Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(2):163-171. doi:10.7150/ijbs.6.163 This issue Cite
Research Paper
The ulcerative colitis marker protein WAFL interacts with accessory proteins in endocytosis
1. Genome Institute of Singapore, Singapore 138672
2. Strategic Research Center IRIS, Karolinska Institute, Stockholm, Sweden
3. Department of Microbiology, Tumor and Cell Biology, Karolinska Institute, Stockholm, Sweden
4. Okinawa Institute of Science and Technology, Onna, Okinawa 904-0412, Japan
Received 2009-11-11; Accepted 2010-3-22; Published 2010-3-29
Abstract
Ulcerative colitis (UC) is one of the major forms of inflammatory bowel disease with unknown cause. A molecular marker, WAFL, has recently been found to be up-regulated in the inflamed colonic mucosa of UC patients. Towards understanding biological function of WAFL, we analyzed proteins interacting with WAFL in HEK-293 cells by immunoprecipitation and mass spectrometry. Among four proteins found to specifically interact with WAFL, both KIAA0196 and KIAA1033 bind to α-appendage of the adaptor protein complex 2 (AP2), which acts as an interaction hub for accessory proteins in endocytosis mediated by clathrin-coated vesicle (CCV). The specific interaction between WAFL and KIAA0196 was also confirmed in human colorectal carcinoma HCT-116 cells by co-immunoprecipitation with specific antibodies. Meta-analyses of the databases of expressed genes suggest that the three genes are co-expressed in many tissues and cell types, and that their molecular function may be classified in the category of 'membrane traffic protein'. Therefore, these results suggest that WAFL may play an important role in endocytosis and subsequent membrane trafficking by interacting with AP2 through KIAA0196 and KIAA1033.
Keywords: Inflammatory bowel disease, Proteomics, WAFL/FKBP15/FKBP133/KIAA0674, KIAA1033, KIAA0196/strumpellin, Wiskott-Aldrich syndrome protein
1. Introduction
Inflammatory bowel disease (IBD) is a complex polygenic chronic inflammatory disease of the gastrointestinal tract with unknown cause, and is in need of additional biomarkers in order to subgroup different forms of IBD. In its current setting, IBD is classified into two major forms, Crohn's disease (CD) and UC [1-4]. Although CD and UC are two major clinically different diseases, there is correlation between their genetic backgrounds, and diagnosis is made difficult when disease manifestations overlap [5-7]. Nine IBD susceptibility loci have been identified and confirmed by linkage studies [8]. The CARD15 gene coding for NOD2, an intracellular peptidoglycan receptor, has been identified to be linked to CD [9,10]. A recent genome-wide association study has identified UC-risk loci encompassing genes, PLA2G2E, IFNG, IL26 and IL22, as well as associations between UC and the MHC region, and between UC and IL23R [11].
Through a differential gene expression screen using subtractive suppression hybridization, it has recently been found that the WAFL (WASP and FKBP-like protein; also known as FKBP15/FKBP133/KIAA0674 [12,13]) gene, which encodes a novel protein with homologies to WASP and FKBP (FK506-binding protein), was up-regulated in the inflamed colonic mucosa of UC patients, but not of CD [14]. The WAFL protein has recently been found to associate with early endosomes, and to move along microtubule tracks. In addition, WAFL also interacts with WASP-interacting protein (WIP) and actin [15]. The mouse homolog of WAFL was reported to partially co-localize with F-actin in the growth cones of dorsal root ganglion neurons [13]. These results suggest that WAFL may be involved in the transport of early endosomes by interacting with endosomes, microtubules and actin filaments. However, how WAFL interacts with endosomes and microtubules is unknown.
To explore biological and pathological roles of WAFL, we have investigated the interactome of WAFL in HEK-293 cells by co-immunoprecipitation, PAGE and LC-MS/MS, and have found that WAFL specifically interacts with accessory proteins involved in endocysis. This result was further supported by a meta-analysis of gene expression profiles of the interacting partners in the Oncomine database [16,17] as well as by gene ontology analysis of the PANTHER database [18], and was confirmed by co-immunoprecipitation of human colorectal carcinoma HCT-116 cells with specific antibodies.
2. Materials and Methods
Cell lines and transfection
Chemicals and reagents used in this study were purchased from Sigma (St. Louis, MO) except for those otherwise specified. HEK-293 cells and HCT-116 cells were cultured in minimum essential medium (MEM; Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS; Invitrogen) and 1.0% (v/v) penicillin/streptomycin (Invitrogen) at 37ºC in a 5% (v/v) CO2 humidified atmosphere. The full-length WAFL cDNA (3757 bp) was cloned into pCMV·SPORT6 plasmid (Invitrogen) between Sal I and Apa I sites. The insert also encoded the WAFL protein fused with a FLAG tag at its C-terminus.
Co-immunoprecipitation and Western blotting
HEK-293 cells expressing the full-length WAFL protein with a FLAG tag were lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.0 mM EDTA, 1.0 mM EGTA, 1% (v/v) Triton X-100, 2.5 mM sodium pyrophosphate, 1.0 mM beta-glycerophosphate, 1.0 mM Na3VO4, 1.0 µg/ml leupeptin, and protease inhibitor cocktail (x 1) (Roche Singapore)) for 1.0 h at 4°C. After centrifuging for 10 min at 13,000 rpm, the supernatant was immunoprecipitated with anti-FLAG M2 affinity gel and eluted according to the manufacturer's instructions (Sigma).
The eluted protein samples were separated by 10% SDS-PAGE, and were stained with Coomassie Brilliant Blue G-250 (CBB; Thermo Fisher, Pittsburgh, PA) or transferred to a nitrocellulose filter. The membrane was incubated with primary antibody (anti-FLAG M2 antibody; F3165) at a concentration of 2.5 μg/ml and then secondary antibody (goat anti-mouse immunoglobulin G antibody; A9044) at a concentration of 0.5 μg/ml. The blot was stained by using ECL reagents (GE Healthcare, Little Chalfont, UK), and was exposed to X-ray film.
Human HCT-116 cell line, purchased from the American Type Culture Collection (ATCC; Manassas, VA), was cultivated in MEM as described above, and was used for co-immunoprecipitation and Western blotting analyses using antibodies, HPA007979 (Sigma) and sc-87442 (Santa Cruz Biotechnology, Santa Cruz, CA), specific for WAFL and KIAA0196, respectively. In brief, HCT-116 cell lysates (~107 cells in 1.0 ml each) prepared as described above were precleared with 30 μl protein A-Sepharose 4B (101142; Invitrogen) for 3 h at 4oC, and then after removing the beads by centrifugation, the supernatants were incubated with antibody, 3 μg, at 4oC overnight. Resulting antibody-protein complexes were collected by incubating the solution with 30 μl of protein A-Sepharose beads for 2 h at 4oC, and then after centrifugation, the beads were washed three times with TBS (50 mM Tris-HCl, pH 7.4, and 150 mM NaCl) buffer, 1.5 ml, for 10 min at 4oC. Proteins were eluted by 70 μl of 0.1 M glycine buffer, pH 2.5, for 20 min at room temperature, and were neutralized with 10 μl of 1.0 M Tris-HCl, pH 8.5. Rabbit serum IgG (sc-2027; Santa Cruz) was used as a negative control. Western blotting was performed as described above.
LC-MS/MS
Cell lysates and protein samples were separated by 10% SDS-PAGE. After staining with CBB G-250 or with a silver stain plus kit (BIO-RAD, Hercules, CA), protein bands were excised out and were then transferred to polypropylene tubes. The excised gels were washed three times with a 1:1 mixture of acetonitrile and 50 mM ammonium bicarbonate for 15 min, dried and then reduced with 10 mM dithiothreitol for 1.0 h at 55ºC. After 55 mM iodoacetamide treatment followed by incubation in dark for 45 min at room temperature, the gel pieces were washed with 100 mM ammonium bicarbonate for 5 min with vortexing, and then with a 1:1 mixture of acetonitrile and 50 mM ammonium bicarbonate for 5 min. The resulting gel pieces were dehydrated with acetonitrile, and were dried up.
In-gel digestion of proteins was performed by adding 50 mM ammonium bicarbonate containing 2.6 ng/µl modified trypsin (Promega, Madison, WI) and incubated in a water bath for 16 h at 37°C. Tryptic peptides were extracted twice in extraction buffer (0.1% (v/v) trifluoroacetic acid, 50% acetonitrile, pH 2.5) by sonicating for 20 min. The peptide solution was dried in a Speed-Vac (Thermo Savant, Marietta, OH) to reduce the volume and was adjusted to 100 µl with 100 mM ammonium bicarbonate before loading onto LC-MS/MS.
The peptide samples were analyzed with a Finnigan Surveyor HPLC system coupled online to a LTQ ion trap mass spectrometer (Thermo Electron, San Jose, CA) equipped with a nano-ESI source. The MS scan range was 200-2000 m/z and mass spectra were collected in data-dependent MS/MS mode in which five of the highest intensity peaks in each MS scan were chosen for collision-induced dissociation (CID), with an isolation width of 3 Da.
LC-MS/MS data analysis
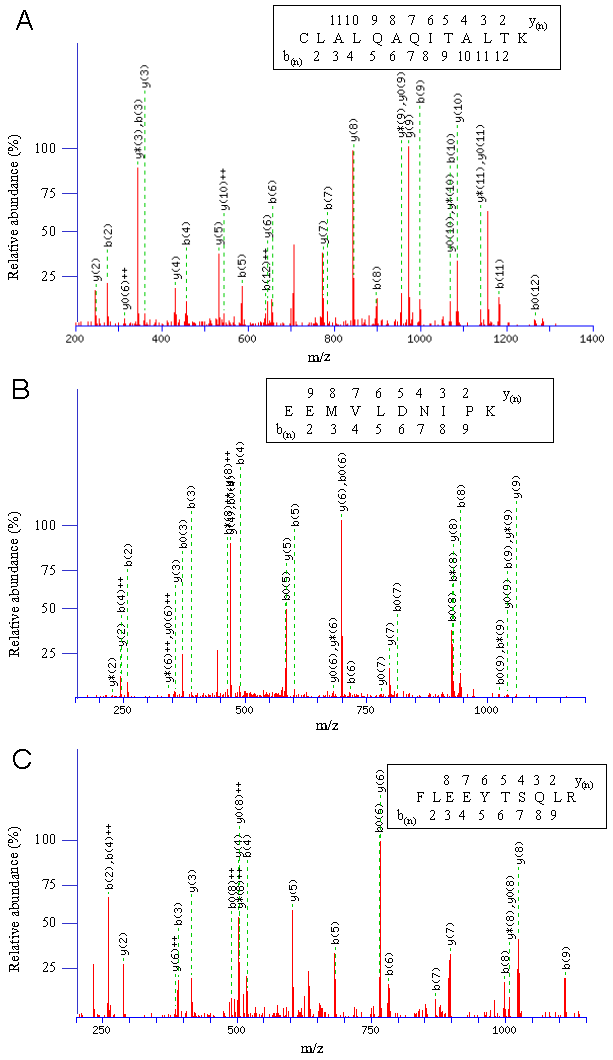
MS/MS data were analyzed by using Mascot (Matrix Science, London, UK). Database search was performed with a carbaminomethylation (Cys) set as fixed modification, oxidized methionine (+16 Da) as variable modifications. Missed cleavages of trypsin were set as one. Peptide charges were confined to 2+ and 3+, and mass tolerance was set to 2.0 Da for parent ions and 0.8 Da for product ions, respectively. Identifications were accepted with at least three consecutive y- and b- ions pair and a minimum ions score of 40. Details of ions score and nomenclature for 'b' and 'y' ions were previously described [19]. Three independent experiments were carried out. One representative result was selected and the interacting proteins detected in at least two experiments are shown.
Meta-analysis of WAFL- interacting partners
Meta-analysis of expression profiles of WAFL-interacting partner genes was carried out by retrieving studies from the Oncomine database [16,17]. Similar meta-analysis has been successfully applied for the identification of signaling pathway components mediated by the estrogen receptor after retrieving top 21 studies from the database [20,21]. Co-occurrence of two factors (co-expression of two genes) for each study was compared among a set of co-expressed genes after removing duplicates and ESTs. The correlation network of co-expressed genes was constructed by assuming that two genes co-expressed at higher frequencies would be more functionally correlated and would more closely interact with each other.
Gene ontology analysis of WAFL and its interacting partners was carried out using the PANTHER database as previously described [18,22]. Top 1,000 genes highly co-expressed were selected as input data from the database, and were compared with the NCBI reference gene set. Statistically significantly (p < 0.01) enriched PANTHER gene ontology categories were identified.
3. Results and Discussion
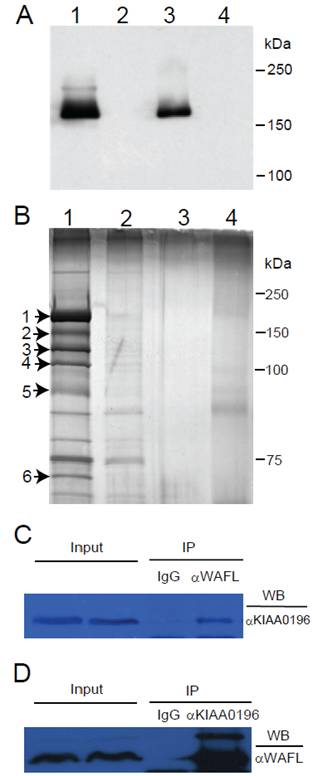
After transfection of an expression plasmid encoding the full-length WAFL with a FLAG tag into HEK-293 cells, the WAFL protein was partially purified by immunoprecipitation with anti-FLAG antibody, and analyzed by SDS-PAGE. When the gel was stained with CBB, a single protein band was observed (data not shown). When analyzed by Western blotting with anti-FLAG antibody, furthermore, a single band was also observed in the immunoprecipitate or total lysate of HEK-293 cells harboring the expression plasmid encoding the WAFL-FLAG fusion protein (Fig. 1A, lanes #1 and #3); the faint band above the main band in lane #1 was also observed as an unknown protein when exposed for a longer period of time. These results indicated that the main band corresponds to the WAFL protein with a FLAG tag. Therefore, we analyzed the single band in the CBB-stained gel by LC-MS/MS after being excised out from the gel. The results confirmed that the band was indeed the WAFL protein (data not shown). When the gel was stained with more sensitive silver staining, many other bands were also detected (Fig. 1B, lane #1). Among these bands, six bands were specifically detected only in the immunoprecipitate from HEK-293 cells expressing the WAFL-FLAG fusion protein, but not in cells untransfected. These six bands were excised out and analyzed by LC-MS/MS. The results are summarized in Table 1.
The band #1 in Fig. 1B showed the highest intensity, and was identified with the highest Mascot score, 1495, as the WAFL protein itself (Fig. 2A). The WAFL protein was found in all the bands #1 through #6, and smaller bands than the band #1 might be cleaved products since their sequence coverage was less than that of the band #1, which covered 35% of the full length by the largest number, 110, of unique peptides. Band #3 contained a hypothetical protein, KIAA1033 (Fig. 2C), and tau tubulin kinase 2 (TTBK2) in addition to WAFL. The KIAA0196 protein (Fig. 2B) was found as a major component of band #4 in addition to WAFL and TTBK2. Polyubiquitin encoded by UBC was also identified in band #6 in addition to WAFL.
(A) Western blot analysis of the WAFL protein fused with a FLAG tag. HEK-293 cells were transfected with an expression plasmid encoding the WAFL-FLAG fusion. The cells were lysed, and then immunoprecipitated with anti-FLAG antibody. Proteins before and after the immuntoprecipitation were resolved by SDS-PAGE. After the electrophoresis, proteins were transferred onto a membrane filter and detected with anti-FLAG antibody. Lane #1: immunoprecipitate from cells transfected; #2: immunoprecipitate from cells untransfected; #3: total proteins from cells transfected; #4: total proteins from cells untransfected. Positions of molecular standards are also shown on the right margin. (B) Proteins co-immunoprecipitated with the WAFL-FLAG fusion protein by using immobilized anti-FLAG antibody. After separating eluates from the resin by SDS-PAGE, proteins were silver-stained. Lane #1: immunopriciptate from cells transfected; #2: immunopriciptate from cells untransfected; #3: immunoprecipitate from lysis buffer (negative control); #4: eluate from anti-FLAG antibody resin used for the immunoprecipitation (negative control). Protein bands on the lane #1 that were not observed in lane #2 are indicated by arrows with numbers on the left margin. Positions of molecular standards are also shown on the right margin. (C) Co-immunoprecipitation (IP) of the KIAA0196 protein with antibody specific for WAFL. Human HCT-116 cell lysate was reacted with antibody specific for WAFL, and the resulting complex was precipitated with protein A-Sepharose. The protein complex was eluted from the Sepharose beads, was resolved by SDS-PAGE, and was analyzed by Western blotting (WB) with antibody specific for KIAA0196. (D) Co-immunoprecipitation of WAFL with antibody specific for the KIAA0196 protein.
Proteins co-immunoprecipitated with WAFL/KIAA0674.
| Band# | Access#a | Identification | Mr (Da) | Score | Uniq. peptide# | Seq. Cover. (%) |
|---|---|---|---|---|---|---|
| 1 | T00363 | WAFL | 135609 | 1495 | 110 | 35 |
| 2 | T00363 | WAFL | 135609 | 1051 | 102 | 29 |
| 3 | T00363 | WAFL | 135609 | 933 | 81 | 22 |
| Q2M389 | KIAA1033 | 137357 | 488 | 46 | 22 | |
| Q8IWY7 | TTBK2 | 183893 | 62 | 7 | 3 | |
| 4 | Q53EL1 | KIAA0196 | 135670 | 1407 | 98 | 36 |
| T00363 | WAFL | 135609 | 175 | 16 | 14 | |
| Q8IWY7 | TTBK2 | 183893 | 50 | 4 | 4 | |
| 5 | T00363 | WAFL | 135609 | 383 | 40 | 17 |
| 6 | T00363 | WAFL | 135609 | 163 | 3 | 5 |
| BAA23486 | Polyubiquitin/UBC | 68448 | 248 | 2 | 4 | |
| Q8IWY7 | TTBK2 | 183893 | 51 | 3 | 4 |
a UniProt accession#
The results shown above indicated that KIAA1033, KIAA0196, polyubiquitin, and TTBK2 interacted with WAFL in HEK-293 cells. Among these proteins, KIAA0196 (also known as strumpellin [23]) had the highest Mascot score, 1407, and 36% of its entire sequence was covered by the largest number, 98, of unique peptides (Table 1). This specific interaction between WAFL and KIAA0196 was also confirmed in human colorectal carcinoma HCT-116 cells under physiological conditions by co-immunoprecipitation with specific antibodies for WAFL and KIAA0196. When HCT-116 cell lysate was incubated with rabbit serum IgG or rabbit anti-WAFL antibody (αWAFL), KIAA0196 was specifically co-immunoprecipitated with WAFL, but not by using the reference serum IgG (Fig. 1C). Similarly, WAFL was specifically co-immunoprecipitated with KIAA0196 by using anti-KIAA0196 antibody (αKIAA0196; Fig. 1D). As observed in Fig. 1A, a minor band with a higher molecular mass than WAFL was again observed when the Western blot was exposed for a longer period of time (Fig. 1D). This may be an alternative form of the WAFL protein that was post-translationally modified since this faint band was detected with two distinct, anti-FLAG and anti-WAFL, antibodies. The KIAA0196 gene was mutated in patients with hereditary spastic paraplegia, a progressive upper-motor neurodegenerative disease [22]. KIAA1033 also had a high Mascot score, 488, and 22% of its entire sequence was covered by a large number, 46, of unique peptides (Table 1).
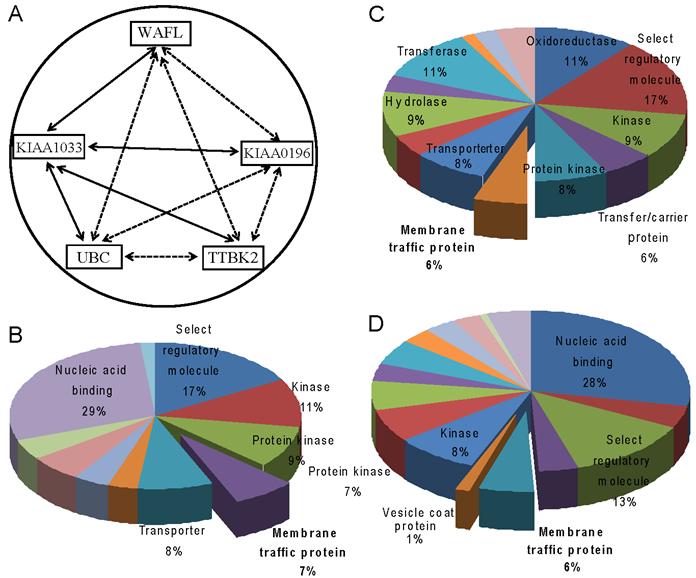
To further understand the relationship between these interacting partners identified by mass spectrometry, we retrieved 20 studies from the Oncomine database [16,17] for each of the interacting partner genes, and analyzed the co-occurrence of the gene with other interacting partner genes. The two-factor analysis showed that WAFL co-occurred most frequently with KIAA1033 (40%); KIAA0196 with KIAA1033 (35%) and UBC (35%); KIAA1033 with UBC (45%); UBC with KIAA1033 (35%); and TTBK2 with KIAA1033 (40%). Results of the three-factor co-occurrence analysis are also consistent with those of the two-factor analysis (data not shown). Fig. 3A summarizes these results by joining with solid lines two genes that showed the highest frequency of co-occurrence among all the combinations, and with dashed lines all the other combinations with less frequent co-occurrences. The significantly frequent co-occurrences of WAFL/FKBP15, KIAA0196, KIAA1033, UBC and TTBK2 are consistent with the results of the study above using mass spectrometry. Among the co-occurrences, KIAA1033 had the highest frequencies of co-occurrences with all the other genes.
We also performed the PANTHER analysis [18] of top 1,000 co-expressed genes with the five genes WAFL, KIAA0196, KIAA1033, UBC and TTBK2 in order to explore their physiological functions by elucidating functional categories in that the genes are commonly expressed. In addition to the categories 'kinase' and 'protein kinase', the category 'membrane traffic protein' as 'molecular function' was commonly found for the three genes WAFL, KIAA0196 and KIAA1033 (Fig. 3B-D). These results suggest that WAFL may be involved in membrane trafficking by interacting with KIAA0196 and KIAA1033. Indeed, Schmid et al [24] reported that KIAA0196 and KIAA1033 are interacting partners of α-appendage, but not β-appendage, of AP2. The AP2 α- and β-appendages act as interaction hubs for accessory proteins which are involved in CCV formation. Thus, these meta-analyses also suggest functional interactions among WAFL, KIAA1033 and KIAA0196, consistent with the results in the present study using co-immunoprecipitation followed by mass spectrometry. Therefore, WAFL may play a role in endocytosis and subsequent membrane trafficking through interaction with KIAA0196, KIAA1033 and other proteins. The WAFL protein consists of at least four domains, WH1 (WASP homology 1; amino-acid residues 69-171), FKBP (aa 178-290), coiled coil (aa ~500-~950) and C-terminal acidic end (aa 1185-1219) [14]. The WH1 domain and the acidic C-terminus are also found in the WASP family of actin polymerization regulators, which activate an actin-related protein (Arp2/3) complex and consequently stimulate actin filament assembly [25]. It seems that WAFL interacts with endosomes via its central coiled-coil domain [15]. Therefore, it is possible that the WAFL coiled-coil domain may bind to KIAA0196 and/or KIAA1033. It is also reasonable that polyubiquitin (band #6 in Fig. 1B) was co-immunoprecipitated with WAFL, since polyubiquitination is often required for endocytosis of cell-surface receptors [26], the proper delivery of proteins into the lumen of late endosomal multivesicular bodies (MVBs), and the formation and scission of cargo-filled intralumenal vesicles [27].
Representative CID fragmentation profiles are given for interacting proteins of WAFL/KIAA0674 identified by LC-MS/MS. (A) CID fragmentation profile of CLALQAQITALTK derived from WAFL/KIAA0674 (T00363). The monoisotopic mass of the neutral peptide is 1429.80 Da. Ions score is 59. b(2)-b(12) and y(2)-y(11) are matched with the peptide shown in the insert. (B) CID fragmentation profile of EEMVLDNIPK derived from KIAA0196 (Q53EL1_HUMAN). The monoisotopic mass of the neutral peptide is 1186.59 Da. Ions score is 39. b(2)-b(9) and y(2)-y(9), except b(7), are matched with the peptide shown in the insert. (C) CID fragmentation profile of FLEEYTSQLR derived from KIAA1033 (Q2M389). The monoisotopic mass of the neutral peptide is 1284.63 Da. Ions score is 44. b(2)-b(9) and y(2)-y(8) are continuously detected and matched with the peptide shown in the insert.
Meta-analysis of WAFL and its interacting proteins. (A) Correlation network of WAFL and its interacting partners. Solid lines indicate the strongest correlation among all the combinations with partner genes, based on co-expression frequencies of corresponding genes in the database, and dashed lines imply statistically significant, but less frequent than that of the strongest, correlations. (B-D) Gene ontology analysis of WAFL and its interacting partners. Top 1,000 genes co-expressed were selected from the PANTHER database, and statistically significantly (p < 0.01) enriched ontology categories were identified as molecular functions. The following three gene sets were analyzed: (B) WAFL, (C) KIAA0196, and (D) KIAA1033. Note that the 'membrane traffic protein' category is commonly enriched.
WAFL has recently been implicated to associate with microtubule and actin cytoskeletons [15]. Indeed, TTBK2, which plays a role in cytoskeleton rearrangement during endocytosis, was also found in the multiple protein bands #3, #4 and #6, although the Mascot score was not as good as other proteins (Table 1). TTBK2 is a member of the casein kinase group of eukaryotic protein kinases, as indicated by its ability to phosphorylate tau and tubulin in vitro [28]. The two TTBK2 phosphorylation sites in tau (Ser208 and Ser210) are priming sites for the phosphorylation of tau by GSK-3b, which is known to influence tau pathology [29]. Mutations of TTKB2 cause spinocerebellarataxia type 11, and affected brain tissues show substantial cerebellar degeneration and tau deposition [30]. Therefore, it is likely that TTBK2 regulates cytoskeleton functions by phosphorylating tau and tubulin. It was also reported that WAFL interacts with actin and WIP [13,15]. In the present mass spectrometry study, however, we failed to detect these interactions. If WAFL specifically interacts with F-actin, not monomeric actin molecules, as shown previously [15], it might be difficult to detect the interaction by using the co-immunoprecipitation approach. The WAFL-WIP interaction was previously detected using transfected cells expressing these proteins from plasmids. If the expression level of endogenous WIP was very low or the affinity between the two proteins was weak, it may also be difficult to detect the interaction.
We have previously found that the gene expression of WAFL was consistently up-regulated in all UC patient biopsies investigated, although individual patient expression levels varied [14]. Furthermore, we have also previously found WAFL to be subject to conditional up-regulation of its mRNA expression in differentiating macrophages. As monocytes differentiated into mature macrophages, interestingly, there was a 2.5-fold induction of WAFL [14]. This suggests that the expression of WAFL is conditionally regulated and becomes elevated as the myeloid cell acquires the characteristics of immuno-competent cells, which are active in endocytosis for processing antigens. However, it is not ruled out that endocytosis is also important in other cells, such as B cells and T cells, involved in gut inflammation.
4. Conclusion
The results in the present study suggest that the WAFL protein, a molecular marker for UC, may function as part of the CCV-mediated endocytosis machinery by interacting with AP2 through KIAA0196 and KIAA1033. The gene expression of WAFL may be up-regulated when monocytes are activated and differentiate into macrophages, which are active in endocytosis. WAFL may also play a role in transport of early endosomes from the cell periphery to the perinuclear region by interacting with actin filaments and microtubules. Further analysis of interacting network of WAFL, KIAA0196 and KIAA1033 should elucidate their physiological and pathological roles in the processes of endocytosis and subsequent endosome trafficking.
Acknowledgements
We thank Low Teck Yew, Liu Jining and Zhao Yan for helpful discussion and technical support for LC-MS/MS and co-immunoprecipitation; and Michael Roy, Ingo Lehman, and members of the laboratory for their critical reading of the manuscript. This work was supported by the Agency for Science, Technology, and Research of the Republic of Singapore, The Swedish Medical Research Council (to SP), and Okinawa Institute of Science and Technology.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Tysk C, Lindberg E, Järnerot G, Flodérus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990-996
2. Thompson NP, Driscoll R, Pounder RE, Wakefield AJ. Genetics versus environment in inflammatory bowel disease: results of a British twin study. B M J. 1996;312:95-96
3. Lapidus A, Bernell O, Hellers G. et al. Incidence of Crohn's disease in Stockholm County 1955-1989. Gut. 1997;41:480-486
4. Orholm M, Binder V, Sørensen TI. et al. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol. 2000;35:1075-1081
5. Orholm M, Munkholm P, Langholz E. et al. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84-88
6. Monsén U, Bernell O, Johansson C, Hellers G. Prevalence of inflammatory bowel disease among relatives of patients with Crohn's disease. Scand J Gastroenterol. 1991;26:302-306
7. Probert CS, Jayanthi V, Hughes AO. et al. Prevalence and family risk of ulcerative colitis and Crohn's disease: an epidemiological study among Europeans and south Asians in Leicestershire. Gut. 1993;34:1547-1551
8. Gaya DR, Russell RK, Nimmo ER, Satsangi J. New genes in inflammatory bowel disease: lessons for complex diseases? Lancet. 2006;367:1271-1284
9. Hugot JP, Chamaillard M, Zouali H. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603
10. Ogura Y, Bonen DK, Inohara N. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603-606
11. Silverberg MS, Cho JH, Rioux JD. et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216-220
12. Kikuno R, Nagase T, Nakayama M. et al. HUGE: a database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE. Nucl Acids Res. 2004;32:D502-D504
13. Nakajima O, Nakamura F, Yamashita N. et al. FKBP133: A novel mouse FK506-binding protein homolog alters growth cone morphology. Biochem Biophys Res Commun. 2006;346:140-149
14. Viklund IM, Kuznetsov NV, Löfberg R. et al. Identification of a new WASP and FKBP-like (WAFL) protein in inflammatory bowel disease: a potential marker gene for ulcerative colitis. Int J Colorectal Dis. 2008;23:921-930
15. Viklund IM, Aspenström P, Meas-Yedid V. et al. WAFL, a new protein involved in regulation of early endocytic transport at the intersection of actin and microtubule dynamics. Exp Cell Res. 2009;315:1040-1052
16. Rhodes DR, Yu J, Shanker K. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1-6
17. Oncomine. http://www.oncomine.org
18. PANTHER. http://www.pantherdb.org
19. Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699-711
20. Brian JW, Vincent G. Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer. 2008;7:1-8
21. Alles MC, Gardiner-Garden M, Nott DJ. et al. Meta-Analysis and gene set enrichment relative to ER status reveal elevated activity of MYC and E2F in the ''basal'' breast cancer subgroup. PLoS ONE. 2009;4:e4710
22. Mi H, Betty L-U, Rozina L. et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucl Acids Res. 2005;31:334-341
23. Valdmanis PN, Meijer IA, Reynolds A. et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet. 2007;80:152-161
24. Schmid EM, Ford MGJ, Burtey A. et al. Role of the AP2 α-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4:1532-1548
25. Millard TH, Sharp SJ, Machesky LM. Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem J. 2004;380:1-17
26. Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: The network at work. Exp Cell Res. 2009;315:1610-1618
27. Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519-547
28. Takahashi M, Tomizawa K, Sato K. et al. A novel tau-tubulin kinase from bovine brain. FEBS Lett. 1995;372:59-64
29. Noble W, Planel E, Zehr C. et al. Inhibition of glycogen synthase kinase-3 by lithium correlates with reduced tauopathy and degeneration in vivo. Proc Natl Acad Sci USA. 2005;102:6990-6995
30. Houlden H, Johnson J, Gardner-Thorpe C. et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet. 2007;39:1434-1436
Author contact
![]() Corresponding author: Dr. Ichiro Maruyama, Okinawa Institute of Science and Technology, 1919-1 Tancha, Onna, Kunigami, Okinawa 904-0412, Japan. Tel: +81-98-966-8496; Fax: +81-98-966-8521. E-mail: ichijp
Corresponding author: Dr. Ichiro Maruyama, Okinawa Institute of Science and Technology, 1919-1 Tancha, Onna, Kunigami, Okinawa 904-0412, Japan. Tel: +81-98-966-8496; Fax: +81-98-966-8521. E-mail: ichijp

 Global reach, higher impact
Global reach, higher impact