10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(1):1-14. doi:10.7150/ijbs.7624 This issue Cite
Research Paper
Variant Ionotropic Receptors Are Expressed in Olfactory Sensory Neurons of Coeloconic Sensilla on the Antenna of the Desert Locust (Schistocerca gregaria)
1. China Agricultural University, Key Lab for Biological Control of the Ministry of Agriculture, Beijing 100193, China
2. University of Hohenheim, Institute of Physiology, 70593 Stuttgart, Germany
3. Department of Evolutionary Neuroethology, Max Planck Institute for Chemical Ecology, Jena, Germany
Received 2013-9-9; Accepted 2013-10-31; Published 2013-12-1
Abstract
The behaviour of the desert locust, Schistocera gregaria, is largely directed by volatile olfactory cues. The relevant odorants are detected by specialized antennal sensory neurons which project their sensory dendrites into hair-like structures, the sensilla. Generally, the responsiveness of the antennal chemosensory cells is determined by specific receptors which may be either odorant receptors (ORs) or variant ionotropic receptors (IRs). Previously, we demonstrated that in locust the co-receptor for ORs (ORco) is only expressed in cells of sensilla basiconica and sensilla trichodea, suggesting that cells in sensilla coeloconica may express different types of chemosensory receptors. In this study, we have identified the genes of S. gregaria which encode homologues of co-receptors for the variant ionotropic receptors, the subtypes IR8a and IR25a. It was found that both subtypes, SgreIR8a and SgreIR25a, are expressed in the antennae of all five nymphal stages and in adults. Attempts to assign the relevant cell types by means of in situ hybridization revealed that SgreIR8a and SgreIR25a are expressed in cells of sensilla coeloconica. Double fluorescence in situ hybridization experiments disclosed that the two IR-subtypes are co-expressed in some cells of this sensillum type. Expression of SgreIR25a was also found in some of the sensilla chaetica, however, neither SgreIR25a nor SgreIR8a was found to be expressed in sensilla basiconica and sensilla trichodea. This observation was substantiated by the results of double FISH experiments demonstrating that cells expressing SgreIR8a or SgreIR25a do not express ORco. These results support the notion that the antenna of the desert locust employs two different populations of OSNs to sense odors; cells which express IRs in sensilla coeloconica and cells which express ORs in sensilla basiconica and sensilla trichodea.
Keywords: locust, olfaction, ionotropic receptors, in situ hybridization
Introduction
The desert locust, Schistocerca gregaria, is a dreaded pest in afro-asian regions, due to its potential to form huge swarms, which invade and crop complete agricultural areas thus causing tremendous economical damage. Many aspects of locust behavior, including aggregation, feeding, mating and oviposition are triggered and guided by volatile chemical cues emitted from con-specifics, food plants or oviposition sites [1]. Insects sense volatile chemicals by olfactory sensory neurons (OSNs) on antennae, which extend their dendrites into hair-like structures. On locust antennae three types of morphologically identifiable olfactory sensilla are distinguished housing different number of OSNs: sensilla basiconica (up to 50 OSNs), sensilla trichodea (1-3 OSNs) and sensilla coeloconica (1-4 OSNs) [2]. Single sensillum recordings from antennae of Schistocerca gregaria and the closely related Locusta migratoria have provided some first insight into the response spectrum of OSNs in the different sensilla types. It was found that basiconic OSNs responded to nymphal as well as to adult aggregation pheromones, while OSNs in s. trichodea responded to odorants from locust feces and to a putative sex pheromone [3, 4]. Finally, OSNs in s. coeloconica responded to organic acids, plant volatiles and nymphal odors; but were inhibited by putative aggregation pheromones [3].
In the past decades significant progress has been made to unravel the molecular mechanisms mediating the odorant-responses of insect OSNs [5-8]. Distinct receptor types residing in the dendritic membrane of OSNs are considered as key elements in odorant detection. Originally in Drosophila [9, 10] and subsequently in many species from various insect orders, large families of highly diverse olfactory receptors have been identified [11-13]. Interestingly, recent studies have provided evidence that two classes of chemosensory receptors may exist in the olfactory system of insects [14-17]. Members of the large and diverse family of odorant receptors (ORs) are expressed in OSNs housed in sensilla trichoidea and sensilla basiconica from flies [18, 19], mosquitoes [20] or moths [21, 21, 22]. These seven transmembrane domain receptor proteins confer ligand-specificity to the OSN [18, 23-25] and most probably heteromerize with a common OR-coreceptor (ORco) to form a receptor-complex which is activated by appropriate odorants leading to a depolarization of OSNs through ionotropic [26] and/or metabotropic mechanisms [27].
The second type of olfactory receptors, named “variant ionotropic receptors” (IRs) due to their sequence relation and structural similarity to ionotropic glutamate receptors (iGluRs) [8, 14] was found to be expressed in OSNs housed in sensilla coeloconica of Drosophila. In Drosophila, each coeloconic OSN appears to express combinations of several IRs from a repertoire of antennal IR genes. IRs are considered to mediate responsiveness of OSNs to organic acids, amines and alcohols [14, 28]. Generally, several variable IRs appear to be co-expressed with one or both IR-subtypes, IR8a and IR25a [29]. These two subtypes are phylogenetically highly conserved and are considered to function as co-receptors [30, 31], thus resembling the functional role of ORco protein in basiconic and trichoid OSNs.
In a previous study exploring the expression of ORco in the antenna of S. gregaria and L. migratoria [32] we could assign the expression of ORco to OSNs located in sensilla basiconica and sensilla trichodea, but found no expression in OSNs of sensilla coeloconica. In addition, for some ORs expression in ORco-positive sensilla types was demonstrated [33]. Together, these results imply that olfactory receptors of the OR-class are involved in odorant responses of basiconic and trichoid OSNs, while s. coeloconica likely express different receptor types. In this study, we set out to explore whether variant ionotropic receptors may be expressed in the antennae of the locust S. gregaria. Towards this goal attempts were made to identify the genes encoding the IR co-receptors IR8a and IR25a and to visualize their expression in the antenna.
Materials and Methods
Insect rearing and tissue collection
Locusts, Schistocerca gregaria, were obtained from local suppliers (Zoo&Co, Filderstadt, Germany). Body parts (antennae, mouth parts, tarsi and brains) of adult animals and antennae of different nymphal stages were dissected from cold anaesthetized insects. Tissues were collected in liquid N2 and subsequently used to isolate total RNA. For in situ hybridization experiments antennae were directly embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Europe, Zoeterwoude, The Netherlands) and stored at -70°C until sectioning.
Identification of IR sequences (SgreIRs) from the antennal transcriptome of Schistocerca gregaria
We used a collection of IR sequences reported in Croset et al, 2010 to generate a BLAST database in Geneious 6 (Biomatters, Auckland, New Zealand), and carried out tblastx queries with a cut off of 10-5 against this database using S. gregaria antennal transcriptome data, kindly provided by Heiko Vogel (Department for Entomology, MPI for Chemical Ecology Jena, Germany) and Andreas Vilcinskas (Institute of Phytopathology and Applied Zoology, Justus-Liebig-University of Giessen, Germany). Identified hits indicating candidate SgreIR sequences were used to re-tblastx the NCBI nr (non-redundant) database to verify identity. This identified several sequences annotated as ionotropic glutamate receptors or variant ionotropic receptors, which were used as queries to perform tblastx again with the Schistocerca gregaria transcriptome database. Finally, identified and extracted contig sequences were assembled to yield putative IR sequences of S. gregaria (SgreIRs).
Amplification of SgreIRs sequences
Total RNA was extracted from frozen male and female antennae using Trizol reagent (Invitrogen, Germany) according to the supplier´s protocol. Poly A+ RNA was purified from 100 μg total RNA using oligo (dT)25 magnetic dynabeads (Invitrogen) following recommended protocols. cDNAs were synthesized from 50 ng mRNA using the Smarter Race cDNA Amplication Kit (Takara, Japan). In order to amplify the 5’ terminal and 3’ terminal sequences of SgreIR8a and SgreIR25a coding sequence specific primers (Supplementary Material: Table S1) were used in PCR reaction with Fermentas High Fidelity Taq (Fisher Scientific, Germany). To overcome GC rich regions in the 5’ part of the SgreIR8a sequence a Taq(R) high GC enhancer (New England Biolabs, USA) was added to the standard PCR reaction. PCR conditions used in SgreIR8a 5’ part were: 95°C for 5 min, then 35 cycles with 94°C for 30 s, 68°C for 30 s and 72°C for 2 min, followed by incubation for 10 min at 72°C. PCR conditions used in SgreIR8a 3’ part were: 95°C for 5 min, then 20 cycles with 94°C for 30s, 70°C for 30 s and 72°C for 1 min 30 s, decreasing the annealing temperature by 0.5°C per cycle. Subsequently, 20 cycles with 60°C annealing temperature were performed followed by incubation for 10 min at 72°C. SgreIR25a sequences (5´ and 3´ parts) were amplified using the following conditions: 94°C for 1 min 40 s, then 20 cycles with 94°C for 30 s, 48°C for 30 s and 72°C for 1 min 30 s, with decreasing the annealing temperature by 0.5°C per cycle. This was followed by 20 further cycles with 38°C annealing temperature and a final incubation for 10 min at 72°C. PCR products were gel-purified using the Geneclean kit (MP Biomedicals, Germany) and adenine nucleotide overhangs were added by incubation with 10 mM dATP and 5U Taq polymerase (Gennaxxon, Germany) at 72°C for 20 min. The resulting A-tailed PCR products were cloned using the pGEM-T vector system (Promega, USA) and sequenced on an ABI310 automatic sequencer employing the BIG dye cycle sequencing kit (v3.1; Applied Biosystems, Foster City, Ca, USA) with vector and gene specific primers.
Tissue and stage-specific expression of IRs
Total RNA was extracted from different adult tissues and nymphal stages using Trizol reagent (Invitrogen) following recommended protocols. Male and female cDNAs were transcribed from 1 μg of total RNA using 4 μl first strand buffer (250 mM Tris pH 8.3, 375 mM KCl, 15 mM MgCl2), 1 μl 10 mM dNTP mix, 1 μl RNaseout, 2 μl DTT (0.1M), 1 μl oligo-dT18 primer (500 ng μl-1) and 1 μl Superscript II reverse transcriptase (Invitrogen) in a total volume 20 μl. Synthesis of cDNA was performed at 50°C for 50 min followed by incubation for 15 min at 70 °C. Non-quantitative RT-PCR was performed using IR-specific sense and anti-sense primers (Supplementary Material: Table S1). PCR conditions used for SgreIR8a were: 94°C for 1 min 30 s, then 20 cycles with 94°C for 30s, 55°C for 30 s and 72°C for 1 min 30 s, with decreasing the annealing temperature by 0.5°C per cycle. Subsequently, 20 cycles at 45°C annealing temperature were performed followed by incubation for 10 min at 72°C. PCR conditions for SgreIR25a were: 94°C for 1 min 30 s, then 40 cycles with 94°C for 30 s, 45°C for 30 s and 72°C for 1 min, followed by incubation for 10 min at 72°C. Primers matching the actin gene of S. gregaria (Supplementary Material: Table S1) were used to verify the quality of the cDNA preparations. PCR conditions for actin were: 94°C for 1 min 30 s, then 40 cycles with 94°C for 30 s, 45°C for 30 s and 72°C for 1 min, followed by incubation for 10 min at 72°C. PCR products were run on 1.2% agarose gels and visualized by ethidium bromide staining.
Sequence analysis and comparison
Sequence alignments shown for IR8a and IR25a sequences, respectively, were conducted using ClustalW [34] and further arranged using the BioEdit program (www.mbio.ncsu.edu/BioEdit/bioedit.html). For SgreIR8a and SgreIR25a structure domain annotation was added according to the DmelIR8a definitions reported in [31]. An unrooted neighbour joining tree comparing the relationship of IR8a and IR25a amino acid sequences from various insect species was calculated based on a ClustalW alignment using the MEGA5 program [35].
In situ hybridization
Digoxigenin (Dig)-labeled or biotin-labeled anti-sense and sense probes were synthesized from linearized pGEM-T vectors containing partial cDNA of SgreIRs or the coding sequence of SgreORco [32] using the T7/Sp6 RNA transcription system (Roche, Germany) following the protocol recommended by the manufacturer. For SgreIR8a riboprobes were transcribed from two plasmids containing 1906 nucleotides of the 5’ part and 1283 nucleotides of the 3’ part, respectively. Accordingly, for SgreIR25a plasmids containing either the 5’ part (1438 nucleotides) or the 3’ part (1669 nucleotides) were used. In ISH experiments 1:1 mixtures of 5’ part and 3’ part riboprobes were used for both SgreIRs. Antennae (embedded in Tissue-Tek) of male and female locusts were used to make 12 μm sections with a Jung CM300 cryostat at -21°C. Sections were thaw mounted on Super Frost Plus slides (Menzel-Gläser, Braunschweig, Germany) and stored at -70°C until use. In situ hybridization was performed using the protocol described in detail previously [32] with few modifications. Briefly, sections were taken out from the -70°C freezer and immediately transferred to 4% PFA for 20 min at 4°C. This was followed by a wash in PBS for 1 min, incubation in 0.2 M HCl for 10 min and two washes in PBS for 1 min each. Then sections were incubated for 10 min in acetylation solution (25% acetic anhydride freshly added in 0.1 M triethanolamine) followed by three 3 min washes in PBS. Pre-hybridization was for 1 hour at 65°C for SgreIR8a and 60°C for SgreIR25a. Hybridization with labeled probes was performed at the same temperatures for 24 hours.
Visualization of Dig-labeled probe hybridizations using color substrate was performed as described earlier [32] using an anti-Dig alkaline phosphatase (AP) conjugated antibody (1:500, Roche) and NBT/BCIP substrate. To increase the signal intensity, polyvinyl alcohol (PVA, MW: 89-98K, Sigma) (1% for SgreIR8a; 2.5% for SgreIR25a) was added to the developing buffer containing NBT/BCIP substrate. Tissue sections were analyzed on a Zeiss Axioskope2 microscope (Zeiss, Oberkochen, Germany) equipped with Axiovision software.
Single and double fluorescent RNA in situ hybridization (FISH) with Dig- and/or biotin-labeled probes was conducted in the same way. Visualization of labeled probes was performed as described earlier [33, 36]. In short, Dig-labeled probes were visualized by the anti-Dig AP-conjugated antibody in combination with HNPP/Fast Red (Roche). For biotin-labeled probes the TSA kit (Perkin Elmer, MA, USA), including an anti-biotin strepavidin horse radish peroxidase-conjugate and fluorescein-tyramides as substrate was used. Sections were analysed for hybridization signals (epifluorescence) using a Zeiss LSM510 Meta laser scanning microscope (Zeiss, Oberkochen, Germany). Confocal image stacks were recorded from antennal segments in the red and green fluorescence channel as well as the transmitted-light channel. Pictures presented are projections of selected optical planes. The red and green fluorescence channels have been overlaid with the transmitted-light channel or are shown separately.
Results
Identification of IRs from the locust, Schistocerca gregaria
In order to identify olfactory ionotropic receptors from S. gregaria (SgreIRs) we have bioinformatically screened transcriptome sequence data from the antenna using a collection of reported IR sequences [29]. These approaches provided nine overlapping contigs with significant similarity to DmelIR8a or putative IR8a sequences of other insects. An assembly of the locust sequences resulted in a putative SgreIR8a sequence which comprises 3719 nucleotides and encodes a protein of 902 amino acids (Fig. 1). The correct assembly of the full-length SgreIR8a sequence was verified by RT-PCR amplification of 5’ and 3’ parts from locust antennal mRNA and sequencing of the PCR products. Similarly, three non-overlapping antennal transcriptome sequences were identified which showed high similarity to LmigIR25a or other candidate insect IR25a sequences. The gaps between stretches of partial sequences were closed by RT-PCR amplification employing gap-spanning primer pairs and sequencing of the PCR products. These efforts led to a SgreIR25a sequence of 2505 nucleotides which encoded a protein of 834 amino acids; sequence comparison suggested that part of the N-terminus is missing (Fig. 2).
To explore the similarity of candidate IR8a sequences from different insect orders we aligned the orthopteran SgreIR8a sequence to lepidopteran, dipteran, coleopteran and hymenopteran sequences (Fig. 1) and calculated the pair-wise identity. This revealed an overall sequence identity between 42.7 and 68.6 %; for certain protein domains a high degree of conservation is particularly evident (Fig. 1). The highest identity across species was found in the region between transmembrane (M) segments M1 – M3, in especially in M2 and the pore loop (P). Fewer identical amino acids are present in the binding domain loops S1 and S2; and very little similarity exists in the amino terminal domain (ATD) and the C-terminus (C) of the proteins. When comparing the sequence of SgreIR25a with the sequences of IR25a from other insects a similar pattern of sequence conservation emerged (Fig. 2). The pair-wise sequence identity ranged from 50.1% to 69.9% between species and the same domains were conserved as in the IR8a proteins, except for the amino terminal domain which was more conserved in the IR25a proteins (Fig. 2). Overall, SgreIR8a and Sgre25a share 29.2% of their amino acids. To further analyze the phylogenetic relationship of the locust IR8a and IR25a with representatives from other insects, a sequence similarity tree was calculated using the MEGA5 program [35]. The resulting neighbor joining tree (Fig. 3) shows that SgreIR8a and SgreIR25a cluster into clearly separated branches comprising insect IR8a and IR25a sequences, respectively. Within the IR8a branch as well as in the IR25a branch, the sequences cluster in an order-specific manner, reflecting that the highest similarity exists between sequences of insects belonging to the same orders.
Alignment of the SgreIR8a amino acid sequence with IR8a sequences from other insects. Positions of the amino terminal domain (ATD), the binding domain lobes (S1 and S2), the pore loop (P) and the transmembrane segments (M1, M2, M3) are marked by bars of different colors referring to their position in DmelIR8a [14]. The positions of key ligand binding residues in iGluRs are marked by asterisks above the sequences. Numbers on the right refer to the number of the last amino acid in the line. Amino acids with at least 50% identity or similarity between sequences are shaded black and grey, respectively. The IR8a amino acid sequences from Agam = Anopheles gambiae, Amel = Apis melifera, Apis = Acyrthosiphon pisum, Bmor = Bombyx mori, Dmel = Drosophila melanogaster, and Tcas = Tribolium castaneum were taken from [29].

Alignment of the SgreIR25a amino acid sequence with IR25a sequences identified in other insects. Numbers on the right refer to the number of the last amino acid in the line. Black and grey shadings indicate amino acids which show at least 70% identity, respectively similarity, between sequences. Labeling of protein domains, abbreviations and origin of sequences are the same as indicated in figure 1. LmigIR25a = Locusta migratoria IR25a (GenBank: AFP33229.1)
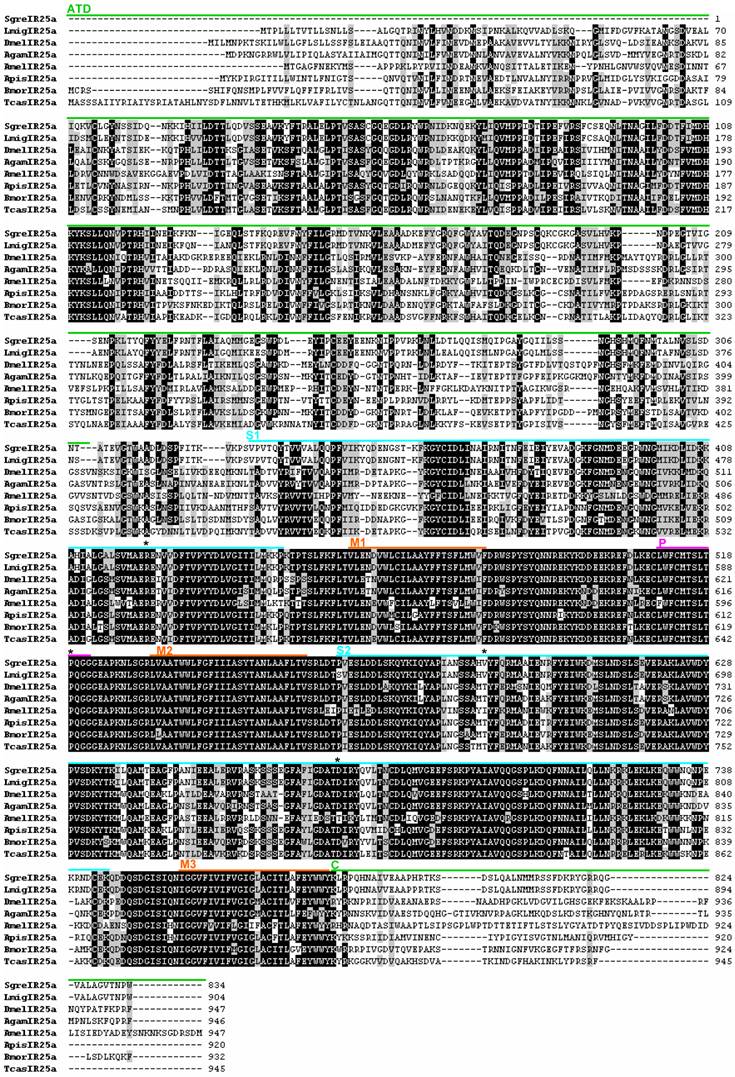
Phylogenetic relationship of IR8a and IR25a sequences from S. gregaria and species belonging to various insect orders. A neighbour-joining tree was constructed using MEGA5 [35] based on a ClustalW alignment of the IRs indicated in figures 1 and 2, as well as sequences from Aaeg = Aedes aegypti, Cpom = Cydia pomonella, Cqui = Culex quinquefasciatus, Dgri = D. grimshawi, Dpse= D. pseudoobscura, Msex = Manduca sexta, Nvit = Nasonia vitripennis and Phum = Pedeculus humanus, [16, 29, 43]. Bootstrap support values are based on 1000 replicates, only support values above 80% are shown. Branch lengths are proportional.
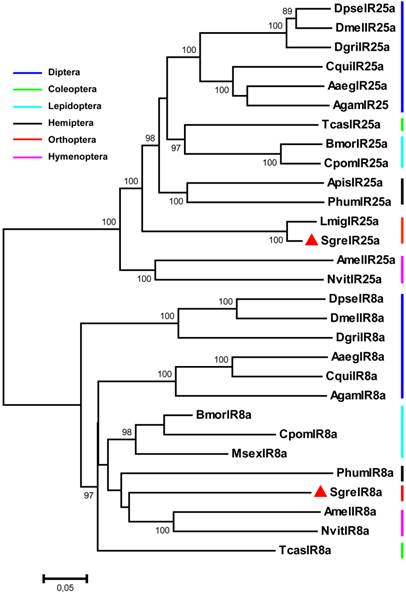
Spatiotemporal expression patterns of SgreIR8a and SgreIR25a
To determine the level of expression for SgreIR8a and SgreIR25a in male and female antenna and to assess if the two IRs may also be expressed in other parts of the body, RT-PCR experiments were performed using IR-specific primers and cDNA prepared from chemosensory (male and female antennae, mouthpart, tarsi) and non-chemosensory (brain) tissues of locust. With primer pairs specific for SgreIR8a and SgreIR25a, respectively, bands of the expected size were found in the antennae of both sexes, with no obvious differences in the expression level. In addition, transcripts for SgreIR25a were also found in cDNAs from mouthparts and at a low level from tarsi. This result indicates that SgreIR25a is expressed not only in the antennae. No PCR products were obtained with cDNA preparations of brain (Fig. 4A).
To explore the onset and time course of SgreIR8a and SgreIR25a expression during development different stages were compared. cDNAs prepared from antennae of different nymphal stages (1st to 5th instars) and adult animals were analysed with IR-specific primers (Fig. 4B). With templates from all stages, PCR products were obtained with slightly different intensities, especially for SgreIR8a. Together the results indicate that both IR-subtypes are expressed in antennae throughout development from the first instar stage to adult.
Identification of the IR-expressing cells on the antenna
Four morphological distinct sensilla types housing sensory cells have been identified on the antenna of the desert locusts: olfactory sensilla basiconica, sensilla trichodea and sensilla coeloconica, while the sensilla chaetica are supposed to serve a gustatory/mechanosensory function [2]. To visualize the cells which express SgreIR8a and SgreIR25a in antennae in situ hybridization (ISH) experiments were performed. Sections through the antennae were incubated with IR-specific anti-sense RNA probes and positive cells visualized employing colour substrates.
Experiments with a SgreIR8a specific Dig-labeled anti-sense RNA probe led to the labeling of several cells within an antennal segment (Fig. 5A and B). Control experiments with a corresponding sense RNA probe did not result in any labeled cells; thus confirming the specificity of the ISH signals (Fig. 5H). More detailed analysis revealed that within a section the SgreIR8a anti-sense RNA probe visualized either individual cells (Fig. 5C and F), two adjacent cells (Fig. 5D) and in some cases even clusters of three cells (Fig. 5E). SgreIR8a-positive cells could clearly be assigned to s. coeloconica (Fig. 5C - F), but were not found under any s. basiconica (Fig. 5C), s. trichodea (Fig. 5F) or s. chaetica (Fig. 5E). For comparison we performed ISH with a SgreORco-specific probe resulting in a complementary labeling pattern, thus confirming our previous results [32] that ORco is expressed in the 20-30 OSNs housed in s. basiconica (Fig. 5G) as well as in the 2-3 OSNs in the s. trichodea (not shown). Together, these results suggest that expression of SgreIR8a is restricted to s. coeloconica.
ISH-experiments with a SgreIR25a-specific anti-sense RNA probe resulted in a labeling pattern quite similar to SgreIR8a (Fig. 6), with either single cells or with two or three adjacent cells on a single section (Fig. 6A - C). In addition with the SgreIR25a probe occasionally clusters of four labeled cells were found (Fig. 6D). While no labeled cells were seen under s. trichodea (Fig. 6E) or s. basiconica (Fig. 6F), we regularly found labeled cells under some of the s. chaetica (Fig. 6G and C). These results indicate that SgreIR25a is more broadly expressed; both in s. coeloconica as well as in a subpopulation of s. chaetica.
Since both IR-subtypes are predominantly expressed in the s. coeloconica, the possibility exists that they could be co-expressed in the same cell. To scrutinize this view, double FISH experiments were performed employing differentially labeled SgreIR8a- and SgreIR25a-specific probes. The results are depicted in (Fig. 7); cells containing IR transcripts were visualized by red or green fluorescence for, respectively, SgreIR25a and SgreIR8a. Although the experiments were hampered by the relatively low FISH signal intensities, we regularly visualized cells which were clearly co-labeled by both probes indicating co-expression of SgreIR8a and SgreIR25a (Fig. 7A - C). In addition, we found cells that appear to express only one of the two receptors (Fig. 7D - I). Together our results indicate heterogeneous expression of SgreIR8a and SgreIR25a in distinct but partly overlapping populations of OSNs.
To support the specific expression of SgreIR8a and SgreIR25a in OSNs of s. coeloconica but not in OSNs of s. basiconica or s. trichodea we performed double FISH with SgreIR- and SgreORco-specific probes. The results for the combination SgreIR8/SgreORco are shown in (Fig. 8). On longitudinal sections through the antenna cells labeled with SgreIR8a (green) and labeled with SgreORco (red) are clearly separated (Fig. 8C and G). The SgreIR8a probe labeled 1-3 cells under s. coeloconica (Fig. 8A and D), while SgreORco probe labeled a cluster of many cells (Fig. 8B and F) thus confirming the results obtained with single probes (Fig. 5). Analysis of more horizontal sections of the antenna revealed that the SgreIR8a-positive cells (Fig. 8E) are intermingled but clearly separated from the clusters of SgreORco-positive cells (Fig. 8G). This labeling pattern is reminiscent of the mixed topography described for s. coeloconica and s. basiconica on the locust antenna [2]. Two-color FISH experiments using the combination SgreIR25a/SgreORco probes gave a similar labeling pattern (Fig. 9). Areas labeled with a SgreIR25a-probe (Fig. 9A) or with a SgreORco-probe (Fig. 9B) were clearly separated (Fig. 9C), indicating that ORco is not expressed in SgreIR25a-positive cells of sensilla coeloconica. In accordance with the result obtained with the single probe ISH and a chromogenic visualization, double FISH experiments demonstrated that a SgreIR25a probe labeled cells also under some s. chaetica (Fig. 9D - F). Also, the SgreIR25a-positive cells of sensilla chaetica (Fig. 9D) did not co-express SgreORco (Fig. 9E) but were well separated from the ORco-expressing cells (Fig. 9F).
Tissue specificity and developmental expression of SgreIR8a and SgreIR25a. RT-PCR experiments were performed using cDNAs prepared from the tissues indicated and primer pairs specific for SgreIR8a and SgreIR25a, respectively. Primers to actin were used as control for the integrity of the cDNA preparations. A, Expression of the IRs in different locust tissues. FA, female antenna; MA, male antenna; M, mouthparts (maxillary and labial palps); B, brain; T, tarsi. B, Comparison of the IR expression in the antennae of different nympal stages (1st to 5th instar) and adults. Amplification products were analysed on agarose gels and visualized by UV illumination after ethidium bromide staining.
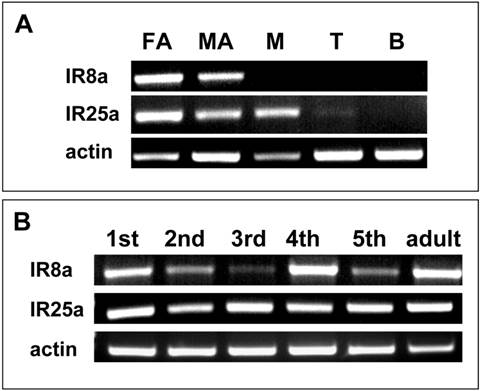
Topography of SgreIR8a gene expression in the antenna S. gregaria. In situ hybridization using SgreIR8a-specific Dig-labeled sense and anti-sense riboprobes and visualization with color substrates. A, Labeling of cells by the SgreIR8a anti-sense RNA probe in two antennal segments of the desert locust. B, Higher magnification of the area boxed in A. C - F, The SgreIR8a anti-sense RNA probe labeled one to three cells under sensilla coeloconica (s.co), but never cells under sensilla basiconica (s.ba, C) , sensilla chaetica (s.ch, E) or sensilla trichodea (s.tr, F). G, Labeling of cells by a Dig-labeled SgreORco-specific anti-sense RNA probe. H, No hybridization signals were observed with the SgreIR8a sense probe. A, B, D, E: female antennae; C, F, G, H: male antennae. Scale bars: 100 μm in A; 50 μm in B, G, H; 20 μm in C, D, E, F.
Localisation of SgreIR25a gene expression in the antenna of S. gregaria. In situ hybridization using SgreIR25a-specific Dig-labeled sense and anti-sense riboprobes and chromogenic visualization. A - D, The SgreIR25a anti-sense RNA probe labeled one (A), two (B), three (C) or four (D) cells under sensilla coeloconica (s.co). No cells under sensilla trichodea (s.tr, E) and sensilla basiconica (s.ba, F) were labeled. For sensilla chaetica (s.ch), cases of no labeled cells (C) and SgreIR25a-positive cells (G) were found. H, No labeling of cells were obtained with the SgreIR25a sense riboprobe. B, C, E, H: female antennae; A, D, G, F: male antennae. Scale bars: 20 μm in A - G; 50 μm in H.
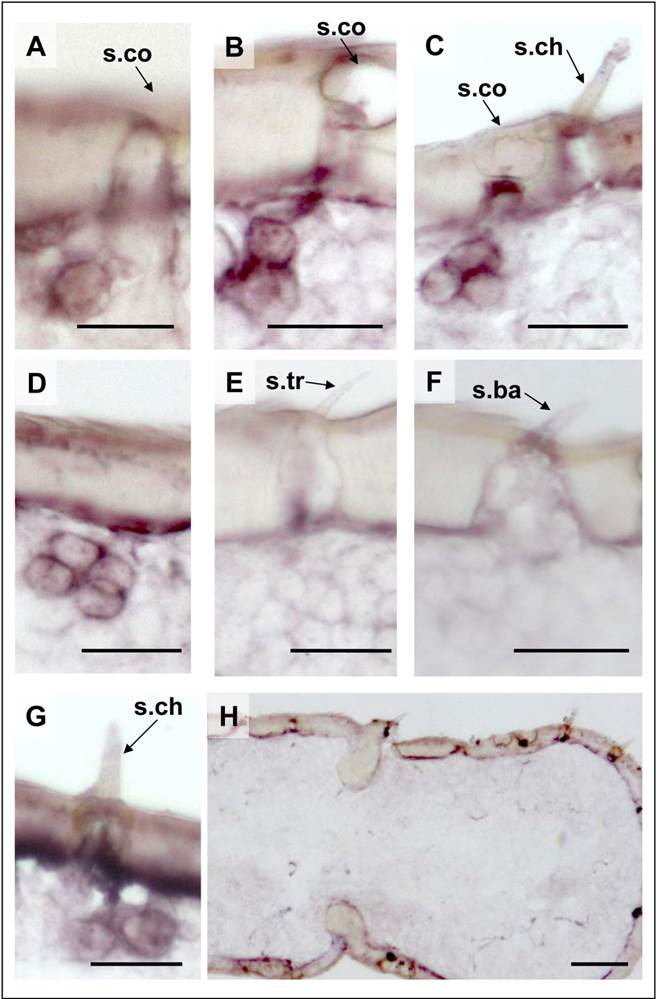
Partial overlap of SgreIR8a and SgreIR25a expression. Double FISH on antennal sections using Dig-labeled SgreIR25a and biotin-labeled SgreIR8a probes with visualization of FISH signals in red (SgreIR25a) and green (SgreIR8a). A - C, Cluster of three cells labeled by both (C), the SgreIR25 probe (red, A) and the SgreIR8a probe (green, B). D - F, Distinct cells that only express SgreIR25a (red, D) or SgreIR8a (green, E) without overlap (F). G - I, Cluster of three cells, with one cell co-expressing SgreIR8a and SgreIR25a (I), the other two cells express SgreIR25a (red, G) but not SgreIR8a (H, green). A - C, G - I: female antennae; D - F: male antennae. Scale bars: 20 μm.
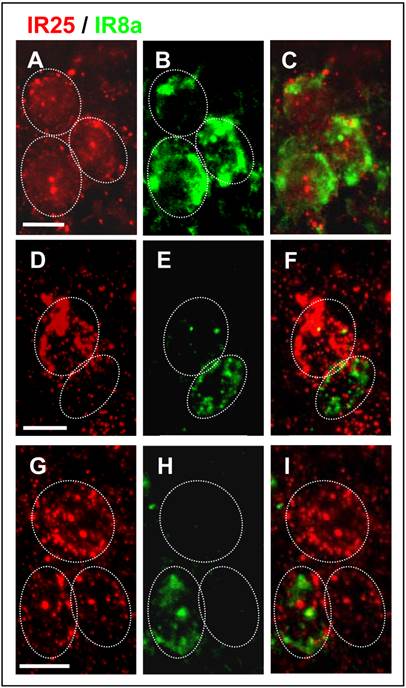
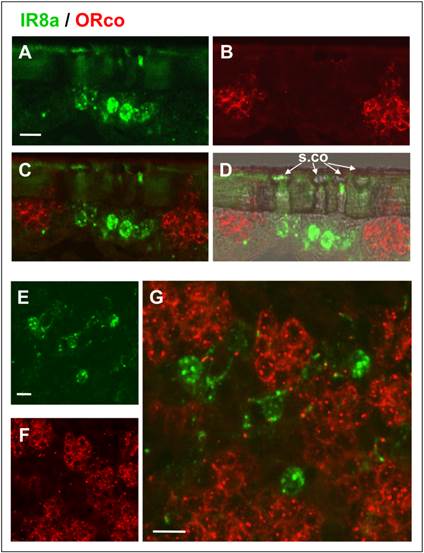
SgreIR8a is not expressed in cells expressing SgreORco. Two-color FISH on antennal sections of male locust using a biotin-labeled SgreIR8a and a Dig-labeled SgreORco probe and detection by green (SgreIR8a) and red (SgreORco) fluorescence. A - D, Hybridization signals on a sagital section of the antenna. The IR8a anti-sense RNA probe labeled cells under sensilla coeloconica which are not labeled by the ORco probe. Pictures show projections of confocal image stacks showing the separated (A, green; B, red) or overlaid (C) fluorescence channels. To better show the morphology of the section the transmitted light channel has been overlaid with the fluorescence channel in D. E - G, Two-color FISH on a more horizontal section of the antenna section confirming the expression of SgreIR8a (green) and SgreORco (red) in different cells. Clusters of ORco-positive cells are intermingled with SgreIR8a-positive cells. Pictures show the separated green (E) and red (F) fluorescence channels and the overlay at higher magnification in G. Scale bars: 20 μm.
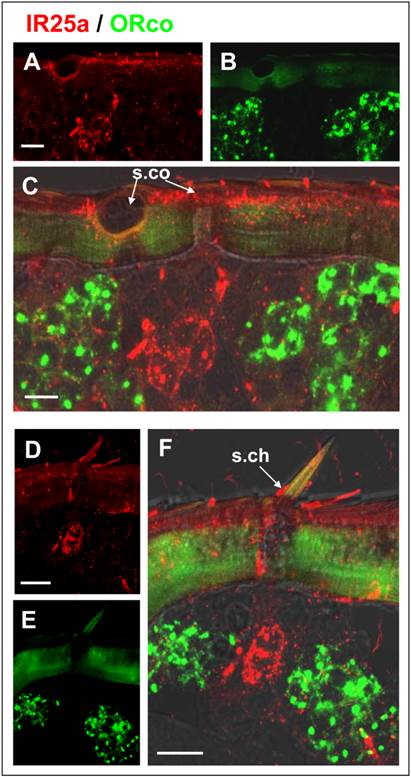
Expression of SgreIR25a and SgreORco locate in different cells. Double FISH on female antennal sections was performed employing Dig-labeled SgreIR25a and biotin-labeled SgreORco probes. Positive cell were visualized by red (SgreIR25a) and green (SgreORco) fluorescence. A - C, Hybridization signals on a longitudinal section of the antenna. The IR25a anti-sense RNA probe labeled cells under sensilla coeloconica which are not labeled by the ORco probe. D - F, The SgreIR25a anti-sense RNA probe labeled cells under sensilla cheatica which do not express SgreORco. Pictures are projections of confocal image stacks showing the separated (A, D, red; B, E, green) or overlaid fluorescence channels (C, F with transmitted light channel). Scale bars: 20 μm.
Discussion
In this study we identified two genes which encode putative co-receptors of variant ionotropic receptors, the subtypes, IR8a and IR25a, of the locust Schistocera gregaria. The identification is based on the pronounced sequence similarity of SgreIR8a and SgreIR25a with the respective sequences from other insect species. Originally the IR8a and IR25a receptors were identified in the fruitfly Drososophila melanogaster as members of a novel type of olfactory receptor family [14]. By means of bioinformatic genome screenings and transcriptome sequence analyses orthologs of IR8a and IR25a have been identified in a number of insect species as well as in other arthropods, such as crustaceans [16, 17, 29, 37]. Most remarkably, IR25a homologs were also found in the nematode Caenorabditis elegans, the mollusc Aplysia californica and the annelid Capitella capitata. Thus, these receptor types seem to be under high selective pressure to maintain the primary structure of the protein [29] suggesting an important functional role of the proteins.
The in situ hybridization experiments have shown that both receptor types, IR8a and IR25a, are mainly expressed in sensory neurons located in sensilla coeloconica. They are not expressed in OSNs of the sensilla basiconica and sensilla trichodea. This notion was substantiated by the results of double FISH experiments, demonstrating that both SgreIRs are not co-expressed with ORco (Figs. 8 and 9), which is selectively expressed in OSNs of s. basiconica and s. trichodea of locust [32]. A similar distribution pattern of ORco was also found in other insects [38, 39]. Recent studies suggest IR8a and IR25a may act as co-receptors and may form heteromers with another variant ionotropic receptor [30, 31] thus resembling the role of ORco for the ligand-specific OR-subtypes. Consequently, the expression of ORco is considered as an indicator for the expression of other OR-types and similarly IR8a and IR25a may be indicators for the expression of IR-subtypes. This view would imply that the sensory cells housed in s. coeloconica of the locust antenna express the variant ionotropic receptor and they are only present in this sensilla type.
Previous electron microscopic studies have identified two morphological distinguishable types of sensilla coeloconica on the antenna of S. gregaria [2]. The double wall type is penetrated by radial pores and contains one to three unbranched sensory neurons (type I), while the non-porous wall type (type II) contains four sensory neurons [2]. The in situ hybridization experiments have shown that SgreIR8a and SgreIR25a are apparently expressed in both types of s. coeloconica; in most cases the number of labeled cells varied from 1 to 3 (Figs. 5 and 6). Clusters of four labeled cells were only obtained using the probes for SgreIR25a (Fig. 6D), suggesting that only SgreIR25a may be expressed in all neurons of type II sensilla. The results of double-labeling studies showed that SgreIR8a and SgreIR25a are co-expressed in a subpopulation of cells, but there are also cells which express only one of the two subtypes. This expression pattern is reminiscent of that in Drosophila, where immunohistochemical studies with specific antibodies have demonstrated that IR8a and IR25 are expressed in distinct but partially overlapping populations of neurons [31]. Although we cannot exclude that in some cases SgreIR co-expression was not detected due to transcript levels below the detection limit, the data indicate a heterogeneous expression pattern of SgreIR8a and SgreIR25a in the sensilla coeloconica of the locust.
SgreIR25a-positive cells were also found in some sensilla chaetica which are supposed to serve gustatory/mechanosensory functions [2]. The notion that IR25a may be present in gustatory chemosensory cells was supported by the result of RT-PCR experiments indicating expression of SgreIR25a in mouths parts, which carry hundreds of s. chaetica (labial palps) and peg-like sensilla (maxillary palps); these sensilla are supposed to have a primary gustatory function [40]. The concept that locust gustatory neurons may co-express SgreIR25a and other ligand binding IRs is in line with some recent studies demonstrating that in Drosophila IR25a is co-expressed with IR7a in gustatory cells on the labellum [29] and that IR76b is involved in the detection of salt [41].
The results of our in situ hybridization experiments that there are no obvious gender differences in the number of SgreIR8a- or SgreIR25a-expressing cells (Figs. 5 and 6) as well as in the levels of SgreIR8a and SgreIR25a transcripts in male and female antennae (Fig. 4) are in agreement with similar numbers of s. coeloconica on the antenna of male and female animals [2, 42]. Overall these data suggest that the two co-receptors are of similar importance in the male and female olfactory system. The presence of SgreIR8a and SgreIR25a transcripts in the antennae of all five nymphal stages is in accordance with the observation that s. coeloconica exist already in first instar stage and are maintened till the adult stage [2]. This may further underline the importance of the variant ionotropic receptor for chemoreception of Schistocera gregaria throughout the entire locust lifespan.
Supplementary Material
Table S1.
Acknowledgements
We are grateful to Heidrun Froß for excellent technical assistance. Furthermore, we thank Heiko Vogel (Department for Entomology, MPI for Chemical Ecology Jena, Germany) and Andreas Vilcinskas (Institute of Phytopathology and Applied Zoology, Justus-Liebig-University of Giessen, Germany) for providing access to the transcriptome data.
Funding
This work was supported by the DFG (German Research Foundation), the NSFC (Natural Science Foundation of China) Project No. 30730012 and the China Scholarship council.
Data deposition
The IR sequences reported in this paper have been deposited in Genbank under accession numbers: KF528686 (SgreIR25a) and KF528687 (SgreIR8a).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hassanali A, Njagi PG, Bashir MO. Chemical ecology of locusts and related acridids. Annu Rev Entomol. 2005;50:223-45
2. Ochieng SA, Hallberg E, Hansson BS. Fine structure and distribution of antennal sensilla of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae). Cell Tissue Res. 1998;291:525-36
3. Ochieng SA, Hansson BS. Responses of olfactory receptor neurones to behaviourally important odours in gregarious and solitarious desert locust, Schistocerca gregaria. Physiol Entomol. 1999;24:28-36
4. Cui X, Wu C, Zhang L. Electrophysiological response patterns of 16 olfactory neurons from the trichoid sensilla to odorant from fecal volatiles in the locust, Locusta migratoria manilensis. Arch Insect Biochem Physiol. 2011;77:45-57
5. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373-91
6. Su CY, Menuz K, Carlson JR. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139:45-59
7. Nakagawa T, Vosshall LB. Controversy and consensus: noncanonical signaling mechanisms in the insect olfactory system. Curr Opin Neurobiol. 2009;19:284-92
8. Rytz R, Croset V, Benton R. Ionotropic Receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 2013
9. Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725-36
10. Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327-38
11. Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72:698-711
12. Jacquin-Joly E, Merlin C. Insect olfactory receptors: contributions of molecular biology to chemical ecology. J Chem Ecol. 2004;30:2359-97
13. Sanchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208-16
14. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149-62
15. Liu C, Pitts RJ, Bohbot JD, Jones PL, Wang G, Zwiebel LJ. Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS Biol. 2010 8
16. Bengtsson JM, Trona F, Montagne N, Anfora G, Ignell R, Witzgall P, Jacquin-Joly E. Putative Chemosensory Receptors of the Codling Moth, Cydia pomonella, Identified by Antennal Transcriptome Analysis. PLoS ONE. 2012;7:e31620
17. Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E. Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol. 2011;20:189-99
18. Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965-79
19. Ha TS, Smith DP. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci. 2006;26:8727-33
20. Schultze A, Pregitzer P, Walter MF, Woods DF, Marinotti O, Breer H, Krieger J. The Co-Expression Pattern of Odorant Binding Proteins and Olfactory Receptors Identify Distinct Trichoid Sensilla on the Antenna of the Malaria Mosquito Anopheles gambiae. PLoS ONE. 2013;8:e69412
21. Gohl T, Krieger J. Immunolocalization of a candidate pheromone receptor in the antenna of the male moth, Heliothis virescens. Invert Neurosci. 2006;6:13-21
22. Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, Nishioka T. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci U S A. 2004;101:16653-8
23. Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418-23
24. Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66-71
25. Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143-60
26. Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002-6
27. Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007-11
28. Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GS. Acid sensing by the Drosophila olfactory system. Nature. 2010;468:691-5
29. Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064
30. Ai M, Blais S, Park JY, Min S, Neubert TA, Suh GS. Ionotropic Glutamate Receptors IR64a and IR8a Form a Functional Odorant Receptor Complex In Vivo in Drosophila. J Neurosci. 2013;33:10741-9
31. Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44-60
32. Yang Y, Krieger J, Zhang L, Breer H. The olfactory co-receptor Orco from the migratory locust (Locusta migratoria) and the desert locust (Schistocerca gregaria): identification and expression pattern. Int J Biol Sci. 2012;8:159-70
33. Xu H, Guo M, Yang Y, You Y, Zhang L. Differential expression of two novel odorant receptors in the locust (Locusta migratoria). BMC Neurosci. 2013;14:50
34. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673-80
35. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-9
36. Krieger J, Raming K, Dewer YME, Bette S, Conzelmann S, Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci. 2002;16:619-28
37. Corey EA, Bobkov Y, Ukhanov K, Ache BW. Ionotropic crustacean olfactory receptors. PLoS ONE. 2013;8:e60551
38. Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703-14
39. Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc Natl Acad Sci U S A. 2004;101:5058-63
40. Blaney WM, Chapman RF. The fine structure of the terminal sensilla on the maxillary palps of Schistocerca gregaria (Forskal) (Orthoptera, Acrididae). Z Zellforsch Mikrosk Anat. 1969;99:74-97
41. Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334-8
42. Greenwood M, Chapman RF. Differences in numbers of sensilla on the antennae of solitarious abd gregarious Locusta migratoria L. (Orthoptera: Acrididae). Int J Morphol and Embryol. 1984;13:295-301
43. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci U S A. 2011;108:7449-54
Author contact
![]() Corresponding authors: Prof. Dr. H. Breer, University of Hohenheim, Institute of Physiology, Garbenstr. 30, 70599 Stuttgart, Germany. e-mail: breerde. Prof. Long Zhang, Ph. D., China Agricultural University, Key Lab for Biological Control, Ministry of Agriculture, Beijing 100193, China, e-mail: locustedu.cn
Corresponding authors: Prof. Dr. H. Breer, University of Hohenheim, Institute of Physiology, Garbenstr. 30, 70599 Stuttgart, Germany. e-mail: breerde. Prof. Long Zhang, Ph. D., China Agricultural University, Key Lab for Biological Control, Ministry of Agriculture, Beijing 100193, China, e-mail: locustedu.cn

 Global reach, higher impact
Global reach, higher impact