10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(15):4883-4897. doi:10.7150/ijbs.83474 This issue Cite
Research Paper
SRSF1-mediated alternative splicing is required for spermatogenesis
1. Guangdong and Shenzhen Key Laboratory of Reproductive Medicine and Genetics, The Center of Reproductive Medicine, Peking University Shenzhen Hospital, Shenzhen 518000, China.
2. Department of Urology & Andrology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310016, China.
3. State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, 100101, China.
4. School of Artificial Intelligence, University of Chinese Academy of Sciences, Beijing, 100049, China.
5. Guangzhou Key Laboratory of Metabolic Diseases and Reproductive Health, Guangdong-Hongkong Metabolism & Reproduction Joint Laboratory, Reproductive Medicine Center, Guangdong Second Provincial General Hospital, Guangzhou, 510317, China.
6. Department of Veterinary Pathobiology, University of Missouri, Columbia, MO 65211, USA.
7. Beijing Institute for Stem Cell and Regenerative Medicine, Beijing, 100101, China.
8. CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, 518055, China.
†These authors contributed equally to this work.
Received 2023-2-12; Accepted 2023-8-25; Published 2023-9-11
Abstract
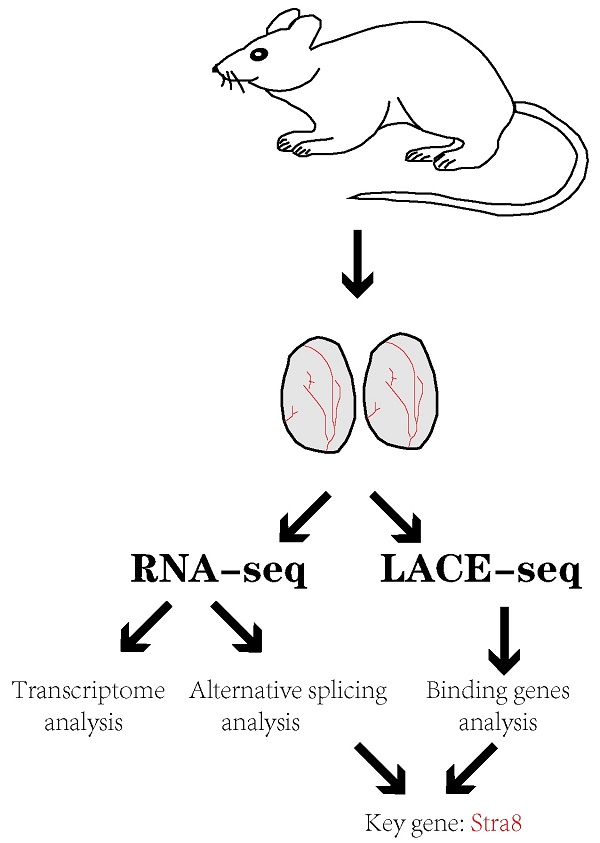
Alternative splicing (AS) plays significant roles in a multitude of fundamental biological activities. AS is prevalent in the testis, but the regulations of AS in spermatogenesis is only little explored. Here, we report that Serine/arginine-rich splicing factor 1 (SRSF1) plays critical roles in alternative splicing and male reproduction. Male germ cell-specific deletion of Srsf1 led to complete infertility by affecting spermatogenesis. Mechanistically, by combining RNA-seq data with LACE-seq data, we showed that SRSF1 affected the AS of Stra8 in a direct manner and Dazl, Dmc1, Mre11a, Syce2 and Rif1 in an indirect manner. Our findings demonstrate that SRSF1 has crucial functions in spermatogenesis and male fertility by regulating alternative splicing.
Keywords: SRSF1, male infertility, alternative splicing, spermatogenesis
Introduction
In mammalian species, the male germline stem cells are initiated to enter meiosis, continuously producing haploid male gametes after puberty [1]. Spermatogenesis consists of three sequential processes: mitosis, meiosis and spermiogenesis [2]. In the first phase of spermatogenesis, spermatogonia stem cells undergo mitotic self-renewal and differentiation into primary spermatocytes. Then, spermatocytes commence two constitutive meiotic divisions without an interval of DNA synthesis to yield haploid round spermatids in the second phase of spermatogenesis. Finally, before being released into seminiferous tubular lumen as spermatozoa, haploid round spermatids undergo a series of stepwise morphogenetic changes [2]. Dysfunction in any of these processes can cause the failure of spermatogenesis, further resulting in severe consequences including infertility [3].
It is well known that alternative splicing (AS) is one of the most important transcriptional and post-transcriptional regulatory mechanisms to enrich the amount of mRNA and protein isoforms from a single gene, and these different protein isoforms always have different structural characteristics and functions [4-6]. Generally, AS occurs in about 95% of human genes and in about 60% of mouse genes, and AS is most prevalent in the testis and brain when compared to other tissues [7, 8]. There are numerous AS events during many developmental processes. Recently, it has been shown that several RNA binding proteins have functions in AS events during spermatogenesis [9-11], indicating the importance of AS events during spermatogenesis. However, understanding the splicing factors that control the AS of key genes in spermatogenesis remains ambiguous.
The critical regulators of alternative splicing are members of the serine/arginine (SR)-rich splicing factor family, which bind to splicing enhancer sequences to promote alternative splicing [12, 13]. In humans, the SR family has at least 12 members that share a conserved SR domain [14]. It has been reported that these SR splicing factors are critical for development. The SR splicing factors can identify the splicing components of precursor RNA, then recruit and assemble spliceosomes to promote or inhibit the occurrence of alternative splicing events [15]. There are large amounts of studies suggesting that SR splicing factors have functions in nearly every step of spliceosome assembly, mRNA export, mRNA stability and genomic stability [16, 17]. Serine/arginine-rich splicing factor 1 (SRSF1), also known as SF2/ASF, is a prototype member of the SR protein family [18]. So far, several studies have suggested that SRSF1 regulates post-transcriptional gene expression via pre-mRNA alternative splicing, mRNA stability, and translation [19, 20]. While SRSF1 is an important regulator of gene expression, very little is known of its functions in the male germ cells.
In this study, by crossing Srsf1Floxed/Floxed (Srsf1F/F) mice with Stra8-Cre mice to generate mutant mice with specific deletion of the Srsf1 gene in male germ cells, we found that SRSF1 knockout caused complete infertility and germ cells were drastically lost during spermatogenesis. We further verified that deletion of the Srsf1 gene in germ cells led to meiotic arrest at the pachytene stage. By combining advanced linear amplification of complementary DNA ends and sequencing (LACE-seq) and RNA-seq with bioinformatics analysis, we unbiasedly mapped the binding sites of SRSF1 at single-nucleotide resolution and revealed the changes of the transcriptome and transcripts splicing in SRSF1-null testes. Our data suggest that SRSF1-mediated alternative splicing is required for spermatogenesis.
Results
Germ cell-specific Srsf1 knockout results in complete male infertility
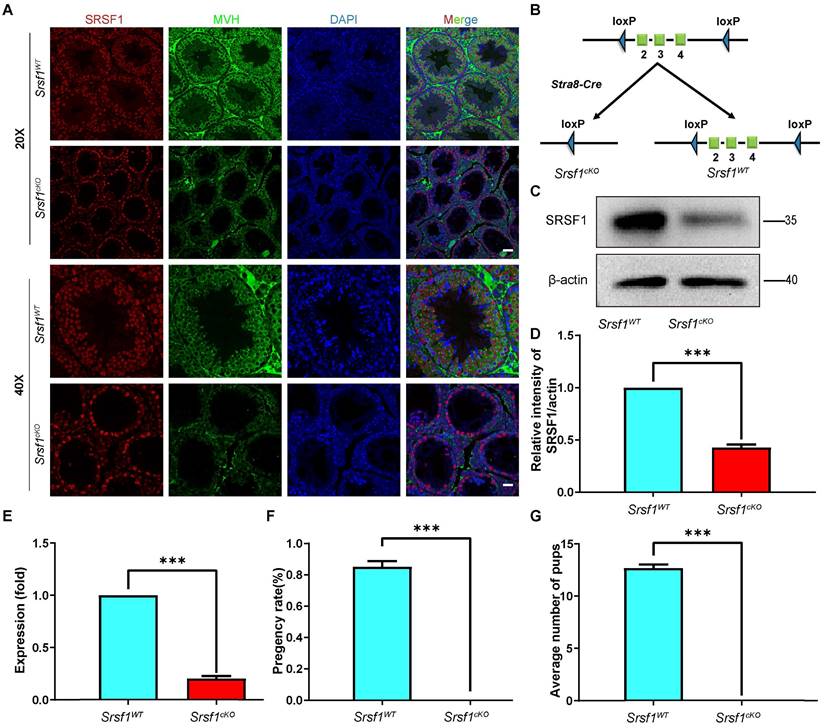
To explore the function of SRSF1 in spermatogenesis, we first analyzed the expression of SRSF1 in the testis by using the anti-SRSF1 antibody. As a well-known splicing factor, co-staining of cross-sections of seminiferous tubules in the adult mouse testis showed that SRSF1 was expressed in both germ cells and somatic cells of the testis (Fig. 1A), implying its potential role in spermatogenesis.
Then, we generated Srsf1 conditional knockout mice (referred to as Srsf1cKO) by crossing Srsf1Floxed/Floxed (Srsf1F/F) mice in which the exons 2-4 were floxed by loxP sites [21], and Stra8-Cre mice in which cre activity is initiated at 3 days after birth [22]. Srsf1 was specifically deleted (Fig. 1B), and the knockout efficiency of SRSF1 was confirmed by using quantitative RT-PCR and Western blotting. The mRNA and protein level of Srsf1 gene was significantly decreased in testes of Srsf1cKO mice (Fig. 1C,1D and Fig. 1E). Also, SRSF1 expression was barely detected in Mouse Vasa Homologue (MVH)-positive germ cells of Srsf1cKO mice (Fig. 1A). Thus, we successfully generated male germ cell-specific knockout mice for SRSF1. Next, we analyzed the phenotypes and found that compared to their littermate controls, the Srsf1cKO male mice were completely infertile (Fig. 1F and Fig. 1G). Although copulatory plugs were routinely observed, no pups were obtained when adult Srsf1cKO males were mated with normal fertile females.
Srsf1 knockout leads to severe defects in spermatogenesis
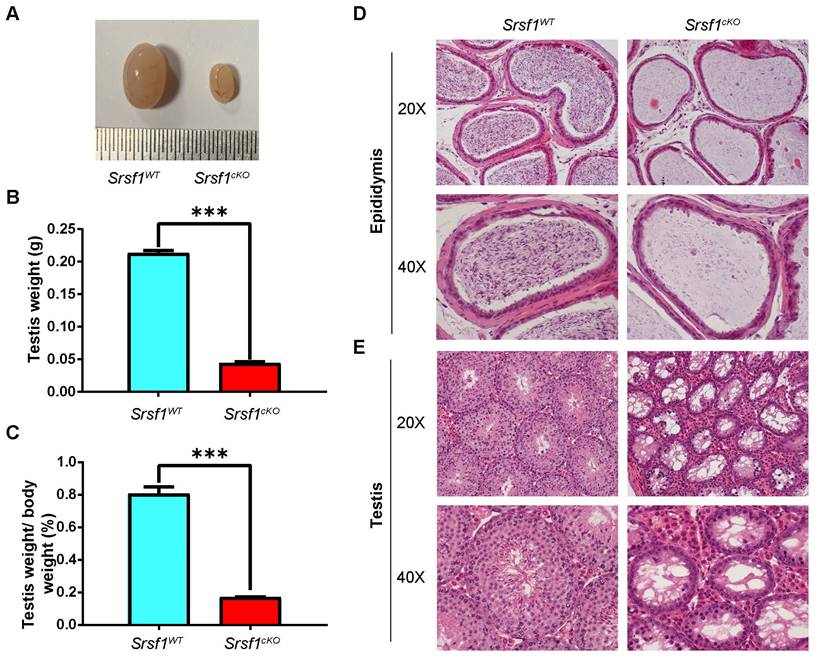
To determine the reasons of infertility in Srsf1cKO male mice, we firstly analyzed the phenotypes. Compared with controls, Srsf1cKO male mice had much smaller testes (Fig. 2A). The testis weight and the testis weight to body weight ratio of Srsf1cKO mice were significantly lower (Fig. 2B and Fig. 2C). Then we analyzed the histology of the epididymes and testes by Hematoxylin and Eosin (H&E) staining. Consistent with this phenotype, hematoxylin staining results showed that no mature spermatozoa were found in the epididymal lumen of Srsf1cKO mice (Fig. 2D). The seminiferous tubules of Srsf1WT testes contained a basal population of spermatogonia, several types of spermatocytes and spermatids. However, germ cells were severely reduced in number, almost no spermatids were present in the seminiferous tubules of Srsf1cKO testes (Fig. 2E). These findings collectively suggested that germ cell-specific Srsf1 knockout results in spermatogenesis failure and thus male infertility.
Srsf1-deficient male germ cells arrest at the pachytene stage
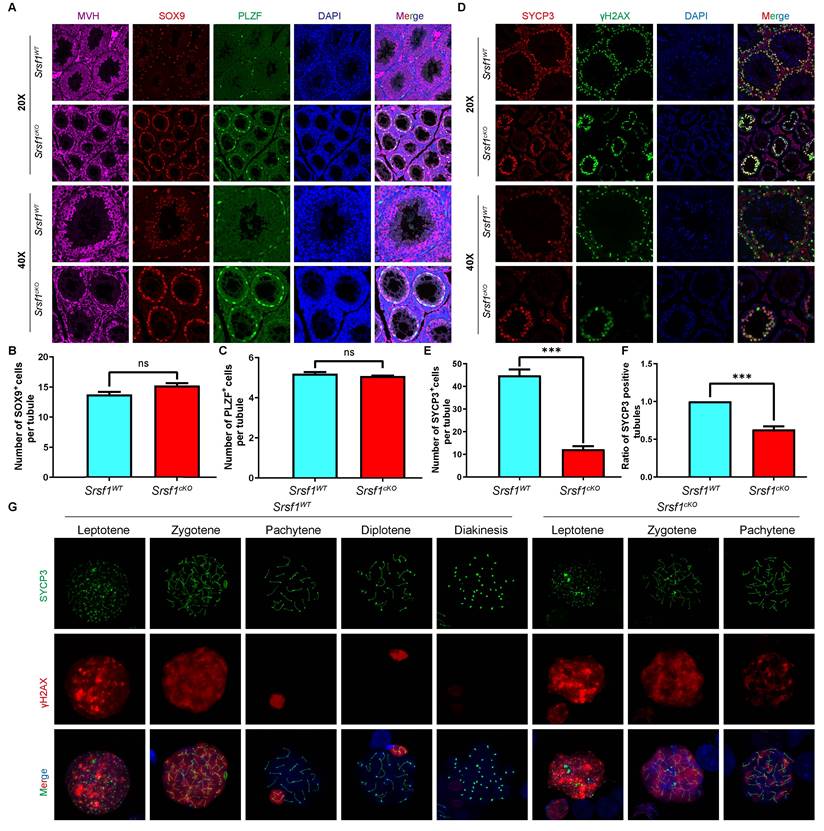
To validate the above results, we performed immunofluorescent co-staining by using antibodies against MVH, SOX9 and PLZF, markers for the germ cells, Sertoli cells, and undifferentiated spermatogonia, respectively. Immunofluorescence results indicated that the number of MVH positive signals was significantly reduced in cKO testicular sections compared with those in the control (Fig. 3A). Sertoli cells marker SOX9 staining and undifferentiated spermatogonia marker PLZF did not show an obvious change (Fig. 3B and 3C).
Germ cell-specific Srsf1 knockout results in complete male infertility. (A) Representative images of localization of SRSF1 in the control and Srsf1cKO testes of 8-week-old mice. The DNA was stained with DAPI, and SRSF1 (red) and MVH (green) were stained with corresponding antibodies. Scale bar: (top) 50 μm; (bottom) 20 μm. (B) Schematic diagram of deletion of Srsf1 exons 2, 3 and 4 and generation of Srsf1 Δ allele by Stra8- Cre-mediated recombination in male germ cells. (C) Western blotting analysis of SRSF1 protein in Srsf1WT and Srsf1cKO total testes of 8-week-old mice. β-actin was detected as an internal control. (D) The relative intensity of SRSF1/actin. β-actin was detected as an internal control. (E) Quantitative RT-PCR analyses showing Srsf1 mRNA level was decreased. β-actin was used as the internal control. Data are presented as the mean ± SEM. P<0.05(*), 0.01(**) or 0.001(***). (F) Pregnancy rates (%) of plugged wild-type females after mating with Srsf1WT and Srsf1cKO 8-week-old males. (G) Average litter size of plugged wild-type females after mating with Srsf1WT and Srsf1cKO 8-week-old males. For this part, at least 3 mice (8-week-old) of each genotype were used for the analysis. Data are presented as the mean ± SEM. P<0.05(*), 0.01(**) or 0.001(***).
Meiosis is a unique process for the differentiation of germ cells. Meiotic recombination and homologous chromosome synapsis are two pivotal events in meiotic progression. Next, we examined meiotic progression by immunostaining the axial element component of the synaptonemal complex with SYCP3 and double-strand break (DSB) marker γH2AX. Similarly, immunofluorescence results indicated that SYCP3 andγH2AX double-positive cells were significantly decreased in Srsf1cKO mice compared with controls (Fig. 3D, 3E and 3F). To further evaluate the detailed phenotype, we observed spermatocytes using mouse germ cell surface spreading by co-immunostaining with antibodies against synaptonemal complex protein 3 (SYCP3) and phosphorylated histone H2AX (γH2AX) (Fig. 3G). The result indicated that loss of SRSF1 led to meiotic arrest at the pachytene stage.
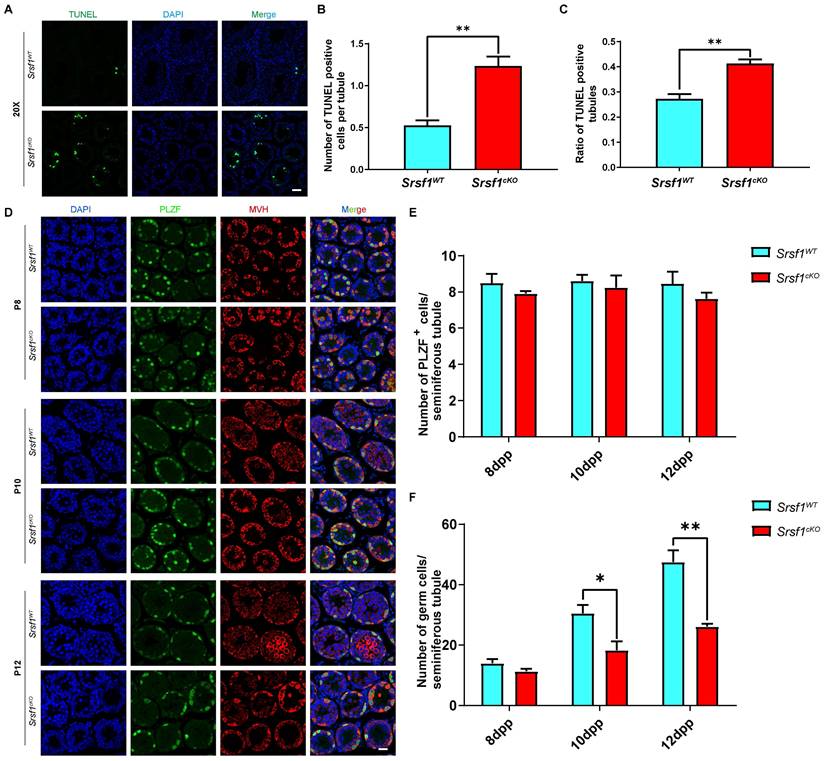
The number of germ cells was sharply decreased in Srsf1cKO mice, probably because the germ cells were undergoing apoptosis. To test this possibility, TUNEL assay was performed, and the results showed that germ cells underwent apoptosis in the Srsf1cKO mice (Fig. 4A, 4B and 4C). Then, to further identify which stage of spermatogenesis was firstly impaired in SRSF1-deficient mice, we performed immunofluorescence staining of the PLZF protein and the germ cell marker MVH to characterize the first wave of spermatogenesis in mice at postnatal day 8 (P8), P10, and P12. Undifferentiated spermatogonia marker PLZF staining showed that the number and location of undifferentiated spermatogonia did not display an obvious change (Fig. 4D and Fig. 4E). However, the results revealed that the numbers of germ cells of Srsf1cKO mice were similar to Srsf1WT mice at P8, but these numbers started to decrease at P10 and sharply dropped at P12 (Fig. 4Dand Fig. 4F). Altogether, these results again supported that Srsf1 has functions during spermatogenesis.
SRSF1 is required for spermatogenesis. (A) The testes of Srsf1cKO were smaller than those of the control (8-week-old, the same as below). (B) Testis weight of Srsf1WT and Srsf1cKO 8-week-old male mice (n=3). (C) Testis weight to body weight ratio of Srsf1WT and Srsf1cKO 8-week-old male mice (n=3). Data are presented as the mean ± SEM. P<0.05(*), 0.01(**) or 0.001(***). (D) Histological analysis of the caudal epididymes of the Srsf1WT and Srsf1cKO mice. Scale bar: (top) 100 μm; (bottom) 50 μm. (E) Histological analysis of the seminiferous tubules of the Srsf1WT and Srsf1cKO mice. Scale bar: (top) 100 μm; (bottom) 50 μm. For this part, at least 3 mice (8-week-old) of each genotype were used for the analysis.
Srsf1 deficiency causes the loss of germ cells. (A) MVH (a marker of germ cells, purple), SOX9 (a marker of Sertoli cells, red) and PLZF (a marker of undifferentiated spermatogonia, green), immunofluorescence analysis of the Srsf1WT and Srsf1cKO 8-week-old male mice. Scale bar: (top) 50 μm; (bottom) 20 μm. (B) Quantification of the SOX9 positive signal cells in the seminiferous tubules of the Srsf1WT and Srsf1cKO mice. (C) Quantification of the PLZF positive signal cells in the seminiferous tubules of the Srsf1WT and Srsf1cKO mice. (D) SYCP3 (red) and γH2AX (green) immunofluorescence analysis of the Srsf1WT and Srsf1cKO 8-week-old male mice. Scale bar: (top) 50 μm; (bottom) 20 μm. (E) Quantification of the SYCP3 positive signal cells in the seminiferous tubules of the Srsf1WT and Srsf1cKO mice. (F) Statistics of the ratio of SYCP3-positive tubule cross-sections in Srsf1WT and Srsf1cKO mice. (G) Srsf1WT and Srsf1cKO mice chromosome spreads of spermatocytes were immunostained with antibodies against SYCP3 (green) and γH2AX (red). γH2AX marked DSB in spermatocytes(8-weekold). Scale bar: 10 μm. In this part, at least 3 mice of each genotype were used for the analysis.
Genome-wide analysis of SRSF1-binding genes in mouse testes
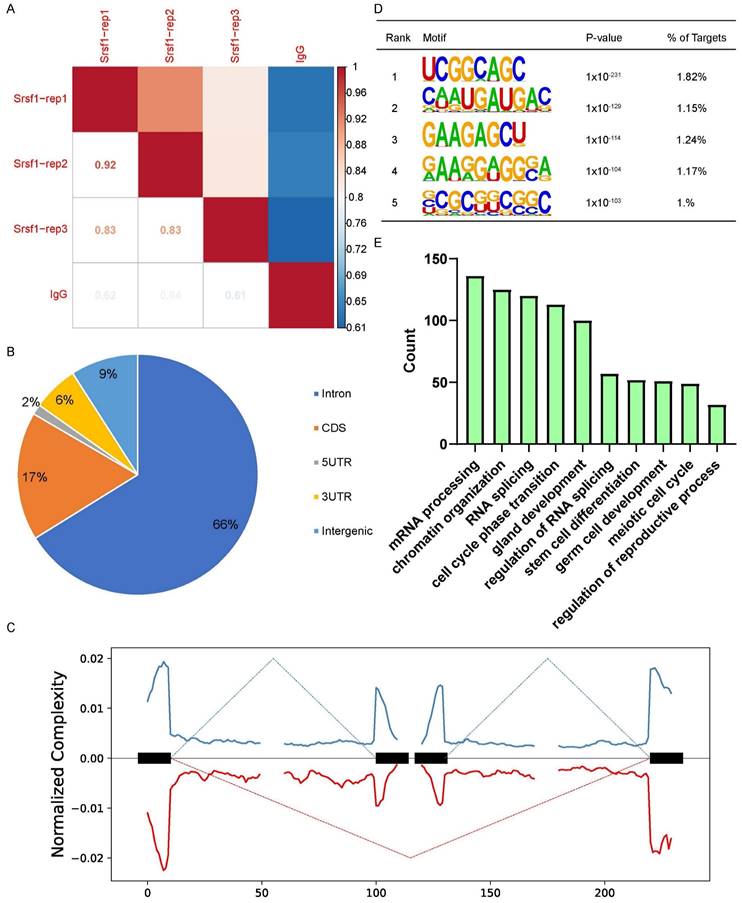
To further investigate the molecular mechanisms by which SRSF1 causes the failure of spermatogenesis, we performed LACE-seq analysis of testes collected at P10 to profile SRSF1-binding sites. Three independent replicates with a high correlation in read counts were pooled together for the subsequent analysis (Fig. 5A). Among these SRSF1 clusters, more than half of them were derived from intergenic regions, while others were aligned to intron, CDS (coding sequence), UTR3 (3′ untranslated region), and UTR5 (5′ untranslated region) (Fig. 5B). We also found that SRSF1 “preferentially” bound to exons and enriched between 0 and 50 nt of the 5′ and 3′ exonic sequences flanking the constitutive splice sites as revealed by analyzing the distributions of SRSF1-binding peaks within 200 nucleotides (nt) upstream or downstream of the constitutive splice site (Fig. 5C). Among these SRSF1 peaks, most of them were significantly enriched for UC-rich consensus motif (Fig. 5D). GO analysis showed that these SRSF1-binding genes were involved in the regulation of mRNA processing, RNA splicing, germ cell development, stem cell differentiation, meiotic cell cycle, and regulation of the reproductive process (Fig. 5E). Together, these analyses suggested that SRSF1 functions as a crucial regulator by directly binding its targets in the reproductive development.
SRSF1 depletion results in the apoptotic death of germ cells. (A) TUNEL immunofluorescence staining of the testes of Srsf1WT and Srsf1cKO. (Scale bar: 50 μm) Green: TUNEL positive signal; Blue: DAPI. At least 3 mice (8-week-old) of each genotype were used for analysis. (B) Number of TUNEL positive cells per tubule section. (C) Quantification of TUNEL positive seminiferous tubules (8-week-old). (D) PLZF (green) and MVH (red) immunofluorescence analysis of the Srsf1WT and Srsf1cKO male mice at P8, P10 and P12. Scale bar, 20 μm. In this part, at least 3 mice of each genotype were used for the analysis. (E) Quantification of the PLZF positive signal cells in the seminiferous tubules of the juvenile Srsf1WT and Srsf1cKO mice. For each time point, at least 3 mice of each genotype were used for the analysis. Data are presented as the mean ± SEM. P<0.05(*), 0.01(**) or 0.001(***). (F) Quantification of the germ cells in the seminiferous tubules of the juvenile Srsf1WT and Srsf1cKO mice. For each time point, at least 3 mice of each genotype were used for the analysis. Data are presented as the mean ± SEM. P<0.05(*), 0.01(**) or 0.001(***).
Changes in transcriptome and splicing of transcripts in SRSF1-null testes
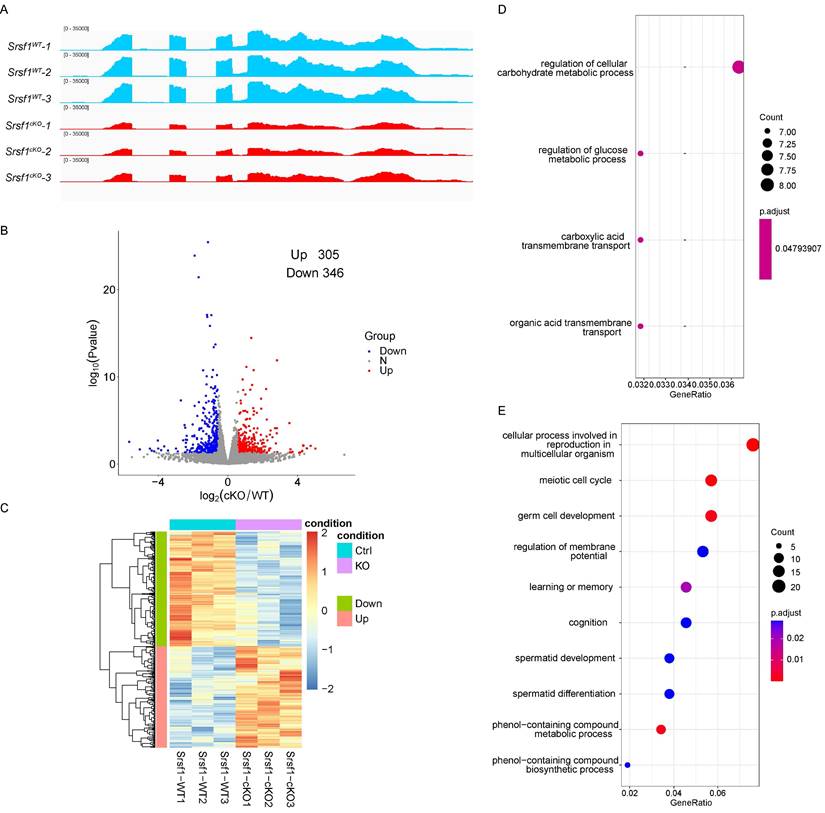
According to the above-presented data, SRSF1 cKO mice displayed defects in spermatogenesis. To explore a comprehensive perspective of the mechanisms of SRSF1 deletion in male germ cells, we isolated mRNA from Srsf1WT and Srsf1cKO testes at P10 and then performed RNA sequencing (RNA-seq). RNA-seq results firstly showed the reduction of Srsf1 RNA in Srsf1cKO mice testes (Fig. 6A). A total of 305 genes were upregulated, and 346 genes were downregulated in Srsf1cKO testes (P value of <0.05, |log2FoldChange| ≥ 0.58) (Fig. 6B). Heatmap analysis showed hierarchical clustering of differential expression genes (DEGs) of Srsf1WT and Srsf1cKO testes (Fig. 6C). To obtain more comprehensive information, we then performed Gene Ontology (GO) annotation. GO analysis showed that these upregulated genes were involved in carbohydrate metabolic processes and carboxylic acid transmembrane transport, (Fig. 6D). Meiotic cell cycle, germ cell development, and cellular processes involved in reproduction in multicellular organisms were significantly enriched among these downregulated genes (Fig. 6E). In short, these differential expression genes may account for the SRSF1-null phenotypes in spermatogenesis.
Global landscape of SRSF1-binding sites in mouse testes as revealed by using LACE-seq. (A) The reproducibility of the LACE-seq data. (B) Genomic distribution of SRSF1 binding sites in testes. CDS, coding sequence. UTR3, 3′ untranslated region. UTR5, 5′ untranslated region. (C) Schematic analysis showing the distribution of SRSF1-binding sites in the vicinity of the 5′ exon-intron and the 3′ intron-exon boundaries. (D) Enriched hexamer motifs bound by SRSF1. The top five enriched motifs are shown. (E) GO enrichment map of SRSF1-binding genes.
Transcriptome changes in SRSF1-null testes. (A) RNA-seq results showing the reduction of Srsf1 RNA in Srsf1cKO mice testes. Three independent RNA-seq experiments are shown. (B) Volcano plot showing transcriptome difference between Srsf1WT and Srsf1cKO testes. (C) Heatmap showing hierarchical clustering of differential expression genes of Srsf1WT and Srsf1cKO male mice testes. (D) GO term enrichment analysis of upregulated genes. (E) GO term enrichment analysis of downregulated genes.
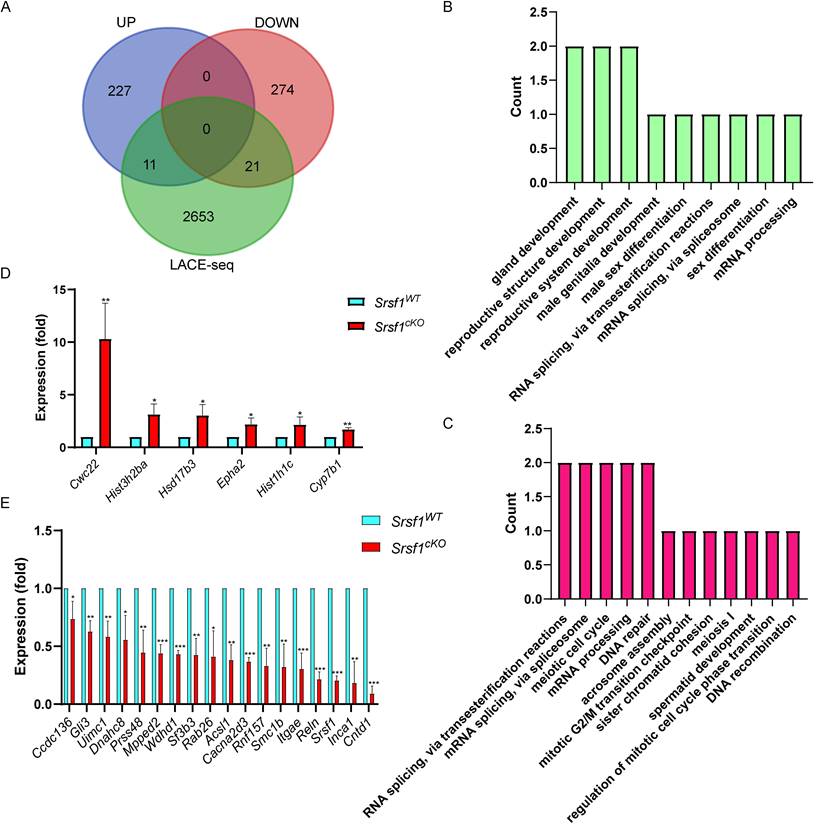
Furthermore, by combining RNA-seq data with LACE-seq identified peaks, we only identified 21 downregulated, and 11 upregulated transcripts as direct targets of SRSF1 in testes (Fig. 7A). To obtain more comprehensive information, similarly, we then performed GO annotation. GO analysis showed that both significantly upregulated genes and SRSF1-binding genes were involved in reproductive development, male sex differentiation, mRNA processing and mRNA splicing (Fig. 7B). And RNA splicing, meiotic cell cycle, meiosis I, spermatid development, DNA repair, and DNA recombination were significantly enriched among both of these significantly downregulated genes and SRSF1-binding genes (Fig. 7C). We next validated both of these significantly DEGs and SRSF1-binding genes which were involved in spermatogenesis by using quantitative polymerase chain reaction (qPCR) to check the mRNA abundance (Fig. 7D and Fig. 7E). These data reflected that deletion of SRSF1 directly affects the expression levels of critical genes involved in spermatogenesis. Nevertheless, most of the changes in gene expression might be regulated indirectly by SRSF1.
Because SRSF1 is an SR protein and played critical roles in AS, we then analyzed the five different types of AS events between Srsf1WT and Srsf1cKO testes by using the rMATS computational tool. Compared with the Srsf1WT group, a total of 1044 AS events were identified as significantly changed in the Srsf1cKO group (|Diff| > 0.05, FDR < 0.001). Among these 1044 changed AS events, most (924) of AS events were skipped exons (SE). Moreover, there were 14 alternative 3′ splice sites (A3SS), 34 alternative 5′ splice sites (A5SS), 55 mutually exclusive exons (MXE), and 17 retained introns (RI) (Fig. 8A and Fig. S1). Importantly, we successfully verified that loss of SRSF1 in testes resulted in differential splicing in Dazl, Dmc1, Mre11a, Syce2 and Rif1 (Fig. 8B), which were critical for spermatogenesis.
The expressions of key SRSF1-binding genes involved in the spermatogenesis change after Srsf1 KO. (A) Venn diagram showing the overlapped genes between differentially expressed genes and SRSF1-binding genes. (B) Correlation analysis between the RNA-seq and LACE-seq. GO analysis of the significantly upregulated genes and SRSF1-binding genes. (C) Correlation analysis between the RNA-seq and LACE-seq. GO analysis of the significantly downregulated genes and SRSF1-binding genes. (D) Quantitative RT-PCR validation of the expression of genes involved in (B). β-actin was used as the internal control. Data are presented as the mean ± SEM. P<0.05(*), 0.01(**) or 0.001(***). (E) Quantitative RT-PCR validation of the expression of genes involved in (C). β-actin was used as the internal control. Data are presented as the mean ± SEM. P<0.05(*), 0.01(**) or 0.001(***).
Srsf1 is required for normal splicing of functional genes during spermatogenesis. (A) The five different types of alternative splicing (AS) events. The numbers of abnormal AS events were counted between Srsf1WT and Srsf1cKO testes by rMATS software. (B) A magnified view showing RNA-seq signals of the selected candidate genes. IgG, immunoglobulin G. Semiquantitative RT-PCR analysis of AS patterns of the changed spliced genes in Srsf1WT and Srsf1cKO testes at P10. (C) Venn diagram shows the correlation between SRSF1-binding genes and AS genes. (D) GO analysis of the AS genes and SRSF1-binding genes. (E) A magnified view showing RNA-seq and LACE-seq signals of the Stra8 genes. IgG, immunoglobulin G. Semiquantitative RT-PCR analysis of AS patterns of the changed spliced genes in Srsf1WT and Srsf1cKO testes at P10. PCR primers are listed in Table S1.
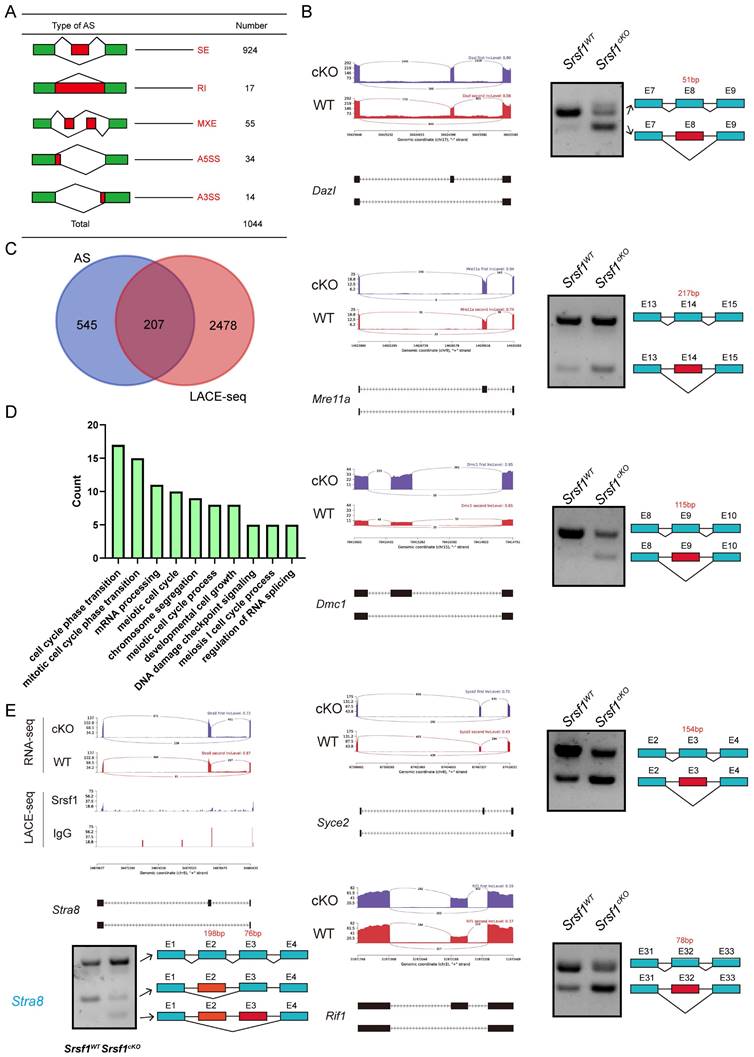
We then asked how these differentially spliced genes were regulated by SRSF1. Venn diagram showed that among these differentially spliced genes, 207 genes were bound by SRSF1 as identified in LACE-seq (Fig. 8C), indicating that these genes are probably the direct targets of SRSF1. GO analysis showed that both AS genes and SRSF1-binding genes were involved in meiotic cell cycle, mRNA processing, chromosome segregation and developmental cell growth (Fig. 8D). Importantly, the data showed that the splicing of Stra8 mRNA was changed after SRSF1 cKO (Fig. 8E). We also performed semiquantitative reverse transcription PCR to confirm the above results (Fig. 8E). Together, these results suggest that SRSF1 is essential for RNA splicing during spermatogenesis.
Discussion
In higher eukaryotes, most genes express primary transcripts that undergo AS regulation in the nucleus to generate different functional mRNAs [23-25]. It is important for spermatogenesis to regulate AS processes, and splicing factors and AS is regulated in a stage-specific manner during spermatogenesis [26, 27]. The SR proteins are well-known splicing factors that are critical for the regulation of AS. As members of the SR protein family, SRs which include 12 members in mammalian systems (SRSF1-12) are well-known for their regulatory function of splicing [28]. The first SRs identified were SRSF1 (previously known as SF2/ASF) [14]. Like other SR splicing factors, several studies in recent years have shown that SRSF1, as a crucial RNA-binding protein (RBP), has well-documented functions as posttranscriptional regulator for mRNA splicing and translation during a multitude of physiological processes and multiple cancer genesis events [29-31].
Recently, it also has been found that RBPs have important functions during germline and early embryo development. As an RBP, SRSF1 is also expressed in testis, however, its functions in male germ cells are still unknown. In this study, by crossing Srsf1F/F mice with Stra8-Cre mice to generate mutant mice, we found that SRSF1 is essential for spermatogenesis and fertility in males.
Our findings showed that the number of germ cells was sharply decreased in Srsf1cKO mice, and Srsf1-deficient male germ cells arrest at the pachytene stage, probably because the pachytene checkpoint-arrested spermatocytes and other abnormal germ cells were undergoing apoptosis. To test this possibility, TUNEL assay was performed, and the results showed that abnormal germ cells underwent apoptosis in the Srsf1cKO mice. To gain a comprehensive perspective of the mechanisms of SRSF1 depletion in male germ cells, we isolated testes from wildtype mouse at P10 and systematically profiled binding landscapes of SRSF1 proteins by using LACE-seq. The results showed that SRSF1 proteins could bind numerous genes in a direct manner. Then, our analysis showed that these SRSF1-binding genes were closely involved in the regulation of mRNA processing, RNA splicing, germ cell development, stem cell differentiation, meiotic cell cycle, and regulation of reproductive processes. In addition, RNA-seq analysis further showed that transcriptome and splicing of transcripts change in SRSF1-null testes. By combining RNA-seq and LACE-seq data, we found that deletion of SRSF1 directly or indirectly affects the expression levels of critical genes involved in spermatogenesis.
Spermatogonial stem cell differentiation and the mitosis-meiosis transition are important steps for spermatogenesis. Retinoic acid (RA) is a crucial factor of spermatogenesis, with functions on spermatogonial stem cell differentiation and subsequent initiation of meiosis [32, 33]. RA has two certain targets, Stra8 and Kit. There is already some evidence suggesting that Stra8 has two different functions in spermatogenesis. On one hand, under the influence of RA, Stra8 functions as a transcriptional repressor of the pluripotency program during differentiation of spermatogonia. When differentiating spermatogonia are near the end of their mitotic phase, Stra8 switches to the second role and acts as a transcription activator of genes involved in meiosis initiation [34-36]. In addition to RA signaling, Dazl is a key intrinsic factor for initiating meiosis [37]. The switch from mitosis to meiosis is a critical step of germ cell development that requires Dazl. In meiosis I, the Mre11 complex is essential for meiotic recombination. During the meiotic prophase, numerous DNA double-strand breaks (DSB) are formed in the genome in order to initiate recombination between homologous chromosomes. The conserved Mre11 complex has functions in mitotic cells for sensing and repairing DSB [38, 39]. In most eukaryotic cells, two recombinases, Rad51 and Dmc1, are responsible for the homologous recombination process. Dmc1, which is essential for both mitotic and meiotic recombination, is present only in meiosis while Rad51 is expressed in both meiotic and mitotic cells [40-42]. Also, Syce2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination [43]. The surveys indicated that Rif1 had crucial functions in DNA double-strand repair by its interaction with TP53BP1 (tumor protein p53 binding protein) [44].Here, we crossed Stra8-Cre mice with Srsf1flox/flox (Srsf1cKO) mice to study the functions of Srsf1 in spermatogenesis. We found that the number of c-KIT+ cells was obviously reduced in cKO testes compared with that in the control. SYCP3 and γH2AX positive cells were also significantly decreased in Srsf1cKO testes compared with controls. These results indicated that Srsf1 had critical roles during spermatogenesis. Of particular note, one of our critical findings was that we identified the binding motif and splicing targets of Srsf1 during spermatogenesis through combined analysis of RNA-seq and LACE-seq data. The two omics data indicated that SRSF1 affects the AS of Stra8 in a direct manner and Dazl, Dmc1, Mre11a, Syce2 and Rif1 in an indirect manner. These above genes are critical for the male germ cell developmental process. However, limitation of the study is that transcriptome and GO data provided here may only due to a lack of germ cells in testis from SRSF1 cKO mice.
In summary, our findings suggest that SRSF1 is critical for male fertility and spermatogenesis. Mechanistic analyses indicate that SRSF1 has functions in posttranscriptional regulation by specifically adjusting the gene expression and AS in direct or indirect manners during spermatogenesis. Specifically, we verified Stra8 as one of the splicing targets of SRSF1. Our investigation suggests that SRSF1-mediated AS is important for spermatogenesis.
Methods
Mice
Mice lacking Srsf1 in male germ cells (referred to as Srsf1cKO) were generated by crossing Srsf1Floxed/Floxed (Srsf1F/F) mice with Stra8-Cre mice. All transgenic mouse lines had C57BL/6J genomic background. Genotyping PCR for Srsf1 was performed using the following primers: forward: GGGACTAATGTGGGAAGAATG, and reverse: AACCTAAACTATTGCTCCCATCTG. The PCR conditions were as follows: 94 °C for 5 min; 35 rounds of 94 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 30 sec; and 72 °C for 5 min. Genotyping PCR for Stra8-Cre was performed using the following primers: forward: ACTCCAAGCACTGGGCAGAA, wildtype reverse: GCCACCATAGCAGCATCAAA and reverse: CGTTTACGTCGCCGTCCAG. The PCR conditions were as follows: 94 °C for 5 min; 35 rounds of 94 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 30 sec; and 72 °C for 5 min. Four genotypes in the progeny, including Srsf1F/+, Srsf1F/-, Srsf1F/+; Stra8-Cre and Srsf1F/-; Stra8-Cre were identified. The Srsf1F/+ male mice were used as control group.
The mice were maintained under specific-pathogen-free (SPF) conditions and housed under controlled environmental conditions with free access to water and food. All animal operations were approved by the Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences (CAS). The assigned accreditation number of the laboratory was IOZ20160033.
Antibodies
β-actin antibody (mouse, sc-47778; Santa Cruz); SYCP3 (mouse, sc-74569; Santa Cruz); γH2AX (rabbit, 9718; Cell Signaling Technology, Inc.); MVH (mouse, ab27591; Abcam); SOX9 antibody (rabbit, AB5535, Sigma-Aldrich); PLZF antibody (goat, AF2944, R&D Systems); SFRS1 polyclonal antibody (rabbit, 12929-2-AP, Proteintech); c-Kit antibody (goat, AF1356, R&D Systems). Horseradish peroxidase-conjugated secondary antibodies were purchased from Zhongshan Golden Bridge Biotechnology Co, LTD (Beijing). Alexa Fluor 488-conjugated antibody, 594-conjugated antibody and Alexa Fluor 647-conjugated antibody were purchased from Life Technologies.
Breeding assay
Males of different genotypes (8 weeks) were used for the breeding assay. Each male mouse was caged with two wild-type ICR (Institute of Cancer Research) females (7 weeks), and their vaginal plugs were checked every morning. The number of pups in each cage was counted within a week of birth. Each male underwent at least eight cycles of the above breeding assay.
Immunoblotting
To prepare protein extracts, testes were homogenized in RIPA lysis buffer supplemented with protease and phosphatase inhibitor cocktail (Roche Diagnostics). After transient ultrasound treatment, the testis lysates were incubated on ice for 30 min and then centrifuged at 4 ℃, 12000 rpm for 20 min. The supernatant was transferred to a new tube and quantified using a BCA reagent kit (Beyotime, P0012-1). Then equal volume loading buffer was added. After being boiled at 95 ℃ for 10 min, the protein lysates were used for immunoblotting analysis. Immunoblotting was performed as described previously [45]. Briefly, the separated proteins in SDS-PAGE were electrically transferred to a polyvinylidene fluoride membrane. After incubation with primary and secondary antibodies, the membranes were scanned with Bio-Rad ChemiDoc XRS+.
Tissue collection and histological analysis
For histological analysis, at least three adult mice for each genotype were analyzed. Testes and caudal epididymides were dissected immediately following euthanasia. The tissues were then fixed in Bouin's fixative (saturated picric acid: 37% formaldehyde: glacial acetic acid= 15: 5: 1) overnight at room temperature, dehydrated in an ethanol series, and embedded in paraffin wax. Then, 5μm sections were cut with a microtome. After 48 °C overnight drying, the sections were deparaffinized in xylene, hydrated by a graded alcohol series and stained with Hematoxylin and Eosin for histological analysis. Images were collected with a Nikon inverted microscope with a charge coupled device (CCD) (Nikon, Eclipse Ti-S, Tokyo, Japan).
Immunofluorescence
Testes used for immunostaining were fixed in 4% paraformaldehyde (pH 7.4) overnight at 4 °C, dehydrated, and embedded in paraffin. Paraffin-embedded testes were cut into sections of 5μm thickness. Then, the sections were deparaffinized, immersed in sodium citrate buffer (pH 6.0) and heated for 15 min in a microwave for antigen retrieval. After blocking with 5% donkey serum albumin, sections were incubated with primary antibodies at 4 °C overnight. Then the sections were incubated with an appropriate FITC-conjugated secondary antibody. The nuclei were stained with DAPI. Images were captured using a laser scanning confocal microscope LSM880 (Carl Zeiss, Germany).
TUNEL assay
TUNEL assay was carried out in accordance with the DeadEndTM Fluorometric TUNEL System (Promega BioSciences, Madison, WI, USA). Images were captured using a laser scanning confocal microscope LSM880 (Carl Zeiss, Germany).
RNA extraction and gene expression analysis
Total RNA was extracted from whole testes using TRNzol Universal Reagent (cat. # DP424, Tiangen, China) according to the manufacturer's instructions. Then reverse transcription (RT) was performed using the 5X All-In-One RT MasterMix (cat. # G490, Abm, Canada). RT-PCR was performed using the UltraSYBR Mixture (cat. # CW0957, Cowin Bio, China) on a LightCycler 480 instrument (Roche). The results were analyzed based on the 2-ΔΔCt method to calculate the fold changes. β-actin was used as an internal control. At least three independent experiments were analyzed. All primer sequences are listed in Table S1.
Semiquantitative PCR experiment was carried out with primers (listed in Table S1) amplifying endogenous transcripts. Then the PCR products were detected on 2% agarose gels. Gapdh was used as an internal control.
RNA sequencing and data analysis
Total testes samples were used from P10 Srsf1WT and Srsf1cKO male mice according to three individual collections. One Total RNA was extracted from whole testes using TRNzol Universal Reagent (cat. # DP424, Tiangen, China) according to the manufacturer's instructions. The quality of RNA samples was examined by NanoDrop 2000&8000 and Agilent 2100 Bioanalyzer, Agilent RNA 6000 Nano Kit. High-quality RNAs were used to prepare the libraries, followed by high-throughput sequencing on an Illumina NovaSeq 6000. The RNA sequencing experiment was supported by Annoroad BioLabs.
After trimming adaptor sequence and rRNA, the retained reads from Srsf1 control and cKO samples were aligned to the mouse genome (mm9) using HISAT2 with default parameters. Only non-RCR duplicate and uniquely mapped reads were used for subsequent analysis. Significantly changed genes were screened using DESeq2 with |log2FC| > 0.58 and p<0.05. Alternative splicing events were identified by rMATS with default parameters. Only events with FDR<0.001 and splicing difference > 0.05 were regarded as significant.
LACE-sequencing and data analysis
Total testes samples were used from P10 WT male mice for LACE-seq. LACE-seq method was performed as described recently by us [46]. Briefly, the samples were firstly irradiated twice with UV-C light on ice at 400 mJ. Then RNA immunoprecipitation of the samples was performed. The immunoprecipitated RNAs were then fragmented by MNase and dephosphorylated. Then a series of steps was performed to include, reverse transcription, first-strand cDNA capture by streptavidin beads, poly(A) tailing, pre-PCR, IVT, RNA purification, RT, PCR barcoding and deep sequencing.
The adapter sequences and poly(A) tails at the 3′ end of raw reads were removed using Cutadapt (v.1.15) with two parameters: -f fastq -q 30,0 -a ATCTCGTATGCCGTCTTCTGCTT -m 18 --max-n 0.25 --trim-n., and -f fastq -a A -m 18 -n 2. Clean reads were first aligned to mouse pre-rRNA using Bowtie, and the remaining unmapped reads were then aligned to the mouse (mm9) reference genome. For LACE-seq data mapping, two mismatches were allowed (Bowtie parameters: -v 2 -m 10 --best -strata; -v 2 -k 10 --best -strata). Peaks were identified by Piranha with parameters: -s -b 20 -p 0.01. Peaks without IgG signal were selected for further usage. For motif analysis, LACE-seq peaks/clusters were first extended 30 nt to 5′ upstream, and overrepresented hexamers in the extended sequences were identified as previously described [47]. The consensus motifs were generated from the top-10 enriched hexamers using WebLogo.
Statistical analysis
All of the experiments were performed at least three times independently. Paired two-tailed Student's t-test was used for statistical analysis. Data analyses were carried out via GraphPad Prism 8.00 (GraphPad Software, Inc.) and presented as mean ± SEM and P<0.05(*), 0.01(**) or 0.001(***) was considered statistically significant.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank Dr Xiang-Dong Fu for providing the Srsf1F/F mice. We appreciate and acknowledge Shiwen Li and Xili Zhu for their technical assistance. We thank all members of the Sun lab for their help and discussion.
Funding sources
This study was supported by National Key R&D Program of China (2021YFA1100302, 2019YFA0109900), National Natural Science Foundation of China (82301806, 31970509), Guangdong Basic and Applied Basic Research Foundation (2021A1515111118, 2020A1515011414), Natural Science Foundation of Shandong Province (ZR2021ZD33) and Basic and Applied Basic Research Foundation of Guangzhou City (202201020292).
Ethics approval and consent to participate
All experiments were conducted following the guidelines and with the approval of the Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences (CAS) (No. IOZ20160033).
Author contributions
Conceptualization was performed by W.L., Q.S., W.Q., F.S., and Z.W.; methodology and investigation by W.L., Z.D., T.M., R.S., Y.L.; visualization by G.H., and Y.N.; supervision by H.S. and Z.H.; writing—original draft by W.L., and Y.L.; and writing—review and editing by W.Q., F.S., and Q.S. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tournaye H, Krausz C, Oates RD. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017;5:544-53
2. Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263-86
3. Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13:493-504
4. Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015;84:291-323
5. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457-63
6. Song H, Wang L, Chen D, Li F. The Function of Pre-mRNA Alternative Splicing in Mammal Spermatogenesis. Int J Biol Sci. 2020;16:38-48
7. de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38:2825-38
8. Zhang X, Ameer FS, Azhar G, Wei JY. Alternative Splicing Increases Sirtuin Gene Family Diversity and Modulates Their Subcellular Localization and Function. Int J Mol Sci. 2021 22
9. Iwamori N, Tominaga K, Sato T, Riehle K, Iwamori T, Ohkawa Y. et al. MRG15 is required for pre-mRNA splicing and spermatogenesis. Proceedings of the National Academy of Sciences. 2016;113:E5408-E15
10. Zagore LL, Grabinski SE, Sweet TJ, Hannigan MM, Sramkoski RM, Li Q. et al. RNA Binding Protein Ptbp2 Is Essential for Male Germ Cell Development. Mol Cell Biol. 2015;35:4030-42
11. Barsh GS, Bao J, Tang C, Li J, Zhang Y, Bhetwal BP. et al. RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility. PLoS Genet. 2014 10
12. Yu T, Cazares O, Tang AD, Kim HY, Wald T, Verma A. et al. SRSF1 governs progenitor-specific alternative splicing to maintain adult epithelial tissue homeostasis and renewal. Dev Cell. 2022
13. Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242
14. Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010;24:1073-4
15. Zheng X, Peng Q, Wang L, Zhang X, Huang L, Wang J. et al. Serine/arginine-rich splicing factors: the bridge linking alternative splicing and cancer. Int J Biol Sci. 2020;16:2442-53
16. Huang Y, Gattoni R, Stévenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837-43
17. Savisaar R, Hurst LD. Purifying Selection on Exonic Splice Enhancers in Intronless Genes. Mol Biol Evol. 2016;33:1396-418
18. Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res. 2014;12:1195-204
19. Howard JM, Sanford JR. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA. 2015;6:93-110
20. Katsuyama T, Moulton VR. Splicing factor SRSF1 is indispensable for regulatory T cell homeostasis and function. Cell Rep. 2021;36:109339
21. Wang H-Y, Xu X, Ding J-H, Bermingham JR, Fu X-D. SC35 Plays a Role in T Cell Development and Alternative Splicing of CD45. Mol Cell. 2001;7:331-42
22. Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738-42
23. Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336
24. Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741-54
25. Liu Y, Luo Y, Shen L, Guo R, Zhan Z, Yuan N. et al. Splicing Factor SRSF1 Is Essential for Satellite Cell Proliferation and Postnatal Maturation of Neuromuscular Junctions in Mice. Stem Cell Reports. 2020;15:941-54
26. Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437-51
27. Schmid R, Grellscheid SN, Ehrmann I, Dalgliesh C, Danilenko M, Paronetto MP. et al. The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res. 2013;41:10170-84
28. Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1-12
29. Zhou X, Wang R, Li X, Yu L, Hua D, Sun C. et al. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J Clin Invest. 2019;129:676-93
30. Maslon MM, Heras SR, Bellora N, Eyras E, Cáceres JF. The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife. 2014;3:e02028
31. Qi Z, Wang F, Yu G, Wang D, Yao Y, You M. et al. SRSF1 serves as a critical posttranscriptional regulator at the late stage of thymocyte development. Science Advances. 2021 7
32. Huang HF, Hembree WC. Spermatogenic response to vitamin A in vitamin A deficient rats. Biol Reprod. 1979;21:891-904
33. Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A. 2006;103:2474-9
34. Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, Page DC. Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc Natl Acad Sci U S A. 2015;112:E2347-56
35. Ishiguro KI, Matsuura K, Tani N, Takeda N, Usuki S, Yamane M. et al. MEIOSIN Directs the Switch from Mitosis to Meiosis in Mammalian Germ Cells. Dev Cell. 2020;52:429-45 e10
36. Sinha N, Whelan EC, Tobias JW, Avarbock M, Stefanovski D, Brinster RL. Roles of Stra8 and Tcerg1l in retinoic acid induced spermatogonial differentiation in mousedagger. Biol Reprod. 2021;105:503-18
37. Lin Y, Gill ME, Koubova J, Page DC. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Sci. 2008;322:1685-7
38. Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551-63
39. Paiano J, Wu W, Yamada S, Sciascia N, Callen E, Paola Cotrim A. et al. ATM and PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nat Commun. 2020;11:857
40. Crickard JB, Greene EC. The biochemistry of early meiotic recombination intermediates. Cell Cycle. 2018;17:2520-30
41. Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439-56
42. Lan WH, Lin SY, Kao CY, Chang WH, Yeh HY, Chang HY. et al. Rad51 facilitates filament assembly of meiosis-specific Dmc1 recombinase. Proc Natl Acad Sci U S A. 2020;117:11257-64
43. Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R, De Rooij DG. et al. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol. 2007;176:741-7
44. Blasiak J, Szczepańska J, Sobczuk A, Fila M, Pawlowska E. RIF1 Links Replication Timing with Fork Reactivation and DNA Double-Strand Break Repair. Int J Mol Sci. 2021 22
45. Lei WL, Han F, Hu MW, Liang QX, Meng TG, Zhou Q. et al. Protein phosphatase 6 is a key factor regulating spermatogenesis. Cell Death Differ. 2020;27:1952-64
46. Su R, Fan L-H, Cao C, Wang L, Du Z, Cai Z. et al. Global profiling of RNA-binding protein target sites by LACE-seq. Nat Cell Biol. 2021;23:664-75
47. Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS. et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996-1006
Author contact
![]() Corresponding authors: Department of Urology & Andrology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, #3 Qingchun East Road, Shangcheng District, Hangzhou, 310016, China. E-mail: sunfeisrrshedu.cn (Fei Sun); Guangdong and Shenzhen Key Laboratory of Reproductive Medicine and Genetics, The Center of Reproductive Medicine, Peking University Shenzhen Hospital, 1120 Lianhua Rd, Futian District, Shenzhen 518000, China. Tel/Fax: +8675583923333-5213; E-mail: qianweipingszcom (Wei-Ping Qian); CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, 1068 Xueyuan Avenue, Nanshan District, Shenzhen 518055, China. Tel/Fax: +86755 86392288; E-mail: cl.liuac.cn (Chenli Liu); Reproductive Medicine Center, Guangdong Second Provincial General Hospital, #466 Xin-Gang-Zhong-Lu, Haizhu District, Guangzhou 510317, China. Tel: +862089169839; E-mail: sunqyorg.cn (Qing-Yuan Sun).
Corresponding authors: Department of Urology & Andrology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, #3 Qingchun East Road, Shangcheng District, Hangzhou, 310016, China. E-mail: sunfeisrrshedu.cn (Fei Sun); Guangdong and Shenzhen Key Laboratory of Reproductive Medicine and Genetics, The Center of Reproductive Medicine, Peking University Shenzhen Hospital, 1120 Lianhua Rd, Futian District, Shenzhen 518000, China. Tel/Fax: +8675583923333-5213; E-mail: qianweipingszcom (Wei-Ping Qian); CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, 1068 Xueyuan Avenue, Nanshan District, Shenzhen 518055, China. Tel/Fax: +86755 86392288; E-mail: cl.liuac.cn (Chenli Liu); Reproductive Medicine Center, Guangdong Second Provincial General Hospital, #466 Xin-Gang-Zhong-Lu, Haizhu District, Guangzhou 510317, China. Tel: +862089169839; E-mail: sunqyorg.cn (Qing-Yuan Sun).

 Global reach, higher impact
Global reach, higher impact