10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(13):5185-5206. doi:10.7150/ijbs.72600 This issue Cite
Review
The role of intestinal stem cell within gut homeostasis: Focusing on its interplay with gut microbiota and the regulating pathways
1. Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China.
2. Cell Therapy & Cell Drugs of Luzhou Key Laboratory, Luzhou 646000, Sichuan, China.
3. South Sichuan Institute of Translational Medicine, Luzhou 646000, Sichuan, China.
4. State Key Laboratory of Quality Research in Chinese Medicine, University of Macau, Macao, China.
5. Department of Oncology, Affiliated Hospital of Southwest Medical University, Luzhou 646000, Sichuan, China.
# These authors contributed equally to the work.
Received 2022-3-4; Accepted 2022-7-29; Published 2022-8-8
Abstract
Intestinal stem cells (ISCs) play an important role in maintaining intestinal homeostasis via promoting a healthy gut barrier. Within the stem cell niche, gut microbiota linking the crosstalk of dietary influence and host response has been identified as a key regulator of ISCs. Emerging insights from recent research reveal that ISC and gut microbiota interplay regulates epithelial self-renewal. This article reviews the recent knowledge on the key role of ISC in their local environment (stem cell niche) associating with gut microbiota and their metabolites as well as the signaling pathways. The current progress of intestinal organoid culture is further summarized. Subsequently, the key challenges and future directions are discussed.
Keywords: Intestinal stem cell, Intestinal barrier, Gut microbiota, Intestinal homeostasis, Signaling pathway
1. Introduction
As a natural defense system, the intestinal luminal surface is covered by just one layer of epithelium. However, that layer is an effective barrier that guards against pathogens and toxins that may be harmful to the host [1-3]. There are several mechanisms that may participate in the disruption of barriers in the gastrointestinal (GI) tract, including inflammation, infection, etc., and lead to dysregulation of intestinal homeostasis. Intestinal homeostasis is maintained by a combination of the GI luminal epithelium, gut microbiota, and intestinal stem cells (ISCs). Recent research has focused on the mechanisms of controlling intestinal homeostasis both in health, injury and recovery conditions. Fortunately, recent technological advancements such as intestinal organoids and organ cultures of the gut have allowed us to gain a deeper understanding of this system.
ISCs in the crypts of the intestines are rare, non-quiescent, undifferentiated cells, which play a crucial role in regulating the homeostasis and regeneration of tissues. Each crypt contains four to six pluripotent stem cells [4, 5]. Nevertheless, a key question is how to coordinate the mechanisms of ISC renewal and differentiation. The microbiota found in the GI tract is made up of various microbial communities which interact with the host at the intestinal ecotone. Gut microbiota influences the intestinal epithelial development by interfering with ISCs through the abundance of commensal bacteria and invasive organisms, as well as through metabolites produced by microorganisms. A balance is assumed to be achieved among ISCs, gut microbiota and other factors within the gut homeostasis.
The aim of this review is thus to review the recent knowledge on the key role of ISC in their local environment (stem cell niche) associating with gut microbiota and their metabolites as well as the signaling pathways. The current development of intestinal organoid culture is further summarized. The key challenges and future directions are further discussed.
2. ISCs in maintaining intestinal barrier
2.1 Intestinal structure and intestinal homeostasis
The GI tract, which has a surface area of approximately 250 cm2, is not only the primary site of nutrient absorption and processing, but also the body's largest endocrine organ [1]. Mucosal surfaces of the GI tract are surrounded by epithelial cells (ECs), creating a natural barrier of intercellular connections that separates the internal and external environments. This barrier selectively absorbs nutrients and electrolytes while simultaneously prevents the passage of potentially harmful substances (antigens, toxins, and microbial by-products) into the body. For the epithelial barrier to remain intact, many defense systems are involved, that the immune system develops normally, and that tolerance to dietary antigens and intestinal microbiota is maintained. This allows the GI tract to function as a robust barrier against potential hazardous infections and toxins [2, 3].
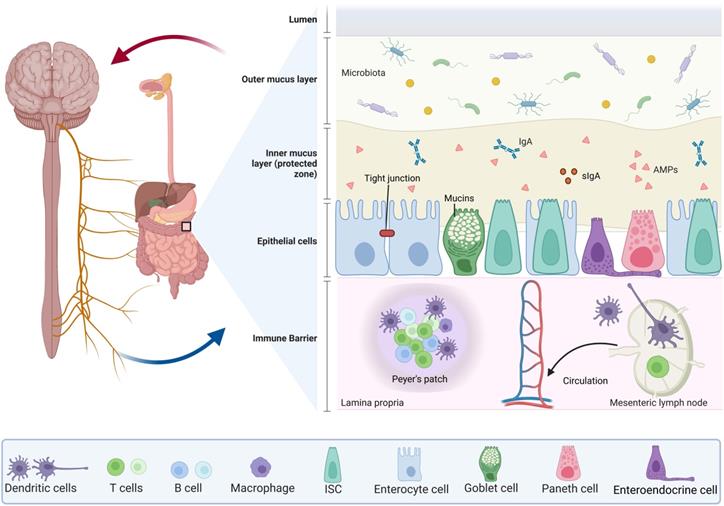
The intestinal barrier is composed of two layers (Figure 1): the outer layer serves as a physical barrier, preventing bacteria from adhering and regulating decidual cell spread into the host tissue; and the internal layer provides a chemical barrier, preventing the spread of bacteria to the host tissue. The intestinal microbiota, the mucus layer, and ECs work together to form the physical barrier against infection. For example, there is competition between intestinal flora and vector pathogens for resources in the GI tract. These colonies of bacteria can process molecules that are essential for mucosal integrity, and regulating immune activity in the deep barrier, among other things. In addition to assisting in the retention of antimicrobial peptide-rich mucus, the mucus layer also helps to prevent mucosal adhesion and subsequent microbial invasion across the epithelium [6, 7]. Furthermore, the mucus layer contains secretory immunoglobulin A (sIgA) produced by a type of plasma cell as well as antimicrobial products produced by the Paneth cells which include phospholipids, negatively charged mucins, and peptides such as trilobal factor family (TFF) peptides and antimicrobial peptides that are active against bacteria, yeast, fungi, viruses, and tumor cells. IgA and other antimicrobial products are essential to maintain gut homeostasis as they are necessary for preventing gut inflammation to occur [8]. In addition, the intestine contains tight junctions (TJs), which are composed of a network of transmembrane protein chains that include intact membrane proteins (claudin family proteins, which are involved in selective ion permeation as the primary mode of transmembrane epithelial transport, and another transmembrane proteins called occludins), binding complex proteins, as well as intracellular signaling molecules [3, 9]. The TJs are controlled and regulated by extracellular signaling molecules. Upon the occurrence of pathogens and commensal microorganisms within the inner layer, TJs act as an immune barrier, organizing immune tolerance and pathogen response [10]. In addition, the gut-associated lymphoid tissue (GALT) is a key immunological system in the gut which is comprised of Peyer's patches, interdigitating lymphocytes, plasma cells and lymphocytes present in the lamina propria, and mesenteric lymph nodes. GALT is divided into two types: organized and diffuse GALT. Organized GALT initiates the immune response to the massive antigen exposure, whereas diffuse GALT is the effector. Diffuse GALT is made up of numerous leukocyte populations that are dispersed on either side of the basement membrane [11].
It is common for intestinal homeostasis to be dysregulated when barrier integrity is disrupted. Maintaining gut homeostasis is achieved through a delicate interplay among ECs, gut microbiome, immune system, as well as ISCs [11-13]. Several findings have shown that ISCs are regulated and influenced by the microenvironment (stem cell niche) derived from specialized epithelial and mesenchymal cells. ISCs are involved in tissue homeostasis and regeneration after injury or inflammation [4, 13, 14]. As expected, intestinal ECs, commensal bacteria, immune system and ISCs work together to maintain the luminal environment in the intestine. Notably, disruption of intestinal homeostasis can result in GI inflammation, such as inflammatory bowel disease (IBD) [8]. The epithelial barrier breach associated with IBD is characterized by an increase in EC death, a decrease in TJ protein expression, and a deficiency in immune response [15], which also contributes to initiation of diseases of other systems.
The components in intestinal structure for maintaining intestinal barrier. The intestinal barrier consists of two layers: the outer layer consists of the intestinal microbiota (including bacteria, viruses, and pathogens), the mucus layer and the ECs, working together to form a physical barrier against infection. The mucus layer contains sIgA produced by Paneth cells and mucins produced by goblet cells, in addition to intercellular TJs. The inner layer provides a chemical barrier to prevent the spread of bacteria into the host tissue. The GALT is an important immune system in the gut and is composed of the Peyer's patches, the interdigitating lymphocytes, plasma cells and lymphocytes presented in the lamina propria, and mesenteric lymph nodes. All of these are closely linked to the blood circulation in the body.
2.2 The role in ISCs in maintaining gut barrier
ISCs in the crypts are undifferentiated and are involved in tissue homeostasis and regeneration after injury or inflammation [4, 13, 14]. ISCs near the crypt base is the source of all putative mitotically developed cells. Back in 2009, Walker et al. pointed out that stem cells that maintain tissue homeostasis were regulated and supported by the surrounding microenvironment, known as the stem cell niche [16]. In 2017, Meran et al. formally introduced the concept of the ISC niche, which includes nearby Paneth cells, neuronal cells, smooth muscle cells and stromal cells, as well as the extracellular matrix together [17]. Moreover, the intestinal epithelium is a site of rapid cell renewal and is highly in need of regulation by the ISCs located at the base of the crypts. The maintenance of ISC activity is influenced by different paracrine ecotropic factors and a number of cellular signals such as Wnt, R-spondin, Notch and bone morphogenetic protein (BMP), as well as by inflammation, gut microbiota and diet. Indeed, following injury, the ISC niche stimulates EC regeneration [18]. When Noggin (a BMP inhibitor) and downstream Wnt signaling targets (e.g. EPhB and C-Myc) are overexpressed in the embryonic intestinal epithelium, exceptional crypts are formed with the assistance of villi that are perpendicular to the crypt-villi axis [19, 20]. For example, each intestinal crypt contains four to six long-lived pluripotent stem cells, which are actively dividing during the day, rather than being quiescent [4, 5].
ISCs replenish themselves by mitosis, dividing into equal stem mobile clones over time. Such division ensures constant stem cell diversification and differentiation. ISCs are required to sustain themselves for an extended period while also provide upward movement to all differentiated types of tissue in question; this is referred to as their "stemness". Stem cells must also be able to provide upward movement to all differentiated mobile phone types of tissue in question. According to Barker et al., the direct progeny of stem cells, known as TA cells (transit-amplifying cells), would unexpectedly dilute any integrated DNA markers used to demonstrate their stemness on remote adult stem cells. A common method of achieving "stemness," therefore, is the long-term storage of DNA labels. When tissue has been severely damaged, stem cells go through mitosis to maintain DNA markers in the tissue. On the contrary, because they are unable to proliferate, DNA labels may continue to be retained by terminally differentiated cells longer than they would in stem cells [4, 14].
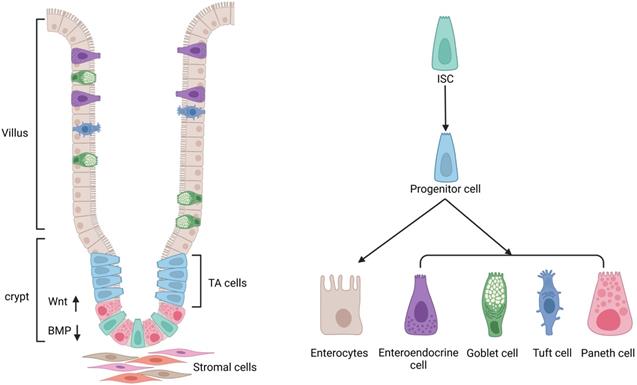
The small intestinal epithelium is made up of one type of absorptive cell and four types of secretory cells (Goblet cells, enteroendocrine cells, tuft cells, and Paneth cells), which are differentiated from stem cells and generated from the endoderm [4, 13, 14, 21] (Figure 2). Most ECs are goblet cells, which account for 10-15 percent of small intestine ECs and 50 percent of colonic ECs. Goblet cells are involved in the production and secretion of mucus, creating a protective barrier for ECs. A recent study showed that ST6GALNAC1 (ST6), a saline acid transferase specifically expressed in goblet cells is essential for mucus integrity and protecting from microbial degradation [22]. It is estimated that enteroendocrine cells account for 1% of the epithelium of the intestine. These cells are dispersed among the mucus as character cells, which are responsible for the production and release of hormones. Tuft cells are a type of secretory cell that has only recently been discovered in the intestines of newborn mice and are thought to be responsible for their secretion [23]. Only tuft cells categorize cyclooxygenase and express the style protein (α-gustducin, Trpm5) in response to vitamins in the intestinal lumen [23]. They are also the only ECs that secrete opioids in response to vitamins [23]. In comparison with other ECs, which appear later and coincide with the appearance of the crypt [13], Paneth cells synthesize and exude antimicrobial peptides into the intestinal lumen. To preserve their capacity to proliferate and self-renew, ISCs rely on the niche cells surround them (Figure 2). When ISCs continually self-renew, they produce progenitor-transporting amplifying cells, and progenitor cells undergo additional mobile divisions before maturation and differentiation. Under normal circumstances, the regeneration of the ECs takes 1-2 weeks in drosophila, whereas in mice it takes only 5-7 days [24, 25].
Since ISCs allow for the continuous replacement of intestinal ECs, they are critical for maintaining the mucosal barrier, which is a mechanism that protects the intestinal barrier from pathogen invasion and bodily damage in pathological or physiological conditions that activate chronic or acute inflammation. Irritable bowel syndrome is a disorder related to the gut, involving abnormal GI motility, abnormal absorption or secretion, and visceral hypersensitivity in the presence of a stressful environment. Actually, these symptoms may also be associated with a low density of enteroendocrine cells, which is caused by a low density of ISCs [21]. Potton et al. had previously discovered that Musashi-1 in mice intestinal tissue was an early marker of stem cells and progenitor cells, and is expressed only in damaged tissue (e.g. tumors) [5]. A recent study demonstrated that deficiency of SETDB1, a histone methyltransferase that mediates the trimethylation of histone H3 at lysine 9, in ISCs led to irreversible disruption of epithelial barrier homeostasis, promoting intestinal inflammation and causing severe IBD [26]. Additionally, Zhang et al. discovered that ISCs controlled the stability in the GI environment through tumor-suppressive autophagy [27].
ISC niche and its differentiation. The regulation of the activity of ISCs is dependent on the stem cell niche, including surrounding stromal cells and signaling molecules. The intestinal epithelium is made up of one type of absorptive cell and four types of secretory cells. Transit-amplifying (TA) cells act as direct descendants of ISCs, including secretory and absorptive progenitors, which can give rise to stem-like cells following stem cell injury. Secretory progenitors are differentiated into Paneth cells, goblet cells, tuft cells, and enteroendocrine cells, while absorptive progenitors are differentiated into enterocytes.
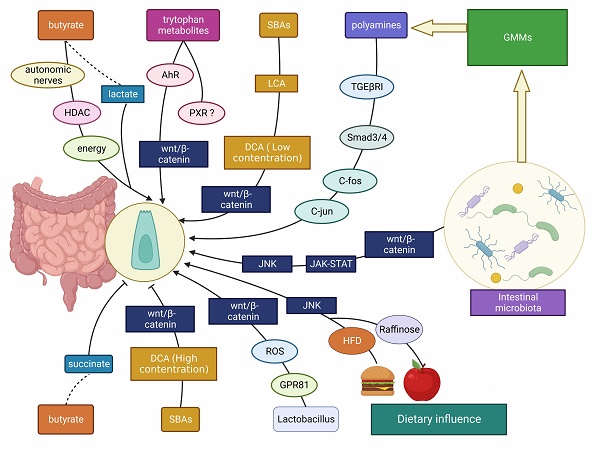
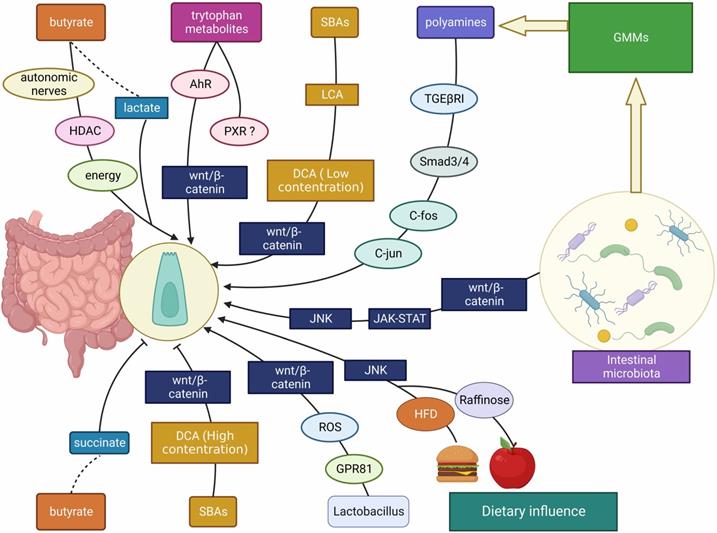
The interplay between gut microbes/ microbial metabolites/ dietary factors and ISCs. The intestinal microbiota, gut microbial metabolites (including SCFAs, SBAs, tryptophan metabolites, etc.) and dietary influences (including HFD, polysaccharides, etc.) affect the development of the intestinal mucosa mainly through Wnt/β-catenin, JNK, JAK-STAT and other signaling molecules to promote/ inhibit ISC proliferation.
3. Interplay of gut microbiota and ISCs
Keeping the intestinal lumen stable can be a complex task, which necessitates interaction among the ECs lining the lumen, ISCs, and gut microbiota. The intestinal microbiota comprises a variety of microbial communities, which include viruses, bacteria, and fungi, which is essential for regulating GI function and immune homeostasis [28, 29].
In healthy state, microbiota variability, as reflected in intestinal bacterial diversity and microbial growth rates, is mainly correlated with genetic, environmental and individual differences [30-34]. Notably, intestinal bacteria imbalance contributes significantly not only to intestinal diseases such as IBD, but also to some systemic chronic conditions such as obesity, cardio-metabolic disease (CMD), type 2 diabetes (T2D), metabolic syndrome, and malnutrition [35-37]. Changes in the abundance of microbiota in healthy and diseased states are shown in Table 1. Firstly, intestinal microbiota has been shown to be abnormally grown under diseases. For instance, Liu et al. found that the abundance of Bacteroides thetaiotaomicron was significantly reduced in obese patients and inversely related to serum glutamate concentrations [38]. Allin et al. discovered a decrease in the abundance of Clostridium spp. and Akkermansia muciniphila, whereas the abundance of Ruminococcus, Sutterella and Streptococcus increased in the gut of T2D patients [39]. Moreover, microbiota was demonstrated as one of main contributors of disease onset and progression. Altered intestinal microbiota profile led to excess endogenous alcohol production, and increased pro-inflammatory factors which contributed to non-alcoholic fatty liver disease (NAFLD) [40-42]. It should be noted that dysbiosis (imbalance of intestinal microbiota) has been regarded as onset of most chronic diseases, due to unhealthy diet or environmental changes. Both gain and loss of function dysbiosis are expected to cause intestinal injury and chronic inflammation, which, coordinating with other factors or solely, contributes to disease.
Throughout the last decade, researchers have discovered that gut bacteria show a complex interplay with ISCs, which plays an important role in gut health. Therefore, this section aims to summarize the recent developments in regulating intestinal microbiota/ derived metabolites-ISC axis (Figure 3). The effects of gut microbes and gut microbial metabolites (GMMs) on ISCs in different gut states are displayed in Table 2.
Alteration of gut microbiota in diseases and the associated mechanisms.
| Disease | Characteristics or Alterations | Effects | References |
|---|---|---|---|
| Healthy subject | High gut bacterial diversity and microbial growth rates; High microbial gene richness; Stable microbiome functional cores | Related to gene, environmental and individual differences | [30-34] |
| Obesity | Decreased abundance: Bacteroides thetaiotaomicron; Oscillospira; Methanobrevibacter smithii; Faecalibacterium prausnitzii Elevated abundance: Eubacterium ventriosum; Roseburia intestinalis | Diet/ high BMI-related | [38, 209-211] |
| Type 2 diabetes (T2D) | Decreased abundance: Clostridium spp; Akkermansia muciniphila; Bacteroides spp.; Butyrate-producing bacteria Elevated abundance: Dorea; Ruminococcus; Sutterella; Streptococcus; Akkermansia muciniphila | Functional alterations in the metagenomes as causes or progression factor | [39, 212-214] |
| Atherosclerotic cardiovascular disease (ACVD) | Decreased abundance: Prevotella Elevated abundance: Streptococcus; Escherichia | Unknown | [215] |
| Cardio-metabolic diseases (CMD) | Decreased abundance: Bacteroides spp.; Faecalibacterium prausnitzii | Their metabolites blocking NF-κB activation and IL-8 secretion | [216] |
| Chronic heart failure (CHF) | Decreased abundance: Alistipes; Faecalibacterium; Oscillibacter spp. Elevated abundance: Ruminococcus; Acinetobacter; Veillonella spp. | Associated with altered fecal and plasma metabolic patterns | [217] |
| Non-alcoholic fatty liver disease (NAFLD) | Decreased abundance: Oscillibacter; Flavonifaractor; Odoribacter; Alistipes spp. Elevated abundance: Clostridium; Anaerobacter; Streptococcus; Escherichia; Lactobacillus; Klebsiella pneumoniae | Excess endogenous alcohol production; An elevated level of proinflammatory cytokines | [40, 42] |
| Non-alcoholic steatohepatitis (NASH) | Decreased abundance: Oscillospira spp. Elevated abundance: Proteobacteria; Enterobacteriaceae; Escherichia spp.; Dorea; Ruminococcus spp. | Excess endogenous alcohol production; Higher fecal concentrations of 2-butanone and 4-methyl-2-pentanone that cause hepatocellular toxicity | [218, 219] |
| Liver cirrhosis | Elevated abundance: Proteobacteria; Fusobacteria | An increase of microbial haem biosynthesis and phosphotransferase systems | [220] |
| Malnutrition, Severe acute malnutrition (SAM) | Decreased abundance: Bifidobacterium longum; Bifidobacterium pseudolongum | Related with not breastfeeding and unhealthy diet | [221] |
The effects of the gut microbiota/ its metabolites on ISCs in different intestinal states.
| Gut microbiota/ metabolites | Model | Effects on ISCs | Mechanisms | References |
|---|---|---|---|---|
| Preterm infant gut microbiota | Humanized microbiome gnotobiotic mouse model | Promoting cell proliferation | Upregulating the expression of Cryptdin 5, Muc3 and Lyz1 | [29] |
| Lactic acid producing bacteria | Normal mouse/ Drosophila | Promoting proliferation | Stimulating the Wnt/β-catenin pathway through GPR81; Nox-mediated generation of ROS | [48, 89, 91, 92] |
| Gut damage in mice caused by radiation and methotrexate; Gut damage in drosophila (Lactobacillus plantarum overgrowth) | Promoting proliferation and enhancing regeneration | GPR81 activation and downstream regulation on Wnt3; Activating production of ROS by the intestinal Nox | [48, 90] | |
| Lactobacillus reuteri | Normal mouse intestinal organoid | Promoting proliferation | Stimulating the Wnt/β-catenin pathway through increase in R-spondins | [49] |
| TNF-induced intestinal organoid damage; C. rodentium-induced intestinal inflammation in mice | Promoting proliferation and differentiation | Activating the Wnt/β-catenin pathway and upregulating Wnt3 and Lrp5 expression | [49] | |
| Erwinia carotovora | E. carotovora infection in Drosophila | Promoting proliferation and division | Initiating the JAK-STAT signaling | [222] |
| Bacillus subtilis | DSS-induced mouse colitis model | Protecting ISCs from inflammatory injury and inducing proliferation | Rebalancing the intestinal flora | [52] |
| SCFAs (acetate, butyrate, propionate) | Crypt culture | Butyrate: Promoting proliferation | Unknown | [57] |
| Mice treated with vancomycin; Intestinal organoids | Valproic acid, Acetate, Propionate, and Butyrate: Promoting proliferation | Notch activating; MEK-ERK signaling; Inhibition of histone deacetylases by butyrate | [58-61] | |
| Primary colonic ISCs | Butyrate: Inhibitory effect Acetate, Propionate: No effect | Foxo3-dependent | [62, 63] | |
| Hydrochloric acid induced rat epithelial injury model | Butyrate: No effect | Unknown | [64] | |
| Lactate (SCFAs intermediate metabolites) | Normal mouse; Mouse small intestine organoid; Gut damage caused by radiation or methotrexate | Promoting proliferation and regeneration | GPR81 stimulates of the Wnt/β-catenin signal pathway | [48] |
| Succinate (SCFAs intermediate metabolites) | Normal rat | Inhibiting proliferation | Unknown | [69] |
| Tryptophan metabolite | AhR-/- mouse | Indoleacetic acid (IAA): Inhibiting proliferation | Suppresses β-catenin signals through AhR | [77] |
| AhR-/- mouse, VillinCreAhrfl/fl mouse; Mouse intestinal organoid | Indole-3-carbinol (I3C): Inhibiting proliferation | Activating AhR | [78, 80] | |
| DSS-induced mouse IBD model | Indoleacrylic acid (IA): Promoting differentiation | Activates AhR to elevate expression of Ki67 | [81] | |
| TNF-α-induced intestinal organoid injury; Candida albicans infected mouse model | Indolealdehyde (IAld): Promoting differentiation | Induction of IL-22 | [82, 83] | |
| Secondary bile acids: lithocholic acid (LCA), deoxycholic acid (DCA) | Human colon cancer cell model; Rat small intestinal crypt cells | Promoting proliferation at low dose, and inhibition at high dose | Promotion: Possible involvement of Wnt/β-catenin signaling Inhibition: EGFR and FXR signaling | [86-88] |
| Polyamines | Rat small intestinal crypt cells; Phytohaemagglutinin induced gut growth | Promoting proliferation | Inducing TGF-βRI; Increasing c-fos, c-myc, and c-jun expression; Acting as energy source | [93-95] |
3.1 Intestinal microbiota and ISCs
Many studies have established a link between the gut microbiota and ISCs, revealing clear communication at the niche level. ISCs are often used as a tool, which is due to its ability to promote a continuous renewal of the intestinal ECs. It has been shown that the intestinal microbiota has an impact on how the intestinal ECs grows and develops. To discover whether early intestinal microbiota contributes to the development of the intestinal tract, Yu and colleagues examined the differentiation and formation of TJs in four different epithelial cell lines [29]. Early intestinal bacteria have been demonstrated to be a cause of villi height and crypt formation [29]. Particularly, early microbes from a preterm infant with normal weight gain mediated increased villus height and crypt depth, elevated cell proliferation, high numbers of goblet cells and Paneth cells, and enhanced TJs [29].
Remarkably, intestinal microbiota exerts varied influences on ISCs during different intestinal states (healthy, injury and regenerative conditions). In a steady-state condition, normal gut microbiome, featuring with balanced beneficial and potential harmful bacteria, helps to maintain gut health. It seems that gut microbiota under healthy condition probably does not give specific advantage to ISCs. Schoenborn et al. demonstrated that no differences in ISC numbers or cycling activity were observed in the small intestine of germ-free and conventionally raised mice, which suggested that enteric microbiota did not impact ISCs under a normal condition [43]. Another study compared the transcriptome/proteome expression phenotypes of primary organoids established from germ-free and microbiota-associated mice, indicating there were no significant differences [44].
In contrast, accumulated evidence confirms that intestinal microbiota is essential for ISC activity during injury and repair states. The cytosolic bacterial peptidoglycan sensor Nod2 was found highly expressed in ISCs linking microbes to gut epithelial regeneration [45]. Nod2 knockout mice were viable and did not show any particular difference compare to wide-type mice [45]. However, under stress conditions, the presence of microbiota made ISCs more prone to respond to microbiota-secreted metabolites in a Nod2 dependent manner, protecting from injury. The results indicate that gut microbiota may not influence ISCs in normal condition, but robustly exerts effect on ISCs function and EC renewal and repair.
Alteration of intestinal microbiota is characterized as either loss of function and/or gain of function. Opportunistic pathogens and their activities could be obtained or overgrow to promote diseases such as infectious diseases, which is termed gain of function dysbiosis. Loss of function dysbiosis is often due to the suppression of beneficial bacteria and their activities, which can be linked to diseases like IBD and obesity. Changes in the composition of the intestinal microbiota can cause abnormal reprogramming of ISCs via a variety of mechanisms including genetic and epigenetic failure, impaired metabolism, ISC stability, and abnormal immune activation [46]. The reciprocal symbiotic connection between the host and microbiota under physiological conditions could be lost as a result of changes in the microbial composition and/or the host's genetic susceptibility to infection [46]. Tao et al. found that the intestine's relative quantity of beneficial and invading bacteria was directly related with the production and expression of inflammatory cytokines, which interacted to regulate immune function [47]. Notably, it is speculated that bacteria, through their contact with the gut epithelium, have an impact on the production and assembly of TJs and affecting host genome and immunity, thereby controlling the permeability and reprogramming of the epithelium [47]. Mucin-degrading bacteria (such as Akkermansia muciniphila) have been shown to be critical for ISC-mediated epithelial growth and intestinal environment stability in a recent study [48]. Wu et al. showed that Lactobacillus reuteri induced intestinal epithelial cell proliferation to repair epithelial damage through an increase in R-spondins and reduced intestinal pro-inflammatory cytokine secretion and serum lipopolysaccharides (LPS) concentrations, thereby activating the Wnt/β-catenin pathway [49]. Furthermore, Buchon et al. demonstrated that both invasive bacteria and commensal bacteria could affect adult ECs renewal, demonstrating the critical role of ISCs in maintaining the GI environment's stability and host defense. However, the gut microbiota-intestinal epithelium interaction is still largely unknown due to unknown variables and processes. This is potentially due to the fact that a large number of microbes exist in the gut and are involved in complex regulations.
Given the role of gut microbiota and ISCs in gut health, it has been suggested that manipulation of gut microbiome via colonization of specific beneficial bacteria strains or dietary supplementation of prebiotics may be beneficial in certain intestinal diseases. Chen et al. demonstrated that Clostridium butyricum inhibited the growth of high-fat diet (HFD)-induced intestinal tumor development in Apcmin/+ mice through modulation of Wnt signaling and alteration of intestinal microbiome [51]. The researchers showed that Bacillus subtilis not only attenuated the inflammatory response, stimulated the proliferation of ISCs, facilitated the repair of the intestinal barrier, but also restored intestinal flora balance. Ferments of B. subtilis relieved IBD [52]. Moreover, Chrysanthemum morifolium polysaccharides have been shown to be useful in the treatment of ulcerative colitis (UC) by enhancing the establishment of healthy gut bacteria, restoring balance to the intestinal micro-ecological environment, and regenerating the immune system [47]. Although it is demonstrated that manipulating gut microbiota presents a promising strategy for maintaining gut health, the methods for precision regulation of gut microbiota are still lacking. The homeostasis of gut is maintained by balance of different microbes. Besides, there are great individual differences among subjects. Supplementation of specific bacterial strains or prebiotics has its limitations and may not benefit all individuals. Thus, it is of interest to develop novel methods that precisely regulate gut microbiota.
3.2 GMMs and ISCs
Gut microbiota is capable of biotransforming either host-derived components such as bile acids or dietary substances to generate various metabolites. Several studies have revealed that GMMs had effects on the host's gut environment as well as the immunological balance.
3.2.1 Short-chain fatty acids
Short-chain fatty acids (SCFAs) as the fermentation products of indigestible carbohydrates, primarily comprising acetate, propionate, and butyrate, are the most studied metabolites of gut bacteria [53]. SCFAs are important energy source of colonocytes. Besides, they directly or indirectly impact on host physiology, as some of SCFAs also serve as the substrate and regulator for intestinal and hepatic gluconeogenesis.
The role of SCFAs on ISCs has been intensively investigated. Since the effect of SCFAs on ISC is confounded by multiple factors, such as diet, gut microbiota, and gut physiology, opposing conclusions have been drawn from recent decades of research [54]. One part of the studies suggests that SCFAs have a proliferative effect on ISC, while conclusion from the other part supports an ineffective or even an inhibitory effect.
In earlier years, Sakata et al. found that SCFAs stimulate EC proliferation through the autonomic nervous system in association with dosage of SCFAs, the uptake of intestinal microorganisms and fermentable fibers [55, 56]. In vitro experiments have demonstrated that SCFAs promoted mucosal proliferation [57, 58]. It is reported that, in small intestinal organoids, HDAC inhibition by butyrate resulted in an enhanced LGR5+ ISC pool [59]. Donohoe et al. suggest that microbial production of butyrate, the main source of energy for colon cells, may provide energy for ISCs and promote its value-added [60]. De Vadder et al. demonstrated that SCFAs promoted intestinal gluconeogenesis through a cAMP-dependent mechanism to control glucose metabolism [61]. Therefore, these studies have suggested a promotive role of SCFAs on ISC proliferation.
However, Boffa et al. illustrated that feeding wheat bran significantly reduced the level of butyrate in the colon and inhibited the proliferation of colon cells [62]. Kaiko and colleagues showed that butyrate inhibited the proliferation of ISCs upon mucosal injury or in crypt-less host organism [63]. In addition, Scheppach demonstrated that butyrate has no effect on the growth of crypt cells [64].
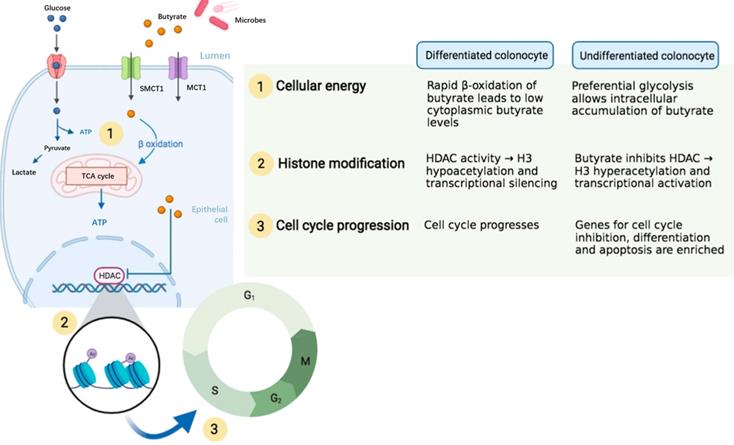
Despite the disagreement regarding the role of SCFAs on ISCs, most of studies have indicated that SCFAs particularly the butyrate enhanced ISC proliferation. The difference in experimental design and conditions across research groups may contribute to the discrepancy of results. For the in vivo studies, the gut microbiota of animals in different houses may be varied in response to dietary fibers or SCFAs. For the in vitro studies, the concentration and culture conditions used may significantly lead to bias in results. In addition, SCFAs have diverse effects on ISCs through not only influencing ISC proliferation but also modulating genetic pathways that might or might not be associated with ISC proliferation. Some researchers have proposed that the distinct metabolic pathways for energy use associating with epigenetics in differentiated and the undifferentiated colonocytes may contribute to the “butyrate paradox” (Figure 4) [65]. Therefore, more studies are advocated to investigate the exact role of SCFAs on ISCs.
Besides the SCFAs, several studies have demonstrated that SCFA intermediate metabolites such as lactate and succinate are regulators of ISCs. Fevr et al. showed that lactate led to larger size of small intestine organoids and enhanced expression of target genes in Wnt/β-catenin pathway which modulates stem cell self-renewal [66, 67]. On the contrary, succinate is reported to mediated superoxide production and mucosa damage in the colon [68]. It is showed that succinate through colonic infusion inhibited colonic epithelial growth in rats [69].
3.2.2 Tryptophan metabolites
Tryptophan as an essential amino acid is readily metabolized by microorganisms to a variety of biologically active amino acid derivatives, such as indole, indoleacetic acid (IAA), indole aldehyde (IAld), indole acrylic acid (IA), indole lactic acid, indole ethanol, indole propionic acid, tryptamine, 3-methylindole, etc. [70-73]. These metabolites are mostly present as ligands for the aryl hydrocarbon receptor (AhR) and the pregnane X receptor (PXR) [74-76]. Although both AhR and PXR participate in immune regulation, intestinal barrier function and cell proliferation, current knowledge mainly supports a role of the microbial metabolites of indole derivatives on ISC via AhR signaling. How tryptophan metabolites influence ISC through PXR remains further studies.
Intestinal microbe-derived indole metabolites influence ISCs through acting as ligand of AhR. It is reported that the expression level of β-catenin was unexpectedly high in the cecum of Ahr‑/- mice, which was decreased after treatment of IAA. IAA suppressed β-catenin signals through AhR-mediated β-catenin degradation [77]. The stem cell niche is maintained by Rnf43 and Znrf3 by blocking Wnt/catenin signaling and limiting its proliferation, and the gut barrier is restored by AhR signals in response to dietary ligands [78]. Recently, Metidji et al. reported that indole-3-carbinol (I3C) inhibited ISC proliferation through activating AhR, which led to the increased expression of Znfr3 and Rnf43 genes for degrading Wnt [79]. In addition, Park et al. also showed that I3C could inhibit ISC proliferation [80].
The microbial indole metabolites also affect ISCs activity through interaction indirectly with other cells. Wlodarska showed that oral administration of Peptostreptococcus russellii to mice resulted in increased production of IA which activated AhR in colonic organoids and macrophage and contributed to elevated expression of Ki67 and proliferation of goblet cells in DSS-induced colitic mice [81]. Previous studies also demonstrated that Lactobacillus reuteri enhanced mouse intestinal EC proliferation through generating IAld metabolite which activated AhR in lamina propria lymphocytes to upregulate IL22 and promote ISC proliferation [82, 83].
A proposed epigenetic configuration involving distinct metabolic pathways that contributes to “butyrate paradox”. Figure reused with permission from Ref. [65]. Copyright @ 2021 by Pooja S. Salvi and Robert A. Cowles.
3.2.3 Secondary bile acids
Since high levels of diet-induced secondary bile acids (SBAs), such as lithocholic acid (LCA) and deoxycholic acid (DCA), may contribute to the development of colorectal cancer, it can be expected that SBAs have a role in the proliferation of ISCs [54, 84]. The direct evidence has been limited. Kozoni et al. reported that LCA increased proliferating cells in colonic crypt of mice [85]. Other findings in cancer cells showed that DCA increased Wnt/β-catenin signaling and promoted colon cancer cell proliferation at low concentrations. However, the effect was reversed at high concentrations [86, 87]. This phenomenon was also reported by Dossa and Milovic's works [87, 88]. These results may suggest a potential role of SBAs on ISCs. However, till now, the direct effect of SBAs on ISCs both in vitro and in vivo is not fully understood.
3.2.4 Other microbial metabolites
A study by Jones showed that Lactobacillus plantarum caused alterations in gut epithelial cell growth by promoting physiological amounts of reactive oxygen species (ROS) from Nox enzyme [89]. In 2018, Latsenko et al. reported that a zero mutation in the Drosophila PGRP-SD gene was related to increased overgrowth of L. plantarum and its metabolite lactate through activation and generation of ROS [90]. In the same year, Lee and colleagues demonstrated that GPR81 on panthenol and stromal cells recognized lactic acid derived from lactobacilli and induced regeneration of intestinal ECs via WNT3/β-catenin [91]. Reedy et al. then corroborated the findings of the aforementioned experiments [92].
Polyamines are a type of compounds that can be either from dietary source or produced by gut microbiota. It was reported that polyamines may serve as energy source or influence intracellular signaling pathways such as the TGF-βRI/Smad3/4, c-fos and c-jun, etc. to promote ISCs proliferation [93-95].
3.3 Dietary influence
Dietary variables may have a direct impact on gut homeostasis either directly or indirectly through regulating the composition of the intestinal microbiota and/or microbial metabolites.
Frieling and colleagues have found that HFD increased ISCs activity in the Drosophila fruit fly. Specifically, the intestinal microbiota and the JNK signaling pathway in intestinal cells were both required for this action to occur [96]. Hou et al. identified diet-microbial metabolism feedforward loop that showed a remarkable role in modulating ISC proliferation in the stressed gut. They found that dietary raffinose promoted Lactobacillus reuteri growth which increased metabolism of raffinose to fructose for enhancing glycolysis to support ISC proliferation and renewal [97].
Gut microbiota mediated dietary metabolites showed potential effects on ISCs. Singh et al. demonstrated that urolithin A, a major microbial metabolite of phenolics from berries and pomegranate fruits, exerted barrier activity by upregulating epithelial TJ proteins in intestinal ECs via the AhR-nuclear factor erythroid 2-related factor 2 (Nrf2) pathway in organoid culture [98], suggesting a potential role in regulating ISCs.
4. Signaling pathways associated with regulation of ISCs
4.1 Notch signaling pathway
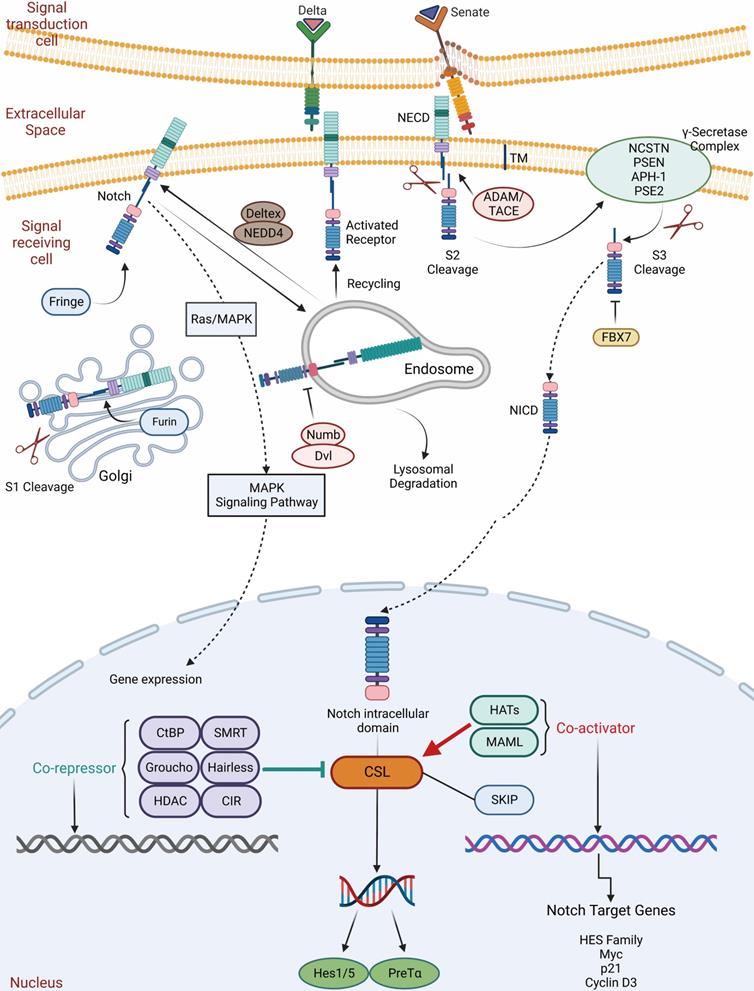
During development and in adult tissues homeostasis as well, the Notch pathway plays an essential role in determining cell fate, which is a highly conserved mechanism. The Notch system regulates pro-secretory cell signaling by influencing both signaling and receiving cells through ligand-receptor interactions. The Notch signaling receptor is a single transmembrane protein with functional extracellular (NECD), transmembrane (TM) and intracellular (NICD) structural domains. Delta-like and Senate (Jagged1, Jagged2) families are ligands for Notch. First, the Notch receptor is sheared and glycosylated within the Golgi of the recipient cell, followed by S1 cleavage with TM-NICD. Endosomes transfer processed receptors to the plasma membrane, where it binds to the ligand via Deltex regulation and NUMB inhibition. Secondly, TACE (TNF-ADAM: metalloproteinase convertase) shears NECD away from the TM-NICD structural domain when it binds to the ligand (S2 cleavage). Lastly, NECD-linked ligands form a complex that is dependent on endocytosis and cycling via Mib ubiquitination in signaling cells. In signal-receiving cells, NICD is released from the TM by-secretase (S3 cleavage), resulting in nuclear translocation and binding to the CSL transcription factor complex, allowing transcription of traditional Notch target genes (Myc, p21, and HES family members) (Figure 5) [99-103].
The Notch signaling pathway in ISC regulation. The Notch signaling receptor is a single transmembrane protein with functional extracellular (NECD), transmembrane (TM) and intracellular (NICD) structural domains. Delta and Senate families are ligands for Notch. First, the Notch receptor is sheared and glycosylated within the Golgi of the recipient cell, followed by S1 cleavage with TM-NICD. Endosomes transfer processed receptors to the plasma membrane, where it binds to the ligand via Deltex regulation and NUMB inhibition. Secondly, TACE (TNF-ADAM: metalloproteinase convertase) shears NECD away from the TM-NICD structural domain when it binds to the ligand (S2 cleavage). Lastly, NECD-linked ligands form a complex. In signal-receiving cells, NICD is released from the TM by-secretase (S3 cleavage), resulting in nuclear translocation, binding to the CSL transcription factor complex and allowing transcription of traditional Notch target genes.
Notch signaling includes Notch receptors, CSL DNA binding proteins, and Notch ligands (DSL proteins), which determines biological cell development by regulating local intercellular interactions and are essential for the maintenance of ISCs [104]. Researchers found that activation of Notch signaling increased ISCs while inhibiting cell differentiation, suggesting that the Notch pathway regulates early intestinal cells [105]. Notch, in contrast to other receptors, receives signals from the surface of the cell and regulates gene expression inside the nucleus. Notch signaling regulates intestinal cell types, and ISCs differentiate into two types: secretory intestinal enteroendocrine cells and nutrient-absorbing intestinal ECs [106]. Ectopic overexpression of NICD decreased the frequency of secretory cells in the mouse intestinal epithelium [107-109]. Inhibiting the Notch pathway, on the other hand, resulted in increased secretory cell lines and fewer absorptive enterocytes and colitis cells [106, 110-112]. Guo and Ohlstein also demonstrated that ISCs with high Delta levels strongly activated Notch signaling in their progeny, resulting in ECs, whereas Notch signaling inhibition or loss resulted in stem cell loss and premature formation and appreciation of enteroendocfine cells; thus, the regulation of Notch signaling on ISC differentiation is bidirectional [113]. To regulate ISC behavior, activity oscillates within a limited threshold. Increased Notch activity promotes ISC development, resulting in the creation of more daughter cells, with increased Notch activity leading to intestinal stem cell differentiation [114]. These data indicate that Notch signaling is crucial to determining how intestinal epithelium develops [13, 108, 114-117].
Recent research has indicated that Notch's influence on the ISC spectrum was mediated through its action on Atoh1, with Hes1 and Atoh1 expression being mutually inhibited [118]. Thus, the degree of Notch activity controls the reciprocal regulation of Hes1 and Atoh1, which is critical for balancing the destiny of cells between uptake and secretion [119, 120]. Furthermore, Atoh1 is controlled by Wnt signaling, whose activity affects GSK3β, a protein kinase that mediates phosphorylation and protein degradation and regulates Atoh1 via ubiquitination [120-122]. Moreover, Gfi1, a transcription factor that is reliant on Atoh1 expression in the intestine, is essential for the differentiation of Atoh1-specific secretory precursors into enteroendocrine and cupped/pan precursors. An investigation has shown that defective Neurog3 suppression at an early stage was required for sub-stasis to occur in Paneth and mucus cell lines, and that sub-stasis resulted in alterations in the cell type ratio in Gif1-deficient mice. Thus, analogous to Hes1/Atoh1, the relative activity of Gfi1/Neurog3 is implicated in the partitioning of cells into secretory lineages [123].
ISC self-renewal is maintained and modulated by Notch signaling. Kapuria et al. found that inhibition of Notch-mediated Tuberous Sclerosis Complex 2 (TSC2) helped to promote the differentiation of ECs [124]. Qu et al. revealed that dual cortisol-like kinase 1 (Dclk1) may act as ISCs marker and that restricting the Notch signaling pathway decreased ISC survival, implying that the Notch pathway is critical for ISC-mediated saphenous fossa regeneration. Dclk1 expression in saphenous ECs may potentially be used as a measure of ISC survival after injury [125]. Bmi1 expression is dynamically controlled and depends on synergistic regulation by Notch, MaM co-activators and β-catenin. Bmi1 regulates mouse ISC proliferation and self-renewal downstream of Notch [126]. Srinivasan et al. then proved that Notch signaling modulated the Bmi1/Lgr5+ ISCs ratio, with suppression of Notch signaling decreasing this ratio and vice versa increasing this ratio. This confirms that Notch signaling can induce asymmetric division in response to intestinal inflammation, establishing a direct link between slow and fast-cycle ISCs [127]. Kwak et al. observed that Ghrelin rehabilitates gut function after intestinal lesions by activating Notch signaling [128]. Jones et al. established that Defacre-expressing Paneth Cells were malleable and that the Notch signaling pathway may induce differentiation of Defacre-expressing Paneth Cells into pluripotent stem cells expressing Lgr5+ crypt base columnar and facilitated acute intestinal regeneration following damage [129].
Recently, the laboratory of Joshua V. Troll demonstrated for the first time that the MyD88-dependent signal induced by the microflora inhibited the Notch signal, thereby promoting the fate of secretory cells. These results link the activity of the microflora with the Notch pathway. In conclusion, Notch signaling is essential for intestinal homeostasis and repair, which when damaged may lead to chronic inflammation and cancer [130].
4.2 Wnt signaling pathway
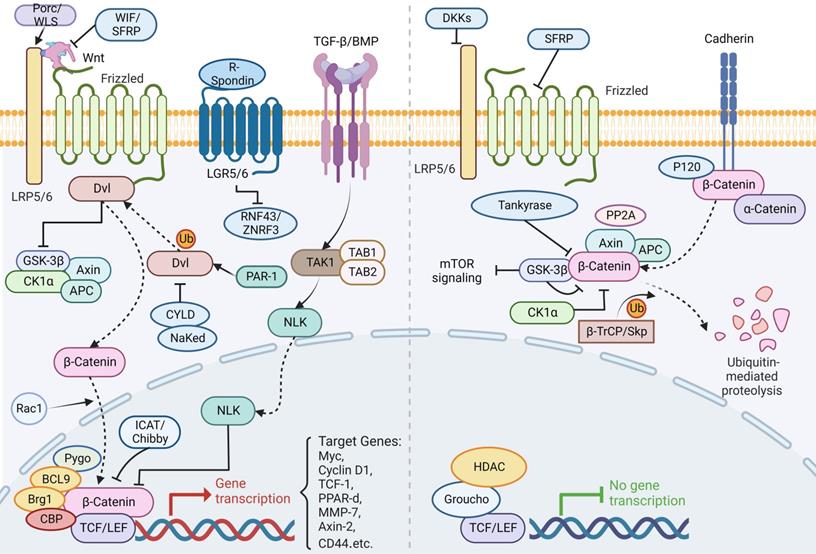
The Wnt pathway is crucial to early development, organogenesis, tissue regeneration and other physiological processes in the animal embryo, which is a highly conserved class of signaling pathway. It's a collection of several downstream signaling pathways that are activated when the ligand protein Wnt binds to membrane protein receptors. The Wnt signaling pathway enables two distinct modes of communication between cells: intercellular (paracrine) communication and autophagic communication (autocrine). The canonical pathway is comprised of Dvl, GSK3, the Wnt family of secretory proteins, APC, Axin, the Frizzled family of transmembrane receptor proteins, β-catenin, and TCF/LEF transcriptional regulators. The planar cell polarity route is involved in cytoskeletal rearrangement and JNK activation; the Wnt/Ca+ pathway is involved in triggering PLC and PKC; additionally, the intracellular pathway is involved in spindle orientation and asymmetric cell division [131]. The following section will discuss the canonical Wnt/β-catenin pathway in further detail. It regulates ISC pluripotency and determines the differentiation fate of cells throughout development. Moreover, it incorporates signals including retinoic acid, fibroblast growth factor (FGF), transforming growth factor beta (TGF-β) and BMP. A ligand for Wnt attaches to Frizzled receptors then assembles a complex on the surface of the cell together with LRP5/6. The binding of R-spondin to LGR5/6 inhibits the frizzled receptor's activity by ubiquitinating ZNRF3 and RNF43. When the Wnt receptor complex is activated, GSK-3β is dissociated from APC/Axin/GSK-3β. Dvl is activated by sequential phosphorylation, polymerization and polyubiquitination in response to PAR-1 and is inhibited by CYLD and NaKed. Stable β-catenin enters the nucleus through Rac1, contacts LEF/TCF, displaces co-repressors, then recruits co-activators in Wnt target genes. Without Wnt signaling, β-catenin acts as a transduction co-regulatory molecule and an intercellular adhesion junction protein, which is phosphorylated by CK1α and the APC/Axin/GSK-3β complex, resulting in ubiquitination and degradation by the proteasome via the β-TrCP/Skp pathway. In addition, β-catenin works in concert with several different transcription factors to control specific targets (Figure 6) [132-141].
The canonical Wnt/β-catenin signaling pathway in ISC regulation. A ligand for the canonical Wnt/β-catenin attaches to Frizzled receptors then assembles a complex on the surface of the cell together with LRP5/6. The binding of R-spondin to LGR5/6 inhibits the frizzled receptor's activity by ubiquitinating ZNRF3 and RNF43. When the Wnt receptor complex is activated, GSK-3β is dissociated from APC/Axin/GSK-3β. Dvl is activated by sequential phosphorylation, polymerization and polyubiquitination in response to PAR-1 and is inhibited by CYLD and NaKed. Stable β-catenin enters the nucleus through Rac1, contacts LEF/TCF, displaces co-repressors, then recruits co-activators in Wnt target genes. On the other hand, TGF-β/BMP regulates NLK into the nucleus through TAK1 to inhibit β-catenin expression. Without Wnt signaling, β-catenin acts as a transduction co-regulatory molecule and an intercellular adhesion junction protein, which is phosphorylated by CK1α and the APC/Axin/GSK-3β complex, resulting in ubiquitination and degradation by the proteasome via the β-TrCP/Skp pathway. In addition, β-catenin works in concert with several different transcription factors to control specific targets.
Not only is Wnt the main force in ISC proliferation, but its mutagenesis is also a major factor in colon cancer [4, 142]. Its activity contributes to intestinal differentiation as well as the stability of ISCs, and either an increase or decrease in β-catenin activity has a significant effect on intestinal epithelial cell generation and differentiation, ultimately leading to the development of GI tumors [143]. Numerous studies have indicated that mutations that activate the Wnt pathway abnormally could accelerate the proliferation of undifferentiated progenitor cells and ultimately resulted in cancer [144]. Wnt/β-catenin signaling activation is often related to carcinogenesis, most notably in colorectal cancer. Zhang et al. revealed a previously unknown method of β-catenin activation, the MST4-p-catenin signaling axis. A MST4 deficiency led to a reduction of ISCs and slowed colon cancer formation. Secondly, the MST4-p-catenin axis was elevated, which has been associated with a poor outcome in human colorectal cancer. In conclusion, this study broadens avenues for colon cancer targeted treatment [145].
Several studies have found that changes in the Wnt pathway and microbial composition regulated the proliferation of intestinal ECs through Myd88 [146-148]. Hirohito Abo et al. showed that early microbiota regulation of Erdr1 stimulated the Wnt pathway in ECs, boosted Lgr5+ stem cell production, and promotes colonic mucosal repair, which may be utilized to treat cancer and mucosal ulcers [149].
Nevertheless, several protein ligands and cytokines have the ability to regulate the Wnt signaling. Strubberg et al. noted that inhibition of proliferation of ISC in patients with cystic fibrosis (CF) by cystic fibrosis transmembrane conductance regulator (Cftr) may promote Wnt/β-catenin signaling, thereby increasing the risk of gut neoplasia [150]. Johansson et al. proved that RALs are required for effective ISC regeneration downstream of Wnt signaling. ISC function and Lgr5 positivity decreased, resulting in fast crypt mortality and insensitivity to Wnt inhibition, and impaired tissue regeneration [151]. Han et al. reported that YTHDF1, which is distinctly expressed in ISCs, enhanced β-catenin activity by increasing the translation of Wnt signaling. It contributes to the maintenance of ISCs during regeneration and tumorigenesis [152]. Moreover, Li et al. proved that deletion of Lats1/2 (the core Hippo kinase) inhibited the Wnt pathway, leading to deficiency of ISC, which is based on transcriptional enhanced associate domain (TEAD). Their team recognized that inhibition of TEAD palmitoylation could prevent the above finding [153]. Wei et al. showed that Erk1/2 deficiency resulted in activation of the Ras/Raf cascade through activation of Wnt signaling, which then transduced Akt activity to enhance Wnt/β-catenin pathways as well as the mTOR. This was associated with a decrease in mesenchymal Bmp4, which is a Wnt repressor [154].
To effectively treat chronic intestinal illnesses, it's necessary to completely understand the developmental mechanisms of ISC. However, there's a dispute about the various ISC lineage hierarchy and segregation theories. Anika Böttcher and colleagues demonstrated a new ecological signal Wnt/PCP pathway in the development of ISC [155]. While Wnt signaling governs ISC self-renewal by inducing Lgr5+, little is known about Lgr5 post-translational regulation. Novellasdemunt et al. indicated that silencing NEDD4 and NEDD4L increased Wnt activation and the amount of ISCs, hence enhancing tumor predisposition and progression [156].
In addition, a study found that increased zinc activity maintained gut integrity through activation of Wnt/β-catenin signaling, shedding insight into the efficient preventative approach of ISC-based exogenous zinc preparations [157]. Dectin-1 (a C-type lectin receptor) could signal to intestinal ECs to activate the Wnt pathway and induced intestinal ECs to proliferate, thereby promoting colorectal cancer (CRC) development [158].
4.3 BMP signaling pathway
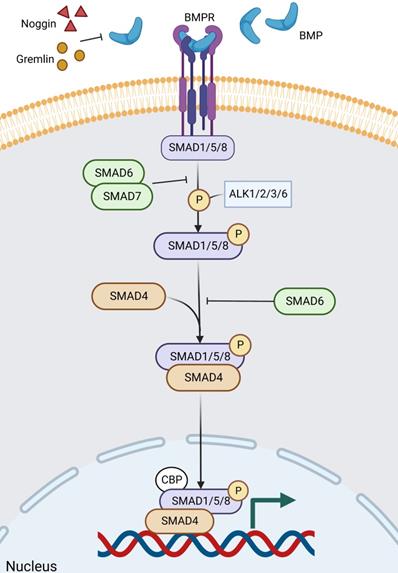
BMP is a growth factor of the TGF-β family, and BMP signaling contains a complex formation of different type 1 and type 2 receptors for serine/threonine kinase activity. Ligand recognition can be obtained from both type 1 and type 2 receptors [159]. During epithelial cell differentiation, SMAD1/5/8, a molecule downstream of BMP signaling, is phosphorylated by ALK2/3/6 and ALK1, followed by BMP signaling highly activated by phosphorylated SMAD1/5/8 [160, 161]. SMAD6/7 is involved in the inhibition of phosphorylation and signaling, particularly SMAD6, which inhibits ALK3/6-mediated signaling (Figure 7) [161, 162]. The main BMPs (both BMP2 and BMP4) in the gut are recognized by the type 1 receptor BMPR1A, which is expressed in mature epithelial cells and mesenchymal cells [163-165]. BMP is negatively regulated by ISCs in the gut [166, 167], and its inhibitors, Noggin and Gremlin (Grem1 and Grem2) are expressed in the crypt [164], not only keeping ISCs and progenitor cells in a BMP-low niche [166, 168-172], but also inducing stem cell proliferation and enhancing Wnt activity [168]. Among other things, there is a negative crosstalk between Wnt and BMP/TGF-β [165].
It has been shown that BMPR1A and SMAD4 mutations cause the human polyposis. If BMPR1A is deleted or overexpressed in mice, ectopic crypt formation is stimulated [161, 164, 168]. In addition, abnormalities in Grem1 or Noggin can also lead to human polyposis, which is related to mutations in BMP pathway genes and a high-risk propensity for CRC [173, 174]. Mechanistically, SMAD/HDAC1-mediated repression of stem cell-associated genes (e.g. Lgr5, Sox9) is driven by BMP signaling, which prevents premature ISC expansion and polyp formation in Wnt-rich niche [175]. Recently, a study has revealed how oncogenes convert the niche environment into a beneficial one - niche remodeling. Secretion of BMP ligands by crypt expressing oncogenic KRAS or PI3K, inhibition of ISC activity, and alteration of Wnt signaling by PDGFRloCD81+ stromal cells induced by crypt with oncogenic PI3K [176]. In conclusion, BMP signaling sustains homeostatic balance by suppressing Lgr5+ ISC proliferation at the base of the crypt while promoting the allocation of secretory cell lines [177].
The BMP signaling pathway in ISC regulation. BMP signaling contains a complex formation of different type 1 and type 2 receptors for serine/threonine kinase activity. Noggin and Gremlin are extracellular inhibitors of BMP. After ligand binding to the receptor and entry into the cell, firstly, the downstream molecules of BMP signaling, SMAD1/5/8, are phosphorylated by ALK2/3/6 and ALK1, and then BMP signaling is highly activated by the phosphorylated SMAD1/5/8. Of these, SMAD6/7, particularly SMAD6, is involved in inhibiting phosphorylation and signaling, which inhibits ALK3/6-mediated signaling. Finally, phosphorylated SMAD1/5/8 aggregates with Smad4 and continues to signal, accumulating in the nucleus and regulating target gene transcription.
4.4 Other pathways
Likewise, other pathways and factors affect intestinal homeostasis and the progression of GI cancers. JAK-STAT signaling has been proven to control the proliferation of ISC and is negatively regulated by Notch [178]. A crucial question is how the intestinal epithelium interacts with the bacterial flora and microbial metabolites and what the mechanism is at the molecular level. Smarcad1 is a member of the Smarcad family of chromatin remodeling factors and is highly expressed in the epithelial region of the gut. Kazakevych and colleagues discovered that Smarcad1 mediated microbially caused inflammatory reactions in mice and coordinated the expression of intestinal epithelial cell gene transcription [179].
Vitamin D/vitamin D receptor (VDR) inadequacy is a major risk factor for colon cancer. Zhang et al. postulated that the intestinal VDR protected mice against dysbiosis by modulating the JAK/STAT system. VDR insufficiency is a major risk factor for colon cancer. They identified potential targets for cancer prevention, the VDR, which protected against the effects of dysbiosis by regulating the JAK/STAT system [180]. TNFAIP8 is a regulator of the Akt/β-catenin signaling axis for intestinal injury repair, which regulates the homeostasis and regeneration of mouse ISC through modulation of microbiota-induced Akt signaling pathways [181].
In addition, YAP1 activity is implicated in the maintenance of ISCs proliferation which acts as an inhibitor of Hippo pathway downstream effectors and/or Wnt signaling, contributing to ISC maintenance and differentiation in transit-amplified cells. The structural integrity of intestinal EC junctions is the primary signal regulating intestinal homeostasis via the Hippo pathway [182]. A recent study demonstrated that suppression of Hippo signaling reprogramed Lgr5+ ISCs to a Klf6+ wound-healing cell state, and YAP overexpression led to inhibition of tumor formation [148].
Finally, autophagy and its regulatory mechanisms are involved in the homeostasis and repair of the gut by regulating TJs and influencing the metabolism and the proliferative and regenerative capacity of ISCs [183]. Defective Atg16l1 autophagy in intestinal ECs led to abnormal morphology of Paneth cells, as evidenced by dysfunction in the secretion of protective antimicrobial peptides [184-186], and resulted in impaired ISC capacity as well [187]. This was because ISCs relied on Wnt and EGF signals to maintain stemness, and these signals are partially provided by Paneth cells. In addition, autophagy-deficient ISCs were more susceptible to excess ROS [188, 189], which made repair of intestinal damage much more difficult. Besides, it was found that deletion of Atg7 led to increased oxidative stress, which promoted P53-mediated apoptosis in Lgr5+ ISCs [190].
5. Current development of intestinal organoid culture
In 2009, Sato et al. successfully cultured crypt extracts from the murine small intestine into self-renewing and differentiating organoids [25]. The successful establishment of the first miniature intestinal organoid opens a new chapter in organoid research and is rapidly becoming a new research hotspot. The intestinal organoid is three-dimensional (3D) cell culture grown from pure ISCs. In 3D culture systems, these organoids can provide an in vitro model that more closely resembles the in vivo environment than traditional two-dimensional (2D) cell culture technique. ISCs replicate the self-renewal seen in vivo in the mature gut, resulting in forming a crypt-villi structure, epithelial polarization and functional lumen [25, 191, 192].
The 3D organoids have many applications in disease modeling, gene function analysis, and regenerative medicine. UC is characterized by chronic inflammation in the colon, and prolonged exposure to an inflammatory environment predisposes to CRC [193]. To understand the genetic mechanisms of inflammation, Nanki et al. analyzed somatic mutations in the epithelium of UC using organoids [194]. In the same year, a parallel study sequenced UC and non-UC crypt foci isolated directly from patients and reported frequent mutations in UC on the NFKBIZ pathway [195]. Pleguezuelos-Manzano et al. showed by organoid that pks+ E. coli induced double-stranded DNA breaks and interstrand crosslinks, and that individuals carrying genotoxic pks+ E. coli individuals may be at higher risk of developing CRC [196]. Notably, several studies showed that transplanted Lgr5+ stem cells led to recovery of damaged colonic lesions and the repaired colonic structures displays appropriate epithelial barrier function [197].
Although organoids have been vastly superior to normal 2D cell cultures, there are still drawbacks, such as a lack of endothelial cells, immune cells and gut microbes, which limits the study of cell-cell and cell-microbe interactions. To better study the interaction of intestinal cells with other types of cells or gut microbe, the recent development of a human intestinal microfluidic organ-on-a-chip model has been engaged to overcome these challenges. Organ chips can integrate different types of cells including ECs, microvascular endothelium, nerve cells, immune cells as well as their interplay with gut microbes [198-200]. Some studies have used immortalized cell lines such as Caco-2 to generate gut chip [201, 202]. However, these cell lines are usually originated from tumor origin and are not suitable for studying intestinal tumorigenesis and normal physiology. The emerging primary culture based on combing organoid and organ chip has made much progress, which enables fluid flow and peristalsis-like deformations and allows the intestinal ECs undergo villus histogenesis with multilineage differentiation that mimics the growth and host defense response of original tissue [203, 204].
As the 3D organoids have enclosed lumens that may limit their value for transport and coculture studies, recently the microinjection technique shows some advantages. For instance, microinjection of fecal microbes into colon organoids demonstrated that the complex microbiota in feces survived and grew in the colon with little change in complexity [205, 206].
In addition, Meran et al. constructed autologous jejunal mucosal grafts using biomaterials, which showed that this biological scaffold can be efficiently expanded in vitro and has highly similar biochemical properties to those in vivo, becoming a boon for patients with intestinal failure [207]. The current shortcomings and problems with this technology include the difficulty of constructing some macroscopic organ functions, technical robustness, production materials, etc. [198].
6. Conclusions and future directions
The digestive tract has an exceptional capacity for homeostasis, as demonstrated by the efficient absorption and secretion of nutrients. As well, it has a strong mucosal barrier that prevents the invasion of pathogenic bacteria and the excessive renewal of epithelial cells and tumor overgrowth, and generates anti-inflammatory components such as IgA and antimicrobial peptides against GI inflammation. Numerous GI and immunometabolic diseases are closely linked to disturbances in gut homeostasis; therefore, intestinal ECs must repair themselves after various pathogenic lesions. The renewal of the intestinal ECs depends on the regulation and support of the ISC and stem cell niches. To maintain dynamic stability, the niche needs to be continuously reshaped and adapted. Notably, the maintenance of ISC activity, which is tightly regulated by several signaling pathways, is influenced by inflammation, gut microbiota, and GMMs.
Numerous evidence has demonstrated that gut microbiota or their GMMs help maintain gut health during homeostasis and, of particular note, they play a critical role in regulating ISC activity to protect against injury. Specific bacteria (e.g., Bacillus and Lactobacillus) and GMMs (e.g., SCFAs) have been identified to directly influence ISC proliferation, renewal and differentiation. Given that the gut microbiota is comprised of a large community of microbes that can be either healthy or pathogenic, it is assumed that the interaction between gut microbes and ISCs is still largely unknown. Thus, a critical future direction for the field of ISCs is to investigate the variables and processes underlying the interaction between the intestinal microbiota and the intestinal epithelium, and more precisely, which signaling pathways or ligand factors are involved, by utilizing finer and more precise molecular markers.
It should be noted that gut microbiota may indirectly impact the stem cell niche via modulating other cells to regulate ISCs. A latest study revealed complex regulation between the gut microbiota, gut neurons, immune cells and ISCs [208]. The metabolite valeric acid from the gut microbiota promoted Tph2 expression in intestinal serotonergic neurons by blocking recruitment of the NuRD complex to the Tph2 promoter. The 5-hydroxytryptamine activated PGE2 production in the PGE2 macrophage subpopulation through its receptor HTR2A/3 A, and PGE2 promoted Wnt/β-catenin signaling by binding to EP1/EP4 to promote ISC self-renewal. A new level of ISC regulation by niche cells and gut microbiota is uncovered [208]. Therefore, it is essential to explore the complex interactions among gut microbiota, niche cells and ISCs.
In addition, the proper functioning of ISC is dependent on the influence of various signaling pathways such as Wnt, Notch, BMP, autophagy and so on. For example, the Notch pathway plays an influential role in determining cell fate. Wnt/β-catenin activity contributes to ISC differentiation as well as stability, and mutations that aberrantly activate the Wnt pathway may accelerate the proliferation of undifferentiated progenitor cells, ultimately leading to cancer. In contrast, BMP signaling inhibits the proliferation of Lgr5+ ISC at the base of the crypt. Defects in autophagy-related genes not only affect the proliferation and differentiation of ISC, but also cause abnormal secretion of antimicrobial peptides by Paneth cells, leading to diseases with impaired intestinal barrier. Other pathways such as YAP, VDR and JAK/STAT also regulate ISC activity. Indeed, in order to understand the role of ISCs in gut homeostasis, it is essential to investigating the tight regulation of ISC maintenance, proliferation, differentiation and stability.
Further, most previous studies on ISCs used drosophila or mouse models of the gut. However, in recent years, there has been a growing interest in human intestinal organoids, achieving an increasing number of exciting achievements. The interaction between ISC and other type of cells or gut microbe could be explored. The new development in primary human organoid culture and organ chip will largely facilitate basic research in precision medicine, bridging the gap between traditional in vitro high-throughput screening and in vivo studies used in disease modeling and drug development, and has potential for regenerative medicine research.
Abbreviations
AhR: aryl hydrocarbon receptor; BMP: bone morphogenetic protein; CMD: cardio-metabolic disease; CF: cystic fibrosis; Cftr: cystic fibrosis transmembrane conductance regulator; CRC: colorectal cancer; DCA: deoxycholic acid; Dclk1: dual cortisol-like kinase 1; EC: epithelial cell; FGF: fibroblast growth factor; GI: gastrointestinal; GALT: gut-associated lymphoid tissue; GMMs: gut microbial metabolites; HFD: high-fat diet; ISC: intestinal stem cell; IBD: inflammatory bowel disease; IAA: indoleacetic acid; IA: indoleacrylic acid; Iald: indole aldehyde; I3C: indole-3-carbinol; LPS: lipopolysaccharides; LCA: lithocholic acid; NAFLD: non-alcoholic fatty liver disease; Nrf2: nuclear factor erythroid 2-related factor 2; NECD: Notch extracellular domain; NICD: Notch intracellular domain; PXR: pregnane X receptor; ROS: reactive oxygen species; SCFAs: short-chain fatty acids; SBAs: secondary bile acids; TJ: tight junction; TA cells: transit-amplifying cells; T2D: type 2 diabetes; TM: transmembrane; TEAD: transcriptional enhanced associate domain; TSC2: Tuberous sclerosis complex 2; TGF-β: transforming growth factor beta; 2D: tow-dimensional; 3D: three-dimensional; UC: ulcerative colitis; VDR: vitamin D receptor.
Acknowledgements
Figures were created in BioRender.com. This study was supported by the Open Research Project Programme of the State Key Laboratory of Quality Research in Chinese Medicine (University of Macau) (Ref. No.: SKL-QRCM-OF2021006), grant from Southwest Medical University (Ref. No. 2021ZKZD017), and grant from Science and Technology Programme of Luzhou, China (Ref. No. 21CGZHPT0001).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411-20
2. Salvo Romero E, Alonso Cotoner C, Pardo Camacho C, Casado Bedmar M, Vicario M. The intestinal barrier function and its involvement in digestive disease. Rev Esp Enferm Dig. 2015;107:686-96
3. Camilleri M, Madsen K, Spiller R, Greenwood-Van Meerveld B, Verne GN. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol Motil. 2012;24:503-12
4. Barker N, Clevers H. Tracking down the stem cells of the intestine: strategies to identify adult stem cells. Gastroenterology. 2007;133:1755-60
5. Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P. et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28-41
6. Antoni L, Nuding S, Weller D, Gersemann M, Ott G, Wehkamp J. et al. Human colonic mucus is a reservoir for antimicrobial peptides. Journal of Crohn's & colitis. 2013;7:e652-64
7. Bevins C, Salzman N. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nature reviews Microbiology. 2011;9:356-68
8. Bloemendaal AL, Buchs NC, George BD, Guy RJ. Intestinal stem cells and intestinal homeostasis in health and in inflammation: A review. Surgery. 2016;159:1237-48
9. Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut Microbiota and Intestinal Trans-Epithelial Permeability. Int J Mol Sci. 2020 21
10. Francescangeli F, De Angelis ML, Zeuner A. Dietary Factors in the Control of Gut Homeostasis, Intestinal Stem Cells, and Colorectal Cancer. Nutrients. 2019 11
11. Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333-44
12. Castellazzi AM, Valsecchi C, Caimmi S, Licari A, Marseglia A, Leoni MC. et al. Probiotics and food allergy. Ital J Pediatr. 2013;39:47
13. Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp Cell Res. 2011;317:2702-10
14. Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856-64
15. Markandey M, Bajaj A, Ilott NE, Kedia S, Travis S, Powrie F. et al. Gut microbiota: sculptors of the intestinal stem cell niche in health and inflammatory bowel disease. Gut Microbes. 2021;13:1990827
16. Walker MR, Patel KK, Stappenbeck TS. The stem cell niche. J Pathol. 2009;217:169-80
17. Meran L, Baulies A, Li VSW. Intestinal Stem Cell Niche: The Extracellular Matrix and Cellular Components. Stem Cells Int. 2017;2017:7970385
18. Santos AJM, Lo YH, Mah AT, Kuo CJ. The Intestinal Stem Cell Niche: Homeostasis and Adaptations. Trends Cell Biol. 2018;28:1062-78
19. Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ. et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684-6
20. Madison B, Braunstein K, Kuizon E, Portman K, Qiao X, Gumucio D. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development (Cambridge, England). 2005;132:279-89
21. El-Salhy M. Possible role of intestinal stem cells in the pathophysiology of irritable bowel syndrome. World J Gastroenterol. 2020;26:1427-38
22. Yao Y, Kim G, Shafer S, Chen Z, Kubo S, Ji Y. et al. Mucus sialylation determines intestinal host-commensal homeostasis. Cell. 2022;185:1172-88.e28
23. Gerbe F, van Es JH, Makrini L, Brulin B, Mellitzer G, Robine S. et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192:767-80
24. Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475-9
25. Sato T, Vries R, Snippert H, van de Wetering M, Barker N, Stange D. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262-5
26. Wang R, Li H, Wu J, Cai ZY, Li B, Ni H. et al. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature. 2020;580:386-90
27. Zhang P, Holowatyj AN, Ulrich CM, Edgar BA. Tumor suppressive autophagy in intestinal stem cells controls gut homeostasis. Autophagy. 2019;15:1668-70
28. Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J. et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5:24
29. Yu Y, Lu L, Sun J, Petrof EO, Claud EC. Preterm infant gut microbiota affects intestinal epithelial development in a humanized microbiome gnotobiotic mouse model. Am J Physiol Gastrointest Liver Physiol. 2016;311:G521-32
30. Falony G, Vieira-Silva S, Raes J. Richness and ecosystem development across faecal snapshots of the gut microbiota. Nature microbiology. 2018;3:526-8
31. Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science (New York, NY). 2015;349:1101-6
32. Zeevi D, Korem T, Godneva A, Bar N, Kurilshikov A, Lotan-Pompan M. et al. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43-8
33. Structure function, diversity of the healthy human microbiome. Nature. 2012; 486: 207-14.
34. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210-5
35. Shen S, Zhong T, Peng C, Fang J, Lv B. Structural modulation of gut microbiota during alleviation of non-alcoholic fatty liver disease with Gynostemma pentaphyllum in rats. BMC complementary medicine and therapies. 2020;20:34
36. Rinninella E, Cintoni M, Raoul P, Lopetuso LR, Scaldaferri F, Pulcini G. et al. Food Components and Dietary Habits: Keys for a Healthy Gut Microbiota Composition. Nutrients. 2019 11
37. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55-71
38. Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y. et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nature medicine. 2017;23:859-68
39. Allin K, Tremaroli V, Caesar R, Jensen B, Damgaard M, Bahl M. et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61:810-20
40. Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X. et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Scientific reports. 2015;5:8096
41. Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C. et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787-94
42. Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X. et al. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell metabolism. 2019;30:675-88.e7
43. Schoenborn A, von Furstenberg R, Valsaraj S, Hussain F, Stein M, Shanahan M. et al. The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut microbes. 2019;10:45-58
44. Hausmann A, Russo G, Grossmann J, Zünd M, Schwank G, Aebersold R. et al. Germ-free and microbiota-associated mice yield small intestinal epithelial organoids with equivalent and robust transcriptome/proteome expression phenotypes. Cellular microbiology. 2020;22:e13191
45. Nigro G, Rossi R, Commere P, Jay P, Sansonetti P. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell host & microbe. 2014;15:792-8
46. Marzano M, Fosso B, Piancone E, Defazio G, Pesole G, De Robertis M. Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer. Cancers (Basel). 2021 13
47. Tao J, Duan J, Jiang S, Feng N, Qiu W, Ling Y. Chrysanthemum morifoliumPolysaccharides from Ramat ameliorate colitis rats by modulating the intestinal microbiota community. Oncotarget. 2017;8:80790-803
48. Kim S, Shin YC, Kim TY, Kim Y, Lee YS, Lee SH. et al. Mucin degrader Akkermansia muciniphila accelerates intestinal stem cell-mediated epithelial development. Gut Microbes. 2021;13:1-20
49. Wu H, Xie S, Miao J, Li Y, Wang Z, Wang M. et al. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut microbes. 2020;11:997-1014
50. Peck BC, Mah AT, Pitman WA, Ding S, Lund PK, Sethupathy P. Functional Transcriptomics in Diverse Intestinal Epithelial Cell Types Reveals Robust MicroRNA Sensitivity in Intestinal Stem Cells to Microbial Status. J Biol Chem. 2017;292:2586-600
51. Chen D, Jin D, Huang S, Wu J, Xu M, Liu T. et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer letters. 2020;469:456-67
52. Zhang X, Tong Y, Lyu X, Wang J, Wang Y, Yang R. Prevention and Alleviation of Dextran Sulfate Sodium Salt-Induced Inflammatory Bowel Disease in Mice With Bacillus subtilis-Fermented Milk via Inhibition of the Inflammatory Responses and Regulation of the Intestinal Flora. Front Microbiol. 2020;11:622354
53. Louis P, Hold G, Flint H. The gut microbiota, bacterial metabolites and colorectal cancer. Nature reviews Microbiology. 2014;12:661-72
54. Xing P, Pettersson S, Kundu P. Microbial Metabolites and Intestinal Stem Cells Tune Intestinal Homeostasis. Proteomics. 2020;20:e1800419
55. Sakata T. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. The British journal of nutrition. 1987;58:95-103
56. Sakata T, von Engelhardt W. Stimulatory effect of short chain fatty acids on the epithelial cell proliferation in rat large intestine. Comparative biochemistry and physiology A, Comparative physiology. 1983;74:459-62
57. Bartram H, Scheppach W, Schmid H, Hofmann A, Dusel G, Richter F. et al. Proliferation of human colonic mucosa as an intermediate biomarker of carcinogenesis: effects of butyrate, deoxycholate, calcium, ammonia, and pH. Cancer research. 1993;53:3283-8
58. Park J, Kotani T, Konno T, Setiawan J, Kitamura Y, Imada S. et al. Promotion of Intestinal Epithelial Cell Turnover by Commensal Bacteria: Role of Short-Chain Fatty Acids. PloS one. 2016;11:e0156334
59. Yin X, Farin H, van Es J, Clevers H, Langer R, Karp J. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nature methods. 2014;11:106-12
60. Donohoe D, Garge N, Zhang X, Sun W, O'Connell T, Bunger M. et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell metabolism. 2011;13:517-26
61. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A. et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84-96
62. Boffa L, Lupton J, Mariani M, Ceppi M, Newmark H, Scalmati A. et al. Modulation of colonic epithelial cell proliferation, histone acetylation, and luminal short chain fatty acids by variation of dietary fiber (wheat bran) in rats. Cancer research. 1992;52:5906-12
63. Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ. et al. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell. 2016;165:1708-20
64. Scheppach W, Dusel G, Kuhn T, Loges C, Karch H, Bartram H. et al. Effect of L-glutamine and n-butyrate on the restitution of rat colonic mucosa after acid induced injury. Gut. 1996;38:878-85
65. Salvi P, Cowles R. Butyrate and the Intestinal Epithelium: Modulation of Proliferation and Inflammation in Homeostasis and Disease. Cells. 2021 10
66. Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Molecular and cellular biology. 2007;27:7551-9
67. Lee Y, Kim T, Kim Y, Lee S, Kim S, Kang S. et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell host & microbe. 2018;24:833-46.e6
68. Fukui S, Shimoyama T, Tamura K, Yamamura M, Satomi M. Mucosal blood flow and generation of superoxide in rat experimental colitis induced by succinic acid. Journal of gastroenterology. 1997;32:464-71
69. Inagaki A, Ichikawa H, Sakata T. Inhibitory effect of succinic acid on epithelial cell proliferation of colonic mucosa in rats. Journal of nutritional science and vitaminology. 2007;53:377-9
70. Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell host & microbe. 2018;23:716-24
71. Alkhalaf L, Ryan K. Biosynthetic manipulation of tryptophan in bacteria: pathways and mechanisms. Chemistry & biology. 2015;22:317-28
72. Roager H, Licht T. Microbial tryptophan catabolites in health and disease. Nature communications. 2018;9:3294
73. Nicholson J, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W. et al. Host-gut microbiota metabolic interactions. Science (New York, NY). 2012;336:1262-7
74. Postler T, Ghosh S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell metabolism. 2017;26:110-30
75. Stockinger B, Di Meglio P, Gialitakis M, Duarte J. The aryl hydrocarbon receptor: multitasking in the immune system. Annual review of immunology. 2014;32:403-32
76. Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet A. et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296-310
77. Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J. et al. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13481-6
78. Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E. et al. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity. 2018;49:353-62.e5
79. Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E. et al. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity. 2018;49:353-62.e5
80. Park J, Lee J, Lee E, Hwang W, Kim D. Indole-3-Carbinol Promotes Goblet-Cell Differentiation Regulating Wnt and Notch Signaling Pathways AhR-Dependently. Molecules and cells. 2018;41:290-300
81. Wlodarska M, Luo C, Kolde R, d'Hennezel E, Annand J, Heim C. et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell host & microbe. 2017;22:25-37.e6
82. Hou Q, Ye L, Liu H, Huang L, Yang Q, Turner J. et al. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell death and differentiation. 2018;25:1657-70
83. Zelante T, Iannitti R, Cunha C, De Luca A, Giovannini G, Pieraccini G. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372-85
84. Martinez-Augustin O, Sanchez de Medina F. Intestinal bile acid physiology and pathophysiology. World journal of gastroenterology. 2008;14:5630-40
85. Kozoni V, Tsioulias G, Shiff S, Rigas B. The effect of lithocholic acid on proliferation and apoptosis during the early stages of colon carcinogenesis: differential effect on apoptosis in the presence of a colon carcinogen. Carcinogenesis. 2000;21:999-1005
86. Pai R, Tarnawski A, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Molecular biology of the cell. 2004;15:2156-63
87. Milovic V, Teller I, Faust D, Caspary W, Stein J. Effects of deoxycholate on human colon cancer cells: apoptosis or proliferation. European journal of clinical investigation. 2002;32:29-34
88. Dossa A, Escobar O, Golden J, Frey M, Ford H, Gayer C. Bile acids regulate intestinal cell proliferation by modulating EGFR and FXR signaling. American journal of physiology Gastrointestinal and liver physiology. 2016;310:G81-92
89. Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW. et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. Embo j. 2013;32:3017-28
90. Iatsenko I, Boquete JP, Lemaitre B. Microbiota-Derived Lactate Activates Production of Reactive Oxygen Species by the Intestinal NADPH Oxidase Nox and Shortens Drosophila Lifespan. Immunity. 2018;49:929-42 e5
91. Lee YS, Kim TY, Kim Y, Lee SH, Kim S, Kang SW. et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe. 2018;24:833-46 e6
92. Reedy AR, Luo L, Neish AS, Jones RM. Commensal microbiota-induced redox signaling activates proliferative signals in the intestinal stem cell microenvironment. Development. 2019 146
93. Rao J, Li L, Bass B, Wang J. Expression of the TGF-beta receptor gene and sensitivity to growth inhibition following polyamine depletion. American journal of physiology Cell physiology. 2000;279:C1034-44
94. Wang J, McCormack S, Viar M, Wang H, Tzen C, Scott R. et al. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. The American journal of physiology. 1993;265:G331-8
95. Bardócz S, Grant G, Brown D, Pusztai A. Putrescine as a source of instant energy in the small intestine of the rat. Gut. 1998;42:24-8
96. von Frieling J, Faisal MN, Sporn F, Pfefferkorn R, Nolte SS, Sommer F. et al. A high-fat diet induces a microbiota-dependent increase in stem cell activity in the Drosophila intestine. PLoS Genet. 2020;16:e1008789
97. Hou Y, Wei W, Guan X, Liu Y, Bian G, He D. et al. A diet-microbial metabolism feedforward loop modulates intestinal stem cell renewal in the stressed gut. Nat Commun. 2021;12:271
98. Singh R, Chandrashekharappa S, Bodduluri SR, Baby BV, Hegde B, Kotla NG. et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10:89
99. Ables J, Breunig J, Eisch A, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nature reviews Neuroscience. 2011;12:269-83
100. Andersson E, Lendahl U. Therapeutic modulation of Notch signalling-are we there yet? Nature reviews Drug discovery. 2014;13:357-78
101. Ntziachristos P, Lim J, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer cell. 2014;25:318-34
102. Weinmaster G, Fischer J. Notch ligand ubiquitylation: what is it good for? Developmental cell. 2011;21:134-44
103. Yuan J, Kousis P, Suliman S, Visan I, Guidos C. Functions of notch signaling in the immune system: consensus and controversies. Annual review of immunology. 2010;28:343-65
104. Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H. et al. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Letters. 2003;535:131-5
105. Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964-8
106. van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1:18
107. Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci U S A. 2005;102:12443-8
108. Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155-8
109. Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev. 2005;19:1686-91
110. Kazanjian A, Noah T, Brown D, Burkart J, Shroyer NF. Atonal homolog 1 is required for growth and differentiation effects of notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology. 2010;139:918-28 28.e1-6
111. Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U. et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377-83
112. van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H. et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959-63
113. Guo Z, Ohlstein B. Stem cell regulation. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science. 2015 350
114. Perdigoto CN, Schweisguth F, Bardin AJ. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development. 2011;138:4585-95
115. Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T. et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877-81
116. Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M. et al. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36-44
117. Lee CS, Perreault N, Brestelli JE, Kaestner KH. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488-97
118. Kim TH, Shivdasani RA. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J Biol Chem. 2011;286:11427-33
119. Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412-7
120. Peignon G, Durand A, Cacheux W, Ayrault O, Terris B, Laurent-Puig P. et al. Complex interplay between β-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 2011;60:166-76
121. Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC. et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238-42
122. Tsuchiya K, Nakamura T, Okamoto R, Kanai T, Watanabe M. Reciprocal targeting of Hath1 and beta-catenin by Wnt glycogen synthase kinase 3beta in human colon cancer. Gastroenterology. 2007;132:208-20
123. Bjerknes M, Cheng H. Cell Lineage metastability in Gfi1-deficient mouse intestinal epithelium. Dev Biol. 2010;345:49-63
124. Kapuria S, Karpac J, Biteau B, Hwangbo D, Jasper H. Notch-mediated suppression of TSC2 expression regulates cell differentiation in the Drosophila intestinal stem cell lineage. PLoS Genet. 2012;8:e1003045
125. Qu D, May R, Sureban SM, Weygant N, Chandrakesan P, Ali N. et al. Inhibition of Notch signaling reduces the number of surviving Dclk1+ reserve crypt epithelial stem cells following radiation injury. Am J Physiol Gastrointest Liver Physiol. 2014;306:G404-11
126. López-Arribillaga E, Rodilla V, Pellegrinet L, Guiu J, Iglesias M, Roman AC. et al. Bmi1 regulates murine intestinal stem cell proliferation and self-renewal downstream of Notch. Development. 2015;142:41-50
127. Srinivasan T, Than EB, Bu P, Tung KL, Chen KY, Augenlicht L. et al. Notch signalling regulates asymmetric division and inter-conversion between lgr5 and bmi1 expressing intestinal stem cells. Sci Rep. 2016;6:26069
128. Kwak SY, Shim S, Park S, Kim H, Lee SJ, Kim MJ. et al. Ghrelin reverts intestinal stem cell loss associated with radiation-induced enteropathy by activating Notch signaling. Phytomedicine. 2021;81:153424
129. Jones JC, Brindley CD, Elder NH, Myers MG Jr, Rajala MW, Dekaney CM. et al. Cellular Plasticity of Defa4(Cre)-Expressing Paneth Cells in Response to Notch Activation and Intestinal Injury. Cell Mol Gastroenterol Hepatol. 2019;7:533-54
130. Troll JV, Hamilton MK, Abel ML, Ganz J, Bates JM, Stephens WZ. et al. Microbiota promote secretory cell determination in the intestinal epithelium by modulating host Notch signaling. Development. 2018 145
131. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50-60
132. Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012 4
133. Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468-77
134. Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192-205
135. de Lau W, Peng W, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes & development. 2014;28:305-16
136. Fearon E. PARsing the phrase "all in for Axin"- Wnt pathway targets in cancer. Cancer cell. 2009;16:366-8
137. MacDonald B, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17:9-26
138. Niehrs C. The complex world of WNT receptor signalling. Nature reviews Molecular cell biology. 2012;13:767-79
139. Metcalfe C, Bienz M. Inhibition of GSK3 by Wnt signalling-two contrasting models. J Cell Sci. 2011;124:3537-44
140. Petersen C, Reddien P. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056-68
141. Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. The EMBO journal. 2012;31:2714-36
142. Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473-81
143. Kurokawa K, Hayakawa Y, Koike K. Plasticity of Intestinal Epithelium: Stem Cell Niches and Regulatory Signals. Int J Mol Sci. 2020 22
144. Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26:570-9
145. Zhang H, Lin M, Dong C, Tang Y, An L, Ju J. et al. An MST4-pβ-Catenin Signaling Axis Controls Intestinal Stem Cell and Tumorigenesis. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2021: e2004850.
146. Cordero JB, Sansom OJ. Wnt signalling and its role in stem cell-driven intestinal regeneration and hyperplasia. Acta Physiol (Oxf). 2012;204:137-43
147. Moossavi S. Location-specific effect of microbiota and MyD88-dependent signaling on Wnt/beta-catenin pathway and intestinal stem cells. Gut Microbes. 2014;5:11-4
148. Cheesman S, Neal J, Mittge E, Seredick B, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proceedings of the National Academy of Sciences of the United States of America. 2011:4570-7
149. Abo H, Chassaing B, Harusato A, Quiros M, Brazil JC, Ngo VL. et al. Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration. Nat Commun. 2020;11:513
150. Strubberg AM, Liu J, Walker NM, Stefanski CD, MacLeod RJ, Magness ST. et al. Cftr Modulates Wnt/β-Catenin Signaling and Stem Cell Proliferation in Murine Intestine. Cell Mol Gastroenterol Hepatol. 2018;5:253-71
151. Johansson J, Naszai M, Hodder MC, Pickering KA, Miller BW, Ridgway RA. et al. RAL GTPases Drive Intestinal Stem Cell Function and Regeneration through Internalization of WNT Signalosomes. Cell Stem Cell. 2019;24:592-607.e7
152. Han B, Yan S, Wei S, Xiang J, Liu K, Chen Z. et al. YTHDF1-mediated translation amplifies Wnt-driven intestinal stemness. EMBO Rep. 2020;21:e49229
153. Li Q, Sun Y, Jarugumilli GK, Liu S, Dang K, Cotton JL. et al. Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription. Cell Stem Cell. 2020;26:675-92.e8
154. Wei G, Gao N, Chen J, Fan L, Zeng Z, Gao G. et al. Erk and MAPK signaling is essential for intestinal development through Wnt pathway modulation. Development. 2020 147
155. Böttcher A, Büttner M, Tritschler S, Sterr M, Aliluev A, Oppenländer L. et al. Non-canonical Wnt/PCP signalling regulates intestinal stem cell lineage priming towards enteroendocrine and Paneth cell fates. Nat Cell Biol. 2021;23:23-31
156. Novellasdemunt L, Kucharska A, Jamieson C, Prange-Barczynska M, Baulies A, Antas P. et al. NEDD4 and NEDD4L regulate Wnt signalling and intestinal stem cell priming by degrading LGR5 receptor. Embo j. 2020;39:e102771
157. Zhou JY, Lin HL, Wang Z, Zhang SW, Huang DG, Gao CQ. et al. Zinc L-Aspartate enhances intestinal stem cell activity to protect the integrity of the intestinal mucosa against deoxynivalenol through activation of the Wnt/β-catenin signaling pathway. Environ Pollut. 2020;262:114290
158. Wang Y, Ren Y, Huang Y, Yu X, Yang Y, Wang D. et al. Fungal dysbiosis of the gut microbiota is associated with colorectal cancer in Chinese patients. American journal of translational research. 2021;13:11287-301
159. Wang R, Green J, Wang Z, Deng Y, Qiao M, Peabody M. et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes & diseases. 2014;1:87-105
160. Heldin C, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465-71
161. Date S, Sato T. Mini-gut organoids: reconstitution of the stem cell niche. Annu Rev Cell Dev Biol. 2015;31:269-89
162. Lönn P, Morén A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell research. 2009;19:21-35
163. Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol. 2021;22:39-53
164. Haramis A, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus G. et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science (New York, NY). 2004;303:1684-6
165. Batts L, Polk D, Dubois R, Kulessa H. Bmp signaling is required for intestinal growth and morphogenesis. Developmental dynamics: an official publication of the American Association of Anatomists. 2006;235:1563-70
166. Kosinski C, Li V, Chan A, Zhang J, Ho C, Tsui W. et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15418-23
167. VanDussen K, Carulli A, Keeley T, Patel S, Puthoff B, Magness S. et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development (Cambridge, England). 2012;139:488-97
168. He X, Zhang J, Tong W, Tawfik O, Ross J, Scoville D. et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nature genetics. 2004;36:1117-21
169. Aoki R, Shoshkes-Carmel M, Gao N, Shin S, May C, Golson M. et al. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cellular and molecular gastroenterology and hepatology. 2016;2:175-88
170. Reynolds A, Wharton N, Parris A, Mitchell E, Sobolewski A, Kam C. et al. Canonical Wnt signals combined with suppressed TGFβ/BMP pathways promote renewal of the native human colonic epithelium. Gut. 2014;63:610-21
171. Stzepourginski I, Nigro G, Jacob J, Dulauroy S, Sansonetti P, Eberl G. et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E506-E13
172. McCarthy N, Manieri E, Storm E, Saadatpour A, Luoma A, Kapoor V. et al. Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell stem cell. 2020;26:391-402.e5
173. Davis H, Irshad S, Bansal M, Rafferty H, Boitsova T, Bardella C. et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nature medicine. 2015;21:62-70
174. Howe J, Bair J, Sayed M, Anderson M, Mitros F, Petersen G. et al. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nature genetics. 2001;28:184-7
175. Qi Z, Li Y, Zhao B, Xu C, Liu Y, Li H. et al. BMP restricts stemness of intestinal Lgr5 stem cells by directly suppressing their signature genes. Nature communications. 2017;8:13824
176. Yum M, Han S, Fink J, Wu S, Dabrowska C, Trendafilova T. et al. Tracing oncogene-driven remodelling of the intestinal stem cell niche. Nature. 2021;594:442-7
177. Sphyris N, Hodder M, Sansom O. Subversion of Niche-Signalling Pathways in Colorectal Cancer: What Makes and Breaks the Intestinal Stem Cell. Cancers. 2021 13
178. Liu W, Singh SR, Hou SX. JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J Cell Biochem. 2010;109:992-9
179. Kazakevych J, Denizot J, Liebert A, Portovedo M, Mosavie M, Jain P. et al. Smarcad1 mediates microbiota-induced inflammation in mouse and coordinates gene expression in the intestinal epithelium. Genome Biol. 2020;21:64
180. Zhang YG, Lu R, Wu S, Chatterjee I, Zhou D, Xia Y. et al. Vitamin D Receptor Protects Against Dysbiosis and Tumorigenesis via the JAK/STAT Pathway in Intestine. Cell Mol Gastroenterol Hepatol. 2020;10:729-46
181. Goldsmith JR, Spitofsky N, Zamani A, Hood R, Boggs A, Li X. et al. TNFAIP8 controls murine intestinal stem cell homeostasis and regeneration by regulating microbiome-induced Akt signaling. Nat Commun. 2020;11:2591
182. Hong AW, Meng Z, Guan KL. The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol. 2016;13:324-37
183. Foerster E, Mukherjee T, Cabral-Fernandes L, Rocha J, Girardin S, Philpott D. How autophagy controls the intestinal epithelial barrier. Autophagy. 2022;18:86-103
184. Cadwell K, Liu J, Brown S, Miyoshi H, Loh J, Lennerz J. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259-63
185. Cadwell K, Patel K, Maloney N, Liu T, Ng A, Storer C. et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135-45
186. Adolph T, Tomczak M, Niederreiter L, Ko H, Böck J, Martinez-Naves E. et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272-6
187. Lassen K, Kuballa P, Conway K, Patel K, Becker C, Peloquin J. et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7741-6
188. Asano J, Sato T, Ichinose S, Kajita M, Onai N, Shimizu S. et al. Intrinsic Autophagy Is Required for the Maintenance of Intestinal Stem Cells and for Irradiation-Induced Intestinal Regeneration. Cell reports. 2017;20:1050-60
189. Levy A, Stedman A, Deutsch E, Donnadieu F, Virgin H, Sansonetti P. et al. Innate immune receptor NOD2 mediates LGR5 intestinal stem cell protection against ROS cytotoxicity via mitophagy stimulation. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:1994-2003
190. Trentesaux C, Fraudeau M, Pitasi CL, Lemarchand J, Jacques S, Duche A. et al. Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity. Proc Natl Acad Sci U S A. 2020;117:11136-46
191. Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190-4
192. Sato T, Stange D, Ferrante M, Vries R, Van Es J, Van den Brink S. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762-72
193. Li V. Modelling intestinal inflammation and infection using 'mini-gut' organoids. Nature reviews Gastroenterology & hepatology. 2021;18:89-90
194. Nanki K, Fujii M, Shimokawa M, Matano M, Nishikori S, Date S. et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature. 2020;577:254-9
195. Kakiuchi N, Yoshida K, Uchino M, Kihara T, Akaki K, Inoue Y. et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature. 2020;577:260-5
196. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood H, Nomburg J. et al. Mutational signature in colorectal cancer caused by genotoxic pks E. coli. Nature. 2020;580:269-73
197. Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X. et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5⁺ stem cell. Nature medicine. 2012;18:618-23
198. Bhatia S, Ingber D. Microfluidic organs-on-chips. Nature biotechnology. 2014;32:760-72
199. Bein A, Shin W, Jalili-Firoozinezhad S, Park M, Sontheimer-Phelps A, Tovaglieri A. et al. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cellular and molecular gastroenterology and hepatology. 2018;5:659-68
200. Steinway S, Saleh J, Koo B, Delacour D, Kim D. Human Microphysiological Models of Intestinal Tissue and Gut Microbiome. Frontiers in bioengineering and biotechnology. 2020;8:725
201. Kim H, Huh D, Hamilton G, Ingber D. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a chip. 2012;12:2165-74
202. Kim H, Ingber D. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integrative biology: quantitative biosciences from nano to macro. 2013;5:1130-40
203. Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A. et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Scientific reports. 2018;8:2871
204. Ma L, Barker J, Zhou C, Li W, Zhang J, Lin B. et al. Towards personalized medicine with a three-dimensional micro-scale perfusion-based two-chamber tissue model system. Biomaterials. 2012;33:4353-61
205. Williamson I, Arnold J, Samsa L, Gaynor L, DiSalvo M, Cocchiaro J. et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cellular and molecular gastroenterology and hepatology. 2018;6:301-19
206. Puschhof J, Pleguezuelos-Manzano C, Martinez-Silgado A, Akkerman N, Saftien A, Boot C. et al. Intestinal organoid cocultures with microbes. Nature protocols. 2021;16:4633-49
207. Meran L, Massie I, Campinoti S, Weston A, Gaifulina R, Tullie L. et al. Engineering transplantable jejunal mucosal grafts using patient-derived organoids from children with intestinal failure. Nature medicine. 2020;26:1593-601
208. Zhu P, Lu T, Wu J, Fan D, Liu B, Zhu X. et al. Gut microbiota drives macrophage-dependent self-renewal of intestinal stem cells via niche enteric serotonergic neurons. Cell research. 2022;32:555-69
209. Gophna U, Konikoff T, Nielsen H. Oscillospira and related bacteria - From metagenomic species to metabolic features. Environmental microbiology. 2017;19:835-41
210. Miller T, Wolin M, Conway de Macario E, Macario A. Isolation of Methanobrevibacter smithii from human feces. Applied and environmental microbiology. 1982;43:227-32
211. Tims S, Derom C, Jonkers D, Vlietinck R, Saris W, Kleerebezem M. et al. Microbiota conservation and BMI signatures in adult monozygotic twins. The ISME journal. 2013;7:707-17
212. Zhong H, Ren H, Lu Y, Fang C, Hou G, Yang Z. et al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. EBioMedicine. 2019;47:373-83
213. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262-6
214. Bryrup T, Thomsen C, Kern T, Allin K, Brandslund I, Jørgensen N. et al. Metformin-induced changes of the gut microbiota in healthy young men: results of a non-blinded, one-armed intervention study. Diabetologia. 2019;62:1024-35
215. Jie Z, Xia H, Zhong S, Feng Q, Li S, Liang S. et al. The gut microbiome in atherosclerotic cardiovascular disease. Nature communications. 2017;8:845
216. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán L, Gratadoux J. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16731-6
217. Cui X, Ye L, Li J, Jin L, Wang W, Li S. et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Scientific reports. 2018;8:635
218. Zhu L, Baker S, Gill C, Liu W, Alkhouri R, Baker R. et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology (Baltimore, Md). 2013;57:601-9
219. Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D. et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology (Baltimore, Md). 2017;65:451-64
220. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64
221. Smith M, Yatsunenko T, Manary M, Trehan I, Mkakosya R, Cheng J. et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science (New York, NY). 2013;339:548-54
222. Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200-11
Author contact
![]() Corresponding authors: Xu Wu, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-13882770623. E-mail: wuxulzedu.cn. Zhangang Xiao, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-18308330263. E-mail: zhangangxiaoedu.cn.
Corresponding authors: Xu Wu, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-13882770623. E-mail: wuxulzedu.cn. Zhangang Xiao, PhD, Laboratory of Molecular Pharmacology, Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou 646000, Sichuan, China. Tel: +86-18308330263. E-mail: zhangangxiaoedu.cn.

 Global reach, higher impact
Global reach, higher impact