Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(4):1146-1162. doi:10.7150/ijbs.80233 This issue Cite
Review
tRNA Modifications and Modifying Enzymes in Disease, the Potential Therapeutic Targets
1. Department of Thoracic Surgery, Xiangya Hospital, Central South University, Xiangya Road 87th, Changsha, 410008, Hunan, PR China.
2. Hunan Engineering Research Center for Pulmonary Nodules Precise Diagnosis & Treatment, Changsha, 410008, Hunan, PR China.
3. National Clinical Research Center for Geriatric Disorders, Changsha, 410008, Hunan, PR China.
4. Institute of Medical Sciences, Xiangya Lung Cancer Center, Xiangya Hospital, Central South University, Changsha 410008, Hunan, PR China.
5. Hunan Key Laboratory of Oncotarget Gene, Hunan Cancer Hospital & The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha 410008, Hunan, PR China.
Received 2022-10-27; Accepted 2023-1-26; Published 2023-2-13
Abstract
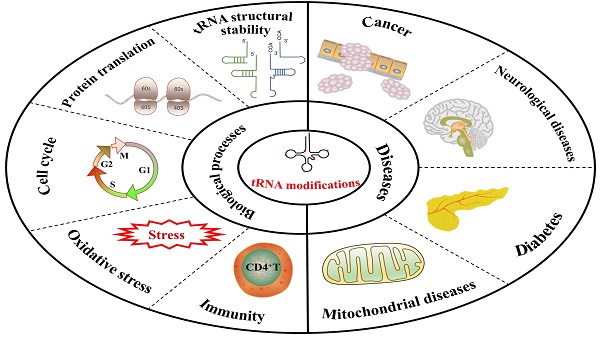
tRNA is one of the most conserved and abundant RNA species, which plays a key role during protein translation. tRNA molecules are post-transcriptionally modified by tRNA modifying enzymes. Since high-throughput sequencing technology has developed rapidly, tRNA modification types have been discovered in many research fields. In tRNA, numerous types of tRNA modifications and modifying enzymes have been implicated in biological functions and human diseases. In our review, we talk about the relevant biological functions of tRNA modifications, including tRNA stability, protein translation, cell cycle, oxidative stress, and immunity. We also explore how tRNA modifications contribute to the progression of human diseases. Based on previous studies, we discuss some emerging techniques for assessing tRNA modifications to aid in discovering different types of tRNA modifications.
Keywords: tRNA modification, tRNA modifying enzyme, signaling pathways, biomarkers
Introduction
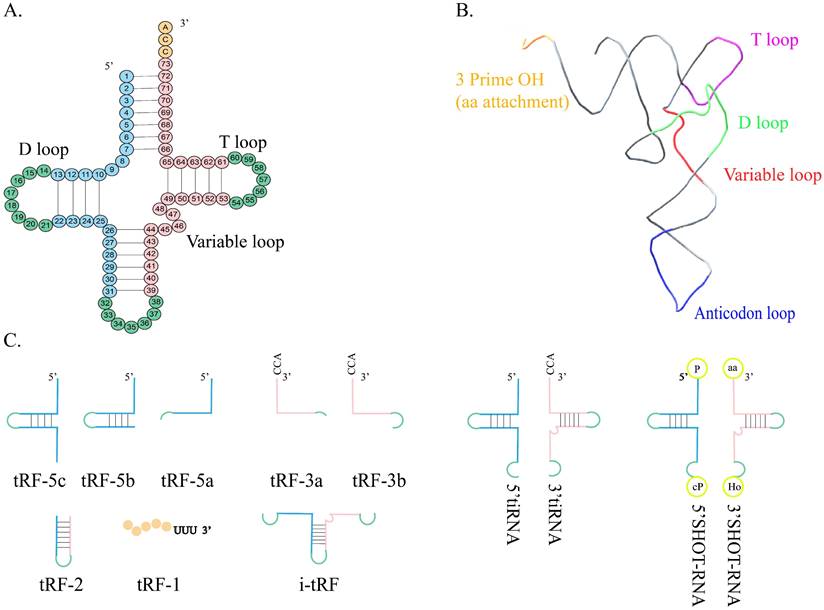
With the rapid development of high-throughput sequencing technology, tRNA-related studies are becoming more common. The classical role of tRNA is to synthesize proteins from different amino acids in the ribosome via the guidance of mRNA. In the process of biogenesis, tRNA is cleaved into different small fragments by specific enzymes including angiogenin (ANG), Dicer, and other RNA enzymes[1, 2]. These fragments are called tRNA-derived small RNA fragments (tsRNA) and can generally be divided into the following two subtypes: tRNA-half, also known as tRNA-derived stress-induced RNA (tiRNA), and tRNA-derived fragments (tRFs) (Figure 1). tsRNA is very abundant and has been identified in all three life domains, including archaea, bacteria, and certain unicellular organisms[3, 4]. tsRNA is not a randomly degraded tRNA fragment, but its production is related to tRNA chemical modification and has a precise sequence and rich biological functions[5-8]. At present, some public databases can continuously identify new tsRNA and predict their downstream targets, which helps the understanding of tRNA[9].
So far, more than 200 RNA modifications are known in all areas of life[10], about half of which are present in tRNAs, including methylation, acetylationdeamination, isomerization, glycosylation, thiolation reactions and pseudouridylation[11], and their frequency and distribution depend on the organism or tRNA species[12].
Among the modifications, methylation of tRNAs is one of the most prominent post-transcriptional modifications, occurring on nitrogen rings of almost all bases[13], mainly including N-methyladenosine/guanosine, N-methylcytidine, and 5-Methyluridine (m5U). tRNA methylation is crucial for its maturation and function execution. Methylation of tRNAs is catalyzed by methyltransferase, including the TRM10, NSUN families, and METTL families, with S-adenosylmethionine as the methyl donor, While ALKB family members catalyze demethylation[14, 15].
N4-acetylcytidine (ac4C) is usually thought of as a conservative chemically modified nucleoside present on tRNAs and rRNAs. According to studies, ac4C can only exist at position 12 in eukaryotic tRNA[16, 17]. The ac4C of tRNA is produced by N-acetyltransferase 10 with the help of THUMPD1, which is combined with tRNA[18].
Pseudouridine (Ψ) is by far the most abundant type of tRNA modification known. In eukaryotes, Ψ synthases are known as PUS, divided into 10 different types, called PUS1 to PUS10[19]. They modify uridine into Ψ by recognizing the secondary structural elements and sequence of the target uridine-containing RNA.
tRNA is a nice model for researching RNA modifications, this is because of the high cellular abundance of tRNA and the existence of a mass of modifications on tRNA. Base modifications can affect many aspects of tRNA functions, such as ensuring correct folding and stable tertiary structure of tRNA, ensuring accurate and efficient translation[20], and affecting cell heat resistance and stress[21]. Due to the importance of tRNA modifying enzymes in protein synthesis, a large number of human diseases are shown to be related to aberrant expression and dysfunction of tRNA modifying enzymes[22], including neurological disorders[23], mitochondrial diseases[24, 25], cancer[26] and diabetes mellitus[27]. Therefore, a comprehensive understanding of tRNA modifications is so necessary for us that we can better understand the physiological and pathological processes of tRNA-mediated human diseases.
Structure and classification of tRNA. A. The shamrock-like secondary structure of tRNA. B. The L-shaped tertiary structure of tRNA. C. tRF-5 is formed by the Dicer cleaving in the D loop or between the D loop and the anticodon loop of mature tRNA. tRF-3 is the 3' terminal portion of mature tRNA, containing the CCA sequence. tRF-2 contains anticodon stem-loop sequences but lacks the 5' end and 3' end of tRNA. tRF-1is derived from the cleavage of the 3' terminal of the pre-tRNA. i-tRF is generated from the interior of mature tRNA spanning anticodon regions. 5'tiRNA is from the 5' terminal of the mature tRNA to the end of the anticodon loop, and 3'-tiRNA is from the 3' end to the end of the anticodon loop. SHOT-RNA is produced in response to sex hormone stimulation and engaged by ANG.
Studying the biological function of tRNA modifications relies on continuous advances in technology to detect modifications. However, due to the complexity and specificity of RNA modifications, the detection, localization, and quantification of RNA modifications have always been a technical challenge, which has resulted in limited research on tRNA modifications in the past[28]. New technologies for RNA modification detection are constantly evolving, opening the door to a new world of tRNA modifications, but there are still some unavoidable limitations[29].
In this review, we focus on the relationship between tRNA modifications and tRNA structural stability, protein translation, cell cycle, oxidative stress, and immunity. In addition, we summarize the common types of tRNA modifications (Figure 2) and assess their possibility as novel diagnostic markers and clinical therapeutic targets in human diseases. Finally, we discuss the current limitations of this field and look ahead to where it was going in the future.
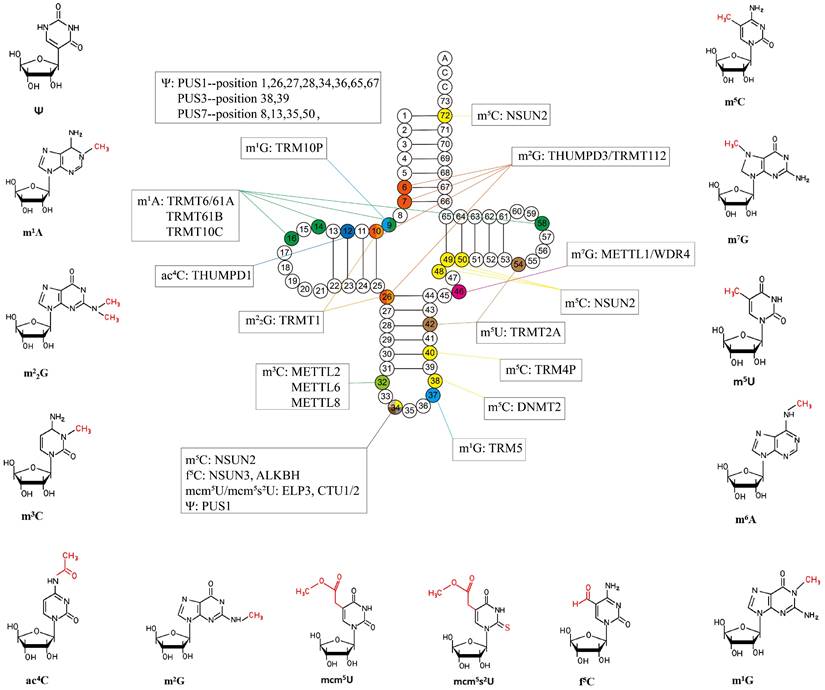
Common tRNA modifications and their chemical structures discussed in this paper. Ψ: Pseudouridine; m1A: N1-methyladenosine; m2G: N2, N2-dimethylguanosine; m5C: 5-methylcytodine; ac4C: N4-acetylcytidine; m2G: N2-methylguanosine; mcm5U: 5-methoxycarbonylmethyluridine; mcm5s2U: 5- methoxycarbonylmethyl-2-thiouridine; f5C: 5-formylcytidine; m3C: 3-methylcytodine; m7G: N7- methylguanosine; m1G: N1-methylguanosine; m6A: N6-methyladenosine; m5U: 5-methyluridine.
tRNA modifications and Biological processes
tRNA modifications and tRNA structural stability
The disruption of Watson-crick base pairing consequently affects the structure of RNAs and thereby influences gene expression and their functions[30]. Similarly, tRNA modifications can affect the secondary and tertiary structure of tRNA by disrupting Watson-crick base pairing. In tRNAs, most of the base modification sites are concentrated in two regions. One is located at the core region of the tertiary structure of tRNA and the other is in the anticodon domain[11]. The effect of modifications on the structure relies on their type and location in the tRNA, including the effects on the hydrophobic character of a base, base pairing and stacking, and charge stabilization of nucleotides[21, 31].
Modifications that occur at the core region of the tRNA structure, that is in the D-loop or T-loop, are necessary for tRNAs to maintain secondary and tertiary structural stability, particularly methylation modifications[31]. The m1A modifications occurring at site 9 (m1A9) of human mt-tRNALys damage the formation of the Watson-Crick base pair within the stem. Similar to m1A, m1G disrupts canonical base pairing by forming methyl groups on tRNA bases and thus blocking the formation of the Watson-Crick face, thereby interfering with secondary structure formation[32]. In addition, tRNA modifications can stabilize the structure of tRNAs by providing a hydrophobic or hydrophilic environment for the bases of the local structures. The m5C48 strengthens the hydrophobicity between the bases, and contributes to the stacking of bases, thus stabilizing the L-shaped tertiary structure of the tRNAs[33]. The m5C40 promotes the binding of Mg2+ to tRNA[34], which is known to stabilize the tertiary structure of tRNAs[35]. By disrupting Watson-Crick base pairing and stabilizing the core of L-shaped structure, m2G10 and m2G26 promote the secondary and tertiary folding of tRNAs[31].
In summary, methylation modifications that occur on tRNAs generally deny Watson-Crick and canonical base pairing. Furthermore, the side chain of N6-threonylcarbamoyladenosine (t6A) by intramolecular hydrogen bonds to make its planar ring be extended, by enhancing π-π stacking with neighboring bases and preventing pairing with the base at position 33 to stabilize the anticodon loop structure[36]. Ψ is widely distributed on the tRNA structure and also helps maintain the correct shape of the tRNA, which is related to its increased hydrophobicity on the one hand, and H bond formation in the anticodon ring in the tRNA on the other hand[19, 37].
Positive charges at the TΨC domain and the D-loop are related to the tertiary folding of tRNAs[38]. There is evidence that both m7G and m1A can influence the formation of non-Watson/Crick hydrogen bonds by carrying positive charges, thereby affecting the stability of tRNA structures[39]. In mitochondrial tRNA, the m3C32 modifications are associated with the folding of mt-tRNASer/Thr(UCN), this is because the m3C32 brings in a positive charge to strengthen electrostatic stability[40].
Furthermore, hypomodified tRNAs become more sensitive to the RNA degradosome and prone to rapid degradation[41]. Therefore, modified nucleotides affect the folding and structure of tRNA in a variety of ways, which is conducive to the construction of the most efficient ASL conformation for effective translation. It is worth noting that although some modifications do not affect the overall structure, they can modulate local dynamics, make tRNA's secondary and tertiary structure harmonized[42].
tRNA modifications at anticodon and Protein translation
Most post-transcriptional modifications occur at the anticodon loop of tRNA, which is crucial for the accurate synthesis of proteins at different translation steps, such as aminoacylation, decoding and translocation[43], especially at positions 34 and 37.
According to the classical wobble hypothesis in 1966, position 34 at the first anticodon is known as the “wobble” position[44]. Nucleosides at this site can bind to non-standard bases, which is called the degeneracy of codons. The modified wobble hypothesis was proposed in 1991, suggesting that specific base modifications select particular codons[45]. Since then, increasing evidence has shown that nucleoside modifications at tRNA positions 34 and 37 are crucial for the accurate and effective translation of genetic code, and modification at position 34 can limit or extend the decoding ability of tRNA[46-48]. The low modification state of the 34th position is not conducive to the combination of codons and anticodons and affects the fidelity of translation[49]. In addition, modification of position 34 was found to directly affect the stability of the anticodon ring plane base and thus affect translation[50].
Hypomodified tRNAs, such as lack of mcm5s2U34, cannot efficiently decode their cognate codons, resulting in ribosome suspension, thereby impinging protein homeostasis[51]. Q34, which occurs on mitochondrial tRNA, is thought to promote tyrosine translation in mitochondria[52]. The ac4C at the tRNAMet wobble position can improve the accuracy of tRNAs reading noninitiating AUG codons and weaken the affinity between tRNA and the codon AUG[53], as a result, the translation of the codon is reduced and ultimately affect protein synthesis. Moreover, the "distal" conformation of ac4C can hinder the misreading of the AUA codon during protein translation[54].
In addition, I34, f5C, and m5C also extend the decoding capability of tRNA[55]. In tRNA, m5C has been demonstrated to optimize codon-anticodon pairing and control translation efficiency and accuracy[56]. The m7G for the corresponding tRNAs might affect ribosome translocation. The lack of METTL1 promotes the movement of ribosomes on mRNAs, resulting in a higher frequency of m7G-tRNA decoding, indicating that m7G-tRNA modification and expression of related modifying enzymes affect translation efficiencies[57-59].
The type of modification at tRNA position 37 depends on the base at position 36[60], mainly to improve the stability of the codon and anti-codon coupling via cross-stacking with the first base of the codon[61, 62]. t6A is a generally conserved modification located at position 37 of tRNA and can promote tRNA binding to the A-site codon and efficient translocation, ensuring the efficiency and correctness of translation[36].
Studies of pseudouridine synthetase (Pus1) in S. cerevisiae have shown that Pus1-dependent pseudouridylation is very important for particular decoding events in vivo. Pus1 deletion markedly increased the codon misreading of CGC (Arg) by tRNAHis[63].
These modifications collectively affect the binding of codons to anticodons and thus affect the protein translation process.
tRNA modifications and Cell cycle
Modification of tRNA is related to the cell cycle. The level of tRNA modifications affected the aggregation status of Cdc13 and cell division[64]. It has been found that irreversible cell cycle arrests both in G1 and G2 in tad3-1 mutant cells because of an impairment in the A to I conversion at position 34 of tRNA[65]. Interestingly, there was an increase in the S phase, relative to G1 and G2, in Trm9 mutant cells due to the lack of Trm9-dependent tRNA modification (mcm⁵U)[66]. Further research has revealed that mcm⁵U affecting the cell cycle may have been achieved through the reciprocal regulation of TORC signaling and tRNA modifications by Elongator[67]. The percentage of G2 phase is increased when METTL1 knockout in mouse embryonic stem cells and intrahepatic cholangiocarcinoma (ICC) cell, lead to slower cell proliferation and colony formation ability is impaired, revealing the key role of METTL1-mediated m7G tRNA modification for the regulation of cell cycle[57, 58]. Besides, TRMT2A, which is involved in the 5-methylation of uracil located at position 54 (m5U54) of tRNA, is a potential regulator of the cell cycle in mammals[68].
tRNA modifications and Oxidative stress
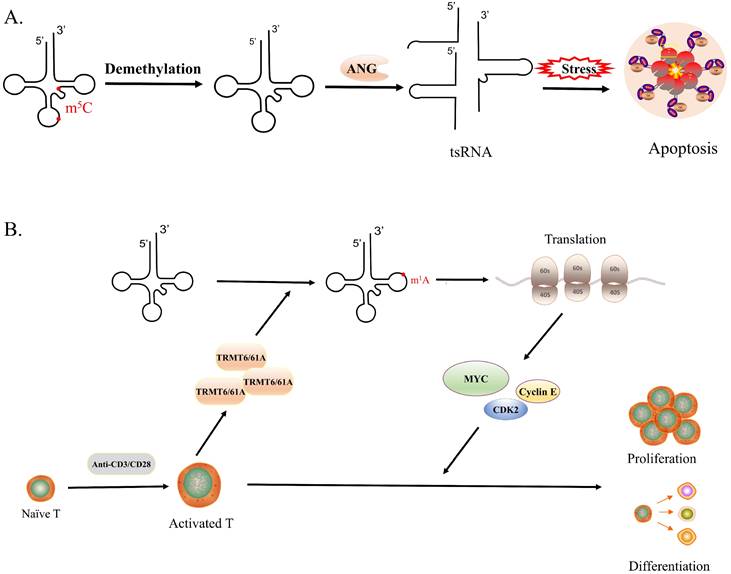
Quantitative mass spectrometry allows one to assess and compare changes in tRNA modifications abundance, which provides clear evidence that tRNA modifications, especially methylation, have a relationship with stress[69]. Specific m5C regulates the cellular stress response. Under oxidative stress, the amount of m5C at site 48 in tRNA decreased and the modification of m5C at the oscillating position increased[31]. The loss of NSUN3-mediated m5Cs in the anti-codon ring of mitochondrial tRNAMet results in reduced induction of mitochondrial ROS under stress[70]. Deletion of NSUN2-mediated tRNA methylation makes cells more sensitive to oxidative stress stimulation, while NSUN2-overexpressing cells had higher cell viability under stress (Figure 3A)[71]. In addition, after heat shock DNMT2 relocalizes to stress particles, and stress-induced cleavage of tRNAs depends on DNMT2[72]. These observations suggest that tRNA methylation is related to oxidative stress in cells.
Other studies have suggested that some rare types of tRNA modifications are also involved in oxidative stress. Biochemical experiments revealed that 3-(3-amino-3-carboxypropyl) uridine U47 (acp3U47) confers thermal stability on tRNA. When tRNA aminocarboxypropyltransferase, which is in charge of the formation of acp3U in tRNA, is knocked out of the Escherichia coli, it causes genome instability in E. coli. under sustained heat stress[73]. It has been reported that genetic analysis showing MNMA, known as the enzyme that catalyzes sulfuration of uridine at position 34 in tRNAs, is important for sustaining oxidative stress[74].
tRNA modifications and Immunity
Certain tRNA modifications are associated with infection and regulate immune function[75]. With the development of new methods, the regulation of cellular immunity mediated by tRNA modifications will become the focus of research. Guanosine 2′-O-methylation located at position 18 (Gm18) in bacterial tRNA has been revealed to antagonize tRNA-induced TLR7/8 activation, indicating the modulation of 2′-O-methylations in tRNAs participates in immune escape[76]. Similarly, data from Adeline Galvanin et al further suggest that under starvation and antibiotic stress conditions, Gm18 methylation in tRNA selectively increases, which inhibits the host immune response by the RNA/TLR7 axis[77]. However, human tRNALys3 was immunosilent despite lacking Gm18, it is due to the 2′-O-methylthymidine modification at position 54 which limits TLR7 activation[78]. The level of tRNA modification is altered during T-cell activation. It has been found that yW and ms2t6A modifications are significantly reduced during T cell proliferation, which may be to reduce the occurrence of ribosomal frameshifting and improve translational fidelity[79]. A recent study revealed that TRMT61A-mediated m1A modification of the tRNA 58 promotes the translation of multiple key proteins such as MYC by modulating codon decoding, to ensure a rapid immune response in CD4+ T cells (Figure 3B). This study, which for the first-time links tRNA-m1A58 modifications to functional changes in T cells, will provide new RNA epigenetic strategies for improving CD4+ T cell-mediated inflammatory responses and enhancing tumor immunotherapy efficacy[80]. T cell proliferation and activation involve complex molecular mechanisms, and the relationship between T cell homeostasis and tRNA modification is still largely unknown. It will be very interesting to explore how tRNA expression regulates immune response in the future, including not only tRNA modification and immune response, but also the relationship between the level of some tRFs and immune response, which would be a very valuable area of research for human health[8, 81, 82].
tRNA modifications and Diseases
The pathological consequences resulting from defects in tRNA modifications, known as 'RNA modopathies', occur in many tissues and cells[83]. Abnormal tRNA modifications affect normal translation by destabilizing tRNA, causing various diseases, including cancer, neurological diseases, diabetes, mitochondrial diseases, and so forth (Table 1). Studying the molecular mechanisms behind diseases is an essential process for diagnosing and treating diseases, but the complexity of genes requires us to make great efforts to keep exploring.
Cancer
There is increasing evidence that some tRNA modifications and the expression of the enzymes they modify are associated with cancer progression. Here we mainly introduce the relationship between several common types of tRNA modifications and cancer, including m7G, m6A, m1A, mcm5U34, m5C, and Ψ. Since there is little evidence that other types of cancer-related RNA modifications are associated with tRNAs, we will not summarize them here.
tRNA modifications and biological processes. A. The lack of m5C modification would increase the affinity between tRNA and ANG, leading to the production of large amounts of tsRNA, which would make cells more sensitive to oxidative stress and promote cell apoptosis. B. The expression of TRMT6/61 increases rapidly in the short term after T cell activation, which generates a large number of tRNA-m1A58, promoting translation and synthesizing a large number of functional proteins timely, in turn, driving the rapid proliferation and differentiation of activated T cells.
Summary of tRNA modifications in human diseases
| tRNA modifications | Diseases | References |
|---|---|---|
| m7G | GBM, LPS, Melanoma, AML, ICC, HCC, Cervical cancer, HNSCC, BC, LC, Microcephalic primordial dwarfisms, NPC, NBL, ESCC | [58, 59, 87-89, 92-95, 97-99] |
| m6A | GBM, Pancreatic cancer | [103] |
| m1A | Cervical cancer, Prostate cancer, HCC, BC, GBM, Alzheimer's disease, Mitochondrial disorders | [105, 109-111, 138, 163] |
| mcm5U34/mcm5s2U34 | BC, Breast cancer, Ovarian cancer, Colon cancer, Melanoma | [116-118, 120] |
| m5C | HNSCC, Ovarian cancer, ID, depression-like behavior, Dubowitz-like syndrome | [124, 125, 127, 132, 135] |
| m3C | Developmental delay and early-onset epileptic encephalopathy | [140] |
| ac4C | Syndromic intellectual disability | [143] |
| PUS | GBM, Neurodevelopmental disorder, Mental disorder, ALS, MLASA | [129, 145, 147, 149, 162] |
| m1G | Childhood diabetes | [156] |
| ms2t6A37 | Type 2 diabetes | [152] |
| τm5U | MELAS | [167] |
Abbreviations: GBM: glioblastoma multiforme; LPS: liposarcoma; AML: acute myeloid leukemia; HCC: hepatocellular carcinoma; ICC: intrahepatic cholangiocarcinoma; HNSCC: head and neck squamous cell carcinoma; NPC: nasopharyngeal carcinoma; ESCC: esophageal squamous cell carcinoma. LC: Lung cancer; BC: bladder cancer; NBL: neuroblastoma. ID: Intellectual Disabilities; ALS: amyotrophic lateral sclerosis; MLASA: mitochondrial myopathy with lactic acidosis and sideroblastic anemia; MELAS: mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes.
m7G in cancer
m7G located at position 46 is one of the most common tRNA modifications and has been found in eukaryotes, prokaryotes, and some archaea. Several recent studies have shown that the widespread m7G tRNA methylomes in mammals are related to the carcinogenesis and development of tumors[84-86]. The loss of METTL1 results in decreased m7G tRNA methylation and expression, inhibition of cell cycle and global translation, as well as the growth of tumors cell in many types, such as melanoma, liposarcoma (LPS), glioblastoma multiforme (GBM), and acute myeloid leukemia (AML). METTL1 mediates altered m7G-modified tRNAs and makes specific tRNAs enriched, especially tRNAArg-TCT-4-1, which increases the translation of mRNAs with the AGA codon, thus regulating the cell cycle[87, 88].
METTL1/WDR4 mediated m7G tRNA modification levels are increased in Intrahepatic cholangiocarcinoma (ICC) and are linked to poor prognosis, this may be because that METTL1/WDR4-mediated m7G tRNA modification in a selective manner affect the translation of oncogenic mRNAs, which have a higher frequency of m7G-related codons in ICC, such as genes involved in cell cycle and EGFR signaling pathway[58]. In the same way, the levels of METTL1/WDR4 are increased in hepatocellular carcinoma (HCC) and related to advanced clinical TNM stages and poor overall survival, indicating METTL1/WDR4 mediated m7G tRNA modification enhances the translation of target mRNAs through a codon-frequency-dependent mechanism[59]. A recent study linked m7G tRNA modification to radiotherapy resistance in HCC revealed the high level of METTL1 in tumor tissue is significantly related to poor prognosis in radiotherapy-treated patients with HCC[89]. Insufficient radiofrequency ablation is associated with a high recurrence of HCC. It has shown that m7G tRNA modification promotes the translation of SLUG/SNAIL under sublethal heat stress, and further makes the malignancy of METTL1 knockout HCC cells restored after sublethal heat exposure. This research elucidates that the METTL1-m7G-SLUG/SNAIL axis has the potential to be a therapeutic target for preventing metastasis of HCC after radiofrequency thermal ablation[90]. In addition, a recent study demonstrated that m7G tRNA modification is critical for enhancing lenvatinib resistance in vivo[91], and METTL1 is implicated in 5-FU sensitivity in HeLa cells[92]. These studies suggest that m7G modification is associated with drug resistance of tumor cells, and provide a promising diagnostic marker and therapeutic target for drug resistance.
Not only in digestive tumors, but the METTL1 mediated m7G tRNA was also shown to enhance the development and malignancy of head and neck squamous cell carcinoma (HNSCC) via adjusting global mRNA translation of the PI3K/AKT/mTOR molecular axis, and shown to alter immune landscape[93]. Xiaoling Ying et al showed the pathological significance of METTL1 which is an oncogene in the development of bladder cancer (BC) through the METTL1-m7G-EGFR/EFEMP1 pathway[94]. In lung cancer, METTL1 mediated m7G tRNA modification and m7G tRNA decoded codon usage, enhances the translation of mRNA and in turn promotes lung cancer progression[95]. Further study found that METTL1 may be involved in the autophagy of A549 cells, and may act via the AKT/mTORC1 pathway in LUAD cells[96]. In nasopharyngeal carcinoma (NPC), METTL1/WDR4 and m7G tRNA modification enhance the EMT process of NPC cells through the WNT/β-catenin pathway to promote NPC progression, and METTL1 is linked to the chemosensitivity of NPC cells to cisplatin and docetaxel[97]. A recent study revealed METTL1-mediated tRNA m7G modification was an independent risk factor for neuroblastoma (NBL) patients and regulated mRNA translation of MTDH and PDCD10 in the form of m7G -related codon dependent manner to promote NBL progression in mechanism[98]. In esophageal squamous cell carcinoma (ESCC), a finding provides a novel translational regulatory mechanism mediated by m7G tRNA modification, which linked autophagy with translational machinery[99]. It is confirmed that METTL1 overexpression in the ESCC cells is closely related to RPTOR/ULK1 axis, and it could be an effective strategy to treat METTL1 overactive ESCC by targeting mTOR/ULK1/autophagy signaling pathway[100]. Taken together, these data demonstrated that m7G tRNA modification can impact the carcinogenesis and development of cancer in a variety of ways, METTL1 can be used as a marker for diagnosis and prognosis and therapeutic target in cancer.
m6A in cancer
tRNA m6A methylation occurs at the nitrogen-6 position of adenine and is another important post-transcriptional modification[101]. This methylation is a dynamically reversible tRNA modification and can be reversed by demethylases, such as ALKBH5[102], which promotes the development of GBM by enhancing the proliferation and self-renewal of tumor stem cells[103]. It is reported that tRNA modified by ALKBH3 demethylase improves protein translation efficiency in cells, which is essential for tumor proliferation[104] and has been proposed as a possible therapeutic target for human pancreatic cancer[13]. But the m6A demethylase activity of ALKBH3 remains uncertain[104, 105]. In colorectal cancer, mutations of m6A regulators affect patient prognosis and may be associated with immune cell infiltration in colon tissues[106]. Besides, m6A RNA methylation regulators, as prognostic factors for colon and prostate cancer, have potential value for the treatment of related tumors and show high prospects in clinical cancer prognostic models[107, 108]. Of note, some studies of m6A in cancer require further exploration of whether they are tRNA modifications.
m1A in cancer
m1A modification has been implicated in many cancers. It can enhance the migration, proliferation, and colony formation in cervical cancer and ovarian cancer cells[109]. The high-level expression of tRNA modifying enzyme hTrm6p/hTrm61p in BC tissue causes a large release of m1A in the urine, further promotes the proliferation of cells and inhibits cell apoptosis by influencing the modification of tRNAMet in BC[110], which consistent with findings in highly aggressive GBM[111]. The m1A on tRNA can be demethylated by ALKBH3, which promotes ribosome assembly and prevent apoptosis via regulating the biogenesis of tsRNA in HeLa, DU145, and PC3 cells[105]. Notably, m1A has been found to regulate tumor development through different pathways in non-small cell lung cancer, pancreatic cancer, prostate cancer, and liver cancer[112-115], however, whether it is related to mRNA modifications or tRNA modifications needs further exploration.
mcm5U34 /mcm5s2U34 in cancer
The 5-carbonylmethyluridine (cm5U) was methylated by human TRM9L and ALKBH8 to generate 5-methoxycarbonylmethyluridine (mcm5U) at U34 of tRNA. According to the report, the level of ALKBH8 expressed in BC is high, and ALKBH8 knockout promotes apoptosis by decreasing the protein expression of anti-apoptotic factors[116]. Another study reported that human TRM9L encodes a negative regulator of tumor growth that is often suppressed in a variety of cancers, including testicular, cervical, colorectal, bladder, breast, lung, and ovarian cancer[117]. In colon cancer, phosphorylated TRM9L links oxidative stress and cell cycle control and proliferation by interacting with 14-3-3 proteins, revealing TRM9L is a crucial downstream effector of the ERK molecular axis[117]. In ovarian cancer, TRM9L regulates the expression of LIN9 to activate pRB and p53 signaling pathways and thus inhibits the proliferation of ovarian cancer cells[118]. ELP3 and CTU1/2, which are tRNA modifying enzymes acting at position 34, are essential for the invasiveness and metastases of breast cancer and linked the tRNA-dependent translation of an ITAF to the IRES-dependent translation of a key oncogenic protein[119]. In addition, U34 tRNA modification is associated with drug resistance in melanoma. BRAFV600E -expressing melanoma cells rely on U34 tRNA modifying enzymes for survival, and activation of tRNA modifying enzymes at position 34 promote the synthesis of HIF1α protein, resulting in enhanced glycolysis and conferring mTORC2‐dependent resistance to targeted BRAF inhibition. Thus, high levels of U34 tRNA modifying enzymes and HIF1α are related to the acquired resistance to anti-BRAF therapy[120, 121].
m5C in cancer
Similar to m1A, m5C is a powerful mechanism for regulating physiological processes in post-transcriptionally[122]. Dysregulation of m5C expression levels is common in various human cancers, but most current studies focus on mRNA, lncRNA or still unknown[123], while m5C is the most abundant in tRNA and rRNA[124]. Studies have shown that m5C tRNA methyltransferases are associated with drug resistance of cancer cells. Co-overexpression of NSUN2 and METTL1 can enhance the cancer cell resistance to 5-FU by stabilizing tRNA and preventing rapid tRNA degradation (RTD) pathways[92]. The study found that the tRNA methyltransferase NSUN2 in HNSCC was significantly upregulated and increased the risk of death in HNSCC patients, indicating that NSUN2 is a potential therapeutic target and prognostic marker in HNSCC[125]. In addition, the interaction between NSUN2 expression and T cell activation status affects patient survival in HNSCC, suggesting that NSUN2 is a potential immune-related marker or therapeutic target, but the molecular mechanism behind it still needs to be further explored[126]. In ovarian cancer, NSUN2/IGF-II is associated with patient survival, and the subgroup of NSUN2highIGF-IIlow has superior survival, whereas NSUN2lowIGF-IIhigh has the worst[127]. Recently, Michaela Frye's group found that m5C and f5C modifications at mitochondrial tRNAMet are required for the dynamic regulation of translation rate, and regulate the metabolic reprogramming of tumor cells, which is required for tumor invasion and metastasis from the primary site[128].
Ψ in cancer
Ψ is the most common epitranscriptomic modification. However, the biological role of Ψ that occurs on tRNA is largely unknown, especially in cancer. Studies have found PUS7 is highly expressed in GBM and predicts poor survival, moreover, PUS7 inhibitors can efficiently suppress GSC-derived tumor progression. Mechanistically, PUS7 regulates the codon-specific translational in GSCs by tRNA pseudouridylation, and regulates GSC growth via downregulating the TYK2-STAT1 pathway[129]. Currently, the inhibitor for pus7 has been identified to reduce Ψ levels of tRNA and inhibit GBM tumorigenesis and growth, providing potential therapeutic drugs for targeting PUS7 in GBM and other cancers[130].
Overall, the study of tRNA modification is a new frontier in cancer research, which not only complements new content in cancer epigenetic regulation, provides new insights into tumor development, and molecular mechanisms of drug resistance, but also it promotes the development of new tumor treatments and may be ideal targets for cancer therapy. In this chapter, we summarize the function of common tRNA modifications in cancer (Table 2) and discuss the relevant regulatory mechanisms (Figure 4), which will contribute to the further study of cancer in the future.
Overview of the regulatory network of tRNA modifications in cancer. Notes: Orange frames represent tRNA modifying enzymes. Gray frames represent protein molecules. EMT: epithelial-to-mesenchymal transition.
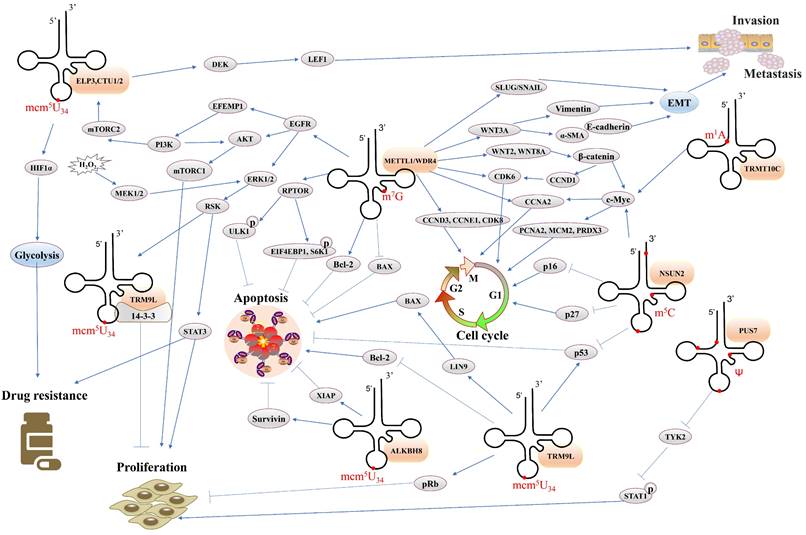
Function of tRNA modifications in cancer
| tRNA modifications | Cancers | tRNA modifying enzymes | Molecular axis | Function | References |
|---|---|---|---|---|---|
| m7G | GBM, Melanoma, Liposarcoma, AML | METTL1 (↑) | Cell cycle related proteins | Change the cell cycle, promote tumor growth | [87, 88] |
| HCC | METTL1/WDR4 (↑) | EGFR, CCNA2 | Promote cell proliferation, migration and invasion | [59] | |
| HCC | METTL1/WDR4 (↑) | SLUG/SNAIL | Promote HCC metastasis under sublethal heat exposure | [90] | |
| HCC | METTL1/WDR4 (↑) | EGFR-ERK-STAT3 | Promote cell proliferation and inhibit apoptosis | [91] | |
| ICC | METTL1/WDR4 (↑) | EGFR, AKT-mTOR, Cell cycle related proteins | Promote cell growth and colony formation, inhibit apoptosis, promote migration and invasion | [58] | |
| HNSCC | METTL1/WDR4 (↑) | PI3K-AKT-mTOR | Promote cell proliferation, migration and invasion, promote cell colony formation, inhibit apoptosis | [93] | |
| Bladder cancer | METTL1 (↑) | EGFR/EFEMP1 | Promote cell proliferation, migration and invasion | [94] | |
| Lung cancer | METTL1 (↑) | AKT/mTORC1 | Promote cell proliferation and colony formation, promote migration and invasion | [95, 96] | |
| NPC | METTL1 (↑) | ANRT/METTL1/WNT/EMT | Promote cell proliferation and colony formation, inhibit apoptosis, promote migration and invasion, and promote drug resistance | [97] | |
| Neuroblastoma | METTL1 (↑) | MTDH, PDCD10 | Promote cell proliferation and inhibit apoptosis | [98] | |
| ESCC | METTL1 (↑) | RPTOR/ULK1 | Promote cell proliferation and colony formation, inhibit apoptosis | [99, 100] | |
| m6A | Pancreatic cancer | ALKBH3 (↑) | Global translation | Promote cell growth | [104] |
| Cervical cancer | TRMT10C (↑) | c-Myc related pathway | Promote cell proliferation, migration and colony formation | [109] | |
| m1A | Cervical cancer, Prostate cancer, HCC | ALKBH3 (↑) | Enhance the sensitivity of tRNA to ANG and promote the binding of tDR and Cyt-c | Promote cell proliferation and inhibit apoptosis | [105] |
| GBM | hTRM6p/61p (↑) | PKCα related pathway | Promote cell colony formation and pelleting ability, and promote invasion | [111] | |
| mcm5U34 /mcm5s2U34 | Bladder cancer | ALKBH8 (↑) | Survivin/XIAP | Promote cell proliferation and inhibit apoptosis | [116] |
| Colon cancer | TRM9L (↓) | H2O2/ERK/RSK/14-3-3 | Inhibit cell proliferation | [117] | |
| Ovarian cancer | TRM9L (↓) | LIN9/Bax/Bcl-2 | Promote cell apoptosis and inhibit cell proliferation | [118] | |
| Breast cancer | ELP3, CTU1/2 (↑) | ELP3-CTU1/2-DEK-IRES-LEF1 | Promote cell invasion and metastasis | [119] | |
| Melanoma | ELP1, ELP3, CTU1/2 (↑) | PI3K/mTORC2 | Promote drug resistance | [120, 121] | |
| m5C | HNSCC | NSUN2 (↑) | c-Myc related pathway | Promote tumor growth | [125] |
| Ψ | GBM | PUS7 (↑) | TYK2/STAT1 | Promote cell growth and self-renewal | [129, 130] |
Notes: ↑, upregulated; ↓, downregulated.
Abbreviations: EMT: epithelial-mesenchymal transition; ANG: angiotensin; GBM: glioblastoma multiforme; AML: acute myeloid leukemia; HCC: hepatocellular carcinoma; ICC: intrahepatic cholangiocarcinoma; HNSCC: head and neck squamous cell carcinoma; NPC: nasopharyngeal carcinoma; ESCC: esophageal squamous cell carcinoma.
Neurological diseases
The effect of tRNA modifications on the nervous system is an emerging aspect of neuroscience. More and more genes involved in tRNA modifications have been proved to be related to neurodevelopmental disorders[131], suggesting the increasing importance of tRNA modifications in human neural development. It is essential for us to know the functions of these tRNA modifying genes or enzymes, which will help us to realize their exact biological roles, and to develop new therapeutic strategies by controlling the expression of tRNA modifications related genes.
Methylation modification and neurological diseases
In patients with mutations in the NSUN2 gene, the cells lacked the corresponding protein, resulting in the deficiency of site-specific 5-cytosine methylation at C47 and C48 of the tRNAAsp, and the patients present with moderate to severe intellectual impairment, facial deformities, and distal myopathy[124, 132, 133]. NSUN2-mediated tRNA methylation is indispensable for the migration and differentiation of intermediate progenitors in the ventricular zone toward the upper-layer neurons. In neural development, deficiency of NSUN2-mediated methylation increases the sensitivity of tRNA to ANG, resulting in the accumulation of more tRF-5 in the brain, which will interfere with differentiation of neuroepithelial stem cells, and reduce response sensitivity to growth factors and decrease the number of upper neurons, eventually resulting in neurodevelopmental defects[134]. Loss of NSUN2-mediated tRNA methylation causes a selective enrichment of tRF-5 in mice, impairs survival of striatal, cortical and hippocampal neurons by triggering cellular stress responses and cell death[71]. Recently, NSUN2-mediated tRNA cytosine methylation was found to be related to emotion. NSUN2 overexpression in the cerebral cortex produces depression-like behavior, and NSUN2 deletion produces an anti-depressant phenotype[135]. Patients with NSUN3 mutations exhibit early-onset mitochondrial encephalopathy and seizures, possibly due to impaired mt-tRNAMet methylation affecting mitochondrial translation[136, 137].
Recent studies have shown heterotropic m1A methylation in the mitochondrial tRNA of these mice by mapping the abundant m1A tRNA modification in the cerebral cortex of mouse models of Alzheimer's disease[138]. Silzer TK et al also reported significant hypermethylation of specific sites at position 9 of mt-tRNA in the cerebellar tissue of individuals with progressive supranuclear palsy and Alzheimer's disease[139], suggesting that hypo m1A modification of mt-tRNA is linked to neurodegenerative diseases.
In addition, the lack of DARLD3 in human cells blocks the m3C formation of tRNAArg. It will suffer from early-onset epileptic encephalopathy and severe developmental delay if human individuals with a deficiency of the DALRD3 gene, indicating that m3C-mediated tRNA modification in mammalian tRNAArg is related to nervous system function[140].
Studies have shown that m7G tRNA modification is involved in neural differentiation and brain development. Mutations in WDR4, which interacts with METTL1 to form a complex, result in defective m7G-mediated tRNA modification and interfere with the correct expression of genes on the neural spectrum[57]. Biallelic variants occurring in the WDR4 gene lead to microcephalic primordial dwarfisms, characterized by severe growth retardation and microcephaly[141].
The latest case report shows biallelic variant in the domain of ALKBH8 causes syndromic intellectual disability, adding a new variant to the ever-expanding list of tRNA modifications related to ALKBH8[142].
ac4C and the nervous system
THUMPD1 is participated in regulating ac4C modification of tRNA. In a recent study, Martin Broly et al reported that 13 individuals from 8 families had a rare mutation of THUMPD1 dysfunction that resulted in a deficiency of ac4C modification of individually purified tRNASer-CGA[143]. The results of research show that the loss of tRNA acetylation due to lack of THUMPD1 function leads to syndromic intellectual disability, this is because the lack of tRNA modification may lead to impaired protein synthesis and thereby affecting the proteostasis at important stages of neural development.
Ψ and the nervous system
PUS3 mutations can cause a rare neurodevelopmental disorder[144]. Recently, a novel homozygous truncating mutation in PUS3 was found to be associated with intellectual impairment, patients with this mutation had dropped levels of uracil isomerization at tRNA positions 38 and 39[145]. Patients with the PUS7 mutation showed delayed speech and aggressive behavior, and experiments in fruit flies demonstrated that this behavioral deficit was caused by the PUS7 mutation[146].
Furthermore, the adenosine deaminase tRNA-specific 2/3 (ADAT2/ADAT3) complex can catalyze adenosine deamination, called inosine. It can lead to an autosomal recessive genetic mental disorder if ADAT2/3 is mutated[147, 148]. The EPL3, a subunit of the elongator complex that modifies the uridine at the tRNA wobble position, is low expressed in the motor cortex of amyotrophic lateral sclerosis (ALS), which may be related to mcm5s2U-modified tRNA levels[149].
Type 2 diabetes
The study of diabetes genetics has changed due to rapid advances in sequencing technologies. Among the risk factors associated with diabetes, some disorders of tRNA modifications are thought to be related to type 2 diabetes[27].
CDKAL1, a tRNA methylthiotransferase, is located at position 37 of cytoplasmic tRNALys[150, 151]. Genetic variations in the CDKAL1 gene are linked to type 2 diabetes in different ethnic groups. It showed hypertrophy of islets, reduced insulin secretion, and impaired glycemic control, which are typical phenotypes related to type 2 diabetes in CDKAL1 knockout mice[150]. Misreading of the Lys codon in CDKAL1-deficient β cells leads to reduced glucose-stimulated proinsulin synthesis, this may be related to the 2-methylthio-modification (ms2-) of N6-threonylcarbonyladenosine at position-37 (ms2t6A37) in tRNAUUULys3[152]. Insulin levels in vivo are regulated by ferrimodulin 2 (Irp2) through iron-mediated CDKAL1-catalyzed tRNA modification. The loss of Irp2 leads to impaired iron-sulfur cluster biosynthesis and functional iron deficiency in the β-cells, and reduces the activity of ferrithionein CDKAL1, resulting in decreased insulin synthesis[153, 154]. These findings suggest that a fully modified tRNALys (UUU) may be required for the correct translation of insulin mRNA.
Interestingly, TRMT10A, a tRNA methyltransferase, methylates guanosine at position 9 of tRNA, has been proved that lack of TRMT10A sensitizes β-cells to apoptosis[155]. Recently, several cases have been reported that TRMT10A is associated with childhood diabetes. In addition to diabetes, the patients also suffered from microcephaly and intellectual disability, and genotype testing suggested homozygous mutations in the TRMT10A gene[156-158].
Mitochondrial disease
Mitochondrial disease, also known as mitochondrial encephalopathy, is a disorder caused by mitochondrial dysfunction with diverse clinical phenotypes, including blindness, deafness, dyskinesia and myopathy[159, 160]. The absence of tRNA modification in mitochondria often leads to pathological consequences[161]. For example, a missense mutation in PUS1 is associated with the disorder mitochondrial myopathy with lactic acidosis and sideroblastic anemia (MLASA) in humans[162]. Besides, mutations in TRMT10C affect mt-tRNA processing and mitochondrial protein synthesis, leading to mitochondrial diseases including deafness in newborns[163].
NSUN3 is an m5C methyltransferase of mt-tRNA, specifically for the 'wobble' 34th position of mt-tRNAMet[137]. Most mt-tRNAMet-C34 forms m5C and further oxidized by ALKBH1 to generate f5C[164, 165]. Deficiency of NSUN3 or ALKBH1 affects mitochondrial translation, results in reduced cell proliferation and may be linked to early-onset mitochondrial encephalomyopathy and seizures[136].
Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) and Myoclonic Epilepsy with Ragged Red Fibers (MERRF)[166] are a group of mitochondrial diseases caused by the lack of taurine modification at the first anticodon nucleotide of mt-tRNALeu(UUR) [167]. Due to the absence of naturally occurring modified nucleoside 5-taurinomethyluridine (τm5U) located at the wobble position (C34) of tRNALeu, resulting in errors in codon translation[168]. High dose of taurine can improve mitochondrial tRNALeu (UUR) modification defect in peripheral blood leukocytes and inhibit stroke in MELAS patients[169]. Mtu1(Trmu) is a kind of highly conserved tRNA modifying enzyme, and associated with the modification of τm5s2U at the 'wobble' 34th position of tRNALys, tRNAGlu and tRNAGln[170]. These abnormal modifications affect the translation function by affecting mitochondrial respiration and thus lead to auditory defects in zebrafish[170] and suppressed osteogenic differentiation in mice[171].
Conclusion & perspective
In summary, the researches on the biological functions of tRNA modifications have made great progress, and become a hot topic in the field of RNA modifications. But there are still many problems to be explored.
The detection technologies of tRNA modifications have limited the development of research. Due to the rich variety of tRNA modification types and their elusive function, the detection, quantification and localization of these tRNA modifications are crucial for understanding the elusive function of tRNA modifications. For the traditional high-throughput RNA sequencing technology, on the one hand, they will affect the preparation of the RNA-seq library, and on the other hand, they cannot implement deep sequencing for some highly modified tsRNA. To solve the problems of existing sequencing technologies, new technologies have been developed and applied, such as PANDORA-Seq[172], mim-tRNA seq[173] and Absolute Quantification RNA-seq[174]. These new techniques alleviate the bias in traditional tRNA sequencing, accurately measure the abundance of tRNA in cells, and reveal the quantitative map of tRNA modifications. However, there are still some unavoidable limitations, and there is no general and precise method to quantify the modification in tRNA. We urgently need to develop more sensitive and reliable detection methods to rapidly and quantitatively detect tRNA modifications, and open new avenues for exploration of more types of RNA modifications in the future.
tRNA modification is driven by various modifying enzymes, and a variety of modifying enzymes directly affect the biological function of tRNA. The existence of tRNA modification enhances the stability of tRNA structure, improves the accuracy of decoding and the efficiency of translation during protein synthesis, and plays an important role in cell cycle and stress. But our understanding of how tRNA modifications facilitate these processes remains incomplete. Considering that the spatiotemporal order of tRNA modifications plays an integral role in its function[175, 176], but related studies are still very rare, it would be interesting to investigate the synergistic mechanism of different modifying enzymes. The tRNA acetylation has also been reported to play an essential role in antibiotic resistance[177], and other types of tRNA modifications were mentioned above for chemoresistance in tumor patients. Therefore, the mechanism of tRNA modifications in drug resistance and immune escape is also worthy of further exploration. In recent years, with the deepening of related studies on tRNA modifications, its effects on human diseases are gradually being revealed[22], especially the effect of tRNA modifying enzymes on cancer. Although there is evidence that aberrant expression of some modifying enzymes is significantly associated with human pathological processes[178], the specific molecular pathways and interactions between tRNA modifications and other noncoding RNA modifications should be further explored. All in all, these studies will help us understand the function of tRNA modifications in epigenetics.
Clinical potential of tRNA modifying enzymes in cancer
| Modifying enzymes | Cancer | Clinical potential | References |
|---|---|---|---|
| METTL1 | HCC | Promote radiotherapy resistance of HCC and is significantly associated with poor prognosis of HCC after radiotherapy | [89] |
| METTL1 | HCC | Promote lenvatinib resistance in HCC | [91] |
| METTL1/WDR4 | HCC | Overexpressed in HCC, correlated with tumor stage and patient survival | [59] |
| METTL1/WDR4 | HCC | Insufficient radiofrequency ablation promotes HCC metastasis | [90] |
| METTL1/WDR4 | ICC | Overexpressed in ICC, and patients with high expression have poor survival and are more likely to relapse | [58] |
| NSUN2, METTL1 | Cervical cancer | Combined knockdown of NSUN2 and METTL1 significantly enhanced the sensitivity of Hela cells to 5-FU | [92] |
| METTL1/WDR4 | HNSCC | Overexpressed in HNSCC and correlates with tumor stage and overall survival | [93] |
| NSUN2 | HNSCC | Overexpressed in HNSCC, leads to short overall survival and a high risk of death | [125] |
| NSUN2 | HNSCC | Interaction with T cell activation, affects the survival of HNSCC patients | [126] |
| NSUN2 | Ovarian cancer | Overexpressed in ovarian cancer, and it has the worst survival in the low NSUN2 and high IGF-II subgroups | [127] |
| METTL1 | BC | Overexpressed in BC, and patients with high expression have poor disease-free survival | [94] |
| hTRM6p/61p | BC | Promote the high discharge of m1A in the urine of BC patients | [110] |
| ALKBH8 | BC | Overexpressed in advanced bladder cancer | [116] |
| METTL1/WDR4 | LUAD | Overexpressed in LUAD and leads to poor survival | [95, 96] |
| METTL1 | NPC | Overexpressed in NPC, which is related to TNM stage and patient survival, and promotes drug resistance | [97] |
| METTL1 | NBL | Overexpressed in advanced NBL and leads to poor prognosis | [98] |
| METTL1 | ESCC | Overexpressed in ESCC, correlates with patient survival and tumor TNM stage | [99] |
| ALKBH3 | Pancreatic cancer | Overexpressed in pancreatic cancer and knockdown inhibits tumor cell growth | [104] |
| ELP1, ELP3, CTU1/2 | Melanoma | Promote drug resistance in melanoma | [120, 121] |
| PUS7 | GBM | Overexpressed in GBM and leads to poor survival | [129, 130] |
Abbreviations: GBM: glioblastoma multiforme; HCC: hepatocellular carcinoma; ICC: intrahepatic cholangiocarcinoma; HNSCC: head and neck squamous cell carcinoma; NPC: nasopharyngeal carcinoma; ESCC: esophageal squamous cell carcinoma. LUAD: Lung adenocarcinoma; BC: bladder cancer; NBL: neuroblastoma.
What is more, the clinical potential of tRNA modifying enzymes has also attracted widespread attention. Studies have revealed that levels of tRNA modification are related to tumor progression, and some high levels of tRNA modifications can be detected in the urine or blood of cancer patients, suggesting that tRNA-modifying enzymes may be new markers for cancer diagnosis and prognosis (Table 3). In addition, tRNA-modifying enzymes and their inhibitors can promote or suppress tumor cell phenotypes, implying their potential as new therapeutic targets. With the development of precision medicine, it will be an attractive strategy for human cancer treatment that targeting tRNA modification systems. However, this kind of research is still in its infancy, what we know about the basic biological functions of tRNA modifications still has a large gap, and there are still limitations in the development of tRNA modifying enzyme inhibitors. We still have a long way to go before it can be applied to clinical practice.
Our ultimate goal is to solve human pain, so it is the top priority to explore the mechanism of action of various modifications and modified enzymes in various diseases, so as to develop targeted drugs to provide new possibilities for disease treatment or to provide new markers for disease diagnosis. In conclusion, we need to reveal the occurrence and function of tRNA modifications from different perspectives.
Abbreviations
tRNA: transfer RNA; mRNA: messenger RNA; tsRNA: tRNA-derived small RNA fragments; tiRNA: tRNA-derived stress-induced RNA; tRF: tRNA-derived fragment; m5U: 5-Methyluridine; ac4C: N4-acetylcytidine; m1A: N1-methyladenosine; m5C: 5-methylcytodine; m2G: N2-methylguanosine; m22G: N2, N2-dimethylguanosine; t6A: N6-Threonylcarbamoyladenosine; m7G: N7-methylguanosine; m1G: N1-methylguanosine; m3C: 3-methylcytosine; mcm5U: 5-methoxycarbonylmethyluridine; mcm5s2U: 5-methoxycarbonylmethyl-2-thiouridine; Q34: Queuosine 34; m5U: 5-methyluridine; acp3U47: 3-(3-amino-3-carboxypropyl) uridine U47; f5C: 5-formylcytidine; m6A: N6-methyladenosine; ms2t6A: 2-methylthio-modification of N6-threonylcarbonyladenosine; Gm: Guanosine 2′-O-methylation; DNMT2: DNA methyltransferase2; TRM10: transfer RNA methyltransferase 10; NSUN: NOP2/Sun RNA methyltransferase; METTL1: methyltransferase like 1; WDR4: WD Repeat Domain 4; THUMPD1: THUMP Domain Containing 1; PUS: Pseudouridine Synthase; TRM9L: transfer RNA methyltransferase 9-like gene; DARLD3: domain containing 3; ALKBH: ALKB homolog; THUMPD1: THUMP Domain Containing 1; CDKAL1: CDK5 Regulatory Subunit Associated Protein 1 Like 1; TRMT10A: TRNA Methyltransferase 10 Homolog A; TRMT61A: TRNA Methyltransferase 61 Homolog A; TRMU: tRNA mitochondrial 2-thiouridylase; TRMT2A: TRNA Methyltransferase 2 Homolog A; ROS: reactive oxygen species; MELAS: mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes; ESCs: embryonic stem cells; ICC: intrahepatic cholangiocarcinoma; GBM: glioblastoma multiforme; LPS: liposarcoma; AML: acute myeloid leukemia; HCC: hepatocellular carcinoma; HNSCC: head and neck squamous cell carcinoma; BC: bladder cancer; LC: lung cancer; NPC: nasopharyngeal carcinoma; ESCC: esophageal squamous cell carcinoma; ID: Intellectual Disabilities; ALS: amyotrophic lateral sclerosis; MLASA: mitochondrial myopathy with lactic acidosis and sideroblastic anemia; MERRF: Myoclonic Epilepsy with Ragged Red Fibers; NBL: neuroblastoma; LUAD: Lung adenocarcinoma; ANG: angiotensin; RTD: rapid tRNA degradation; EMT: epithelial-mesenchymal transition.
Acknowledgements
Funding
This work was supported by the National Natural Science Foundation of China (No. 81974367, No. 82003065, No. 82172655), the National Natural Science Foundation of Hunan Province (No. 2020JJ4132, No. 2021JJ70023), and National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases (Lung Cancer, No. z027002).
Author Contributions
CD designed the study. WC and CD drafted the manuscript. WC, DZ, JJ, CZ and CD collected the data and conducted the picture processing. WC, FT and CD revised the manuscript. All authors have read and approved the final version of manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Akiyama Y, Lyons SM, Fay MM, Tomioka Y, Abe T, Anderson PJ. et al. Selective Cleavage at CCA Ends and Anticodon Loops of tRNAs by Stress-Induced RNases. Front Mol Biosci. 2022;9:791094
2. Di Fazio A, Schlackow M, Pong SK, Alagia A, Gullerova M. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic Acids Res. 2022;50:1734-52
3. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78
4. Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909
5. Zong T, Yang Y, Zhao H, Li L, Liu M, Fu X. et al. tsRNAs: Novel small molecules from cell function and regulatory mechanism to therapeutic targets. Cell Prolif. 2021;54:e12977
6. Liu B, Cao J, Wang X, Guo C, Liu Y, Wang T. Deciphering the tRNA-derived small RNAs: origin, development, and future. Cell Death Dis. 2021;13:24
7. Su Z, Wilson B, Kumar P, Dutta A. Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annu Rev Genet. 2020;54:47-69
8. Shan N, Li N, Dai Q, Hou L, Yan X, Amei A. et al. Interplay of tRNA-Derived Fragments and T Cell Activation in Breast Cancer Patient Survival. Cancers (Basel). 2020 12
9. Li N, Shan N, Lu L, Wang Z. tRFtarget: a database for transfer RNA-derived fragment targets. Nucleic Acids Res. 2021;49:D254-D60
10. Amalric A, Bastide A, Attina A, Choquet A, Vialaret J, Lehmann S. et al. Quantifying RNA modifications by mass spectrometry: a novel source of biomarkers in oncology. Crit Rev Clin Lab Sci. 2022;59:1-18
11. Grosjean H, Edqvist J, Stråby KB, Giegé R. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J Mol Biol. 1996;255:67-85
12. Krutyhołowa R, Zakrzewski K, Glatt S. Charging the code - tRNA modification complexes. Curr Opin Struct Biol. 2019;55:138-46
13. Huang H, Li H, Pan R, Wang S, Liu X. tRNA modifications and their potential roles in pancreatic cancer. Arch Biochem Biophys. 2021;714:109083
14. Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst). 2007;6:429-42
15. Motorin Y, Helm M. RNA nucleotide methylation: 2021 update. Wiley Interdiscip Rev RNA. 2022;13:e1691
16. Kowalski S, Yamane T, Fresco JR. Nucleotide sequence of the "denaturable" leucine transfer RNA from yeast. Science. 1971;172:385-7
17. Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139-40
18. Sharma S, Langhendries JL, Watzinger P, Kötter P, Entian KD, Lafontaine DL. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242-58
19. Rintala-Dempsey AC, Kothe U. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;14:1185-96
20. Han L, Phizicky EM. A rationale for tRNA modification circuits in the anticodon loop. RNA. 2018;24:1277-84
21. Lorenz C, Lünse CE, Mörl M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules. 2017 7
22. Chujo T, Tomizawa K. Human transfer RNA modopathies: diseases caused by aberrations in transfer RNA modifications. FEBS J. 2021;288:7096-122
23. Ramos J, Fu D. The emerging impact of tRNA modifications in the brain and nervous system. Biochim Biophys Acta Gene Regul Mech. 2019;1862:412-28
24. Asano K, Suzuki T, Saito A, Wei FY, Ikeuchi Y, Numata T. et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 2018;46:1565-83
25. Bohnsack MT, Sloan KE. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell Mol Life Sci. 2018;75:241-60
26. Rapino F, Delaunay S, Zhou Z, Chariot A, Close P. tRNA Modification: Is Cancer Having a Wobble? Trends Cancer. 2017;3:249-52
27. Wei FY, Tomizawa K. tRNA modifications and islet function. Diabetes Obes Metab. 2018;20(Suppl 2):20-7
28. Sarkar A, Gasperi W, Begley U, Nevins S, Huber SM, Dedon PC. et al. Detecting the epitranscriptome. Wiley Interdiscip Rev RNA. 2021;12:e1663
29. Yolu Y, Ammann G, Barraud P, Jora M, Limbach PA, Motorin Y. et al. Instrumental analysis of RNA modifications. Crit Rev Biochem Mol Biol. 2021;56:178-204
30. Lu L, Katsaros D, Mayne ST, Risch HA, Benedetto C, Canuto EM. et al. Functional study of risk loci of stem cell-associated gene lin-28B and associations with disease survival outcomes in epithelial ovarian cancer. Carcinogenesis. 2012;33:2119-25
31. Väre VY, Eruysal ER, Narendran A, Sarachan KL, Agris PF. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules. 2017 7
32. Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574-85
33. Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes (Basel). 2019 10
34. Chen Y, Sierzputowska-Gracz H, Guenther R, Everett K, Agris PF. 5-Methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem-loop of yeast phenylalanine tRNA. Biochemistry. 1993;32:10249-53
35. Xu Y, MacKerell AD Jr, Nilsson L. Structural effects of modified ribonucleotides and magnesium in transfer RNAs. Bioorg Med Chem. 2016;24:4826-34
36. Lin H, Miyauchi K, Harada T, Okita R, Takeshita E, Komaki H. et al. CO(2)-sensitive tRNA modification associated with human mitochondrial disease. Nat Commun. 2018;9:1875
37. Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492-501
38. Nobles KN, Yarian CS, Liu G, Guenther RH, Agris PF. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002;30:4751-60
39. Agris PF, Sierzputowska-Gracz H, Smith C. Transfer RNA contains sites of localized positive charge: carbon NMR studies of [13C]methyl-enriched Escherichia coli and yeast tRNAPhe. Biochemistry. 1986;25:5126-31
40. Kleiber N, Lemus-Diaz N, Stiller C, Heinrichs M, Mai MM, Hackert P. et al. The RNA methyltransferase METTL8 installs m(3)C(32) in mitochondrial tRNAs(Thr/Ser(UCN)) to optimise tRNA structure and mitochondrial translation. Nat Commun. 2022;13:209
41. Kimura S, Waldor MK. The RNA degradosome promotes tRNA quality control through clearance of hypomodified tRNA. Proc Natl Acad Sci U S A. 2019;116:1394-403
42. Biedenbänder T, de Jesus V, Schmidt-Dengler M, Helm M, Corzilius B, Fürtig B. RNA modifications stabilize the tertiary structure of tRNAfMet by locally increasing conformational dynamics. Nucleic Acids Res. 2022;50:2334-49
43. Zhou JB, Wang ED, Zhou XL. Modifications of the human tRNA anticodon loop and their associations with genetic diseases. Cell Mol Life Sci. 2021;78:7087-105
44. Crick FH. Codon-anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548-55
45. Agris PF. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie. 1991;73:1345-9
46. Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R. et al. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem. 2002;277:16391-5
47. Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1-13
48. Agris PF, Narendran A, Sarachan K, Väre VYP, Eruysal E. The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity. Enzymes. 2017;41:1-50
49. Patil A, Chan CT, Dyavaiah M, Rooney JP, Dedon PC, Begley TJ. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 2012;9:990-1001
50. Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y. et al. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J Biol Chem. 2008;283:18801-11
51. Nedialkova DD, Leidel SA. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 2015;161:1606-18
52. Suzuki T, Yashiro Y, Kikuchi I, Ishigami Y, Saito H, Matsuzawa I. et al. Complete chemical structures of human mitochondrial tRNAs. Nat Commun. 2020;11:4269
53. Kawai G, Hashizume T, Miyazawa T, McCloskey JA, Yokoyama S. Conformational characteristics of 4-acetylcytidine found in tRNA. Nucleic Acids Symp Ser. 1989:61-2
54. Kumbhar BV, Kamble AD, Sonawane KD. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at "wobble" 34th position in the anticodon loop of tRNA. Cell Biochem Biophys. 2013;66:797-816
55. Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. 2016;12:546-51
56. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H. et al. Biological roles of RNA m(5)C modification and its implications in Cancer immunotherapy. Biomark Res. 2022;10:15
57. Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-Mediated m(7)G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol Cell. 2018;71:244-55.e5
58. Dai Z, Liu H, Liao J, Huang C, Ren X, Zhu W. et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell. 2021;81:3339-55.e8
59. Chen Z, Zhu W, Zhu S, Sun K, Liao J, Liu H. et al. METTL1 promotes hepatocarcinogenesis via m(7) G tRNA modification-dependent translation control. Clin Transl Med. 2021;11:e661
60. Anton BP, Russell SP, Vertrees J, Kasif S, Raleigh EA, Limbach PA. et al. Functional characterization of the YmcB and YqeV tRNA methylthiotransferases of Bacillus subtilis. Nucleic Acids Res. 2010;38:6195-205
61. Murphy FVT, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol. 2004;11:1186-91
62. Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005;44:8078-89
63. Khonsari B, Klassen R. Impact of Pus1 Pseudouridine Synthase on Specific Decoding Events in Saccharomyces cerevisiae. Biomolecules. 2020 10
64. Arias L, Martínez F, González D, Flores-Ríos R, Katz A, Tello M. et al. Modification of Transfer RNA Levels Affects Cyclin Aggregation and the Correct Duplication of Yeast Cells. Front Microbiol. 2020;11:607693
65. Tsutsumi S, Sugiura R, Ma Y, Tokuoka H, Ohta K, Ohte R. et al. Wobble inosine tRNA modification is essential to cell cycle progression in G(1)/S and G(2)/M transitions in fission yeast. J Biol Chem. 2007;282:33459-65
66. Patil A, Dyavaiah M, Joseph F, Rooney JP, Chan CT, Dedon PC. et al. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle. 2012;11:3656-65
67. Candiracci J, Migeot V, Chionh YH, Bauer F, Brochier T, Russell B. et al. Reciprocal regulation of TORC signaling and tRNA modifications by Elongator enforces nutrient-dependent cell fate. Sci Adv. 2019;5:eaav0184
68. Chang YH, Nishimura S, Oishi H, Kelly VP, Kuno A, Takahashi S. TRMT2A is a novel cell cycle regulator that suppresses cell proliferation. Biochem Biophys Res Commun. 2019;508:410-5
69. Yolu Y, van de Logt E, Kellner-Kaiser S. The Stress-Dependent Dynamics of Saccharomyces cerevisiae tRNA and rRNA Modification Profiles. Genes (Basel). 2021 12
70. Trixl L, Amort T, Wille A, Zinni M, Ebner S, Hechenberger C. et al. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell Mol Life Sci. 2018;75:1483-97
71. Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020-39
72. Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M. et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590-5
73. Takakura M, Ishiguro K, Akichika S, Miyauchi K, Suzuki T. Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat Commun. 2019;10:5542
74. Zhou J, Lénon M, Ravanat JL, Touati N, Velours C, Podskoczyj K. et al. Iron-sulfur biology invades tRNA modification: the case of U34 sulfuration. Nucleic Acids Res. 2021;49:3997-4007
75. Koh CS, Sarin LP. Transfer RNA modification and infection - Implications for pathogenicity and host responses. Biochim Biophys Acta Gene Regul Mech. 2018;1861:419-32
76. Freund I, Buhl DK, Boutin S, Kotter A, Pichot F, Marchand V. et al. 2'-O-methylation within prokaryotic and eukaryotic tRNA inhibits innate immune activation by endosomal Toll-like receptors but does not affect recognition of whole organisms. RNA. 2019;25:869-80
77. Galvanin A, Vogt LM, Grober A, Freund I, Ayadi L, Bourguignon-Igel V. et al. Bacterial tRNA 2'-O-methylation is dynamically regulated under stress conditions and modulates innate immune response. Nucleic Acids Res. 2020;48:12833-44
78. Keller P, Freund I, Marchand V, Bec G, Huang R, Motorin Y. et al. Double methylation of tRNA-U54 to 2'-O-methylthymidine (Tm) synergistically decreases immune response by Toll-like receptor 7. Nucleic Acids Res. 2018;46:9764-75
79. Rak R, Polonsky M, Eizenberg-Magar I, Mo Y, Sakaguchi Y, Mizrahi O. et al. Dynamic changes in tRNA modifications and abundance during T cell activation. Proc Natl Acad Sci U S A. 2021 118
80. Liu Y, Zhou J, Li X, Zhang X, Shi J, Wang X. et al. tRNA-m(1)A modification promotes T cell expansion via efficient MYC protein synthesis. Nat Immunol. 2022
81. Gao Z, Jijiwa M, Nasu M, Borgard H, Gong T, Xu J. et al. Comprehensive landscape of tRNA-derived fragments in lung cancer. Mol Ther Oncolytics. 2022;26:207-25
82. Chen Y, Shen J. Mucosal immunity and tRNA, tRF, and tiRNA. J Mol Med (Berl). 2021;99:47-56
83. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol. 2021;22:375-92
84. Enroth C, Poulsen LD, Iversen S, Kirpekar F, Albrechtsen A, Vinther J. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. 2019;47:e126
85. Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G. et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol Cell. 2019;74:1304-16.e8
86. Liu Y, Yang C, Zhao Y, Chi Q, Wang Z, Sun B. Overexpressed methyltransferase-like 1 (METTL1) increased chemosensitivity of colon cancer cells to cisplatin by regulating miR-149-3p/S100A4/p53 axis. Aging (Albany NY). 2019;11:12328-44
87. Katsara O, Schneider RJ. m(7)G tRNA modification reveals new secrets in the translational regulation of cancer development. Mol Cell. 2021;81:3243-5
88. Orellana EA, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W. et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81:3323-38.e14
89. Liao J, Yi Y, Yue X, Wu X, Zhu M, Chen Y. et al. Methyltransferase 1 is required for nonhomologous end-joining repair and renders hepatocellular carcinoma resistant to radiotherapy. Hepatology. 2022
90. Zhu S, Wu Y, Zhang X, Peng S, Xiao H, Chen S. et al. Targeting N(7)-methylguanosine tRNA modification blocks hepatocellular carcinoma metastasis after insufficient radiofrequency ablation. Mol Ther. 2022
91. Huang M, Long J, Yao Z, Zhao Y, Zhao Y, Liao J. et al. METTL1-mediated m7G tRNA modification promotes lenvatinib resistance in hepatocellular carcinoma. Cancer Res. 2022
92. Okamoto M, Fujiwara M, Hori M, Okada K, Yazama F, Konishi H. et al. tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells. PLoS Genetics. 2014;10:e1004639
93. Chen J, Li K, Chen J, Wang X, Ling R, Cheng M. et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun (Lond). 2022;42:223-44
94. Ying X, Liu B, Yuan Z, Huang Y, Chen C, Jiang X. et al. METTL1-m(7) G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin Transl Med. 2021;11:e675
95. Ma J, Han H, Huang Y, Yang C, Zheng S, Cai T. et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol Ther. 2021;29:3422-35
96. Wang C, Wang W, Han X, Du L, Li A, Huang G. Methyltransferase-like 1 regulates lung adenocarcinoma A549 cell proliferation and autophagy via the AKT/mTORC1 signaling pathway. Oncol Lett. 2021;21:330
97. Chen B, Jiang W, Huang Y, Zhang J, Yu P, Wu L. et al. N(7)-methylguanosine tRNA modification promotes tumorigenesis and chemoresistance through WNT/β-catenin pathway in nasopharyngeal carcinoma. Oncogene. 2022;41:2239-53
98. Huang Y, Ma J, Yang C, Wei P, Yang M, Han H. et al. METTL1 promotes neuroblastoma development through m(7)G tRNA modification and selective oncogenic gene translation. Biomark Res. 2022;10:68
99. Han H, Zheng S, Lin S. N(7)-methylguanosine (m(7)G) tRNA modification: a novel autophagy modulator in cancer. Autophagy. 2022:1-3
100. Han H, Yang C, Ma J, Zhang S, Zheng S, Ling R. et al. N(7)-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun. 2022;13:1478
101. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
102. Ru W, Zhang X, Yue B, Qi A, Shen X, Huang Y. et al. Insight into m(6)A methylation from occurrence to functions. Open Biol. 2020;10:200091
103. Dong Z, Cui H. The Emerging Roles of RNA Modifications in Glioblastoma. Cancers (Basel). 2020 12
104. Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K. et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7:42271
105. Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J. et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533-45
106. Zhang Q, Cai Y, Kurbatov V, Khan SA, Lu L, Zhang Y. et al. Gene Alterations of N6-Methyladenosine (m(6)A) Regulators in Colorectal Cancer: A TCGA Database Study. Biomed Res Int. 2020;2020:8826456
107. Liu T, Li C, Jin L, Li C, Wang L. The Prognostic Value of m6A RNA Methylation Regulators in Colon Adenocarcinoma. Med Sci Monit. 2019;25:9435-45
108. Xu J, Liu Y, Liu J, Xu T, Cheng G, Shou Y. et al. The Identification of Critical m(6)A RNA Methylation Regulators as Malignant Prognosis Factors in Prostate Adenocarcinoma. Front Genet. 2020;11:602485
109. Wang Q, Zhang Q, Huang Y, Zhang J. m(1)A Regulator TRMT10C Predicts Poorer Survival and Contributes to Malignant Behavior in Gynecological Cancers. DNA Cell Biol. 2020;39:1767-78
110. Shi L, Yang XM, Tang DD, Liu G, Yuan P, Yang Y. et al. Expression and significance of m1A transmethylase, hTrm6p/hTrm61p and its related gene hTrm6/hTrm61 in bladder urothelial carcinoma. Am J Cancer Res. 2015;5:2169-79
111. Macari F, El-Houfi Y, Boldina G, Xu H, Khoury-Hanna S, Ollier J. et al. TRM6/61 connects PKCα with translational control through tRNAi(Met) stabilization: impact on tumorigenesis. Oncogene. 2016;35:1785-96
112. Zhao M, Shen S, Xue C. A Novel m1A-Score Model Correlated With the Immune Microenvironment Predicts Prognosis in Hepatocellular Carcinoma. Front Immunol. 2022;13:805967
113. Zheng Q, Yu X, Zhang Q, He Y, Guo W. Genetic characteristics and prognostic implications of m1A regulators in pancreatic cancer. Biosci Rep. 2021 41
114. Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br J Cancer. 2011;104:700-6
115. Konishi N, Nakamura M, Ishida E, Shimada K, Mitsui E, Yoshikawa R. et al. High expression of a new marker PCA-1 in human prostate carcinoma. Clin Cancer Res. 2005;11:5090-7
116. Ohshio I, Kawakami R, Tsukada Y, Nakajima K, Kitae K, Shimanoe T. et al. ALKBH8 promotes bladder cancer growth and progression through regulating the expression of survivin. Biochem Biophys Res Commun. 2016;477:413-8
117. Gu C, Ramos J, Begley U, Dedon PC, Fu D, Begley TJ. Phosphorylation of human TRM9L integrates multiple stress-signaling pathways for tumor growth suppression. Sci Adv. 2018;4:eaas9184
118. Chen HM, Wang J, Zhang YF, Gao YH. Ovarian cancer proliferation and apoptosis are regulated by human transfer RNA methyltransferase 9-likevia LIN9. Oncol Lett. 2017;14:4461-6
119. Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M. et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213:2503-23
120. McMahon M, Ruggero D. A wobbly road to drug resistance in melanoma: tRNA-modifying enzymes in translation reprogramming. EMBO J. 2018 37
121. Rapino F, Delaunay S, Rambow F, Zhou Z, Tharun L, De Tullio P. et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature. 2018;558:605-9
122. García-Vílchez R, Sevilla A, Blanco S. Post-transcriptional regulation by cytosine-5 methylation of RNA. Biochim Biophys Acta Gene Regul Mech. 2019;1862:240-52
123. Li M, Tao Z, Zhao Y, Li L, Zheng J, Li Z. et al. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J Transl Med. 2022;20:214
124. Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856-63
125. Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA Transferase NSUN2 Gene Expression is Associated with Poor Prognosis in Head and Neck Squamous Carcinoma. Cancer Invest. 2018;36:246-53
126. Lu L, Gaffney SG, Cannataro VL, Townsend J. Transfer RNA methyltransferase gene NSUN2 mRNA expression modifies the effect of T cell activation score on patient survival in head and neck squamous carcinoma. Oral Oncol. 2020;101:104554
127. Yang JC, Risch E, Zhang M, Huang C, Huang H, Lu L. Association of tRNA methyltransferase NSUN2/IGF-II molecular signature with ovarian cancer survival. Future Oncol. 2017;13:1981-90
128. Delaunay S, Pascual G, Feng B, Klann K, Behm M, Hotz-Wagenblatt A. et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature. 2022
129. Cui Q, Yin K, Zhang X, Ye P, Chen X, Chao J. et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat Cancer. 2021;2:932-49
130. Zhang DY, Ming GL, Song H. PUS7: a targetable epitranscriptomic regulator of glioblastoma growth. Trends Pharmacol Sci. 2021;42:976-8
131. Blaze J, Akbarian S. The tRNA regulome in neurodevelopmental and neuropsychiatric disease. Mol Psychiatry. 2022
132. Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S. et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet. 2012;49:380-5
133. Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S. et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:847-55
134. Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S. et al. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and?Motility. Stem Cell Reports. 2017;8:112-24
135. Blaze J, Navickas A, Phillips HL, Heissel S, Plaza-Jennings A, Miglani S. et al. Neuronal Nsun2 deficiency produces tRNA epitranscriptomic alterations and proteomic shifts impacting synaptic signaling and behavior. Nat Commun. 2021;12:4913
136. Paramasivam A, Meena AK, Venkatapathi C, Pitceathly RDS, Thangaraj K. Novel Biallelic NSUN3 Variants Cause Early-Onset Mitochondrial Encephalomyopathy and Seizures. J Mol Neurosci. 2020;70:1962-5
137. Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA. et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039
138. Shafik AM, Zhou H, Lim J, Dickinson B, Jin P. Dysregulated mitochondrial and cytosolic tRNA m1A methylation in Alzheimer's disease. Hum Mol Genet. 2021
139. Silzer TK, Pathak GA, Phillips NR. Mitochondrial tRNA methylation in Alzheimer's disease and progressive supranuclear palsy. BMC Med Genomics. 2020;13:71
140. Lentini JM, Alsaif HS, Faqeih E, Alkuraya FS, Fu D. DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. Nat Commun. 2020;11:2510
141. Trimouille A, Lasseaux E, Barat P, Deiller C, Drunat S, Rooryck C. et al. Further delineation of the phenotype caused by biallelic variants in the WDR4 gene. Clin Genet. 2018;93:374-7
142. Waqas A, Nayab A, Shaheen S, Abbas S, Latif M, Rafeeq MM. et al. Case Report: Biallelic Variant in the tRNA Methyltransferase Domain of the AlkB Homolog 8 Causes Syndromic Intellectual Disability. Front Genet. 2022;13:878274
143. Broly M, Polevoda BV, Awayda KM, Tong N, Lentini J, Besnard T. et al. THUMPD1 bi-allelic variants cause loss of tRNA acetylation and a syndromic neurodevelopmental disorder. Am J Hum Genet. 2022;109:587-600
144. Nøstvik M, Kateta SM, Schönewolf-Greulich B, Afenjar A, Barth M, Boschann F. et al. Clinical and molecular delineation of PUS3-associated neurodevelopmental disorders. Clin Genet. 2021;100:628-33
145. Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM. et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet. 2016;135:707-13
146. de Brouwer APM, Abou Jamra R, Körtel N, Soyris C, Polla DL, Safra M. et al. Variants in PUS7 Cause Intellectual Disability with Speech Delay, Microcephaly, Short Stature, and Aggressive Behavior. Am J Hum Genet. 2018;103:1045-52
147. Ramos J, Proven M, Halvardson J, Hagelskamp F, Kuchinskaya E, Phelan B. et al. Identification and rescue of a tRNA wobble inosine deficiency causing intellectual disability disorder. RNA. 2020;26:1654-66
148. Ramos J, Han L, Li Y, Hagelskamp F, Kellner SM, Alkuraya FS. et al. Formation of tRNA Wobble Inosine in Humans Is Disrupted by a Millennia-Old Mutation Causing Intellectual Disability. Mol Cell Biol. 2019 39
149. Bento-Abreu A, Jager G, Swinnen B, Rué L, Hendrickx S, Jones A. et al. Elongator subunit 3 (ELP3) modifies ALS through tRNA modification. Hum Mol Genet. 2018;27:1276-89
150. Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A. et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest. 2011;121:3598-608
151. Arragain S, Handelman SK, Forouhar F, Wei FY, Tomizawa K, Hunt JF. et al. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J Biol Chem. 2010;285:28425-33
152. Vangaveti S, Ranganathan SV, Agris PF. Physical Chemistry of a Single tRNA-Modified Nucleoside Regulates Decoding of the Synonymous Lysine Wobble Codon and Affects Type 2 Diabetes. J Phys Chem B. 2022;126:1168-77
153. Santos M, Anderson CP, Neschen S, Zumbrennen-Bullough KB, Romney SJ, Kahle-Stephan M. et al. Irp2 regulates insulin production through iron-mediated Cdkal1-catalyzed tRNA modification. Nat Commun. 2020;11:296
154. Shigi N. Biosynthesis and Degradation of Sulfur Modifications in tRNAs. Int J Mol Sci. 2021 22
155. Cosentino C, Toivonen S, Diaz VE, Atta M, Ravanat JL, Demine S. et al. Pancreatic β-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 2018;46:10302-18
156. Stern E, Vivante A, Barel O, Levy-Shraga Y. TRMT10A mutation in a child with diabetes, short stature, microcephaly and hypoplastic kidneys. J Clin Res Pediatr Endocrinol. 2021
157. Şıklar Z, Kontbay T, Colclough K, Patel KA, Berberoğlu M. Expanding the Phenotype of TRMT10A Mutations: Case Report and a Review of the Existing Cases. J Clin Res Pediatr Endocrinol. 2021
158. Brener A, Zeitlin L, Wilnai Y, Birk OS, Rosenfeld T, Chorna E. et al. Looking for the skeleton in the closet-rare genetic diagnoses in patients with diabetes and skeletal manifestations. Acta Diabetol. 2022;59:711-9
159. Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R. et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080
160. Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359-407
161. Suzuki T, Nagao A, Suzuki T. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip Rev RNA. 2011;2:376-86
162. Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet. 2004;74:1303-8
163. Metodiev MD, Thompson K, Alston CL, Morris AAM, He L, Assouline Z. et al. Recessive Mutations in TRMT10C Cause Defects in Mitochondrial RNA Processing and Multiple Respiratory Chain Deficiencies. Am J Hum Genet. 2016;98:993-1000
164. Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C. et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104-19
165. Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45:7401-15
166. Helm M, Florentz C, Chomyn A, Attardi G. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR). Nucleic Acids Res. 1999;27:756-63
167. Meseguer S, Navarro-González C, Panadero J, Villarroya M, Boutoual R, Sánchez-Alcázar JA. et al. The MELAS mutation m.3243A>G alters the expression of mitochondrial tRNA fragments. Biochim Biophys Acta Mol Cell Res. 2019;1866:1433-49
168. Kamble AS, Fandilolu PM, Sambhare SB, Sonawane KD. Idiosyncratic recognition of UUG/UUA codons by modified nucleoside 5-taurinomethyluridine, τm5U present at 'wobble' position in anticodon loop of tRNALeu: A molecular modeling approach. PLoS One. 2017;12:e0176756
169. Ohsawa Y, Hagiwara H, Nishimatsu SI, Hirakawa A, Kamimura N, Ohtsubo H. et al. Taurine supplementation for prevention of stroke-like episodes in MELAS: a multicentre, open-label, 52-week phase III trial. J Neurol Neurosurg Psychiatry. 2019;90:529-36
170. Zhang Q, Zhang L, Chen D, He X, Yao S, Zhang Z. et al. Deletion of Mtu1 (Trmu) in zebrafish revealed the essential role of tRNA modification in mitochondrial biogenesis and hearing function. Nucleic Acids Res. 2018;46:10930-45
171. He Q, Zhao Q, Li Q, Pan R, Li X, Chen Y. Mtu1 defects are correlated with reduced osteogenic differentiation. Cell Death Dis. 2021;12:61
172. Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y. et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol. 2021;23:424-36
173. Behrens A, Rodschinka G, Nedialkova DD. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell. 2021;81:1802-15.e7
174. Hu JF, Yim D, Ma D, Huber SM, Davis N, Bacusmo JM. et al. Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat Biotechnol. 2021;39:978-88
175. Li J, Zhu WY, Yang WQ, Li CT, Liu RJ. The occurrence order and cross-talk of different tRNA modifications. Sci China Life Sci. 2021;64:1423-36
176. Sokołowski M, Klassen R, Bruch A, Schaffrath R, Glatt S. Cooperativity between different tRNA modifications and their modification pathways. Biochim Biophys Acta Gene Regul Mech. 2018;1861:409-18
177. Hou YM, Masuda I, Foster LJ. tRNA methylation: An unexpected link to bacterial resistance and persistence to antibiotics and beyond. Wiley Interdiscip Rev RNA. 2020;11:e1609
178. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303-22
Author contact
![]() Corresponding author: Department of Thoracic Surgery, Xiangya Hospital, Central South University, Xiangya Road 87th, Changsha, 410008, Hunan, PR China. Email: duancjxyedu.cn
Corresponding author: Department of Thoracic Surgery, Xiangya Hospital, Central South University, Xiangya Road 87th, Changsha, 410008, Hunan, PR China. Email: duancjxyedu.cn

 Global reach, higher impact
Global reach, higher impact