10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(3):1127-1143. doi:10.7150/ijbs.102431 This issue Cite
Research Paper
TTC7B triggers the PI4KA-AKT1-RXRA-FTO axis and inhibits colon cancer cell proliferation by increasing RNA methylation
Key Laboratory of Carcinogenesis and Translational Research (MOE/Beijing), Division of Etiology, Peking University Cancer Hospital and Institute, Beijing, 100142, China.
# Equal first authors.
Received 2024-8-16; Accepted 2024-12-31; Published 2025-1-13
Abstract
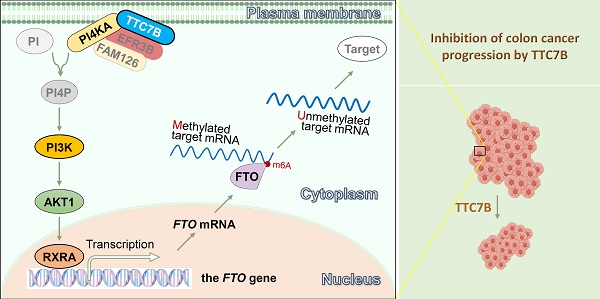
TTC7B is the PI4KA-binding protein. The upstream regulatory network associated with the expression of genes involved in RNA N6-adenine (m6A) methylation is not clear. Bioinformatics analysis revealed that the expression levels of TTC7B, PI4KA, and FTO are positively correlated with each other across human tissues. These genes are consistently downregulated in many cancers. We initially confirmed the correlation of the expression of these genes in colon cancer tissues from patients (n=105) and reported that TTC7B downregulation was significantly associated with poor prognosis. We subsequently performed a series of biological experiments and demonstrated that TTC7B upregulated RXRA expression probably through the PI4KA-mediated AKT1 pathway and that RXRA was a transcription factor for the FTO gene. TTC7B inhibited the proliferation of colon cancer cells by increasing the recruitment of RXRA to the FTO promoter, increasing FTO expression, and decreasing the total RNA m6A level. Ablation of FTO demethylase activity completely abolished the inhibitory effect of TTC7B on the proliferation of cancer cells in vitro and in vivo. In conclusion, our study demonstrated for the first time that TTC7B triggers the RXRA-FTO axis through PI4KA binding, which leads to a decrease in total RNA m6A modification and the inhibition of colon cancer progression.
Keywords: TTC7B, FTO, m6A, RXRA, PI4KA, AKT1, colon cancer
Introduction
RNA modifications are recognized as rapid adaptive responses of cells to environmental factors. As the most abundant type of RNA modification, N6-methyladenosine (m6A) plays important roles in RNA metabolism, cell fate determination, and cancer development [1-4]. It is unclear how cells sense environmental factors and coordinately modulate the expression levels of genes encoding RNA m6A modification enzymes, including the core components of methyltransferase complexes (METTL3, METTL14 and WTAP) and the demethylases FTO and ALKBH5 [5].
The tetratricopeptide repeat (TPR) domain is common in a variety of proteins and is associated with multiple protein complexes. Abnormalities in TPR family members are related to dysfunctions in cell motility, mitochondrial respiration, and intestinal development [6-8]. In our recent work, we reported that TTC22, a member of the TPR family, directly interacts with the RPL4 protein and WTAP pre-mRNA to increase WTAP expression and promote colon cancer metastasis through the upregulation of SNAI1 [9]. Tetratricopeptide repeat domain 7B (TTC7B) is another TPR protein that physically interacts with PI4KA on the cellular membrane [10, 11]. The level of TTC7B transcription is not only inversely correlated with that of the TTC22 gene but also positively correlated with those of the FTO and PI4KA genes in numerous normal/cancer tissues and cancer cell lines according to The Cancer Genome Atlas (TCGA), Genotype Tissue Expression (GTEx), and Cancer Cell Line Encyclopedia (CCLE) datasets (Figures S1-S3) [12-14]. However, the effects of the TTC7B gene on RNA modifications and the underlying mechanisms are largely unknown.
Here, we report for the first time that TTC7B upregulated the expression of the RXRA gene, probably through PI4KA binding, and increased the recruitment of the RXRA protein to the FTO promoter, which subsequently upregulated FTO expression and downregulated total RNA m6A modification (TRm6A) and cancer cell proliferation.
Results
TTC7B expression is correlated with FTO expression and colon cancer metastasis
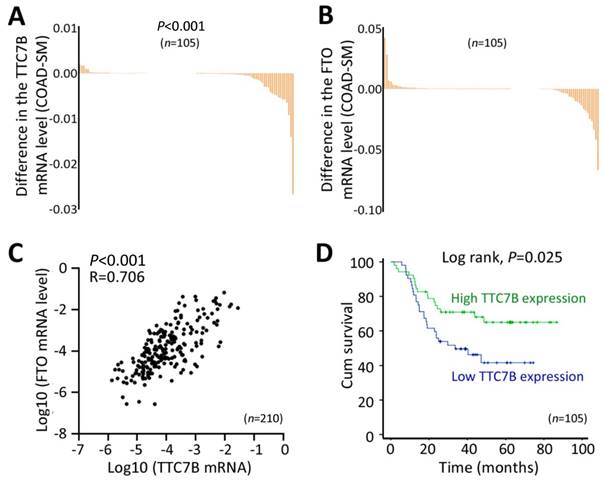
Because TTC7B, FTO, and PI4KA expression is frequently downregulated in many cancers in the TCGA project, including colon adenocarcinoma (COAD) (Figure S4), we performed a validation experiment using COAD and paired surgical margin (SM) samples from 105 patients. Our quantitative RT‒PCR (qRT‒PCR) confirmed the results of the above bioinformatics analyses. The TTC7B mRNA level was significantly lower in the COAD tissues than in the SM tissues from these patients (Figure 1A), and the FTO mRNA level was also lower in the COAD samples than in the SM samples, but the difference was not significant (Figure 1B). In addition, the level of TTC7B mRNA in COAD tissues at advanced invasion stages was lower than that in COAD tissues at early invasion stages (P=0.038; Table S1). Similar differences in the mRNA levels of the TTC7B and FTO genes were also detected in the SM samples (P=0.036 and 0.037; Tables S1 and S2). Consistent with the above bioinformatics analyses, the mRNA levels of the TTC7B and FTO genes were strongly correlated with each other in the COAD samples (R=0.706, P<0.001; Figure 1C). Kaplan‒Meier analysis revealed that the overall survival (OS) of patients with low TTC7B expression was much shorter than that of patients with high TTC7B expression (log rank test, P=0.025; Figure 1D). A significant difference in the OS of patients with low and high FTO expression was not observed (log rank test, P=0.567; data not shown). Thus, we further investigated whether the TTC7B gene regulates FTO expression and TRm6A levels in detail, as described below.
Changes in the expression of the TTC7B gene in cancer tissues. (A and B) Comparison of the mRNA levels of the TTC7B and FTO genes in colon adenocarcinoma (COAD) and paired surgical margin (SM) tissues. (C) Relationships between the mRNA levels of the TTC7B and FTO genes in COAD and paired SM tissue samples (n=210) from 105 patients. The correlation coefficient (R) and P value are also labeled. (D) Kaplan‒Meier survival curves of patients with COAD with high and low TTC7B expression levels.
TTC7B upregulates FTO expression and downregulates TRm6A
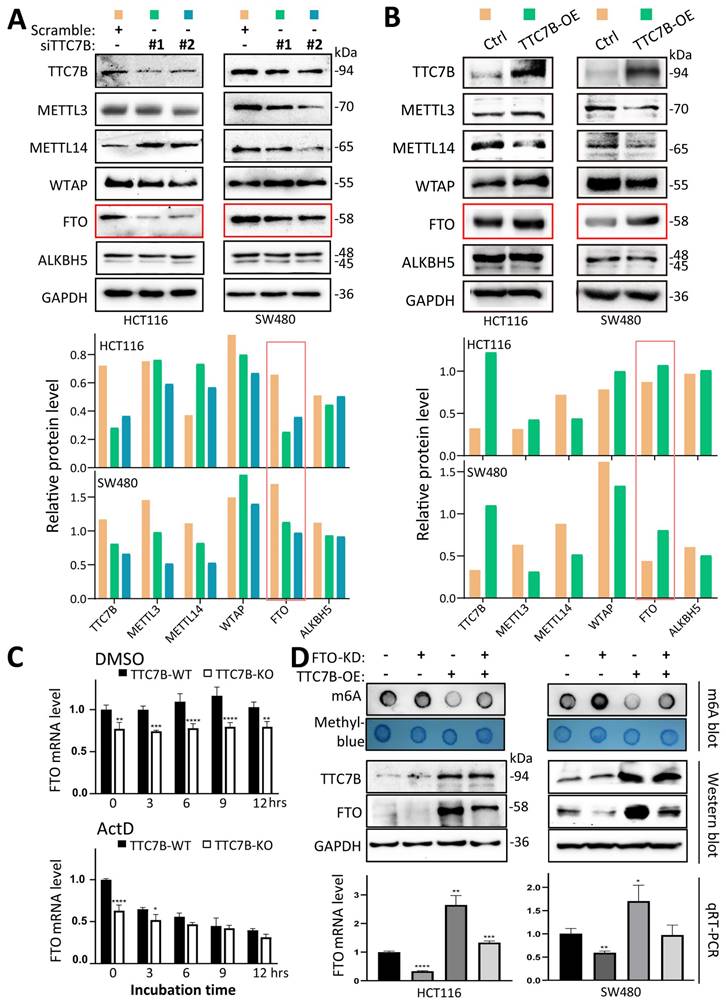
According to the qRT‒PCR analysis, baseline TTC7B mRNA was almost detectable in most of the 15 tested human cancer cell lines, and a medium expression level of TTC7B was detected in the colon cancer cell lines HCT116, LoVo, and SW480 (Figure 2A). Thus, we downregulated TTC7B expression (siTTC7B) in these colon cancer cells via two siRNAs targeting TTC7B mRNA (siTTC7B#1 and siTTC7B#2). m6A-specific dot blot analysis revealed that siTTC7B significantly increased TRm6A abundance in these siTTC7B cells 48 hr posttransfection (Figure 2B). A similar difference in the TRm6A level was induced in these cell lines by TTC7B knockout (TTC7B-KO) (Figure 2C). This was confirmed by liquid chromatography coupled with mass/mass spectrometry (LC‒MS/MS) and ELISA analyses. The ratio of m6A/rA in TTC7B-KO HCT116 cells was greater than that in TTC7B wild-type (TTC7B-WT) cells (Figure 2D). In contrast, TTC7B overexpression (TTC7B-OE) significantly decreased the TRm6A level in these cells, as determined by m6A-specific ELISA and dot blotting (Figure 2E and 2F). In the rescue experiment, the restoration of TTC7B expression mostly mitigated the TTC7B-KO-induced increase in TRm6A (Figure 2G). These results indicate that TTC7B inhibited TRm6A modification.
In our RNA-seq experiments, the GO analysis results revealed that the genes upregulated in TTC7B-KO cells were enriched in mRNA processing and splicing in HCT116 cells, whereas the genes downregulated in TTC7B-OE cells were enriched in DNA replication. In contrast, both the TTC7B-KO downregulated genes and the TTC7B-OE upregulated genes were enriched in cell adhesion binding molecules in these cells (Figure S5). Our m6A-antibody-immunoprecipitated RNA sequencing (meRIP-seq) data further revealed that TTC7B-KO downregulated the mRNA m6A levels of genes related to endocytosis, infection, and colorectal cancer, according to the results of the mRNA pathway analysis (Figure S6 and Supplementary Data File 1). These data support the hypothesis that TTC7B might be a cancer-related gene.
Combined analysis of these RNA-seq and meRIP-seq datasets revealed that the proportion of differentially downregulated transcripts among differentially m6A-modified transcripts (or genes, n=1725) was significantly greater than that among transcripts (n=8292) without differential m6A modifications in TTC7B-KO HCT116 cells (63.9% vs. 51.8%, P<0.001; Figure S7A and Supplementary Data File 2). No difference was detected between differentially m6A-modified transcripts with and without differential expression (37.6% vs. 37.5%, respectively). These 1725 genes are enriched mainly in transcription processes via DAVID functional annotation bioinformatics analysis [15]. Using the publicly available RNA-seq dataset for FTO-KO HEK293T cells [16], we compared the proportion of differentially expressed transcripts in TTC7B-KO HCT116 and FTO-KO HEK293T cells. We found that the proportions of both differentially upregulated and downregulated transcripts were the same among the overlapping gene set (n=1015) in these two cell models (Figure S7B). These findings suggest that FTO and RNA m6A modifications may contribute to TTC7B-KO-induced downregulation.
To determine the mechanisms by which TTC7B decreases TRm6A levels, we examined the changes in the expression of five core m6A modification enzymes in the siTTC7B and TTC7B-OE HCT116 and SW480 cell lines. We found that the level of the FTO protein was consistently decreased in the two siTTC7B cell lines via Western blot analysis (Figure 3A), whereas it was markedly increased in the two TTC7B-OE cell lines (Figure 3B). No consistent changes in the expression of the other four m6A modifiers (METTL3, METTL14, WTAP, and ALKBH5) were observed. Similar differences in the transcript levels of these m6A modifier genes were also observed in the RNA-seq datasets (Figure S8). Notably, according to the qRT‒PCR analysis, the level of FTO mRNA was consistently decreased in TTC7B-KO cells, whereas the inhibition of RNA polymerase activity by ActD (at a final concentration of 5 μg/mL) abolished the decrease in FTO mRNA expression induced by TTC7B-KO. The results of the bioinformatics analysis confirmed the positive correlation between TTC7B and FTO transcripts in both normal and cancer tissues of the gut (Figure S2A and S2B). These phenomena strongly indicate that TTC7B upregulated FTO transcription (Figure 3C).
We further evaluated the importance of changes in FTO expression in the TTC7B-mediated regulation of TRm6A modification. While the TRm6A level was obviously decreased by TTC7B-OE in scramble control cells, it was only weakly decreased by TTC7B-OE in FTO stable knockdown (FTO-KD) cells (Figure 3D).
RXRA is essential for TTC7B-mediated upregulation of FTO expression
To screen possible genes contributing to TTC7B-upregulated FTO transcription, differentially expressed genes (DEGs) induced by TTC7B-OE and TTC7B-KO in HCT116 cells were screened via RNA-seq (Figure S9A).
Effects of functional changes in the TTC7B gene on total RNA m6A levels in human cancer cells. (A) The expression status of the TTC7B and FTO genes in a set of cancer cell lines according to qRT‒PCR analysis. (B) Effect of siRNA-mediated knockdown of TTC7B expression on total RNA m6A levels in 3 cancer cell lines according to m6A immunoblot analysis. The extent of TTC7B downregulation by siTTC7B#1 and siTTC7B#2 was monitored via qRT‒PCR and Western blotting. (C) Effect of knockout of TTC7B exon-11 (TTC7B-KO) via CRISPR/Cas9 on total RNA m6A levels in HCT116, SW480, and LoVo cells. The knockout status of TTC7B exon 11 was confirmed by PCR sequencing. (D and E) Results of LC‒MS/MS and ELISA analyses for detecting total RNA m6A levels in colon cancer cells with and without TTC7B-KO or TTC7B-OE. (F) Effects of TTC7B overexpression (TTC7B-OE) on total RNA m6A levels in HCT116, SW480 and LoVo cells. (G) Effect of TTC7B-OE on the TTC7B-KO-induced increase in total RNA m6A levels in colon cancer HCT116 and SW480 cells in the rescue experiment. These experiments were performed 72 hr after FTO-OE or TTC7B-OE. */**/***/****: P<0.05/0.01/0.001/0.0001, respectively, according to Student's t test.

As mentioned above, the TTC22 gene is another upstream regulator of TRm6A in HCT116 cells [9]; thus, we selected TTC7B-DEGs that overlapped with TTC22-DEGs (n=1980) to reduce the number of candidate genes that may contribute to m6A-upstream regulatory networks (Figure S9B, left). We further excluded m6A-downstream genes from the list of these 1980 DEGs via publicly available RNA-seq datasets [17-20], including DEGs induced by the knockdown of FTO, ALKBH5, METTL3&14, and WTAP expression. Finally, 421 DEGs that did not overlap with these four groups of genes were considered TTC7B&TTC22-regulated m6A-upstream candidates (Supplementary Data File 3) and were used in further bioinformatics analysis (Figure 4B, middle and right). KEGG/Gene Ontology (GO) analysis revealed that these 421 candidates were significantly enriched in 13 molecular function clusters [15]. Retinoid X receptor (RXR) isoform α and β genes (RXRA and RXRB) and ESR2 were enriched in multiple DNA binding clusters (Figure S9C, red letters highlighted).
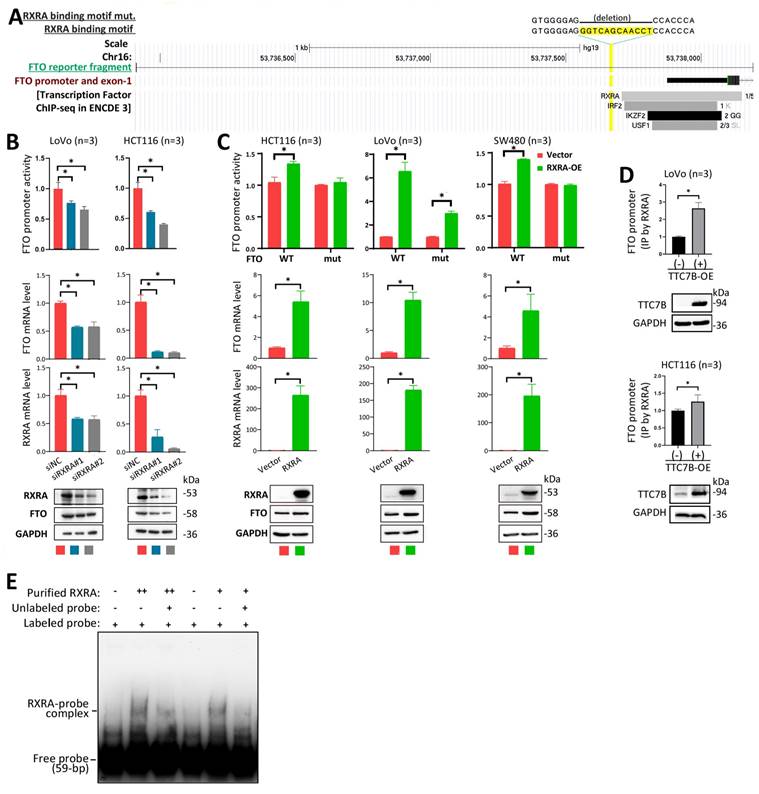
RXRA generally forms homoduplexes with itself or heteroduplexes with RXRB, retinoid acid receptors (RARs) or other NRs to bind promoters and regulate the transcription of its target genes [21]. According to the ChIP-seq datasets in the Encyclopedia of DNA Elements (ENCODE) [22], four (RXRA, IKZF2, IRF2, and USF1) of these 421 genes overlapped with 143 transcription factors that were bound to the FTO promoter (Table S3). Our qRT‒PCR analyses revealed that only the RXRA gene was downregulated in TTC7B-KD cells and upregulated in TTC7B-OE cells, whereas the RXRB, IKZF2, IRF2, and USF1 genes were downregulated in both TTC7B-KD and TTC7B-OE cells (Figure S10). In addition, the transcript levels of the TTC7B, RXRA, and FTO genes were positively correlated with each other in gut cancer tissues in the TCGA project (Figure S2C and S2D). Therefore, the RXRA gene was selected as the top candidate for further study.
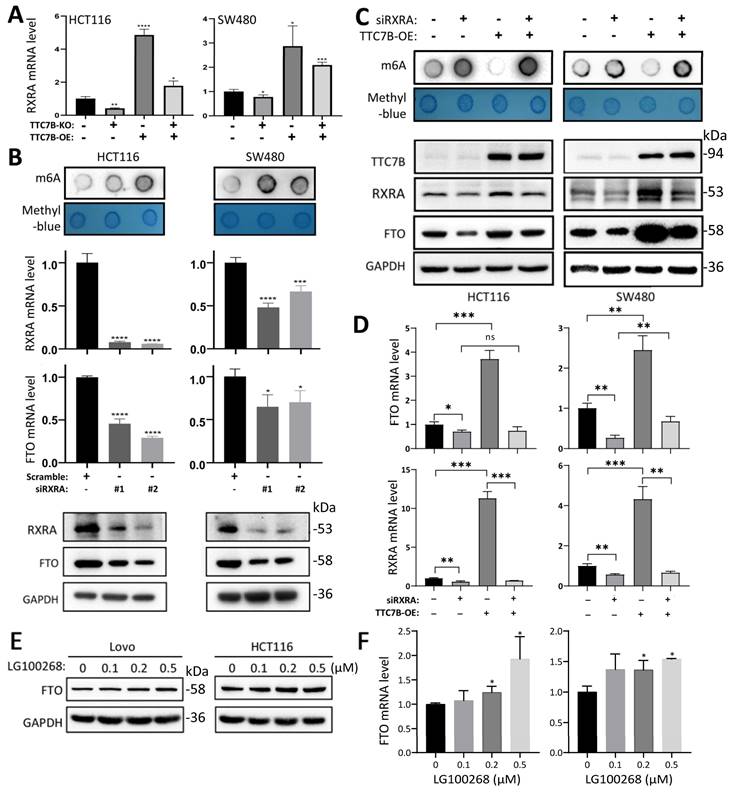
The results of qRT‒PCR analysis confirmed that the level of RXRA mRNA was significantly increased in TTC7B-OE cells and decreased in TTC7B-KO cells (Figure 4A). siRNA-mediated knockdown of RXRA expression (siRXRA) not only decreased the mRNA and protein levels of the FTO gene according to qRT‒PCR and Western blot analyses but also increased the TRm6A level in HCT116 and SW480 cells according to dot blot analysis (Figure 4B). Notably, while TTC7B-OE greatly increased FTO levels and decreased TRm6A levels in scramble control cells, TTC7B-OE only weakly affected FTO or TRm6A levels in siRXRA cells (Figure 4C). Similar alterations in FTO mRNA levels were also observed via qRT‒PCR analysis (Figure 4D). In addition, the expression level of FTO gradually increased in a dose-dependent manner 12 hr after treatment with LG100268, an agonist of retinoid X receptors [23], according to qRT‒PCR and Western blot analyses (Figure 4E and 4F). These phenomena demonstrate that RXRA may be essential for TTC7B-mediated upregulation of FTO expression and TRm6A modification.
The binding of the transcription factor RXRA to the FTO promoter increases gene transcription
To investigate whether RXRA directly regulates the transcription of the FTO gene as a transcription factor in detail, we analyzed the JASPAR database of transcription factor binding profiles [24] and identified one potential RXRA binding motif (5'-gggtcagcaacct-3') within the FTO promoter (Figure 5A, highlighted in yellow). We subsequently constructed an FTO luciferase reporter plasmid by inserting an FTO promoter fragment (2290 bp) and its RXRA binding motif-deleted mutant into the pGL3-basic vector and used the resulting plasmids to transfect HCT116, LoVo, and SW480 colon cancer cell lines. We found that siRXRA not only significantly decreased the activity of the FTO promoter in the reporter analysis but also decreased the endogenous mRNA and protein levels of the FTO gene in HCT116 and LoVo cells according to the qRT‒PCR and Western blot analyses (Figure 5B). Notably, RXRA overexpression (RXRA-OE) increased the activity of the wild-type FTO promoter but not the activity of the FTO promoter mutant in HCT116 and SW480 cells (Figure 5C). Although the activity of the FTO promoter mutant was still increased by RXRA-OE in LoVo cells, the fold change in the activity of the FTO promoter mutant was only half that of the wild-type FTO promoter. qRT‒PCR and Western blot analyses revealed that endogenous FTO expression levels were also increased by RXRA-OE in the cells used in the reporter analysis. In the ChIP‒PCR analysis, the amount of RXRA-enriched FTO promoter DNA was significantly increased by TTC7B-OE in HCT116 and LoVo cells (Figure 5D). The results of the EMSA also revealed that the purified GST-RXRA protein directly bound the biotin-labeled FTO promoter probe and that the unlabeled probe inhibited RXRA-FTO promoter binding (Figure 5E). Taken together, the above data strongly support the hypothesis that RXRA is a transcription factor for the FTO gene.
Characterization of the FTO gene as a TTC7B target that inhibits total RNA m6A levels. (A and B) Comparison of the protein levels of various m6A modifiers in HCT116 and SW480 cells with those in siTTC7B or TTC7B-OE cells by Western blot analysis. The protein levels relative to those of GAPDH are displayed below the lane images. FTO bands with consistent alterations by siTTC7B and TTC7B-OE are highlighted with red frames. (C) Effect of ActD treatment on TTC7B-KO-induced changes in FTO mRNA levels in HCT116 cells. (D) Effect of shRNA-mediated stable knockdown of FTO expression (FTO-KD) on total RNA m6A levels in two cell lines with and without TTC7B-OE, as determined by m6A dot blot analysis. */**/***/****: P<0.05/0.01/0.001/0.0001, respectively, according to Student's t test. These experiments were performed 48 hr after FTO-OE or TTC7B-OE.
Effects of the RXRA gene on TTC7B-induced changes in FTO expression and RNA m6A modification. (A) qRT‒PCR analysis of the effects of TTC7B-KO and TTC7B-OE on RXRA expression in HCT116 and SW480 cells 48 hr posttransfection. (B) qRT‒PCR analysis of the effects of siRNA-mediated knockdown of RXRA on total RNA m6A and the protein and mRNA levels of the FTO gene in HCT116 and SW480 cells. (C and D) qRT‒PCR analysis of the effects of siRXRA (#1) on TTC7B-induced total RNA m6A and the protein and mRNA levels of the FTO gene in HCT116 and SW480 cells 72 hr and 48 hr after siRNA and TTC7B expression vector transfection, respectively. (E and F) The protein and mRNA levels of the FTO gene in LoVo and HCT116 cells 12 hr after treatment with the retinoid X receptor agonist LG100268 at various concentrations; */**/***/****: P<0.05/0.01/0.001/0.0001, respectively, according to Student's t test.
TTC7B inhibits cancer cell proliferation in an FTO-dependent manner
We further studied the effect of TTC7B on the proliferation of colon cancer cells and found that TTC7B-OE significantly inhibited the proliferation of HCT116, LoVo, and SW480 cells (Figure S11A), whereas TTC7B-KO significantly increased the proliferation and colony formation of HCT116 and SW480 cells (Figure S11B and S11C). Furthermore, the restoration of TTC7B expression completely mitigated the effect of TTC7B-KO on the proliferation of these cells (Figure S11D). These experimental results support the hypothesis that TTC7B may inhibit COAD progression.
Next, we evaluated the role of the FTO gene on the TTC7B-mediated inhibition of colon cancer cell proliferation using FTO-KO cell models. Notably, we found that stable TTC7B-OE did not affect the proliferation of the four FTO-KO cell lines but did significantly inhibit the proliferation of their FTO-WT counterparts (Figure 6A). These in vitro experimental results were confirmed by animal experiments. Stable TTC7B-OE did not inhibit the growth of FTO-KO LoVo cells transplanted into nude mice (s.c.), whereas it significantly inhibited the growth of FTO-WT LoVo cells (Figure 6B; P<0.001). The proportion of Ki67-expressing cells was inhibited by TTC7B-OE only in xenografts derived from FTO-WT LoVo cells (70% to 53%) but not in those derived from FTO-KO cells (31% to 42%). In contrast, TTC7B-KO significantly increased the growth of HCT116 cells (Figure 6C). These results confirmed that TTC7B inhibited the proliferation of colon cancer cells in an FTO-dependent manner.
Interestingly, the proliferation rate of FTO-KO HCT116 and SW480 cells was lower than that of cells harboring wild-type FTO alleles (FTO-WT) (Figure S12A). Consistent with these findings, the restoration of FTO expression by FTO-OE mitigated the effect of FTO-KO on LoVo and RKO cell proliferation (Figure S12B). The inhibitory effect of FTO KO on the growth of LoVo cells was also detected in the nude mouse model (Figure 6B; P=0.046).
Regulation of FTO expression by RXRA as a transcription factor. (A) Schematic diagram of the FTO promoter. The locations of one predicted (by JASPAR) RXRA binding motif, the binding fragments for four DEGs in the ENCODE 3 database, and the FTO promoter fragment integrated into the reporter are displayed. (B) Effects of RXRA knockdown on FTO promoter activity and endogenous FTO expression, as determined by a luciferase reporter assay, qRT‒PCR, and Western blot 72 hr posttransfection. (C) Activity of the wild-type FTO promoter (WT) and RXRA binding motif-deleted FTO promoter (mut) in cell lines with and without RXRA-OE for 72 hr. The level of endogenous FTO expression was also determined via qRT‒PCR and Western blotting. (D) ChIP‒PCR showing whether RXRA bound to the FTO promoter in cells with and without TTC7B-OE. (E) The results of the EMSA for detecting the binding of the biotin-labeled FTO promoter probe with the purified RXRA protein. *: P<0.05 according to Student's t test.
Effect of the FTO gene on TTC7B-mediated inhibition of colon cancer cell proliferation. (A) Effects of TTC7B-OE on the proliferation of four colon cancer cell lines with and without FTO KO. (B) Effects of TTC7B-OE on the growth of LoVo cells with and without FTO KO in nude mice. The weights of the xenografts on the 23rd experimental day and the constituent ratios of Ki67(+) and Ki67(-) cell populations in representative xenografts derived from LoVo cells with different gene managements are displayed on the below. The P values obtained via Student's t test are listed. (C) Effect of TTC7B-KO on the growth of HCT116 cells. The weights of the xenografts on the 21st experimental day are displayed on the right. The P value obtained via the Mann-Whitney test is listed. (D) Comparison of total RNA m6A levels in FTO-KO cells with and without restoration of FTO expression. The wild-type FTO expression vector containing the FTO promoter (FTO-wt) and its dioxygenase mutant (FTO-mut) are illustrated. (E) Effect of stable TTC7B-OE on the proliferation of HCT116 and LoVo cells with and without FTO KO and/or the restoration of FTO expression, as determined with the IncuCyte long-term live-cell observation platform. TTC7B and FTO expression in these cells was determined by Western blotting, and the results are displayed on the right.
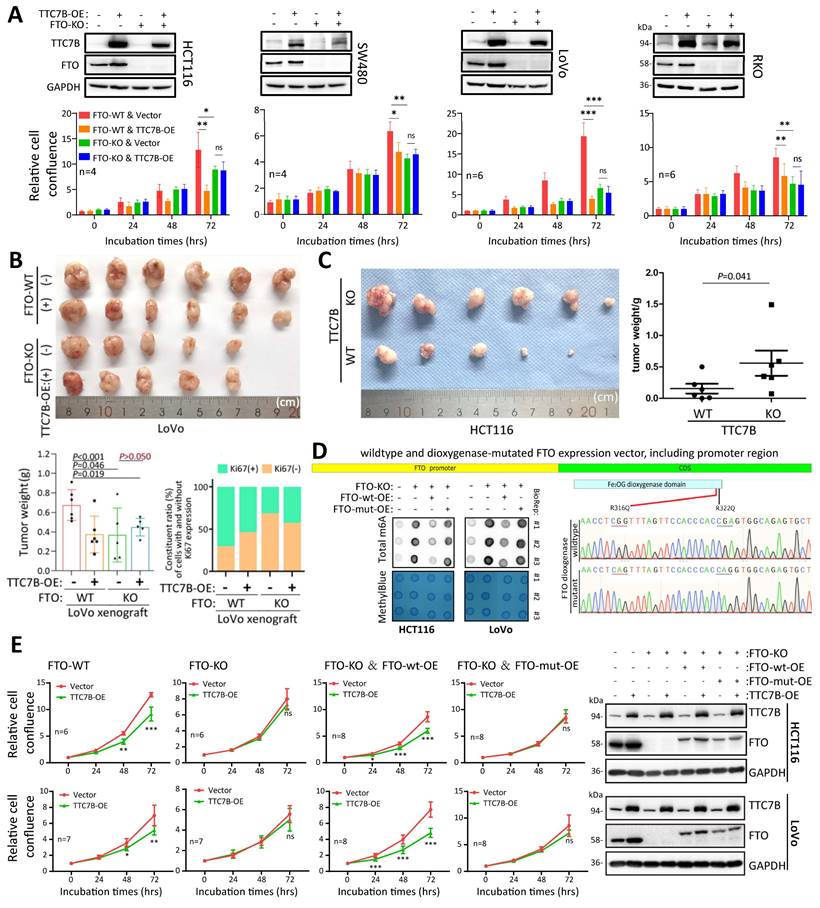
To further evaluate whether the effect of TTC7B on the proliferation of colon cancer cells is dependent on the demethylase activity of FTO, we constructed a special wild-type FTO expression vector containing the FTO promoter (FTO-wt), which is responsible for the regulatory effect of the TTC7B-RXRA axis on FTO transcription. A demethylase activity-null FTO mutant expression vector with R316Q and R322Q mutations (FTO-mut) was also constructed according to a previous report [25]. As expected, FTO-wt overexpression (FTO-wt-OE) mostly mitigated the impact of FTO-KO on the TRm6A level, and this effect was not induced by FTO-mut overexpression (FTO-mut-OE) (Figure 6D). While FTO-KO abolished the inhibitory effect of TTC7B-OE on the proliferation of HCT116 and LoVo cells, the restoration of FTO expression by FTO-wt-OE mitigated the effect of FTO-KO on TTC7B-OE. However, this effect was not observed in cells in which FTO expression was restored by FTO-mut-OE (Figure 6E). Together, these results indicate not only that FTO plays an essential role in the inhibition of colon cancer cell proliferation by TTC7B but also that the TTC7B-mediated inhibition of cancer cell proliferation might be dependent on the demethylase activity of FTO.
TTC7B may upregulate RXRA expression through binding the PI4KA protein
TTC7B is a given PIK4A-binding protein, and the TTC7B-PIK4A interaction activates both the PI3K-AKT and MEK-ERK signaling pathways, two crucial pathways for cell proliferation [26-28]. We found that inhibition of the TTC7B-PI4KA interaction via siRNA-mediated knockdown of PI4KA (PI4KA-KD) or inhibition of PI4KA by GSK-A1 abolished TTC7B-induced RXRA upregulation at both the mRNA and protein levels (Figure 7A and 7B). In addition, the repression of PI3K/AKT1 activity by treatment with the AKT1 inhibitor MK2206 or the PI3K inhibitor BYL719 completely blocked the effect of TTC7B on RXRA expression in colon cancer cells, whereas the repression of MEK/ERK activity by treatment with the MEK1/2 inhibitor U0126 or selumetinib did not (Figure 7C). These results suggest that TTC7B increases RXRA expression through binding to PIK4A and subsequently activating the PI3K‒AKT1 pathway (Figure 7D).
TTC7A is an important paralog of TTC7B, and their codisruption results in synthetic lethal interactions [29]. We found that the mRNA level of TTC7A is inversely and significantly associated with that of TTC7B in human cancer cell lines according to CCLE datasets (n=1037; r=-0.41). Interestingly, siRNA-mediated knockdown of TTC7A also increased the TRm6A levels in colon cancer cell lines (Figure S13). However, no significant alteration in the amount of FTO was detected in these cells. These results suggest that there are functional differences between TTC7A and TTC7B.
Discussion
RNA modifications play crucial roles in host cellular adaptive responses and the development of diseases, including cancer and obesity [30-32]. It is not clear how environmental factors trigger cellular adaptive responses and how cells coordinate the expression levels of m6A modifier genes to maintain TRm6A dynamics. Our recent work indicated that TTC22 is an upstream regulator of TRm6A and colon cancer metastasis in an RPL4-WTAP-YTHDF1 axis-dependent manner [9]. TTC7B expression is inversely correlated with TTC22 expression and is positively correlated with FTO expression. In the present study, we reported for the first time that TTC7B is another upstream regulator of TRm6A and colon cancer cell proliferation. TTC7B increased RXRA expression through the PI4KA-AKT1 signaling pathway, subsequently increasing FTO transcription and decreasing TRm6A levels. TTC7B inhibits the proliferation of colon cancer cells in an RXRA-FTO axis-dependent manner.
There are two RNA demethylases: FTO and ALKBH5. The regulatory mechanisms of these genes have rarely been reported. Our series of experiments indicated that RXRA is a transcription factor of the FTO gene. As a master regulator, RXRA regulates the transcriptional activity of target genes through the formation of duplexes with many partners, including RARA itself, RXRB, and thyroid hormone receptor [21]. The partners that cooperate with RXRA to regulate FTO expression are worthy of further study. Interestingly, RARA is an FTO target gene in AML, and the regulation of RARA expression by FTO depends on its m6A demethylase activity [33]. Our work and that of others indicate that there may be a negative feedback loop between RXRA and FTO expression. FTO posttranscriptionally downregulates the expression of RXRA's partner, whereas RXRA upregulates FTO transcription.
The functions of FTO in the development of cancers remain unclear. FTO has been reported to repress the growth of cancers in the colon, liver, and stomach, while it has also been reported to promote cancer cell proliferation [34-40]. Compared with that in paired normal tissues, FTO was downregulated in breast carcinoma (BRCA), cervical carcinoma (CESC), COAD, lung carcinoma (LUAD and LUSC), and uterine cancer (UCEC and UCS) tissues but upregulated in esophageal carcinoma (ESCA), pancreatic adenocarcinoma (PAAD), and stomach adenocarcinoma (STAD) tissues, according to TCGA pancancer RNA-seq datasets [40-42]. These phenomena suggest that the impact of the FTO gene on cancer development may be cancer specific. We did not observe a significant difference in FTO mRNA levels between COAD and paired normal tissues from 105 patients by qRT‒PCR. Although FTO-KO (or FTO-OE) inhibited (or enhanced) colon cancer cell proliferation, unexpectedly, we found that TTC7B-OE, which upregulates FTO expression, inhibited cell proliferation in an FTO-dependent manner. Our in vivo experiment fully confirmed these in vitro results. Both FTO-KO and TTC7B-OE significantly inhibited the growth of LoVo cells in nude mice, and the loss of FTO abolished the effect of TTC7B-OE, suggesting a dominant effect of TTC7B over FTO on colon cancer cell proliferation. In other words, the effect of FTO on cell proliferation may be weaker than that of TTC7B alone or in combination. FTO is a potential therapeutic target for cancer, and several small-molecule FTO inhibitors are under development [43, 44]. Our findings described above suggest that TTC7B and RXRA may be closely associated with the sensitivity of colon cancer cells to FTO inhibitors because of their crucial regulation of FTO expression.
Effect of PI4KA-AKT activity on RXRA upregulation by TTC7B. (A) Effects of siRNA-mediated knockdown of PI4KA (PI4KA-KD) and inhibition of PI4KA by GSK-A1 on the abundance of RXRA protein in cells with and without TTC7B-OE. (B) Effect of PI4KA-KD on the abundance of RXRA mRNA in cells with and without TTC7B-OE. (C) Effects of treatment with the PI3K inhibitor BLY719, the AKT inhibitor MK2206, and MERK1/2 inhibitors (U0126 and selumetinib) on the TTC7B-induced increase in RXRA expression in cells. (D) Graphical summary of the ability of the TTC7B gene to upregulate the PI3K-AKT-RXRA-FTO axis and to decrease the proliferation of colon cancer cells. The RXRA protein levels relative to those of GAPDH (A) or ACTIN and Tubulin (C) are displayed below the lane images.
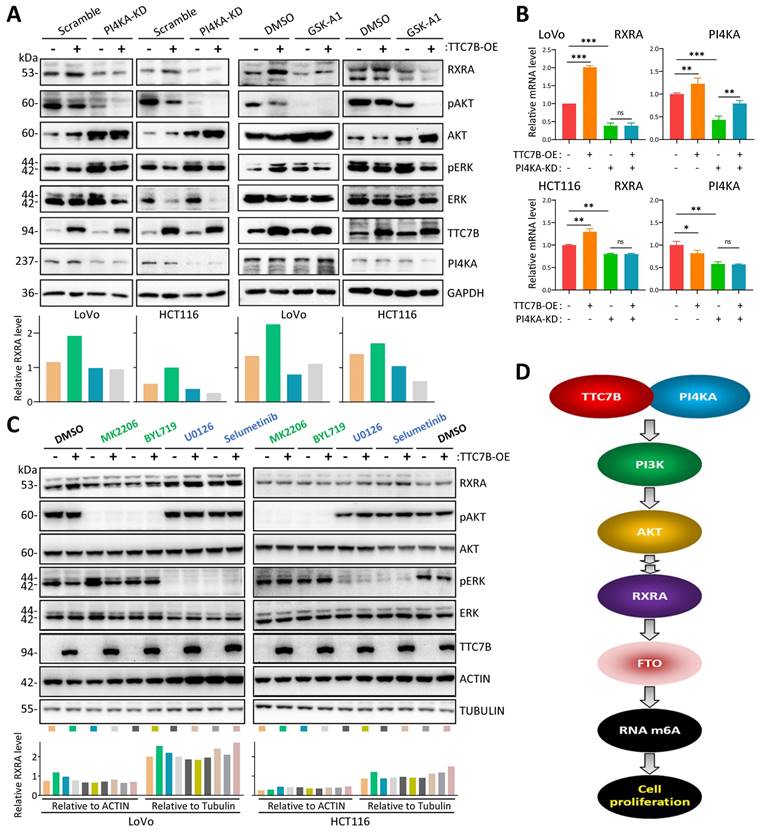
The TTC7B protein mainly localizes to the cytoplasm and plasma membrane and physically interacts with PI4KA, which activates several key signaling pathways, including the PI3K/AKT and MEK/ERK pathways, which promote cancer cell proliferation [26-28]. We found that inhibition of the TTC7B-PI4KA interaction with PI4KA-KD and the PI4KA inhibitor GSK-A1 blocked the RXRA upregulation induced by TTC7B. Repression of PI3K/AKT1 activity with AKT1/PI3KA inhibitors completely abolished TTC7B-upregulated RXRA expression, whereas repression of MEK/ERK activity with MEK inhibitors did not. These phenomena indicate that TTC7B may increase RXRA expression through the PI4KA-AKT1 pathway. The detailed mechanism by which RXRA is upregulated by AKT1 needs to be further studied.
Retinoic acid, a natural agonist of retinoic acid receptor (RAR) and retinoic acid X receptor (RXR), plays important roles in cell growth, differentiation, and organogenesis [45]. Some RXRA agonists have been used to treat cancers. For example, bexarotene combined with cisplatin and vinorelbine resulted in a 25% response rate with better-than-expected survival rates in patients with non-small cell lung cancer [46]. The combination of all-trans retinoic acid (ATRA) and arsenic trioxide (ATO) safely cures acute promyelocytic leukemia. In addition, the synergistic targeting of Pin1 by ATO and ATRA is an attractive approach for combating breast cancer and other cancers [47]. Whether RXRA upregulation by TTC7B might enhance the therapeutic efficacy of ATRA- and RXRA agonist-mediated treatments for cancers is worthy of study.
In conclusion, the present study indicated that TTC7B inhibits colon cancer progression in an FTO-dependent manner both in vitro and in vivo. Our findings also revealed that TTC7B upregulates the transcription factor RXRA, which subsequently binds to the FTO promoter and activates FTO transcription. Our study revealed a novel TTC7B-PI4KA-AKT1-RXRA-FTO axis in colon cancer cells, which may be useful for the development of anticancer RNA demethylase inhibitors.
Methods
Human tissue samples
Colon carcinomas (n=105) and paired surgical margin samples were obtained from patients at Beijing Cancer Hospital who were enrolled in our previous studies [48]. The Institutional Review Board of Peking University Cancer Hospital & Institute approved this study, which was carried out in accordance with the principles outlined in the Declaration of Helsinki. Informed consent was obtained from each patient prior to their inclusion in the study.
Cell culture
The human HCT116 and SW480 cancer cell lines were kindly provided by Professor Yuanjia Chen at Peking Union Medical College Hospital. The RKO cell line was kindly provided by Professor Guoren Deng at the University of California. The HCT-8 and U87 cell lines were obtained from Professor W Du and W Fang, Peking Union Medical College Hospital. The HeLa, SGC7901, and MGC803 cell lines were kindly provided by Professor Yang Ke. The H1299 and LoVo cell lines were kindly provided by Professor Chengchao Shou. The A549 and MCF-7 cell lines were kindly provided by Professors Zhiqian Zhang and Yuntao Xie, respectively, at Peking University Cancer Hospital. The H661, H292, and MKN45 cell lines were purchased from the National Laboratory Cell Resource Sharing Platform (Beijing, China). HCT116, SW480, RKO, LoVo, HCT-8, H661, H1299, A549, H292, SGC7901, MKN45, and MGC803 cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA). HeLa, U87, and MCF-7 cells were cultured in DMEM (Gibco). All the cell lines were cultured in the corresponding medium supplemented with 10% fetal bovine serum (FBS) and a 1% penicillin (10 kU/mL)/streptomycin mixture (10 mg/mL) (C0222, Beyotime, Beijing, China).
Treatment with small-molecule inhibitors
To inhibit RNA polymerase activity, HCT116 cells were treated with ActD (HY-17559; MedChemExpress, NJ, USA) at a final concentration of 5 μg/mL. The RXRA agonist LG100268 (HY-15340, MedChemExpress, NJ, USA) was used at 0/0.1/0.2/0.5 μM to treat HCT116 and LoVo cells for 12 hr. To inhibit PI4KA activity, GSK-A1 (HY-125118, MedChemExpress, NJ, USA) was used at final concentration of 200 nM to treat cells for 12 hr. To block related cell signal pathways, the following drugs were used to treat cells for 6 hr at different final concentrations: MK2206 (HY-10358, MedChemExpress, NJ, USA) at 10 μM; BYL719 (S2814, Shelleck, TX, USA) at 10 μM; U0126 (S1901, Beyotime, Shanghai, China) at 10 μg/mL; Selumetinib (SD5933, Beyotime, Shanghai, China) at 10 μM.
Construction of the TTC7B, RXRA, and FTO expression vectors
The pCMV-MCS-3Flag-TTC7B expression vector was purchased from Vigene Bio (c14_5685, Shandong, China). To generate cells with transient TTC7B overexpression (TTC7B-OE), HCT116, SW480 and LoVo cells were transfected with pCMV-TTC7B-3Flag or an empty control vector with XtremeGENE HP DNA Transfection Reagent (Roche, Mannheim, Germany). To generate cells with stable TTC7B overexpression, the abovementioned cells were transfected with pCMV-TTC7B-3Flag or empty control vector and selected with G418. The pReceiver-Lv242-RXRA expression vector was purchased from GeneCopoeia (EX-A0529-Lv242, Guangzhou, China). The plasmid pCMV-hFTO-3xFLAG for FTO overexpression was kindly provided by Prof. Yun-Gui Yang of the Chinese Academy of Sciences. To generate a wild-type FTO expression vector (FTO-wt) that can be regulated by endogenous transcription factors, we replaced the CMV promoter in the pCMV-hFTO-3xFLAG vector with a DNA fragment (-1454 ~ +29 bp) in the FTO promoter. To generate FTO mutants without RNA demethylase activity, we replaced both R316 and R322 with glutamine (R316Q&R322Q) via the Fast Mutagenesis System (FM111-01; TransGen Biotech, Beijing, China) according to previously published methods [25].
Knockdown of gene expression by siRNA/shRNA
For the knockdown of gene expression, synthesized duplex RNAi oligos targeting human TTC7B, FTO, and RXRA were purchased from Gene Pharma (Shanghai, China; Table S4). The LV3(H1/GFP&Puro)-shFTO, TTC7B, and empty vector plasmids were also purchased from Gene Pharma. When the cells reached 70-80% confluence, they were transfected with siRNAs or plasmids by PEI MAX (24765-100; Polysciences, PA, USA) according to the manufacturer's instructions. Successful knockdown of the target gene was confirmed by Western blotting and qRT‒PCR. Scramble siRNAs were used as the negative control.
Knockout of the TTC7B and FTO genes via CRISPR/Cas9
To generate TTC7B-KO and FTO-KO cells, the CRISPR/Cas9 editing system was used according to a published protocol [49]. Briefly, cells transfected with plasmids containing CRISPR/Cas9 and TTC7B single guide RNA (sgRNA; Table S4) were selected with puromycin (final concentration, 1 μg/mL) for 2 weeks, whereas CRISPR/Cas9- and FTO-sgRNA-transfected cells were selected by flow cytometry (BD Biosciences, Franklin Lakes, USA) at 48 hr posttransfection. The surviving cells were harvested and seeded into 96-well plates for the formation of cell clones and further expansion. The expanded monoclonal cell populations were pooled and designated TTC7B-KO or FTO-KO cells. Two weeks later, monoclonal cells with good growth conditions were selected. The status of target gene knockout was determined by PCR sequencing. Three to five subclones with failed target gene knockout were pooled and used as TTC7B-WT or FTO-WT cells.
RNA extraction and quantitative RT‒PCR (qRT‒PCR)
Total RNA was isolated with TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. The quality and concentration of the RNA samples were determined with a NanoDrop 2000 system (Thermo Fisher Scientific, Inc.). cDNA was synthesized from the qualified RNA samples with TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Quantitative RT‒PCR (qRT‒PCR) was performed using a StepOne Real-time PCR System (Applied Biosystems, Foster City, CA, USA) and SYBR Green PCR master mix reagents (FastStart Universal SYBR Green Master, Roche, Mannheim, Germany). The primer sequences are listed in Table S4.
m6A dot blot
The m6A dot blot assay was performed in accordance with a previously reported protocol with slight modifications [50]. Briefly, the indicated amount of total cellular RNA was denatured in a 3-fold volume of RNA incubation buffer (46.2% formaldehyde and 53.8% 20 × SSC) at 95 °C for 5 min, followed by chilling on ice. Next, RNA samples (50-200 ng) were spotted directly onto a positively charged nylon membrane (RPN303B, GE Healthcare, Chicago, USA) and air-dried for 5 min. The membrane was then UV cross-linked in an ultraviolet crosslinker, blocked with 1% blocking reagent (11096176001, Roche) in maleic acid (pH 7.5), and then incubated with a m6A antibody (1:3000, ab151230, Abcam, Cambridge, UK) overnight at 4 °C. HRP-conjugated anti-rabbit IgG secondary antibody was added to the membrane for 1 hr at room temperature with gentle shaking, after which the membrane was developed with enhanced chemiluminescence. Methylene blue staining was used to verify that equal amounts of RNA were spotted on the membrane.
LC‒MS/MS for detection of the m6A/rA ratio
LC‒MS/MS quantification of m6A was performed by Cloudseq Biotech Inc. (Shanghai, China) following the recommended protocol. Total RNA from TTC7B-KO and control HCT116 cells was isolated with TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA). One microgram of total RNA was digested with 4 µL of nuclease P1 (Sigma, St. Louis, MO) in 40 µL of buffer solution (10 mM Tris-HCl pH 7.0, 100 mM NaCl, 2.5 mM ZnCl2) at 37 °C for 12 hr, followed by incubation with 1 µL of alkaline phosphatase (Sigma‑Aldrich, Merck KGaA, Darmstadt, Germany) at 37 °C for 2 hr. The RNA mixture was diluted to 100 µL and injected into the LC‒MS/MS instrument. The nucleosides were separated by reversed-phase high-performance liquid chromatography on an Agilent C18 column (Agilent Technologies, San Diego, CA) coupled with mass spectrometry detection with an AB SCIEX QTRAP 5500 (AB Sciex LLC, Framingham, MA). Pure nucleosides were used to generate standard curves, from which the concentrations of adenosine (A) and m6A in the sample were calculated. The level of m6A was then calculated as a percentage of total adenosine in RNA (rA) [51].
Quantification of the m6A abundance via m6A-specific ELISA
The total RNA m6A level was quantified with an EpiQuik m6A RNA Methylation Quantification Kit (P-9005; Epigentek Group, NY, USA) according to the manufacturer's instructions [52]. Briefly, 200 ng of RNA sample or 1 ng of m6A positive control RNA standard was added to the assay wells and incubated at 37 ºC for 90 min, followed by incubation with the capture antibody for 1 hr and then incubation with the detection antibody mixture for 30 min at room temperature. The absorbance at 450 nm was detected with a Synergy H1 Microplate Reader (Biotek, VT, USA). The relative level of m6A RNA methylation in total RNA was calculated according to the following formula: m6A% = [(ODSample- ODNC) ÷ S]/[(ODPC - ODNC) ÷ P] ×100% (S is the amount of input sample RNA in ng; P is the amount of input positive control in ng).
Western blot
For Western blot analysis, cells at approximately 80% confluence were harvested. Proteins were extracted from these cells and diluted in 1× cell lysis buffer. The cell lysates were separated by SDS‒PAGE and electroblotted onto nitrocellulose membranes. Nonspecific binding was blocked with 5% nonfat milk in PBS overnight at 4 °C, and the membranes were rinsed twice with PBST. Then, the membranes were incubated with the indicated primary antibodies at room temperature for 1.5 hr, washed six times with PBST, incubated with horseradish peroxidase-labeled secondary antibodies for 45 min and washed again as described above. The protein bands were visualized with an enhanced chemiluminescence system (Thermo Scientific, Rockford, IL) (Supplemental Material). The primary antibodies used for TTC7B and other proteins are listed in Table S5.
RNA sequencing and m6A-antibody-immunoprecipitated RNA sequencing (meRIP-seq)
For whole-genome transcriptome profiling, four libraries were generated from total RNA samples extracted from TTC7B-KO, control, TTC7B-OE, and vector control HCT116 cells by sequencing on an Illumina NovaSeq platform, and 150 bp paired-end reads were generated. HISAT2 (version 2.0.5) was used to map the cleaned reads to the human hg38 reference genome. Upregulated and downregulated genes (P-adj<0.05) were identified to determine significant functions and significant signaling pathways on the basis of the Gene Ontology database (GO-Analysis) or KEGG database (Pathway-Analysis). The differentially expressed genes are listed in Supplementary Data File 4. The raw data were deposited under the Gene Expression Omnibus accession number GSE247502.
meRIP-seq was performed as described in our previous report [9]. The differentially methylated mRNAs between TTC7B-KO and TTC7B-WT HCT116 cells are listed in Supplementary Data File 5. The raw data have been deposited, and the Gene Expression Omnibus accession number is GSE266858.
Selection of m6A-upstream differentially expressed genes (DEGs) with RNA-seq datasets
Our RNA-seq datasets for HCT116 cells with and without TTC7B-KO/-OE (GSE247502) and TTC22-KD/-OE (GSE189982) were used to identify TTC7B&TTC22-related DEGs (n=1980), as illustrated in Figure S9B. RNA-seq datasets for shALKBH5 cells (GSE144968), shFTO cells (GSE136204), siWTAP cells (GSE142386), and siMETTL3&14 cells (GSE146806) were downloaded from the Gene Expression Omnibus, a publicly available database [17-20], and used to exclude m6A-downstream genes (n=1559) among the TTC7B&TTC22-related DEGs (Supplementary Data File 3).
Dual-luciferase reporter assay
The wild-type FTO gene promoter (2290 bp) was amplified via PCR, digested with the restriction enzymes KpnI and XhoI, inserted into the pGL3-basic vector and used to transfect HCT116, SW480, and LoVo cells in 24-well plates. siRNAs for RXRA, scrambled siRNA, the TTC7B plasmid or the control vector were cotransfected with the Renilla vector (3 wells/group). The activities of both Renilla and firefly luciferases were measured with the Dual-Luciferase Reporter Assay System (Yeasen, Shanghai, China). The results are presented after normalization to the measured values of Renilla luciferase.
Chromatin immunoprecipitation assay (ChIP)
LoVo and HCT116 cells were transfected with pCMV-MCS-TTC7B-3Flag and the vector, respectively. These cells were then collected and used in the ChIP assay as previously described [53]. Briefly, a rabbit antibody against RXRA was used to precipitate the bound DNA. Normal rabbit IgG was used as a negative control. The precipitated DNA was subsequently purified with a DNA, RNA, and protein purification kit (740609--250; Macherey-Nagel, Düren, Germany). The FTO promoter DNA fragment was amplified via quantitative PCR.
Electrophoresis mobility shift assay (EMSA)
The 59-nt FTO promoter sense-strand and biotin-labeled antisense-strand DNA fragments were synthesized by Thermo Fisher Scientific (Waltham, MA, USA; Table S4) and annealed as EMSA hot probes. DNA fragments with the same sequence but without biotin labeling were used as unlabeled cold probes. To purify the RXRA protein, the coding DNA sequence for full-length RXRA from the pReceiver-Lv242-RXRA vector (EX-A0529-Lv242, GeneCopoeia, Guangzhou, China) was inserted into the pGEX-4T1 vector. The GST-RXRA protein was induced with IPTG and purified with glutathione Sepharose beads (17-0756-01, GE Healthcare, Sweden). A chemiluminescent EMSA kit (GS009, Beyotime, Shanghai, China) was used for the shift assay according to the manufacturer's instructions [54].
Assessment of cell proliferation with IncuCyte, the CCK-8 kit and colony formation assays
For long-term dynamic observation of cell proliferation, colon cancer cells were seeded into 96-well plates (3000 cells/well, n=4/6 wells/group) and cultured for at least 96 hr to generate proliferation curves. The cells were photographed every 24 hr on a long-term dynamic observation platform (IncuCyte, Essen, MI, USA). Cell confluence was analyzed with IncuCyte ZOOM software (Essen, Ann Arbor, MI, USA).
For the CCK-8 assay, 100 μL of cell suspension (30000 cells/mL) was added to each well of a 96-well plate with 6 wells/treatment. Cell proliferation was analyzed via a cell counting kit-8 (A311-01/02; Vazyme, Nanjing, China). Briefly, 10 μL of CCK-8 solution was added to each well of the plate, and the plate was incubated for 1 hr in a 37 °C incubator. The absorbance at 450 nm was subsequently measured with a Bio-Rad microplate reader. The cells were measured daily for 3 continuous days. The average absorbance was calculated.
For colony formation analysis, approximately 1000 cells in 2 mL of RPMI 1640 medium supplemented with 10% FBS were cultured in 6-well plates (3 wells/treatment) at 37 °C with 5% CO2. Two weeks later, the cell colonies were fixed with methanol, stained with crystal violet solution, photographed, and counted.
Xenografts in BALB/c nude mice
LoVo or HCT116 cells resuspended in 0.1 mL of PBS (2×107 cells/mL) were inoculated subcutaneously into the bilateral inguinal region of 6-week-old female nude mice (5/6 mice per group, 2×106 cells per injection; purchased from Beijing Huafukang Biotech) [55]. The mice were sacrificed by spine dislocation on the 23rd or 21st inoculation day, and the xenografts were separated, weighed, and photographed. The tumor samples were processed into formalin-fixed and paraffin-embedded (FFPE) sections.
Statistical analyses
All the statistical analyses were performed with SPSS 16.0 software. Student's t test was used for the determination of mRNA expression and relative luciferase activity. Differences in the TTC7B expression level in tissue samples were assessed via the Mann‒Whitney U test. The correlation between the levels of TTC7B and FTO expression was measured with a nonparametric correlation test and curvilinear regression model. For the Kaplan‒Meier survival analysis, CC patients were classified into high and low TTC7B expression groups, with the median mRNA level (0.0623) used as the cutoff value. Statistical significance was assigned at P < 0.05 (*), P < 0.01 (**) or P < 0.001 (***). All experiments were performed at least three times with triplicate samples.
Supplementary Material
Supplementary figures and tables.
Supplementary Data File 1.
Supplementary Data File 2.
Supplementary Data File 3.
Supplementary Data File 4.
Supplementary Data File 5.
Acknowledgements
This work was supported by grants from the Beijing Hospital Authority Mission Plan (SML20191101) to DD and from the Science Foundation of Peking University Cancer Hospital (grant #2022--8) to TW. The authors gratefully acknowledge Prof. Yun-Gui Yang from the Chinese Academy of Sciences for kindly sending the FTO overexpression plasmids, and they also gratefully acknowledge Prof. Pietro DeCamilli at Yale University for the gift of the TTC7B plasmid.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zheng G, Dahl JA, Niu Y. et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol Cell. 2013;49(1):18-29
2. Agarwala SD, Glitzblau HG, Hochwagen A. et al. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8(6):e1002732
3. Lin S, Choe J, Du P. et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62(3):335-345
4. Ding N, You A, Tian W. et al. Chidamide increases the sensitivity of Non-small Cell Lung Cancer to Crizotinib by decreasing c-MET mRNA methylation. Int J Biol Sci. 2020;16(14):2595-2611
5. Jaafa C, Aguiiar RCT. Dynamic multilayered control m6A RNA demethylase activity. PNAS. 2024;121(46):e2317847121
6. Boehlke C, Janusch H, Hamann C. et al. A Cilia Independent Role of Ift88/Polaris during Cell Migration. PLoS One. 2015;10(10):e0140378
7. Bottani E, Cerutti R, Harbour ME. et al. TTC19 Plays a Husbandry Role on UQCRFS1 Turnover in the Biogenesis of Mitochondrial Respiratory Complex III. Mol Cell. 2017;67(1):96-105.e4
8. Neves JF, Afonso I, Borrego L. et al. Missense mutation of TTC7A mimicking tricho-hepato-enteric (SD/THE)syndrome in a patient with very-early onset inflammatory bowel disease. Eur J Med Genet. 2018;61(4):185-188
9. You A, Tian W, Gu LK. et al. TTC22 promotes m6A-mediated WTAP expression and colon cancer metastasis in an RPL4 binding-dependent pattern. Oncogene. 2022;41(32):3925-3938
10. Ulengin-Talkish I, Parson MAH, Jenkins ML. et al. Palmitoylation targets the calcineurin phosphatase to the phosphatidylinositol 4-kinase complex at the plasma membrane. Nat Commun. 2021;12(1):6064
11. Dornan GL, Dalwadi U, Hamelin D. et al. Probing the Architecture, Dynamics, and Inhibition of the PI4KIIIα/TTC7/FAM126 Complex. J Mol Biol. 2018;430(18 Pt B):3129-3142
12. Cancer Cell Line Encyclopedia Consortium, Genomics of Drug Sensitivity in Cancer Consortium. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528:84-87
13. GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580-585
14. Tang Z, Li Chenwei, Kang B. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98-W102
15. Sherman BT, Hao M, Qiu J. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022 doi:10.1093/nar/gkac194
16. Bartosovic M, Molares HC, Gregorova P. et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017;45(19):11356-11370
17. Shen C, Sheng Y, Zhu AC. et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell. 2020;27(1):64-80.e9
18. Su R, Dong L, Li Y. et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38(1):79-96.e11
19. Wang LJ, Xue Y, Li H. et al. Wilms' tumor 1-associating protein inhibits endothelial cell angiogenesis by m6A-dependent epigenetic silencing of desmoplakin in brain arteriovenous malformation. J Cell Mol Med. 2020;24(9):4981-4991
20. Müller S, Bley N, Busch B. et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, posttranscriptional superesuper-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 2020;48(15):8576-8590
21. Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841-850
22. Wang J, Zhuang J, Iyer S. et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013;41(Database issue):D171-D176
23. Boehm MF, Zhang L, Zhi L. et al. Design and Synthesis of Potent Retinoid X Receptor Selective Ligands That Induce Apoptosis in Leukemia Cells. J Med Chem. 1995;38(16):3146-3155
24. Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I. et al. A JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50(D1):D165-D173
25. Jia G, Fu Y, Zhao X. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885-887
26. Yoshida S, Ohya Y, Goebl M. et al. A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J Biol Chem. 1994;269(2):1166-1172
27. Dornan GL, Mcphail JA, Burke JE. Type III phosphatidylinositol 4 kinases: structure, function, regulation, signalling and involvement in disease. Biochem Soc Trans. 2016;44(1):260-266
28. Adhikari H, Kattan WE, Kumar S. et al. Oncogenic KRAS is dependent upon an EFR3A-PI4KA signaling axis for potent tumorigenic activity. Nat Commun. 2021;12(1):5248
29. Thompson NA, Ranzani M, Weyden L. et al. Combinatorial CRISPR screen identifies fitness effects of gene paralogues. Nat Commun. 2021;12:1302
30. Zhou J, Wan J, Gao X. et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591-594
31. Yang S, Wei J, Cui Y-H. et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782
32. Wang X, Wu R, Liu Y. et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16(7):1221-1235
33. Li Z, Weng H, Su R. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N-Methyladenosine RNA demethylase. Cancer Cel.l. 2017;31:127-141
34. Yue C, Chen J, Li Z. et al. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J Exp Clin Cancer Res. 2020;39(1):240
35. Zuidhof HR, Calkhoven CF. Oncogenic and Tumor-Suppressive Functions of the RNA Demethylase FTO. Cancer Res. 2022;82(12):2201-2212
36. Tan Z, Shi S, Xu J. et al. RNA N6-methyladenosine demethylase FTO promotes pancreatic cancer progression by inducing the autocrine activity of PDGFC in an m6A-YTHDF2-dependent manner. Oncogene. 2022;41(20):2860-2872
37. Relier S, Ripoll J, Guillorit H. et al. FTO-mediated cytoplasmic m(6)Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat Commun. 2021;12:1716
38. Nan YB, Liu S, Luo Q. et al. m6A demethylase FTO stabilizes LINK-A to exert oncogenic roles via MCM3-mediated cell-cycle progression and HIF-1α activation. Cell Rep. 2023;42(10):113273
39. Sun Z, Sun X, Qin G. et al. FTO promotes proliferation and migration of bladder cancer via enhancing stability of STAT3 mRNA in an m6A-dependent manner. Epigenetics. 2023;18(1):2242688
40. Xiang MY, Liu WS, Tian W. et al. RNA N-6-methyladenosine enzymes and resistance of cancer cells to chemotherapy and radiotherapy. Epigenomics. 2020;12(9):801-809
41. Cerami E, Gao J, Dogrusoz U. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404
42. Gao J, Aksoy BA, Dogrusoz U. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1
43. Qing Y, Dong L, Gao L. et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol Cell. 2021;81(5):922-939.e9
44. Huang Y, Su R, Sheng Y. et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35(4):677-691.e10
45. Kadison A, Kim J, Maldonado T. et al. Retinoid signaling directs secondary lineage selection in pancreatic organogenesis. J Pediatr Surg. 2001;36(8):1150-1156
46. Khuri FR, Rigas JR, Figlin RA. et al. Multi-institutional phase I/II trial of oral bexarotene in combination with cisplatin and vinorelbine in previously untreated patients with advanced non-small-cell lung cancer. J Clin Oncol. 2001;19(10):2626-2637
47. Kozono S, Lin Y-M, Seo H-S. et al. Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells. Nat Commun. 2018;9(1):3069
48. Yuan H, Ren QW, Du Y. et al. LncRNA miR663AHG represses the development of colon cancer in a miR663a-dependent manner. Cell Death Discov. 2023;9(1):220
49. Joung J, Konermann S, Gootenberg JS. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828-863
50. Shen L, Liang Z, Yu H. Dot Blot Analysis of N6-methyladenosine RNA Modification Levels. Bio Protoc. 2017;7(1):e2095
51. Yuan BF. Liquid Chromatography-Mass Spectrometry for Analysis of RNA Adenosine Methylation. Methods Mol Biol. 2017;1562:33-42
52. Xu Y, Zhou J, Li L. et al. FTO-mediated autophagy promotes progression of clear cell renal cell carcinoma via regulating SIK2 mRNA stability. Int J Biol Sci. 2022;18(15):5943-5962
53. Li Z, Qiao J, Ma W. et al. P14AS upregulates gene expression in the CDKN2A/2B locus through competitive binding to PcG protein CBX7. Front Cell Dev Biol. 2022;10:993525
54. Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2(8):1849-61
55. Li P, Zhang X, Gu L. et al. P16 methylation increases the sensitivity of cancer cells to the CDK4/6 inhibitor palbociclib. PLoS One. 2019Oct25;14(10):e0223084
Author contact
![]() Corresponding authors: Wei Tian or Dajun Deng, Key Laboratory of Carcinogenesis and Translational Research (MOE/Beijing), Division of Cancer Etiology, Peking University Cancer Hospital and Institute, Fu-Cheng-Lu #52, Haidian District, Beijing, 100142, China, Tel.: 86-10-88196752, Fax: 86-10-88122437, E-mail: tianweiedu.cn or dengdajunedu.cn.
Corresponding authors: Wei Tian or Dajun Deng, Key Laboratory of Carcinogenesis and Translational Research (MOE/Beijing), Division of Cancer Etiology, Peking University Cancer Hospital and Institute, Fu-Cheng-Lu #52, Haidian District, Beijing, 100142, China, Tel.: 86-10-88196752, Fax: 86-10-88122437, E-mail: tianweiedu.cn or dengdajunedu.cn.

 Global reach, higher impact
Global reach, higher impact