10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2024; 20(5):1652-1668. doi:10.7150/ijbs.92988 This issue Cite
Review
The Burgeoning Significance of Liquid-Liquid Phase Separation in the Pathogenesis and Therapeutics of Cancers
1. Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University, Changsha 410011, China.
2. Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410078, China.
3. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha 410078, China.
4. Department of Pathology, the Second Xiangya Hospital, Central South University, Changsha 410011, China.
5. Hunan key laboratory of the research and development of novel pharmaceutical preparations, Changsha Medical University, Changsha 410219, China.
* These authors contributed equally to this work.
Received 2023-12-6; Accepted 2024-2-1; Published 2024-2-12
Abstract
Liquid-liquid phase separation (LLPS) is a physiological phenomenon that parallels the mixing of oil and water, giving rise to compartments with diverse physical properties. Biomolecular condensates, arising from LLPS, serve as critical regulators of gene expression and control, with a particular significance in the context of malignant tumors. Recent investigations have unveiled the intimate connection between LLPS and cancer, a nexus that profoundly impacts various facets of cancer progression, including DNA repair, transcriptional regulation, oncogene expression, and the formation of critical membraneless organelles within the cancer microenvironment. This review provides a comprehensive account of the evolution of LLPS from the molecular to the pathological level. We explore the mechanisms by through which biomolecular condensates govern diverse cellular physiological processes, encompassing gene expression, transcriptional control, signal transduction, and responses to environmental stressors. Furthermore, we concentrate on potential therapeutic targets and the development of small-molecule inhibitors associated with LLPS in prevalent clinical malignancies. Understanding the role of LLPS and its interplay within the tumor milieu holds promise for enhancing cancer treatment strategies, particularly in overcoming drug resistance challenges. These insights offer innovative perspectives and support for advancing cancer therapy.
Keywords: liquid-liquid phase separation, biomolecular condensates, cancer, super enhancer, disease
Introduction
In both human society and business, the effective collaboration of diverse groups often necessitates their organization into functional systems. A parallel concept exists within the realm of cellular biology, where human cells employ a process known as liquid-liquid phase separation (LLPS) to partition their constituents. This in vivo physiological mechanism is believed to underlie the formation of membrane-free structures within cells, such as nucleoli [1]. The resulting regionalization of cellular activities, often referred to as "biomolecular condensates," arises from the coalescence of various components of membraneless cellular structures via LLPS [2]. These condensates play key roles in numerous cellular functions, including the regulation of gene expression, control of nucleic acid transcription, and in vivo responses to environmental stimuli [2, 3]. This physiological phenomenon, LLPS, has gained considerable attention in recent research [1]. It is integral to our understanding of how cells compartmentalize and optimize their functions. Previous research has extensively investigated the formation of condensates in the context of amyotrophic lateral sclerosis [4]. As our comprehension of LLPS continues to evolve, pathological processes related to neurodegenerative diseases and certain cancers are also being viewed anew as manifestations of LLPS-driven processes [5, 6]. A profound grasp of these biomolecular condensates assumes paramount importance in the exploration of diverse biological phenomena and the formulation of therapeutic interventions.
This comprehensive review aims to elucidate the physiological and biochemical underpinnings of LLPS and biomolecular condensates, thereby facilitating a more profound comprehension of the intricate relationship between LLPS and cancer. Our focus centers on elucidating the mechanisms through which phase separation orchestrates the progression of select malignancies, offering valuable insights into the realm of targeted therapies and strategies to overcome drug resistance challenges. By delving into these intricacies, this review endeavors to provide researchers with a robust foundation for further inquiry and advancement in the field.
Development of LLPS
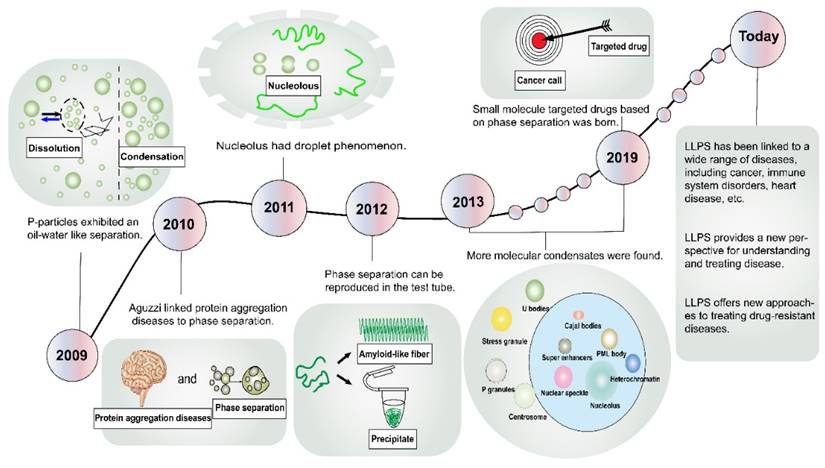
Development History
In the domain of physical chemistry, the term “phase separation” conventionally denotes the spontaneous demixing of two coexisting fluids [7]. Remarkably, the literature has harbored descriptions of membraneless organelles for over a century, yet within the biological community, few researchers have paid them much heed [8]. A critical turning point transpired in 2009 when Brangwynne et al. discerned “oil-water” separation-like behaviors within P granules in Caenorhabditis elegans (Figure 1) [9]. Subsequent to this, in 2010, Aguzzi et al. probed the pathogenesis of protein aggregation diseases, revealing an intimate linkage to phase separation dynamics [10]. In 2011, Brangwynne et al. extended their explorations to nucleosomes, uncovering analogous fluidic traits [11]. The year 2012 witnessed Li and Kato et al. harnessing molecular methodologies to simulate phase separation phenomena within the laboratory setting [12, 13]. Subsequent endeavors unearthed an array of molecular condensates, encompassing Cajal bodies [14], nuclear speckles [15], stress granules (SGs) [16], signaling puncta [17], promyelocytic leukemia (PML) bodies [18], super-enhancers (SEs) [19], heterochromatin [20], and more. These advancements in cell structure elucidation have precipitated innovative avenues for disease intervention. In a landmark development in 2019, Wheeler et al. identified lipoamide as the first small molecule capable of targeting the phase separation of the fused-in sarcoma (FUS) protein, thereby paving the way for the treatment of neurodegenerative disorders [21]. In recent years, the frontiers of LLPS research have expanded exponentially, culminating in a burgeoning body of literature linking LLPS to a spectrum of diseases, including cancer, infectious ailments, cardiovascular disorders, and neurological maladies [22-25].
Formation Mechanisms of LLPS
The fundamental physical principles that underpin the phenomenon of LLPS in polymers have gained considerable attention and have shed light on the understanding of LLPS in biological systems [26]. The concentration of molecules within a limited space involves an energy cost, yet numerous weak interactions can offset the entropic cost associated with LLPS [27].
LLPS is thought to be initiated by weak, multivalent interactions, often provided by intrinsically disordered regions and structured domains in proteins, which aid in specific molecular recognition [28]. For example, aromatic residues such as tyrosine, with their side chains containing delocalized π electrons, contribute to π-π stacking interactions that facilitate LLPS [29]. Notably, it has been observed that π-π interactions are also present in non-aromatic amino acids, as demonstrated in the fragile X mental retardation protein [29]. Positively charged amino acids, such as lysine and arginine, participate in cation-π interactions with electron-rich aromatic groups, further promoting LLPS [30]. Interestingly, these cation-π interactions are strong enough to overcome the repulsive effects typically seen in cation-anion interactions, thus aiding in phase separation among similarly charged molecules [31]. Charge-charge interactions are increasingly recognized as key contributors to LLPS. The aggregation of polymers with opposing charges, resulting in charge neutralization, can lead to the formation of droplets, as observed in mixtures of RNA and cationic peptides [32, 33]. Moreover, hydrophobic interactions and dipole-dipole interactions, which are independent of the amino acid composition of low complexity domains, have been suggested as mechanisms driving LLPS [26, 34].
History of the discovery and development of LLPS. Representative milestone findings promoting the development of LLPS are enumerated in the figure. LLPS, liquid-liquid phase separation; PML, promyelocytic leukemia.
The role of LLPS in the regulation of cellular responses is a highly intricate and precisely controlled process, involving not only weak intermolecular interactions within cells but also significantly influenced by intra- and extracellular environmental factors. Key environmental variables include temperature, ion concentrations, and pH levels, each capable of modulating the physicochemical properties of cellular molecules, thereby impacting the occurrence and characteristics of LLPS [35-37]. Additionally, in certain pathological states, such as some neurological disorders, the aberrant aggregation of proteins is closely associated with alterations in the LLPS process, suggesting a potential role of this mechanism in disease onset and progression [38]. The energy state of the cell, particularly ATP levels, also plays a crucial role in influencing the dynamics of LLPS [39]. Overall, LLPS as a mechanism for regulating cellular responses relies on the interplay of multiple protein interactions and a nuanced response to the cellular environment. This dynamic separation mechanism contributes to the organization and functional regulation of intracellular spaces, ensuring the optimal execution of cellular responses.
Physiological Functions of LLPS
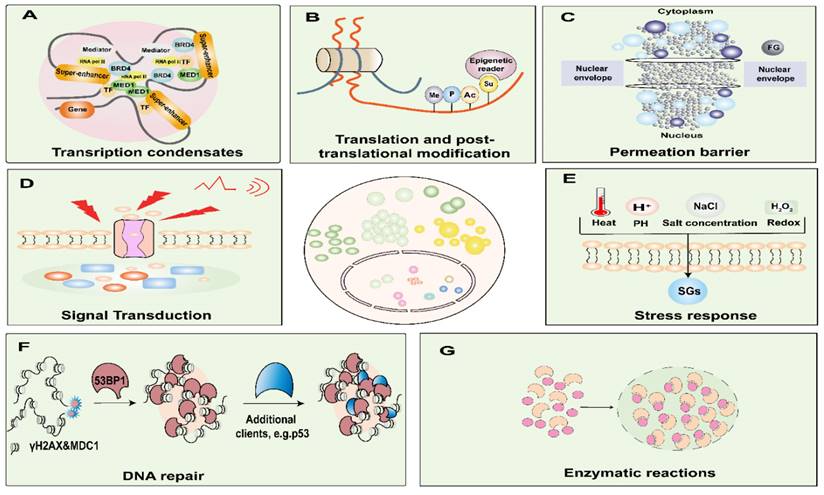
LLPS exerts intricate regulatory control over a spectrum of biological processes and functions, encompassing but not limited to gene expression, signal transduction, enzymatic responses, and stress response (Figure 2) [40]. It is imperative to acknowledge that the aberrant occurrence of LLPS, either temporally or spatially, has the potential to culminate in the formation of recalcitrant and irreversibly structured entities. Such untoward events can precipitate obstructive phenomena or pathological aggregations, thereby underpinning the onset of various maladies [41]. Consequently, a comprehensive comprehension of the molecular underpinnings of LLPS in the context of cancer assumes paramount significance, constituting a critical prerequisite for the development of efficacious therapeutic strategies.
Involvement in the function of the nucleus
LLPS in transcriptional regulation
Transcription factors (TFs) exert meticulous control over gene expression by virtue of their ability to bind to cis-acting regulatory elements, known as enhancers, and by orchestrating the recruitment of coactivators alongside RNA polymerase II (Pol II) [42]. Remarkably, SEs, comprised of several hundred clusters of enhancers, wield the authority to initiate transcription of vital genes that underpin cell identity, thanks to the phase-separating attributes residing within the intrinsically disordered regions of TFs and cofactors [43]. Emerging evidence has illuminated the key role of phase-separated condensates in steering transcriptional regulation [44]. Notably, TFs such as the Octamer-binding transcription factor 4 (OCT4) governing embryonic stem cell pluripotency and the yeast TF GCN4 have been shown to form phase-separated condensates alongside Mediator, an indispensable coactivator complex [45]. Sabari et al. have eloquently demonstrated that transcriptional coactivators enriched in SEs, namely, bromodomain-containing protein 4 (BRD4) and mediator complex subunit 1 (MED1), manifest as nuclear puncta, exhibiting liquid-like properties [43].
Intriguingly, Pol II exhibits colocalization with mediators within stable condensates while diligently executing the transcription of both messenger RNAs and various non-coding RNAs, all of which display hallmarks of phase-separated condensates (e.g., rapid fluorescence recovery post-photobleaching and susceptibility to 1,6-hexanediol, an inhibitor of LLPS) [46, 47]. Furthermore, Pol II initiates cluster formation by interacting with the intrinsically disordered carboxy-terminal domain and activators, and the subsequent phosphorylation of this domain leads to its release from the clusters, thereby triggering the transcription of the target gene [48]. Recent investigations have unveiled that biomolecular condensation augments the recruitment of the negative elongation factor to promoters and activates the positive transcription elongation factor b, thereby facilitating Pol II elongation [49, 50]. This orchestrated process orchestrates the transition from promoter-proximal pausing to transcription elongation [49, 50]. Interestingly, the products of transcription, i.e., RNAs, are not mere passive spectators but actively partake in influencing the formation and characteristics of condensates through a feedback mechanism. Henninger et al. have illuminated this intricate interplay by revealing that transcriptional regulation incorporates a feedback loop wherein low levels of RNA serve as promoters of transcriptional condensate formation, whereas elevated RNA levels can dismantle transcription condensates, consequently downregulating transcription [51].
LLPS in DNA damage
DNA damage possesses the potential to instigate genomic instability, a prominent hallmark of cancerous cells. One of the initial cellular responses to DNA damage entails the enzymatic synthesis of poly(ADP-ribose) (PAR) polymers, orchestrated by the enzymes of PAR polymerase 1 [52]. The cellular rejoinder to DNA damage is notably characterized by the escalation of PAR levels, driven by the hyperactivation of PAR polymerase enzymes, which are proficient at discerning DNA breaks within the chromatin structure [53]. Consequently, this instigates the swift accumulation of proteins bearing low-complexity sequence domains [53].
Functions of biomolecular condensates. Biomolecular condensates are involved in nuclear functions and are critical for DNA damage, nuclear translocation, enzymatic reactions, amplification, transcription, translation, and post-translational modification processes. Condensates are involved in signal transduction, stress sensing, and transport processes. A. Model of a phases-separated complex at gene regulatory elements. At the super-enhancer locus, transcriptional regulators with extensive interactions, including TFs, BRD4, MED1, and RNA pol II, are enriched to form a phase-separated condensate, which is separated from others. B. Post-translational modification processes such as phosphorylation, acetylation, ubiquitination, and SUMOylation. C. Among the FG-nucleoporins form the central pore of the nuclear pore complexes, which governs the nucleocytoplasmic transport through the pores. D. Condensates are involved in signal transduction. E. Condensates are involved in stress response. F. After DNA damage sites cause the formation of γH2AX and recruitment of MDC1 for nucleation, 53BP1 accumulates and phase separates, and P53 acts as a scaffold for client molecules, interacting transiently with 53BP1, where they find an environment permissive for their activation. G. Phase separation significantly accelerates the efficiency of multienzyme biocatalysis. BRD4, bromodomain-containing protein; RNA Pol II, RNA polymerase II; MED1, mediator complex subunit 1; TF, transcription factors. Me, methylation; P, phosphorylation; Ac, acetylation; Su, and small ubiquitin-like modifier; FG, Phenylalanine-glycine-rich; SGs, stress granules; 53PB1, p53 binding protein 1; γH2AX, phosphorylated histone H2AX; MDC1, Mediator of DNA damage checkpoint protein 1.
Remarkably, entities such as FUS/translocated in sarcoma, Ewing sarcoma (EWS), and TATA-box binding protein associated factor 15 emerge as prominent facilitators of phase separation, thus contributing significantly to this intricate landscape [54]. The phenomenon of FUS-driven LLPS demonstrates remarkable efficacy in recruiting and sequestering splicing factors at sites of DNA damage [55]. Furthermore, the phase separation of tumor protein p53 binding protein 1 (TP53BP1), a critical tumor suppressor protein intricately involved in orchestrating the balance between cell division and cell cycle arrest, imparts profound influence upon the fluid-like behavior governing DNA repair processes [56, 57].
Another compelling study underscores that the Rad52 DNA repair protein undergoes accumulation within distinct droplets, effectively collaborating with DNA damage-inducible intranuclear microtubule filaments. This collaboration enhances the aggregation of DNA damage sites, thereby contributing to the preservation of genomic stability [58]. Additionally, the activity of cytoplasmic nucleic acid exonuclease three prime repair exonuclease 1 (EXO1) experiences modulation through the process of cyclic GMP-AMP synthase-driven phase separation, culminating in the formation of molecular condensates that effectively curtail DNA deterioration [59]. Thus, a more profound understanding of the intricate mechanisms underpinning how LLPS regulates DNA repair holds substantial promise in unveiling novel therapeutic opportunities.
LLPS in chromatin organization
In most eukaryotes, a significant portion of the genome is ensconced within heterochromatin, a multifaceted scaffold pivotal for nuclear architecture, DNA repair, genome stability, and the quelling of transposons and gene expression [60, 61]. It is posited that heterochromatin's gene-silencing activity arises, in part, from the capability of heterochromatin protein 1 (HP1), the “reader” of histone H3 lysine 9 methylation (H3K9me), to form complexes through increased multivalent interactions with H3K9me-modified chromatin [20]. Considering heterochromatin organization from the perspective of phase separation provides a foundation for uncovering genomic conformational disturbances and their links to diseases. For example, methyl-CpG-binding protein 2 (MECP2), a chromatin organizer governing gene expression, competes with linker histone H1 to form distinct chromatin condensates in vitro and heterochromatin foci in vivo [62].
LLPS is intricately linked to chromatin modifications which can modulate phase-separation properties, thereby influencing chromatin organization and function. Post-translational modifications (PTMs) are well-established as pivotal regulators of biomolecular condensate formation [63].
Phosphorylation stands out as a highly characterized PTM that exerts plays a central role in signal transduction pathways [64]. For instance, the activation of dual-specificity tyrosine-phosphorylation-regulated kinase 3 (DYRK3) facilitates the dissolution of SGs, releasing the mechanistic target of rapamycin complex 1 (mTOR1) for signaling and enhancing its activity by direct phosphorylation of the mTOR1 inhibitor 40-kDa proline-rich Akt substrate (PRAS40) [65]. Intriguingly, phosphorylation promotes the pathological aggregation of microtubule-associated protein Tau by fostering electrostatic interactions conducive to phase separation, shedding light on the connection between LLPS and neurodegeneration in tauopathies [66].
Arginine methylation, a common PTM with broad implications across various cellular processes (e.g., chromatin remodeling, RNA processing, and cell signaling), directly interferes with phase separation by disrupting cation-π interactions, as exemplified in FUS [67].
Furthermore, small ubiquitin-like modifier (SUMOylation) represents an additional PTM that modifies cohorts of functionally related proteins through covalent attachment to lysine residues in a multitude of proteins [68]. PML proteins, for instance, undergo extensive SUMOylation, subsequently recruiting client proteins into PML nuclear bodies through SUMO-interacting motifs/SUMO interactions [69].
LLPS in the permeation barrier
Extensive evidence suggests that multivalent protein assemblies are crucial for phase separation. Li et al. suggested a mechanism through which multivalent interactions could result in sharp transitions between physically and functionally distinct states and connect disparate length scales in the cell. This process potentially contributes to forming the structure and function of the cellular body [12]. The primary pathway for nuclear-cytoplasmic communication is facilitated by nuclear pore complexes. These complexes are comprised of closely packed, inherently disordered nucleoporins rich in phenylalanine and glycine. They create a selective barrier that regulates permeability [70]. These phenylalanine-glycine-rich nucleoporins form the core pore of the nuclear pore complexes and are instrumental in managing nucleocytoplasmic transport [71, 72]. Their unique sequences allow them to undergo LLPS, forming hydrogel-like structures crucial for the regulation of cargo transport into and out of the nucleus.
Involvement in enzymatic reactions
Evidence suggests that the net effect of crowding because of complex coacervation of multivalent macromolecules is to increase reaction rates. For example, LLPS may achieve precise control over the rate of enzymatic reactions by finely modulating the local concentration of reactants [73]. Furthermore, LLPS compartmentalizes the mixed solution to produce a condensate with a particular material concentration., LLPS facilitates the formation of specific affinity interaction domains within cells, involving enzymes, substrates, and other biomolecules. This phenomenon contributes to the organized assembly and regulation of enzymatic reactions [74].
Involvement in signal transduction
Intracellular signaling networks are essential for regulating cellular behavior and homeostasis. Recent research indicates that biomolecular condensates enhance the presence and control the function of signaling molecules through multivalent interactions [75, 76]. These interactions, common in membrane-associated signaling pathways, suggest a broader applicability for such regulatory mechanisms [77].
LLPS is instrumental in the assembly of membrane receptors and associated signaling entities, evident in processes like T-cell receptor (TCR) signaling in immune cells and the activation of cyclic GMP-AMP synthase (cGAS) and stimulator of interferon genes [17, 78, 79]. TCR phosphorylation, for instance, leads to the spontaneous formation of liquid-like clusters of downstream signaling proteins like GRB2, phospholipase C, and others, enhancing actin polymerization and contributing to TCR signaling [17]. In the context of innate immunity, cGAS activation by cytosolic DNA triggers a phase transition, facilitating immune signaling [78]. In the context of innate immunity, cGAS activation by cytosolic DNA triggers a phase transition, facilitating immune signaling. This activation results in a cascade of events, including the production of type I interferons and proinflammatory cytokines [80]. Additionally, recent findings show that stimulator of interferon genes forms structured condensates, attracting TANK-binding kinase 1 (TBK1) and regulating innate immune responses [79]. These results demonstrated that protein phase separation can create a distinct physical and biochemical compartment that facilitates signaling.
Involvement in stress response to external stimuli
Cellular adaptation mechanisms to diverse physicochemical environmental conditions (like temperature, pH, salinity, redox states) involve the dynamic formation or dissociation of biomolecular condensates under stress. In response to abrupt environmental changes, these condensates can rapidly assemble or disperse.
During heat shock, cells compartmentalize protein aggregates, impacting mRNA expression under stress [81]. Adverse conditions can also trigger the formation of SGs, which safeguard cells against various stressors [82]. For instance, under heat stress, SGs containing mRNA and proteins influence mRNA localization, translation, and degradation, as well as regulate signaling pathways and antiviral responses [35]. This dynamic aggregation and disaggregation of proteins is an adaptive, self-regulating process that may enhance organismal resilience during stress.
Zlotorynski et al. demonstrated that changes in pH can influence phase separation. The yeast prion protein Sup35 forms liquid-like condensates at pH 6, which solidify into a gel-like state and dissolve upon pH increase, aiding in translation resumption and enhancing cellular fitness post-stress [36, 83].
Furthermore, salt concentration and charge variations can induce cellular stress. Cummings et al. observed that green fluorescent protein undergoes LLPS more readily at higher salt concentrations, forming liquid condensates from solid precipitates [37]. Also, macromolecular crowding under hyperosmotic stress leads to the formation of liquid-demixing condensates of apoptosis signal-regulating kinase 3 (ASK3), influencing its activity [84]. PolyA binding protein-binding protein 1 (Pbp1), a regulator of target of rapamycin complex 1 (TORC1) signaling and autophagy, interacts with TORC1 during respiratory growth, triggering TORC1-mediated autophagy. This process is crucial for eliminating SGs and damaged organelles, thus protecting cells from oxidative stress due to redox imbalances [85, 86].
Transcription factors activated by LLPS and possible potential targets.
| Tumor model | Complex | Cellular mechanisms (Transcription factors) | Reference |
|---|---|---|---|
| Prostate Cancer | SPOP | SPOP mutations enhance autophagy and NFE2L2 activation by directly modulating SQSTM1 LLPS and ubiquitination. | [96] |
| MED1 | MED1 is required for androgen receptor-mediated transcription. | [97] | |
| OCT4 | OCT4 is recruited to specific genomic loci to activate other TFs. | [98] | |
| AR-SV | AR-SVs promote tumor growth by orchestrating transcriptional reprogramming. | [99] | |
| LSD1 | LSD1 function as a transcriptional corepressor and coactivator. | [100] | |
| BRD4 | BRD4 participates in super-enhancers organization and oncogenes expression regulation. | [175] | |
| rIGSRNA | Tumor growth repression. | [176] | |
| Osteosarcoma | MYC | MYC regulates the SEs containing genes and mediates the transcriptional amplification of its target genes. | [102] |
| HOXB8 and FOSL1 | HOXB8 and FOSL1 form condensates to regulate chromatin accessibility and oncogenic transcription. | [103] | |
| Multiple Myeloma | SRC-3 | SRC-3 LLPS enhanced recruitment of NSD2 to the condensate. | [106] |
| BRD4 | BRD4 participates in super-enhancers organization and oncogenes expression regulation. | [107] | |
| Lung Cancer | EML4-ALK | EML4-ALK LLPS activate STAT3 phosphorylation and downstream pathways of tumor transformation. | [111] |
| MNX1-AS1 | MNX1-AS1 binds to IGF2BP1 and drives its phase separation to maintain mRNA stability of c-Myc and E2F1 | [112] | |
| EZH2 | Myristoylation-mediated LLPS of EZH2 activates STAT3 signaling. | [113] | |
| YAP/TAZ | YAP/TAZ activates target genes and is directly involved in the control of S-phase entry and mitosis. | [114] | |
| MELTF-AS1 | MELTF-AS1 regulates tumorigenesis by driving phase separation of YBX1 to activate ANXA8. | [115] | |
| USP42 | USP42 controls phase separation of spliceosome component PLRG1. | [116] | |
| Breast Cancer | NUDT5 | NUDT5 affects the phase separation process by regulating ATP synthesis in the nucleus of breast cancer cells. | [177] |
| Cationic polymers | Blocks the translation of TGFβ1 mRNA in tumor cells by inducing RNA LLPS. | [122] | |
| Par3 | Par3 acts as a mechanical mediator of breast cancer aggressiveness. | [123] | |
| TAZ-NANOG | TAZ-NANOG LLPS promotes the transcription of SOX2 and OCT4. | [124] | |
| Pancreatic Cancer | SRPK2 | SRPK2 mediates SG formation through overactivation driven by the IGF1/PI3K/mTOR/S6K1 pathway. | [127] |
| KMT2D | KMT2S low-complexity domains creates an LLPS environment that helps stabilize WDR5 protein and complex formation. | [129] | |
| Leukemia | YY1 | YY1 binds to HDAC1 / 3 and regulates the high expression of METTL3. | [130] |
| YTHDC1 | YTHDC1 binds to m6A-mRNA to form nYACs, protecting the stability of its target gene mRNA from degradation. | [132] | |
| PML/RARα | Deneddylation of PML/RARα restores its phase separation process to reconstruct functional nuclear bodies and activates RARα signaling | [133] | |
| NUP98 | NUP98 results in the transcriptional activation of leukemogenic genes. | [135] | |
| Myxoid liposarcoma | FUS-CHOP | FUS-CHOP recruits BRD4 to carcinogenic condensates. | [141] |
| Ewing sarcoma | EWS-FLI1 | EWS-FLI1 binds to the protein chaperone network and regulates the transcription gene targets. | [139] |
| Synovial sarcoma | SS18-SSX | SS18 enriches BRG1 into the condensate through the interaction between SS1 and BRG18. | [143] |
| Glioblastoma | BYSL | BYSL is involved in the malignant progression of glioblastoma through the GSK-3β/β-linked protein signaling pathway. | [178] |
| NONO | NONO depletion reduces nuclear TAZ LLPS, while ectopic NONO expression promotes the LLPS. | [145] | |
| Ovarian cancer | BRD4 | BRD4 participates in super-enhancers organization and oncogenes expression regulation. | [152] |
| Esophageal Cancer | ILF3 | ILF3 encodes two double-stranded RNA -binding proteins to regulate gene transcription and protein translation. | [155] |
| BRD4 | BRD4 participates in super-enhancers organization and oncogenes expression regulation. | [179] | |
| Colorectal cancer | NONO | NONO LLPS enhances DNA damage repair by accelerating nuclear EGFR-induced DNA-PK activation. | [157] |
| BRD4 | BRD4 participates in super-enhancers organization and oncogenes expression regulation | [158] | |
| NOP53 | NOP53 inhibits p53 activation and enhances radio-resistance in colorectal cancer cells | [159] |
LLPS contributes to the development of cancer
LLPS has been implicated in the epigenetic dysregulation that contributes to carcinogenesis and tumor progression [87-89]. Recent studies suggest that phase-separated condensates may play a role in modulating drug distribution and concentration, thereby affecting drug efficacy and resistance in cancer cells [90]. Table 1 presents an overview of potential targets and emerging strategies for treating malignancies linked to LLPS, offering a theoretical framework for the identification of effective target molecules.
LLPS in prostate cancer
Prostate cancer, a prevalent malignancy in men worldwide, is effectively treated in its early stages with surgery or radiation, often combined with androgen-deprivation therapy in more advanced cases [91, 92]. However, the response to this treatment varies, commonly leading to castration-resistant prostate cancer (CRPC), a lethal stage of the disease [93]. Addressing the resistance to antiandrogen therapies in CRPC patients is a critical aspect of ongoing clinical research [94].
Recent studies have identified the tumor suppressor speckle-type POZ protein (SPOP) as frequently mutated in solid tumors, particularly prostate cancer. These mutations disrupt phase separation and co-localization in membraneless organelles [95]. Notably, prostate cancer-associated SPOP mutations have been found to enhance autophagy and activate the NFE2L2/NRF2 pathway, crucial in managing oxidative stress, by directly modulating sequestosome1 (SQSTM1) LLPS and ubiquitination [96]. This finding supports the possibility that this oncogenic pathway may guide targeted therapy toward SPOP-mutated cancers. A recent study showed that androgen receptor (AR) forms dynamic AR-rich, liquid-like foci with coactivator MED1 to SEs in cellular prostate cancer models, promoting an oncogenic transcriptional program [97]. This discovery suggests that targeting the SPOP mutation could be a promising therapeutic approach. In cellular models of prostate cancer, the AR forms dynamic, liquid-like foci with MED1 in SEs, driving an oncogenic transcriptional program [96]. OCT4 recruitment to specific genomic loci activates other TFs at SEs, indicating that disrupting these interactions could be a novel therapeutic strategy for advanced tumors [98].
Xie et al. discovered that the AR LLPS inhibitor ET516 potentially disrupts the AR feedback mechanism. This compound acts as a precursor to increased resistance by specifically targeting AR LLPS, countering resistance mechanisms arising from AR mutations or splicing events [94]. Additionally, UT-143, an AR-selective irreversible covalent antagonist, was found to hinder LLPS formation and mutagenesis, leading to chromatin condensation and disassembly of the AR splice variant 7 (AR-V7) interactome, resulting in a transcriptionally inactive complex [99]. Lastly, research has shown that lysine-specific demethylase 1 (LSD1), an AR coactivator in prostate cancer, drives disease progression through SE-mediated oncogenic programs, which could be countered using LSD1 inhibitors to suppress CRPC growth [100].
LLPS in osteosarcoma
Osteosarcoma, a highly aggressive primary bone malignancy prevalent in pediatric and adolescent populations, is frequently associated with unfavorable prognosis and low survival rates, particularly in cases involving metastasis and recurrent disease [101]. Substantial evidence from prior investigations underscores the crucial role of the MYC-driven SE signaling pathway in osteosarcoma tumorigenesis [102]. The administration of SE inhibitors, such as THZ1 and JQ1, has demonstrated marked success in abrogating the proliferative, migratory, and invasive capacities of osteosarcoma cells. Thus, the therapeutic targeting of the MYC/SE axis emerges as a promising avenue for the management of osteosarcoma patients [102].
In a study conducted by Lu et al., it was elucidated that core regulatory circuit components, namely homeobox B8 (HOXB8) and fos-like antigen 1 (FOSL1), residing proximal to the SE locus in osteosarcoma, exhibit the ability to form dense and dynamically phase-separated droplets in vitro and liquid-like puncta within cell nuclei. These observations suggest a potential role in supporting SE-driven transcriptional processes [103]. Furthermore, the utilization of the histone H3 lysine 27 (H3K27) demethylase inhibitor GSK‐J4 disrupts the phase separation dynamics of core regulatory circuit factors. This disruption results in reduced chromatin accessibility within SE regions and the subsequent inhibition of aberrant oncogenic transcriptional programs. As such, GSK‐J4 emerges as a prospective therapeutic candidate for patients afflicted with metastatic and chemoresistant osteosarcoma, offering promising prospects for improving clinical outcomes [103].
LLPS in multiple myeloma
Multiple myeloma progression is linked to genetic and epigenetic anomalies [104]. Recent studies highlight the impact of altered epigenetic mechanisms in these cancer cells, leading to immature, drug-resistant phenotypes [105]. Notably, steroid receptor coactivator-3 (SRC-3), characterized by its LLPS properties, is integral to drug resistance in multiple myeloma [106]. Research indicates that SRC-3 LLPS enhances histone methyltransferase nuclear receptor binding SET domain protein 2 (NSD2) recruitment, influencing histone H3 at lysine 36 (H3K36me) and apoptosis, thereby facilitating drug resistance. Furthermore, the SRC-3 inhibitor SI-2 shows potential in sensitizing cells to bortezomib by disrupting NSD2 [106]. Regulating gene expression to decrease LLPS may increase drug efficacy, and targeting SRC-3 could reduce bortezomib resistance. Additionally, small-molecule bromodomain inhibitors like JQ1 in xenograft multiple myeloma models demonstrate significant antiproliferative effects, indicating bromodomain and extra-terminal domain (BET) inhibition as a promising cancer treatment strategy [107].
LLPS in lung cancer
Lung cancer, notably with a high incidence and mortality rate, is a leading cause of cancer deaths [108]. Despite advances, resistance to therapy remains a challenge in advanced non-small cell lung cancer (NSCLC) [109]. A significant contributor to NSCLC oncogenesis is the mesenchymal lymphoma kinase (anaplastic lymphoma receptor tyrosine kinase [ALK]) fusion mutations, often associated with accelerated cancer progression [110]. Qin et al. identified that the echinoderm microtubule-associated protein-like 4/ALK (EML/ALK) variant undergoes phase separation in tumors, enhancing STAT3 phosphorylation and tumor transformation [111]. Additionally, long non-coding RNA (lncRNA) MNX1-AS1, upregulated by gene amplification and c-Myc, stabilizes TF1 mRNA in association with insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) by LLPS, exhibiting potent anti-tumor effects in xenograft models, thus presenting a potential biomarker and therapeutic target [112]. Zhang et al. observed that myristoylation induces phase-separated liquid droplets of enhancer of zeste homolog 2 (EZH2) in lung cancer cells, compartmentalizing and activating STAT3, and promoting cell growth [113]. Targeting EZH2 myristoylation emerges as a novel lung cancer treatment strategy.
Further, steroid receptor coactivator-1 (SRC-1) forms phase-separated condensates with Yes-associated protein/TEA domain, enhancing gene expression. This process is disruptible by elvitegravir, an anti-HIV drug, inhibiting YAP's oncogenic activity [114]. Lu et al. demonstrated that lncRNA MELTF-AS1 binds to and induces phase separation of Y-box binding protein 1 (YBX1), activating ANXA8 transcription and NSCLC tumorigenesis [115]. Moreover, ubiquitin-specific peptidase 42 (USP42) regulates phase separation of spliceosome component PLRG1, facilitating mRNA splicing and tumorigenesis [116]. Treatment with BRD4-targeting JQ1, combined with chemoradiotherapy and anti-PD-L1 antibody, enhances anti-tumor immunity in NSCLC [117].
LLPS in breast cancer
Breast cancer ranks as the second most prevalent cancer among women globally, with significant mortality implications. Despite advancements in screening and treatment, high rates of invasion and metastasis contribute to low survival rates [118]. Research shows that tumor suppressor SPOP mutants, by impairing phase separation, can induce breast cancer through mutation-caused mislocalization [95]. In the cellular context, ATP, acting as an estrogen cofactor, is involved in the dynamic phase separation process, influencing the pathophysiology of breast cancer [119, 120]. Nudix hydrolase 5 (NUDT5), crucial for ATP synthesis in breast cancer cell nuclei, is targeted by TH5427, a specific inhibitor that disrupts nuclear ATP generation, impacting hormone-induced chromatin remodeling and progestin-dependent gene regulation [121]. Studies using a mouse breast cancer model have highlighted that cationic polymers, like cDex and DETA-Dex, can impede RNA LLPS, thereby inhibiting TGFβ1 mRNA translation in tumor cells [122]. Moreover, Hu et al. identified that the cell-polarity protein Par3 facilitates the phase separation of junctional adhesion molecule-A during breast cancer metastasis, altering osmotic pressure and mechanical properties [123]. Inhibiting atypical protein kinase C (aPKC) can reduce Par3's mechanical transmission, thus hindering breast cancer invasion, migration, and potentially improving survival [123]. Additionally, drug resistance in breast cancer is closely linked to cancer stem cells. The phase separation of TAZ-Nanog homeobox enhances the transcription of SOX2 and OCT4. Targeting Nanog homeobox or TAZ could therefore elevate chemosensitivity by diminishing cancer stem cell prevalence in breast cancer cells [124].
LLPS in pancreatic cancer
Pancreatic cancer, characterized by its aggressive nature and ranking foremost among asymptomatic malignancies, exhibits a dismal prognosis, with significant survival improvement only achievable through resection achieving macroscopic tumor clearance [125]. Consequently, precision medicine emerges as crucial for enhancing therapeutic outcomes and prognoses for this disease. Notably, 1,6-hexanediol, by disrupting protein-mediated abnormal LLPS, substantially reduces MYC expression, effectively curtailing pancreatic cancer growth [126]. The serine/arginine-rich protein kinase 2 (SRPK2), identified as a key regulator of stress granule (SG) formation in obesity-associated pancreatic ductal adenocarcinoma, modulates SG assembly through the hyperactivation of the IGF1/PI3K/mTOR1/S6K1 pathway. Furthermore, SRPK2, as a substrate of S6K1, is implicated in mRNA stability, splicing, and lipid metabolism [127, 128]. The S6K1 inhibitor PF4708671 shows preferential efficacy in diminishing SGs and inhibiting growth in obesity-related pancreatic cancer [127]. Additionally, the low-complexity sequence domains of histone lysine (K)-specific methyltransferase 2D (KMT2D) facilitate a LLPS environment, which is instrumental in regulating histone monomethylation and transcription, thus promoting pancreatic cancer progression [129]. Therefore, targeting the LLPS microenvironment emerges as a promising strategy in pancreatic cancer therapy.
LLPS in leukemia
Leukemia is a malignant hematologic cancer with a long history of targeted therapy development. TF Yin-Yang 1 (YY1) is a new target for treating acute myeloid leukemia. It binds to histone deacetylase 1/3 (HDAC1/3) and controls the high production of methyltransferase-like protein 3 (METTL3) through moderate LLPS [130]. Treatment with an HDAC inhibitor treatment significantly reduces the binding of YY1 to HDAC 1/3, resulting in an excessive LLPS state, thereby reducing the expression of METTL3 and the proliferation of acute myeloid leukemia cells [130]. RNA-binding protein YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) is highly expressed in many cases of acute myeloid leukemia and is a new potential target for treating this disease [131]. Cheng et al. reported that YTHDC1 binds m6A-mRNA by phase separation to form nuclear YTHDC1-m6A condensates, which prevent the degradation of target gene mRNA, thereby allowing acute myelocytic leukemia cells to survive and maintain their undifferentiated state [132].
A study revealed that deneddylation of PML /retinoic acid receptor α restores phase separation to reconstitute functional PML nuclear bodies and activates retinoic acid receptor α, the eradication of acute promyelocytic leukemia. Hence, PML proteins in leukemia are promising targets [133, 134]. In addition, it was found that nucleoporin 98 (NUP98) fusion oncoproteins drive LLPS with the help of homo- and hetero-typic interactions, thereby forming aberrant nuclear loci and affecting gene expression in leukemia [135]. Therefore, therapeutic modulation of aberrant LLPS using fusion oncoproteins is a compelling prospect for cancer treatment. In T-cell acute lymphoblastic leukemia, heterozygous somatic mutations of T-cell acute lymphocytic leukemia protein 1 (TAL1) oncogene are acquired, which introduce binding motifs for the TF myeloblastosis oncogene and creates a SE upstream of the TAL1 oncogene. In this way, the aberrantly formed SE drives monoallelic TAL1 expression, promoting the progression of leukemia [136].
Ubiquitously transcribed tetratricopeptide repeat, X chromosome (UTX) (a critical tumor suppressor) regulates genome-wide histone modifications and high-order chromatin interactions in a condensation-dependent manner [137]. UTX encodes the H3K27 demethylase; thus, GSK-J4 is a novel, highly promising epigenetic therapy option against TAL1-positive T-cell acute lymphoblastic leukaemia [138].
LLPS in sarcoma
The therapeutic targeting of translocation-driven sarcomas presents significant challenges. Recent focus on fusion proteins has opened avenues for identifying novel therapeutic targets, exemplified by FUS-C/EBP HOmologous protein (FUS-CHOP) in myxoid liposarcoma and EWS RNA-binding protein 1/friend leukemia virus integration 1 (EWSR1/FLI1) in Ewing's sarcoma [139, 140]. FUS-CHOP is observed to form phase-separated condensates that co-localize with BRD4 within SEs, impacting chromatin remodeling and transcription, thus offering a potential therapeutic avenue for myxoid liposarcoma [141]. The prion-like domain in EWS-FLI1 is associated with aberrant phase separation events, leading to the activation of numerous gene targets in Ewing's sarcoma [139]. Hsp104, a hexameric AAA+ protein disaggregase from yeast, demonstrates efficacy in mitigating the toxicity and aggregation of FUS-CHOP and EWS-FLI, suggesting its potential utility in counteracting the deleterious effects of abnormal fusion proteins in sarcomas [142]. Additionally, the oncoprotein SS18-SSX, a distinctive marker of synovial sarcoma, has been scrutinized by Chang et al., who identified that the self-association of its intrinsically disordered QPGY domain leads to LLPS, contributing to its oncogenic activity in synovial sarcoma [143]. Consequently, targeting phase-separated structures of SS18-SSX1 with small molecules emerges as a promising therapeutic strategy for synovial sarcoma.
LLPS in glioblastoma
Glioblastoma, the most lethal form of primary brain cancer, presents a formidable clinical challenge. Current treatment strategies, encompassing surgery, chemotherapy, and adjuvant radiotherapy, yield a median survival duration of merely 18 months for afflicted patients [144]. Research led by Wei et al. has highlighted the critical role of non-POU-domain-containing octamer-binding protein (NONO), a TAZ‐binding protein, in facilitating the TAZ-driven oncogenic transcriptional mechanism and promoting TAZ-mediated LLPS, thereby driving the transcriptional program of glioblastoma [145]. The suppression of NONO expression has been shown to inhibit TAZ-driven tumorigenesis, suggesting the modulation of NONO as a novel therapeutic approach against TAZ-driven glioblastoma [145]. Furthermore, Gene Set Enrichment Analysis has identified a strong association of fatty acid-binding protein 5 (FABP5), implicated in LLPS, with key signaling pathways in glioblastoma, including nuclear factor-kappa B (NF-κB) signaling [146, 147]. A negative correlation between BRD4 expression and overall survival in glioma patients has been observed, with the novel BRD4 inhibitor GNE987 targeting c-Myc expression and modulating transcription through BRD4 downregulation [148]. Additionally, recent studies have unveiled that the nucleoprotein AHNAK impedes TP53BP1 oligomerization and phase separation, thereby curtailing glioma cell proliferation and inducing apoptosis [56, 149].
LLPS in ovarian cancer
Ovarian cancer remains one of the leading causes of death in women worldwide, although the prognosis of early-stage disease is good [150]. Research indicates that genes related to LLPS are aberrantly expressed in epithelial ovarian cancer, influencing the cell cycle and DNA replication, and thus presenting as potential prognostic indicators for this malignancy [151]. Despite the theoretical efficacy of BET bromodomain inhibitors in the treatment of epithelial ovarian cancer, initial clinical trials have yielded suboptimal results [152]. Zhang et al. have suggested a novel approach, wherein the combined use of phase separation of BRD4 droplets and aurora kinase inhibitors effectively counteracts the antagonistic impact of BET bromodomain inhibitors, thereby targeting and eradicating JQ1-resistant ovarian cancer cells [153].
LLPS in esophageal cancer
Esophageal squamous cell carcinoma, classified among the deadliest forms of human cancer, is notorious for its treatment resistance and high recurrence rate, with a notable absence of approved targeted therapies currently available [154]. In this context, the macrophage-associated lncRNA (MALR) is significantly overexpressed in esophageal squamous cell carcinoma. It is hypothesized that MALR interacts with the double-stranded RNA-binding domain 3 of interleukin-enhancing factor 1 (ILF3), consequently inducing ILF3 LLPS and activating hypoxia-inducible factor 1α (HIF1α) signaling pathways [155]. Therefore, the MALR-ILF3-HIF1α axis presents itself as a potential therapeutic target in the treatment of esophageal squamous cell carcinoma.
LLPS in colorectal cancer
Colorectal cancer ranks as one of the most fatal diseases globally, with patients frequently facing high recurrence, metastasis, diagnostic challenges, and poor prognostic outcomes [156]. In colorectal cancer tissues, the expression of NONO is notably elevated. NONO-mediated LLPS recruits DNA-dependent protein kinase and nuclear epidermal growth factor receptor, enhancing their phosphorylation and consequently accelerating DNA repair in tumor cells [157]. Kondo et al. identified a novel compound, aminocyclopropenone 1n (ACP-1n), that impedes SE-driven MYC expression by inhibiting BRD4 functionality in the nucleus of colorectal cancer HCT4 cells in vitro, thereby suppressing oncogenic transcription [158]. Additionally, the nucleolar protein53 (Nop53) is overexpressed in colorectal cancer and correlates with poor prognosis. Nop53 forms cohesions in the nucleolus and exhibits sensitivity to 1,6-hexanediol treatment [159]. It inhibits p53 activation and enhances radioresistance in colorectal cancer cells, making it a potential target for increasing radiosensitivity in cancer patients [159]. Moreover, DExD-box helicase 21 (DDX21), a notable RNA-binding protein, is highly expressed in colorectal cancer. The phase-separated condensates of DDX21 target and activate minichromosome maintenance complex component 5 (MCM5), facilitating colorectal cancer cell migration and invasion, and triggering the activation of the Epithelial-mesenchymal transition pathway, which is crucial in the metastatic regulatory circuitry [160].
LLPS in Wilms tumor
Nephroblastoma is one of the most common solid tumors in children; particularly, Wilms tumor is the most common type of pediatric kidney tumors [161]. Within this context, AF9 and ENL, both YEATS domain-containing proteins and readers of histone acetylation, play pivotal roles in chromatin modification and transcription regulation [162, 163]. Furthermore, mutations in the YEATS domain of ENL have been identified as functionally significant in Wilms tumor. These mutations augment phase separation and transcription, leading to an aberrant gene expression profile [164]. This emerging understanding offers novel insights into potential mechanism-based strategies to mitigate the oncogenic impacts of ENL mutations.
LLPS in liver cancer
The liver ranks as the sixth most common primary cancer site in humans, with liver cancer patients often facing low survival rates unless the disease is detected early [165]. A circular RNA known as VAMP3 has been identified to facilitate the phase separation of cytoplasmic activation/proliferation-associated protein 1 (CAPRIN1), thereby promoting stress granule formation in cells. This process, in turn, inhibits c-Myc translation, regulating cell proliferation and metastasis [166]. Furthermore, glycogen accumulation has been recognized as a critical oncogenic initiator in malignant liver transformation. This accumulation disrupts Hippo signaling through glycogen phase separation, thereby exacerbating tumor development [167].
Regulatory ethods of applying LLPS in cancer treatment
Given that LLPS can impact tumorigenesis through various pathways, there is a need to develop practical strategies for modulating the formation of condensates and targeting cancer-associated proteins. Recent advancements have revealed several regulatory methods for biomolecular condensates, including molecule drug treatment, RNA interference, and DNA editing.
As anticipated, evidence suggests that small molecule drug treatment can effectively modulate phase separation (Table 2). For instance, 1,6-Hexanediol disrupts hydrophobic interactions, thereby compromising the 3-dimensional genome organization in living cells and inhibiting LLPS [47]. JQ1 inhibits osteosarcoma, multiple myeloma, and ovarian cancer cell proliferation, migration, and invasion by suppressing SEs genes as inhibitors targeting transcriptional activators (Figure 3) [102, 107, 153]. Additionally, HDACi (NCT03564704) and TSA (NCT03185429) are currently being evaluated in clinical trials to determine their efficacy and safety in treating patients with leukemia and esophageal tumors.
RNA interference to modulate LLPS can be a potential regulatory method. RNA interference relies on the cell creating small RNA molecules that can target and inhibit the harmful RNA sequences that need to be silenced [168]. For example, the suppressor of gene silencing 3 (SGS3) LLPS drives RNA-dependent RNA polynerase 6 (RDR6) to form small interfering RNA bodies in cytoplasm, which is essential for small interfering RNA production and gene silencing [169].
Small molecule inhibitors related to LLPS in prevalent clinical malignancies. These diseases include glioblastoma, breast cancer, sarcoma, colorectal cancer, ovarian cancer, leukemia, lung cancer, pancreatic cancer, prostate cancer, multiple myeloma, and osteosarcoma cancer. ACP-1n, aminocyclopropenone 1n; EVG, elvitegravir; HDACi, histone deacetylase inhibitor; LSD 1i, lysine-specific histone demethylase 1 inhibitor; SARICA, small-molecule-selective AR-irreversible covalent antagonists.
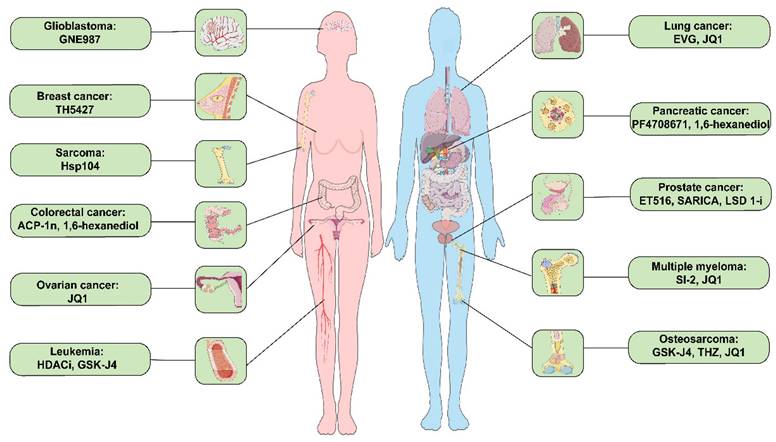
Small molecule inhibitors related to LLPS in malignant tumors and their mechanisms of action.
| Tumor model | Inhibitors | Mechanisms | Reference |
|---|---|---|---|
| Prostate Cancer | ET516 | ET516 blocks AR transcription and inhibits the growth of castration-resistant prostate cancer. | [94] |
| SARICA | Interfering with LLPS formation or mutagenesis resulted in chromatin condensation and dissociation of the AR-V7 interactome. | [99] | |
| LSD 1-i | Target multiple oncogenic pathways in CRPC and that disrupting MYC signaling. | [100] | |
| Osteosarcoma | THZ | THZ inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing SEs genes. | [102] |
| JQ1 | JQ1 inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing SEs genes. | [102] | |
| GSK-J4 | GSK-J4 binds directly to HOXB8-IDR and breaks the loop CRC condensate. | [103] | |
| Multiple Myeloma | SI-2 | SI-2 disrupts SRC-3 LLPS and sensitizes Bortezomib treatment in myeloma cells. | [106] |
| JQ1 | JQ1 preferentially disrupts the transcription process. | [107] | |
| Lung Cancer | EVG | regulates YAP transcriptional activity by reducing H3K27ac mark levels at YAP target genes | [114] |
| Breast Cancer | TH5427 | TH5427 blocked nuclear ATP synthesis, chromatin remodeling, and gene regulation. | [121] |
| Pancreatic Cancer | 1,6-hexanediol | disrupt protein-mediated abnormal LLPS and significantly reduce MYC expression. | [126] |
| PF-4708671 | S6K1 inhibition selectively attenuates SGs and impairs obesity-associated pancreatic ductal adenocarcinoma development. | [127] | |
| Leukemia | HDACi | HDACi inhibits METTL3 expression and proliferation of acute myeloid leukemia cells. | [130] |
| Glioblastoma | 1-AZ | 1-AZ inhibition of GSK-1β partially reverses BYSL down-regulation of β-linked protein transcriptional activity. | [178] |
| Ovarian cancer | JQ1 | JQ1 preferentially disrupts the transcription process. | [153] |
| Esophageal Cancer | TSA | TSA promotes esophageal squamous cell carcinoma migration and Epithelial-mesenchymal-transition through two signaling pathways of BRD4/ERK1/2. | [179] |
| Colorectal cancer | ACP-1n | ACP-1n blocks BRD4 in the nucleus of colorectal cancer HCT4 cells to suppress SE-driven MYC expression. | [158] |
Furthermore, the gene editing technique known as clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR associated (Cas) nuclease 9 (CRISPR-Cas9) offers another avenue for LLPS regulation. This technique is derived from the natural defense system of bacteria and archaea against viral invasion [170]. Notably, Shin et al. recently reported CasDrop, a novel optogenetic technology developed based on CRISPR-Cas9, enables controlled liquid condensation at specific genomic loci [171].
Conclusions
Over the past decade, LLPS has emerged as a burgeoning and captivating concept in cellular biology. Despite its nascent stage, research in phase separation holds considerable potential. Cancer, often associated with dire prognoses and challenging treatments, also incurs substantial economic burdens on individuals and societies. The selective drug distribution and concentration abilities within phase-separated structures can influence drug efficacy and resistance in cancer cells. For example, Mitrea et al. have described a previously unexplored drug discovery approach based on identifying condensate-modifying therapeutics [172]. Consequently, targeting the regulation of phase separation is becoming pivotal in both the diagnosis and treatment of diseases. Clinical trials exploring inhibitors that target transcriptional activators have reported reductions in tumor growth, underlining their therapeutic potential.
However, the understanding of LLPS in cancer remains incomplete. The direct link between the anticancer activity of transcription-related inhibitors and the regulation of biomolecular cohesions is not fully established. This gap in knowledge indicates the need for more comprehensive research. Moreover, most existing studies rely on cultured cancer cells, leaving the comparability of phase separation phenomena and biomolecular cohesions in vitro versus in vivo environments uncertain. Such differences cast doubt on whether current research findings can be translated into clinical settings. Additionally, the intercommunication mechanisms among these biomolecular condensates have yet to be elucidated. To address these critical issues, a multidisciplinary approach is essential. Molecular biologists and biochemists, for instance, are focusing on understanding the molecular mechanisms underlying LLPS. They study how specific proteins and nucleic acids interact to form phase-separated structures and how these structures influence cellular functions in cancer. Moreover, computational biologists play a crucial role in simulating these complex interactions and predicting how changes in LLPS could lead to cancerous states. They use computational tools to simulate the LLPS process, providing insights to guide experimental design and hypothesis testing. Clinical researchers and oncologists translate these findings into potential therapeutic strategies. Finally, this collaboration extends to chemists and drug developers, who leverage knowledge from these studies to design and synthesize new compounds that can modulate LLPS in cancer cells.
Furthermore, the predominant use of fluorescence recovery after photobleaching (FRAP) in previous studies to demonstrate LLPS has faced skepticism [173]. The critics, including Thompson et al. [174], believe that FRAP relies on morphological assessment rather than quantitative data, which limits the study of LLPS. This critique underscores the urgent need for “new tools” to yield more precise data. Herein, we propose that by integrating techniques such as FRAP, Förster Resonance Energy Transfer, and super-resolution microscopy, researchers can gain a more comprehensive understanding of LLPS phenomena in cancer cells. These methods can provide quantitative data and deeper insights into the molecular interactions within phase-separated structures. We look forward to ongoing research that integrates multiple technologies, as it is expected to further clarify the relationship between cancer and LLPS potentially leading to breakthroughs in cancer treatment.
Acknowledgements
We thank the grants supported by the National Natural Science Foundation of China (No. 82073944), Scientific Research Project of Hunan Health Commission (No. 202113010170), Hunan Provincial Natural Science Foundation of China (No. 2022JJ30834, 2022JJ40679 and 2022JJ80119), Degree & Postgraduate Education Reform Project of Central South University (2023JGB123), Natural Science Foundation of Changsha (No. kq2202386), and the Fundamental Research Funds for the Central Universities of Central South University (No. 2022XQLH154).
Funding
This research was supported by the National Natural Science Foundation of China (No. 82073944), Scientific Research Project of Hunan Health Commission (No. 202113010170), Hunan Provincial Natural Science Foundation of China (No. 2022JJ30834, 2022JJ40679 and 2022JJ80119), Degree & Postgraduate Education Reform Project of Central South University (2023JGB123), Natural Science Foundation of Changsha (No. kq2202386), and the Fundamental Research Funds for the Central Universities of Central South University (No. 2022XQLH154).
Author contributions
J.Q. and Q.Q. conceived of the article. W.-X.X., and J.Q. performed the literature search and data collection. W.-X.X., J.Q., Q.Q. and X.-Q.T. drafted the manuscript. J.Q., Y-W.W., J.L., and Y.-H.D. critically revised the work. All authors modified and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 2019;176:419-34
2. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285-98
3. Cable J, Brangwynne C, Seydoux G, Cowburn D, Pappu RV, Castañeda CA. et al. Phase separation in biology and disease-a symposium report. Ann N Y Acad Sci. 2019;1452:3-11
4. Fernandopulle M, Wang G, Nixon-Abell J, Qamar S, Balaji V, Morihara R. et al. Inherited and Sporadic Amyotrophic Lateral Sclerosis and Fronto-Temporal Lobar Degenerations arising from Pathological Condensates of Phase Separating Proteins. Hum Mol Genet. 2019;28:R187-r96
5. Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727-36
6. Alberti S, Carra S. Quality Control of Membraneless Organelles. J Mol Biol. 2018;430:4711-29
7. Aota A, Mawatari K, Takahashi S, Matsumoto T, Kanda K, Anraku R. et al. Phase separation of gas-liquid and liquid-liquid microflows in microchips. Microchimica Acta. 2009;164:249-55
8. Hirose T, Ninomiya K, Nakagawa S, Yamazaki T. A guide to membraneless organelles and their various roles in gene regulation. Nat Rev Mol Cell Biol. 2023;24:288-304
9. Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729-32
10. Aguzzi A, O'Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9:237-48
11. Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334-9
12. Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336-40
13. Kato M, Han TW, Xie S, Shi K, Du X, Wu LC. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753-67
14. Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653
15. Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011 3
16. Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041-56
17. Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595-9
18. Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R. et al. Compositional Control of Phase-Separated Cellular Bodies. Cell. 2016;166:651-63
19. Sengupta S, George RE. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer. 2017;3:269-81
20. Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241-5
21. Wheeler RJ, Lee HO, Poser I, Pal A, Doeleman T, Kishigami S. et al. Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv. 2019: 721001.
22. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123-33
23. Tanaka T, Ishimoto C, Chylack LT Jr. Phase separation of a protein-water mixture in cold cataract in the young rat lens. Science. 1977;197:1010-2
24. Mo Y, Feng Y, Huang W, Tan N, Li X, Jie M. et al. Liquid-Liquid Phase Separation in Cardiovascular Diseases. Cells. 2022 11
25. Zhang X, Zheng R, Li Z, Ma J. Liquid-liquid Phase Separation in Viral Function. J Mol Biol. 2023: 167955.
26. Brangwynne Clifford P, Tompa P, Pappu Rohit V. Polymer physics of intracellular phase transitions. Nature Physics. 2015;11:899-904
27. Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017 357
28. Ren J, Zhang Z, Zong Z, Zhang L, Zhou F. Emerging Implications of Phase Separation in Cancer. Adv Sci (Weinh). 2022;9:e2202855
29. Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P. et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018 7
30. Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ. et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell. 2018;173:720-34.e15
31. Kim S, Huang J, Lee Y, Dutta S, Yoo HY, Jung YM. et al. Complexation and coacervation of like-charged polyelectrolytes inspired by mussels. Proc Natl Acad Sci U S A. 2016;113:E847-53
32. Aumiller WM Jr, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem. 2016;8:129-37
33. Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A. et al. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol Cell. 2017;65:1044-55.e5
34. Yeo GC, Keeley FW, Weiss AS. Coacervation of tropoelastin. Adv Colloid Interface Sci. 2011;167:94-103
35. Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26:668-79
36. Adame-Arana O, Weber CA, Zaburdaev V, Prost J, Jülicher F. Liquid Phase Separation Controlled by pH. Biophys J. 2020;119:1590-605
37. Cummings CS, Obermeyer AC. Phase Separation Behavior of Supercharged Proteins and Polyelectrolytes. Biochemistry. 2018;57:314-23
38. Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY. et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066-77
39. Kang J, Lim L, Song J. ATP enhances at low concentrations but dissolves at high concentrations liquid-liquid phase separation (LLPS) of ALS/FTD-causing FUS. Biochem Biophys Res Commun. 2018;504:545-51
40. Sheu-Gruttadauria J, MacRae IJ. Phase Transitions in the Assembly and Function of Human miRISC. Cell. 2018;173:946-57.e16
41. Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S. et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018 37
42. Fukaya T, Lim B, Levine M. Enhancer Control of Transcriptional Bursting. Cell. 2016;166:358-68
43. Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018 361
44. Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13-23
45. Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV. et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018;175:1842-55.e16
46. Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412-5
47. Ulianov SV, Velichko AK, Magnitov MD, Luzhin AV, Golov AK, Ovsyannikova N. et al. Suppression of liquid-liquid phase separation by 1,6-hexanediol partially compromises the 3D genome organization in living cells. Nucleic Acids Res. 2021;49:10524-41
48. Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H. et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25:833-40
49. Rawat P, Boehning M, Hummel B, Aprile-Garcia F, Pandit AS, Eisenhardt N. et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol Cell. 2021;81:1013-26.e11
50. Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A. et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558:318-23
51. Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng MM. et al. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell. 2021;184:207-25.e24
52. Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18:610-21
53. Chong S, Graham TGW, Dugast-Darzacq C, Dailey GM, Darzacq X, Tjian R. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol Cell. 2022;82:2084-97.e5
54. Zuo L, Zhang G, Massett M, Cheng J, Guo Z, Wang L. et al. Nat Commun. 2021; 12: 1491.
55. Levone BR, Lenzken SC, Antonaci M, Maiser A, Rapp A, Conte F. et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J Cell Biol. 2021 220
56. Ghodke I, Remisova M, Furst A, Kilic S, Reina-San-Martin B, Poetsch AR. et al. AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation. Mol Cell. 2021;81:2596-610.e7
57. Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R. et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. Embo j. 2019;38:e101379
58. Oshidari R, Huang R, Medghalchi M, Tse EYW, Ashgriz N, Lee HO. et al. DNA repair by Rad52 liquid droplets. Nat Commun. 2020;11:695
59. Zhou W, Mohr L, Maciejowski J, Kranzusch PJ. cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol Cell. 2021;81:739-55 e7
60. Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18:204-11
61. Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732-44
62. Wang L, Hu M, Zuo MQ, Zhao J, Wu D, Huang L. et al. Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell Res. 2020;30:393-407
63. Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol. 2022;23:329-49
64. Martin IM, Aponte-Santamaría C, Schmidt L, Hedtfeld M, Iusupov A, Musacchio A. et al. Phosphorylation tunes elongation propensity and cohesiveness of INCENP's intrinsically disordered region. J Mol Biol. 2022;434:167387
65. Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791-805
66. Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun. 2017;8:275
67. Guccione E, Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol. 2019;20:642-57
68. Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325-36
69. Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F. et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol. 2014;204:931-45
70. Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D. et al. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol. 2016;215:57-76
71. Celetti G, Paci G, Caria J, VanDelinder V, Bachand G, Lemke EA. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J Cell Biol. 2020 219
72. Vovk A, Gu C, Opferman MG, Kapinos LE, Lim RY, Coalson RD. et al. Simple biophysics underpins collective conformations of the intrinsically disordered proteins of the Nuclear Pore Complex. Elife. 2016 5
73. Stroberg W, Schnell S. Do Cellular Condensates Accelerate Biochemical Reactions?. Lessons from Microdroplet Chemistry. Biophys J. 2018;115:3-8
74. Liu M, He S, Cheng L, Qu J, Xia J. Phase-Separated Multienzyme Biosynthesis. Biomacromolecules. 2020;21:2391-9
75. Huang WYC, Alvarez S, Kondo Y, Lee YK, Chung JK, Lam HYM. et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science. 2019;363:1098-103
76. Case LB, Zhang X, Ditlev JA, Rosen MK. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 2019;363:1093-7
77. Strzyz P. Phase separation tunes signal transduction. Nat Rev Mol Cell Biol. 2019;20:263
78. Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704-9
79. Yu X, Zhang L, Shen J, Zhai Y, Jiang Q, Yi M. et al. The STING phase-separator suppresses innate immune signalling. Nat Cell Biol. 2021;23:330-40
80. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630
81. Wallace Edward WJ, Kear-Scott Jamie L, Pilipenko Evgeny V, Schwartz Michael H, Laskowski Pawel R, Rojek Alexandra E. et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 2015;162:1286-98
82. Cinar H, Cinar S, Chan HS, Winter R. Pressure-Induced Dissolution and Reentrant Formation of Condensed, Liquid-Liquid Phase-Separated Elastomeric α-Elastin. Chemistry. 2018;24:8286-91
83. Zlotorynski E. Stress responses: Reversible sequestration over irreversible aggregation. Nat Rev Mol Cell Biol. 2018;19:73
84. Watanabe K, Morishita K, Zhou X, Shiizaki S, Uchiyama Y, Koike M. et al. Cells recognize osmotic stress through liquid-liquid phase separation lubricated with poly(ADP-ribose). Nat Commun. 2021;12:1353
85. Kato M, Yang YS, Sutter BM, Wang Y, McKnight SL, Tu BP. Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell. 2019;177:711-21.e8
86. Yang YS, Kato M, Wu X, Litsios A, Sutter BM, Wang Y. et al. Yeast Ataxin-2 Forms an Intracellular Condensate Required for the Inhibition of TORC1 Signaling during Respiratory Growth. Cell. 2019;177:697-710.e17
87. Nozawa RS, Yamamoto T, Takahashi M, Tachiwana H, Maruyama R, Hirota T. et al. Nuclear microenvironment in cancer: Control through liquid-liquid phase separation. Cancer Sci. 2020;111:3155-63
88. Peng PH, Hsu KW, Wu KJ. Liquid-liquid phase separation (LLPS) in cellular physiology and tumor biology. Am J Cancer Res. 2021;11:3766-76
89. Tong X, Tang R, Xu J, Wang W, Zhao Y, Yu X. et al. Liquid-liquid phase separation in tumor biology. Signal Transduct Target Ther. 2022;7:221
90. Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall'Agnese A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386-92
91. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34
92. Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6:76-85
93. Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501-11
94. Xie J, He H, Kong W, Li Z, Gao Z, Xie D. et al. Targeting androgen receptor phase separation to overcome antiandrogen resistance. Nat Chem Biol. 2022;18:1341-50
95. Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N. et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol Cell. 2018;72:19-36.e8
96. Gao K, Shi Q, Liu Y, Wang C. Enhanced autophagy and NFE2L2/NRF2 pathway activation in SPOP mutation-driven prostate cancer. Autophagy. 2022;18:2013-5
97. Zhang F, Biswas M, Massah S, Lee J, Lingadahalli S, Wong S. et al. Dynamic phase separation of the androgen receptor and its coactivators key to regulate gene expression. Nucleic Acids Res. 2023;51:99-116
98. Takayama KI, Kosaka T, Suzuki T, Hongo H, Oya M, Fujimura T. et al. Subtype-specific collaborative transcription factor networks are promoted by OCT4 in the progression of prostate cancer. Nat Commun. 2021;12:3766
99. Thiyagarajan T, Ponnusamy S, Hwang DJ, He Y, Asemota S, Young KL. et al. Inhibiting androgen receptor splice variants with cysteine-selective irreversible covalent inhibitors to treat prostate cancer. Proc Natl Acad Sci U S A. 2023;120:e2211832120
100. Li M, Liu M, Han W, Wang Z, Han D, Patalano S. et al. LSD1 inhibition disrupts super-enhancer driven oncogenic transcriptional programs in castration-resistant prostate cancer. Cancer Res. 2023
101. Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705-18
102. Chen D, Zhao Z, Huang Z, Chen DC, Zhu XX, Wang YZ. et al. Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res. 2018;6:11
103. Lu B, Zou C, Yang M, He Y, He J, Zhang C. et al. Pharmacological Inhibition of Core Regulatory Circuitry Liquid-liquid Phase Separation Suppresses Metastasis and Chemoresistance in Osteosarcoma. Adv Sci (Weinh). 2021;8:e2101895
104. Abramson HN. The Multiple Myeloma Drug Pipeline-2018: A Review of Small Molecules and Their Therapeutic Targets. Clin Lymphoma Myeloma Leuk. 2018;18:611-27
105. De Smedt E, Lui H, Maes K, De Veirman K, Menu E, Vanderkerken K. et al. The Epigenome in Multiple Myeloma: Impact on Tumor Cell Plasticity and Drug Response. Front Oncol. 2018;8:566
106. Liu J, Xie Y, Guo J, Li X, Wang J, Jiang H. et al. Targeting NSD2-mediated SRC-3 liquid-liquid phase separation sensitizes bortezomib treatment in multiple myeloma. Nat Commun. 2021;12:1022
107. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904-17
108. Collins LG, Haines C, Perkel R, Enck RE. Lung cancer: diagnosis and management. Am Fam Physician. 2007;75:56-63
109. Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med. 2021;27:1345-56
110. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069-75
111. Qin Z, Sun H, Yue M, Pan X, Chen L, Feng X. et al. Phase separation of EML4-ALK in firing downstream signaling and promoting lung tumorigenesis. Cell Discov. 2021;7:33
112. Zhu Q, Zhang C, Qu T, Lu X, He X, Li W. et al. MNX1-AS1 Promotes Phase Separation of IGF2BP1 to Drive c-Myc-Mediated Cell-Cycle Progression and Proliferation in Lung Cancer. Cancer Res. 2022;82:4340-58
113. Zhang J, Zeng Y, Xing Y, Li X, Zhou L, Hu L. et al. Myristoylation-mediated phase separation of EZH2 compartmentalizes STAT3 to promote lung cancer growth. Cancer Lett. 2021;516:84-98
114. Zhu G, Xie J, Fu Z, Wang M, Zhang Q, He H. et al. Pharmacological inhibition of SRC-1 phase separation suppresses YAP oncogenic transcription activity. Cell Res. 2021;31:1028-31
115. Lu X, Wang J, Wang W, Lu C, Qu T, He X. et al. Copy number amplification and SP1-activated lncRNA MELTF-AS1 regulates tumorigenesis by driving phase separation of YBX1 to activate ANXA8 in non-small cell lung cancer. Oncogene. 2022;41:3222-38
116. Liu S, Wang T, Shi Y, Bai L, Wang S, Guo D. et al. Cell Death Differ. 2021; 28: 2482-98.
117. Wang J, Xu Y, Rao X, Zhang R, Tang J, Zhang D. et al. BRD4-IRF1 axis regulates chemoradiotherapy-induced PD-L1 expression and immune evasion in non-small cell lung cancer. Clin Transl Med. 2022;12:e718
118. Waks AG, Winer EP. Breast Cancer Treatment: A Review. Jama. 2019;321:288-300
119. Chu XY, Xu YY, Tong XY, Wang G, Zhang HY. The Legend of ATP: From Origin of Life to Precision Medicine. Metabolites. 2022 12
120. Wright RH, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J. et al. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science. 2016;352:1221-5
121. Page BDG, Valerie NCK, Wright RHG, Wallner O, Isaksson R, Carter M. et al. Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells. Nat Commun. 2018;9:250
122. Xing Z, Xue J, Ma X, Han C, Wang Z, Luo S. et al. Intracellular mRNA phase separation induced by cationic polymers for tumor immunotherapy. J Nanobiotechnology. 2022;20:442
123. Hu Y, Xie Q, Chen S, Zhao W, Zhao X, Ruan Q. et al. Par3 promotes breast cancer invasion and migration through pull tension and protein nanoparticle-induced osmotic pressure. Biomed Pharmacother. 2022;155:113739
124. Liu X, Ye Y, Zhu L, Xiao X, Zhou B, Gu Y. et al. Niche stiffness sustains cancer stemness via TAZ and NANOG phase separation. Nat Commun. 2023;14:238
125. Gillen S, Schuster T, Meyer Zum Büschenfelde C, Friess H, Kleeff J. Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010;7:e1000267
126. Ming Y, Chen X, Xu Y, Wu Y, Wang C, Zhang T. et al. Targeting liquid-liquid phase separation in pancreatic cancer. Transl Cancer Res. 2019;8:96-103
127. Fonteneau G, Redding A, Hoag-Lee H, Sim ES, Heinrich S, Gaida MM. et al. Stress Granules Determine the Development of Obesity-Associated Pancreatic Cancer. Cancer Discov. 2022;12:1984-2005
128. Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM. et al. Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell. 2017;171:1545-58.e18
129. Li W, Wu L, Jia H, Lin Z, Zhong R, Li Y. et al. The low-complexity domains of the KMT2D protein regulate histone monomethylation transcription to facilitate pancreatic cancer progression. Cell Mol Biol Lett. 2021;26:45
130. Li M, Li M, Xia Y, Li G, Su X, Wang D. et al. HDAC1/3-dependent moderate liquid-liquid phase separation of YY1 promotes METTL3 expression and AML cell proliferation. Cell Death Dis. 2022;13:992
131. Yan H, Zhang L, Cui X, Zheng S, Li R. Roles and mechanisms of the m(6)A reader YTHDC1 in biological processes and diseases. Cell Death Discov. 2022;8:237
132. Cheng Y, Xie W, Pickering BF, Chu KL, Savino AM, Yang X. et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39:958-72 e8
133. Shao X, Chen Y, Xu A, Xiang D, Wang W, Du W. et al. Deneddylation of PML/RARα reconstructs functional PML nuclear bodies via orchestrating phase separation to eradicate APL. Cell Death Differ. 2022;29:1654-68
134. Li Y, Ma X, Wu W, Chen Z, Meng G. PML Nuclear Body Biogenesis, Carcinogenesis, and Targeted Therapy. Trends Cancer. 2020;6:889-906
135. Chandra B, Michmerhuizen NL, Shirnekhi HK, Tripathi S, Pioso BJ, Baggett DW. et al. Phase Separation Mediates NUP98 Fusion Oncoprotein Leukemic Transformation. Cancer Discov. 2022;12:1152-69
136. Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373-7
137. Shi B, Li W, Song Y, Wang Z, Ju R, Ulman A. et al. UTX condensation underlies its tumour-suppressive activity. Nature. 2021;597:726-31
138. Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F. et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30:508-21
139. Ahmed NS, Harrell LM, Wieland DR, Lay MA, Thompson VF, Schwartz JC. Fusion protein EWS-FLI1 is incorporated into a protein granule in cells. Rna. 2021;27:920-32
140. Riggi N, Cironi L, Provero P, Suvà ML, Stehle JC, Baumer K. et al. Expression of the FUS-CHOP fusion protein in primary mesenchymal progenitor cells gives rise to a model of myxoid liposarcoma. Cancer Res. 2006;66:7016-23
141. Owen I, Yee D, Wyne H, Perdikari TM, Johnson V, Smyth J. et al. The oncogenic transcription factor FUS-CHOP can undergo nuclear liquid-liquid phase separation. J Cell Sci. 2021 134
142. Ryan JJ, Sprunger ML, Holthaus K, Shorter J, Jackrel ME. Engineered protein disaggregases mitigate toxicity of aberrant prion-like fusion proteins underlying sarcoma. J Biol Chem. 2019;294:11286-96
143. Cheng Y, Shen Z, Gao Y, Chen F, Xu H, Mo Q. et al. Phase transition and remodeling complex assembly are important for SS18-SSX oncogenic activity in synovial sarcomas. Nat Commun. 2022;13:2724
144. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93-108
145. Wei Y, Luo H, Yee PP, Zhang L, Liu Z, Zheng H. et al. Paraspeckle Protein NONO Promotes TAZ Phase Separation in the Nucleus to Drive the Oncogenic Transcriptional Program. Adv Sci (Weinh). 2021;8:e2102653
146. Tang Q, Mao X, Chen Z, Ma C, Tu Y, Zhu Q. et al. Liquid-liquid phase separation-related gene in gliomas: FABP5 is a potential prognostic marker. J Gene Med. 2023: e3517.
147. Wang Y, Wahafu A, Wu W, Xiang J, Huo L, Ma X. et al. FABP5 enhances malignancies of lower-grade gliomas via canonical activation of NF-κB signaling. J Cell Mol Med. 2021;25:4487-500
148. Ma L, Li G, Yang T, Zhang L, Wang X, Xu X. et al. An inhibitor of BRD4, GNE987, inhibits the growth of glioblastoma cells by targeting C-Myc and S100A16. Cancer Chemother Pharmacol. 2022;90:431-44
149. Zhao Z, Xiao S, Yuan X, Yuan J, Zhang C, Li H. et al. AHNAK as a Prognosis Factor Suppresses the Tumor Progression in Glioma. J Cancer. 2017;8:2924-32
150. Wei Y, Erfani S, Schweer D, de Gouvea R, Qadir J, Shi J. et al. Targeting receptor tyrosine kinases in ovarian cancer: Genomic dysregulation, clinical evaluation of inhibitors, and potential for combinatorial therapies. Mol Ther Oncolytics. 2023;28:293-306
151. Qiu Y, Pan M, Chen X. A Liquid-Liquid Phase Separation-Related Gene Signature as Prognostic Biomarker for Epithelial Ovarian Cancer. Front Oncol. 2021;11:671892
152. Baudin A, Moreno-Romero AK, Xu X, Selig EE, Penalva LOF, Libich DS. Structural Characterization of the RNA-Binding Protein SERBP1 Reveals Intrinsic Disorder and Atypical RNA Binding Modes. Front Mol Biosci. 2021;8:744707
153. Zhang Z, Ma P, Jing Y, Yan Y, Cai MC, Zhang M. et al. BET Bromodomain Inhibition as a Therapeutic Strategy in Ovarian Cancer by Downregulating FoxM1. Theranostics. 2016;6:219-30
154. Reichenbach ZW, Murray MG, Saxena R, Farkas D, Karassik EG, Klochkova A. et al. Clinical and translational advances in esophageal squamous cell carcinoma. Adv Cancer Res. 2019;144:95-135
155. Liu J, Liu ZX, Li JJ, Zeng ZL, Wang JH, Luo XJ. et al. The macrophage-associated lncRNA MALR facilitates ILF3 liquid-liquid phase separation to promote HIF1α signaling in esophageal cancer. Cancer Res. 2022
156. Zhang Y, Li H, Lv L, Lu K, Li H, Zhang W. et al. Autophagy: Dual roles and perspective for clinical treatment of colorectal cancer. Biochimie. 2023;206:49-60
157. Fan XJ, Wang YL, Zhao WW, Bai SM, Ma Y, Yin XK. et al. NONO phase separation enhances DNA damage repair by accelerating nuclear EGFR-induced DNA-PK activation. Am J Cancer Res. 2021;11:2838-52
158. Kondo H, Mishiro K, Iwashima Y, Qiu Y, Kobayashi A, Lim K. et al. Discovery of a Novel Aminocyclopropenone Compound That Inhibits BRD4-Driven Nucleoporin NUP210 Expression and Attenuates Colorectal Cancer Growth. Cells. 2022 11
159. Shi J, Chen SY, Shen XT, Yin XK, Zhao WW, Bai SM. et al. NOP53 undergoes liquid-liquid phase separation and promotes tumor radio-resistance. Cell Death Discov. 2022;8:436
160. Gao H, Wei H, Yang Y, Li H, Liang J, Ye J. et al. Phase separation of DDX21 promotes colorectal cancer metastasis via MCM5-dependent EMT pathway. Oncogene. 2023;42:1704-15
161. Hohenstein P, Pritchard-Jones K, Charlton J. The yin and yang of kidney development and Wilms' tumors. Genes Dev. 2015;29:467-82
162. Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem Cell Biol. 2009;87:65-75
163. Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L. et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature. 2020;577:121-6
164. Kabra A, Bushweller J. The Intrinsically Disordered Proteins MLLT3 (AF9) and MLLT1 (ENL) - Multimodal Transcriptional Switches With Roles in Normal Hematopoiesis, MLL Fusion Leukemia, and Kidney Cancer. J Mol Biol. 2022;434:167117
165. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6
166. Chen S, Cao X, Zhang J, Wu W, Zhang B, Zhao F. circVAMP3 Drives CAPRIN1 Phase Separation and Inhibits Hepatocellular Carcinoma by Suppressing c-Myc Translation. Adv Sci (Weinh). 2022;9:e2103817
167. Liu Q, Li J, Zhang W, Xiao C, Zhang S, Nian C. et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell. 2021;184:5559-76.e19
168. Manage KI, Rogers AK, Wallis DC, Uebel CJ, Anderson DC, Nguyen DAH. et al. A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans. Elife. 2020 9
169. Tan H, Luo W, Yan W, Liu J, Aizezi Y, Cui R. et al. Phase separation of SGS3 drives siRNA body formation and promotes endogenous gene silencing. Cell Rep. 2023;42:111985
170. Wang SW, Gao C, Zheng YM, Yi L, Lu JC, Huang XY. et al. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer. 2022;21:57
171. Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P. et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell. 2018;175:1481-91.e13
172. Mitrea DM, Mittasch M, Gomes BF, Klein IA, Murcko MA. Modulating biomolecular condensates: a novel approach to drug discovery. Nat Rev Drug Discov. 2022;21:841-62
173. Wu W, Tan Y, Yin H, Jiang M, Jiang Y, Ma X. et al. Phase separation is required for PML nuclear body biogenesis and function. Faseb j. 2023;37:e22986
174. Muzzopappa F, Hummert J, Anfossi M, Tashev SA, Herten DP, Erdel F. Detecting and quantifying liquid-liquid phase separation in living cells by model-free calibrated half-bleaching. Nat Commun. 2022;13:7787
175. Li X, Baek G, Ramanand SG, Sharp A, Gao Y, Yuan W. et al. BRD4 Promotes DNA Repair and Mediates the Formation of TMPRSS2-ERG Gene Rearrangements in Prostate Cancer. Cell Rep. 2018;22:796-808
176. Esposito M, Fang C, Cook KC, Park N, Wei Y, Spadazzi C. et al. TGF-β-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat Cell Biol. 2021;23:257-67
177. Tong XY, Quan Y, Zhang HY. NUDT5 as a novel drug target and prognostic biomarker for ER-positive breast cancer. Drug Discov Today. 2021;26:620-5
178. Sha Z, Zhou J, Wu Y, Zhang T, Li C, Meng Q. et al. BYSL Promotes Glioblastoma Cell Migration, Invasion, and Mesenchymal Transition Through the GSK-3β/β-Catenin Signaling Pathway. Front Oncol. 2020;10:565225
179. Liu D, Liu Y, Qi B, Gu C, Huo S, Zhao B. Trichostatin A promotes esophageal squamous cell carcinoma cell migration and EMT through BRD4/ERK1/2-dependent pathway. Cancer Med. 2021;10:5235-45
Author contact
![]() Corresponding author: Jian Qu, MD, Ph.D., Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University, No. 139 Middle Renmin Road, Changsha, China, 410011; Tel.: +86-15973190614; Fax number: +86-0731-85292072; E-mail: qujianstanleyedu.cn.
Corresponding author: Jian Qu, MD, Ph.D., Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University, No. 139 Middle Renmin Road, Changsha, China, 410011; Tel.: +86-15973190614; Fax number: +86-0731-85292072; E-mail: qujianstanleyedu.cn.

 Global reach, higher impact
Global reach, higher impact