Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(7):978-991. doi:10.7150/ijbs.7.978 This issue Cite
Research Paper
PTPBR7 Binding Proteins in Myelinating Neurons of the Mouse Brain
1. Department of Cell Biology, Nijmegen Centre for Molecular Life Sciences, Radboud University Nijmegen Medical Centre, Geert Grooteplein 28, 6525 GA Nijmegen, The Netherlands
2. Department of Biochemistry and Microbiology, Ghent University, K.L. Ledeganckstraat 35, 9000 Gent, Belgium
3. Department of Biochemistry, Nijmegen Centre for Molecular Life Sciences, Radboud University Nijmegen Medical Centre, Geert Grooteplein 28, 6525 GA Nijmegen, The Netherlands
4. Neural Development Unit, UCL, Institute of Child Health, 30 Guildford Street, London, UK
Received 2011-4-18; Accepted 2011-7-21; Published 2011-8-9
Abstract
Mouse protein tyrosine phosphatase PTPBR7 is a receptor-like, transmembrane protein that is localized on the surface of neuronal cells. Its protein phosphatase activity is reduced upon multimerization, and PTPBR7-deficient mice display motor coordination defects. Extracellular molecules that may influence PTPBR7 activity, however, remain to be determined. We here show that the PTPBR7 extracellular domain binds to highly myelinated regions in mouse brain, in particular the white matter tracks in cerebellum. PTPBR7 deficiency does not alter this binding pattern, as witnessed by RAP in situ staining of Ptprr-/- mouse brain sections. Additional in situ and in vitro experiments also suggest that sugar moieties of heparan sulphate and chondroitin sulphate glycosaminoglycans are not critical for PTPBR7 binding. Candidate binding proteins were affinity-purified exploiting the PTPBR7 extracellular domain and identified by mass spectrometric means. Results support the suggested link between PTPRR isoforms and cerebellar calcium ion homeostasis, and suggest an additional role in the process of cell-cell adhesion.
Keywords: cerebellum, RAP in situ, extracellular domain, ligand protein, PTPRR, protein tyrosine phosphatase, mass spectrometry.
Introduction
Molecular and cellular mechanisms that underlie the development and function of the vertebrate nervous system include signalling events by cell adhesion molecules (CAMs) and Receptor Tyrosine Kinases [1, 2]. Receptor Protein Tyrosine Phosphatases (RPTPs) regulate phosphotyrosine signalling by counteracting the tyrosine kinases [3]. RPTPs show strong developmental expression in the central (CNS) and peripheral nervous system, coinciding with significant events such as axogenesis, target contact, synaptogenesis and plasticity [4, 5]. The structural resemblance of many RPTPs to neural CAMs (for example Immunoglobulin-like domains and Fibronectin type III domains) has made them candidate receptors for contact-mediated cues encountered by growth cones. Indeed in vivo roles for RPTPs in neural development and function have been shown [3, 4].
Although numerous downstream effectors of RPTP signalling have been identified, the ligands have been discovered for only a small number of RPTPs [6]. The CAM-like subfamily members RPTPμ, RPTPκ, RPTPλ and RPTPδ all display homophilic interactions that are important in cell adhesion processes [6, 7]. Although RPTPσ and PTP-LAR have an extracellular segment that is similar to these homophilic RPTPs, they rather bind to multiple other extracellular proteins. RPTPδ, RPTPσ and PTP-LAR all three bind netrin-G ligand-3 (NGL-3) [8]. RPTPσ binds nucleolin, alpha latroxin, contactin, the heparan sulphate (HS) proteoglycans agrin and collagen ΧVІІІ, and chondroitin-sulphate (CS) produced by astroglia [9-13]. A splice form of human PTP-LAR binds the laminin-nidogen complex, a major component of the extracellular matrix that modulates neurite outgrowth, proliferation and differentiation [14]. The Drosophila homolog DLAR influences the development of synapses through the binding of two HS proteoglycans, syndecan and dallylike, that have positive and negative effects, respectively, on its PTP activity [7]. The homologous RPTPγ and RPTPζ each bind to distinct contactin family members [15]. In addition, RPTPζ can interact with tenascin and pleiothropin but, intriguingly, only for the latter an effect on activity was reported [16]. Pleiothropin binding resulted in RPTPζ dimerisation and inactivation, as had been found for several other RPTPs upon the artificial induction of dimers [6].
PTPBR7, a receptor-type isoform that is encoded by the mouse Ptprr gene, appears on the cell surface as a homomultimeric protein that displays a much reduced phosphatase activity when compared to the monomeric Ptprr-encoded, cytosolic variants [17]. Prenatally, PTPBR7 is expressed in spinal ganglia and in Purkinje precursor cells within the developing cerebellum. After birth, PTPBR7 transcripts gradually decrease in the maturing Purkinje cells (PCs) and remain expressed throughout all other brain regions [18], suggestive of a neurodevelopmental role. Unexpectedly, Ptprr knockout mice did not display overt brain malformations but rather performed quite poorly in various locomotive tests [19], reminiscent of findings in ataxic animal models with deficits in cerebellar calcium ion homeostasis [20].
Here we report on the ligand binding potential of the PTPBR7 extracellular segment, which does not contain any known CAM-like or protein interaction motifs. Using Receptor Alkaline Phosphatase in situ (RAP in situ) methodology [21, 22] we demonstrate PTPBR7 ectodomain binding to myelinated regions in mouse brain. Affinity purification and identification of associating proteins supports a role for PTPBR7 in cell-cell adhesion and Ca2+-regulated processes in the mouse brain.
Materials and methods
RT-PCR and cloning
C57BL/6 mouse brain RNA preparations were subjected to RT-PCR to obtain cDNA fragments encoding the extracellular domain of PTPBR7, including its signal peptide. Approximately 5 µg of total RNA was reverse-transcribed by random hexamers priming and the SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA). Resulting cDNA was subsequently used in a standard PCR reaction with primers bearing restriction sites for EcoRI or KpnI to facilitate cloning. Primer sequences were: BR7ecto forward, 5'- ATGAATTCGCGGCCGCCACCATGAGGAGAGCGGTCGGC - 3'; BR7ecto226 reverse, 5'-AGGGTACCTCCTTCTTTGCTCCAGATC-3'; BR7ecto225 reverse, 5'- AGGGTACCTTCTTTGCTCCAGATCTTG-3'; BR7ecto224 reverse, 5'- AGGGTACCTTTGCTCCAGATCTTGTC-3'; BR7ecto223 reverse, 5'- AGGGTACCGCTCCAGATCTTGTCTGC-3'; BE7ecto222 reverse, 5'- AGGGTACCCCAGATCTTGTCTGCTTC-3'; BR7ecto221 reverse, 5'- AGGGTACCGATCTTGTCTGCTTCGTG-3'; BR7ecto220 reverse, 5'- TGGGTACCCTTGTCTGCTTCGTGCTG-3'.
The mammalian expression plasmid pBG-flexilinker was generated from pBG [11] by inserting a linker encoding GGGGSGGGGSP upstream of the PLAP open reading frame (Supplementary Material: Fig. S1). pBG-flexilinker was linearized using HindIII and ClaI in order to introduce an adapter sequence containing EcoRI and KpnI sites (a heteroduplex of oligonucleotides 5'- AGCTGAATTCTTAAGGTACCT-3' and 5'- CGAGGTACCTTAAGAATTC-3'). The mammalian expression plasmid pHLsec [23] and a version containing the extracellular domain of RPTPµ (eRPTPµ) [24] were kindly provided by Dr. Aricescu. The unique EcoRI and KpnI restriction sites allowed insertion of the BR7ecto226 PCR product into empty pHLsec to generate plasmid pHLsec-BR7ecto226-His.
Antibodies, Glycosaminoglycans (GAGs) and GAGs hydrolytic enzymes
Rabbit polyclonal antiserum α-BR7 directed towards the extracellular domain of PTPBR7, anti-VSV-G tag monoclonal antibody P5D4, and single chain variable fragment antibodies to HS (HS4C3), CS-A and -C (IO3H10), and CS-E (GD3G7) are described elsewhere [25-29]. Monoclonal antibody 11-5B to CNPase, goat anti-mouse Alexa-Fluor488 antibody, and rabbit anti-mouse alkaline phosphatase-conjugated antibody were from Dakopatts (Glostrup, Denmark), Millipore (Billerica, MA) and Invitrogen, respectively. Heparanase III (Flavobacterium heparinum) was from Ibex Technologies Inc. (Quebec). Bovine kidney HS, shark cartilage CS-A and CS-C and chondroitinase ABC (Proteus vulgaris) were from Sigma-Aldrich (St. Louis, MO). Shark cartilage CS-D and squid cartilage CS-E were from Seikagaku (Tokyo, Japan).
Cell culture and transient transfection
HEK293T cells (ATCC nr. CRL-11268) were cultured in humidified chambers at 37°C and 5% CO2 using Dulbecco's Modified Eagle's Medium (DMEM) reconstituted with pyruvate and glutamine and supplemented with 10% foetal calf serum. When cells in T75 cm2 flasks reached 60-70% confluency, they were transfected using the DEAE-dextran method [26] and cultured in DMEM, 10% foetal calf serum. After 48 hours conditioned culture media were collected, sterile-filtered and buffered with 20 mM Hepes (pH 7.4) [30]. Alkaline phosphatase (AP) activity in the medium was determined using pNPP (Eastman Kodak, Rochester, NY) [30].
Brain tissue sections and lysates
Wild type and Ptprr knockout mice of 9 - 12 months of age were anesthetized and perfused with PBS. Whole brains were extracted, snap frozen in liquid nitrogen-cold isopentane, and stored at -80°C or used for sagittal and coronal cryosectioning [30]. The 10 μm cryosections were dried under air flow at RT.
To prepare brain lysates, frozen brains were thawed and homogenized at 4°C in buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1mM PMSF, 10 mM NaF, 1 mM Na3VO4 and protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany).
Receptor alkaline phosphatase (RAP) in situ staining
For RAP in situ applications [30] cryosections were thawed at RT and processed using published methods [31]. The conditioned media containing the various AP-fused PTPBR7 extracellular domains were used as probe and PLAP-containing medium served as background control. Each assay employed 200 µl/section of conditioned culture medium containing the same AP activity. For some applications conditioned media were first concentrated using Vivaspin 15 membranes (Sartorius Stedim Biotech GmbH, Germany). The staining was at RT overnight using NBT-BCIP (Sigma-Aldrich, St. Louis, MO) as substrate [30].
After the RAP in situ procedure sections were briefly washed in milliQ, embedded in a water-soluble embedding and examined on a Leica DM LB microscope with HC PL Fluotar objective (Leica Microsystem GmbH, Germany). In each experiment minimally three sections per condition were analyzed. Per section three digital images of cerebellar white matter were taken and for each the mean gray value in three representative areas was determined using ImageJ [32]. Statistical analyses involved Student's t-test.
For some experiments tissue sections received a pre-treatment with chondroitinase, involving an overnight incubation at RT in 1 IU/ml chondroitinase ABC digestion mix (Sigma-Aldrich) or in chondroitinase buffer alone (50 mM Tris-HCl pH 8.0, 60 mM sodium acetate, 0.02% BSA) as control. Activity of the enzyme under these conditions was assessed independently in a GAG ELISA (see below).
Immunohistochemistry and immunoblotting
Cryosections were briefly rinsed at RT in TBS and fixed for 10 minutes in acetone at -20°C. After 2 washes, endogenous mouse immunoglobulins were blocked using the VECTOR M.O.M Immunodetection Kit (VECTOR Laboratories, Inc. Burlingame, CA). After blocking, sections were washed twice in TBS and pre-incubated 10 minutes in primary antibody M.O.M. Diluent in TBS supplemented with 2% Goat Serum. Sections were then incubated for 2 hours at RT with anti CNPase (1:100 dilution), washed twice in TBS and incubated for 1 hour at RT with goat-anti-mouse-conjugated Alexa-Fluor488 (1:500 dilution). Finally, sections were washed twice in TBS, embedded in Mowiol (Sigma-Aldrich) and analyzed by fluorescence microscopy (Leica DMRA).
For immunoblotting, conditioned medium was mixed with sample buffer, subjected to SDS-PAGE and blotted onto PVDF membranes by electro-transfer. Membranes were incubated with rabbit polyclonal antiserum α-BR7 (1:3000 dilution) in TBS containing 0.5% Tween 20 and 3% BSA, followed by Alexa-Fluor680- conjugated goat-anti-rabbit antibody (1:10000 dilution) in the same buffer. Immunoreactive bands were visualized on an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Affinity purification using His-tagged PTPBR7 ectodomain
His-Select Nickel affinity gel (Sigma-Aldrich) was washed in washing buffer (50 mM Tris-HCl, pH 8.0; 300 mM NaCl; 1mM PMSF) and incubated on an orbital shaker for 2 hours at 4˚C with conditioned medium from cells expressing BR7ecto226-His, the His-tagged RPTPµ extracellular domain [24], or with medium from untransfected control cells. Beads were then collected and washed 3 times with washing buffer containing 10 mM imidazole. Subsequently, beads were incubated with brain lysate overnight at 4°C on an orbital shaker. Beads were again collected and washed in washing buffer with 1% Triton X-100 and 150 mM NaCl. Bound proteins were eluted in the same buffer containing 250 mM imidazole. Eluates were mixed with sample buffer, boiled and subjected to SDS-PAGE. Following staining with Coomassie Brilliant Blue, relevant parts were sliced out and used for LC-MS analysis (see below).
GAG ELISA
GAG binding to the extracellular domain of PTPBR7 was tested using a modified ELISA set-up [25]. 100 µl of HS, CS-A, CS-C, CS-D or CS-E (all 10 μg/ml) were coated in 96-well microplates at RT overnight. Excess GAGs was removed by washing 6 times in ELISA buffer (0.2 M NaCl in PBST). Wells were blocked at RT for 1 hour using 3% cold water fish skin gelatin in ELISA buffer. After 6 washes different dilutions of conditioned medium containing BR7ecto226 AP-tagged protein were added to the wells and incubation was for 1.5 hours at RT in the dark. Following another 6 washes, 100 μl of phosphatase reaction mix (1 mg/ml pNPP in SEAP buffer [30]) per well were added. After incubating for 2 hours at RT absorbance at 405 nm was recorded using a Wallac Victor 3 reader (Perkin Elmer, Waltham, MA).
Some of the CS-A, CS-C and CS-E -coated microplate wells were used to check chondroitinase ABC enzyme activity. Following an overnight digestion with 1 IU/ml chondroitinase ABC and several washes with PBST, these wells were incubated with 100 μl of VSV-tagged CS-specific single-chain variable fragment antibodies (1:10 dilution). Subsequent incubations were done using anti-VSV monoclonal antibody P5D4 (1:10 dilution) followed by AP-conjugated rabbit anti-mouse antibody (1:1000). After 6 washes, 100 µl of 1 mg/ml pNPP in SEAP buffer [30] was added per well and absorbance at 405 nm was recorded.
In-Gel digestion
Coomassie Brilliant Blue-stained gel bands were minced and destained by 3 consecutive washes in 750 µl buffer containing 200 mM ammonium bicarbonate in 50% (v/v) acetonitrile for 30 min at 30°C. After drying the gel pieces in a Speedvac (Thermo Savant, Holbrook, USA), 200 µl trypsin solution (0.002 µg/µl in a 50 mM ammonium bicarbonate buffer solution, pH 8.0) was added and allowed to be absorbed by the gel for 45 min on ice. Gel bands were completely immersed by adding additional 800 µl buffer solution and incubated at 37°C overnight. The supernatant was recovered, and the resulting peptides were extracted twice with 500 µl of 60% (v/v) acetonitrile, 0.1% (v/v) formic acid. The pooled peptide extracts were dried in a SpeedVac and resuspended in 15 µl 2% (v/v) acetonitrile, 0.1% (v/v) formic acid for analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS).
LC-ESI-FT-MS/MS
A volume of 5 µl of each fraction was analyzed independently using a fully automated LC-MS/MS setup. Peptides were first separated on an Agilent 1200 chromatographic system (Agilent, Santa Clara, CA, USA) and on-line measured on a LTQ-FT Ultra mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The samples were first loaded and desalted on a (5 mm x 0.3 mm) Zorbax 300SB-C18 trapping column at a 4 µl/min flow rate using a 2% (v/v) acetonitrile, 0.1% formic acid buffer, and then separated on a (150 mm x 75 µm) Zorbax 300SB-C18 analytical column (Agilent) by a 50 min linear gradient ranging from 2% (v/v) to 80% (v/v) acetonitrile, 0.1% formic acid at a 0.3 µl/min flow rate. The LC-effluent was directly coupled to a Triversa NanoMate ESI source (Advion, Ithaca, NY, USA), working in nano-LC mode and equipped with a D-chip whereon a 1.55 kV voltage was supplied. During the LC-separation the FT-ICR mass analyzer acquired MS scans at 100,000 resolution, the 3 most intense precursor peptides for each MS scan were automatically selected and fragmented by the LTQ ion trap mass analyzer.
Mass spectrometry data handling
Raw LC-ESI-FT-MS/MS data were analyzed using Mascot Daemon v2.2.2 (Matrix Science. Londen, UK). The processed spectra were searched against the SwissProt Mus musculus database (Swiss-Prot release 57.15). Carbamidomethyl (C) and oxidation (HMW) were selected as variable modifications, and two missed cleavage were allowed with trypsin as the cleaving agent. Decoy database searches were done with a tolerance of 10 ppm for the precursor ion and 0.5 Da in the MS/MS mode. Keratins were excluded from the final protein list. Only protein identifications with a calculated protein probability higher than 0.99 and with MS/MS ionscore ≥ 30 were retained.
Results
PTPBR7 ectodomain probes for RAP in situ
To investigate whether the Ptprr-encoded receptor-like isoform PTPBR7 displays ligand binding potential, we generated a panel of fusion proteins to be used in the RAP in situ technique [30]. Briefly, seven cDNA fragments encoding the extracellular domain of PTPBR7 were fused in-frame with the PLAP coding sequence via an intervening, flexible peptide linker (Supplementary Material: Fig. S1). All started at PTPBR7 amino acid (aa) codon 1 and ended at codon 220, 221, 222, 223, 224, 225 or 226, respectively (Fig. 1A). This variety of probes serves selection on the basis of optimal fusion protein yield and conformational flexibility required for the binding assay.
After transfection of HEK293T cells, expression levels and sizes of the ectodomain probes were monitored on western blots using antiserum α-BR7 [26]. All probes except BR7ecto225 were properly secreted in the culture medium and displayed the expected apparent molecular weight of 83 kDa (Fig. 1B). Correct folding was assessed by measuring alkaline phosphatase (AP) activity towards 4-nitrophenyldisodium orthophosphate, pNPP (Fig. 1C). All conditioned media, except for BR7ecto225, met the minimal AP specific activity requirements for RAP in situ purposes [30]. In all subsequent experiments specific AP activities were equalized and unfused PLAP-containing conditioned medium served as control.
PTPBR7 extracellular domain binds to myelinated tracts in mouse brain
The different PTPBR7 ectodomain-AP fusion probes, except for BR7ecto225, were used on sagittal sections of adult mouse brain. Positive staining was observed in regions of the corpus callosum, alveus, fimbria fornix, anterior commisure of olfactory bulb, hippocampal area in the cerebrum and in cerebellar-related fiber tracts (Fig. 2), all regions associated with high myelination [33]. These RAP in situ-positive regions of the cerebrum only partially coincide with the areas where the Ptprr gene is expressed as determined by RNA in situ hybridization [18]. No qualitative differences in the binding patterns were observed for the various ectodomain probes, indicating comparable binding abilities (data not shown). The patterns are distinct from those obtained using RPTPσ extracellular probes [11, 31], underscoring specificity.
PTPBR7 extracellular domain binds to white matter in mouse cerebellum
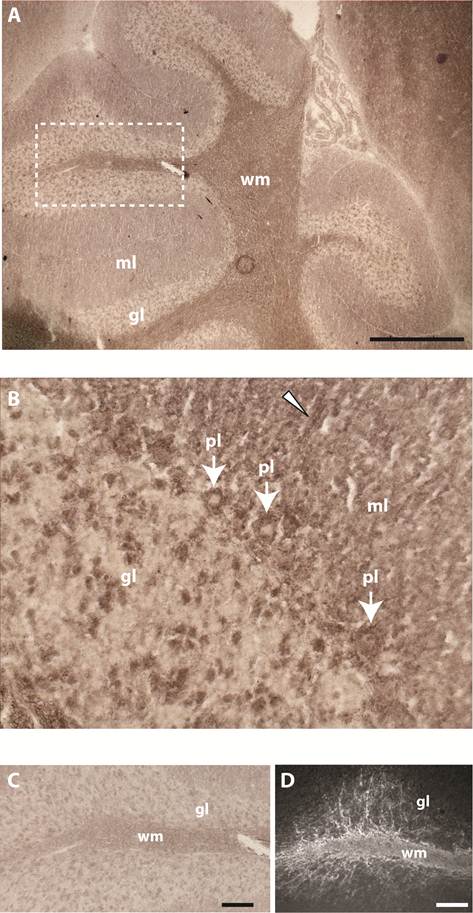
Past studies showed that gene Ptprr displays highest expression levels in granular layer and PCs in the adult mouse cerebellum [18]. RAP in situ staining using the various PTPBR7 ectodomain probes resulted in positive signals for white matter of the cerebellum. Some staining was also present in PCs, single neurons of the granular layer and tracts of the molecular layer (Fig. 3). The pattern observed in the white matter is reminiscent of that of myelinating oligodendrocytes, as revealed by 2'-3'-cyclic-nucleotide 3'-phosphodiesterase (CNPase) immunoreactivity (Fig. 3D).
We next examined whether PTPRR deficiency may have an impact on the localization and level of expression of putative PTPBR7 ligands. No apparent difference in the BR7ecto226 binding patterns was observed when comparing wild type and Ptprr knock-out cerebellar sections (data not shown).
An attempt to reduce the staining intensity of the BR7ecto226 probe by adding increasing amounts of a 6-Histidine tagged version of this PTPBR7 extracellular segment (BR7ecto226-His) was not successful (Supplementary Material: Fig. S2), leading us to conclude that either saturation of binding sites could not be achieved under these conditions or that AP-mediated dimerization [30] of the BR7ecto226 probe is favouring ligand binding over that of the monomeric His-tagged competitor. It has been shown that monomeric RPTPσ binds less well to ligands under RAP conditions as compared to the dimeric probe [34].
Assessment of binding to heparan sulphate and chondroitin sulphate GAGs
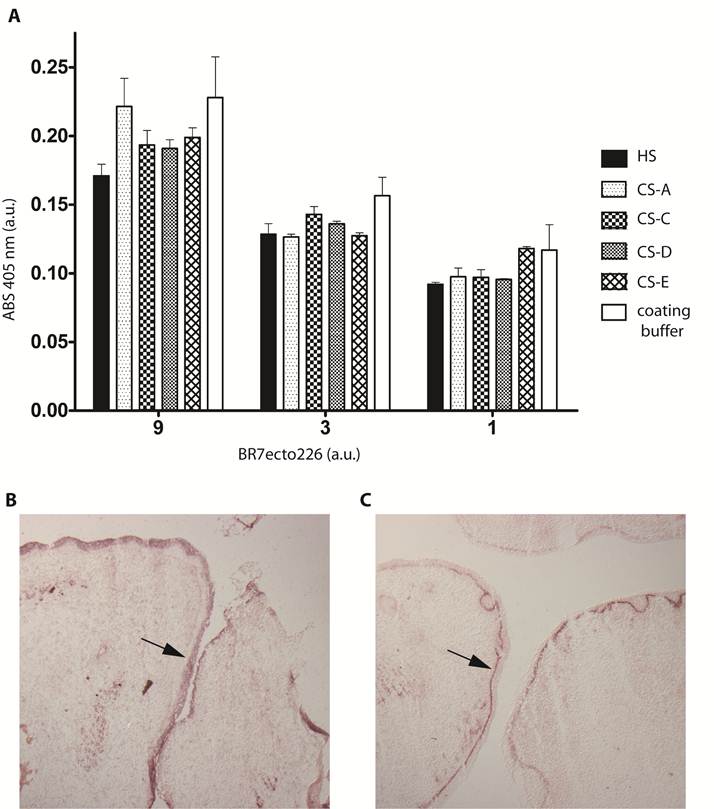
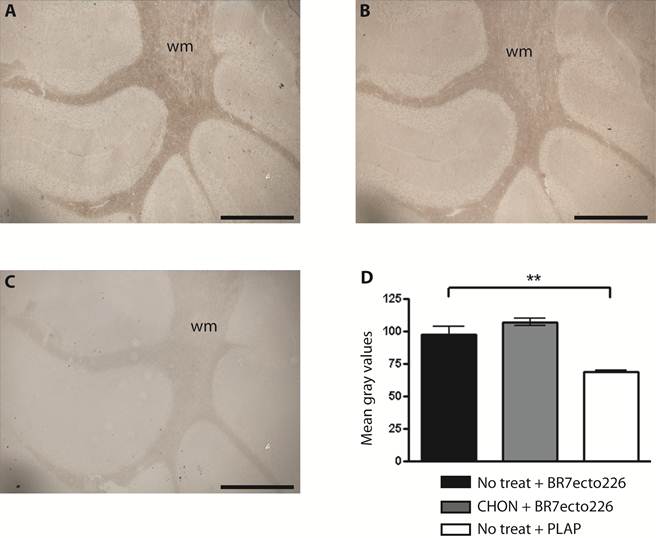
Sulphated glycosaminoglycans (GAGs) have important roles in development and plasticity in the postnatal brain [35] and some have turned out to be RPTP ligands [6]. We therefore tested whether the PTPBR7 ectodomain could directly bind to a panel of GAG modifications using a tailored ELISA protocol [25]. Successful coating of purified HS and CS-A, -C, -D and -E chains onto microtiter plate wells was confirmed by the application of single chain GAG-specific antibodies in parallel control wells (data not shown). Direct binding of probe BR7ecto226 to the HS and CS molecules, as determined by measuring AP activity in the wells, could not be demonstrated in this set-up (Fig. 4). This may also reflect an essential contribution of the GAGs protein moiety to the binding and, therefore, we performed a complementary experiment using the RAP in situ assay. Removal of the most abundant GAG modification in the CNS, CS, by chondroitinase ABC treatment did not alter the staining pattern or the staining intensity of the BR7ecto226 probe on cerebellar tissue sections (Fig. 5). Successful chondroitinase ABC treatment could be confirmed independently using the aforementioned ELISA assay (data not shown). Finally, RAP in situ staining patterns showed GAG-mediated basal lamina binding for RPTPσ [11] but not for PTPRR probes (Figure 4). These findings imply that HS and CS are not the major binding sites for the PTPBR7 extracellular domain.
Characterization of PTPBR7 ectodomain fusion proteins used for RAP in situ assays on mouse brain sections. (A) Schematic representation of mouse PTPBR7 (72 kDa) and the various BR7ecto probes (83 kDa). Signal peptide (SP), hydrophobic (HR) and transmembrane (TM) regions, kinase-interacting motif (KIM), catalytic protein tyrosine phosphatase domain (PTP), flexible Glycine linker (Gly linker), Placental alkaline phosphatase enzyme (PLAP) and the segment recognised by the α-BR7 antiserum are indicated. The seven amino acid residues (position 220, 221, 222, 223, 224, 225 and 226 in PTPBR7) that form the seven different junctions between the extracellular domain and the Glycine linker are highlighted in bold. Resulting probes are named 'BR7ecto' followed by the fusion residue number. (B) Expression and secretion of BR7ecto probes as detected on immunoblots. Conditioned culture media of HEK293T cells transiently transfected with pBG-flexilinker-based expression constructs for the various BR7ecto probes (indicated by the respective fusion residue number on the top of each lane) were subjected to SDS-PAGE and immunoblotted using rabbit polyclonal α-BR7 antiserum. The arrowhead points at the position of the BR7ecto probes. Molecular size markers (in kDa) are indicated on the left. (C) Graph reporting the alkaline phosphatase activity within the different BR7ecto probe-containing conditioned media as determined spectrophotometrically using pNPP as a substrate.
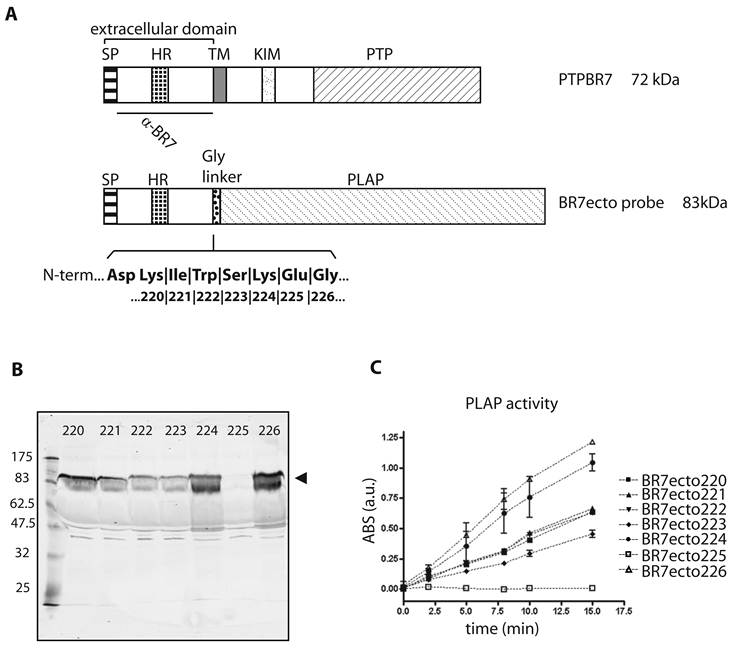
The PTPBR7 ectodomain binds specifically to highly myelinated regions in mouse brain. RAP in situ assays using BR7ecto226 (A), BR7ecto224 (B) and control PLAP protein (C) as a probe were performed on adult mouse brain cryosections. Reddish-purple staining was identified in corpus callosum (cc), alveus (alv), fimbria (fi), columns of the fornix (fx), anterior commissure of olfactory bulb (aco), hippocampal fissure (hf) in the cerebrum and cerebellar-related fiber tracts (wm). Probes BR7ecto220, BR7ecto221, BR7ecto222 and BR7ecto223 gave identical results. Bar = 1 mm.

Binding pattern of the BR7ecto226 probe in adult mouse cerebellum. Low (A) and high (B) magnification of the cerebellum show strong staining concentrated in white matter (wm). Less intense staining is observed in Purkinje cells (pl, arrows), in single cells of the granular layer (gl) and in tracts (arrowhead) of the molecular layer (ml). (C) Enlargement of the region in panel A (dashed rectangle) showing detailed staining of fiber tracts at the white matter branching. D) Using monoclonal antibody 11-5B immuno-reactivity for CNPase was detected in white matter (wm), resembling the BR7ecto226 staining pattern (C), with fiber tracts elongating from the white matter into the granular layer (gl). Bars represent 1 mm (A) and 0.1 mm (C,D).
PTPBR7 ectodomain does not bind to heparan sulphate and chondroitin sulphate moieties. (A) Medium containing the BR7ecto226 probe was added to microplate wells previously coated with one of the indicated GAG chains, or with coating buffer as a control. Binding was assessed by addition of pNPP as substrate and measuring absorption (ABS) at 405 nm. Successful coating of the wells was confirmed separately, exploiting GAG-specific antibodies. Intensities did not significantly differ in comparison with the uncoated control. Results (n=3) are presented as Mean ± S.D. (B) RAP in situ staining of the epidermis (arrow) in mouse embryonic cryosections using BR7ecto226 as a probe. (C) The RPTPσ ectodomain probe displays distinct binding to the basal laminae (arrow), whereas BR7ecto226 does not.
Removal of CS chains does not alter PTPBR7 ectodomain binding to cerebellar white matter. BR7ecto226 probe-containing medium was incubated on Chondroitinase ABC-treated coronal sections of mouse cerebellum (B), and on non-treated control sections (A), according to the RAP in situ procedure. Low magnification images do not show overt differences in binding. Chondroitinase ABC activity was confirmed separately, exploiting GAG-specific antibodies (data not shown). Sections stained with unfused PLAP-containing medium (C) served as control. (D) Quantification of staining intensities within the white matter. Mean gray values are shown. The experiment was done in triplicate and BR7ecto226 and PLAP staining intensities were consistently found to be significantly different. Results (n=3) are presented as Mean ± S.D. (p=0.0013). CHON= chondroitinase ABC. No treat = no treatment. Bar = 1 mm.
Candidate proteins that bind to the PTPBR7 extracellular domain
As an approach to directly identify candidate protein molecules that can bind to the PTPBR7 extracellular segment, we exploited the BR7ecto226-His fusion protein to affinity-purify such proteins from mouse brain lysates. Two independent purifications were undertaken to address reproducibility. Furthermore, a similar procedure was followed using the His-tagged RPTPμ extracellular domain [24] as affinity reagent, to verify specificity of the procedure. His-tagged ectodomains were produced in transfected HEK293T cells, purified using nickel beads and subsequently used to capture mouse brain proteins. The ectodomain-displaying beads were extensively washed and the specifically bound proteins were then eluted and size-separated on SDS-PAGE. Relevant gel lanes were then excised and subjected to enzymatic digestion. Retrieved peptides were subsequently identified by means of nanoLC coupled to mass spectrometry.
Peptides of three different proteins were retrieved reproducibly in the duplicate BR7ecto226-purified samples (Table 1): the guanine nucleotide-binding protein G(o) subunit alpha (Gnao1), the IgLON family cell adhesion molecule Neurotrimin (Ntm), and α/β subunits of Calcium/calmodulin-dependent kinase II (Camk2a/b). These peptides did not overlap with those affinity-purified using RPTPμ extracellular domain or nickel beads alone as controls and generated protein Mascot scores above 60, minimizing false positive rates. It is of note that PSD-95, the most abundant membrane-associated protein of the post-synaptic density fraction, was not present in our isolates. Both findings support the specificity of the applied procedure. At first sight it may come as a surprise to find cytoplasmic candidates like Gαo and CaMKII when using extracellular protein segments as bait but under the conditions used there might still be multi-subunit protein complexes that could explain co-purification. Future experiments should aim at the validation and functional testing of the identified binding proteins.
Proteins identified by LC/MS-MS and Mascot Search that were affinity-purified from brain lysates using BR7ecto226-His as bait. Proteins that had Mascot scores above 60 and did not purify in isolates using an unrelated His-tagged bait or nickel beads alone are listed. Data from two independent isolates are shown. A full list of proteins identified in this way is provided as (Supplementary Material: Table S1). *CaMKII protein was detectable in control samples but Mascot scores (38) were below threshold. Subunits α and β of CaMKII have high sequence identity and could not be distinguished in the analysis.
| Protein name | Protein mnemonic identifier | Uniprot Accession number | Mass (kDa) | Mascot scores | Queries matched | Function |
|---|---|---|---|---|---|---|
| Guanine nucleotide-binding protein G(o) subunit alpha | Gnao1 | GNAO_MOUSE | 40 | 85 | 2 | Modulator / transducer of GPCR signaling. G(o) protein function is not clear. GNAO1 mutation implicated in breast cancer |
| 139 | 5 | |||||
| Neurotrimin | Ntm | NTRI_MOUSE | 38 | 77 | 5 | Neural cell adhesion molecule of the IgLON subfamily of immunoglobulins |
| 31 | 1 | |||||
| Calcium/calmodulin-dependent protein kinase type II | Camk2a Camk2b | KCC2A_MOUSE KCC2B_MOUSE | 54 60 | 153* | 4 | Prominent kinase in the central nervous system, implicated in long-term potentiation and neurotransmitter release |
| 200* | 5 |
Discussion
The mouse gene Ptprr encodes a receptor-like transmembrane protein isoform, PTPBR7, which is expressed on many cells of neuronal origin [18]. Our previous work has shown that PTPBR7 phosphatase activity is negatively regulated through homo-multimerisation [17]. Combined with the phenotype displayed by Ptprr knock-out mice [19] this prompted us to search for putative PTPBR7 ligands. Exploiting the RAP in situ strategy we localised binding sites for the extracellular domain of PTPBR7 to highly myelinated areas in mouse brain, including white matter of the cerebellum. While highly sulphated GAGs proved to be ligands for RPTPσ and PTP-LAR [7, 9, 11], we found no evidence for an interaction of the PTPBR7 ectodomain with HS and CS in vitro or in situ. In parallel, we used the PTPBR7 extracellular domain to affinity-purify candidate protein ligands from brain lysates and used mass spectrometry for identification. The obtained associating proteins (Gαo, Neurotrimin and CaMKII) point to Ca2+-regulated processes, cell-cell communication and adhesion. Immunopurification of endogenous protein complexes containing any of these three proteins and PTPBR7, exploiting brain lysates from wildtype and Ptprr-/- animals, was unsuccessful (our unpublished results). This may be due to aspecificity or lack of sensitivity of the antibodies used, but could also reflect that only a small subset of the proteins is involved in the interaction or that their encounter is transient. It is of note that also ERK1/2 MAP kinases, well-established PTPBR7 interactors, were not detected in these experiments, and reports on ERK-PTP complexes indeed are based on ectopic expression.
We found that highly myelinated tracts in adult brain tissue represent preferred sites of PTPBR7 ectodomain binding. These regions do not mirror Ptprr expression patterns as revealed by RNA in situ hybridisation [18], which may reflect PTPBR7 localization and interactions on neuronal processes far away from the cell body. The concordance of PTPBR7 ligand staining with myelinating oligodendrocytes in cerebellar white matter may even point to a role for PTPBR7 in the process of myelination. A direct role in remyelination has been demonstrated for another RPTP, RPTPζ/β, whose expression is induced within remyelinating oligodendrocytes in multiple sclerosis lesions [36]. The locomotive impairment observed in Ptprr-/- animals [19], however, is not accompanied by obvious myelination defects arguing against such a role for PTPBR7.
Myelination, neurite outgrowth and synaptic plasticity are strongly based on cell adhesion mechanisms [1, 2]. Intriguingly, a cell-cell adhesion molecule presented as a candidate interactor of the PTPBR7 extracellular domain; Neurotrimin (Table 1). Neurotrimin is a member of the IgLON family of neural cell adhesion molecules that contains members like Neuronal growth regulator 1, LSAMP and OBCAM/OPCML. In vitro and in vivo studies indicate that IgLON members either enhance or inhibit neurite outgrowth and synaptogenesis [37]. Neurotrimin is expressed in cerebellar granule and Purkinje cells and its developmental pattern of expression points to roles in axon fasciculation and synaptogenesis [38]. PTPBR7 is expressed at all developmental stages in mouse cerebellar granular cells. In contrast, its expression in Purkinje cells gradually decreases over the first weeks after birth [18]. The putative PTPBR7 - Neurotrimin interaction may well contribute to Purkinje cell maturation and parallel fiber synaptogenesis, processes that are essential for cerebellar functioning in locomotive processes.
The identification of cytosolic CaMKII α/β subunits as a candidate interactor that is reproducibly purified using the extracellular domain of PTPBR7 first came as a surprise. However, we also found CaMKII binding to the tyrosine phosphatase domain of PTPBR7 in an independent glutathione S-transferase pull-down experiment (unpublished data). Since PTPBR7 is able to form homo-multimers [17] it might be that the PTPBR7 ectodomain stably associates with full-length PTPBR7 and its associating proteins from mouse brain lysate. This may involve cell surface-exposed PTPBR7 but could also address PTPBR7-containing complexes following its internalisation and subsequent vesicular trafficking. Alternatively, other CaMKII-associated transmembrane proteins may mediate the interaction with the PTPBR7 ectodomain probe. A similar type of reasoning may underlie the identification of Gαo in BR7ecto226-His purified material.
CaMKII is highly expressed in brain and its serine/threonine kinase activity is dependent on increased calcium levels. The enzyme is concentrated in the post synaptic density (PSD) fraction and is critically involved in the synaptic plasticity that underlies processes of learning and memory [39]. Mice lacking CaMKIIα display limbic epilepsy [40]. Also Gαo functioning has been linked to neuronal calcium signalling processes and locomotion. The study of ionic currents in mouse hippocampal CA3 neurons revealed that modulation of Ca2+ currents by G protein-coupled receptors is changed and much slower recovery kinetics are displayed when Gαo isoforms are lacking [41]. Gαo deficient animals have normal brain morphology but suffer from tremors and occasional seizures, display severe motor control impairment and have a reduced life span of only two months [42, 43]. Intriguingly, locomotive impairment combined with normal brain morphology was also observed in Ptprr-/- mice [19] and in several mouse models that lack cerebellar calcium binding proteins [20]. A functional interaction between PTPBR7 and Gαo or CaMKII containing complexes could well explain such in vivo parallels.
In conclusion, our work puts forward highly myelinated areas in brain as a source of PTPBR7 ligands. Using the PTPBR7 extracellular domain as bait, we identified protein candidates that lend support to PTPBR7 involvement in two major processes. On the one hand as part of cell-cell adhesion complexes during cerebellar development; on the other hand in calcium ion-regulated events that are instrumental in neuronal development and plasticity. This provides a working hypothesis to start testing the functional implications of the observed interactions in cell and animal models.
Supplementary Material
Figure S1: Schematic representation of the pBG-flexilinker vector used to generate RAP in situ probes. Figure S2: The binding of BR7ecto226 probe to the mouse cerebellum is not altered by the PTPBR7 extracellular domain itself. Table S1: Mascot scores for proteins identified by LC‐ESI‐FT‐MS/MS that were affinity‐purified from brain lysates.
Abbreviations
AP: alkaline phosphatase; CAM: cell adhesion molecule; CaMKII: calcium/calmodulin-dependent kinase II; CNPase: 2'-3'-cyclic-nucleotide 3'-phosphodiesterase; CNS: central nervous system; CS: chondroitin sulphate; HR: hydrophobic region; HS: heparan sulphate; GAG: glycosaminoglycan; Goα: guanine nucleotide-binding protein G(o) subunit alpha; Ntm: neurotrimin; PLAP: placental alkaline phosphatase; pNPP: 4-nitrophenyldisodium orthophosphate; RPTP: receptor protein tyrosine phosphatase.
Acknowledgements
We thank Mirthe Erkens, Nick van Bakel and Catharina E. van der Zee for technical assistance and Radu Aricescu (Oxford, UK) for generously providing pHLsec-based constructs. Frank Böhmer is thanked for supportive guidance and critical reading of the manuscript. This work was supported in part by a Marie Curie Research Training Network (PTPNET/MRTN-CT-2006-035830) to AS and WH.
Conflict of Interests
The authors declare that no conflict of interest exists.
References
1. Dityatev A, Schachner M, Sonderegger P. The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat Rev Neurosci. 2010;11:735-746
2. Togashi H, Sakisaka T, Takai Y. Cell adhesion molecules in the central nervous system. Cell Adh Migr. 2009;3:29-35
3. Hendriks WJ, Elson A, Harroch S, Stoker AW. Protein tyrosine phosphatases: functional inferences from mouse models and human diseases. Febs J. 2008;275:816-830
4. Johnson KG, Van Vactor D. Receptor protein tyrosine phosphatases in nervous system development. Physiol Rev. 2003;83:1-24
5. Stoker AW. Receptor tyrosine phosphatases in axon growth and guidance. Curr Opin Neurobiol. 2001;11:95-102
6. den Hertog J, Ostman A, Bohmer FD. Protein tyrosine phosphatases: regulatory mechanisms. Febs J. 2008;275:831-847
7. Johnson KG, Tenney AP, Ghose A, Duckworth AM, Higashi ME, Parfitt K, Marcu O, Heslip TR, Marsh JL, Schwarz TL, Flanagan JG, Van Vactor D. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 2006;49:517-531
8. Kwon SK, Woo J, Kim SY, Kim H, Kim E. Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J Biol Chem. 2010;285:13966-13978
9. Shen Y, Tenney AP, Busch SA, Horn KP, Cuascut FX, Liu K, He Z, Silver J, Flanagan JG. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326:592-596
10. Krasnoperov V, Bittner MA, Mo W, Buryanovsky L, Neubert TA, Holz RW, Ichtchenko K, Petrenko AG. Protein-tyrosine phosphatase-sigma is a novel member of the functional family of alpha-latrotoxin receptors. J Biol Chem. 2002;277:35887-35895
11. Aricescu AR, McKinnell IW, Halfter W, Stoker AW. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase sigma. Mol Cell Biol. 2002;22:1881-1892
12. Alete DE, Weeks ME, Hovanession AG, Hawadle M, Stoker AW. Cell surface nucleolin on developing muscle is a potential ligand for the axonal receptor protein tyrosine phosphatase-sigma. Febs J. 2006;273:4668-4681
13. Fry EJ, Chagnon MJ, Lopez-Vales R, Tremblay ML, David S. Corticospinal tract regeneration after spinal cord injury in receptor protein tyrosine phosphatase sigma deficient mice. Glia. 2010;58:423-433
14. O'Grady P, Thai TC, Saito H. The laminin-nidogen complex is a ligand for a specific splice isoform of the transmembrane protein tyrosine phosphatase LAR. J Cell Biol. 1998;141:1675-1684
15. Bouyain S, Watkins DJ. The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules. Proc Natl Acad Sci U S A. 2010;107:2443-2448
16. Fukada M, Fujikawa A, Chow JP, Ikematsu S, Sakuma S, Noda M. Protein tyrosine phosphatase receptor type Z is inactivated by ligand-induced oligomerization. FEBS Lett. 2006;580:4051-4056
17. Noordman YE, Augustus ED, Schepens JT, Chirivi RG, Rios P, Pulido R, Hendriks WJ. Multimerisation of receptor-type protein tyrosine phosphatases PTPBR7 and PTP-SL attenuates enzymatic activity. Biochim Biophys Acta. 2008;1783:275-286
18. van den Maagdenberg AM, Bachner D, Schepens JT, Peters W, Fransen JA, Wieringa B, Hendriks WJ. The mouse Ptprr gene encodes two protein tyrosine phosphatases, PTP-SL and PTPBR7, that display distinct patterns of expression during neural development. Eur J Neurosci. 1999;11:3832-3844
19. Chirivi RG, Noordman YE, Van der Zee CE, Hendriks WJ. Altered MAP kinase phosphorylation and impaired motor coordination in PTPRR deficient mice. J Neurochem. 2007;101:829-840
20. Hendriks WJ, Dilaver G, Noordman YE, Kremer B, Fransen JA. PTPRR Protein Tyrosine Phosphatase Isoforms and Locomotion of Vesicles and Mice. Cerebellum. 2009;8:80-88
21. Flanagan JG, Leder P. The kit ligand: a cell surface molecule altered in steel mutant fibroblasts. Cell. 1990;63:185-194
22. Flanagan JG, Cheng H-J, Feldheim DA, Hattori M, Lu Q, Vanderhaeghen P. Alkaline phosphatase fusions of ligands or receptors as in situ probes for staining of cells, tissues, and embryos. Methods Enzymol. 2000;327:19-35
23. Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243-1250
24. Aricescu AR, Siebold C, Choudhuri K, Chang VT, Lu W, Davis SJ, van der Merwe PA, Jones EY. Structure of a Tyrosine Phosphatase Adhesive Interaction Reveals a Spacer-Clamp Mechanism. Science. 2007;317:1217-1220
25. van Kuppevelt TH, Dennissen MA, van Venrooij WJ, Hoet RM, Veerkamp JH. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J Biol Chem. 1998;273:12960-12966
26. Dilaver G, van de Vorstenbosch R, Tarrega C, Rios P, Pulido R, van Aerde K, Fransen J, Hendriks W. Proteolytic processing of the receptor-type protein tyrosine phosphatase PTPBR7. Febs J. 2007;274:96-108
27. Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931-941
28. Smetsers TFCM, van de Westerlo EMA, ten Dam GB, Overes IM, Schalkwijk J, van Muijen GNP, van Kuppevelt TH. Human Single-Chain Antibodies Reactive with Native Chondroitin Sulfate Detect Chondroitin Sulfate Alterations in Melanoma and Psoriasis. J Invest Dermatol. 2004Mar;122(3):707-16
29. ten Dam GB, van de Westerlo EM, Purushothaman A, Stan RV, Bulten J, Sweep FC, Massuger LF, Sugahara K, van Kuppevelt TH. Antibody GD3G7 selected against embryonic glycosaminoglycans defines chondroitin sulfate-E domains highly up-regulated in ovarian cancer and involved in vascular endothelial growth factor binding. Am J Pathol. 2007;171:1324-1333
30. Stoker A. Methods for identifying extracellular ligands of RPTPs. Methods. 2005;35:80-89
31. Sajnani-Perez G, Chilton JK, Aricescu AR, Haj F, Stoker AW. Isoform-specific binding of the tyrosine phosphatase PTPsigma to a ligand in developing muscle. Mol Cell Neurosci. 2003;22:37-48
32. Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36-42
33. Franklin RJM, ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci. 2008Nov;9(11):839-55
34. Lee S, Faux C, Nixon J, Alete D, Chilton J, Hawadle M, Stoker AW. Dimerization of Protein Tyrosine Phosphatase {sigma} Governs both Ligand Binding and Isoform Specificity. Mol Cell Biol. 2007;27:1795-1808
35. Kwok JC, Afshari F, Garcia-Alias G, Fawcett JW. Proteoglycans in the central nervous system: plasticity, regeneration and their stimulation with chondroitinase ABC. Restor Neurol Neurosci. 2008;26:131-145
36. Harroch S, Furtado GC, Brueck W, Rosenbluth J, Lafaille J, Chao M, Buxbaum JD, Schlessinger J. A critical role for the protein tyrosine phosphatase receptor type Z in functional recovery from demyelinating lesions. Nat Genet. 2002;32:411-414
37. Hashimoto T, Maekawa S, Miyata S. IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem Funct. 2009;27:496-498
38. Chen S, Gil O, Ren YQ, Zanazzi G, Salzer JL, Hillman DE. Neurotrimin expression during cerebellar development suggests roles in axon fasciculation and synaptogenesis. J Neurocytol. 2001;30:927-937
39. Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914-931
40. Butler LS, Silva AJ, Abeliovich A, Watanabe Y, Tonegawa S, McNamara JO. Limbic epilepsy in transgenic mice carrying a Ca2+/calmodulin-dependent kinase II alpha-subunit mutation. Proc Natl Acad Sci U S A. 1995;92:6852-6855
41. Greif GJ, Sodickson DL, Bean BP, Neer EJ, Mende U. Altered regulation of potassium and calcium channels by GABA(B) and adenosine receptors in hippocampal neurons from mice lacking Galpha(o). J Neurophysiol. 2000;83:1010-1018
42. Jiang M, Gold MS, Boulay G, Spicher K, Peyton M, Brabet P, Srinivasan Y, Rudolph U, Ellison G, Birnbaumer L. Multiple neurological abnormalities in mice deficient in the G protein Go. Proc Natl Acad Sci U S A. 1998;95:3269-3274
43. Valenzuela D, Han X, Mende U, Fankhauser C, Mashimo H, Huang P, Pfeffer J, Neer EJ, Fishman MC. G alpha(o) is necessary for muscarinic regulation of Ca2+ channels in mouse heart. Proc Natl Acad Sci U S A. 1997;94:1727-1732
Author contact
![]() Corresponding author: Tel: +31-24-3614329, Fax: +31-24-3615317. E-mail: w.hendriksru.nl (W.J.A.J. Hendriks)
Corresponding author: Tel: +31-24-3614329, Fax: +31-24-3615317. E-mail: w.hendriksru.nl (W.J.A.J. Hendriks)

 Global reach, higher impact
Global reach, higher impact