10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(12):4731-4743. doi:10.7150/ijbs.72482 This issue Cite
Review
SARS-CoV-2 and the Nucleus
Department of Biomedical Sciences, Faculty of Health Sciences, University of Macau, Taipa, Macau, China
Received 2022-2-28; Accepted 2022-6-20; Published 2022-7-11
Abstract
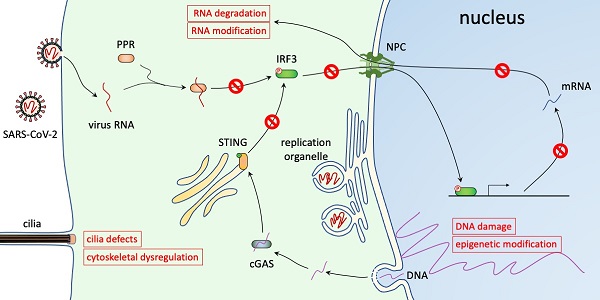
The ongoing COVID-19 pandemic is caused by an RNA virus, SARS-CoV-2. The genome of SARS-CoV-2 lacks a nuclear phase in its life cycle and is replicated in the cytoplasm. However, interfering with nuclear trafficking using pharmacological inhibitors greatly reduces virus infection and virus replication of other coronaviruses is blocked in enucleated cells, suggesting a critical role of the nucleus in virus infection. Here, we summarize the alternations of nuclear pathways caused by SARS-CoV-2, including nuclear translocation pathways, innate immune responses, mRNA metabolism, epigenetic mechanisms, DNA damage response, cytoskeleton regulation, and nuclear rupture. We consider how these alternations contribute to virus replication and discuss therapeutic treatments that target these pathways, focusing on small molecule drugs that are being used in clinical studies.
Keywords: SARS-CoV-2, nuclear transport, innate immunity, epigenetics, cilia, DNA damage
Introduction
The ongoing coronavirus disease 2019 (COVID-19) is one of the deadliest infectious diseases in history. To date, it has caused more than 536 million confirmed cases and over 6.31 million deaths (Weekly epidemiological update on COVID-19, retrieved from https://www.who.int on 24 June 2022). COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the Coronaviridae family of enveloped RNA viruses (1). The Coronaviridae family includes the Letovirinae and Orthocoronavirinae subfamilies and members of the latter are commonly known as coronaviruses (2). The coronaviruses are categorized into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (2). SARS-CoV-2 belongs to the genus Betacoronavirus. Betacoronavirus can be further divided into four lineages, or subgroups (3,4). Common cold-causing HKU1 and OC43 belong to subgroup A, SARS-CoV and SARS-CoV-2 belong to subgroup B, and MERS-CoV is a member of subgroup C. All beta-coronaviruses are enveloped with a lipid bilayer that contains transmembrane proteins: membrane (M), envelope (E), and spike (S) structural proteins (5-7). The M and E proteins are essential for virion assembly and budding and determine the shape and size of the virus particles. The S protein forms surface projections and mediates viral entry into the host cells. It has been the center for anti-viral research and the major target for vaccine and therapeutic development. Inside of the envelope is the viral genome, a 26-32 kilobase (29.8-29.9 kilobase for SARS-CoV-2) positive-sense single-stranded RNA. The RNA genome is organized and protected by the nucleocapsid (N) structural protein. More than two-thirds of the SARS-CoV-2 genome encodes 16 nonstructural proteins (Nsp1-Nsp16), while the rest encodes S, E, M, N, and other open reading frames (ORFs) (6,7).
The binding of the S protein to its receptor, angiotensin-converting enzyme 2 (ACE2) for SARS-CoV-2, triggers membrane fusion and the viral genome is released into the cytosol. S protein contains an RGD tripeptide which can facilitate its binding to multiple integrins and viral entry into ACE2-negative cells (8-11). The virus can also enter monocytes using an antibody-Fcγ receptor-mediated mechanism (12). In the cytosol, the viral genome directs the expression of viral proteins to hijack cellular machinery (13,14). These include the transcription and translation machinery that are necessary for viral production and the innate immune responses that fight against the virus. In addition, the viruses also hijack cellular pathways to reduce cellular metabolism or repurpose them to optimize viral production. The exact pathways that are taken over vary greatly among viruses, and heavily depend on their life cycle. One of the most important factors contributing to pathway alternation is the site of replication, especially if the virus genome is replicated in the cytoplasm or in the nucleus (15). For viruses that are replicated in the nucleus, they must hijack nuclear import machinery to allow the viral genome to enter the nucleus and nuclear export machinery to transport the viral ribonucleoproteins out of the nucleus. Viruses that are replicated in the cytoplasm, including all positive-sense RNA viruses of eukaryotes, encode their own DNA or RNA polymerase, allowing the synthesis of their genomes outside of the nucleus.
Although coronavirus genome replication and transcription occur in the cytoplasm, the nucleus is important for their replication (16,17). A number of the viral proteins contain nuclear localization signals (NLSs) and/or display nuclear localization (18). Some also contain nuclear exporting signals (18). Several non-structural proteins were shown to alter the nuclear import and export functions to impair the translocation of transcription factors involved in immune responses (18). Here, we focus on SARS-CoV-2 and discuss the relationships between this coronavirus and the nucleus and their therapeutic implications.
The nucleus and nuclear transportation
The life cycle of coronavirus doesn't show clear dependency on the nucleus, but virus clearance is accelerated by blocking nuclear entry and virus infection is reduced by inhibiting nuclear export (15,19,20), suggesting a role of the nucleus in virus replication. Virus replication was also shown to be greatly reduced in enucleated cells. The avian infectious bronchitis virus, a gamma-coronavirus, cannot replicate in enucleated cells (16). The murine hepatitis virus, a beta-coronavirus, can replicate in enucleated cells, but viral production is greatly decreased (down to 0.6 - 15% of control nucleated cells, dependent on the virus strains) (17). Replication of SARS-CoV-2 in enucleated cells has not been tested, but the above results suggest that the nucleus may similarly contribute to SARS-CoV-2 viral production.
The nucleus hosts almost all the cell's genomic material (except for mitochondrial DNAs) and contains nucleolus and a number of nuclear bodies involved in RNA synthesis, RNA processing, and ribosome assembly. It is the site of gene regulation, as transcription factors must enter the nucleus to activate or inhibit gene expression. The nucleus is bounded by the nuclear envelope consisting of the outer nuclear membrane (ONM), the inner nuclear membrane (INM), and the nuclear lamina. Between the ONM and the INM is the nuclear lumen, or the perinuclear space. Because the ONM is continuous with the endoplasmic reticulum (ER), the nuclear lumen is connected to the ER lumen. The INM and ONM are also continuous, but they contain different sets of proteins (21). Unanchored proteins diffuse freely on the planes of INM and ONM, but diffusion between these two layers is tightly regulated. The INM and ONM are connected by two types of protein complexes. First, they are connected by the linker of nucleoskeleton and cytoskeleton (LINC) complexes consisting of nesprins on the ONM and SUN proteins on the INM (22,23). The KASH motif of nesprins and the SUN domain of SUN proteins interact in the lumen and bridge the ONM and the INM. The LINC complexes couple the nucleoskeleton (the nuclear lamina) to the cytoskeleton and play important functions in mechanical signaling and force transduction. Second, they are connected by the nuclear pore complexes (NPC) (24). In fact, the ONM and the INM are continuous at the NPCs (21,24). The NPCs are large multi-protein pores on the nuclear envelope (24). The pores are open to both the cytoplasm and the nucleoplasm and are the sole gateway of transportation across the nuclear envelope. The nuclear lamina is composed of lamins (lamin A, B1, B2, and C) (25). These intermediate filament proteins form a meshwork underneath the INM and provide structural support to the nucleus. The lamina also provides attaching points for the LINC complexes, interacts with the NPCs, and through anchoring chromatids, regulates gene expression (25,26).
Nuclear transportation relies on the NPC and the gradient of Ran (Ras-related nuclear protein) small GTPase. The NPCs are giant pore complexes with eightfold rotational symmetry (24,27). Mammalian NPCs are about 110 MDa and are composed of multiple copies of ~30 different nucleoporins (Nups) (24,27). The inner diameter of an NPC is about 50 nm. However, the nucleoporins within the central channels contain repeating sequences of phenylalanine and glycine (FG repeats) that form disordered projections into the lumen (24). These projections create a permeability barrier and exclude the passage of large proteins. In general, small proteins (<50 kDa) can pass the NPC by passive diffusion while larger proteins require the transport receptors (24). This size limitation is approximate because the barrier is not stiff and surface residues of the cargo proteins can greatly alter the kinetics of their NPC passage (28,29). It can also be affected by post-translational modification. For example, glycosylation was shown to facilitate nuclear entry of BSA and neuroD1 (30,31). In addition, as shown for YAP, mechanical stress on the nuclear membrane can stretch the nuclear pore to increase nuclear import (32).
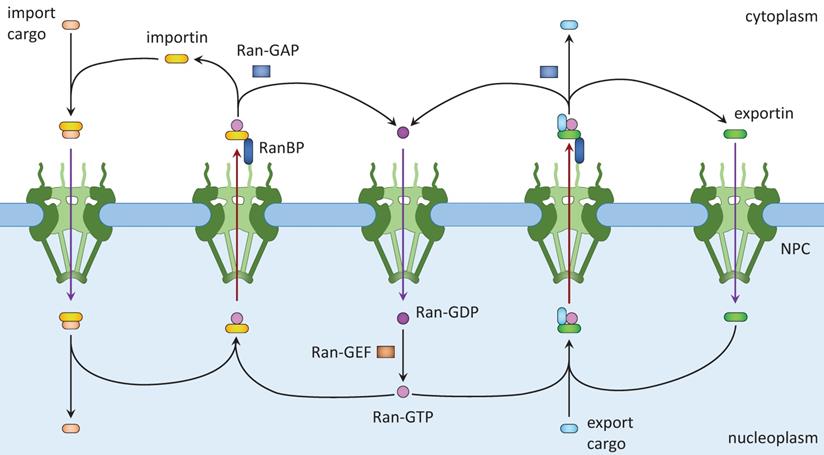
Active nuclear transport requires the Ran system. Ran is a member of the Ras family of small GTPases (15,18). Ran exists in two, GTP-bound and GDP-bound, forms. Ran binding proteins and Ran GTPase activating protein (GAP) in the cytoplasm facilitate the conversion of Ran-GTP into Ran-GDP, whereas RCC1, the nucleotide exchange factor (GEF) for Ran, converts Ran-GDP into Ran-GTP in the nucleus. Together, a Ran-GTP/Ran-GDP gradient is formed across the nuclear envelope. Nuclear import (Fig. 1, left) starts with the binding of NLS-containing cargos to importins, which allows them to shuttle between the cytoplasm and the nucleoplasm. Inside the nucleus, Ran-GTP binds to importin and releases the cargo, resulting in its nuclear accumulation. Ran-GTP and importin move together to the cytoplasm, in which Ran-GTP is converted to Ran-GDP and importin dissociates. Ran-GDP enters the nucleus and is converted to Ran-GTP there. Released importin and Ran-GTP can then participate in another round of nuclear importation. Nuclear export (Fig. 1, right) starts with nuclear export signal (NES)-mediated cargo (protein, ribonucleoprotein complex, or RNA) binding to Ran-GTP and an exportin. After passing through the NPC, the complex dissociates when Ran-GTP is hydrolyzed to Ran-GDP. Ran-GDP and exportin then diffuse into the nucleus for reuse.
Nuclear import and export. Left: Cargo to be imported binds to cytoplasmic importin and enters the nucleus, where it is released from importin by Ran-GTP. The Ran-GTP/importin complex moves to the cytoplasm and binds RanBP. Ran-GAP activates the GTPase activity of Ran to convert Ran-GTP to Ran-GDP and release importin. Ran-GDP enters the nucleus and is recharged by a Ran-GEF. Right: Cargo to be exported binds to exportin and Ran-GTP in the nucleus. The complex moves to the cytoplasm where Ran-GTP is converted to Ran-GDP, leading to the disassembly of the complex. Both Ran-GDP and exportin then enter the nucleus for another round of nuclear export.
Viral replication and the nucleus
Upon membrane fusion, the coronavirus genomic RNA is uncoated and released into the cytosol. The released RNA first serves as an mRNA and is translated into polypeptides that are processed to form the replication and transcription complexes (RTC) (6,7). Some of the RTC subunits of SARS-CoV, namely Nsp3, Nsp4, and Nsp6, work together to remodel intracellular membranes, leading to the formation of convoluted membranes (CMs) and double-membrane vesicles (DMVs) (33). The SARS-CoV-2 Nsp3, 4, and 6 contain similar transmembrane domains as their SARS-CoV counterparts and are expected to function likewise in membrane rearrangement (34,35). Virus-induced membrane structures (CMs and DMVs) are collectively called the replication organelles, as they are believed to be the specialized sites of viral RNA synthesis and protect viral RNAs from being detected by the innate immunity (6). Cryotomographic studies revealed that the membrane of DMVs induced by the murine hepatitis coronavirus contains pore complexes that allow the export of synthesized RNAs to the cytosol (36). Viral infection also induces the formation of annulate lamellae, parallel stacks of ER-derived membranes. Annulate lamellae contain NPC proteins, suggesting a link to the nuclear envelope (37). The formation and functions of annulate lamellae are still poorly understood.
The genomic RNA contains a nested set of subgenomic mRNAs, which encode the M, E, S, and N structural proteins and additional accessory proteins (5). The N protein is expressed by cytoplasmic ribosomes and binds to replicated RNAs after they exit the replication organelles. The M, E, and S proteins are transmembrane proteins and, expectedly, are synthesized by rough ER-associated ribosomes and co-translationally translocated into the ER. Vesicles containing these proteins are transported to the ER-to-Golgi intermediate compartment (ERGIC). The ERGIC clusters, concentrated with viral proteins, then assemble around the newly formed N-encapsulated genomic RNAs, resulting in assembled virus particles in the lumen of secretory vesicles (5). The virus particles then go through the secretory pathway before being released via a lysosomal-dependent pathway (38).
The nucleus may contribute to virus production due to its connection to the replication organelles. Notably, the replication organelles are usually located in the perinuclear region of the cells. These organelles are interconnected and connect to the ER, so, in principle, material exchange can occur between the nuclear lumen and the intermembrane space of the DMVs. However, given that synthesized RNAs are protected by the inner membrane and enter the cytosol through the pore complex, it is unlikely that luminal factors can considerably affect virus replication. Similarly, transmembrane proteins synthesized in the ER can diffuse to the ONM. The E protein of SARS-CoV-2, when overexpressed alone, was indeed shown to localize to the nucleus (and the cytoplasm) (39). However, the functional significance of this localization is unknown.
The nucleus is also in close proximity to the ERGIC and the Golgi complexes. Two proteins involved in nuclear transportation, RanBP1 and importin-α, regulate the dynamics of the Golgi complexes (40-42). A pool of RanBP1 localized to the trans-Golgi network in neurons and mediates the nuclear export of proteins that regulate Golgi condensation (40). Importin-α directly interacts with GM130 on Golgi and functions in Golgi disassembly and spindle assembly during mitosis (41,42). Interestingly, importin-α may promote Golgi disassembly independent of Ran (41,43). The Golgi complexes are highly fragmented in SARS-CoV-2 infected cells (44). Whether this is due to altered nuclear transportation, however, has not been tested.
Viral proteins in the nucleus
Supporting a role of the nucleus in viral production, several viral proteins were shown to contain NLS and/or NES and localize to the nucleus. The SARS-CoV N protein, which is 90% identical to SARS-CoV-2 N protein, contains multiple NLSs and a signal for nucleolar localization (18). Nucleolar localization was indeed observed for N protein of SARS-CoV (45). N protein of SARS-CoV-2, however, is mostly cytoplasmic (39). The S protein of SARS-CoV-2 was also shown to be present in the nucleus (39,46). In addition to the N and S proteins, many other viral proteins, when overexpressed, are detectable in the nucleus. These include E, ORF9a, Nsp1, Nsp3N, Nsp5 to Nsp7, Nsp9, Nsp10, and Nsp12 to Nsp16 (39). Among them, only Nsp13 exhibits foci localization and enriches in the splicing compartment (39). Many of these proteins are small and probably enter the nucleus by passive diffusion.
Because viral assembly occurs in the cytoplasm, nuclear localization of some of the viral proteins may not be desirable and they need to be exported from the nucleus. Pharmacological inhibition of nuclear export leads to nuclear accumulation of viral proteins and significantly decreases viral infection (20). Several of the SARS-CoV-2 proteins, including N, S, M, E, Nsp9, Nsp12, and ORF3a, contain predicted NESs (18). These NESs may promote the nuclear export of these proteins or allow them to interact with exportins to hijack nuclear transportation (see below). The mechanism of nuclear export is best understood for SARS-CoV N protein. Phosphorylated N protein binds to adaptor protein 14-3-3 for nuclear export (47). SARS-CoV-2 N protein also binds to 14-3-3 in a phosphorylation-dependent manner (48).
Targeting nuclear translocation of immune regulators
Pathogenic coronaviruses commonly interfere with the interferon (IFN) pathway but trigger an aberrant immune response by producing excessive cytokines and chemokines (cytokine storm) (14). IFNs are a family of antiviral cytokines produced and released by host cells upon detecting various pathogen-associated molecular patterns. There are three types of IFNs: Type I IFNs (including α and β) are anti-viral. The only type II IFN (γ) fights against both bacterial and virus infections. Whereas type III IFNs (λ) are produced after virus or fungal infections. All three types of IFNs are targets of coronaviruses. The expression of type I and III IFNs induced by SARS-CoV-2 is affected by many factors, like the tissue location, severity, and age (49). Type II IFN is also downregulated by SARS-CoV-2 (50,51).
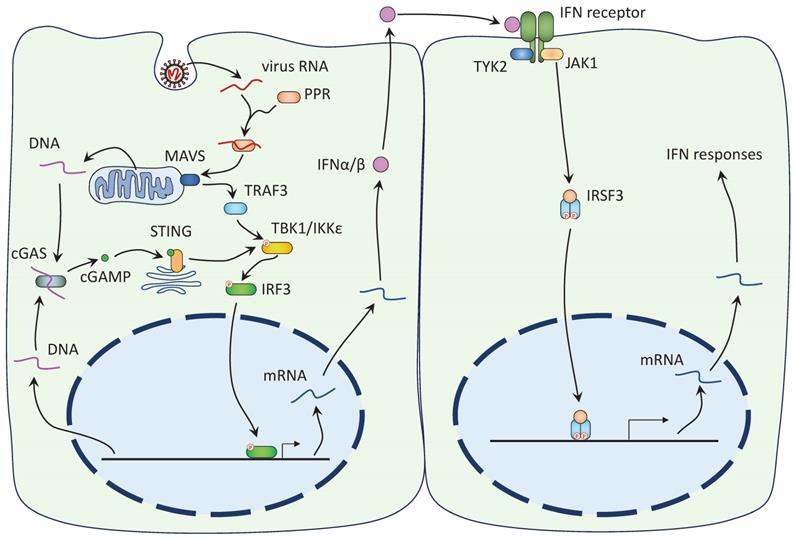
The mechanisms of IFN antagonism have been extensively studied for type I IFNs (52,53). Type I IFN response begins with the recognition of virus RNA by host cell pattern recognition receptors (PPRs), like RIG-I and MDA5 (Figure 2). PPRs then bind to mitochondrial-associated adaptor MAVS and recruit and activate TRAF3. TRAF3 activates the interferon regulatory factors (IRF) kinases, TBK1 and IKKε, which phosphorylate IRF3 and IRF7. Phosphorylated IRF3/7 enter the nucleus to initiate the expression of the IFN genes. TBK1 and IKKε also phosphorylate IκB and facilitate the nuclear translocation of NF-κB, another important regulator of innate immune response. Expressed IFNs are released by the host cells and bind to the IFN receptors on the nearby cells to activate the Janus kinase (JAK)-STAT signaling cascade (Figure 2). Receptor-associated JAK phosphorylates and activates both STAT1 and STAT2, which, together with IRF9, form the IFN-stimulated gene factor 3 (ISGF3). The ISGF3 enters the nuclear with the help of importins α1/β1 and triggers the expression of antiviral genes (19).
SARS-CoV-2 antagonizes IFN with many, likely redundant, mechanisms targeting both the pre-IFN IRF3 pathway and the downstream JAK-STAT pathway (Table 1). Regulations on the mRNA level also exist (see below). In three studies, many of the viral proteins, including Nsp1, Nsp3, Nsp6, Nsp7, Nsp12, Nsp13, Nsp14, Nsp15, ORF3a, ORF6, ORF7a, ORF7b, and M, were shown to suppress IFN-α/β signaling, mostly by preventing the phosphorylation and subsequent nuclear entry of IRF3 (54-56). In addition, Nsp5 was also reported to inhibit IFN response by targeting IRF3 (57,58). Two viral proteins, Nsp2 and S, however, enhance IFN signaling (54). Several of these proteins, including Nsp1, Nsp3, Nsp6, Nsp13, ORF3a, ORF6, ORF7, and M, also inhibit nuclear translocation of STAT1/STAT2 (54,55,59). Moreover, N protein was shown to inhibit the phosphorylation and nuclear translocation of STAT1/STAT2 (60). It should be noted that these studies mostly relied on the ectopic expression of individual protein, and have identified overlapped but not identical sets of viral proteins (54-56).
SARS-Cov-2 induces the IFN-I response. Left: 1) The viral RNA is recognized by host cell PPRs and together they activate MAVS on mitochondria. MAVS recruits TRAF3 to activate TBK1/IKKε, which in turn phosphorylates IRF3. IRF3 Phosphorylation leads to its nuclear translocation and the expression of the IFN-I genes. 2) DNA released from damaged mitochondria or ruptured nucleus is recognized by cGAS, leading to the synthesis of 2'3'-cGAMP. cGAMP binds to ER-localized STING and promotes its dimerization and translocation to the Golgi complex. STING activates TBK1 to induce IFN response. Right: Expressed IFNs are released and bind to the IFN receptors on the nearby cells. Receptor-associated JAK phosphorylates both STAT1 and STAT2, allowing them to dimerize and interact with IRF9 to form the ISFG3. The ISGF3 enters the nucleus and induces the expression of the IFN response genes.
Mechanisms of IFN down-regulation by SARS-CoV-2 proteins
| TBK1 phosphorylation | IRF3 phosphorylation | IRF3 translocation | STAT phosphorylation | ISGF3 translocation | |
|---|---|---|---|---|---|
| Nsp1 | Yes (55) | Yes (55) | Yes (55) | ||
| Nsp3 | Yes (59) | Yes (59) | |||
| Nsp5 | No (57) | Yes (57) | |||
| Nsp6 | No (55) | Yes (55) | Yes (55) | Yes (55) | Yes (55) |
| Nsp7 | |||||
| Nsp12 | No (67) | Yes (67) | |||
| Nsp13 | Yes (55) | Yes (55) | Yes (55,56) | Yes (55) | Yes (55) |
| Nsp14 | Yes (56) | ||||
| Nsp15 | Yes (56) | ||||
| ORF3a | Yes (55) | Yes (55) | |||
| ORF6 | No (54,55) | Yes (54-56) | Yes (55,63-65) | ||
| ORF7a | Yes (55) | Yes (55) | |||
| ORF7b | Yes (55) | Yes (55) | |||
| M | Yes (55) | Yes (55) | |||
| N | Yes (60) | Yes (60) |
* Yes/No: whether the process (indicated by the column heading) is inhibited by the protein (indicated by the row label). The numbers in parentheses refer to the references.
The underlying mechanism of IFN downregulation is best understood for ORF6. SARS-CoV-2 ORF6 was shown to interact with importin α2 and inhibit importin-dependent nuclear translocation of both IRF3 and ISGF3 (55). Interestingly, ORF6 of SARS-CoV also interacts with importin α2 and β1 and sequesters them on the ER and Golgi membrane (61). ORF6 additionally interacts with Nup98, an NPC component, and Rae1, a nuclear mRNA export factor, and interferes with nuclear trafficking, including STATs translocation (62-65). Inhibitors of importins are effective drugs for COVID-19, but the mechanism of action is still unclear (15,19,66).
The protease domain of Nsp3 (PLPro) can downregulate IFN signaling by cleaving a ubiquitin-like protein, ISG15 (59). Nsp3 prevents ISGylation of IRF3 by ISG15 and decreases both phosphorylation and nuclear translocation of IRF3 (59). Directed cleavage of IRF3 by PLPro can also occur (58). Nsp5 also contains a protease domain and blocks nuclear translocation of phosphorylated IRF3, but this activity is independent of its protease domain (57). Nsp12 blocks the nuclear translocation of IRF3 without affecting its phosphorylation (67). Therefore, Nsp5 and Nsp12 may interact with the nuclear transport machinery. Both Nsp6 and Nsp13 bind to TBK1 (55,68), but function differently to inhibit IRF3 phosphorylation. Nsp13 inhibits the phosphorylation of TBK1, whereas Nsp6 binds to phosphorylated TBK1 and inhibits its kinase activity (55,68).
In addition to the IFN responses, the virus also interferes with other anti-viral innate immune pathways. However, the inhibitions are not as tight as those of the IFN responses and whether nuclear entry of the transcription factors is altered by the virus is not well understood. Nevertheless, SARS-CoV-2 ORF9c interacts with proteins involved in NF-kB signaling (62) and NF-κB nuclear translocation is affected by the loss of Nup62 induced by Nsp9 expression (69), demonstrating that SARS-CoV-2 can alter NF-kB signaling through regulating its nuclear entry. The NF-κB pathways are critical for the cytokine storm (70). In the inactive state, the NF-κB heterodimer is sequestered in the cytoplasm by its inhibitor IκB. Inflammatory stimuli activate the IκB kinase, an enzyme complex consisting of IKKα kinase, IKKβ kinase, and a regulatory protein NEMO. Active IκB kinase phosphorylates IκB and causes its ubiquitinylation and proteasome degradation. Freed NF-κB then enters the nucleus to trigger the expression of inflammatory genes. The NF-κB pathways play dual roles in SARS-CoV-2 biology. On one hand, both Nsp5 and Nsp13 were shown to inhibit NF-κB (68,71). Nsp5 cleaves NEMO to reduce IFN signaling (71). On the other hand, Nsp5 was shown to activate NF-κB in another study (72). In addition, many viral proteins, including ORF3a, ORF7a, M, N, and S, activate NF-κB (11,73-76). Surprisingly, the NF-κB pathways may be essential for SARS-CoV-2, as its depletion results in a complete inhibition of virus replication (77).
Upstream of IFN and NF-κB, the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway has a direct connection to the nucleus. Damage to the nuclear envelope or mitochondria releases DNA to the cytoplasm (Fig 2). Cytoplasmic DNAs are sensed by cGAS and induce the production of cyclic GMP-AMP, which binds to STING to activate TBK1 (78). TBK1 then phosphorylates IRF3 to induce IFN production. Similar to the NF-κB pathways, cGAS-STING seems to play dual roles in COVID-19. ORF3a, ORF9b, N, and 3CLPro (protease domain of Nsp5) of SARS-CoV-2 were shown to inhibit STING (79,80) and pharmacological activation of cGAS-STING reduces virus infection (81,82). However, elevated cGAS-STING also contributes to the cytokine storm and inflammation (83,84).
Targeting host mRNAs
SARS-CoV-2 cuts down cellular protein synthesis to inhibit immune responses and optimize viral replication. This is in part achieved by targeting host mRNAs, including inhibiting mRNA release after transcription, blocking nuclear trafficking of mRNAs, and accelerating degradation of mRNAs (85). SARS-CoV-2 induces the transcription of IFN mRNAs but most of these mRNAs are retained at the sites of transcription and degraded eventually in the nucleus (85). Escaped mRNAs can be transported to the cytoplasm, but global mRNA export is also inhibited by viral proteins. ORF6 interacts with Nup98 and Rae1 and impairs bidirectional nuclear transport, including host mRNA export (64,65). Nsp1, which suppresses host gene expression, also blocks mRNA export (86,87). The N-terminus of Nsp1 binds to NXF1, a subunit of the mRNA export receptor NXF1-NXT1. Nsp1 binding does not interfere with NXF1 binding to RNAs but impairs its interaction with nuclear export factors and the NPC (87). In the cytoplasm, exported host mRNAs face increased degradation mediated by Nsp1 (85,86). Together, these studies demonstrated that SARS-CoV-2 targets multiple mRNA pathways to dampen the translation of host proteins.
Coronavirus, cilia, and ciliary trafficking
Cilia are microtubule-dependent organelles projected from the cell body (88,89). Two types of cilia exist, the motile cilia that beat in coordinated waves to move extracellular material relative to the cell and the immotile primary cilia that act as cellular antennas and perform highly specialized sensory functions. Motile cilia play critical roles in the defense against airway infections (90). The respiratory tract is lined by epithelial cells that are mostly ciliated. Each ciliated epithelial cell has about two hundred cilia that beat in a coordinated fashion to propel the mucus layer and inhaled particles and pathogens out of the airways. SARS-CoV-2 preferentially targets ciliated cells and causes a rapid loss of the ciliated epithelium (90). Motile cilia in SARS-CoV-2 infected cells display structural abnormalities, both in shapes and sizes (90). The expression of Foxj1, a ciliogenesis regulator, is reduced by virus infection and may contribute to some of these cilia defects (90). Primary cilia are sensory organelles found in most non-blood cells in our bodies. With some exceptions, most cells possess one single primary cilium. The loss of smell and taste is a common symptom of SARS-CoV-2 infection and is related to cilia damage (91,92). ORF10 interacts with ZYG11B, an adapter protein of the CUL2 E3 ligase, and induces proteasomal degradation of proteins involved in ciliogenesis or cilium structure (93). Overexpression of ORF10 inhibits primary cilium assembly (93).
The machinery of nuclear trafficking has a surprising role in ciliogenesis and translocation of cilia proteins (88,89). At least ten Nups as well as importin-β1 and β2 are found to be present in cilia (89,94). The Nups localize to the base of the cilia and form a diffusion barrier to regulate protein translocation between the cilia and the cytoplasm (88,89). The structural organization of these Nups is poorly understood but is expected to share some features of the NPC. Similar to nuclear trafficking, ciliary transport depends on the FG repeats of the Nups and a ciliary Ran-GTP, cytoplasmic Ran-GDP gradient (88,89). Importins are also involved in ciliary transport. Importin-β1 interacts with CRB3-CLPI at the cilia and overexpression of an importin β1 mutant leads to strong cilia abnormalities (94). Importin β2 recognizes ciliary localization signals and mediates translocation of KIF17, an important component of the intraflagellar transport (95). Given that both SARS-CoV and SARS-CoV-2 ORF6s are reported to interact with and inhibit importins (α2 (55) and β1 (61)), it is possible that ORF6 may affect ciliogenesis and cilia maintenance.
In a SARS‐CoV‐2-human protein interactomic study, Nsp13 is found to interact with 12 centrosomal components (62). The centrosome and the basal body of cilia are closely related microtubule-organizing centers and both contain centriole (62,96). Nsp6 of SARS‐CoV, when overexpressed alone, induces vesicle formation at the centrosome. Vesicle formation, however, does not occur when Nsp4 is coexpressed (33). The functional importance of these interactions is unknown.
Epigenetic alternations by coronavirus
Epigenetic mechanisms, such as DNA/RNA methylation and histone modification, are important regulators of gene expression. Differences in epigenetic modifications contribute greatly to the susceptibility to SARS-CoV-2 and the prevalence of comorbidities (97,98). Epigenetic regulations of ACE2 expression and their effects on COVID-19 outcomes have been extensively studied and have great therapeutic potential (97-99).
Not surprisingly, SARS-CoV-2 also induces epigenetic changes to optimize viral infection and limit immunity. First, mRNA methylation is affected by virus infection. Epitranscriptomic studies using blood samples of COVID-19 patients and healthy controls have revealed significant differences in their N6-methylation of adenosine (m6A) modification profiles (100,101). METTL3, a subunit of the N6-adenosine-methyltransferase, is essential for m6A modification of SARS-CoV-2 RNAs and helps them to evade the RIG-I immune response (102-104). Depletion of METTL3 reduces virus infection in human lung fibroblasts but, surprisingly, enhances infection in human hepatocarcinoma cells (104,105). In infected cells, the expression of METTL3 is increased while the expression of FTO, a demethylase, is decreased (103,106). An increase in METTL3 expression relies on its interaction with the viral RNA-dependent RNA polymerase (106). Interestingly, while METTL3 and FTO are normally nuclear, cytoplasmic localization is observed in infected cells (106).
Second, DNA methylation is altered by SARS-CoV-2 and shows significant differences between COVID-19 patients and healthy controls. Profiles of DNA methylation can be used to calculate epigenetic ages and aging in COVID-19 patients was shown to be accelerated (107-109). Differentially methylated regions have been identified using blood, kidney, and heart samples, and many of these regions are close to genes involved in viral defense and interferon signaling (110,111).
Third, histone modification is altered by SARS-CoV-2. An interactomic study has identified direct interactions between viral proteins (E, Nsp5, Nsp8, and Nsp13) and epigenetic regulators (62). Inhibition of BRD2, a BET family reader of histone acetylation inhibition, reduces ACE2 expression and blocks SARS-CoV-2 (112). Genome-wide studies of H3K4me3 and H3K27me3 modifications in peripheral blood mononuclear cells have demonstrated that genes related to immune response are upregulated after infection (113). NETosis, the formation of neutrophil extracellular traps (NET), is an antimicrobial cell death pathway (114). NETs contain modified chromatin and are characterized by the presence of citrullinated histone H3. Cell-free DNA and citrullinated histone H3 are increased in sera from patients with COVID-19 and correlate with the severity of the syndromes (115).
Lastly, chromatin organization contributes to viral susceptibility and is affected by the virus. Chromatin accessibility regulates the expression profiles of ACE2 and related genes and modulates differential virus entry across tissues (116). In a genome-wide CRISPR screen, the SWI/SNF chromatin remodeling complex was found to promote SARS-CoV-2 infection, suggesting a role of chromatin remodeling in virus life cycle (117). Single-cell transposase-accessible chromatin with sequencing experiments with peripheral blood mononuclear cells showed that COVID-19 induces significant chromatin remodeling (118,119). Changes in chromatin organization are enriched on genes of the inflammatory pathways and facilitate long-term adaptive immune responses (118,119). Drastic chromatin remodeling also occurs in olfactory sensory neurons and leads to significant downregulation of the olfactory receptor genes and, potentially, anosmia (117).
Genetic alternations by coronavirus
One of the nuclear processes that is frequently triggered by virus infection is the DNA damage response, even for mRNA viruses (77,120,121). Proteins involved in DNA damage response are essential for virus infection as a small molecule inhibitor of the ATR kinase blocks SARS-CoV-2 replication (121). DNA damage response is quickly induced after SARS-CoV-2 infection but is swiftly suppressed in less than a day (77). This can be due to increased DNA damage caused by virus-induced nuclear rupture (81,122). Increased DNA damage and changes in the DNA damage response may lead to genomic changes or even genomic instability.
Besides DNA damage, SARS-CoV-2 was reported to affect two other aspects of the host genome. First, accelerated telomere shortening was found in infected Vero E6 cells and COVID-19 survivors (120,107). A telomere is the end region of a chromosome containing repetitive DNA sequences. Shortening of telomeres occurs during aging and increases the severity of COVID-19 (123). Accelerated telomere shortening implies that aging is accelerated in COVID-19 patients. However, it should be noted that contradictory results, i.e. the lack of accelerated telomere shortening and accelerated aging, have also been reported (124). Second, virus genome and vaccine mRNA were reported to be reverse-transcribed and integrated into the host genome (125-127). These findings are highly debated due to the artificial expression of long interspersed nuclear element-1, a reverse transcriptase, and the concern of artifacts during library preparation (128-131).
Other nuclear pathways affected by coronavirus
In addition to gating nuclear trafficking and altering epigenetic mechanisms, several nuclear pathways can also play roles in the virus life cycle or host cell anti-viral activities.
The nucleus may contribute to the virus life cycle indirectly through the cytoskeleton. All three classes of cytoskeletal elements (actin, intermediate filament, and microtubule) have been reported to involve in virus entry (132). The actin cytoskeleton plays several essential roles in viral entry. A recent report showed that myosin IIA directly interacts with the S protein of SARS-CoV-2 and facilitates virus infection (43). SARS-CoV-2 infection also stimulates casein kinase II and p38 MAP kinase pathways to induce actin polymerization (77). Nsp2 interacts with strumpellin, a component of the Wiskott-Aldrich syndrome protein and scar homology (WASH) complex, which regulates actin assembly and may contribute to virus egress (62). Intermediate filaments participate in virus replication and assembly (132). Vimentin intermediate filaments were shown to form a ''cage-like'' network around the DMVs and Withaferin A, a pharmacological inhibitor of vimentin, blocks viral replication (44). Lastly, the microtubule cytoskeleton is involved in ER-to-Golgi transportation of the virus particles (132). The nucleus senses mechanical forces and reorganizes the cytoskeleton through triggering mechanosignaling pathways, like the YAP/TAZ and serum response pathways (133). Lamin and INM protein emerin were shown to regulate actin dynamics (134). The LINC complexes couple the nucleus to the cytoskeleton and regulate Rho activity (135,136). The Rho family of small GTPases are master regulators of the cytoskeleton. SARS-CoV-2 may regulate Rho activity through Nsp7 binding to RhoA (62). S protein was also shown to activate RhoA (137). Alternatively, ORF3a interacts with SUN2 and may thus regulate the many functions of the LINC complexes, including Rho regulation (62). In addition, actin dynamics and the serum response can regulate NF-κB activity (138-140). MRTF, an activator of the serum response factor, binds to p65 in the nucleus and suppresses NF-κB activity (140). Similarly, lamin A/C deficiency reduces NF-κB-regulated transcription (138).
A recent study links the nucleus and the nuclear lamina to DNA-induced IFN response during SARS-CoV-2 infection (81). The nuclear lamina provides structural support to the nuclear envelope and protects it against mechanical stress. Defects in the nuclear lamina increase the frequency of nuclear envelope rupture, which exposes genomic DNA to cytoplasmic nucleases, damages DNA, and leads to genome instability (141). SARS-CoV-2 was shown to induce the cGAS-STING pathways because of the presence of cytoplasmic genomic DNAs, which are greatly increased by cell fusion (81,122). Interaction between the S protein and ACE2 can not only mediate virus entry, but also induce cell-cell fusion, resulting in the formation of syncytia - cells with multiple nuclei (142). Nuclei in the fused cells have lower levels of lamin A/C, the major contributor of the mechanical property of the nucleus, and have evident nuclear membrane blebs containing DNA (81). These blebs protrude from the nuclear lamina and rupture, leading to the release of DNA to the cytoplasm. Consistently, DNA damage foci, stained by γH2AX, are found to accumulate in infected cells with lower lamin A/C (81). diABZI, a potent STING activator, greatly reduces SARS-CoV-2 infection (81). These results suggest that nuclear integrity modulates host cell immune response during virus infection.
Therapeutic implications
Small molecule drugs targeting the above pathways have therapeutic potential and some are being used in clinical studies. Epigenetic drugs have been reviewed and are not discussed here (143,144).
Because the IFN-I response is down-regulated by coronaviruses, many studies have been conducted to evaluate the effects of interferon treatments (145). In a study involving 11,330 subjects, the World Health Organization has found no benefit of IFN-β1 and recommends against using IFN-I for the treatment of COVID-19 (146). The virus not only reduces IFN expression by blocking upstream signaling like the nuclear entry of IRF3, but also blocks the downstream JAK-STAT signaling by preventing nuclear translocation of ISGF3. Therefore, targeting nuclear trafficking may be more effective in fighting the virus and is a tempting alternative to IFN administration. The uses of inhibitors of nuclear transport in clinical studies have been previously summarized (15,18,19). Notably, Ivermectin, an FDA-approved importin α inhibitor, is included in almost 80 ongoing clinical trials for SARS-CoV-2 and has been shown to significantly improve viral clearance in several studies (15,19). Drugs against the nuclear export machinery are also shown to be beneficial for treating SARS-CoV-2. Selinexor and Verdinexor, drugs of the selective inhibitors of nuclear export (SINE) family, are FDA-approved and have been in clinical trials for treating COVID-19 (18). Selinexor and Verdinexor target exportin-1 (also known as XPO1 and CRM1) and treatment of Selinexor inhibits the nuclear export of ORF3b, ORF9b, and N protein and induces anti-viral response (20). Selinexor treatment also leads to ACE2 accumulation in the nucleus (20).
Cytoplasmic DNAs can activate the cGAS-STING pathway and induce antiviral responses (81). Activators of the cGAS-STING pathway have been clinically tested for cancer therapy. One of which, diABZI, is an activator of STING and was shown to effectively block SARS-CoV-2 replication (81,82). However, STING activation is associated with SARS-CoV-2-induced inflammation and inhibition of cGAS-STING also exhibits beneficial effects (83,84,147). Compared to manipulating cGAS-STING, pharmacological inhibition of NF-κB may be more promising, as it can inhibit virus replication and dampen cytokine storm (70,148). Several NF-κB inhibitors have been used in clinical/preclinical trials (149).
DNA damage repair pathways play poorly understood roles in SARS-CoV-2 replication (121). In an antiviral drug screen, berzosertib, an inhibitor of the ATR kinase, exhibited potent anti-SARS-CoV-2 activity (121). Many pharmacological inhibitors of DNA damage repair have been identified and are used in clinical studies for cancer therapy (150). It is worth testing the effects of these drugs on SARS-CoV-2.
Conclusion
We have summarized the evidence demonstrating the importance of the nucleus in SARS-CoV-2 infection. The nucleus plays multiple roles in the virus life cycle, but these roles are complicated and sometimes counteract each other. Some of the apparent incompatibilities are due to the differences between the in vivo and in vitro settings used in the studies. Or, it can be due to the differential effects of the virus on different cell types or different stages of infection. A better understanding of these roles of the nucleus requires more mechanistic studies.
Nuclear biology is understudied in COVID-19 research. The importance of the nucleus and its many interactions with the virus merit more studies in this area and promise fruitful results. Further works are expected to uncover new roles of the nucleus in SARS-CoV-2 infection and COVID-19 pathogenesis. They will also provide mechanical details of the SARs-CoV-2-nucleus interactions and fuel the discovery of new treatments.
Abbreviations
COVID-19: Coronavirus disease 2019; SARS-CoV: Severe acute respiratory syndrome coronavirus; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; M: membrane protein; E: envelope protein; S: spike protein; N: nucleocapsid; Nsp: nonstructural protein; ORF: open reading frame; ACE2: Angiotensin-converting enzyme 2; NLS: nuclear localization signals; NES: nuclear export signal; ONM: outer nuclear membrane; INM: inner nuclear membrane; ER: endoplasmic reticulum; ERGIC: ER-to-Golgi intermediate compartment; LINC: linker of nucleoskeleton and cytoskeleton; NPC: nuclear pore complex; Nup: nucleoporin; Ran: Ras-related nuclear protein; GAP: GTPase activating protein; GEF: nucleotide exchange factor; RTC: replication and transcription complexes; CM: convoluted membrane; DMV: double-membrane vesicle; IFN: Interferon; PPR: pattern recognition receptor; IRF: interferon regulatory factor; ISGF3: IFN-stimulated gene factor 3; cGAS: cyclic GMP-AMP synthase; STING: simulator of interferon genes; NF-κB: nuclear factor kappa B; NET: neutrophil extracellular traps; m6A: N6-methylation of adenosine.
Acknowledgements
This work was supported by funding from the Science and Technology Development Fund, Macau SAR (file 0077/2020/A2) and from the University of Macau (file: SRG2020-00015-FHS).
Author Contributions
M.C., Y.M. and W.C. wrote and edited the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021Mar;19(3):141-54
2. Zmasek CM, Lefkowitz EJ, Niewiadomska A, Scheuermann RH. Genomic evolution of the Coronaviridae family. Virology. 2022May1;570:123-33
3. Woo PCY, Huang Y, Lau SKP, Yuen K-Y. Coronavirus Genomics and Bioinformatics Analysis. Viruses. 2010Aug24;2(8):1804-20
4. Li X, Chang J, Chen S, Wang L, Yau TO, Zhao Q. et al. Genomic Feature Analysis of Betacoronavirus Provides Insights Into SARS and COVID-19 Pandemics. Front Microbiol. 2021;12:614494
5. V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021Mar;19(3):155-70
6. Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat Rev Mol Cell Biol. 2022Jan;23(1):21-39
7. Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020Dec1;41(12):1100-15
8. Nader D, Fletcher N, Curley GF, Kerrigan SW. SARS-CoV-2 uses major endothelial integrin αvβ3 to cause vascular dysregulation in-vitro during COVID-19. PloS One. 2021;16(6):e0253347
9. Park EJ, Myint PK, Appiah MG, Darkwah S, Caidengbate S, Ito A. et al. The Spike Glycoprotein of SARS-CoV-2 Binds to β1 Integrins Expressed on the Surface of Lung Epithelial Cells. Viruses. 2021Apr9;13(4):645
10. Simons P, Rinaldi DA, Bondu V, Kell AM, Bradfute S, Lidke DS. et al. Integrin activation is an essential component of SARS-CoV-2 infection. Sci Rep. 2021Oct14;11(1):20398
11. Robles JP, Zamora M, Adan-Castro E, Siqueiros-Marquez L, Escalera GM de la, Clapp C. The spike protein of SARS-CoV-2 induces endothelial inflammation through integrin α5β1 and NF-κB signaling. J Biol Chem. 2022Mar1;298(3):101695
12. Junqueira C, Crespo Â, Ranjbar S, de Lacerda LB, Lewandrowski M, Ingber J. et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022Apr6;606(7914):576-84
13. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022Jan;23(1):3-20
14. Suryawanshi RK, Koganti R, Agelidis A, Patil CD, Shukla D. Dysregulation of Cell Signaling by SARS-CoV-2. Trends Microbiol. 2021Mar1;29(3):224-37
15. Sajidah ES, Lim K, Wong RW. How SARS-CoV-2 and Other Viruses Build an Invasion Route to Hijack the Host Nucleocytoplasmic Trafficking System. Cells. 2021Jun;10(6):1424
16. Evans MR, Simpson RW. The coronavirus avian infectious bronchitis virus requires the cell nucleus and host transcriptional factors. Virology. 1980Sep1;105(2):582-91
17. Wilhelmsen KC, Leibowitz JL, Bond CW, Robb JA. The replication of murine coronaviruses in enucleated cells. Virology. 1981Apr15;110(1):225-30
18. Uddin MdH, Zonder JA, Azmi AS. Exportin 1 inhibition as antiviral therapy. Drug Discov Today. 2020Oct1;25(10):1775-81
19. Martin AJ, Jans DA. Antivirals that target the host IMPα/β1-virus interface. Biochem Soc Trans. 2021Jan13;49(1):281-95
20. Kashyap T, Murray J, Walker CJ, Chang H, Tamir S, Hou B. et al. Selinexor, a novel selective inhibitor of nuclear export, reduces SARS-CoV-2 infection and protects the respiratory system in vivo. Antiviral Res. 2021Aug1;192:105115
21. De Magistris P, Antonin W. The Dynamic Nature of the Nuclear Envelope. Curr Biol. 2018Apr23;28(8):R487-97
22. Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol. 2015Jan5;208(1):11-22
23. Luxton GG, Starr DA. KASHing up with the nucleus: Novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol. 2014;28(1):69-75
24. Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Structure and Assembly of the Nuclear Pore Complex. Annu Rev Biophys. 2019May6;48(1):515-36
25. Karoutas A, Akhtar A. Functional mechanisms and abnormalities of the nuclear lamina. Nat Cell Biol. 2021Feb;23(2):116-26
26. Janota CS, Calero-Cuenca FJ, Gomes ER. The role of the cell nucleus in mechanotransduction. Curr Opin Cell Biol. 2020Apr1;63:204-11
27. Raices M, D'Angelo MA. Nuclear pore complexes and regulation of gene expression. Curr Opin Cell Biol. 2017Jun1;46:26-32
28. Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D. et al. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol. 2016Oct3;215(1):57-76
29. Frey S, Rees R, Schünemann J, Ng SC, Fünfgeld K, Huyton T. et al. Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell. 2018Jun28;174(1):202-217.e9
30. Andrali SS, Qian Q, Ozcan S. Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J Biol Chem. 2007May25;282(21):15589-96
31. Rondanino C, Bousser M-T, Monsigny M, Roche A-C. Sugar-dependent nuclear import of glycosylated proteins in living cells. Glycobiology. 2003Jul1;13(7):509-19
32. Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ. et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell. 2017;171(6):1397-1410.e14
33. Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. mBio. 2013Aug13;4(4):e00524-13
34. Thomas S. Mapping the Nonstructural Transmembrane Proteins of Severe Acute Respiratory Syndrome Coronavirus 2. J Comput Biol. 2021Sep;28(9):909-21
35. Santerre M, Arjona SP, Allen CN, Shcherbik N, Sawaya BE. Why do SARS-CoV-2 NSPs rush to the ER? J Neurol. 2021Jun1;268(6):2013-22
36. Wolff G, Limpens RWAL, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM. et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020Sep11;369(6509):1395-8
37. Eymieux S, Blanchard E, Uzbekov R, Hourioux C, Roingeard P. Annulate lamellae and intracellular pathogens. Cell Microbiol. 2021;23(8):e13328
38. Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M. et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell. 2020Dec10;183(6):1520-1535.e14
39. Zhang J, Cruz-cosme R, Zhuang M-W, Liu D, Liu Y, Teng S. et al. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct Target Ther. 2020Nov17;5(1):1-3
40. Mencarelli C, Nitarska J, Kroecher T, Ferraro F, Massey K, Riccio A. et al. RanBP1 Couples Nuclear Export and Golgi Regulation through LKB1 to Promote Cortical Neuron Polarity. Cell Rep. 2018Sep4;24(10):2529-2539.e4
41. Chang C-C, Chen C-J, Grauffel C, Pien Y-C, Lim C, Tsai S-Y. et al. Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-α. Nat Commun. 2019Sep20;10(1):4307
42. Wei J-H, Zhang ZC, Wynn RM, Seemann J. GM130 Regulates Golgi-Derived Spindle Assembly by Activating TPX2 and Capturing Microtubules. Cell. 2015Jul16;162(2):287-99
43. Chen J, Fan J, Chen Z, Zhang M, Peng H, Liu J. et al. Nonmuscle myosin heavy chain IIA facilitates SARS-CoV-2 infection in human pulmonary cells. Proc Natl Acad Sci. 2021Dec14;118(50):e2111011118
44. Cortese M, Lee J-Y, Cerikan B, Neufeldt CJ, Oorschot VMJ, Köhrer S. et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe. 2020Dec9;28(6):853-866.e5
45. Timani KA, Liao Q, Ye L, Zeng Y, Liu J, Zheng Y. et al. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005Dec1;114(1):23-34
46. Eymieux S, Rouillé Y, Terrier O, Seron K, Blanchard E, Rosa-Calatrava M. et al. Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell Mol Life Sci. 2021Apr;78(7):3565-76
47. Surjit M, Kumar R, Mishra RN, Reddy MK, Chow VTK, Lal SK. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein Is Phosphorylated and Localizes in the Cytoplasm by 14-3-3-Mediated Translocation. J Virol. 2005Sep;79(17):11476-86
48. Tugaeva KV, Hawkins DEDP, Smith JLR, Bayfield OW, Ker D-S, Sysoev AA. et al. The Mechanism of SARS-CoV-2 Nucleocapsid Protein Recognition by the Human 14-3-3 Proteins. J Mol Biol. 2021Apr16;433(8):166875
49. Sposito B, Broggi A, Pandolfi L, Crotta S, Clementi N, Ferrarese R. et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell. 2021Sep16;184(19):4953-4968.e16
50. Geng H, Subramanian S, Wu L, Bu H-F, Wang X, Du C. et al. SARS-CoV-2 ORF8 Forms Intracellular Aggregates and Inhibits IFNγ-Induced Antiviral Gene Expression in Human Lung Epithelial Cells. Front Immunol. 2021;12:679482
51. Zhang Y, Chen Y, Li Y, Huang F, Luo B, Yuan Y. et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc Natl Acad Sci. 2021Jun8;118(23):e2024202118
52. Satarker S, Tom AA, Shaji RA, Alosious A, Luvis M, Nampoothiri M. JAK-STAT Pathway Inhibition and their Implications in COVID-19 Therapy. Postgrad Med. 2021Jul4;133(5):489-507
53. Park A, Iwasaki A. Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 2020Jun10;27(6):870-8
54. Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z. et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020Jul30;11(1):3810
55. Xia H, Cao Z, Xie X, Zhang X, Chen JY-C, Wang H. et al. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020Oct6;33(1):108234
56. Yuen C-K, Lam J-Y, Wong W-M, Mak L-F, Wang X, Chu H. et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020Jan1;9(1):1418-28
57. Fung S-Y, Siu K-L, Lin H, Yeung ML, Jin D-Y. SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3. Int J Biol Sci. 2021;17(6):1547-54
58. Moustaqil M, Ollivier E, Chiu H-P, Van Tol S, Rudolffi-Soto P, Stevens C. et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg Microbes Infect. 2021Jan1;10(1):178-95
59. Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A. et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020Nov;587(7835):657-62
60. Mu J, Fang Y, Yang Q, Shu T, Wang A, Huang M. et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020Sep15;6(1):1-4
61. Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol. 2007Sep;81(18):9812-24
62. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020Jul;583(7816):459-68
63. Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS. et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci. 2020Nov10;117(45):28344-54
64. Addetia A, Lieberman NAP, Phung Q, Hsiang T-Y, Xie H, Roychoudhury P. et al. SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98. mBio. 2021Apr13;12(2):e00065-21
65. Kato K, Ikliptikawati DK, Kobayashi A, Kondo H, Lim K, Hazawa M. et al. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem Biophys Res Commun. 2021Jan15;536:59-66
66. Bryant A, Lawrie TA, Dowswell T, Fordham EJ, Mitchell S, Hill SR. et al. Ivermectin for Prevention and Treatment of COVID-19 Infection: A Systematic Review, Meta-analysis, and Trial Sequential Analysis to Inform Clinical Guidelines. Am J Ther. 2021Aug;28(4):e434
67. Wang W, Zhou Z, Xiao X, Tian Z, Dong X, Wang C. et al. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol Immunol. 2021Apr;18(4):945-53
68. Vazquez C, Swanson SE, Negatu SG, Dittmar M, Miller J, Ramage HR. et al. SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon activation through distinct mechanisms. PLOS ONE. 2021Jun24;16(6):e0253089
69. Makiyama K, Hazawa M, Kobayashi A, Lim K, Voon DC, Wong RW. NSP9 of SARS-CoV-2 attenuates nuclear transport by hampering nucleoporin 62 dynamics and functions in host cells. Biochem Biophys Res Commun. 2022Jan1;586:137-42
70. Attiq A, Yao LJ, Afzal S, Khan MA. The triumvirate of NF-κB, inflammation and cytokine storm in COVID-19. Int Immunopharmacol. 2021Dec;101(Pt B):108255
71. Chen J, Li Z, Guo J, Xu S, Zhou J, Chen Q. et al. SARS-CoV-2 nsp5 Exhibits Stronger Catalytic Activity and Interferon Antagonism than Its SARS-CoV Ortholog. J Virol. 2022Apr27;96(8):e0003722
72. Li W, Qiao J, You Q, Zong S, Peng Q, Liu Y. et al. SARS-CoV-2 Nsp5 Activates NF-κB Pathway by Upregulating SUMOylation of MAVS. Front Immunol. 2021;12:750969
73. Khan S, Shafiei MS, Longoria C, Schoggins JW, Savani RC, Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. eLife. 2021Dec6;10:e68563
74. Su C-M, Wang L, Yoo D. Activation of NF-κB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci Rep. 2021Jun29;11(1):13464
75. Umar S, Palasiewicz K, Meyer A, Kumar P, Prabhakar BS, Volin MV. et al. Inhibition of IRAK4 dysregulates SARS-CoV-2 spike protein-induced macrophage inflammatory and glycolytic reprogramming. Cell Mol Life Sci. 2022May19;79(6):301
76. Xia J, Tang W, Wang J, Lai D, Xu Q, Huang R. et al. SARS-CoV-2 N Protein Induces Acute Lung Injury in Mice via NF-ĸB Activation. Front Immunol. 2021;12:791753
77. Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Marrero MC. et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020Aug6;182(3):685-712.e19
78. Hopfner K-P, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. 2020Sep;21(9):501-21
79. Han L, Zhuang M-W, Deng J, Zheng Y, Zhang J, Nan M-L. et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J Med Virol. 2021;93(9):5376-89
80. Rui Y, Su J, Shen S, Hu Y, Huang D, Zheng W. et al. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduct Target Ther. 2021Mar15;6(1):1-11
81. Zhou Z, Zhang X, Lei X, Xiao X, Jiao T, Ma R. et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduct Target Ther. 2021Nov3;6(1):1-13
82. Li M, Ferretti M, Ying B, Descamps H, Lee E, Dittmar M. et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci Immunol. 2021May18;6(59):eabi9007
83. Messaoud-Nacer Y, Culerier E, Rose S, Maillet I, Rouxel N, Briault S. et al. STING agonist diABZI induces PANoptosis and DNA mediated acute respiratory distress syndrome (ARDS). Cell Death Dis. 2022Mar25;13(3):269
84. Neufeldt CJ, Cerikan B, Cortese M, Frankish J, Lee J-Y, Plociennikowska A. et al. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. Commun Biol. 2022Jan12;5(1):1-15
85. Burke JM, Clair LAS, Perera R, Parker R. SARS-CoV-2 infection triggers widespread host mRNA decay leading to an mRNA export block. RNA. 2021Nov1;27(11):1318-29
86. Finkel Y, Gluck A, Nachshon A, Winkler R, Fisher T, Rozman B. et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature. 2021Jun;594(7862):240-5
87. Zhang K, Miorin L, Makio T, Dehghan I, Gao S, Xie Y. et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci Adv. 2021Feb;7:eabe7386
88. Lu L, Madugula V. Mechanisms of ciliary targeting: entering importins and Rabs. Cell Mol Life Sci. 2018Feb1;75(4):597-606
89. Johnson CA, Malicki JJ. The Nuclear Arsenal of Cilia. Dev Cell. 2019Apr22;49(2):161-70
90. Robinot R, Hubert M, de Melo GD, Lazarini F, Bruel T, Smith N. et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat Commun. 2021Jul16;12(1):4354
91. Bryche B, St Albin A, Murri S, Lacôte S, Pulido C, Ar Gouilh M. et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav Immun. 2020Oct1;89:579-86
92. Shelton JF, Shastri AJ, Fletez-Brant K, Stella Aslibekyan, Auton A. The UGT2A1/UGT2A2 locus is associated with COVID-19-related loss of smell or taste. Nat Genet. 2022Feb;54(2):121-4
93. Wang L, Liu C, Yang B, Zhang H, Jiao J, Zhang R. et al. SARS-CoV-2 ORF10 impairs cilia by enhancing CUL2ZYG11B activity. J Cell Biol. 2022Jun8;221(7):e202108015
94. Fan S, Fogg V, Wang Q, Chen X-W, Liu C-J, Margolis B. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J Cell Biol. 2007Jul30;178(3):387-98
95. Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW. et al. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12(7):703-10
96. Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q. et al. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 2015Dec3;163(6):1484-99
97. Saksena N, Bonam SR, Miranda-Saksena M. Epigenetic Lens to Visualize the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Infection in COVID-19 Pandemic. Front Genet. 2021;12:581726
98. Sen R, Garbati M, Bryant K, Lu Y. Epigenetic mechanisms influencing COVID-19. Genome. 2021Apr;64(4):372-85
99. Kaneko S, Takasawa K, Asada K, Shinkai N, Bolatkan A, Yamada M. et al. Epigenetic Mechanisms Underlying COVID-19 Pathogenesis. Biomedicines. 2021Sep;9(9):1142
100. Meng Y, Zhang Q, Wang K, Zhang X, Yang R, Bi K. et al. RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes. Cell Death Dis. 2021Jul23;12(8):1-10
101. Qiu X, Hua X, Li Q, Zhou Q, Chen J. m6A Regulator-Mediated Methylation Modification Patterns and Characteristics of Immunity in Blood Leukocytes of COVID-19 Patients. Front Immunol. 2021;12:774776
102. Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A. et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol. 2020Apr;5(4):584-98
103. Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG. et al. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 2021May11;35(6):109091
104. Burgess HM, Depledge DP, Thompson L, Srinivas KP, Grande RC, Vink EI. et al. Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev. 2021Jul1;35(13-14):1005-19
105. Liu J, Xu Y-P, Li K, Ye Q, Zhou H-Y, Sun H. et al. The m6A methylome of SARS-CoV-2 in host cells. Cell Res. 2021Apr;31(4):404-14
106. Zhang X, Hao H, Ma L, Zhang Y, Hu X, Chen Z. et al. Methyltransferase-like 3 Modulates Severe Acute Respiratory Syndrome Coronavirus-2 RNA N6-Methyladenosine Modification and Replication. mBio. 2021Jul6;12(4):e0106721
107. Mongelli A, Barbi V, Gottardi Zamperla M, Atlante S, Forleo L, Nesta M. et al. Evidence for Biological Age Acceleration and Telomere Shortening in COVID-19 Survivors. Int J Mol Sci. 2021Jan;22(11):6151
108. Cao X, Li W, Wang T, Ran D, Davalos V, Planas-Serra L. et al. Accelerated biological aging in COVID-19 patients. Nat Commun. 2022Apr19;13(1):2135
109. Corley MJ, Pang APS, Dody K, Mudd PA, Patterson BK, Seethamraju H. et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J Leukoc Biol. 2021;110(1):21-6
110. Balnis J, Madrid A, Hogan KJ, Drake LA, Chieng HC, Tiwari A. et al. Blood DNA methylation and COVID-19 outcomes. Clin Epigenetics. 2021May25;13(1):118
111. Li S, Ma F, Yokota T, Garcia G, Palermo A, Wang Y. et al. Metabolic reprogramming and epigenetic changes of vital organs in SARS-CoV-2-induced systemic toxicity. JCI Insight. 2021Jan25;6(2):e145027
112. Samelson AJ, Tran QD, Robinot R, Carrau L, Rezelj VV, Kain AM. et al. BRD2 inhibition blocks SARS-CoV-2 infection by reducing transcription of the host cell receptor ACE2. Nat Cell Biol. 2022Jan;24(1):24-34
113. Yang X, Rutkovsky AC, Zhou J, Zhong Y, Reese J, Schnell T. et al. Characterization of Altered Gene Expression and Histone Methylation in Peripheral Blood Mononuclear Cells Regulating Inflammation in COVID-19 Patients. J Immunol. 2022Apr15;208(8):1968-77
114. Saunders CA, Parent CA. Emerging roles for the nucleus during neutrophil signal relay and NETosis. Curr Opin Cell Biol. 2020Feb1;62:135-43
115. Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA. et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020Jun4;5(11):e138999
116. Muus C, Luecken MD, Eraslan G, Sikkema L, Waghray A, Heimberg G. et al. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat Med. 2021Mar;27(3):546-59
117. Zazhytska M, Kodra A, Hoagland DA, Frere J, Fullard JF, Shayya H. et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell. 2022Mar17;185(6):1052-1064.e12
118. You M, Chen L, Zhang D, Zhao P, Chen Z, Qin E-Q. et al. Single-cell epigenomic landscape of peripheral immune cells reveals establishment of trained immunity in individuals convalescing from COVID-19. Nat Cell Biol. 2021Jun;23(6):620-30
119. Li S, Wu B, Ling Y, Guo M, Qin B, Ren X. et al. Epigenetic Landscapes of Single-Cell Chromatin Accessibility and Transcriptomic Immune Profiles of T Cells in COVID-19 Patients. Front Immunol. 2021;12:625881
120. Victor J, Deutsch J, Whitaker A, Lamkin EN, March A, Zhou P. et al. SARS-CoV-2 triggers DNA damage response in Vero E6 cells. Biochem Biophys Res Commun. 2021Nov19;579:141-5
121. Garcia G, Sharma A, Ramaiah A, Sen C, Purkayastha A, Kohn DB. et al. Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication. Cell Rep. 2021Apr6;35(1):108940
122. Liu X, Wei L, Xu F, Zhao F, Huang Y, Fan Z. et al. SARS-CoV-2 spike protein-induced cell fusion activates the cGAS-STING pathway and the interferon response. Sci Signal. 2022Apr12;15(729):eabg8744
123. Sanchez-Vazquez R, Guío-Carrión A, Zapatero-Gaviria A, Martínez P, Blasco MA. Shorter telomere lengths in patients with severe COVID-19 disease. Aging. 2021Jan15;13(1):1-15
124. Franzen J, Nüchtern S, Tharmapalan V, Vieri M, Nikolić M, Han Y. et al. Epigenetic Clocks Are Not Accelerated in COVID-19 Patients. Int J Mol Sci. 2021Jan;22(17):9306
125. Yin Y, Liu X, He X, Zhou L. Exogenous Coronavirus Interacts With Endogenous Retrotransposon in Human Cells. Front Cell Infect Microbiol. 2021;11:609160
126. Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci. 2021May25;118(21):e2105968118
127. Aldén M, Olofsson Falla F, Yang D, Barghouth M, Luan C, Rasmussen M. et al. Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line. Curr Issues Mol Biol. 2022Mar;44(3):1115-26
128. Briggs E, Ward W, Rey S, Law D, Nelson K, Bois M. et al. Assessment of potential SARS-CoV-2 virus integration into human genome reveals no significant impact on RT-qPCR COVID-19 testing. Proc Natl Acad Sci. 2021Nov2;118(44):e2113065118
129. Kazachenka A, Kassiotis G. SARS-CoV-2-Host Chimeric RNA-Sequencing Reads Do Not Necessarily Arise From Virus Integration Into the Host DNA. Front Microbiol. 2021;12:676693
130. Yan B, Chakravorty S, Mirabelli C, Wang L, Trujillo-Ochoa JL, Chauss D. et al. Host-Virus Chimeric Events in SARS-CoV-2-Infected Cells Are Infrequent and Artifactual. J Virol. 2021Jul12;95(15):e00294-21
131. Parry R, Gifford RJ, Lytras S, Ray SC, Coin LJM. No evidence of SARS-CoV-2 reverse transcription and integration as the origin of chimeric transcripts in patient tissues. Proc Natl Acad Sci. 2021Aug17;118(33):e2109066118
132. Wen Z, Zhang Y, Lin Z, Shi K, Jiu Y. Cytoskeleton—a crucial key in host cell for coronavirus infection. J Mol Cell Biol. 2020Dec1;12(12):968-79
133. Cho S, Irianto J, Discher DE. Mechanosensing by the nucleus: From pathways to scaling relationships. J Cell Biol. 2017Feb;216(2):305-15
134. Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497(7450):507-11
135. Porter L, Minaisah R-M, Ahmed S, Ali S, Norton R, Zhang Q. et al. SUN1/2 Are Essential for RhoA/ROCK-Regulated Actomyosin Activity in Isolated Vascular Smooth Muscle Cells. Cells. 2020Jan6;9(1):132
136. Thakar K, May CK, Rogers A, Carroll CW. Opposing roles for distinct LINC complexes in regulation of the small GTPase RhoA. Weis K, editor. Mol Biol Cell. 2017Jan1;28(1):182-91
137. DeOre BJ, Tran KA, Andrews AM, Ramirez SH, Galie PA. SARS-CoV-2 Spike Protein Disrupts Blood-Brain Barrier Integrity via RhoA Activation. J Neuroimmune Pharmacol. 2021Dec1;16(4):722-8
138. Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD. et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004Feb1;113(3):370-8
139. Jain N, Iyer KV, Kumar A, Shivashankar GV. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci. 2013Jul9;110(28):11349-54
140. Hayashi K, Murai T, Oikawa H, Masuda T, Kimura K, Muehlich S. et al. A novel inhibitory mechanism of MRTF-A/B on the ICAM-1 gene expression in vascular endothelial cells. Sci Rep. 2015May29;5(1):10627
141. Cho S, Vashisth M, Abbas A, Majkut S, Vogel K, Xia Y. et al. Mechanosensing by the Lamina Protects against Nuclear Rupture, DNA Damage, and Cell-Cycle Arrest. Dev Cell. 2019;49(6):920-35
142. Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM. et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020Dec1;39(23):e106267
143. El Baba R, Herbein G. Management of epigenomic networks entailed in coronavirus infections and COVID-19. Clin Epigenetics. 2020Aug5;12(1):118
144. Atlante S, Mongelli A, Barbi V, Martelli F, Farsetti A, Gaetano C. The epigenetic implication in coronavirus infection and therapy. Clin Epigenetics. 2020Oct21;12(1):156
145. Lu L-Y, Feng P-H, Yu M-S, Chen M-C, Lin AJ-H, Chen JL. et al. Current utilization of interferon alpha for the treatment of coronavirus disease 2019: A comprehensive review. Cytokine Growth Factor Rev. 2022Jan13;63:34-43
146. WHO Solidarity Trial Consortium, Pan H, Peto R, Henao-Restrepo A-M, Preziosi M-P, Sathiyamoorthy V. et al. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N Engl J Med. 2021Feb11;384(6):497-511
147. Domizio JD, Gulen MF, Saidoune F, Thacker VV, Yatim A, Sharma K. et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022Mar;603(7899):145-51
148. Nilsson-Payant BE, Uhl S, Grimont A, Doane AS, Cohen P, Patel RS. et al. The NF-κB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication. J Virol. 2021Nov9;95(23):e0125721
149. Kandasamy M. NF-κB signalling as a pharmacological target in COVID-19: potential roles for IKKβ inhibitors. Naunyn Schmiedebergs Arch Pharmacol. 2021Mar;394(3):561-7
150. Li L, Guan Y, Chen X, Yang J, Cheng Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front Pharmacol. 2021;11:629266
Author contact
![]() Corresponding author: Wakam Chang, University of Macau, E12-3016, Avenida da Universidade, Taipa, Macau, China. Phone: (853) 88224986. Email: wakamchangedu.mo
Corresponding author: Wakam Chang, University of Macau, E12-3016, Avenida da Universidade, Taipa, Macau, China. Phone: (853) 88224986. Email: wakamchangedu.mo

 Global reach, higher impact
Global reach, higher impact