ1 positive feedback loop increases the cisplatin resistance of non-small cell lung cancer by inducing G6PD expression. Am J Transl Res. 2019;11:6860-6876
149. Tan Y, Xu F, Xu L, Cui J. Long non-coding RNA LINC01748 exerts carcinogenic effects in non-small cell lung cancer cell lines by regulating the microRNA-520a-5p/HMGA1 axis. Int J Mol Med. 2022;49:22
150. Quintavalle C, Burmeister K, Piscuoglio S, Quagliata L, Karamitopoulou E, Sepe R. et al. High mobility group A1 enhances tumorigenicity of human cholangiocarcinoma and confers resistance to therapy. Mol Carcinog. 2017;56:2146-2157
151. Chang H, Yao Y. lncRNA TMPO antisense RNA 1 promotes the malignancy of cholangiocarcinoma cells by regulating let-7g-5p/ high-mobility group A1 axis. Bioengineered. 2022;13:2889-2901
152. D'Angelo D, Mussnich P, Rosa R, Bianco R, Tortora G, Fusco A. High mobility group A1 protein expression reduces the sensitivity of colon and thyroid cancer cells to antineoplastic drugs. BMC Cancer. 2014;14:851
153. Loria R, Laquintana V, Bon G, Trisciuoglio D, Frapolli R, Covello R. et al. HMGA1/E2F1 axis and NFkB pathways regulate LPS progression and trabectedin resistance. Oncogene. 2018;37:5926-5938
154. Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913-30
155. Sharma K, Le N, Alotaibi M, Gewirtz DA. Cytotoxic autophagy in cancer therapy. Int J Mol Sci. 2014;15:10034-51
156. Conte A, Paladino S, Bianco G, Fasano D, Gerlini R, Tornincasa M. et al. High mobility group A1 protein modulates autophagy in cancer cells. Cell Death Differ. 2017;24:1948-1962
157. Wu QQ, Liu C, Cai Z, Xie Q, Hu T, Duan M. et al. High-mobility group AT-hook 1 promotes cardiac dysfunction in diabetic cardiomyopathy via autophagy inhibition. Cell Death Dis. 2020;11:160
158. Li G, Luo W, Wang B, Qian C, Ye Y, Li Y. et al. HMGA1 Induction of miR-103/107 Forms a Negative Feedback Loop to Regulate Autophagy in MPTP Model of Parkinson's Disease. Front Cell Neurosci. 2021;14:620020
159. Wei H, Qu L, Dai S, Li Y, Wang H, Feng Y. et al. Structural insight into the molecular mechanism of p53-mediated mitochondrial apoptosis. Nat Commun. 2021;12:2280
160. Pierantoni GM, Rinaldo C, Mottolese M, Di Benedetto A, Esposito F, Soddu S. et al. High-mobility group A1 inhibits p53 by cytoplasmic relocalization of its proapoptotic activator HIPK2. J Clin Invest. 2007;117:693-702
161. Esposito F, Tornincasa M, Chieffi P, De Martino I, Pierantoni GM, Fusco A. High-mobility group A1 proteins regulate p53-mediated transcription of Bcl-2 gene. Cancer Res. 2010;70:5379-88
162. Lau KM, Chan QK, Pang JC, Ma FM, Li KK, Yeung WW. et al. Overexpression of HMGA1 deregulates tumor growth via cdc25A and alters migration/invasion through a cdc25A-independent pathway in medulloblastoma. Acta Neuropathol. 2012;123:553-71
163. Baldassarre G, Belletti B, Battista S, Nicoloso MS, Pentimalli F, Fedele M. et al. HMGA1 protein expression sensitizes cells to cisplatin-induced cell death. Oncogene. 2005;24:6809-19
164. Contrepois K, Thuret JY, Courbeyrette R, Fenaille F, Mann C. Deacetylation of H4-K16Ac and heterochromatin assembly in senescence. Epigenetics Chromatin. 2012;5:15
165. Doubleday PF, Fornelli L, Kelleher NL. Elucidating Proteoform Dynamics Underlying the Senescence Associated Secretory Phenotype. J Proteome Res. 2020;19:938-948
166. Satou W, Suzuki T, Noguchi T, Ogino H, Fujii M, Ayusawa D. AT-hook proteins stimulate induction of senescence markers triggered by 5-bromodeoxyuridine in mammalian cells. Exp Gerontol. 2004;39:173-9
167. De Martino M, Palma G, Arra C, Chieffi P, Fusco A, Esposito F. Characterization of HMGA1P6 transgenic mouse embryonic fibroblasts. Cell Cycle. 2020;19:2281-2285
168. Parry AJ, Hoare M, Bihary D, Hänsel-Hertsch R, Smith S, Tomimatsu K. et al. NOTCH-mediated non-cell autonomous regulation of chromatin structure during senescence. Nat Commun. 2018;9:1840
169. Yanagisawa BL, Resar LM. Hitting the bull's eye: targeting HMGA1 in cancer stem cells. Expert Rev Anticancer Ther. 2014;14:23-30
170. Xian L, Georgess D, Huso T, Cope L, Belton A, Chang YT. et al. HMGA1 amplifies Wnt signalling and expands the intestinal stem cell compartment and Paneth cell niche. Nat Commun. 2017;8:15008
171. Wang J, Liu J, Sun G, Meng H, Wang J, Guan Y. et al. Glioblastoma extracellular vesicles induce the tumour-promoting transformation of neural stem cells. Cancer Lett. 2019;466:1-12
172. Wu Y, Chen X, Zhao Y, Wang Y, Li Y, Xiang C. Genome-wide DNA methylation and hydroxymethylation analysis reveal human menstrual blood-derived stem cells inhibit hepatocellular carcinoma growth through oncogenic pathway suppression via regulating 5-hmC in enhancer elements. Stem Cell Res Ther. 2019;10:151
173. Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J. et al. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One. 2012;7:e48533
174. Palmieri D, Valentino T, D'Angelo D, De Martino I, Postiglione I, Pacelli R. et al. HMGA proteins promote ATM expression and enhance cancer cell resistance to genotoxic agents. Oncogene. 2011;30:3024-35
175. Akhter MZ, Luthra K, Rajeswari MR. Molecular aspects on adriamycin interaction with hmga1 regulatory region and its inhibitory effect on HMGA1 expression in human cervical cancer. J Biomol Struct Dyn. 2016;34:877-91
176. Leung YK, Chan QK, Ng CF, Ma FM, Tse HM, To KF. et al. Hsa-miRNA-765 as a key mediator for inhibiting growth, migration and invasion in fulvestrant-treated prostate cancer. PLoS One. 2014;9:e98037
177. Baron RM, Lopez-Guzman S, Riascos DF, Macias AA, Layne MD, Cheng G. et al. Distamycin A inhibits HMGA1-binding to the P-selectin promoter and attenuates lung and liver inflammation during murine endotoxemia. PLoS One. 2010;5:e10656
178. Scala S, Portella G, Fedele M, Chiappetta G, Fusco A. Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc Natl Acad Sci U S A. 2000;97:4256-61
179. Kitchen MO, Yacqub-Usman K, Emes RD, Richardson A, Clayton RN, Farrell WE. Epidrug mediated re-expression of miRNA targeting the HMGA transcripts in pituitary cells. Pituitary. 2015;18:674-84
180. Maasch C, Vater A, Buchner K, Purschke WG, Eulberg D, Vonhoff S, Klussmann S. Polyetheylenimine-polyplexes of Spiegelmer NOX-A50 directed against intracellular high mobility group protein A1 (HMGA1) reduce tumor growth in vivo. J Biol Chem. 2010;285:40012-8
181. Grant MA, Baron RM, Macias AA, Layne MD, Perrella MA, Rigby AC. Netropsin improves survival from endotoxaemia by disrupting HMGA1 binding to the NOS2 promoter. Biochem J. 2009;418:103-12
182. Akhter MZ, Rajeswari MR. Triplex forming oligonucleotides targeted to hmga1 selectively inhibit its expression and induce apoptosis in human cervical cancer. J Biomol Struct Dyn. 2017;35:689-703
183. Beckerbauer L, Tepe JJ, Cullison J, Reeves R, Williams RM. FR900482 class of anti-tumor drugs cross-links oncoprotein HMG I/Y to DNA in vivo. Chem Biol. 2000;7:805-12
184. Huso TH, Resar LM. The high mobility group A1 molecular switch: turning on cancer - can we turn it off? Expert Opin Ther Targets. 2014;18:541-53
185. Chiappetta G, Ottaiano A, Vuttariello E, Monaco M, Galdiero F, Gallipoli A. et al. HMGA1 protein expression in familial breast carcinoma patients. Eur J Cancer. 2010;46:332-9
Author contact
![]() Corresponding authors: Jing Zhong (zhongjing2002edu.cn) and Xuyu Zu (zuxuyu0108com)
Corresponding authors: Jing Zhong (zhongjing2002edu.cn) and Xuyu Zu (zuxuyu0108com)

 Global reach, higher impact
Global reach, higher impact