Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(4):1316-1335. doi:10.7150/ijbs.81518 This issue Cite
Review
Functional roles, regulatory mechanisms and theranostics applications of ncRNAs in alcohol use disorder
1. Department of Pharmacy, Affiliated Psychological Hospital of Anhui Medical University, Hefei, 230000, China
2. Department of Pharmacy, First Affiliated Hospital of Anhui Medical University, Hefei, 230022, China
3. Department of Pharmacy, Hefei Fourth People's Hospital, Hefei, 230000, China
4. Psychopharmacology Research Laboratory, Anhui Mental Health Center, Hefei, 230000, China
5. Anhui Clinical Research Center for Mental Disorders, Hefei,230000, China
6. The Grade 3 Pharmaceutical Chemistry Laboratory of State Administration of Traditional Chinese Medicine, Hefei, 230022, China
7. Anhui Province Key Laboratory of Major Autoimmune Diseases, Anhui Institute of Innovative Drugs, School of Pharmacy, Anhui Medical University, Hefei, 230032, China
# These authors contributed equally to this work.
Received 2022-12-4; Accepted 2023-2-2; Published 2023-2-21
Abstract
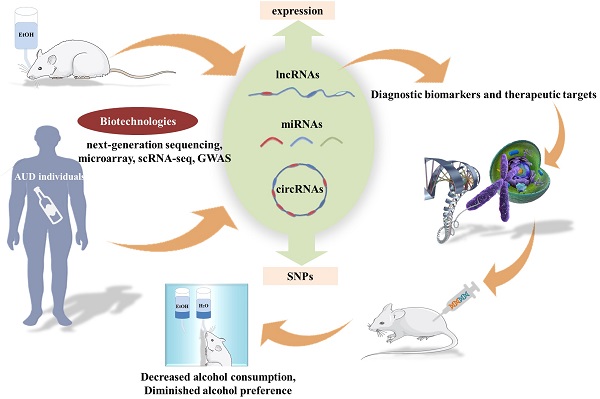
Alcohol use disorder (AUD) is one of the most prevalent neuropsychological disorders worldwide, and its pathogenesis is convoluted and poorly understood. There is considerable evidence demonstrating significant associations between multiple heritable factors and the onset and progression of AUD. In recent years, a substantial body of research conducted by emerging biotechnologies has increasingly highlighted the crucial roles of noncoding RNAs (ncRNAs) in the pathophysiology of mental diseases. As in-depth understanding of ncRNAs and their mechanisms of action, they have emerged as prospective diagnostic indicators and preclinical therapeutic targets for a variety of psychiatric illness, including AUD. Of note, dysregulated expression of ncRNAs such as circRNAs, lncRNAs and miRNAs was routinely found in AUD individuals, and besides, exogenous regulation of partial ncRNAs has also been shown to be effective in ameliorating alcohol preference and excessive alcohol consumption. However, the exact molecular mechanism still remains elusive. Herein, we systematically summarized current knowledge regarding alterations in the expression of certain ncRNAs as well as their-mediated regulatory mechanisms in individuals with AUD. And finally, we detailedly reviewed the potential theranostics applications of gene therapy agents targeting ncRNAs in AUD mice. Overall, a deeper comprehension of functional roles and biological mechanisms of ncRNAs may make significant contributions to the accurate diagnosis and effective treatment of AUD.
Keywords: alcohol use disorder, noncoding RNAs, emerging biotechnology, gene therapy agents, regulatory mechanisms, theranostics applications
Introduction
Alcohol use disorder (AUD) is a kind of chronic recurrent psychiatric illness characterized by an increased tolerance to alcohol, compulsive drinking regardless of physical and mental health, and loss of control over alcohol intake, ultimately leading to the progressive emergence of withdrawal syndrome, grand mal seizures, Wernicke encephalopathy (WE) and even death [1]. The consequences of AUD concerned considerable disability, comorbidity and mortality, as well as substantial medical, caregiver and socioeconomic burdens [2, 3]. On account of the easy availability and rapidly increasing consumption of alcohol, AUD has become one of the most pervasive mental diseases globally, with a male to female ratio of approximately 5:1 [4, 5]. Epidemiological studies showed that the prevalence of AUD was highest in developed countries (8.4%, 95% CI 8.0-8.9), followed by middle-income countries (5.4%, 5.0-6.0), and lowest in less developed countries [1]. Although tools such as the Diagnostic and Statistical Manual of Mental Disorders (DSM) and International Classification of Disease (ICD) criteria, questionnaires and blood tests played an indispensable role in the screening and diagnosis of AUD, less than 15% of AUD patients were diagnosed and treated in a timely manner [6]. Owing to evident contraindications and poor therapeutic effect, the clinical application of anti-craving drugs and alcohol-sensitive drugs was extremely limited, accounting for a persistently high recurrent incidence of AUD [7]. What's worse, the early diagnosis and treatment of AUD still face great challenges as the number of drinkers increases and the abstinence rate declines year by year [4].
AUD is a complicated multifactorial illness whose etiology and pathogenesis have yet been completely understood. It is estimated that 40-60% of AUD risk can be attributed to hereditary factors, with the remainder being linked to individual physiopsychological features, social contextual variables and gene-environment interactions [6]. Long-lasting changes in the expression of brain protein-coding genes have been the most intensively studied in AUD patients and rodents [8, 9]. However, these genes only accounted for less than 2% of the mammalian genome sequence, and the rest of genomic transcripts are noncoding RNAs (ncRNAs) that were previously assumed to be incapable of coding for proteins [10]. More specifically, ncRNAs are a diverse class of bioactive regulatory molecules that are broadly present in organisms, mediating multiple biological processes such as chromatin and histone remodeling, mRNA splicing, transcription, translation, post-transcriptional modification and signal transduction [11]. Moreover, it has recently been shown that ncRNAs possessed the hidden potential to encode peptides and proteins [12]. Emerging biotechnologies such as next-generation sequencing, microarray, single cell RNA sequencing (scRNA-seq) and genome-wide association studies (GWAS) have outperformed traditional genetic techniques in terms of detection efficiency, accuracy and automation [13]. In practice, a large number of studies based on cutting-edge biotechnologies have revealed that ncRNAs were abundant in central nervous system (CNS) and played a regulatory role in brain function and homeostasis, as well as the pathological processes of mental diseases through a range of genetic and epigenetic mechanisms [14, 15]. As a consequence, it was hypothesized that the regulatory network in which ncRNAs participate may affect potential molecular targets and signaling pathways that drive specific cellular biological responses and fates, eventually leading to the occurrence and development of AUD.
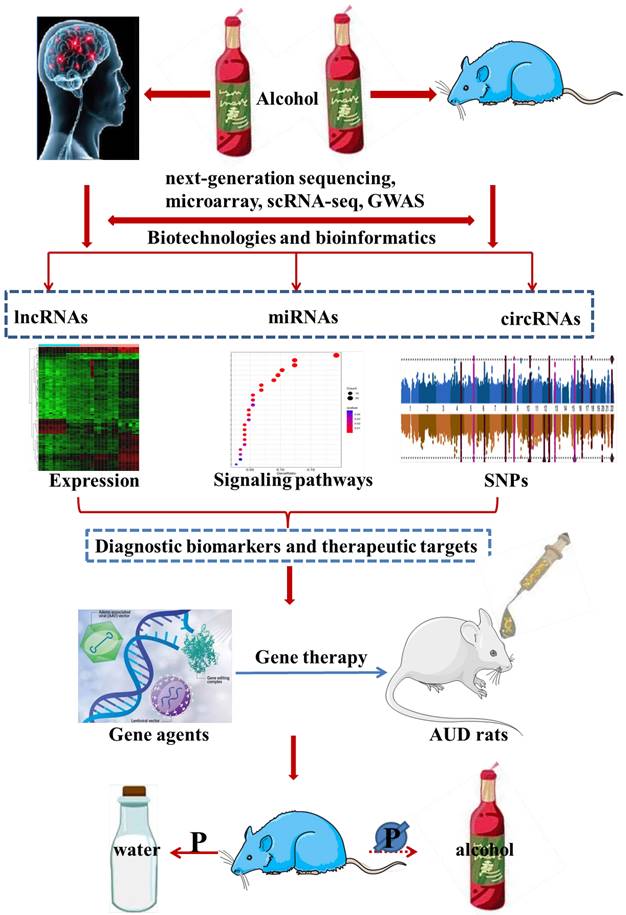
The current situation of AUD diagnosis and treatment is not encouraging, and the urgency to shed light on the etiology and pathophysiology of AUD is becoming more and more prominent. Notably, ncRNAs have been widely used as prospective biomarkers for many different types of diseases, and fortunately, considerable advance has been made in the treatment of a variety of disease models by simulating or silencing the AUD-related ncRNAs [16-19]. More recently, a great deal of studies have assessed the expression of ncRNAs as well as their relevant molecular functions and biological mechanisms in AUD individuals. Despite many exciting advances, reliable diagnostic biomarkers for AUD and their relationship with AUD pathogenesis remain to be further explored. In this review, we mainly focused on the dysregulated expression of ncRNAs in AUD patients and animal models, and then, introduced their diagnostic value and prospects for future research and theranostics application. In addition, we systematically summarized current knowledge regarding the pathogenic ncRNAs-mediated biological mechanisms and functions in AUD individuals. Finally, we discussed the therapeutic effect of ncRNAs-targeting gene therapy agents on AUD animal models in detail. In summary, this review presented a comprehensive description of significant relationships between the expression and single nucleotide polymorphisms (SNPs) of ncRNAs and the progression of AUD, as well as insights into the underlying mechanisms of AUD, which may be of significant implications for the exploitation of diagnostic biomarkers and gene therapy agents for AUD. The content outline of our present review was shown in Figure 1.
Overview of ncRNAs
Thousands of unique ncRNAs sequences exist within cells, which constitute a sizeable portion of the transcriptional landscape [20, 21]. According to the differences in size, structure, subcellular localization and biological properties, ncRNAs are generally categorized as regulatory ncRNAs and housekeeping ncRNAs [22]. More specifically, housekeeping ncRNAs are the most fundamental and abundant RNAs that play important roles in the genomic transcription and translation, including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), etc., whereas regulatory ncRNAs are the functional components of gene expression with heterogeneous molecular groups, which can be further divided into microRNAs (miRNAs), small interfering RNAs (siRNAs), piwi interacting RNAs (piRNAs), long non coding RNAs (lncRNAs), circular RNAs (circRNAs) and natural antisense transcripts [23]. These ncRNAs have long been regarded as transcriptional “junks RNA” when their expression, mechanism and function are unknown [24]. In recent years, with the continuous development of emerging biotechnology and in-depth research on ncRNAs, scientific researchers have increasingly understood that ncRNAs are engaged in a variety of developmental and pathological associated cellular processes and pathways in vivo. Insights into the potential roles of ncRNAs in development and diseases, particularly in brain diseases, have made ncRNAs prospective tools and targets for novel therapeutic approaches. Dysregulation of these ncRNAs, especially lncRNAs, circRNAs and miRNAs, was consistently observed in AUD individuals, which may be associated with the onset and progression of AUD.
CircRNAs and alcohol use disorder
CircRNAs are a kind of ncRNA with covalently closed loops and no polyadenylate tail and cap structure, allowing them to resist exonuclease degradation and so have a prolonged half-life [25]. CircRNAs, originally discovered in pathogens, were subsequently found to be ubiquitously expressed in eukaryotes and have been closely linked to a variety of cellular and physiological processes through multiple mechanisms of actions, including competing endogenous RNAs (ceRNAs) network and alternative splicing [26]. Although the majority of circRNAs were expressed at modest levels in most tissues, individual circRNAs can gradually accumulate to high levels in certain cell types and body fluids, particularly in brain and blood [27-29]. Inspired by the characteristics of high abundance and evolutionary conservation, a growing body of research has highlighted the intriguing prospects of circRNAs as diagnostic markers and therapeutic targets for AUD.
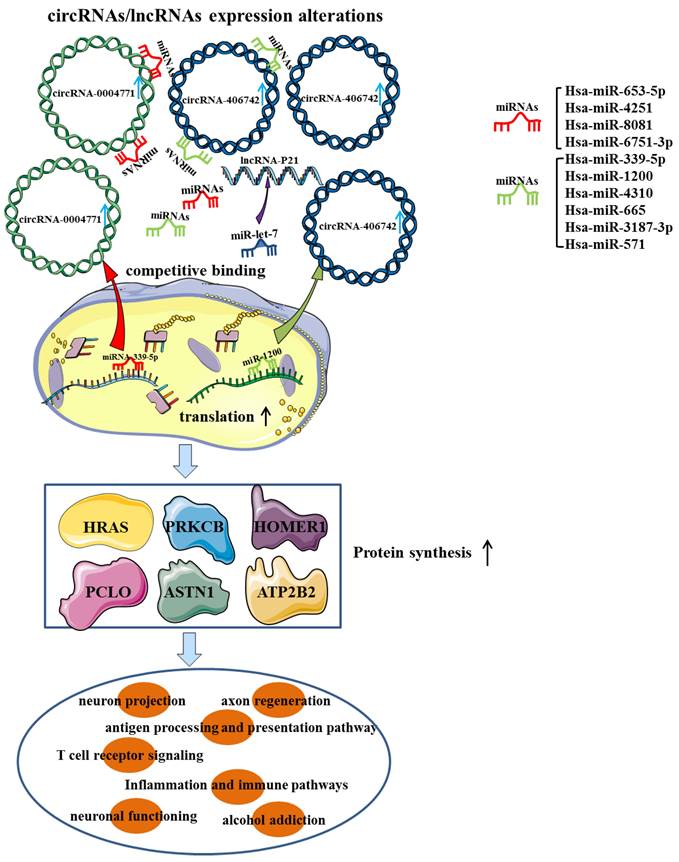
High-throughput sequencing (HTS) identified a total of 399 improperly expressed circRNAs in brain samples from mice exposed to chronic intermittent ethanol (CIE), including 150 up-regulated circRNAs and 249 down-regulated circRNAs. Furthermore, bioinformatic analysis of circRNAs has demonstrated that these dyregulated circRNAs were mainly enriched in GABAergic synapse, retrograde endocannabinoid signaling and morphine addiction [30]. CircRNAs within the exosomes are not easily degraded and can pass through the blood-brain barrier, making circulating exosomal circRNAs abundant and also of great research value in brain diseases [31]. CircRNA-sequencing and quantitative real time PCR (qRT-PCR) results suggested that the expression of serum exosomal hsa-circ-0002130, 0000896, 0004771, 0000825 and 0007177 in AUD patients were significantly higher than those in healthy controls. Among them, hsa-circ-0004771 may be a sensitive diagnostic biomarker that was related to the severity of AUD and can effectively distinguish AUD patients from controls, with an area under curve (AUC) value of 0.874. Mechanistically, differentially expressed (DE) circRNAs may interact with several AD-related miRNAs and then affect infectious and inflammatory pathways [32]. In another study of genome-wide expression of circRNAs in the nucleus accumbens (NAc) of 32 AUD cases/controls, circRNA-406742 expression was found to be decreased in AUD patients and negatively correlated with miR-1200 expression. The ceRNAs networks (circRNA-406742/ miR-1200/HRAS, HOMER1, PRKCB and PCLO) were thought to participate in the pathogenesis of AUD (Figure 2). Besides, the cis-eQTLs of circRNA-000480, 001675, 104942 and 406742 were highly enriched in GWAS & Sequencing Consortium of Alcohol and Nicotine Use and Psychiatric Genetics Consortium AUD GWAS [33, 34]. In conclusion, circRNAs functioned as miRNAs sponge to affect synaptic transmission and neural function through the regulation of target genes, providing a fundamental basis for future studies on the underlying mechanisms of AUD. (Table 1).
LncRNAs and alcohol use disorders
LncRNAs are one of the most abundant classes of ncRNAs in the brain with a length of more than 200 nucleotides and no open reading frames [35]. It is well acknowledged that lncRNAs are of great biological significance in brain function and dysfunction, and that most of which are expressed in a tissue- and temporal- specific manner in the CNS [36]. Evidence has increasingly accumulated that lncRNAs are not only dysregulated in a wide range of neurological illnesses, but also play regulatory roles in gene expression, cellular functions and pathological processes [37, 38]. With the extensive use of modern biotechnologies and bioinformatics, dysregulated lncRNAs were frequently found in the brain of individuals with AUD and have also been related to the pathophysiology of AUD.
The lncRNAs expression and their-related mechanisms in AUD individuals
LncRNA MALAT-1 was shown to be dramatically up-regulated in the cerebellum, hippocampus and brain stem of AUD individuals. Similarly, substantial upregulation of MALAT-1 was observed in the hippocampus and brain stem, as well as the cortex of AUD rats following alcohol withdrawal [39]. There existed a positive correlation between MALAT-1 and neuropeptide 1, and both of them were noticeably enhanced in excitatory synapses and participated in the brain's reward response, which may increase alcohol craving and initiate AUD progression by forming complexes with PSD-95 and NMDA NR2 receptors [40]. It was found that the stability of lncRNA-P21 was decreased in AUD rat model. The possible mechanism was that lncRNA P21 bound to the RNA-binding protein HuR and recruited let-7/AGO2, thereby targeting mRNA translation and boosting AUD development [40, 41] (Figure 2).
The content outline of our present review.
The potential ceRNAs (circRNAs/lncRNAs-miRNAs-mRNAs) network may be involved in the pathogenesis of alcohol use disorder.
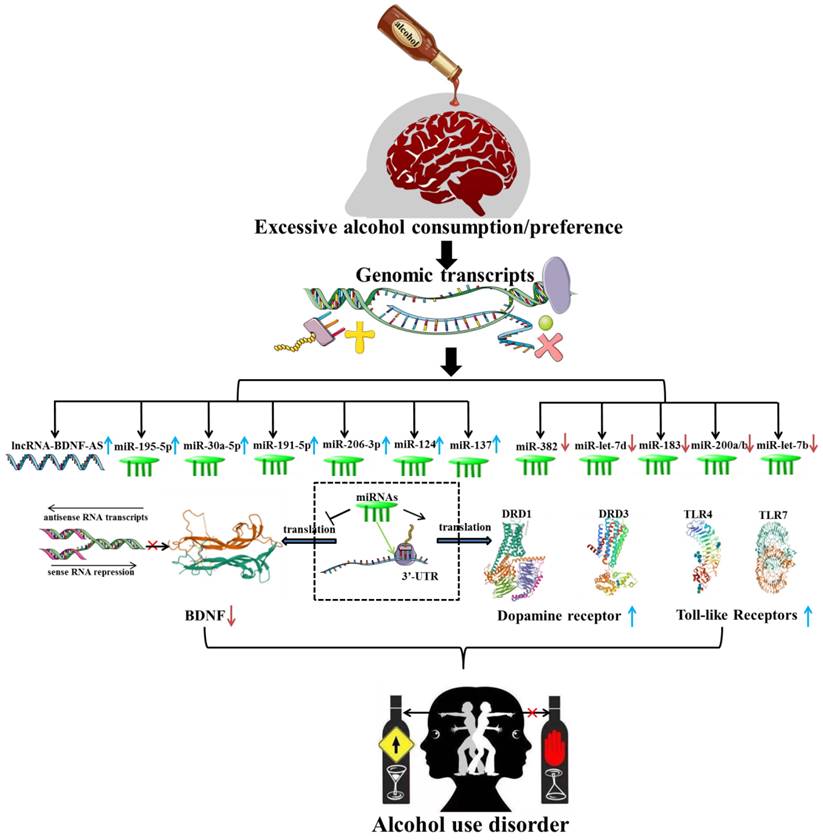
In recent years, several investigations have verified the aberrant expression of brain-derived neurotrophic growth factor (BDNF) in AUD patients and animal models [42, 43]. LncRNA BDNF-AS, a naturally occurring BDNF antisense, was robustly elevated in early onset AUD amygdala (AMY), which appeared to account for decreased BDNF expression and increased EZH2 recruitment. Bohnsack et al. emphasized the importance of enhanced lncRNA BDNF-AS expression for synaptic plasticity and epigenetic reprogramming in the human AMY region [44] (Figure 3). A recent transcriptome analysis has revealed the aberrant expression of lncRNAs, including lincRNA (SNORD3C, RP11-403B2.6, SNORD3C, RP11-638F5.1, RP4-723E3.1, SNORD3C), pseudogene (RP11-252O2.2, HSPA7, RP5-1033K19.2, KRT16P2, RP11-299H22.5) and antisense (RP13-126C7.1, RP11-543H23.2, RP5-1185I7.1) in different brain regions of AUD patients. Further mechanistic studies have shown that these lncRNAs were related to splicing factors dysfunction and may be involved in the course of AUD by regulating post-transcriptional processes and mature mRNA production [45]. In an original study, Farri et al. indicated that several lncRNAs, including NCRNA-00092, 00174 and 00284, may have a cooperative role in cellular plasticity, ultimately leading to the formation of a fixed pattern of chronic alcohol abuse [46]. Furthermore, RNA-seq has demonstrated that eleven lncRNAs (NCRNA-00051, 00256A, 00250A, 00256B, 00245, 00176, 00116, 00175, 00173, 00230B, 00107) were expressed substantially higher in the prefrontal cortex (PFC) of AUD individuals than in controls. While the expression of three lncRNAs (NCRNA-00169, 00161 and 00290) were significantly decreased in AUD individuals [47]. Drake et al. found that 36 lncRNAs modules were obviously associated with AUD and were mostly abundant in alcohol-related processes such as microglia, postsynaptic membrane, glutamatergic synapse, basal neuron and immune-related systems [48]. Saba et al. first revealed an unannotated transcript resembling a lncRNA, which was then termed as “Long noncoding RNA for alcohol preference” (Lrap). Genetic studies revealed that the disrupting Lrap may result in an escalation of alcohol consumption and a pronounced preference for alcohol through controlling gene expression and splicing of a large number of brain transcripts [49]. Given the overlapping changes seen in different types of substance dependence [50], NEAT1, MIAT, MEG3 and EMX2OS, which were previously identified to be up-regulated in heroin/cocaine individuals, were proposed to be involved in the process of developing alcohol addition by modulating synaptic function, synaptogenesis and neuroadaptative mechanisms [51, 52]. In conclusion, all these findings highlighted the potentially regulatory functions of lncRNAs in AUD.
Although numerous studies have identified brain transcriptomic patterns in AUD patients and animal models, the expression levels of transcripts in unique cell types have been rarely investigated. A recent scRNA-seq revealed that lncRNA FP700111.1, AC079793.1, AL807742.1, AC008957.2, AL022068.1, AL512598.2, AC098588.2, AC023421.2 AC104123.1, TMEM72-AS1, LINC01006 was elevated in GABAergic neurons, excitatory neurons, astrocytes, oligodendrocyte progenitor cells (OPCs), oligodendrocytes and microglia of AUD individuals, respectively. Ingenuity pathway analysis (IPA) showed that the identified DElncRNAs were predominately associated with neuroinflammatory signaling, GNRH signaling and neuroimmune response [53]. (Table 2).
CircRNAs in alcohol use disorder
| CircRNAs | Source | Species | Expression | Target | Enriched pathways | References |
|---|---|---|---|---|---|---|
| circ_0002130 | Serum, Exosomes | Human | Up | miR-6074/miR-6514-5p/miR-662/miR-326/miR-4482-3p | ||
| circ_0000896 | Serum, Exosomes | Human | Up | miR-4498/miR-3692-5p/miR-1270/miR-5001-5p/miR-4492 | ||
| circ_0004771 | Serum, Exosomes | Human | Up | miR-8081/miR-6751-3p/ miR-339-5p/miR-4251/miR-653-5p | ||
| circ_0000825 | Serum, Exosomes | Human | Up | miR-340-3p/miR-30a-5p/ miR-4686/miR-4724-3p/miR-5195-3p | T cell receptor signaling, Antigen processing, presentation pathways | [32] |
| circ_0007177 | Serum, Exosomes | Human | Up | miR-224-5p/miR-514a-5p/miR-6891-3p/miR-670-3p/miR-153-5p | ||
| circ_002910 | Serum, Exosomes | Human | Down | miR-1258/miR-6812-5p/miR-6751-5p/miR-6855-5p/miR-6803-5p | ||
| circ_004794 | Serum, Exosomes | Human | Down | miR-6782-5p/miR-6894-5p/miR-30b-3p/miR-6751-5p/miR-6796-5p | ||
| circ_406742 | Nucleus accumbens | Human | Down | miR-1200 | ||
| circ_000390 | Nucleus accumbens | Human | Down | miR-361-5p | ||
| circ_065645 | Nucleus accumbens | Human | Down | miR-571 | synaptic transmission, cellular localization, | [33, 34] |
| circ_405170 | Nucleus accumbens | Human | Down | miR-4310 | neural development | |
| circ_101134 | Nucleus accumbens | Human | Down | miR-665 | ||
| circ_001072 | Nucleus accumbens | Human | Down | miR-3187-3p | ||
| 150 circRNAs | Brain samples | Mice | Up | NM | morphine addiction GABAergic synapse | |
| 249 circRNAs | Brain samples | Mice | Down | NM | Endocannabinoid signaling | [30] |
NM: Not mentioned
Several candidate miRNAs specifically target Brain-derived neurotrophic factor (BDNF), Dopamine receptor (DR) and Toll-like receptor (TLR), which are widely acknowledged to be associated with the occurrence and development of alcohol use disorder.
The lncRNAs risk locus associated with AUD
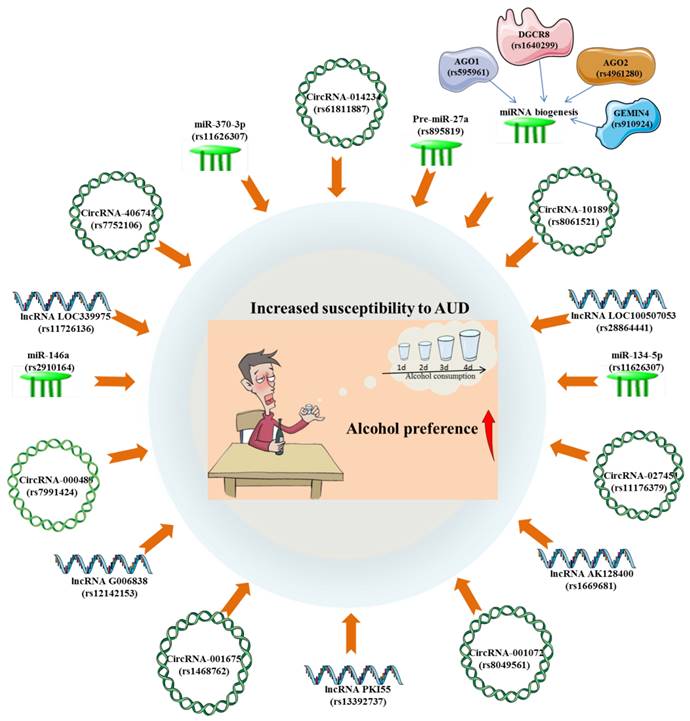
The GWAS of AUD has revealed the possible biological functions of the risk variants of related lncRNAs and their potential regulatory roles on gene expression. A prior GWAS observed an evident connection between lncRNA LOC100507053 locus (rs28864441) and AUD in African-Americans. Mechanistically, LOC100507053, as part of the larger ADH gene cluster, has the capacity to control numerous ADH genes through antisense function [54]. Surprisingly, LOC339975 is classified as an enhancer lncRNA on chromosome 4 and may contribute to the increased ability of the chromosome to translate ADH [55]. It was found that the associated allele of rs11726136 may be responsible for the decreased lncRNA LOC339975 expression in NAc, thereby participating in the pathogenesis of AUD through functional consequence [56]. The cis-eQTLs analysis has demonstrated that the alternative allele of rs1669681 confers a lower lncRNA AK128400 expression and rs12142153 confers a higher lncRNA G006838 expression in AUD individuals, which might be important for elucidating the putative etiological mechanisms of AUD [48]. Besides, there is experimental evidence that eQTLs for a lncRNA PKI55 locus (rs13392737) in the NAc were significantly enriched in AUD GWAS signals [57]. (Figure 4).
MiRNAs and alcohol use disorder
MiRNAs are short endogenous single-stranded RNA molecules of 16-25 nucleotides that comprehensively regulate a variety of physiological and pathological processes in eukaryotic organisms [58]. In terms of mechanisms of action, miRNAs primarily function in posttranscriptional regulation of target genes by inhibiting messenger RNA (mRNA) translation or promoting mRNA degradation [59]. MiRNAs, highly abundant in the brain, are involved in multiple biological functions such as cell proliferation, differentiation, neurodegeneration, programmed death, DNA repair, self-renewal, synapse formation and plasticity, and show great potential as diagnostic markers and therapeutic targets for brain diseases [60]. Over the past decades, genomics and genetics studies have identified a number of genes that may influence drinking behavior in humans and animal models. Notably, dysregulation of miRNAs lead to alterations in biological functions and cellular mechanisms, which may further contribute to the neuropathogenesis of AUD.
LncRNAs in alcohol use disorder
| LncRNAs | Source | Species | Expression | Target | Pathways | References |
|---|---|---|---|---|---|---|
| MALAT-1 | Cerebellum Hippocampus Brain stem | Human | Up | Neuropeptide 1 | Excitatory synapses Brain reward response | [39] |
| P21 | Brain tissue | Rats | Down | miR-let-7 | Neuropeptides activation, Neurotoxicity | [40, 41] |
| BDNF-AS | Amygdala | Human | Up | BDNF/E2H2 | Synaptic plasticity, Epigenetic reprogramming | [44] |
| SNORD3C/ RP11-403B2.6/ SH3RF3-AS1/ HSPA7 | Superior frontal cortex | Human | Up | Specific splicing factors | Dysregulation of splicing factors | [45] |
| RP11-252O2.2 | Nucleus accumbens | Human | Up | |||
| RP11-543H23.2/ RP11-258F1.1/ TBL1XR1-AS1/ AFAP1-AS1/RP11-61I13.3/ AC137932.6/ SNORD3C/RP11-638F5.1/ FAM225B/ LINC00313/RP4-723E3.1/ HSPA7/ RPLP0P2/ RP5-1033K19.2/KRT16P2/ CTD-2575K13.6/AC000367.1/RP11-299H22.5 | Basolateral amygdala | Human | Up | |||
| RP11-543H23.2/ RP11-258F1.1/ RP5-1185I7.1/ RP11-350G8.5/RP13-126C7.1/ SNORD3C/ HSPA7/RPLP0P2/MTND6P6 | Central nucleus of the amygdala | Human | Up | |||
| NCRNA00092/NCRNA00174/NCRNA00284 | Brain tissue | Human | NM | NM | Cellular plasticity | [46] |
| NCRNA-00051/00256A/00250A/00256B/00245/00176/00116/ 00175/00173/00230B/00107 | Prefrontal cortex | Human | Up | NM | NM | [47] |
| NCRNA00169/NCRNA00161/NCRNA00290 | Prefrontal cortex | Human | Down | |||
| Lrap | Brain tissues | Rats | Down | P2rx4 | Dysregulation of splicing factors | [49] |
| MEG3/MIAT/NEAT1/EMX2OS | Nucleus accumbens | Human | Up | NM | Synaptic function, Synaptogenesis, Neuroadaptative | [51, 52] |
| FP700111.1 | GABAergic neurons | Human | Up | NM | Neuroinflammatory, GNRH signaling, Neuroimmune response | [53] |
| AC079793.1, AL807742.1 | Excitatory neurons | Human | Up | NM | ||
| AC008957.2, AL022068.1, AL512598.2, AC098588.2, AC023421.2, AC104123.1 | Astrocytes | Human | Up | SLC1A3 | ||
| TMEM72-AS1, LINC01006 | Oligodendrocytes | Human | Up | NM | ||
| AP002453.1 | Excitatory neurons | Human | Down | NM | ||
| AC069228.1 | Oligodendrocytes | Human | Down | NM | ||
| AC011586.2 | microglia | Human | Down | NM |
NM: Not mentioned.
The miRNAs expression and their-related mechanisms in AUD patients
The miRNAs expression profile and functional analysis revealed that several miRNAs were aberrantly expressed and involved in the progression of AUD through a variety of biological mechanisms. Lewohl et al. reported that about 35 miRNAs (hsa-miR-553/369-3p/18a/339-5p/1/7/196a/301a/144/let-7g/153/let-7f/203/34c-5p/101/376c/665/152/194/423-5p/515-3p/374b/140/519b-3p/586/135b/92a/15b/580/146a/454-3p/380/652/802/196b) were significantly up-regulated in the PFC of AUD individuals when compared with controls. It was then revealed that the predicted target genes of these up-regulated miRNAs have a great overlap with the mRNAs expression profile, thereby leading to the neuronal plasticity, deterioration and concomitant adaptation of neuronal functions [61]. Besides, an original research emphasized that the expression of miR-34a and -34c in hippocampal postmortem tissue of AUD subjects were remarkably higher than those of controls [62]. MiRNA expression profiling conducted by the Affymetrix GeneChip miRNA 2.0 revealed that 12 miRNAs (miR-493/29b/377/375/516a-2/299-3p/488/379/149/767-5p/105/3065-5p) were significantly up-regulated and 4 miRNAs (miR-3162/572/1227/2355) were down-regulated in the PFC of AUD subjects. Among them, 4 up-regulated miRNAs was located on chromosome 14q32, whose predicted gene targets are mainly involved with oligodendrocyte growth, differentiation and signaling [63].
The significant associations between alcohol use disorder (susceptibility, consumption and preference) and SNPs of circRNAs, lncRNAs and miRNAs.
The development of reliable, minimally invasive biomarkers in readily available body fluids may be beneficial for early prediction and diagnosis of advanced diseases. Small RNA-seq analysis identified that 7 miRNAs (miR-451a, miR-10a-5p, miR-100-5p, miR-3613-5p, miR-7704, miR-1290 and miR-4488) and 5 miRNAs (miR-126-3p, miR-10a-5p, miR-1290, miR-4488 and miR-1273h-5p) were differently expressed in the saliva of 56 African-Americans (AA) participants (28 AUD patients and 28 controls) and 64 European-Americans (EA) participants (32 AUD patients and 32 controls), respectively. These saliva miRNAs may be involved in DNA binding, alternative splicing or calcium-dependent cell-cell adhesion, and can serve as easily accessible biomarkers for prediction of AUD with the accuracy of 79.1% in AAs and 72.2% in EAs [64]. Plasma miRNAs profiling revealed that the plasma concentrations of miR-122-5p, miR-3937, miR-193b-3p and miR-4507 were significantly associated with alcohol consumption and may play a mediatory role in the pathogenesis of AUD [65]. Next generation sequencing identified that several serum miRNAs (miR-92b, miR-96, miR-24 and miR-136) were abnormally expressed in AUD individuals, which appeared to be synchronized with the changes in brain miRNAs expression. Ignacio et al. further suggested that these biomarkers may be responsible for alterations in CNS structure and function by consistently affecting cell death, cell proliferation and cell cycle processes [66]. The extracellular release of miRNAs has been found to be one of the mechanisms by which ethanol modulates immunological activation, highlighting the important role of miRNAs in the pathogenesis of AUD. A prior study reported that miR-let-7b released from microglia-derived microvesicles may be responsible for microglial activation and increased Toll-like receptor 7 (TLR7) expression, which further contribute to the ethanol-induced neurotoxicity [67] (Figure 3). MiRNA-Seq determined a number of upregulated circulating extracellular vesicle-bound miRNAs (EV-miRNAs) (miR-155/154/34c/450a/204/124a/211/96) and downregulated (EV-miRNAs) (miR-1224/625) as candidate biomarkers for chronic heavy drinking (CHD), whose expressions were found to be significantly associated with alcohol dose. Additional functional studies suggested that overexpression of miR-155, miR-154, miR-34c, miR-450a, and miR-204 could result in a heightened inflammatory response, while overexpression of miR-625 could lead to increased production of the inflammatory cytokines TNFα or IL-6 in CHD peripheral blood mononuclear cells (PBMC) followed by stimulation with PMA/ionomycin [68]. The stress system has long been considered to play an essential role in motivating compulsive drinking, continued alcohol use and relapse. More recently, it has been revealed that up-regulation of psychological stress-related miR-10a, miR-21 and their downstream genes (transactivation responsive (TAR)-RNA-binding protein associated complex) may lead to increased alcohol intake in binge and alcohol abuse [69].
A large number of studies highlighted the critical roles of miRNAs-dependent repression of target genes and miRNAs-mRNAs regulatory networks in the pathogenesis of AUD. Small RNA-seq analysis determined 19 mature DEmiRNAs (miR-10a-5p, miR-182-5p, miR-1246, miR-126b-5p, miR-4485-3p, miR-486-3p, miR-486-5p, miR-144-5p, miR-1248, miR-5100, miR-1231, miR-144-3p, miR-122-5p, miR-412-5p, miR-4326, miR-302a-5p, miR-6868-3p, miR-5196-3p and miR-412-5p) and 97 DEmRNAs in reward-related or alcohol-responsive brain regions of AUD subjects and controls. It was then revealed that the miRNAs-mRNAs regulatory networks constructed using differential expression and negative correlation pairs could regulate AUD-related pathways, including CREB signaling, IL-8 signaling and axon guidance signaling, which might contribute to the onset and progression of AUD [70]. A transcriptome analysis by microarray-based assay revealed the differential expression of miR-130a in postmortem PFC of 23 AUD subjects and 23 matched controls. Further functional annotation clustering analysis showed that the hub target genes (ITPR2 and ATP1A2) of miR-130a were responsible for the control of ion channel function, which may be involved in the etiological process of AUD [71]. Although the miRNAs alterations in AUD patients showed great consistency, some controversies still existed. A recent meta-analysis of 6 clinical studies revealed that most miRNAs related to dopamine and gamma-aminobutyric acid receptor subunit were significantly up-regulated in AUD patients, while approximately 9% of miRNAs were down-regulated, including miR-567, miR-126, miR-1, miR-432, and miR-15 [72]. Undoubtedly, large-scale studies and more clinical work are needed if the screened specific miRNAs are to be clinically applied as potential biomarkers for AUD (Table 3).
MiRNAs in primates with alcohol use disorder
| MiRNAs | Source | Species | Expression | Target | Pathways | References |
|---|---|---|---|---|---|---|
| miR-553/369-3p/18a/339-5p/1/7/196a/301a/144/let-7g/153/let-7f/203/34c-5p/101/376c/665/152/194/423-5p/515-3p/374b/140/519b-3p/586/135b/92a/15b/580/146a/454-3p/380/652/802/196b | Prefrontal cortex | Human | Up | CXCR4/DICER1/BIRC6/SCARB2/EDIL3/SLAIN1/SSR1/GART/FAM108B1/RNF103/FBXL3/TEX2/FRYL/PAPD4/SESTD1/RIPK2/CDKN1B | Neuronal plasticity, Neuronal deterioration, Neuronal adaptation | [61] |
| miR-34a/miR-34c | Hippocampal tissue | Human | Up | NM | Neuronal physiology, pathology | [62] |
| miR-451a/10a-5p/100-5p/3613-5p/126-3p | Saliva | Human | Up | NM | DNA binding, Alternative splicing, Calcium-dependent cell-cell adhesion | [64] |
| miR-1290/1273h-5p/ 7704/4488 | Saliva | Human | Down | NM | DNA binding, Alternative splicing, Calcium-dependent cell-cell adhesion | [64] |
| miR-96/320b-1/1976/24-1/30a/96-5p/127/136/320b-2/421/671/3615/3676 | Serum | Human | Up | p53 and TNF | Cell proliferation, Cell cycle processes, CNS structure, function | [66] |
| miR-let-7b | Hippocampal tissue | Human | Up | TLR7 | Neurotoxicity | [67] |
| miR-10a/ miR-21 | Peripheral blood | Human | Up | TRBP | Autoimmune, HPA axis reactivity | [69] |
| miR-10a-5p/182-5p/1246/126b-5p/ 4485-3p/486-3p/486-5p/144-5p/124/5100/1231/144-3p/ | Brain tissues | Human | Up | GPRC5C/PTH1R/PIM1/GPR75/CDKN1A/VEGFA/ADRB1/IFNLR1 THBS1/GPR88/JAK3/ITGA9/KCNJ12/VIPR1 | CREB pathways, IL-8 pathways, Axon guidance, neuroplasticity | [70] |
| miR-122-5p/412-5p/4326/302a-5p/6868-3p/5196-3p/412-5p | Brain tissues | Human | Down | PLD1/FNBP1/CDH1/MMP15/TUBB6/GIPR/PDE8A/GPR27 | Opioid pathways, G-Protein receptor | [70] |
| miR-193b-3p/122-5/3937 | Plasma | Human | Up | ALDH2/ FLI/SMAD3/ XPO6/SLC7A11/ FOXP1 | Transmembrane receptor protein Serine/Threonine kinase | [65] |
| miR-4507 | Plasma | Human | Down | ADAM19/C16orf95 | Cell surface receptor | [65] |
| miR-493/29b/377/375/516a-2/299-3p/488/379/149/767-5p/105/3065-5p | Frontal cortex | Human | Up | THBS2/CHN2/NDE1/UGT8/CNP/ENPP2/SEMA4D1 | Oligodendrocyte growth, Differentiation, Neuronal migration | [63] |
| miR-3162/572/1227/2355 | Frontal cortex | Human | Down | |||
| miR-130a | Prefrontal cortex | Human | Down | ITPR2/ATP1A2/ JARID2 | ion channel function, Neuroadaptations | [71] |
| miR-155/154/34c/450a/204/124a/211/96 | Extracellular vesicle, Plasma | Macaque | Up | TNFα or IL-6 | Inflammation, Immune activation | 68] |
| miR-1224/625 | Extracellular vesicle, Plasma | Macaque | Down |
NM: Not mentioned.
The miRNAs expression and their-related mechanisms in AUD animal models
Altered genes expression in the brains of alcohol addicts underlies the neural adaptations for alcoholism and compulsive drinking. The next-generation sequencing identified 14 known alcohol-sensitive miRNAs and 13 putative novel miRNAs in Drosophila repeatedly exposed to alcohol. Specifically, the overexpression of miR-6 and miR-310 were correlated with the increased ethanol sensitivity [73]. Ethanol exposure may lead to upregulation of miR-153a/725/30d/let-7k/100/738/732 in Zebrafish embryos, which may further account for ethanol toxicology. Of course, whether alcohol exposure of Zebrafish embryos can be applied as an AUD model remains to be further validated [74]. miR-7, miR-153, miR-152, miR-15B, miR-203 and miR-144 were predicted to target key genes implicated in the pathophysiology of AUD. The alterations in the expression of these mentioned miRNAs in different cell lines (HEK293T, SH SY5Y and 1321 N1 cells) following exposure to alcohol were consistent with those identified in postmortem brain of AUD patients. Therefore, Steenwyk et al. proposed that cellular models of AUD can be applied to illustrate the underlying mechanisms of gene expression changes that occurred in AUD patients, and that the exploration of effector cells and intercellular cascade mechanism for AUD is indispensable [75].
An original study supported that the expression of miR-411, miR-203, miR-137 and miR-92a were obviously reduced in the mPFC of AUD mice [76]. It was widely acknowledged that the calcium- and voltage-gated BK channel was significantly associated with alcohol tolerance, which may lead to the development of AUD [77]. Pietrzykowski et al. reported that chronic alcohol exposure lead to an increased expression of miR-9, which may participate in the alternative splicing and neuronal adaptation by down-regulating the expression of its target gene, the alpha subunit of the BK channel [78]. miR-382, a critical novel gene, was found to be significantly down-regulated in both alcohol-treated neuronal cells and the NAc of AUD rats, and played a regulatory role in alcohol preference and voluntary intake through the negative regulation of the dopamine D1 receptor (DRD1) and DeltaFosB pathway [79]. A recent research also reported that the expression of miR-382 was significantly reduced in the PFC area, which accompanied with an increase in DeltaFosB expression [80]. The miR-lethal-7 (let-7) family are widely expressed in neurons throughout the brain and has been implicated in several neurophysiological functions and neurobiological responses to drugs abuse [81-83]. Dreyer et al. previously found that miR-let-7d had a positive effect on reducing cocaine-induced conditioned place preference (CPP) and improving measures of anxiety- and depression-like behaviors in mice [84]. Consistently, it was found that miR-let-7d expression was robustly reduced in AUD rats and inversely correlated with the alcohol intake and preference. Bahi et al. also hypothesized that the down-regulation of miR-let-7d might result in the elevated dopamine D3 receptor expression, which in turn enhance alcohol-related behavior responses [85] (Figure 3).
It was generally accepted that the expression of BDNF was decreased in most neuropsychiatric disorders and negatively correlated with the severity of the diseases. Ehinger et al. indicated an inverse correlation between blood serum BDNF levels and alcohol intake in rapid-onset and late-onset AUD rats. In contrast, the expression of miR-30a-5p, miR-195-5p, miR-191-5p and miR-206-3p, which were known to target BDNF, were robustly elevated in the serum of rapid-onset AUD rats [86]. It was found that the expression of miR-206 was significantly up-regulated in medial prefrontal cortex (mPFC) of rats with a history of AUD. Tapocik et al. also speculated that long-lasting induction of miR-206 expression may alter the neural circuits functions and synaptic plasticity through indirectly targeting BDNF 3'-UTR, which may contribute to escalated alcohol consumption [87]. Another research article demonstrated that the expression of miR-206 was robustly increased, while the BDNF expression was reduced in mPFC, CeA and hippocampus of mice exposed to repeated cycles of CIE [88]. It was also reported that the expression of miR-30a-5p in the mPFC of mice with excessive alcohol consumption was robustly up-regulated, which was accompanied by a decrease in BDNF expression. Moreover, the overexpression of miR-30a-5p conducted by a stereotaxic infusion of Ad-miR-30a-5p in the mPFC resulted in an escalation of alcohol intake and a preference for water in the two-bottle choice model, which may be attributed to the decreased BDNF expression [89]. miR-124, which has also been reported to target BDNF, was found to be remarkably decreased after alcohol withdrawal in AUD mice, accompanied by a significant increase in the target gene CDC42, which may be responsible for the drinking relapse [90]. A previous study reported that miR-124a was downregulated, while BDNF was upregulated in the dorsolateral striatum (DLS) of rats following voluntary alcohol intake. Surprisingly, exogenous overexpression of miR-124a mediated by lentiviral (LV)-miR-124a enhanced about fivefold ethamol-induced CPP and alcohol consumption, which may be induced by decreased BDNF expression [91]. Ureña-Peralta et al. observed a significant down-regulation of miR-183 Cluster (miR-96/-182/-183) and miR-200a/b expression and an up-regulation of miR-125b in long-term chronic alcohol exposure WT mice. These miRNAs may lead to increased alcohol intake by regulating the voltage-gated sodium channel, neuron hyperexcitability, innate immune TLR4 signaling pathway, which eventually cause alcohol abuse [92] (Figure 3).
Adolescence is a vulnerable period of neurodevelopment, and besides, alcohol exposure during pubertal development can lead to long-term and widespread neurobiological dysfunction, which may further be involved in the development of AUD. Binge drinking in adolescent is a potential risk factor for AUD and comorbid anxiety in adulthood. Kyzar et al. found that miR-137 was significantly upregulated and its target genes Lsd1 and Lsd1+8a were downregulated in the AMY of adolescent intermittent ethanol (AIE) adult, which may be caused by epigenetic reprogramming. As a result, they highlighted that miR-137 may be applied as a prospective predictive biomarker and therapeutic target for AUD-associated anxiety and AUD susceptibility in adulthood following adolescent alcohol exposure [93]. Prins et al. indicated that peripubertal binge alcohol exposure has long term effects on the miRNAs (miR-10a-5p/26a/103/495) expression and further account for changes in hippocampal function, which may be mediated by the regulation of BDNF and sirtuin-1 (SIRT1) expression [94]. MiRNAs within the EVs can cross the blood-brain barrier and are very stable in the peripheral circulation, which are widely used as reliable biomarkers for brain diseases. It has been confirmed that long-term ethanol exposure reduced the levels of miR-146a-5p and miR-21-5p, thereby triggering the high expression of inflammatory target genes (Traf6, Stat3 and Camk2a). These findings also supported the notion that circulating miRNAs may be involved in the early pathological process of adult AUD patients, such as ethanol-induced neuroinflammation and brain injury in adolescents [95]. It has also been demonstrated that 6 miRNAs (miR-19a-3p/19b-3p/29a-3p/29c-3p/34a/488-3p) were significantly altered in rats exposed to repeated binge alcohol compared to control-treated counterparts [96].
Based on the emerging biotechnology and bioinformatics approaches, a great deal of studies revealed the coordinated dysregulation of miRNAs and associated mRNAs network in the AUD rats brain, which may be of great biological implications for the course of AUD. miRNA microarrays analysis showed that 41 rat miRNAs were significantly altered in the mPFC of rats after a history of AUD. Bioinformatics analysis further identified 33 miRNAs putatively targeting 89 mRNAs, which in turn affect neurotransmission, neuroadaptation, synaptic plasticity, axonal guidance and neurotransmitter signaling [97]. Besides, integration analysis of ex vivo miRNAs and protein expression by Gorini et al. revealed that miRNAs (miR-488-3p/410-3p/3084-3p/let-7a-2-3p/200a-3p/140-3p/96-5p/3107-3p/34b-5p/141-3p/410-3p) were predominantly upregulated and mRNAs were downregulated in CIE-exposed dependent mice. Functional analysis suggested that the coordinated, synergistic regulation of miRNAs and proteins may be responsible for multiple neuroadaptations, driving the behavioral transition from alcohol consumption to dependence [98]. Nunez et al. first reported a positive correlation between increased miRNAs expression and increased mRNA expression in the early stage of AUD, shedding novel light on the current understanding of AUD pathogenesis. They also speculated that the early activation of miRNAs (let-7, miR-7, miR-15, miR-101, miR-140, miR-152, miR-17, miR-34, miR-135, miR-144, miR-146, miR-301, miR-339 and miR-368) may be an adaptive regulation in the initial stage of AUD [99]. It was also demonstrated that long-term ethanol exposure can lead to temporal- and brain area-dependent dysregulation of complex gene networks in C57BL/6J mice. Surprisingly, three DEmiRNAs (miR-187-3p, miR-2137 and miR-7b-3p) were generally detected in AMY, NAc and PFC regions [100]. The expression profiles revealed 11 miRNAs (miR-182, miR-382, miR-9, miR-543, miR-483, miR-206, miR-130a, miR-16, miR-30a and miR27a) and 9 mRNAs (BDNF, SATB2, NRD4A2, GALR1, CREB, SLC17A7-8, ARC) whose expressions were significantly altered after alcohol dependency and withdrawal in rats. Integrated variations of miRNAs-mRNAs expressions in rat brain were linked to pathways, such as reward pathways, synaptic plasticity, neuron differentiation, neurotransmission and chromatin organization, thereby reconstituting the neural circuit function required for the development of AUD [101]. A genome-wide analysis of miRNAs expression identified that AUD-associated miRNAs modules showed significant enrichment for both neuronal, astrocyte and glial marker genes. Among them, miR-449a/b were found to have additional functions related to cellular proliferation and neurotoxic effects in the brain, and may be involved in the pathogenesis of AUD through the negative regulation of hub genes (ELAVL4, DPYSL3, and KCNJ6) in the NAc [102]. A recent study by Martinez reported that the expressions of miR-9-3p, 15b-5p, 16-5p and 222-3p were upregulated in the ethanol-drinking rats model and were almost unchanged in the groups with simultaneous ingestion of ethanol and caffeine. These findings only appeared to indicate that greater caffeine consumption in rats may result in weak changes in serum miRNAs that do not counteract the genetic changes caused by AUD [81] (Table 4).
The miRNAs risk locus associated with AUD
More and more studies have emphasized that there is a close relationship between genotype polymorphism and phenotypic diversity, all of which are of significant importance for understanding the pathogenesis of diseases and predicting the outcome of diseases. A genetic association study described that individuals who are allele C carriers of the miR-146a G>C polymorphism (rs2910164) were susceptible to AUD [103]. Besides, it was found that the Pre-miR-27a rs895819 A>G polymorphism were significantly associated with heavy alcohol consumption in a Mediterranean population, which may account for the occurrence of AUD [104]. Weighted gene co-expression network analyses (WGCNA) on miRNAs expression in NAc of AUD identified 3 significant miRNAs modules and 25 miRNAs hub genes, including hsa-miR-34b-5p/34c-5p/34c-3p/375/34b-3p/4652-3p/4423-3p/383-5p/212-3p/377-5p/132-3p/1912/1180-3p/382-5p/370-3p/361-5p/4760-3p/3189-5p/134-5p/523-3p/4720-3p/4633-5p/4762-5p/4311/555. It was then observed that the significant eQTLs for these 25 miRNA hubs were predominantly enriched in alcohol related GWAS [57]. Among them, hsa-miR-34 family miRNAs have been shown to possibly exhibit trans-eQTL effects with variants associated with NNAT and PSMB5, which may be implicated in the pathogenesis of AUD [57, 105]. There existed a significant association between genes variants of miRNA biogenesis and AUD risk. Specifically, it was found that AGO1 rs595961, AGO2 rs4961280 and DGCR8 rs1640299 have significantly altered the risk for AUD [106]. These findings highlighted the potential relevance of polymorphisms within miRNAs genes as prospective biomarkers to illustrate the susceptibility to AUD (Figure 4).
The potential therapeutic effects of miRNA-relevant biological agents on AUD
Gene therapy is considered to be the third revolution in the field of medicine and pharmacy because it can fundamentally prevent, cure or alleviate various genetic diseases [107]. Detecting gene mutations and comprehending the specific molecular mechanism of hereditary disorders, the type of genetic modifications and delivery methods are critical procedures to perform gene therapy. Gene therapy drugs, including DNA drugs based on DNA modification (e.g., in vivo gene therapy drugs based on viral vectors, in vitro gene therapy drugs, naked plasmid drugs, etc.) and RNA drugs (e.g., antisense oligonucleotide (ASO) drugs, RNA interference drugs, mRNA gene therapy, etc.), are promising therapeutic tools [108]. After years of tortuous development, a total of about 43 gene therapy drugs have been approved for clinical application and more than 3,000 preclinical and clinical trials of gene therapy are under way or have been completed worldwide [109]. Beyond that, in-vivo gene therapy based on viral vectors and small nucleic acid drugs (ASO, siRNA and miRNA) have made significant progresses in preclinical and clinical trials of AUD, which have greatly revolutionized the field and are garnering fresh interest.
MiRNAs in animals with alcohol use disorders
| MiRNAs | Source | Species | Expression | Target | Pathways | References |
|---|---|---|---|---|---|---|
| miR-411/ 203/ 137/ 92a | Prefrontal cortex | Mice | Down | GluA2/Faah/Ppard | Neuroadaptations | [76] |
| miR-9 | Brain tissue | Rat | Up | BK channel | Alternative splicing, Neuronal adaptation, Neuronal plasticity | [78] |
| miR-382 | Nucleus accumbens | Rat | Down | DRD1 | DeltaFosB pathways | [79] |
| miR-let-7d | Hippocampus | Mice | Down | DRD3 | Epigenetic regulation, neuropathophysiology | [85] |
| miR-30a-5p/195-5p/ 191-5p/206-3p | Serum | Rat | Up | BDNF | Neuronal differentiation, Synapse formation, Synaptic plasticity | [86] |
| miR-206 | Prefrontal cortex | Rat | Up | BDNF | Neuroadaptations | [87] |
| miR-30a-5p | Prefrontal cortex | Rat | Up | BDNF | Neuronal proliferation, differentiation, survival, Synaptic plasticity | [89] |
| miR-124 | Brain tissue | Mouse | Down | Cdc42 | Neuronal development | [90] |
| miR-488-3p/410-3p/3084-3p/let-7a-2-3p/200a-3p/140-3p/96-5p/3107-3p/34b-5p/141-3p/410-3p | Brain tissue | Mice | Up | DNM1L/HS90A/DNM1L/DPYL3/DYN1 and so on | Neuroadaptations | [98] |
| miR-let-7/7/15/101/140/152/17/34/135/144/146/301/339/368 | Frontal cortex | Mouse | Up | Aak1/Alcam/Cdc42/Scd1/Mlf2/Gtdc1/Kifc2/Mat2b and so on | Adaptive response | [99] |
| miR-187-3p/2137/7b-3p | Brain tissue | Mice | Down | Mrpl49/Eml5/Bicd2/Acta2/Arhgap44/Dusp/Ccdc149/Fbxl8 Polr2d and so on | Neurotoxicity, Neuronal function | [100] |
| miR-182/382/9/543/483/206/130a/16/30a/27a | Brain tissue | Mice | Up/Down | BDNF/SATB2/NRD4A2/GALR1/CREB/SLC17A7-8/ARC | Reward pathways, Synaptic plasticity, Neurotransmission | [101] |
| miR-449a/b | Nucleus accumbens | Mice | Up | ELAVL4/DPYSL3/KCNJ6 | Neurobiological processes | [102] |
| miR-9-3p/15b-5p/16-5p/222-3p | Serum | Rat | Up | Caspase-3/XIAP/IGF-1R | Neurogenesis, Neuroprotector effects | [81] |
| miR-96/182/183/200a/200b | Cerebral cortex | Mice | Down | Nav1.3/Trpv1/Smad3/PP1-γ/Il1r1/Mapk14/Sirt1/Lrp6/BDNF | Neuroinflammatory pathways | [92] |
| miR-125b | Cerebral cortex | Mice | Up | |||
| miR-137 | Amygdala | Rats | Up | LSD1/BDNF | Chromatin remodeling, H3K9 dimethylation | [93] |
| miR-124a | Dorsolateral striatum | Rats | Down | BDNF | TrkB signaling, DRD3 pathways | [91] |
| miR-10a-5p/26a/103/495 | Hippocampus | Rats | Up | BDNF/SIRT1 | Neuronal development, Neurogenesis | [94] |
| miR-146a-5p/ miR-21-5p | Extracellular vesicles, Plasma | Rats | Down | Traf6/Stat3/Camk2a | Neuroinflammation, Neurotoxicity | [95] |
| miR-19a-3p/19b-3p/29a-3p/29c-3p/34a/488-3p | Hippocampus | Rats | Down | ATXN1/KCNC3/VAMP2/VDAC1 | Synaptic plasticity, Memory formation | [96] |
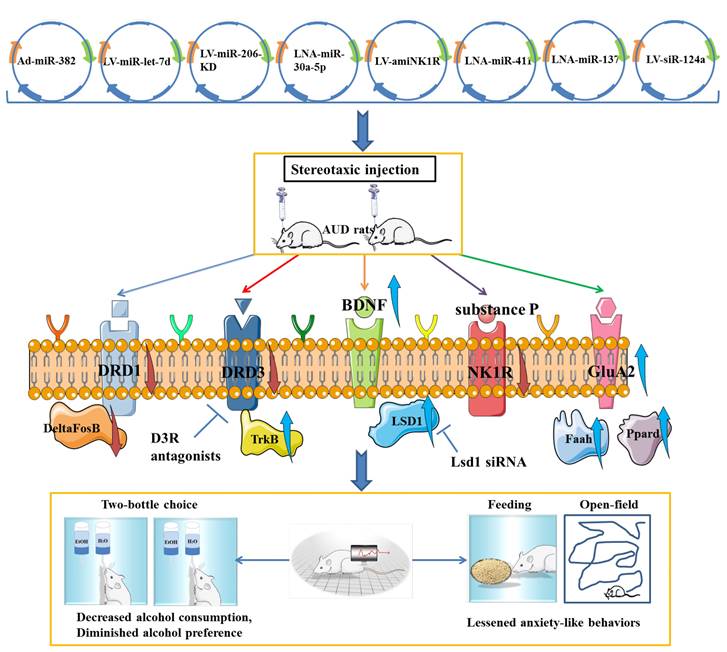
It was confirmed that overexpression of miR-382 via adenovirus (Ad)-miR-382 could effectively affect the drinking behavior and responses, such as lowering voluntary alcohol intake and the preference for alcohol in rats under the intermittent access two-bottle choice drinking paradigm [79]. LV-mediated up-regulation of miR-let-7d in accumbal could effectively decrease alcohol preference and intake by directly targeting the dopamine D3 receptor to decrease its expression [85]. There is experimental evidence that viral-mediated overexpression of miR-206 in the mPFC was sufficient to induce the escalation of alcohol self-administration of rats. Given the causal role of low expression of miR-206 in the induction of BDNF expression and recovery of neural function, it can be speculated that miR-206 could be a potential therapeutic target for AUD [87]. Tapocik et al. have not indicated the potential effect of miRNA-206 knockdown on alcohol intake and alcohol preference in AUD rats, which probably due to the complexity of knockout techniques in vivo. Furthermore, Darcq et al. indicated that inhibition of miR-30a-5p in the mPFC using a locked nucleic acid sequence could decrease excessive alcohol intake and restore the BDNF expression, while overexpression of miR-30a-5p could produce an escalation of alcohol intake and a preference for alcohol [89]. The neuropeptide substance P and neurokinin-1 receptor (NK1R) are widely acknowledged to be involved in the stress response and drug reward systems. The artificial miRNA (amiRNA) carried by lentiviral vector that target the NK1R effectively decreased the voluntary alcohol consumption of AUD rats and NK1R expression in the hippocampus, positively supporting the potential use of amiRNA as a therapeutic agent for the treatment of AUD [110]. It was confirmed that inhibition of miR-137 through direct infusion of antagomiR-137 into the AMY could effectively rescue AIE-induced alcohol intake and anxiety-like behaviors. Further mechanistic studies showed that silencing of miR-137 has an effect on normalizing the decreased LSD1 expression, decreased LSD1 occupancy and decreased BDNF IV expression via increasing H3K9 dimethylation in AIE adult rats [93]. Interestingly, knockdown of miR-124a expression via LV-siR-124a infusion in the DLS positively attenuate ethanol-induced CPP and voluntary alcohol consumption, which was similar to the regulatory effect of LV-BDNF infusion [91]. Although miR-411 was found to be reduced in mice exposed to a chronic alcohol drinking paradigm, silencing of miR-411 by infusion of antagomiR-411 into the mPFC could decrease voluntary alcohol consumption and preference of mice with a history of AUD, which may be related to the elevated levels of glutamate receptor AMPA-2 subunit (GluA2), fatty acid amide hydrolase (Faah) and peroxisome proliferator activated receptor delta (Ppard) [76]. In conclusion, all these findings positively supported the involvement of miRNAs in AUD pathogenesis and may represent novel therapeutic targets for AUD (Figure 5).
Taken together, gene therapy agents can effectively ameliorate AUD-related symptoms by mimicking or silencing AUD-related miRNAs, providing theoretical guidance and practical reference for exogenous regulation of miRNAs in the treatment of AUD. Once the effectiveness of the miRNAs-target agents is experimentally verified, their safety and delivery efficiency can be further optimized. Notably, multiple significant advancements in preclinical models are required to convert preclinical research into clinical practice. Firstly, the safety of gene transfection system, such as host immune response and long-term toxicity, deserved to be great concern. The second issue that needs to be addressed is the effectiveness of the objective genes. The current reality is that most gene therapies are non-specific. The stability and targeting of gene transmission systems are other important restrictions that prevent gene therapy techniques from prospering. In addition, the development and optimization of efficient, safe and targeted gene delivery vectors are also indispensable. The last and most critical constraint is the species difference between humans and model animals, which may result in dramatically different treatment outcomes. In the near future, gene therapy will eventually shine in the treatment of refractory diseases with the continuous development of emerging biotechnologies and materials.
Conclusions and future perspective
Alcohol use disorder (AUD) is an intractable global public health issue with high morbidity, disability and mortality. The current situation of AUD diagnosis and treatment is alarming because of the low rate of consultation and treatment success. Genetic alterations and epigenetic modifications have been associated with the susceptibility to AUD and are involved in the pathogenesis of AUD. Concretely, γ-aminobutyric acid (GABA) transporter 3 (GAT-3) was confirmed to be selectively reduced in the central AMY of AUD patients, accompanied by the impaired GABA clearance [111]. An elevated level of TLR4 signal was found in the ventral tegmental area (VTA) of alcohol-preferring (P) rats in a prior research [112]. In addition, RNA methylation analysis by Arraystar has revealed 29 mRNAs, 5 lncRNAs, and 3 miRNAs that were differently methylated in postmortem NAc of AUD patients and controls [113]. Moonat et al. emphasized the crucial roles of aberrant histone deacetylase 2-mediated histone modifications and synaptic plasticity within the AMY in the pathogenesis of AUD [114]. It has been widely reported that biological processes such as oxidative stress, GABAergic neurotransmission and neuroimmune were strongly related to the onset and progression of AUD [115-117]. Nevertheless, there is still a lack of research on the relationships between ncRNAs and genetic and epigenetic mechanisms, as well as the possible involvement of ncRNAs-mediated cellular functions and biological processes in the course of AUD. Most noteworthy, whether these biological mechanisms are governed by the upstream ncRNAs is another study avenue.
The positive effects of miRNAs-associated biotechnological agents on attenuating alcohol preference, decreasing voluntary alcohol intake and lessening anxiety-like behaviors, as well as the involvement of potential regulatory mechanisms mediated by the gene therapy agents in alcohol use disorder.
A large number of ncRNAs have been implicated in the onset and progression of drug addiction [118]. Whether these ncRNAs can be used as diagnostic indicators and therapeutic targets for AUD are also worthy of further study. Given the recent finding of peptides and proteins encoded by ncRNAs in tumors [119], it is worth investigating whether peptides and proteins encoded by ncRNAs might potentially serve as prospective diagnostic biomarkers and pharmacological targets for AUD. Once validated, these findings will provide a wealth of opportunities for biomarkers, targets and gene therapy drugs. Exosomes participated in cell-to-cell transmission of information and substances through particular mechanisms, and have shown great application advantages and prospects in the diagnosis and treatment of various diseases [120]. Owing to the good stability, biological activity, targeting specificity, and easy crossing of the blood-brain barrier, circulating exosomes are thought to be a valuable model for understanding the pathogenesis of brain disorders [121]. In fact, studies on the potential roles and regulatory mechanisms of exosomal ncRNAs in AUD are particularly lacking, which may point to a weak avenue for understanding the pathophysiology of AUD. More recently, we have performed plasma exosomal miRNA-seq analysis on 25 clinical samples and identified a large number of differentially expressed circulating exosomal miRNAs that could effectively distinguish AUD from normal controls.
Advances in the field of gene therapy have raised the prospect of curing resistant diseases. As we reviewed, the favorable effects of miRNAs-associated gene therapy agents on alleviating alcohol preference and intake have suggested that miRNAs may be prospective therapeutic targets for AUD. However, it is worth evaluating whether therapeutic agents delivered by adenovirus, lentivirus vector and stereotaxic injection might cause substantial damage to rats and affect their alcohol self-administration. Exosomes, as favorable non-viral gene delivery carriers, can protect gene material from the host immune response, and has the advantages of high biocompatibility, low clearance rate, strong permeation retention effect, and cell-targeted delivery. In the future, engineered exosomes can be designed for tissue- and cell-specific delivery, such as specific recognition sequences installed outside exosomes or protein-protein interactions designed by exosomal proteins to enrich exosomes, thereby improving the targeting effect of exosomes as well as loading and delivery efficiency, which may represent a reliable strategy for AUD therapy. Encouragingly, exosomes engineering is gradually being applied preclinical studies, which has shown great clinical benefits and a broad applicability promise. It's also worth noting that neither an escalation nor an inhibition of alcohol intake and preference experiments were performed in transgenic mice. Furthermore, the current reality is that therapeutic effects have only been studied in animal models of AUD, and have not risen to the height of human trials, which deserved to be further investigated. To our knowledge, some other types of ncRNAs such as piRNAs, snRNAs and snoRNAs have been identified to be significantly associated with the progression of certain diseases, which may also play an unforeseen role in the pathogenesis of AUD.
Stereotactic injection may have higher selectivity and regulatory efficiency than systemic injection, which is conducive to safer and more efficient role of gene therapy agents in the treatment of AUD. The pathophysiological processes of brain diseases such as AUD may be precisely pinpointed to a certain brain region. It may lead to poor therapeutic efficiency and significant disparities in therapeutic outcomes among different species if gene therapy agents delivered by exosomes cannot precisely navigate to specific brain regions. But fortunately, systemic administration of EV-derived miR-124 has been shown to attenuate radiation-induced brain injury and secondary neuroinflammation [122]. Another study has also demonstrated that systemic administration of rabies virus glycoprotein (RVG)-exosomes loaded with miR-124 effectively promoted cortical neurogenesis and protected against ischemic injury [123]. In addition, it was found that systemic antimiR-337-3p delivery has a favorable rescue effect on cerebral ischemia-mediated damage [124]. Lai et al. reported that systemic exosomal miR-193b-3p delivery dramatically alleviated neuroinflammation in early brain damage following subarachnoid hemorrhage in mice [125]. Although some other studies have also shown that systemic delivery of miRNA-related gene therapy agents has a favorable effect on brain diseases, the existence of blood-brain barrier may have a slight impact on the therapeutic effect of systemic delivery. Due to its capacity to quickly cross the blood-brain barrier, exosomes delivery vectors can be carefully designed to localize to the induced region of AUD, thereby providing a unique therapeutic benefit.
Despite substantial efforts, only a few gene therapy agents have been approved for clinical use due to many challenges such as potential genotoxicity, inefficiencies in gene transfer or editing, immune responses to repeated drug delivery vectors, and a lack of a societal consensus on controversial issues. Over the past few years, the scientific advances and clinical successes have demonstrated the potential for lasting human health benefits gained from gene therapy, so we have good grounds to remain optimistic and step up efforts to make this therapy part of our standard approach to treating major human diseases. In conclusion, with the emergence and widespread application of modern biotechnology, such as next generation sequencing, microarray, scRNA-seq, GWAS and engineered exosomes, we will finally make breakthroughs in the investigation of the pathogenesis of AUD and the development of therapeutic drugs. Gene therapy could one day upend current treatment strategies for genetic diseases by the use of precise genetic manipulation and delivery tools.
Abbreviations
AUD: alcohol use disorder; ncRNAs: noncoding RNAs; circRNAs: circular RNAs; lncRNAs: long noncoding RNAs; miRNAs: microRNAs; SNPs: single nucleotide polymorphisms; WE: Wernicke encephalopathy; DSM: Diagnostic and Statistical Manual of Mental Disorders; ICD: International Classification of Disease; GWAS: genome-wide association studies; DE: differently expressed; CIE: chronic intermittent ethanol; CNS: central nervous system; ceRNAs: competing endogenous RNAs; AUC: area under curve; WGCNA: weighted gene co-expression network analyses; HTS: High-throughput sequencing; NAc: nucleus accumbens; BDNF: brain-derived neurotrophic growth factor; AMY: amygdala; PFC: prefrontal cortex; scRNA-seq: single-cell RNA sequencing; DR: dopamine receptor; TLR: Toll-like receptor; NK1R: neurokinin-1 receptor; EV: extracellular vesicle; CHD: chronic heavy drinking; PBMC: peripheral blood mononuclear cells; amiRNA: artificial miRNA; GABA: γ-aminobutyric acid; VTA: ventral tegmental area; P: preferring; Ad: adenovirus; LV: lentiviral; CPP: conditioned place preference.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 82104298); Scientific Research Project of Anhui Education Department (Grant No. 2022AH050652); Anhui Province Medical and Health Key Specialty Construction Project; Applied Medicine Research Project of Hefei (Grant No. Hwk2022yb027) and Hefei Fourth People's Hospital project (Grant No. HFSY2022ZD02).
Author Contributions
J.Q. W, Y.R. L and J. Li conceived the original idea. J.Q. W and Y.R. L wrote main manuscript and accomplished the figures and tables. J. Li polished the original manuscript. Q.R. X, J. Liang and J.L. W revised the figures and tables. All authors read and approved the final version of manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Carvalho AF, Heilig M, Perez A, Probst C, Rehm J. Alcohol use disorders. The Lancet. 2019;394:781-92
2. Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. The lancet. 2009;373:2223-33
3. Rehm J, Shield KD. Global burden of disease and the impact of mental and addictive disorders. Current psychiatry reports. 2019;21:1-7
4. Organization WH. Global status report on alcohol and health 2018: World Health Organization; 2019
5. Hasin DS, Wall M, Witkiewitz K, Kranzler HR, Falk D, Litten R. et al. Change in non-abstinent WHO drinking risk levels and alcohol dependence: a 3 year follow-up study in the US general population. The Lancet Psychiatry. 2017;4:469-76
6. Schuckit MA. Alcohol-use disorders. The Lancet. 2009;373:492-501
7. Kranzler HR, Soyka M. Diagnosis and pharmacotherapy of alcohol use disorder: a review. Jama. 2018;320:815-24
8. Xu M, Lin Z. Genetic influences of dopamine transport gene on alcohol dependence: a pooled analysis of 13 studies with 2483 cases and 1753 controls. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2011;35:1255-60
9. Thapa KS, Chen AB, Lai D, Xuei X, Wetherill L, Tischfield JA. et al. Identification of Functional Genetic Variants Associated With Alcohol Dependence and Related Phenotypes Using a High-Throughput Assay. Alcoholism: Clinical and Experimental Research. 2020;44:2494-518
10. Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nature Reviews Genetics. 2010;11:559-71
11. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033-55
12. Wang J, Zhu S, Meng N, He Y, Lu R, Yan G-R. ncRNA-encoded peptides or proteins and cancer. Molecular Therapy. 2019;27:1718-25
13. Guarnaccia M, Gentile G, Alessi E, Schneider C, Petralia S, Cavallaro S. Is this the real time for genomics? Genomics. 2014;103:177-82
14. Wu Y-Y, Kuo H-C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. Journal of Biomedical Science. 2020;27:1-23
15. Varela MA, Roberts TC, Wood MJ. Epigenetics and ncRNAs in brain function and disease: mechanisms and prospects for therapy. Neurotherapeutics. 2013;10:621-31
16. Tang X-H, Guo T, Gao X-Y, Wu X-L, Xing X-F, Ji J-F. et al. Exosome-derived noncoding RNAs in gastric cancer: functions and clinical applications. Molecular Cancer. 2021;20:1-15
17. Qureshi IA, Mehler MF. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics. 2013;10:632-46
18. Li M, Yang Y, Wang Z, Zong T, Fu X, Aung LHH. et al. Piwi-interacting RNAs (piRNAs) as potential biomarkers and therapeutic targets for cardiovascular diseases. Angiogenesis. 2021;24:19-34
19. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q. et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588
20. Esteller M. Non-coding RNAs in human disease. Nature reviews genetics. 2011;12:861-74
21. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nature Reviews Cancer. 2018;18:5-18
22. Hongkuan Z, Karsoon T, Shengkang L, Hongyu M, Huaiping Z. The functional roles of the non-coding RNAs in molluscs. Gene. 2021;768:145300
23. Zhang P, Wu W, Chen Q, Chen M. Non-coding RNAs and their integrated networks. Journal of integrative bioinformatics. 2019 16
24. Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cellular and molecular life sciences. 2010;67:217-37
25. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature biotechnology. 2014;32:453-61
26. Kristensen LS, Andersen MS, Stagsted LV, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics. 2019;20:675-91
27. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nature reviews Clinical Oncology. 2022;19:188-206
28. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS genetics. 2013;9:e1003777
29. Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Progress in neurobiology. 2020;186:101746
30. Gong Z, Rong X, Li X, Wang H, Liu D, He L. et al. Male mice exposed to chronic intermittent ethanol exposure exhibit significant upregulation or downregulation of circular RNAs. The American Journal of Drug and Alcohol Abuse. 2022;48:562-72
31. Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PloS one. 2015;10:e0141214
32. Liu Y, Li J, Bu H, Wang H, Zhang Y, Shen Q. et al. Circular RNA expression alteration identifies a novel circulating biomarker in serum exosomal for detection of alcohol dependence. Addiction Biology. 2021;26:e13031
33. Vornholt E, Drake J, Mamdani M, McMichael G, Taylor ZN, Bacanu SA. et al. Identifying a novel biological mechanism for alcohol addiction associated with circRNA networks acting as potential miRNA sponges. Addiction biology. 2021;26:e13071
34. Vornholt ES. TRANSCRIPTOMIC PROFILING OF POSTMORTEM PREFRONTAL CORTEX AND NUCLEUS ACCUMBENS FROM CHRONIC ALCOHOL ABUSERS. 2020.
35. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393-407
36. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775-89
37. Beermann J, Piccoli M-T, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiological reviews. 2016
38. Puvvula PK. LncRNAs regulatory networks in cellular senescence. International journal of molecular sciences. 2019;20:2615
39. Kryger R, Fan L, Wilce PA, Jaquet V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol. 2012;46:629-34
40. Zhu S, Wu J, Hu J. Non-coding RNA in alcohol use disorder by affecting synaptic plasticity. Experimental Brain Research. 2022:1-15
41. Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S. et al. LincRNA-p21 suppresses target mRNA translation. Molecular cell. 2012;47:648-55
42. Logrip ML, Barak S, Warnault V, Ron D. Corticostriatal BDNF and alcohol addiction. Brain research. 2015;1628:60-7
43. Pandey SC. A critical role of brain-derived neurotrophic factor in alcohol consumption. Biological psychiatry. 2016;79:427-9
44. Bohnsack JP, Teppen T, Kyzar EJ, Dzitoyeva S, Pandey SC. The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Translational psychiatry. 2019;9:1-11
45. Van Booven D, Li M, Sunil Rao J, Blokhin IO, Dayne Mayfield R, Barbier E. et al. Alcohol use disorder causes global changes in splicing in the human brain. Translational psychiatry. 2021;11:1-9
46. Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Molecular psychiatry. 2015;20:1438-47
47. Farris SP, Mayfield RD. RNA-Seq reveals novel transcriptional reorganization in human alcoholic brain. International review of neurobiology. 2014;116:275-300
48. Drake J, McMichael GO, Vornholt ES, Cresswell K, Williamson V, Chatzinakos C. et al. Assessing the role of long noncoding RNA in nucleus accumbens in subjects with alcohol dependence. Alcoholism: Clinical and Experimental Research. 2020;44:2468-80
49. Saba LM, Hoffman PL, Homanics GE, Mahaffey S, Daulatabad SV, Janga SC. et al. A long non-coding RNA (Lrap) modulates brain gene expression and levels of alcohol consumption in rats. Genes, Brain and Behavior. 2021;20:e12698
50. Farris SP, Harris RA, Ponomarev I. Epigenetic modulation of brain gene networks for cocaine and alcohol abuse. Frontiers in neuroscience. 2015;9:176
51. Mayfield RD. Emerging roles for ncRNAs in alcohol use disorders. Alcohol. 2017;60:31-9
52. Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. Journal of neurochemistry. 2011;116:459-66
53. Brenner E, Tiwari GR, Liu Y, Brock A, Mayfield RD. Single cell transcriptome profiling of the human alcohol-dependent brain. bioRxiv. 2019: 780304.
54. Gelernter J, Kranzler H, Sherva R, Almasy L, Koesterer R, Smith A. et al. Genome-wide association study of alcohol dependence: significant findings in African-and European-Americans including novel risk loci. Molecular psychiatry. 2014;19:41-9
55. Denham AN, Drake J, Gavrilov M, Taylor ZN, Bacanu S-A, Vladimirov VI. Long Non-Coding RNAs: The New Frontier into Understanding the Etiology of Alcohol Use Disorder. Non-coding RNA. 2022;8:59
56. Adkins AE, Hack LM, Bigdeli TB, Williamson VS, McMichael GO, Mamdani M. et al. Genomewide association study of alcohol dependence identifies risk loci altering ethanol-response behaviors in model organisms. Alcoholism: Clinical and Experimental Research. 2017;41:911-28
57. Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A. et al. Integrating mRNA and miRNA weighted gene co-expression networks with eQTLs in the nucleus accumbens of subjects with alcohol dependence. PloS one. 2015;10:e0137671
58. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2010;11:597-610
59. Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Current opinion in cell biology. 2009;21:452-60
60. Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Molecular cancer. 2014;13:1-18
61. Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-regulation of microRNAs in brain of human alcoholics. Alcoholism: Clinical and Experimental Research. 2011;35:1928-37
62. Santos-Bezerra DP, Cavaleiro AM, Santos AS, Suemoto CK, Pasqualucci CA, Jacob-Filho W. et al. Alcohol Use Disorder is Associated with Upregulation of MicroRNA-34a and MicroRNA-34c in Hippocampal Postmortem Tissue. Alcoholism: Clinical and Experimental Research. 2021;45:64-8
63. Manzardo A, Gunewardena S, Butler M. Over-expression of the miRNA cluster at chromosome 14q32 in the alcoholic brain correlates with suppression of predicted target mRNA required for oligodendrocyte proliferation. Gene. 2013;526:356-63
64. Rosato AJ, Chen X, Tanaka Y, Farrer LA, Kranzler HR, Nunez YZ. et al. Salivary microRNAs identified by small RNA sequencing and machine learning as potential biomarkers of alcohol dependence. Epigenomics. 2019;11:739-49
65. Karabegović I, Abozaid Y, Maas SC, Labrecque J, Bos D, De Knegt RJ. et al. Plasma microRNA signature of alcohol consumption: the Rotterdam Study. The Journal of Nutrition. 2022
66. Ignacio C, Hicks SD, Burke P, Lewis L, Szombathyne-Meszaros Z, Middleton FA. Alterations in serum microRNA in humans with alcohol use disorders impact cell proliferation and cell death pathways and predict structural and functional changes in brain. BMC neuroscience. 2015;16:1-18
67. Coleman LG, Zou J, Crews FT. Microglial-derived miRNA let-7 and HMGB1 contribute to ethanol-induced neurotoxicity via TLR7. Journal of neuroinflammation. 2017;14:1-15
68. Lewis SA, Doratt B, Sureshchandra S, Pan T, Gonzales SW, Shen W. et al. Profiling of extracellular vesicle-bound miRNA to identify candidate biomarkers of chronic alcohol drinking in nonhuman primates. Alcoholism: Clinical and Experimental Research. 2022;46:221-31
69. Beech RD, Leffert JJ, Lin A, Hong KA, Hansen J, Umlauf S. et al. Stress-Related Alcohol Consumption in Heavy Drinkers Correlates with Expression of mi R-10a, mi R-21, and Components of the TAR-RNA-Binding Protein-Associated Complex. Alcoholism: Clinical and Experimental Research. 2014;38:2743-53
70. Lim Y, Beane-Ebel JE, Tanaka Y, Ning B, Husted CR, Henderson DC. et al. Exploration of alcohol use disorder-associated brain miRNA-mRNA regulatory networks. Translational psychiatry. 2021;11:1-10
71. Wang F, Gelernter J, Zhang H. Differential expression of miR-130a in postmortem prefrontal cortex of subjects with alcohol use disorders. Journal of addiction research & therapy. 2013 4
72. Mohebbati R, Sadeghnia HR. The Role of microRNAs in Alcoholism: A Meta-analytic Review. Current Pharmaceutical Design. 2022;28:1926-31
73. Ghezzi A, Zomeno M, Pietrzykowski AZ, Atkinson NS. Immediate-early alcohol-responsive miRNA expression in Drosophila. Journal of neurogenetics. 2016;30:195-204
74. Soares AR, Pereira PM, Ferreira V, Reverendo M, Simoes J, Bezerra AR. et al. Ethanol exposure induces upregulation of specific microRNAs in zebrafish embryos. Toxicological sciences. 2012;127:18-28
75. Van Steenwyk G, Janeczek P, Lewohl JM. Differential effects of chronic and chronic-intermittent ethanol treatment and its withdrawal on the expression of miRNAs. Brain sciences. 2013;3:744-56
76. Most D, Salem NA, Tiwari GR, Blednov YA, Mayfield RD, Harris RA. Silencing synaptic MicroRNA-411 reduces voluntary alcohol consumption in mice. Addiction biology. 2019;24:604-16
77. Treistman SN, Martin GE. BK Channels: mediators and models for alcohol tolerance. Trends in neurosciences. 2009;32:629-37
78. Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM. et al. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274-87
79. Li J, Li J, Liu X, Qin S, Guan Y, Liu Y. et al. Micro RNA expression profile and functional analysis reveal that mi R-382 is a critical novel gene of alcohol addiction. EMBO molecular medicine. 2013;5:1402-14
80. Shahvaroughi E, Ebadi M. Investigation of the Changes in the Expression of Mir-382 and FOSBΔ Genes in the Prefrontal Area Following Alcohol Addiction in Male Wistar Rats
81. Martinez M, Rossetto IM, Arantes RM, Lizarte F, Tirapelli L, Tirapelli D. et al. Serum miRNAs are differentially altered by ethanol and caffeine consumption in rats. Toxicology Research. 2019;8:842-9
82. Maffioletti E, Cattaneo A, Rosso G, Maina G, Maj C, Gennarelli M. et al. Peripheral whole blood microRNA alterations in major depression and bipolar disorder. Journal of affective disorders. 2016;200:250-8
83. He Y, Wang ZJ. Let-7 microRNAs and opioid tolerance. Frontiers in Genetics. 2012;3:110
84. Bahi A, Dreyer J-L. Lentiviral-mediated let-7d microRNA overexpression induced anxiolytic-and anti-depressant-like behaviors and impaired dopamine D3 receptor expression. European Neuropsychopharmacology. 2018;28:1394-404
85. Bahi A, Dreyer J-L. Lentiviral-mediated up-regulation of let-7d microRNA decreases alcohol intake through down-regulating the dopamine D3 receptor. European Neuropsychopharmacology. 2020;37:70-81
86. Ehinger Y, Phamluong K, Darevesky D, Welman M, Moffat JJ, Sakhai SA. et al. Differential correlation of serum BDNF and microRNA content in rats with rapid or late onset of heavy alcohol use. Addiction Biology. 2021;26:e12890
87. Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A. et al. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. Journal of Neuroscience. 2014;34:4581-8
88. Solomon MG, Griffin WC, Lopez MF, Becker HC. Brain regional and temporal changes in BDNF mRNA and microRNA-206 expression in mice exposed to repeated cycles of chronic intermittent ethanol and forced swim stress. Neuroscience. 2019;406:617-25
89. Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Molecular psychiatry. 2015;20:1240-50
90. Mizuo K, Katada R, Okazaki S, Tateda K, Watanabe S, Matsumoto H. Epigenetic regulation of MIR-124 under ethanol dependence and withdrawal. Nihon Arukoru Yakubutsu Igakkai zasshi= Japanese journal of alcohol studies & drug dependence. 2012;47:155-63
91. Bahi A, Dreyer JL. Striatal modulation of BDNF expression using micro RNA 124a-expressing lentiviral vectors impairs ethanol-induced conditioned-place preference and voluntary alcohol consumption. European Journal of Neuroscience. 2013;38:2328-37
92. Ureña-Peralta J, Alfonso-Loeches S, Cuesta-Diaz C, García-García F, Guerri C. Deep sequencing and miRNA profiles in alcohol-induced neuroinflammation and the TLR4 response in mice cerebral cortex. Scientific reports. 2018;8:1-17
93. Kyzar EJ, Bohnsack JP, Zhang H, Pandey SC. MicroRNA-137 drives epigenetic reprogramming in the adult amygdala and behavioral changes after adolescent alcohol exposure. Eneuro. 2019 6
94. Prins SA, Przybycien-Szymanska MM, Rao YS, Pak TR. Long-term effects of peripubertal binge EtOH exposure on hippocampal microRNA expression in the rat. PLoS One. 2014;9:e83166
95. Ibáñez F, Ureña-Peralta JR, Costa-Alba P, Torres J-L, Laso F-J, Marcos M. et al. Circulating micrornas in extracellular vesicles as potential biomarkers of alcohol-induced neuroinflammation in adolescence: Gender differences. International journal of molecular sciences. 2020;21:6730
96. Asimes A, Kim CK, Rao YS, Bartelt K, Pak TR. microRNA expression profiles in the ventral hippocampus during pubertal development and the impact of peri-pubertal binge alcohol exposure. Non-coding RNA. 2019;5:21
97. Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank J. et al. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. The pharmacogenomics journal. 2013;13:286-96
98. Gorini G, Nunez YO, Mayfield RD. Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain. PLoS One. 2013;8:e82565
99. Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA. et al. Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC genomics. 2013;14:1-21
100. Osterndorff-Kahanek EA, Tiwari GR, Lopez MF, Becker HC, Harris RA, Mayfield RD. Long-term ethanol exposure: Temporal pattern of microRNA expression and associated mRNA gene networks in mouse brain. PloS one. 2018;13:e0190841
101. Sinirlioglu Z, Coskunpinar E, Akbas F. miRNA and mRNA expression profiling in rat brain following alcohol dependence and withdrawal. Cellular and Molecular Biology. 2017;63:49-56
102. Vornholt E, Drake J, Mamdani M, McMichael G, Taylor ZN, Bacanu S-A. et al. Network preservation reveals shared and unique biological processes associated with chronic alcohol abuse in NAc and PFC. PloS one. 2020;15:e0243857
103. Novo-Veleiro I, González-Sarmiento R, Cieza-Borrella C, Pastor I, Laso F-J, Marcos M. A genetic variant in the microRNA-146a gene is associated with susceptibility to alcohol use disorders. European Psychiatry. 2014;29:288-92
104. Barragán R, Coltell O, Asensio EM, Francés F, Sorlí JV, Estruch R. et al. MicroRNAs and drinking: Association between the Pre-miR-27a rs895819 polymorphism and alcohol consumption in a Mediterranean population. International journal of molecular sciences. 2016;17:1338
105. Mamdani M. A Systems Biology Approach to Detect eQTLs Associated with miRNA and mRNA Co-expression Networks in the Nucleus Accumbens of Chronic Alcoholic Patients. 2014.
106. Gedik H, Erdal ME, Yilmaz SG, Sengul C, Sengul CB, Herken H. Association of microRNA biogenesis pathway gene variants and alcohol dependence risk. DNA and cell biology. 2015;34:220-6
107. Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. Gene therapy comes of age. Science. 2018;359:eaan4672
108. Nedorezova DD, Dubovichenko MV, Belyaeva EP, Grigorieva ED, Peresadina AV, Kolpashchikov DM. Specificity of oligonucleotide gene therapy (OGT) agents. Theranostics. 2022;12:7132
109. Shahryari A, Burtscher I, Nazari Z, Lickert H. Engineering gene therapy: advances and barriers. Advanced Therapeutics. 2021;4:2100040
110. Baek MN, Jung KH, Halder D, Choi MR, Lee B-H, Lee B-C. et al. Artificial microRNA-based neurokinin-1 receptor gene silencing reduces alcohol consumption in mice. Neuroscience letters. 2010;475:124-8
111. Augier E, Barbier E, Dulman RS, Licheri V, Augier G, Domi E. et al. A molecular mechanism for choosing alcohol over an alternative reward. Science. 2018;360:1321-6
112. Aurelian L, Warnock K, Balan I, Puche A, June H. TLR4 signaling in VTA dopaminergic neurons regulates impulsivity through tyrosine hydroxylase modulation. Translational psychiatry. 2016;6:e815-e
113. Liu Y, Zhang H. RNA m6A Modification Changes in Postmortem Nucleus Accumbens of Subjects with Alcohol Use Disorder: A Pilot Study. Genes. 2022;13:958
114. Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC. Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biological psychiatry. 2013;73:763-73
115. Yang M, Zhou X, Tan X, Huang X, Yuan L, Zhang Z. et al. The Status of Oxidative Stress in Patients with Alcohol Dependence: A Meta-Analysis. Antioxidants. 2022;11:1919
116. Warden A, Erickson E, Robinson G, Harris RA, Mayfield RD. The neuroimmune transcriptome and alcohol dependence: potential for targeted therapies. Pharmacogenomics. 2016;17:2081-96
117. Gatta E, Guidotti A, Saudagar V, Grayson DR, Aspesi D, Pandey SC. et al. Epigenetic regulation of GABAergic neurotransmission and neurosteroid biosynthesis in alcohol use disorder. International Journal of Neuropsychopharmacology. 2021;24:130-41
118. Zhao Y, Qin F, Han S, Li S, Zhao Y, Wang H. et al. MicroRNAs in drug addiction: Current status and future perspectives. Pharmacology & Therapeutics. 2022: 108215.
119. Zhu S, Wang J, He Y, Meng N, Yan G-R. Peptides/proteins encoded by non-coding RNA: a novel resource bank for drug targets and biomarkers. Frontiers in Pharmacology. 2018;9:1295
120. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977
121. Ranganathan M, Rahman M, Ganesh S, D'Souza DC, Skosnik PD, Radhakrishnan R. et al. Analysis of circulating exosomes reveals a peripheral signature of astrocytic pathology in schizophrenia. The World Journal of Biological Psychiatry. 2022;23:33-45
122. Leavitt RJ, Acharya MM, Baulch JE, Limoli CL. Extracellular Vesicle-Derived miR-124 Resolves Radiation-Induced Brain InjuryExtracellular Vesicle Therapy for the Irradiated Brain. Cancer research. 2020;80:4266-77
123. Yang J, Zhang X, Chen X, Wang L, Yang G. Exosome mediated delivery of miR-124 promotes neurogenesis after ischemia. Molecular Therapy-Nucleic Acids. 2017;7:278-87
124. Wang X, Suofu Y, Akpinar B, Baranov SV, Kim J, Carlisle DL. et al. Systemic antimiR-337-3p delivery inhibits cerebral ischemia-mediated injury. Neurobiology of disease. 2017;105:156-63
125. Lai N, Wu D, Liang T, Pan P, Yuan G, Li X. et al. Systemic exosomal miR-193b-3p delivery attenuates neuroinflammation in early brain injury after subarachnoid hemorrhage in mice. Journal of Neuroinflammation. 2020;17:1-13
Author contact
![]() Corresponding authors: Ya-ru Liu, E-mail: liuyaruedu.cn; Jun Li, E-mail: lijunedu.cn
Corresponding authors: Ya-ru Liu, E-mail: liuyaruedu.cn; Jun Li, E-mail: lijunedu.cn

 Global reach, higher impact
Global reach, higher impact