10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(3):1065-1080. doi:10.7150/ijbs.104460 This issue Cite
Review
The Role of mRNA Modifications in Bone Diseases
Division of Spine Surgery, Department of Orthopaedics, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong 510515, China; The First School of Clinical Medicine, Southern Medical University, Guangzhou, Guangdong 510515, China.
# These authors contributed equally to this work.
Received 2024-9-30; Accepted 2024-12-24; Published 2025-1-13
Abstract
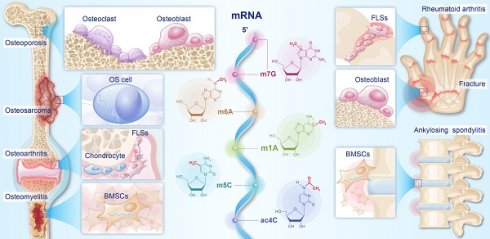
As a type of epigenetic modifications, mRNA modifications regulate the metabolism of mRNAs, thereby influencing gene expression. Previous studies have indicated that dysregulation of mRNA modifications is closely associated with the occurrence and progression of bone diseases (BDs). In this study, we first introduced the dynamic regulatory processes of five major mRNA modifications and their effects on the nucleus export, stability, and translation of mRNAs. We then summarized the mechanisms of mRNA modifications involved in the development of osteoporosis, osteoarthritis, rheumatoid arthritis, ankylosing spondylitis, fractures, osteomyelitis, and osteosarcoma. Finally, we reviewed therapeutic strategies for BDs based on the above mechanisms, focusing on regulating osteoblast and osteoclast differentiation, inhibiting cellular senescence and injury, and alleviating inflammation. This review identified mRNA modifications as potential targets for treating BDs and proposes perspectives on the diversity, targetability, and safety of mRNA-modifying therapies.
Keywords: messenger RNA, RNA modifications, bone diseases
1. Introduction
Gene expression is influenced by epigenetic modifications without altering nucleotide sequences [1]. With the rapid advancement of epigenetics, research has expanded beyond traditional DNA and histone modifications to encompass RNA modifications—a diverse and complex class of epigenetic modifications with over 170 types identified to date [2]. Among all RNAs, mRNA carries genetic information from DNA and serves as a template for protein synthesis. mRNA modifications regulate mRNA metabolism and affect gene expression. Recently, mRNA modifications have become a research hotspot for a wide range of diseases, especially cancer, neurodegenerative diseases, and autoimmune diseases [3-5].
An increasing number of studies have focused on mRNA modifications that regulate bone diseases (BDs). m6A, ac4C, and m5C modifications are shown to regulate the metabolism of mRNAs related to osteoclastic and osteogenic differentiation, and they then participate in the occurrence and development of osteoporosis (OP) [6-8]. In rheumatoid arthritis (RA), abnormalities in m5C and m6A modifications of related mRNAs have been reported to exacerbate joint inflammation and the proliferation of fibroblast-like synoviocytes (FLSs) [9, 10]. The proliferation and metabolic abnormalities observed in osteosarcoma (OS) cells can be linked to dysregulated mRNA modifications, specifically in m6A, m5C, and ac4C [11-13]. In addition, numerous studies have shown that mRNA modifications are also involved in the development of fractures, osteoarthritis (OA), osteomyelitis (OM), and ankylosing spondylitis (AS). Thus, abnormal mRNA modifications disrupt normal cellular functions, contributing to the development and progression of BDs.
Based on the above mechanisms of mRNA modifications, researchers have proposed corresponding therapeutic strategies for BDs. Viruses and certain drugs have been used to regulate mRNA modification regulators [6, 8, 14]. Specifically, adenovirus, lentivirus, and adeno-associated virus are used as vectors to elevate the levels of METTL14 (an m6A writer) and YTHDF1 (an m6A reader) in bone marrow mesenchymal stem cells (BMSCs), which contribute to increased osteoblast differentiation by enhancing the stability and translation of autophagy genes such as Beclin-1, ultimately alleviating bone loss in OP and accelerating bone healing in fractures [6, 14]. The m5C reader Y-box binding protein 1 (YBX1) is activated by sciadopitysin, resulting in increased bone mass in ovariectomized (OVX) mice by bolstering the stability of bone morphogenetic protein 4 (BMP4) [8]. These studies suggest that therapies targeting mRNA modifications could correct abnormal cellular functions and prevent the progression of BDs.
In this review, we introduced the dynamic regulatory mechanisms of five major mRNA modifications and their effects on mRNA metabolism (nucleus export, stability, translation, etc.). We then summarized the pathogenesis of the mRNA modifications involved in BDs, including fracture, OP, OA, RA, AS, OM, and OS. Finally, we concluded the strategies of mRNA-modifying therapies for BDs, mainly including the regulation of osteoblast and osteoclast differentiation, as well as the inhibition of cell senescence, injury, and inflammation. This review describes the cross-talk between mRNA modifications and BDs, which may uncover novel insights into BD mechanisms and pave the way for the development of epigenetic-based therapeutic approaches for BDs.
2. Overview of mRNA modifications (Fig. 1)
The understanding of mRNA modifications is gradually deepening with advanced detection technologies. Currently, over 170 types of RNA modifications have been discovered across various RNA species. Chemical modifications play important roles in the metabolism and function of mRNA, thus widely regulating human diseases. The regulators of mRNA modifications primarily consist of writers, erasers, and readers. Writers install chemical modifications on RNAs, while erasers remove these modifications to ensure the dynamic reversibility of modifications. Readers, on the other hand, recognize modification sites, bind to the modified RNAs, and regulate their metabolism and function. In this review, we focused on five major mRNA modifications closely related to BDs and summarized how these modifications regulated the metabolism of mRNAs through regulators.
2.1 N6-methyladenosine (m6A) (Fig. 1a)
m6A, a chemical modification resulting from the addition of a methyl group to the sixth nitrogen atom of adenosine, was initially discovered by Saneyoshi et al. in the valine tRNAs of Escherichia coli in 1969 [15]. With the emergence of technologies such as MeRIP-seq [16], m6A modifications have been discovered to be prevalent in mammalian mRNAs, and their abundance undergoes dynamic changes during mammalian development [17]. The m6A modifications of mRNA are primarily catalyzed by the methyltransferase complex (MTC), with METTL3, METTL14, and WTAP serving as core subunits [18]. METTL3 exhibits methyltransferase activity, whereas METTL14 and WTAP exert pivotal influences on m6A levels through their supporting functions [19]. METTL14 promotes the allosteric activation of METTL3, facilitating the recognition of RNA substrates [20]. WTAP mediates the localization of the METTL3-METTL14 complex to nuclear speckles and significantly influences their accumulation within these nuclear speckles [21]. In mammalian mRNAs, m6A modifications cluster near the stop codon and 3' untranslated region (3' UTR), often within the GGACU sequence [22]. This localization is likely related to the recruitment of the MTC to specific mRNA regions by VIRMA, RBM15, and RBM15B [23]. Moreover, the occurrence of m6A modifications in different regions of mRNA may influence mRNA metabolism in varying ways, contributing to diverse effects.
Currently, the identified m6A erasers primarily consist of fat mass and obesity-associated protein (FTO) and ALKBH5, which achieve the reversibility of m6A modifications [24]. FTO has been shown to remove m6A modifications from nuclear mRNAs by catalyzing their demethylation using α-ketoglutarate [25]. ALKBH5, on the other hand, catalyzes the demethylation of m6A through a Fe2+-dependent mechanism [26].
Typical m6A readers are members of the YTH domain-containing protein family, which directly recognizes m6A sites on mRNAs through their YTH domains, thus regulating mRNA metabolism [27]. YTHDC1 resides in the nucleus, regulating mRNA splicing and promoting nuclear export [28]. The YTHDC2 protein is localized in both the nucleus and cytoplasm, where it plays a role in mRNA degradation by recruiting the 5'→3' exoribonuclease XRN1 [29]. However, another study demonstrated that YTHDC2 has a relatively modest effect on mRNA stability and, more notably, can enhance translation efficiency [30]. The mechanism of YTHDC2 in regulating mRNA translation and stability needs to be further explored. It has been reported that YTHDF2 removes mRNAs from the translatable pool, thereby promoting their degradation [31], while YTHDF1 promotes translation by recruiting translation initiation factors and facilitating ribosome loading onto mRNA [32]. In addition, YTHDF3 promotes protein synthesis in synergy with YTHDF1 and affects the decay of methylated mRNAs mediated by YTHDF2 [33]. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) and heterogeneous nuclear ribonucleoproteins (hnRNPs) also serve as m6A readers [34, 35]. hnRNPs mainly regulate the processing of pri-miRNAs and pre-mRNAs [36, 37], while IGF2BPs enhance mRNA stability and translation by recruiting RNA stability factors and eukaryotic translation initiation factor (eIF) proteins [34, 38]. In summary, the differential expression of m6A readers in various biological contexts may lead to distinct outcomes of mRNA modifications, thereby playing opposing roles in the onset and progression of diseases.
The regulatory processes of mRNA modifications. Formation and erasure mechanisms of mRNA modifications, along with their impacts on mRNA.
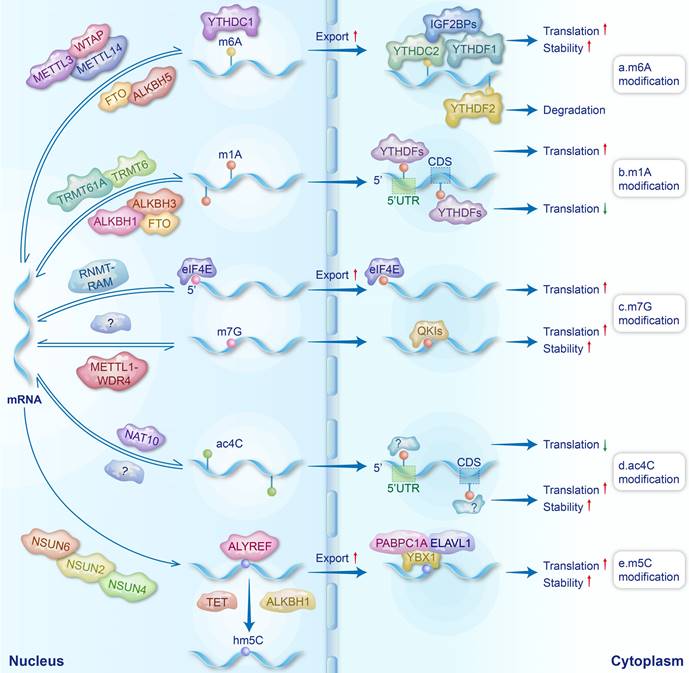
2.2 N1-methyladenosine (m1A) (Fig. 1b)
m1A is a methylation modification that occurs on the first nitrogen atom of RNA adenine. Since its initial discovery in 1961, m1A modifications have primarily been identified in the tRNAs and rRNAs of various fungi and bacteria [39, 40]. In 2016, Dominissini et al. demonstrated the existence of m1A modifications in eukaryotic mRNAs using MeRIP-seq technology [41]. In eukaryotic mRNAs, the ratio of m1A/A is shown to be lower compared to m6A/A and Ψ/U in the same sample. m1A is enriched near the start codon within the 5' untranslated region (5' UTR) and within the coding sequence (CDS) [42]. As m1A writers, RNA methyltransferase 61A (TRMT61A) exhibits enzymatic activity, while tRNA methyltransferase 6 (TRMT6) is responsible for substrate selection [43]. They form an α2β2 heterotetramer complex, TRMT6/61A, which catalyzes the m1A modifications in mRNAs [44]. Additionally, TRMT61B and TRMT10C have also been reported to be involved in the m1A modifications of mRNAs [45, 46]. The erasers of m1A include the AlkB family, namely ALKBH1, ALKBH3, and FTO [46]. It has been reported that m6A readers (YTHDF proteins) can also bind to m1A modifications and function as m1A readers [47]. The effects of m1A modifications on mRNA metabolism are position dependent. Specifically, m1A located at the 5' UTR, with its positive charge, influences the secondary/tertiary structure of mRNA around the translation initiation site and disrupts the formation of hydrogen bonds, thus interfering with Watson-Crick base pairing and promoting the translation initiation and the recruitment of elongation factors [41, 42]. Moreover, m1A modifications within CDS have been reported to affect translation elongation and potentially reduce translation efficiency [42].
2.3 N7-methylguanosine (m7G) (Fig. 1c)
m7G, the methylation of the seventh nitrogen atom on guanine in RNAs, can be categorized into m7G cap modifications and internal m7G modifications. As early as 1975, Muthukrishnan et al. discovered m7G modifications in the 5'-cap structure of mRNAs in viruses and erythroblast cells [48]. The methylation of the guanine cap in mammalian mRNAs is catalyzed by RNA guanine-N7 methyltransferase (RNMT) and RNMT-activating miniprotein [49]. The 5' cap modified with m7G can protect RNAs from hydrolysis by nucleases and regulate the stability, nuclear export, splicing, and translation of mRNAs by eIF4E and cap-binding complexes [50-53]. As the “writer” of internal m7G modifications in mRNAs, the METTL1-WDR4 complex mainly catalyzes the m7G modifications in 5' UTRs, CDS, and 3' UTRs [54]. The m7G modifications within mRNA have been shown to promote the translation process [54]. Quaking proteins have been identified as readers of mRNA internal m7G modifications. These proteins shuttle m7G-modified transcripts into stress granules to enhance mRNA stability and reduce translational efficiency [55]. To date, the erasers responsible for removing m7G modifications have yet to be discovered. Identifying these erasers is essential for elucidating the comprehensive regulatory mechanisms through which m7G modifications influence mRNAs.
2.4 N4-acetylcytosine (ac4C) (Fig. 1d)
ac4C is an acetylation modification that occurs on the fourth nitrogen atom of cytosine [56], and its abundance is much lower than other RNA modifications [57]. Early research on ac4C primarily focused on tRNAs and rRNAs in bacteria and fungi [58, 59]. In 2018, ac4C was first discovered in the mRNAs of HeLa cells [60]. The comprehension of ac4C remains elementary, and so far, NAT10 stands as the sole identified writer of ac4C, primarily localized within the nucleus [61, 62]. It is noteworthy that ac4C in different regions of mRNAs has diverse effects on translation. ac4C modifications in CDS significantly enhance the stability of mRNAs, leading to improved translation efficiency [60]. Furthermore, ac4C modifications at the codon wobble position were found to facilitate the recognition of cognate tRNAs to mRNAs, resulting in the upregulation of translation efficiency [63]. Conversely, acetylation in the 5' UTR can suppress translation initiation [64]. Due to limited research on ac4C, the identification of ac4C erasers and readers has not yet been reported. Notably, remodelin has been identified as an inhibitor of NAT10 [65].
2.5 5-methylcytosine (m5C) (Fig. 1e)
The m5C modifications of RNAs involve the methylation of the fifth carbon atom of cytosine [66]. m5C is particularly prominent in tRNAs and rRNAs but relatively rare in mammalian mRNAs, accounting for only 0.02%-0.09% of the total cytosine content [67, 68]. The majority of m5C modifications in mRNAs are mediated by NSUN2 and NSUN6, while the remaining modifications are attributed to NSUN4 [69-71]. Unlike other RNA modifications, the erasers of m5C do not directly remove the methyl group but rather oxidize m5C to products such as 5-hydroxymethylcytosine [72]. The erasers of m5C include the ten-eleven translocation (TET) family of demethylases (TET1, TET2, and TET3), as well as ALKBH1, which prevents the binding of m5C reader proteins by oxidizing m5C [73, 74].
Currently identified m5C readers include Aly/REF export factor (ALYREF) [69], YBX1 [75], as well as YTHDF2 and SRSF2 [76, 77]. ALYREF regulates the nuclear export of mRNAs [69], while YBX1 modulates mRNA translation and enhances mRNA stability by recruiting poly(A) binding protein 1 and ELAV-like RNA binding protein 1 to mRNAs [72, 78, 79].
3. Role of mRNA modifications in BDs (Fig. 2)
BDs, encompassing diverse bone, joint, and soft tissue issues, are a major disability cause globally, with significant economic implications. Many studies have emphasized that chemical modifications within mRNAs affect the physiological and pathological processes of cells involved in BDs, thus playing a crucial role in BDs, as shown in Table 1. This review introduces the important roles of mRNA modifications in the following seven BDs.
mRNA modifications in bone diseases.
| Modification | Regulator | Disease | Expression (sample) | Mechanism/function | Refs. |
|---|---|---|---|---|---|
| m6A | METTL14 | osteoporosis | Downregulated (human and mouse) | METTL14-m6A-NFATc1, SIRT1, Beclin-1, SMAD1, GPX4 and TCF1/RUNX2 mRNA modulating osteoblast and osteoclast differentiation | [6,85-89] |
| ankylosing spondylitis | Downregulated (human) | METTL14-m6A-FOXO3a mRNA modulating autophagy activity and inflammation | [144] | ||
| METTL14-m6A-ELMO1 mRNA modulating the directed migration of MSCs | [145] | ||||
| rheumatoid arthritis | Downregulated (human) | METTL14-m6A-TNFAIP3 mRNA modulating the progression of inflammation | [9] | ||
| Upregulated (rat) | METTL14-m6A-LASP1 mRNA modulating the activation of FLSs and related inflammatory reactions | [119] | |||
| osteosarcoma | Upregulated (human) | METTL14-m6A-MN1 mRNA modulating tumor progression and ATRA chemotherapy resistance | [11] | ||
| METTL3 | osteoporosis | Upregulated (mouse) | METTL3-m6A-CHI3L1,Atp6v0d2,Traf6 and Pth1r modulating osteoclast differentiation, bone resorption and the osteogenic differentiation of BMSCs | [90-92] | |
| rheumatoid arthritis | ATT inhibits METTL3 (mouse) | METTL3-m6A-ICAM2 mRNA modulating apoptosis of RA-FLS cells, inhibiting proliferation, migration, and invasion | [121] | ||
| —— (mouse) | METTL3-m6A-TTC4 mRNA modulating inflammation and oxidative stress | [120] | |||
| osteoarthritis | Upregulated (human and rat) | METTL3-m6A-ACSL4,NEK7 mRNA regulating chondrocyte ferroptosis and pyroptosis | [106, 108] | ||
| METTL3-m6A-ATG7 mRNA regulating autophagy and senescence in FLSs | [107] | ||||
| osteomyelitis | Upregulated (human and rat) | —— | [153, 154] | ||
| osteosarcoma | Upregulated (human) | METTL3-ATG5 mRNA modulating oncogenic autophagy | [130] | ||
| WTAP | osteoarthritis | Upregulated (human) | WTAP-m6A-FRZB mRNA regulating extracellular matrix degradation, inflammatory response, and oxidative stress in osteoarthritis chondrocytes | [109] | |
| osteosarcoma | Upregulated (human) | WTAP-m6A-HMBOX1 mRNA regulating osteosarcoma growth and metastasis via PI3K/AKT pathway | [129] | ||
| FTO | osteoporosis | Downregulated (human) | FTO-m6A-PPARG and protective protein mRNAs modulating osteoblast differentiation and bone formation; safeguarding osteoblasts from genotoxic | [93,94] | |
| osteomyelitis | Downregulated (rat) | FTO-m6A-MDM2 mRNA modulating SA-triggered ferroptosis in BMSCs | [155] | ||
| osteoarthritis | Upregulated (human) | —— | [112] | ||
| ALKBH5 | osteoporosis | Upregulated in osteogenic process (rat) | ALKBH5-m6A-Runx2 mRNA modulating osteoblast differentiation | [96] | |
| osteoarthritis | Downregulated (mouse) | ALKBH5-m6A-CYP1B1 mRNA, leading to mitochondrial dysfuncton and promoting MSC senescence | [110] | ||
| rheumatoid arthritis | Upregulated (human) | ALKBH5-m6A-JARID2 mRNA modulating the proliferation, migration, and invasion of RA FLSs | [122] | ||
| osteosarcoma | Downregulated (human) | ALKBH5-m6A-SOCS3 mRNA modulating cell proliferation and apoptosis | [131] | ||
| YTHDF1 | osteoporosis | Downregulated (human) | YTHDF1-m6A-Zfp839 mRNA modulating BMSC osteogenesis | [97] | |
| osteoarthritis | Downregulated (human) | —— | [113] | ||
| YTHDC1 | osteoporosis | Downregulated (human and mouse) | YTHDC1-m6A-HUR-PTPN6 mRNA modulating osteoclast differentiation | [98] | |
| IGFBP2 | osteoarthritis | Upregulated (human) | —— | [113] | |
| ac4C | NAT10 | osteoporosis | Downregulated (human and mouse) | NAT10-ac4C-RUNX2 mRNA modulating osteogenic differentiation of bone BMSCs | [7] |
| osteosarcoma | Upregulated (human) | NAT10-ac4C-YTHDC1mRNA-m6A-LDHA/PFKM mRNA modulating the glycolysis of cancer cells | [13] | ||
| m5C | NSUN2 | osteosarcoma | Upregulated (human) | NSUN2-m5C-FABP5 mRNA and anti-apoptotic mRNA modulating fatty acid metabolism and apoptosis in osteosarcoma cells | [12,132] |
| rheumatoid arthritis | Upregulated (human and rat) | NSUN2-m5C-SFRP1 mRNA modulating Wnt/β-catenin signaling pathway and proliferation of RA FLS | [10] | ||
| YBX1 | osteoporosis | Downregulated (mouse) | YBX1-m5C-BMP4 mRNA modulating osteogenic differentiation of BMSCs | [8] | |
| m7G | METTL1 | osteosarcoma | Upregulated (human) | —— | [133] |
| osteoarthritis | Downregulated (human) | —— | [115] | ||
| eIF4E2 | osteoarthritis | Upregulated (human) | —— | [114] | |
| m1A | TRMT10C | osteosarcoma | —— | ACAT1 was identified as an m1A methylation-related metabolic gene | [134] |
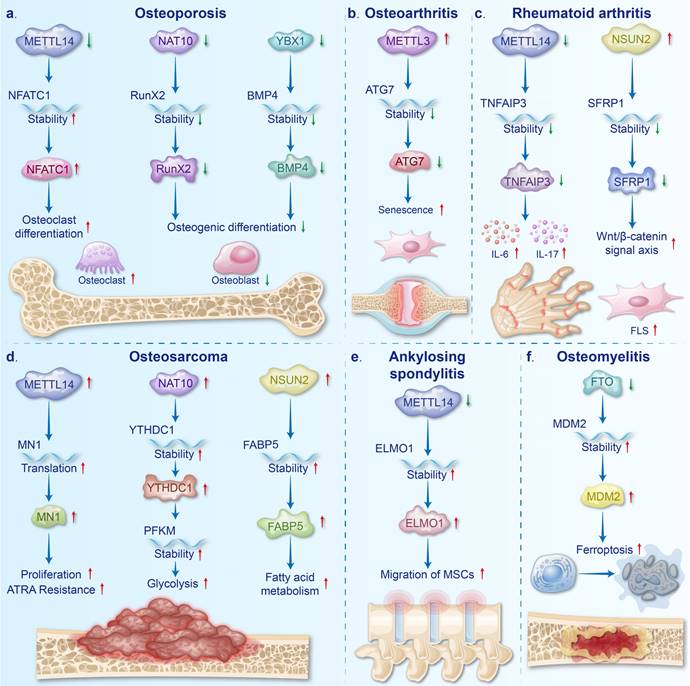
Mechanisms of mRNA modifications in the BDs. a. Abnormalities of METTL14 (m6A), NAT10 (ac4C) and YBX1 (m5C) affected the mRNA stability of NFATC1, RUNX2 and BMP4, respectively. These processes contributed to disorders of osteoclast differentiation and osteoblast differentiation leading to OP. b. The mRNA stability of ATG7 is reduced by the upregulation of METTL3 (m6A), which contributes to the senescence of FLSs in OA. c. Dysregulation of METTL14 (m6A) and NSUN2 (m5C) reduced mRNA stability of TNFAIP3 and SFRP1, respectively, and promoted inflammation and proliferation of FLS in RA. d. The mRNA translation of MN1 and the mRNA stability of YTHDC1 and FABP5 were regulated by METTL14 (m6A), NAT10 (ac4C), and NSUN2 (m5C), respectively, which resulted in the progression of OS. e. The downregulation of METTL14 (m6A) enhanced the mRNA stability of ELMO1, and promoted the migration and osteogenesis differentiation of MSCs in AS. f. mRNA stability of MDM2 was increased by FTO (m6A) downregulation, which promoted BMSCs ferroptosis in OM.
3.1 Osteoporosis (OP) (Fig. 2a)
OP is a systemic BD resulting from an imbalance in bone cell function characterized by enhanced bone fragility, decreased bone density, and degradation of bone tissue microstructure [1, 80]. OP can be divided into two categories: primary and secondary, with the primary type being more common [81]. Primary OP comprises mainly senile OP and postmenopausal OP, while secondary OP can be triggered by a variety of factors, including diseases, medications, and nutritional imbalances [82, 83]. As a common metabolic BD, OP poses a major challenge to global public health [84].
Downregulation of METTL14 in the bone tissues of OP patients and OVX mice is believed to promote osteoclast differentiation and inhibit osteoblast differentiation. Increased osteoclast differentiation is attributed to decreased SIRT1 mRNA stability and increased NFATc1 mRNA stability, both of which result from reduced m6A modification [85, 86]. Deng et al. demonstrated that overexpression of METTL14 inhibits osteoclast differentiation by enhancing the stability of GPX4 mRNA, further reinforcing the concept that METTL14 deficiency facilitates osteoclast differentiation [87]. METTL14 inhibits osteoclastogenesis and bone resorption via enhancing GPX4 stability through an m6A-HuR dependent mechanism. Additionally, the inhibition of osteoblast differentiation is linked to the reduced stability of Beclin-1 and SMAD1 mRNA by downregulating METTL14 [6, 88]. Overexpression of METTL14 promotes osteoblast differentiation by enhancing the stability of TCF1 mRNA, supporting the notion that METTL14 deficiency inhibits osteoblast differentiation [89].
Upregulated METTL3 in the bone tissues of OP patients and OVX mice is deemed to enhance osteoclast differentiation and bone resorption [90]. This viewpoint is further supported by the finding that silencing METTL3 inhibits osteoclast differentiation [91]. Mechanistically, METTL3 promotes osteoclast differentiation by stabilizing CHI3L1 mRNA, while its silencing reduces Atp6v0d2 expression and impairs Traf6 nuclear export, leading to decreased osteoclast differentiation [90, 91]. However, METTL3 also facilitates osteoblast differentiation and mitigates osteoporosis progression by enhancing the translation efficiency of Pth1r mRNA [92]. The reported data suggest that the upregulation of METTL3 promotes osteoblast and osteoclast differentiation. High levels of METTL3 in bone tissue ultimately result in low bone mass, potentially due to the stronger promotion of osteoclast differentiation.
FTO is upregulated during osteoblast differentiation and is thought to promote this process by reducing the stability of PPARG mRNA and increasing the stability of protective protein mRNAs, such as Hspa1a [93, 94]. Additionally, downregulation of FTO promotes osteoclast differentiation in diabetes-induced OP, further indicating that upregulated FTO plays a crucial role in counteracting OP [95]. ALKBH5, another m6A eraser, is also upregulated during osteoblast differentiation [96]. Knockdown of ALKBH5 inhibits osteoblast differentiation by reducing the stability of Runx2 mRNA, further confirming the essential role of ALKBH5 upregulation in osteoblast differentiation [96].
Studies have demonstrated that the m6A readers play crucial roles in OP prevention by promoting osteoblast differentiation and inhibiting osteoclast differentiation, respectively. Specifically, YTHDF1 enhances the translation of Zfp839, which promotes osteoblast differentiation through its interaction with Runx2 [97]. Additionally, YTHDC1 inhibits osteoclast differentiation by stabilizing PTPN6 mRNA in an m6A-HUR-dependent manner [98]. Moreover, aberrant expression of METTL3 and METTL14 leads to abnormal m6A modification levels on mRNAs related to osteoclast and osteoblast differentiation. This affects the recognition of these m6A-modified mRNA by m6A readers, resulting in abnormal biological effects by readers. Specifically, in OP, the downregulation of METTL14 reduces m6A modifications on Beclin-1 and SMAD1 mRNA, weakening the recognition of these mRNAs by IGF2BPs and the stabilizing effects on mRNAs. This leads to decreased stability of Beclin-1 and SMAD1 mRNAs, thereby inhibiting osteoblast differentiation [6, 88]. Additionally, supplementation of METTL14 increases m6A modifications on NFATc1, enhancing its recognition by YTHDF2 and diminishing its stabilization, ultimately suppressing osteoclast differentiation [85]. The upregulation of METTL3 in osteoporosis increases m6A modifications on Atp6v0d2, facilitating its recognition by YTHDF2, which reduces its stability. This process results in the differentiation of smaller osteoclasts with enhanced bone resorption activity [91].
The downregulation of the ac4C writer NAT10 and the m5C reader YBX1 contributes to the pathogenesis of OP by inhibiting osteoblast differentiation. Specifically, downregulated NAT10 reduces the stability of RUNX2 mRNA, leading to suppressed osteoblast differentiation [7]. Ybx1 downregulation diminishes the stability of BMP4 mRNA and blocks its release from the endothelium, further inhibiting osteoblast differentiation [8]. Additionally, research on m1A modifications in OP remains limited, with preliminary findings identifying 28 m1A-SNPs associated with bone mineral density [99].
3.2 Osteoarthritis (OA) (Fig. 2b)
OA is the most common joint disease that involves damage to the articular cartilage, alteration of the subchondral bone, and affecting the ligaments and muscles surrounding the joint [100]. Age serves as the primary risk factor for OA, with gender, genetics, and obesity contributing as well [101]. OA has been shown to increase the risk of diabetes in older adults, hinder the prevention of serious complications in diabetic patients, and negatively impact both cardiovascular and mental health in the elderly [102-105]. Notably, chondrocytes and FLSs play pivotal roles in OA, and the chemical modifications of mRNAs can affect their physiological and pathological processes.
Upregulation of METTL3 is considered to be involved in the progression of OA. Elevated METTL3 levels not only promote chondrocyte pyroptosis by increasing NEK mRNA expression but also impair autophagy in OA-FLSs by reducing the stability of ATG7 mRNA [106, 107]. Additionally, METTL3 downregulation has been shown to inhibit chondrocyte ferroptosis and alleviate OA progression, further underscoring the critical role of upregulated METTL3 in OA pathogenesis [108]. WTAP, another m6A writer, is upregulated in clinical osteoarthritic cartilage and TNF-α-induced chondrocytes. Elevated WTAP expression causes extracellular matrix degradation, inflammation, and oxidative stress in osteoarthritic chondrocytes by reducing FRZB levels and activating the Wnt/β-catenin pathway [109].
Aberrant expression of m6A erasers and readers significantly influences the progression of OA by modulating complex biological processes, such as senescence and inflammatory responses. Specifically, downregulation of ALKBH5 in OA enhances the stability of CYP1B1 mRNA, which leads to mitochondrial dysfunction and accelerates the aging of mesenchymal stem cells (MSCs) [110]. Moreover, upregulation of IGF2BP3 in OA promotes macrophage polarization and the expression of inflammatory cytokines [111]. In patients with OA, the expression levels of FTO and IGFBP2 are also elevated, while YTHDF2 expression is decreased [112, 113]. However, the precise mechanisms by which these molecules contribute to OA pathogenesis remain to be further elucidated.
In recent years, the role of m7G has attracted attention in OA research. Studies have shown that the m7G reader EIF4E2 is significantly upregulated in OA synovial tissues, while the m7G writer METTL1 is notably downregulated [114, 115]. These findings suggest a potential involvement of m7G in OA pathogenesis, although the specific mechanisms remain largely unexplored and require further investigation.
3.3 Rheumatoid arthritis (RA) (Fig. 2c)
RA is a chronic inflammatory autoimmune disease characterized by immune cell infiltration, synovial hyperplasia, and destruction of the articular cartilage and bone [116, 117]. Patients with RA can suffer from progressive joint ankylosis, destruction, deformity, and disability if left untreated [117]. Approximately 1% to 2% of the world's population suffers from RA, with women at a higher risk of developing the disease [118].
The m6A writers play critical roles in the pathogenesis of RA by regulating inflammation and FLS activation. Studies have demonstrated that both METTL14 expression and m6A modifications are significantly downregulated in the peripheral blood mononuclear cells of RA patients [9]. This reduction enhances the stability of TNFAIP3 mRNA, leading to the increased expression of inflammatory cytokines, such as IL-6 and IL-17 [9]. However, another study indicated that METTL14 expression was upregulated in synovial tissues, which stabilized LASP1 mRNA and exacerbated FLS activation and inflammation via the LASP1/SRC/AKT signaling pathway [119]. These findings suggest that the expression of modified regulators may vary across different organ tissues in the same disease, and that the specific mechanisms need further investigation. As another m6A writer, METTL3 has been implicated in promoting oxidative stress and inflammation in arthritic tissues by destabilizing TTC4 mRNA [120]. The downregulation of METTL3 inhibits the ICAM2/PI3K/AKT/p300 signaling pathway, effectively slowing RA progression and further supporting the critical role of METTL3 in RA pathogenesis [121].
Aberrant m6A and m5C modifications are pivotal in the pathogenesis of RA. Specifically, upregulation of the m6A eraser ALKBH5 significantly promotes the proliferation, migration, and invasion of RA FLSs by increasing the stability of JARID2 mRNA [122]. On the other hand, elevated expression of the m5C writer NSUN2 in RA decreases the stability of SFRP1 mRNA, leading to activation of the Wnt/β-catenin signaling pathway and thereby accelerating RA progression [10].
3.4 Osteosarcoma (OS) (Fig. 2d)
OS, the primary bone tumor mainly affecting children and adolescents, is best treated with a combination of surgery and chemotherapy [123-125]. Although chemotherapy has raised the five-year survival rate to nearly 80%, the overall survival rate remains in the 60%-70% range due to challenges such as metastasis, drug resistance, and high recurrence rates [126-128].
The abnormal elevation of m6A writers has been reported to promote the proliferation of OS cells and the formation of drug resistance. Specifically, upregulation of WTAP reduces the stability of HMBOX1 mRNA and activates the PI3K/AKT signaling pathway, thereby promoting tumor cell proliferation [129]. Additionally, overexpression of METTL14 has been shown to increase the translation efficiency of MN1 mRNA, leading to all-trans retinoic acid (ATRA) chemotherapy resistance and accelerating tumor progression [11]. Furthermore, the upregulation of METTL3 stabilizes ATG5 mRNA, enhancing autophagy, which is another key mechanism driving OS progression [130]. Additionally, clinical data have revealed that the downregulation of the m6A eraser ALKBH5 contributes to OS development [131]. This finding is further supported by subsequent experiments, where overexpression of ALKBH5 effectively inhibits OS progression by stabilizing SOCS3 mRNA and activating the STAT3 signaling pathway [131].
Upregulation of the m5C writer NSUN2 and the ac4C writer NAT10 is implicated in tumor progression through the regulation of cellular metabolic mechanisms. Specifically, elevated NSUN2 expression significantly enhances fatty acid metabolism in tumor cells by stabilizing FABP5 mRNA [12]. Notably, chemotherapy-induced downregulation of NSUN2 expression can effectively trigger apoptosis in tumor cells, further reinforcing the link between NSUN2 upregulation and poor prognosis in OS [132]. Additionally, increased NAT10 expression promotes glycolysis in tumor cells by stabilizing YTHDC1, PFKM, and LDHA mRNA, thus providing essential energy support for tumor growth [13].
Current research on m7G and m1A modifications in OS remains in the early exploratory stages. Upregulation of the m7G writer complex METTL1/WDR4 in OS has been shown to enhance the m7G methylation of tRNA, thereby promoting OS progression by increasing the translation efficiency of oncogenic mRNAs [133]. However, it has not yet been demonstrated that METTL1/WDR4 directly modifies mRNA to contribute to OS development. Additionally, the m1A writer TRMT10C has been observed to potentially promote epithelial mesenchymal transition and increase sensitivity to chemotherapeutic drugs in tumor cells by regulating the expression of ACAT1 [134].
3.5 Others (Fig. 2e-f)
Fractures represent disruptions in the structural integrity and continuity of bone, often resulting from trauma or pathological conditions [135]. Since about 10% of fractures failed to heal properly under traditional therapy (surgery and conservative treatment), it is important to explore the roles of mRNA modifications in fracture healing to find new therapeutic strategies [136].
METTL3 expression is markedly downregulated during the early phases of fracture healing and is thought to facilitate this reparative process [137]. In vivo studies demonstrate that local administration of the METTL3 plasmid at the fracture site significantly delays healing, reinforcing the notion that METTL3 downregulation is conducive to fracture repair [137]. Mechanistic investigations have revealed that METTL3 impairs osteoblast differentiation by inhibiting the maturation of miR-7212-5p [137]. However, whether METTL3 exerts its inhibitory effects on fracture healing directly through m6A-mediated mRNA regulation remains to be elucidated. Concurrently, the upregulation of YTHDF1 during osteoblast differentiation appears to enhance fracture healing, potentially through the activation of autophagy signaling pathways [14].
AS is a chronic rheumatic disease characterized by persistent inflammation and heterotopic bone formation [138]. Clinically, it presents as inflammation at the points of attachment, primarily affecting the spine and sacroiliac joints [139]. The disease is more prevalent among young and middle-aged males, increasing the risk of spinal skeletal dysfunction and disability [140, 141]. The etiology of AS is complex, involving environmental triggers, infections, genetic susceptibility, and immune dysregulation [142, 143]. Recent studies have identified the roles of m6A and ac4C modifications in AS pathogenesis. Specifically, the downregulation of METTL14 has been shown to destabilize FOXO3a mRNA, leading to impaired autophagy and exacerbated inflammation [144]. Concurrently, decreased METTL14 levels enhance the migratory capacity of MSCs by increasing the stability of ELMO1 mRNA [145]. The restoration or overexpression of METTL14 following therapeutic intervention further underscores the critical role of METTL14 downregulation in AS development [144]. Additionally, a significant reduction in FTO expression has been observed in interspinous ligament samples from patients with AS, although the precise mechanism by which FTO contributes to AS pathology remains to be elucidated [146]. Moreover, existing research has established a close association between reduced NAT10 expression and AS disease activity, and NAT10 levels can be used to improve the sensitivity and specificity of early AS diagnosis [147].
OM, an inflammatory BD primarily caused by bacterial, fungal, and mycobacterial infections, typically emerges following bone injury or vascular insufficiency [148-150]. In bone tissue, limited blood flow and the presence of bacterial biofilms impede antibiotic delivery, making treatment challenging [151, 152]. METTL3 is significantly upregulated in both OM animal models and in bone marrow from OM patients, and is believed to play a role in the inflammatory response [153, 154]. This viewpoint is further supported by findings that inhibition of METTL3 with STM2457 in OM cell models leads to a marked reduction in the release of IL-6 and TNF-α [153]. Additionally, the deletion of the m6A eraser FTO in Staphylococcus aureus-exposed BMSCs has been shown to induce ferroptosis by stabilizing MDM2 mRNA [155].
4. RNA modification-based therapeutics for BDs (Fig. 3)
Therapeutic strategies based on mRNA modifications refer to the modulation of the chemical modifications of mRNAs to effectively inhibit the occurrence and progression of diseases. As shown in Table 2. These strategies can regulate cell differentiation, senescence, and death while suppressing inflammatory responses, ultimately leading to significant improvements in the occurrence and progression of BDs.
Therapeutic strategies based on mRNA modifications.
| Modification | Disease | Intervention | Target mRNA/axis | Functions/mechanisms | Refs. |
|---|---|---|---|---|---|
| m6A | osteoporosis | LV-METTL14, AV-METTL14, METTL14 exosomes injection | SIRT1, SMAD1, NFATC1, Beclin-1 | Promoting osteogenic differentiation of BMSCs and inhibiting osteoclast differentiation | [6,85-87] |
| fracture | Cell sheets of BMSCs overexpressing YTHDF1 | Autophagy signaling pathway | Promoting osteogenic differentiation of BMSCs | [14] | |
| osteoarthritis | AAV-siWTAP injection | FRZB | Inhibiting inflammation and extracellular matrix degradation, suppressing oxidative stress | [109] | |
| sh-METTL3 injection | NEK7 | Inhibiting chondrocyte pyroptosis and suppressing inflammation | [106] | ||
| MSCs overexpressing ALKBH5 | CYP1B1 | Mitigating cellular senescence and reducing inflammation | [110] | ||
| AAV9.HAP-1-si-METTL3 injection | ATG7 | Suppressing cellular senescence and ameliorated cartilage destruction | [107] | ||
| osteomyelitis | LV-pcDNA-FTO vector injection | MDM2/TLR4/SLC7A11 axis | Suppressing iron death | [155] | |
| STM2457 injection | METTL3-MyD88 | Inhibiting METTL3 to alleviate inflammation | [153] | ||
| rheumatoid arthritis | Artemisitene injection | METTL3-ICAM2 | Inhibiting METTL3 to impede the migration and invasion of RA-FLSs | [121] | |
| sh-ALKBH5 injection | JARID2 | Inhibiting proliferation, migration, and invasion of RA FLSs | [122] | ||
| sh-METTL14 injection | LASP1/SRC/AKT axis | Inhibiting the activation, proliferation, and invasion of FLSs | [119] | ||
| ankylosing spondylitis | AdV-shELMO1 injection | ELMO1 | Counteracting the upregulation of ELMO1 expression caused by METTL14, thereby ameliorating ectopic ossification | [145] | |
| osteosarcoma | Spautin-1 | USP13/METTL3/ATG5 | Inhibiting proliferation and metastasis of OS | [130] | |
| m5C | osteoporosis | sciadopitysin | YBX1 | Improving osteogenic differentiation of BMSCs | [8] |
| ac4C | osteoporosis | AdV-NAT10 injection | RUNX2 | Promoting osteogenic differentiation of BMSCs | [7] |
| Mijiao | NAT10 | Promoting osteogenic differentiation of BMSCs by boosting NAT10 expression | [158] |
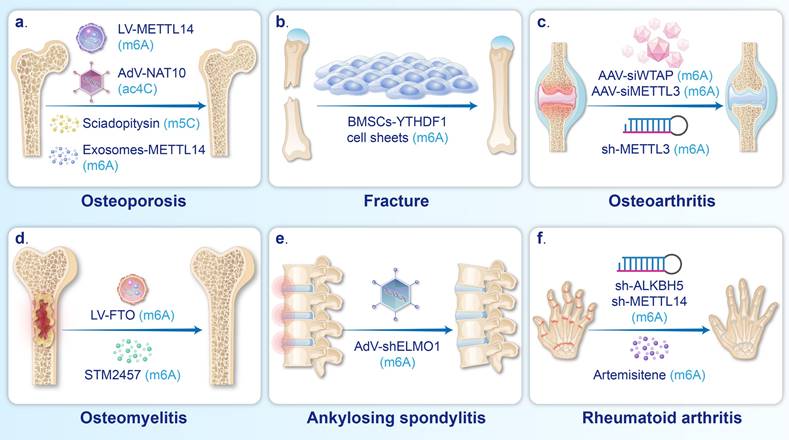
Therapeutic strategies for BDs based on mRNA modifications. a. AdV-NAT10, LV-METTL14, Sciadopitysin, and METTL14 exosomes were used to alleviate OP. b. Cell sheets of BMSCs overexpressing YTHDF1 promoted fracture healing. c. Intra-articular injection of AAV-siWTAP, AAV-siMETTL3, and sh-METTL3 could mitigate the progression of OA. d. OM can be relieved by injection of LV-FTO and STM2457. e. Injection of AdV-shELMO1 could alleviated ectopic ossification in AS. f. The progression of Rheumatoid arthritis could be suppressed by the treatment of Artemisitene, sh-METTL14, and sh-ALKBH5.
4.1 Regulating bone homeostasis in OP and fractures (Fig. 3a-b)
Osteoblasts are derived from MSCs in bone marrow and blood [156]. Stimulating MSCs to migrate and differentiate into osteoblasts, as well as enhancing their ability to synthesize the bone matrix, is key to treating OP and fractures [157].
Numerous studies have demonstrated that the regulation of m6A deposition in mRNAs plays a critical role in enhancing osteoblast differentiation. Injecting LV-METTL14 can enhance the m6A level and stability of SIRT1, SMAD1, and Beclin-1 mRNA, in turn promoting the osteogenic differentiation of BMSCs and alleviating bone loss in OVX mice [6, 86, 88]. Transplantation of BMSC sheets overexpressing the m6A reader YTHDF1 around the fracture site has been shown to enhance autophagy, thus promoting osteoblast differentiation and ultimately facilitating fracture healing in rats [14].
Emerging evidence suggests that ac4C and m5C are also important targets in treatment strategies aimed at promoting osteoblast differentiation. It has been reported that intramuscular injection of adenovirus overexpressing NAT10 can upregulate the acetylation and stability of RUNX2 mRNA, which enhances osteoblast differentiation and increases the trabecular volume fraction and trabecular number in OVX mice [7]. Traditional Chinese medicine, Mijiao, has been shown to increase the stability and expression of RUNX2 by upregulating NAT10, which further improves osteoblast differentiation and mitigates bone loss in OVX mice [158]. In addition, the m5C reader YBX1 can be upregulated by injection of sciadopitysin, resulting in increased vascular-dependent osteogenic differentiation via enhancing the stability of BMP4 mRNA [8].
Osteoclasts, originating from mononuclear/macrophage lineage progenitor cells, play a crucial role in maintaining bone homeostasis by mediating bone resorption [159], yet excessive activation of osteoclasts can lead to pathological bone loss [160]. In such cases, inhibiting osteoclast differentiation through the regulation of m6A serves as a strategy for treating OP. For example, injection of METTL14 exosomes into the bone marrow cavity or intravenous administration of LV-METTL14 has been shown to reduce the stability of NFATc1 mRNA and increase the stability of SIRT1 mRNA, thus effectively inhibiting osteoclast differentiation and alleviating bone loss in OVX mice [85, 86].
4.2 Inhibiting inflammatory response, cell loss, and cellular senescence in OA and OM (Fig. 3c-d)
Inflammation, cellular senescence, and cellular damage are common causes of the development of BDs, such as OA and OM [161, 162]. Regulating these pathological processes through the modulation of m6A serves as a therapeutic strategy for BDs. For example, intra-articular injection of AAV-siWTAP can improve the OARSI score in a mouse model of OA [109]. According to X et al., the downregulation of WTAP reduced the m6A modifications of FRZB mRNA, leading to increased FRZB expression, inhibiting the Wnt/β-catenin pathway and ultimately suppressing the inflammatory cascade in OA [109]. In addition, an intraperitoneal injection of STM2457 (a METTL3 inhibitor) in an OM mouse model effectively suppresses inflammation by downregulating MyD88 and NF-κB-related inflammatory molecules in macrophages [153].
Regarding the suppression of cellular senescence, intra-articular injection of MSCs overexpressing ALKBH5 attenuates many types of cellular senescence and reduces inflammation induced by senescent cells, which in turn alleviates cartilage defects and OARSI scores in OA mice [110]. The increased expression of ALKBH5 facilitates CYP1B1 mRNA degradation via m6A demethylation, thus attenuating senescence in MSCs [110]. Additionally, intra-articular injection of METTL3 siRNA can increase the stability of ATG7 mRNA, leading to upregulation of autophagy in OA-FLSs, suppression of MSC senescence, and improvement of cartilage degradation [107]. The pyroptosis of chondrocytes and the expression of inflammatory factors in OA mice can be inhibited by the injection of sh-METTL3 [106]. Xiong et al. demonstrated that the m6A modifications of NEK7 mRNA were reduced by the downregulation of METTL3, leading to decreased NEK7 expression and reduced chondrocyte pyroptosis [106]. Furthermore, the intravenous injection of lentiviruses carrying pcDNA-FTO vectors reduces the methylation and stability of MDM2 mRNA, further downregulating the MDM2/TLR4/SLC7A11 signaling pathway, which subsequently inhibits ferroptosis in BMSCs and reduces histopathological damage in bone [155].
4.3 Others (Fig. 3e-f)
It has been reported that inhibiting the proliferation and migration of FLSs while promoting their apoptosis can reduce synovial hyperplasia and cartilage degeneration in RA [163, 164]. Intraperitoneal injection of artemisitene in an RA mouse model inhibits METTL3, which in turn reduces the expression of ICAM2 mRNA, ultimately preventing the migration and invasion of RA-FLSs and inducing apoptosis [121]. Intra-articular injection of sh-METTL14 in rat models of RA has been found to prevent the activation, proliferation, and invasion of FLSs by reducing the methylation and stability of LASP1 mRNA, thus inhibiting the LASP1/SRC/AKT signaling axis [119]. In addition, the proliferation, migration, and invasion of FLSs are inhibited by intra-articular injection of sh-ALKBH5, which reduces m6A modification levels and the stability of JARID2 mRNA [122].
Suppressing the proliferation, metastasis, and drug resistance of OS represents a central challenge in the treatment of this disease. Wang et al. reveal that injecting Spautin-1 into mice bearing xenografts effectively inhibits the USP13-enhanced stabilization of METTL3, subsequently reducing the stability of ATG5 mRNA and thereby curbing tumor proliferation and metastasis [130]. Additional researches indicate that knockdown of METTL14, NAT10, and NSUN2 in OS cell lines significantly inhibits tumor growth in nude mice [11-13]. Overexpression of ALKBH5 has also been found to suppress the proliferation of U2OS cells in nude mice [131]. Moreover, Li et al. further elucidate that knocking down METTL14 and NSUN2 in OS cells can respectively potentiate the inhibitory effects of ATRA and doxorubicin on tumors, thereby suppressing tumor drug resistance [11, 132].
Abnormally increased osteogenic differentiation of MSCs has been shown to play a significant role in pathological bone formation in AS [165], and inhibiting this differentiation has been identified as a strategy to alleviate spinal joint stiffness. Xie et al. reported that the injection of AV-ELMO1 into the tail vein counteracted METTL14-induced upregulation of ELMO1 expression, thus ameliorating heterotopic ossification [145].
5. Conclusions and future perspectives
Scientists have made significant progress in revealing the epigenetic mechanisms underlying the occurrence and progression of BDs. As an important component of epigenetics, mRNA modifications have emerged as new focuses and novel therapeutic targets against BDs. In this review, we have summarized the regulatory mechanisms of five mRNA modifications and explored their involvement in the occurrence and development of BDs. Current research has highlighted that the cooperation between writers and erasers achieves dynamic reversibility of modifications, and that different modification types, modification sites, and readers will have different effects on the processes of mRNA export, translation, and degradation. We found that abnormal mRNA modification levels were involved in the development of BDs in multiple aspects. Abnormal modifications can affect mRNA stability and translation efficiency, disrupt the balance between osteogenic and osteoclastic differentiation, promote inflammation and cytopathia, and induce metabolic reprogramming and drug resistance in tumors. In addition, targeting mRNA modification regulators shows promise in diagnosing and treating BDs. Taking m6A modification as an example, Deng et al. demonstrated that METTL14 is downregulated in serum samples from postmenopausal women with OP and positively correlates with bone mineral density [87]. This finding suggests that METTL14 has potential as a diagnostic biomarker for OP. Furthermore, animal studies have confirmed that modulating abnormal levels of m6A regulators, such as through viral injection methods, can effectively treat OP. Therefore, developing safe and clinically validated drugs targeting m6A modification regulators holds significant clinical value for the treatment of OP.
The targetability and safety of mRNA-modified therapeutic strategies need to be improved. Viral vectors, agonists, and inhibitors have achieved good therapeutic effects in animal models by regulating mRNA modifications. However, mRNA modification regulators may regulate multiple mRNAs simultaneously, which can lead to the occurrence of other diseases while treating BDs. Moreover, mRNA modification regulators can exert opposing effects when acting on different cell types. Therefore, accurately targeting diseased cells and specific mRNAs in BDs is essential for improving the therapeutic efficacy of BDs and avoiding the occurrence of side effects. Nanomaterials, particularly bone-targeted nanotechnology, may be ideal candidates for achieving this goal [166]. Furthermore, given the critical role of mRNA modifications in biological processes, further exploration of unknown mRNA modification regulators—such as specific readers of m1A—and their molecular mechanisms underlying biological effects is essential for advancing our understanding of this field. Building on this foundation, in-depth investigations into mRNA modifications associated with BDs, as well as the mechanisms by which pathogenic factors lead to dysregulation of these modification regulators, hold significant scientific value and clinical relevance. Although this review discusses mRNA modifications in some BDs, whether mRNA modifications are involved in the development of certain metabolic BDs, such as rickets, hyperthyroidism, and hypothyroidism, and their underlying mechanisms, remains to be further investigated.
Future studies will aim to elucidate the complex networks and mechanisms of action between specific lncRNAs and DNA/RNA modifying enzymes in BDs. In these future studies, two models of modification synergistic effects are particularly worthy of further elucidation. The first model involves one type of mRNA modification regulating the expression of the regulatory factors of another mRNA modification, thereby influencing the occurrence and progression of BDs through the latter modification. The other modification interaction is reflected in different mRNA modifications jointly regulating the same mRNA, collectively determining the progression of the disease. These complex networks and mechanisms will go beyond mere descriptions of associations to portray deep molecular mechanism atlases. The discovery of these epigenetic regulatory mechanisms may lead to potential therapeutic approaches. As our understanding of interactions deepens, epigenetic biomarkers may be utilized for the early diagnosis, prognostic assessment, and monitoring of the therapeutic response in BDs. Future treatments for BDs are likely to evolve in a more personalized direction. By targeting specific aberrant mRNA modifications in individual patients, interventions are expected to improve the efficacy and precision of treatment.
Acknowledgements
This study was supported by GuangDong Basic and Applied Basic Research Foundation (Grant No. 2023A1515111003), the China Postdoctoral Science Foundation (Grant No. 2024M751327), and the President Foundation of Nanfang Hospital, Southern Medical University (Grant No. 2023B020).
Author contributions
Zehui Li and Keyu Meng wrote the manuscript and designed the figures. Shanwei Lan, Zhengda Ren, Zhongming Lai, Xiang Ao, and Zhongyuan Liu contributed to collect the references and design the tables. Jiajia Xu, Xiaoyi Mo and Zhongmin Zhang conceived the project, reviewed and revised the manuscript. All authors participated in the manuscript and approved the final version.
Abbreviations
BDs: bone diseases
OP: osteoporosis
RA: rheumatoid arthritis
OS: osteosarcoma
OA: osteoarthritis
OM: osteomyelitis
AS: ankylosing spondylitis
FLSs: fibroblast-like synoviocytes
BMSCs: bone marrow mesenchymal stem cells
MSCs: mesenchymal stem cells
OVX: ovariectomized
m6A: N6-methyladenosine
m1A: N1-methyladenosine
m7G: N7-methylguanosine
ac4C: N4-acetylcytosine
m5C: 5-methylcytosine
3' UTR: 3'untranslated region
5' UTR: 5'untranslated region
CDS: coding sequence
MTC: methyltransferase complex
FTO: fat mass and obesity-associated protein
IGF2BPs: insulin-like growth factor 2 mRNA-binding proteins
hnRNPs: heterogeneous nuclear ribonucleoproteins
eIF: eukaryotic translation initiation factor
TRMT61A: tRNA methyltransferase 61A
TRMT6: tRNA methyltransferase 6
RNMT: RNA guanine-N7 methyltransferase
TET: ten-eleven translocation
ALYREF: Aly/REF export factor
YBX1: Y-Box binding protein 1
BMP4: bone morphogenetic protein 4
ATRA: all-trans retinoic acid
Competing Interests
The authors have declared that no competing interest exists.
References
1. Letarouilly JG, Broux O, Clabaut A. New insights into the epigenetics of osteoporosis. Genomics. 2019;111:793-8
2. Cappannini A, Ray A, Purta E, Mukherjee S, Boccaletto P, Moafinejad SN. et al. MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 2024;52:D239-D44
3. Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124
4. Yen YP, Chen JA. The m(6)A epitranscriptome on neural development and degeneration. J Biomed Sci. 2021;28:40
5. Zhou H, Mao L, Xu H, Wang S, Tian J. The functional roles of m(6)A modification in T lymphocyte responses and autoimmune diseases. Cytokine Growth Factor Rev. 2022;65:51-60
6. He M, Lei H, He X, Liu Y, Wang A, Ren Z. et al. METTL14 Regulates Osteogenesis of Bone Marrow Mesenchymal Stem Cells via Inducing Autophagy Through m6A/IGF2BPs/Beclin-1 Signal Axis. Stem Cells Transl Med. 2022;11:987-1001
7. Yang W, Li HY, Wu YF, Mi RJ, Liu WZ, Shen X. et al. ac4C acetylation of RUNX2 catalyzed by NAT10 spurs osteogenesis of BMSCs and prevents ovariectomy-induced bone loss. Mol Ther Nucleic Acids. 2021;26:135-47
8. Li YJ, Guo Q, Ye MS, Cai G, Xiao WF, Deng S. et al. YBX1 promotes type H vessel-dependent bone formation in an m5C-dependent manner. JCI Insight. 2024;9:e172345
9. Tang J, Yu Z, Xia J, Jiang R, Chen S, Ye D. et al. METTL14-Mediated m6A Modification of TNFAIP3 Involved in Inflammation in Patients With Active Rheumatoid Arthritis. Arthritis Rheumatol. 2023;75:2116-29
10. Huang Y, Xu P, Liao F, Ca H, Wang X, Wang X. et al. Fat mass and obesity-associated protein inhibit the pathology of rheumatoid arthritis through the NSUN2/SFRP1/Wnt/beta-catenin signal axis. J Pharm Pharmacol. 2024;76:283-94
11. Li HB, Huang G, Tu J, Lv DM, Jin QL, Chen JK. et al. METTL14-mediated epitranscriptome modification of MN1 mRNA promote tumorigenicity and all-trans-retinoic acid resistance in osteosarcoma. EBioMedicine. 2022;82:104142
12. Yang M, Wei R, Zhang S, Hu S, Liang X, Yang Z. et al. NSUN2 promotes osteosarcoma progression by enhancing the stability of FABP5 mRNA via m(5)C methylation. Cell Death Dis. 2023;14:125
13. Mei Z, Shen Z, Pu J, Liu Q, Liu G, He X. et al. NAT10 mediated ac4C acetylation driven m(6)A modification via involvement of YTHDC1-LDHA/PFKM regulates glycolysis and promotes osteosarcoma. Cell Commun Signal. 2024;22:51
14. Gao X, Wang J, Wang Y, Li W, Pan Z. The m(6)A Reader YTHDF1 Accelerates the Osteogenesis of Bone Marrow Mesenchymal Stem Cells Partly via Activation of the Autophagy Signaling Pathway. Stem Cells Int. 2023;2023:5563568
15. Saneyoshi M, Harada F, Nishimura S. Isolation and characterization of N6-methyladenosine from Escherichia coli valine transfer RNA. Biochim Biophys Acta. 1969;190:264-73
16. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
17. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-46
18. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-5
19. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233-47
20. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z. et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575-8
21. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177-89
22. Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A. et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 2015;29:2037-53
23. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z. et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10
24. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
25. Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19:88
26. Li W, Hao Y, Zhang X, Xu S, Pang D. Targeting RNA N(6)-methyladenosine modification: a precise weapon in overcoming tumor immune escape. Mol Cancer. 2022;21:176
27. Deng X, Qing Y, Horne D, Huang H, Chen J. The roles and implications of RNA m(6)A modification in cancer. Nat Rev Clin Oncol. 2023;20:507-26
28. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412
29. Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m(6)A Transcripts by the 3'->5' RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol Cell. 2017;68:374-87 e12
30. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y. et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115-27
31. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-99
32. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-20
33. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315-28
34. Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M. et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657-75
35. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
36. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR. et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9:420
37. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299-308
38. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
39. Dunn DB. The occurrence of 1-methyladenine in ribonucleic acid. Biochim Biophys Acta. 1961;46:198-200
40. Srivastava R, Gopinathan KP. Ribosomal RNA methylation in Mycobacterium smegmatis SN2. Biochem Int. 1987;15:1179-88
41. Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441-6
42. Sun H, Li K, Liu C, Yi C. Regulation and functions of non-m6A mRNA modifications. Nat Rev Mol Cell Biol. 2023Oct;24(10):714-731
43. Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:5173-8
44. Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281-90
45. Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551:251-5
46. Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J. et al. Base-Resolution Mapping Reveals Distinct m(1)A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol Cell. 2017;68:993-1005 e9
47. Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH Domain-Containing Proteins as the Readers for N1-Methyladenosine in RNA. Anal Chem. 2018;90:6380-4
48. Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33-7
49. Gonatopoulos-Pournatzis T, Dunn S, Bounds R, Cowling VH. RAM/Fam103a1 is required for mRNA cap methylation. Mol Cell. 2011;44:585-96
50. Bueren-Calabuig JA, M GB, Cowling VH, Pisliakov AV. Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: insights from accelerated molecular dynamics simulations. Nucleic Acids Res. 2019;47:8675-92
51. Furuichi Y, LaFiandra A, Shatkin AJ. 5'-Terminal structure and mRNA stability. Nature. 1977;266:235-9
52. Osborne MJ, Volpon L, Memarpoor-Yazdi M, Pillay S, Thambipillai A, Czarnota S. et al. Identification and Characterization of the Interaction Between the Methyl-7-Guanosine Cap Maturation Enzyme RNMT and the Cap-Binding Protein eIF4E. J Mol Biol. 2022;434:167451
53. Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657-68
54. Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G. et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol Cell. 2019;74:1304-16 e8
55. Zhao Z, Qing Y, Dong L, Han L, Wu D, Li Y. et al. QKI shuttles internal m(7)G-modified transcripts into stress granules and modulates mRNA metabolism. Cell. 2023;186:3208-26 e27
56. Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020;583:638-43
57. Wang L, Tao Y, Zhai J, Xue M, Zheng C, Hu H. The emerging roles of ac4C acetylation "writer" NAT10 in tumorigenesis: A comprehensive review. Int J Biol Macromol. 2024;254:127789
58. Zachau HG, Dutting D, Feldmann H. The structures of two serine transfer ribonucleic acids. Hoppe Seylers Z Physiol Chem. 1966;347:212-35
59. Stern L, Schulman LH. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J Biol Chem. 1978;253:6132-9
60. Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G. et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell. 2018;175:1872-86 e24
61. Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T. et al. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem. 2014;289:35724-30
62. Xie L, Zhong X, Cao W, Liu J, Zu X, Chen L. Mechanisms of NAT10 as ac4C writer in diseases. Mol Ther Nucleic Acids. 2023;32:359-68
63. Wang C, Hou X, Guan Q, Zhou H, Zhou L, Liu L. et al. RNA modification in cardiovascular disease: implications for therapeutic interventions. Signal Transduct Target Ther. 2023;8:412
64. Arango D, Sturgill D, Yang R, Kanai T, Bauer P, Roy J. et al. Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol Cell. 2022;82:2912
65. Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527-32
66. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971-5
67. Nachtergaele S, He C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017;14:156-63
68. Huber SM, van Delft P, Mendil L, Bachman M, Smollett K, Werner F. et al. Formation and abundance of 5-hydroxymethylcytosine in RNA. Chembiochem. 2015;16:752-5
69. Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY. et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606-25
70. Liu J, Huang T, Zhang Y, Zhao T, Zhao X, Chen W. et al. Sequence- and structure-selective mRNA m(5)C methylation by NSUN6 in animals. Natl Sci Rev. 2021;8:nwaa273
71. Yang L, Ren Z, Yan S, Zhao L, Liu J, Zhao L. et al. Nsun4 and Mettl3 mediated translational reprogramming of Sox9 promotes BMSC chondrogenic differentiation. Commun Biol. 2022;5:495
72. Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639
73. Arguello AE, Li A, Sun X, Eggert TW, Mairhofer E, Kleiner RE. Reactivity-dependent profiling of RNA 5-methylcytidine dioxygenases. Nat Commun. 2022;13:4176
74. Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282-5
75. Wang Y, Wei J, Feng L, Li O, Huang L, Zhou S. et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol Cancer. 2023;22:81
76. Dai X, Gonzalez G, Li L, Li J, You C, Miao W. et al. YTHDF2 Binds to 5-Methylcytosine in RNA and Modulates the Maturation of Ribosomal RNA. Anal Chem. 2020;92:1346-54
77. Ma HL, Bizet M, Soares Da Costa C, Murisier F, de Bony EJ, Wang MK. et al. SRSF2 plays an unexpected role as reader of m(5)C on mRNA, linking epitranscriptomics to cancer. Mol Cell. 2023;83:4239-54 e10
78. Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL. et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol Cell. 2019;75:1188-202 e11
79. Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X. et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978-90
80. Genant HK, Cooper C, Poor G, Reid I, Ehrlich G, Kanis J. et al. Interim report and recommendations of the World Health Organization Task-Force for Osteoporosis. Osteoporos Int. 1999;10:259-64
81. Guo Y, Jia X, Cui Y, Song Y, Wang S, Geng Y. et al. Sirt3-mediated mitophagy regulates AGEs-induced BMSCs senescence and senile osteoporosis. Redox Biol. 2021;41:101915
82. Guan Z, Xuanqi Z, Zhu J, Yuan W, Jia J, Zhang C. et al. Estrogen deficiency induces bone loss through the gut microbiota. Pharmacol Res. 2023;196:106930
83. Sobh MM, Abdalbary M, Elnagar S, Nagy E, Elshabrawy N, Abdelsalam M. et al. Secondary Osteoporosis and Metabolic Bone Diseases. J Clin Med. 2022;11:2382
84. NIH Consensus Development Panel on Osteoporosis Prevention, Diagnosis, Therapy. Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285:785-95
85. Yang JG, Sun B, Wang Z, Li X, Gao JH, Qian JJ. et al. Exosome-targeted delivery of METTL14 regulates NFATc1 m6A methylation levels to correct osteoclast-induced bone resorption. Cell Death Dis. 2023;14:738
86. Wang C, Chen R, Zhu X, Zhang X, Lian N. METTL14 alleviates the development of osteoporosis in ovariectomized mice by upregulating m(6)A level of SIRT1 mRNA. Bone. 2023;168:116652
87. Deng M, Luo J, Cao H, Li Y, Chen L, Liu G. METTL14 represses osteoclast formation to ameliorate osteoporosis via enhancing GPX4 mRNA stability. Environ Toxicol. 2023;38:2057-68
88. Huang C, Wang Y. Downregulation of METTL14 improves postmenopausal osteoporosis via IGF2BP1 dependent posttranscriptional silencing of SMAD1. Cell Death Dis. 2022;13:919
89. Wang X, Zou C, Li M, Hou C, Jiang W, Bian Z. et al. METTL14 upregulates TCF1 through m6A mRNA methylation to stimulate osteogenic activity in osteoporosis. Hum Cell. 2023;36:178-94
90. Wang C, Zhang X, Chen R, Zhu X, Lian N. EGR1 mediates METTL3/m(6)A/CHI3L1 to promote osteoclastogenesis in osteoporosis. Genomics. 2023;115:110696
91. Li D, Cai L, Meng R, Feng Z, Xu Q. METTL3 Modulates Osteoclast Differentiation and Function by Controlling RNA Stability and Nuclear Export. Int J Mol Sci. 2020;21:1660
92. Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y. et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772
93. Chen LS, Zhang M, Chen P, Xiong XF, Liu PQ, Wang HB. et al. The m(6)A demethylase FTO promotes the osteogenesis of mesenchymal stem cells by downregulating PPARG. Acta Pharmacol Sin. 2022;43:1311-23
94. Zhang Q, Riddle RC, Yang Q, Rosen CR, Guttridge DC, Dirckx N. et al. The RNA demethylase FTO is required for maintenance of bone mass and functions to protect osteoblasts from genotoxic damage. Proc Natl Acad Sci U S A. 2019;116:17980-9
95. Shen X, Lan C, Lin Y, Zhang F, Zhang Y, Chen M. et al. Suppression of TLR4 prevents diabetic bone loss by regulating FTO-mediated m(6)A modification. Int Immunopharmacol. 2023;122:110510
96. Feng L, Fan Y, Zhou J, Li S, Zhang X. The RNA demethylase ALKBH5 promotes osteoblast differentiation by modulating Runx2 mRNA stability. FEBS Lett. 2021;595:2007-14
97. Liu T, Zheng X, Wang C, Wang C, Jiang S, Li B. et al. The m(6)A "reader" YTHDF1 promotes osteogenesis of bone marrow mesenchymal stem cells through translational control of ZNF839. Cell Death Dis. 2021;12:1078
98. Zhang M, Guan J, Yu S, Zhang Y, Cheng L, Zhang Y. YTHDC1 inhibits osteoclast differentiation to alleviate osteoporosis by enhancing PTPN6 messenger RNA stability in an m6A-hUR-dependent manner. J Leukoc Biol. 2024;115:1154-64
99. Han L, Wu J, Wang M, Zhang Z, Hua D, Lei S. et al. RNA Modification-Related Genetic Variants in Genomic Loci Associated with Bone Mineral Density and Fracture. Genes (Basel). 2022;13:1892
100. Hutton CW. Osteoarthritis: the cause not result of joint failure? Ann Rheum Dis. 1989;48:958-61
101. Motta F, Barone E, Sica A, Selmi C. Inflammaging and Osteoarthritis. Clin Rev Allergy Immunol. 2023;64:222-38
102. Park D, Park YM, Ko SH, Choi YH, Min DU, Ahn JH. et al. Association between knee osteoarthritis and the risk of cardiovascular disease and the synergistic adverse effects of lack of exercise. Sci Rep. 2023;13:2777
103. Botha-Scheepers S, Riyazi N, Kroon HM, Scharloo M, Houwing-Duistermaat JJ, Slagboom E. et al. Activity limitations in the lower extremities in patients with osteoarthritis: the modifying effects of illness perceptions and mental health. Osteoarthritis Cartilage. 2006;14:1104-10
104. Jeon CY, Lokken RP, Hu FB, van Dam RM. Physical activity of moderate intensity and risk of type 2 diabetes: a systematic review. Diabetes Care. 2007;30:744-52
105. Hawker GA, Croxford R, Bierman AS, Harvey P, Ravi B, Kendzerska T. et al. Osteoarthritis-related difficulty walking and risk for diabetes complications. Osteoarthritis Cartilage. 2017;25:67-75
106. Xiong X, Xiong H, Peng J, Liu Y, Zong Y. METTL3 Regulates the m(6)A Modification of NEK7 to Inhibit the Formation of Osteoarthritis. Cartilage. 2023: 19476035231200336.
107. Chen X, Gong W, Shao X, Shi T, Zhang L, Dong J. et al. METTL3-mediated m(6)A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann Rheum Dis. 2022;81:87-99
108. Cheng S, Xu X, Wang R, Chen W, Qin K, Yan J. Chondroprotective effects of bone marrow mesenchymal stem cell-derived exosomes in osteoarthritis. J Bioenerg Biomembr. 2024;56:31-44
109. An X, Wang R, Lv Z, Wu W, Sun Z, Wu R. et al. WTAP-mediated m(6)A modification of FRZB triggers the inflammatory response via the Wnt signaling pathway in osteoarthritis. Exp Mol Med. 2024;56:156-67
110. Ye G, Li J, Yu W, Xie Z, Zheng G, Liu W. et al. ALKBH5 facilitates CYP1B1 mRNA degradation via m6A demethylation to alleviate MSC senescence and osteoarthritis progression. Exp Mol Med. 2023;55:1743-56
111. Lu Y, Zhang H, Pan H, Zhang Z, Zeng H, Xie H. et al. Expression pattern analysis of m6A regulators reveals IGF2BP3 as a key modulator in osteoarthritis synovial macrophages. J Transl Med. 2023;21:339
112. Chen Z, Wang W, Hua Y. Expression patterns of eight RNA-modified regulators correlating with immune infiltrates during the progression of osteoarthritis. Front Immunol. 2023;14:1019445
113. Liu Z, Liu H, Li D, Ma L, Lu T, Sun H. et al. Comprehensive analysis of m6A RNA methylation modification patterns and the immune microenvironment in osteoarthritis. Front Immunol. 2023;14:1128459
114. Hao L, Shang X, Wu Y, Chen J, Chen S. Construction of a Diagnostic m(7)G Regulator-Mediated Scoring Model for Identifying the Characteristics and Immune Landscapes of Osteoarthritis. Biomolecules. 2023;13:539
115. Chen Z, Hua Y. Identification of m7G-related hub biomarkers and m7G regulator expression pattern in immune landscape during the progression of osteoarthritis. Cytokine. 2023;170:156313
116. Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS. et al. Rheumatoid arthritis. Nat Rev Dis Primers. 2018;4:18001
117. Huang J, Fu X, Chen X, Li Z, Huang Y, Liang C. Promising Therapeutic Targets for Treatment of Rheumatoid Arthritis. Front Immunol. 2021;12:686155
118. Itoh Y. Metalloproteinases: potential therapeutic targets for rheumatoid arthritis. Endocr Metab Immune Disord Drug Targets. 2015;15:216-22
119. Li X, Xu X, Zhang Q, Ling M, Li X, Tan X. METTL14 promotes fibroblast-like synoviocytes activation via the LASP1/SRC/AKT axis in rheumatoid arthritis. Am J Physiol Cell Physiol. 2023;324:C1089-C100
120. Lu H, Lu X, Xie Q, Wan H, Sun Y. TTC4 inhibits NLRP3 inflammation in rheumatoid arthritis by HSP70. Int J Rheum Dis. 2023;26:1751-9
121. Chen J, Lin X, He J, Liu D, He L, Zhang M. et al. Artemisitene suppresses rheumatoid arthritis progression via modulating METTL3-mediated N6-methyladenosine modification of ICAM2 mRNA in fibroblast-like synoviocytes. Clin Transl Med. 2022;12:e1148
122. Kuang Y, Li R, Wang J, Xu S, Qiu Q, Lin S. et al. ALKBH5-Mediated RNA m(6) A Methylation Regulates the Migration, Invasion, and Proliferation of Rheumatoid Fibroblast-Like Synoviocytes. Arthritis Rheumatol. 2024;76:192-205
123. Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18:609-24
124. Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83-103
125. Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18:39-50
126. Link MP, Goorin AM, Miser AW, Green AA, Pratt CB, Belasco JB. et al. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. N Engl J Med. 1986;314:1600-6
127. Miller BJ, Cram P, Lynch CF, Buckwalter JA. Risk factors for metastatic disease at presentation with osteosarcoma: an analysis of the SEER database. J Bone Joint Surg Am. 2013;95:e89
128. Jiang J, Pan H, Li M, Qian B, Lin X, Fan S. Predictive model for the 5-year survival status of osteosarcoma patients based on the SEER database and XGBoost algorithm. Sci Rep. 2021;11:5542
129. Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y. et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6)A-dependent manner. Cell Death Dis. 2020;11:659
130. Wang C, Meng Y, Zhao J, Ma J, Zhao Y, Gao R. et al. Deubiquitinase USP13 regulates glycolytic reprogramming and progression in osteosarcoma by stabilizing METTL3/m(6)A/ATG5 axis. Int J Biol Sci. 2023;19:2289-303
131. Yang Z, Cai Z, Yang C, Luo Z, Bao X. ALKBH5 regulates STAT3 activity to affect the proliferation and tumorigenicity of osteosarcoma via an m6A-YTHDF2-dependent manner. EBioMedicine. 2022;80:104019
132. Shao D, Liu C, Wang Y, Lin J, Cheng X, Han P. et al. DNMT1 determines osteosarcoma cell resistance to apoptosis by associatively modulating DNA and mRNA cytosine-5 methylation. FASEB J. 2023;37:e23284
133. Wang Z, Yu P, Zou Y, Ma J, Han H, Wei W. et al. METTL1/WDR4-mediated tRNA m(7)G modification and mRNA translation control promote oncogenesis and doxorubicin resistance. Oncogene. 2023;42:1900-12
134. Wang G, Wang H, Cheng S, Zhang X, Feng W, Zhang P. et al. N1-methyladenosine methylation-related metabolic genes signature and subtypes for predicting prognosis and immune microenvironment in osteosarcoma. Front Genet. 2022;13:993594
135. Xiao B, Liu Y, Chandrasiri I, Adjei-Sowah E, Mereness J, Yan M. et al. Bone-Targeted Nanoparticle Drug Delivery System-Mediated Macrophage Modulation for Enhanced Fracture Healing. Small. 2024;20:e2305336
136. Einhorn TA, Gerstenfeld LC. Fracture healing: mechanisms and interventions. Nat Rev Rheumatol. 2015;11:45-54
137. Mi B, Xiong Y, Yan C, Chen L, Xue H, Panayi AC. et al. Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J Cell Mol Med. 2020;24:6385-96
138. Schett G, Lories RJ, D'Agostino MA, Elewaut D, Kirkham B, Soriano ER. et al. Enthesitis: from pathophysiology to treatment. Nat Rev Rheumatol. 2017;13:731-41
139. Liu L, Yuan Y, Zhang S, Xu J, Zou J. Osteoimmunological insights into the pathogenesis of ankylosing spondylitis. J Cell Physiol. 2021;236:6090-100
140. Ward MM, Deodhar A, Gensler LS, Dubreuil M, Yu D, Khan MA. et al. 2019 Update of the American College of Rheumatology/Spondylitis Association of America/Spondyloarthritis Research and Treatment Network Recommendations for the Treatment of Ankylosing Spondylitis and Nonradiographic Axial Spondyloarthritis. Arthritis Rheumatol. 2019;71:1599-613
141. Calin A, Brophy S, Blake D. Impact of sex on inheritance of ankylosing spondylitis: a cohort study. Lancet. 1999;354:1687-90
142. Fang Y, Liu J. Novel regulatory role of non-coding RNAs in ankylosing spondylitis. Front Immunol. 2023;14:1131355
143. Lai NS, Yu HC, Chen HC, Yu CL, Huang HB, Lu MC. Aberrant expression of microRNAs in T cells from patients with ankylosing spondylitis contributes to the immunopathogenesis. Clin Exp Immunol. 2013;173:47-57
144. Chen Y, Wu Y, Fang L, Zhao H, Xu S, Shuai Z. et al. METTL14-m6A-FOXO3a axis regulates autophagy and inflammation in ankylosing spondylitis. Clin Immunol. 2023;257:109838
145. Xie Z, Yu W, Zheng G, Li J, Cen S, Ye G. et al. TNF-alpha-mediated m(6)A modification of ELMO1 triggers directional migration of mesenchymal stem cell in ankylosing spondylitis. Nat Commun. 2021;12:5373
146. Sun X, Zhou C, Zhu J, Wu S, Liang T, Jiang J. et al. Identification of clinical heterogeneity and construction of a novel subtype predictive model in patients with ankylosing spondylitis: An unsupervised machine learning study. Int Immunopharmacol. 2023;117:109879
147. Luo Q, Zhu J, Wang S, Fu P, Fu B, Huang Z. et al. Decreased expression of NAT10 in peripheral blood mononuclear cells from new-onset ankylosing spondylitis and its clinical significance. Arthritis Res Ther. 2024;26:7
148. Ma X, Xia W, Zong Y, Jiang C, Shan H, Lin Y. et al. Tumor necrosis factor-alpha promotes Staphylococcus aureus-induced osteomyelitis through downregulating endothelial nitric oxide synthase. J Microbiol Immunol Infect. 2021;54:1018-27
149. Dayan GH, Mohamed N, Scully IL, Cooper D, Begier E, Eiden J. et al. Staphylococcus aureus: the current state of disease, pathophysiology and strategies for prevention. Expert Rev Vaccines. 2016;15:1373-92
150. Berendt AR, Peters EJ, Bakker K, Embil JM, Eneroth M, Hinchliffe RJ. et al. Diabetic foot osteomyelitis: a progress report on diagnosis and a systematic review of treatment. Diabetes Metab Res Rev. 2008;24(Suppl 1):S145-61
151. Zhou Z, Pan C, Lu Y, Gao Y, Liu W, Yin P. et al. Combination of Erythromycin and Curcumin Alleviates Staphylococcus aureus Induced Osteomyelitis in Rats. Front Cell Infect Microbiol. 2017;7:379
152. Noskin GA, Rubin RJ, Schentag JJ, Kluytmans J, Hedblom EC, Smulders M. et al. The burden of Staphylococcus aureus infections on hospitals in the United States: an analysis of the 2000 and 2001 Nationwide Inpatient Sample Database. Arch Intern Med. 2005;165:1756-61
153. Hu CY, Jiao Y. A METTL3 Inhibitor Alleviates the Onset of Osteomyelitis in a Mouse Model by Targeting MyD88. Jpn J Infect Dis. 2023;76:191-6
154. Shi X, Ni H, Wu Y, Guo M, Wang B, Zhang Y. et al. Diagnostic signature, subtype classification, and immune infiltration of key m6A regulators in osteomyelitis patients. Front Genet. 2022;13:1044264
155. Song M, Lv K, Xu Z, Li J, Sun J, Shi J. et al. N6 methyladenosine eraser FTO suppresses Staphylococcus aureus-induced ferroptosis of bone marrow mesenchymal stem cells to ameliorate osteomyelitis through regulating the MDM2/TLR4/SLC7A11 signaling pathway. Cell Biol Int. 2024;48:450-60
156. Sims NA, Martin TJ. Coupling Signals between the Osteoclast and Osteoblast: How are Messages Transmitted between These Temporary Visitors to the Bone Surface? Front Endocrinol (Lausanne). 2015;6:41
157. Mazziotta C, Lanzillotti C, Iaquinta MR, Taraballi F, Torreggiani E, Rotondo JC. et al. MicroRNAs Modulate Signaling Pathways in Osteogenic Differentiation of Mesenchymal Stem Cells. Int J Mol Sci. 2021;22:2362
158. Xiao D, Huang S, Tang Z, Liu M, Di D, Ma Y. et al. Mijiao formula regulates NAT10-mediated Runx2 mRNA ac4C modification to promote bone marrow mesenchymal stem cell osteogenic differentiation and improve osteoporosis in ovariectomized rats. J Ethnopharmacol. 2024;330:118191
159. Charles JF, Aliprantis AO. Osteoclasts: more than 'bone eaters'. Trends Mol Med. 2014;20:449-59
160. Zaidi M, Turner CH, Canalis E, Pacifici R, Sun L, Iqbal J. et al. Bone loss or lost bone: rationale and recommendations for the diagnosis and treatment of early postmenopausal bone loss. Curr Osteoporos Rep. 2009;7:118-26
161. McLean RR. Proinflammatory cytokines and osteoporosis. Curr Osteoporos Rep. 2009;7:134-9
162. He X, Hu W, Zhang Y, Chen M, Ding Y, Yang H. et al. Cellular senescence in skeletal disease: mechanisms and treatment. Cell Mol Biol Lett. 2023;28:88
163. Nygaard G, Firestein GS. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat Rev Rheumatol. 2020;16:316-33
164. Wang Q, Chu P, Yu X, Li J, Zhang W, Gong M. ZFAS1 knockdown inhibits fi broblast-like synoviocyte proliferation, migration, invasion and inflammation, and promotes apoptosis via miR-3926/FSTL1 in rheumatoid arthritis. Exp Ther Med. 2021;22:914
165. Xie Z, Wang P, Li Y, Deng W, Zhang X, Su H. et al. Imbalance Between Bone Morphogenetic Protein 2 and Noggin Induces Abnormal Osteogenic Differentiation of Mesenchymal Stem Cells in Ankylosing Spondylitis. Arthritis Rheumatol. 2016;68:430-40
166. Zheng L, Zhuang Z, Li Y, Shi T, Fu K, Yan W. et al. Bone targeting antioxidative nano-iron oxide for treating postmenopausal osteoporosis. Bioact Mater. 2022;14:250-61
Author contact
![]() Corresponding authors: Email: zzmzzcedu.cn (Zhongmin Zhang), 269549364com (Xiaoyi Mo), xujiajiaedu.cn (Jiajia Xu).
Corresponding authors: Email: zzmzzcedu.cn (Zhongmin Zhang), 269549364com (Xiaoyi Mo), xujiajiaedu.cn (Jiajia Xu).

 Global reach, higher impact
Global reach, higher impact