10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(7):2991-3010. doi:10.7150/ijbs.109593 This issue Cite
Review
Crosstalk between RNA-binding proteins and non-coding RNAs in tumors: molecular mechanisms, and clinical significance
1. Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China.
2. Hunan Engineering Research Center of Tumor organoid Technology and application, Public Service Platform of Tumor organoids Technology, 283 Tongzipo Road, Changsha, 410013, Hunan, China.
3. University of South China, Hengyang, 421001, Hunan, China.
* The authors contribute equally to the work.
Received 2024-12-30; Accepted 2025-4-5; Published 2025-4-21
Abstract
RNA-binding proteins, integral in regulating RNA metabolism and gene expression, collaborate closely with non-coding RNAs, which are pivotal in post-transcriptional gene regulation. Both elements are essential for the progression of tumors. While recent research has increasingly illuminated their individual mechanisms, the intricate network interplay between them still requires further exploration. This article has provided a comprehensive review of the roles played by RNA-binding proteins and their associated non-coding RNAs in tumor biology. It delves into the intricate functions of various RNA-binding proteins in tumors, including their involvement in alternative splicing, m6A modification, alternative polyadenylation, and phase separation. Furthermore, it highlights the diverse and significant roles of different non-coding RNAs, such as microRNAs, long non-coding RNAs, and circRNAs, in tumor progression. The interaction between RNA-binding proteins and regulated non-coding RNAs is also explored, providing insights into their collective impact on metabolic reprogramming, immunity, drug resistance, metastasis, and ferroptosis. This in-depth exploration not only deepens our understanding of the mechanisms underlying tumorigenesis but also lays a foundation for developing innovative therapeutic strategies.
Keywords: RNA-binding proteins, non-coding RNAs, phase separation, splicing, tumor immunity
Introduction
Tumors, a disease caused by genetic and epigenetic changes that cause cells to lose control over normal growth and differentiation, represent one of the most significant public health challenges today [1, 2]. The progression of tumors involves a myriad of molecular mechanisms, with the regulation of gene expression being a crucial aspect. Gene expression goes through processes from transcription and translation to post-transcriptional and post-translational modifications, all of which are regulated at different levels [3, 4]. Among them, RNA-binding proteins (RBPs) and non-coding RNAs (ncRNAs) are two types of molecules that play an important role at the gene transcription level, and there are complex interactions between them, which affect the biological behavior of tumor cells [5, 6]. For example, RBPs can modulate the stability, transport, degradation, and functional performance of ncRNAs by recognizing and binding to their specific sequences or structures [7]. Conversely, ncRNAs can also interact with RBPs to affect the expression, localization, activity, and phase separation of RBPs. These interactions take place in various cellular compartments, including the nucleus, cytoplasm, and cell membrane [8]. Furthermore, these interactions manifest in a variety of physiological or pathological conditions, including hypoxia, inflammation, and tumor environments [9-11]. As a result of these interactions, gene expression is finely regulated, thereby impacting tumor cell behaviors such as proliferation, migration, invasion, apoptosis, autophagy, differentiation, stemness, and immune evasion [12-16]. For instance, recent studies have demonstrated that in metabolic disorders such as obesity-associated periodontitis, the long non-coding RNA AC018926.2 modulates the ITGA2/FAK/AKT signaling axis by binding to PARP1, thereby regulating stem cell differentiation [17].
This article aims to review the roles of RBPs and their associated ncRNAs in tumors, alongside the molecular mechanisms of their interactions. On one hand, the study of RBPs and ncRNAs has emerged as a highly active and promising research area in life sciences, encompassing not only the fundamental principles of gene expression regulation but also their significant implications for human health and disease [18]. In gallbladder carcinoma, exosome-derived lncRNA TRPM2-AS activates the NOTCH1 signaling pathway through direct interaction with PABPC1, significantly enhancing tumor angiogenesis and metastatic dissemination [19]. Particularly in oncology, RBPs and ncRNAs are identified as key players in numerous tumor-related signaling pathways and cellular processes, impacting critical aspects of tumor cell behavior such as proliferation, apoptosis, migration, invasion, and angiogenesis [20]. On the other hand, gaining a comprehensive understanding of the roles and interaction mechanisms of RBPs and ncRNAs in tumors not only aids in elucidating the molecular mechanisms of tumor genesis and progression, but also facilitates the identification of novel targets and strategies for tumor diagnosis, prognosis, and therapy. Recent investigations have further revealed that circRNF13 stabilizes ITGB1 mRNA by enhancing the phase separation capacity of IGF2BP1, thereby promoting cisplatin resistance in oral squamous cell carcinoma [21].
Currently, there is considerable research on the roles of RBPs and ncRNAs in tumors, as well as their interaction mechanisms. However, most of these studies focus on one or a few classes of RBPs or ncRNAs, lacking a comprehensive and systematic overview of the field as a whole. In addition, these studies tend to ignore the interactions between RBPs and ncRNAs, focusing only on their respective functions. Hence, a comprehensive review of this field is necessary, aiming to analyze the roles and interaction mechanisms of RBPs and ncRNAs in tumors from multiple perspectives and levels, thereby offering a structured framework and guidance for future research.
Initially, we will discuss the functional attributes of various RBPs in tumors, encompassing aspects like alternative splicing, m6A modification, APA, and phase separation. Second, we will review the role of different types of ncRNAs in tumors and the interactions between RBPs and their regulated ncRNAs, including miRNAs, lncRNAs, and circRNAs. Subsequently, we will summarize the roles of RBPs and ncRNAs in tumor-related aspects such as metabolism, immunity, drug resistance, metastasis, and ferroptosis. Finally, we will look forward to future research directions and challenges in this field.
Exploring the functional properties of RBPs in tumors
Regulating of alternative splicing
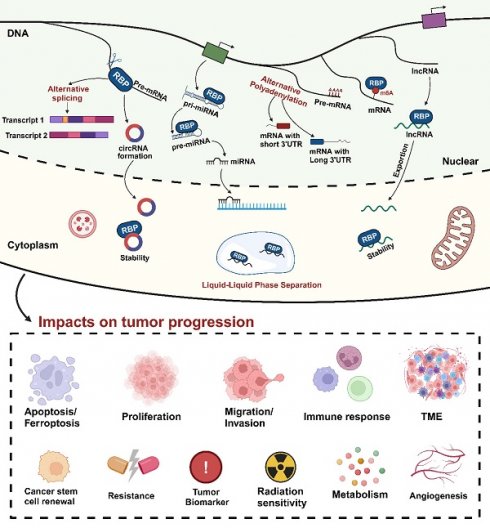
Alternative splicing mediated by RBPs is a critical post-transcriptional regulatory mechanism that promotes mRNA stability and ensures protein diversity. In tumors, abnormal splicing events mainly include exon skipping (ES), alternative 5' splice site selection (A5SS), alternative 3' splice site selection (A3SS), mutually exclusive exons (MXE), and intron retention (RI) [22, 23]. Among these, splicing abnormalities or splicing errors are a major factor in cancer development, and its occurrence is frequently linked to RBPs in tumor [24]. RBPs are involved in alternative splicing in tumors in a variety of roles. Some RBPs are able to form complexes with splicing-associated core proteins that work together to control splicing molecules in tumor cells [25-28]. Furthermore, certain RBPs modulate spliceosome activity by binding to the splicing sites [29, 30].
SR proteins, comprising 12 members (SRSF1-12) in mammals, share conserved domain architectures [31]. These proteins regulate splice site selection and exon recognition by binding to pre-mRNA cis-elements and facilitating spliceosome assembly, thereby governing the production of alternative splice isoforms [32]. Notably, SRSF10 exerts oncogenic functions through dual regulation of RNA splicing and metabolic reprogramming. In hepatocellular carcinoma (HCC), SRSF10 suppresses MDM4 exon skipping to downregulate p53 protein levels, concurrently reducing CD8+ T-cell infiltration, inhibiting IFNα/γ signaling, and inducing HIF1α-mediated PD-L1 upregulation, collectively driving tumor progression and immune evasion [33]. In gliomas, SRSF10 serves as an independent prognostic marker by modulating BCLAF1 exon 5a splicing to dysregulate cell cycle progression, with its overexpression correlating with enhanced sensitivity to immune checkpoint therapy [34]. Mechanistically, SRSF10 stabilizes MYB transcripts to upregulate glycolytic enzymes (GLUT1, HK1, LDHA), fostering lactate accumulation and histone H3K18 lactylation [35]. This metabolic reprogramming establishes a feedforward loop that promotes M2 macrophage polarization and impairs CD8+ T-cell function, ultimately conferring resistance to anti-PD-1 therapy. Pharmacological inhibition of SRSF10 (e.g., compound 1C8) reverses immunosuppressive microenvironments and synergizes with immune checkpoint blockade, highlighting its potential as a pan-cancer therapeutic target [35].
Heterogeneous nuclear RNA (hnRNA) is the primary transcriptional product of RNA polymerase II in eukaryotes [36]. Newly synthesized hnRNAs form complexes with various proteins in a non-selective manner during transcription, and heterogeneous nuclear ribonucleoproteins (hnRNPs) are indispensable protein components of these complexes [36]. HnRNPs consist of RNA-binding domains (RBDs) and auxiliary domains, which guide hnRNPs to interact with target genes or other proteins by recognizing specific nucleic acid sequences. The hnRNP protein family includes several members, such as hnRNPH1, E1, A1, K, and PTBP1. Unlike SR proteins, the hnRNP family generally plays an inhibitory role in alternative splicing [37]. SR proteins, such as SRSF1, typically promote spliceosome assembly and enhance exon inclusion by binding to exonic enhancers, whereas hnRNPs tend to bind to silencer elements, leading to exon skipping or intron retention through spatial hindrance, RNA structural remodeling, or competitive inhibition of SR protein activity. For example, hnRNPH1 regulates the alternative splicing of meiosis-related genes in germ cells by recruiting PTBP2 and SRSF3 [38]. Its loss results in chromosome synapsis defects and disrupted germ cell-support cell communication, ultimately leading to infertility. HnRNP E1, through binding to the nucleic acid structure of PNUTS pre-mRNA, inhibits its splicing to form lncRNA-PNUTS [39]. When hnRNP E1 is silenced or undergoes nucleocytoplasmic shuttling, the inhibition is released, promoting the generation of oncogenic lncRNAs, which, in turn, enhance epithelial-mesenchymal transition (EMT) and breast cancer metastasis by sequestering miR-205. Additionally, PTBP1 (a member of the hnRNP family) accelerates exon skipping by forming intronic RNA loops or directly inhibits splicing activity through trans-exonic loops, thereby regulating splicing events of cancer-related genes [25]. In apoptosis regulation, hnRNP A1 competes with SR protein SRSF1 for binding to Bcl-x pre-mRNA, inhibiting the splicing of the pro-apoptotic Bcl-xS isoform while promoting the expression of the anti-apoptotic Bcl-xL isoform, thereby enhancing tumor cell survival [37]. This antagonistic regulation is particularly prominent in cancer, where the dynamic balance between hnRNPs and SR proteins determines the final outcome of splicing events, influencing tumor progression, metastasis, and chemoresistance.
The RNA Binding Motif Protein (RBM) family also plays a pivotal role in the regulation of alternative splicing. For instance, RBM22 is mainly involved in pre-mRNA splicing and is crucial for maintaining the conformation of the catalytic core of spliceosome, serving as a bridge between the catalytic core and other essential spliceosomal proteins [40]. Additionally, RBM25 regulates alternative splicing by binding to the AMOTL1 pre-mRNA exon splicing enhancer motif 5'-CGGGCA-3' motif. This binding leads to increased expression of CircAMOTL1L, which in turn influences epithelial-mesenchymal transition (EMT) and suppresses the proliferation and metastasis of prostate cancer cells [41]. In lung cancer cells, mutations in RBM10 have been identified as inducers of alternative splicing of key genes such as EIF4H, CD44, UBAP2L, and KDM6A, which ultimately promoting cell growth [42].
Additional regulators of alternative splicing orchestrate the occurrence of splicing events in cells. Notably, the knockdown of PRPF19 leads to the conversion of the MDM4 splice isoform from the stable full-length MDM4-fl to the unstable MDM4-s, which lacks exon 6 [43]. The splicing factor ESRP1 interacts with the exon flanking regions involved in forming Circ-BIRC6 through alternative splicing, thereby facilitating the formation of Circ-BIRC6 in human embryonic stem cells [44].
Recent studies have demonstrated that dysregulation of eIF4E can extensively reprogram alternative splicing in a mutation-independent manner. Specifically, eIF4E selectively promotes the nuclear export and translation of mRNAs encoding splicing factors (e.g., SF3B1 and U2AF1) via its nuclear RNA export activity, thereby markedly increasing the protein levels of spliceosomal components [45]. Mechanistically, eIF4E directly binds to splicing factor transcripts (such as SF3B1 and U2AF1) and enhances their translational efficiency through polysome loading, while also physically interacting with the spliceosomal complex and specific precursor mRNAs to directly influence splice site selection. This dual action results in altered splicing patterns in approximately 800 transcripts in cell lines and around 4,600 transcripts in high-eIF4E acute myeloid leukemia (AML) patients. Further studies revealed that this process involves: (1) translation-splicing coupling regulation, whereby the RNA-binding protein HuR/ELAVL1 synergistically enhances splicing reprogramming by stabilizing splicing factor mRNAs [46]; (2) the MNK-eIF4E signaling axis, wherein the oncogenic isoform MNK2b—generated by the selective splicing of MNK2—promotes splicing factor translation by sustaining eIF4E phosphorylation while circumventing p38-MAPK-mediated tumor suppression [47]; and (3) an evolutionarily conserved mechanism, as evidenced by eIF4E's regulation of Sxl gene splicing in Drosophila, which affects sex determination [48]. These findings reveal that eIF4E globally modulates the abundance of splicing factors and spliceosomal function, leading to widespread reprogramming of the splicing landscape that far exceeds the impact of single splicing factor mutations, thereby offering a novel mechanistic framework for splicing dysregulation in malignancies such as AML.
Given the crucial role of RBPs in alternative splicing, they have emerged as potential targets for therapeutic drugs. In recent years, scientists have developed various drugs that target RBPs or their mediated alternative splicing, including small molecule compounds and oligonucleotides [49]. These drugs work by disrupting the interactions between RBPs and RNA or proteins, or by altering the expression level or activity of RBPs. This affects specific or a wide range of alternative splicing events, leading to therapeutic effects such as inhibition of tumor cell proliferation or induction of apoptosis. For instance, deletion of RBM10 increases the sensitivity of EGFR-mutated LUAD cells to spliceosome inhibitors. Combining spliceosome inhibitors with osimertinib, an EGFR tyrosine kinase inhibitor, enhances therapeutic efficacy, especially in RBM10-deficient LUAD, and overcome drug resistance [42]. Therefore, a comprehensive understanding of the molecular mechanisms and networks by which RBPs regulate alternative splicing, as well as the development of drugs targeting RBPs or their mediated splicing events, is significant for clinical diagnosis and therapy [50].
Regulation of alternative polyadenylation
Alternative polyadenylation (APA) is an RNA processing mechanism that generates transcripts with distinct 3' ends by selecting different polyadenylation sites within the mRNA's 3' untranslated region (3'UTR) or coding region [51, 52]. This process regulates mRNA stability, translational efficiency, and protein function. Dysregulation of APA is widespread in cancer; for example, the prevalent shortening of the 3'UTR in cancer cells can lead to upregulation of oncogenes (such as NQO1) or inactivation of tumor suppressor genes, thereby promoting tumor proliferation, metastasis, and metabolic abnormalities [53-55]. Recent findings indicate that various processing factors at the 3' end, along with RBPs like hnRNP C, CPSF6, and Ppn1 are significantly associated with the APA process [53, 56, 57]. These proteins are usually involved in the APA process by interacting with complexes formed at the 3' end of pre-mRNAs, including CPSF, CSTF, CFI, CFII, CTD, and RNAPII (Fig.1B) [51]. CPSF6, cleavage and polyadenylation specificity factor 6, is a vital member of the SR protein superfamily, whose structure and function are inextricably to the processing at the 3' end of mRNA. CPSF6 facilitates the use of proximal polyadenylation signals (PAS) by binding to the U-rich region on mRNA precursors. In hepatocellular carcinoma cells, CPSF6 significantly contributes to HCC progression by upregulating NQO1 expression through APA [53]. Additionally, CPSF6 promotes the assembly of mRNA 3' end processing complexes and influences mRNA maturation and function by recruiting other processing factors like factor interacting with APOLA and CPSF1 (FIP1L1) [54]. Interestingly, CPSF6 regulates APA in cancer cells through liquid-liquid phase separation (LLPS), independent of its expression level. This regulation leads to the shortening of the 3'UTR in cell cycle-related genes, thereby accelerating cell proliferation [54].
The hnRNP family includes a variety of proteins, such as hnRNP A1, A2/B1, C1/C2, H1, etc., which plays a multifaceted role in the regulation of alternative polyadenylation [58]. For example, hnRNPC contributes to cancer progression in metastatic colon cancer cells by altering the selection of APA sites and affecting MTHFD1L and NAP1L1 expression [56]. Furthermore, it is hypothesized that the shortening of the 3'UTR in lncRNA DSCAM-AS1 is mainly linked to the splicing factor hnRNPL [59]. In addition, Hematopoietic- and neurologic-expressed sequence 1 (HN1) is associated with the senescence phenotype of cancer cells, and low expression of hnRNPA1 in cancer cells contributes to the prolongation of the 3'UTR on HN1, which regulates cancer cell senescence [55].
Other RBPs also contribute to gene expression regulation via APA. For instance, PPN1, a component of the DPS subcomplex, regulates the localization and stability of the CPSF complex in the nucleus through interactions with proteins like Dis2 and Swd22. This in turn affects the expression of phosphate homeostasis genes (e.g., pho1 and pho84) via an APA mechanism [57]. Moreover, ELAV/Hu family proteins are known for regulating APA of pre-mRNAs in the drosophila nervous system, where they facilitate the extensive expression of mRNA isoforms with a long 3' UTRs [60].
In summary, APA represents a distinct type of alternative splicing, yet the precise mechanisms by which RBPs regulate APA remain an emerging area of research. Significantly, a growing number of studies have highlighted the impact of alternative polyadenylation on gene expression and disease progression [61]. Consequently, deepening the understanding of the APA process has vital clinical implications and may help identify novel biomarkers and therapeutic targets.
Regulation of m6A modification
In addition to alternative splicing, RBPs can also affect ncRNA function and expression through m6A modification [62]. N6-methyladenosine (m6A) modification, a prevalent form of eukaryotic mRNA modification, comprises approximately 0.1%-0.4% of all adenosine residues in mRNAs [63]. Modification of m6A involves three key components: the “writer” that add m6A modification to specific RNA sequences, the “eraser” that remove existing m6A modifications, and a “reader” that recognize and bind to m6A-modified RNA [64]. The m6A modification exerts its functional effects by dynamically regulating RNA translation, stability, and processing.
The "writer" components (such as the METTL3/METTL14/WTAP complex) catalyze m6A modification. Methylation of targeted RNA modulates its secondary structure or binding sites, thereby affecting the recruitment of RNA-binding proteins (RBPs). For example, RBM10 can inhibit the m6A methylation of lncRNA MALAT1, a member of the lncRNA family, by recruiting METTL3. It also reduces MALAT1 expression by binding and regulating it, thereby affecting the phosphorylation of the downstream PI3K/AKT/mTOR pathway, and ultimately impacting the invasion and migration of NSCLC [65].
The "eraser" components (such as ALKBH5 and FTO) dynamically regulate m6A modification levels through demethylation. The removal of m6A modifications can directly impact RNA stability and processing efficiency. The m6A modification is continuously converted under the action of “writer” and “eraser”, dynamically regulating the transcription and translation of eukaryotic ncRNA. For instance, ALKBH5 can decrease the m6A methylation level of lncRNA-NEAT1, leading to the upregulation of NEAT1 expression. Elevated NEAT1 then acts as a scaffold, influencing the expression of EZH2 and consequently promoting the invasion and metastasis of gastric cancer (GC) cells [66]. Furthermore, ALKBH5 demethylates pri-miR-194-2, an effect that relies on m6A modification, thereby inhibiting the biogenesis of miR-194-2 and consequently reducing distant metastasis of esophageal cancer. Additionally, ALKBH5 enhances the stability of circCCDC134 and increases its expression through demethylation of m6A modification, a process that subsequently facilitates the growth and metastasis of cervical cancer [67, 68].
The "reader" components (such as YTHDF1/2/3) regulate RNA fate by recognizing m6A modification sites. For example, YTHDF2 binds to m6A-modified RNAs and recruits the CCR4-NOT complex, thereby promoting the degradation of target RNAs [69]; YTHDF1 enhances the translation efficiency of target RNAs by associating with ribosomal complexes; and the YTHDF family members (YTHDF1/2/3) competitively bind to the same target, thereby regulating its functional balance. For instance, in non-small cell lung cancer (NSCLC), YTHDF3 preferentially binds to the m6A sites on YAP pre-mRNA, while YTHDF1 and YTHDF2 competitively bind to respectively promote YAP translation or degradation, ultimately determining YAP protein levels and influencing tumor progression [68].
Phase separation
RBPs coordinate the distribution and function of RNA within specific cellular regions, a process that is essential to ensure the timely and spatially accurate expression and functioning of RNA molecules [70]. Consequently, the aggregation of specific molecules at precise cellular locations within membraneless organelles is essential for establishing order. Proteins that facilitate LLPS often possess intrinsically disordered regions (IDRs). These IDRs potentially mediate the initiation of LLPS through weak and non-specific affinity interactions with various targets [71].
RBPs have been shown to influence gene expression and function via LLPS [72, 73]. For example, DDX21 is an RNA helicase containing a DEAD domain and is known to be involved in ribosomal RNA processing, RNA polymerase II-mediated transcription, and can affect gene expression regulation and cell differentiation, but the role of DDX21 in LLPS is still unclear. Consequently, Han et al. discovered that DDX21 forms a phase-separated condensate in colorectal cancer cells. This phase-separated DDX21 shows a high affinity for binding to the MCM5 gene locus, thereby modulating the expression of MCM5 (Fig.1D) [74]. DDX3X and DDX3Y, encoded by genes on the X and Y chromosomes, are sexually dimorphic RNA helicases. Studies have shown that DDX3Y demonstrates a greater tendency for LLPS compared to DDX3X. This is attributed to variations in their N-terminal intrinsically disordered regions and ATPase activities. Such differences crucially influence mRNA translation repression and the aggregation of the FUS protein [75].
Furthermore, phase separation acts as a dynamic platform for responding to various cellular signals and stresses [76]. Phase separation also facilitates the swift transport and distribution of RNA-binding proteins and RNA, thereby regulating RNA stability and availability. As an example, FUS, a protein involved in RNA metabolism and DNA repair, responds to DNA damage signals via phase separation. It forms a subnuclear foci at the DNA damage site and interacts with other DNA damage response (DDR) factors to promote the repair of DNA double-strand breaks (DSBs) [77]. Additionally, the malfunction of FUS in amyotrophic lateral sclerosis (ALS) is associated with the development of this disease. Upon mutation of FUS, the protein accumulates in the cytoplasm and exhibits phase separation capabilities. This leads to the formation of LLPS condensates that disrupt the phase separation equilibrium of FMRP, thereby inhibiting protein translation [78].
Furthermore, RBPs can modulate the differential expression and function of RNA via the selective barrier created by phase separation. For instance, TDP-43 selectively binds to and regulates RNA containing long motif clusters via phase separation. Mutations in FUS can disrupt the phase separation equilibrium of TDP-43, leading to alterations in the aggregation number and size within TDP-43's binding regions and changes in TDP-43's RNA binding profile [79].
In summary, RBPs are pivotal in modulating the biological functions of RNA via phase separation mechanisms. Research in this field not only deepened our understanding of the mechanisms of intracellular RNA processing and expression, but also opened up avenues for novel therapeutic strategies. However, studies on the regulation of non-coding RNAs by RBPs through phase separation are still limited, and further exploration in this emerging field is necessary.
Regulatory Functions of RNA-Binding Proteins in Tumor Biology. A. RBPs and the spliceosome machinery synergistically regulate alternative splicing. B. The mechanism of APA regulation by RBPs is mainly through the regulation of the length of mRNA 3'UTRs. C. RBPs affects mRNA translation, stability, and degradation through dynamic regulation of N(6)-methyladenosine (m6A). D. RBPs perform biological functions through membrane-less organelles generated by liquid-liquid phase separation (LLPS).
The role of non-coding RNAs in tumors
CircRNA
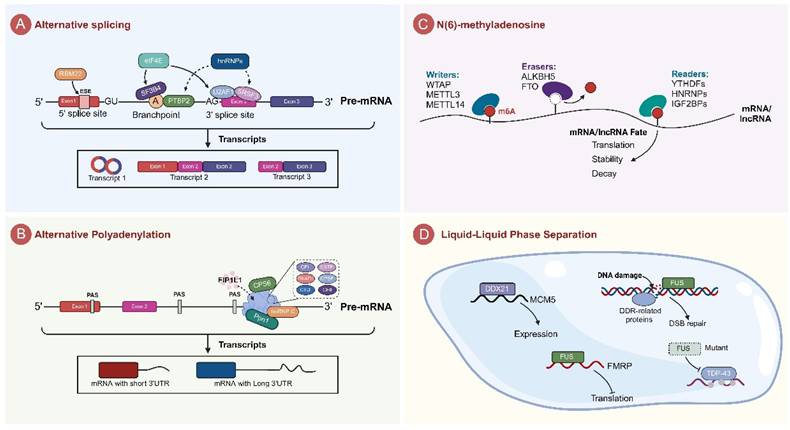
Circular RNA (circRNA) is a novel type of non-coding RNA that is formed by the covalent closure of the 5' and 3' ends of the precursor RNA. They are mostly localized to the cytoplasm, are highly sequence conserved, are not easily degraded by nucleases, are very stable in vivo, and are specific to tissue or developmental stage. CircRNA is an important regulator of tumorigenesis and development, and recent studies have shown that circRNA can regulate the proliferation, apoptosis, migration and invasion of tumor cells [80]. For example, circRNA WHSC1 is significantly overexpressed in uterine cancer tissues, which substantially enhances the proliferation, migration, and invasion capabilities of uterine cancer cells while concurrently reducing apoptosis (Fig.2A) [81]. It is well known that elevated levels of circSEPT9 and circCDYL significantly boost the proliferation of tumor cells and simultaneously decrease apoptosis rates [82, 83]. Moreover, circRNAs like circRILPL1 and circZNF215 hold promise as potential cancer biomarkers. Their expression levels in certain cancers are closely linked to disease severity and prognosis, making them useful tools for cancer diagnosis and prognostic [84, 85]. Furthermore, certain circRNAs affect the responses of tumor cells to chemotherapeutic agents and play a role in tumor drug resistance by modulating drug metabolism or apoptosis pathways. As an example, the circRNA cDOPEY2 was found to be significantly down-regulated in cisplatin-resistant esophageal squamous cell carcinoma (ESCC) cells. The reintroduction of cDOPEY2 substantially improved the efficacy of cisplatin against ESCC cells [86]. CircRNAs not only regulate intracellular processes but also play a significant role in modulating the tumor microenvironment. For instance, circRNA circHIPK3 is significantly upregulated in breast cancer, which contributes to tumor progression by affecting angiogenesis in endothelial cells in the tumor microenvironment [87]. Additionally, circRNAs are closely related to cancer stem cells, which are crucial in tumor biology due to their ability to self-renewal and multi-directional differentiation. It has been shown that that the circRNA circIPO11 enhances the self-renewal ability of hepatocellular carcinoma stem cells, which in turn promotes their proliferation [88]. It must be acknowledged that the study of circRNAs is still at an early stage and most of their mechanistic role in tumors remains elusive. Nevertheless, with technological advancements and deepening the understanding of circRNA functions, they demonstrate substantial potential for early diagnosis, prognosis and treatment of tumors.
lncRNA
Long non-coding RNAs (lncRNAs) constitute a class of RNA molecules that do not encode for proteins and whose transcripts are more than 200 nucleotides in length. Initially, lncRNAs were considered as “transcriptional noise” because of their low sequence conservation, extremely low expression levels, and limited detectability in genetic screenings [89]. In recent years, with the development of new technologies, an increasing number of studies uncovering the significant role of lncRNA in tumorigenesis, progression, prognosis, and treatment [90]. For instance, the overexpression of lncRNA HOTAIR, which is commonly observed in various tumors including liver, breast, and lung cancers, is frequently associated with tumor progression, increased malignancy, and poor prognosis (Fig.2B) [91-93]. Additionally, some lncRNAs function as tumor suppressors, such as lncRNA MEG3, whose diminished expression is closely associated with cancer progression and poor prognosis [94]. LncRNAs are involved in tumor regulation via diverse mechanisms. One major way is to act as molecular sponges to absorb and modulate miRNA activity, which in turn influences the expression of miRNA target genes. For example, lncRNA LINC00667, which is overexpressed in liver cancer, acts as a molecular sponge for miR-130a-3p. This interaction exacerbates liver cancer progression by regulating androgen receptor expression [95]. Secondly, lncRNAs can affect tumor cell activity by regulating protein function or localization through interaction with proteins. Particularly in breast cancer, SP1-induced overexpression of lncRNA AGAP2-AS1 upregulates MyD88 expression and activates the NF-κB signaling pathway by binding to the transcriptional coactivator CBP, which promotes breast cancer growth and enhances trastuzumab resistance [96]. Additionally, certain lncRNAs play a role in regulating the tumor microenvironment. For instance, through the miR-361 regulatory network, lncRNA NEAT1 influences the expression of STAT3 and other genes pivotal to the tumor microenvironment, such as MEF2D, ROCK1, WNT7A, and VEGF-A, thereby altering the immune microenvironment of the tumor and facilitating the progression of aggressive endometrial cancer [97]. LncRNAs also contribute to the development of tumor resistance. The down-regulation of lncRNA SNHG15 increases tumor cell sensitivity to 5-FU, whereas its overexpression promotes chemotherapeutic drug resistance [98]. In terms of diagnosis and therapy, the expression patterns of certain lncRNAs are closely associated with tumorigenesis, progression and prognosis, and thus they may serve as effective cancer biomarkers. For example, the increased expression of lncRNA LINC00853 in serum small extracellular vesicles may serve as a novel biomarker for early-stage hepatocellular carcinoma, particularly in AFP-negative hepatocellular carcinoma (HCC) cases [99]. Furthermore, therapeutic approaches targeting lncRNAs, including small interfering RNAs (siRNAs) and nucleic acid drugs, have demonstrated promise in cancer treatment. However, these strategies encountered significant challenges in terms of specificity, effective delivery, and tolerability [100].
MiRNA
Micro RNAs (miRNAs) are a class of small non-coding RNAs, typically 19-24 nucleotides long, whose primary function is to regulate gene expression by inhibiting mRNA translation or inducing mRNA degradation through binding to the 3' untranslated region of mRNA. It is well-recognized that miRNAs have multiple biological functions. They can regulate key genes involved in cell proliferation, apoptosis, migration, and angiogenesis, thereby affecting cell growth, invasion, and metastasis. Furthermore, miRNAs play a significant role in the occurrence and progression of various diseases [101]. The role of miRNAs in tumors is particularly noteworthy, and it is widely accepted that they have important regulatory roles in tumorigenesis and progression. Generally, miRNAs are classified into two types based on their effects on tumor progression: tumor-inhibitory miRNAs and tumor-promoting miRNAs [102]. Tumor-inhibitory miRNAs play a pivotal role in hindering tumorigenesis and progression by specifically targeting and suppressing the activity of certain oncogenes or signaling pathways. For instance, the miR-34a and miR-200 families are often downregulated in various cancers. These miRNA classes specifically target and inhibit several critical tumor-promoting factors, including key components of signaling pathways like Notch, Wnt, and MAPK (Fig.2C). In this way, they effectively inhibit the proliferation, invasion, and migration of cancer cells [103, 104].
Mode of action of non-coding RNAs in tumors. A. circRNAs affect tumor cell apoptosis, proliferation and invasion, drug resistance, angiogenesis, stemness and other malignant progression. B. lncRNAs act in tumors through different mechanisms, including triggering apoptosis, regulating the tumor microenvironment (TME), affecting drug sensitivity, and serving as biomarkers. C. miRNAs in tumors influence drug sensitivity and radiosensitivity by regulating downstream signaling pathways.
Conversely, the expression of certain miRNAs is elevated in tumors and are therefore classified as tumor-promoting miRNAs. For example, miR-21 and miR-155, are frequently upregulated in various cancers. They target and suppress key tumor suppressor genes such as PTEN and TP53, or elevate the expression of VEGF, which enhances angiogenesis and further promotes tumor development and progression [105, 106].
Recent studies have revealed that miRNAs not only regulate tumor development and progression, but also affect tumor response to treatment, particularly chemo-radiotherapy sensitivity. For instance, miR-29a can target the abnormal expression of MDM2, thereby enhancing the sensitivity of glioma cells to temozolomide treatment [107]. Additionally, Studies have shown that elevated levels of miR-449b in tumors are positively correlated with the sensitivity of tumor cells to radiotherapy. Furthermore, eEF-2 kinase functions as a key intermediary in the radiation-sensitizing effect exerted by miR-449b [108]. This finding highlights the potential of augmenting cancer therapy efficacy through the regulation of miRNA expression.
The regulation of non-coding RNAs by RBPs in tumors
Recent research highlights the significant role of RBPs in the biogenesis, functionality, and stability of non-coding RNAs, extending beyond their interaction with mRNA (Fig.3A).
RBPs regulation of circRNAs
Despite circRNAs have fewer RBP-binding sites than their linear mRNA counterparts, there is strong evidence that they interact with RBPs, which play a crucial role in various aspects of circRNA biology, including their generation, post-transcriptional regulation, functional execution, translation, specific modifications, and potential involvement in extracellular transport pathways [80].
Roles of RBPs in the biogenesis of circRNAs
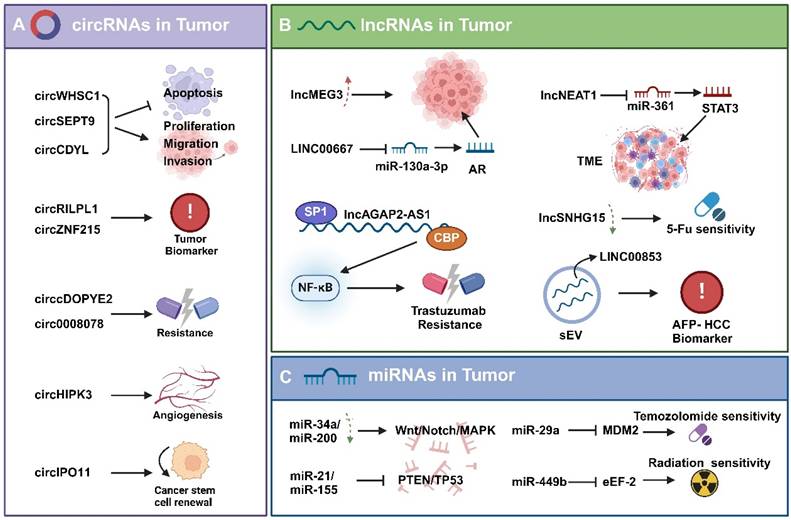
RBPs play an essential role in the regulation of circRNA production. For example, the quaking protein (QKI) significantly enhances circRNA production. QKI binds to specific RNA sequences in circRNA-producing precursor mRNAs and aligns these sequences closely, thus enabling splicing reactions between them. Specifically, QKI typically binds to intronic regions designated for reverse splicing. This binding acts as a “bridge” in circRNA generation, causing the RNA molecule to loop tightly enough for reverse splicing to occur [109-111]. Besides QKI, numerous other RBPs, including FUS, ADAR1, MBNL1 have been recognized to have an effect on circRNA production [112-114].
RBPs regulate the stability of circRNAs
In circRNAs, m6A modifications are usually catalyzed by RNA-binding proteins, including METTL3, METTL14, and WTAP. For instance, METTL14 can inhibit the progression of gastric cancer by affecting the stability of circORC5 through m6A modification, which in turn affects the miR-30c-2-3p/AKT1S1 signaling axis. YTHDF1, as an m6A-modified “reader protein” enhances the stability of circALG1 and its binding affinity to miR-342-5p, thus significantly enhancing the function of circALG1 as a sponge. These proteins identify specific RNA sequences and introduce m6A modifications to them [115, 116].
Roles of RBPs in the regulation of lncRNAs
RBPs regulates lncRNA stability, translocation, and transcription through various mechanisms such as alternative splicing, alternative polyadenylation, and m6A modification, thereby altering lncRNA function and expression (Fig.3B).
Roles of RBPs in the regulation of lncRNA stability
For instance, PTBP1 binds to lncRNA MACC1-AS1, thereby enhancing its sponging effect on miRNAs [117]. Similarly, HNRNPL forms a stabilizing complex with lncRNA SChLAP1, which subsequently enhances the interaction of SChLAP1 with ACTN4 [118]. This binding to lncRNAs not only alters the stability of lncRNAs, but also affects their interactions with other molecules. LINREP, a long-stranded non-coding RNA that is prominently expressed in glioblastoma (GBM), has an m6A modification site and is recognized by HuR, a mechanism that protects it from RNase L degradation. When the m6A modification site of LINREP is mutated or the m6A-writing enzyme METTL3 is knocked down, both the stability and function of LINREP are affected [119].
Roles of RBPs in transcription and localization of lncRNAs
M6A modifications are also crucial in regulating the transcription of the lncRNA XIST, especially in the nucleus. For instance, YTHDC1, is a nuclear m6A “reader protein” that recognizes the m6A site on XIST and recruits the PRC2 complex to silence genes on the X chromosome [120]. It has been observed that p53 acts directly on the promoters of lncRNAs, including NEAT1, whose transcription is regulated by the lncRNA ST7-AS1 [121, 122].
RBPs can also influence the intracellular localization of lncRNAs. The RBPs HuR and GRSF1 can impact the stability and export of their target lncRNA RMRP, subsequently controlling RMRP's localization in the cytoplasm and mitochondria [123]. Additionally, PTBP1 and hnRNPK can regulate the subcellular distribution of lncRNA SININEUP and the assembly of translation initiation complexes, thereby enhancing the translation of target mRNAs [124].
Roles of RBPs in the regulation of miRNAs
RBPs can affect miRNA processing, stability, translocation, and function by binding to either miRNA precursors or mature forms (Fig.3C). The RBPs drosha and DGCR8 form a microprocessor complex that generates short-chain pre-miRNAs by binding to long-stranded pri-miRNAs [125]. Furthermore, it has been shown that the efficiency of the activity of this complex varies depending on the type of miRNAs [126]. Dicer, another key miRNA processing enzyme, cleaves pre-miRNAs into mature double-stranded miRNAs, where one strand is degraded and the other strand binds to the RNA-induced silencing complex (RISC) to fulfill miRNA functions [127]. RBPs can also inhibit the miRNA maturation process. For instance, LIN28 is an RBP that is prominently expressed in embryonic development and stem cells and binds to the let-7 family of pre-miRNAs, thereby preventing cleavage by Dicer and inhibiting let-7 maturation [128].
The regulation of non-coding RNAs by RBPs in Tumors. A. Some RBPs (e.g., QKI, FUS, ADAR1, and MBNL1) regulate the biogenesis of circRNAs, while others (e.g., METTL3 and YTHDF1) affect the stability of circRNAs. B. Some RBPs (e.g., PTBP1, hnRNPL, and HuR) control the stability and degradation of lncRNAs, others (e.g., YTHDC1 and p53) inhibit the transcription of lncRNAs, and some others (e.g., HuR, GRSF1, PTBP1, and hnRNPK) regulate the subcellular localization of lncRNAs. C. RBPs such as Drosha, DGCR8, Dicer, LIN28 bind to pri-miRNA/pre-mRNA and affect the processing and maturation of miRNAs; while RBPs such as Ago2, TARBP2, HuR bind to miRNA and affect their stability, translocation, and function.
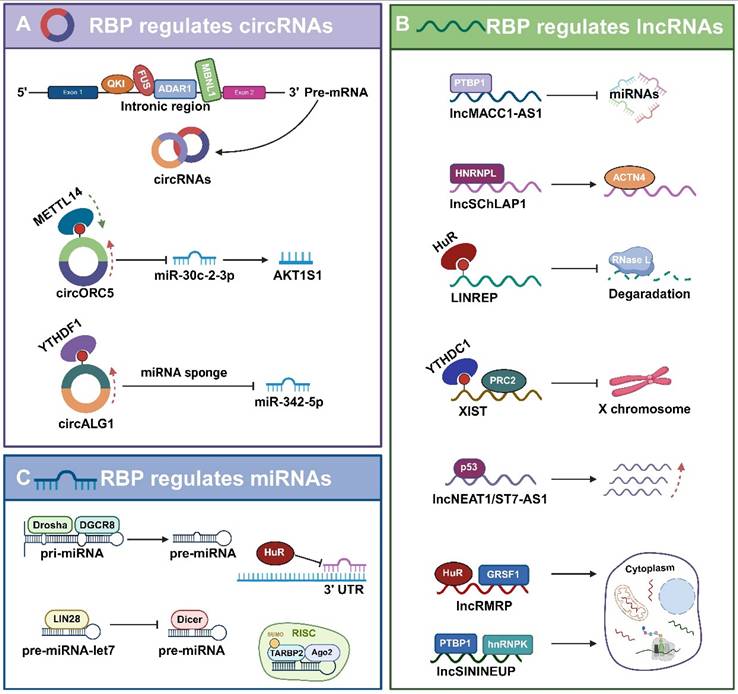
RBPs also bind to mature miRNAs, thereby affecting their functional roles. HuR protein, which are commonly expressed member of the ElaV family, interacts with miRNAs, particularly inhibiting miRNA binding to the 3'UTR of mRNAs, and promoting the dissociation of miRISC from the target mRNAs, thereby diminishing the function of miRNAs [129]. As previously mentioned, mature miRNAs processed by Dicer are incorporated into the RISC. Argonaute (Ago) protein, the core component of RISC, binds to mature miRNAs and recognizes the target mRNAs to inhibit their translation or degradation [101]. TAR (HIV-1) RNA binding protein 2 (TARBP2) is also a component of RISC. SUMOylated TARBP2 undergoes a post-translational modification process that forms a RISC-loading complex by recruiting Ago2 and facilitates increased loading of pre-miRNAs into the RLC [130].
Collectively, RBPs play a crucial role in miRNA regulation, including binding to pri-miRNAs, pre-miRNAs, or mature miRNAs, as well as facilitating or inhibiting their processing, localization, stability, or targeting efficiency. These findings provide important insights into the miRNA regulatory network.
Roles of non-coding RNAs regulated by RBP in tumors
Tumor metastasis
Tumor metastasis involves the process by which tumor cells detach from their primary site and colonize other parts of the body through the blood or lymphatic system, or directly invade adjacent tissues to form new tumors [131, 132]. Regarding the pattern of tumor cell metastasis, Douglas Hanahan proposed the metastatic cascade model. The model suggests that tumor cell metastasis requires six steps: invasion, migration, traversal between vascular endothelial cells, circulation, colonization, and regrowth. These steps, in turn, are closely associated with factors like cellular plasticity, tumor heterogeneity, the tumor microenvironment, and the regulation of gene expression and signaling pathways [133]. Current research is increasingly focusing on the interactions between tumor cells and their surrounding microenvironment, as well as on the epigenetic regulation of tumor cells, including the role of RBPs and their regulated ncRNAs [134, 135]. During cancer metastasis, dysfunction of RBPs leads to aberrant expression or malfunction of mRNAs or ncRNAs linked to cellular plasticity, thereby impacting the epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET) processes in cancer cells [136-138]. RBPs act as oncogenes or tumor suppressors by modulating the stability and processing of lncRNAs and circRNAs (Fig.4A). HNRNPC can specifically bind to the lncRNA DDX11-AS1 and promotes the Wnt/β-catenin and AKT pathways as well as the EMT process, thus facilitating glioma metastasis [139]. The RBP FUS binds to the lncRNA MALAT1 and activates STAT3 signaling, which in turn promotes the proliferation and metastasis of lung cancer cells. RBM25 activates the EMT program in prostate cancer cell metastasis by inducing its biogenesis through binding to circAMOLT1 and subsequently regulating the circAMOTL1L/miR-193a-5p/Pcdha signaling pathway [41]. The overexpression of ALKBH5 in cervical cancer decreases the expression and stability of circCCDC134, which in turn promotes the growth and metastatic of cancer cells by influencing HIF1A transcription [67]. CIB-3b disrupts TRBP-Dicer interactions by binding to TRBP, leading to disrupted maturation of miRNAs (e.g., miR-181, miR-320, miR-106, and let-7) in hepatocellular carcinoma cells. This disruption affects EMT-associated signaling pathways and thus regulates metastasis [140]. It has been demonstrated that HIF1 activates the expression of LIN28, which binds to the precursor of let-7 and prevents its cleavage by Dicer, thus inhibiting let-7 maturation. Increased activity of HIF1 and reduced expression of let-7 can enhance the invasion and migration of breast cancer cells, particularly in the case of brain metastasis. This correlation is evident in the modulation of the PDGF/PDGFR signaling pathway in breast cancer cells, which impairs the anti-metastatic effect of LK-99 against breast cancer [141]. Overall, RBPs can promote or inhibit tumor cell metastasis by influencing the processing, stability, and subcellular localization of ncRNAs by regulating processes such as EMT and MET. RBPs can also modulate signaling pathways involved in growth, apoptosis, angiogenesis, and immune escape in tumor cells. Consequently, the role of RBPs and their regulated ncRNAs in tumor metastasis unveils new mechanisms and targets for cancer therapy and provide new perspectives and research avenues in the field of cancer biology.
Metabolic reprogramming
One of the hallmarks of cancer is the reprogramming of energy production that favor increased glycolysis even in the presence of oxygen, a phenomenon known as the Warburg effect. This phenomenon, observed in various tumor types, coincides with the maintenance of the malignant phenotype of tumor cells through metabolic pathways involving fatty acids, cholesterol, and glutamine [142]. RBPs and their regulated ncRNAs play a key role in the metabolic reprogramming of tumors, and can affect various metabolism-related pathways, including those of glucose, amino acids, and fatty acids, which can promote tumorigenesis and progression (Fig.4B).
Specific RBPs have been identified as regulators of glycolysis and glycolytic pathways. ENO1 protein is both a glycolytic enzyme enolase and an RNA-binding site, has been identified as an RNA-binding protein [143]. ENO1 acts as an RBP that binds and stabilizes YAP1 mRNA, thereby promoting the growth of hepatocellular carcinoma cells by activating intracellular arachidonic acid metabolism [144]. LIN28b enhances aerobic glycolysis and lactate secretion in tumor cells via the LIN28b/MYC/miR-34a pathway [145]. In terms of amino acid metabolism, the interactions between these RBPs and ncRNAs can influence the synthesis and catabolism of essential amino acids in tumor cells, which may affect cell proliferation and survival [146]. Besides upregulating GPT2 expression, LncRNA UCA1 also regulates inosine monophosphate dehydrogenase 1 and 2 (IMPDH1/2) expression via TWIST1, which alters metabolite levels and promotes guanine nucleotide de novo synthesis, thereby reprogramming the metabolism of bladder cancer cells [147]. Circ_0062682 promotes PHGDH expression and activity by interacting with miR-940 to increase cellular serine metabolism and enhance proliferation, migration, invasion, and drug resistance in colorectal cancer cells [148]. Moreover, RBPs and ncRNAs significantly regulate fatty acid metabolism, which in turn influences the energy homeostasis and signaling of tumor cells by modulating the synthesis and oxidation of fatty acids. Triglyceride lipase (ATGL) is a major enzyme in lipolysis and is regulated by NEAT1, which controls ATGL expression by interacting with miR-124-3p to modulate aberrant lipid metabolism and promote the proliferation of hepatocellular carcinoma cells [149]. NEAT1 stabilizes the RPRD1B protein and enhances fatty acid uptake and synthesis via the c-Jun/c-Fos/SREBP1 signaling axis to promote primary tumor metastasis in lymph nodes [150]. CircPRKAA1 binds to sterol regulatory elements binding protein 1 (mSREBP-1), which in turn increases fatty acid synthesis by upregulating its transcription, thereby promoting tumor growth [151]. In summary, RNA-binding proteins and ncRNAs emerge as novel therapeutic targets due to their critical role in tumor metabolic reprogramming. The development of small molecule inhibitors or antagonists targeting molecules like ENO1, LIN28b, UCA1, and NEAT1 may be effective in inhibiting tumor growth and metastasis. Thus, exploring the mechanisms by which RBPs and ncRNAs contribute to the metabolic reprogramming of tumors remains a vital field of research in future cancer therapies.
Tumor immunity
RBPs are crucial for the function of immune cells, mainly through the post-transcriptional regulation of RNA metabolism and function. The dysfunction of RBPs and aberrant RNA metabolism are closely associated with a variety of autoimmune and autoinflammatory diseases [152, 153]. Various RBPs play pivotal roles in auto-reactive inflammatory responses by orchestrating a complex regulatory network of DNA, RNA, and proteins in immune cells (Fig.4C) [153].
Many human cancers have the ability to evade the adaptive immune system as certain cancer cells can achieve immune escape by expressing or modulating RBPs. These proteins affect the metabolism and function of self or viral-derived RNAs, thus inhibiting the expression or activity of immune checkpoint molecules and blocking the activation and destruction of immune cells [154, 155]. FMRP is an RNA-binding protein that regulates the expression of various immune-related genes in tumor cells, thereby influencing the ability of tumor to evade immune surveillance. Elevated expression of FMRP in tumor cells promotes the secretion of immune-suppressive factors, including IL-33, Protein S, and exosomes, as well as the recruitment of regulatory T-cells and immune-suppressive macrophages [156]. This would create an immune-suppressive tumor microenvironment, as well as depletion and clearance of cytotoxic CD8 T cells. Conversely, reduced or absent expression of FMRP in tumor cells leads to the secretion of pro-inflammatory factors, including CCL7, CCL9, CXCL9, and CXCL10, which recruits more cytotoxic CD8 T cells and enhances the anti-tumor response of lymphocytes [157].
Furthermore, ncRNAs regulated by RBPs play a crucial role in tumor immunity. NEAT1 enhances the expression of PTRF by interacting with the PTRF/Cavin-1, thereby stabilizing its mRNA, and PTRF activates NF-κB signaling by inhibiting the expression of UBXN1, thereby upregulating the transcription of PD-L1 and promoting the immune escape of tumor cells [158]. In lung adenocarcinoma, mutations in KRAS can activate the PI3K-STAT3 signaling pathway, thereby inhibiting the expression of miR-34a and leading to elevated expression of CD47, which allows tumor cells to evade immune system surveillance [159].
Multifaceted roles of RBP-regulated non-coding RNAs in tumors. A. RBPs promote the invasion and metastasis of tumor cells by affecting mRNAs and ncRNAs associated with cellular plasticity and regulating EMT and MET processes. B. RBPs modulate ncRNA to affect genes and enzyme activities related to metabolism, impacting various metabolic pathways such as glucose metabolism, amino acid metabolism, and fatty acid metabolism, influencing tumor progression. C. RBPs regulate the number of CD8+ T cells in the TME by directly controlling the activity of immunosuppressive and proinflammatory factors. Additionally, some RBPs regulate the transcription of ncRNAs affecting PD-L1 and CD47, suppressing the RIG-I-MAVS signaling pathway, thus facilitating tumor immune evasion. D. RBPs disrupt the conduction of related signaling pathways such as the apoptosis signaling pathway, PTEN pathway, and PKR/eIF2α pathway by affecting the expression levels of ncRNAs, regulating the drug sensitivity of tumor cells. E. RBPs and their regulated ncRNAs mainly affect the binding of ferroptosis-related molecules such as NCOA4 binding to FTH1, promoting the expression of ferroptosis-related genes DMT1 and TFR1, leading to excessive accumulation of iron ions, and ultimately inducing cell death by ferroptosis.
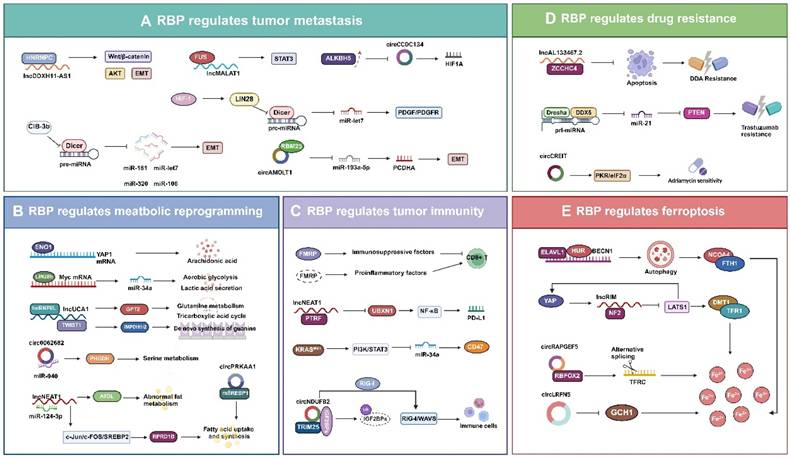
CircNDUFB2 is lowly expressed in non-small cell lung cancer (NSCLC) and negatively correlates with the malignancy degree of NSCLC. CircNDUFB2 serves as a bridge to enhance the interaction between TRIM25 and IGF2BPs, subsequently promoting the ubiquitin-mediated degradation of IGF2BPs. Additionally, circNDUFB2 activates the RIG-I-MAVS pathway by being recognized by the RIG-I signaling mechanism, thereby recruiting immune cells into the tumor microenvironment and participating in the immune response of tumor cells [160].
Drug resistance
Tumor cell resistance is a major obstacle to the treatment of patients with tumors, limiting the efficacy of chemotherapy, targeted therapy, radiotherapy, immunotherapy, and other treatments for a wide range of solid cancers. This resistance is also one of the leading causes of tumor-related deaths worldwide [161]. Therapeutic resistance in cancer can be classified into natural and acquired resistance based on the timing of resistance development. Natural resistance that exists or rapidly develops in tumor cells before treatment may arise from genetic abnormalities, tumor heterogeneity, or intrinsic defense mechanisms. Conversely, acquired resistance which develops gradually in tumor cells post-treatment may result from modifications in driver oncogenes, activation of tumor-associated signaling pathways, or adaptation of the tumor microenvironment [162]. Growing evidence indicates that epigenetic regulation plays a crucial role in tumor drug resistance, and interest in the mechanisms involving RBPs and their interacting is escalating [163]. RBPs can influence the expression, function, and silencing of tumor-associated genes through various mechanisms, particularly by impacting ncRNAs, thus altering the drug response of tumor cells (Fig.4D). ZCCHC4 inhibits apoptotic signaling in hepatocellular carcinoma cells by interacting with lncRNA AL133467.2, thereby suppressing its pro-apoptotic function and thus promoting the chemoresistance to DNA-damaging agents (DDAs) in these cells [164]. In 17q23-amplified breast cancer, the RNA-binding protein DDX5 interacts with the Drosha complex and affects the maturation of miR-21, which directly inhibits PTEN, an important mechanism for trastuzumab resistance in HER2+ breast cancers [165]. Overexpression of circRNA-CREIT enhances the sensitivity of triple-negative breast cancer cells to adriamycin, which is linked to the inhibition of the PKR/eIF2α signaling axis [166]. Numerous therapeutic strategies have been developed targeting RBPs and their regulated ncRNAs to overcome tumor resistance. Responsiveness to sorafenib treatment can be restored in sorafenib-resistant patient-derived xenograft mice by subcutaneous treatment with siRNAs targeting circRNA-SORE [167]. The overexpression of circRNA17 in a mouse model, transplanted with enzalutamide-resistant cells demonstrated restoration of enzalutamide sensitivity in prostate cancer treatment [168]. However, how to safely and effectively deliver siRNA or shRNA to the target site without side effects remains a major challenge in clinical applications.
Ferroptosis
Ferroptosis is a mode of cell death induced by iron ions and lipid peroxidation that plays a significant role in tumorigenesis, progression, and metastasis. Ferroptosis is regulated through diverse cellular metabolic pathways, including redox homeostasis, iron metabolism, mitochondrial activity, and amino acids, lipids, and glucose metabolism [169]. Recent evidence suggests that RBPs and their regulatory ncRNAs are instrumental in controlling key aspects of ferroptosis, including the glutathione-GPX4 pathway, glutamate/cystine transport, and the metabolism of both iron and lipids (Fig.4E) [170-172]. ELAVL1/HuR interacts with BECN1 mRNA to stabilize its expression, thereby increasing autophagy levels and causing NCOA4 to bind to FTH1. This interaction results in the degradation of ferritin and an excessive release of iron ions, culminating in the ferroptosis of hepatic stellate cells [173]. LncRIM is a lncRNA activated by YAP, which inhibits the kinase activity of LATS1 by binding to NF2. This inhibition enhances the transcriptional activity of YAP, promotes the expression of iron metabolism-related genes DMT1 and TFR1, and increases the level of intracellular iron ions. lncRIM also promotes its own expression by forming a feedback loop, thereby further enhancing YAP activity. This results in the excessive accumulation of iron ions, leading to lipid peroxidation and ultimately inducing ferroptosis in cells [174]. The expression of lncRIM is regulated by various RBPs. HuR can bind to lncRIM and stabilizes its expression, thereby enhancing YAP activation. Conversely, some RBPs (e.g.YTHDF3) negatively regulate lncRNA GAS5, which reduces its activating effect on YAP [175, 176]. Although the mechanism of circRNA role in ferroptosis remains unclear, it has been demonstrated that in endometrial cancer, circRAPGEF5 affects ferroptosis by regulating the alternative splicing of the transferrin receptor (TFRC) through interacting with the RNA-binding protein fox-1 homologue 2 (RBFOX2) [177]. Furthermore, it has been demonstrated that circLRFN5 can cause elevated iron levels, increased lipid peroxidation, and the development of ferroptosis in GBM cells by reducing the expression of GCH1, a molecule that inhibits ferroptosis through the production of the antioxidant tetrahydrobiopterin (BH4) [178]. Many studies have reported that miRNAs regulate the occurrence of ferroptosis by modulating genes involved in iron metabolism, antioxidants, and lipid metabolism [122, 179]. Overall, although the mechanism of RBP-regulated ncRNAs in ferroptosis remains unclear, an increasing number of studies are aimed at unraveling this mystery. The role of RBPs and their regulated ncRNAs in ferroptosis is becoming increasingly significant.
Conclusion and outlook
RBPs and their regulated ncRNAs play central roles in tumorigenesis and progression through mechanisms including alternative RNA splicing, m6A modification, APA processing, and phase separation [1, 18, 20]. For instance, the RBP ZCCHC4 suppresses DNA damage-induced apoptosis by interacting with the long non-coding RNA AL133467.2, thereby promoting chemoresistance in hepatocellular carcinoma cells [164]. Meanwhile, the ncRNA circRTN4 facilitates tumor growth and hepatic metastasis in pancreatic ductal adenocarcinoma via the circRTN4-miR-497-5p-HOTTIP axis while simultaneously stbilizing RAB11FIP1 to drive epithelial-mesenchymal transition [137]. These findings not only elucidate the molecular basis of tumor heterogeneity but also highlight novel therapeutic opportunities targeting RNA regulatory networks.
Emerging studies have validated the therapeutic potential of targeting RBP/ncRNA interactions [180]. Small-molecule inhibitors such as curaxin CBL0137 activate ZBP1 to induce necroptosis in cancer-associated fibroblasts and restore immune checkpoint blockade sensitivity in melanoma models [181]. RNA-targeting therapies, including the miR-122 antagonist Miravirsen, have progressed to Phase II clinical trials [182]. CRISPR/dCas13-based RNA editing platforms further enable precise modulation of oncogenic ncRNAs [183]. Notably, serum exosomal miRNA panels (miR-181b, miR-193b, miR-195, miR-411) demonstrate clinical utility for preoperative prediction of lymph node metastasis in T1 colorectal cancer, with risk stratification models reducing overtreatment rates by 76% without compromising diagnostic accuracy [184], warranting further clinical validation.
Breakthroughs in combination therapies reveal synergistic effects between RNA-targeting agents and immunotherapies. The FMRP-targeting PROTAC degrader sc1-VHLL, delivered via lipid nanoparticles, specifically ubiquitinates FMRP in tumor cells, remodeling the tumor microenvironment by enhancing CD8+ T cell infiltration and reducing Treg populations in CT26-bearing mice, thereby converting immunologically "cold" tumors to "hot" lesions [185]. Parallel studies show that ADAR1 ablation activates PKR/MDA5 pathways through reduced RNA editing, overcoming PD-1 resistance caused by antigen presentation defects [186]. Additionally, radiotherapy-induced expansion of immunosuppressive MDSCs via the NF-κB-YTHDF2 feedback loop can be reversed by YTHDF2 inhibition, potentiating synergy with PD-L1 blockade. These findings collectively establish RNA processing proteins as pivotal regulators of immunomodulation and temporal therapeutic targets [187].
Future investigations should prioritize three directions: (1) mapping dynamic RBP-ncRNA interactomes across tumor subclones using spatial transcriptomics and RNA in situ imaging; (2) optimizing RNA structure-targeted drug design through AI-driven screening platforms; (3) accelerating clinical translation via interdisciplinary frameworks integrating organoid models and PDX platforms. As the RNA epigenetics landscape becomes increasingly deciphered, RBP/ncRNA-centered therapeutic paradigms promise to redefine precision oncology.
Abbreviations
RBP, RNA-binding protein; ncRNA, non-coding RNA; AS, Alternative splicing; PPT, polypyrimidine tract; L-3′ SS, 3′ splice site of exon 3a; S-3′ SS, original 3′ splice site of exon 3; AML, acute myeloid leukemia; SR proteins, serine/arginine-rich proteins; SF3B1, Splicing factor 3b subunit 1; U2AF1, U2 small nuclear RNA auxiliary factor 1; SF3B4, Splicing factor 3b subunit 4; hTERT, human telomerase reverse transcriptase; NSCLC, non-small cell lung cancer; DR8, Death receptor 8; hnRNP, heterogeneous nuclear ribonucleoprotein; SE, skipped exons; A5SS, alternative 5' splice site selection; A3SS, alternative 3' splice site selection; MXE, mutually exclusive exons; RI, intron retention; PTBP2, Polypyrimidine tract-binding protein 2; SRSF3, Serine/arginine-rich splicing factor 3; RBM, RNA binding motif protein; EMT, epithelial-mesenchymal transition; EIF4H, Eukaryotic translation initiation factor 4H; CD44, Cluster of Differentiation 44; UBAP2L, Ubiquitin-associated protein 2-like; KDM6A, Lysine demethylase 6A; PRPF19, Pre-mRNA processing factor 19; MDM4, MDM4 regulator of p53; ESRP1, Epithelial splicing regulatory protein 1; EGFR, Epidermal growth factor receptor; LUAD, Lung adenocarcinoma; APA, Alternative Polyadenylation; 3'UTR, 3' untranslated region; hnRNP C, Heterogeneous nuclear ribonucleoprotein C; CPSF6, Cleavage and polyadenylation specificity factor subunit 6; Ppn1, Protein phosphatase 1; CPSF, Cleavage and polyadenylation specificity factor; CSTF, Cleavage stimulation factor; CFI, Cleavage factor I; CFII, Cleavage factor II; CTD, C-terminal domain; RNAPII, RNA polymerase II; PAS, polyadenylation signals; NQO1, NAD (P)H quinone dehydrogenase 1; FIP1L1, Factor interacting with APOLA and CPSF1; LLPS, liquid-liquid phase separation; MTHFD1L, Methylenetetrahydrofolate dehydrogenase 1-like; NAP1L1, Nucleosome assembly protein 1-like 1; HN1, Hematopoietic- and neurologic-expressed sequence 1; DPS, Developmental pluripotency associated 2; Dis2, Dis2 phosphatase; Swd22, Snf2-related CREBBP activator protein; ELAV/Hu, ELAV like RNA binding protein/Hu protein; m6A, N6-methyladenosine; WTAP, Wilms tumor 1 associated protein; METTL, Methyltransferase like; PI3K, Phosphoinositide 3-kinase; AKT, AKT serine/threonine kinase; mTOR, Mechanistic target of rapamycin kinase; ALKBH, AlkB homolog; FTO, Fat mass and obesity-associated protein; EZH2, Enhancer of zeste 2 polycomb repressive complex 2 subunit; YTHDF, YTH N6-methyladenosine RNA binding protein; YAP, Yes-associated protein; IDRs, intrinsically disordered regions; DDR, DNA damage response; DSBs, DNA double-strand breaks; ALS, amyotrophic lateral sclerosis; circRNA, Circular RNA; ESCC, esophageal squamous cell carcinoma; LncRNA, Long non-coding RNAs; NF-κB, Nuclear factor kappa-light-chain-enhancer of activated B cells; HCC, hepatocellular carcinoma; siRNAs, small interfering RNA; miRNA, Micro RNA; QKI, quaking protein; HNRNPL, Heterogeneous nuclear ribonucleoprotein L; PRC2, Polycomb repressive complex 2; HuR, Human antigen R; GRSF1, G-rich RNA sequence binding factor 1; DGCR8, DiGeorge syndrome critical region gene 8; RISC, RNA-induced silencing complex; Ago, Argonaute; TARBP2, TAR (HIV-1) RNA binding protein 2; SUMO, Small Ubiquitin-like Modifier; RLC, RISC Loading Complex; HIF1, Hypoxia-inducible factor 1; PDGF/PDGFR, Platelet-derived growth factor/Platelet-derived growth factor receptor; ENO1, Enolase 1; LIN28b, Lin-28 Homolog B; IMPDH1/2, Inosine monophosphate dehydrogenase 1 and 2; TWIST1, Twist family bHLH transcription factor 1; ATGL, Adipose triglyceride lipase; mSREBP-1, Membrane-bound sterol regulatory element-binding protein 1; KRAS, Kirsten rat sarcoma virus; IGF2BP, Insulin-like growth factor 2 mRNA binding protein; DDA, DNA-damaging agents; shRNA, Short hairpin RNA; GPX4, Glutathione peroxidase 4; ELAVL1, ELAV like RNA binding protein 1; HuR, Human antigen R; NCOA4, Nuclear receptor coactivator 4; FTH1, Ferritin heavy chain 1; NF2, Neurofibromin 2; DMT1, Divalent metal transporter 1; TFR1, Transferrin receptor 1; TFRC, Transferrin receptor; RBFOX2, RNA-binding protein fox-1 homologue 2; GBM, Glioblastoma multiforme; BH4, Tetrahydrobiopterin.
Acknowledgements
All figures created by biorender.com.
Funding
This work was supported in part by grants from the following sources: the National Natural Science Foundation of China (82302987, 82303534, 82203233, 82202966, 82173142), the Natural Science Foundation of Hunan Province (2023JJ60469, 2023JJ40413, 2023JJ30372, 2023JJ30375, 2022JJ80078, 2020JJ5336), Key Research and Development Program of Hunan Province (2022SK2051), Science and Technology Innovation Program of Hunan Province (2023RC3199, 2023SK4034, 2023RC1073), the Research Project of Health Commission of Hunan Province (202203034978, 202202055318, 202109031837, 202109032010, 20201020), the Changsha Science and Technology Board (kh2201054), Ascend Foundation of National cancer center (NCC201909B06), and by Hunan Cancer Hospital Climb Plan (ZX2020001-3, YF2020002, 2023NSFC-A001, 2023NSFC-A002, 2023NSFC-A004).
Author contributions
CSW and PQ contributed to drafting and editing of the manuscript, shared the first authorship. LQJ and ZYJ designed, revised and finalized the manuscript. MQF, XXM, ZWL, JXJ and TSM participated in the drafting and editing manuscript. YWJ, HYQ, OYLD, and LSZ participated in the revision and coordination. LJG, WJW, XLZ and PMJ contributed to literature search. WNYY and TYY participated in the conception and coordination. All authors contributed toward data analysis, drafting and revising the paper and agreed to be accountable for all aspects of the work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X. et al. RNA-binding proteins in tumor progression. J Hematol Oncol. 2020;13:90
2. Xu X, Peng Q, Jiang X, Tan S, Yang Y, Yang W. et al. Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential. Exp Mol Med. 2023;55:1357-70
3. Zhang Y, Mao Q, Xia Q, Cheng J, Huang Z, Li Y. et al. Noncoding RNAs link metabolic reprogramming to immune microenvironment in cancers. J Hematol Oncol. 2021;14:169
4. Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345-54
5. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 2021;41:109-20
6. Kameda S, Ohno H, Saito H. Synthetic circular RNA switches and circuits that control protein expression in mammalian cells. Nucleic Acids Res. 2023;51:e24
7. Li L, Miao H, Chang Y, Yao H, Zhao Y, Wu F. et al. Multidimensional crosstalk between RNA-binding proteins and noncoding RNAs in cancer biology. Semin Cancer Biol. 2021;75:84-96
8. Li X, Liang QX, Lin JR, Peng J, Yang JH, Yi C. et al. Epitranscriptomic technologies and analyses. Sci China Life Sci. 2020;63:501-15
9. Han L, Huang C, Wang X, Tong D. The RNA-binding protein GRSF1 promotes hepatocarcinogenesis via competitively binding to YY1 mRNA with miR-30e-5p. J Exp Clin Cancer Res. 2022;41:17
10. Lee J, Kang H. Nucleolin Regulates Pulmonary Artery Smooth Muscle Cell Proliferation under Hypoxia by Modulating miRNA Expression. Cells. 2023;12:817
11. Yoon DS, Lee KM, Choi Y, Ko EA, Lee NH, Cho S. et al. TLR4 downregulation by the RNA-binding protein PUM1 alleviates cellular aging and osteoarthritis. Cell Death Differ. 2022;29:1364-78
12. Liu H, Luo J, Luan S, He C, Li Z. Long non-coding RNAs involved in cancer metabolic reprogramming. Cell Mol Life Sci. 2019;76:495-504
13. Brownmiller T, Juric JA, Ivey AD, Harvey BM, Westemeier ES, Winters MT. et al. Y Chromosome LncRNA Are Involved in Radiation Response of Male Non-Small Cell Lung Cancer Cells. Cancer Res. 2020;80:4046-57
14. Hu R, Zhu X, Chen C, Xu R, Li Y, Xu W. RNA-binding protein PUM2 suppresses osteosarcoma progression via partly and competitively binding to STARD13 3'UTR with miRNAs. Cell Prolif. 2018;51:e12508
15. Ji H, Kim TW, Lee WJ, Jeong SD, Cho YB, Kim HH. Two circPPFIA1s negatively regulate liver metastasis of colon cancer via miR-155-5p/CDX1 and HuR/RAB36. Mol Cancer. 2022;21:197
16. Xu H, Jiang Y, Xu X, Su X, Liu Y, Ma Y. et al. Inducible degradation of lncRNA Sros1 promotes IFN-gamma-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat Immunol. 2019;20:1621-30
17. Qu HL, Sun LJ, Li X, Liu F, Sun HH, He XT. et al. Long non-coding RNA AC018926.2 regulates palmitic acid exposure-compromised osteogenic potential of periodontal ligament stem cells via the ITGA2/FAK/AKT pathway. Cell Prolif. 2023;56:e13411
18. Kang D, Lee Y, Lee JS. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers (Basel). 2020;12:2699
19. He Z, Zhong Y, Regmi P, Lv T, Ma W, Wang J. et al. Exosomal long non-coding RNA TRPM2-AS promotes angiogenesis in gallbladder cancer through interacting with PABPC1 to activate NOTCH1 signaling pathway. Mol Cancer. 2024;23:65
20. Murphy JJ, Surendranath K, Kanagaraj R. RNA-Binding Proteins and Their Emerging Roles in Cancer: Beyond the Tip of the Iceberg. Int J Mol Sci. 2023;24:9612
21. Xu X, Peng Q, Ren Z, Han Y, Jiang X, Wu Z. et al. CircRNF13 enhances IGF2BP1 phase separation-mediated ITGB1 mRNA stabilization in an m6A-dependent manner to promote oral cancer cisplatin chemoresistance. Mol Cancer. 2025;24:36
22. Eymin B. Targeting the spliceosome machinery: A new therapeutic axis in cancer? Biochem Pharmacol. 2021;189:114039
23. Wang BD, Ceniccola K, Hwang S, Andrawis R, Horvath A, Freedman JA. et al. Alternative splicing promotes tumour aggressiveness and drug resistance in African American prostate cancer. Nat Commun. 2017;8:15921
24. Cortes-Lopez M, Chamely P, Hawkins AG, Stanley RF, Swett AD, Ganesan S. et al. Single-cell multi-omics defines the cell-type-specific impact of splicing aberrations in human hematopoietic clonal outgrowths. Cell Stem Cell. 2023;30:1262-81 e8
25. Ye R, Hu N, Cao C, Su R, Xu S, Yang C. et al. Capture RIC-seq reveals positional rules of PTBP1-associated RNA loops in splicing regulation. Mol Cell. 2023;83:1311-27 e7
26. Han J, An O, Ren X, Song Y, Tang SJ, Shen H. et al. Multilayered control of splicing regulatory networks by DAP3 leads to widespread alternative splicing changes in cancer. Nat Commun. 2022;13:1793
27. Moss ND, Wells KL, Theis A, Kim YK, Spigelman AF, Liu X. et al. Modulation of insulin secretion by RBFOX2-mediated alternative splicing. Nat Commun. 2023;14:7732
28. Rezvykh AP, Ustyugov AA, Chaprov KD, Teterina EV, Nebogatikov VO, Spasskaya DS. et al. Cytoplasmic aggregation of mutant FUS causes multistep RNA splicing perturbations in the course of motor neuron pathology. Nucleic Acids Res. 2023;51:5810-30
29. Kang HS, Sanchez-Rico C, Ebersberger S, Sutandy FXR, Busch A, Welte T. et al. An autoinhibitory intramolecular interaction proof-reads RNA recognition by the essential splicing factor U2AF2. Proc Natl Acad Sci U S A. 2020;117:7140-9
30. Guo H, Xu J, Xing P, Li Q, Wang D, Tang C. et al. RNA helicase DHX15 exemplifies a unique dependency in acute leukemia. Haematologica. 2023;108:2029-43
31. Zheng X, Peng Q, Wang L, Zhang X, Huang L, Wang J. et al. Serine/arginine-rich splicing factors: the bridge linking alternative splicing and cancer. Int J Biol Sci. 2020;16:2442-53
32. Tao Y, Zhang Q, Wang H, Yang X, Mu H. Alternative splicing and related RNA binding proteins in human health and disease. Signal Transduct Target Ther. 2024;9:26
33. Luo X, Zhang Z, Li S, Wang Y, Sun M, Hu D. et al. SRSF10 facilitates HCC growth and metastasis by suppressing CD8 (+)T cell infiltration and targeting SRSF10 enhances anti-PD-L1 therapy. Int Immunopharmacol. 2024;127:111376
34. An W, Yang Q, Xi Y, Pan H, Huang H, Chen Q. et al. Identification of SRSF10 as a promising prognostic biomarker with functional significance among SRSFs for glioma. Life Sci. 2024;338:122392
35. Cai J, Song L, Zhang F, Wu S, Zhu G, Zhang P. et al. Targeting SRSF10 might inhibit M2 macrophage polarization and potentiate anti-PD-1 therapy in hepatocellular carcinoma. Cancer Commun (Lond). 2024;44:1231-60
36. Lu Y, Wang X, Gu Q, Wang J, Sui Y, Wu J. et al. Heterogeneous nuclear ribonucleoprotein A/B: an emerging group of cancer biomarkers and therapeutic targets. Cell Death Discov. 2022;8:337
37. Kedzierska H, Piekielko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 2017;396:53-65
38. Feng S, Li J, Wen H, Liu K, Gui Y, Wen Y. et al. hnRNPH1 recruits PTBP2 and SRSF3 to modulate alternative splicing in germ cells. Nat Commun. 2022;13:3588
39. Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK. et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105-15
40. Soubise B, Jiang Y, Douet-Guilbert N, Troadec MB. RBM22, a Key Player of Pre-mRNA Splicing and Gene Expression Regulation, Is Altered in Cancer. Cancers (Basel). 2022;14:643
41. Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC. et al. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38:2516-32
42. Bao Y, Zhang S, Zhang X, Pan Y, Yan Y, Wang N. et al. RBM10 Loss Promotes EGFR-Driven Lung Cancer and Confers Sensitivity to Spliceosome Inhibition. Cancer Res. 2023;83:1490-502
43. Yano K, Takahashi RU, Shiotani B, Abe J, Shidooka T, Sudo Y. et al. PRPF19 regulates p53-dependent cellular senescence by modulating alternative splicing of MDM4 mRNA. J Biol Chem. 2021;297:100882
44. Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY. et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149
45. Ghram M, Morris G, Culjkovic-Kraljacic B, Mars JC, Gendron P, Skrabanek L. et al. The eukaryotic translation initiation factor eIF4E reprograms alternative splicing. EMBO J. 2023;42:e110496
46. Chang SH, Elemento O, Zhang J, Zhuang ZW, Simons M, Hla T. ELAVL1 regulates alternative splicing of eIF4E transporter to promote postnatal angiogenesis. Proc Natl Acad Sci U S A. 2014;111:18309-14
47. Maimon A, Mogilevsky M, Shilo A, Golan-Gerstl R, Obiedat A, Ben-Hur V. et al. Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell Rep. 2014;7:501-13
48. Graham PL, Yanowitz JL, Penn JK, Deshpande G, Schedl P. The translation initiation factor eIF4E regulates the sex-specific expression of the master switch gene Sxl in Drosophila melanogaster. PLoS Genet. 2011;7:e1002185
49. Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers GJL, Giovannetti E. et al. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resist Updat. 2020;53:100728
50. Bashari A, Siegfried Z, Karni R. Targeting splicing factors for cancer therapy. RNA. 2023;29:506-15
51. Zhang Y, Liu L, Qiu Q, Zhou Q, Ding J, Lu Y. et al. Alternative polyadenylation: methods, mechanism, function, and role in cancer. J Exp Clin Cancer Res. 2021;40:51
52. Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18-30
53. Tan S, Zhang M, Shi X, Ding K, Zhao Q, Guo Q. et al. CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression. J Exp Clin Cancer Res. 2021;40:85
54. Liu S, Wu R, Chen L, Deng K, Ou X, Lu X. et al. CPSF6 regulates alternative polyadenylation and proliferation of cancer cells through phase separation. Cell Rep. 2023;42:113197
55. Jia Q, Nie H, Yu P, Xie B, Wang C, Yang F. et al. HNRNPA1-mediated 3' UTR length changes of HN1 contributes to cancer- and senescence-associated phenotypes. Aging (Albany NY). 2019;11:4407-37
56. Fischl H, Neve J, Wang Z, Patel R, Louey A, Tian B. et al. hnRNPC regulates cancer-specific alternative cleavage and polyadenylation profiles. Nucleic Acids Res. 2019;47:7580-91
57. Benjamin B, Sanchez AM, Garg A, Schwer B, Shuman S. Structure-function analysis of fission yeast cleavage and polyadenylation factor (CPF) subunit Ppn1 and its interactions with Dis2 and Swd22. PLoS Genet. 2021;17:e1009452
58. Mo L, Meng L, Huang Z, Yi L, Yang N, Li G. An analysis of the role of HnRNP C dysregulation in cancers. Biomark Res. 2022;10:19
59. Elhasnaoui J, Miano V, Ferrero G, Doria E, Leon AE, Fabricio ASC. et al. DSCAM-AS1-Driven Proliferation of Breast Cancer Cells Involves Regulation of Alternative Exon Splicing and 3'-End Usage. Cancers (Basel). 2020;12:1453
60. Wei L, Lee S, Majumdar S, Zhang B, Sanfilippo P, Joseph B. et al. Overlapping Activities of ELAV/Hu Family RNA Binding Proteins Specify the Extended Neuronal 3' UTR Landscape in Drosophila. Mol Cell. 2020;80:140-55 e6
61. Masuda A, Takeda J, Okuno T, Okamoto T, Ohkawara B, Ito M. et al. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 2015;29:1045-57
62. Zou S, Gou X, Wen K. Advances in the role of long non-coding RNAs and RNA-binding proteins in regulating DNA damage repair in cancer cells. Int J Mol Med. 2023;52:93
63. Zheng F, Du F, Zhao J, Wang X, Si Y, Jin P. et al. The emerging role of RNA N6-methyladenosine methylation in breast cancer. Biomark Res. 2021;9:39
64. Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell. 2019;74:640-50
65. Zhuang Y, Li T, Hu X, Xie Y, Pei X, Wang C. et al. MetBil as a novel molecular regulator in ischemia-induced cardiac fibrosis via METTL3-mediated m6A modification. FASEB J. 2023;37:e22797
66. Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY. et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379-89
67. Liang L, Zhu Y, Li J, Zeng J, Wu L. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J Exp Clin Cancer Res. 2022;41:261
68. Jin D, Guo J, Wu Y, Yang L, Wang X, Du J. et al. m (6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40
69. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M. et al. YTHDF2 destabilizes m (6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626
70. Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711-9
71. Peng Q, Tan S, Xia L, Wu N, Oyang L, Tang Y. et al. Phase separation in Cancer: From the Impacts and Mechanisms to Treatment potentials. Int J Biol Sci. 2022;18:5103-22
72. Elguindy MM, Mendell JT. NORAD-induced Pumilio phase separation is required for genome stability. Nature. 2021;595:303-8
73. Villanueva E, Smith T, Queiroz RML, Monti M, Pizzinga M, Elzek M. et al. Efficient recovery of the RNA-bound proteome and protein-bound transcriptome using phase separation (OOPS). Nat Protoc. 2020;15:2568-88
74. Gao H, Wei H, Yang Y, Li H, Liang J, Ye J. et al. Phase separation of DDX21 promotes colorectal cancer metastasis via MCM5-dependent EMT pathway. Oncogene. 2023;42:1704-15
75. Shen H, Yanas A, Owens MC, Zhang C, Fritsch C, Fare CM. et al. Sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA metabolism through phase separation. Mol Cell. 2022;82:2588-603 e9
76. Hoefig KP, Reim A, Gallus C, Wong EH, Behrens G, Conrad C. et al. Defining the RBPome of primary T helper cells to elucidate higher-order Roquin-mediated mRNA regulation. Nat Commun. 2021;12:5208
77. Levone BR, Lenzken SC, Antonaci M, Maiser A, Rapp A, Conte F. et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J Cell Biol. 2021;220:e202008030
78. Birsa N, Ule AM, Garone MG, Tsang B, Mattedi F, Chong PA. et al. FUS-ALS mutants alter FMRP phase separation equilibrium and impair protein translation. Sci Adv. 2021;7:eabf8660
79. Hallegger M, Chakrabarti AM, Lee FCY, Lee BL, Amalietti AG, Odeh HM. et al. TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell. 2021;184:4680-96 e22
80. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188-206
81. Liu Y, Chen S, Zong ZH, Guan X, Zhao Y. CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the development of endometrial cancer. J Cell Mol Med. 2020;24:6898-907
82. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-4
83. Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R. et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19:73
84. Liao W, Du J, Li L, Wu X, Chen X, Feng Q. et al. CircZNF215 promotes tumor growth and metastasis through inactivation of the PTEN/AKT pathway in intrahepatic cholangiocarcinoma. J Exp Clin Cancer Res. 2023;42:125
85. Wu P, Hou X, Peng M, Deng X, Yan Q, Fan C. et al. Circular RNA circRILPL1 promotes nasopharyngeal carcinoma malignant progression by activating the Hippo-YAP signaling pathway. Cell Death Differ. 2023;30:1679-94
86. Liu Z, Gu S, Wu K, Li L, Dong C, Wang W. et al. CircRNA-DOPEY2 enhances the chemosensitivity of esophageal cancer cells by inhibiting CPEB4-mediated Mcl-1 translation. J Exp Clin Cancer Res. 2021;40:361
87. Shi P, Liu Y, Yang H, Hu B. Breast cancer derived exosomes promoted angiogenesis of endothelial cells in microenvironment via circHIPK3/miR-124-3p/MTDH axis. Cell Signal. 2022;95:110338
88. Gu Y, Wang Y, He L, Zhang J, Zhu X, Liu N. et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol Cancer. 2021;20:132
89. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24:430-47
90. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-63
91. Porman AM, Roberts JT, Duncan ED, Chrupcala ML, Levine AA, Kennedy MA. et al. A single N6-methyladenosine site regulates lncRNA HOTAIR function in breast cancer cells. PLoS Biol. 2022;20:e3001885
92. Topel H, Bagirsakci E, Comez D, Bagci G, Cakan-Akdogan G, Atabey N. lncRNA HOTAIR overexpression induced downregulation of c-Met signaling promotes hybrid epithelial/mesenchymal phenotype in hepatocellular carcinoma cells. Cell Commun Signal. 2020;18:110
93. Zhang C, Xu L, Deng G, Ding Y, Bi K, Jin H. et al. Exosomal HOTAIR promotes proliferation, migration and invasion of lung cancer by sponging miR-203. Sci China Life Sci. 2020;63:1265-8
94. Xu J, Wang X, Zhu C, Wang K. A review of current evidence about lncRNA MEG3: A tumor suppressor in multiple cancers. Front Cell Dev Biol. 2022;10:997633
95. Qin Z, Liu X, Li Z, Wang G, Feng Z, Liu Y. et al. LncRNA LINC00667 aggravates the progression of hepatocellular carcinoma by regulating androgen receptor expression as a miRNA-130a-3p sponge. Cell Death Discov. 2021;7:387
96. Dong H, Wang W, Mo S, Chen R, Zou K, Han J. et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37:202
97. Dong P, Xiong Y, Yue J, Xu D, Ihira K, Konno Y. et al. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J Exp Clin Cancer Res. 2019;38:295
98. Saeinasab M, Bahrami AR, Gonzalez J, Marchese FP, Martinez D, Mowla SJ. et al. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J Exp Clin Cancer Res. 2019;38:172
99. Kim SS, Baek GO, Ahn HR, Sung S, Seo CW, Cho HJ. et al. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol Oncol. 2020;14:2646-59
100. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629-51
101. Shang R, Lee S, Senavirathne G, Lai EC. microRNAs in action: biogenesis, function and regulation. Nat Rev Genet. 2023;24:816-33
102. Otmani K, Rouas R, Lewalle P. OncomiRs as noncoding RNAs having functions in cancer: Their role in immune suppression and clinical implications. Front Immunol. 2022;13:913951
103. Li S, Wei X, He J, Cao Q, Du D, Zhan X. et al. The comprehensive landscape of miR-34a in cancer research. Cancer Metastasis Rev. 2021;40:925-48
104. Xue B, Chuang CH, Prosser HM, Fuziwara CS, Chan C, Sahasrabudhe N. et al. miR-200 deficiency promotes lung cancer metastasis by activating Notch signaling in cancer-associated fibroblasts. Genes Dev. 2021;35:1109-22
105. Cao LQ, Yang XW, Chen YB, Zhang DW, Jiang XF, Xue P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer. 2019;18:148
106. Deng T, Zhang H, Yang H, Wang H, Bai M, Sun W. et al. Exosome miR-155 Derived from Gastric Carcinoma Promotes Angiogenesis by Targeting the c-MYB/VEGF Axis of Endothelial Cells. Mol Ther Nucleic Acids. 2020;19:1449-59
107. Chen Q, Wang W, Chen S, Chen X, Lin Y. miR-29a sensitizes the response of glioma cells to temozolomide by modulating the P53/MDM2 feedback loop. Cell Mol Biol Lett. 2021;26:21
108. Ji C, Xu Q, Guo L, Wang X, Ren Y, Zhang H. et al. eEF-2 Kinase-targeted miR-449b confers radiation sensitivity to cancer cells. Cancer Lett. 2018;418:64-74
109. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
110. Han J, Meng J, Chen S, Wang X, Yin S, Zhang Q. et al. YY1 Complex Promotes Quaking Expression via Super-Enhancer Binding during EMT of Hepatocellular Carcinoma. Cancer Res. 2019;79:1451-64
111. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
112. Liu S, Yang N, Jiang X, Wang J, Dong J, Gao Y. FUS-induced circular RNA ZNF609 promotes tumorigenesis and progression via sponging miR-142-3p in lung cancer. J Cell Physiol. 2021;236:79-92
113. Omata Y, Okawa M, Haraguchi M, Tsuruta A, Matsunaga N, Koyanagi S. et al. RNA editing enzyme ADAR1 controls miR-381-3p-mediated expression of multidrug resistance protein MRP4 via regulation of circRNA in human renal cells. J Biol Chem. 2022;298:102184
114. Zhao Y, Song J, Dong W, Liu X, Yang C, Wang D. et al. The MBNL1/circNTRK2/PAX5 pathway regulates aerobic glycolysis in glioblastoma cells by encoding a novel protein NTRK2-243aa. Cell Death Dis. 2022;13:767
115. Fan HN, Chen ZY, Chen XY, Chen M, Yi YC, Zhu JS. et al. METTL14-mediated m (6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer. 2022;21:51
116. Lin C, Ma M, Zhang Y, Li L, Long F, Xie C. et al. The N (6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol Cancer. 2022;21:80
117. Zhang X, Zhou Y, Chen S, Li W, Chen W, Gu W. LncRNA MACC1-AS1 sponges multiple miRNAs and RNA-binding protein PTBP1. Oncogenesis. 2019;8:73
118. Ji J, Xu R, Ding K, Bao G, Zhang X, Huang B. et al. Long Noncoding RNA SChLAP1 Forms a Growth-Promoting Complex with HNRNPL in Human Glioblastoma through Stabilization of ACTN4 and Activation of NF-kappaB Signaling. Clin Cancer Res. 2019;25:6868-81
119. Ji X, Liu Z, Gao J, Bing X, He D, Liu W. et al. N (6)-Methyladenosine-modified lncRNA LINREP promotes Glioblastoma progression by recruiting the PTBP1/HuR complex. Cell Death Differ. 2023;30:54-68
120. Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM. et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580-6
121. Sheng J, He X, Yu W, Chen Y, Long Y, Wang K. et al. p53-targeted lncRNA ST7-AS1 acts as a tumour suppressor by interacting with PTBP1 to suppress the Wnt/beta-catenin signalling pathway in glioma. Cancer Lett. 2021;503:54-68
122. Zhang Y, Luo M, Cui X, O'Connell D, Yang Y. Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 2022;29:1850-63
123. Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R. et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016;30:1224-39
124. Toki N, Takahashi H, Sharma H, Valentine MNZ, Rahman FM, Zucchelli S. et al. SINEUP long non-coding RNA acts via PTBP1 and HNRNPK to promote translational initiation assemblies. Nucleic Acids Res. 2020;48:11626-44
125. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne). 2018;9:402
126. Nogami M, Miyamoto K, Hayakawa-Yano Y, Nakanishi A, Yano M, Okano H. DGCR8-dependent efficient pri-miRNA processing of human pri-miR-9-2. J Biol Chem. 2021;296:100409
127. Wei X, Ke H, Wen A, Gao B, Shi J, Feng Y. Structural basis of microRNA processing by Dicer-like 1. Nat Plants. 2021;7:1389-96
128. Mayr F, Schutz A, Doge N, Heinemann U. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res. 2012;40:7492-506
129. Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088-100
130. Chen C, Zhu C, Huang J, Zhao X, Deng R, Zhang H. et al. SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat Commun. 2015;6:8899
131. Zeng Q, Michael IP, Zhang P, Saghafinia S, Knott G, Jiao W. et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature. 2019;573:526-31
132. Terry AR, Nogueira V, Rho H, Ramakrishnan G, Li J, Kang S. et al. CD36 maintains lipid homeostasis via selective uptake of monounsaturated fatty acids during matrix detachment and tumor progression. Cell Metab. 2023;35:2060-76 e9
133. Michael IP, Saghafinia S, Hanahan D. A set of microRNAs coordinately controls tumorigenesis, invasion, and metastasis. Proc Natl Acad Sci U S A. 2019;116:24184-95
134. Zheng Z, Zeng X, Zhu Y, Leng M, Zhang Z, Wang Q. et al. CircPPAP2B controls metastasis of clear cell renal cell carcinoma via HNRNPC-dependent alternative splicing and targeting the miR-182-5p/CYP1B1 axis. Mol Cancer. 2024;23:4
135. Wang H, Huo X, Yang XR, He J, Cheng L, Wang N. et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136
136. Thompson EW, Haviv I. The social aspects of EMT-MET plasticity. Nat Med. 2011;17:1048-9
137. Wong CH, Lou UK, Fung FK, Tong JHM, Zhang CH, To KF. et al. CircRTN4 promotes pancreatic cancer progression through a novel CircRNA-miRNA-lncRNA pathway and stabilizing epithelial-mesenchymal transition protein. Mol Cancer. 2022;21:10
138. Zhu Y, Huang C, Zhang C, Zhou Y, Zhao E, Zhang Y. et al. LncRNA MIR200CHG inhibits EMT in gastric cancer by stabilizing miR-200c from target-directed miRNA degradation. Nat Commun. 2023;14:8141
139. Xiang Z, Lv Q, Zhang Y, Chen X, Guo R, Liu S. et al. Long non-coding RNA DDX11-AS1 promotes the proliferation and migration of glioma cells by combining with HNRNPC. Mol Ther Nucleic Acids. 2022;28:601-12
140. Peng T, He Y, Wang T, Yu J, Ma X, Zhou Z. et al. Discovery of a Novel Small-Molecule Inhibitor Disrupting TRBP-Dicer Interaction against Hepatocellular Carcinoma via the Modulation of microRNA Biogenesis. J Med Chem. 2022;65:11010-33
141. Wyss CB, Duffey N, Peyvandi S, Barras D, Martinez Usatorre A, Coquoz O. et al. Gain of HIF1 Activity and Loss of miRNA let-7d Promote Breast Cancer Metastasis to the Brain via the PDGF/PDGFR Axis. Cancer Res. 2021;81:594-605
142. Thompson CB, Vousden KH, Johnson RS, Koppenol WH, Sies H, Lu Z. et al. A century of the Warburg effect. Nat Metab. 2023;5:1840-3
143. Huppertz I, Perez-Perri JI, Mantas P, Sekaran T, Schwarzl T, Russo F. et al. Riboregulation of Enolase 1 activity controls glycolysis and embryonic stem cell differentiation. Mol Cell. 2022;82:2666-80 e11
144. Sun L, Suo C, Zhang T, Shen S, Gu X, Qiu S. et al. ENO1 promotes liver carcinogenesis through YAP1-dependent arachidonic acid metabolism. Nat Chem Biol. 2023;19:1492-503
145. Chen C, Bai L, Cao F, Wang S, He H, Song M. et al. Targeting LIN28B reprograms tumor glucose metabolism and acidic microenvironment to suppress cancer stemness and metastasis. Oncogene. 2019;38:4527-39
146. Zhao H, Wu W, Li X, Chen W. Long noncoding RNA UCA1 promotes glutamine-driven anaplerosis of bladder cancer by interacting with hnRNP I/L to upregulate GPT2 expression. Transl Oncol. 2022;17:101340
147. Liu SS, Li JS, Xue M, Wu WJ, Li X, Chen W. LncRNA UCA1 Participates in De Novo Synthesis of Guanine Nucleotides in Bladder Cancer by Recruiting TWIST1 to Increase IMPDH1/2. Int J Biol Sci. 2023;19:2599-612
148. Sun S, Li C, Cui K, Liu B, Zhou M, Cao Y. et al. Hsa_circ_0062682 Promotes Serine Metabolism and Tumor Growth in Colorectal Cancer by Regulating the miR-940/PHGDH Axis. Front Cell Dev Biol. 2021;9:770006
149. Liu X, Liang Y, Song R, Yang G, Han J, Lan Y. et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer. 2018;17:90
150. Jia Y, Yan Q, Zheng Y, Li L, Zhang B, Chang Z. et al. Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. J Exp Clin Cancer Res. 2022;41:287
151. Li Q, Yao H, Wang Y, Wu Y, Thorne RF, Zhu Y. et al. circPRKAA1 activates a Ku80/Ku70/SREBP-1 axis driving de novo fatty acid synthesis in cancer cells. Cell Rep. 2022;41:111707
152. Bill R, Wirapati P, Messemaker M, Roh W, Zitti B, Duval F. et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science. 2023;381:515-24
153. Liu J, Cao X. RBP-RNA interactions in the control of autoimmunity and autoinflammation. Cell Res. 2023;33:97-115
154. Zhang X, Smith SM, Wang X, Zhao B, Wu L, Hu X. Three paralogous clusters of the miR-17~92 family of microRNAs restrain IL-12-mediated immune defense. Cell Mol Immunol. 2021;18:1751-60
155. Daher M, Basar R, Gokdemir E, Baran N, Uprety N, Nunez Cortes AK. et al. Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells. Blood. 2021;137:624-36
156. Thomas S, Fayet OM, Truffault F, Fadel E, Provost B, Hamza A. et al. Altered expression of fragile X mental retardation-1 (FMR1) in the thymus in autoimmune myasthenia gravis. J Neuroinflammation. 2021;18:270
157. Zeng Q, Saghafinia S, Chryplewicz A, Fournier N, Christe L, Xie YQ. et al. Aberrant hyperexpression of the RNA binding protein FMRP in tumors mediates immune evasion. Science. 2022;378:eabl7207
158. Yi K, Cui X, Liu X, Wang Y, Zhao J, Yang S. et al. PTRF/Cavin-1 as a Novel RNA-Binding Protein Expedites the NF-kappaB/PD-L1 Axis by Stabilizing lncRNA NEAT1, Contributing to Tumorigenesis and Immune Evasion in Glioblastoma. Front Immunol. 2021;12:802795
159. Hu H, Cheng R, Wang Y, Wang X, Wu J, Kong Y. et al. Oncogenic KRAS signaling drives evasion of innate immune surveillance in lung adenocarcinoma by activating CD47. J Clin Invest. 2023;133:e153470
160. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H. et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12:295
161. Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:299-309
162. Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther. 2022;7:121
163. Cabral LKD, Tiribelli C, Sukowati CHC. Sorafenib Resistance in Hepatocellular Carcinoma: The Relevance of Genetic Heterogeneity. Cancers (Basel). 2020;12:1576
164. Zhu H, Chen K, Chen Y, Liu J, Zhang X, Zhou Y. et al. RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal Transduct Target Ther. 2022;7:240
165. Liu Y, Xu J, Choi HH, Han C, Fang Y, Li Y. et al. Targeting 17q23 amplicon to overcome the resistance to anti-HER2 therapy in HER2+ breast cancer. Nat Commun. 2018;9:4718
166. Wang X, Chen T, Li C, Li W, Zhou X, Li Y. et al. CircRNA-CREIT inhibits stress granule assembly and overcomes doxorubicin resistance in TNBC by destabilizing PKR. J Hematol Oncol. 2022;15:122
167. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X. et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5:298
168. Sang Y, Chen B, Song X, Li Y, Liang Y, Han D. et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol Ther. 2019;27:1638-52
169. Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266-82
170. Li J, Liu J, Zhou Z, Wu R, Chen X, Yu C. et al. Tumor-specific GPX4 degradation enhances ferroptosis-initiated antitumor immune response in mouse models of pancreatic cancer. Sci Transl Med. 2023;15:eadg3049
171. Lee J, You JH, Roh JL. Poly (rC)-binding protein 1 represses ferritinophagy-mediated ferroptosis in head and neck cancer. Redox Biol. 2022;51:102276
172. Wang Q, Guo Y, Wang W, Liu B, Yang G, Xu Z. et al. RNA binding protein DAZAP1 promotes HCC progression and regulates ferroptosis by interacting with SLC7A11 mRNA. Exp Cell Res. 2021;399:112453
173. Zhang Z, Yao Z, Wang L, Ding H, Shao J, Chen A. et al. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14:2083-103
174. He XY, Fan X, Qu L, Wang X, Jiang L, Sang LJ. et al. LncRNA modulates Hippo-YAP signaling to reprogram iron metabolism. Nat Commun. 2023;14:2253
175. Jonas K, Calin GA, Pichler M. RNA-Binding Proteins as Important Regulators of Long Non-Coding RNAs in Cancer. Int J Mol Sci. 2020;21:2969
176. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A. et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m (6)A reader YTHDF3. Mol Cancer. 2019;18:143
177. Zhang J, Chen S, Wei S, Cheng S, Shi R, Zhao R. et al. CircRAPGEF5 interacts with RBFOX2 to confer ferroptosis resistance by modulating alternative splicing of TFRC in endometrial cancer. Redox Biol. 2022;57:102493
178. Jiang Y, Zhao J, Li R, Liu Y, Zhou L, Wang C. et al. CircLRFN5 inhibits the progression of glioblastoma via PRRX2/GCH1 mediated ferroptosis. J Exp Clin Cancer Res. 2022;41:307
179. Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D. et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19:43
180. Wu P. Inhibition of RNA-binding proteins with small molecules. Nat Rev Chem. 2020;4:441-58
181. Zhang T, Yin C, Fedorov A, Qiao L, Bao H, Beknazarov N. et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature. 2022;606:594-602
182. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K. et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-94
183. Liang WW, Muller S, Hart SK, Wessels HH, Mendez-Mancilla A, Sookdeo A. et al. Transcriptome-scale RNA-targeting CRISPR screens reveal essential lncRNAs in human cells. Cell. 2024;187:7637-54 e29
184. Miyazaki K, Wada Y, Okuno K, Murano T, Morine Y, Ikemoto T. et al. An exosome-based liquid biopsy signature for pre-operative identification of lymph node metastasis in patients with pathological high-risk T1 colorectal cancer. Mol Cancer. 2023;22:2
185. Peng R, Huang Q, Wang L, Qiao G, Huang X, Jiang J. et al. G-Quadruplex RNA Based PROTAC Enables Targeted Degradation of RNA Binding Protein FMRP for Tumor Immunotherapy. Angew Chem Int Ed Engl. 2024;63:e202402715
186. Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A. et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565:43-8
187. Wang L, Dou X, Chen S, Yu X, Huang X, Zhang L. et al. YTHDF2 inhibition potentiates radiotherapy antitumor efficacy. Cancer Cell. 2023;41:1294-308 e8
Author contact
![]() Corresponding authors: Yujuan Zhou or Qianjin Liao, Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China, 283 Tongzipo Road, Changsha 410013, Hunan, China. Tel.: 86-731-88651681; Fax: 86-731-88651999; Email: yujany_zhoucom or march-oncom.
Corresponding authors: Yujuan Zhou or Qianjin Liao, Hunan Key Laboratory of Cancer Metabolism, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China, 283 Tongzipo Road, Changsha 410013, Hunan, China. Tel.: 86-731-88651681; Fax: 86-731-88651999; Email: yujany_zhoucom or march-oncom.

 Global reach, higher impact
Global reach, higher impact