10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(7):728-742. doi:10.7150/ijbs.6475 This issue Cite
Review
Glutamate Signaling in Benign and Malignant Disorders: Current Status, Future Perspectives, and Therapeutic Implications
Departments of Cancer Genetics and Urology, Center for Genetics and Pharmacology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY.
Received 2013-4-15; Accepted 2013-6-15; Published 2013-8-9
Abstract
Glutamate, a nonessential amino acid, is the major excitatory neurotransmitter in the central nervous system. As such, glutamate has been shown to play a role in not only neural processes, such as learning and memory, but also in bioenergetics, biosynthetic and metabolic oncogenic pathways. Glutamate has been the target of intense investigation for its involvement not only in the pathogenesis of benign neurodegenerative diseases (NDDs) such as Parkinson's disease, Alzheimer's disease, schizophrenia, multiple sclerosis, and amyotropic lateral sclerosis (ALS), but also in carcinogenesis and progression of malignant diseases. In addition to its intracellular activities, glutamate in secreted form is a phylogenetically conserved cell signaling molecule. Glutamate binding activates multiple major receptor families including the metabotropic glutamate receptors (mGluRs) and ionotropic glutamate receptors (iGluRs), both of which have been implicated in various signaling pathways in cancer. Inhibition of extracellular glutamate release or glutamate receptor activation via competitive or non-competitive antagonists decreases growth, migration and invasion and induces apoptosis in breast cancer, melanoma, glioma and prostate cancer cells. In this review, we discuss the current state of glutamate signaling research as it relates to benign and malignant diseases. In addition, we provide a synopsis of clinical trials using glutamate antagonists for the treatment of NDD and malignant diseases. We conclude that in addition to its potential role as a metabolic biomarker, glutamate receptors and glutamate-initiated signaling pathways may provide novel therapeutic opportunities for cancer.
Keywords: Glutamate, mGluR, iGluR, GRM1a, mGluR1a, signaling, breast, cancer, melanoma, prostate, brain, glioma.
Introduction
Recent studies have implicated glutamate (Glu) signaling in the development and progression of malignant diseases. Glu, a non-essential amino acid, is the major excitatory neurotransmitter in the central and peripheral nervous systems (CNS, PNS). In addition, it functions as a source of energy for rapidly dividing cells such as lymphocytes and cancer cells (reviewed in [1]). Many decades of research have demonstrated the importance of glutamatergic signaling in the CNS [2, 3]. Processes such as synaptic plasticity and learning and memory which rely on long-term potentiation are known to require functional Glu receptors [4].
Glu initiates signaling cascades upon binding to both ionotropic (iGluR) and metabotropic Glu receptors (mGluR) (reviewed in [2, 3, 5, 6]). The two receptor families differ in their mechanisms of activation and downstream effectors: iGluRs are voltage-gated ion channels that initiate Ca2+ and/or K+ influx and downstream signaling while mGluRs are atypical G-protein-coupled receptors (GPCRs) which activate second messenger pathways such as phospholipase C (PLC), phosphoinositide 3 kinase/retrovirus AK thymoma/mTOR (PI3K/AKT/mTOR) and mitogen activated protein kinase (MAPK) signaling (Table 1, Figure 1 & 2). Both receptor families are further classified into subgroups based on amino acid sequence homology, pharmacology and other signaling characteristics. First, the iGluRs are segregated into three subfamilies based on agonist binding: the N-methyl-D-aspartate (NMDAR), a-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPAR), and kainate receptor (KR) subfamilies Table 1, Figure 1). Second, the mGluR family contains three subfamilies known as groups I, II and III (Table 1, Figure 2). The mGluR subfamilies differ in the second messenger systems that are activated upon ligand binding. Group I contains mGluR1 and 5 and initiates signaling via the PLC/Inositol 1,4,5-triphosphate/diacylglycerol (PLC/IP3/DAG) pathway (Figure 2). Group II (mGluR 2 and 3) and III (mGluR4, 6, 7, 8) activates the inhibitory G-protein, Gi, resulting in decreased cAMP levels (Table 1). Studies have shown that these are not rigid rules: some mGluRs are known to bind to other excitatory or inhibitory G-proteins (see [2] for review).
Ionotropic and metabotropic glutamate receptor families.
| Glutamate receptor family | Subgroup | # of genes | Second messenger signaling system |
|---|---|---|---|
| iGluR | NMDAR | 7 | Ca2+, K+ influx |
| AMPAR | 4 | Ca2+, K+ influx | |
| Kainate | 5 | Ca2+, K+ influx | |
| mGluR | Group I | 2 | Ca2+, coupling to K+ channels, PLC/IP3/DAG, PI3K/AKT/mTOR, MAPK |
| Group II | 2 | Inhibition of adenylyl cyclase; shutdown of cAMP signaling | |
| Group III | 4 | Inhibition of adenylyl cyclase; shutdown of cAMP signaling |
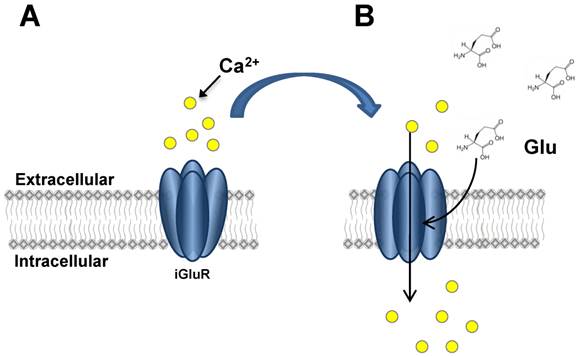
iGluR activation. This figure illustrates the activation and opening of iGluR ion channels. In Panel A, the closed channel does not allow ion influx. Upon Glu binding (Panel B), conformational change in iGluR subunits allows Ca2+ influx to occur by diffusion through the open channel.
mGluR activation. This figure depicts the mGluR1a receptor before (Panel A) and after (Panel B) Glu binding. Relevant domains are noted, such as: amino terminal domain (ATD), cysteine-rich domain (CRD), transmembrane domain (TMD), cytoplasmic tail domain (CTD). Binding of Glu activates G-protein signaling and downstream second messenger pathways including PKC, PLC and Ca2+ signaling.
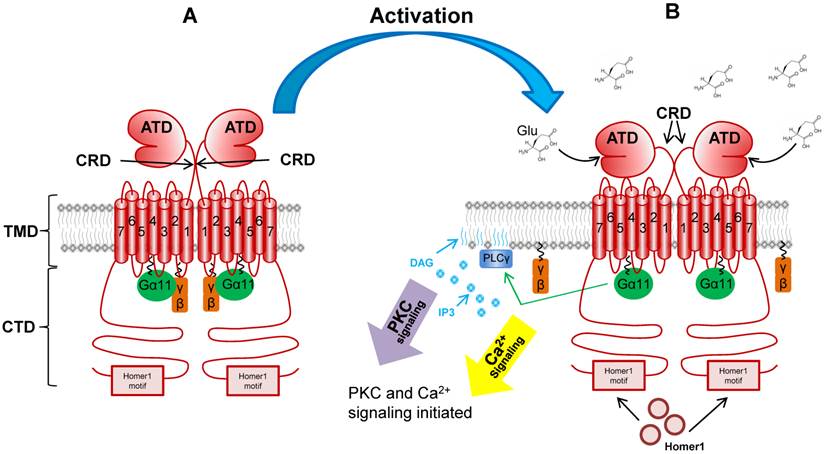
The eight mGluR genes are alternatively spliced to encode 12 isoforms (reviewed in [2]). As is the case for all GPCRs, all mGluR isoforms contain the typical seven transmembrane domains (TMD) as well as an amino-terminal (N-terminal) ligand-binding domain (ATD) and a carboxy-terminal (C-terminal) cytoplasmic domain (CTD) (Figure 2) [7]. The mGluRs are referred to as atypical GPCRs for a number of reasons. First, the ATD is far larger than other GPCR family members at approximately 55 kDa [8]. The unique two-lobed shape of this domain in which Glu binds between the two lobes is referred to as the “venus fly trap” domain [9]. Second, the receptors differ from classic GPCRs and further the isoforms differ from one another at the level of the CTD. The mGluR1a and mGluR5a isoforms contain a long proline-rich CTD which has been shown to participate in multiple protein-protein interactions. Downstream of the proline-rich region is the Homer1 binding motif. This domain facilitates the interaction of the active receptor with Homer1a, b and c; an event which mediates the coupling of mGluR1a/5a to multiple critical downstream signaling components [10-14]. Finally, the mGluR family differs from other GPCRs in that it exists as a functional homodimer. Disulfide bridging between the two receptor monomers occurs in the short cysteine rich domain (CRD) between the ATD and the TMD which contains a number of cysteine residues (Figure 2) [7, 15-17].
The protein-protein interactions that occur at the Homer1 binding motif of mGluR1a/5a can modulate the activity of the receptor in multiple critical ways. Characterized by the consensus sequence PPxxFR (where x is any amino acid), the Homer1 binding motif is known to recruit Homer1 proteins via the Homer Ena/VASP Homology 1 domain (EVH1) located at the N-terminus of all Homer1 isoforms [18-20]. The Homer1 gene encodes multiple isoforms of varying length; the best studied of which are Homer1a, b and c [reviewed in 21]. Homer1b and c (known as “long form” isoforms) also contain coiled-coil and leucine zipper domains which facilite homodimerization, allowing long form Homer1 isoforms to couple the active receptor to other Homer1-bound downstream signaling components, such as Insitol triphosphate receptor (IP3R), PI3K and ERK1/2 [19, 21, 22-26]. In many cases, Shank scaffolding proteins are involved in these protein-protein interactions which, in neural cells, are referred to as postsynaptic densities and are critical for proper neurotransmission [19, 27-28]. In contrast to the function of other isoforms, Homer1a is shorter in length and lacks the coiled-coil domains allowing for scaffolding activity, lending to it's classification as a dominant-negative isoform [reviewed in 21; 29-30] . Since it retains the EVH1 domain, but lacks coiled-coil and leuzine zippers, Homer1a simply occupies the Homer1 binding domain without coupling to other proteins. The dynamics of Homer1a and long form isoform expression in neural cells is one mechanism of generating synpatic plasticity and modulating the interaction of mGluR1a/5a with signaling components [see 24, 31 for example]. Finally, Homer1 isoform binding to mGluR1a/5a can influence its trafficking between the cytoplasm and plasma membrane compartments - a fact which influences the stability of the protien [24].
In recent years, experimental data have implicated both iGluRs and mGluRs in malignant disease. Furthermore, many studies have indicated that mGluRs are the predominant mediators of glutamatergic signaling in cancer. Importantly, mGluR1 was recently found to be overexpressed in human prostate tumors and further, serum Glu levels correlated with primary prostate cancer (PCa) aggressiveness [31, 32]. In addition, rodent mGluR1 protein expression has been shown to cause phenotypes associated with cellular transformation in cultured melanocytes [33, 34]. These data indicate that mGluR1 can function as an oncogene in certain cell types and that Glu and mGluR1 may be important markers of disease progression in some cancers. In this review, we discuss the current state of Glu signaling research in cellular transformation and in the translational application of these studies to preclinical studies and clinical trials. We focus on the best-studied cancers in which Glu signaling has been implicated: glioma, melanoma and breast and prostate carcinoma. We conclude that there may be similarities between these cancers with respect to glutamatergic signaling and that mGluR1 is an important new target in anti-cancer drug discovery.
Glutamate blockade for the treatment of benign neurological and psychiatric disorders
Aberrant Glu signaling is implicated in the etiology and progression of many neurological and psychiatric disorders such as neurodegenerative disease (NDD), severe autism, Fragile X syndrome (mental retardation), schizophrenia, long-term depression, bipolar disorder and recovery after traumatic CNS injury (for reviews, see [6, 35-43]). In some cases (i.e. schizophrenia), a lack of mGluR signaling is thought to be a contributor to disease severity and mGluR agonists have been successful in providing some symptomatic relief [44, 45]. However, this research is plagued by the high side effects associated with mGluR agonists (reviewed in [46]). Current efforts are focused on developing a molecule with better tolerance and a higher penetrance for symptomatic relief in schizophrenia patients [46]. In many other cases, overstimulation of GluRs by excessive Glu release or persistence of Glu in the synaptic cleft is correlated with disease onset or symptomatic severity - a phenomenon called excitotoxicity [47]. In this situation, the excitatory amino acid transporter (EAAT) system which is usually responsible for removing Glu from the synaptic cleft after signal transduction is either unable to keep up with the flood of excess Glu, or is rendered nonfunctional. Therefore, blockade of Glu release, receptor antagonism or inhibition of downstream signaling has been used extensively in the treatment of NDDs, psychiatric maladies and to protect against permanent neural damage post-injury [48].
Many Glu signaling antagonists have been described in the literature and have had success in the clinic as treatments for neurological disorders [2, 3, 49-54]. Research into GluR antagonists as treatments for major depressive disorder (MDD) have shown that iGluR signaling blockade can be highly successful (reviewed in [55]). Ketamine, a noncompetitive NMDAR antagonist, provides symptomatic relief in MDD patients within hours of infusion [56, 57]. This finding was especially significant because patients that were resistant to antidepressants still responded well to ketamine, therefore opening the possibility that Glu signaling blockade may be a viable treatment option for other drug-resistant mood disorders such as lithium-resistant bipolar disorder [58]. mGluRs may also be targets for therapeutic intervention in patients with mood disorders. Indeed, the mGluR5 antagonists 2-methyl-6-(phenylethynyl)-pyridine (MPEP) and 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl] pyridine (MTEP) have been shown to decrease depression-related behaviors in rats [58, 59]. A new mGluR5 antagonist with potential activity against MDD and other mood disorders, BCI-838, is currently being tested for safety in the clinic (Clinicaltrials.gov #NCT01548703).
Perhaps one of the most successful uses of Glu-targeted therapeutics is represented by the treatment of the NDD amyotrophic lateral sclerosis (ALS) with the drug riluzole [60]. Marketed by Sanofi-Aventis as Rilutek, FDA-approval for this drug as a treatment for ALS was achieved in 1995 [61, 62]. Since then, it has been suggested as a therapy for bipolar or major depressive disorders, but has not been granted FDA-approval for these indications [63]. In fact, Clinicaltrials.gov reveals that this drug is under investigation in the clinic for a number of other neurological disorders such as obsessive-compulsive disorder (Clinicaltrials.gov #NCT00523718), MDD (#NCT01204918), multiple sclerosis (#NCT00501943), autism (#NCT01661855), Huntington's disease (#NCT00277602), Parkinson's disease (#NCT00013624), Cervical spondylotic myelopathy (#NCT01257828), neural protection after traumatic spinal cord injury (#NCT01597518), as well as in various combination therapies and as treatment for other psychiatric disorders.
Classified as an “anti-excitotoxic” drug, riluzole blocks the release of Glu from the cell and may also block the activity of mGluR1a [64]. Although riluzole appears to antagonize NMDA and kainate receptors from electrophysiology data (reviewed in [65]), in vitro studies have been unsuccessful in demonstrating direct binding of the molecule to these receptors. In fact, it is still not completely clear exactly which Glu receptors are affected by riluzole [64-67]. As will be discussed in future sections of this review, riluzole shows promise as an anti-cancer therapeutic [67]. The drug is well-tolerated at high doses in long-term treatment paradigms with low side effects, making it an ideal medicine.
Glutamate signaling in malignant diseases
Throughout the decades, members of the GluR family have been studied for their potential role in cellular transformation, in cancer progression, and as diagnostic and therapeutic targets. Indeed, expression of at least one subunit of iGluR or mGluR has been reported in carcinoma of the breast, prostate, thyroid and lung as well as in oral squamous cell carcinoma, colorectal cancer, melanoma, osteosarcoma, neuroblastoma, astrocytoma, multiple myeloma, glioma, medulloblastoma and some leukemias [34, 68-70]. This list is likely not all-inclusive since studies implicating Glu signaling in malignant disease are rapidly being published. This literature is confounded by a general lack of studies on the human orthologues of Glu receptors and further by the fact that in most studies, only one subunit of a particular GluR is examined.
In addition to expression differences in iGluRs in benign versus malignant cells, somatic mutations in iGluR genes have been described. First, one report proposes that iGluRs are upregulated in an androgen-stimulated manner in the PCa cell lines LNCaP and PC-3 [71]. Second, current work by Endele, et al. has identified somatic mutations in the NMDAR family members Glu receptor subunit zeta-2A and -2B (GRIN2A and GRIN2B) that impart susceptibility to not only schizophrenia but also to melanoma [72]. The mechanism for the involvement of iGluR signaling in melanoma seems to be related to the receptor's inability to allow Ca2+ influx and the downstream signaling repercussions [72]. Further, Wei, et al. recently catalogued somatic mutations in the iGluR superfamily in melanoma patients where they are found with high mutation frequency [73]. Interestingly, the expression of iGluRs may be regulated by DNA methylation since at least one study reported that not only is the promoter of Glu receptor ionotropic kainate 2 (GRIK2) differentially methylated depending on the tissue type analyzed but also that the gene is silenced by hypermethylation in gastric cancer cell lines [74]. Moreover, GRIK2 may have tumor suppressor activity in normal gastric epithelium since expression of the gene in normal cells results in decreased colony formation and invasion [74]. These data indicate that there may be a role for iGluR signaling in the development and progression of some cancers. Follow up studies will be needed to determine whether these effects are due to Glu binding directly to iGluRs or whether these effects are mediated by coupling to mGluRs.
In addition to the mutations uncovered in iGluR genes, somatic mutations in the mGluRs (e.g., mGluR1, mGluR3, mGluR5) have also been reported to promote breast cancer, melanoma and renal cell carcinoma growth and progression in vivo [75-80]. Using high throughput studies in cancer genomics, a relatively high number of missense and nonsense mutations were identified in human mGluR1a in a wide variety of cancers such as: adenocarcinoma and squamous cell carcinoma of lung, triple-negative breast cancer, basal cell and ductal carcinoma of the breast, adenocarcinoma of the large intestine, lymphoma and leukemia, high-grade astrocytoma, squamous cell carcinoma of skin and upper aerodisgestive tract, and serous carcinoma of ovary [81-83]. This work has led to the discovery of multiple mutations in the ligand binding domain (LBD), TMD and CTD in mGluR1a (Table 2) [81-88]. Interestingly, most of the base changes reported thus far have been missense mutations which may suggest that truncation of the protein is not favorable for cellular transformation. Moreover, all of the mutations discussed in Table 2 have been uncovered in various types of cancer. Future studies that determine the penetrance of these somatic mutations within specific cancer patient populations will be extremely useful.
Understanding the biological relevance or significance of domain-specific mutations in the mGluR1a gene is essential to define their translational potentials in cancers. An important study by Esseltine and colleagues has elucidated the functional consequences of a number of somatic mutations in mGluR1a [84]. As shown in Table 2, a large number of somatic mutations are clustered in the LBD of mGluR1a. A number of these LBD mutations, including A168V, R375G an d G396V, result in a reduction in the basal activity of the receptor [84]. Since it has been demonstrated that mGluR1a signaling pathways have a high basal level of activity as measured by G-protein activation [89], it is tempting to speculate that there is a selective advantage to reducing basal G-protein activity in cancer cells. However, since not all mutations follow this paradigm, the link between these LBD mutations, basal G-protein activity and mGluR1a signaling in cancer cells requires further investigation [84]. As discussed previously, Homer1 binding to mGluR1a can influence the active receptor at the level of subcellular localization and interaction with signaling components such as ERK1/2 [90]. The P1148L mutation, which is located in the Homer1 binding motif, results in not only mislocalization of the receptor, but also in reduced Homer1b binding - two consequences which have a major impact on receptor activity [84]. The P11148L and R696W mutations (located in the Homer1 binding motif and the TMD, respectively) were shown to influence mGluR1a-dependent phosphorylation of ERK1/2 without influencing the activation of lipid signaling [84]. These data indicate somatic mutations in mGluR1a can have second messenger-dependent effects on receptor signaling. Future studies may lead to not only more clues as to how mGluR1a functions in cancer cells, but also in possible diagnostic and prognostic tools.
Somatic mutations identified in the mGluR1a gene in cancer patients.
| Domain | Mutation | Type | Reference |
|---|---|---|---|
| LBD ( n = 19 ) | S33L | Missense | 85 |
| D44E | Missense | 81 | |
| R71K | Missense | 85 | |
| R78H | Missense | 138 | |
| D87H | Missense | 138 | |
| A91T | Missense | TCGA* | |
| K153N | Missense | TCGA | |
| A168V | Missense | 81 | |
| A184T | Missense | 86 | |
| W224C | Missense | 138 | |
| A229S | Missense | 138 | |
| R275H | Missense | TCGA | |
| R297 - STOP | Nonsense | 84 | |
| R375G | Missense | 81 | |
| E386 - STOP | Nonsense | 81 | |
| G396V | Missense | 81 | |
| I414V | Missense | TCGA | |
| P444L | Missense | 88 | |
| C547F | Missense | 85 | |
| TMD loops ( n = 10 ) | D619A | Missense | 87 |
| S626C | Missense | 137 | |
| T655N | Missense | TCGA | |
| R661H | Missense | TCGA | |
| R684C | Missense | 82 | |
| G688V | Missense | 81 | |
| R696W | Missense | 83 | |
| Q706 - STOP | Nonsense | 85 | |
| N782I | Missense | TCGA | |
| S783I | Missense | 87 | |
| CTD ( n = 4 ) | R967H | Missense | 137 |
| E1006K | Missense | 85 | |
| D1096N | Missense | 138 | |
| P1148L | Missense | 84 |
*TCGA: The cancer genome atlas. http://cancergenome.nih.gov.
Glutamate receptor expression and effective antagonists in different cancer cell lines.
| Cancer type | Cell line | GluR genes detected | Glu antagonists with growth inhibitory effect | Refs |
|---|---|---|---|---|
| Glioma | U87-MG, U343, C6 | mGluR1-8, NR2-3, GluR2-7, KA1-2 | Memantine, MK801 | [33, 91] |
| Neuroblastoma | SK-N-AS | mGluR2-8, NR1-3, GluR2-4, 6-7, KA1-2 | Dizocilpine, GYKI 52466 | [91] |
| Rhabdomyosarcoma & Medulloblastoma | TE671 | mGluR2-4, 6-8, NR1-3, KA1-2, GluR2, 4, 6, 7 | Dizocilpine, GYKI 52466 | [91] |
| Astrocytoma | MOGGCCM | mGluR1-8, NR1-3, KA1-2, GluR1-4; GluR6-7 | Dizocilpine, GYKI 52466 | [91] |
| Lung carcinoma | SK-LU-1 | mGluR1-2, 4-8, NR1-3, KA1-2, GluR1-2, 4, 6 | N.D. | [91] |
| Colon adenocarcinoma | HT29, LS180 | mGluR1-8, NR1-3, KA1-2, GluR1-2, 4, 6-7 | Dizocilpine, GYKI 52466 | [91] |
| T-cell leukemia | Jurkat E6.1 | mGluR1-7, NR2-3, KA1-2, GluR2-4, 6-7 | N.D. | [91] |
| Multiple myeloma | RPMI 8226 | mGluR2-7, NR1-2, KA1-2, GluR4, 6 | N.D. | [91] |
| Breast carcinoma | T47D | mGluR1-7, NR1-3, KA1-2, GluR2, 4-7 | Dizocilpine, BAY36-7620, riluzole, GYKI 52466 | [91, 133] |
| Thyroid carcinoma | FTC 238 | mGluR2, 4, 7, NR1-3, KA1-2, GluR2-4, 6-7 | Dizocilpine, GYKI 52466 | [91] |
| Prostate carcinoma | PC-3, LNCaP, DU145, VCaP, 22RV1, E006AA, MDA-PCa2B, | mGluR1a, mGluR1-8, NR1-3 | Riluzole, BAY36-7620 | [32, 61,71, 91] |
| Melanoma | C8161, UACC903, HT144, SKMEL2 | mGluR1 | Riluzole, BAY36-7620 | [130] |
Confirming at the transcript level that multiple GluR groups are involved in malignant disease, Stepulak, et al. conducted a study in which a panel of human tumor-derived cell lines were assayed for the expression of several iGluR and all eight mGluR genes [91]. Using semi-quantitative reverse transcription PCR (RT PCR), this work illustrated that multiple mGluRs are expressed in each of the 12 cell lines that were investigated. It should be noted that for mGluR1-8, the RT-PCR primers chosen were not able to differentiate between isoforms. The following human cell lines were interrogated for mGluR expression: rhabdomyosarcoma/medulloblastoma (TE671), neuroblastoma (SK-N-AS), thyroid carcinoma (FTC 238), lung carcinoma (SK-LU-1 and A549), astrocytoma (MOGGCCM), multiple myeloma (RPMI 8226), glioma (U87-MG and U343), colon adenocarcinoma (HT 29), T cell leukemia cells (Jurkat E6.1), breast carcinoma (T47D) and colon adenocarcinoma (LS180) [91]. These data are summarized in Table 3.
In addition to the growing number of studies reporting GluR expression in malignant disease, Glu antagonists have been shown to have anti-proliferative and anti-migratory effects both in vitro and in vivo [92]. In a study designed to test the effect of Glu antagonists on migration and proliferation, Rzeski and colleagues exposed astrocytoma (MOGGCCM), rhabdomyosarcoma/medullobastoma (TE671), neuroblastoma (SK-NA-S) and thyroid carcinoma (FTC238) cells to the Glu antagonists dizocilpine and GYK152466 [92]. Interestingly, differential responses to the two drugs were seen in subsets of the cell lines. These effects were attributable to the ability of both drugs to block Ca2+ influx since growing the cells in a reduced-Ca2+ environment mitigated the effect [92]. Dizocilpine is widely known to cause a variety of unpleasant neural side effects including brain lesions and cognitive disruption which are likely due to both the drug's potent blockage of Ca2+ influx in NMDAR-expressing neurons and the pleiotropic role for Glu signaling in the nervous system [93]. Due to these side effects, the drug is not suitable for cancer therapy and is therefore not used in the clinic. However, these data are important in that they establish what is emerging as a general phenomenon: Glu antagonists and blockade of Glu release from cells has anti-tumor effects. Recalling that NMDARs are often coupled to Group I mGluRs, the mechanism for NMDAR involvement in tumor cell growth and migration may lie in mGluR signaling, after all. The mechanism of the effect of Glu antagonists on cancer cells is under-investigated.
Glutamate excitotoxicity and glioma
The role of glutamatergic signaling in the progression of glioma is well established (reviewed in [70, 94, 95]). Glioma represents the family of malignant tumors comprised of cells of glial origin and can contain ependymal cells, oligodendrocytes, astrocytes or a mixture of cell types. Making up 80% of all malignant brain diseases, gliomas can be either fast-growing or slow-growing, and that classification trends with treatability: slow-growing gliomas are more treatable and vice versa [96]. Importantly, glioma explants have been shown to continuously secrete Glu and therefore maintain a higher level of intra- and extracellular Glu than normal glial cells [97]. Tumors that secrete Glu are highly resistant to chemotherapy and standard apoptosis-inducing therapeutics - a phenomenon that may be mediated by the same mechanism whereby Glu functions as a survival factor at normal physiological doses in the developing nervous system [98]. However, too much Glu is known to be toxic, and although the malignant glioblastoma cells are themselves resist to excitotoxicity, neighboring neurons are not. Local apoptosis of neurons is commonly observed in glioma patients and is often a source of extensive brain tissue damage despite successful tumor resection [99]. In fact, it has been postulated that a selective infiltrative capacity for certain gliomas lies in their abilities to create more space for growth by killing neighboring cells [100]. Research has shown that excitotoxic levels of Glu arise from a shift in the balance between Glu transporter activity and secretion through the amino acid antiporter, cysteine-glutamine exchanger (X-c) (reviewed in [101] and below). The mechanisms by which normal cells secrete and remove Glu from the extracellular space are critically altered in transformed glial cells (reviewed in [70, 94]).
The role of EAAT in glioma-induced excitotoxicity has been well-studied. Normally, after stimulation of GluRs, Glu is emptied from the synaptic cleft or intracellular space by EAATs, of which there are multiple genes and splice variants in humans [33]. The role of EAATs in the growth and progression of glioma was studied in detail using the in vitro rat glioma cell line, C6 [33]. C6 cells normally express excitatory amino acid carrier 1 (EAAC1, an EAAT); however, clonal populations of the cell line have been shown not only to release Glu into the medium, but also to have selective loss of EAAC1 expression [33]. In animal models, Glu-releasing C6 clones showed a distinct growth advantage over those that did not secrete Glu. In addition, co-culture of Glu-secreting clones of C6 with neurons resulted in apoptosis of the neural populations [33]. It is counter-intuitive that gliomas such as astrocytoma release high levels of Glu into the extracellular space since a main function of normal astrocytes is to remove Glu post-signaling in the normal CNS. Immunohistochemical studies on human astrocytomas show a complete loss of the glutamate transporter-1 (GLT1) splice variant and aberrant localization of a second EAAT, glutamate/aspartate transporter (GLAST) [97]. It is likely not a coincidence that GLT1 and GLAST are the predominantly-expressed EAATs by astrocytes in the rat brain [102]. Together, the aforementioned studies indicate that impaired Glu transporter function and consequential constitutive Glu signaling is an important part of cellular transformation in glial cells. The observed reduction in EAAT activity in glioma is attributable to mGluR signaling (reviewed in [94, 103]). Expression of mGluR2, 3 and 5 but not 1 has been detected in normal glial cell types including astrocytes. Activation of mGluR3 or 5 with chemical agonists resulted in decreased EAAT (GLT1 and GLAST) expression in astrocytes in culture, demonstrating a direct link between mGluRs and EAAT expression. Importantly, glioblastoma cells display increased mGluR5 expression in vivo [104]. These data are suggestive of a direct role for mGluR in the fundamental change between normal astrocyte function and the transformed phenotype of reduced EAAT expression. Another important part in establishing an excitotoxic phenotype lies in the Glu secretion mechanism.
The method by which glioma cells release Glu into the extracellular space is mediated by the amino acid exchange antiporter, X-c [105]. This heterodimer catalyzes the exchange of extracellular cysteine for intracellular Glu at a 1:1 stoichiometric ratio. Many gliomas were found to have higher than normal levels of X-c - a significant reason for the accumulation of extracellular Glu [106]. As predicted given the results discussed thus far, silencing of X-c expression using small interfering RNA (siRNA) resulted in marked reduction in local apoptosis and brain swelling as well as reduced tumor growth rates [101, 107]. It is tempting, therefore, to assume that anti-X-c drugs would be potent treatments for glioma. Since the X-c antiporter has critical functions in the protection against oxidative stress outside of the nervous system, and since the loss of glutathione via X-c inhibition would therefore be cytotoxic, it is unlikely that anti-X-c therapies will be a viable option for glioma treatment [101, 107].
Downstream of the GluR family, phosphatase and tensin homolog (PTEN) and AKT signaling have been heavily implicated in glioma progression, resistance to chemotherapy and excitotoxicity. Loss of heterozygosity of PTEN has been shown for many cancers including glioma (as well as prostate, see [108] for a review). It is significant that the highly aggressive and most common form of glioma, glioblastoma multiforme, shows a higher penetrance of loss of PTEN function than the lower-grade astrocytomas [109]. The loss of PTEN-mediated inhibition of PI3K activity has important consequences for the cell: First, unchecked AKT activation leads to protection from excitotoxicity [103, 110, 111]. Second, since PTEN is also responsible for inhibiting Tumor necrosis factor-α (TNFα) signaling which activates nuclear factor κ enhancer of activated B cells (NFκB), elimination of TNFα inhibition results in overactive NFκB. This signaling situation is a well-known survival factor in many cancers [112]. It is clear from these data that glioma cells develop unique signaling adaptations that allow them to survive in an excitotoxic microenvironment. Treatments for glioma must involve shutting down these anti-apoptosis signals and removing toxic levels of Glu from the tumor microenvironment.
Glu antagonists have been extensively employed in the treatment of glioma. In recent years, there have been multiple significant failures of clinical trials utilizing other drugs families such as the receptor tyrosine kinase inhibitor family (reviewed in [113]). This may be partially due to the fact that inhibition of RTKs does not mitigate the release of Glu from glioma cells. Differential effects have been documented for iGluR and mGluR antagonists. First, the AMPAR/KR inhibitors, ZK 200775 and GYKI 52466, while having little effect on glioma growth in vitro, have pronounced anti-proliferative and anti-excitotoxic effects in rat hippocampal glioma models. Similar results were obtained for the NMDAR antagonists, norketamine [72] and MK801 (memantine) [33]. In fact, memantine has been employed in a Phase II clinical trial to determine its safety/efficacy in glioma patients; however results are not yet available (Clinicaltrials.gov #NCT01260467). Similarly, the effectiveness of talampanel (an AMPAR antagonist) against glioma has been explored in the clinic. This study was completed with results that were not encouraging: no significant activity against glioma was observed (reviewed in [33, 94]).
Despite all of the preclinical work and clinical trials in progress or completed involving iGluR antagonists, studies targeting mGluR in glioma treatment are less commonly found. Perhaps the major discovery that gliomas release Glu and create excitotoxic local conditions within the brain will prompt new studies of this type. Indeed, a review by Roesler and colleagues in 2010 defines the family of mGluRs as “emerging targets” in glioma [114]. So far, no clinical trials are available using mGluR antagonists in glioma patients on Clinicaltrials.gov. Future studies in this field are clearly needed to determine the efficacy of mGluR antagonists and Glu release inhibitors such as riluzole against glioma.
mGluR expression causes cellular transformation in melanocytes
Although many reports of receptor expression in cell lines have been published, few population studies in human patient samples are available and few have established a mechanism for human mGluR in either carcinogenesis or disease progression. Certainly the first direct evidence for the oncogenic activity of the mGluRs was shown in melanocytes by Chen and colleagues [34, 115]. Overexpression of mGluR1 in non-cancerous melanocytes was sufficient to induce transformed phenotypes, indicating that mGluR1 functions as a proto-oncogene (reviewed in [33, 34]). Indeed, mGluR1 expression is detected in human melanoma samples and transformed melanoma cell lines, but not in benign nevi or normal melanocytes. In the pioneering study in the field, Pollock, et. al found that mGluR1 protein was expressed in 7 of 19 human melanoma biopsy samples and 12 of 18 human melanoma cell lines. As is the case in many studies, these data were obtained using western blot with an undescribed antibody and the resulting figure shows bands without any size marker or indication of the size of the protein [79]. In addition, due to the lack of mGluR isoform-specific antibodies, differential expression of mGluR1 isoforms could not be demonstrated. In agreement with these data, transgene insertion into an intron in the mouse mGluR1 gene resulted in increased susceptibility to melanoma - furthering the model that mGluR1 may induce oncogenic transformation [79, 116-118]. Subsequent work demonstrated that the ectopic expression of murine mGluR1 in normal melanocytes in vitro was sufficient to induce rapid and aggressive tumor growth in nude mouse models. Angiogenesis and metastasis were readily apparent in this model, suggesting that the normal melanocytes were completely transformed by murine mGluR1 expression. Further, murine mGluR1 overexpression was found to be sufficient to cause malignant phenotypes in murine melanocytes both in vitro and in vivo [120]. Interestingly, constitutive expression of mGluR1 is required to maintain the transformed phenotype. Using a unique conditional expression system in transgenic mice, Ohtani, et al. showed that long-term expression of mGluR1 stimulated melanocytes growth and eventually led to melanoma, while silencing mGluR1 expression was sufficient to abrogate melanoma development [78]. This requirement for constitutive mGluR1 expression may indicate that mGluR1 is an ideal drug target [78].
Although the exact downstream signaling involved in the ability of mGluR1 to transform melanocytes is not yet known, some clues have been found using both genome-wide sequencing and candidate approach techniques. First, stimulation of Glu signaling in melanoma cell lines with mGluR agonists results in activation of MAPK; specifically ERK1/2 via active PKC [119]. Downstream activation of AKT2, but not AKT3, is found in mGluR1 overexpressed lines and this effect is enough to cause transformation in other cell lines [120, 121].
Whole-exome sequencing of melanoma and normal matched patient samples has revealed an accumulation of mutations in Glu signaling components including GRIN2A - an NMDAR subunit [70, 122]. Further studies have similarly implicated a number of other Glu signaling components: Mutations were found in the other NMDAR subunits GRIN1 and GRIN3, as well as in PLCβ4, mGluR3, PYK2 and receptor tyrosine-protein kinase erbB-4 (ERBB4) [70, 73]. PYK2 has been shown to interact directly with GRIN2A via PH-domain binding - further indicating a role for aberrant GluR signaling in cellular transformation [123]. Similarly, PLCβ4 is activated by mGluR signaling and produces the second messengers DAG and IP3.
Interestingly, mutations in mGluR3 were found to be selectively activating. Mutational analysis of mGluR3 in vitro demonstrated that recapitulation of the somatic mutations (found in patients) imparted increased activation of mitogen activated protein kinase kinase 1/2 (MEK 1/2). Phosphorylated MEK1/2 resulted in transformed phenotypes including loss of anchorage-dependency in growth regulation and increased migration [70]. Further, cells carrying the same mGluR3 mutations formed increased micrometastases in vivo [70]. Finally, ERBB4 is a well-known oncogene and its mutations are associated with increased risk not only for certain types of cancer, but also for schizophrenia [124]. As a downstream effector of mGluR1 signaling, these data represent an important link between GluRs and second messenger signaling mechanisms in melanoma. Follow-up studies have illustrated a link between constitutively active ERBB4 and increased invasion and migration in melanoma cells [125].
Drugs that block Glu release from the cell or otherwise disrupt Glu signaling have shown great promise in melanoma treatment in preclinical trials. It has been clearly demonstrated that transformed melanocytes release high levels of Glu into the medium. Similarly, glioma cells have also been shown to secrete Glu [33, 70]. Blockade of Glu release from melanocytes by riluzole results in decreased cell proliferation, suggesting an autocrine-like loop whereby mGluR1 expression is linked with increased extracellular Glu and proliferation. However, a direct causal relationship has not been established between mGluR activation and Glu release [126, 127]. Riluzole is able to suppress the growth of human melanoma xenografts in vivo by 50% compared to vehicle-treated controls [127]. Further, the Glu antagonists LY367385 and BAY36-7620 are also able to suppress proliferation and Glu release in vitro [127]. Taken together, these data lend further evidence to the hypothesis that riluzole, or other GluR antagonists, may have great potential in the treatment of melanoma. Indeed, a phase 0 clinical trial of riluzole in patients with advanced melanoma demonstrated a 34% success rate (n=12 patients, all expressing mGluR1) in decreasing tumor load [129]. Importantly, treated tumors showed decreased phospho-AKT (p-AKT) and/or p-ERK, indicating that the anti-tumor effect was due to inhibition of mGluR signaling [128]. Unfortunately, the follow-up phase II study with riluzole was not as successful, with no change in response evaluation criteria in solid tumors (RECIST) measurements. 42% of treated patients did not exhibit disease progression, however, implying that riluzole was at least able to suspend tumor growth in this population [129, 130]. Recent work has focused on combination therapies such as riluzole and the tyrosine kinase inhibitor, sorafenib [129]. Marketed by Bayer as Nexavar, sorafenib is a small molecule that targets the tyrosine kinases PDGF and VEGF as well as Raf kinases such as B- and C-Raf [131]. In preclinical studies using a combination of riluzole and sorafenib, significant inhibition of tumor growth was observed. These data were confirmed in cancer cell line investigated and were shown to be coincident with a reduction in p-ERK activity levels. A clinical trial to confirm these data in human melanoma patients is currently in progress (Clinicaltrials.gov #NCT01303341).
In summary, a role for mGluR signaling in melanomagenesis has been clearly established. This heterogeneous malignancy is not simply cured by Glu blockade, however, given the less-than-impressive results of the phase II trial that was conducted on riluzole in late-stage mGluR1-expressing melanoma patients. These results may be explainable by the mechanism of action of riluzole. It is likely that blocking the release of Glu from the cell is not enough to exert an anti-tumor effect in human patients. Combination therapy involving riluzole and either tyrosine kinase inhibitors like sorafenib or radiation treatment may provide some relief for patients suffering from this aggressive tumor. The next few years will be critical for this field: If riluzole is not the key to block Glu signaling in transformed melanocytes, then new drugs as Glu-release inhibitor or competitive or non-competitive Glu receptor inhibitor will need to be developed.
Preliminary studies suggest that riluzole is effective against breast carcinoma
Breast cancer, a “steroid hormone dependent” carcinoma, represents a group of malignancies with characteristic steroid receptor expression and steroid hormone responsiveness. Despite the decades of research and successes in treating estrogen receptor (ER)-positive, progesterone receptor (PR)-positive and human epidermal growth factor receptor 2 (HER2)-positive breast cancers, patients that are negative for these markers (triple negative breast cancers; TNBCs) have been forced to rely on broad-spectrum cytotoxic agents [132]. Research that identifies hormone-independent markers and therapeutic targets for these specific breast cancers are needed.
Following the recent explosion of interest in Glu signaling in melanomagenesis and treatment, Speyer and colleagues undertook a pilot study to determine the role of mGluR signaling in breast cancer [133]. First, mGluR1 expression was assayed in a number of TNBC breast cancer cell lines. Including normal breast epithelial cells, all of the cell lines examined were found to express mGluR1 protein [133]. Furthermore, Glu signaling was active in these cells since phospho-AKT2 (p-AKT) was stimulated by the AMPA/mGluR1 Group 1 agonist, quisqualic acid and inhibited by the non-competitive mGluR1 antagonist, BAY36-7620 [133]. Similar to the findings discussed above for melanoma cells in culture, TNBC cell growth is inhibited by mGluR1 antagonists and shRNA knockdown of mGluR1 resulted in a statistically significant reduction in proliferation [133]. As was the case in other studies of this nature and for the same reasons described before, no differentiation among isoforms of mGluR1 was made and it is still unknown whether one isoform is more important than another in the regulation of cancer cell proliferation. In fact, knockdown was assayed at the level of mRNA expression using primers that are homologous to both the mGluR1a and mGluR1b mRNAs, indicating that both isoforms may have been knocked down using this shRNA (determination made by BLASTn search). Nevertheless, these results are significant in that they establish that for TNBCs, mGluR1 may play a similar role in tumor growth and possibly even transformation as has been shown for melanomagenesis. Interestingly, another group has reported that breast cancer cells secrete Glu into the medium, indicating that glioma, melanoma and breast cancer may have some intriguing similarities [134, 135].
In a TNBC xenograft model, doses of riluzole that were found to be effective in treating ALS patients showed up to 80% decrease in tumor volume [133], further indicating that mGluR and Glu signaling are emerging therapeutic targets in breast cancer treatment. We postulate that multiple studies are underway at the time of the writing of this review and that the coming years in the field of Glu signaling in TNBC research will be very interesting.
Glutamate and glutamate receptors in prostate cancer cells and tissues
The role of Glu signaling in prostate carcinogenesis and progression has not been investigated until very recently. In 2009, one study was published which suggested that subsets of mGluR genes are expressed in the PCa cell lines PC-3 and LNCaP [71]. It is interesting to note that different subsets of NMDARs and mGluRs were detected in the two cell lines - but the functional significance of these data remains unknown [71].
We recently reported that serum Glu levels correlate with primary PCa aggressiveness and that Glu antagonists decrease PCa cell proliferation [31]. Most importantly, the clinical significance of Glu signaling in PCa was determined by measuring serum-Glu levels in normal adult males and patients with primary untreated organ-confined or metastatic castrate-resistant tumors [31]. Using Gleason score (GS) as a pathologic grading pattern (reflecting PCa aggressiveness or lethality) and other clinical parameters (PSA and clinical stage) to divide primary PCa into the clinically low, mid, or high aggressiveness subgroups, it was found that higher serum Glu levels correlated with higher GS (>8) and more aggressive disease [31]. Further, racial disparity was found: African American (AA) PCa patients had higher serum Glu levels than Caucasian PCa patients. In AAs, serum-Glu levels in metastatic castrate-resistant PCa (mCRPCa) were also found to be higher than in the primary PCa (early stage and less aggressive) or normal research subjects [31]. Although the source of increased circulating Glu is unknown at this time, it is tempting to speculate that PCa cells release Glu into the serum - as has been shown for breast cancer cells in culture and glioma in human or rodent cell systems. Furthermore, we have also detected extracellular Glu release in Glu-depleted conditioned media collected from different PCa cell lines (Koochekpour et al, unpublished data). In addition, our descriptive pilot study demonstrated mGluR1a (also known as GRM1a) overexpression in primary and metastatic PCa tissues [31, 32].
The clinical significance of these studies lies in the fact that Glu has the potential to serve as a metabolic serum biomarker for primary PCa in Caucasians and/or for primary and metastatic PCa in African Americans. In addition, mGluR1a may also serve as a molecular target for therapeutic interventions in both African Americans and Caucasians. In fact, microarray studies have previously demonstrated that targeting the AKT/mTOR pathway with anti-mTOR or anti-MAPK/ERK drugs resulted in decreased growth of PCa cell lines in vitro [136]. Taken together, these data prompt a model in which glutamatergic signaling presents itself as a critical druggable target for PCa.
In PCa cell lines, mGluR1 is detected and Glu signaling plays a role in growth regulation [31]. As we previously published, not only is mGluR1 expression is detected at high levels in high-grade human PCa tumors, but also, in PCa cell lines, multiple bands of varying size are detected on western blot using a commercial anti-mGluR1 antibody (1:1000; Proteintech Group, Chicago, IL. Catalog #19955) [31]. Interestingly, the predominant band (i.e. ~75 KDa) that is detected with this antibody (which was raised against an N-terminal peptide) is significantly smaller than the predicted monomeric molecular weight of ~125 KDa for mGluR1a, which may indicate that the protein is processed in some way in PCa cells. This phenomenon was global: All 7 PCa cell lines tested displayed this smaller band, regardless of their androgen responsiveness or the clinical aggressiveness of their tumors of origin [31]. Glu signaling in these lines is functional, since inhibition with the antagonists BAY36-7620 or riluzole and deprivation of Glu were both able to significantly decrease PCa cells growth. In fact, riluzole induced apoptosis in a time- and dose-dependent manner in PCa cells [31]. This effect was mediated by activation of caspases, since levels of cleaved caspase-3, -7 and -9 and -PARP were increased with increased doses of riluzole. Finally, both antagonists were also able to suppress two of the most important malignancy-associated phenotypes: migration and invasion. This study prompts the model that high serum Glu levels create a favorable growth environment or microenvironment for PCa. It will be critical to determine whether this is the case for other cancers (e.g. breast, melanoma and glioma) where Glu antagonists have shown anti-proliferative effects. Large scale clinical studies will certainly be necessary to define clinical and histopathological significance of Glu (ligand) and mGLuR1 (receptor) expression and signaling in PCa. It is noteworthy a non-invasive serum Glu test either alone or in combination with other PCa biomarkers [prostate cancer 3 (PCA3) for example; see [108] for review] for PCa diagnosis could potentially aid in early-stage diagnosis and treatment planning. Finally. Glu antagonists may serve as novel therapeutic agents for PCa [32].
Concluding remarks
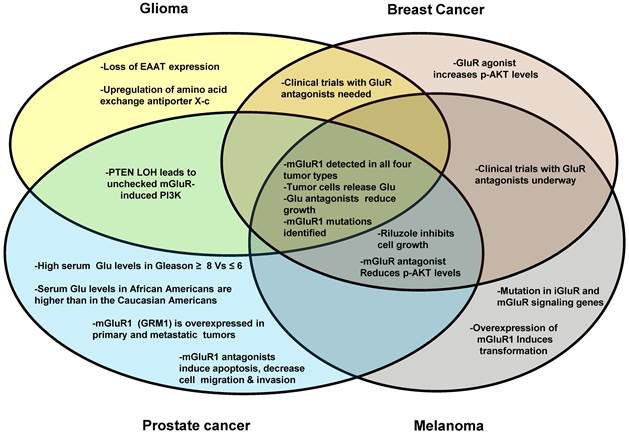
Research into the role of Glu signaling in cancer development and progression is still in its infancy; however, important progress has been made in recent years. The discovery that rodent mGluR overexpression is sufficient for cellular transformation in cultured melanocytes has had a tremendous impact on the field. Further, the ability of glioma, and possibly advanced TNBCs, to secrete Glu into the tumor microenvironment has prompted the question of whether this feature is common to other types of cancers [94, 134, 135]. It is tempting to speculate that both glioma and melanoma originate from cells of the ectodermal germ layer or simply share a common neuroendocrine origin given the similarities between the malignancies. Interestingly, neuroendocrine cells also exist in both benign and malignant prostate tissues and the neuroendocrine differentiation pathway is believed to be one of the most important mechanisms for the development of castrate-resistant PCa. In Figure 3, we present the similarities and differences among the cancers that have been discussed in this review. It is clear that more research is needed to define the clinical and histopathological significance of Glu-mGlUR expression and signaling in glioma, melanoma, breast cancer, and PCa. Given these preliminary studies, it will be extremely interesting to follow the field of glutamatergic signaling in cancer in future years.
GluR involvement in different cancer types. This Venn diagram illustrates the similarities and differences among the four cancer types with respect to the involvement of glutamatergic signaling. Glioma, breast cancer, prostate cancer and melanoma are represented by the circles. Overlapping areas indicate the expressional and functional similarities for GluR genes among different cancer types investigated. For example, mGluR proteins have been detected in all four cancer types, therefore this phrase is in an area where all four circles overlap.
Acknowledgements
This work was supported by Roswell Park Cancer Institute and National Cancer Institute (NCI) grant #P30 CA016056. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or National Cancer Institute.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kelly A, Stanley CA. Disorders of glutamate metabolism. Ment Retard Dev Disabil Res Rev. 2001;7:287-95
2. Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997;37:205-37
3. Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK. et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405-96
4. Kullmann DM. The Mother of All Battles 20 years on: is LTP expressed pre- or postsynaptically? J Physiol. 2012;590:2213-6
5. Julio-Pieper M, Flor PJ, Dinan TG, Cryan JF. Exciting times beyond the brain: metabotropic glutamate receptors in peripheral and non-neural tissues. Pharmacol Rev. 2011;63:35-58
6. Ribeiro FM, Paquet M, Cregan SP, Ferguson SS. Group I metabotropic glutamate receptor signalling and its implication in neurological disease. CNS Neurol Disord Drug Targets. 2010;9:574-95
7. Seebahn A, Sticht H, Enz R. Expression, purification, and structural analysis of intracellular C-termini from metabotropic glutamate receptors. Methods Enzymol. 2013;520:257-79
8. Iwazumi T. Myofibril tension fluctuations and molecular mechanisms of contraction. Adv Exp Med Biol. 1988;226:595-608
9. Beqollari D, Kammermeier PJ. Venus fly trap domain of mGluR1 functions as a dominant negative against group I mGluR signaling. J Neurophysiol. 2010;104:439-48
10. Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP, Worley PF. et al. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 2000;26:143-54
11. Brauner-Osborne H, Wellendorph P, Jensen AA. Structure, pharmacology and therapeutic prospects of family C G-protein coupled receptors. Curr Drug Targets. 2007;8:169-84
12. Fagni L, Chavis P, Ango F, Bockaert J. Complex interactions between mGluRs, intracellular Ca2+ stores and ion channels in neurons. Trends Neurosci. 2000;23:80-8
13. Luo P, Li X, Fei Z, Poon W. Scaffold protein Homer 1: implications for neurological diseases. Neurochem Int. 2012;61:731-8
14. Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol. 2007;8:206
15. El Moustaine D, Granier S, Doumazane E, Scholler P, Rahmeh R, Bron P. et al. Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc Natl Acad Sci U S A. 2012;109:16342-7
16. Muto T, Tsuchiya D, Morikawa K, Jingami H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2007;104:3759-64
17. Romano C, Yang WL, O'Malley KL. Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J Biol Chem. 1996;271:28612-6
18. Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP. et al. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 2000;26:143-54
19. Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717-726
20. Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707-716
21. Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol. 2007;8(2):206.1-206.12
22. Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137(1):159-71
23. Mao L, Yang L, Tang Q, Samdani S, Zhang G, Wang JQ. The scaffold protein Homer1b/c links metabotropic glutamate receptor 5 to extracellular signal-regulated protein kinase cascades in neurons. J Neurosci. 2005;25:2741-2752
24. Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543-547
25. Rong R, Ahn JY, Huang H, Nagata E, Kalman D, Kapp JA, Tu J, Worley PF, Snyder SH, Ye K. PI3 kinase enhancer-Homer complex couples mGluRI to PI3 kinase, preventing neuronal apoptosis. Nat Neurosci. 2003;6:1153-1161
26. Xiao B, Tu JC, Worley PF. Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol. 2000;10:370-374
27. Brandstätter JH, Dick O, Boeckers TM. The postsynaptic scaffold proteins ProSAP1/Shank2 and Homer1 are associated with glutamate receptor complexes at rat retinal synapses. J Comp Neurol. 2004;475:551-563
28. Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113:1851-1856
29. Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: A protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284-288
30. Kato A, Ozawa F, Saitoh Y, Hirai K, Inokuchi K. Vesl, a gene encoding VASP/Ena family related protein, is upregulated during seizure, long-term potentiation and synaptogenesis. FEBS Lett. 1997;412:183-189
31. Koochekpour S, Majumdar S, Azabdaftari G, Attwood K, Scioneaux R, Subramani D. et al. Serum glutamate levels correlate with Gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin Cancer Res. 2012;18:5888-901
32. Koochekpour S. Glutamate, a metabolic biomarker of aggressiveness and a potential therapeutic target for prostate cancer. Asian J Androl. 2013;15:212-3
33. Takano T, Lin JH, Arcuino G, Gao Q, Yang J, Nedergaard M. Glutamate release promotes growth of malignant gliomas. Nat Med. 2001;7:1010-5
34. Teh JL, Chen S. Glutamatergic signaling in cellular transformation. Pigment Cell Melanoma Res. 2012;25:331-42
35. Kim K, Lee SG, Kegelman TP, Su ZZ, Das SK, Dash R. et al. Role of excitatory amino acid transporter-2 (EAAT2) and glutamate in neurodegeneration: opportunities for developing novel therapeutics. J Cell Physiol. 2011;226:2484-93
36. Mehta A, Prabhakar M, Kumar P, Deshmukh R, Sharma PL. Excitotoxicity: bridge to various triggers in neurodegenerative disorders. Eur J Pharmacol. 2013;698:6-18
37. Bailey AR, Giunta BN, Obregon D, Nikolic WV, Tian J, Sanberg CD. et al. Peripheral biomarkers in Autism: secreted amyloid precursor protein-alpha as a probable key player in early diagnosis. Int J Clin Exp Med. 2008;1:338-44
38. Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370-7
39. Carlson GC. Glutamate receptor dysfunction and drug targets across models of autism spectrum disorders. Pharmacol Biochem Behav. 2012;100:850-4
40. Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012;37:4-15
41. Ray B, Long JM, Sokol DK, Lahiri DK. Increased secreted amyloid precursor protein-alpha (sAPPalpha) in severe autism: proposal of a specific, anabolic pathway and putative biomarker. PLoS One. 2011;6:e20405
42. Sokol DK, Maloney B, Long JM, Ray B, Lahiri DK. Autism, Alzheimer disease, and fragile X: APP, FMRP, and mGluR5 are molecular links. Neurology. 2011;76:1344-52
43. Yerys BE, Wallace GL, Sokoloff JL, Shook DA, James JD, Kenworthy L. Attention deficit/hyperactivity disorder symptoms moderate cognition and behavior in children with autism spectrum disorders. Autism Res. 2009;2:322-33
44. Lisman JE, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S. et al. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008;31:234-42
45. Patil ST, Zhang L, Martenyi F, Lowe SL, Jackson KA, Andreev BV. et al. Activation of mGlu2/3 receptors as a new approach to treat schizophrenia: a randomized Phase 2 clinical trial. Nat Med. 2007;13:1102-7
46. Kirkpatrick B. Understanding the physiology of schizophrenia. J Clin Psychiatry. 2013;74:e05
47. Dong XX, Wang Y, Qin ZH. Molecular mechanisms of excitotoxicity and their relevance to pathogenesis of neurodegenerative diseases. Acta Pharmacol Sin. 2009;30:379-87
48. Park E, Velumian AA, Fehlings MG. The role of excitotoxicity in secondary mechanisms of spinal cord injury: a review with an emphasis on the implications for white matter degeneration. J Neurotrauma. 2004;21:754-74
49. Coyle JT, Basu A, Benneyworth M, Balu D, Konopaske G. Glutamatergic synaptic dysregulation in schizophrenia: therapeutic implications. Handb Exp Pharmacol. 2012:267-95
50. Duty S. Targeting glutamate receptors to tackle the pathogenesis, clinical symptoms and levodopa-induced dyskinesia associated with Parkinson's disease. CNS Drugs. 2012;26:1017-32
51. Goeldner C, Ballard TM, Knoflach F, Wichmann J, Gatti S, Umbricht D. Cognitive impairment in major depression and the mGlu2 receptor as a therapeutic target. Neuropharmacology. 2013;64:337-46
52. Pin JP, Bockaert J. Get receptive to metabotropic glutamate receptors. Curr Opin Neurobiol. 1995;5:342-9
53. Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34:1-26
54. Schwarz S, Froelich L, Burns A. Pharmacological treatment of dementia. Curr Opin Psychiatry. 2012;25:542-50
55. Mathews DC, Henter ID, Zarate CA. Targeting the glutamatergic system to treat major depressive disorder: rationale and progress to date. Drugs. 2012;72:1313-33
56. Berman RM, Cappiello A, Anand A, Oren DA, Heninger GR, Charney DS. et al. Antidepressant effects of ketamine in depressed patients. Biol Psychiatry. 2000;47:351-4
57. Zarate CA Jr, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA. et al. A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry. 2006;63:856-64
58. Pilc A, Chaki S, Nowak G, Witkin JM. Mood disorders: regulation by metabotropic glutamate receptors. Biochem Pharmacol. 2008;75:997-1006
59. Cryan JF, Mombereau C, Vassout A. The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neurosci Biobehav Rev. 2005;29:571-625
60. Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7:639-49
61. Miller RG. Carrell-Krusen Symposium invited lecture. Clinical trials in motor neuron diseases. J Child Neurol. 1999;14:173-9
62. Owen RT. Glutamatergic approaches in major depressive disorder: focus on ketamine, memantine and riluzole. Drugs Today (Barc). 2012;48:469-78
63. Zarate C Jr, Machado-Vieira R, Henter I, Ibrahim L, Diazgranados N, Salvadore G. Glutamatergic modulators: the future of treating mood disorders? Harv Rev Psychiatry. 2010;18:293-303
64. Malgouris C, Daniel M, Doble A. Neuroprotective effects of riluzole on N-methyl-D-aspartate- or veratridine-induced neurotoxicity in rat hippocampal slices. Neurosci Lett. 1994;177:95-9
65. Doble A. The pharmacology and mechanism of action of riluzole. Neurology. 1996;47:S233-41
66. Akamatsu K, Shibata MA, Ito Y, Sohma Y, Azuma H, Otsuki Y. Riluzole induces apoptotic cell death in human prostate cancer cells via endoplasmic reticulum stress. Anticancer Res. 2009;29:2195-204
67. McDonnell ME, Vera MD, Blass BE, Pelletier JC, King RC, Fernandez-Metzler C. et al. Riluzole prodrugs for melanoma and ALS: design, synthesis, and in vitro metabolic profiling. Bioorg Med Chem. 2012;20:5642-8
68. Cavalheiro EA, Olney JW. Glutamate antagonists: deadly liaisons with cancer. Proc Natl Acad Sci U S A. 2001;98:5947-8
69. Kalariti N, Pissimissis N, Koutsilieris M. The glutamatergic system outside the CNS and in cancer biology. Expert Opin Investig Drugs. 2005;14:1487-96
70. Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18:4240-6
71. Pissimissis N, Papageorgiou E, Lembessis P, Armakolas A, Koutsilieris M. The glutamatergic system expression in human PC-3 and LNCaP prostate cancer cells. Anticancer Res. 2009;29:371-7
72. Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I. et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021-6
73. Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J. et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442-6
74. Wu CS, Lu YJ, Li HP, Hsueh C, Lu CY, Leu YW. et al. Glutamate receptor, ionotropic, kainate 2 silencing by DNA hypermethylation possesses tumor suppressor function in gastric cancer. Int J Cancer. 2010;126:2542-52
75. Abdel-Daim M, Funasaka Y, Komoto M, Nakagawa Y, Yanagita E, Nishigori C. Pharmacogenomics of metabotropic glutamate receptor subtype 1 and in vivo malignant melanoma formation. J Dermatol. 2010;37(7):635-46
76. Choi KY, Chang K, Pickel JM, Badger JD 2nd, Roche KW. Expression of the metabotropic glutamate receptor 5 (mGluR5) induces melanoma in transgenic mice. Proc Natl Acad Sci. 2011;108(37):15219-24
77. Martino JJ, Wall BA, Mastrantoni E, Wilimczyk BJ, La Cava SN, Degenhardt K, White E, Chen S. Metabotropic glutamate receptor 1 (Grm1) is an oncogene in epithelial cells. Oncogene. 2012 epub ahead of print
78. Ohtani Y, Harada T, Funasaka Y, Nakao K, Takahara C, Abdel-Daim M. et al. Metabotropic glutamate receptor subtype-1 is essential for in vivo growth of melanoma. Oncogene. 2008;27:7162-70
79. Pollock PM, Cohen-Solal K, Sood R, Namkoong J, Martino JJ, Koganti A. et al. Melanoma mouse model implicates metabotropic glutamate signaling in melanocytic neoplasia. Nat Genet. 2003;34:108-12
80. Prickett TD, Wei X, Cardenas-Navia I, Teer JK, Lin JC, Walia V. et al. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet. 2011;43:1119-26
81. Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869-73
82. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807-12
83. Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268-74
84. Esseltine JL, Willard MD, Wulur IH, Lajiness ME, Barber TD, Ferguson SS. Somatic mutations in GRM1 in cancer alter metabotropic glutamate receptor 1 intracellular localization and signaling. Mol Pharmacol. 2013;83:770-80
85. Durinck S, Ho C, Wang NJ, Liao W, Jakkula LR, Collisson EA, Pons J, Chan SW, Lam ET, Chu C, Park K, Hong SW, Hur JS, Huh N, Neuhaus IM, Yu SS, Grekin RC, Mauro TM, Cleaver JE, Kwok PY, LeBoit PE, Getz G, Cibulskis K, Aster JC, Huang H, Purdom E, Li J, Bolund L, Arron ST, Gray JW, Spellman PT, Cho RJ. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1(2):137-43
86. Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, López-Guerra M, Colomer D, Navarro A, Baumann T, Aymerich M, Rozman M, Delgado J, Giné E, Hernández JM, González-Díaz M, Puente DA, Velasco G, Freije JM, Tubío JM, Royo R, Gelpí JL, Orozco M, Pisano DG, Zamora J, Vázquez M, Valencia A, Himmelbauer H, Bayés M, Heath S, Gut M, Gut I, Estivill X, López-Guillermo A, Puente XS, Campo E, López-Otín C. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47-52
87. Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395-9
88. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareño C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157-60
89. Dale LB, Bhattacharya M, Anborgh PH, Murdoch B, Bhatia M, Nakanishi S, Ferguson SS. G protein-coupled receptor kinase-mediated desensitization of metabotropic glutamate receptor 1A protects against cell death. J Biol Chem. 2000;275(49):38213-20
90. Rozengurt E. Mitogenic signaling pathways induced by G protein-coupled receptors. J Cell Physiol. 2007;213(3):589-602
91. Stepulak A, Luksch H, Gebhardt C, Uckermann O, Marzahn J, Sifringer M. et al. Expression of glutamate receptor subunits in human cancers. Histochem Cell Biol. 2009;132:435-45
92. Rzeski W, Turski L, Ikonomidou C. Glutamate antagonists limit tumor growth. Proc Natl Acad Sci U S A. 2001;98:6372-7
93. Lim AL, Taylor DA, Malone DT. Consequences of early life MK-801 administration: long-term behavioural effects and relevance to schizophrenia research. Behav Brain Res. 2012;227:276-86
94. de Groot J, Sontheimer H. Glutamate and the biology of gliomas. Glia. 2011;59:1181-9
95. Sontheimer H. Ion channels and amino acid transporters support the growth and invasion of primary brain tumors. Mol Neurobiol. 2004;29:61-71
96. Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205:613-21
97. Ye ZC, Sontheimer H. Glioma cells release excitotoxic concentrations of glutamate. Cancer Res. 1999;59:4383-91
98. Beier D, Schulz JB, Beier CP. Chemoresistance of glioblastoma cancer stem cells--much more complex than expected. Mol Cancer. 2011;10:128
99. Steinbach JP, Weller M. Apoptosis in gliomas: molecular mechanisms and therapeutic implications. J Neurooncol. 2004;70:245-54
100. Sontheimer H. Malignant gliomas: perverting glutamate and ion homeostasis for selective advantage. Trends Neurosci. 2003;26:543-9
101. Savaskan NE, Seufert S, Hauke J, Trankle C, Eyupoglu IY, Hahnen E. Dissection of mitogenic and neurodegenerative actions of cystine and glutamate in malignant gliomas. Oncogene. 2011;30:43-53
102. Lehre KP, Danbolt NC. The number of glutamate transporter subtype molecules at glutamatergic synapses: chemical and stereological quantification in young adult rat brain. J Neurosci. 1998;18:8751-7
103. Noch E, Khalili K. Molecular mechanisms of necrosis in glioblastoma: the role of glutamate excitotoxicity. Cancer Biol Ther. 2009;8:1791-7
104. Aronica E, Gorter JA, Ijlst-Keizers H, Rozemuller AJ, Yankaya B, Leenstra S. et al. Expression and functional role of mGluR3 and mGluR5 in human astrocytes and glioma cells: opposite regulation of glutamate transporter proteins. Eur J Neurosci. 2003;17:2106-18
105. Baker DA, Xi ZX, Shen H, Swanson CJ, Kalivas PW. The origin and neuronal function of in vivo nonsynaptic glutamate. J Neurosci. 2002;22:9134-41
106. Sontheimer H. A role for glutamate in growth and invasion of primary brain tumors. J Neurochem. 2008;105:287-95
107. Savaskan NE, Heckel A, Hahnen E, Engelhorn T, Doerfler A, Ganslandt O. et al. Small interfering RNA-mediated xCT silencing in gliomas inhibits neurodegeneration and alleviates brain edema. Nat Med. 2008;14:629-32
108. Willard SS, Koochekpour S. Regulators of gene expression as biomarkers for prostate cancer. Am J Cancer Res. 2012;2:620-57
109. Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J Natl Cancer Inst. 1999;91:1922-32
110. Haas-Kogan D, Shalev N, Wong M, Mills G, Yount G, Stokoe D. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8:1195-8
111. Sonoda Y, Ozawa T, Aldape KD, Deen DF, Berger MS, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61:6674-8
112. Koul D, Takada Y, Shen R, Aggarwal BB, Yung WK. PTEN enhances TNF-induced apoptosis through modulation of nuclear factor-kappaB signaling pathway in human glioma cells. Biochem Biophys Res Commun. 2006;350:463-71
113. Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK. et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061-83
114. Roesler R, Brunetto AT, Abujamra AL, de Farias CB, Brunetto AL, Schwartsmann G. Current and emerging molecular targets in glioma. Expert Rev Anticancer Ther. 2010;10:1735-51
115. Shin SS, Martino JJ, Chen S. Metabotropic glutamate receptors (mGlus) and cellular transformation. Neuropharmacology. 2008;55:396-402
116. Chen S, Zhu H, Wetzel WJ, Philbert MA. Spontaneous melanocytosis in transgenic mice. J Invest Dermatol. 1996;106:1145-51
117. Zhu H, Reuhl K, Botha R, Ryan K, Wei J, Chen S. Development of early melanocytic lesions in transgenic mice predisposed to melanoma. Pigment Cell Res. 2000;13:158-64
118. Zhu H, Reuhl K, Zhang X, Botha R, Ryan K, Wei J. et al. Development of heritable melanoma in transgenic mice. J Invest Dermatol. 1998;110:247-52
119. Marin YE, Namkoong J, Cohen-Solal K, Shin SS, Martino JJ, Oka M. et al. Stimulation of oncogenic metabotropic glutamate receptor 1 in melanoma cells activates ERK1/2 via PKCepsilon. Cell Signal. 2006;18:1279-86
120. Shin SS, Wall BA, Goydos JS, Chen S. AKT2 is a downstream target of metabotropic glutamate receptor 1 (Grm1). Pigment Cell Melanoma Res. 2010;23:103-11
121. Shin S, Palis BE, Phillips JL, Stewart AK, Perry RR. Cutaneous melanoma in Asian-Americans. J Surg Oncol. 2009;99:114-8
122. Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP. et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006-14
123. Seabold GK, Burette A, Lim IA, Weinberg RJ, Hell JW. Interaction of the tyrosine kinase Pyk2 with the N-methyl-D-aspartate receptor complex via the Src homology 3 domains of PSD-95 and SAP102. J Biol Chem. 2003;278:15040-8
124. Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH. et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824-8
125. Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR. et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127-32
126. Namkoong J, Martino JJ, Chen S. From existing therapies to novel targets: a current view on melanoma. Front Biosci. 2006;11:2081-92
127. Namkoong J, Shin SS, Lee HJ, Marin YE, Wall BA, Goydos JS. et al. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Res. 2007;67:2298-305
128. Yip D, Le MN, Chan JL, Lee JH, Mehnert JA, Yudd A. et al. A phase 0 trial of riluzole in patients with resectable stage III and IV melanoma. Clin Cancer Res. 2009;15:3896-902
129. Lee HJ, Wall BA, Wangari-Talbot J, Shin SS, Rosenberg S, Chan JL. et al. Glutamatergic pathway targeting in melanoma: single-agent and combinatorial therapies. Clin Cancer Res. 2011;17:7080-92
130. Le MN, Chan JL, Rosenberg SA, Nabatian AS, Merrigan KT, Cohen-Solal KA. et al. The glutamate release inhibitor Riluzole decreases migration, invasion, and proliferation of melanoma cells. J Invest Dermatol. 2010;130:2240-9
131. Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7:3129-40
132. Bilici A, Arslan C, Altundag K. Promising therapeutic options in triple-negative breast cancer. J BUON. 2012;17:209-22
133. Speyer CL, Smith JS, Banda M, DeVries JA, Mekani T, Gorski DH. Metabotropic glutamate receptor-1: a potential therapeutic target for the treatment of breast cancer. Breast Cancer Res Treat. 2012;132:565-73
134. Sharma MK, Seidlitz EP, Singh G. Cancer cells release glutamate via the cystine/glutamate antiporter. Biochem Biophys Res Commun. 2010;391:91-5
135. Seidlitz EP, Sharma MK, Saikali Z, Ghert M, Singh G. Cancer cell lines release glutamate into the extracellular environment. Clin Exp Metastasis. 2009;26:781-7
136. Kinkade CW, Castillo-Martin M, Puzio-Kuter A, Yan J, Foster TH, Gao H. et al. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J Clin Invest. 2008;118:3051-64
137. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061-8
138. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609-15
Author contact
![]() Corresponding author: Dr. Shahriar Koochekpour, Departments of Cancer Genetics and Urology, Center for Genetics and Pharmacology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA, Telephone: 716-845-3345; Fax: 716-845-1698; Email: Shahriar.Koochekpourorg.
Corresponding author: Dr. Shahriar Koochekpour, Departments of Cancer Genetics and Urology, Center for Genetics and Pharmacology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA, Telephone: 716-845-3345; Fax: 716-845-1698; Email: Shahriar.Koochekpourorg.

 Global reach, higher impact
Global reach, higher impact