10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(15):4834-4848. doi:10.7150/ijbs.87028 This issue Cite
Review
The bidirectional interplay between ncRNAs and methylation modifications in gastrointestinal tumors
1. Department of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China.
2. Henan Liver Transplantation Centre, China.
3. Henan Organ Transplantation Quality Control Centre, China.
4. Open and Key Laboratory for Hepatobiliary & Pancreatic Surgery and Digestive Organ Transplantation at Henan Universities, China.
5. Henan Innovative Research Group for Hepatobiliary & Pancreatic Surgery and Digestive Organ Transplantation, China.
Received 2023-6-11; Accepted 2023-8-26; Published 2023-9-11
Abstract
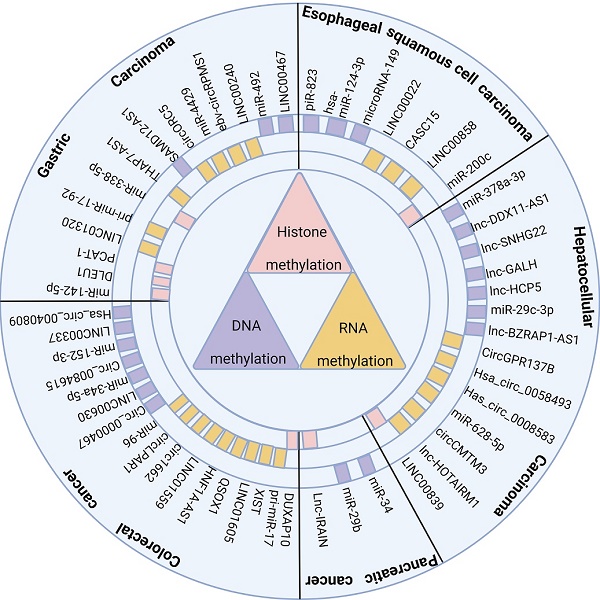
The aberrant expression of methylation and ncRNAs, two crucial regulators of epigenetic modifications, has been widely demonstrated in cancer. The complex interplay between them is essential in promoting malignant phenotype, poor prognosis, and drug resistance in GI tumors (including esophageal, gastric, colorectal, liver, and pancreatic cancers). Therefore, we summarize the interrelation process between ncRNAs and methylation modifications in GI tumors, including the detailed mechanism of methylation enzyme regulation of ncRNAs, the molecular mechanism of ncRNAs regulation of methylation modifications, and the correlation between the interactions between ncRNAs and methylation modifications and clinical features of tumors. Finally, we discuss the potential value of ncRNAs and methylation modifications in clinical diagnosis and therapy.
Keywords: ncRNAs, methylation, GI tumors, Mechanism, Biomarker and Therapy
1. Introduction
Cancer remains a serious global public health issue, with digestive system cancers accounting for a large portion of all cancers. According to statistics, it is estimated that there will be 4.82 million new cancer cases in China in 2022, with digestive system cancers accounting for as much as 42.5% [1]. In 2021, digestive system cancers were also the most common cancer type in the United States, accounting for 25% of all new cancer cases [2]. Digestive system cancers include esophageal cancer, gastric cancer, colorectal cancer, liver cancer, pancreatic cancer, and gallbladder cancer. In addition to their high incidence, the prognosis for digestive system tumors is often poor [3]. According to global cancer statistics in 2020, the 5-year survival rate for pancreatic cancer is less than 10%, while the 5-year survival rate for esophageal and liver cancer is less than 20%, indicating an abysmal prognosis [4]. The high incidence and mortality of digestive system tumors are mainly due to the lack of currently available effective diagnostic and treatment methods [5]. For example, there is a lack of widely available screening methods for esophageal cancer, and patients are often diagnosed with cancer in the late stages [6]. Therefore, understanding the pathogenesis of cancer, identifying biomarkers that aid in the early diagnosis of digestive tract tumors, and finding therapeutic targets are of great significance in reducing the incidence of digestive tract tumors and improving prognosis.
NcRNAs are a class of RNA molecules that do not encode proteins, and they play an important role in regulating cellular transcription and post-transcriptional processes [7]. Based on their length, structure, and function, ncRNAs can be divided into several types, including miRNA, lncRNA, circRNA, and piRNA [8, 9]. NcRNAs play a significant role in regulating gene expression in digestive system tumors. They use their complex epigenetic regulatory mechanisms to act as upstream regulators of downstream oncogenes or tumor suppressor genes [10]. For example, in liver cancer, circMDK upregulates ATG16L1 by inhibiting miR-346 and miR-874-3p, which promotes the occurrence and development of liver cancer [11]. LncRNA AGPG has been shown to play a role in esophageal cancer by binding to and stabilizing PFKB3, which increases glycolytic flux and cell cycle progression in esophageal cancer cells. [12]. Moreover, ncRNAs are also subject to epigenetic regulation and play a role in cancer. For instance, in colorectal cancer, m6A methyltransferase catalyzes the maturation of miR-124, which promotes colorectal cancer metastasis through the miR-124/SPRED2/MAPK axis [13]. These studies highlight the crucial role of ncRNAs in regulating digestive system tumors, as they can act as intermediate molecules to link many cancer-related molecules together.
In recent years, there have been increasing studies on epigenetic modifications [14]. Epigenetic modifications refer to a genetic regulatory mechanism that affects gene expression without changing DNA sequence [15]. In cancer, epigenetic modifications can affect gene expression and function by modifying DNA, histones, RNA, ncRNAs, etc., thereby affecting tumor development [16]. There are many epigenetic modifications currently known, including methylation [17], acetylation [18], ubiquitination [19], and others. Among them, methylation is the most studied, as it is a common modification in cell activity and can change the expression level or structural function of DNA, RNA, and proteins [20]. Many studies have found that methyltransferases can regulate tumor progression and drug resistance in the digestive system by catalyzing methylation or demethylation of cancer-related genes [21].
NcRNAs and methylation modifications are crucial factors that affect the occurrence and development of digestive system cancer, and exploring their interaction is valuable. Thus, this paper summarizes the common methylases and their interacting ncRNAs in recent years in digestive system tumors (Figure 1), examines the mechanism of interaction between them, and their impact on tumor occurrence and development, including the clinical features of tumors and their biological processes. Hopefully, this review will provide novel insights for studying ncRNAs associated with methylation modification and identify new clinical diagnostic markers and therapeutic targets.
ncRNAs regulate cancer progression through DNA methylation, RNA methylation, and histone methylation in digestive system tumors
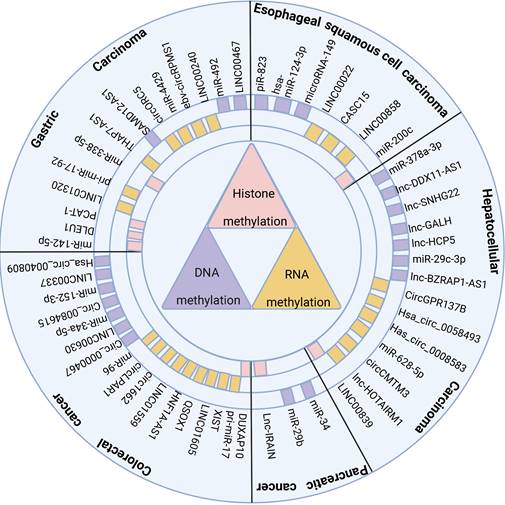
2. Methyltransferases
Methylation is an essential modification of nucleic acids and proteins. Specific sites on nucleic acids and proteins are methylated by the catalysis of methyltransferases, resulting in changes in gene expression or protein structure and function [22]. In this section, we will introduce several methylesterases commonly found in digestive tumors, which are classified into three categories according to the methylation modification substrate: DNA, histone, and RNA methyltransferases.
2.1 DNA methyltransferases
DNA methylation is adding a methyl group to the fifth carbon atom of cytosine on DNA, producing 5-methylcytosine (5-mC) [23]. The DNA methyltransferases (DNMTs) involved in the association between digestive system tumors and ncRNAs include DNMT1, DNMT3A, and DNMT3B. DNMT1 is the most abundant DNA methyltransferase, responsible for maintaining the existing DNA methylation pattern. It is often overexpressed in digestive system tumors, resulting in a highly methylated global or local state [24, 25]. On the other hand, DNMT3A and DNMT3B are often mutated or deleted in tumor cells, leading to a globally or locally hypomethylated state [26].
2.2 Histone methyltransferases
Histone methylation usually refers to the addition or removal of methyl groups on lysine residues of histones, which affects the structure and function of chromatin [27]. Common histone methyltransferases associated with ncRNAs in digestive system tumors include EZH2, RIZ1, and LSD1. EZH2 is often overexpressed or mutated in cancer, and it silences some tumor suppressor genes by methylating H3K27 to H3K27me3 [28]. H3K27 is a lysine residue and the 27th amino acid of histone H3 [29]. RIZ1 can methylate H4R3 to H4R3me1/2; its low expression or loss in cancer is a principal element in cancer cell biology [30]. LSD1 is slightly different from the first two methyltransferases. A histone demethylase can remove methyl groups from H3K4me1/2 or H3K9me1/2. It is often overexpressed in cancer and is associated with poor prognosis in multiple cancers [31-33].
2.3 RNA methyltransferases
RNA methylation mainly involves the methylation modification of bases or sugar rings on RNA molecules, which can alter the structure and function of RNA [34]. The RNA methyltransferases associated with ncRNAs in digestive system tumors are METTL3, METTL14, WTAP, and FTO. METTL3 and METTL14 are m6A writers, which can transfer methyl groups to adenosine on RNA molecules. However, their expression and functions vary in tumor tissues [35]. WTAP does not have a methylation function, but it is an essential auxiliary factor of m6A methyltransferase and can form a complex with METTL3 and METTL14 to regulate m6A modification [36]. FTO, on the other hand, is a demethylase that can remove m6A methylation modification on RNA molecules. FTO is overexpressed in various cancers and promotes biological processes such as tumor cell proliferation, angiogenesis, and drug resistance by reducing m6A levels [37-39].
3. Methylation to ncRNAs
In this section, we will mainly introduce the mechanism by which methyltransferases actively regulate ncRNAs involved in the occurrence and development of digestive system tumors. We will also discuss the impact of this regulation on the biological functions of tumor cells (Table 1).
3.1 DNA methylation modifies ncRNAs
Research has found that many ncRNA promoters contain abundant CpG sites, which makes these ncRNAs susceptible to regulation by DNA methyltransferases [40, 41]. In esophageal cancer, it has been shown that miR-149 can inhibit the growth and invasiveness of ESCC cells by suppressing the RNF2/Wnt/β-catenin pathway. However, DNMT3B methylates the miR-149 promoter and suppresses miR-149 expression, reversing its anti-cancer effect [42]. Additionally, DNMT1 indirectly regulates BCAT1 by methylating miR-124-3, which directly targets BCAT1 to affect the proliferation and migration of esophageal cancer cells [43]. BCAT1 is an enzyme that converts the α-amino group of branched-chain amino acids to a-KG and is highly correlated with the occurrence and development of various cancers [44]. DNMT1 indirectly regulates BCAT1 by methylating miR-124-3. In liver cancer, it was found that DNMT1 methylates miR-16-5p to play a carcinogenic role. Interestingly, as a lncRNA, SNHG22 recruits DNMT1 to the promoter region of miR-16-5p through interaction with EZH2. In this axis, ncRNAs actively regulate methyltransferases while also being catalyzed by them [45]. Additionally, it has been found that DNMT1 silences miR-378a-3p by methylating it, thereby promoting the expression of TRAF1 and activating the NF-κB signaling pathway. The p65 in the NF-κB signaling pathway can promote the upregulation of DNMT1, thereby forming a positive feedback loop [46]. Furthermore, in colorectal cancer, miR-152-3p was found to upregulate and inhibit TMSB10, while upregulation of DNMT1 can reverse the effect of miR-152-3p on CRC tumor growth [47]. DNA methyltransferases mainly regulate the occurrence and development of digestive system tumors by indirectly affecting various oncogenes by regulating miRNA promoter methylation.
3.2 Histone methylation modifies ncRNAs
Many studies have found that histone methylation plays a vital role in regulating ncRNA expression [48]. In esophageal cancer, EZH2, as a histone methyltransferase, promotes esophageal cancer cells' invasion and EMT process by upregulating miR-200c. Although the mechanism of regulating cells by EZH2 and miR-200c is not yet precise, they are still promising biomarkers for treating esophageal cancer patients [49]. In addition, in gastric cancer, embryonic ectoderm development protein (EED) can suppress the expression of miR-338-5p by promoting histone methylation. EED has been confirmed to be responsible for the methylation of histone H3K27 [50]. Knockdown of EED promotes miR-338-5p expression, inhibiting the proliferation and invasion of GC cells. Interestingly, miR-338-5p can inhibit the expression of METTL3 and regulate the methylation level of CDCP1. EED can promote CDCP1 expression and GC development through this axis [51]. Another study found that histone methyltransferase RIZ1 can mediate the enrichment of H3K9me1 on the promoter of lncRNA HOTAIRM1, promoting the growth and metastasis of HCC cells [30]. In addition to histone methyltransferases, histone demethylases also have research on regulating ncRNAs. LSD1 is an enzyme that regulates the demethylation of H3K4me1/2 and H3K9me1/2 [52]. Studies have found that high expression of LSD1 significantly promotes the migration and EMT process of GC cells. Mechanistically, LSD1 promotes miR-142-5p expression and downregulates CD9 through its demethylation function [53]. Overall, histone methylation plays an essential role in digestive system tumors by regulating the expression of ncRNAs, and targeting various histone methyltransferases is a promising therapeutic approach.
The molecular mechanisms and biological processes involved in the regulation of ncRNAs by methylases
| Type | methylase | ncRNA | Trend of regulation | Axis/pathway | Biological activities | Reference |
|---|---|---|---|---|---|---|
| DNA methylation | DNMT3B | miR-149 | down-regulated | DNMT3B/miR-149/RNF2/Wnt/β-catenin | promote migration, invasiveness and EMT | [42] |
| DNMT1 | miR-124-3p | down-regulated | DNMT1/miR-124-3p/BCAT1 | promote proliferation and migration | [43] | |
| DNMT1 | miR-152-3p | down-regulated | DNMT1/miR-125-3p/TMSB10 | promote proliferation, migration, invasion and inhibit apoptosis | [47] | |
| DNMT1 | miR-378a-3p | down-regulated | DNMT1/miR-378a-3p/TRAF1/NF-κB positive feedback loop | promote angiogenesis | [46] | |
| DNMT1 | miR-16-5p | down-regulated | SNHG22/EZH2/DNMT1/miR-16-5p | promote proliferation, migration, invasion, and angiogenesis | [45] | |
| DNMT1 | miR-34a | down-regulated | DNMT1/miR-34a/Notch | [41] | ||
| Histone methylation | EZH2 | miR-200c | up-regulated | EZH2/miR-200c | promote migration, EMT | [49] |
| LSD1 | miR-142-5p | up-regulated | LSD1/miR-142-5p/CD9 | promote migration, EMT | [53] | |
| R1ZI | HOTAIRM1 | up-regulated | R1ZI/HOTAIRM1 | promote proliferation, migration and Invasion | [30] | |
| EED | miR-338-5p | down-regulated | EED/miR-3385p/METTL3/CDCP1 | promote proliferation and invasion | [51] | |
| RNA methylation | FTO | LINC00022 | up-regulated | FTO/LINC00022/p21 | promote cell-cycle and proliferation | [65] |
| FTO | CASC15 | up-regulated | FTO/CASC15/SIM2 | promote proliferation and inhibit apoptosis | [103] | |
| FTO | circGPR137B | up-regulated | circGPR137B/miR-4739/FTO feedback loop | promote growth and invasion | [66] | |
| WTAP | circCMTM3 | up-regulated | WTAP/circCMTM3/IGF2BP1/PARK7 | inhibit ferroptosis | [64] | |
| METTL14 | circORC5 | down-regulated | METTL14/circORC5/miR-30c-2-3p/AKT1S1 | inhibit proliferation, invasion | [111] | |
| METTL14 | LINC01320 | up-regulated | METTL14/LINC01320/miR-495-5p/RAB19 | promote proliferation, migration, and invasion | [67] | |
| METTL14 | XIST | down-regulated | METTL14/YTHDF2/XIST | inhibit proliferation and invasion | [62] | |
| METTL14 | miR-17-5p | down-regulated | METTL14/miR-17-5p/MFN2 | promote apoptosis | [140] | |
| METTL14 | circFUT8 | up-regulated | miR-628-5p/METTL14/circFUT8/miR-552-3p/CHMP4B | promote proliferation and inhibit apoptosis | [63] | |
| METTL3 | circ1662 | up-regulated | METTL3/circ1662/YAP1 | promote invasion, migration and EMT | [60] | |
| METTL3 | THAP7-AS1 | up-regulated | SP1/METTL3/THAP7-AS1/CLUB4/ miR-22-3p,miR-320a/ PI3K/AKT | promote proliferation, migration and invasion | [57] | |
| METTL3 | miR-17-92 | up-regulated | METTL3/miR-17-92/PTEN, TMEM127/AKT/mTOR | promote proliferation, migration and invasion | [56] | |
| METTL3 | LINC01559 | down-regulated | METTL3/LINC01559/miR-106b-5p/PTEN | promote proliferation and metastasis | [58] | |
| METTL3 | HNF1A-AS1 | up-regulated | METTL3/HNF1A-AS1/IGF 2BP2/CCND1 | promote migration, invasion, cell cycle and angiogenesis | [117] | |
| METTL3 | circ QSOX1 | up-regulated | METTL3/circ QSOX1/miR-326 and miR-330-5p/PGAM1 | promote proliferation, migration and invasion | [59] | |
| METTL3 | circ_0058493 | up-regulated | METTL3/circ_0058493/YTHDC1 | promote proliferation and metastasis | [61] |
3.3 RNA methylation modifies ncRNAs
RNA methylation modification occurring on ncRNAs mainly involves m6A methylation. The m6A methyltransferases METTL3 and METTL14 play an important role in regulating ncRNAs in digestive system tumors [54, 55]. In gastric cancer, METTL3 was found to catalyze m6A in an m6A/DGCR8-dependent manner to process pri-miR-1792 into a mature form. MiR-1792 activates the AKT/mTOR pathway to promote gastric cancer cell resistance by targeting PTEN or TMEM127 [56]. LncRNA THAP7-AS1 was shown to promote GC cell progression. At the same time, THAP7-AS1 was upregulated by METTL3-mediated m6A modification, which enabled CUL4B protein entry into the nucleus by facilitating the interaction of nuclear localization signal (NLS) with input protein α1. CUL4B indirectly regulates the PI3K/Akt pathway in the nucleus by regulating miR22-3P and miR-320a. In this axis, METTL3 and the methylation regulation of ncRNAs by CLU4B play an important role in the development of gastric cancer [57]. In addition, METTL3 upregulates the expression of LINC01559 and circQSOX1 by methylating them, and these two ncRNAs also promote colorectal cancer progression by sequestering miRNAs [58, 59]. Additionally, METTL3 upregulates the expression of circ 1662 by binding to its flanking sequences and adding m6A modifications, while circ1662 promotes CRC invasion and migration by upregulating the YAP1 protein [60]. METTL3 promotes cell proliferation and migration in liver cancer through the circ_0058493/YTHDC1 axis [61]. The role of METTL14 is similar to METTL3, as studies have shown that METTL14 promotes the proliferation and invasion of CRC cells by regulating the m6A modification of lncRNA XIST. It was also found that XIST can be recognized and degraded by the m6A reader protein YTHDF2. Upregulation of XIST is achieved through METTL14-mediated RNA degradation inhibition of YTHDF2 [62]. Furthermore, in HCC, it was found that METTL14 upregulates the expression of circFUT8 through m6A modification. CircFUT8 promotes HCC progression through the miR-552-3p/CHMP4B axis. However, M1 macrophages can reverse this process by secreting miR-628-5p into HCC cells to target and inhibit METTL14 [63]. Additionally, WTAP plays an important role in m6A modification by forming a complex with methyltransferases. Research has found that WTAP regulates m6A modification of circCMTM3 in liver cancer. The silencing of CircCMTM3 inhibits PARK7 expression by binding to IGF2BP1, inducing iron death in liver cancer cells [64]. FTO, as an m6A demethylase, is also important. In esophageal cancer, FTO reduces the m6A methylation level of LINC00022 and promotes its upregulation. The mechanism is that FTO slows down the decay rate of LINC00022 in a YTHDF2-dependent manner. LINC00022 directly binds to the p21 protein, promoting its degradation and inducing esophageal cancer cell proliferation and progression [65]. Additionally, in liver cancer, a study found that FTO indirectly inhibits miR-4739 by demethylating circGPR137B, while miR-4739 can inhibit FTO expression, forming a positive feedback loop [66].
In summary, RNA methylation modification of ncRNAs plays a vital role in regulating digestive system tumors [67]. However, current research has mainly focused on m6A methylation, and it is believed that more types of RNA methylation modifications regulating ncRNAs will be discovered in the future (Figure 2).
4. ncRNAs to methylation
In this section, we will discuss the mechanism by which ncRNAs actively regulate methylation modification and their impact on the biological processes of cancer cells. We will divide ncRNAs into miRNA, lncRNA, and circRNA and describe their roles in regulating methylation in digestive system tumors (Table 2).
4.1 miRNA regulates methylation modification
MiRNAs are a class of non-coding RNAs approximately 21-23 nucleotides long, which regulate gene expression by binding specifically to the target messenger RNA (mRNA) and inhibiting its transcription [68]. In digestive system tumors, miRNAs mainly regulate cancer progression by inhibiting methyltransferases. In gastric cancer, miR-492 promotes gastric cancer cells' stemness and invasion ability by directly inhibiting DNMT3B [69]. In addition, miR-4429 directly targets and inhibits METTL3, regulating gastric cancer progression. METTL3 can stabilize SEC62 in liver cancer cells by m6a modification to promote cell proliferation [70]. Research has shown that miR-338-5p inhibits METTL3 expression to suppress the proliferation and migration of GC cells. However, miR-338-5p is inhibited by LINC00240 [71]. In colorectal cancer, miR-515-5p directly inhibits DNMT1, while circ_0040809 competes with miR-515-5p to restore DNMT1 expression [72]. MiR-34a-5p can be transported to CRC cells through extracellular vesicles derived from mesenchymal stem cells (MSC-EVs) and directly inhibits c-MYC to regulate DNMT3a expression. DNMT3a can regulate CRC progression by promoting PTEN methylation [73]. In addition, miR-96 promotes FTO upregulation by downregulating AMPKα2 in CRC, and FTO inhibits MYC expression through demethylation modification. The inhibition of miR-96 in CRC cells' malignant phenotype is achieved by regulating FTO demethylation modification [74]. In HCC, miR-29c-3p directly inhibits DNMT3B, while DNMT3B regulates HCC progression by methylating LATS1. LATS1 has been identified as a tumor suppressor gene, and its inhibition by DNMT3B promotes HCC cell proliferation and migration [75, 76]. Furthermore, LINC00839 upregulates WTAP to promote liver cancer cell proliferation and invasion through the sponge-like effect of miR-144-3p. MiR-144-3p can directly inhibit WTAP from exerting its effect [77]. In pancreatic cancer, miR-29b targets and inhibits DNMT3B to suppress pancreatic cancer cell apoptosis [78]. In summary, in digestive system tumors, miRNAs play a role by directly targeting the mRNA of methyltransferases. Additionally, miRNAs often act as tumor suppressors to regulate the pro-cancer effect of methyltransferases.
Three methylation-modified enzymes regulate the progress of related cell biology by actively regulating ncRNAs. DNA methylase DNMT1 and DNMT3B promote cancer progression through functional up-regulation of downstream microRNA. In contrast, RNA methylase mainly promotes cancer progression through the up-regulation of downstream oncogenic microRNA, lncRNA, and circRNA. Histone methylase promotes cancer progression by down-regulating the expression of related cancer suppressor microRNA.
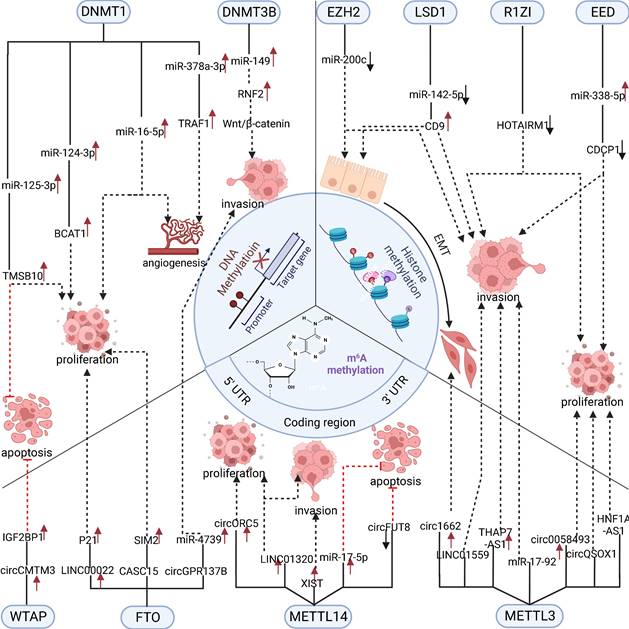
4.2 lncRNA regulates methylation modification
LncRNAs are ncRNAs with a length greater than 200 bp, and their mechanism of action is complex [79]. Compared to miRNAs that directly bind to methyltransferase mRNA to regulate its expression level, lncRNAs can affect the occurrence and development of digestive system tumors by regulating the expression or activity of methyltransferase in multiple ways. For example, lncRNA HCP5 indirectly promotes the expression of DNMT3A by sponging miR-29b-3p in HCC cells, and DNMT3A promotes the migration and invasion ability of HCC cells by methylating and activating the AKT pathway [80]. In addition to indirectly regulating the expression level of methylation enzymes, lncRNAs can also regulate methylation enzymes in other ways. In ESCC, LINC00858 upregulates FTO expression by recruiting ZNF184. FTO promotes the proliferation, invasion, and migration characteristics of ESCC cells by demethylating MYC and upregulating its expression [81]. Besides indirectly regulating the expression of methylesterases, lncRNAs can also act by recruiting methylesterases to the binding sites of target genes, and lncRNAs can directly catalyze epigenetic modifications of methylesterases to alter their activities. For example, in gastric cancer, lncRNAs SAMD12-AS1 and LINC00467 can directly interact with DNMT1 to promote DNMT1-catalyzed p53 and Reprimo promoter methylation, respectively, to regulate gastric carcinogenesis and development [82, 83]. Furthermore, lncRNA DLEU1 can promote gastric cancer cell proliferation by upregulating KLF2 by recruiting LSD1 to the KLF2 promoter region [84]. In colorectal cancer, the knockdown of LINC00337 inhibited angiogenesis and proliferation of CRC cells because LINC00337 inhibited CNN1 expression by recruiting DNMT1. In contrast, CNN1, a critical oncogenic factor in colorectal cancer, was upregulated by the knockdown of LINC003377 [85, 86]. Comparatively, LINC01605 can promote m6A modification of SPTBN2 by binding to METTL3, and LINC01605 itself is regulated by SMYD2-EP300-mediated histone methylation modifications [87]. In hepatocellular carcinoma, lncRNA BZRAP1-AS1 promotes the malignant phenotype of HCC cells by interacting with DNMT3b to downregulate THBS1[88]. According to another study, lncRNA DDX11-AS1 promoted LATS2 methylation by interacting with EZH2 and DNMT1. Knockdown of DDX11-AS1 promoted LATS2 expression and thus inhibited the proliferation and invasion of HCC cells [89]. Moreover, besides the recruitment of methylesterase, it was found that lncRNA DUXAP10 could also inhibit its action by binding to LSD1. DUXAP10 directly inhibits the methylation of p21 and PTEN by LSD1, thereby promoting CRC cell proliferation [90]. In pancreatic cancer, the lncRNA IRAIN inhibits the expression of the downstream target KLF2 by binding to LSD1 and EZH2[91]. Eventually, lncRNAs can also influence the activity of methylation enzymes by regulating their epigenetic modifications. Linc-GALH indirectly regulates the methylation status and expression of Gankyrin by promoting DNMT1 degradation by regulating the ubiquitination status of DNMT1 in HCC cells. Gankyrin is closely associated with the development and metastasis of HCC [92, 93]. To summarize, lncRNAs play a role in gastrointestinal tumors by indirectly influencing the methylation of downstream targets by regulating methylation enzyme expression and activity. However, the way lncRNAs regulate methylation enzymes is complex and more research is required to understand their mechanisms of effect.
4.3 circRNA regulates methylation modification
CircRNAs are a class of circular ncRNAs that are more stable than linear RNAs and are rich in miRNA binding sites [94]. CircRNAs, therefore, act mainly as miRNA sponges, and in addition, circRNAs can regulate the translation and post-translational modifications of many proteins [95, 96]. CircRNAs have been found to regulate the expression of methylesterase through miRNA sponges in a variety of digestive tumors. For example, in colorectal cancer, Circ_0084615 upregulated DNMT3A expression through the sponge miR-599 to promote the proliferation, migration, and invasion of colorectal cancer cells [97]. Similarly, Circ_0000467 inhibited miR-651-5p, and DNMT3B was a direct target of miR-651-5p. Knockdown of circ_0000467 upregulated DNMT3B expression and suppressed the malignant phenotype of colorectal cancer cells [98]. In HCC, circ_0008583 also promoted HCC cancer progression by inhibiting miR-1301-3p and up-regulating METTL3 [99]. In addition to acting as miRNA sponges, circRNAs can also regulate digestive tumor progression in other ways. In EBV-associated gastric cancer, circPRMS1, produced by EBV, promotes the proliferation, migration, and invasion of gastric cancer cells. The mechanism is that circPRMS1 binds directly to Sam68 and activates the recruitment of Sam68 to the promoter region of METTL3 to promote its transcription [100]. In addition, it was found that circPAR1 encapsulated by exosomes could directly bind to eIF3h and inhibit its interaction with METTL3. The oncogene BRD4's translation depends on the complex of METTL3 and eIF3h. circPAR1 indirectly downregulated the expression of BRD4 to inhibit the development of colorectal cancer [101]. In conclusion, there are few studies on the regulation of methylation enzymes by circRNAs in gastrointestinal tumors, and circRNAs mainly regulate methylation enzymes through sponge miRNAs. Further mechanisms of CircRNA regulation of methylesterase remain to be explored.
The molecular mechanisms and biological processes involved in the regulation of methylases by ncRNAs
| Type | ncRNA | Methylase | Trend of regulation | Axis/pathway | Biological activities | Reference |
|---|---|---|---|---|---|---|
| miRNA | miR-492 | DNMT3B | repress | miR-492/DNMT3B | promote proliferation, metastasis and stemness | [69] |
| miR-338-5p | METTL3 | repress | LINC00240/miR-338-5p/METTL3 | inhibit proliferation, migration, invasion and promote apoptosis | [71] | |
| miR-4429 | METTL3 | repress | miR-4429/METTL3/SEC62 | inhibit proliferation and promote apoptosis | [70] | |
| miR-515-5p | DNMT1 | repress | circ_0040809/miR-515-5p/DNMT1 | inhibit proliferation, migration and promote apoptosis | [72] | |
| miR-34a-5p | DNMT3A | repress | miR-34a-5p/cMYC/DNMT3a/PTEN | inhibit proliferation, migration, invasion and promote apoptosis | [73] | |
| miR-96 | FTO | repress | miR-96/AMPKα2/FTO/MYC | promote proliferation, migration, invasion and inhibit apoptosis | [74] | |
| miR-29c-3p | DNMT3B | repress | miR-29c-3p/DNMT3B/LATS1 | inhibit proliferation, migration, invasion and promote apoptosis | [75] | |
| miR-144-3p | WTAP | repress | LINC00839/miR-144-3p/WTAP | promote proliferation, invasion and migration | [77] | |
| miR‑29b | DNMT3B | repress | miR‑29b/DNMT3B | inhibit proliferation and promote apoptosis | [78] | |
| LncRNA | LINC00858 | FTO | promote | LINC00858/ZNF184/FTO/MYC | promote proliferation, apoptosis, cell cycle, migration and invasion | [81] |
| LINC00467 | DNMT1 | promote | LINC00467/DNMT1/Reprimo | proliferation, apoptosis, migration and invasion | [83] | |
| SAMD12-AS1 | DNMT1 | promote | SAMD12-AS1/DNMT1/p53 | promote proliferation and cell cycle | [82] | |
| DLEU1 | LSD1 | promote | DLEU1/LSD1/KLF2 | promote proliferation and inhibit apoptosis | [84] | |
| LINC00337 | DNMT1 | promote | LINC00337/DNMT1/CNN1 | promote tumorigenesis and angiogenesis | [85] | |
| LINC01605 | METTL3 | promote | SMYD2-EP300/LINC01605/METTL3/SPTBN2 | promote proliferation and metastasis | [87] | |
| DDX11-AS1 | DNMT1, EZH2 | promote | DDX11-AS1/DNMT1, EZH2/LATS2 | promote proliferation, migration, and invasion | [89] | |
| Linc-GALH | DNMT1 | repress | Linc-GALH/DNMT1/Gankyrin | promote migration and invasion | [92] | |
| HCP5 | DNMT3A | repress | HCP5/miR-29b-3p/DNMT3A/AKT | promote proliferation, migration, and invasion | [80] | |
| BZRAP1-AS1 | DNMT3B | promote | BZRAP1-AS1/DNMT3B/THBS1 | promote proliferation, migration and angiogenesis | [88] | |
| DUXAP10 | LSD1 | promote | DUXAP10/LSD1/p21, PTEN | promote proliferation, apoptosis and cell cycle | [90] | |
| IRAIN | LSD1, EZH2 | promote | IRAIN/LSD1, EZH2/KLF2,P15 | promote cell cycle and inhibit apoptosis | [91] | |
| CircRNA | circRPMS1 | METTL3 | promote | circRPMS1/Sam68/METTL3 | promote proliferation, migration, invasion and inhibit apoptosis | [100] |
| Circ_0084615 | DNMT3A | promote | Circ_0084615/miR-599/DNMT3A | promote proliferation, migration and invasion | [97] | |
| Circ_0000467 | DNMT3B | promote | Circ_0000467/miR-651-5p/DNMT3B | promote growth, migration and invasion | [98] | |
| circLPAR1 | METTL3 | promote | circLPAR1/eIF3h/METTL3/BRD4 | promote proliferation, migration and invasion | [101] | |
| circ_0008583 | MTTTL3 | promote | circ_0008583/miR-1301-3p/METTL3 | promote proliferation, migration and invasion | [99] |
5. Association of ncRNAs with methylesterases in various gastrointestinal tumors
In gastrointestinal tumors, a variety of ncRNAs modified by methylation and methylation enzymes regulated by ncRNAs are profoundly associated with tumor prognosis and clinical features. This section summarizes the relationship between ncRNAs and associated methylesterases and their clinical characteristics and prognosis in various GI tumors (Table 3). This suggests that ncRNAs and methylesterases may serve as key diagnostic markers or therapeutic targets in the following tumors.
5.1 Esophageal Cancer
The incidence and mortality rate of esophageal cancer remains high, with more than 600,000 people diagnosed worldwide each year [102]. Increasingly, ncRNAs and methylation modifications have been found to play an important role in esophageal cancer. Zeng et al. found that low expression of miR-124-3p was highly correlated with the TNM stage and differentiation of ESCC. The methylation of miR-124-3p by DNMT1 played a key role in the regulation of miR-124-3p expression [43]. This suggests that miR-124-3p and DNMT1 may be important prognostic markers for ESCC. In addition, upregulation of LINC00022 was significantly associated with poorer OS in ESCC patients, implying that LINC00022 is a poor prognostic factor. And LINC00022 was upregulated by m6A methylation of FTO [65]. In 45 patients with esophageal cancer, lncRNA CASC15 was highly expressed in tumor tissues, and its expression was significantly associated with low OS, TNM stage, and lymphatic metastasis in patients. CASC15 was also upregulated by m6A methylation of FTO [103]. In conclusion, ncRNAs modified by methylation are closely related to the poor prognosis of esophageal cancer. The selection of targeted drugs targeting methylating enzyme-associated ncRNAs may be effective in treating esophageal cancer.
5.2 Gastric cancer
Gastric cancer has the fifth highest incidence of all malignancies and has a poor prognosis, with a 5-year survival rate of less than 30% [104, 105]. The development of gastric cancer is associated with many factors, including diet [106], H. pylori infection [107], and epigenetic changes [108]. Because there are no apparent symptoms in the early stages of gastric cancer, patients are often diagnosed in the late stages of gastric cancer and therefore have a low 5-year survival rate [109]. Currently, the main treatments for gastric cancer are surgical resection and chemotherapy, but the results are still poor [110]. Therefore, there is a need to find an effective therapeutic target or early diagnostic marker. METTL14 was found to be upregulated in GC and significantly correlated with poor OS, TNM stage, and lymphatic metastasis in GC patients. The oncogenic effect of METTL14 was achieved by methylation of circORC5 [111]. In addition, high expression of METTL3 was found to be associated with lymphatic metastasis in gastric cancer patients, and METTL3 could act through the methylation of miR-17-92 [56]. In addition to methylesterases, many ncRNAs regulating methylation modifications have been linked to GC prognosis. LINC00240 was significantly upregulated in gastric cancer and was associated with poor patient survival, TNM stage, and distant metastasis [71]. And LINC00240 could indirectly upregulate METTL3. These two studies suggest that the mutual regulation of ncRNAs and METTL3 has an important impact on the prognosis of gastric cancer patients. In addition, the upregulation of lncRNA DLEU1 was also associated with lymphatic metastasis and TNM stage of gastric cancer patients. As well as, DLEU1 could promote the methylation of LSD1 [84].
5.3 Colorectal cancer
Colorectal cancer is the second most common cancer worldwide, and its incidence is expected to more than double by 2035 [112, 113]. The high incidence of colorectal cancer is mainly attributed to its asymptomatic early stage and the lack of practical screening tools [114]. Current screening methods detect only 40% of cancer cases [113]. Currently, the treatment options for colorectal cancer include surgery, chemotherapy, immunotherapy, and targeted therapy [115]. The primary treatment for patients with advanced colorectal cancer is still chemotherapy, but the current resistance rate to chemotherapy is high [116]. Therefore, there is an urgent need to identify markers and therapeutic targets that can diagnose colorectal cancer at an early stage. Many ncRNAs involved in methylation modifications were found to be closely related to the clinical prognosis of colorectal cancer. For example, LINC01605 is highly expressed in colorectal cancer, and its expression correlates with tumor stage, lymph node metastasis, and distant metastasis of colorectal cancer patients. High expression of LINC01605 predicts poor overall survival, and LINC01605 can promote the function of METTL3 to exert oncogenic effects [87]. Furthermore, METTL3 downregulated LINC01559 expression, and low LINC01559 expression was also associated with TNM stage, lymph node metastasis, and distant metastasis in colorectal cancer patients [58]. Moreover, HNF1A-AS1, circQSOX1, and circ1622 are all highly expressed in colorectal cancer and associated with poor prognosis in colorectal cancer patients, and they are all upregulated by METEL3 methylation [59, 60, 117]. High expression of METTL3 has been shown to be highly expressed in colorectal cancer and correlated with low OS in patients. These results confirm that METTL3 and its mutually regulated ncRNA molecules are strongly associated with the prognosis of colorectal cancer patients and have promise as diagnostic markers and therapeutic targets for CRC.
5.4 Liver cancer
Liver cancer is the third most common cause of cancer deaths. In 2020 a total of 83,000 people died from liver cancer worldwide [118]. Ninety percent of all primary liver cancers are hepatocellular carcinoma (HCC) [119]. Risk factors for HCC include hepatitis B virus, hepatitis C virus, and alcohol consumption [120]. It was found that ncRNAs and their associated methylation enzymes are aberrantly expressed in hepatocellular carcinoma and correlated with patient prognosis. MiR-378a-3p was downregulated by DNMT1 methylation in HCC. Low expression of MiR-378a-3p was associated with poor OS, tumor thrombosis, and (microvascular density) MVD in patients [46]. As a highly vascular tumor, high MVD in hepatocellular carcinoma represents enhanced angiogenesis, and its prognostic significance in HCC is significant [121, 122]. Alternatively, LncRNAs SNHG22, DDX11-AS1, and Linc-GALH were found to be upregulated in HCC and associated with poor prognosis and clinical features of hepatocellular carcinoma. All three lncRNAs can act by regulating DNMT1 methylation oncogenic factor [45, 89, 92]. LncRNA BZRAP1-AS1 is highly expressed in HCC and strongly correlates with tumor size, microvascular invasion, and TNM stage. DNMT3B is upregulated by BZRAP1-AS1 [88]. Besides, miR-29c-3p can inhibit DNMT3B, and low expression of miR-29c-3p is associated with intrahepatic metastasis and tumor multiplicity in HCC patients [75]. Many ncRNAs regulating methylesterase are associated with poor prognosis of HCC, and the discovery of more ncRNAs in relation to methylesterase regulation may help to find appropriate diagnostic markers for HCC.
Expression and clinical features of ncRNAs or methylases in patients with GI cancer
| Cancer type | ncRNAs/methylase | Expression | Clinical features | Reference |
|---|---|---|---|---|
| ESCC | miR-124-3p | down | TNM stage and differentiation grade | [43] |
| ESCC | LINC00022 | up | OS | [65] |
| ESCC | CASC15 | up | OS, TNM stage and lymphatic metastasis | [103] |
| GC | METTL14 | down | OS | [111] |
| GC | LINC01320 | up | OS | [67] |
| GC | THAP7-AS1 | up | OS | [57] |
| GC | METTL3 | up | lymph node metastasis | [56] |
| GC | LINC00240 | up | OS, TNM stage,distant metastasis and lymph nodes metastasis | [71] |
| GC | SAMD12-AS1 | up | OS | [82] |
| GC | DLEU1 | up | lymph node metastasis, TNM stage | [84] |
| CRC | METTL14 | up | RFS, larger tumor size, lymphatic invasion, remote metastasis and TNM stage | [62] |
| CRC | circ1662 | up | OS, lymph node metastasis and vascular invasion | [60] |
| CRC | LINC01559 | down | TNM stage, lymphatic metastasis and distant metastasis | [58] |
| CRC | HNF1A-AS1 | up | OS, higherpathological stage (III/IV), lymph node metastasis and distant metastasis | [117] |
| CRC | circQSOX1 | up | OS | [59] |
| CRC | circ_0040809 | up | OS | [72] |
| CRC | LINC01605 | up | OS, lymph node metastases, distal metastases and advanced tumor stage | [87] |
| CRC | DUXAP10 | up | tumor sizes, TNM stages and lymph node metastasis | [90] |
| CRC | Circ_0084615 | up | OS | [97] |
| CRC | circLPAR1 | down | OS | [101] |
| HCC | miR-378 a-3p | down | OS, thrombosis of tumor blood vessel and MVD | [46] |
| HCC | SNHG22 | up | OS | [45] |
| HCC | circGPR137B | down | OS | [66] |
| HCC | CircCMTM3 | up | OS | [64] |
| HCC | hsa_circ_0058493 | up | OS | [61] |
| HCC | miR-29c-3p | down | OS, tumor size, multiplicity, and intrahepatic metastasis | [75] |
| HCC | LINC00839 | up | OS, tumor size, lymph node metastasis and poor tumor differentiation | [77] |
| HCC | DDX11-AS1 | up | serum AFP levels and tumor stage | [89] |
| HCC | Linc-GALH | up | intrahepatic metastasis, vascular invasion and distant metastasis | [92] |
| HCC | HCP5 | up | large tumors, metastasis, high histological grade tissues, and recurrence | [80] |
| HCC | BZRAP1-AS1 | up | tumor size, microvascular invasion and TNM stage | [88] |
| PC | IRAIN | up | advanced pathological stage, larger tumor size, and lymph node metastasis | [91] |
5.5 Pancreatic cancer
Pancreatic cancer has the worst prognosis of all cancers, although its incidence is slightly lower than that of other digestive system tumors [123]. Pancreatic cancer's mortality to incidence ratio is as high as 94% [124]. The mortality rate of most other tumors has been decreasing year by year, but the mortality rate of pancreatic cancer has remained almost unchanged [125]. There is still a lack of effective treatment for pancreatic cancer [126]. The LncRNA IRAIN was found to be highly expressed in pancreatic cancer, and its high expression predicted advanced pathological stage, larger tumor size, and lymph node metastasis. target genes by binding to LSD1 and EZH2 [91]. Targeting IRAIN, LSD1, or EZH2 may be a potential approach to treat pancreatic cancer. There are few studies on the interaction between ncRNAs and methylation modifications in PC. More studies are needed to explore the potential value of ncRNAs and methylation modifications in the clinical prognosis of pancreatic cancer.
6. Potential clinical value of ncRNAs involved in methylation modifications
With the continuous research on the clinical application of ncRNA, we found that ncRNAs have a considerable prospect in gastrointestinal cancer, which is expected to be used as clinical tumor biomarkers, therapeutic targets, and targeted targets for anti-drug resistance (Figure 3).
6.1 Serve as biomarkers
The discovery of more effective diagnostic markers can help diagnose patients with gastrointestinal tumors early so that patients can receive early treatment [127]. At the same time, effective prognostic markers can reflect the malignancy of patients' tumors and determine the recurrence and metastasis of tumors [128]. With the in-depth study of ncRNAs and related methylation enzymes, both of them are expected to be effective diagnostic and prognostic markers in gastrointestinal tumors. In colorectal cancer, circLPAR1, which is encapsulated by exosomes, was found to have reduced expression in the plasma of patients. Its low expression was associated with poor OS in colorectal cancer patients. More importantly, it was found that exosomal circLPAR1 was stable in plasma, and its expression did not change significantly after repeated freeze-thawing or long-term storage at room temperature. CircLPAR1 was used to differentiate colorectal patients from normal controls with better efficiency than CEA and CA19-9 [101]. CEA and CA19-9 are currently and popularly used diagnostic markers for colorectal cancer [129]. This suggests that circLPAR1 holds promise as an effective diagnostic and prognostic marker for colorectal cancer. The mechanism is that circLPAR1 exerts its oncogenic effect by inhibiting METTL3. And METTL3 was also found to be highly expressed in the plasma of CRC patients [130]. Similarly, increased expression of circ_0038138 was found in plasma exosomes of gastric cancer patients. Circ_0038138 indirectly downregulated EZH2 through miR-198 [131]. In another study, EZH2 was found to be present in small extracellular vesicles (sEV) of gastric cancer cells, and its expression was significantly higher in the plasma of patients than in healthy subjects [132]. On the other hand, METTL3 can also be used as a marker to detect drug sensitivity to guide drug administration. It was found that gastric cancer patients with high METTL3 were more sensitive to everolimus. This may be since everolimus, as an mTOR inhibitor, blocks the METTL3/miR-17-92/PTEN/AKT/mTOR axis. And high expression of METTL3 suggested activation of the AKT/mTOR pathway [56]. Marker assays against plasma are noninvasive and easily promoted screening tool, and assays against specific ncRNAs and methylesterases in serum are expected to be an effective screening tool [133]. It is believed that with more in-depth studies on ncRNAs and related methylesterases, more effective diagnostic and prognostic markers will be found.
6.2 Serve as therapeutic targets
Gastrointestinal tumors are solid tumors, and the current treatment is still based on surgery, chemotherapy, and radiotherapy [134-136]. In recent years, with the continuous exploration of molecular biology, targeted therapies targeting specific genes and cells have been developed. Among them, the delivery of siRNA to tumor cells through nanomaterials has become a promising approach for cancer therapy [137]. Du et al. demonstrated that delivery of circMDK siRNA via poly (β-amino esters) (PAEs) to a liver cancer model could effectively inhibit tumor progression. CircMDK is upregulated in hepatocellular carcinoma by m6A methylation modifications [11]. However, the mechanism of how circMDK is modified by methylation is unclear. Further investigation of the mechanism of action of circMDK on m6A methylation-modified enzymes by demethylating circMDK may be a new approach to target circMDK for the treatment of HCC.
NcRNAs can be used as biomarkers for patients and targets for drug resistance therapy in clinical practice. circRNA in plasma exosomes has high stability, sensitivity, and specificity, which can replace the existing tumor markers. lncRNA PCAT-1, miR-17-5p, LINC00630, and circQSOX1 play important drug resistance roles in anti-chemotherapy, anti-radiotherapy, and anti-immunotherapy, respectively.
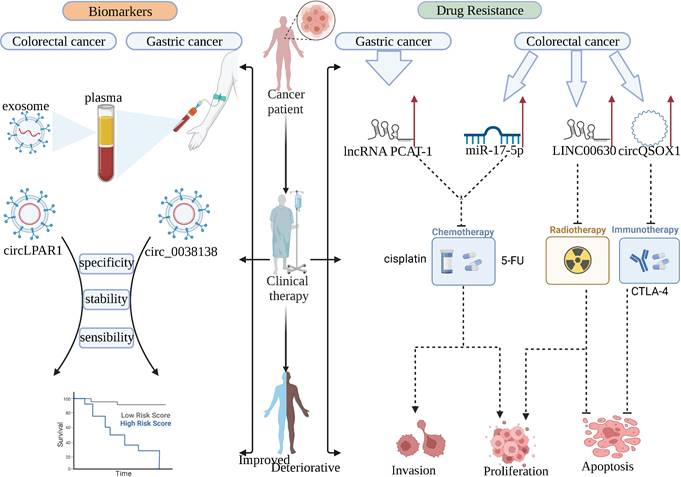
6.3 Serve as a target for improving drug resistance
Drug resistance has been a significant challenge in the treatment of tumors, and eliminating tumor resistance through a combination of drugs is a promising strategy for the treatment of tumors [138]. It was found that the knockdown of lncRNA PCAT-1 could re-sensitize drug-resistant GC cells to cisplatin. And PACT-1 is resistant to GC cells by inhibiting PTEN through binding to EZH2. Targeting PACT-1 is a promising therapeutic approach for patients with cisplatin-resistant gastric cancer [139]. Similarly, in colorectal cancer, overexpression of miR-17-5p regulated by METTL14 methylation leads to 5-FU resistance in CRC cells. Knockdown of miR-17-5p could improve the sensitivity of CRC cells to 5-FU [140]. In addition to chemoresistance, radioresistance is also an important factor affecting patient outcomes [141]. High expression of LINC00630 was found to be associated with radioresistance in CRC. This was mainly achieved by the inhibition of BEX1 by LINC00630 through EZH2. Targeted inhibition of LINC00630 significantly improved the radioresistance of CRC cells [142]. Immunotherapy is also an emerging therapeutic strategy in recent years, as CTLA-4 is an immune checkpoint that is expressed mainly on Treg cells and mediates their immunosuppressive capacity [143]. Anti-CTLA-4 therapy is highly effective in cancer patients who are sensitive to it, but unfortunately, most patients do not respond to this therapy. Therefore, addressing CTLA-4 resistance in tumor cells is an urgent issue at present [144]. It was found that circQSOX1 upregulation in colorectal cancer inhibited the anti-CTLA-4 treatment response in colorectal cancer cells. Knockdown of circQSOX1 could effectively improve the sensitivity of colorectal cancer tumors to CTLA-4. Mechanistically, METTL3 upregulates circQSOX1 via m6A methylation, and circQSOX1 indirectly promotes PGAM1 expression through sponge miR-326 and miR-330-5p to make colorectal cancer cells exhibit resistance to immunotherapy [59]. Further exploration of the regulatory mechanisms between ncRNAs and methylation modifications is expected to address the drug resistance problem in clinical treatment and help identify more effective therapeutic targets.
7. Conclusion
As methylation modifications of DNA, histones, and RNA have been intensively studied, there is increasing evidence for the important role of methylation modifications in cancer regulation. With the rapid development of transcriptomics, the role of ncRNAs in gastrointestinal tumors has been gradually revealed. As two essential regulators of epigenetic modifications, the interplay between ncRNAs and methylation modifications has been shown to play an important role in regulating the malignant phenotype of GI tumors, influencing the prognosis of tumor patients and drug resistance. DNA and histone methylation modifications can directly regulate the transcription of ncRNAs, and RNA methylation can regulate the degradation, synthesis, and processing of ncRNAs. In addition, ncRNAs can also directly or indirectly regulate the activity of methylation enzymes and further regulate the expression level of cancer-related genes in various ways. At the same time, ncRNAs and methylation modifications can also form a feedback loop to promote tumor cell proliferation and metastasis. For example, the oncogenic effect of CircGPR137B in HCC is achieved by upregulating FTO to develop a positive feedback loop. In clinical applications, some ncRNAs that can be stably expressed in plasma have shown potential value as effective diagnostic markers. In addition, siRNAs and methylation enzyme inhibitors targeting ncRNAs have shown great promise in tumor therapy. It is believed that further exploration of the regulatory mechanisms of ncRNAs and methylation modification will lead to the discovery of more effective diagnostic markers and therapeutic targets for eventual clinical application.
Acknowledgements
This work was supported by the Province Medical Science and Technology Research Program (Jointly Co-construction) Project of Henan (LHGJ20210324), and the National Natural Science Foundation of China (82170648).
Author contributions
Wenzhi Guo designed and guided the review. M.K conceived of this review manuscript and drafted the manuscript. R.G helped to draft and revised the manuscript. X.Y reviewed and revised this manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Xia C. et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135(5):584-590
2. Siegel RL. et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7-33
3. Wang D. et al. Cyclooxygenases and Prostaglandins in Tumor Immunology and Microenvironment of Gastrointestinal Cancer. Gastroenterology. 2021;161(6):1813-1829
4. Siegel RL. et al. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7-30
5. Ameli Mojarad M. et al. piRNA: A promising biomarker in early detection of gastrointestinal cancer. Pathol Res Pract. 2022;230:153757
6. Rogers JE. et al. Esophageal cancer: emerging therapeutics. Expert Opin Ther Targets. 2022;26(2):107-117
7. Cai Z. et al. Role of traditional Chinese medicine in ameliorating mitochondrial dysfunction via non-coding RNA signaling: Implication in the treatment of neurodegenerative diseases. Front Pharmacol. 2023;14:1123188
8. Rayford KJ. et al. Trypanosoma cruzi dysregulates expression profile of piRNAs in primary human cardiac fibroblasts during early infection phase. Front Cell Infect Microbiol. 2023;13:1083379
9. Kielbowski K. et al. The role of selected non-coding RNAs in the biology of non-small cell lung cancer. Adv Med Sci. 2023;68(1):121-137
10. Lin L. et al. Analysis of autophagy-related genes and associated noncoding RNAs and transcription factors in digestive system tumors. Future Oncol. 2019
11. Du A. et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. 2022;21(1):109
12. Liu J. et al. Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nat Commun. 2020;11(1):1507
13. Peng W. et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38(1):393
14. Ang HL. et al. Mechanism of epithelial-mesenchymal transition in cancer and its regulation by natural compounds. Med Res Rev. 2023
15. Zong X. et al. Longitudinal multi-omics alterations response to 8-week risperidone monotherapy: Evidence linking cortical thickness, transcriptomics and epigenetics. Front Psychiatry. 2023;14:1127353
16. Huang E, Chen L. RNA N(6)-methyladenosine modification in female reproductive biology and pathophysiology. Cell Commun Signal. 2023;21(1):53
17. Stevens C. et al. Genomic, epigenomic, and transcriptomic signatures of prostate cancer between African American and European American patients. Front Oncol. 2023;13:1079037
18. Li S. et al. PROTACs: Novel tools for improving immunotherapy in cancer. Cancer Lett. 2023:216128.
19. Song T. et al. TRIM28 represses renal cell carcinoma cell proliferation by inhibiting TFE3/KDM6A-regulated autophagy. J Biol Chem. 2023:104621.
20. Zheng X. et al. Effect of N6-methyladenosine methylation-related gene signature for predicting the prognosis of hepatocellular carcinoma patients. Front Surg. 2023;10:1052100
21. Liu WJ. et al. Epigenetic modifications in esophageal cancer: An evolving biomarker. Front Genet. 2022;13:1087479
22. Sutopo NC. et al. Role of histone methylation in skin cancers: Histone methylation-modifying enzymes as a new class of targets for skin cancer treatment. Biochim Biophys Acta Rev Cancer. 2023;1878(3):188865
23. Rotimi OA. et al. Early-life AFB1 exposure: DNA methylation and hormone alterations. Vitam Horm. 2023;122:237-252
24. Ren W. et al. Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation. Genes (Basel). 2018 9(12)
25. Bhandari SK. et al. Unchanged PCNA and DNMT1 dynamics during replication in DNA ligase I-deficient cells but abnormal chromatin levels of non-replicative histone H1. Sci Rep. 2023;13(1):4363
26. Zhou Z. et al. DNA Methyltransferase Inhibitors and their Therapeutic Potential. Curr Top Med Chem. 2018;18(28):2448-2457
27. Ozair A. et al. DNA Methylation and Histone Modification in Low-Grade Gliomas: Current Understanding and Potential Clinical Targets. Cancers (Basel). 2023 15(4)
28. Entezari M. et al. The pharmacological and biological importance of EZH2 signaling in lung cancer. Biomed Pharmacother. 2023;160:114313
29. Boileau RM. et al. Loss of MLL3/4 decouples enhancer H3K4 monomethylation, H3K27 acetylation, and gene activation during embryonic stem cell differentiation. Genome Biol. 2023;24(1):41
30. Li YQ. et al. Inactivation of lncRNA HOTAIRM1 caused by histone methyltransferase RIZ1 accelerated the proliferation and invasion of liver cancer. Eur Rev Med Pharmacol Sci. 2020;24(17):8767-8777
31. Liu HM. et al. Discovery of acridine-based LSD1 inhibitors as immune activators targeting LSD1 in gastric cancer. Eur J Med Chem. 2023;251:115255
32. Liu C. et al. LSD1 Stimulates Cancer-Associated Fibroblasts to Drive Notch3-Dependent Self-Renewal of Liver Cancer Stem-like Cells. Cancer Res. 2018;78(4):938-949
33. Hou G. et al. LSD1 regulates Notch and PI3K/Akt/mTOR pathways through binding the promoter regions of Notch target genes in esophageal squamous cell carcinoma. Onco Targets Ther. 2019;12:5215-5225
34. Liu Y. et al. Involvement of RNA methylation modification patterns mediated by m7G, m6A, m5C and m1A regulators in immune microenvironment regulation of Sjogren's syndrome. Cell Signal. 2023:110650.
35. Zhao NN. et al. Controllable assembly of dendritic DNA nanostructures for ultrasensitive detection of METTL3-METTL14 m(6)A methyltransferase activity in cancer cells and human breast tissues. Biosens Bioelectron. 2023;228:115217
36. Huang L. et al. WTAP regulates autophagy in colon cancer cells by inhibiting FLNA through N6-methyladenosine. Cell Adh Migr. 2023;17(1):1-13
37. Cheng S. et al. FTO-mediated m(6)A modification promotes malignant transformation of gastric mucosal epithelial cells in chronic Cag A(+) Helicobacter pylori infection. J Cancer Res Clin Oncol. 2023
38. Wu J. et al. N6-methyladenosine methylation regulator FTO promotes oxidative stress and induces cell apoptosis in ovarian cancer. Epigenomics. 2022;14(23):1509-1522
39. Gui S. et al. Effects of Helicobacter pylori on the expression of the FTO gene and its biological role in gastric cancer. Oncol Lett. 2023;25(4):143
40. Symeonidis A. et al. Contingent Synergistic Interactions between Non-Coding RNAs and DNA-Modifying Enzymes in Myelodysplastic Syndromes. Int J Mol Sci. 2022 23(24)
41. Ma Y. et al. DNA methyltransferase mediates the hypermethylation of the microRNA 34a promoter and enhances the resistance of patient-derived pancreatic cancer cells to molecular targeting agents. Pharmacol Res. 2020;160:105071
42. Yang J. et al. DNA methyltransferase 3 beta regulates promoter methylation of microRNA-149 to augment esophageal squamous cell carcinoma development through the ring finger protein 2/Wnt/beta-catenin axis. Bioengineered. 2022;13(2):4010-4027
43. Zeng B. et al. The role of DNMT1/hsa-miR-124-3p/BCAT1 pathway in regulating growth and invasion of esophageal squamous cell carcinoma. BMC Cancer. 2019;19(1):609
44. Ding Y. et al. BCAT1, as a prognostic factor for HCC, can promote the development of liver cancer through activation of the AKT signaling pathway and EMT. J Mol Histol. 2023;54(1):25-39
45. Zhang Y. et al. Long non-coding RNA SNHG22 facilitates hepatocellular carcinoma tumorigenesis and angiogenesis via DNA methylation of microRNA miR-16-5p. Bioengineered. 2021;12(1):7446-7458
46. Zhu B. et al. DNMT1-induced miR-378a-3p silencing promotes angiogenesis via the NF-kappaB signaling pathway by targeting TRAF1 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2021;40(1):352
47. Wang C. et al. DNMT1 maintains the methylation of miR-152-3p to regulate TMSB10 expression, thereby affecting the biological characteristics of colorectal cancer cells. IUBMB Life. 2020;72(11):2432-2443
48. Costa P. et al. Epigenetic reprogramming in cancer: From diagnosis to treatment. Front Cell Dev Biol. 2023;11:1116805
49. Nourmohammadi F. et al. EZH2 regulates oncomiR-200c and EMT markers in esophageal squamous cell carcinomas. Sci Rep. 2022;12(1):18290
50. Wang J. et al. Inhibition of EED activity enhances cell survival of female germline stem cell and improves the oocytes production during oogenesis in vitro. Open Biol. 2023;13(1):220211
51. Zhang F. et al. Methylation of microRNA-338-5p by EED promotes METTL3-mediated translation of oncogene CDCP1 in gastric cancer. Aging (Albany NY). 2021;13(8):12224-12238
52. Zhang Y. et al. Phosphoproteomic analysis on ovarian follicles reveals the involvement of LSD1 phosphorylation in Chicken follicle selection. BMC Genomics. 2023;24(1):109
53. Zhao LJ. et al. LSD1 deletion represses gastric cancer migration by upregulating a novel miR-142-5p target protein CD9. Pharmacol Res. 2020;159:104991
54. Huang L. et al. METTL3 promotes colorectal cancer metastasis by promoting the maturation of pri-microRNA-196b. J Cancer Res Clin Oncol. 2022
55. Liu J. et al. N(6) -methyladenosine-modified lncRNA ARHGAP5-AS1 stabilises CSDE1 and coordinates oncogenic RNA regulons in hepatocellular carcinoma. Clin Transl Med. 2022;12(11):e1107
56. Sun Y. et al. N(6)-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. 2020;11(10):836
57. Liu HT. et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2022;29(3):627-641
58. Shi K. et al. RNA methylation-mediated LINC01559 suppresses colorectal cancer progression by regulating the miR-106b-5p/PTEN axis. Int J Biol Sci. 2022;18(7):3048-3065
59. Liu Z. et al. N6-methyladenosine-modified circular RNA QSOX1 promotes colorectal cancer resistance to anti-CTLA-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Updat. 2022;65:100886
60. Chen C. et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021;11(9):4298-4315
61. Wu A. et al. Methyltransferase-Like 3-Mediated m6A Methylation of Hsa_circ_0058493 Accelerates Hepatocellular Carcinoma Progression by Binding to YTH Domain-Containing Protein 1. Front Cell Dev Biol. 2021;9:762588
62. Yang X. et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19(1):46
63. Wang L. et al. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol Lett. 2022;27(1):106
64. Chen S. et al. WTAP-mediated m6A modification on circCMTM3 inhibits hepatocellular carcinoma ferroptosis by recruiting IGF2BP1 to increase PARK7 stability. Dig Liver Dis. 2022
65. Cui Y. et al. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40(1):294
66. Liu L. et al. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer. 2022;21(1):149
67. Hu N. and H. Ji, N6-methyladenosine (m6A)-mediated up-regulation of long noncoding RNA LINC01320 promotes the proliferation, migration, and invasion of gastric cancer via miR495-5p/RAB19 axis. Bioengineered. 2021;12(1):4081-4091
68. Rivera J. et al. Mitochondria Localized microRNAs: An Unexplored miRNA Niche in Alzheimer's Disease and Aging. Cells. 2023 12(5)
69. Wu S. et al. miR-492 promotes chemoresistance to CDDP and metastasis by targeting inhibiting DNMT3B and induces stemness in gastric cancer. Biosci Rep. 2020 40(3)
70. He H. et al. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem Biophys Res Commun. 2019;517(4):581-587
71. Wang G. et al. Long non-coding RNA LINC00240 promotes gastric cancer progression via modulating miR-338-5p/METTL3 axis. Bioengineered. 2021;12(2):9678-9691
72. Mao G. et al. Hsa_circ_0040809 regulates colorectal cancer development by upregulating methyltransferase DNMT1 via targeting miR-515-5p. J Gene Med. 2021;23(12):e3388
73. Zhao J. et al. Mesenchymal Stem Cell-derived Extracellular Vesicles Transmitting MicroRNA-34a-5p Suppress Tumorigenesis of Colorectal Cancer Through c-MYC/DNMT3a/PTEN Axis. Mol Neurobiol. 2022;59(1):47-60
74. Yue C. et al. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKalpha2-FTO-m6A/MYC axis. J Exp Clin Cancer Res. 2020;39(1):240
75. Wu H. et al. miR-29c-3p regulates DNMT3B and LATS1 methylation to inhibit tumor progression in hepatocellular carcinoma. Cell Death Dis. 2019;10(2):48
76. Hong SH. et al. Inhibition of EZH2 exerts antitumorigenic effects in renal cell carcinoma via LATS1. FEBS Open Bio. 2023
77. Zhou X. et al. LINC00839/miR-144-3p/WTAP (WT1 Associated protein) axis is involved in regulating hepatocellular carcinoma progression. Bioengineered. 2021;12(2):10849-10861
78. Wang L.H. et al. Downregulation of miR-29b targets DNMT3b to suppress cellular apoptosis and enhance proliferation in pancreatic cancer. Mol Med Rep. 2018;17(2):2113-2120
79. Cusenza VY. et al. The lncRNA epigenetics: The significance of m6A and m5C lncRNA modifications in cancer. Front Oncol. 2023;13:1063636
80. Zhou Y. et al. Long non-coding RNA HCP5 functions as a sponge of miR-29b-3p and promotes cell growth and metastasis in hepatocellular carcinoma through upregulating DNMT3A. Aging (Albany NY). 2021;13(12):16267-16286
81. Ke S. et al. Long intergenic non-protein coding RNA 00858 participates in the occurrence and development of esophageal squamous cell carcinoma through the activation of the FTO-m6A-MYC axis by recruiting ZNF184. Genomics. 2023;115(3):110593
82. Lu GH. et al. LncRNA SAMD12-AS1 Promotes the Progression of Gastric Cancer via DNMT1/p53 Axis. Arch Med Res. 2021;52(7):683-691
83. Wu Y, Du J. Downregulated Reprimo by LINC00467 participates in the growth and metastasis of gastric cancer. Bioengineered. 2022;13(5):11893-11906
84. Li X. et al. Long non-coding RNA DLEU1 predicts poor prognosis of gastric cancer and contributes to cell proliferation by epigenetically suppressing KLF2. Cancer Gene Ther. 2018;25(3-4):58-67
85. Xu X. et al. LINC00337 promotes tumor angiogenesis in colorectal cancer by recruiting DNMT1, which suppresses the expression of CNN1. Cancer Gene Ther. 2021;28(12):1285-1297
86. Zhang Z. et al. CNN1 Represses Bladder Cancer Progression and Metabolic Reprogramming by Modulating HIF-1alpha Signaling Pathway. Front Oncol. 2022;12:859707
87. Yue M. et al. LINC01605, regulated by the EP300-SMYD2 complex, potentiates the binding between METTL3 and SPTBN2 in colorectal cancer. Cancer Cell Int. 2021;21(1):504
88. Wang W. et al. Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation. J Transl Med. 2019;17(1):421
89. Li Y. et al. Long noncoding RNA DDX11-AS1 epigenetically represses LATS2 by interacting with EZH2 and DNMT1 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2019;514(4):1051-1057
90. Lian Y. et al. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci Rep. 2017;7(1):7312
91. Lian Y. et al. Long non-coding RNA IRAIN suppresses apoptosis and promotes proliferation by binding to LSD1 and EZH2 in pancreatic cancer. Tumour Biol. 2016;37(11):14929-14937
92. Xu X. et al. The long non-coding RNA Linc-GALH promotes hepatocellular carcinoma metastasis via epigenetically regulating Gankyrin. Cell Death Dis. 2019;10(2):86
93. Peng G. et al. Gankyrin-mediated interaction between cancer cells and tumor-associated macrophages facilitates prostate cancer progression and androgen deprivation therapy resistance. Oncoimmunology. 2023;12(1):2173422
94. Wu Z. et al. Mechanism underlying circRNA dysregulation in the TME of digestive system cancer. Front Immunol. 2022;13:951561
95. Shoda K. et al. circRNA: A New Biomarker and Therapeutic Target for Esophageal Cancer. Biomedicines. 2022 10(7)
96. Wang P. et al. The function and regulation network mechanism of circRNA in liver diseases. Cancer Cell Int. 2022;22(1):141
97. Zhang B. et al. Circ_0084615 is an oncogenic circular RNA in colorectal cancer and promotes DNMT3A expression via repressing miR-599. Pathol Res Pract. 2021;224:153494
98. Zou L. et al. Circ_0000467 modulates malignant characteristics of colorectal cancer via sponging miR-651-5p and up-regulating DNMT3B. Nucleosides Nucleotides Nucleic Acids. 2023;42(2):134-150
99. Wang F. et al. Has_circ_0008583 modulates hepatocellular carcinoma progression through the miR-1301-3p/METTL3 pathway. Bioengineered. 2022;13(1):1185-1197
100. Zhang JY. et al. ebv-circRPMS1 promotes the progression of EBV-associated gastric carcinoma via Sam68-dependent activation of METTL3. Cancer Lett. 2022;535:215646
101. Zheng R. et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer. 2022;21(1):49
102. Zhang X. et al. Combining serum inflammation indexes at baseline and post treatment could predict pathological efficacy to anti-PD-1 combined with neoadjuvant chemotherapy in esophageal squamous cell carcinoma. J Transl Med. 2022;20(1):61
103. Qin B. et al. Long non-coding RNA CASC15 facilitates esophageal squamous cell carcinoma tumorigenesis via decreasing SIM2 stability via FTO-mediated demethylation. Oncol Rep. 2021;45(3):1059-1071
104. Dos Santos EC. et al. Iroquois Family Genes in Gastric Carcinogenesis: A Comprehensive Review. Genes (Basel). 2023 14(3)
105. Ayub A. et al. Gastric Linitis Plastica: Clinical Characteristics and Outcomes from the National Cancer Database. Anticancer Res. 2023;43(4):1543-1548
106. Narmcheshm S. et al. Association between gastric cancer and the intake of different types of iron and meats. BMC Nutr. 2023;9(1):53
107. Usui Y. et al. Helicobacter pylori, Homologous-Recombination Genes, and Gastric Cancer. N Engl J Med. 2023;388(13):1181-1190
108. Wang X. et al. The N(6)-methyladenine DNA demethylase ALKBH1 promotes gastric carcinogenesis by disrupting NRF1 binding capacity. Cell Rep. 2023;42(3):112279
109. Asif B. et al. Cancer surveillance as an alternative to prophylactic total gastrectomy in hereditary diffuse gastric cancer: a prospective cohort study. Lancet Oncol. 2023;24(4):383-391
110. Pucher PH. et al. Diagnosis and treatment for gastro-oesophageal cancer in England and Wales: analysis of the National Oesophago-Gastric Cancer Audit (NOGCA) database 2012-2020. Br J Surg. 2023
111. Fan HN. et al. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer. 2022;21(1):51
112. Papamichael D. et al. Treatment of colorectal cancer in older patients: International Society of Geriatric Oncology (SIOG) consensus recommendations 2013. Ann Oncol. 2015;26(3):463-76
113. Hossain MS. et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers (Basel). 2022 14(7)
114. Linke C. et al. Quantification of mitochondrial cfDNA reveals new perspectives for early diagnosis of colorectal cancer. BMC Cancer. 2023;23(1):291
115. Chen Y. et al. The Role of the Tumor Microenvironment and Treatment Strategies in Colorectal Cancer. Front Immunol. 2021;12:792691
116. Al Bitar S. et al. Molecular mechanisms targeting drug-resistance and metastasis in colorectal cancer: Updates and beyond. World J Gastroenterol. 2023;29(9):1395-1426
117. Bian Y. et al. m(6)A Modification of Long Non-Coding RNA HNF1A-AS1 Facilitates Cell Cycle Progression in Colorectal Cancer via IGF2BP2-Mediated CCND1 mRNA Stabilization. Cells. 2022 11(19)
118. Kuok C. et al. Inhibitory Effect of Hernandezine on the Proliferation of Hepatocellular Carcinoma. Biol Pharm Bull. 2023;46(2):245-256
119. Foerster F. et al. Emerging immunotherapy for HCC: A guide for hepatologists. Hepatology. 2022;75(6):1604-1626
120. Huang DQ. et al. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18(4):223-238
121. Sun R. et al. Identification of subtypes of hepatocellular carcinoma and screening of prognostic molecular diagnostic markers based on cell adhesion molecule related genes. Front Genet. 2022;13:1042540
122. Zhang Q. et al. Evaluation of Intra-Tumoral Vascularization in Hepatocellular Carcinomas. Front Med (Lausanne). 2020;7:584250
123. Mattiuzzi C. and G. Lippi, Cancer statistics: a comparison between World Health Organization (WHO) and Global Burden of Disease (GBD). Eur J Public Health. 2020;30(5):1026-1027
124. Bray F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424
125. Khalaf N. et al. Burden of Pancreatic Cancer: From Epidemiology to Practice. Clin Gastroenterol Hepatol. 2021;19(5):876-884
126. Choi SI. et al. Complexation of drug and hapten-conjugated aptamer with universal hapten antibody for pancreatic cancer treatment. J Control Release. 2023
127. de Mello RA. et al. Current and potential biomarkers in gastric cancer: a critical review of the literature. Future Oncol. 2021;17(25):3383-3396
128. Yamamoto T. et al. Inflammation-Related Biomarkers for the Prediction of Prognosis in Colorectal Cancer Patients. Int J Mol Sci. 2021 22(15)
129. Hu M. et al. Circulating tumor cells in colorectal cancer in the era of precision medicine. J Mol Med (Berl). 2022;100(2):197-213
130. Zhou D. et al. METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Mol Oncol. 2021;15(8):2172-2184
131. Zheng Y. et al. Cancer-derived exosomal circ_0038138 enhances glycolysis, growth, and metastasis of gastric adenocarcinoma via the miR-198/EZH2 axis. Transl Oncol. 2022;25:101479
132. Zhao LJ. et al. Lysine demethylase LSD1 delivered via small extracellular vesicles promotes gastric cancer cell stemness. EMBO Rep. 2021;22(8):e50922
133. Yu W. et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32(4):466-477
134. Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71(3):264-279
135. Llovet JM. et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18(5):293-313
136. Patel SG. et al. Early age onset colorectal cancer. Adv Cancer Res. 2021;151:1-37
137. Coradduzza D. et al. Role of Nano-miRNAs in Diagnostics and Therapeutics. Int J Mol Sci. 2022 23(12)
138. Jin H. et al. Rational combinations of targeted cancer therapies: background, advances and challenges. Nat Rev Drug Discov. 2023;22(3):213-234
139. Li H. et al. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem. 2020;121(2):1353-1361
140. Sun K. et al. METTL14-dependent maturation of pri-miR-17 regulates mitochondrial homeostasis and induces chemoresistance in colorectal cancer. Cell Death Dis. 2023;14(2):148
141. Cabrera-Licona A. et al. Deciphering the epigenetic network in cancer radioresistance. Radiother Oncol. 2021;159:48-59
142. Liu F. et al. Long noncoding RNA LINC00630 promotes radio-resistance by regulating BEX1 gene methylation in colorectal cancer cells. IUBMB Life. 2020;72(7):1404-1414
143. Yao F. et al. Novel nanotherapeutics for cancer immunotherapy by CTLA-4 aptamer-functionalized albumin nanoparticle loaded with antihistamine. J Cancer Res Clin Oncol. 2023
144. Bagchi S. et al. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu Rev Pathol. 2021;16:223-249
Author contact
![]() Corresponding authors: fccguowzedu.cn (Wenzhi Guo); ranguo16com (Ran Guo).
Corresponding authors: fccguowzedu.cn (Wenzhi Guo); ranguo16com (Ran Guo).

 Global reach, higher impact
Global reach, higher impact