Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(4):1178-1191. doi:10.7150/ijbs.79430 This issue Cite
Review
The role of gut microbiota in T cell immunity and immune mediated disorders
1. Department of Anatomy, Pusan National University School of Medicine, Yangsan 50612, Republic of Korea
2. PNU GRAND Convergence Medical Science Education Research Center, Pusan National University School of Medicine, Yangsan 50612, Republic of Korea
Received 2022-9-30; Accepted 2023-1-23; Published 2023-2-13
Abstract
Gut microbiota was only considered as a commensal organism that aids in digestion, but recent studies revealed that the microbiome play a critical role in both physiological and pathological immune system. The gut microbiome composition is altered by environmental factors such as diet and hygiene, and the alteration affects immune cells, especially T cells. Advanced genomic techniques in microbiome research defined that specific microbes regulate T cell responses and the pathogenesis of immune-mediated disorders. Here, we review features of specific microbes-T cell crosstalk and relationship between the microbes and immunopathogenesis of diseases including in cancers, autoimmune disorders and allergic inflammations. We also discuss the limitations of current experimental animal models, cutting-edge developments and current challenges to overcome in the field, and the possibility of considering gut microbiome in the development of new drug.
Keywords: microbiome, T cell, immune system, autoimmune disease, fecal microbiota transplant
Introduction
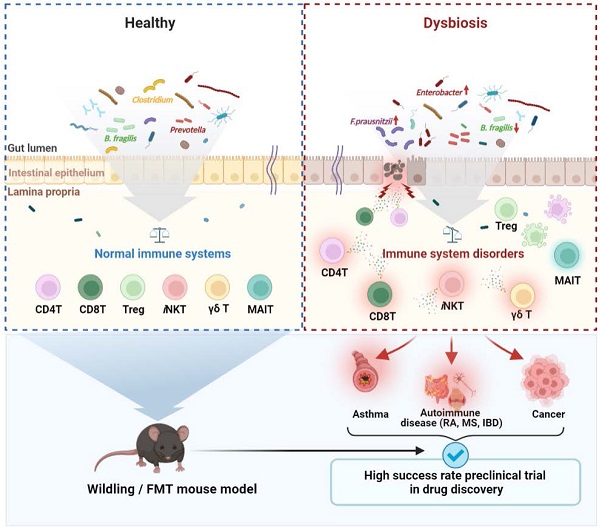
The microbiota, which has been studied for decades, forms a complex network evolving with humans [1-3]. The gut microbiota influences the development, homeostasis, and function of the immune system [4, 5].
The gut microbiota interacts to T cells with antigen-specific recognition [6], or signals via Toll-like receptors and Nod-like receptors. These signals mediate cell induction and function, thus ensuring homeostasis in the human immune system [7]. The microbiota species has been shown to be associated with the differentiation of T cells such as helper T (Th) 1, Th2, Th17, and regulatory T (Treg) cells [8-10]. Also, short-chain fatty acids (SCFAs), metabolites of the gut microbiota, regulate T cell differentiation and activation [11]. In addition, germ-free (GF) mice have reduction of Treg cells, the absence of Th17 cells, and imbalance of Th1/Th2 cells, which is skewed into the Th2 response [12]. The immune phenotype in GF mice was modulated to Th1 skewed-responses by reconstitution of specific microbe [8]. In recent mass cytometry by time-of-flight (CyTOF®) analysis, immune landscape in specific pathogen-free (SPF) mice was significantly distinct from wildling mice which has wildling bacterial microbiome and wildling mice was more similarly phenocopied the immune response of humans than SPF mice [13]. Therefore, these studies indicate that gut microbiota can regulate immune landscape and responses including differentiation, homeostasis and development of T cells, and alternation of gut microbiota may be closely linked to the pathogenesis of immune-mediated diseases.
Gut microbiota dysbiosis is caused by a variety of mechanisms, including microbiome imbalance, immune dysregulation, proinflammatory mechanisms, and metabolic activities. Dysbiosis lead to various T cell-related diseases, including rheumatoid arthritis (RA), type 1 and type 2 diabetes, asthma, cardiovascular disease, inflammatory bowel disease (IBD), cancer, liver disease, and psychiatric disorders [14-20].
Despite the importance of understanding microbiome-T cell interactions, most immunology experiments have been performed with limited microbiome composition such as SPF or GF mice. Thus, experimental results using gnotobiotic mouse models in the drug development stage cannot fully represent humans [21]. To address these limitations, a fecal microbiota transplant (FMT) mouse model with a gut microbiota similar to that of humans was developed using human or wild mouse feces [22-24]. The FMT mouse model has an abundant and more diverse gut microbiome than the current experimental animal models [23]. In addition, mice implanted with the wild mouse or human microbiota not only exhibit different degrees of microbial diversity but their systemic immunity is also affected.
Here, we discuss the specific microbes-T cell crosstalk, and distribution of the gut microbiota according to T cell-mediated diseases. In addition, we discuss the need to use the FMT mouse model to overcome the limitations of the current experimental mouse model, and to consider the human gut microbiota in drug discovery.
Microbiota and T cells
Numerous studies have been conducted on the crosstalk between the gut microbiota and immune cells. Commensal microbiota affects the function, development, and differentiation of T cells to maintain host immune homeostasis.
A recent report that the microbiome acts as an antigen for T cells, specific microbes, such as segmented filamentous bacteria (SFB), contribute to the development of microbiota-specific T cells in the thymus [25]. T cells mainly regulate adaptive immune responses and can be divided into CD4+ T cells and CD8+ T cells. Specific commensal microbiota induces Th cell polarization and cytotoxic CD8+ T cell activity. In addition, unconventional T cells including natural killer T (NKT), γδ T, and mucosal-associated invariant T (MAIT) cells recognize microbe groups based on their microbe-derived metabolites.
CD4+ T cells
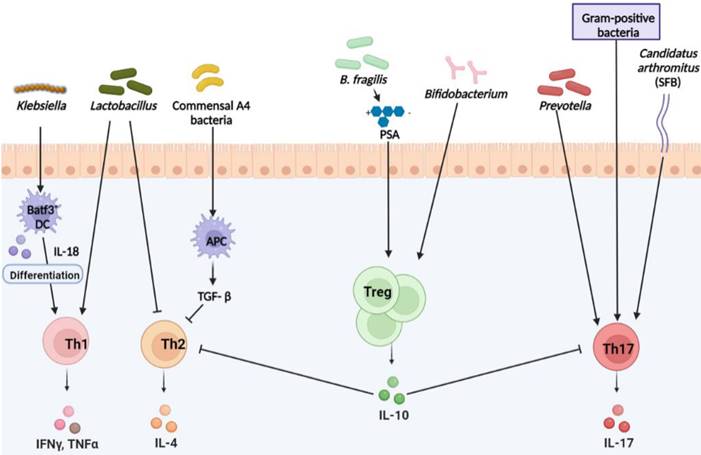
CD4+ T cells differentiate into various Th lineages with different effector functions. Several studies have demonstrated the role of the microbiome in Th cell differentiation and homeostasis. Klebsiella (K.) genera, such as K. aeromobilis and K. pneumoniae, induce Th1 cell responses in the gut (Fig. 1). Colonized Klebsiella in GF mouse intestines enhances Th1 cell proliferation, leading to Th1 cell augmentation in the intestine [26]. Probiotic bacteria have also been shown to modulate Th1 cell activity. Representative probiotic Lactobacillus strains are closely related to Th1 cells. Lactobacillus (L.) plantarum [27, 28] and L. salivarius [29] enhanced the production of Th1 cytokines, tumor necrosis factor alpha (TNFα), and interferon gamma (IFNγ). Lactobacillus strains isolated from fermented foods also upregulated TNFα secretion via macrophage activation, simultaneously, the amount of the Th2 cytokine interleukin (IL)-4 decreased [30]. Based on these data, the gut microbiota could be involved in both Th1 and Th2 cell activities, and other Lactobacillus studies support this hypothesis [28, 31]. Mazmanian et al. reported that GF mice appeared more biased toward Th2 cell responses than Th1 cell. However, colonization of GF mice with polysaccharides (PSA) produced by Bacteroides (B.) fragilis corrected the imbalance by skewing Th2 cell to Th1 cell, resulting in decreased IL-4 and increased IFNγ production [8]. This indicated that the microbiota could concurrently affect Th1 and Th2 cells. Klebsiella-induced Th1 cell differentiation is mediated by basic leucine zipper ATF-like transcription factor 3 (Batf3)-dependent dendritic cells (DCs) and TLR signaling pathway with IL-18 signaling [26]. Under homeostatic control, Klebsiella-specific Th1 cells can be regulated without induction of severe gut inflammation. Once Klebsiella are dominant during dysbiosis, severe gut inflammation can be elicited following induction of Th1 cell differentiation. Indeed, enrichment of Klebsiella strains in the fecal of IBD patients than healthy controls has been reported [32].
Th2 cell secreting IL-4, IL-5, and IL-13 significant role in humoral immunity and defense against helminth infections and contribute to chronic inflammatory diseases, such as asthma and allergy [33]. Lactobacillus strains and B. fragilis were found to inhibit Th2 activity by positively influencing Th1 activity [8, 28, 30, 31] (Fig. 1). The influence of the microbiota on Th2 activity appears during the IBD phase. Bamias et al. reported that SAMP1/YitFc mouse models of Crohn's disease (CD) -like ileitis indicated that commensal bacteria induced Th2 response characteristic of the chronic phase of SAMP1/YitFc ileitis, and symptoms also exacerbated [34]. Studies have shown that Lachnospiraceae strains are Th2 inhibiting microbes. Commensal A4 bacteria, a member of the Lachnospiraceae family, induce transforming growth factor-beta (TGF-β) production from DCs, which results in the inhibition of Th2 cell differentiation and activity [35].
Th17 cells produce IL-17, a potent proinflammatory cytokine that causes tissue damage and is involved in the pathogenesis of inflammatory and autoimmune diseases [36]. Also, Th17 cells are absent in GF mice and are inducible upon microbial colonization [12] Studies have reported that SFB and gram-positive bacteria induce Th17 cell differentiation (Fig. 1). Atarashi et al. reported that SFB or commensal bacteria-derived ATP activates a specific subset of lamina propria cells, and promotes inducing Th17 cell differentiation [37, 38]. In addition, Ivanov et al. reported that SPF mouse treatment with vancomycin, a gram-positive bacterial antibiotic, reduced the frequency of Th17 cells in the small intestine, implicating specific bacteria in Th17 cell differentiation [39]. According to Koji and Ivanov, Th17 cell differentiation is determined by gut microbes. Subsequent studies revealed that SFB or 'Candidatus Arthromitus' is a gram-positive bacterium that promotes Th17 cell differentiation and secretion of several cytokines, such as IL-17 and IL-22 secretions [37, 40-42]. Also, Prevotella is another bacterium responsible for robust Th17 cell and cytokine secretion in the mouse colon [43]. Hang et al. reported that a microbial-produced secondary bile acid negatively affected Th17 cell differentiation. Among 30 different primary and secondary bile acids, 3-oxolithocholic acid blocked Th17 transcription factor RORγt, and Th17 cell differentiation was reduced in SPF mice [44]. Although how intestinal Th17 differentiation is induced by specific bacteria was fully undefined, many reports have clearly shown that the microbiome directly and indirectly contributes to Th17 cell differentiation and function. Therefore, Th17 differentiation by the microbiota colonization is linked across diverse autoimmune diseases, such as RA, multiple sclerosis (MS) and IBD, discussed in more detail in the 'Autoimmune diseases' section.
Microbiota and CD4+ T cells. Microbiota mediates T cell differentiation in homeostatic and pathogenic conditions. Klebsiella activates T helper (Th) 1 cell in the intestinal via antigen presenting cells (APCs) to secrete IFNγ and TNFα. Lactobacillus activates Th1 cells but inhibits Th2 cells. Symbiotic A4 bacteria of the Lachnospiraceae family inhibit the production of IL-4 secreted by Th2 cells by APCs. Th17 cells are activated by Prevotella, gram-positive bacteria, and SFB, secreting IL-17 and causing inflammation. Bacteroides fragilis (B. fragilis) produces PSA, which activates regulatory T (Treg) cells. Bifidobacterium also activates Treg cells, which relieve inflammation by inhibiting or balancing Th1, Th2, and Th17 cells. * PSA: Polysaccharide A, a capsular carbohydrate from the commensal gut bacteria B. fragilis * Batf3: basic leucine zipper ATF-like transcription factor 3
Treg cells play a crucial role in preventing autoimmune diseases through the modulation of immune responses, maintaining immune homeostasis, and providing immune tolerance to both self and non-self-innocuous antigens [45]. B. fragilis is a well-known microbe capable of robust Treg cell populations by producing PSA. This microbial product leads to the development of Foxp3+ Treg cells with IL-10 production [46] (Fig. 1). Bifidobacterium strains also affect Treg cells, and it was confirmed that Bifidobacterium (Bi.) infantis and Bi. bifidum act to promote Treg cell generation [47, 48]. Lactobacillus strains are involved in Treg cell differentiation and activity, and Lacticaseibacillus casei induces Treg cell development and IL-10 secretion [49]. Forsythe et al. reported that L. reuteri attenuated allergic airway responses in BALB/c mice [50], and that this microbe promotes Foxp3 expression [51]. Thus, L. reuteri, affects Treg cell development and the severity of autoimmune diseases. L. acidophilus strain L-92 (L-92) showed similar effects in BALB/c mice. Under allergic contact dermatitis (ACD) conditions, oral treatment with L-92 resulted in increased Foxp3, IL-10, and TGF-β expression in mice. They concluded that L-92 mediates ACD by enhancing Treg production and Th1 and Th2 responses [52]. Another Lactobacillus strain, L. murinus, has been shown to regulates RORγt+ Treg cells in the small intestine, which attenuates lung inflammation associated with Mycobacterium tuberculosis infection [53]. Microbial bile acid metabolites are involved in Treg cell differentiation and homeostasis. Song et al. observed that SPF mice fed a nutrient-rich diet produced more primary bile acids than GF mice, and this affected the restoration of RORγt+ Treg and Foxp3+ Treg cells [54]. In contrast, lithocholic acid, a secondary bile acid synthesized by bacteria, isoallolithocholic acid, inhibits Treg cell differentiation [44]. Zhang et al. reported that treatment with the penicillin antibiotic, ampicillin, reduced Treg cell proliferation and deregulated the Th1 response to bacterial infection. It can be seen that dysbiosis by antibiotics affects Treg cell production [55]. Likewise, microbiota can be altered by various factors, and this change also affects Treg cell generation and activities. Although the mechanism of intestinal Treg cell differentiation by specific commensal bacteria was not clearly defined, intestinal Treg cell colonization is clearly connected to specific bacteria and autoimmune diseases, for example, decreased abundance of Clostridia strains in RA and ameliorative effect of B. fragilis in IBD, discussed in more detail in the 'Autoimmune diseases' section.
CD8+ T cells
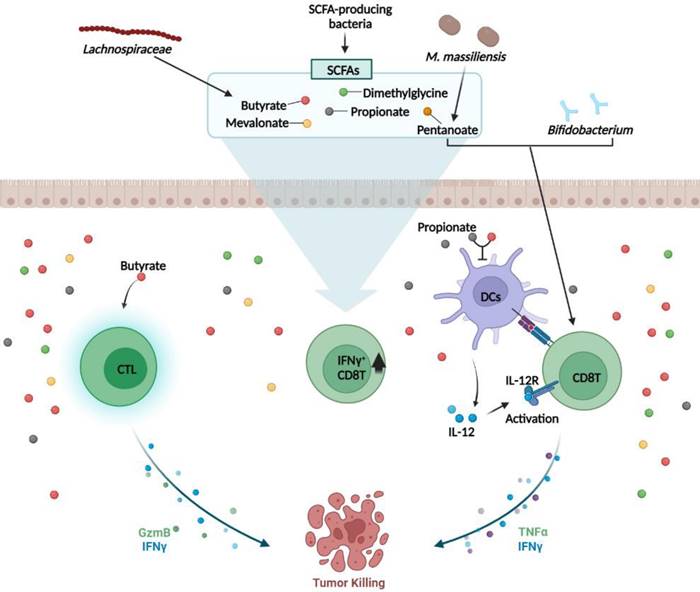
CD8+ T cells are critical for immune defense against intracellular pathogens, including viruses and bacteria, and for tumor surveillance [56]. Sivan et al. showed that probiotic species (Bifidobacteria) could determine the anti-tumor efficacy of CD8+ T cells [57] (Fig. 2). Vétizou et al. showed that particular microbes can influence cytotoxic T lymphocyte antigen-4 (CTLA-4) blockage for cancer immunotherapy. They found that B. fragilis, B. thetaiotaomicron, and Burkholderiales were the major microbes that restored the anti-tumor efficacy of αCTLA-4 treatment [58]. Recent studies have demonstrated that microbial byproducts mediate CD8+ T cell function. Butyrate and propionate, two major SCFAs, inhibit CD8+ T cell activation by controlling IL-12 production by antigen presenting cell (APC) [59]. Another study confirmed that butyrate directly increases CD8 T cell activity by upregulating IFNγ and granzyme B expression [60]. Luu et al. demonstrated that Megasphaera (M.) massiliensis produced pentanoate promotes effector CD8 T cells activity. In the presence of M. massiliensis, they detected higher IFNγ and TNFα expression, and this positively influenced the efficacy of adoptive T cell therapy [61] (Fig. 2). As a result, Microbiota-derived SCFAs modulate CD8 T cell responses during cancer immunotherapy. In contrast, Yang et al. discovered that butyrate derived from Lachnospiraceae species inhibited IFNγ secreting CD8 T cells. They elucidated that butyrate restrained the stimulator of the IFN gene (STING) activation in DCs, which is associated with CD8 T cell responses, resulting in lower radiotherapy efficacy [62]. Induction of IFNγ+CD8 T cells may be associated to specific bacterial strains-derived metabolites, such as mevalonate, dimethylglycine [63] and SCFAs [64], which enter circulation and induce systemic activation of CD8 T cells (Fig. 2). Thus, the microbiota can modulate CD8 T cell function and immunotherapy efficacy.
Unconventional T cell subsets
Natural Killer T cells
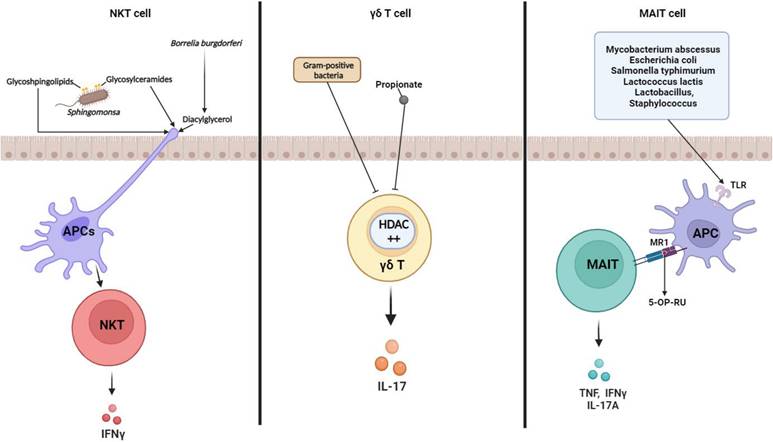
CD1d-restricted NKT cells regulate the broad range of immune responses between the innate and adaptive immune systems. They have the capacity to kill target cells and modulate immune response-secreting cytokines [65, 66]. Several studies have revealed that commensal microbiota regulate NKT cells homeostasis. Sphingomonas are gram-negative bacteria that are mainly found in the natural environment and are representative NKT cell stimulators. Since glycosphingolipids and glycosylceramides from Sphingomonas act as microbial antigens, they stimulate NKT cell activation and IFNγ secretion [67, 68] (Fig. 3). In the immune cells of GF mice, a lower frequency of iNKT cells was detected than that in SPF mice, but intragastric administration of Sphingomonas increased iNKT cell frequency [69]. Therefore, Sphingomonas exposure affects NKT-cell-modulated diseases. Olszak et al. found that commensal microbiota reduced mucosal iNKT cell accumulation. They observed a higher frequency and number of colon iNKT cells in GF mice than in that for SPF mice, implicating IBD and allergic asthma morbidity [70]. Sphingolipid produced by B. fragilis regulates homeostasis with suppression of NKT cell proliferation in neonatal mice [71], indicating that NKT cell-microbiota networking is critical in maintaining of balance between protective responses and excessive inflammation. CXCL16 expression is mediated by microbial bile acids, which affect hepatic NKT cell accumulation. Clostridium species modified bile acids promote the CXCL16 level in liver sinusoidal endothelial cells. Moreover, recruiting hepatic NKT cells has shown impressive anti-tumor responses against EL4 lymphoma tumors [72]. According to these findings, gut microbiota significantly affects NKT cells and mediates several diseases.
γδ T cells
γδ T cells are a small population of T cells that promote inflammatory responses and are particularly important for initial inflammatory and immune responses [69]. Similar to other T cell subsets, γδ T cells are also influenced by the microbiome. Under SPF conditions, microbes activate interleukin-1 receptor 1 (IL-1R1) expression on γδ T cells in the small intestine, thereby maintaining IL-17 production [73]. Li et al. showed that IL-17A-producing γδ T (γδ T17) cell homeostasis is mediated by the commensal microbiota in the liver. Commensal microbiota affects γδ T17 cell activation and IL-17 cytokine secretion, which accelerates non-alcoholic fatty liver disease (NAFLD) progression [74]. Microbial byproducts also downregulated γδ T17 cell activity. Vancomycin treatment increased γδ T17 cells in conventional mice, indicating that gram-positive bacteria repress γδ T cell function. Dupraz et al. confirmed that propionate is the main factor modulating IL-17 secretion by γδ T cells [75] (Fig. 3). Another study showed that commensal microbiota is associated with γδ T cell expansion in the mouse lung, which can promote particulate matter-induced neutrophilia [76]. As previously reported, γδ T cells that recognize lipids and phosphoantigens, as intermediates produced by microbiota via metabolic pathway, undergo various immune responses and epigenetic modification. This modification is indirectly induced through microbiota-epithelial cell interaction [77]. Although the interrelationship between intestinal γδ T cell and specific commensal bacteria was not clearly defined, localization and motility of γδ T cells are microbiota-dependently regulated in the epithelial layer [78] which is critical in protective immunosurveillance of the gut surface, indicating that commensal microbiota play an important role in the protective function of γδ T cells in diseases.
Microbiota and CD8+ T cells. Bifidobacterium and Mobilicoccus massiliensis (M. massiliensis)-derived pentanoate activates CD8+ T cells and increases the secretion of IFNγ and TNFα, thereby enhancing the anti-tumor CD8+ T cell response. Lachnospiraceae species-derived butyrate and propionate, the two major short-chain fatty acids (SCFAs), decrease APC-induced IL-12 production, thereby inhibiting CD8+ T cell activation and IFNγ secretion. In addition, butyrate directly activates cytotoxic T lymphocyte (CTL) cells via other pathways to upregulate the secretion of IFNγ and granzyme B (GzmB). Microbiota-derived SCFAs mediate anti-tumor responses by modulating CD8 T cell responses. SCFAs such as mevalonate and dimethylglycine increase IFNγ+CD8 T cells in the intestinal. *TNF: tumor necrosis factor
Microbiota and unconventional T cells. Modulation of unconventional T cell subset functions by microbe-derived metabolites and their own microbiota. Glycosphingolipids and glycosylceramides from Sphingomonas act as microbial antigens to activate NKT cells and secrete IFNγ (left). Gut microbiota represses IL-17 production by cecal γδ T cells. Gram-positive bacteria and short-chain fatty acids (SCFAs; ex. propionate) repress IL-17 producing γδ T cells (middle). Various bacterial antigens (ex.5-OP-RU) presented by APC are presented to MR1 (ligand), bind to the TCR of MAIT cells, activate MAIT cells, and secrete cytokines such as TNF, IFNγ, and IL17A (right). TCR in MAIT cells interacts with MR-1 in APC and is semi-constant. * TCR: T cell receptor; HDAC: Histone deacetylases; TLR: Toll Like Receptors; MR: MHC class I related-1 molecule; 5-OP-RU: MAIT cell ligand
Mucosal-associated invariant T cells
MAIT cells are MHC Ib-restricted innate-like lymphocytes and play a critical role in mucosal homeostasis. Interest in MAIT cell-microbiota interaction has increased since some studies have reported that MAIT cells can recognize bacteria and protect against microbial infection [79, 80]. Koay et al. showed that the number of MAIT cells in GF mice was noticeably lower than that in SPF mice. They found that the MAIT cell development in the thymus is impaired as the microbiome does not exist in GF mice [81]. Subsequent studies corroborated the significance of the microbiome on MAIT cell development and generation [72, 82]. Moreover, MAIT cell has an anti-microbial function, and some microbiome infections trigger MAIT cell function enhancement. For example, Francisella tularensis [80], Mycobacteroides abscessus, Escherichia (E.) coli [83], Salmonella typhimurium [84], and Lactococcus lactis [85] act as MAIT cell activators. Activation of MAIT cells recognizes various bacterial antigens (eg, 5-OP-RU) presented by MHC class 1b protein MR1 and secretes cytokines such as TNF, IFNγ, and IL-17A (Fig. 3). However, the use of mouse models for MAIT cell studies is much more limited than the use of human MAIT cells. SPF and GF mice lack microbiome complexity and richness, and this microbial environment is not sufficient to develop mouse models for MAIT cell studies. Indeed, MAIT cells are found in human blood and peripheral tissues, including the liver and gut lamina propria [86, 87], and their frequency is higher than that in mice [88]. Furthermore, microbiome comparisons between laboratory mice and humans are strikingly different [89]; these factors may influence several aspects of host immune system and diseases.
T cell-mediated inflammatory disease and microbiota
The etiology of autoimmune diseases can mainly be explained by genetic mutations and environmental factors [90, 91]. In particular, individual genetic susceptibility to diseases and subsequent environmental triggers cause changes in the composition of symbiotic microorganisms that interact with the immune system. Recent studies have highlighted the importance of microorganisms in autoimmune diseases such as RA [92], experimental autoimmune encephalomyelitis (EAE) [93], asthma, and IBD [94]. In this context, several reports have identified that intestinal microbes and their metabolites regulate T cell differentiation and function [63, 95]. In addition, disease progression and severity are affected by the composition of an individual's microbiota [94, 96] (Fig. 4).
Alteration of gut microbiota in autoimmune disease. Altered abundance of the gut microbiota in patients with Rheumatoid arthritis (RA), Multiple sclerosis (MS), and Inflammatory bowel disease (IBD) is indicated. * Red: high; blue: low; number: reference
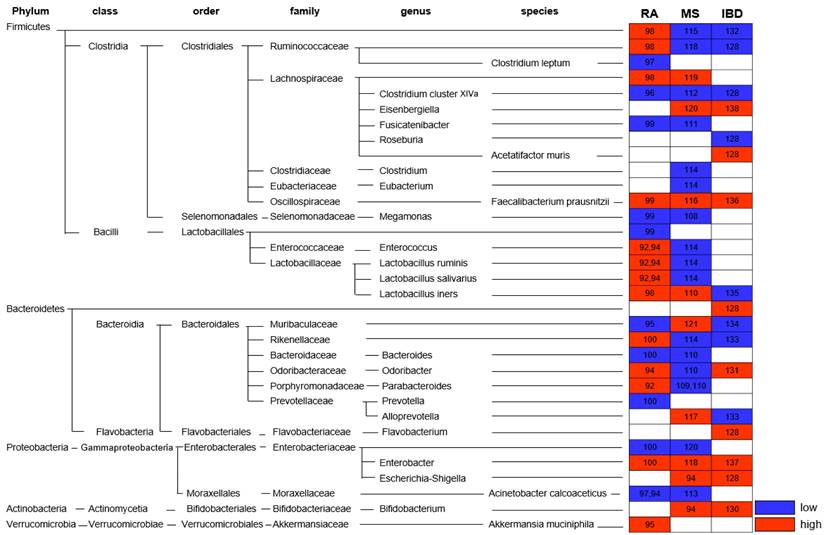
Autoimmune diseases
Rheumatoid arthritis
RA is a systemic chronic inflammatory disease that mainly affects the joints and is known to be an autoimmune disease accompanied by bone and cartilage destruction. Although the pathogenesis remains unclear, the microbiome, along with genetic and environmental factors, is also involved in the pathogenesis of RA. Numerous studies have investigated the gut microbiome abundance in patients with RA (Fig. 4). Among them, the abundance of Firmicutes, Ruminococcaceae, Bacteroides, Prevotella, Lactobacillus group, and Faecalibacterium (F.) prausnitzii increased, whereas that of Rikenellaceae, Alloprevotella, Bifidobacterium, Clostridium (C.) cluster XIVa, Enterobacter, Enterococcus, Fusicatenibacter, Megamonas, Odoribacter, and C. leptum decreased [97-105]. Scher et al. found that in the gut of patients with recent-onset RA, an increased abundance of Prevotella (P.) copri and Bacteroides and decreased abundance of C. leptum was observed [98].
In the RA mouse model, collagen-induced arthritis (CIA), the Lactobacillaceae lineage was significantly more abundant. The Lactobacillus genus was significantly more abundant after collagen-mediated induction in CIA-sensitive mice than in CIA-resistant mice [92, 106]. Ivanov et al. found that Candidatus Arthromitus or SFB colonization induces Th17 cells, rapid onset autoimmune arthritis, and exacerbates CIA [41]. Conversely, commensal bacteria-derived butyrate (C. cluster XIVa, including Lachnospiraceae, which are major butyrate producers) suppresses the development or improves symptoms of autoimmune arthritis [107]. Numerous studies have shown that SFB and prevotella, which are known to promote the secretion of Th17 cells and inflammatory cytokines, are greatly increased in autoimmune RA. Thus, the differential abundance of the gut microbiota suggests that it may orchestrate the development and symptoms of autoimmune arthritis through a Th cell-mediated pathway.
Multiple sclerosis
MS is an autoimmune and chronic inflammatory demyelinating disease of the central nervous system (CNS) caused by proinflammatory Th1 and Th17 cells [108]. Although Th17 cells have been reported to have a significant impact on MS pathogenesis, it has been suggested that microbiota alterations may also be factors in MS initiation and severity [99, 109, 110]. According to Koji and Ivanov, recruitment and activation of myelin-specific CD4+ T cells from immune processes depends on the availability of the target commensal gut microbiota [109]. Lee et al. suggested that GF mice harboring only SFB develop EAE, as colonization with SFB induces Th17 (IL-17) in the CNS, and that gut bacteria may influence neuroinflammation [111]. Additionally, SFB affects the disease by increasing cytokine production by Th1 (IFNγ) and Th17 (IL-17) cells in EAE (mouse model of MS) [111-113]. In patients with MS, Akkermansia (Ak.) muciniphila and Acinetobacter (Ac.) calcoaceticus both increased (induced proinflammatory responses), but Parabacteroides distasonis was reduced (stimulated anti-inflammatory IL-10-expressing) [99]. In addition, the abundance of Bacteroidetes, Lachnospiraceae, Rikenellaceae, Eisenbergiella, Escherichia-Shigella, F. prausnitzii, and Flavobacterium increased in patients with MS, whereas Firmicutes, Ruminococcaceae, Bacteroides, Bifidobacterium, Clostridium, L. salivarius, L. iners, L. ruminis, Megamonas, Odoribacter, Parabacteroides, and Prevotella decreased [99, 114-128] (Fig. 4). These results are similar to those of EAE. EAE onset and severity showed differences based on the composition of the microbiota community. At the onset of EAE, Acetatifactor (A.) muris, C. leptum, Turicibacter sanguinis, C. cluster XlVa, and Erysipelotrichaceae families increased, whereas Lactobacillus decreased [129]. In addition, Bifidobacterium, Prevotella, and Lactobacillus were negatively correlated with EAE severity, whereas Anaeroplasma, Rikenellaceae, and Clostridium were positively correlated with disease severity [119, 130]. Oral administration of L. murinus and L. reuteri [119, 131] is effective in reducing Th1/Th17 cells and related cytokines IFNγ/IL-17 and in alleviating and preventing EAE symptoms [109, 110, 132]. Altogether, data from MS patients and the murine EAE model found the composition of microbiota correlated with disease onset and clinical severity. Since the immune responses are activated or suppressed depending on the type of gut bacteria, the immunoregulatory effect on MS patients can be applied differently. Therefore, the diverse gut microbiota is expected to provide new preventive and therapeutic approaches targeting autoimmune response in patients with MS.
Inflammatory bowel disease
IBD is an autoimmune disease that occurs in the digestive tract owing to chronic inflammation and is divided into CD and ulcerative colitis (UC) [133]. Chronic inflammation leads to the production of reactive oxygen species (ROS), which impair DNA. It also controls proinflammatory mediators that promote cellular survival and growth. Consequently, patients with IBD have an increased risk of developing CRC [94]. In IBD patients, Enterobacteriaceae, Muribaculaceae, and A. muris, which are commensal proinflammatory bacterial taxa, were overgrown, and the Firmicutes family, especially the Clostridia class and Roseburia, were decreased [134]. Numerous studies have shown that the intestinal microbial abundance in IBD patients of Eisenbergiella, Escherichia-Shigella, F. prausnitzii, Parabacteroides, Ac. calcoaceticus, and Ak. muciniphila were higher, whereas Bacteroidetes, Rikenellaceae, Ruminococcaceae, C. cluster XIVa, and Flavovobacterium showed low abundance [134-144] (Fig. 4). A higher frequency of cells was observed in the gut microbiome of the IBD mouse model compared to mice colonized with healthy donor microbiotas [145]. In addition, SPF mice develop colitis with conventional microbiota but not under GF conditions [146]. The administration of Clostridium has been reported to prevent inflammation by increasing induced iTreg cells that secrete TGF-β and IL-10 [10]. In addition, Lactobacillus (especially L. murinus) induces Treg cell expansion by secreting IL-10 and TGF-β in colonic DCs and macrophages, and B. fragilis also relieves inflammation by inducing Treg cells [133, 147, 148]. IBD is influenced by the composition of the gut microbiota; therapeutic supplementation with probiotics, prebiotics, synbiotics, and FMT is also being investigated to suppress inflammation [149-151]. Therefore, it is expected that specific bacterial strains such as Clostridium and B. fragilis that induce Treg cells in the intestinal environment can be a novel treatment targeting IBD patients with intestinal bacterial dysbiosis.
Allergic airway inflammation (AAI) and Asthma
Asthma is a chronic airway inflammation disease characterized by airway hyper-responsiveness (AHR), airflow obstruction, airway inflammation, and airway remodeling [152]. In addition, eosinophil infiltration and higher concentrations of Th2 cytokines (IL-4, IL-5, and IL-13) and immunoglobulin E (IgE) were detected in the serum and bronchoalveolar lavage fluid of patients with asthma [152, 153]. According to current research, maternal or pediatric intestinal microbial clusters affect the immune system and are involved in allergic diseases, such as asthma. Herbst et al. observed that the total number of infiltrating lymphocytes and eosinophils was elevated in the airways of allergic GF mice compared to that in control SPF mice [154]. Oral treatment with live L. reuteri in a mouse model of allergic airway inflammation could attenuate the symptoms of an asthmatic response by inducing Treg cell production and increasing IL-10 secretion. This microbe also reduces the secretion of inflammatory cytokines, such as monocyte chemoattractant protein1 (MCP1), IL-5, and IL-13 including T lymphocytes and macrophages [50]. In addition, robust L. murinus Treg cells in the mesenteric lymph nodes and lung and decreased proinflammatory cytokine secretion are related to AAI severity. These data demonstrate that AAI and asthma can also be attenuated by strain-specific probiotics, such as in patients with RA and MS. Antibiotics are known to induce changes in the intestinal microbiota, and among them, azithromycin is reported to alter the gut microbiome and attenuate airway inflammation in allergic asthma [155]. In summary, altered gut microbiome may influence the onset and severity of asthma or disease with AAI. In addition, it will provide potential new biomarkers and suggest the possibility of more targeted therapies for autoimmune diseases by regulating the gut bacteria strains.
Cancers and microbiota
Interactions between the gut microbiome and immune system are thought to influence cancer immune surveillance. The human immune system plays an important role in tumor suppression, as tumor cells express immune checkpoints, such as programmed death (PD) 1/PD-ligand 1 and CTLA-4, to avoid the immune system response and suppress anti-tumor immunity [156, 157]. In addition, PD-1 and CTLA-4 expression increased by CD8+ tumor-infiltrating T lymphocytes in patients with pancreatic ductal adenocarcinoma (PDAC) [158, 159]. Blocked CTLA-4 induces a dramatic decrease in Bacteroidales and Burkholderiales, with a relative increase in Clostridiales [58]. Several studies have reported that PD-1 blockade in cancer patients is associated with gut microbiota, including Akkermansia, Bifidobacterium, and Faecalibacterium [57, 160-162]. In addition, cancers, such as CRC and PDAC, may be caused by dysbiosis of the gut microbiome. Numerous studies have analyzed and compared the gut microbiome abundance of different cancer patients and healthy donors. When the gut microbiota was analyzed in patients with various types of cancers such as PDAC, CRC, breast cancer, ovarian cancer, and cervical cancer, Proteobacteria, Firmicutes phyla and Actinobacteria abundance was very high compared to that in a healthy donor [163-166]. Conversely, Bacteroidetes are more abundant in healthy donors than in cancer patients with PDAC, CRC, ovarian cancer, or breast cancer [167]. Dysbiosis in patients with CRC causes increased Ak. muciniphila, E. coli, P. copri, Alistipes putredinis, Ruminococcus torques, and Prevotella [94, 168]. Microbiota-produced SCFA, such as acetate, butyrate, and propionate, have great effects on disease prevention and health promotion. Butyrate is also known to play an important role in cancer prevention [169]. In particular, fecal samples of patients with PDAC are depleted in the phylum Firmicutes, which includes beneficial bacteria including F. prausnitzii, Eubacterium rectale, and Roseburia intestinalis [170]. Butyrate secreted by Firmicutes suppresses histone deacetylase activity and downregulates proinflammatory cytokines in intestinal epithelial cells and immune cells to alleviate CRC symptoms [171, 172]. Zhang et al. reported that as a result of analyzing the intestinal microflora in lung cancer, lower levels of Kluyvera, Escherichia-Shigella, Dialister, Faecalibacterium, and Enterobacter were found in patients with lung cancer, whereas Veillonella, Fusobacterium, and Bacteroides were significantly higher than in healthy individuals [173]. In breast cancer patients, the anti-tumor effect was higher in patients with a high abundance of specific gut microbiomes, such as Ak. muciniphila, Bi. longem, Collinsella aerofaciens, and F. prausnitizi [174]. Cheng et al. reported that the gut microbiota in patients with ovarian cancer can negatively regulate estrogen levels [175]. Taken together, comparing the abundance of gut microbiota could be a useful biomarker for certain cancer patients and that the gut microbiome influences the occurrence, development, treatment, and prognosis of cancers; it will also provide new strategies for the prevention, diagnosis, and treatment of cancers.
Study of wildling or humanized mice by fecal microbiota transfer and its necessity
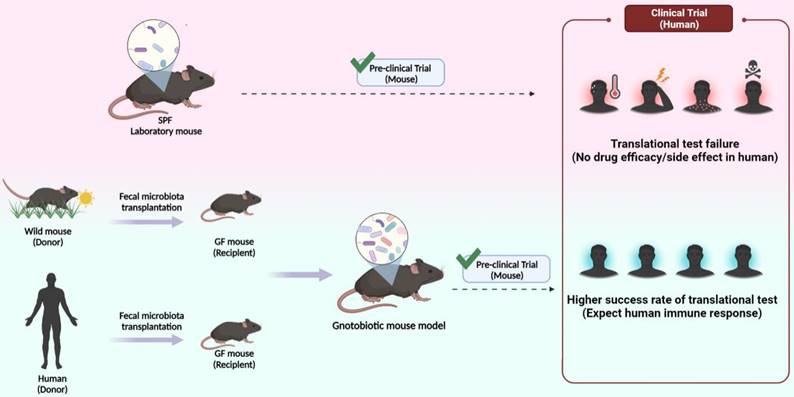
As a representative experimental animal, the mouse is essential for immunologists and biomedical research because it has more than 90% genetic similarity to humans [176] as well as advantages such as its small size, low cost, and ease of handling. SPF and GF mice with a limited microbiome managed in an ultra-hygienic system to prevent pathogen invasion in the 1960s [177] were used. Ironically, the limited microbiota in laboratory mice is a growing concern in human immunology and clinical research. Unlike laboratory mice, humans have been exposed to external pathogens from birth, which naturally leads to a higher microbiota complexity and abundance in humans than in laboratory mice [89]. Furthermore, in terms of the immune system, the microbial challenge from natural habitats matures the human immune system, whereas laboratory mice have an immature immune system similar to that of neonates. According to a study by Beura et al., laboratory mice lacked memory CD8+ T cell subsets that experienced protection against pathogen invasion [178]. This difference has limited the translational research value from laboratory mice to humans. Especially in the drug development and testing stages, preclinical testing using laboratory mouse model does not always show the same value as clinical testing in humans. The failure rate of translational tests is approximately 90%, mainly because drug candidate toxicity cannot be predicted at the clinical stage [179]. Therefore, to increase the success rate of clinical trials, preclinical trials must be reinforced. Therefore, immunologists have proposed the use of wild mice. As wild mice live in natural habitats similar to humans, using wild mice could provide the same environmental conditions as laboratory mice in terms of gut microbiota and immune system aspects. Recent studies have supported the importance of investigating wild mice. Rosshart et al. captured wild mice from Maryland, USA, and compared the gut microbiota with SPF mice. Interestingly, the gut microbiota composition differed between the groups. Relatively higher levels of Proteobacteria and Bacteroidetes were detected in wild mice, whereas Firmicutes levels were lower than in laboratory mice [22]. Ericsson et al. also consistently demonstrated that the wild mice microbiota differed from laboratory mice [180]. In addition, wild mice showed distinct immune system differences compared with laboratory mice. Wild mice had higher proportions of effector and memory T cells and higher cytokine production [178, 181]. Therefore, utilizing the wild mice-microbiome that influences immunological properties could be an option to improve the value of preclinical findings. Recently, wild mouse-based studies have been conducted using various wild microbiota-transplantation methods. It has been reported that the rewilded laboratory mouse via wild microbiota colonization showed similar microbial community [13, 22] and immune system fitness [13, 178, 182-184] and enhanced disease resistance [22]. Additionally, mice implanted with human microbiota have been studied for decades to determine their roles in health and disease. Burz et al. reported that when microbiota from a patient with non-alcoholic fatty liver disease (NAFLD) and healthy individuals were transplanted into mice, they displayed a microbiome composition similar to that of a human donor. When a high-fructose, high-fat diet was administered to mice transplanted with intestinal microbes from patients with a fatty liver, NAFLD characteristics, including increased body weight, steatosis, and plasma cholesterol, were observed. These results suggest that gut bacteria play a role in the development of obesity and steatosis and that targeting the gut microbiota could be a preventive or therapeutic strategy for managing NAFLD [24]. Another study identified a group of gut microbes that either promoted or inhibited tumorigenesis after transplantation of fecal samples from patients with CRC into GF mice. Baxter et al. suggested that a better understanding of the gut microbiota could lead to the development of prebiotic or probiotic therapies to prevent or delay the development and progression of CRC [185]. Britton et al. suggested that the fecal microbiota of humans with IBD alters intestinal CD4+ T cell homeostasis in mice and induces more Th2 and Th17 cells. In addition, it has been reported that mice colonized with IBD microbiota in a colitis model suffer from more severe diseases [145]. Based on current studies, fecal transfer of microbiota can be utilized to alter the laboratory mouse microbiome community and immune fitness; this process can facilitate the prediction of human immune responses. Therefore, FMT models such as the wildling or humanized model by fecal transfer would enhance the reliability of preclinical tests by providing accurate data in drug efficacy tests (Fig. 5). This would lead to the holistic development of the pharmaceutical industry.
Conclusion
This review discusses the relationship between gut microbiota and the immune system. In recent decades, significant advances have been made in immunology and microbiology using microbiota and germ-free animals and next-generation sequencing. Many such studies have revealed the correlation between gut microbiota and the immune system. Based on this, we have discussed the role of gut microbiome in activating T cells and the development and promotion of autoimmune diseases and various cancers. FMT mouse models of Human and wild mice are expected to increase the success rate of preclinical studies. Furthermore, it will be possible to overcome the limitations of using GF or SPF mice by gut microbiome research which will open new avenues for disease treatment and prevention based on the distribution of the gut microbiome.
Acknowledgements
We thank the members of the Hong lab for critical review of this manuscript. This work was supported by a 2-Year Research Grant of Pusan National University.
Author Contributions
All authors researched data for the article, made substantial contribution to discussion of content, and wrote, reviewed and edited the manuscript before submission.
The necessity for laboratory mouse expressing wild and human microbiome. SPF experimental mouse currently used a low success rate due to translational failure in the clinical stage despite success in preclinical tests (top). A humanized or wildling mouse model in which feces of the human and wild mouse are transplanted is predicted to show high translational test success rate in clinical trials after successful preclinical tests (bottom).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Berg G, Rybakova D, Fischer D, Cernava T, Vergès MC, Charles T. et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:103
2. Derrien M, Alvarez AS, de Vos WM. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019;27:997-1010
3. Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y. et al. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588:303-7
4. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H. et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296-302
5. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A. et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 2017;168:928-43.e11
6. Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL. et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152-6
7. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA. et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974-7
8. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107-18
9. Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677-89
10. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337-41
11. Kim CH, Park J, Kim M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune Netw. 2014;14:277-88
12. Lee N, Kim WU. Microbiota in T-cell homeostasis and inflammatory diseases. Exp Mol Med. 2017;49:e340
13. Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH. et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019 365
14. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. Bmj. 2018;361:k2179
15. Karlsson F, Tremaroli V, Nielsen J, Bäckhed F. Assessing the human gut microbiota in metabolic diseases. Diabetes. 2013;62:3341-9
16. Ananthakrishnan AN, Bernstein CN, Iliopoulos D, Macpherson A, Neurath MF, Ali RAR. et al. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. 2018;15:39-49
17. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800-12
18. Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59:328-39
19. Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G. et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65:830-9
20. Cenit MC, Sanz Y, Codoñer-Franch P. Influence of gut microbiota on neuropsychiatric disorders. World J Gastroenterol. 2017;23:5486-98
21. Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD. et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med. 2006;355:1018-28
22. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K. et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015-28.e13
23. Lundberg R, Toft MF, Metzdorff SB, Hansen CHF, Licht TR, Bahl MI. et al. Human microbiota-transplanted C57BL/6 mice and offspring display reduced establishment of key bacteria and reduced immune stimulation compared to mouse microbiota-transplantation. Sci Rep. 2020;10:7805
24. Burz SD, Monnoye M, Philippe C, Farin W, Ratziu V, Strozzi F. et al. Fecal Microbiota Transplant from Human to Mice Gives Insights into the Role of the Gut Microbiota in Non-Alcoholic Fatty Liver Disease (NAFLD). Microorganisms. 2021 9
25. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu WH, Saldana-Morales FB. et al. Thymic development of gut-microbiota-specific T cells. Nature. 2021;594:413-7
26. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S. et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science. 2017;358:359-65
27. Matsusaki T, Takeda S, Takeshita M, Arima Y, Tsend-Ayush C, Oyunsuren T. et al. Augmentation of T helper type 1 immune response through intestinal immunity in murine cutaneous herpes simplex virus type 1 infection by probiotic Lactobacillus plantarum strain 06CC2. Int Immunopharmacol. 2016;39:320-7
28. Takeda S, Takeshita M, Kikuchi Y, Dashnyam B, Kawahara S, Yoshida H. et al. Efficacy of oral administration of heat-killed probiotics from Mongolian dairy products against influenza infection in mice: alleviation of influenza infection by its immunomodulatory activity through intestinal immunity. Int Immunopharmacol. 2011;11:1976-83
29. Dayong Ren DW, Hongyan Liu, Minghao Shen, Hansong Yu. Two strains of probiotic Lactobacillus enhance immune response and promote naive T cell polarization to Th1. Food and Agricultural Immunology. 2019;30:281-95
30. Won TJ, Kim B, Song DS, Lim YT, Oh ES, Lee DI. et al. Modulation of Th1/Th2 balance by Lactobacillus strains isolated from Kimchi via stimulation of macrophage cell line J774A.1 in vitro. J Food Sci. 2011;76:H55-61
31. Shiro Takeda MH, Hiroki Yoshida, Masahiko Takeshita, Yukiharu Kikuchi, Chuluunbat Tsend-Ayush, Bumbein Dashnyam, Satoshi Kawahara, Michio Muguruma, Wataru Watanabe, Masahiko Kurokawa. Antiallergic activity of probiotics from Mongolian dairy products on type I allergy in mice and mode of antiallergic action. Journal of Functional Foods. 2014;9:60-9
32. Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655-62
33. Walker JA, McKenzie ANJ. T(H)2 cell development and function. Nat Rev Immunol. 2018;18:121-33
34. Bamias G, Okazawa A, Rivera-Nieves J, Arseneau KO, De La Rue SA, Pizarro TT. et al. Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J Immunol. 2007;178:1809-18
35. Wu W, Liu HP, Chen F, Liu H, Cao AT, Yao S. et al. Commensal A4 bacteria inhibit intestinal Th2-cell responses through induction of dendritic cell TGF-β production. Eur J Immunol. 2016;46:1162-7
36. Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677-88
37. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S. et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367-80
38. Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M. et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808-12
39. Ivanov II, Frutos Rde L Manel N, Yoshinaga K Rifkin DB, Sartor RB et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337-49
40. Farkas AM, Panea C, Goto Y, Nakato G, Galan-Diez M, Narushima S. et al. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J Immunol Methods. 2015;421:104-11
41. Ivanov II, Atarashi K Manel N, Brodie EL Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485-98
42. Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G. et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99-103
43. Huang Y, Tang J, Cai Z, Zhou K, Chang L, Bai Y. et al. Prevotella Induces the Production of Th17 Cells in the Colon of Mice. J Immunol Res. 2020;2020:9607328
44. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J. et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. 2019;576:143-8
45. Dominguez-Villar M, Hafler DA. Regulatory T cells in autoimmune disease. Nat Immunol. 2018;19:665-73
46. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204-9
47. O'Mahony C, Scully P, O'Mahony D, Murphy S, O'Brien F, Lyons A. et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4:e1000112
48. Verma R, Lee C, Jeun EJ, Yi J, Kim KS, Ghosh A. et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci Immunol. 2018 3
49. Smits HH, Engering A, van der Kleij D, de Jong EC, Schipper K, van Capel TM. et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J Allergy Clin Immunol. 2005;115:1260-7
50. Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am J Respir Crit Care Med. 2007;175:561-9
51. Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med. 2009;179:186-93
52. Shah MM, Saio M, Yamashita H, Tanaka H, Takami T, Ezaki T. et al. Lactobacillus acidophilus strain L-92 induces CD4(+)CD25(+)Foxp3(+) regulatory T cells and suppresses allergic contact dermatitis. Biol Pharm Bull. 2012;35:612-6
53. Bernard-Raichon L, Colom A, Monard SC, Namouchi A, Cescato M, Garnier H. et al. A Pulmonary Lactobacillus murinus Strain Induces Th17 and RORγt(+) Regulatory T Cells and Reduces Lung Inflammation in Tuberculosis. J Immunol. 2021;207:1857-70
54. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W. et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. 2020;577:410-5
55. Sorini C, Cardoso RF, Gagliani N, Villablanca EJ. Commensal Bacteria-Specific CD4(+) T Cell Responses in Health and Disease. Front Immunol. 2018;9:2667
56. Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161-8
57. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-9
58. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-84
59. Nastasi C, Fredholm S, Willerslev-Olsen A, Hansen M, Bonefeld CM, Geisler C. et al. Butyrate and propionate inhibit antigen-specific CD8(+) T cell activation by suppressing IL-12 production by antigen-presenting cells. Sci Rep. 2017;7:14516
60. Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S. et al. Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Sci Rep. 2018;8:14430
61. Luu M, Riester Z, Baldrich A, Reichardt N, Yuille S, Busetti A. et al. Microbial short-chain fatty acids modulate CD8(+) T cell responses and improve adoptive immunotherapy for cancer. Nat Commun. 2021;12:4077
62. Yang K, Hou Y, Zhang Y, Liang H, Sharma A, Zheng W. et al. Suppression of local type I interferon by gut microbiota-derived butyrate impairs antitumor effects of ionizing radiation. J Exp Med. 2021 218
63. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600-5
64. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K. et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8(+) T Cells. Immunity. 2019;51:285-97.e5
65. Krijgsman D, Hokland M, Kuppen PJK. The Role of Natural Killer T Cells in Cancer-A Phenotypical and Functional Approach. Front Immunol. 2018;9:367
66. Brailey PM, Lebrusant-Fernandez M, Barral P. NKT cells and the regulation of intestinal immunity: a two-way street. Febs j. 2020;287:1686-99
67. Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD. et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520-5
68. Mattner J, Debord KL, Ismail N, Goff RD, Cantu C 3rd, Zhou D. et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525-9
69. Girardi M. Immunosurveillance and immunoregulation by gammadelta T cells. J Invest Dermatol. 2006;126:25-31
70. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489-93
71. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D. et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123-33
72. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M. et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018 360
73. Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7:140-50
74. Li F, Hao X, Chen Y, Bai L, Gao X, Lian Z. et al. The microbiota maintain homeostasis of liver-resident γδT-17 cells in a lipid antigen/CD1d-dependent manner. Nat Commun. 2017;7:13839
75. Dupraz L, Magniez A, Rolhion N, Richard ML, Da Costa G, Touch S. et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. 2021;36:109332
76. Yang C, Kwon DI, Kim M, Im SH, Lee YJ. Commensal Microbiome Expands Tγδ17 Cells in the Lung and Promotes Particulate Matter-Induced Acute Neutrophilia. Front Immunol. 2021;12:645741
77. Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL. et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A. 2011;108:8743-8
78. Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, Mucida D. Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell. 2017;171:783-94.e13
79. Meierovics AI, Cowley SC. MAIT cells promote inflammatory monocyte differentiation into dendritic cells during pulmonary intracellular infection. J Exp Med. 2016;213:2793-809
80. Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci U S A. 2013;110:E3119-28
81. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF. et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. 2016;17:1300-11
82. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019 366
83. Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M. et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701-8
84. Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. 2012;80:3256-67
85. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361-5
86. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C. et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54
87. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D. et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250-9
88. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B. et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. 2015;212:1095-108
89. Nagpal R, Wang S, Solberg Woods LC, Seshie O, Chung ST, Shively CA. et al. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-human Primate, and Human Feces. Front Microbiol. 2018;9:2897
90. Khan MF, Wang H. Environmental Exposures and Autoimmune Diseases: Contribution of Gut Microbiome. Front Immunol. 2019;10:3094
91. Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat Rev Genet. 2016;17:160-74
92. Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H. et al. Role of the Gut Microbiome in Modulating Arthritis Progression in Mice. Sci Rep. 2016;6:30594
93. Johanson DM 2nd, Goertz JE, Marin IA, Costello J, Overall CC, Gaultier A. Experimental autoimmune encephalomyelitis is associated with changes of the microbiota composition in the gastrointestinal tract. Sci Rep. 2020;10:15183
94. Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S. et al. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep. 2020;31:107471
95. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121-41
96. Lu D, Zhang JB, Wang YX, Geng ST, Zhang Z, Xu Y. et al. Association between CD4(+) T cell counts and gut microbiota and serum cytokines levels in HIV-infected immunological non-responders. BMC Infect Dis. 2021;21:742
97. Salem F, Kindt N, Marchesi JR, Netter P, Lopez A, Kokten T. et al. Gut microbiome in chronic rheumatic and inflammatory bowel diseases: Similarities and differences. United European Gastroenterol J. 2019;7:1008-32
98. Rodrigues GSP, Cayres LCF, Gonçalves FP, Takaoka NNC, Lengert AH, Tansini A. et al. Detection of Increased Relative Expression Units of Bacteroides and Prevotella, and Decreased Clostridium leptum in Stool Samples from Brazilian Rheumatoid Arthritis Patients: A Pilot Study. Microorganisms. 2019 7
99. Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA. et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114:10713-8
100. Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B. et al. Brief Report: Intestinal Dysbiosis in Ankylosing Spondylitis. Arthritis Rheumatol. 2015;67:686-91
101. Maeda Y, Takeda K. Role of Gut Microbiota in Rheumatoid Arthritis. J Clin Med. 2017 6
102. Lange L, Thiele GM, McCracken C, Wang G, Ponder LA, Angeles-Han ST. et al. Symptoms of periodontitis and antibody responses to Porphyromonas gingivalis in juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2016;14:8
103. Rogier R, Evans-Marin H, Manasson J, van der Kraan PM, Walgreen B, Helsen MM. et al. Alteration of the intestinal microbiome characterizes preclinical inflammatory arthritis in mice and its modulation attenuates established arthritis. Sci Rep. 2017;7:15613
104. Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J. et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43
105. Sun Y, Chen Q, Lin P, Xu R, He D, Ji W. et al. Characteristics of Gut Microbiota in Patients With Rheumatoid Arthritis in Shanghai, China. Front Cell Infect Microbiol. 2019;9:369
106. Dorożyńska I, Majewska-Szczepanik M, Marcińska K, Szczepanik M. Partial depletion of natural gut flora by antibiotic aggravates collagen induced arthritis (CIA) in mice. Pharmacol Rep. 2014;66:250-5
107. Takahashi D, Hoshina N, Kabumoto Y, Maeda Y, Suzuki A, Tanabe H. et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine. 2020;58:102913
108. Iwakura Y, Ishigame H. The IL-23/IL-17 axis in inflammation. J Clin Invest. 2006;116:1218-22
109. Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C. et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538-41
110. Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J. et al. High frequency of intestinal T(H)17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. 2017;3:e1700492
111. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615-22
112. Calcinotto A, Brevi A, Chesi M, Ferrarese R, Garcia Perez L, Grioni M. et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat Commun. 2018;9:4832
113. Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S. et al. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041-50
114. Takewaki D, Yamamura T. Gut microbiome research in multiple sclerosis. Neurosci Res. 2021;168:28-31
115. Cantoni C, Lin Q, Dorsett Y, Ghezzi L, Liu Z, Pan Y. et al. Alterations of host-gut microbiome interactions in multiple sclerosis. EBioMedicine. 2022;76:103798
116. Yadav M, Ali S, Shrode RL, Shahi SK, Jensen SN, Hoang J. et al. Multiple sclerosis patients have an altered gut mycobiome and increased fungal to bacterial richness. PLoS One. 2022;17:e0264556
117. Shah S, Locca A, Dorsett Y, Cantoni C, Ghezzi L, Lin Q. et al. Alterations of the gut mycobiome in patients with MS. EBioMedicine. 2021;71:103557
118. Toghi M, Bitarafan S, Kasmaei HD, Ghafouri-Fard S. Bifidobacteria: A probable missing puzzle piece in the pathogenesis of multiple sclerosis. Mult Scler Relat Disord. 2019;36:101378
119. He B, Hoang TK, Tian X, Taylor CM, Blanchard E, Luo M. et al. Lactobacillus reuteri Reduces the Severity of Experimental Autoimmune Encephalomyelitis in Mice by Modulating Gut Microbiota. Front Immunol. 2019;10:385
120. Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T. et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10:e0137429
121. Ling Z, Cheng Y, Yan X, Shao L, Liu X, Zhou D. et al. Alterations of the Fecal Microbiota in Chinese Patients With Multiple Sclerosis. Front Immunol. 2020;11:590783
122. Picchianti-Diamanti A, Panebianco C, Salemi S, Sorgi ML, Di Rosa R, Tropea A. et al. Analysis of Gut Microbiota in Rheumatoid Arthritis Patients: Disease-Related Dysbiosis and Modifications Induced by Etanercept. Int J Mol Sci. 2018 19
123. Schepici G, Silvestro S, Bramanti P, Mazzon E. The Gut Microbiota in Multiple Sclerosis: An Overview of Clinical Trials. Cell Transplant. 2019;28:1507-27
124. Tremlett H, Fadrosh DW, Faruqi AA, Zhu F, Hart J, Roalstad S. et al. Gut microbiota in early pediatric multiple sclerosis: a case-control study. Eur J Neurol. 2016;23:1308-21
125. Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep. 2013;3:2191
126. Cox LM, Maghzi AH, Liu S, Tankou SK, Dhang FH, Willocq V. et al. Gut Microbiome in Progressive Multiple Sclerosis. Ann Neurol. 2021;89:1195-211
127. Troci A, Zimmermann O, Esser D, Krampitz P, May S, Franke A. et al. B-cell-depletion reverses dysbiosis of the microbiome in multiple sclerosis patients. Sci Rep. 2022;12:3728
128. Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM. et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484
129. Montgomery TL, Künstner A, Kennedy JJ, Fang Q, Asarian L, Culp-Hill R. et al. Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc Natl Acad Sci U S A. 2020;117:27516-27
130. Bellone M, Brevi A, Huber S. Microbiota-Propelled T Helper 17 Cells in Inflammatory Diseases and Cancer. Microbiol Mol Biol Rev. 2020 84
131. Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H. et al. Salt-responsive gut commensal modulates T(H)17 axis and disease. Nature. 2017;551:585-9
132. Hofstetter HH, Ibrahim SM, Koczan D, Kruse N, Weishaupt A, Toyka KV. et al. Therapeutic efficacy of IL-17 neutralization in murine experimental autoimmune encephalomyelitis. Cell Immunol. 2005;237:123-30
133. Zenewicz LA, Antov A, Flavell RA. CD4 T-cell differentiation and inflammatory bowel disease. Trends Mol Med. 2009;15:199-207
134. Alam MT, Amos GCA, Murphy ARJ, Murch S, Wellington EMH, Arasaradnam RP. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020;12:1
135. Glover JS, Browning BD, Ticer TD, Engevik AC, Engevik MA. Acinetobacter calcoaceticus is Well Adapted to Withstand Intestinal Stressors and Modulate the Gut Epithelium. Front Physiol. 2022;13:880024
136. Gu ZY, Pei WL, Zhang Y, Zhu J, Li L, Zhang Z. Akkermansia muciniphila in inflammatory bowel disease and colorectal cancer. Chin Med J (Engl). 2021;134:2841-3
137. Gomez-Nguyen A, Basson AR, Dark-Fleury L, Hsu K, Osme A, Menghini P. et al. Parabacteroides distasonis induces depressive-like behavior in a mouse model of Crohn's disease. Brain Behav Immun. 2021;98:245-50
138. Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47-55
139. Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S. et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7:9523
140. Gao Z, Chen KY, Mueller O, Zhang H, Rakhilin N, Chen J. et al. Microbiota of Inflammatory Bowel Disease Models. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:2374-7
141. Nomura K, Ishikawa D, Okahara K, Ito S, Haga K, Takahashi M. et al. Bacteroidetes Species Are Correlated with Disease Activity in Ulcerative Colitis. J Clin Med. 2021 10
142. Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroenterol Res Pract. 2014;2014:872725
143. Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin Microbiol Rev. 2019 32
144. Gryaznova MV, Solodskikh SA, Panevina AV, Syromyatnikov MY, Dvoretskaya YD, Sviridova TN. et al. Study of microbiome changes in patients with ulcerative colitis in the Central European part of Russia. Heliyon. 2021;7:e06432
145. Britton GJ, Contijoch EJ, Mogno I, Vennaro OH, Llewellyn SR, Ng R. et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity. 2019;50:212-24.e4
146. Hudcovic T, Stĕpánková R, Cebra J, Tlaskalová-Hogenová H. The role of microflora in the development of intestinal inflammation: acute and chronic colitis induced by dextran sulfate in germ-free and conventionally reared immunocompetent and immunodeficient mice. Folia Microbiol (Praha). 2001;46:565-72
147. Goldenberg MM. Multiple sclerosis review. P t. 2012;37:175-84
148. Blumberg RS, Strober W. Prospects for research in inflammatory bowel disease. Jama. 2001;285:643-7
149. Singh R, Nieuwdorp M, ten Berge IJ, Bemelman FJ, Geerlings SE. The potential beneficial role of faecal microbiota transplantation in diseases other than Clostridium difficile infection. Clin Microbiol Infect. 2014;20:1119-25
150. Wasilewski A, Zielińska M, Storr M, Fichna J. Beneficial Effects of Probiotics, Prebiotics, Synbiotics, and Psychobiotics in Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21:1674-82
151. Akutko K, Stawarski A. Probiotics, Prebiotics and Synbiotics in Inflammatory Bowel Diseases. J Clin Med. 2021 10
152. Agrawal DK, Shao Z. Pathogenesis of allergic airway inflammation. Curr Allergy Asthma Rep. 2010;10:39-48
153. Broide DH. Molecular and cellular mechanisms of allergic disease. J Allergy Clin Immunol. 2001;108:S65-71
154. Herbst T, Sichelstiel A, Schär C, Yadava K, Bürki K, Cahenzli J. et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198-205
155. Park HK, Choi Y, Lee DH, Kim S, Lee JM, Choi SW. et al. Altered gut microbiota by azithromycin attenuates airway inflammation in allergic asthma. J Allergy Clin Immunol. 2020;145:1466-9.e8
156. Qin S, Xu L, Yi M, Yu S, Wu K, Luo S. Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer. 2019;18:155
157. Buchbinder EI, Desai A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am J Clin Oncol. 2016;39:98-106
158. Wartenberg M, Cibin S, Zlobec I, Vassella E, Eppenberger-Castori S, Terracciano L. et al. Integrated Genomic and Immunophenotypic Classification of Pancreatic Cancer Reveals Three Distinct Subtypes with Prognostic/Predictive Significance. Clin Cancer Res. 2018;24:4444-54
159. Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47-52
160. Derosa L, Routy B, Thomas AM, Iebba V, Zalcman G, Friard S. et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med. 2022;28:315-24
161. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-7
162. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP. et al. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017;19:848-55
163. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-8
164. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A. et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018;8:403-16
165. Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N. et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138-47
166. Zhou B, Sun C, Huang J, Xia M, Guo E, Li N. et al. The biodiversity Composition of Microbiome in Ovarian Carcinoma Patients. Sci Rep. 2019;9:1691
167. Wu AH, Tseng C, Vigen C, Yu Y, Cozen W, Garcia AA. et al. Gut microbiome associations with breast cancer risk factors and tumor characteristics: a pilot study. Breast Cancer Res Treat. 2020;182:451-63
168. Ma Y, Zhang Y, Xiang J, Xiang S, Zhao Y, Xiao M. et al. Metagenome Analysis of Intestinal Bacteria in Healthy People, Patients With Inflammatory Bowel Disease and Colorectal Cancer. Front Cell Infect Microbiol. 2021;11:599734
169. Liu L, Li L, Min J, Wang J, Wu H, Zeng Y. et al. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell Immunol. 2012;277:66-73
170. Zhou W, Zhang D, Li Z, Jiang H, Li J, Ren R. et al. The fecal microbiota of patients with pancreatic ductal adenocarcinoma and autoimmune pancreatitis characterized by metagenomic sequencing. J Transl Med. 2021;19:215
171. Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012-7
172. Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485s-93s
173. Zhang WQ, Zhao SK, Luo JW, Dong XP, Hao YT, Li H. et al. Alterations of fecal bacterial communities in patients with lung cancer. Am J Transl Res. 2018;10:3171-85
174. Vitorino M, Baptista de Almeida S, Alpuim Costa D, Faria A, Calhau C, Azambuja Braga S. Human Microbiota and Immunotherapy in Breast Cancer - A Review of Recent Developments. Front Oncol. 2021;11:815772
175. Cheng H, Wang Z, Cui L, Wen Y, Chen X, Gong F. et al. Opportunities and Challenges of the Human Microbiome in Ovarian Cancer. Front Oncol. 2020;10:163
176. Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GL, Wides R. et al. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science. 2002;296:1661-71
177. Betts AO. Pathogen-free pigs for industry and research. Proc R Soc Med. 1962;55:259-62
178. Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA. et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512-6
179. Van Norman GA. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is it Time to Rethink Our Current Approach? JACC Basic Transl Sci. 2019;4:845-54
180. Ericsson AC, Montonye DR, Smith CR, Franklin CL. Modeling a Superorganism - Considerations Regarding the Use of "Dirty" Mice in Biomedical Research Yale J Biol Med. 2017; 90: 361-71.
181. Abolins S, King EC, Lazarou L, Weldon L, Hughes L, Drescher P. et al. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat Commun. 2017;8:14811
182. Lin JD, Devlin JC, Yeung F, McCauley C, Leung JM, Chen YH. et al. Rewilding Nod2 and Atg16l1 Mutant Mice Uncovers Genetic and Environmental Contributions to Microbial Responses and Immune Cell Composition. Cell Host Microbe. 2020;27:830-40.e4
183. Yeung F, Chen YH, Lin JD, Leung JM, McCauley C, Devlin JC. et al. Altered Immunity of Laboratory Mice in the Natural Environment Is Associated with Fungal Colonization. Cell Host Microbe. 2020;27:809-22.e6
184. Leung JM, Budischak SA, Chung The H, Hansen C, Bowcutt R, Neill R. et al. Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biol. 2018;16:e2004108
185. Baxter NT, Zackular JP, Chen GY, Schloss PD. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome. 2014;2:20
Author contact
![]() Corresponding author: Changwan Hong, Department of Anatomy, Pusan National University School of Medicine Room 504, 49 Busandaehak-ro, Yangsan, Gyeongsangnam-do, 50612, South Korea. Tel: +82-51-510-8041; Fax.: +82-51-510 -8049; E-mail: chongac.kr
Corresponding author: Changwan Hong, Department of Anatomy, Pusan National University School of Medicine Room 504, 49 Busandaehak-ro, Yangsan, Gyeongsangnam-do, 50612, South Korea. Tel: +82-51-510-8041; Fax.: +82-51-510 -8049; E-mail: chongac.kr

 Global reach, higher impact
Global reach, higher impact