Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(3):253-260. doi:10.7150/ijbs.7.253 This issue Cite
Research Paper
Oxidative Stress Can Affect the Gene Silencing Effect of DOTAP Liposome in an In Vitro Translation System
Division of Chemical Engineering, Graduate School of Engineering Science, Osaka University, Osaka, Japan
Received 2011-1-13; Accepted 2011-3-11; Published 2011-3-15
Abstract
Oxidative stress can affect in vitro GFP expression through its control of the gene silencing effect of the liposome prepared by 1,2-dioleoyl-3-trimethyl-ammonium propane (DOTAP). The gene silencing effect of cationic DOTAP liposome in in vitro GFP expression, especially focusing on its translation process, and the effects of oxidative stress on its silencing effect were investigated. GFP expression, initiated by mRNA, was found to be thoroughly inhibited in the presence of DOTAP liposome at concentration of more than 2.5 mM, though its inhibitory effect was reduced in the presence of hydrogen peroxide. The analyses of (i) the interaction of mRNA with DOTAP, (ii) the chemical structure of DOTAP, and (iii) the membrane fluidity of DOTAP liposome imply the possible role of gene expression by the liposome membrane and stress conditions.
Keywords: Oxidative stress, GFP expression, gene silencing, liposome
Introduction
In recent years, attempts have been made to apply gene delivery techniques to various medical and industrial uses (RNA interfere etc.).1 In the conventional gene delivery technique, the nucleic acid is usually delivered inside the cell as a complex with a vector (liposome etc.) through an endocytosis pathway of the biological cell.2 Among the possible gene vectors, DOTAP (1,2-dioleoyl-3-trimethyl-ammonium-propane) is quite popular because of its novel usability.3 The interaction between DOTAP and nucleic acids is very strong in comparison with other lipids, and it can offer high transfection efficiency inside a host cell. However, due to these strong interactions, nucleic acids in the complex sometimes cannot be expressed.2 The effect of the vector on gene repression (silencing) inside a host cell should be minimized in the ideal gene delivery technique.
Oxidative stresses caused by reactive oxygen species (ROS) (e.g., superoxide, hydrogen peroxide, and hydroxyl radical) are inevitable stresses of biological systems on earth. Various diseases (i.e. cancer, Alzheimer's disease) have been reported to be related to oxidative stress through the oxidation of biomolecules such as proteins or lipids.4 It has been reported that the ROS are abundantly generated in the cancer cells as a self-defense in contrast to the normal cell system.5 A novel strategy for a drug delivery system based on the characteristics of cancer cells has also been reported, in which a tandem catalyst of artificial SOD (superoxide dismutase) and natural catalase conjugate has been developed for induction of the apoptosis of cancer cell through the elimination of oxidative stress.6 It seems rational to consider a novel gene delivery system that can be selectively induced and, also, regulated in response to the oxidative stress generated from cancer cells.
Biomembranes are commonly known to act as physical boundaries to separate cellular biomolecules from the environment. The potential functions of the biomembrane, that produce a dynamic changes in the environmental conditions (stresses), has recently been revealed through basic research on a model biomembrane. A model biomembrane, liposome, has previously been reported to induce a variety of potential functions under variable environmental conditions, including a molecular chaperone-like function,7 the translocation of proteins across membranes,8 membrane fusion,9 and LIPOzyme functions10-14 under appropriate stress conditions. In our series of works, it has been previously reported that liposomes can affect the in vitro gene expression system15-20,31 including a series of steps (i.e. (i) transcription (DNA to RNA), (ii) translation (mRNA to polypeptide), and (iii) folding (conformational change of polypeptide), herewith defined as “Biomembrane Interference.” Especially in the case of oxidative stress, it has also been reported that variations in the characteristics of the liposome membrane can occur through the oxidation of the unsaturated part inside a phospholipid bilayer by a reactive oxygen species.21 It is expected that the gene expression affected by the membrane (or liposome) could also be affected by oxidative stress.
The final emphasis of this study was an attempt to achieve On-Off control of in vitro gene expression based on “Biomembrane Interference” phenomena (Supplementary Material: Fig.S1). As the first step for the final goal, the possibility of regulating gene expression by oxidative stress was investigated, especially focusing on the “translation” of GFP mRNA, in this study through the investigation of the “gene silencing effect” of DOTAP liposome in in vitro gene expression of GFP.
Experimental Materials
1,2-Dioleoyl-3-trimethyl-ammonium-propane (DOTAP) and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) were purchased from Avanti Polar Lipids (Alabaster, AL). A Rapid Translation System RTS 100 E. coli HY Kit (RTS-Kit) was purchased from Roche Diagnostics (Indianapolis, IN). The SV Total RNA Isolation System was purchased from Promega (Madison, WI). Other chemicals of commercially guaranteed reagent grade were purchased from Wako Pure Chemicals (Osaka, Japan) and used without further purification.
Liposome Preparation
A solution of DOTAP and DOPC in chloroform was dried in a round-bottom flask by rotary evaporation under reduced pressure. The lipid films obtained were dissolved in chloroform twice, and the solvent was evaporated. The lipid film was kept under a high vacuum for at least 3 hours, and then hydrated with distilled water at room temperature to form multilamellar vesicles (MLVs). The vesicle suspension was frozen at -80 oC and thawed in a water bath at 50 oC to enhance the transformation of small MLVs into larger ones (LUVs). This freeze-thaw cycle was performed five times. MLVs were used to prepare small unilamellar vesicles (SUVs) by extruding the MLV suspension 11 times through two layers of polycarbonate membranes with mean pore diameters of 100 nm using an extruding device (Liposofast; Avestin Inc., Ottawa, Canada). Final liposome concentration is 20 mM. The liposome size was set at 100 nm in all the experiment based on the results of the preliminary experiments which performed at different sizes (50-200nm).
In vitro Expression of Green Fluorescent Protein (GFP) with Liposome
The Rapid Translation System (RTS-Kit), an in vitro translation system based on E. coli lysate, was used for cell-free protein synthesis. The pIVEX control vector GFP, containing the GFP gene, was used as an expression vector. The reaction solution (9.6 μl) contained the following components: 2 μl E.coli lysate, 2 μl reaction mix, 2.4 μl amino acid solution, 0.2 μl methionine solution, 1 μl reconstitution buffer, and recovery mRNA in 2.5 μl of nuclease-free water. The reaction was initiated by mixing the reaction solutions with or without liposome (final lipid concentration is 0 - 4 mM) at 30 oC for 6 hours and was kept at 4 oC for 24 hours. The amount of GFP synthesized using an RTS-Kit was evaluated from the fluorescence of GFP (Ex = 395 nm, Em = 509 nm). The total GFP expression in the presence or absence of liposomes was evaluated based on the relative GFP fluorescence intensity compared with control (without liposome), using a fluorescence spectrophotometer (FP-6500; JASCO, Tokyo, Japan) with a slit width of 5 nm at excitation and emission of the light path.
Evaluation of Membrane Fluidity of Liposome
To measure the outer membrane fluidity, a fluorescent probe, 1,3,5-hexatriene (DPH), was added to the liposome solution (molar ratio; liposome/DPH 250/1). The fluorescence polarization of DPH (Ex=360 nm, Em=430 nm) was measured after incubation at 30 °C for an hour using a fluorescence spectrophotometer (FP-6500; JASCO, Tokyo, Japan). After the sample was excited with vertically polarized light (360 nm), the emission intensities (430 nm) both parallel (I//) and perpendicular (I⊥) to excited light were recorded. The polarization of DPH was then calculated based on the following equation (P).
P = (I// - I⊥) / (I// + I⊥)
The membrane fluidity was evaluated based on the reciprocal of polarization (1/P).
UV and Mass Spectra Analysis of Oxidized DOTAP
The structural change of oxidative DOTAP was investigated by UV and Mass analysis. DOTAP was treated for 1 hour in 5 mM H2O2 solution. After the oxidized DOTAP was extracted by chloroform/water system, it was recovered by evaporation under a vacuum. The ethanol solution of the oxidized DOTAP was analyzed using a UV spectrophotometer (UV-1800; SHIMADZU, Kyoto, Japan) and mass spectra. (JEOL JMS-700)
Transcription and purification of GFP mRNA
As the plasmid DNA, pIVEX control vector GFP (Roche) was used. The plasmid DNA was once treated with restriction enzyme ApaL I for 1 h incubation at 37 oC, in order to cleave the AmpR gene so as to obtain line DNA fragments harboring the GFP gene before transcription. The transcription of mRNA encoding GFP gene (861 bp) was carried out using T7 RiboMAXTM Expression Large Scale RNA Production System (Promega, Madison, WI, USA), withch includes T7 RNA polymerase as a transcriptional enzyme. Transcription was achieved for 30 min at 37 oC. The obtained mRNA was recovered and purified with the SV total RNA Isolation Kit (Promega, Madison, WI, USA). The mRNA products were quantified from the absorbance at 260 nm and electrophoresis on 1% agarose gel.
Results and Discussion
Effects of Oxidative Stress on in vitro GFP Expression with DOTAP Liposome
The effects of the addition of DOTAP liposome on the in vitro gene expression of GFP especially translation process were first investigated. After the mRNA was transcribed from GFP gene on plasmid DNA, it was recovered and used for the GFP expression in an in vitro gene expression system according to the previous method.16,17,31 Figure 1(a) shows the dependence of GFP expression on the concentrations of DOTAP liposome. The amount of GFP expression was reduced with increases in the DOTAP concentrations and was completely inhibited at DOTAP concentrations of more than 2.5 mM. There have been some previous reports of the inhibitory role of cationic DOTAP liposome on the gene expression of GFP in an E.coli cell-free translation system.31,16,17,18 It has been reported that the DOTAP liposome can neutralize the negatively-charged mRNA through strong electrostatic interactions between the cationic quaternary ammonium groups on liposome and the negatively-charged phosphate groups of mRNA, resulting in a silencing of translation.16,17 It has also been reported that, due to this electrostatic interaction, the conformational change of RNAs occurs on the DOTAP liposome, resulting in an inhibitory output of GFP expression.16-18 It is considered that the above results regarding the GFP silencing could be induced by the entrapment of the mRNA molecule by DOTAP liposome through their electrostatic interaction.
Effects of (a) DOTAP concentration, (b) H2O2 concentration, and (c) H2O2 with 2.5 mM DOTAP on in vitro GFP Expression Translated from Recovered mRNA(n=3).
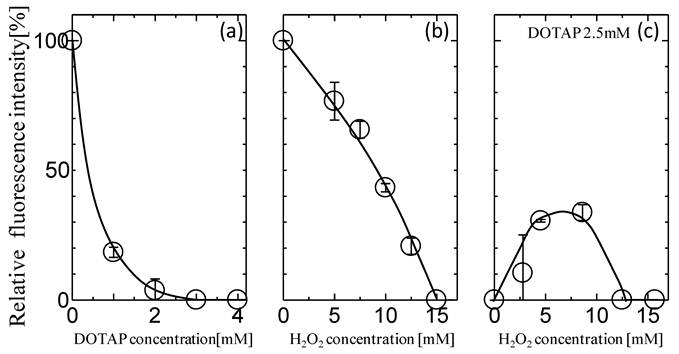
Oxidative stress has been reported to play a role in maintaining the homeostasis of biological systems.22 It has been reported that protein expression can also be modulated in a biological cell under the oxidative stress condition.22,23 Figure 1(b) shows the typical behaviors of GFP expression as a function of H2O2 concentration. The GFP expression decreased with increases in the H2O2 concentrations and its value became zero at more than 10 mM. This decrease could be caused by the oxidation of various biomolecules at high H2O2 Concentrations. It has been reported that the concentration level of H2O2 in the biological cell is a few mM at maximum4,5, implying the possibility of the partial expression of GFP in the biological system. In order to investigate the effects of oxidative stress on the GFP expression with DOTAP, both H2O2 and DOTAP liposome were added together into the in vitro translation system by setting the DOTAP concentration at 2.5 mM, which completely inhibited the GFP expression. It is worth noting that the GFP expression was observed at a specific concentration of H2O2. At higher H2O2 concentrations (more than 15 mM), the GFP was not expressed, probably, due to the oxidation of molecules occurring at high H2O2 concentrations. On the other hand, at lower H2O2 concentrations (less than 10 mM), GFP expression was observed and its value reached a plateau of 30%. Although both DOTAP and H2O2 were shown to inhibit GFP expression (Fig.1(a) and (b)), GFP was found to be expressed in the presence of both. Although the DOTAP liposome plays a role in the gene silencing effect in in vitro protein expression systems, the oxidative stress was shown to reduce the gene silencing effect of the DOTAP liposome.
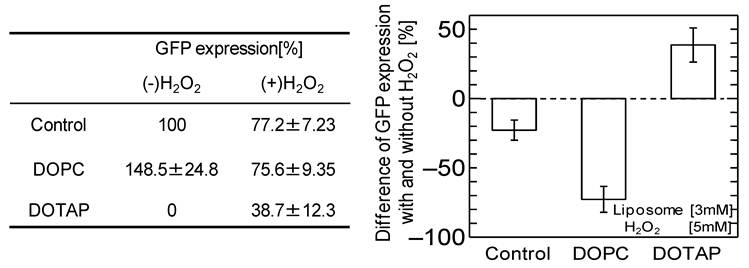
It has been reported that adding neutral liposome can enhance GFP expression by more than 1.2-1.7 times. 19,31 The effects of the addition of the DOPC liposome, harboring the same acyl-chains and a neutral head group, on the GFP expression were investigated with and without oxidative stress, as shown in Fig.2. Without oxidative stress, the GFP expression was enhanced by the addition of neutral DOPC liposome, similar to the previously-reported case. However, the GFP expression was inhibited by adding the H2O2. The above results imply that oxidative stress negatively affects not only in vitro cell free systems but also that with neutral liposome, resulting in the down-regulation of GFP expression. The results also indicate that oxidative stress varies the effects of cationic DOTAP liposome and neutral DOPC liposome on in vitro GFP expression. In the case of cationic DOTAP liposome, it is expected that the oxidative stress could be a key factor in regulating in vitro gene expression through the weakened interaction between the liposome and mRNA.
Percentage of GFP expression decreases by oxidation stress addition with or without DOPC liposome and DOTAP liposome. [Liposome 3 mM, H2O2 5 mM]
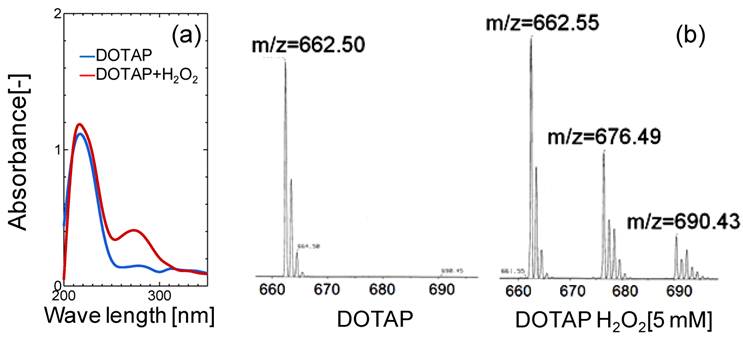
(a) UV and (b) Mass Spectra Analyses of DOTAP Molecule before and after H2O2 treatment.
Oxidative Stress Can Vary the Interaction of DOTAP Liposomes and mRNA
The interactive behaviors of DOTAP liposome and mRNA were further studied in order to investigate a possible mechanism of the above phenomena on the oxidative stress-induced regulation of GFP expression. The mRNA/DOTAP liposome solution was analyzed by using agarose gel electrophoresis (Supplementary Material: Fig.S2). The DOTAP liposome and mRNA at various concentration ratios were mixed and incubated for one hour at 30 oC. The bands at 861 bp and 1722 bp represent free mRNA and its dimer, respectively, indicating mRNA was free from DOTAP liposome. The amount of free mRNA was reduced with an increase in the DOTAP concentrations since mRNA that formed complexes with DOTAP was neutralized and was not able to move during the electrophoresis. Especially at higher concentrations (more than 2 mM), the band for free mRNA was found to disappear. The relative concentrations of the mRNA bands in electrophoresis were determined by using densitometer analysis, and the adsorbed amount of mRNA on the DOTAP liposome was calculated (Supplementary Material: Fig.S2) at various ratios of mRNA/DOTAP. Considering the balance of the total negative charge of the mRNA and the total positive charge of DOTAP, mRNA can interact with the DOTAP liposome with more than equivalent molar ratios of the above charged groups. It has been reported that mRNA and DOTAP cannot interact because DOTAP has enough area to interact with mRNA.24 It is considered that some parts of an mRNA molecule could interact with the positively-charged surface of the DOTAP liposome. The mRNA structure with DOTAP liposome was analyzed by using CD spectra and FT-IR to investigate the interaction between mRNA and DOTAP liposome, resulting that the conformational change of mRNA occurred followed by the interaction between DOTAP liposome and nucleobases of mRNA (data not shown).
In order to evaluate the lipid oxidation, the UV and mass spectra of DOTAP molecules were measured before and after the oxidation treatment of DOTAP by 5 mM H2O2. As shown in Fig.3(a), a new peak in the UV spectra was observed at 275 nm after the H2O2 treatment, indicating that the DOTAP molecule was oxidized and its structure was changed. It is known that the band at 275 nm originated from the keto-group or aldehyde group.25 In order to examine the chemical structure of oxidized DOTAP in more detail, the mass spectra of the DOTAP were analyzed before and after oxidation (Fig.3(b)). After H2O2-oxidation, two m/z peaks were newly observed in addition to that of normal DOTAP, showing that some oxidized DOTAP species were generated. Based on the above results, possible molecular structures of DOTAP after the oxidation are shown in Supplementary Material: Fig.S3. Oxidized DOTAP includes carboxyl or aldehyde groups, although it is uncertain where the carbonyl oxygen is added.
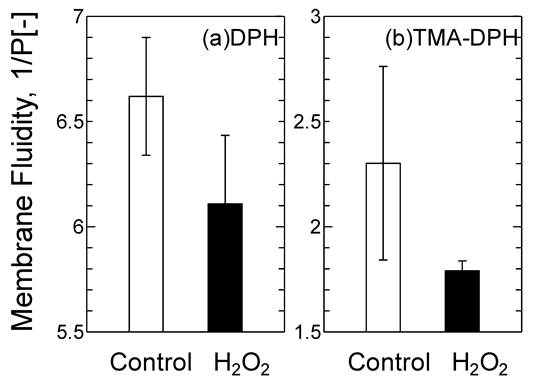
The oxidized DOTAP molecule may cause drastic changes in the surface properties of the DOTAP liposome as a “molecular assembly”. The membrane fluidity of DOTAP liposome was investigated by using DPH and TMA-DPH as molecular probes of the interior and interface of the membrane, respectively (Fig.4). Membrane fluidity has been reported to correspond with the hydrophobicity of lipid membranes and is one of the essential parameters to show the dynamic interaction between mRNA and DOTAP liposome. The membrane fluidities of both the interior and the interface of DOTAP liposome were found to be reduced in response to H2O2 addition. It was reported that the membrane fluidity decreased when the oxidized lipid contents increased.26 The interaction between proteins and liposome has been reported to be correlated with the membrane fluidity, which also corresponds to the hydrophobicity.26 It has recently been reported in our series of papers that the peptide fragment of the SOD can be recruited on the liposome surface through the combined effects of the electrostatic interaction, hydrophobic interaction, and hydrogen bond. 19,27,31 It is considered that the interaction between DOTAP liposome and mRNA could similarly be governed, first by the electrostatic interaction (surface charge), secondly by the hydrophobic interaction (membrane fluidity), and finally by the hydrogen bond stabilization (i.e. interaction of the side residue of the RNA and the oxidized group in the acyl chain). The decrease in membrane fluidity of the DOTAP liposome could be a key to weaken the above combined interaction of the mRNA with DOTAP liposome.
In conclusion, although DOTAP liposome inhibited in vitro GFP expression though its “gene silencing effect,” oxidative stress was found to reduce its effect. The above Off-On effect of DOTAP liposome and oxidative stress could be investigated based on DOTAP liposome's recognition of the GFP mRNA on its surface. Based on phenomena related to the up-regulation of target product under stress conditions in vivo16, 17, 18, 19, 20 and in vitro,28, 29, 30 gene expression can usually be achieved by modulation of the possible recognition of biomolecules by the “(stressed) biomembrane” as described in our series of reports. If the DOTAP liposome-mRNA complex approached to the cancer cell generating oxidative stress, the target gene will be expressed through the regulation of the liposome mRNA interaction. After the mRNA/DOTAP liposome could be delivered onto the cell surface or into the cytoplasm of the cell, the response of mRNA- liposome complex could happen on the surface of biomembrane of the target cancer cell or at the inside of the cell. Although further investigation is needed, we now show a possibility of a new conceptual gene delivery system considering the characteristic of the liposome membrane through the careful discussion. It is expecting that the above phenomena could be utilized for “biomembrane”-based gene regulation although further investigation is needed.
Variation of the Membrane Fluidity of DOTAP liposome before and after H2O2 [5 mM] Treatment.
Supplementary Material
Fig.S1 - Conceptual illustration of the present study. Fig.S2 - Adsorption of mRNA on DOTAP liposome. Fig.S3 - Expected chemical Structure of oxidized DOTAP.
Acknowledgements
The fundamental concept was supported by the Research Group of “Membrane Stress Biotechnology.” The study was supported by a Funding Program for Next Generation World-Leading Researchers of the Council for Science and Technology Policy (CSTP)(No.GR0660), a Grant-in-Aid for Scientific Research (No. 19656203, 19656220, and 20360350) from the MEXT of Japan, a grant from the 21COE and GCOE programs (Osaka Univ.) of JSPS, and a grant from MRDL (Grad. Sch. of Eng. Sci., Osaka Univ.). The authors are grateful to the Research Center for Solar Energy Chemistry and the Gas hydrate Analyzing System of Osaka University.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Hannon G J. RNA interference. Nature. 2002;418:244-251
2. Yuhoung X, Szoka F C. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochem. 1996;35:5616-5623
3. Templeton N S, Lasic D D, Frederik P M, Strey H H, Roberts D D, Pavlakis G N. Improved DNA: Liposome complexes for increased systemic delivery and gene expression. Nature Biotech. 1997;15:647-652
4. Stadtman E R, Berlett B S. Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metabol. Rev. 1998;30:225-243
5. Behrend L, Henderson G, Zwacka R M. Reactive oxygen species in oncogenic transformation. Mol. Mech. Signal. 2003;31:1441-1444
6. Naruta Y, Sasayama M, Ichikawa K. Functional modeling of manganese-containing O2 evolution enzymes with manganese porphyrin dimmers. J. Mol. Catalysis A:. 1997;117:115-121
7. Kuboi R, Yoshimoto M, Walde P, Luisi P L. Refolding of carbonic anhydrase assisted by 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine liposomes. Biotech. Progress. 1997;13:828-836
8. Umakoshi H, Yoshimoto M, Shimanouchi T, Kuboi R, Komasawa I. Model system for heat-induced translocation of cytoplasmic β-galactosidase across phospholipid bilayer membrane. Biotech. Progress. 1998;4:218-226
9. Yoshimoto M, Walde P, Umakoshi H, Kuboi R. Conformationally changed cytochrome c-mediated fusion of enzyme- and substrate-containing liposomes. Biotech. Progress. 1999;15:689-696
10. Tuan LQ, Umakoshi H, Shimanouchi T, Kuboi R. Liposome-recruited activity of oxidized and fragmented superoxide dismutase. Langmuir. 2008;24:350-354
11. Tuan L Q, Umakoshi H, Shimanouchi T, Kuboi R. Liposome membrane can act like molecular and metal chaperones for oxidized and fragmented superoxide dismutase. Enzyme Microb. Tech. 2009;44:101-106
12. Umakoshi H, Morimoto K, Ohama Y, Nagami H, Shimanouchi T, Kuboi R. Liposome modified with Mn-porphyrin complex can simultaneously induce antioxidative enzyme-like activity of both superoxide dismutase and peroxidase. Langmuir. 2008;24:4451-4455
13. Umakoshi H, Tuan L Q, Shimanouchi T, Kuboi R. Role of liposome on recognition and folding of oxidized and fragmented superoxide dismutase for its re-activation. Biochem. Eng. J. 2009;46:313-319
14. Umakoshi H, Morimoto K, Yasuda N, Shimanouchi T, Kuboi R. Development of liposome-based mimics of superoxide dismutase and peroxidase based on the "LIPOzyme" concept. J. Biotech. 2010;147:59-63
15. Bui H T, Umakoshi H, Suga K, Nishida M, Shimanouchi T, Kuboi R. Negatively charged liposome as a potent inhibitor of post-translation during in vitro synthesis of green fluorescent protein. Biochem. Eng. J. 2009;46:154-160
16. Bui H T, Umakoshi H, Suga K, Tanabe T, Shimanouchi T, Kuboi R. Cationic liposome inhibits gene expression in E.coli cell-free translation system. Membrane. 2009;34:146-151
17. Bui H T, Umakoshi H, Suga K, Tanabe T, Ngo K X, Shimanouchi T, Kuboi R. Cationic liposome can interfere mRNA translation in an E.coli cell-free translation system. Biochem. Eng. J. 2010;52:38-43
18. Suga K, Umakoshi H, Tomita H, Tanabe T, Shimanouchi T, Kuboi R. Liposomes destabilize tRNA during heat stress. Biotech. J. 2010;5:526-529
19. Umakoshi H, Suga K, Bui H T, Nishida M, Shimanouchi T, Kuboi R. Charged liposome affects the translation and folding steps of in vitro expression of green fluorescent protein. J. Biosci. Bioeng. 2009;108:450-454
20. Umakoshi H, Nishida M, Suga K, Ishii H, Bui H T. Characterization of green fluorescent protein using aqueous two-phase systems. Solv. Extr. Res. Dev. Japan. 2009;16:145-150
21. Gilbert O, Fruhwirth A, Hermetter L A. Oxidized phospholipids: From molecular properties to disease. Biochim. Biophys. Acta. 2007;1772:718-736
22. Wulf D. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47-95
23. Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nature Immun. 2002;3:1129-1134
24. Cristina M, Maura M, Augusto A, Daniela P, Ruggero C, Giulio C. Surface area of lipid membranes regulates the DNA-binding capacity of cationic liposomes. Phys. Letters. 2009;94:33903-33906
25. Braude BA. Ultraviolet and visible light absorption. In: (ed.) Braude B A, Nachod FC. Determination of organic structures by physical methods. New York: Academic Press. 1955:131
26. Jan W, Nina V V, Olga K, Antonie J W G V. Oxidation of unsaturated phospholipids in membrane bilayer mixtures is accompanied by membrane fluidity changes. Biochim. Biophys. Acta. 2000;1487:61-73
27. Kuboi R, Umakoshi H. Analysis and separation of amyloid β-peptides using aqueous two-phase systems under stress conditions - From aqueous two-phase system to liposome membrane system. Solv. Extr. Res. Dev. Japan. 2006;13:9-21
28. Ngo K X, Umakoshi H, Shimanouchi T, Bui H T, Kuboi R. Enhanced release of chitosanase from Streptomyces griseus through direct interaction of liposome with cell membrane under heat stress. J. Biosci. Bioeng. 2008;106:602-605
29. Ngo K X, Umakoshi H, Sugaya H, Shimanouchi T, Kuboi R. Chitosanase displayed on liposome can increase its activity and stability. J. Biotechnol. 2010;146:105-113
30. Umakoshi H, Fukuta Y, Kuboi R. Utilization of cell response under heat, chemical, and combined stresses for selective recovery of cytoplasmic β-galactosidase from Escherichia coli cells. Biotechnol. Prog. 1998;14:909-912
31. Bui H T, Umakoshi H, Ngo K X, Nishida M, Shimanouchi T, Kuboi R. Liposome membrane itself can affect gene expression in the Escherichia coli cell-free translation system. Langmuir. 2008;24:10537-10542
Author contact
![]() Corresponding author: 1-3 Machikaneyama-cho, Toyonaka, Osaka 560-8531, Japan. E-mail: umakoshies.osaka-u.ac.jp; Tel: +81-(0)6-6850-6287; Fax: +81-(0)6-6850-6286
Corresponding author: 1-3 Machikaneyama-cho, Toyonaka, Osaka 560-8531, Japan. E-mail: umakoshies.osaka-u.ac.jp; Tel: +81-(0)6-6850-6287; Fax: +81-(0)6-6850-6286

 Global reach, higher impact
Global reach, higher impact