Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(13):5103-5122. doi:10.7150/ijbs.75410 This issue Cite
Review
Phase separation in Cancer: From the Impacts and Mechanisms to Treatment potentials
1. Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China.
2. Hunan Key Laboratory of Translational Radiation Oncology, 283 Tongzipo Road, Changsha 410013, Hunan, China.
* Contributed equally
Received 2022-5-23; Accepted 2022-7-16; Published 2022-8-1
Abstract
Cancer is a public health problem of great concern, and it is also one of the main causes of death in the world. Cancer is a disease characterized by dysregulation of diverse cellular processes, including avoiding growth inhibitory factors, avoiding immune damage and promoting metastasis, etc. However, the precise mechanism of tumorigenesis and tumor progression still needs to be further elucidated. Formations of liquid-liquid phase separation (LLPS) condensates are a common strategy for cells to achieve diverse functions, such as chromatin organization, signal transduction, DNA repair and transcriptional regulation, etc. The biomolecular aggregates formed by LLPS are mainly driven by multivalent weak interactions mediated by intrinsic disordered regions (IDRs) in proteins. In recent years, aberrant phase separations and transition have been reported to be related to the process of various diseases, such as neurodegenerative diseases and cancer. Herein, we discussed recent findings that phase separation regulates tumor-related signaling pathways and thus contributes to tumor progression. We also reviewed some tumor virus-associated proteins to regulate the development of virus-associated tumors via phase separation. Finally, we discussed some possible strategies for treating tumors by targeting phase separation.
Keywords: phase separation, cancer, biomolecular condensates, tumor-related signaling pathways, virus-associated tumors
Introduction
Cancer is a complex disease that is based on several “cancer hallmarks” described by Hanahan and Weinberg in 2011. This includes self-sufficiency in growth signals, genomic instability and mutation, resistance to cell death and avoidance of immune surveillance, and others [1-3]. Tumor progression is mainly driven by tumor suppressor gene inactivation and oncogene over activation, such as P53, MYC, RAS, EGFR, etc. [4-8]. Some tumor-associated signaling pathways are evolutionarily conserved in mammals, which plays a key role in tumor cell development and differentiation. Increasing evidence has shown that aberrant activation of tumor-associated signaling pathways in many different tumors can induce tumor cells proliferation, metastasis and epithelial-mesenchymal transition, such as Wnt/beta-catenin signaling, Hippo signaling and mTOR signaling [9-11].
In addition, tumor associated virus is also one of the important causes of cancer, which is related to about 20% of human tumors. These tumor viruses can affect various cell activities and lead to the occurrence of human malignant tumors [12, 13]. At present, several recognized human tumor viruses have been involved in the development of human cancer, including hepatitis B virus (HBV) and hepatitis C virus (HCV) in liver cancer, EB virus (EBV) in lymphoma, human papillomavirus (HPV) in cervical cancer, Kaposi sarcoma herpesvirus (KSHV) in Kaposi sarcoma [14]. Although great progress has been made in identifying driving mutations and related oncogenic signaling pathways of oncogenes or tumor suppressor genes, the exact pathological mechanism of tumorigenesis or tumor development is still largely unknown.
Phase separation is a relatively unfamiliar concept in biology, but it is a very common phenomenon in the field of physical chemistry. It describes the dynamic concentration of biomolecules from a homogeneous environment into a relatively dense phase to form a sparse phase and a dense phase [15]. The formation of cellular compartmentalization and membrane-less organelles in cells can be explained by LLPS theory. LLPS occurs when multivalent biopolymers interact instantaneously to coalesce into a dense membrane-less condensate [16-18]. The characteristics of LLPS include the liquid properties of the formed condensate droplets, such as spherical, fusion and fission, and then relaxation into a sphere [19]. More and more evidences show that LLPS is the basis for the formation of various subcellular membrane-less compartments, such as stress granules(SGs) [20-22], Cajal bodies[23], nucleolus [24, 25], splicing speckles [26-28], and processing bodies (P-bodies) [29]. The proteins involved in the formation of LLPS aggregates usually have intrinsically disordered regions (IDRs). These IDRs may mediate weak-affinity and non-specifically interactions of multiple targets to trigger LLPS [30-32]. Many functions of IDR depend on their structural properties, such as spacers, flexible linkers or entropic springs [33, 34].
Growing evidence suggests that LLPS condensates are related to the pathogenesis of neurodegenerative diseases [35]. In the past few years, LLPS condensates in cells have been known to be associated with several proteins that accumulate in neurodegeneration, including FUS [36, 37], TDP-43 [38, 39], HNRNPA1 [40], and DDX [41], as well as Tau [42, 43]. Some evidence suggested that there is a closely link between the development of cancer and the formation of phase separation condensate [44-48], such as transcriptional condensates, PRC1 condensates, super enhancers, DNA repair condensates, stress granules, Paraspeckles, SPOP/DAXX bodies and PML foci[49-54]. Here, we summarize the latest findings on the function and mechanism of LLPS-related condensates. We discuss recent results that biomolecular condensate is involved in regulating tumor-related signaling pathways, thereby contributing to cancer cell survival. We also summarize the function and mechanisms of LLPS condensates in virus-associated proteins to promote the progression of virus-associated tumors. Finally, we discuss how this LLPS condensate affects cancer treatments.
Characteristics of LLPS condensates
LLPS is a thermodynamic process that divides the mixture into dense phase and dilute phase to achieve the lowest free energy state [19, 55, 56]. Phase separation is the characteristics of many macromolecules, such as proteins, RNA/DNA or their complexes like chromatin [57, 58]. LLPS is involved in the assembly process of many membrane-less condensates (Table 1), such as Cajal bodies, nucleoli and nuclear bodies, which is conducive to the efficient or orderly regulation of various complex biochemical reactions in cells [59-62]. The weak intramolecular and intermolecular interactions, including electrostatic, cation-pi and pi-pi interactions, hydrophobic, are the driving forces for the formation of LLPS membrane-less organelles [31, 63, 64]. The assembly of biomolecular aggregates can be promoted by these weak interactions. Special interactions between the condensates can promote the formation of different compartments [19, 65].
Membraneless organelles formed by phase separation
| Name | Location | Functions | References |
|---|---|---|---|
| Nucleoli | Nucleus | The site of ribosomal RNA (rRNA) production and ribosome subunit assembly | [219] |
| P-bodies | Cytoplasm | Associated with translation repression and 5'-to-3'mRNA decay | [220] |
| Cajal bodies | Nucleus | Involved in the formation of ribonucleoproteins including small nuclear RNPs | [221] |
| PML bodies | Nucleus | Involved in a wide variety of biological processes ranging from senescence to viral infections or stemness | [222] |
| Stress granules | Cytoplasm | Play an important in the stress response and may contribute to some degenerative diseases | [223] |
| U-bodies | Cytoplasm | Involved in mRNA decay and translational repression | [224] |
| Paraspeckles | Nucleus | Mediate the nuclear retention of some A-to-I hyper-edited mRNAs, gene transcription, RNA splicing, and RNA stability | [225] |
| GW/P bodies | Cytoplasm | Translational repressors of mRNA through Ago2-mediated RNA silencing | [226] |
| Polycomb bodies | Nucleus | Mediate down-regulation of target genes | [227] |
| Nuclear speckle | Nucleus | Inhibition of mRNA splicing | [228] |
The governing mechanism of phase separation condensates in the cell is a multivalent interaction [66-68]. Intrinsic disordered regions (IDRs) and low complexity regions (LCRs) can promote the multivalent interaction of proteins [69]. LCRs lack a stable three-dimensional structure, and often serve as a scaffold that interacts with short and flexible interacting motifs [30, 70]. Some specific amino acids are often highly enriched in IDRs, such as hydrophilic residues (serine, arginine, glutamine, glutamate, and lysine), aromatic residues (tyrosine, phenylalanine, and tryptophan) and charged residues. These residues contribute to form electrostatic interactions, pi-pi interaction and cation-pi interactions, respectively. In contrast, aliphatic residues are less frequently observed in low-complexity domains, such as leucine, valine and isoleucine [71-73].
Proteins and nucleic acids (DNA and RNA) are the main components and mediators of LLPS. Their biophysical properties and phase separation behaviors can vary them to form a highly multi-component system in condensates [74, 75]. Many RNA-binding proteins (RBPs) have IDRs and LCRs, referred to as prion-like domains (PLDs), so that phase separation condensates can be formed in an overcrowded nuclear environment [37, 76-78]. The protein containing PLDs is initially concerned because they can be assembled into a self-template protein aggregate. These LLPS aggregates may be infectious because they can spread between individuals [46, 79]. Approximately 70 human RBPs contain a PLDs via some database of LLPS-related proteins (Table 2), including TDP-43 (transactivation response element DNA-binding protein 43), FUS (fused in sarcoma), EWSR1 (Ewing sarcoma breakpoint region 1), TAF15 (TATA-binding protein-associated factor 15) and hnRNPA1/A2 (heterogeneous nuclear ribo-nucleoproteins A1/A2) [80]. Indeed, Wang et al. predicted the saturation concentration of a class of proteins with domain similar length to PLD and RBD of FUS family proteins by using a specific model, and identified some proteins that may provide key scaffold functions for many biochemical compartments in cells [76]. Other proteins containing repetitive sequences of Src homology 3 (SH3) domain and proline-rich motifs (PRMs) can also be used as scaffold proteins. Phase separation can be driven by these multivalent SH3/ RPM domains in a concentration dependent manner [67, 74].
Biological macromolecular phase separation is not limited to proteins. RNA is another important component of phase separation condensates. In fact, RNA is widely involved in the formation of RNA/protein-rich membrane-less aggregates in cells by promoting the LLPS [44, 81]. For example, the two cytoplasmic RNA particles, SG and P body driven via LLPS are responsible for different main functions, but they also exchange mRNA in each other and share many RBPs [82, 83]. Additionally, RNA is an ideal scaffold element for its single-stranded, multivalent, and flexible structures. For example, long non-coding RNAs (lncRNAs) participate in the formation of membrane-less organelles as scaffolds, such as NEAT1 and HSATIII binding for many specific proteins in nuclear body, and keep the dynamic shuttle of proteins and RNAs in nucleoplasm.[49, 84]. The formation of phase separation is closely related to the type and concentration of RNA. High RNA/protein ratio can inhibit the formation of phase separation droplets, while low RNA/protein ratio can promote the formation of phase separation droplets. The decrease of nuclear RNA level or genetic alteration of RNA binding leads to excessive phase separation in cells, which promotes the formation of cytotoxic solid like aggregates [37].
Phase separation related databases
| Name | URL | Functions |
|---|---|---|
| IUPred | https://iupred2a.elte.hu/plot | Prediction of disordered protein regions |
| PLAAC | http://plaac.wi.mit.edu/ | Prediction of prion-like region |
| PONDR | http://www.pondr.com | Predictor of natural disordered regions |
| MobiDB | https://mobidb.org | Provides information about intrinsically disordered regions and related features |
| CIDER | http://pappulab.wustl.edu/CIDER/ | Calculation of many different parameters associated with disordered protein sequences |
| ZipperDB | https://services.mbi.ucla.edu/zipperdb/ | Predictions of fibril-forming segments within protein |
| D2P2 | http://d2p2.pro/ | Database of disordered protein predictions |
| Metadisorder | http://iimcb.genesilico.pl/metadisorder/ | Prediction of protein disorder |
| Expasy | https://web.expasy.org/compute_pi/ | Computation of the theoretical pI (isoelectric point) and Mw (molecular weight) |
| AMYCO | http://bioinf.uab.es/amycov04/ | Evaluation of mutation impact on prion-like proteins aggregation propensity |
| RPS | http://rps.renlab.org/#/Home | A comprehensive database of RNAs involved in liquid-liquid phase separation |
| RNAPhaSep | http://www.rnaphasep.cn/#/Home | A resource of RNAs undergoing phase separation |
| LLPSDB | http://biocomp.ucas.ac.cn/llpsdb/home.aspx | A database of proteins undergoing liquid-liquid phase separation in vitro |
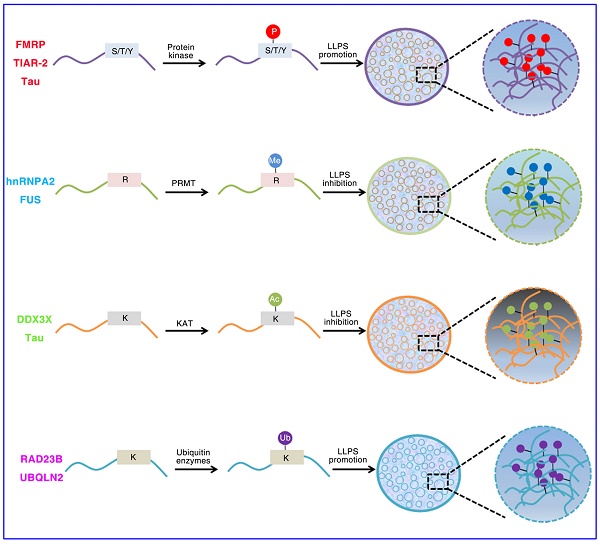
Role of Post-translational modifications (PTMs) of intrinsically disordered proteins regulates LLPS. (A). Phosphorylation of FMRP, TIAR-2 and Tau changes the intermolecular interactions and thus promotes FUS/RNA phase separation. (B). Methylation of hnRNPA2 and FUS inhibits the phase separation by weakening the cation-π interactions. (C). DDX3X and Tau IDRs acetylated by lysine acetyltransferase results in impaired phase separation. (D). The RAD23B and UBQLN2 formed LLPS by triggering the multivalent interactions between ubiquitin-associated domains and ubiquitin chains of ubiquitinated proteins.
The regulation of LLPS condensates
The phase separation of the protein is strictly controlled by various mechanisms. The multivalent affinity of intermolecular and intramolecular can be regulated by physical conditions such as pH, temperature, ion concentration and osmotic pressure, thereby changing the phase separation behaviors of the biomolecular system [19, 44, 85]. Recent results suggest that ParB phase separation condensate needs the ATPase activity of para to maintain. Further experiments show that motor protein can participate in the control of LLPS droplet number and subcellular localization [86]. Interestingly, the size, number and subcellular localization of some nucleolar compartments, such as stress granules, heterochromatin domain or P granules, can be controlled by ATP by the similar mechanisms, and after ATP deletion, the fluidity of stress granules was significantly inhibited[61, 87, 88].
IDRs are enriched in post-translational modifications (PTMs) sites. These modifications can result in changes in secondary or tertiary structure and can create or destroy interaction sites [32]. PTMs can have a strong effect on the charge state and/or binding motifs of proteins, and thus are primary regulators of LLPS (Figure 1). Phosphorylation, acetylation, methylation, sumoylation and ubiquitination are the most common PTMs [58, 89-92]. For example, both phosphorylation and phosphomimetic variants in low complexity domain inhibit its prion like characteristics and aggregation tendency. The aggregation tendency of FUS can be significantly reduced when the phase separation of FUS is destroyed by the presence of RNA or salt [93]. The occurrence of DDX4 phase separation is driven by its N-terminal RGG-rich domain in vitro, and it can also form liquid condensates in cells. The cation-π interactions between repeated FG and RG motifs can promote the formation of DDX4 LLPS droplets. However, PRMT1 expression can inhibit the formation of DDX4 droplets, mainly because PRMT1 mediates the asymmetric dimethylation of DDX4 [41, 94]. The phase separation of DDX3X can be driven by the N-terminal IDRs, and the formation of DDX3X droplets can be destroyed by acetylation of multiple lysine residues. HDAC6 can enhance DDX3X phase separation by deacetylating IDRs, which is also necessary to promote SG maturation [95]. The IDR of C. elegans PcG protein SOP-2 can mediate phase separation, which can be regulated by sumoylation [91]. The p62 can form phase separation droplets with liquid properties in vivo which can be induced by adding k63 ubiquitin chain, so that a large number of ubiquitin signals are enriched in p62 droplets [90].
LLPS has been found to be regulated by post transcriptional modifications (Figure 2) [74]. RNA N6-methyladenosine (m6A), as the most common RNA modification, has been reported to be related to the progression of a variety of life activities and diseases [96]. YTHDF1-3 is a cytoplasmic m6A binding protein, which has been proved to undergo phase separation both in cells and in vitro. YTHDF proteins can bind to methylated mRNA, resulting in phase separation [97]. Similarly, Wang, et al. also reported that m6A enhanced the phase separation ability of YTHDF2 through experiments in vitro and in vivo. In cells, YTHDF2 itself has a weak ability to undergo LLPS, and its phase separation ability is significantly enhanced after binding with m6A mRNA. Although YTHDF2 protein itself can form phase separation droplets in vitro, the addition of m6A modified RNA significantly promotes its LLPS ability [98]. Recent studies have reported that highly active enhancer RNA (eRNA) can be modified by m6A to recruit YTHDC1 to form a LLPS condensate, which depends on its C-terminal disordered region and arginine residues. The formation of BRD4 coactivator LLPS condensates can be promoted by YTHDC1/m6A-eRNA phase separation condensate co-mixes [99]. In addition, mRNA degradation can be promoted by YTHDF1 phase separation and the interaction of YTHDF1-AGO2 [100]. These studies showed that the composition of intracellular LLPS transcriptome can be regulated by the distribution and number of m6A sites in mRNA, which indicates that phase separation can control the cellular characteristics of m6A modified mRNA.
RNA G-quadruplex is a secondary structure of nucleic acids formed in guanine rich sequences. It can interact multivalent with RNA binding proteins and RNA, which also makes it a favorable scaffold for RNA-driven phase separation (Figure 2) [84]. SHR mRNA has RNA G-quadruplex structure, which can undergo LLPS under physiological conditions. Under the condition of more G-quadruplex, the ability of G-quadruplex to trigger phase separation will be significantly enhanced. In addition, the formation of phase separation is closely related to the number of G-quadruplex and the length of loops [101]. Recently, Liu, et al. reported that single stranded DNA with parallel G-quadruplex structure can functionally cooperate with G-quadruplex binding protein to form phase separation droplets by using specific giant membrane vesicles as a protocell model [102]. In addition, the LLPS of FUS condensate formation is significantly enhanced through the interaction between FUS and G-quadruplex-RNA [103]. These studies have indicated that special RNA secondary structures may have an important role in forming different phase separation condensates in a cellular environment.
The role of LLPS in oncogenic signaling
Since “phase separation” is involved in various life activities of cells, its abnormal state will inevitably lead to the occurrence of many diseases. Neurodegenerative diseases including frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) have been linked to disruption of the components and properties of LLPS condensates [104, 105]. The relationship between the LLPS stress particles and these ALS/FTD-associated proteins, including HNRNPA1/HNRNPA2, TDP-43 and FUS, has always been the main focus to correlate LLPS particles to neurodegeneration [106]. More and more evidences show that LLPS are involved in many major cellular processes, such as heterochromatin and genome organization, transcription, and stress responses [58, 107-110]. Some studies have reported that LLPS can also occur on many carcinogenic signaling molecules [111, 112]. Here, we will elucidate the relationship between oncogenic signaling pathways and mis-regulated phase separation (Figure 3). Additionally, we also discuss the mechanisms of phase separation of oncogenic signaling molecules and its potential significance.
LLPS in p53 signaling
The p53 protein and its cellular pathway are highly conserved in evolution, which can lead to cell death, mediate tumor inhibition or maintain cell homeostasis through a group of regulatory, informed and comprehensive responses to environmental disturbances [113]. p53 can act as an internal monitor of many cellular stresses and DNA damage response, such as telomere shedding, mitochondrial and ribosomal biological changes, spindle poisoning, starvation, hypoxia or oncogene activation. Depending on the degree of cell damage, p53 can induce cell cycle arrest or cell death, aging and DNA repair [114, 115]. In 50% of human cancers, p53 mutations hinder their binding to the specific target sequence. Therefore, many studies have been carried out on the function of p53 and the consequences caused by its loss of function [116].
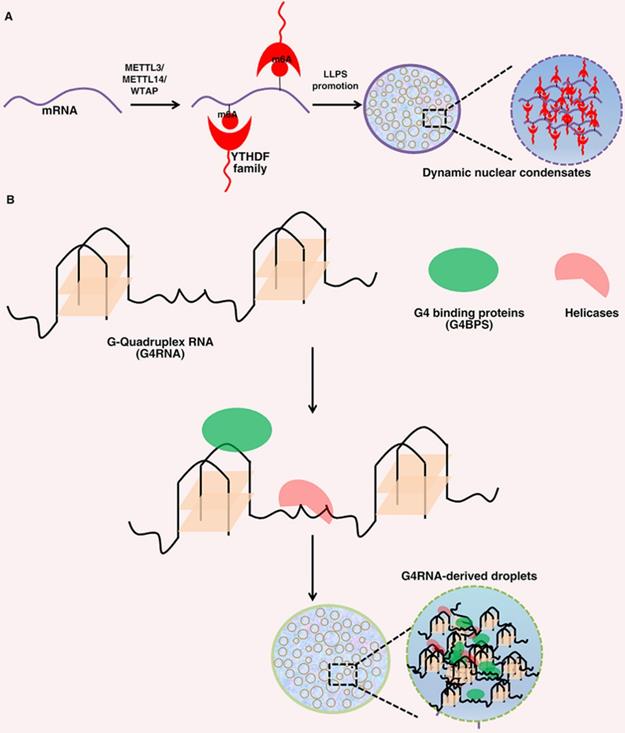
Roles of N6-methyladenosine(m6A) and G-quadruplex Structures of RNA in LLPS. (A). The METTL3/ METTL14/WTAP writer complex co-transcriptionally methylates mRNAs. A set of YTHDF family ''reader'' proteins bind directly or indirectly to m6A-mRNAs and thus promotes YTHDF-m6A-mRNAs phase separation. (B). As a scaffold, G4RNAs can interact with G4BP and RNA helicase to promote phase separation.
Recently, another important aspect of p53 that has been reported is that it can play its function by participating in the formation of LLPS liquid-like condensates. For example, p53 is involved in the formation of PML and Cajal bodies under stress response conditions [117, 118]. P53 itself has the potential to form phase separation liquid-like condensates, which can be regulated by post-translational modification and other cellular molecules [116]. Some studies have found that p53 protein amyloid formation exists in human cancer tissues. The formation of p53 amyloid protein in cells can lead to its functional inactivation and promote its transformation into oncoprotein. Cancer-associated mutation of p53 can accelerate the protein aggregation and amyloid formation by destroying the folding of p53 core domain [116]. In addition, the formation of aggregation structure in cancer may be caused by the formation of p53 phase separation condensate, such as mutant amyloid oligomer [119]. P53 signal transduction and DNA damage response can be regulated by p53 binding protein 1 (53BP1) [120]. 53BP1 can form phase separation droplets, which enrich tumor suppressor protein p53. The expression of p53 target gene and 53BP1-dependent induction of p53 can be inhibited by destroying the phase separation of 53BP1 [51]. The scaffold protein AHNAK can regulate the phase separation potential of 53BP1 by binding to its oligomeric domain. The excessive accumulation of 53BP1 in chromatin, enhancement of its LLPS, increasing of p53 response, destruction of the survival of cancer cells, and the aging of non-transformed cells are closely related to the loss of AHNAK [121]. These studies suggest that one of the reasons for the loss of p53 function may be the phase separation of p53 and the formation of amyloid protein. Lemos, et al. found that the small molecule compound aminothiazole can destroy p53 condensate by interacting with p53, which indicates that the compound changes the condensation behavior of p53 according to the type of p53 mutation. Furthermore, the compound does not cause reactivation of mutant p53 and is active on p53 phase separation condensate. These results provide evidence for p53 phase separation condensation of mutations in cells and provide tools to regulate this process [122].Therefore, targeting the formation of p53 phase separation may become an important way of cancer treatment.
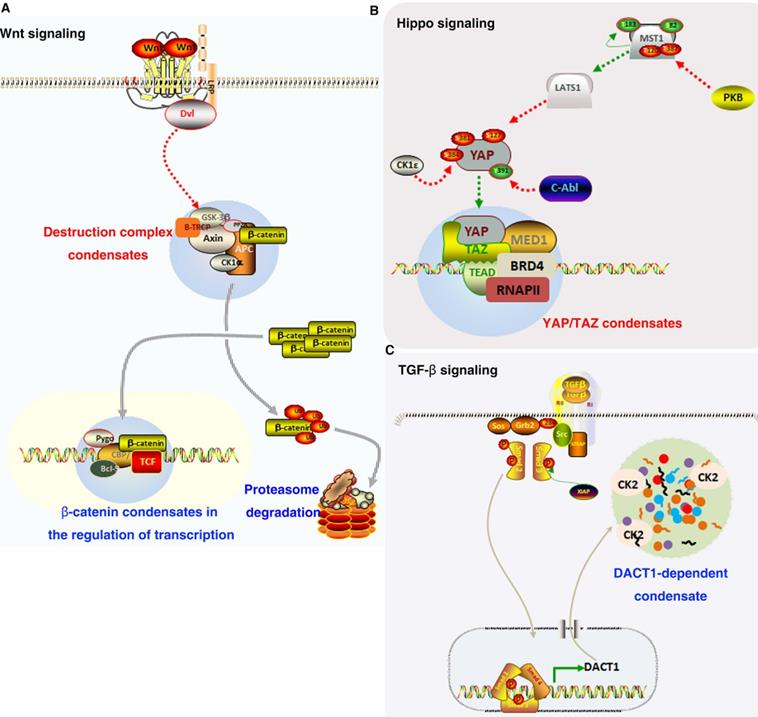
Role of LLPS in oncogenic signaling. (A). In Wnt/β-catenin signaling pathway, Axin, and APC assemble into a destruction complex condensate that recruits other members such as GSK3 and CKI. β-catenin accumulates and is transported to the nucleus, where it may localize to condensates at super-enhancers to activate the expression of target genes. (B). YAP/TAZ condensates co-localize with TEAD and recruit RNA Pol II to promote the expression of downstream target gene. (C). TGF-β promotes the expression of DACT1, which through LLPS is required for compartmentalising hundreds of proteins including CK2.
LLPS in Wnt/β-catenin signaling
Wnt/β-catenin signaling is one of the key pathways controlling stemness and development, and is closely related to cancer [123]. Wnt/β-catenin signaling participates in a variety of tumor physiological processes such as proliferation, migration/invasion and apoptosis [124-126]. Wnt/β-catenin signaling pathway generally refers to the canonical Wnt signaling, which can be divided into three main components: β-catenin protein, degradation complex and membrane protein three main components [127]. The localization of Wnt protein receptors low-density lipoprotein receptor related protein group (LRP5/6) and frizzled (Fzd2) receptors on the cell membrane [128, 129]. The Axin, glycogen synthase kinase 3β (Gsk3β), adenomatous colorectal polyps (APC), casein kinase 1α (CK1α), and dishevelled (DVL) protein to form the β-catenin destruction complex. CK1α and GSK3β can promote β-catenin ubiquitination and subsequent proteasome degradation by controlling its phosphorylation successively [130, 131]. Un-phosphorylated β-catenin gradually accumulates in cytoplasm and transports to the nucleus to activate Wnt downstream target genes by interacting with lymphoid enhancer-binding factor (LEF) and T cell-specific factor (TCF) co-activators [132].
Recent studies have shown that LLPS can occur in some signaling molecules in Wnt signaling pathway, which is very important for the regulation of Wnt functions [133]. IDR in Axin can drive its phase separation and promote the formation of destruction complex. Phase separation phenomenon has also been found in APC molecules, which can enhance the dynamic of Axin phase separation droplets in vitro. The assembly of β-catenin destruction complex and β-catenin phosphorylation by GSK3β/CK1α are driven by phase separation, which then maintain β-catenin protein stability and regulate Wnt/β-catenin signal transduction [133]. In vitro, IDRs of APC undergoes phase separation. In colorectal cells, β‑catenin degradation and Axin puncta formation can be promoted by expressing IDR of APC [134]. Dvl is a multivalent protein interacting with other Wnt signaling proteins. Dvl was also observed to form puncta in cells, and its DIX domain is important for the puncta formation. Although Dvl protein has been proposed to have an ability to undergo LLPS, there is no experimental evidence that has been reported by far [133, 135, 136]. Recent studies have shown that coactivator, mediators and some transcription factors enrich on super enhancers to form phase separation condensates [110, 137]. Interestingly, β-catenin interacts with DNA binding factors and selectively occupies the super enhancer to form a phase separated condensate [138]. These studies provide the possibility that phase separation is involved in the assembly of Wnt pathway molecules to regulate the development of cancers.
LLPS in Hippo signaling
The Hippo pathway is a conserved pathway that plays a key role in organ development, immune regulation and tissue regeneration [139]. A variety of cancer progression has been found to be related to the dysregulation of Hippo pathway, such as lung, colorectal, ovarian and pancreatic cancers [140-143]. The Hippo pathway is composed of a huge protein network, which not only regulates the growth of different tissues in the process of regeneration and development, but also controls the occurrence and development of cancer in pathological state [10]. Many of these functions are mediated by transcriptional factors YAP and TAZ, which directly regulates gene expression by controlling the transcription factor TEAD family [139].
Some studies have highlighted that the nuclear cytoplasmic shuttle behavior of Hippo pathway transcription coactivators YAP and TAZ is much more dynamic than previously recognized, and that YAP and TAZ are also regulated by LLPS [144]. TAZ forms nuclear condensates through LLPS to compartmentalize BRD4 and coactivators MED1, the transcription elongation factor CDK9 for transcription, and its DNA-binding cofactor TEAD4. Hippo signaling pathway can negatively regulated the phase separated ability of TAZ via LATS-mediated mediated phosphorylation. In addition, the coiled-coil domain can drive the phase separation of TAZ [145]. Some super enhancer markers such as Oct4, Sox2, H3K27ac and Nanog can co-locate with YAP in mouse embryonic stem cells. In addition, YAP also guides the formation of Med1 labeled aggregates at its binding site through LLPS [146]. Mechanistically, the activation of TAZ in the nucleus and the occurrence of TAZ phase separation are regulated by paraspeckle protein NONO. Overexpression of NONO promoted nuclear TAZ phase separation, while low expression of NONO decreased nuclear TAZ phase separation. Moreover, the low expression of NONO inhibited the interaction of TAZ with enhancers and TEAD [147]. In addition, the phase separation of LATS1 can be promoted by phosphatidic acid-binding lncRNA SNHG9, thereby promoting carcinogenic YAP signaling. These findings have revealed a tumor-associated lncRNA as a key regulator of YAP by facilitating the formation of LATS1 phase separation condensates [148]. Taken together, it can be assumed that Hippo signaling molecules are activated through phase separation and thus play a key role in the occurrence and progression of tumors.
LLPS in TGF-β signaling
Transforming growth factor β (TGF-β) signaling pathway is an evolutionarily conserved pathway and plays a key role in some biological processes, such as cell apoptosis, migration, growth, differentiation, tumorigenesis and development [149]. TGF-β receptor can be activated by its ligand to promote the phosphorylation of serine/threonine residues, and then induce the phosphorylation of intracellular effector SMADs [150]. The activated SMADs Protein can transfer to the nucleus, activate the transcription of its target genes and regulate cellular functions [151]. Smad family consists of many proteins whose main function is to transducing extracellular signals to the nucleus, in which SMAD4 is the key regulator BMP and TGF-β signaling pathway, while SMAD2/3 mainly controls TGF-β signal transduction of subfamily members [149].
Recently, some signaling molecules with IDR in TGF signaling pathway are enriched in super-enhancers condensates by phase separation. TGF-β signaling factors SMAD3 can form nuclear foci when the signaling pathway is activated. The results show that these SMAD3 nuclear foci are condensates formed by phase separation [138]. In addition, TGF-β signal can induce DACT1 to form phase separated condensates in cytoplasm, which can inhibit the function of Wnt signaling pathway. The deletion of IDR in DACT1 can destroy its ability to form phase separation condensate and inhibit Wnt pathway. Moreover, the role of DACT1 in breast cancer and prostate cancer bone metastasis is also dependent on the maintenance of DACT1 aggregates in cellular [152]. These studies suggest that phase separation may influence tumor progression by regulating TGF-β signaling pathway.
LLPS in AMPK signaling
In eukaryotic cells, AMP-activated protein kinase (AMPK) can monitor ATP: ADP: AMP ratio through as an energy sensor [153]. The increase of intracellular ADP/AMP relative level or the decrease of ATP level can activate AMPK, including stress responses triggered by tissue ischemia, hypoxia, muscle resection or glucose deprivation [154]. AMPK has been proved to be closely related to the occurrence and progression of tumors, such as lung, liver and pancreatic cancer [155-157]. A-kinase anchoring protein 1 (AKAP1) can be phosphorylated by AMPK in mitochondria. As a scaffold protein of protein kinase A (PKA), AKAP1 can promote oxidative phosphorylation and mitochondrial fusion by promoting the phosphorylation of dynamin-related protein 1 (DRP1) and mitochondria fusion factor [153, 158].
Recently, Zhang, et al. reported that RIA, a type I regulatory subunit of PKA, can undergo phase separation and form liquid-like biomolecular aggregates enriched in cAMP and PKA, which is essential for cAMP compartmentation. They further showed that RIA phase separation can be effectively inhibited by PKA fusion oncoprotein related to atypical liver cancer, which can induce abnormal cAMP signaling. The destruction of RIA-LLPs in normal cells can promote cell transformation and induce cell proliferation. Their work suggested phase separated as an important assembler of signaling condensates and highlights the pathological significance of this dynamic structural disorder [111].
LLPS in mTOR signaling
Mechanistic target of rapamycin (mTOR) is usually assembled into several complexes, such as mTOR complex 1/2 (mTORC1/2). As a protein kinase, mTOR participates in the regulation of cell survival, growth, immunity and metabolism [159]. A variety of diseases, including tumors, are related to mTOR signaling deregulation [160]. mTORC1 is sensitive to rapamycin and contains mTOR, mLST8 and RAPTOR. mTORC2 is not sensitive to rapamycin and contains mTOR, MAPKAP1, mLST8 and RICTOR[161]. Growth factors and nutrition can activate mTORC1. Different from mTORC1, the activation of mTORC2 only needs growth factor signaling, but its specific molecular mechanism is not completely clear [9]. mTORC1 mediates the expression or phosphorylation of eIF4E, S6K1, 4E-BP1, lipin1, ULK1, TFEB, ATF4, HIF1α, etc., and regulates the nucleotide, lipid and protein synthesis, thereby controlling cell proliferation, growth, metabolism and autophagy. mTORC2 controls the phosphorylation of SGK, PKC and Akt, etc., thus regulating cell apoptosis growth, migration and metabolism[9, 160, 162].
Recently, Zhang, et al. showed that heat stress can promote mTORC1 mediated PGL-1/3 phosphorylation and LLPS of PGL-1/-3 to form PGL particles resistant to autophagy degradation. Moreover, the accumulation of PGL phase separation particles is an adaptive response to thermal stimulation to maintain embryo survival. They found that mTORC1-regulated phase separation of PGL-1/-3 acts as a switching pressure sensor, coupling LLPS to autophagic degradation and stress adaptation [89]. In addition, Schilling, et al. determined that mTOR is a key regulator of survival motor neuron (SMN) phase separation condensation in Cajal body through siRNA-based system. Proteomic analysis revealed that there was TOR dependent phosphorylation in the subunits of SMN complex. They also demonstrated that the ability to condense in Cajal bodies by phase separation can be controlled by phosphorylation of serine 49 and 63 of SMN. Their findings link cellular energy with SMN complex phase separation condensation and UsnRNP biogenesis, and emphasize the regulation of TOR signaling as a reasonable concept for the treatment of SMN-related diseases [163]. These results showed that LLPS plays a key role in the mTOR pathway, but further evidence is needed to determine whether phase separation regulates tumor progression by activating the mTOR signaling pathway.
LLPS in autophagy
Autophagy is an intracellular protective mechanism that can transfer damaged cellular substances to lysosomes for degradation, provide molecular precursors and energy, and allow the basic turnover of cellular components [164]. Autophagy dysregulation are associated with a variety of diseases. For example, in cancer, autophagy can both inhibit tumor initiation and promote cancer progression [165]. Induction of autophagy can be triggered by several intracellular and extracellular stimuli, such as nutrient starvation and serum starvation, oxidative stress and eliminated proteins aggregates, and inhibitors of TOR, e.g., rapamycin [166]. The autophagy process consists of four key steps: initiation, nucleation, maturation and degradation, each of which involves many key proteins. Such as, the autophagy related gene 13 (ATG13), ATG101 Unc-51-like kinase 1 (ULK1) and FIP200 play a role in the initiation step. VPS15, Beclin-1, autophagy and beclin 1 regulator 1 (AMBRA-1), ATG14L and VPS34 are involved in the nucleation step. ATG7, ATG10, ATG5, ATG12, ATG3, LC3 (ATG8), lipid phosphatidylethanolamine (PE) 14, phosphoinositol 3-phosphate and ATG4 are involved in the maturation step. SQSTM1 (p62), neighbor of BRCA1 (NBR1), multiprotein HOPS complex, syntaxin 17 and EPG5 are involved in the degradation step [167].
Role of LLPS in autophagy. (A). p62 interaction with NBR1 proteins and bind to ubiquitin and the polyubiquitin chains of autophagy receptor OPTN to form autophagy receptor condensates; (B). ULK1 complex contains FIP200, ATG13, ULK1 and ATG101. ATG13 interact with FIP200 by the IDR domains multivalent interactions and thus drive LLPS of the ULK1 complex to recruit downstream autophagy proteins for autophagosome formation.
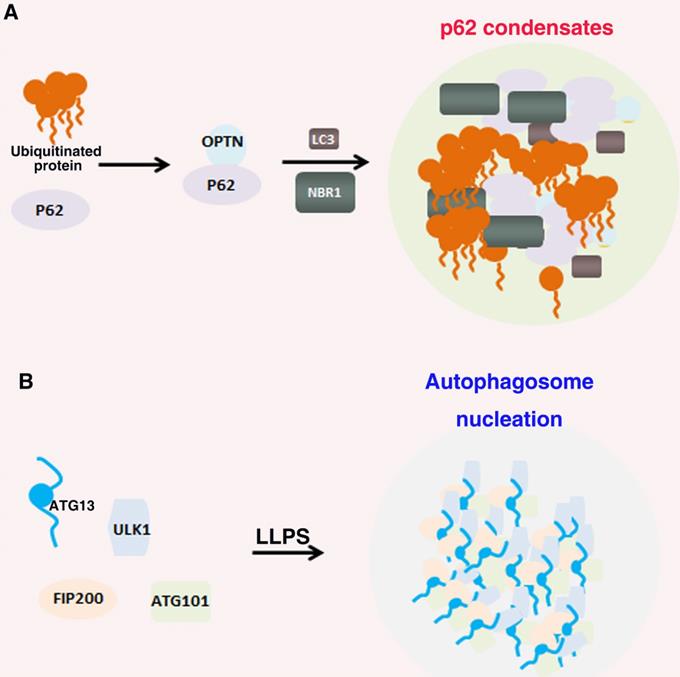
Many biomolecules of autophagy undergo LLPS that regulate many cellular functions (Figure 4)[168]. P62 can form phase separation droplets with liquid properties, such as fusion and fission, micron sized spheres, and recovery rapidly after photobleaching in vivo. In the mechanism, the interaction ubiquitin with p62, p62 polymerization and the multivalent state of ubiquitin chain can significantly promote the LLPS of p62. Furthermore, post translational modifications such as phosphorylation can also regulate the phase separation of p62 [90, 169]. In addition, Agudo-Canalejo, et al. also examined how phase separation condensates containing p62 protein in cellular were isolated by autophagosomes, and proved that autophagosome-like vesicles were formed on the surface of protein-free droplets by partial wetting in vitro [170]. The preautophagosome structure (PAS) is a LLPS condensates containing Atg proteins. The Atg1 complex initiated by autophagy forms droplets through LLPS, and phosphorylation or point mutation can inhibit LLPS and further destroy the formation of PAS [171]. IPMK dysregulation can enhance autophagy activity by activating TFEB, which can form a condensate driven by phase separation with dynamic characteristics. Nuclear TFEB phase separation condensates can participate in the transcription of downstream target genes by interacting with transcriptional condensate MED1 [172]. Wilfling, et al, reported that a selective autophagy pathway is a LLPS condensates formed by endocytic proteins. They found that endocytic protein Ede1 binds Atg8 and mediates LLPS into condensates [173]. These results suggest that the LLPS play a critical, active role in autophagy signaling pathway.
LLPS in immunity
In recent years, a large number of studies are looking for the mechanism of tumor immunity. More and more drugs for immune checkpoint therapy have been applied in a variety of cancers [174]. Cancer immune therapy has shown remarkable benefits in the treatment of many tumors [175]. The innate and adaptive immune system are closely related to tumor development and induction of anti-tumor immune responses [176]. Recently, LLPS has played an increasingly important role in immunology, which also provides a new direction for a deeper understanding of immune response (Figure 5)[15, 177].
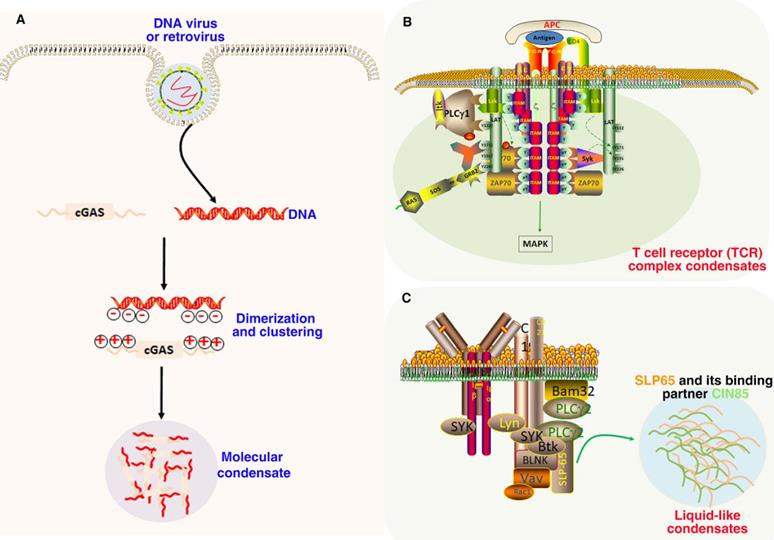
The main function of cGAS-STING signaling pathway is to monitor exogenous DNA and activate innate immune response, such as interferon response after pathogen infection. It is mainly composed of the cyclic GMP-AMP receptor stimulator of interferon genes (STING) and the second messenger cyclic GMP-AMP (cGAS) [178]. The antitumor immune response can be significantly promoted by the activation of cGAS-STING signaling pathway to produce type I interferon [179]. The interaction between DNA and cGAS will strongly induce the formation of phase separation droplets. The LLPS of cGAS-DNA is driven by the increasing of DNA binding valence at the N-terminal of positively charged and disordered cGAS. These findings suggest that DNA promotes the phase separation of cGAS and activates innate immune signaling [180]. Moreover, LLPS of cGAS can not only enhance cytoplasmic DNA sensing, but also inhibit TREX1-regulated DNA degradation. The results revealed a new molecular mechanism, that is, cGAS-DNA activates innate immune response and balances cytoplasmic DNA degradation by phase separation [181]. Mutant tumor suppressor can regulate cGAS-STING pathway by phase separation, and reveal the function and pathogenesis of NF2-associated tumors by controlling antitumor immunity [182]. In addition, in DNA virus infected cells, STING can undergo phase separation to form a biological condensate with an organized membrane structure. The 2'3'-cGAMP can induce the formation of STING phase separation, so as to separate STING-TBK1 from IRF3 and prevent excessive activation of innate immunity [183].
The role of phase separation has also been reported in B cell receptor (BCR) and T cell receptor (TCR) pathways [15]. Once TCR is activated, downstream signaling proteins spontaneously aggregate into phase separated clusters, thereby promoting signal output in Jurkat T cells and in vitro. These results suggest that phase separation is closely related to the reconstruction of T cell signal pathway and the promotion of specific biochemical and signal transduction reactions [112]. In the study of Huang, et al., authors established a foundation for the dynamic proofreading of receptor-mediated Ras activation. They further demonstrated that this kinetic proofreading was modulated by the LAT (linker for activation of T cells)-Grb2-SOS phosphotyrosine-driven phase transition at the membrane [184]. In BCR pathways, cluster BCR is located in the ordered phase-like region, which can sort the key regulatory factors of BCR activation, and a minimum prediction model is proposed, in which cluster receptors stably expand the ordered domain by using super-resolution microscope, resulting in their collective activation [185]. In addition, Effective B cell activation requires the LLPS of CIN85, SLP65 and lipid vesicles into phase separation through the vesicle binding of SLP65 and the hybrid interaction between the proline rich motifs (PRMs) of SLP65 and the SH3 domains of CIN85. The results suggested that LLPS, driven by the transient interaction between vesicle and scaffold protein, is a cellular mechanism of organize signal transducers and aggregation [186]. In consideration of the important role of LLPS in immune signal activation, it may also provide a new perspective for immunotherapy of tumors.
Role of LLPS in immune signaling. (A). Double-stranded DNA binding with cGAS prominently promoted their phase separation and the formation of condensates. In these condensates, cGAS is highly concentrated, which further promotes its catalytic activity by changing the multivalence interaction between cGAS and DNA. (B). The TCR complex is phosphorylated by LCK on ITAM domain, which further recruits the kinase ZAP70. The transmembrane protein LAT is then phosphorylated by ZAP70 and drives LLPS through multivalent interactions with GRB2 and SOS1 for MAPK signaling. (C). SLP65 and its binding partner CIN85 form LLPS condensates in the cytosol of B cells through multivalent interactions between the SLP65 and CIN85.
LLPS in metabolism
Metabolic reprogramming can be widely observed in the occurrence and development of tumors, which endows tumor cells with the ability of malignant proliferation. Even under aerobic conditions, tumor cells like to perform glycolysis to obtain energy and metabolites. This phenomenon was first discovered by Otto Warburg and is now known as “Warburg effect” or “aerobic glycolysis” [187]. Rapid proliferation of cancer cells has been shown to require the support of metabolic reprogramming. The increase of metabolites produced by aerobic glycolysis, such as lactic acid, is related to promoting cancer cell proliferation and metastasis. Therefore, increasing efforts have been invested in trying to develop new therapeutic drugs that target cancer metabolism [188]. Glycolysis plays a central role in metabolism, providing energy as well as carbon feedstocks for anabolic pathways Glycolysis can provide energy and carbon materials for anabolic pathway, and play a core role in metabolism. Recently, under specific stress conditions, some glycolytic enzymes were found to be involved in the formation of phase separation condensates [189]. The glycolytic bodies formed by LLPS are a new RNP particle. The RNA substrates of the glycolytic bodies reside in glycolysis machinery, and the RNA plays key roles in glycolytic body biogenesis and maintenance [190]. Glucose consumption usually increases in cancer cells to support cancer cell proliferation. It was found that Saccharomyces cerevisiae also had “glycolysis” or “G body”. In hypoxia environment, Saccharomyces cerevisiae can concentrate glycolytic enzymes to form a non-membrane bound particle, namely “G body”. Snf1p, a homolog of AMP-activated protein kinase, is necessary for G body formation. The formation of G-bodies can affect cell division by affecting the level of glycolysis [191]. A recent study showed that the accumulated glycogen undergoes LLPS, leading to the assembly of Laforin-Mst1/2 complex, thereby isolating Hippo kinase Mst1/2 in glycogen condensates to reduce its inhibition of YAP. Moreover, deficiency of G6PC or PYGL, glycogenolysis enzymes in both human and mice results in glycogen storage diseases along with liver enlargement and tumorigenesis in a YAP-dependent manner. In addition, in humans and mice, the loss of function of PYGL or G6PC, glycogenolysis enzymes will lead to glycogen storage impairment, resulting in the promotion of tumorigenesis in a YAP-dependent manner [192]. These studies suggest that metabolism can regulate tumor progression through phase separation.
LLPS in tumor virus-associated proteins
Oncogenic viruses can affect various cellular events and lead to the occurrence of human malignant tumors, which is also one of the most important reasons for the occurrence and development of tumors [12]. Approximately 20% of all human oncogenesis is caused by cancer-causing viruses. Oncogenic viruses can cause about 20% of human oncogenesis. Viral infection can cause uncontrolled proliferation, chronic inflammation, and the expression of some key regulatory proteins [13]. oncogenic viruses, such as HBV, HPV, EBV, HCV, HTLV-1 and KSHV are associated with approximately 10%-18% of human tumor worldwide [12]. The viral protein LLPS participates in a series of regulatory steps in the viral replication and lytic cycle (Figure 6). A key function of phase separation driven by viral-encode proteins is the formation of “viral factories” or “viral inclusions”. The formation of LLPS of some viral proteins may not be related to virus replication and assembly, but interferes with the function of the host cell. This interference may depend on the interaction with cellular proteins or changes in host gene transcription, that is, “LLPS mediated host cell function interference” [193-197].
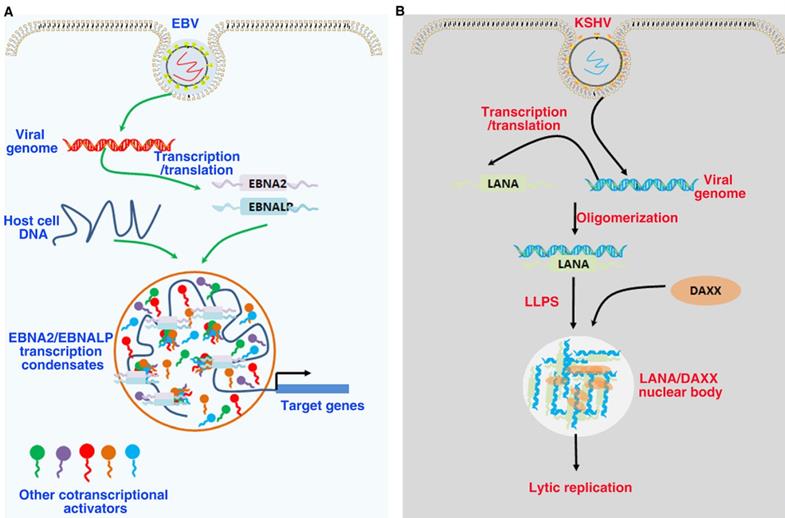
Role of LLPS in tumor virus-associated proteins. (A). EBNA2 and EBNALP recruit other coactivators and transcription factors forming phase-separated condensates at enhancer sites to drive gene activation. These are driven in part by the interactions of IDRs. (B). LANA-associated nuclear bodies structures self-assembly through LLPS to build dynamic structures. DAXX is a component of the latent phase LANA-associated nuclear bodies, and the low complexity, multivalent N-terminal domain interactions driving LLPS.
LLPS in EBV
EBV is the first oncogenic DNA virus found to continuously infect humans and some other primates. As a class of I carcinogens, EBV is associated with a variety of human malignant tumors, such as epithelial and lymphoid tumors, including gastric cancer, nasopharyngeal carcinoma, Burkitt's lymphoma, Hodgkin's lymphoma and extranodal NK/T cell lymphoma[198]. EBV mainly infects the host in the form of latent infection. EBV has three types of latent infection, of which type III expresses all latent infection genes, such as latent membrane proteins LMP1/2, non-coding EBV encoded RNA (EBER1/2), six Epstein-Barr nuclear antigens (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C and EBNA LP), and viral microRNA (miRNA) [199]. EBNA2 and its coactivator EBNALP are the earliest expressed transcription factors after EBV infects B cells. The co-expression of EBNALP and EBNA2 can regulate the expression of specific genes to drive quiescent B cells into the cell cycle, so as to promote the growth and transformation of B cells [200]. EBNA2 and EBNALP can be enriched on super enhancers such as MYC and RUNX3 to form phase separated condensates. Destroying the phase separation of EBNA2 and EBNALP with 1,6 hexanediol can inhibit the expression of their downstream target genes. IDRs and proline specific residues of EBNA2 and EBNALP contribute to the formation phase separation droplet [201]. In addition, EBNA2 can regulate cell alternative splicing events by interacting with components of the splicing mechanism. When the phase separation of EBNA2 is destroyed with 1,6 hexanediol, its ability to regulate splicing can be effectively inhibited [202]. These findings suggest that EBNA2 regulates the transcription of downstream genes as a transcription factor and regulates the splicing of downstream genes as a splicing factor both depend on its phase separation properties. Since 1,6 hexanediol has a wide range of properties of destroying phase separation condensates, further studies are needed to specifically destroy the phase separation ability of EBNA2 by mutating proline residues of EBNA2. These findings provide a basis for understanding the mechanism of phase separation in EBV-host cell interaction and involved in controlling target gene expression. It also provides a new idea for the treatment of EBV-related cancers.
LLPS in KSHV
KSHV belongs to γ-Herpesvirus, a common DNA virus, has been considered to be related to a variety of human malignant tumors, including Kaposi's sarcoma and lymphoma [203]. Immunohistochemistry showed that latency-associated nuclear antigen (LANA) was expressed in almost all KSHV-related tumors, which showed that LANA could be used for diagnosis in KSHV-infected tumors [204]. LANA can recruit host machinery into the viral genome, thus playing a key role in the latency of KSHV [205]. Destroying the phase separation of LANA can change the chromosome conformation of KSHV. During KSHV lytic reactivation to form LANA-related replication compartments, LANA nuclear bodies undergo major morphological transitions. These findings suggest the LANA nucleosome is a dynamic molecular condensates dependent on phase separation, and undergo morphological changes corresponding to different modes of virus replication [206].
LLPS in HPV
Human papillomavirus (HPV) may cause HPV-related tumor, which is mainly transmitted through sexual behavior [207]. About 5% of cancers worldwide are associated with HPV infection, including all cervical cancers and an increasing number of oropharyngeal cancers [208]. The types 16 and 18 HPV are the most common causes of HPV-associated tumors, with about 70% of precancerous cervical lesions and cervical cancer [207]. The genome of HPV encodes for six nonstructural viral proteins (E1, E2, E4, E5, E6, and E7) from the early region of the viral genome. E1 and E2 proteins are mainly involved in viral DNA replication and early transcriptional regulation. As viral oncogenes, E5, E6 and E7 can promote cell transformation and immortalization [209]. HPV infection can induce the formation of HPV E1/E2 foci or replication foci, which contain the main regulatory factors of viral helicase E1 and E2, and DDR protein. These foci have been reported to form by means of phase separation [206]. In addition, super-enhancers condensate has been reported to be regulated by phase separation [137], formation of super-enhancers condensates have been postulated as a new way of HPV-16 integration [210], highlighting the potential role of phase separation in viral tumorigenesis [211].
LLPS in cancer therapy
With the increasing understanding of the role of LLPS in many biological processes, it is gradually recognized that LLPS can regulate some tumor-associated proteins and their downstream and upstream signal pathways, so as to achieve tumor targeted therapy. Recently, Klein found that antitumor drugs are enriched in specific protein droplets in vitro, which occurs via physicochemical characteristic independent of drug targets. This phenomenon has also been found in cancer cells. The distribution of drugs in tumor cells can affect the activity of drugs. Moreover, changing the properties of phase separation condensate affects the concentrations and activity of the drug. These studies showed that the selective distribution and concentration of drugs in the phase separation condensates are helpful to the pharmacodynamics of drugs, and a further understanding of this phenomenon may be helpful to the research of cancer treatment in the future[212, 213].
Anti-PD-L1/PD-1 therapy has shown good clinical effects in a large number of types of tumors, but drug resistance has also appeared in solid tumors, including primary, adaptive and acquired problems [214-218]. Recently, Yu, et al. reported that YAP undergo nuclear translocation and LLPS after IFN promoted anti-PD-1 treatment of cancer cells. YAP interacts with histone acetyltransferase EP300, transcription factor TEAD4 and mediator1 to form phase separation transcriptional condensates, so as to promote target gene transcription. Destroying the LLPS ability of YAP can inhibit cancer cells growth, enhance immune response and make tumor cells sensitive to anti-PD-1 therapy. The prognosis of tumor patients was negatively correlated with YAP activity. These results indicate that YAP regulates the IFN-γ pro-tumor effect via its LLPS ability and shows that YAP can be used as a predictive biomarker and target of anti-PD-1 combination therapy [215]. These studies suggest that the development of drugs regulating LLPS may be a potential way to treat cancer with abnormal condensates and protein aggregates.
Conclusions
Up to now, more and more cancer-related proteins with phase separation function have been discovered and identified, and their regulation through phase separation may affect their cellular domain, function and the life processes of cancer cells. In this review, we have presented recent findings that phase separation regulates tumor-related signaling pathways and reviewed some tumor virus-associated proteins to regulate the development of virus-associated tumors via phase separation; we have also discussed some possible strategies for treating tumors by targeted phase separation. In recent years, the study of phase separation has provided valuable opportunities for in-depth understanding of the pathophysiological process of organisms and the occurrence and development mechanism of various diseases. However, phase separation is a young biological research direction, and there are still many problems to be solved on how to accurately control phase separation. Currently, studies on the role of phase separation in the regulation of cancer mainly focus on the role of condensates formed by phase separation in cancer cells. However, the specific mechanism of the dynamic characteristics of phase separation in the occurrence and development of tumors is still lacking. Therefore, the relationship between phase separation and cancer still needs further exploration. 1,6 hexanediol is the most important chemical to destroy condensates, which can be used to explore the biological function of condensates. However, there is no specificity for 1.6 hexanediol to destroy the condensates in cells, so more specific methods to destroy phase separation need to be urgently explored. For example, gene editing techniques can investigate the function of protein condensates by specifically disrupting the protein phase separation ability by changing the amino acids necessary for phase separation. At present, many studies have verified whether proteins can undergo phase separation by purifying proteins in vitro. However, due to the complexity of the intracellular environment and molecular regulation mechanism, the simulated conditions in vitro cannot completely replace the intracellular environment. Fortunately, with the progress of optogenetics, more and more phase separation studies have been applied to optogenetics. Compared with previous studies on protein and nucleic acid phase separation mainly by means of in vitro reconstruction, optogenetics can be used to dynamically observe the interaction of proteins in living cells, thus realizing the spatiotemporal control of phase separation in living cells. At present, the main methods to study the phase separation of macromolecules are to label them with fluorescence, and then use ordinary optical microscope to detect whether they form droplets or carry out fluorescent recovery after photobleaching experiments. However, ordinary microscope can only observe the state of fluorescent labeled protein or nucleic acid at a fixed time point, and cannot know the dynamic changes of phase separation condensate. Therefore, monomolecular magnetic resonance, atomic force microscopy, cryo-electron microscopy and other techniques should be more used to study the internal structure and dynamic characteristics of biological macromolecular condensates.
Abbreviations
LLPS: liquid-liquid phase separation
IDRs: intrinsically disordered regions
P-bodies: processing bodies
SGs: stress granules
LCRs: low-complexity regions
RBPs: RNA-binding proteins
PLDs: prion-like domains
PRMs: proline-rich motifs
N-WASP: neural Wiskott-Aldrich syndrome protein
SH3: SRC homology 3
FUS: fused in sarcoma
TDP-43: transactivation response element DNA-binding protein 43
EWSR1: Ewing sarcoma breakpoint region 1
TAF15: TATA-binding protein-associated factor 15
HnRNPA: heterogeneous nuclear ribo-nucleoproteins A
LncRNAs: long noncoding RNAs
m6A: RNA N6-methyladenosine
eRNA: enhancer RNAs
ALS: amyotrophic lateral sclerosis
FTD: frontotemporal dementia
53BP1: p53-binding protein 1
PKA: protein kinase A
SMN: survival motor neuron
ATG13: autophagy related gene 13
PAS: pre-autophagosomal structure
cGAS: cyclic GMP-AMP
TCR: T cell receptor
BCR: B cell receptor
LAT: linker for activation of T cells
Funding
This work was supported in part by grants from the following sources: the National Natural Science Foundation of China (82173142, 81972636, 81872281, 81772842), the Natural Science Foundation of Hunan Province (2020JJ5336, 2019JJ40175, 2019JJ40183), the Research Project of Health Commission of Hunan Province (202203034978, 202109031837, 20201020, C2019066), Hunan Provincial Science and Technology Department (2020TP1018), Changsha Science and Technology Project (kh2201054), Ascend Foundation of National cancer center (NCC201909B06, NCC2018b68), and supported by Hunan Cancer Hospital Climb Plan (ZX2020001-3, YF2020002).
Author contributions
Qiu Peng, Shiming Tan, Longzheng Xia, Nayiyuan Wu, Linda Oyang, Yanyan Tang, Min Su, Xia Luo, Ying Wang, Xiaowu Sheng collected the related paper and drafted the manuscript. Yujuan Zhou and Qianjin Liao revised and finalized the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Darwiche N. Epigenetic mechanisms and the hallmarks of cancer: an intimate affair. Am J Cancer Res. 2020;10:1954-78
2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
3. Wang E, Aifantis I. RNA Splicing and Cancer. Trends Cancer. 2020;6:631-44
4. Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer discovery. 2015;5:1024-39
5. Prior IA, Hood FE, Hartley JL. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020;80:2969-74
6. Duffy MJ, Synnott NC, Crown J. Mutant p53 as a target for cancer treatment. Eur J Cancer. 2017;83:258-65
7. Dang CV. MYC on the path to cancer. Cell. 2012;149:22-35
8. Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160-74
9. Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer. 2018;18:744-57
10. Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246-57
11. Bugter JM, Fenderico N, Maurice MM. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat Rev Cancer. 2021;21:5-21
12. Mirzaei H, Ghorbani S, Khanizadeh S, Namdari H, Faghihloo E, Akbari A. Histone deacetylases in virus-associated cancers. Rev Med Virol. 2020;30:e2085
13. Akram N, Imran M, Noreen M, Ahmed F, Atif M, Fatima Z. et al. Oncogenic Role of Tumor Viruses in Humans. Viral Immunol. 2017;30:20-7
14. Gaglia MM, Munger K. More than just oncogenes: mechanisms of tumorigenesis by human viruses. Curr Opin Virol. 2018;32:48-59
15. Xiao Q, McAtee CK, Su X. Phase separation in immune signalling. Nat Rev Immunol. 2021
16. Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol. 2021;22:183-95
17. Langdon EM, Gladfelter AS. A New Lens for RNA Localization: Liquid-Liquid Phase Separation. Annu Rev Microbiol. 2018;72:255-71
18. Alberti S. Phase separation in biology. Curr Biol. 2017;27:R1097-R102
19. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285-98
20. Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. Elife. 2016 5
21. Wang M, Tao X, Jacob MD, Bennett CA, Ho JJD, Gonzalgo ML. et al. Stress-Induced Low Complexity RNA Activates Physiological Amyloidogenesis. Cell Rep. 2018;24:1713-21 e4
22. Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z. et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun. 2019;10:2006
23. Shimobayashi SF, Ronceray P, Sanders DW, Haataja MP, Brangwynne CP. Nucleation landscape of biomolecular condensates. Nature. 2021;599:503-6
24. Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22:165-82
25. Frottin F, Schueder F, Tiwary S, Gupta R, Korner R, Schlichthaerle T. et al. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342-7
26. Liu S, Wang T, Shi Y, Bai L, Wang S, Guo D. et al. USP42 drives nuclear speckle mRNA splicing via directing dynamic phase separation to promote tumorigenesis. Cell Death Differ. 2021;28:2482-98
27. Liao SE, Regev O. Splicing at the phase-separated nuclear speckle interface: a model. Nucleic Acids Res. 2021;49:636-45
28. Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM. et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543-8
29. Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD. et al. Properties of Stress Granule and P-Body Proteomes. Mol Cell. 2019;76:286-94
30. Uversky VN. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18-30
31. Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R. et al. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell. 2016;63:72-85
32. Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr Opin Struct Biol. 2021;67:41-50
33. van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK. et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589-631
34. Piovesan D, Necci M, Escobedo N, Monzon AM, Hatos A, Micetic I. et al. MobiDB: intrinsically disordered proteins in 2021. Nucleic Acids Res. 2021;49:D361-D7
35. Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A. et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015;88:678-90
36. Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY. et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066-77
37. Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918-21
38. Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG. et al. RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron. 2019;102:321-38 e8
39. Gasset-Rosa F, Lu S, Yu H, Chen C, Melamed Z, Guo L. et al. Cytoplasmic TDP-43 De-mixing Independent of Stress Granules Drives Inhibition of Nuclear Import, Loss of Nuclear TDP-43, and Cell Death. Neuron. 2019;102:339-57 e7
40. Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208-19
41. Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936-47
42. Zhang X, Vigers M, McCarty J, Rauch JN, Fredrickson GH, Wilson MZ. et al. The proline-rich domain promotes Tau liquid-liquid phase separation in cells. J Cell Biol. 2020 219
43. Zhang X, Lin Y, Eschmann NA, Zhou H, Rauch JN, Hernandez I. et al. RNA stores tau reversibly in complex coacervates. PLoS Biol. 2017;15:e2002183
44. Zhang H, Ji X, Li P, Liu C, Lou J, Wang Z. et al. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci China Life Sci. 2020;63:953-85
45. Taniue K, Akimitsu N. Aberrant phase separation and cancer. FEBS J. 2021
46. Jiang S, Fagman JB, Chen C, Alberti S, Liu B. Protein phase separation and its role in tumorigenesis. Elife. 2020 9
47. Boija A, Klein IA, Young RA. Biomolecular Condensates and Cancer. Cancer cell. 2021;39:174-92
48. Alberti S, Dormann D. Liquid-Liquid Phase Separation in Disease. Annu Rev Genet. 2019;53:171-94
49. Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH. et al. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol Cell. 2018;70:1038-53 e7
50. Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK. et al. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019;33:799-813
51. Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R. et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38:e101379
52. de The H, Pandolfi PP, Chen Z. Acute Promyelocytic Leukemia: A Paradigm for Oncoprotein-Targeted Cure. Cancer cell. 2017;32:552-60
53. Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N. et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol. 2019;21:1578-89
54. Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N. et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol Cell. 2018;72:19-36 e8
55. Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017 357
56. Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39-58
57. Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R. et al. Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Mol Cell. 2019;76:646-59 e6
58. Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L. et al. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell. 2019;179:470-84 e21
59. Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P. et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell. 2018;175:1481-91 e13
60. Milovanovic D, Wu Y, Bian X, De Camilli P. A liquid phase of synapsin and lipid vesicles. Science. 2018;361:604-7
61. Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334-9
62. Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729-32
63. Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P. et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018 7
64. Laflamme G, Mekhail K. Biomolecular condensates as arbiters of biochemical reactions inside the nucleus. Commun Biol. 2020;3:773
65. Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 2019;176:419-34
66. Weng J, Wang W. Dynamic multivalent interactions of intrinsically disordered proteins. Curr Opin Struct Biol. 2020;62:9-13
67. Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336-40
68. Dao TP, Kolaitis RM, Kim HJ, O'Donovan K, Martyniak B, Colicino E. et al. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol Cell. 2018;69:965-78 e6
69. Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem. 2017;292:19110-20
70. Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553-84
71. Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18-29
72. Wang Z, Zhang H. Phase Separation, Transition, and Autophagic Degradation of Proteins in Development and Pathogenesis. Trends Cell Biol. 2019;29:417-27
73. Hayashi Y, Ford LK, Fioriti L, McGurk L, Zhang M. Liquid-Liquid Phase Separation in Physiology and Pathophysiology of the Nervous System. J Neurosci. 2021;41:834-44
74. Lu J, Qian J, Xu Z, Yin S, Zhou L, Zheng S. et al. Emerging Roles of Liquid-Liquid Phase Separation in Cancer: From Protein Aggregation to Immune-Associated Signaling. Front Cell Dev Biol. 2021;9:631486
75. Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R. et al. Compositional Control of Phase-Separated Cellular Bodies. Cell. 2016;166:651-63
76. Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M. et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell. 2018;174:688-99 e16
77. Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC. et al. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell. 2018;173:677-92 e20
78. Duan Y, Du A, Gu J, Duan G, Wang C, Gui X. et al. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 2019;29:233-47
79. Chakravarty AK, Jarosz DF. More than Just a Phase: Prions at the Crossroads of Epigenetic Inheritance and Evolutionary Change. J Mol Biol. 2018;430:4607-18
80. Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem J. 2017;474:1417-38
81. Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA. et al. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220-30
82. Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ. et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871-84
83. Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803-8
84. Asamitsu S, Shioda N. Potential roles of G-quadruplex structures in RNA granules for physiological and pathological phase separation. J Biochem. 2021;169:527-33
85. Quiroz FG, Fiore VF, Levorse J, Polak L, Wong E, Pasolli HA. et al. Liquid-liquid phase separation drives skin barrier formation. Science. 2020 367
86. Guilhas B, Walter JC, Rech J, David G, Walliser NO, Palmeri J. et al. ATP-Driven Separation of Liquid Phase Condensates in Bacteria. Mol Cell. 2020;79:293-303 e4
87. Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241-5
88. Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487-98
89. Zhang G, Wang Z, Du Z, Zhang H. mTOR Regulates Phase Separation of PGL Granules to Modulate Their Autophagic Degradation. Cell. 2018;174:1492-506 e22
90. Sun D, Wu R, Zheng J, Li P, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405-15
91. Qu W, Wang Z, Zhang H. Phase separation of the C. elegans Polycomb protein SOP-2 is modulated by RNA and sumoylation. Protein Cell. 2020;11:202-7
92. Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M. et al. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell. 2018;173:706-19 e13
93. Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH. et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951-67
94. Hofweber M, Dormann D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem. 2019;294:7137-50
95. Saito M, Hess D, Eglinger J, Fritsch AW, Kreysing M, Weinert BT. et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat Chem Biol. 2019;15:51-61
96. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G. et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121
97. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF. et al. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424-8
98. Wang J, Wang L, Diao J, Shi YG, Shi Y, Ma H. et al. Binding to m(6)A RNA promotes YTHDF2-mediated phase separation. Protein Cell. 2020;11:304-7
99. Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT. et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81:3368-85 e9
100. Li J, Chen K, Dong X, Xu Y, Sun Q, Wang H. et al. YTHDF1 promotes mRNA degradation via YTHDF1-AGO2 interaction and phase separation. Cell Prolif. 2021: e13157.
101. Zhang Y, Yang M, Duncan S, Yang X, Abdelhamid MAS, Huang L. et al. G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res. 2019;47:11746-54
102. Liu X, Xiong Y, Zhang C, Lai R, Liu H, Peng R. et al. G-Quadruplex-Induced Liquid-Liquid Phase Separation in Biomimetic Protocells. J Am Chem Soc. 2021;143:11036-43
103. Ishiguro A, Lu J, Ozawa D, Nagai Y, Ishihama A. ALS-linked FUS mutations dysregulate G-quadruplex-dependent liquid-liquid phase separation and liquid-to-solid transition. J Biol Chem. 2021;297:101284
104. Ryan VH, Fawzi NL. Physiological, Pathological, and Targetable Membraneless Organelles in Neurons. Trends Neurosci. 2019;42:693-708
105. Nedelsky NB, Taylor JP. Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat Rev Neurol. 2019;15:272-86
106. Ryan VH, Perdikari TM, Naik MT, Saueressig CF, Lins J, Dignon GL. et al. Tyrosine phosphorylation regulates hnRNPA2 granule protein partitioning and reduces neurodegeneration. EMBO J. 2021;40:e105001
107. Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL. et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature. 2019;575:390-4
108. Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018 361
109. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123-33
110. Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV. et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018;175:1842-55 e16
111. Zhang JZ, Lu TW, Stolerman LM, Tenner B, Yang JR, Zhang JF. et al. Phase Separation of a PKA Regulatory Subunit Controls cAMP Compartmentation and Oncogenic Signaling. Cell. 2020;182:1531-44 e15
112. Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595-9
113. Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20:471-80
114. Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275-83
115. Stegh AH. Targeting the p53 signaling pathway in cancer therapy - the promises, challenges and perils. Expert Opin Ther Targets. 2012;16:67-83
116. Kamagata K, Kanbayashi S, Honda M, Itoh Y, Takahashi H, Kameda T. et al. Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci Rep. 2020;10:580
117. Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W. et al. The function of PML in p53-dependent apoptosis. Nat Cell Biol. 2000;2:730-6
118. Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105-31
119. Petronilho EC, Pedrote MM, Marques MA, Passos YM, Mota MF, Jakobus B. et al. Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands. Chem Sci. 2021;12:7334-49
120. Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci U S A. 1994;91:6098-102
121. Ghodke I, Remisova M, Furst A, Kilic S, Reina-San-Martin B, Poetsch AR. et al. AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation. Mol Cell. 2021;81:2596-610 e7
122. Lemos C, Schulze L, Weiske J, Meyer H, Braeuer N, Barak N. et al. Identification of Small Molecules that Modulate Mutant p53 Condensation. iScience. 2020;23:101517
123. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461-73
124. Soleas JP, D'Arcangelo E, Huang L, Karoubi G, Nostro MC, McGuigan AP. et al. Assembly of lung progenitors into developmentally-inspired geometry drives differentiation via cellular tension. Biomaterials. 2020;254:120128
125. Salik B, Yi H, Hassan N, Santiappillai N, Vick B, Connerty P. et al. Targeting RSPO3-LGR4 Signaling for Leukemia Stem Cell Eradication in Acute Myeloid Leukemia. Cancer cell. 2020;38:263-78 e6
126. Peng Q, Chen L, Wu W, Wang J, Zheng X, Chen Z. et al. EPH receptor A2 governs a feedback loop that activates Wnt/beta-catenin signaling in gastric cancer. Cell Death Dis. 2018;9:1146
127. Taciak B, Pruszynska I, Kiraga L, Bialasek M, Krol M. Wnt signaling pathway in development and cancer. J Physiol Pharmacol. 2018 69
128. Zeng CM, Chen Z, Fu L. Frizzled Receptors as Potential Therapeutic Targets in Human Cancers. Int J Mol Sci. 2018 19
129. Acebron SP, Niehrs C. beta-Catenin-Independent Roles of Wnt/LRP6 Signaling. Trends Cell Biol. 2016;26:956-67
130. Wiese KE, Nusse R, van Amerongen R. Wnt signalling: conquering complexity. Development. 2018 145
131. Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985-99
132. Zhang Y, Wang X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13:165
133. Shi Q, Kang K, Chen YG. Liquid-liquid phase separation drives the beta-catenin destruction complex formation. Bioessays. 2021;43:e2100138
134. Li TM, Ren J, Husmann D, Coan JP, Gozani O, Chua KF. Multivalent tumor suppressor adenomatous polyposis coli promotes Axin biomolecular condensate formation and efficient beta-catenin degradation. Sci Rep. 2020;10:17425
135. Schwarz-Romond T, Merrifield C, Nichols BJ, Bienz M. The Wnt signalling effector Dishevelled forms dynamic protein assemblies rather than stable associations with cytoplasmic vesicles. J Cell Sci. 2005;118:5269-77
136. Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T. et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781-90
137. Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412-5
138. Zamudio AV, Dall'Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM. et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol Cell. 2019;76:753-66 e6
139. Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19:480-94
140. Park J, Eisenbarth D, Choi W, Kim H, Choi C, Lee D. et al. YAP and AP-1 Cooperate to Initiate Pancreatic Cancer Development from Ductal Cells in Mice. Cancer Res. 2020;80:4768-79
141. Munoz-Galvan S, Felipe-Abrio B, Verdugo-Sivianes EM, Perez M, Jimenez-Garcia MP, Suarez-Martinez E. et al. Downregulation of MYPT1 increases tumor resistance in ovarian cancer by targeting the Hippo pathway and increasing the stemness. Mol Cancer. 2020;19:7
142. Janse van Rensburg HJ, Azad T, Ling M, Hao Y, Snetsinger B, Khanal P. et al. The Hippo Pathway Component TAZ Promotes Immune Evasion in Human Cancer through PD-L1. Cancer Res. 2018;78:1457-70
143. Cheung P, Xiol J, Dill MT, Yuan WC, Panero R, Roper J. et al. Regenerative Reprogramming of the Intestinal Stem Cell State via Hippo Signaling Suppresses Metastatic Colorectal Cancer. Cell Stem Cell. 2020;27:590-604 e9
144. Manning SA, Kroeger B, Harvey KF. The regulation of Yorkie, YAP and TAZ: new insights into the Hippo pathway. Development. 2020 147
145. Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI. et al. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat Cell Biol. 2020;22:453-64
146. Sun X, Ren Z, Cun Y, Zhao C, Huang X, Zhou J. et al. Hippo-YAP signaling controls lineage differentiation of mouse embryonic stem cells through modulating the formation of super-enhancers. Nucleic Acids Res. 2020;48:7182-96
147. Wei Y, Luo H, Yee PP, Zhang L, Liu Z, Zheng H. et al. Paraspeckle Protein NONO Promotes TAZ Phase Separation in the Nucleus to Drive the Oncogenic Transcriptional Program. Adv Sci (Weinh). 2021: e2102653.
148. Li RH, Tian T, Ge QW, He XY, Shi CY, Li JH. et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021;31:1088-105
149. Zhao M, Mishra L, Deng CX. The role of TGF-beta/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14:111-23
150. Syed V. TGF-beta Signaling in Cancer. J Cell Biochem. 2016;117:1279-87
151. Colak S, Ten Dijke P. Targeting TGF-beta Signaling in Cancer. Trends Cancer. 2017;3:56-71
152. Esposito M, Fang C, Cook KC, Park N, Wei Y, Spadazzi C. et al. TGF-beta-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat Cell Biol. 2021;23:257-67
153. Yuan J, Dong X, Yap J, Hu J. The MAPK and AMPK signalings: interplay and implication in targeted cancer therapy. J Hematol Oncol. 2020;13:113
154. Wang Z, Wang N, Liu P, Xie X. AMPK and Cancer. Exp Suppl. 2016;107:203-26
155. Gao L, Xu Z, Huang Z, Tang Y, Yang D, Huang J. et al. CPI-613 rewires lipid metabolism to enhance pancreatic cancer apoptosis via the AMPK-ACC signaling. J Exp Clin Cancer Res. 2020;39:73
156. Fang G, Zhang P, Liu J, Zhang X, Zhu X, Li R. et al. Inhibition of GSK-3beta activity suppresses HCC malignant phenotype by inhibiting glycolysis via activating AMPK/mTOR signaling. Cancer Lett. 2019;463:11-26
157. Ashrafizadeh M, Mirzaei S, Hushmandi K, Rahmanian V, Zabolian A, Raei M. et al. Therapeutic potential of AMPK signaling targeting in lung cancer: Advances, challenges and future prospects. Life Sci. 2021;278:119649
158. Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ. et al. Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise-Regulated Kinases and AMPK Substrates. Cell Metab. 2015;22:922-35
159. Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12:71
160. Huang S. mTOR Signaling in Metabolism and Cancer. Cells. 2020 9
161. Populo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci. 2012;13:1886-918
162. Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183-203
163. Schilling M, Prusty AB, Boysen B, Oppermann FS, Riedel YL, Husedzinovic A. et al. TOR signaling regulates liquid phase separation of the SMN complex governing snRNP biogenesis. Cell Rep. 2021;35:109277
164. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528-42
165. Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307-18
166. Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12
167. Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer discovery. 2019;9:1167-81
168. Noda NN, Wang Z, Zhang H. Liquid-liquid phase separation in autophagy. J Cell Biol. 2020 219
169. Danieli A, Martens S. p62-mediated phase separation at the intersection of the ubiquitin-proteasome system and autophagy. J Cell Sci. 2018 131
170. Agudo-Canalejo J, Schultz SW, Chino H, Migliano SM, Saito C, Koyama-Honda I. et al. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. 2021;591:142-6
171. Fujioka Y, Alam JM, Noshiro D, Mouri K, Ando T, Okada Y. et al. Phase separation organizes the site of autophagosome formation. Nature. 2020;578:301-5
172. Chen D, Wang Z, Zhao YG, Zheng H, Zhao H, Liu N. et al. Inositol Polyphosphate Multikinase Inhibits Liquid-Liquid Phase Separation of TFEB to Negatively Regulate Autophagy Activity. Dev Cell. 2020;55:588-602 e7
173. Wilfling F, Lee CW, Erdmann PS, Zheng Y, Sherpa D, Jentsch S. et al. A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits. Mol Cell. 2020;80:764-78 e7
174. Cortez MA, Anfossi S, Ramapriyan R, Menon H, Atalar SC, Aliru M. et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer. 2019;58:244-53
175. Rogado J, Sanchez-Torres JM, Romero-Laorden N, Ballesteros AI, Pacheco-Barcia V, Ramos-Levi A. et al. Immune-related adverse events predict the therapeutic efficacy of anti-PD-1 antibodies in cancer patients. Eur J Cancer. 2019;109:21-7
176. Candeias SM, Gaipl US. The Immune System in Cancer Prevention, Development and Therapy. Anticancer Agents Med Chem. 2016;16:101-7
177. Xia S, Chen Z, Shen C, Fu TM. Higher-order assemblies in immune signaling: supramolecular complexes and phase separation. Protein Cell. 2021;12:680-94
178. Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. 2020;21:501-21
179. Wang Y, Luo J, Alu A, Han X, Wei Y, Wei X. cGAS-STING pathway in cancer biotherapy. Mol Cancer. 2020;19:136
180. Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704-9
181. Zhou W, Mohr L, Maciejowski J, Kranzusch PJ. cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol Cell. 2021;81:739-55 e7
182. Meng F, Yu Z, Zhang D, Chen S, Guan H, Zhou R. et al. Induced phase separation of mutant NF2 imprisons the cGAS-STING machinery to abrogate antitumor immunity. Mol Cell. 2021;81:4147-64 e7
183. Yu X, Zhang L, Shen J, Zhai Y, Jiang Q, Yi M. et al. The STING phase-separator suppresses innate immune signalling. Nat Cell Biol. 2021;23:330-40
184. Huang WYC, Alvarez S, Kondo Y, Lee YK, Chung JK, Lam HYM. et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science. 2019;363:1098-103
185. Stone MB, Shelby SA, Nunez MF, Wisser K, Veatch SL. Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife. 2017 6
186. Wong LE, Bhatt A, Erdmann PS, Hou Z, Maier J, Pirkuliyeva S. et al. Tripartite phase separation of two signal effectors with vesicles priming B cell responsiveness. Nat Commun. 2020;11:848
187. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73:377-92
188. Peng Q, Zhou Y, Oyang L, Wu N, Tang Y, Su M. et al. Impacts and mechanisms of alternative mRNA splicing in cancer metabolism, immune response, and therapeutics. Mol Ther. 2021
189. Prouteau M, Loewith R. Regulation of Cellular Metabolism through Phase Separation of Enzymes. Biomolecules. 2018 8
190. Fuller GG, Han T, Freeberg MA, Moresco JJ, Ghanbari Niaki A, Roach NP. et al. RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. Elife. 2020 9
191. Jin M, Fuller GG, Han T, Yao Y, Alessi AF, Freeberg MA. et al. Glycolytic Enzymes Coalesce in G Bodies under Hypoxic Stress. Cell Rep. 2017;20:895-908
192. Liu Q, Li J, Zhang W, Xiao C, Zhang S, Nian C. et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell. 2021;184:5559-76 e19
193. Savastano A, Ibanez de Opakua A, Rankovic M, Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun. 2020;11:6041
194. Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudriere-Gesbert C, Gaudin Y. et al. Negri bodies are viral factories with properties of liquid organelles. Nat Commun. 2017;8:58
195. Metrick CM, Koenigsberg AL, Heldwein EE. Conserved Outer Tegument Component UL11 from Herpes Simplex Virus 1 Is an Intrinsically Disordered, RNA-Binding Protein. mBio. 2020 11
196. Guseva S, Milles S, Jensen MR, Schoehn G, Ruigrok RW, Blackledge M. Structure, dynamics and phase separation of measles virus RNA replication machinery. Curr Opin Virol. 2020;41:59-67
197. Brocca S, Grandori R, Longhi S, Uversky V. Liquid-Liquid Phase Separation by Intrinsically Disordered Protein Regions of Viruses: Roles in Viral Life Cycle and Control of Virus-Host Interactions. Int J Mol Sci. 2020 21
198. Cao Y, Xie L, Shi F, Tang M, Li Y, Hu J. et al. Targeting the signaling in Epstein-Barr virus-associated diseases: mechanism, regulation, and clinical study. Signal Transduct Target Ther. 2021;6:15
199. Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer. 2016;16:789-802
200. Sinclair AJ, Palmero I, Peters G, Farrell PJ. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13:3321-8
201. Peng Q, Wang L, Qin Z, Wang J, Zheng X, Wei L. et al. Phase Separation of Epstein-Barr Virus EBNA2 and Its Coactivator EBNALP Controls Gene Expression. J Virol. 2020 94
202. Peng Q, Wang L, Wang J, Liu C, Zheng X, Zhang X. et al. Epstein-Barr virus EBNA2 phase separation regulates cancer-associated alternative RNA splicing patterns. Clin Transl Med. 2021;11:e504
203. Wong EL, Damania B. Linking KSHV to human cancer. Curr Oncol Rep. 2005;7:349-56
204. Mariggio G, Koch S, Schulz TF. Kaposi sarcoma herpesvirus pathogenesis. Philos Trans R Soc Lond B Biol Sci. 2017 372
205. Broussard G, Damania B. Regulation of KSHV Latency and Lytic Reactivation. Viruses. 2020 12
206. Vladimirova O, De Leo A, Deng Z, Wiedmer A, Hayden J, Lieberman PM. Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation. PLoS Pathog. 2021;17:e1009231
207. Szymonowicz KA, Chen J. Biological and clinical aspects of HPV-related cancers. Cancer Biol Med. 2020;17:864-78
208. Berman TA, Schiller JT. Human papillomavirus in cervical cancer and oropharyngeal cancer: One cause, two diseases. Cancer. 2017;123:2219-29
209. Li Y, Xu C. Human Papillomavirus-Related Cancers. Adv Exp Med Biol. 2017;1018:23-34
210. Dooley KE, Warburton A, McBride AA. Tandemly Integrated HPV16 Can Form a Brd4-Dependent Super-Enhancer-Like Element That Drives Transcription of Viral Oncogenes. mBio. 2016 7
211. Lopez N, Camporeale G, Salgueiro M, Borkosky SS, Visentin A, Peralta-Martinez R. et al. Deconstructing virus condensation. PLoS Pathog. 2021;17:e1009926
212. Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall'Agnese A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386-92
213. Howard TP, Roberts CWM. Partitioning of Chemotherapeutics into Nuclear Condensates-Opening the Door to New Approaches for Drug Development. Mol Cell. 2020;79:544-5
214. Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv4
215. Yu M, Peng Z, Qin M, Liu Y, Wang J, Zhang C. et al. Interferon-gamma induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol Cell. 2021;81:1216-30 e9
216. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer cell. 2015;27:450-61
217. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707-23
218. Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L. et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375:1856-67
219. Fulka H, Langerova A. Nucleoli in embryos: a central structural platform for embryonic chromatin remodeling? Chromosome Res. 2019;27:129-40
220. Luo Y, Na Z, Slavoff SA. P-Bodies: Composition, Properties, and Functions. Biochemistry. 2018;57:2424-31
221. Logan MK, McLaurin DM, Hebert MD. Synergistic interactions between Cajal bodies and the miRNA processing machinery. Mol Biol Cell. 2020;31:1561-9
222. Lallemand-Breitenbach V, de The H. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol. 2018;52:154-61
223. Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell. 2017;68:808-20 e5
224. Buckingham M, Liu JL. U bodies respond to nutrient stress in Drosophila. Exp Cell Res. 2011;317:2835-44
225. Li K, Wang Z. Speckles and paraspeckles coordinate to regulate HSV-1 genes transcription. Commun Biol. 2021;4:1207
226. Moser JJ, Chan EK, Fritzler MJ. An SNP in the trinucleotide repeat region of the TNRC6A gene maps to a major TNGW1 autoepitope in patients with autoantibodies to GW182. Adv Exp Med Biol. 2013;768:243-59
227. Sugishita H, Kondo T, Ito S, Nakayama M, Yakushiji-Kaminatsui N, Kawakami E. et al. Variant PCGF1-PRC1 links PRC2 recruitment with differentiation-associated transcriptional inactivation at target genes. Nat Commun. 2021;12:5341
228. Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A. et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol. 2015;210:529-39
Author contact
![]() Corresponding authors: Qianjin Liao or Yujuan Zhou, Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China. Hunan Key Laboratory of Translational Radiation Oncology, 283 Tongzipo Road, Changsha 410013, Hunan, China. Tel: 86-731-88651681; Fax: 86-731-88651999; Email: liaoqianjinorg.cn; or yujany_zhoucom.
Corresponding authors: Qianjin Liao or Yujuan Zhou, Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China. Hunan Key Laboratory of Translational Radiation Oncology, 283 Tongzipo Road, Changsha 410013, Hunan, China. Tel: 86-731-88651681; Fax: 86-731-88651999; Email: liaoqianjinorg.cn; or yujany_zhoucom.

 Global reach, higher impact
Global reach, higher impact