10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2024; 20(8):3126-3139. doi:10.7150/ijbs.96210 This issue Cite
Research Paper
Particulate matter facilitates amphiregulin-dependent lung cancer proliferation through glutamine metabolism
1. Graduate Institute of Biomedical Sciences, China Medical University, Taichung, Taiwan.
2. Department of Respiratory Therapy, Fu-Jen Catholic University, New Taipei City, Taiwan.
3. Department of Life Science, National Taiwan Normal University, Taipei, Taiwan.
4. Center for Molecular Medicine, China Medical University Hospital, Taichung, Taiwan.
5. Department of Medical Research, China Medical University Hsinchu Hospital, Hsinchu, Taiwan.
6. Department of Medical Laboratory Science and Biotechnology, Asia University, Taichung, Taiwan.
7. Division of General Thoracic Surgery, Department of Surgery, Changhua Christian Hospital, Changhua, Taiwan.
8. Department of Physical Therapy, Asia University, Taichung, Taiwan.
9. Department of General Thoracic Surgery, Asia University Hospital, Taichung, Taiwan.
10. Department of Pharmacology, School of Medicine, China Medical University, Taichung, Taiwan.
11. Chinese Medicine Research Center, China Medical University, Taichung, Taiwan.
Received 2024-3-12; Accepted 2024-5-16; Published 2024-5-27
Abstract
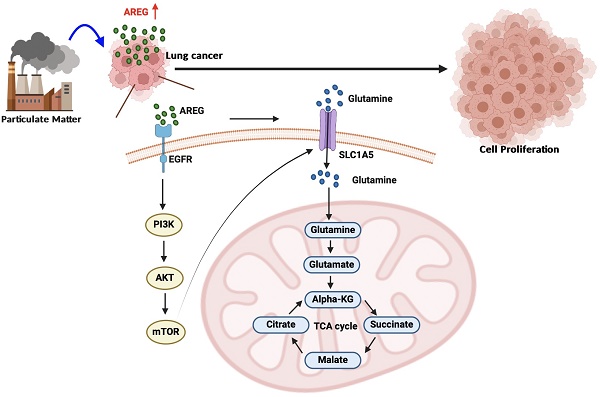
Although many cohort studies have reported that long-term exposure to particulate matter (PM) causes lung cancer, the molecular mechanisms underlying the PM-induced increases in lung cancer progression remain unclear. We applied the lung cancer cell line A549 (Parental; A549.Par) to PM for an extended period to establish a mimic PM-exposed lung cancer cell line, A549.PM. Our results indicate that A549.PM exhibits higher cell growth and proliferation abilities compared to A549.Par cells in vitro and in vivo. The RNA sequencing analysis found amphiregulin (AREG) plays a critical role in PM-induced cell proliferation. We observed that PM increases AREG-dependent lung cancer proliferation through glutamine metabolism. In addition, the EGFR/PI3K/AKT/mTOR signaling pathway is involved in PM-induced solute carrier family A1 member 5 (SLC1A5) expression and glutamine metabolism. Our findings offer important insights into how lung cancer proliferation develops upon exposure to PM.
Keywords: Lung cancer, Particulate matter, Glutamine metabolism, Amphiregulin, SLC1A5
1. Introduction
Air pollution is a global issue that affects people worldwide. In recent years, the impact of air pollution on health has garnered increasing attention [1]. The World Health Organization (WHO) estimated that the combined impacts of indoor and outdoor air pollution result in approximately 7 million premature deaths annually [2]. Particulate matter (PM) is a key indicator of air pollution introduced into the air by various natural and human activities [3]. Research conducted in many regions of the world indicates that air pollution is associated with an increasing number of adverse health effects. Exposure to PM also has been identified as the cause of numerous health effects including respiratory symptoms, exacerbation of chronic respiratory and cardiovascular diseases [4, 5]. The International Agency for Research on Cancer (IARC), in a meta-analysis of findings from 14 studies of outdoor air pollution conducted largely in North America and Europe, reported a statistically significant 9% increase in risk for lung cancer incidence or mortality per each 10 µg/m3 increase in PM2.5 concentrations [6]. PM also induces cell migration, invasion, and epithelial-mesenchymal transition (EMT) in lung cancer cells, thereby promoting tumor metastasis [7]. Although the relationship between PM and lung cancer is established, the underlying mechanism remains unclear.
Long-term exposure to PM2.5 causes DNA damage and activates AhR, epidermal growth factor receptor (EGFR), and the immune system, leading to tumorigenesis [8]. Amphiregulin (AREG), is an epidermal growth factor ligand of EGFR [9], further promoting distant metastasis, anti-apoptosis, and drug resistance in cancer [10]. AREG has been found to play a role in various physiological processes, including breast development, bone formation, cell invasion, and angiogenesis [11-13]. AREG stimulates cell migration and proliferation while reducing lung cancer cell apoptosis [14]. Moreover, AREG is linked to a worse prognosis and shorter survival in lung cancer patients [15]. Therefore, AREG plays a crucial role in regulating lung cancer cell growth, differentiation, and metastasis.
Previous studies have indicated that tumor cells adapt to the tumor microenvironment by altering their metabolic pathways [16]. Tumor cells mainly obtain nutrients through metabolic pathways involving glucose, fatty acids, glutamine, and small-molecule amino acids to support tumor cell growth [17]. Glutamine is considered central to cell growth and metabolism in both normal and cancer cells. Moreover, glutamine participates in cell signaling pathways to promote hallmarks of malignancies, including sustaining proliferation, invasion, and metastasis [18]. Solute carrier family A1 member 5 (SLC1A5) is the primary transporter of glutamine [19]. The SLC1A5 variation is essential for the metabolic reprogramming of cancer because it transports glutamine into mitochondria [20]. In the glutamine metabolic pathway, glutamine enters tumor cells through SLC1A5, where it is converted into glutamate by glutaminase and then enters the mitochondria. Glutamate dehydrogenase converts glutamate in the mitochondria into α-ketoglutarate (α-KG), which enters the tricarboxylic acid (TCA) cycle to provide energy to tumor cells and promote tumor cell growth [21-24]. However, the association of metabolism with PM and lung cancer has not been widely explored.
Our research sought to determine whether long-term exposure to PM through metabolic pathways promotes lung cancer cell proliferation. We applied the lung cancer cell line A549 (Parental; A549.Par) to PM (25 μg/ml) for an extended period to establish a mimic PM-exposed lung cancer cell line (A549.PM). Our in vitro and in vivo evidence showed that long-term exposure to PM promotes lung cancer growth. PM upregulates AREG expression through the EGFR/PI3K/AKT/mTOR signaling pathway, leading to an increase in the expression of the glutamine transporter SLC1A5. This promotion of SLC1A5 enhances mitochondrial respiration via the glutamine-derived TCA cycle metabolites, consequently stimulating the proliferation of lung cancer cells.
2. Materials and Methods
The detailed sources of antibodies, chemicals, cell culture, quantitative real-time polymerase chain reaction (RT-qPCR), western blot, ELISA, Seahorse and immunohistochemistry (IHC) staining assays are all obtainable within Supplementary Materials and Methods.
2.1 PM
The PMs were standard reference material 1649b (SRM1649b) purchased from the National Institute of Standards and Technology NIST (MD, USA). The PMs consisted of urban dust. They were dispersed in ultra-pure water and sonicated for 30 minutes in an ultrasonic bath before being used. All PMs were stored at 4 °C.
2.2 Cell viability assay
Cell proliferation was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and cell counting kit-8 (CCK-8) assay (ab228554, Abcam, Burlingame, CA, USA). Cells were transfected with AREG or SLC1A5 shRNA at 24 hr. In the MTT assay, cells were seeded in 96 well plates at a concentration of 2000 cells per well for 24 hr and 48 hr. After cultures were washed with PBS, 0.5 mg/ml of MTT solution was added and incubated for 1 hr at 37°C. The formazan crystals were dissolved in 50 μl of DMSO and the absorbance was measured at 450 nm [25, 26]. In the CCK-8 assay, cells were seeded in 96 well plates at a concentration of 3000 cells per well for 24 hr and 48 hr. After cultures, the CCK8 regent was added and incubated for 2 hr at 37 °C, the absorbance was measured at 450 nm.
2.3 Colony formation assay
Cells were transfected with AREG or SLC1A5 shRNA. After 24 hr, A549.PM cells were re-plated in six-well plates at a density of 3000 cells per well and cultured with DMEM supplemented with 10% FBS for 7 days. At the end of the incubation period, the cells were washed twice with PBS, fixed in methanol, stained with 5% crystal violet, and manually counted under a microscope [27].
2.4 RNA Sequencing (RNA-Seq) and data analysis
Total RNA of the A549.Par and A549.PM cells were isolated for RNA sequencing. RNA quality and integrity were examined using Bioanalyzer 2100 and RNA 1000 Nano LabChip Kit (Agilent); samples with an RNA integrity number less than 7 were excluded from the subsequent assay. After the mRNA fragmented and cDNA library preparation, RNA sequencing was conducted using the Illumina HiSeq 4000 (paired-end, 150 base pairs, PE150) and mapped by using the HISAT package (http://ccb.jhu.edu/software/hisat2). EdgeR was utilized to estimate the differentially regulated genes of all transcripts by calculating fragments per kilobase per million (FPKM). Differentially expressed genes were determined with a log2 (fold change) >1 or log2 (fold change) <-1 and statistical significance (p-value < 0.05) using the R package [28].
2.5 Meta-analysis of microarray datasets from The Cancer Genome Atlas (TCGA) database
Using the TCGA dataset of lung adenocarcinoma cancers, we identified 48 patients analyzed for AREG gene expression and 57 patients analyzed for SLC1A5 gene expression for each tumor sample.
2.6 Measurement of Oxygen Consumption Rate (OCR)
A549.Par and A549.PM cells (2 × 104 cells/well) were seeded in Seahorse XF24-well microplate (Agilent). After the seeding period, cells were cultured in an assay medium without sodium bicarbonate and HEPES in a non-CO2 incubator at 37 °C for 1 hr. OCR was examined using Seahorse XF Cell Mito Stress Test Kit (103015-100, Agilent, Santa Clara, CA, USA). After baseline measurements, for OCR, oligomycin, the reversible inhibitor of oxidative phosphorylation FCCP (p-trifluoromethoxy carbonyl cyanide phenylhydrazone), and the mitochondrial complex I inhibitor rotenone plus the mitochondrial complex III inhibitor antimycin A (Rote/AA) were sequentially injected. Data were assessed by Seahorse XF24 Analyzer (Agilent, Santa Clara, CA, USA) [29].
2.7 Glutamine levels
The concentration of glutamine was measured using a Glutamine Detection Assay Kit (Abcam, Burlingame, CA, USA) following the manufacturer's instructions after culturing cells (1 × 104/well) in 24-well plates [29].
2.8 Mass spectrometry by carbon-13 tracing experiments
A549.Par or A549.PM cells are cultured for 24 hours in glucose/glutamine-free Dulbecco MEM which is either fully labeled 13C-glutamine or 1,6-13C glucose. After 24 hours, remove the media, and wash the cells with PBS twice then detect the relative amounts of metabolites (from glycolysis or glutamine metabolism) in the cells.
2.9 In vivo tumor growth assay
Mice will be subcutaneously injected with 0.2 mL Matrigel containing 5 × 106 A549. Par or A549.PM cells. After injection, the tumor development was monitored by IVIS imaging system (Xenogen, UK) every week. After 4 weeks, the mice were sacrificed by CO2 inhalation and the tumors were removed, fixed with 4% paraformaldehyde/PBS, and embedded in paraffin. Subsequently, the samples will be processed for AREG and SLC1A5 staining.
2.10 Statistical analysis
All values are expressed as the mean ± standard deviation (SD). Statistical differences between the experimental groups were assessed for significance using the Student's t-test. Statistical comparisons of more than two groups were performed using one-way analysis of variance (ANOVA) with Bonferroni's post hoc test. Between-group differences were significant if the p-value was less than 0.05.
3. Results
3.1 PM-induced lung cancer progression in vitro and in vivo
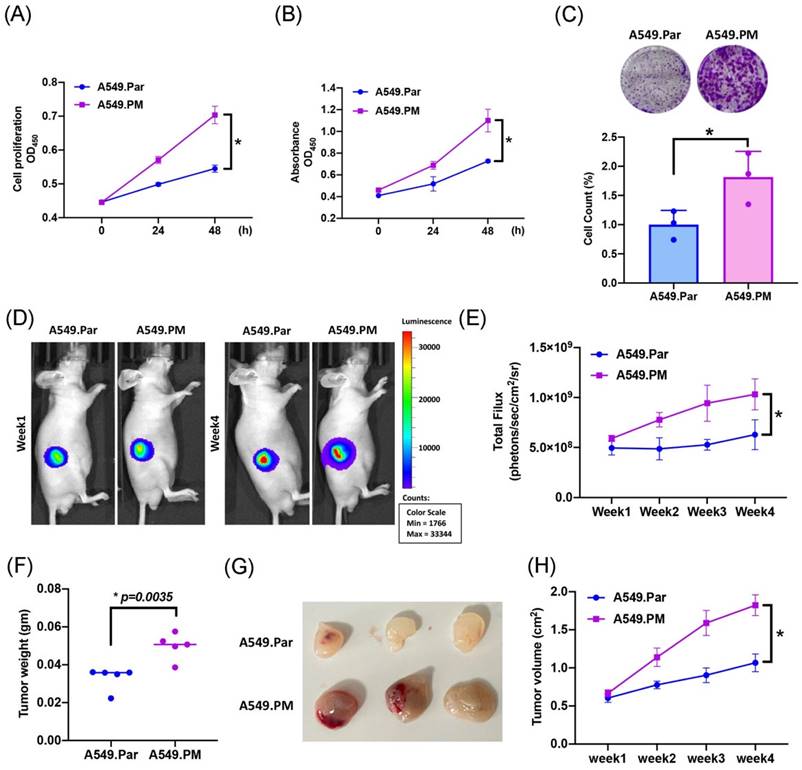
A previous study has shown that PM plays an important role in lung cancer proliferation and metastasis [30]. To validate whether PM induced lung cancer proliferation, we exposed A549 cells (A549.Par) to 25 μg/mL PM for long-term exposure (A549.PM) to mimic PM exposure behavior. The result of the MTT and CCK-8 assay showed that A549.PM cells have a higher cell proliferation ability than A549.Par cells (Figure 1A&B). The growth rate was significantly enhanced in A549.PM cells compared with A549.Par cells, as demonstrated by the colony assay (Figure 1C). In our in vivo data, we found that A549.PM cells exhibit higher tumor growth than A549.Par cells (Figure 1D&E). Furthermore, A549.PM cells also increase tumor weight (Figure 1F) and tumor volume (Figure 1G&H).
3.2 Long-term exposure to PM increases AREG expression in lung cancer
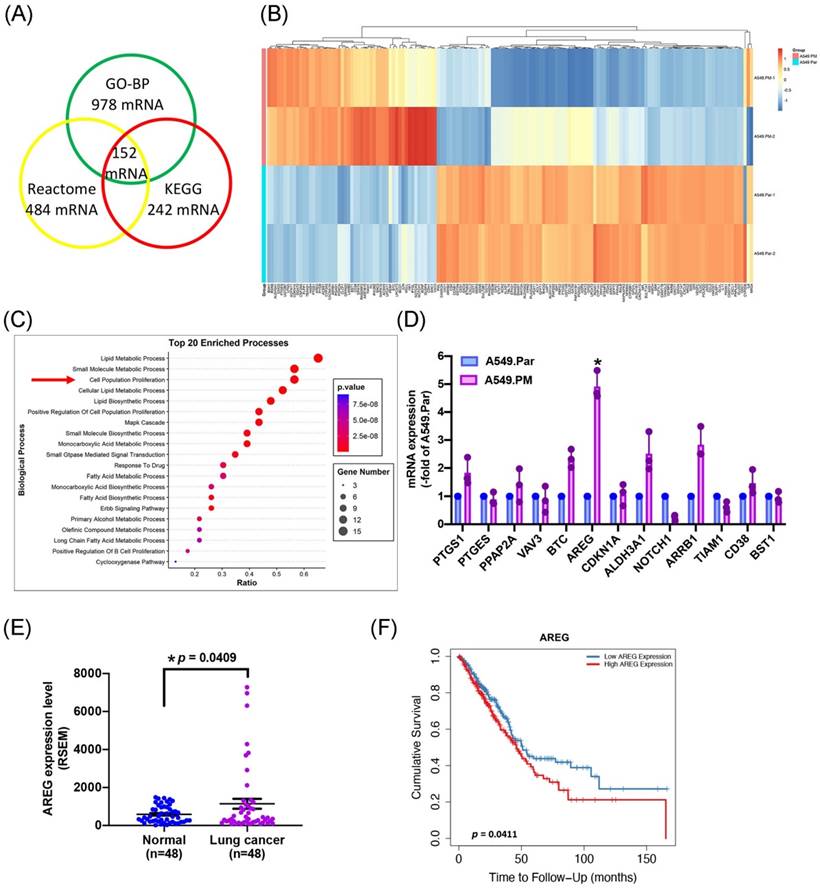
To identify the molecules responsible for the proliferative biological effects of PM, we performed RNA-Seq analysis in A549.Par and A549.PM cells. We found that Gene Ontology Biological Processes (GO-BP) (false discovery rate [FDR] < 0.01, fold change [FC] > 2) altered the expression of 978 genes, Reactome altered 484 genes, and KEGG altered 242 genes, with 152 genes altered under all three conditions (Figure 2A&B). RNA-Seq data also provided strong support for our earlier observations that PM promotes lung cancer cell proliferation (Figure 2C). These data confirm that long-term exposure to PM increases lung cancer cell growth and proliferation.
To determine which gene has the highest expression as predicted from the RNA-seq analysis in the proliferation group, the expression of mRNA was verified by qPCR. The results showed that PTGS1, BTC, AREG, ALDH3A1, and ARRB1 were secreted by A549.PM cells were upregulated compared to A549.Par cells. Among these genes, AREG levels in A549.PM cells were higher than all other up-regulated genes (Figure 2D). To examine the effects of AREG levels on lung cancer progression, we investigated the clinical significance of AREG identified in lung adenocarcinoma samples from the TCGA database. We found higher levels of AREG mRNA expression in tumor tissue than in adjacent normal tissue (Figure 2E). In Timer 2.0 analysis, progressively higher AREG expression among lung cancer patients was associated with correspondingly lower overall survival rates (Figure 2F). These results indicate that PM increases AREG expression in lung cancer cells.
PM increases cell proliferation in lung cancer. (A&B) The cell proliferation ability of A549.Par and A549.PM cells was analyzed by MTT and CCK-8 assays. (C) The cell growth of A549.Par and A549.PM cells was measured by colony assay. (D-F) A549.Par and A549.PM cells were subcutaneously injected into the right flanks of BALB/c-nu mice. Four weeks later, the mice were sacrificed, and the tumors were excised and weighed. (G&H) At week 4, the mice were sacrificed, and the tumors were excised and photographed. *p < 0.05 compared with A549.Par expression.
PM promotes AREG expression in lung cancer cells. (A&B) Overlap of genes in GO-BP, Reactome, and KEGG pathways expressed in the A549.PM cell line. The cluster heatmap shows the expression of 152 genes in the A549.PM cell line compared to the A549.Par cell line. (C) Gene ontology (GO) enrichment analysis of genes from the two groups. (D) The expression levels of selected up-regulated genes were verified by qPCR. (E) AREG mRNA expression in tumor tissue and adjacent normal tissue was analyzed using records from the TCGA database. (F) Timer 2.0 analysis determined the overall survival rates of patients with lung adenocarcinoma and levels of AREG expression. *p < 0.05 compared with A549.Par expression.
3.3 PM-induced increases in AREG expression and facilitates lung cancer proliferation
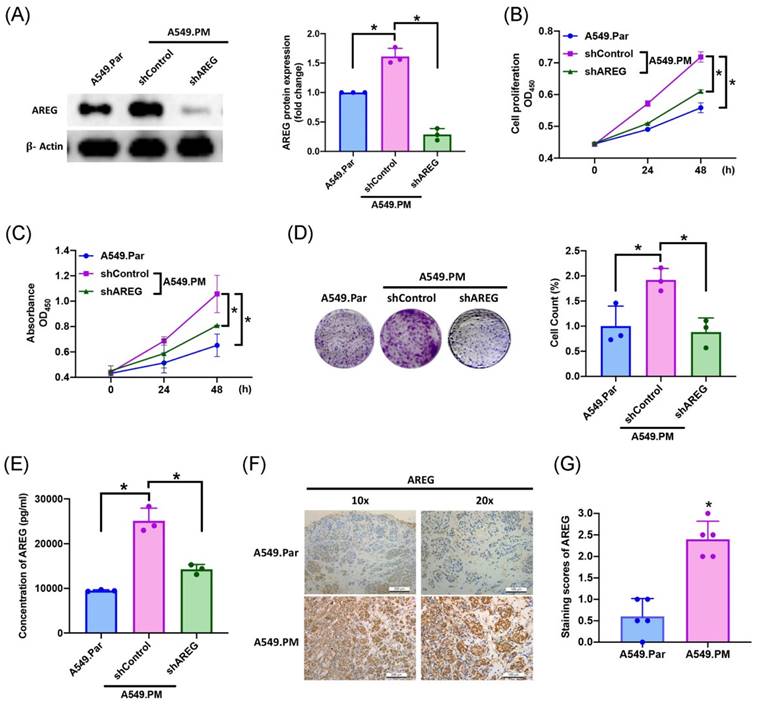
In several cancers (ovarian, breast, and lung cancers), high AREG expression correlates with a worse prognosis [31-33]. To validate whether the amount of AREG secreted from lung cancer cells affects cell proliferation, we found that long-term exposure to PM promotes lung cancer AREG protein expression, which is significantly decreased when transfected with AREG shRNA, as determined by western blot assay (Figure 3A). We also observed a significant decrease in cell proliferation ability as assessed by MTT and CCK-8 assays (Figure 3B&C). Colony assay results also showed that transfection with AREG shRNA significantly decreased cell growth ability (Figure 3D). According to ELISA results, PM exposure increased AREG expression, while transfection with AREG shRNA inhibited this phenomenon (Figure 3E). IHC staining of tumor tissue from in vivo study demonstrated that the expression of AREG significantly increased in the A549.PM group compared to the A549.Par group (Figure 3F&G). These results reveal that PM promotes AREG-dependent lung cancer proliferation.
PM-induced AREG expression enhances lung cancer growth and proliferation. A549.PM cells were transfected with AREG shRNA for 24 hr. (A) Western blot analysis examined levels of AREG and β-actin. The graphs densitometric analysis of protein expression normalized to β-actin. (B&C) Cell proliferation was analyzed by MTT and CCK-8 assays for 2 days. (D) Cell growth ability was measured by colony assay. (E) Levels of AREG expression were examined by ELISA assays. (F&G) Tumors from sacrificed mice were stained with AREG and scored by intensity from 0 to 3 (where 0 = negative; 1 = weak; 2 = moderate; 3 = strong). *p < 0.05 compared with A549.Par or A549.PM expression.
3.4 PM-induced increases in AREG-dependent lung cancer proliferation through glutamine metabolism
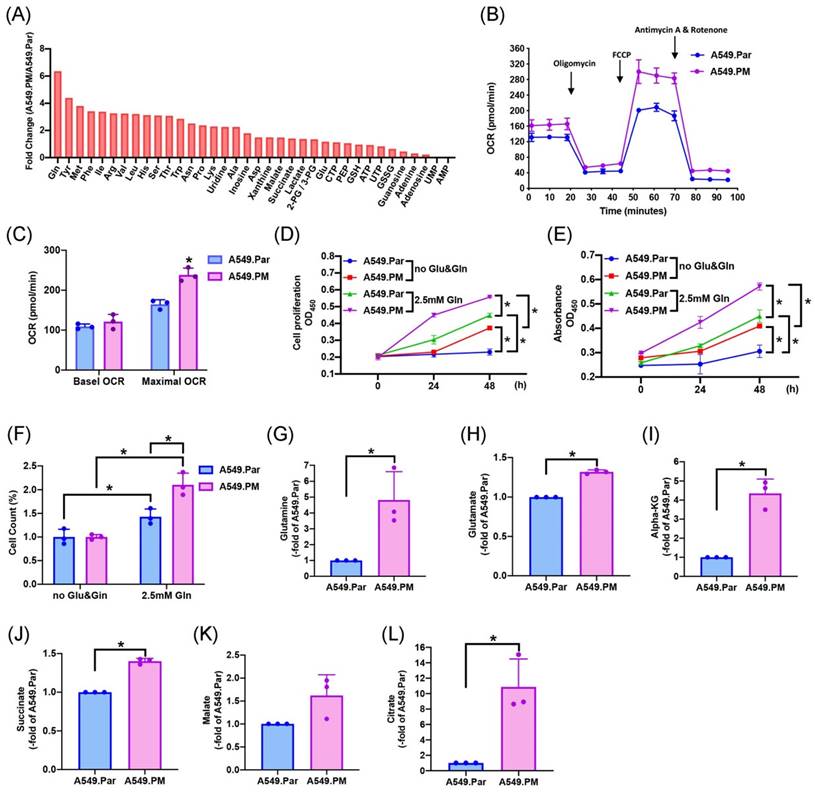
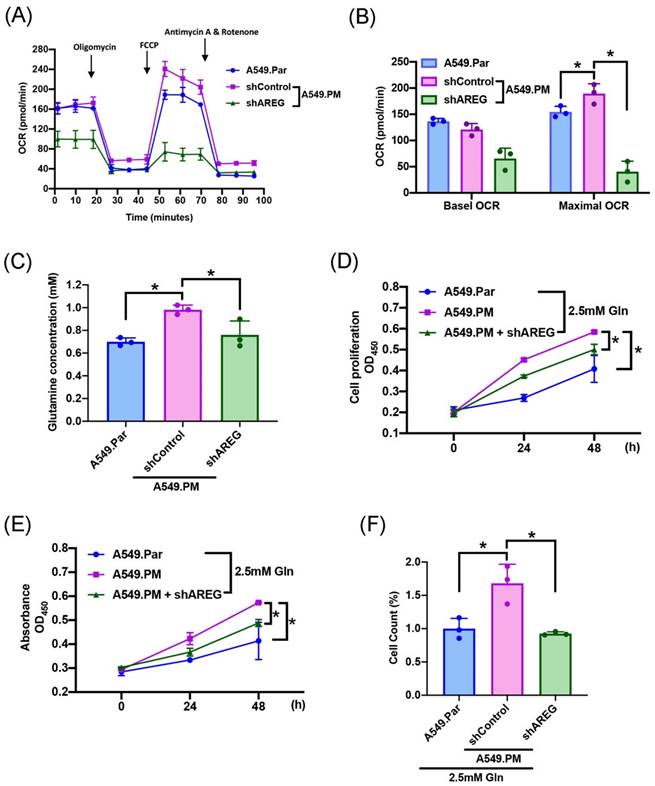
The two main sources of energy for cell development are the metabolism of glucose and glutamine [17]. We sought to determine which metabolic pathway enhances lung cancer progression. Mass spectrometry data showed that glutamine concentration is higher in A549.PM cells than in A549.Par cells (Figure 4A). A549.PM cells also exhibited increased OCR compared to A549.Par cells (Figure 4B&C). In the MTT, CCK-8, and colony assay, cells were seeded in glucose and glutamine-free medium. We found that treating with 2.5 mM glutamine in both A549.Par and A549.PM cells resulted in higher cell growth and proliferation than in the glucose and glutamine-free groups (Figure 4D-F). Additionally, we found that A549.PM cells exhibit increased levels of glutamine-derived TCA cycle metabolites such as glutamine, glutamate, alpha-KG, succinate, malate, and citrate compared to the A549.Par group (Figure 4G-L). Our Seahorse and glutamine concentration assay showed that when A549.PM cells were transfected with AREG shRNA, there was a significant decrease in OCR and glutamine concentration (Figure 5A-C). We also found that when A549.PM cells were transfected with AREG shRNA, there was a significant decrease in cell growth and proliferation ability (Figure 5D-F). This finding implies that PM stimulates AREG expression and glutamine metabolism in lung cancer to encourage tumor cell growth.
PM increases TCA cycle metabolites through mitochondrial glutaminolysis, thereby increasing lung cancer glutamine metabolism and promoting cell proliferation. (A) Metabolite concentration analysis in the mitochondria from A549.Par and A549.PM cells by mass spectrometry assay. (B&C) Oxygen consumption rate (OCR) was measured in A549.Par and A549.PM cells following the addition of oligomycin (1 µM), FCCP (0.5 µM), and the electron transport inhibitor rotenone/antimycin A (0.5 µM). (D&E) A549.Par and A549.PM cells were incubated with 2.5 mM glutamine. Cell proliferation was analyzed by MTT and CCK-8 assays for 2 days. (F) A549.Par and A549.PM cells were incubated with 2.5 mM glutamine for 7 days. Cell growth ability was measured by colony assay. (G-L) The metabolic abundance of glutamine-derived TCA metabolites (glutamine, glutamate, α-KG, succinate, malate, and citrate) in the purified mitochondria from A549.PM cells. *p < 0.05 compared with A549.Par or A549.PM expression.
PM stimulates the expression of AREG and facilitates the glutamine metabolism of lung cancer cells to encourage cell proliferation. A549.PM cells were transfected with AREG shRNA for 24 hr. (A&B) Oxygen consumption rate (OCR) was measured by following the addition of oligomycin (1 µM), FCCP (0.5 µM), and electron transport inhibitor, rotenone/antimycin A (0.5 µM). (C) The concentration of glutamine in A549.Par and A549.PM cells. (D&E) A549.Par and A549.PM cells were incubated with 2.5 mM glutamine. The cell proliferation was analyzed by MTT and CCK-8 assays for 2 days. (F) A549.Par and A549.PM cells were incubated with 2.5 mM glutamine for 7 days. The cell growth ability was measured by colony assay. *p < 0.05 compared with A549.Par or A549.PM expression.
3.5 PM increases AREG-dependent glutamate metabolism and lung cancer proliferation through SLC1A5
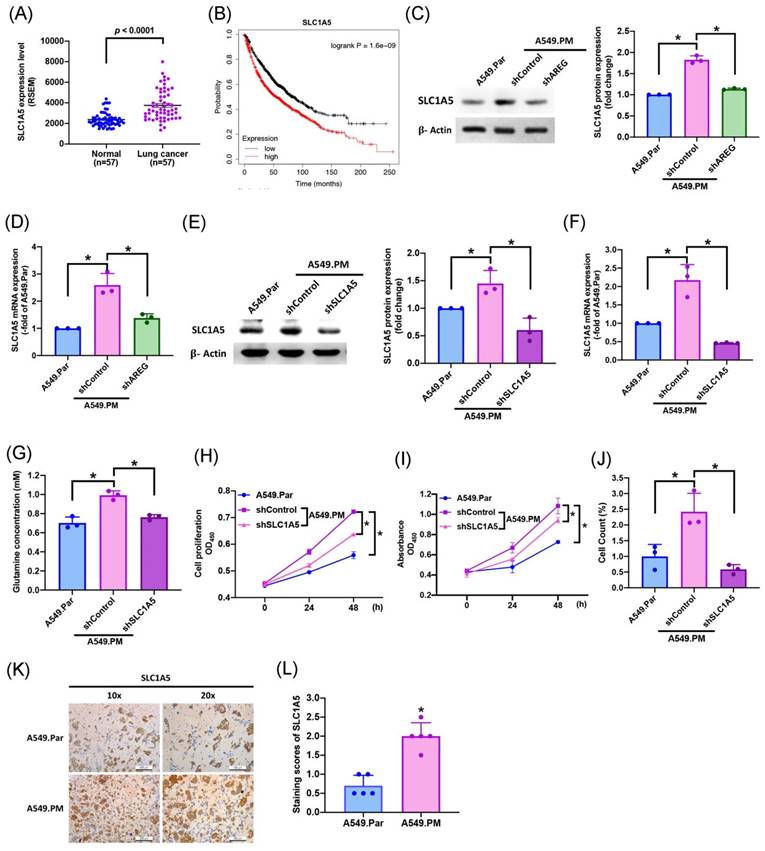
Several previous studies have shown that SLC1A5 is the most important transporter in glutamine metabolism [34, 35]. To identify whether long-term PM exposure is associated with SLC1A5 levels in A549.PM cell lines, our data showed that levels of SLC1A5 mRNA expression were much higher in tumor tissue than in adjacent normal tissue (Figure 6A). In Kaplan-Meier analysis, progressively higher SLC1A5 expression among lung cancer patients was associated with correspondingly lower overall survival rates (Figure 6B). We found that when A549.PM cells were transfected with AREG shRNA, there was a significant decrease in SLC1A5 protein level and mRNA expression (Figure 6C&D). Additionally, long-term exposure to PM increased lung cancer SLC1A5 protein and mRNA expressions, which were significantly decreased when transfected with SLC1A5 shRNA, as determined by western blot and RT-qPCR assay (Figure 6E&F). Our glutamine concentration assay also showed that when A549.PM cells were transfected with AREG shRNA, there was a significant decrease in glutamine concentration (Figure 6G). Next, we aimed to examine whether the level of SLC1A5 promotes long-term PM exposure to lung cancer cell progression. We found that when transfected with SLC1A5 shRNA, there was a significant decrease in cell proliferation and growth by MTT, CCK-8 and colony assays compared with the A549.PM group (Figure 6H-J). In vivo, IHC staining of tumor tissue also demonstrated that the expression of SLC1A5 significantly increased in the A549.PM group compared to the A549.Par group (Figure 6K&L). These results reveal that PM-induced AREG expression through SLC1A5 affects glutamate metabolism and proliferation in lung cancer cells.
PM increases AREG-dependent glutamate metabolism and lung cancer proliferation through SLC1A5. (A) SLC1A5 mRNA expression in tumor tissue and adjacent normal tissue was analyzed using records from The Cancer Genome Atlas (TCGA) database. (B) Kaplan-Meier analysis determined levels of SLC1A5 expression and overall survival rates of patients with lung cancer. (C&D) A549.PM cells were transfected with AREG shRNA for 24 hr. The levels of SLC1A5 and β-actin were examined by Western blot analysis and the graphs densitometric analysis of protein expression normalized to β-actin, while the expression of SLC1A5 was verified using RT-qPCR. (E&F) A549.PM cells were transfected with SLC1A5 shRNA for 24 hr. The levels of SLC1A5 and β-actin were examined by Western blot analysis and the graphs densitometric analysis of protein expression normalized to β-actin, while the expression of SLC1A5 was verified using quantitative RT-qPCR. (G) The concentration of glutamine in A549.Par and A549.PM cells. (H&I) Cell proliferation was analyzed by MTT and CCK-8 assays for 2 days. (J) Cell growth ability was measured by colony assay. (K&L) Tumors from sacrificed mice were stained with SLC1A5 and scored by intensity from 0 to 3 (where 0 = negative; 1 = weak; 2 = moderate; 3 = strong). *p < 0.05 compared with A549.Par or A549.PM expression.
3.6 PM facilitates AREG-dependent glutamine metabolism through EGFR/ PI3K/AKT/mTOR signaling pathway
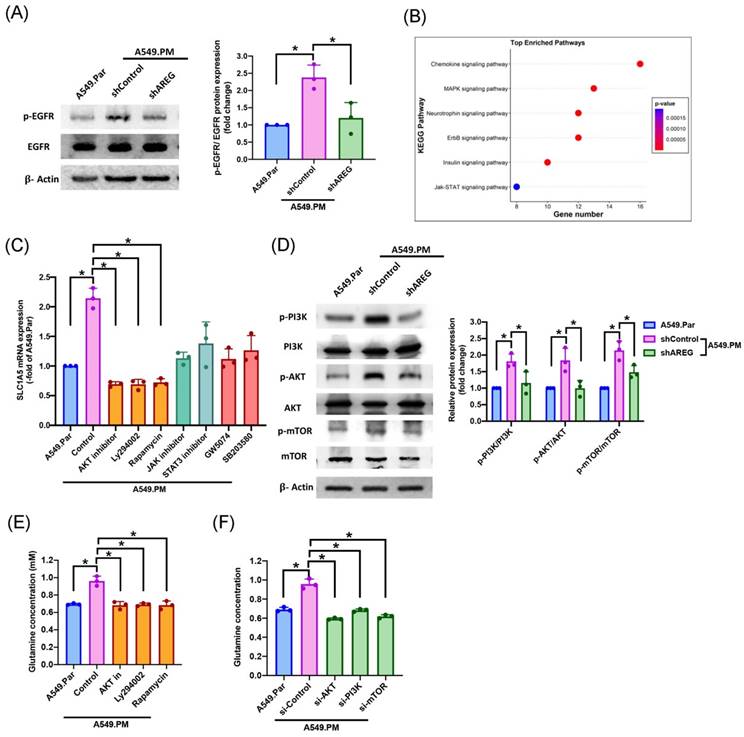
The AREG/EGFR axis induces tumor cell proliferation through multiple signaling pathways [11]. We sought to determine the interaction between AREG and EGFR. Our results showed that when A549.PM cells were transfected with AREG shRNA, there was a significant decrease in p-EGFR level (Figure 7A). Next, we aimed to determine which signaling pathway is involved in PM promoting glutamine metabolism. RNA-Seq data also revealed that A549.PM cells significantly upregulated several EGFR downstream and proliferation signaling pathways such as chemokine, MAPK, neurotrophin, ErbB, insulin, and JAK/STAT signaling pathways (Figure 7B). RT-qPCR data demonstrated that when A549.PM cells were treated with PI3K (Ly294002) /AKT/mTOR (Rapamycin), JAK/STAT, and RAF (GW5074)/p38 (SB203580) inhibitors, the PI3K/AKT/mTOR cascade showed a greater decrease in SLC1A5 mRNA expression compared to the JAK/STAT and RAF/p38 cascades (Figure 7C). Our data also confirmed that transfection with AREG shRNA significantly decreased PI3K/AKT/mTOR phosphorylation expression (Figure 7D). Additionally, we showed that treatment of A549.PM cells with PI3K/AKT/mTOR inhibitors and their respective siRNAs inhibited glutamine concentration (Figure 7E&F). These results suggest that PM induces AREG generation through EGFR/PI3K/AKT/mTOR signaling, thereby increasing SLC1A5 to promote glutamine metabolism in lung cancer.
PM-induced AREG expression promotes glutamine metabolism in lung cancer through the EGFR/PI3K/AKT/mTOR pathway. (A) A549.PM cells were transfected with AREG shRNA for 24 hr. Western blot analysis examined levels of EGFR phosphorylation and β-actin. The graphs demonstrate densitometric analysis of p-EGFR/EGFR. (B) Signaling pathways in A549.PM cells were analyzed by RNA Sequencing. (C) Cells were treated with indicated inhibitors, and the expression of SLC1A5 was verified by qPCR. (D) A549.PM cells were transfected with AREG shRNA for 24 hr. Western blot analysis examined levels of PI3K, AKT, and mTOR phosphorylation and β-actin. The graphs demonstrate densitometric analysis of p-PI3K/PI3k, p-AKT/AKT and p-mTOR/mTOR. A549.PM cells were incubated with PI3K, AKT, and mTOR inhibitor or PI3K, AKT, and mTOR siRNA for 24 h. (E&F) Levels of glutamine expression were examined by glutamine Assay Kit. *p < 0.05 compared with A549.Par or A549.PM expression.
4. Discussion
Long-term exposure to PM is associated with an increased risk of lung cancer [36]. Lung cancer cells stimulated with PM demonstrated higher proliferation, migration, and invasion of cancer cells [37]. We found that long-term exposure to PM in lung cancer promotes the highest cell proliferation. Our in vivo data showed that A549.PM cells have higher tumor growth, tumor weight and tumor volume than A549.Par cells. This study elucidated the functions of PM-promoted lung cell proliferation both in vitro and in vivo.
The presence of AREG is closely linked to the oncogenic process; increasingly higher levels of AREG expression correlate with worse prognosis in several cancers, such as ovarian, glioma, head and neck, breast, and lung cancers [31, 38, 39]. AREG overexpression has been demonstrated in a wide variety of human lung cancer tissues, and AREG is assumed to play an important role in promoting lung cancer proliferation [40]. AREG also promotes tumor growth in pancreatic, colorectal, liver, and lung cancers [41-43]. Our analysis of RNA sequencing data identified that long-term exposure to PM upregulates AREG expression in lung cancer cells. We also found that treatment with AREG shRNA reduces lung cancer tumor growth and cell proliferation compared to the A549.PM group. Our results suggest that targeting AREG could be a valuable therapeutic approach for lung cancer exposed with PM.
Metabolic reprogramming is a hallmark of cancer, and targeting metabolism is a potential therapeutic strategy [44, 45]. Cancer cells mainly obtain nutrients through metabolic pathways involving glucose, fatty acids, glutamine, and small molecule amino acids to support their growth. During cancer cell growth and survival, glucose and glutamine are the main metabolic pathways that significantly influence it. Cancer cells break down glucose into pyruvate through glycolysis and utilize the high-energy compounds (ATP and NADH) and lactate released during this process to fuel their growth [34, 35]. Additionally, cancer cells rely on glutamine-mediated TCA cycle to provide essential metabolites for growth [46]. Our analysis of mass spectrometry data and seahorse assay identified that long-term exposure to PM increases glutamine concentration and OCR in lung cancer cells. Our findings also revealed that long-term exposure to PM increases glutamine metabolism, promoting lung cancer tumor growth and cell proliferation. As air pollution worsens, PM not only accumulates in the cytoplasm but also mitochondria and cell nuclei. This uptake activates cell signaling pathways, further promoting cell proliferation [47, 48]. We also found that long-term exposure to PM increases the concentration of glutamine-derived TCA cycle metabolites such as glutamine, glutamate, alpha-KG, succinate, malate, citrate, and aspartate. PM augments AREG-dependent lung cancer proliferation through increasing glutamine metabolism. Therefore, regulating glutamine metabolism is a novel avenue for developing remedies for lung cancer growth exposed to PM.
SLC1A5 is responsible for transporting glutamine. Previous research has confirmed that SLC1A5 contributes to cancer progression by promoting glutamine metabolism [49]. Many studies have shown that overexpression of SLC1A5 promotes progression in triple-negative basal-like breast cancer [49], prostate cancer [50] and lung cancer [51]. Our study found that long-term exposure to PM increased SLC1A5 expression in lung cancer. We also found that high levels of AREG expression, promoted by PM, can upregulate SLC1A5 expression and enhance lung cancer tumor growth and cell proliferation. Our results suggest that the functional importance of glutamine transport mediated by SLC1A5 in cancer cell proliferation and survival, and SLC1A5 in the glutamine metabolism pathway, is an important therapeutic target for lung cancer.
AREG is a ligand of the epidermal growth factor receptor [9]. Through binding to EGFR, AREG activates multiple downstream signaling pathways, including Ras/MAPK, PI3K/AKT, mTOR, and STAT [52, 53]. These signaling cascades modulate gene expression and induce various cellular responses such as proliferation, survival, invasiveness, and angiogenesis [11]. AREG, through PI3K/Akt signaling, induces its expression to promote epithelial ovarian cancer cell migration and proliferation [54]. Our analysis of RNA sequencing data identified that long-term exposure to PM upregulates the PI3K/AKT/mTOR, JAK/STAT, and RAF/p38 signaling pathways. When A549.PM cells are treated with PI3K/AKT/mTOR, JAK/STAT, and RAF/p38 inhibitors, the PI3K/AKT/mTOR cascade significantly reduces SLC1A5 mRNA expression compared to the JAK/STAT and RAF/p38 cascades. We also found that AREG, via EGFR/PI3K/AKT/mTOR signaling, regulates glutamine concentration and SLC1A5 expression. These results suggest that the EGFR/PI3K/AKT/mTOR pathway plays critical role in PM-mediated glutamate metabolism in lung cancer. Our results illustrate that targeting AREG might offer a novel treatment approach for lung cancer. In the future, whether to use EGFR inhibitors such as erlotinib (Tarceva®) and gefitinib (Iressa®) for targeting AREG and decreasing long-term exposure to PM-induced lung cancer patients is worthy of further study.
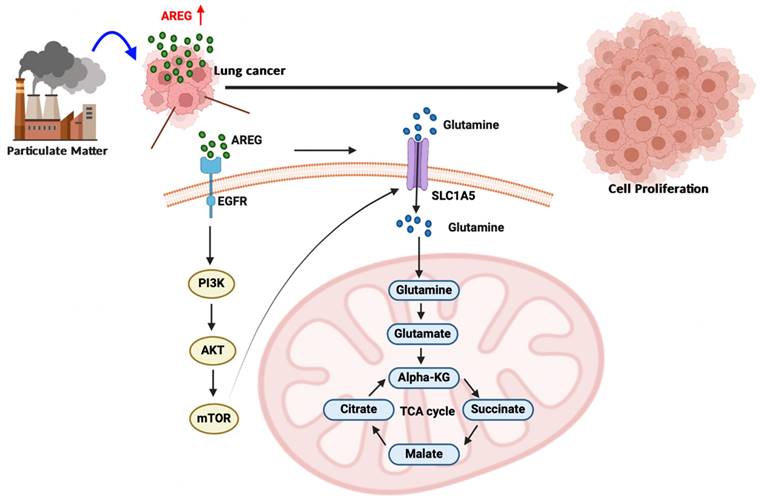
A schematic diagram showing how PM increases AREG-dependent glutamine metabolism and lung cancer proliferation. Long-term exposure to PM leads to increased AREG expression, which in turn induces SLC1A5 expression through the EGFR/PI3K/AKT/mTOR signaling pathway. The upregulation of SLC1A5 facilitates the accumulation of glutamine-derived TCA cycle metabolites, thereby promoting lung cancer cell growth and proliferation.
Previous studies have demonstrated long-term exposure of mice to PM through intratracheal instillation [55, 56]. A limitation of our research is that we didn't have the equipment for intratracheal instillation or the required expertise. Therefore, in our in vivo study, PM powder was utilized for long-term exposure to lung cancer cell lines and subsequently injected subcutaneously into mice. In future studies, using intratracheal instillation in vivo model, which more closely mimics natural exposure to PM, can be employed to more clearly explore how long-term exposure to PM increases the expression of AREG and promotes lung cancer growth through glutamine metabolism.
5. Conclusion
In conclusion, our study results demonstrate that long-term exposure to PM increases AREG expression in lung cancer cells. AREG, in turn, upregulates the glutamine transporter SLC1A5 and promotes the accumulation of glutamine-derived TCA cycle metabolites via the EGFR/PI3K/AKT/mTOR signaling pathway, thereby promoting lung cancer cell growth and proliferation (Figure 8).
Supplementary Material
Supplementary materials and methods, table.
Acknowledgements
Funding
This study was supported by grants from the National Science and Technology Council (NSTC 111-2320-B-468-006-), the Ministry of Science and Technology of Taiwan (MOST 110-2320-B-039-022-MY3), China Medical University Hospital (DMR-113-124), Asia University Hospital (11051008; 11151019), Changhua Christian Hospital (108-CCH-IRP-028).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
Ya-Jing Jiang: Conceptualization, Data curation, Formal analysis, Methodology, Software and Writing - original draft. Trung-Loc Ho: Data curation, Formal analysis and Methodology. Chia-Chia Chao: Investigation and Project administration. Xiu-Yuan He: Formal analysis and Methodology. Po-Chun Chen: Investigation and Project administration. Fang-Ju Cheng: Methodology and Software. Wei-Chien Huang: Project administration and Software. Chang-Lun Huang: Investigation and Resources. Po-I Liu: Resources, Supervision and Writing - review & editing. Chih-Hsin Tang: Conceptualization, Resources, Supervision and Writing - review & editing. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhu F, Ding R, Lei R, Cheng H, Liu J, Shen C. et al. The short-term effects of air pollution on respiratory diseases and lung cancer mortality in Hefei: A time-series analysis. Respir Med. 2019;146:57-65
2. Perez Velasco R, Jarosinska D. Update of the WHO global air quality guidelines: Systematic reviews - An introduction. Environ Int. 2022;170:107556
3. Kim KH, Kabir E, Kabir S. A review on the human health impact of airborne particulate matter. Environ Int. 2015;74:136-43
4. Bazyar J, Pourvakhshoori N, Khankeh H, Farrokhi M, Delshad V, Rajabi E. A comprehensive evaluation of the association between ambient air pollution and adverse health outcomes of major organ systems: a systematic review with a worldwide approach. Environ Sci Pollut Res Int. 2019;26:12648-61
5. Halonen JI, Lanki T, Yli-Tuomi T, Tiittanen P, Kulmala M, Pekkanen J. Particulate air pollution and acute cardiorespiratory hospital admissions and mortality among the elderly. Epidemiology. 2009;20:143-53
6. Hamra GB, Guha N, Cohen A, Laden F, Raaschou-Nielsen O, Samet JM. et al. Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ Health Perspect. 2014;122:906-11
7. Lin H, Zhang X, Feng N, Wang R, Zhang W, Deng X. et al. LncRNA LCPAT1 Mediates Smoking/ Particulate Matter 2.5-Induced Cell Autophagy and Epithelial-Mesenchymal Transition in Lung Cancer Cells via RCC2. Cell Physiol Biochem. 2018;47:1244-58
8. Chen YJ, Roumeliotis TI, Chang YH, Chen CT, Han CL, Lin MH. et al. Proteogenomics of Non-smoking Lung Cancer in East Asia Delineates Molecular Signatures of Pathogenesis and Progression. Cell. 2020;182:226-44 e17
9. Holtan SG, DeFor TE, Panoskaltsis-Mortari A, Khera N, Levine JE, Flowers MED. et al. Amphiregulin modifies the Minnesota Acute Graft-versus-Host Disease Risk Score: results from BMT CTN 0302/0802. Blood Adv. 2018;2:1882-8
10. Kuramochi H, Nakajima GO, Hayashi K, Araida T, Yamamoto M. Amphiregulin/epiregulin mRNA Expression and Primary Tumor Location in Colorectal Cancer. Anticancer Res. 2019;39:4729-36
11. Berasain C, Avila MA. Amphiregulin. Semin Cell Dev Biol. 2014;28:31-41
12. Steponaitis G, Kazlauskas A, Skiriute D, Vaitkiene P, Skauminas K, Tamasauskas A. Significance of Amphiregulin (AREG) for the Outcome of Low and High Grade Astrocytoma Patients. J Cancer. 2019;10:1479-88
13. Zaiss DMW, Gause WC, Osborne LC, Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42:216-26
14. Florentin J, Zhao J, Tai YY, Sun W, Ohayon LL, O'Neil SP. et al. Loss of Amphiregulin drives inflammation and endothelial apoptosis in pulmonary hypertension. Life Sci Alliance. 2022 5
15. Busser B, Sancey L, Josserand V, Niang C, Khochbin S, Favrot MC. et al. Amphiregulin promotes resistance to gefitinib in nonsmall cell lung cancer cells by regulating Ku70 acetylation. Mol Ther. 2010;18:536-43
16. Martinez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669-80
17. Zhao H, Li Y. Cancer metabolism and intervention therapy. Mol Biomed. 2021;2:5
18. Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678-84
19. Hassanein M, Qian J, Hoeksema MD, Wang J, Jacobovitz M, Ji X. et al. Targeting SLC1a5-mediated glutamine dependence in non-small cell lung cancer. Int J Cancer. 2015;137:1587-97
20. Yoo HC, Park SJ, Nam M, Kang J, Kim K, Yeo JH. et al. A Variant of SLC1A5 Is a Mitochondrial Glutamine Transporter for Metabolic Reprogramming in Cancer Cells. Cell Metab. 2020;31:267-83 e12
21. Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:749
22. Kim J, DeBerardinis RJ. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019;30:434-46
23. Ko YH, Lin Z, Flomenberg N, Pestell RG, Howell A, Sotgia F. et al. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance. Cancer Biol Ther. 2011;12:1085-97
24. Chen YF, Lu YH, Tsai HY. Crude extract of Desmodium gangeticum process anticancer activity via arresting cell cycle in G1 and modulating cell cycle-related protein expression in A549 human lung carcinoma cells. BioMedicine. 2022;12:31-9
25. Cheng FJ, Huynh TK, Yang CS, Hu DW, Shen YC, Tu CY. et al. Hesperidin Is a Potential Inhibitor against SARS-CoV-2 Infection. Nutrients. 2021 13
26. Lee KT, Su CH, Liu SC, Chen BC, Chang JW, Tsai CH. et al. Cordycerebroside A inhibits ICAM-1-dependent M1 monocyte adhesion to osteoarthritis synovial fibroblasts. Journal of food biochemistry. 2022: e14108.
27. Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang X. et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16:141
28. Su CM, Tsai CH, Chen HT, Wu YS, Chang JW, Yang SF. et al. Melatonin improves muscle injury and differentiation by increasing Pax7 expression. Int J Biol Sci. 2023;19:1049-62
29. Wu YY, Law YY, Huang YW, Tran NB, Lin CY, Lai CY. et al. Glutamine metabolism controls amphiregulin-facilitated chemoresistance to cisplatin in human chondrosarcoma. Int J Biol Sci. 2023;19:5174-86
30. Chao X, Yi L, Lan LL, Wei HY, Wei D. Long-term PM2.5 exposure increases the risk of non-small cell lung cancer (NSCLC) progression by enhancing interleukin-17a (IL-17a)-regulated proliferation and metastasis. Aging (Albany NY). 2020;12:11579-602
31. Chen JC, Lee IN, Huang C, Wu YP, Chung CY, Lee MH. et al. Valproic acid-induced amphiregulin secretion confers resistance to temozolomide treatment in human glioma cells. BMC Cancer. 2019;19:756
32. Huang YW, Lin CY, Tsai HC, Fong YC, Han CK, Huang YL. et al. Amphiregulin promotes cisplatin chemoresistance by upregulating ABCB1 expression in human chondrosarcoma. Aging (Albany NY). 2020;12:9475-88
33. Chen JC, Huang C, Lee IN, Wu YP, Tang CH. Amphiregulin enhances cell migration and resistance to doxorubicin in chondrosarcoma cells through the MAPK pathway. Mol Carcinog. 2018;57:1816-24
34. Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front Pharmacol. 2011;2:49
35. Bose S, Le A. Glucose Metabolism in Cancer. Adv Exp Med Biol. 2018;1063:3-12
36. Zhang Z, Zhu D, Cui B, Ding R, Shi X, He P. Association between particulate matter air pollution and lung cancer. Thorax. 2020;75:85-7
37. Yang B, Chen D, Zhao H, Xiao C. The effects for PM2.5 exposure on non-small-cell lung cancer induced motility and proliferation. Springerplus. 2016;5:2059
38. Carvalho S, Lindzen M, Lauriola M, Shirazi N, Sinha S, Abdul-Hai A. et al. An antibody to amphiregulin, an abundant growth factor in patients' fluids, inhibits ovarian tumors. Oncogene. 2016;35:438-47
39. Gao J, Ulekleiv CH, Halstensen TS. Epidermal growth factor (EGF) receptor-ligand based molecular staging predicts prognosis in head and neck squamous cell carcinoma partly due to deregulated EGF- induced amphiregulin expression. J Exp Clin Cancer Res. 2016;35:151
40. Hsu YL, Huang MS, Cheng DE, Hung JY, Yang CJ, Chou SH. et al. Lung tumor-associated dendritic cell-derived amphiregulin increased cancer progression. J Immunol. 2011;187:1733-44
41. Yamada M, Ichikawa Y, Yamagishi S, Momiyama N, Ota M, Fujii S. et al. Amphiregulin is a promising prognostic marker for liver metastases of colorectal cancer. Clin Cancer Res. 2008;14:2351-6
42. Yotsumoto F, Fukami T, Yagi H, Funakoshi A, Yoshizato T, Kuroki M. et al. Amphiregulin regulates the activation of ERK and Akt through epidermal growth factor receptor and HER3 signals involved in the progression of pancreatic cancer. Cancer Sci. 2010;101:2351-60
43. Gilmore JL, Gonterman RM, Menon K, Lorch G, Riese DJ 2nd, Robling A. et al. Reconstitution of amphiregulin-epidermal growth factor receptor signaling in lung squamous cell carcinomas activates PTHrP gene expression and contributes to cancer-mediated diseases of the bone. Mol Cancer Res. 2009;7:1714-28
44. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
45. Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267-77
46. Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, Haigis MC. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science. 2017;358:941-6
47. Zheng L, Dong H, Zhao W, Zhang X, Duan X, Zhang H. et al. An Air-Liquid Interface Organ-Level Lung Microfluidics Platform for Analysis on Molecular Mechanisms of Cytotoxicity Induced by Cancer-Causing Fine Particles. ACS Sens. 2019;4:907-17
48. Ferecatu I, Borot MC, Bossard C, Leroux M, Boggetto N, Marano F. et al. Polycyclic aromatic hydrocarbon components contribute to the mitochondria-antiapoptotic effect of fine particulate matter on human bronchial epithelial cells via the aryl hydrocarbon receptor. Part Fibre Toxicol. 2010;7:18
49. van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D. et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35:3201-8
50. Wang Q, Hardie RA, Hoy AJ, van Geldermalsen M, Gao D, Fazli L. et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J Pathol. 2015;236:278-89
51. Xue M, Hong W, Jiang J, Zhao F, Gao X. Circular RNA circ-LDLRAD3 serves as an oncogene to promote non-small cell lung cancer progression by upregulating SLC1A5 through sponging miR-137. RNA Biol. 2020;17:1811-22
52. Berasain C, Castillo J, Perugorria MJ, Prieto J, Avila MA. Amphiregulin: a new growth factor in hepatocarcinogenesis. Cancer Lett. 2007;254:30-41
53. Busser B, Sancey L, Brambilla E, Coll JL, Hurbin A. The multiple roles of amphiregulin in human cancer. Biochim Biophys Acta. 2011;1816:119-31
54. Bolitho C, Moscova M, Baxter RC, Marsh DJ. Amphiregulin increases migration and proliferation of epithelial ovarian cancer cells by inducing its own expression via PI3-kinase signaling. Mol Cell Endocrinol. 2021;533:111338
55. Chen Q, Wang Y, Yang L, Sun L, Wen Y, Huang Y. et al. PM2.5 promotes NSCLC carcinogenesis through translationally and transcriptionally activating DLAT-mediated glycolysis reprograming. J Exp Clin Cancer Res. 2022;41:229
56. Liu J, Li S, Fei X, Nan X, Shen Y, Xiu H. et al. Increased alveolar epithelial TRAF6 via autophagy-dependent TRIM37 degradation mediates particulate matter-induced lung metastasis. Autophagy. 2022;18:971-89
Author contact
![]() Corresponding authors: Chih-Hsin Tang, PhD; Email: chtangcmu.edu.tw. Department of Pharmacology, School of Medicine, China Medical University, Taichung, Taiwan; No. 91, Hsueh-Shih Road, Taichung, Taiwan; Tel: (886) 4-22053366 Ext. 7726. Po-I Liu, MD, PhD; Email: piliu0724com. Department of Thoracic Surgery, Asia University Hospital; No. 222, Fuxin Rd., Wufeng Dist., Taichung City 41354, Taiwan (R.O.C.); Tel: (886) 4-37061668 Ext. 1570.
Corresponding authors: Chih-Hsin Tang, PhD; Email: chtangcmu.edu.tw. Department of Pharmacology, School of Medicine, China Medical University, Taichung, Taiwan; No. 91, Hsueh-Shih Road, Taichung, Taiwan; Tel: (886) 4-22053366 Ext. 7726. Po-I Liu, MD, PhD; Email: piliu0724com. Department of Thoracic Surgery, Asia University Hospital; No. 222, Fuxin Rd., Wufeng Dist., Taichung City 41354, Taiwan (R.O.C.); Tel: (886) 4-37061668 Ext. 1570.

 Global reach, higher impact
Global reach, higher impact