10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(9):2879-2896. doi:10.7150/ijbs.84994 This issue Cite
Review
The potential role and mechanism of circRNA/miRNA axis in cholesterol synthesis
1. Cancer Institute, The Affiliated Hospital of Qingdao University, Qingdao University, Qingdao Cancer Institute, Qingdao, Shandong, 266000, China.
2. Department of Liver Disease Center, The Affiliated Hospital of Qingdao University, Qingdao, Shandong, 266000, China.
3. Interventional Medicine Center, The Affiliated Hospital of Qingdao University, Qingdao, Shandong, 266000, China.
4. Department of Community Health Promotion, Qingdao Municipal Center for Disease Control & Prevention, Qingdao Institute of Preventive Medicine, Qingdao, Shandong, 266033, China.
5. Department of Endocrinology, Qilu Hospital (Qingdao), Cheeloo College of Medicine, Shandong University, Qingdao, Shandong, 266000, China.
6. School of Life Sciences, Tsinghua University, Beijing, 100084, China.
# These authors contributed equally to this study.
Received 2023-4-5; Accepted 2023-5-16; Published 2023-5-29
Abstract
Cholesterol levels are an initiating risk factor for atherosclerosis. Many genes play a central role in cholesterol synthesis, including HMGCR, SQLE, HMGCS1, FDFT1, LSS, MVK, PMK, MVD, FDPS, CYP51, TM7SF2, LBR, MSMO1, NSDHL, HSD17B7, DHCR24, EBP, SC5D, DHCR7, IDI1/2. Especially, HMGCR, SQLE, FDFT1, LSS, FDPS, CYP51, and EBP are promising therapeutic targets for drug development due to many drugs have been approved and entered into clinical research by targeting these genes. However, new targets and drugs still need to be discovered. Interestingly, many small nucleic acid drugs and vaccines were approved for the market, including Inclisiran, Patisiran, Inotersen, Givosiran, Lumasiran, Nusinersen, Volanesorsen, Eteplirsen, Golodirsen, Viltolarsen, Casimersen, Elasomeran, Tozinameran. However, these agents are all linear RNA agents. Circular RNAs (circRNAs) may have longer half-lives, higher stability, lower immunogenicity, lower production costs, and higher delivery efficiency than these agents due to their covalently closed structures. CircRNA agents are developed by several companies, including Orna Therapeutics, Laronde, and CirCode, Therorna. Many studies have shown that circRNAs regulate cholesterol synthesis by regulating HMGCR, SQLE, HMGCS1, ACS, YWHAG, PTEN, DHCR24, SREBP-2, and PMK expression. MiRNAs are essential for circRNA-mediated cholesterol biosynthesis. Notable, the phase II trial for inhibiting miR-122 with nucleic acid drugs has been completed. Suppressing HMGCR, SQLE, and miR-122 with circRNA_ABCA1, circ-PRKCH, circEZH2, circRNA-SCAP, and circFOXO3 are the promising therapeutic target for drug development, specifically the circFOXO3. This review focuses on the role and mechanism of the circRNA/miRNA axis in cholesterol synthesis in the hope of providing knowledge to identify new targets.
Keywords: Cholesterol synthesis, circRNAs, HMGCR, SQLE, miR-122, nucleic acid drugs.
Introduction
Cholesterol is an important component of vertebrate organisms' membrane and plasma lipoproteins and regulates membrane fluidity and permeability. Cholesterol is also a precursor of steroid hormones, bile acids, and vitamin D. However, plasma cholesterol levels have been firmly the initiating factor of atherosclerosis, cardiovascular disease (ASCVD), and cancer, which are the leading causes of disease and death worldwide. Therefore, controlling cholesterol levels is essential for preventing and treating atherosclerosis [1-3]. The human body gets 300-500 mg of cholesterol from the diet every day and produces about 700-900 mg of cholesterol from scratch [4]. Approximately 50% of endogenous cholesterol is synthesized in the liver. HMG-CoA reductase (HMGCR) is the rate-limiting enzyme in cholesterol synthesis. Statins, which are the HMGCR inhibitors, have been widely used for the treatment of ASCVD. Statins also increase survival rates for cancer patients. However, the efficacy of statins was limited by compensatory increases in HMGCR protein. Statins also induced myopathy and hepatotoxicity [5, 6]. Therefore, more research is required to identify new therapeutic targets and agents. Indeed, many genes play a key role in cholesterol synthesis, including HMG-CoA synthetase 1 (HMGCS1), mevalonate kinase (MVK), phosphomevalonate kinase (PMK), and mevalonate diphosphate decarboxylase (MVD, also named MDD), Farnesyl diphosphate farnesyl transferase 1 (FDFT1), squalene epoxidase (SQLE, also known as squalene monooxygenase (SM)), lanosterol synthase (LSS, also known as oxidosqualene cyclase (OSC)), farnesyl diphosphate synthase (FDPS, also named farnesyl pyrophosphate synthase (FPPS)), sterol 14alpha-demethylase (CYP51, also named cytochrome P450 family 51 subfamily A member 1 (CYP51A1)), transmembrane 7 superfamily member 2 (TM7SF2), lamin B receptor (LBR), methylsterol monooxygenase 1 (MSMO1), NAD(P) dependent steroid dehydrogenase-like (NSDHL), hydroxysteroid 17-beta dehydrogenase 7 (HSD17B7), 24-dehydrocholesterol reductase reductase (DHCR24, also known as seladin-1), cholestenol delta-isomerase (EBP), delta7-sterol 5-desaturase (SC5D), 7-Dehydrocholesterol reductase (DHCR7), isopentenyl diphosphate isomerase 1 and 2 (IDI1/2) [5, 7, 8]. However, more studies are still needed to identify the medicinal properties of these targets.
Circular RNAs (circRNAs) are covalently closed-loop single-stranded RNA. CircRNAs have no 5'-3' polarities and a polyadenylated tail, making them much more stable and resistant to RNase R degradation than linear RNA. CircRNAs regulate gene expression by serving as the miRNA sponges, protein scaffolds and sponges, encoding proteins, and regulating splicing and transcription [9-11]. So far, many small nucleic acid drugs and vaccines were approved for market, including Inclisiran (Proprotein convertase subtilisin/kexin-9 (PCSK9) siRNA), Patisiran (transthyretin (TTR) siRNA), Inotersen (TTR antisense oligonucleotide (ASO)), Givosiran (aminolevulinate synthase 1 (ALAS1) siRNA), Lumasiran (hydroxyacid oxidase 1 (HAO1) siRNA), Nusinersen (exon 7 of survival motor neuron 2 (SMN2) ASO), Volanesorsen (Apolipoprotein C3 (APOC3) ASO), Eteplirsen (exon 51 of Duchenne muscular dystrophy (DMD) ASO), Golodirsen (exon 53 of DMD ASO), Viltolarsen (exon 53 of DMD ASO), Casimersen (exon 7 of DMD ASO), Elasomeran (COVID19 Spike glycoprotein mRNA vaccine, also named mRNA-1273), Tozinameran (COVID19 Spike glycoprotein mRNA vaccine, also named BNT162b) [12-23]. There are also multiple nucleic acid agents in preclinical or clinical studies. However, most of these agents are linear RNA drugs. Compared to linear RNA agents, circRNAs may have prolonged half-lives, high stability, low immunogenicity, low production cost, and high delivery efficiency due to the covalently closed structures. CircRNA agents are being developed by several companies, such as Orna Therapeutics, Laronde, CirCode, and Therorna [24, 25]. Interestingly, circRNAs also regulated cholesterol synthesis by serving as miRNA sponges [26, 27]. The formation, classification, and function of circRNAs and miRNAs, please see reviews by other groups [28, 29]. Therefore, we focused on the role and mechanism of the circRNA/miRNA axis in regulating cholesterol synthesis to affect atherosclerosis and provided some potential targets for the diagnosis and treatment of atherosclerosis.
The mechanism of cholesterol synthesis
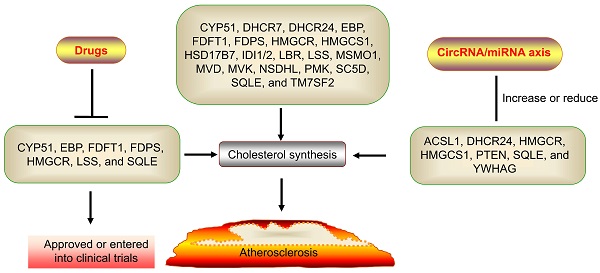
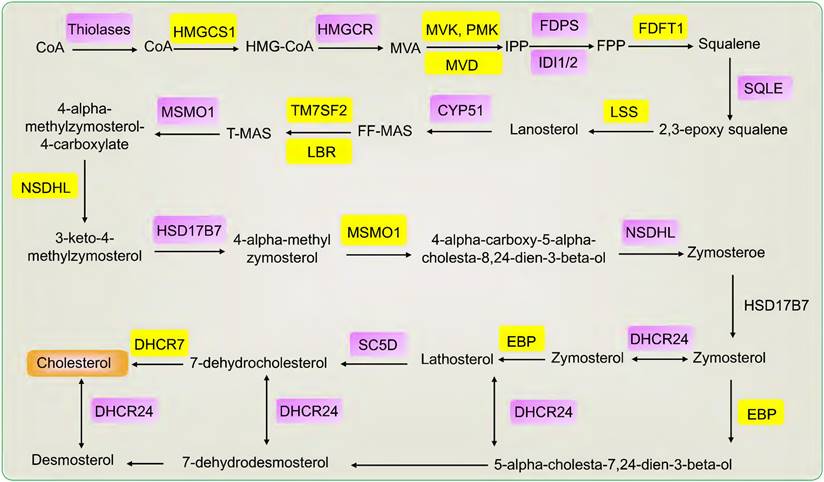
Cholesterol is biosynthesized in three main steps. Firstly, the synthesis of isoprene pyrophosphate (IPP). Acetyl-coenzyme A (CoA) is catalyzed to acetyl-CoA by thiolases and then catalyzed to form 3-hydroxy-3-methylglutaryl CoA (HMG-CoA) by HMGCS1. HMGCR catalyzes HMG-CoA to form mevalonate (MVA). Mevalonate was phosphorylated and decarboxylated to produce IPP by three sequential ATP-dependent Enzymes, including MVK, PMK, and MVD. Secondly, the synthesis of squalene. IPP is catalyzed to form farnesyl pyrophosphate (FPP) and then catalyzed to form squalene by FDFT1. Thirdly, the synthesis of cholesterol. Squalene is catalyzed to form 2,3-epoxy squalene by SQLE and then to form lanosterol by LSS. Lanosterol is catalyzed to form desmosterol and cholesterol after methyl transfer, oxidation, and decarboxylation reaction and then catalyzed to form cholesterol [7, 30, 31]. According to KEGG, many genes are involved in cholesterol synthesis, such as FDPS, CYP51, TM7SF2, LBR, MSMO1, NSDHL, HSD17B7, DHCR24, EBP, SC5D, DHCR7, IDI1/2 [8]. Specifically, FDPS catalyzes the conversion of isopentenyl diphosphate into farnesyl pyrophosphate. CYP51 is a housekeeping gene of the cytochrome P450 that catalyzes the conversion of lanosterol into 4,4-dimethyl-5-alpha-cholesta-8,14,24-trien-3-beta-ol (FF-MAS). TM7SF2 encodes beta-hydroxysterol Delta (14)-reductase (C14SR, DHCR14) that catalyzes the conversion of FF-MAS into 14-demethyllanosterol (T-MAS). LBR and DHCR14 uniquely share the same delta-14 reductase activity in cholesterol biosynthesis. MSMO1 is an intermediate enzyme of cholesterol biosynthesis. NSDHL is a 3beta-hydroxysterol dehydrogenase that catalyzes the conversion of 4-alpha-carboxy-5-alpha-cholesta-8,24-dien-3-beta-ol into zymosterone. HSD17B7 catalyzes the conversion of zymosterone to zymosterol. DHCR24 catalyzes the conversion of desmosterol to cholesterol. EBP catalyzes the conversion of zymostenol into lathosterol. SC5D catalyzes the conversion of lathosterol into 7-dehydrocholesterol. DHCR7 catalyzes the conversion of 7-dehydrocholesterol to form cholesterol and is the final step in cholesterol synthesis. IDI1/2 is the cytoplasmic enzyme involved in cholesterol synthesis [8, 32-35]. Taken together, many genes play a central role in cholesterol synthesis, including HMGCR, SQLE, HMGCS1, FDFT1, LSS, MVK, PMK, MVD, FDPS, CYP51, TM7SF2, LBR, MSMO1, NSDHL, HSD17B7, DHCR24, EBP, SC5D, DHCR7, IDI1/2 (Fig. 1).
The genes and products of cholesterol biosynthesis pathway.
The potential of cholesterol synthesis genes in drug development
As described above, the cholesterol synthesis pathway involves multiple genes. Especially, HMGCR and SQLE are the rate-limiting enzymes in cholesterol synthesis. Statins have been widely used for the treatment of ASCVD by suppressing HMGCR [36-38]. Many studies have shown that statins increase survival rates for cancer patients, including prostate cancer (PCa), lung cancer, gastric cancer (GC), renal cell carcinoma (RCC), breast cancer, colorectal cancer, ovarian cancer, pancreatic cancer, esophageal cancer, endometrial cancer, suggesting that HMGCR is a broad-spectrum anticancer and cardiovascular disease target [39, 40]. Many drugs have entered the stage of market or clinical trials by targeting other cholesterol synthesis genes, such as SQLE, FDFT1, LSS, FDPS, CYP51, and EBP (Table 1). The SQLE inhibitors include terbinafine [41, 42], liranaftate [43], naftifine [44], Butenafine Hydrochloride [44], Amorolfine Hydrochloride [45, 46]. The FDFT1 inhibitors include BPH-652 (also named BMS-188745), S-BPH-652 (also named BMS-188494 or SQ-32709) [47-50], Lapaquistat acetate (also named TAK-475) [51, 52]. The LSS inhibitors include Oxiconazole Nitrate (also named Ro 13-8996) [53, 54] and BIBB-515 (also named BIBB 515 BS) [55]. The FDPS inhibitors include alendronate [56], incadronate (INC, also named cimadronate or YM-175) [56, 57], ibandronate [56, 58, 59], minodronate [56], risedronate [56], pamidronate [56], zoledronate [56]. The CYP51 inhibitors include albaconazole (also named stiefel or UR-9825) [60, 61], arasertaconazole nitrate [62, 63], Bifonazole (also named B3LYP) [64-67], butoconazole (BTZ) [68, 69], clotrimazole [70], dapaconazole [71, 72], eberconazole (EBZ) [73], econazole (also named EcoNai™, SEPA, Spectazole, Ecostatin, or Pevaryl) [74, 75], efinaconazole (also named KP-103 or Jublia) [41, 76, 77], fluconazole [78], flutrimazole [79, 80], fosravuconazole (F-RVCZ, the prodrug of ravuconazole (also named E1224)) [81-86], genaconazole (also named SCH 39304, a racemic mixture that contains 50% of the SCH 42427 and 50% of SCH 42426 enantiomers) [87, 88], HCP002 (a phosphate-modified derivative of voriconazole) [89], IDP113 [90], isavuconazole (ISA, the prodrug of isavuconazole (BAL 4815)) [91, 92], ketoconazole (KTC) [93-95], levoketoconazole [95, 96], itraconazole [97], luliconazole [85, 98], miconazole [99], opelconazole (also named PC945) [100, 101], oteseconazole (also named VT-1161) [102], posaconazole [84, 103], pramiconazole (also named R126638) [104-107], quilseconazole (also named VT-1129) [108, 109], SSY726 [110, 111], voriconazole (VRC) [112, 113], VT-1598 [114, 115]. The EBP inhibitors include DSP-0390 (also named RB55ZW48XG) [116-118]. Thus, EBP, FDFT1, FDPS, HMGCR, LSS, and SQLE are promising targets for drug development.
The drugs in the market and clinical trials targeting cholesterol synthesis genes.
| Name | Structure | Target | Diseases | Status | Refs |
|---|---|---|---|---|---|
| Atorvastatin | 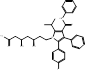 PubChem CID: 60823 | HMGCR | ASCVD | Market | [36-38] |
| Fluvastatin | 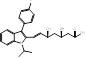 PubChem CID: 446155 | HMGCR | ASCVD | Market | [36-38] |
| Lovastatin | 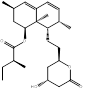 PubChem CID: 53232 | HMGCR | ASCVD | Market | [36-38] |
| Pravastatin | 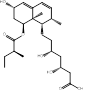 PubChem CID: 54687 | HMGCR | ASCVD | Market | [36-38] |
| Rosuvastatin | 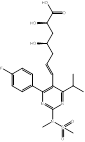 PubChem CID: 446157 | HMGCR | ASCVD | Market | [36-38] |
| Simvastatin | 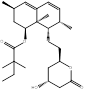 PubChem CID: 54454 | HMGCR | ASCVD | Market | [36-38] |
| Terbinafine |  PubChem CID: 1549008. | SQLE | Onychomycosis and superficial dermatomycoses | Market | [41, 42] |
| Liranaftate |  PubChem CID: 3936. | SQLE | Tinea | Market | [43] |
| Naftifine | 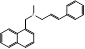 PubChem CID: 47641. | SQLE | Tinea corporis, Tinea cruris, Tinea pedis | Market | [44] |
| Butenafine | 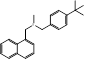 PubChem CID: 2484. | SQLE | Mycoses, onychomycosis, pityriasis versicolor, Tinea corporis, Tinea cruris, Tinea pedis | Market | [44] |
| Amorolfine |  PubChem CID: 54260. | SQLE | Onychomycosis and various local dermal mycoses | Market | [45, 46] |
| BPH-652 | 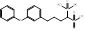 PubChem CID: 10004539. | FDFT1 | Cholesterol-lowering agent | early clinical trials (Completed) | [47-49] |
| S-BPH-652 | 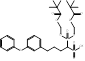 PubChem CID: 154098. | FDFT1 | Hyperlipidaemia | Phase 2 (Discontinued) | [48-50] |
| Lapaquistat Acetate | 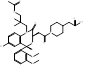 PubChem CID: 9874248. | FDFT1 | Hypercholesterolemia | Phase 3 (Completed) | [51, 52] |
| Oxiconazole Nitrate | 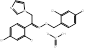 PubChem CID: 9556529. | LSS | Tinea pedis, tinea cruris, and tinea corporis | Market | [53, 54] |
| BIBB-515 |  PubChem CID: 501398. | LSS | Hyperlipidemia | Phase 1 (Completed) | NCT02266498 (ClinicalTrials.gov), NCT02266485, [55] |
| Alendronate | 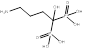 PubChem CID:2088. | FDPS | Corticosteroid-induced osteoporosis; Fracture; Male osteoporosis; Malignant hypercalcaemia; Osteitis deformans; Osteoporosis; Postmenopausal osteoporosis | Market | [56] |
| Incadronate | 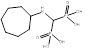 PubChem CID: 3013050. | FDPS | Malignant hypercalcaemia | Market | [56, 57] |
| Ibandronate |  PubChem CID: 6918123. | FDPS | Cancer metastases; Malignant hypercalcaemia; Osteoporosis; Postmenopausal osteoporosis | Market | [56, 58, 59] |
| Minodronate |  PubChem CID: 130956. | FDPS | Osteoporosis | Market | [56] |
| Risedronate |  PubChem CID: 5245. | FDPS | Corticosteroid-induced osteoporosis; Male osteoporosis; Osteitis deformans; Osteoporosis; Postmenopausal osteoporosis | Market | [56] |
| Pamidronate |  PubChem CID: 4674. | FDPS | Osteoporosis | Market | [56] |
| Zoledronate | 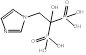 PubChem CID: 68740. | FDPS | Bone metastases; Corticosteroid-induced osteoporosis; Fracture; Male osteoporosis; Malignant hypercalcaemia; Mesothelioma; Multiple myeloma; Osteitis deformans; Postmenopausal osteoporosis | Market | [56] |
| Albaconazole | 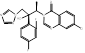 PubChem CID: 208952. | CYP51 | Onychomycosis | Phase 2 (Completed) | [60, 61] |
| Candidiasis Vulvaginitis | Phase 2 (Terminated) | NCT00199264 | |||
| Arasertaconazole nitrate | 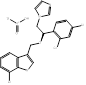 PubChem CID: 9806019 | CYP51 | Vulvovaginal Candidiasis (VVC) | Phase 3 (Planning) | [62, 63] |
| Bifonazole | 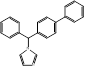 PubChem CID: 2378. | CYP51 | Otomycosis, onychomycos, isseborrhoeic dermatitis of the scalp | Market | [64-67] |
| Butoconazole |  PubChem CID: 47472. | CYP51 | Vulvovaginal candidiasis | Market | [68, 69] |
| Clotrimazole |  PubChem CID: 2812. | CYP51 | Skin, oral and vaginal candida infections | Market | [70] |
| Dapaconazole |  PubChem CID: 51001696. | CYP51 | Tinea Pedis | Phase 3 (completed) | NCT03320486, [71, 72] |
| Eberconazole |  PubChem CID: 72051. | CYP51 | Cutaneous fungal infections | Market | [73] |
| Econazole |  PubChem CID: 3198. | CYP51 | Fungal infections such as tinea pedis and cruris, pityriasis versicolor | Market | [74, 75] |
| Efinaconazole |  PubChem CID: 489181. | CYP51 | Onychomycosis | Market | [41, 76, 77] |
| Fluconazole |  PubChem CID: 3365. | CYP51 | Vulvovaginal candidiasis (RVVC) | Market | [78] |
| Flutrimazole |  PubChem CID: 3401. | CYP51 | Superficial skin fungal infections | Market | [79, 80] |
| Fosravuconazole |  PubChem CID: 9807507. | CYP51 | Onychomycosis | Market | [81-86] |
| Genaconazole |  PubChem CID: 452261, 456001. | CYP51 | Meningitis, Cryptococcal HIV Infections | Phase 1 (Completed) | NCT00000677, [87, 88] |
| HCP002 |  PubChem CID: unknown. | CYP51 | Invasive fungal infections (IFI) | Phase 1 (Recruiting) | [89] |
| IDP113 | Unknown | CYP51 | Tinea capitis (Discontinued) | Phase 2 (ongoing on 30 Aug 2010) | [90] |
| Isavuconazole |  PubChem CID: 6918485. | CYP51 | Invasive aspergillosis (IA) and invasive mucormycosis (IM) | Market | [91, 92] |
| Ketoconazole |  PubChem CID: 456201. | CYP51 | Systemic and superficial mycoses, cushing's syndrome (CS) | Market | [93-95] |
| Levoketoconazole |  PubChem CID: 47576. | CYP51 | CS | Market | [95, 96] |
| Itraconazole |  PubChem CID: 55283. | CYP51 | Broad spectrum antifungal agent | Market | [97] |
| Luliconazole |  PubChem CID: 3003141. | CYP51 | Onychomycosis | Market | [85, 98] |
| Miconazole |  PubChem CID: 4189. | CYP51 | Superficial and cutaneous disease | Market | [99] |
| Opelconazole |  PubChem CID: 121383526. | CYP51 | Pulmonary Aspergillosis | Phase 3 (Recruiting) | [100, 101] |
| Oteseconazole |  PubChem CID: 77050711. | CYP51 | Recurrent Vulvovaginal Candidiasis | Market | [102] |
| Posaconazole |  PubChem CID: 468595. | CYP51 | Broad-spectrum antifungal | Market | [84, 103] |
| Pramiconazole |  PubChem CID: 3013050. | CYP51 | Pityriasis versicolor (PV) | Phase 2 (Completed) | [104-107] |
| Quilseconazole | 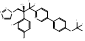 PubChem CID: 91886002. | CYP51 | Systemic Cryptococcus infections | Phase 1 (underway) | [108, 109] |
| SSY726 | 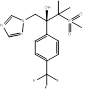 PubChem CID: 486307. | CYP51 | Mycoses (Discontinued) | Phase 2 | [110, 111] |
| Voriconazole | 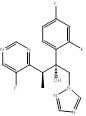 PubChem CID: 71616. | CYP51 | IFI | Market | [112, 113] |
| VT-1598 | 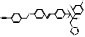 PubChem CID: 126715974. | CYP51 | Coccidioidomycosis | Phase 1 (completed) | NCT04208321, [114, 115] |
| DSP-0390 | 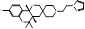 PubChem CID: 154975891. | EBP | Recurrent High-Grade Glioma | Phase 1 (Recruiting) | NCT05023551, [116-118] |
The potential role and mechanism of the circRNA/miRNA axis in cholesterol synthesis
CircRNA/miR-140-3p/HMGCR and HMGCS1 axis
CircRNA_ABCA1
CircRNA_ABCA1 (also named circRNA_36781) is located in the exonic of ABCA1. CircRNA_ABCA1 expression was increased in aortic vessels of HFD-induced apoE-/- mice and H2O2-induced mouse aortic endothelial cells (MAECs) injury model, suggesting that circRNA_ABCA1 is a potential diagnostic biomarker for atherosclerosis. CircRNA_ABCA1 could serve as miR-140-3p sponge increasing vascular endothelial injury and atherosclerosis by regulating the miR-140-3p/MAP2K6 axis [119]. MiR-140-3p also suppressed cholesterol biosynthesis by binding and suppressing the 3'UTR of HMGCR and HMGCS1 [120], suggesting that circRNA_ABCA1 promoted cholesterol biosynthesis by regulating miR-140-3p/HMGCR and HMGCS1 axis. It is worth mentioning that ABCA1 promotes cholesterol efflux to apolipoprotein A-I (apoA-I) to suppress foam cell formation. Previous studies from our laboratory and others have shown that ABCA1 promoted cholesterol efflux to suppress foam cell formation and atherosclerosis development [121-125]. However, the role of circRNA_ABCA1 on ABCA1 expression and cholesterol efflux remains unclear.
CircUGGT2 and circ-PRKCH
CircRNA UDP-glucose glycoprotein glucosyltransferase 2 (circUGGT2, also named hsa_circ_0008274) and circ-protein kinase C eta (circ-PRKCH, also named hsa_circ_0032131) are located in the exonic of UGGT2 and PRKCH (encodes PKCη). UGGT2 is the central hub of the endoplasmic reticulum mate network and regulates the PERK-ATF4-CHOP pathway and IL-8 expression [126]. PRKCH is a member of the PKC family and regulates RGS2, ABCA1, and CTLA-4 expression. Both UGGT2 and PRKCH play an essential role in lipid metabolism and inflammatory response [127-129]. CircUGGT2 and circ-circ-PRKCH could serve as the miR-140-3p sponge [130-132], suggesting that circUGGT2 and circ-PRKCH promoted cholesterol biosynthesis by regulating miR-140-3p/HMGCR and HMGCS1 axis. CircUGGT2 also increased cholesterol efflux by stimulating ABCG1, SR-B1, and miR-186-3p/ABCA1 axis in THP-1 macrophage-derived foam cells [133], suggesting that circUGGT2 not only increased cholesterol synthesis but also cholesterol efflux. Notably, astaxanthin increased the expression of circUGGT2 and then increased cholesterol efflux by stimulating ABCA1, ABCG1, and SR-B1 expression in THP-1 macrophage-derived foam cells. Astaxanthin also suppressed foam cell formation and atherosclerosis development by enhancing ABCA1, ABCG1, and SR-B1 expression in apoE-/- mice [133, 134], suggesting that circUGGT2 may be an anti-atherosclerotic RNA in vivo. However, more studies are needed.
CircRNA/miR-133b and miR-221-5p/SQLE axis
CircRNA/miR-133b/SQLE axis
Zeste homolog 2 (EZH2) could encode circRNAs, including circEZH2 (also named hsa_circ_0006357) and hsa_circ_0008324. Many studies have shown that EZH2 plays a crucial role in cholesterol synthesis and atherosclerosis development. EZH2 siRNA and inhibitors promoted cholesterol synthesis by enhancing multiple genes expression, including HMGCS1, FDFT1, SQLE, LSS, CYP51A1, DHCR7, DHCR24, and HMGCR [135], suggesting that EZH2 suppressed cholesterol synthesis. However, EZH2 promoted atherosclerosis development in vivo. Specifically, myeloid EZH2 deficiency reduced atherosclerosis development by reducing neutrophil migration and macrophage foam cell inflammatory responses, such as nitric oxide (NO), IL-6, and IL-12 [136]. EZH2 reduced ABCA1 expression by promoting triple methylation of lysine 27 (H3K27) in the ABCA1 promoter region and then reduced cholesterol efflux to promote foam cell formation and atherosclerosis development [137, 138]. EZH2 regulated miR-139-5p methylation and its target STAT1 expression through H3K27me3 and then promoted ox-LDL-induced HASMCs apoptosis, plaque formation, and inflammatory response in atherosclerosis mice [139]. EZH2 promoted the expression of MMP2 and MMP9 and their-mediated migration of aortic smooth muscle cells (MASMCs) and atherosclerosis development by promoting the methylation of TIMP2 [140]. As mentioned above, EZH2 is a parental gene of circEZH2 [141, 142]. suggesting that circEZH2 may regulate cholesterol synthesis and atherosclerosis development by regulating EZH2 expression. However, more studies are needed.
It is worth noting that circEZH2 could serve as a sponge of miR-133b [142]. MiR-133b suppressed SQLE expression by targeting SQLE 3'UTR [143, 144], suggesting that circEZH2 promoted cholesterol synthesis by regulating the miR-133b/SQLE axis. In addition, circEZH2 promoted fatty acid uptake by regulating miR-378b/CD36 and the LPL axis. CircEZH2 also promoted fatty acid uptake by promoting Fatty acid desaturase 1 (FADS1) and stearoyl-CoA desaturase 1 (SCD1) expression [145]. Many studies have shown that CD36, LPL, FADS1, and SCD1 promoted atherosclerosis development by regulating lipid metabolism. CD36 promoted cholesterol uptake, foam cell formation, and fatty acid uptake. LPL is responsible for the hydrolysis of triglycerides to glycerol and free fatty acids and is a critical factor in fatty acid uptake. FADS1 and SCD1 mainly promoted unsaturated fatty acid synthesis. Therefore, circEZH2 promoted cholesterol synthesis and uptake to foam cell formation and atherosclerosis development by regulating the miR-133b/SQLE axis and miR-378b/CD36 axis. Indeed, many circRNAs could serve as a sponge of miR-133b, including circ_0005273 [146], circRAB3IP [147], circ_0007031 [148], circ_0006459 [149], circ-HECTD1 [150], circ_0039569 [151], circ_BIRC6_001271 [152], suggesting that these circRNAs promoted cholesterol synthesis by regulating miR-133b/SQLE axis. However, more studies are needed.
CircRNAs/miR-221-5p/SQLE axis
Sterol regulatory element binding protein (SREBP) cleavage activating protein (SCAP) could encode circRNAs, including circRNA-SCAP (also named circSCAP, hsa_circ_0001292), has_circRNA_103352, hsa_circ_0065214, hsa_circ_0007291. These circRNAs are located in the exonic of SCAP. SCAP also regulated cholesterol synthesis. SCAP could bind to SREBPs and form SCAP-SREBP complex. When cholesterol in the endoplasmic reticulum (ER) is too low (below 5 %), SCAP binds to the Coat Protein complex II (COPII) protein and escorts the SCAP-SREBP complex from the ER to the Golgi. After several conformational changes, SREBP2 separates from the SCAP-SREBP2 complex and enters the nucleus. SREBPs promoted cholesterol synthesis genes by binding to HMGCR and SQLE [153]. As mentioned above, circRNA-SCAP is located in SCAP, suggesting that circRNA-SCAP may regulate cholesterol synthesis by regulating SCAP expression and SCAP-SREBP2 complex.
It is worth noting that circRNA-SCAP may be a potential biomarker of atherosclerotic plaque stability. Serum circRNA-SCAP and phosphodiesterase 3B (PDE3B) were upregulated in 25 patients with cerebral atherosclerosis, and ox-LDL-disposed THP-1 foam cells, whereas miR-221-5p level was decreased. CircRNA-SCAP is a miR-221-5p sponge [154]. MiR-221-5p could decrease cholesterol content in the liver by targeting and suppressing SQLE [155], suggesting that circRNA-SCAP promoted cholesterol synthesis by regulating the miR-221-5p/SQLE axis. MiR-221-5p also suppressed PDE3B expression by targeting PDE3B 3'UTR. By regulating the miR-221-5p/PDE3B axis, circRNA-SCAP promoted lipid deposition (total cholesterol (TC) and triglycerides (TG)), apoptosis (increased pro-apoptotic molecule Bax and cleaved-caspase 3 (caspase 3) and decreased anti-apoptotic molecule Bcl-2), inflammation (IL-6, IL-1β, TNFα, and COX-2), and oxidative stress (increased pro-oxidation molecule ROS and malondialdehyde (MDA) level and decreased anti-oxidation molecule superoxide dismutase (SOD) level) [154]. Thus, circRNA-SCAP promoted atherosclerosis development by regulating miR-221-5p/SQLE and PDE3B axis. In addition, circRNA-XPO4 also served as a miR-221-5p sponge [156], suggesting that circRNA-XPO4 promoted cholesterol synthesis by regulating the miR-221-5p/SQLE axis. However, more studies are needed.
CircRNAs/miR-188-5p/HMGCS1 axis
Circ_0001513 increased HMGCS1 expression by serving as a sponge of miR‑188‑5p [157], suggesting that circ_0001513 increased cholesterol synthesis by regulating the miR-188-5p/HMGCS1 axis. In addition, circ-PRMT5 [158] and hsa-circRNA-005843 [159] could also serve as a sponge of miR‑188‑5p, suggesting that these circRNAs increased cholesterol synthesis by regulating miR-188-5p/HMGCS1 axis. In addition, circ-PRMT5 also serves as a sponge for miR-203 [160] and miR-377 [161]. MiR-203 suppressed atherosclerotic plaque formation by binding and suppressing E26 oncogene homolog 2 (Ets2) expression, which promotes intraplaque proinflammatory phenotype [162]. MiR-377 suppressed atherosclerosis development by regulating DNA Methyltransferase 1 (DNMT1)/LPL/GPIHBP1 axis (triglyceride metabolism) and spleen tyrosine kinase (Syk) expression in apoE-/- mice [163, 164]. Circ-PRMT5 may promote cholesterol synthesis, intraplaque proinflammatory phenotype, and triglyceride metabolism by regulating the miR-188-5p/HMGCS1 axis, miR-203/Ets2 axis, miR-377/DNMT1/LPL/GPIHBP1 axis, and miR-377/Syk axis. However, circ-PRMT5 could also serve as a sponge of miR-145 [165]. MiR-145 reduced ABCA1 expression and cholesterol efflux to promote foam cell formation and atherosclerosis development by targeting the ABCA1 3'UTR [125, 166]. Therefore, miR-145/ABCA1 axis may attenuate the pro-atherogenic effect of circ-PRMT5. More studies are needed to confirm the role of circ-PRMT5 on atherosclerosis in vivo.
CircRNAs/miR-34a-5p/ACSL1 axis and miR-141-3p/YWHAG and PTEN axis
HMGCS1 could endcode five circRNAs, including circ-HMGCS1 (also named circHMGCS1, hsa_circ_0072391), hsa_circ_0072387, circHMGCS1-016 (also named hsa_circ_0008621), hsa_circ_0072389, hsa_circ_0072386. These circRNAs are located in the exon 4-6 of HMGCS1, and serve as a sponge of miR‑338‑5p [167]. However, the role of miR-338-5p in atherosclerosis has unclear. Interestingly, circ-HMGCS1 and hsa_circ_0072387 suppress lipid synthesis. CircHMGCS1-016 could increase CD73 and galectin (GAL-8) expression by serving as a sponge of miR-1236-3p [168]. CD73 has a weak anti-atherosclerosis effect in the early stages of the disease. However, as the disease progresses, CD73 promotes the accretion of atherosclerotic plaque by suppressing lipid catabolism [169]. GAL-8 promotes atherosclerosis development by enhancing inflammation, platelet aggregation, and thromboxane generation [170]. Therefore, circHMGCS1-016 may promote atherosclerosis by regulating miR-1236-3p/CD73 and the GAL-8 axis. The role of hsa_circ_0072389, and hsa_circ_0072386 in lipid synthesis and atherosclerosis has unclear. More studies are needed.
CircRNAs/miR-34a-5p/ACSL1 axis
Circ-HMGCS1 could serve as a sponge of miR‑34a‑5p [171], miR-581 [172], miR-892a [172], and miR-503-5p [173]. MiR-34a-5p suppressed long-chain acyl-CoA synthetase 1 (ACSL1) expression by targeting ACSL1 3'UTR, an essential enzyme for the synthesis of fatty acyl-CoA, triglycerides, phospholipids, and cholesterol esters [174, 175]. However, ACSL1 also promotes lipid efflux. MiR-34a-5p increases the level of triglycerides and cholesterol in the liver by suppressing ACSL1 expression [175], suggesting that circ-HMGCS1 may suppress lipid levels although it inhibits lipid synthesis by regulating miR-34a-5p/ACSL1 axis. In addition, miR-34a-5p also increased lipid droplet accumulation by suppressing adipose triglyceride lipase (ATGL) expression which is a key lipolysis gene and enhances adipose tissue lipolysis [176]. MiR-34a-5p suppressed ADAM10 expression by targeting ADAM10 3'UTR [177]. MiR-581 suppressed ABCG1 expression by targeting ABCG1 3'UTR [178]. Many studies have shown that ADAM10 and ABCG1 play a key role in promoting cholesterol efflux, suggesting that circ-HMGCS1 suppressed lipid accumulation by regulating miR-34a-5p/ACSL1, ATGL, ADAM10 axis, and miR-581/ABCG1 axis. MiR-503-5p promoted proinflammatory cytokines and adhesion molecules level and atherosclerosis development by regulating smad family members 1 (smurf1), 2 (smurf2), and 7 (Smad7) in RAW264.7 macrophage-derived foam cells and apoE-/- mice [179], suggesting that circ-HMGCS1 may suppress proinflammatory cytokines by regulating miR-503-5p/smurf1, smurf2, Smad7 axis. Therefore, circ-HMGCS1 promotes lipid synthesis by regulating the miR-34a-5p/ACSL1 axis. Circ-HMGCS1 suppresses lipid accumulation and proinflammatory cytokines by regulating miR-34a-5p/ACSL1, ATGL, ADAM10 axis, miR-581/ABCG1 axis, and miR-503-5p/smurf1, smurf2, Smad7 axis. Circ-HMGCS1 may be an anti-atherosclerotic RNA. However, more studies are needed.
Notable, many circRNAs could serve as a sponge of miR-34a-5p, including circOgdh (also named mmu_circ_0000231) [176], circMED12L [180], circ_FURIN [181], circ_CSNK1E [182], circ0036602 [183], circ-LRP1B [184], circHUWE1 [185], circITGA7 [186], circNFIX [187, 188], circRNA-CIDN [189], circ_0009910 [190], circ_0039569 [191], hsa_circ_0018069 [192], suggesting that these circRNAs may promote lipid synthesis but suppress lipid accumulation by regulating miR-34a-5p/ACSL, ATGL, and ADAM10 axis.
CircRNAs/miR-141-3p/YWHAG and PTEN axis
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma (YWHAG, encoding 14-3-3γ) regulates lipid metabolism and glucose homeostasis by regulating the localization of Lipin1 and GLUT4 [193, 194]. PTEN also regulates lipid metabolism and glucose homeostasis by regulating SREBP-1c and GSK-3β expression [195]. Hsa_circ_0072387 could serve as a sponge of miR-141-3p (also named miR-141) and miR-503-5p [196, 197]. MiR-141-3p increases triglyceride and cholesterol synthesis by upregulating YWHAG and downregulating PTEN expression, respectively [198], suggesting that hsa_circ_0072387 may suppress lipid synthesis by regulating miR-141-3p/YWHAG and PTEN axis. As mentioned above, miR-503-5p promoted proinflammatory response and atherosclerosis development by regulating smurf1, smurf2, and Smad7; thus, hsa_circ_0072387 may suppress lipid synthesis and pro-inflammatory response by regulating miR-141-3p/YWHAG and PTEN axis, miR-503-5p/smurf1, smurf2, Smad7 axis.
Many circRNAs could serve as a sponge of miR-141-3p, including circDLG1 [199], circDIDO1 [200], circ_100395 (also named exo-circ_100395) [201], circ_0075943 [201], circTRPS1 (also named hsa_circ_0085361) [202], circRNA_100338 [203-205], circKEAP1 [206], circ-LRP6 [207], circZEB1 [208], circRNA-SMG1.72 (also named circ-SMG1.72) [209], circSOBP [210], hsa_circRNA_100395 [211], circ_0061140 [212], circATRNL1 [213], circ-GBR10 [214], suggesting that these circRNAs suppress lipid synthesis by regulating miR-141-3p/YWHAG and PTEN axis.
CircRNAs/miR-494-3p/PTEN axis
CircCYP51 (also named circ_0081001) is derived from CYP51 and is a potential biomarker for the diagnosis and prognosis of osteosarcoma (OS) [215]. CircCYP51 could serve as a sponge of miR-494-3p [216]. MiR-494-3p promoted proinflammatory macrophage polarization by suppressing Wnt signaling in atherosclerosis [217]. MiR-494-3p also promoted plasma cholesterol levels by suppressing PTEN [218, 219]. As mentioned above, PTEN was negatively correlated with cholesterol synthesis, suggesting that circCYP51 suppresses proinflammatory macrophage polarization and cholesterol synthesis by regulating miR-494-3p/Wnt and PTEN axis.
CircRNAs/miR-892b and miR-217-5p/DHCR24 axis
CircRNAs/miR-892b/DHCR24 axis
CircPTK2 (also named hsa_circ_0003221) is located in exons 3-7 of protein tyrosine kinase 2 (PTK2). CircPTK2 increased DHCR24 expression by serving as a sponge of miR-892b [220], suggesting that circPTK2 promotes cholesterol synthesis by regulating the miR-892b/DHCR24 axis. CircPTK2 could serve as a sponge of miR-1278 [221], miR-139-3p [222], and miR-758-3p (miR-758) [223], MiR-1278 suppressed cardiomyocyte inflammation in myocardial ischemia by reducing IL-22 and CXCL14 expression [224], suggesting that circPTK2 promotes cholesterol synthesis and inflammation by regulating the miR-892b/DHCR24 axis and miR-1278/IL-22 and CXCL14 axis. However, miR-758-3p suppressed cholesterol efflux and foam cell formation by targeting ABCA1 3'UTR [225]. MiR-758-3p/ABCA1 axis may attenuate the pro-atherogenic effect of circPTK2. More studies are needed to confirm the role of circPTK2 on atherosclerosis in vivo.
CircRNAs/miR-217-5p/KLF5/DHCR24 axis
CircEZH2 enhanced Krüppel-like factor 5 (KLF5) expression by sponging with miR-217-5p [226]. Interestingly, KLF5 increases cholesterol synthesis by activating the DHCR24 promoter [227]. As mentioned earlier, circEZH2 promoted cholesterol synthesis and uptake by regulating the miR-133b/SQLE axis and miR-378b/CD36 axis. Therefore, circEZH2 promoted cholesterol synthesis and uptake to enhance foam cell formation and atherosclerosis development by regulating the miR-217-5p/KLF5/DHCR24 axis, miR-133b/SQLE axis, and miR-378b/CD36 axis.
In addition, many circRNAs could serve as a sponge of miR-217-5p, including circROBO1 [228], circ_0033596 [229], and circ_0002099 [230], suggesting that these circRNAs promoted cholesterol synthesis by regulating miR-217-5p/KLF5/DHCR24 axis.
CircRNAs/miR-122/SREBP-2, HMGCR, and PMK axis
MiR-122 antagonism decreases hepatic lipid metabolism and cholesterol biosynthesis by suppressing several genes expression, including acetyl-CoA carboxylase alpha (ACC1), acetyl-CoA carboxylase beta (ACC2), ATP citrate lyase (ACLY), SCD1, Fatty acid synthase (FASN, also named FAS), SREBP-2, HMGCR, and PMK [10]. MiR-122 antagonism is a promising strategy for the treatment of ASCVD. Many circRNAs could serve as a sponge of miR-122, including ciRS-122 (also named hsa_circ_0005963) [231], circRNA_002581 [232], circCDK17 [233], circ_0007142 [234], circ_0011269 [235], circ-IARS [236], circ_0072995 [237], circFOXO3 (also named hsa_circ_0006404) [238], circ_pleiotrophin (circ_PTN) [239], and circ_1639 [240], suggesting that these circRNAs may suppress cholesterol biosynthesis by serving as a sponge of miR-122 (Table 2). Significantly, inhibits miR-122 with LNA-antagomiR-122 (also named SPC3649 or Miravirsen, was developed by SantarisPharma) and N-acetylgalactosamine-conjugated anti-microRNA-122 oligonucleotide (also named RG-101 was developed by Regulus Therapeutics) for the treatment of hepatitis C virus (HCV) infections has completed the phase II trial [241, 242]. More importantly, circFOXO3 is located in exon 3 of forkhead box O3 (FOXO3). CircFOXO3 rs12196996, a polymorphism at the gene flanking intron, is associated with circFOXO3 levels and the risk of ASCVD in the Chinese Han population [243]. The clinical application potential of circFOXO3 in tumor diagnosis and treatment is immense [244], suggesting that circFOXO3 may be a promising future target in the diagnosis and treatment of cancer and cardiovascular disease.
The potential role and mechanism of circRNAs in cholesterol synthesis.
| CircRNAs | Axis | Refs |
|---|---|---|
| circRNA_ABCA1 | miR-140-3p/HMGCR and HMGCS1 axis | [119, 120] |
| circUGGT2 | miR-140-3p/HMGCR and HMGCS1 axis | [120, 130, 131] |
| circ-PRKCH | miR-140-3p/HMGCR and HMGCS1 axis | [120, 132] |
| circEZH2 | miR-133b/SQLE axis | [142-144] |
| miR-217-5p/KLF5/DHCR24 axis | [226, 227] | |
| circ_0005273 | miR-133b/SQLE axis | [143, 144, 146] |
| circRAB3IP | miR-133b/SQLE axis | [143, 144, 147] |
| circ_0007031 | miR-133b/SQLE axis | [143, 144, 148] |
| circ_0006459 | miR-133b/SQLE axis | [143, 144, 149] |
| circ-HECTD1 | miR-133b/SQLE axis | [143, 144, 150] |
| circ_0039569 | miR-133b/SQLE axis | [143, 144, 151] |
| circ_BIRC6_001271 | miR-133b/SQLE axis | [143, 144, 152] |
| circRNA-SCAP | miR-221-5p/SQLE axis | [154, 155] |
| circRNA-XPO4 | miR-221-5p/SQLE axis | [155, 156] |
| circ_0001513 | miR-188-5p/HMGCS1 axis | [157] |
| circ-PRMT5 | miR-188-5p/HMGCS1 axis | [157, 158] |
| hsa-circRNA-005843 | miR-188-5p/HMGCS1 axis | [157, 159] |
| circHMGCS1 | miR-34a-5p/ACSL1 axis | [171, 175] |
| circOgdh | miR-34a-5p/ACSL1 axis | [175, 176] |
| circMED12L | miR-34a-5p/ACSL1 axis | [175, 180] |
| circ_FURIN | miR-34a-5p/ACSL1 axis | [175, 181] |
| circ_CSNK1E | miR-34a-5p/ACSL1 axis | [175, 182] |
| circ0036602 | miR-34a-5p/ACSL1 axis | [175, 183] |
| circ-LRP1B | miR-34a-5p/ACSL1 axis | [175, 184] |
| circHUWE1 | miR-34a-5p/ACSL1 axis | [175, 185] |
| circITGA7 | miR-34a-5p/ACSL1 axis | [175, 186] |
| circNFIX | miR-34a-5p/ACSL1 axis | [175, 187, 188] |
| circRNA-CIDN | miR-34a-5p/ACSL1 axis | [175, 189] |
| circ_0009910 | miR-34a-5p/ACSL1 axis | [175, 190] |
| circ_0039569 | miR-34a-5p/ACSL1 axis | [175, 191] |
| hsa_circ_0018069 | miR-34a-5p/ACSL1 axis | [175, 192] |
| hsa_circ_0072387 | miR-141-3p/YWHAG and PTEN axis | [196-198] |
| circDLG1 | miR-141-3p/YWHAG and PTEN axis | [198, 199] |
| circDIDO1 | miR-141-3p/YWHAG and PTEN axis | [198, 200] |
| circ_100395 | miR-141-3p/YWHAG and PTEN axis | [198, 201] |
| circ_0075943 | miR-141-3p/YWHAG and PTEN axis | [198, 201] |
| circTRPS1 | miR-141-3p/YWHAG and PTEN axis | [198, 202] |
| circRNA_100338 | miR-141-3p/YWHAG and PTEN axis | [198, 203-205] |
| circKEAP1 | miR-141-3p/YWHAG and PTEN axis | [198, 206] |
| circ-LRP6 | miR-141-3p/YWHAG and PTEN axis | [198, 207] |
| circZEB1 | miR-141-3p/YWHAG and PTEN axis | [198, 208] |
| circRNA-SMG1.72 | miR-141-3p/YWHAG and PTEN axis | [198, 209] |
| circSOBP | miR-141-3p/YWHAG and PTEN axis | [198, 210] |
| hsa_circRNA_100395 | miR-141-3p/YWHAG and PTEN axis | [198, 211] |
| circ_0061140 | miR-141-3p/YWHAG and PTEN axis | [198, 212] |
| circATRNL1 | miR-141-3p/YWHAG and PTEN axis | [198, 213] |
| circ-GBR10 | miR-141-3p/YWHAG and PTEN axis | [198, 214] |
| circ_0081001 | miR-494-3p/PTEN axis | [198, 216, 218, 219] |
| circPTK2 | miR-892b/DHCR24 axis | [220] |
| circROBO1 | miR-217-5p/KLF5/DHCR24 axis | [226-228] |
| circ_0033596 | miR-217-5p/KLF5/DHCR24 axis | [226, 227, 229] |
| circ_0002099 | miR-217-5p/KLF5/DHCR24 axis | [226, 227, 230] |
| ciRS-122 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 231] |
| circRNA_002581 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 232] |
| circCDK17 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 233] |
| circ_0007142 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 234] |
| circ_0011269 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 235] |
| circ-IARS | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 236] |
| circ_0072995 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 237] |
| circFOXO3 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 238] |
| circ_PTN | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 239] |
| circ_1639 | miR-122/SREBP-2, HMGCR, and PMK axis | [10, 240] |
Conclusions and Future Directions
Many genes play a central role in cholesterol synthesis, including CYP51, DHCR7, DHCR24, EBP, FDFT1, FDPS, HMGCR, HMGCS1, HSD17B7, IDI1/2, LBR, LSS, MSMO1, MVD, MVK, NSDHL, PMK, SC5D, SQLE, and TM7SF2. Many circRNAs regulate cholesterol synthesis by regulating ACSL1, DHCR24, HMGCR, HMGCS1, PTEN, SQLE, and YWHAG expression by sponging miRNAs. Some circRNAs were also involved in other atherosclerotic risk factors (Table 3). Notable, CYP51, EBP, FDFT1, FDPS, HMGCR, LSS, and SQLE, are promising therapeutic targets for drug development due to many specific inhibitors have been approved and entered into clinical research by targeting these genes. Many circRNAs regulated cholesterol biosynthesis by regulating HMGCR expression via sponging miR-122. Several drugs targeting miR-122 have completed the phase II trial for the treatment of HCV infections, including Miravirsen and RG-101. Thus, the circRNA/miR-122/HMGCR axis is a promising therapeutic axis for drug development. However, several interesting and critical tasks remain to be explored: (1) The naming of circRNA is not uniform and even a little confusing, such as HMGCS1 could encode five circRNA, including hsa_circ_0072391, hsa_circ_0072387, hsa_circ_0008621, hsa_circ_0072389, and hsa_circ_0072386. However, hsa_circ_0072391 is also named circ-HMGCS1 or circHMGCS1, while hsa_circ_0008621 is also named circHMGCS1-016. SCAP could encode circRNAs, including hsa_circ_0001292, has_circRNA_103352, hsa_circ_0065214, and hsa_circ_0007291. However, only hsa_circ_0001292 is also named circRNA-SCAP or circSCAP. (2) Several circRNAs not only promoted cholesterol biosynthesis but also promoted cholesterol efflux or suppressed proinflammatory cytokines, including circUGGT2, circ-PRMT5, circ-HMGCS1, circOgdh, circMED12L, circ_FURIN, circ_CSNK1E, circ0036602, circ-LRP1B, circHUWE1, circITGA7, circNFIX, circRNA-CIDN, circ_0009910, circ_0039569, hsa_circ_0018069, and circPTK2. The role of these circRNAs in atherosclerosis remains to be investigated in vivo. (3) The state of the disease may affect circRNAs studies, such as circHMGCS1-016. CircHMGCS1-016 may exhibit an anti-atherogenic effect in the early stages of the disease. However, as the disease progresses, circHMGCS1-016 may exhibit a pro-atherogenic effect. The development of drugs and diagnostic reagents must consider the state of disease progression. (4) CircRNAs regulate gene expression through various mechanisms, including sponge miRNA, protein scaffold and sponge, encoding protein, and regulation of splicing and transcription. However, so far, almost all circRNAs regulate cholesterol synthesis genes through sponge miRNA. Whether there are other mechanisms is not clear. (5) Until now, most circRNA's role in cholesterol synthesis has been studied in vitro. However, there are many factors influencing the development of the disease. The effect of circRNAs on the disease still needs to be studied in vivo. (6) Given that inhibits miR-122 completed the phase II trial, circFOXO3 is a promising target for drug research by sponging miR-122. However, more studies are needed. (7) Many drugs have been approved for market by targeting HMGCR and SQLE expression. Several circRNAs may be promising therapeutic targets for drug development by targeting HMGCR and SQLEM, such as circRNA_ABCA1, circ-PRKCH, circEZH2, and circRNA-SCAP. However, more studies are needed. (8) The development of new drugs usually requires preclinical studies in multiple animal models before clinical application to improve drug development's success rate. The development of circRNAs drugs also requires much research. (9) Current methods of circRNA synthesis are limited by low cyclization efficiency and the high cost of enzymes and other reagents. There is an urgent need to address these issues. (10) The current study has shown that circRNAs have many targets. However, it may be caused by different dosing doses. Whether there are multiple targets in vivo still needs much research.
In summary, drugs that target CYP51, EBP, FDFT1, FDPS, HMGCR, LSS, SQLE, and miR-122 have entered the stage of market or clinical trials. CircRNA_ABCA1, circ-PRKCH, circEZH2, circRNA-SCAP, and circFOXO3 are promising therapeutic targets for drug development, specifically circFOXO3. With the progress of science and technology, the deepening of research, and the cooperation of scientific research, we believe there will be the clinical application of circRNAs agents soon.
The role and mechanism of circRNAs that are involved in multiple atherosclerotic risk factors.
| CircRNAs | Axis | Function | Refs |
|---|---|---|---|
| circRNA_ABCA1 | miR-140-3p/HMGCR and HMGCS1 axis | Increased cholesterol synthesis | [119, 120] |
| miR-140-3p/MAP2K6 axis | Increased vascular endothelial injury | [119, 120, 130, 131] | |
| circUGGT2 | miR-140-3p/HMGCR and HMGCS1 axis | Increased cholesterol synthesis | [120, 130, 131] |
| miR-140-3p/MAP2K6 axis | Increased vascular endothelial injury | [119, 120, 130, 131] | |
| miR-186-3p/ABCA1 axis | Increased cholesterol efflux | [133] | |
| circEZH2 | miR-133b/SQLE axis | Increased cholesterol synthesis | [142-144] |
| miR-217-5p/KLF5/DHCR24 axis | Increased cholesterol synthesis | [226, 227] | |
| miR-378b/CD36 axis | Increased cholesterol uptake | [145] | |
| LPL, FADS1 and SCD1 | Increased fatty acid uptake | [145] | |
| circRNA-SCAP | miR-221-5p/SQLE axis | Increased cholesterol synthesis | [154, 155] |
| miR-221-5p/PDE3B axis | Promoted lipid deposition, apoptosis, inflammation, and oxidative stress | [154] | |
| circRNA-XPO4 | miR-221-5p/SQLE axis | Increased cholesterol synthesis | [155, 156] |
| miR-221-5p/PDE3B axis | Promoted lipid deposition, apoptosis, inflammation, and oxidative stress | [154-156] | |
| circ-PRMT5 | miR-188-5p/HMGCS1 axis | Increased cholesterol synthesis | [157, 158] |
| miR-203/Ets2 axis | Increased intraplaque proinflammatory phenotype | [162] | |
| miR-377/DNMT1/LPL/GPIHBP1 axis | Increased triglyceride metabolism | [161, 163, 164] | |
| miR-145/ABCA1 axis | Increased cholesterol efflux | [125, 165, 166] | |
| circ-HMGCS1 | miR-34a-5p/ACSL1 axis | Promoted lipid synthesis but suppressed lipid accumulation | [171, 175] |
| miR-34a-5p/ATGL axis | Promoted lipolysis | [171, 176] | |
| miR-34a-5p/ADAM10 axis | Increased cholesterol efflux | [171, 177] | |
| miR-581/ABCG1 axis | Increased cholesterol efflux | [172, 178] | |
| miR-503-5p/smurf1, smurf2, Smad7 axis | Suppressed proinflammatory cytokines and adhesion molecules level | [173, 179] | |
| hsa_circ_0072387 | miR-141-3p/YWHAG and PTEN axis | Increased triglyceride and cholesterol synthesis | [196, 198] |
| miR-503-5p/smurf1, smurf2, Smad7 axis | Suppressed proinflammatory cytokines and adhesion molecules level | [179, 197] | |
| circCYP51 | miR-494-3p/PTEN axis | Suppressed cholesterol synthesis | [198, 216, 218, 219] |
| miR-494-3p/Wnt axis | Suppressed proinflammatory macrophage polarization | [216, 217] | |
| circPTK2 | miR-892b/DHCR24 axis | Increased cholesterol synthesis | [220] |
| miR-1278/IL-22 and CXCL14 axis | Promoted cardiomyocytes inflammation | [221, 224] | |
| miR-758-3p/ABCA1 axis | Increased cholesterol efflux | [223, 225] |
Acknowledgements
We thank colleagues in Dr. Dongming Xing's laboratory for the technical help and the stimulating discussions provided during this investigation. The authors are grateful for the financial support provided by the Qingdao Major Scientific and Technological Project for Distinguished Scholars (20170103), the Laoshan Major Scientific and Technological Project for Distinguished Scholars (20181030), the Natural Science Foundation of Shandong Province (ZR2020MH369, ZR2021QC088).
Author contributions
WC and JX participated in the writing-original draft. YW, MY, DW, and XH participated in supervision, and resources. BL, CS, LL, and WH participated in formal analysis and investigation. YS and DX participated in conceptualization, writing review & editing, project administration, and funding acquisition. All authors read and approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yang K, Xiao Q, Niu M, Pan X, Zhu X. Exosomes in atherosclerosis: Convergence on macrophages. Int J Biol Sci. 2022;18:3266-81
2. Tan H, Yue T, Chen Z, Wu W, Xu S, Weng J. Targeting FGF21 in cardiovascular and metabolic diseases: from mechanism to medicine. Int J Biol Sci. 2023;19:66-88
3. Li X, Yang Y, Wang Z, Jiang S, Meng Y, Song X. et al. Targeting non-coding RNAs in unstable atherosclerotic plaques: Mechanism, regulation, possibilities, and limitations. Int J Biol Sci. 2021;17:3413-27
4. Yang J, Wang L, Jia R. Role of de novo cholesterol synthesis enzymes in cancer. J Cancer. 2020;11:1761-7
5. Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225-45
6. Wujun Chen YZ, Yang Yuan, Meng Zhu, Wenchao Hu, Ning Liu, Dongming Xing. New insights into the suppression of inflammation and lipid accumulation by JAZF1. Genes Dis. 2022
7. Shi Q, Chen J, Zou X, Tang X. Intracellular Cholesterol Synthesis and Transport. Front Cell Dev Biol. 2022;10:819281
8. Ershov P, Kaluzhskiy L, Mezentsev Y, Yablokov E, Gnedenko O, Ivanov A. Enzymes in the Cholesterol Synthesis Pathway: Interactomics in the Cancer Context. Biomedicines. 2021 9
9. Wang S, Wang Y, Cheng H, Zhang Q, Fu C, He C. et al. The Networks of Noncoding RNAs and Their Direct Molecular Targets in Myocardial Infarction. Int J Biol Sci. 2022;18:3194-208
10. Fernandez-Tussy P, Ruz-Maldonado I, Fernandez-Hernando C. MicroRNAs and Circular RNAs in Lipoprotein Metabolism. Curr Atheroscler Rep. 2021;23:33
11. Chen W, Liu Y, Li L, Liang B, Wang S, Xu X. et al. The potential role and mechanism of circRNAs in foam cell formation. Noncoding RNA Res. 2023;8:315-25
12. Chiriboga CA. Nusinersen for the treatment of spinal muscular atrophy. Expert Rev Neurother. 2017;17:955-62
13. Dhillon S. Viltolarsen: First Approval. Drugs. 2020;80:1027-31
14. Heo YA. Golodirsen: First Approval. Drugs. 2020;80:329-33
15. Hoy SM. Patisiran: First Global Approval. Drugs. 2018;78:1625-31
16. Keam SJ. Inotersen: First Global Approval. Drugs. 2018;78:1371-6
17. Lamb YN. Inclisiran: First Approval. Drugs. 2021;81:389-95
18. Lazarte J, Hegele RA. Volanesorsen for treatment of familial chylomicronemia syndrome. Expert Rev Cardiovasc Ther. 2021;19:685-93
19. Lim KR, Maruyama R, Yokota T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des Devel Ther. 2017;11:533-45
20. Scott LJ. Givosiran: First Approval. Drugs. 2020;80:335-9
21. Scott LJ, Keam SJ. Lumasiran: First Approval. Drugs. 2021;81:277-82
22. Shirley M. Casimersen: First Approval. Drugs. 2021;81:875-9
23. Suzuki Y, Ishihara H. Difference in the lipid nanoparticle technology employed in three approved siRNA (Patisiran) and mRNA (COVID-19 vaccine) drugs. Drug Metab Pharmacokinet. 2021;41:100424
24. Fan X, Yang Y, Chen C, Wang Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat Commun. 2022;13:3751
25. Garber K. Orna Therapeutics: circular logic. Nat Biotechnol. 2022
26. Fasolo F, Di Gregoli K, Maegdefessel L, Johnson JL. Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc Res. 2019;115:1732-56
27. Navarro E, Mallen A, Cruzado JM, Torras J, Hueso M. Unveiling ncRNA regulatory axes in atherosclerosis progression. Clin Transl Med. 2020;9:5
28. Zhang S, Wang W, Wu X, Zhou X. Regulatory Roles of Circular RNAs in Coronary Artery Disease. Mol Ther Nucleic Acids. 2020;21:172-9
29. Huang X, Zhao Y, Zhou H, Li Y. Circular RNAs in atherosclerosis. Clin Chim Acta. 2022;531:71-80
30. Chua NK, Coates HW, Brown AJ. Squalene monooxygenase: a journey to the heart of cholesterol synthesis. Prog Lipid Res. 2020;79:101033
31. Gutierrez-Garcia R, del Pozo T, Suazo M, Cambiazo V, Gonzalez M. Physiological copper exposure in Jurkat cells induces changes in the expression of genes encoding cholesterol biosynthesis proteins. Biometals. 2013;26:1033-40
32. Alphonse PA, Jones PJ. Revisiting Human Cholesterol Synthesis and Absorption: The Reciprocity Paradigm and its Key Regulators. Lipids. 2016;51:519-36
33. Qiu Y, Li D. Inhibition of mevalonate 5-diphosphate decarboxylase by fluorinated substrate analogs. Biochim Biophys Acta. 2006;1760:1080-7
34. Zerenturk EJ, Sharpe LJ, Ikonen E, Brown AJ. Desmosterol and DHCR24: unexpected new directions for a terminal step in cholesterol synthesis. Prog Lipid Res. 2013;52:666-80
35. Seeger MA, Paller AS. The role of abnormalities in the distal pathway of cholesterol synthesis in the Congenital Hemidysplasia with Ichthyosiform erythroderma and Limb Defects (CHILD) syndrome. Biochim Biophys Acta. 2014;1841:345-52
36. CR S. The pharmacology of statins. Pharmacol Res. 2014:3-11
37. Almeida SO, Budoff M. Effect of statins on atherosclerotic plaque. Trends Cardiovasc Med. 2019;29:451-5
38. Zahedipour F, Butler AE, Rizzo M, Sahebkar A. Statins and angiogenesis in non-cardiovascular diseases. Drug Discov Today. 2022;27:103320
39. Huang B, Song BL, Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab. 2020;2:132-41
40. Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer. 2020;1874:188394
41. Gregoriou S, Kyriazopoulou M, Tsiogka A, Rigopoulos D. Novel and Investigational Treatments for Onychomycosis. J Fungi (Basel). 2022 8
42. Gupta AK, Talukder M, Venkataraman M. Review of the alternative therapies for onychomycosis and superficial fungal infections: posaconazole, fosravuconazole, voriconazole, oteseconazole. Int J Dermatol. 2022;61:1431-41
43. Sulaiman A, Wan X, Fan J, Kasimu H, Dong X, Wang X. et al. Analysis on curative effects and safety of 2% liranaftate ointment in treating tinea pedis and tinea corporis & cruris. Pak J Pharm Sci. 2017;30:1103-6
44. Hammoudi Halat D, Younes S, Mourad N, Rahal M. Allylamines, Benzylamines, and Fungal Cell Permeability: A Review of Mechanistic Effects and Usefulness against Fungal Pathogens. Membranes (Basel). 2022 12
45. Gupta AK, Stec N, Summerbell RC, Shear NH, Piguet V, Tosti A. et al. Onychomycosis: a review. J Eur Acad Dermatol Venereol. 2020;34:1972-90
46. Krauss J, Muller C, Klimt M, Valero LJ, Martinez JF, Muller M. et al. Synthesis, Biological Evaluation, and Structure-Activity Relationships of 4-Aminopiperidines as Novel Antifungal Agents Targeting Ergosterol Biosynthesis. Molecules. 2021 26
47. Song J, Shang N, Baig N, Yao J, Shin C, Kim BK. et al. Aspergillus flavus squalene synthase as an antifungal target: Expression, activity, and inhibition. Biochem Biophys Res Commun. 2019;512:517-23
48. Sharma A, Slugg PH, Hammett JL, Jusko WJ. Estimation of oral bioavailability of a long half-life drug in healthy subjects. Pharm Res. 1998;15:1782-6
49. Sharma A, Slugg PH, Hammett JL, Jusko WJ. Clinical pharmacokinetics and pharmacodynamics of a new squalene synthase inhibitor, BMS-188494, in healthy volunteers. J Clin Pharmacol. 1998;38:1116-21
50. AdisInsight. BMS 188494. 2008.
51. Suzuki N, Ito T, Matsui H, Takizawa M. Anti-inflammatory and cytoprotective effects of a squalene synthase inhibitor, TAK-475 active metabolite-I, in immune cells simulating mevalonate kinase deficiency (MKD)-like condition. Springerplus. 2016;5:1429
52. Stein EA, Bays H, O'Brien D, Pedicano J, Piper E, Spezzi A. Lapaquistat acetate: development of a squalene synthase inhibitor for the treatment of hypercholesterolemia. Circulation. 2011;123:1974-85
53. Oxiconazole. Drugs and Lactation Database (LactMed(R)). Bethesda (MD). 2006
54. Jegasothy BV, Pakes GE. Oxiconazole nitrate: pharmacology, efficacy, and safety of a new imidazole antifungal agent. Clin Ther. 1991;13:126-41
55. Eisele B, Budzinski R, Muller P, Maier R, Mark M. Effects of a novel 2,3-oxidosqualene cyclase inhibitor on cholesterol biosynthesis and lipid metabolism in vivo. J Lipid Res. 1997;38:564-75
56. Ohno K, Mori K, Orita M, Takeuchi M. Computational insights into binding of bisphosphates to farnesyl pyrophosphate synthase. Curr Med Chem. 2011;18:220-33
57. Fujita T, Izumo N, Fukuyama R, Meguro T, Yasutomi C, Nakamuta H. et al. Incadronate and etidronate accelerate phosphate-primed mineralization of MC4 cells via ERK1/2-Cbfa1 signaling pathway in a Ras-independent manner: further involvement of mevalonate-pathway blockade for incadronate. Jpn J Pharmacol. 2001;86:86-96
58. Inderjeeth CA, Glendenning P, Ratnagobal S, Inderjeeth DC, Ondhia C. Long-term efficacy, safety, and patient acceptability of ibandronate in the treatment of postmenopausal osteoporosis. Int J Womens Health. 2015;7:7-17
59. Liu XW, Jin HF, Du CQ, Tang LJ. Farnesyl Pyrophosphate Synthase Blocker Ibandronate Reduces Thoracic Aortic Fibrosis in Diabetic Rats. Am J Med Sci. 2019;357:323-32
60. Peyton LR, Gallagher S, Hashemzadeh M. Triazole antifungals: a review. Drugs Today (Barc). 2015;51:705-18
61. Sigurgeirsson B, van Rossem K, Malahias S, Raterink K. A phase II, randomized, double-blind, placebo-controlled, parallel group, dose-ranging study to investigate the efficacy and safety of 4 dose regimens of oral albaconazole in patients with distal subungual onychomycosis. J Am Acad Dermatol. 2013;69:416-25
62. AdiInsight. A phase III trial of Arasertaconazole for the treatment of vulvovaginal candidiasis (VVC). 15 Jan. 2016
63. EMBL-EBI. CHEBI:83692 - arasertaconazole nitrate. 23 July. 2015
64. Baran R, Gupta AK, Pierard GE. Pharmacotherapy of onychomycosis. Expert Opin Pharmacother. 2005;6:609-24
65. Drabinska B, Dettlaff K, Ratajczak T, Kossakowski K, Chmielewski MK, Cielecka-Piontek J. et al. Structural and Spectroscopic Properties of Isoconazole and Bifonazole-Experimental and Theoretical Studies. Int J Mol Sci. 2022 24
66. Naldi L, Diphoorn J. Seborrhoeic dermatitis of the scalp. BMJ Clin Evid. 2015. 2015
67. Vennewald I, Klemm E. Otomycosis: Diagnosis and treatment. Clin Dermatol. 2010;28:202-11
68. Butoconazole. Drugs and Lactation Database (LactMed(R)). Bethesda (MD). 2006
69. Doering PL, Santiago TM. Drugs for treatment of vulvovaginal candidiasis: comparative efficacy of agents and regimens. DICP. 1990;24:1078-83
70. Clotrimazole. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. Bethesda (MD). 2012
71. Antunes NJ, Coombes G, da Cunha KF, Moreira FL, Pilon AC, Lopes NP. et al. In vitro metabolism of the new antifungal dapaconazole using liver microsomes. Drug Metab Pharmacokinet. 2022;47:100475
72. Gobbato AA, Babadopulos T, Gobbato CA, Ilha Jde O, Gagliano-Juca T, De Nucci G. A randomized double-blind, non-inferiority Phase II trial, comparing dapaconazole tosylate 2% cream with ketoconazole 2% cream in the treatment of Pityriasis versicolor. Expert Opin Investig Drugs. 2015;24:1399-407
73. Moodahadu-Bangera LS, Martis J, Mittal R, Krishnankutty B, Kumar N, Bellary S. et al. Eberconazole-pharmacological and clinical review. Indian J Dermatol Venereol Leprol. 2012;78:217-22
74. Firooz A, Nafisi S, Maibach HI. Novel drug delivery strategies for improving econazole antifungal action. Int J Pharm. 2015;495:599-607
75. Kim D, Lim YR, Ohk SO, Kim BJ, Chun YJ. Functional expression and characterization of CYP51 from dandruff-causing Malassezia globosa. FEMS Yeast Res. 2011;11:80-7
76. Saunders J, Maki K, Koski R, Nybo SE. Tavaborole, Efinaconazole, and Luliconazole: Three New Antimycotic Agents for the Treatment of Dermatophytic Fungi. J Pharm Pract. 2017;30:621-30
77. Vlahovic TC, Gupta AK. Efinaconazole topical solution (10%) for the treatment of onychomycosis in adult and pediatric patients. Expert Rev Anti Infect Ther. 2022;20:3-15
78. Sobel JD, Nyirjesy P. Oteseconazole: an advance in treatment of recurrent vulvovaginal candidiasis. Future Microbiol. 2021;16:1453-61
79. Loffler J, Kelly SL, Hebart H, Schumacher U, Lass-Florl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263-8
80. Rigopoulos D, Gregoriou S, Kontochristopoulos G, Ifantides A, Katsambas A. Flutrimazole shampoo 1% versus ketoconazole shampoo 2% in the treatment of pityriasis versicolor. A randomised double-blind comparative trial. Mycoses. 2007;50:193-5
81. Gupta AK, Venkataraman M, Talukder M. Onychomycosis in Older Adults: Prevalence, Diagnosis, and Management. Drugs Aging. 2022;39:191-8
82. Nyuykonge B, Siddig EE, Mhmoud NA, Nyaoke BA, Zijlstra EE, Verbon A. et al. Epidemiological cut-off values for itraconazole and ravuconazole for Madurella mycetomatis, the most common causative agent of mycetoma. Mycoses. 2022;65:1170-8
83. Uehara H, Yamaguchi S, Komatsu K, Miyagi T, Takahashi K. Successful treatment of subcutaneous Purpureocillium lilacinum infection with fosravuconazole. J Dermatol. 2023;50:e104-e5
84. Mazzeti AL, Capelari-Oliveira P, Bahia MT, Mosqueira VCF. Review on Experimental Treatment Strategies Against Trypanosoma cruzi. J Exp Pharmacol. 2021;13:409-32
85. Gupta AK, Wang T, Cooper EA. Dermatophytomas: Clinical Overview and Treatment. J Fungi (Basel). 2022 8
86. Pfarr KM, Krome AK, Al-Obaidi I, Batchelor H, Vaillant M, Hoerauf A. et al. The pipeline for drugs for control and elimination of neglected tropical diseases: 1. Anti-infective drugs for regulatory registration. Parasit Vectors. 2023;16:82
87. Kim H, Radwanski E, Lovey R, Lin CC, Nomeir AA. Pharmacokinetics of the active antifungal enantiomer, SCH 42427 (RR), and evaluation of its chiral inversion in animals following its oral administration and the oral administration of its racemate genaconazole (RR/SS). Chirality. 2002;14:436-41
88. Lamb DC, Kelly DE, Baldwin BC, Gozzo F, Boscott P, Richards WG. et al. Differential inhibition of Candida albicans CYP51 with azole antifungal stereoisomers. FEMS Microbiol Lett. 1997;149:25-30
89. Sun LN, Zhang Y, Fang QC, Shen Y, Li X, Chen C. et al. LC-MS/MS methods to quantify HCP002 in human plasma and urine: applications in a pharmacokinetic study. Bioanalysis. 2022;14:307-16
90. AdisInsight. IDP113. 18 Jul. 2018
91. Ananda-Rajah MR, Kontoyiannis D. Isavuconazole: a new extended spectrum triazole for invasive mold diseases. Future Microbiol. 2015;10:693-708
92. Lewis JS 2nd, Wiederhold NP, Hakki M, Thompson GR 3rd. New Perspectives on Antimicrobial Agents: Isavuconazole. Antimicrob Agents Chemother. 2022;66:e0017722
93. Gupta AK, Lyons DC. The Rise and Fall of Oral Ketoconazole. J Cutan Med Surg. 2015;19:352-7
94. Pivonello R, Simeoli C, Di Paola N, Colao A. Cushing's disease: adrenal steroidogenesis inhibitors. Pituitary. 2022;25:726-32
95. Fleseriu M, Auchus RJ, Pivonello R, Salvatori R, Zacharieva S, Biller BMK. Levoketoconazole: a novel treatment for endogenous Cushing's syndrome. Expert Rev Endocrinol Metab. 2021;16:159-74
96. Gilis-Januszewska A, Boguslawska A, Rzepka E, Ziaja W, Hubalewska-Dydejczyk A. Individualized medical treatment options in Cushing disease. Front Endocrinol (Lausanne). 2022;13:1060884
97. Pierard GE, Arrese JE, Pierard-Franchimont C. Itraconazole. Expert Opin Pharmacother. 2000;1:287-304
98. Shokri A, Abastabar M, Keighobadi M, Emami S, Fakhar M, Teshnizi SH. et al. Promising antileishmanial activity of novel imidazole antifungal drug luliconazole against Leishmania major: In vitro and in silico studies. J Glob Antimicrob Resist. 2018;14:260-5
99. Fothergill AW. Miconazole: a historical perspective. Expert Rev Anti Infect Ther. 2006;4:171-5
100. Logan A, Wolfe A, Williamson JC. Antifungal Resistance and the Role of New Therapeutic Agents. Curr Infect Dis Rep. 2022;24:105-16
101. Murray A, Cass L, Ito K, Pagani N, Armstrong-James D, Dalal P. et al. PC945, a Novel Inhaled Antifungal Agent, for the Treatment of Respiratory Fungal Infections. J Fungi (Basel). 2020 6
102. De SK. Oteseconazole: First Approved Orally Bioavailable and Selective CYP51 Inhibitor for the Treatment of Patients with Recurrent Vulvovaginal Candidiasis. Curr Med Chem. 2023
103. Clark NM, Grim SA, Lynch JP 3rd. Posaconazole: Use in the Prophylaxis and Treatment of Fungal Infections. Semin Respir Crit Care Med. 2015;36:767-85
104. Ausma J, Pennick G, Bohets H, van de Velde V, Borgers M, Fothergill A. Absence of an active metabolite for the triazole antifungal pramiconazole. Acta Derm Venereol. 2007;87:22-6
105. Geria AN, Scheinfeld NS. Pramiconazole, a triazole compound for the treatment of fungal infections. IDrugs. 2008;11:661-70
106. Gupta AK, Foley KA. Antifungal Treatment for Pityriasis Versicolor. J Fungi (Basel). 2015;1:13-29
107. Vanden Bossche H, Ausma J, Bohets H, Vermuyten K, Willemsens G, Marichal P. et al. The novel azole R126638 is a selective inhibitor of ergosterol synthesis in Candida albicans, Trichophyton spp, and Microsporum canis. Antimicrob Agents Chemother. 2004;48:3272-8
108. Trevino-Rangel RJ, Gonzalez GM, Montoya AM, Rojas OC, Elizondo-Zertuche M, Alvarez-Villalobos NA. Recent Antifungal Pipeline Developments against Candida auris: A Systematic Review. J Fungi (Basel). 2022 8
109. Warrilow AG, Parker JE, Price CL, Nes WD, Garvey EP, Hoekstra WJ. et al. The Investigational Drug VT-1129 Is a Highly Potent Inhibitor of Cryptococcus Species CYP51 but Only Weakly Inhibits the Human Enzyme. Antimicrob Agents Chemother. 2016;60:4530-8
110. AdisInsight. SSY726. 12 Dec. 2007
111. Andriole VT. The 1998 Garrod lecture. Current and future antifungal therapy: new targets for antifungal agents. J Antimicrob Chemother. 1999;44:151-62
112. Lakhani P, Patil A, Majumdar S. Challenges in the Polyene- and Azole-Based Pharmacotherapy of Ocular Fungal Infections. J Ocul Pharmacol Ther. 2019;35:6-22
113. Perez-Cantero A, Lopez-Fernandez L, Guarro J, Capilla J. Azole resistance mechanisms in Aspergillus: update and recent advances. Int J Antimicrob Agents. 2020;55:105807
114. Hargrove TY, Garvey EP, Hoekstra WJ, Yates CM, Wawrzak Z, Rachakonda G. et al. Crystal Structure of the New Investigational Drug Candidate VT-1598 in Complex with Aspergillus fumigatus Sterol 14alpha-Demethylase Provides Insights into Its Broad-Spectrum Antifungal Activity. Antimicrob Agents Chemother. 2017 61
115. Wiederhold NP, Lockhart SR, Najvar LK, Berkow EL, Jaramillo R, Olivo M. et al. The Fungal Cyp51-Specific Inhibitor VT-1598 Demonstrates In Vitro and In Vivo Activity against Candida auris. Antimicrob Agents Chemother. 2019 63
116. Reardon DA, Narita Y, Arakawa Y, Goldlust SA, Ansstas G, Mei J. et al. DSP-0390, an oral emopamil binding protein (EBP) inhibitor, in patients with recurrent high-grade glioma: A first-in-human, phase 1 study. J Clin Oncol. 2022 40
117. Reardon DA, Narita Y, Arakawa Y, Goldlust SA, Ansstas G, Mei J. et al. DSP-0390, an oral emopamil binding protein (EBP) inhibitor, in patients with recurrent high-grade glioma: A first-in-human, phase 1 study. Journal of Clinical Oncolog. 2022Journal of Clinical Oncology 40(16_suppl):TPS2077-TPS2077; 40: TPS2077-TPS.
118. AdiInsight. DSP 0390. 18 Jul. 2022
119. Li H, Liu X, Sun N, Wang T, Zhu J, Yang S. et al. Differentially Expressed Circular Non-coding RNAs in Atherosclerotic Aortic Vessels and Their Potential Functions in Endothelial Injury. Front Cardiovasc Med. 2021;8:657544
120. Bhardwaj A, Singh H, Trinidad CM, Albarracin CT, Hunt KK, Bedrosian I. The isomiR-140-3p-regulated mevalonic acid pathway as a potential target for prevention of triple negative breast cancer. Breast Cancer Res. 2018;20:150
121. Chen W, Li L, Wang J, Zhang R, Zhang T, Wu Y. et al. The ABCA1-efferocytosis axis: A new strategy to protect against atherosclerosis. Clin Chim Acta. 2021;518:1-8
122. Chen W, Wang S, Xing D. New Horizons for the Roles and Association of APE1/Ref-1 and ABCA1 in Atherosclerosis. J Inflamm Res. 2021;14:5251-71
123. Chen W, Xing J, Liu X, Wang S, Xing D. The role and transformative potential of IL-19 in atherosclerosis. Cytokine Growth Factor Rev. 2021;62:70-82
124. Chen W, Zhong Y, Feng N, Guo Z, Wang S, Xing D. New horizons in the roles and associations of COX-2 and novel natural inhibitors in cardiovascular diseases. Mol Med. 2021;27:123
125. Zhang S, Li L, Wang J, Zhang T, Ye T, Wang S. et al. Recent advances in the regulation of ABCA1 and ABCG1 by lncRNAs. Clin Chim Acta. 2021;516:100-10
126. Hung HH, Nagatsuka Y, Solda T, Kodali VK, Iwabuchi K, Kamiguchi H. et al. Selective involvement of UGGT variant: UGGT2 in protecting mouse embryonic fibroblasts from saturated lipid-induced ER stress. Proc Natl Acad Sci U S A. 2022;119:e2214957119
127. Torisu K, Zhang X, Nonaka M, Kaji T, Tsuchimoto D, Kajitani K. et al. PKCeta deficiency improves lipid metabolism and atherosclerosis in apolipoprotein E-deficient mice. Genes Cells. 2016;21:1030-48
128. Park DW, Lee HK, Lyu JH, Chin H, Kang SW, Kim YJ. et al. TLR2 stimulates ABCA1 expression via PKC-eta and PLD2 pathway. Biochem Biophys Res Commun. 2013;430:933-7
129. Liu HY, Pedros C, Kong KF, Canonigo-Balancio AJ, Xue W, Altman A. Leveraging the Treg-intrinsic CTLA4-PKCeta signaling pathway for cancer immunotherapy. J Immunother Cancer. 2021 9
130. Zhou GK, Zhang GY, Yuan ZN, Pei R, Liu DM. Has_circ_0008274 promotes cell proliferation and invasion involving AMPK/mTOR signaling pathway in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:8772-80
131. Gao C, Wen Y, Jiang F, Gu X, Zhu X. Circular RNA circ_0008274 upregulates granulin to promote the progression of hepatocellular carcinoma via sponging microRNA -140-3p. Bioengineered. 2021;12:1890-901
132. Zhao J, Li T, Luo W. Silencing of circ-PRKCH protects against lipopolysaccharide (LPS)-evoked chondrocyte damage and extracellular matrix loss by the miR-140-3p/ADAM10 axis. Gen Physiol Biophys. 2021;40:89-101
133. Liu J, Wei Y, Lin Y, Zhang P, Zhang Z, Huang H. et al. Expression of the circular RNAs in astaxanthin promotes cholesterol efflux from THP-1 cells based on RNA-seq. Genes Nutr. 2021;16:13
134. Zou TB, Zhu SS, Luo F, Li WQ, Sun XR, Wu HF. Effects of Astaxanthin on Reverse Cholesterol Transport and Atherosclerosis in Mice. Biomed Res Int. 2017;2017:4625932
135. Xu X, Chen J, Li Y, Yang X, Wang Q, Wen Y. et al. Targeting epigenetic modulation of cholesterol synthesis as a therapeutic strategy for head and neck squamous cell carcinoma. Cell Death Dis. 2021;12:482
136. Neele AE, Chen HJ, Gijbels MJJ, van der Velden S, Hoeksema MA, Boshuizen MCS. et al. Myeloid Ezh2 Deficiency Limits Atherosclerosis Development. Front Immunol. 2020;11:594603
137. Wei X, Zhang Y, Xie L, Wang K, Wang X. Pharmacological inhibition of EZH2 by GSK126 decreases atherosclerosis by modulating foam cell formation and monocyte adhesion in apolipoprotein E-deficient mice. Exp Ther Med. 2021;22:841
138. Meng XD, Yao HH, Wang LM, Yu M, Shi S, Yuan ZX. et al. Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE(-/-) Mice. Mol Ther Nucleic Acids. 2020;19:84-96
139. Zheng X, Zhao X, Han Z, Chen K. Enhancer of zeste homolog 2 participates in the process of atherosclerosis by modulating microRNA-139-5p methylation and signal transducer and activator of transcription 1 expression. IUBMB Life. 2021;73:238-51
140. Miao R, Qi C, Fu Y, Wang Y, Lang Y, Liu W. et al. Silencing of circARHGAP12 inhibits the progression of atherosclerosis via miR-630/EZH2/TIMP2 signal axis. J Cell Physiol. 2022;237:1057-69
141. Zhao X, Ma X, Guo J, Mi M, Wang K, Zhang C. et al. Circular RNA CircEZH2 Suppresses Transmissible Gastroenteritis Coronavirus-induced Opening of Mitochondrial Permeability Transition Pore via Targeting MiR-22 in IPEC-J2. Int J Biol Sci. 2019;15:2051-64
142. Yao B, Zhang Q, Yang Z, An F, Nie H, Wang H. et al. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol Cancer. 2022;21:140
143. Wang S, Dong L, Ma L, Yang S, Zheng Y, Zhang J. et al. SQLE facilitates the pancreatic cancer progression via the lncRNA-TTN-AS1/miR-133b/SQLE axis. J Cell Mol Med. 2022;26:3636-47
144. Qin Y, Zhang Y, Tang Q, Jin L, Chen Y. SQLE induces epithelial-to-mesenchymal transition by regulating of miR-133b in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai). 2017;49:138-48
145. Wang D, Zhao Z, Shi Y, Luo J, Chen T, Xi Q. et al. CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity. Animals (Basel). 2022 12
146. Wu L, Gao J, Liu S, Jia Y, Li C, Duan L. CircRNA circ_0005273 contributes to the cisplatin resistance of cervical cancer cells by sponging miR-133b. J Obstet Gynaecol. 2022:1-8
147. Chen D, Wang Y, Yang F, Keranmu A, Zhao Q, Wu L. et al. The circRAB3IP Mediated by eIF4A3 and LEF1 Contributes to Enzalutamide Resistance in Prostate Cancer by Targeting miR-133a-3p/miR-133b/SGK1 Pathway. Front Oncol. 2021;11:752573
148. He X, Ma J, Zhang M, Cui J, Yang H. Circ_0007031 enhances tumor progression and promotes 5-fluorouracil resistance in colorectal cancer through regulating miR-133b/ABCC5 axis. Cancer Biomark. 2020;29:531-42
149. He J, Ming Y, MinLi Y, Han Z, Jiang J, Zhou J. et al. hsa_circ_0006459 and hsa_circ_0015962 affect prognosis of Dengue fever. Sci Rep. 2019;9:19425
150. Dai Q, Ma Y, Xu Z, Zhang L, Yang H, Liu Q. et al. Downregulation of circular RNA HECTD1 induces neuroprotection against ischemic stroke through the microRNA-133b/TRAF3 pathway. Life Sci. 2021;264:118626
151. Zhao P, Zhang J. Circ_0039569 Competes with MARCKS for miR-133b Binding Sites to Promote the Progression of Renal Cell Carcinoma. Nephron. 2022;146:404-17
152. Yu H, Xu L, Liu Z, Guo B, Han Z, Xin H. Circ_MDM2_000139, Circ_ATF2_001418, Circ_CDC25C_002079, and Circ_BIRC6_001271 Are Involved in the Functions of XAV939 in Non-Small Cell Lung Cancer. Can Respir J. 2019;2019:9107806
153. Brown MS, Radhakrishnan A, Goldstein JL. Retrospective on Cholesterol Homeostasis: The Central Role of Scap. Annu Rev Biochem. 2018;87:783-807
154. He Q, Shao D, Hao S, Yuan Y, Liu H, Liu F. et al. CircSCAP Aggravates Oxidized Low-density Lipoprotein-induced Macrophage Injury by Upregulating PDE3B by miR-221-5p in Atherosclerosis. J Cardiovasc Pharmacol. 2021;78:e749-e60
155. Zhang DD, Wang DD, Wang Z, Wang YB, Li GX, Sun GR. et al. Estrogen Abolishes the Repression Role of gga-miR-221-5p Targeting ELOVL6 and SQLE to Promote Lipid Synthesis in Chicken Liver. Int J Mol Sci. 2020 21
156. Zeng B, Wang H, Luo J, Xie M, Zhao Z, Chen X. et al. Porcine Milk-Derived Small Extracellular Vesicles Promote Intestinal Immunoglobulin Production through pIgR. Animals (Basel). 2021 11
157. Yuan Y, Gong Y, Zhong L, Ding X, Yang Z, Su X. et al. Circular RNA expression profile and competing endogenous RNA regulatory network in preeclampsia. Placenta. 2022;119:32-8
158. Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19:269-79
159. Liu T, Zhang G, Wang Y, Rao M, Zhang Y, Guo A. et al. Identification of Circular RNA-MicroRNA-Messenger RNA Regulatory Network in Atrial Fibrillation by Integrated Analysis. Biomed Res Int. 2020;2020:8037273
160. Zhang LW, Wang B, Yang JX, Yang H. Circ-PRMT5 stimulates migration in esophageal cancer by binding miR-203. Eur Rev Med Pharmacol Sci. 2020;24:9965-72
161. Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099
162. Nie W, Zhang X, Yan H, Li S, Zhu W, Fan F. et al. Xiaoxianggou attenuates atherosclerotic plaque formation in endogenous high Ang II ApoE(-/-) mice via the inhibition of miR-203 on the expression of Ets-2 in endothelial cells. Biomed Pharmacother. 2016;82:173-9
163. Guo Y, Huang S, Ma Y, Zhang J, Wen Y, Zhou L. et al. MiR-377 mediates the expression of Syk to attenuate atherosclerosis lesion development in ApoE(-/-) mice. Biomed Pharmacother. 2019;118:109332
164. Chen LY, Xia XD, Zhao ZW, Gong D, Ma XF, Yu XH. et al. MicroRNA-377 Inhibits Atherosclerosis by Regulating Triglyceride Metabolism Through the DNA Methyltransferase 1 in Apolipoprotein E-Knockout Mice. Circ J. 2018;82:2861-71
165. Du W, Li D, Guo X, Li P, Li X, Tong S. et al. Circ-PRMT5 promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif Cells Nanomed Biotechnol. 2019;47:4120-30
166. Sala F, Aranda JF, Rotllan N, Ramirez CM, Aryal B, Elia L. et al. MiR-143/145 deficiency attenuates the progression of atherosclerosis in Ldlr-/-mice. Thromb Haemost. 2014;112:796-802
167. Liang J, Li X, Xu J, Cai GM, Cao JX, Zhang B. hsa_circ_0072389, hsa_circ_0072386, hsa_circ_0008621, hsa_circ_0072387, and hsa_circ_0072391 aggravate glioma via miR-338-5p/IKBIP. Aging (Albany NY). 2021;13:25213-40
168. Xu YP, Dong ZN, Wang SW, Zheng YM, Zhang C, Zhou YQ. et al. circHMGCS1-016 reshapes immune environment by sponging miR-1236-3p to regulate CD73 and GAL-8 expression in intrahepatic cholangiocarcinoma. J Exp Clin Cancer Res. 2021;40:290
169. Sutton NR, Bouis D, Mann KM, Rashid IM, McCubbrey AL, Hyman MC. et al. CD73 Promotes Age-Dependent Accretion of Atherosclerosis. Arterioscler Thromb Vasc Biol. 2020;40:61-71
170. Romaniuk MA, Tribulatti MV, Cattaneo V, Lapponi MJ, Molinas FC, Campetella O. et al. Human platelets express and are activated by galectin-8. Biochem J. 2010;432:535-47
171. Gerlach BD, Marinello M, Heinz J, Rymut N, Sansbury BE, Riley CO. et al. Resolvin D1 promotes the targeting and clearance of necroptotic cells. Cell Death Differ. 2020;27:525-39
172. Wang F, Xu X, Zhang N, Chen Z. Identification and integrated analysis of hepatocellular carcinoma-related circular RNA signature. Ann Transl Med. 2020;8:294
173. Zhen N, Gu S, Ma J, Zhu J, Yin M, Xu M. et al. CircHMGCS1 Promotes Hepatoblastoma Cell Proliferation by Regulating the IGF Signaling Pathway and Glutaminolysis. Theranostics. 2019;9:900-19
174. Cao Y, Wang S, Liu S, Wang Y, Jin H, Ma H. et al. Effects of Long-Chain Fatty Acyl-CoA Synthetase 1 on Diglyceride Synthesis and Arachidonic Acid Metabolism in Sheep Adipocytes. Int J Mol Sci. 2020 21
175. Tian WH, Wang Z, Yue YX, Li H, Li ZJ, Han RL. et al. miR-34a-5p Increases Hepatic Triglycerides and Total Cholesterol Levels by Regulating ACSL1 Protein Expression in Laying Hens. Int J Mol Sci. 2019 20
176. Liu K, Liu X, Deng Y, Li Z, Tang A. CircRNA-mediated regulation of brown adipose tissue adipogenesis. Front Nutr. 2022;9:926024
177. He J, Zhao H, Liu X, Wang D, Wang Y, Ai Y. et al. Sevoflurane suppresses cell viability and invasion and promotes cell apoptosis in colon cancer by modulating exosomemediated circHMGCS1 via the miR34a5p/SGPP1 axis. Oncol Rep. 2020;44:2429-42
178. Xu Y, Kang P, Leng K, Yao Y, Liao G, Han Y. et al. Circ_ASPH promotes cholangiocarcinoma growth and metastasis through the miR-581/ATP-binding cassette transporter G1 signaling pathway. Cancer Commun (Lond). 2020;40:545-50
179. Wang Y, Xu Z, Wang X, Zheng J, Peng L, Zhou Y. et al. Extracellular-vesicle containing miRNA-503-5p released by macrophages contributes to atherosclerosis. Aging (Albany NY). 2021;13:12239-57
180. Wu B, Sun Y, Hou J. CircMED12L Protects Against Hydrogen Peroxide-induced Apoptotic and Oxidative Injury in Human Lens Epithelial Cells by miR-34a-5p/ALCAM axis. Curr Eye Res. 2022;47:1631-40
181. Zhang S, Guo G. Circ_FURIN promotes trophoblast cell proliferation, migration and invasion in preeclampsia by regulating miR-34a-5p and TFAP2A. Hypertens Res. 2022;45:1334-44
182. Ding L, Liu GL, Lu L, Ge L, Wang JY. circ_CSNK1E modulates airway smooth muscle cells proliferation and migration via miR-34a-5p/VAMP2 axis in asthma. Cell Signal. 2022;95:110340
183. Ye X, Zhu B, Han J, Huang J, Wu Y. Circ-0036602 Acts As a Sponge of MiR-34a-5p and MiR-431-5p to Promote Cervical Cancer Proliferation and Invasion. J Genomics. 2022;10:16-25
184. Zhang S, Luo J, Zeng S. Circ-LRP1B functions as a competing endogenous RNA to regulate proliferation, apoptosis and oxidative stress of LPS-induced human C28/I2 chondrocytes. J Bioenerg Biomembr. 2022;54:93-108
185. Yang X, Sun Q, Song Y, Li W. circHUWE1 Exerts an Oncogenic Role in Inducing DDP-Resistant NSCLC Progression Depending on the Regulation of miR-34a-5p/TNFAIP8. Int J Genomics. 2021;2021:3997045
186. Qi L, Wang W, Zhao G, Jiang H, Zhang Y, Zhao D. et al. Circular RNA circitga7 accelerates glioma progression via miR-34a-5p/VEGFA axis. Aging (Albany NY). 2021;13:13138-52
187. Cheng J, Nie D, Li B, Gui S, Li C, Zhang Y. et al. CircNFIX promotes progression of pituitary adenoma via CCNB1 by sponging miR-34a -5p. Mol Cell Endocrinol. 2021;525:111140
188. Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H. NFIX Circular RNA Promotes Glioma Progression by Regulating miR-34a-5p via Notch Signaling Pathway. Front Mol Neurosci. 2018;11:225
189. Xiang Q, Kang L, Wang J, Liao Z, Song Y, Zhao K. et al. CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine. 2020;53:102679
190. Cao HX, Miao CF, Sang LN, Huang YM, Zhang R, Sun L. et al. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 2020;243:117255
191. Jin C, Shi L, Li Z, Liu W, Zhao B, Qiu Y. et al. Circ_0039569 promotes renal cell carcinoma growth and metastasis by regulating miR-34a-5p/CCL22. Am J Transl Res. 2019;11:4935-45
192. Li M, Wang Y, Liu Y, Zhang X, Liu J, Wang P. Low Expression of hsa_circ_0018069 in Human Bladder Cancer and Its Clinical Significance. Biomed Res Int. 2019;2019:9681863
193. Capobianco V, Nardelli C, Ferrigno M, Iaffaldano L, Pilone V, Forestieri P. et al. miRNA and protein expression profiles of visceral adipose tissue reveal miR-141/YWHAG and miR-520e/RAB11A as two potential miRNA/protein target pairs associated with severe obesity. J Proteome Res. 2012;11:3358-69
194. Ahonen MA, Asghar MY, Parviainen SJ, Liebisch G, Horing M, Leidenius M. et al. Human adipocyte differentiation and composition of disease-relevant lipids are regulated by miR-221-3p. Biochim Biophys Acta Mol Cell Biol Lipids. 2021;1866:158841
195. Deng Q, Ma D, Sun G, Yuan X, Wang Z, Liu G. PTEN influences insulin and lipid metabolism in bovine hepatocytes in vitro. J Dairy Res. 2019;86:73-6
196. Dou Z, Li S, Ren W, Wang Q, Liu J, Kong X. et al. Decreased expression of hsa_circ_0072387 as a valuable predictor for oral squamous cell carcinoma. Oral Dis. 2019;25:1302-8
197. Han L, Cheng J, Li A. hsa_circ_0072387 Suppresses Proliferation, Metastasis, and Glycolysis of Oral Squamous Cell Carcinoma Cells by Downregulating miR-503-5p. Cancer Biother Radiopharm. 2021;36:84-94
198. Fan L, Wang C, Zhan P, Liu Y. miR-141-3p is Poorly Expressed in Polycystic Ovary Syndrome and Correlates with Glucose and Lipid Metabolism. Int J Endocrinol. 2021;2021:2022938
199. Chen DL, Sheng H, Zhang DS, Jin Y, Zhao BT, Chen N. et al. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol Cancer. 2021;20:166
200. Ma L, Wei J, Zeng Y, Liu J, Xiao E, Kang Y. et al. Mesenchymal stem cell-originated exosomal circDIDO1 suppresses hepatic stellate cell activation by miR-141-3p/PTEN/AKT pathway in human liver fibrosis. Drug Deliv. 2022;29:440-53
201. Zhang C, Cao J, Lv W, Mou H. CircRNA_100395 Carried by Exosomes From Adipose-Derived Mesenchymal Stem Cells Inhibits the Malignant Transformation of Non-Small Cell Lung Carcinoma Through the miR-141-3p-LATS2 Axis. Front Cell Dev Biol. 2021;9:663147
202. Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y. et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30:1054-70
203. Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428
204. Cheng X, Tian P, Zheng W, Yan X. Piplartine attenuates the proliferation of hepatocellular carcinoma cells via regulating hsa_circ_100338 expression. Cancer Med. 2020;9:4265-73
205. Huang XY, Huang ZL, Zhang PB, Huang XY, Huang J, Wang HC. et al. CircRNA-100338 Is Associated With mTOR Signaling Pathway and Poor Prognosis in Hepatocellular Carcinoma. Front Oncol. 2019;9:392
206. Wang Y, Ren F, Sun D, Liu J, Liu B, He Y. et al. CircKEAP1 Suppresses the Progression of Lung Adenocarcinoma via the miR-141-3p/KEAP1/NRF2 Axis. Front Oncol. 2021;11:672586
207. Yu Y, Dong G, Li Z, Zheng Y, Shi Z, Wang G. circLRP6 contributes to osteosarcoma progression by regulating the miR1413p/HDAC4/HMGB1 axis. Int J Oncol. 2022 60
208. Chen D, Chou FJ, Chen Y, Tian H, Wang Y, You B. et al. Targeting the radiation-induced TR4 nuclear receptor-mediated QKI/circZEB1/miR-141-3p/ZEB1 signaling increases prostate cancer radiosensitivity. Cancer Lett. 2020;495:100-11
209. Xiao Y, Liu G, Sun Y, Gao Y, Ouyang X, Chang C. et al. Targeting the estrogen receptor alpha (ERalpha)-mediated circ-SMG1.72/miR-141-3p/Gelsolin signaling to better suppress the HCC cell invasion. Oncogene. 2020;39:2493-508
210. Chao F, Song Z, Wang S, Ma Z, Zhuo Z, Meng T. et al. Novel circular RNA circSOBP governs amoeboid migration through the regulation of the miR-141-3p/MYPT1/p-MLC2 axis in prostate cancer. Clin Transl Med. 2021;11:e360
211. Peng N, Shi L, Zhang Q, Hu Y, Wang N, Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS One. 2017;12:e0170287
212. Li X, Lu S, Jiang Y, Xu K, He J, Qiu Z. et al. Circ_0061140 facilitates adenomyosis progression by up-regulating LIN28B in vitro. Gynecol Obstet Invest. 2022
213. Wang D, Luo Y, Wang G, Yang Q. CircATRNL1 promotes epithelial-mesenchymal transition in endometriosis by upregulating Yes-associated protein 1 in vitro. Cell Death Dis. 2020;11:594
214. Guo W, Mu K, Zhang B, Sun C, Zhao L, Li HR. et al. The circular RNA circ-GRB10 participates in the molecular circuitry inhibiting human intervertebral disc degeneration. Cell Death Dis. 2020;11:612
215. Kun-Peng Z, Chun-Lin Z, Jian-Ping H, Lei Z. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14:1513-20
216. Wei W, Ji L, Duan W, Zhu J. Circular RNA circ_0081001 knockdown enhances methotrexate sensitivity in osteosarcoma cells by regulating miR-494-3p/TGM2 axis. J Orthop Surg Res. 2021;16:50
217. van Ingen E, Foks AC, Woudenberg T, van der Bent ML, de Jong A, Hohensinner PJ. et al. Inhibition of microRNA-494-3p activates Wnt signaling and reduces proinflammatory macrophage polarization in atherosclerosis. Mol Ther Nucleic Acids. 2021;26:1228-39
218. Xu F, Liu G, Wang L, Wang X, Jin X, Bo W. miR-494 promotes progression of retinoblastoma via PTEN through PI3K/AKT signaling pathway. Oncol Lett. 2020;20:1952-60
219. Wezel A, Welten SM, Razawy W, Lagraauw HM, de Vries MR, Goossens EA. et al. Inhibition of MicroRNA-494 Reduces Carotid Artery Atherosclerotic Lesion Development and Increases Plaque Stability. Ann Surg. 2015;262:841-7 discussion 7-8
220. Lu P, Jiang Y, Xia Z. Hsa_circ_0003221 facilitates the malignant development of bladder cancer cells via resulting in the upregulation of DHCR24 by targeting miR-892b. Investig Clin Urol. 2022;63:577-88
221. Yang Z, Jin J, Chang T. CircPTK2 (hsa_circ_0003221) Contributes to Laryngeal Squamous Cell Carcinoma by the miR-1278/YAP1 Axis. J Oncol. 2021;2021:2408384
222. Chi X, Gu X, Chen S, Shen X. Circ_0003221 Downregulation Restrains Cervical Cancer Cell Growth, Metastasis and Angiogenesis by Governing the miR-139-3p/S100A14 Pathway. Reprod Sci. 2022;29:1822-35
223. Xie H, Wang J, Wang B. Circular RNA Circ_0003221 Promotes Cervical Cancer Progression by Regulating miR-758-3p/CPEB4 Axis. Cancer Manag Res. 2021;13:5337-50
224. Liu D, Qiao C, Luo H. MicroRNA-1278 ameliorates the inflammation of cardiomyocytes during myocardial ischemia by targeting both IL-22 and CXCL14. Life Sci. 2021;269:118817
225. Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D. et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31:2707-14
226. Liu P, Wang Z, Ou X, Wu P, Zhang Y, Wu S. et al. The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol Cancer. 2022;21:198
227. Daimiel LA, Fernandez-Suarez ME, Rodriguez-Acebes S, Crespo L, Lasuncion MA, Gomez-Coronado D. et al. Promoter analysis of the DHCR24 (3beta-hydroxysterol Delta(24)-reductase) gene: characterization of SREBP (sterol-regulatory-element-binding protein)-mediated activation. Biosci Rep. 2012;33:57-69
228. Wang Z, Yang L, Wu P, Li X, Tang Y, Ou X. et al. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol Cancer. 2022;21:29
229. Jing B, Hui Z. Circular RNA_0033596 aggravates endothelial cell injury induced by oxidized low-density lipoprotein via microRNA-217-5p /chloride intracellular channel 4 axis. Bioengineered. 2022;13:3410-21
230. Fang P, Jiang Q, Liu S, Gu J, Hu K, Wang Z. Circ_0002099 is a novel molecular therapeutic target for bladder cancer. Drug Dev Res. 2022
231. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T. et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539-55
232. Jin X, Gao J, Zheng R, Yu M, Ren Y, Yan T. et al. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis. 2020;11:123
233. Qiu F, Ou D, Tan H, Gao Y, Zi D. The circCDK17/miR-122-5p/ASF1B axis regulates the progression of cervical cancer. Histol Histopathol. 2022: 18527.
234. Yin W, Xu J, Li C, Dai X, Wu T, Wen J. Circular RNA circ_0007142 Facilitates Colorectal Cancer Progression by Modulating CDC25A Expression via miR-122-5p. Onco Targets Ther. 2020;13:3689-701
235. Xu X, Chen Y, Tan B, Wang D, Yuan Z, Wang F. Circular RNA circ_0011269 sponges miR-122 to regulate RUNX2 expression and promotes osteoporosis progression. J Cell Biochem. 2020
236. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K. et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177
237. Huang X, Luo Y, Li X. Circ_0072995 Promotes Ovarian Cancer Progression Through Regulating miR-122-5p/SLC1A5 Axis. Biochem Genet. 2022;60:153-72
238. Yang F, Chen Y, Luo L, Nong S, Li T. circFOXO3 Induced by KLF16 Modulates Clear Cell Renal Cell Carcinoma Growth and Natural Killer Cell Cytotoxic Activity through Sponging miR-29a-3p and miR-122-5p. Dis Markers. 2022;2022:6062236
239. Chen C, Deng L, Nie DK, Jia F, Fu LS, Wan ZQ. et al. Circular RNA Pleiotrophin promotes carcinogenesis in glioma via regulation of microRNA-122/SRY-box transcription factor 6 axis. Eur J Cancer Prev. 2020;29:165-73
240. Lu X, Liu Y, Xuan W, Ye J, Yao H, Huang C. et al. Circ_1639 induces cells inflammation responses by sponging miR-122 and regulating TNFRSF13C expression in alcoholic liver disease. Toxicol Lett. 2019;314:89-97
241. Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: A Promising Therapeutic Target in Cancer. Int J Mol Sci. 2022 23
242. Deng Y, Campbell F, Han K, Theodore D, Deeg M, Huang M. et al. Randomized clinical trials towards a single-visit cure for chronic hepatitis C: Oral GSK2878175 and injectable RG-101 in chronic hepatitis C patients and long-acting injectable GSK2878175 in healthy participants. J Viral Hepat. 2020;27:699-708
243. Zhou YL, Wu WP, Cheng J, Liang LL, Cen JM, Chen C. et al. CircFOXO3 rs12196996, a polymorphism at the gene flanking intron, is associated with circFOXO3 levels and the risk of coronary artery disease. Aging (Albany NY). 2020;12:13076-89
244. Zhang L, Wang Y, Zhang Y, Zhao Y, Li P. Pathogenic mechanisms and the potential clinical value of circFoxo3 in cancers. Mol Ther Nucleic Acids. 2021; 23: 908-17. 1. Yang K, Xiao Q, Niu M, Pan X, Zhu X. Exosomes in atherosclerosis: Convergence on macrophages. Int J Biol Sci. 2022;18:3266-81
Author contact
![]() Corresponding authors: Cancer Institute, The Affiliated Hospital of Qingdao University, Qingdao University, Qingdao Cancer Institute, Qingdao, Shandong, 266071. E-mail addresses: 14268657com (Yingchun Shao), xdm_tsinghuacom (Dongming Xing).
Corresponding authors: Cancer Institute, The Affiliated Hospital of Qingdao University, Qingdao University, Qingdao Cancer Institute, Qingdao, Shandong, 266071. E-mail addresses: 14268657com (Yingchun Shao), xdm_tsinghuacom (Dongming Xing).

 Global reach, higher impact
Global reach, higher impact