10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2007; 3(6):342-348. doi:10.7150/ijbs.3.342 This issue Cite
Research Paper
Differences in Apoptosis and Cell Cycle Distribution between Human Melanoma Cell Lines UACC903 and UACC903(+6), before and after UV Irradiation
1. Department of Biochemistry and Molecular Biology, the George Washington University School of Medicine and Health Sciences, Washington, DC, USA
2. Department of Pathology, Loyola University Medical Center, Maywood, IL, USA
3. Division of Hematology, Center for Biological Evaluation and Research, FDA, Bethesda, MD, USA
Received 2007-6-26; Accepted 2007-7-13; Published 2007-7-16
Abstract
Introduction of human chromosome 6 into malignant melanoma cell line UACC903 resulted in generation of the chromosome 6-mediated suppressed cell subline UACC903(+6) that displays attenuated growth rate, anchorage-dependency, and reduced tumorigenicity. We have showed that overexpression of a chromosome 6-encoded tumor suppressor gene led to partial suppression to UACC903 cell growth. We now describe the differences in apoptosis and cell cycle between UACC903 and UACC903(+6) before and after UV irradiation. MTT assay revealed 86.92±8.24% of UACC903 cells viable, significantly (p<0.01) higher than 48.76±5.31% of UACC903(+6), at 24 hr after 254-nm UV irradiation (40 J/M2). Before UV treatment, flow cytometry analysis revealed 6.06±0.20% apoptosis in UACC903, significantly (p=0.01) lower than 6.67±0.15% in UACC903(+6). The G0/G1, S and G2/M phase cells of UACC903 were, respectively, 54.10±0.59%, 22.31±0.50% and 16.85±0.25%, all significantly (p<0.01) different from the corresponding percentages (58.82±0.35%, 20.48±0.05%, and 13.17±0.45%) of UACC903(+6). After the UV treatment, UACC903 cells in apoptosis, G0/G1, S, and G2/M became 12.59±0.17%, 38.90±0.67%, 19.74±0.70%, and 27.01±0.66%, respectively, while UACC903(+6) cells were 24.16±0.48%, 37.97±0.62%, 19.20±0.52%, and 15.69±0.14%. TUNEL assay revealed 2.31±0.62% apoptosis in UACC903, significantly (p<0.01) lower than 9.60±1.14% of UACC903(+6), and a linear and exponential increase of apoptosis, respectively, in response to the UV treatment. These results indicate that UACC903(+6) cells have a greater tendency to undergo apoptosis and are thus much more sensitive to UV irradiation. Our findings further suggest a novel mechanism for chromosome 6-mediated suppression of tumorigenesis and metastasis, i.e., through increased cell death.
Keywords: Melanoma, UV irradiation, apoptosis, cell cycle arrest
1. Introduction
Cutaneous malignant melanoma (CMM) is the most serious form of skin cancer because of greater likelihood for metastasis and apoptotic resistance to available chemotherapy and biotherapy [1-3]. CMM derives from the transformation of melanocytes in skin due to accumulation of genetic alterations, likely involving in oncogenes and suppressor genes [4]. Consistent with this view, introduction of a neo-tagged chromosome 6 into the different CMM cell lines led to suppression of tumorigenicity [5] and metastasis [6;7]. Molecular analyses of CMM cell lines resulted in identification of the chromosome 6-encoded tumor suppressor gene connexin 43 [8;9] and the chromosome 1-encoded metastasis suppressor gene KiSS-1 [10]. While it is generally assumed that the suppression is due to reduced cell proliferation, apoptotic features of CMM cell lines and the chromosome 6-mediated suppressed cell sublines have not yet been defined.
This study utilized human CMM cell line UACC903 and the chromosome 6-mediated suppressed cell subline UACC903(+6) [5]. UACC903 displays malignant tumorigenic phenotypes in in vitro culture and in mouse xenografts. The malignant phenotypes including a rapid growth rate and anchorage-independent growth in culture and fast tumor formation in athymic nude mice are suppressed in the cell subline UACC903(+6) [5;8;9]. In addition, we observed that 103 of 121 (85%) colony-forming clones selected from the UACC903(+6) cells underwent crisis and eventually cell death at different time points during cell culture [8]. Accordingly, we hypothesize that the UACC903 cell line is resistant, while UACC903(+6) cell line is sensitive, to environmental factor-induced cell death that may be via an apoptotic pathway.
To test this hypothesis, we characterized the differences of apoptosis and cell cycle distribution between these two cell lines before and after UV irradiation. Our results demonstrated significantly higher apoptosis and G0/G1 arrest in the UACC903(+6) cells than in UACC903 before UV treatment. After UV irradiation, the UACC903 cells displayed G2/M arrest and apoptotic resistance compared to UACC903(+6). The results increase our understanding of the effects of UV irradiation on apoptosis and cell cycle control observed in two closely related melanoma cell lines and provide new insight into the cellular mechanisms by which the chromosome 6-mediated tumor suppression operates.
2. MATERIALS and METHODS
Cell culture
UACC903 and UACC903(+6) cell lines were originally obtained from University of Arizona Cancer Center[5]. UACC903 cells were cultured in RPMI1640 with 10% fetal bovine serum, 2 mM L-glutamine. UACC903(+6) cells were cultured in the same medium with 600 μg/ml G418 to select for the pSV2neo-tagged chromosome 6.
UV irradiation
Cells were cultured for triplicate experiments. After removing medium, cells were exposed to UVC (254 nm) generated by UV Crosslinker (Model BLX254, Life Technologies, Gaithersburg, MD) at indicated doses. Medium was added immediately to continue culture until designated time points. Cells at 0 hr were a non-irradiation control.
MTT assay
This was conducted using MTT kit (Promega, Madison, WI) following manufacturer's instructions. Triplicate experiments were performed in 96-well plates. 490-nm absorbance was recorded using spectrophotometer (SPECTRAmax, Molecular Devices, Sunnyvale, California).
DNA ladders
These were detected by previously described methods [11]. Both attached and floating cells were harvested for analysis. DNA samples were fractionated by electrophoresis. Gel images were documented using a PhotoDocIT system (UVP, Upland, CA).
Flow cytometry
This was performed by previously described methods[12;13]. Both attached and floating cells were harvested for cell cycle analysis. For early apoptosis, attached cells were harvested for sorting in annexin-V-fluorescein and PI buffer (Roche, Nonnenwald, Penzberg, Germany). For each experiment, 10,000 cells were analyzed using ELITE Flow Cytometry (Beckman Coulter, Fullerton, CA) and Cell Quest software (Becton Dickinson, Franklin Lakes, NJ). Triplicates were performed in all cases except for the detection of early apoptotic cells.
TUNEL assay
Terminal DNA transferase-mediated dUTP nick end labeling (TUNEL) assay was employed to determine in situ apoptotic DNA breaks [14;15] by using the Detection Kit (Roche Applied Sciences, Germany) following manufacturer' instructions. Cells attached to glass slides in culture dishes were subjected to TUNEL assay and were then analyzed under Olympus BX 60 fluorescent microscope using Evolution CCD System and ImagePro software (Media Cybernetics, Rockville, MD).
Statistical analysis
Calculations of mean, SD, and p values were performed on triplicate experiments using XLSTAT 2006 (New York, NY). The Student T-test was used to calculate p-values for comparison. The significant statistics was set at a p-value <0.05.
3. RESULTS
Dose and time dependent response of UACC903 and UACC903(+6) cells to UV irradiation
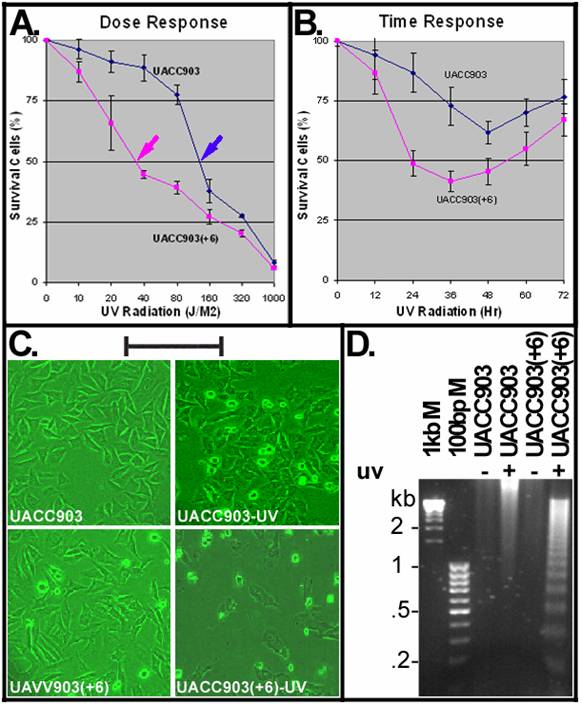
To determine a dose causing 50% cell death (LD50), UACC903 and UACC903(+6) cells were irradiated with UVC (254-nm) at doses including 10, 20, 40, 80, 160, 320 and 1000 J/M2 and viable cells were measured at 24 hr after UV irradiation by MTT assay. The results indicated that UACC903 viable cells were significantly (p<0.05) higher than UACC903(+6) at all doses except for 1000 J/M2 (data not shown). The UV LD50 for UACC903 was 137 J/M2, 3.8-fold higher than that (36 J/M2) to UACC903(+6) (Fig.1A). Thus, 40-J/M2 UVC was the dose chosen for subsequent experiments.
A time dependent response of the UACC903(+6) cells indicated rapid cell death that reached a peak with 41.45±4.14% survival at 36 hr and recovered slowly with 67.08±6.64% survival at 72 hr. In contrast, the UACC903 cells showed attenuated death that reached a peak with 61.67±4.81% survival at 48 hr and recovered rapidly with 76.86±7.17% survival at 72 hr (Fig. 1B). The difference between these two cell lines was highly significant (p=7.62E-11).
Apoptotic DNA ladders before and after UV irradiation
Figure 1C shows morphologies of UACC903 and UACC903(+6) cells before and 24 hr after UV irradiation. The polygon cells were viable and the round became dead. Few round cells were seen before UV irradiation, in contrast to the significant increase after UV treatment. Apoptotic cell death was demonstrated by DNA fragmentation. Figure 1D shows the typical apoptotic DNA ladders (2-fold multiplies of 0.2-kb fragments) in the UV-treated UACC903(+6) samples, as well as strong and weak smears of high molecular weight DNA, respectively, in the UV-treated UACC903 and two untreated samples. The ladders demonstrate apoptosis and the smears are consistent with reports that apoptotic DNA fragments can be as high as 300 kb [11;16].
UV induced apoptosis. (A) Dose-dependent response to UV irradiation. UACC903 and UACC903(+6) cells were exposed to UVC (254 nm) at indicated doses. Viable cells were measured by MTT assay at 24 hr after the UV treatment. Red and blue arrowheads point to the UVC LD50 of the UACC903(+6) (36 J/M2) and UACC903 (137 J/M2) cell lines calculated by fitting trendlines to the data. Means and SD were calculated using the 490-nm absorbance of UV-treated cells divided by that of untreated control. At 40 J/M2, UACC903 viable cells were 88.74±5.54% that is significantly (p=0.0002) higher than UACC903(+6) (44.67±1.12%). (B) Time-dependent response to UV treatment. Viable cells were measured by MTT assay at indicated time after 40-J/M2 UVC irradiation. Samples at 0 hr were not-irradiated controls. (C) Morphology of UACC903 and UACC903(+6) cells before and at 24 hr after exposure to 40-J/M2 UVC. Polygon cells were viable and the round became dead. The density of UACC903(+6) appears lower than that of UACC903 because the former had more floating dead cells than the latter and all floating cells had been removed from the culture dishes before taking images. Bar, 200 nm. (D) Representative gel image of apoptotic DNA ladders. Genomic DNA were extracted from both attached and floating cells before and at 24 hr after 40-J/M2 UVC irradiation using phosphate-citric acid buffer (0.2M Na2HPO4, 4mM citric acid) with a final concentration of 18 units/ml RNase A, 100 μg/ml proteinase K, and 0.1% Triton X-100. One microgram per DNA sample was loaded into a well of 1.5% agarose gel for electrophoresis to fraction DNA ladders. DNA markers and sizes in kilo-base pairs (kb) are indicated. Triplicate experiments were performed.
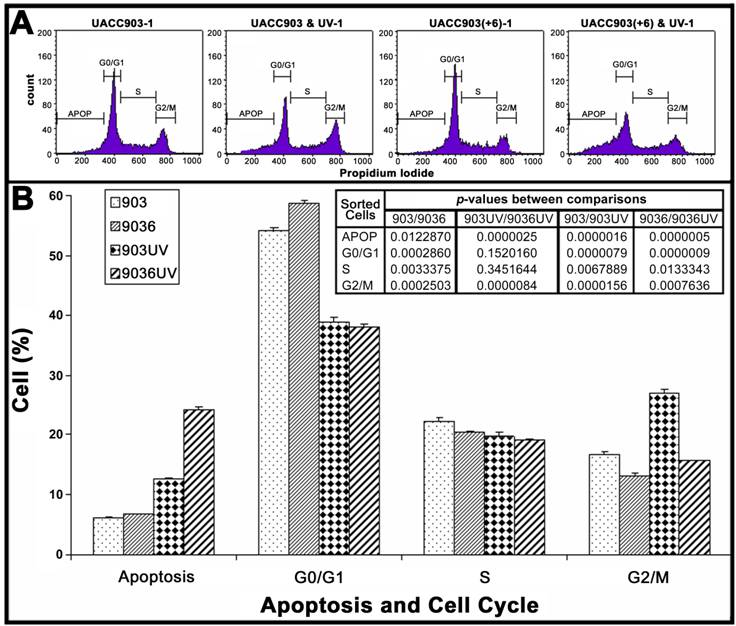
Cell distributions in apoptosis and cell cycle before and after UV irradiation
To quantify cell distributions in apoptosis and cell cycle, both attached and floating cells of UACC903 and UACC903(+6) in culture dishes were analyzed by flow cytometry (Fig. 2A). The results showed that before UV irradiation, the UACC903 cells distributed to 6.06±0.20%, 54.10±0.59%, 22.31±0.50%, and 16.85±0.25% in apoptosis, G0/G1, S, and G2/M, respectively and the corresponding UACC903(+6) cells were 6.67±0.15%, 58.82±0.35%, 20.48±0.05%, and 13.17±0.45%. The differences between UACC903 and UACC903(+6) cells were significant with p-values of 0.01 (apoptosis), 0.0003 (G0/G1), 0.003 (S), and 0.0003 (G2/M). At 24 hr after UV irradiation, the UACC903 cells redistributed to 12.59±0.17% (apoptosis), 38.90±0.67% (G0/G1), 19.74±0.70% (S), and 27.01±0.66% (G2/M), and the corresponding UACC903(+6) cells shifted to 24.16±0.48%, 37.97±0.62%, 19.20±0.52%, and 15.69±0.14%. The differences in apoptosis and G2/M between the UV-treated UACC903 and UACC903(+6) cells were highly significant with p-values of 0.000003 (apoptosis) and 0.000008 (G2/M), whereas the differences in G0/G1 and S were not (G0/G1 p = 0.15, S p = 0.35) (Fig. 2B).
Early apoptosis in UACC903 and UACC903(+6) before and after UV irradiation
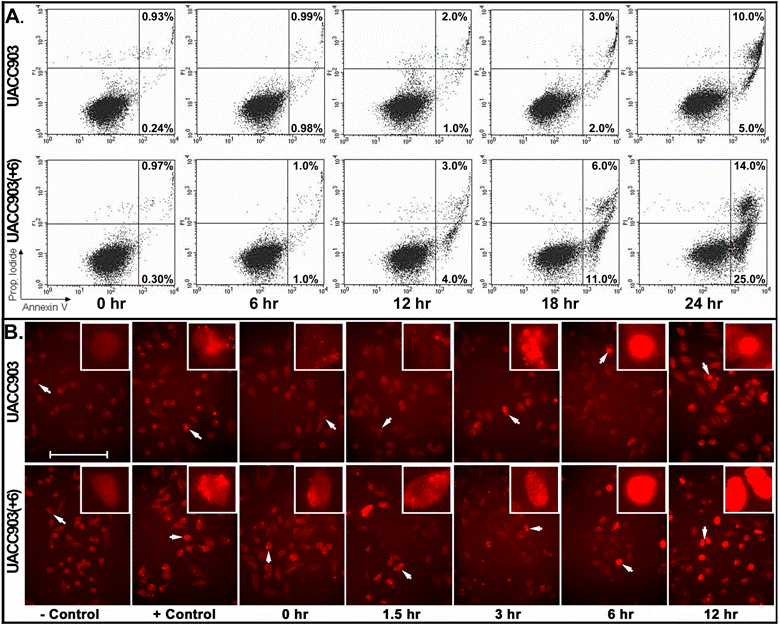
For early apoptosis, cells attached to culture dishes were harvested at 0, 6, 12, 18, and 24 hr after UV irradiation and stained with both annexin-V and PI for flow cytometry analysis. The results showed that early apoptosis in the UACC903 population was 0.24% (0 hr), 0.98% (6 hr), 1% (12 hr), 2% (18 hr) and 5% (24 hr). The corresponding cells in UACC903(+6) were 0.3%, 1%, 4%, 11% and 25%. Few differences between UACC903 and UACC903(+6) cells were observed at 0- and 6-hr time points. At 12-, 18- and 24-hr, early apoptosis in UACC903(+6) were 4-, 5.5- and 5-fold higher than those in UACC903, respectively (Fig. 3A).
Apoptotic DNA breaks in UACC903 and in UACC903(+6) before and after UV irradiation
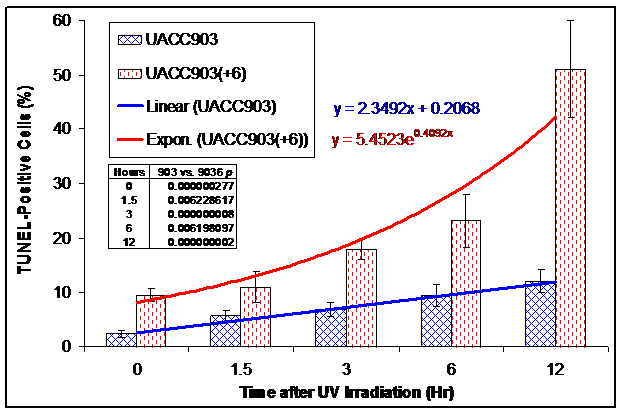
We conducted a TUNEL assay on UACC903 and UACC903(+6) cells to determine the in situ apoptotic DNA breaks before and 1.5, 3, 6, and 12 hr after the UV irradiation. The results showed 2.31±0.62%, 5.59±1.08%, 6.80±1.25%, 9.46±2.10%, and 12.17±2.11% TUNEL-positive cells in the UACC903 cells at 0, 1.5, 3, 6, and 12 hr time points, respectively. The corresponding cells in UACC903(+6) were 9.60±1.14%, 10.93±2.90%, 18.01±1.84%, 23.18±4.82%, and 50.99±12.12%. The corresponding differences between UACC903 and UACC903(+6) cells were highly significant with p-values of 0.0000003, 0.006, 0.000000008, 0.006, and 0.000000002. Trendlines fitted mathematically to the data revealed the linear (y = 2.35x + 0.21) and exponential (y = 5.45e0.41x) increase of the TUNEL-positive cells in UACC903 and UACC903(+6) populations, respectively, where x represents time after the UV treatment and y is a percentage of the TUNEL-positive cells (Fig. 4B).
The differences in apoptosis and cell cycle distribution of UACC903 and UACC903(+6) cells before and 24 hr after the UV treatment. (A) Representative plots of one set of triplicate experiments. Cells in apoptosis (APOP), G0/G1, S, and G2/M of the cell cycle were sorted based on DNA contents after staining, with propidium iodide (PI), UACC903 and UACC903(+6) cells before and after UV treatment. 10,000 cells were sorted using ELITE Flow Cytometry (Beckman Coulter) and analyzed with Cell Quest software (Becton Dickinson). (B) Bar graph of cell distributions. Means and SD were calculated from triplicate experiments. On one hand, the UV treatment induced an increase in apoptosis of UACC903 and UACC903(+6) cells by 6.53% (p = 0.000002) and 17.49% (p = 0.0000005) and in G2/M cells by 10.16% (p = 0.00002) and 2.52% (p = 0.0008), respectively. On the other hand, the same treatment led to a decrease in UACC903 and UACC903(+6) G0/G1 cells by 15.20% (p = 0.000008) and 20.85% (p = 0.0000009) and in S cells by 3.14% (p = 0.007) and 1.28% (p = 0.01), respectively. Interestingly, out of 15.20% reduction of UACC903 cells from G0/G1 phase by UV treatment, 61% cells were redistributed to G2/M and 39% to apoptosis. In contrast, out of 20.85% reduction of the UACC903(+6) G0/G1 cells, 13% were shifted to G2/M and 87% to apoptosis. P-values between all comparisons were calculated by the Student T-tests. 903: UACC903, 9036: UACC903(+6), 903UV: UACC903 treated with UV, 9036UV: UACC903(+6) treated with UV.
Detection of early apoptosis. (A) Plots of sorted apoptotic cells. The cells attached to culture dishes before and at 6, 12, 18, and 24 hr after UV treatment were harvested and stained with annexin-V-FITC plus PI. 10,000 cells of each experiment were sorted by ELITE Flow Cytometry (Beckman Coulter) and analyzed using Cell Quest software (Becton Dickinson). Early apoptotic cells (Annexin-V+ and PI-) were displayed in the lower right quadrant and late apoptotic cells (Annexin-V+ and PI+) were shown in the upper right quadrant. The percentages of apoptotic cells were listed in the quadrants. Because floating cells were excluded from analysis, the percentages in the upper right quadrants underestimate the numbers of late apoptotic cells. (B) Representative images of in situ apoptotic DNA breaks. Cells attached to glass slides in culture dishes before and at 1.5, 3, 6 and 12 hr after the UV irradiation were processed by the terminal deoxynucleotidyl transferase-mediated dUTP biotin nick-end labeling (TUNEL) assay using the In Situ Cell Death Detection Kit (TMR, Roche). The images were acquired with a high sensitivity Evolution MP Charge-Coupled Device (CCD) system and ImagePro software v.5.1 (Media Cybernetics). Cells for negative controls were not treated by UV irradiation and processed under the same TUNEL conditions without adding the terminal transferase. Cells for positive controls were incubated, after permeabilization, in the nicking buffer (500 U/ml DNase I, 50-mM Tris-HCl, pH 7.5, 1 mg/ml BSA) for 10 min at room temperature to induce DNA strand breaks, prior to the TUNEL labeling. Bar: 200 nm. Insert: 4 x 200 nm magnification.
4. DISCUSSION
Resistance to apoptosis and nonstop proliferation are characteristics of malignant cells different from their normal counterparts. Dr. Trent and his colleagues have demonstrated a more rapid proliferation of human malignant melanoma cell line UACC903 in culture and mouse xenografts than that of the chromosome 6-mediated suppressed cell subline UACC903(+6) [5;8]. Previously our laboratory discovered the chromosome 6-encoded connexin 43 (Cx43) suppressing proliferation of these melanoma cells[9]. In the present study, we demonstrate, for the first time, the differences in apoptosis and cell cycle distributions between UACC903 and UACC903(+6) cells, before and after UV irradiation. This discovery provides new insights into cellular mechanisms by which the chromosome 6-mediated tumor suppression operates.
UACC903 and UACC903(+6) cells display different rates in apoptosis and cell cycle distribution before UV treatment, as measured by cell-cycle flow cytometry and TUNEL assays. The cell-cycle flow cytometry detected 10.1% higher apoptotic cells in the UACC903(+6) cell population than in the UACC903 (p = 0.012), while the TUNEL assay demonstrated 4.16-fold higher in the former than in the latter (p <0.01). Because these assays measure different stages of apoptosis, the proportion of apoptotic cells was expected to vary. The TUNEL assay detects the in situ DNA strand breaks [14;15]; whereas cell-cycle flow cytometry cannot detect apoptosis until the integrity of cell membrane is altered and the degraded nuclear DNA leak out of cells [12;13;17]. It is worthy mentioning that the annexin-V flow cytometry data (Fig. 3A) cannot be compared directly with the results derived from the cell-cycle flow cytometry and TUNEL assays for several reasons. First, only attached cells (viable and early apoptotic cells) were used for the annexin-V flow cytometry (Fig. 3A) and TENUL assays (Fig. 3B and Fig. 4); in contrast, both attached and floating (dead) cells were harvested for the cell-cycle flow cytometry (Fig. 2). Second, apoptotic DNA strand breaks detected by the TUNEL assay occur earlier than reversion of phosphatitylserine from the inner leaflet of the cell membrane to the outer leaflet that can be bound by annexin-V [12-15]. Third, the annexin-V flow cytometry aimed at revealing the UV-induced early apoptotic changes which indeed provided a reference time point, that is, prior to 12 hr after the UV treatment for the TUNEL assays. Although 10.1% apoptotic difference seems small, it can be amplified in response to environmental factors, for example, UV irradiation as demonstrated in this study. In vivo, host factors might affect survival and/or apoptosis differentially on these two cell lines, therefore, affecting tumorigenicity. Apparently, fewer apoptotic cells in the UACC903 cell population than those in the UACC903(+6) suggests more viable malignant cells are available for proliferation and tumorigenesis. Additionally, fewer G0/G1 and more S cells in UACC903 than in UACC903(+6) are evidence of more active proliferation of the former than that of the latter.
Bar graph of the TUNEL-positive apoptotic cells before and after UV irradiation. UACC903 and UACC903(+6) cells before and at 1.5, 3, 6 and 12 hr after the UV irradiation were processed by the terminal deoxynucleotidyl transferase-mediated dUTP biotin nick-end labeling (TUNEL) assay using the In Situ Cell Death Detection Kit (TMR, Roche). On average, 700 cells were analyzed for each of triplicate experiments to calculate means and SD of the TUNEL-positive cells. The trendlines and equations on chart fitted mathematically to the data demonstrate the exponential and linear increase in the TUNEL-positive apoptotic cells in UACC903(+6) and UACC903 cells, respectively, in response to the UV treatment. The difference of the TUNEL-positive cells between UACC903 and UACC903(+6) at each time point was highly significant with all p <0.01, as indicated in chart. 903: UACC903, 9036: UACC903(+6)
These results suggest that at least 2 cellular mechanisms, activated apoptosis and G0/G1 arrest in UACC903(+6) cells, underlie the chromosome 6-mediated tumor suppression. This notion is supported by the fact that expression of the chromosome 6-encoded tumor suppressor gene Cx43 was higher in UACC903(+6) than in UACC903 cells, and that transfection and overexpression of Cx43 in UACC903 cells resulted in partial suppression to growth [9]. It has been reported that Cx43 caused G0/G1 arrest of osteosarcoma U2OS cells [18] and sensitized prostate cancer cells to TNFα-induced apoptosis [19]. The partial suppression to UACC903 growth by Cx43 suggests other genes may also play a role in the chromosome 6-mediated G0/G1 arrest and apoptosis.
Genotoxicity of UV irradiation caused redistribution of the melanoma cells in apoptosis and cell cycle. The dose- and time-dependent response demonstrated that the UACC903 cells were resistant, while UACC903(+6) were sensitive, to UV-induced apoptosis. Interestingly, the trendlines fitted mathematically to the TUNEL-positive cells revealed a linear and exponential increase of apoptosis in UACC903 and UACC903(+6) cells, respectively. Apparently, different molecular mechanisms underlie these apoptotic characteristics. Regarding UV-induced cell cycle redistribution, out of 15.20% reduction of UACC903 cells from G0/G1, 61% were redistributed to G2/M and 39% to apoptosis. In contrast, out of 20.85% reduction of the UACC903(+6) G0/G1 cells, 13% were shifted to G2/M and 87% to apoptosis. Therefore, G2/M arrest and apoptotic resistance are the major differences observed between UACC903 and UACC903(+6) cell lines in response to the UVC treatment. The UVC-induced UACC903 G2/M arrest is reminiscent of a recent report that all of UVA (400 – 320 nm), UVB (320 – 290 nm) and UVC (290 – 200 nm) induced G2/M arrest of the G361 human melanoma cell line[20]. The G2/M arrest could be due to UV-induced DNA damage and might provide a mechanism for DNA repair. In this regard, it is known that UVC and UVB induce formation of cyclobutane pyrimidine dimers and (6 – 4) photoproducts which are normally repaired by a nucleotide excision repair mechanism[21]. While UVA and UVB play a role as an etiology in development of skin cancer including malignant melanoma, UVC is the most frequently used UV light in studying UV-induced DNA damage and apoptosis.
In summary, we characterized the differences in apoptosis and cell cycle distribution between human malignant melanoma cell line UACC903 and the chromosome 6-mediated suppressed cell line UACC903(+6), before and after UV irradiation. Without UV treatment, S- and G2/M cells were significantly higher in UACC903 than in UACC903(+6) cell line; whereas the latter had more cells in apoptosis and G0/G1 than the former. After UV irradiation, G2/M arrest and apoptotic resistance become the major differences between UACC903 and UACC903(+6) cell lines. While it is of interest to further study the molecular basis underlying these differences, our current findings provide a novel mechanism by which chromosome 6 suppresses tumorigenesis and metastasis.
Acknowledgements
Supports: NIH-NIDDK-06-925 and the Catherine B McCormick Genomics Center. We thank Dr. Allan Goldstein and AnhThu Nguyen for critical review and discussion of the manuscript and assistance in English grammars.
Conflict of interests
Authors state that there are no any competing financial, professional, or personal conflicts of interest related to this publication.
References
1. Atkins MB. The treatment of metastatic melanoma with chemotherapy and biologics. Curr Opin Oncol. 1997;9(2):205-13
2. Buzaid AC. Management of metastatic cutaneous melanoma. Oncology (Williston Park). 2004;18(11):1443-50
3. Koon HB, Atkins MB. Update on therapy for melanoma: opportunities for patient selection and overcoming tumor resistance. Expert Rev Anticancer Ther. 2007;7(1):79-88
4. Su YA, Trent JM. Genetics of Cutaneous Malignant Melanoma. Cancer Control. 1995;2(5):392-7
5. Trent JM, Stanbridge EJ, McBride HL, Meese EU, Casey G, Araujo DE. et al. Tumorigenicity in human melanoma cell lines controlled by introduction of human chromosome 6. Science. 1990;247(4942):568-71
6. Welch DR, Chen P, Miele ME, McGary CT, Bower JM, Stanbridge EJ. et al. Microcell-mediated transfer of chromosome 6 into metastatic human C8161 melanoma cells suppresses metastasis but does not inhibit tumorigenicity. Oncogene. 1994;9(1):255-62
7. Miele ME, Robertson G, Lee JH, Coleman A, McGary CT, Fisher PB. et al. Metastasis suppressed, but tumorigenicity and local invasiveness unaffected, in the human melanoma cell line MelJuSo after introduction of human chromosomes 1 or 6. Mol Carcinog. 1996;15(4):284-99
8. Su YA, Ray ME, Lin T, Seidel NE, Bodine DM, Meltzer PS. et al. Reversion of monochromosome-mediated suppression of tumorigenicity in malignant melanoma by retroviral transduction. Cancer Res. 1996;56(14):3186-91
9. Su YA, Bittner ML, Chen Y, Tao L, Jiang Y, Zhang Y. et al. Identification of tumor-suppressor genes using human melanoma cell lines UACC903, UACC903(+6), and SRS3 by comparison of expression profiles. Mol Carcinog. 2000;28(2):119-27
10. Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE. et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88(23):1731-7
11. Gong J, Traganos F, Darzynkiewicz Z. A selective procedure for DNA extraction from apoptotic cells applicable for gel electrophoresis and flow cytometry. Anal Biochem. 1994;218(2):314-9
12. Darzynkiewicz Z, Li X, Gong J. Assays of cell viability: discrimination of cells dying by apoptosis. Methods Cell Biol. 1994;41:15-38
13. Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184(1):39-51
14. Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119(3):493-501
15. Ben-Sasson SA, Sherman Y, Gavrieli Y. Identification of dying cells--in situ staining. Methods Cell Biol. 1995;46:29-39
16. Walker PR, Kokileva L, LeBlanc J, Sikorska M. Detection of the initial stages of DNA fragmentation in apoptosis. Biotechniques. 1993;15(6):1032-40
17. Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148(7):2207-16
18. Zhang YW, Morita I, Ikeda M, Ma KW, Murota S. Connexin43 suppresses proliferation of osteosarcoma U2OS cells through post-transcriptional regulation of p27. Oncogene. 2001;20(31):4138-49
19. Wang M, Berthoud VM, Beyer EC. Connexin43 increases the sensitivity of prostate cancer cells to TNFalpha-induced apoptosis. J Cell Sci. 2007;120(Pt 2):320-9
20. Kowalczuk CI, Priestner MC, Pearson AJ, Saunders RD, Bouffler SD. Wavelength dependence of cellular responses in human melanocytes and melanoma cells following exposure to ultraviolet radiation. Int J Radiat Biol. 2006;82(11):781-92
21. Tornaletti S, Pfeifer GP. UV damage and repair mechanisms in mammalian cells. Bioessays. 1996;18(3):221-8
Author contact
![]() Correspondence to: Yan A. Su, MD, PhD, Department of Biochemistry and Molecular Biology, GWUSMHS, Ross Hall, Rm. 555, 2300 EYE Street, NW, Washington, DC 20037. Tel: 202-994-1891; Email: bcmyasedu
Correspondence to: Yan A. Su, MD, PhD, Department of Biochemistry and Molecular Biology, GWUSMHS, Ross Hall, Rm. 555, 2300 EYE Street, NW, Washington, DC 20037. Tel: 202-994-1891; Email: bcmyasedu

 Global reach, higher impact
Global reach, higher impact