10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(10):1097-1107. doi:10.7150/ijbs.9859 This issue Cite
Research Paper
Cationicity-Enhanced Analogues of the Antimicrobial Peptides, AcrAP1 and AcrAP2, from the Venom of the Scorpion, Androctonus crassicauda, Display Potent Growth Modulation Effects on Human Cancer Cell Lines
1. School of Pharmaceutical Sciences, China Medical University, Shenyang 110001, Liaoning, PR China
2. Natural Drug Discovery Group, School of Pharmacy, Queen's University, Belfast BT9 7BL, Northern Ireland, UK
* These authors contributed equally to this work.
Received 2014-6-10; Accepted 2014-8-31; Published 2014-9-21
Abstract
The non disulphide-bridged peptides (NDBPs) of scorpion venoms are attracting increased interest due to their structural heterogeneity and broad spectrum of biological activities. Here, two novel peptides, named AcrAP1 and AcrAP2, have been identified in the lyophilised venom of the Arabian scorpion, Androctonus crassicauda, through “shotgun” molecular cloning of their biosynthetic precursor-encoding cDNAs. The respective mature peptides, predicted from these cloned cDNAs, were subsequently isolated from the same venom sample using reverse phase HPLC and their identities were confirmed by use of mass spectrometric techniques. Both were found to belong to a family of highly-conserved scorpion venom antimicrobial peptides - a finding confirmed through the biological investigation of synthetic replicates. Analogues of both peptides designed for enhanced cationicity, displayed enhanced potency and spectra of antimicrobial activity but, unlike the native peptides, these also displayed potent growth modulation effects on a range of human cancer cell lines. Thus natural peptide templates from venom peptidomes can provide the basis for rational analogue design to improve both biological potency and spectrum of action. The diversity of such templates from such natural sources undoubtedly provides the pharmaceutical industry with unique lead compounds for drug discovery.
Keywords: Scorpion, Venom, Molecular Cloning, Antimicrobial, Peptide, analogue design.
Introduction
Venoms remain an under-utilised resource for drug discovery within the pharmaceutical industry despite a track record over many years of providing lead compounds for the generation of blockbuster drugs such as the ACE inhibitors (captopril) for the treatment of hypertension and the first incretin mimetic (exenatide) for the treatment of type-2 diabetes [1, 2]. Also, the often exquisitely-specific interactions between venom proteins/peptides and their cognate molecular targets, has been an attribute that has provided in many instances, a much fuller understanding of the functional basis of endogenous regulatory systems in both man and animals in areas as diverse as blood clotting and neuronal excitability [3, 4]. Currently, the study of venom components with the view to the discovery of novel therapeutic lead compounds and novel disease-related molecular targets, is undergoing a renaissance due in part to the failure of organic chemistry to fill the fast-emptying pipelines of new products from the pharmaceutical industry [3-5]. Venom proteomes and peptidomes (venomes) thus represent a unique chemical space which can be re-evaluated for drug discovery end-points through the application of contemporary analytical technologies [6, 7]. Although venoms from selected species of venomous animals have been studied for several decades, a systematic approach involving venom gland transcriptome sequencing and proteomic/peptidomic inventory of all encoded toxins, has not been achieved to date for even a single species. Of the total predicted number of venom proteins/peptides occurring in the venoms of all known venomous animals that is conservatively, many hundreds of thousands, only a small fraction of a single percentage has been isolated and structurally/functionally characterised to date [8, 9]. The potential for drug lead discovery of this unique and sizeable library of naturally-selected “smart” molecules, thus remains to be realised. The current study focuses on the venom of the buthid scorpion, Androctonus crassicaudus (literally, the fat-tailed man killer), and more specifically on the group of non-disulphide-bridged peptides (NDBPs) that include the bradykinin-potentiating peptides (BPPs) and the antimicrobial peptides (AMPs) [10]. The AMPs are widely-distributed in scorpion venoms and possess a relatively narrow spectrum of antimicrobial activity with little effects on Gram-negative bacteria [11]. Here we describe the structural and functional characterisation of two similar AMPs initially identified in the venom of A. crassicauda through “shotgun” cloning of their biosynthetic precursor-encoding cDNAs from a lyophilised venom-derived cDNA library. The peptides were C-terminally amidated 18-mers with the amino acid sequences, FLFSLIPH/NAISGLI/LSAFK, and were named in accordance with standard nomenclature as AcrAP1 and AcrAP2, respectively [10]. Synthetic versions of both peptides were found to have potent inhibitory effects against a reference Gram-positive bacterium and a yeast, but to be essentially inactive against a reference Gram-negative bacterium, in keeping with previous observations for structural homologues [12]. Both peptides were inactive against a range of human cancer cell lines at concentrations up to and including 10-4 M. Design, synthesis and biological evaluation of cationicity-enhanced analogues of both peptides, resulted in peptides with a more potent and broad spectrum antimicrobial activity and which possessed growth modulating effects on all human cancer cell lines tested. These data are indicative of the potential of natural venom peptide templates in the generation of analogues with enhanced biological activity and spectrum of actions that may be useful leads in the development of novel therapeutics.
Materials and Methods
Acquisition of scorpion venom
Authentic lyophilised Androctonus crassicauda venom (10 mg) was obtained from Latoxan, Valence, France. Donor scorpions were collected in the field and identified by experts before relocation to the company facility in France. Venom was collected by trained individuals using electrical stimulation (15V) consistent with obtaining sufficient material and with causing a low degree of stress to the animals. Venom was obtained by this procedure at regular intervals over many months (Latoxan, personal communication). All procedures requiring the use of live animals had been approved by appropriate national licensing and local ethics authorities and permits to acquire animals from the wild had been secured from appropriate national bodies where necessary.
Molecular cloning of antimicrobial peptide biosynthetic precursor-encoding cDNAs from a venom-derived cDNA library
A 5 mg sample of Androctonus crassicauda venom was first dissolved in 1ml of cell lysis/mRNA stabilisation buffer (Dynal Biotech, UK). Polyadenylated mRNA was isolated by means of magnetic oligo-dT beads as described by the manufacturer (Dynal Biotech, UK) and subjected to 3′-RACE procedures to obtain full-length antimicrobial peptide biosynthetic precursor nucleic acid sequence data using a SMART-RACE kit (Clontech UK) following manufacturer's instructions. The 3′-RACE reactions employed an NUP primer (supplied with the kit) and a sense primer (S1; 5′-GATGGAAATAAAGTATCTTCTTACCGT-3′) that was designed to a highly- conserved domain of the signal peptide region of previously-characterised antimicrobial peptide precursor cDNAs from closely-related Androctonus scorpion species [12]. The PCR cycling procedure was as follows—initial denaturation step: 60 s at 94 °C; 35 cycles: denaturation 30 s at 94 °C, primer annealing for 30 s at 62 °C; extension for 180 s at 72°C. Resultant PCR products were gel-purified, cloned using a pGEM-T vector system (Promega Corporation) and sequenced using an ABI 3100 automated capillary sequencer.
Isolation and structural characterisation of antimicrobial peptides from lyophilised venom
A second 5 mg sample of lyophilised Androctonus crassicauda venom was dissolved in 0.5ml of trifluoroacetic acid (TFA)/water (0.05/99.95; v/v) and clarified by centrifugation (1100 x g; 5 min). The supernatant was decanted and subjected to reverse-phase HPLC fractionation using a Waters HPLC system fitted with an analytical column (Phenomenex C-5; 250 mm×4.6 mm). This was eluted with a linear gradient formed from TFA/water (0.05/99.95; v/v) to TFA/water/acetonitrile (0.05/19.95/80.00; v/v/v) in 240 min at a flow rate of 1 ml/min. The column effluent was continuously monitored at λ214 nm and fractions (1 ml) were automatically collected at minute intervals. The computed molecular masses of the novel mature peptides, predicted from open-reading frames within the novel cloned cDNAs, were used to interrogate a mass spectral library of venom peptides derived from sequential analysis of each reverse phase HPLC fraction using matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry (MALDI-TOF MS) on a linear time-of-flight Voyager DE mass spectrometer (Perseptive Biosystems, MA, USA) in positive detection mode using α-cyano-4-hydroxycinnamic acid as the matrix. Internal mass calibration of the instrument was achieved using standard peptides that provided a determined accuracy of ±0.1%. The peptides with masses coincident with the novel putative cDNA-encoded peptides were subjected to primary structural analysis by means of MS/MS fragmentation sequencing using an LCQ-Fleet electrospray ion-trap mass spectrometer (Thermo Fisher Scientific, CA, USA).
Prediction of putative AMP secondary structures and physico-chemical properties
Putative peptide secondary structures were predicted using GOR secondary structure prediction software (http://distill.ucd.ie/distill/). Additional physico-chemical properties, including hydrophobicity, hydrophobic moment, net charge at neutral pH and helical wheel plots, were determined using the Heliquest server menu (http://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParamsV2.py) [13].
Peptide synthesis
Each natural antimicrobial peptide and their respective cationicity-enhanced analogues, were separately synthesised by solid phase methodology using Rink amide resin and standard Fmoc chemistry, by means of an automated PS3 peptide synthesiser (Protein Technologies, USA), followed by standardised deprotection and cleavage from resin. Each synthetic peptide was analysed by reverse phase HPLC and MALDI-TOF mass spectrometry to establish both degree of purity and authenticity of structure.
Antimicrobial minimal inhibitory concentration (MIC) and minimum bactericidal (MBC) assays
The antimicrobial activity of each synthetic peptide was assessed by determining their minimal inhibitory concentrations (MICs) against reference strains of a Gram-positive bacterium, Staphylococcus aureus (NCTC 10788), a Gram-negative bacterium, Escherichia coli (NCTC 10418), and a yeast, Candida albicans (NCPF 1467). The reference strains of microorganisms were initially incubated in Mueller-Hinton Broth (MHB) for 16-20 h. Upon achieving their respective logarithmic growth phases, as measured by the optical density (OD) of cultures at 550 nm, each was diluted to 1 × 106 colony-forming units (cfu)/ ml (bacteria) or to 5 × 105 cfu/ ml (yeast). Samples of each were then added to 96-well microtitre plates to which the peptides to be tested were added to achieve final on-plate concentrations of 1 - 250 µM. After 24 h incubation, the OD of each well was measured at 550 nm using a Synergy HT plate reader (BioTek, USA), and the data were analysed using GraphPad Prism 5 software. The MIC was defined as the minimum concentration of peptide that produced an OD similar to that of the negative control (medium minus microorganisms). After this, 10μl of the medium from each well was inoculated onto a Mueller-Hinton agar (MHA) plate and incubated for 24h for measurement of minimum bactericidal concentrations (MBCs) which were defined as the concentration of peptide from the MIC experiments from which no colonies could be grown.
Haemolysis assay
Defibrinated horse blood (TCS Biosciences Ltd, UK) was used to produce a 2% (v/v) suspension of red blood cells in sodium phosphate-buffered saline (PBS) by repeated suspensions and centrifugations until clear supernatants were achieved. Peptide solutions were prepared to achieve final concentrations in the test tubes of 1 - 250 µM. Samples (200 μl) of red blood cell suspensions were incubated with peptides at 37oC for 60 min. The degree of lysis of red blood cells was assessed by haemoglobin release measured by optical density changes of supernatants at λ550 nm using an ELISA plate reader (BioTek EL808). Negative controls employed consisted of a 2% (v/v) red cell suspension and PBS in equal volumes and positive controls consisted of a 2% (v/v) red cell suspension and an equal volume of PBS containing 2% (v/v) of the non-ionic detergent, Triton X-100 (Sigma-Aldrich).
Culture and maintenance of human cancer cell lines
The human prostate carcinoma cell line, PC-3, and the human lung adenocarcinoma cell line, NCI-H460, were cultured using RPMI-1640 culture medium (Invitrogen). Before culturing cells, this medium was supplemented with 10% (v/v) foetal bovine serum (FBS) (Sigma) and 0.1% (w/v) gentamicin (Sigma) and the cells were seeded into 150 cm2 culture flasks (Nunc, UK). The human breast carcinoma cell line, MDA-MB-435s, and the tumourigenic mammary gland cell line, MCF-7, were cultured using a Dulbecco's Modified Eagle's Medium (DMEM) (Sigma), which was supplemented with 10% FBS and 0.1% gentamicin. The cells were seeded into 150 cm2 culture flasks.
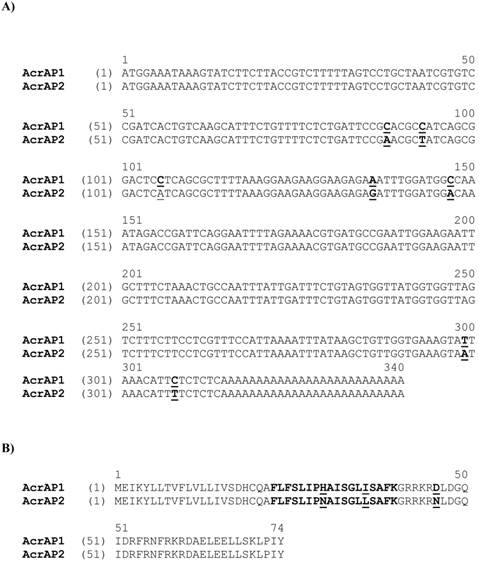
Nucleotide and translated open-reading frame amino acid sequences of cloned cDNAs encoding the biosynthetic precursors of AcrAP1 (A) and AcrAP2 (B). Putative signal peptides are double-underlined, mature peptides are single-underlined and stop codons are indicated by asterisks.

Assessment of growth modulating effects of synthetic natural peptides and their analogues on human cancer cells using the MTT cell viability assay
Yellow-coloured MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) is reduced by the mitochondria of living cells to form a purple-coloured (formazan) crystalline derivative. These crystals can be solubilised by the addition of dimethyl sulphoxide (DMSO) and their concentration determined spectrophotometrically, as detailed below.
The protocol employed has been described in detail previously [14]. Briefly, each cell line was seeded onto 96-well plates at a density of 5 × 103 cells per well. After this, cells were treated with a range of peptide concentrations (10-9 - 10-4M) or with serum-free medium alone (n=8 for each) and were incubated for 24 h at 37oC. This was repeated in triplicate, randomizing lines in which tests and controls were placed. Following this, 10 μl of a 5 mg/ml MTT solution was added to each well and the plates were incubated for an additional 4 h. The growth medium was then removed using a 1 ml syringe fitted with a 21-guage needle and 100 μl of DMSO were added to each well and mixed vigorously to dissolve the formazan crystals that had developed. The absorbance was measured using an EL808™ Absorbance Microplate Reader (Biolise BioTek, USA) at λ550 nm and the statistical analyses were performed using Student's t-test through GraphPad prism software for Windows. The results were considered to be statistically significant if the p value was <0.05.
Results
Molecular cloning of venom precursor cDNAs encoding A crassicauda antimicrobial peptides (AcrAPs)
From the A. crassicauda venom-derived cDNA library, two novel putative antimicrobial peptide precursor-encoding cDNAs were consistently cloned (Figure 1A and B). The translated open-reading frames of both transcripts displayed a high degree of primary structural identity and each contained 74 amino acid residues. Alignments of open-reading frame nucleotide (Figure 2A) and amino acid sequences (Figure 2B), using the AlignX programme of the Vector NTI bioinformatics suite (Informax), revealed that putative antimicrobial peptide-encoding regions were located in identical positions within respective precursors. Both AcrAP-encoding precursors exhibited high degrees of both nucleotide and primary structural similarity with heterogeneity predominantly localised to their C-terminal regions. AcrAP precursor structures were constituted by a signal peptide domain, a single copy of an AcrAP-encoding domain and a C-terminal extension peptide domain. Both AcrAP sequences were flanked C-terminally by glycyl residues that are positioned to act as amide donors to the C-terminal residue of the mature peptides.
Alignment of nucleotide sequences of cloned cDNAs encoding the biosynthetic precursors of AcrAP1 and AcrAP2 (A). Sites of nucleotide sequence differences are shown in bold typeface and are underlined. Alignment of translated open-reading frame amino acid sequences of cloned cDNAs encoding the biosynthetic precursors of AcrAP1 and AcrAP2 (B). Respective mature peptide sequences are shown in bold typeface and sites of amino acid substitutions are underlined.
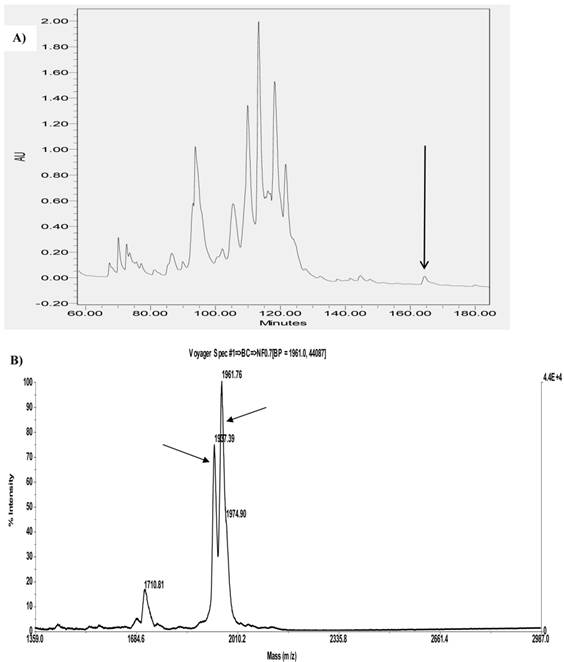
Identification of AcrAP1 and AcrAP2 in reverse phase HPLC fractions of venom and their primary structural confirmation
A single fraction (#168) containing peptides with molecular masses coincident with those of both novel putative antimicrobial peptides (AcrAP1 and AcrAP2) predicted from respective cloned cDNAs, was resolved following reverse phase HPLC fractionation of lyophilised venom (Figure 3A and B). Subsequent MS/MS fragmentation sequencing of doubly-charged ions arising from both peptides, identified in full scan MS spectra and analysed through ion trapping in the LCQ electrospray mass spectrometer, confirmed their predicted primary structures (Figure 4A and B).
Prediction of putative AMP secondary structures and physico-chemical properties
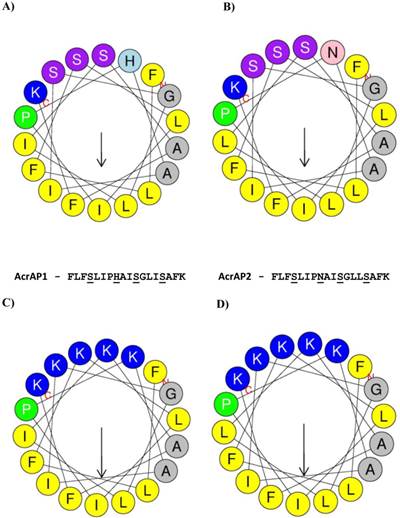
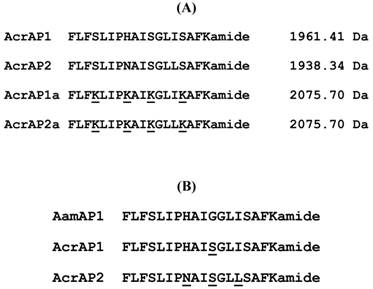
Helical wheel plots of both AcrAP1 and AcrAP2 were constructed and are illustrated in Figure 5A and B. These plots revealed and amphipathic character in both peptides with the hydrophobic amino acid residue (I, F, L, A) side-chains essentially occurring on one face of the molecules and the hydrophilic amino acid residue (K, S, H, N) side-chains, occurring on the other. In view of this arrangement, it was decided to synthesise analogues of each peptide in which the hydrophilic amino acid residues were substituted with lysyl (K) residues to construct a cationic amphipathic peptide in which the hydrophobic amino acid residue side-chains occupied one face of the molecule while the positively-charged (cationic) amino acid residue side-chains occupied the other (Figure 5C and D). Table 1 summarises a sample of the relevant physico-chemical properties of natural AcrAP1 and AcrAP2 and their respective cationicity-enhanced analogues, AcrAP1a and AcrAP2a. The primary structures and molecular masses of each of the 4 peptides used in this study are shown in Figure 6A and the closest structural homologue to the AcrAPs, AamAP1 (Androctonus amoreuxi antimicrobial peptide 1 [12]), identified through NCBI-BLASTp analysis, is aligned with these in Figure 6B.
Minimal inhibitory concentrations (MIC), minimum bactericidal concentrations (MBC) and haemolytic activity of the synthetic natural peptides and their cationicity-enhanced analogues
These data are summarised in Table 2. The replicates of the natural peptides, AcrAP1 and AcrAP2, displayed identical potencies against all three reference microorganisms in both MIC and MBC assays (MIC S. aureus 8 µM, E. coli >250 µM, C. albicans 16 µM; MBC S. aureus 32 µM, E. coli -not tested, C. albicans 64 µM). The cationicity-enhanced analogues of both peptides exhibited a significant increase in potency and spectrum of action and their respective MICs and MBCs were likewise identical with one exception (MBC s of AcrAP1a and AcrAP2a for C. albicans were 4 µM and 8µM, respectively). MICs for S. aureus were 4 µM, E. coli 8 µM and C. albicans 4 µM, while MBCs were 32 µM for both S. aureus and E. coli. The concentrations of peptides effecting 100% lysis of horse red blood cells were 64 µM (AcrAP1 and AcrAP2) and 32 µM (AcrAP1a and AcrAP2a).
Physico-chemical characteristics of the natural A. crassicauda antimicrobial peptides, AcrAP1 and AcrAP2, and their cationicity/amphipathicity enhanced analogues, AcrAP1a and AcrAP2a. C = random coil and H = α-helix.
| Peptides | Secondary structure | α-helix (%) | Hydrophobicity (H) | Hydrophobic moment (μH) | Net charge |
|---|---|---|---|---|---|
| AcrAP1 | FLFSLIPHAISGLISAFK CHHHHHHHHHHHHHHHCC | 83.33 | 0.902 | 0.437 | +2 |
| AcrAP2 | FLFSLIPNAISGLLSAFK CHHHHHHHHHHHHHHHCC | 83.33 | 0.856 | 0.474 | +2 |
| AcrAP1a | FLFKLIPKAIKGLIKAFK CHHHHHHHHHHHHHHHHC | 88.89 | 0.681 | 0.640 | +6 |
| AcrAP2a | FLFKLIPKAIKGLLKAFK CHHHHHHHHHHHHHHHHC | 88.89 | 0.676 | 0.639 | +6 |
Mean inhibitory concentrations (MICs) and and minimum bactericidal concentrations (MBCs) of AcrAP1, AcrAP2, AcrAP1a and AcrAP2a against the test micro-organisms employed and their concentrations effecting 100% lysis of horse erythrocytes.
| Peptides | MIC (µM) | MBC (µM) 100% Haemolysis (µM) | ||||
|---|---|---|---|---|---|---|
| S. aureus | E. coli | C. albicans | S. aureus | E. coli | C. Horse red cells albicans | |
| AcrAP1 | 8 | >250 | 16 | 32 | NT | 64 64 |
| AcrAP2 | 8 | >250 | 16 | 32 | NT | 64 64 |
| AcrAP1a | 4 | 8 | 4 | 32 | 32 | 4 32 |
| AcrAP2a | 4 | 8 | 4 | 32 | 32 | 8 32 |
Region of reverse phase HPLC chromatogram of Androctonus crassicauda venom with an arrow indicating the retention time/elution position of absorbance peak (fraction #168) containing both AcrAP1 and AcrAP2 (A). MALDI-TOF mass spectrum of a sample of fraction #168 indicating major ions (M+H)+ corresponding in m/z values to AcrAP1 (1961.76) and AcrAP2 (1937.39) (B).
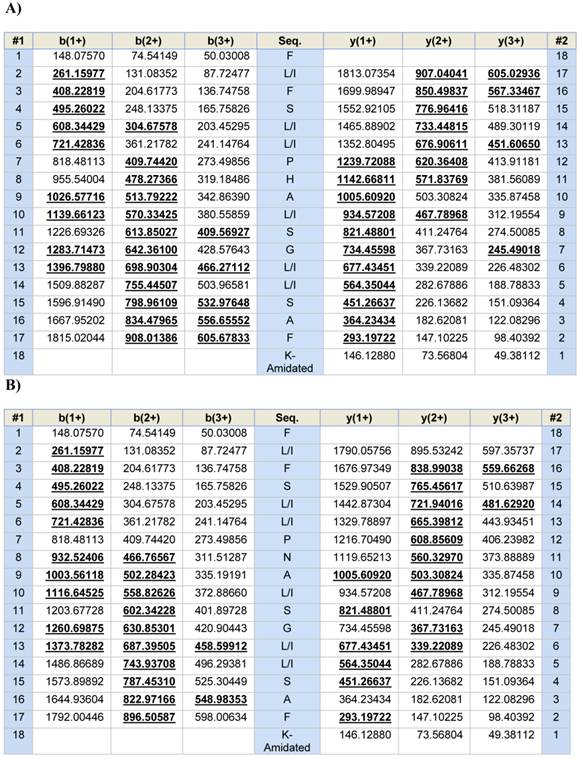
Predicted b- and y-ion series (singly-, doubly- and triply-charged) arising from MS/MS fragmentation of AcrAP1 (A) and AcrAP2 (B). Observed ions are indicated in bold typeface and are underlined.
Assessment of growth modulating effects of synthetic natural peptides and their analogues on human cancer cells
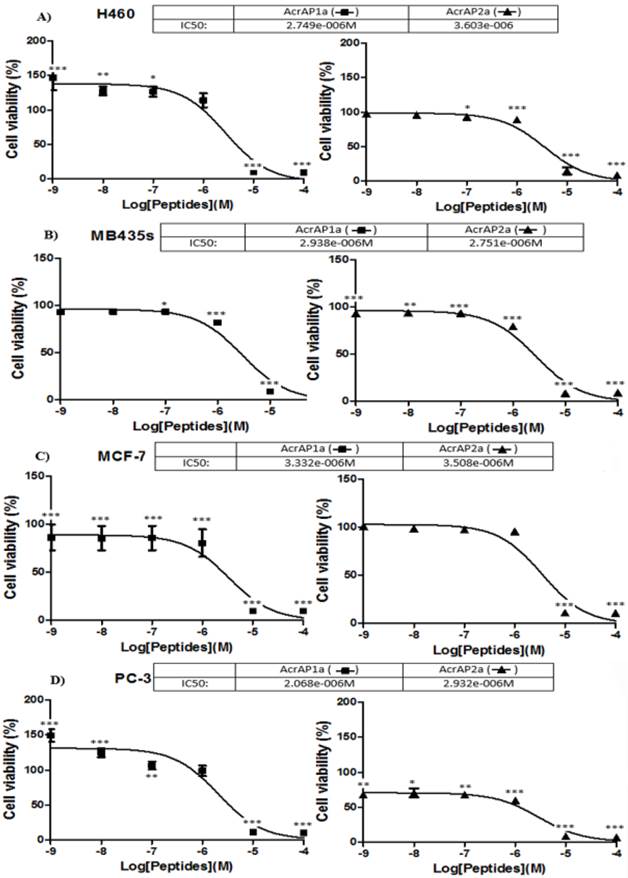
Both synthetic natural peptides, AcrAP1 and AcrAP2, were found to be ineffective in modulating the growth of any of the four human cancer cell lines tested at the concentrations employed (10-4 - 10-9 M) (data not shown). In contrast, both cationicity-enhanced analogues, AcrAP1a and AcrAP2a, displayed potent but subtle differences in their effects on all four cancer cell lines tested (Figure 7A -D). Inhibition of proliferation of all cell lines was observed with both analogue peptides at concentrations above 10-6 M, with IC50 values ranging between 2.068 x 10-6 M and 3.603 x 10-6 M (p<0.001) (Figure 7 A-D). An intriguing observation was that of a significant promotion of growth of both H460 and PC-3 cells with AcrAP1a at concentrations below 10-6 M (Figure 7 A and D). At concentrations of 10-7 M (p<0.05), 10-8 M (p<0.01) and 10-9 M (p<0.001), AcrAP1a caused proliferation of H460 cells (Figure 7A) and at concentrations of 10-8 M (p<0.001) and 10-9 M (p<0.001), caused proliferation of PC-3 cells (Figure 7D). This effect was not observed for AcrAP2a on any of the cancer cell lines tested. At the concentration of AcrAP1a (10-9 M) producing the most significant growth promotion effects on both H460 and PC-3 cancer cell lines (p>0.001), the peptide produced no observable haemolysis.
Helical wheel plots of AcrAP1 (A), AcrAP2 (B), AcrAP1a (C) and AcrAP2a (D). Sites of amino acid substitutions are indicated in each peptide (underlined).
Comparative alignment of the natural AMPs, AcrAP1 and AcrAP2, with their cationicity-enhanced analogues, AcrAP1a and AcrAP2a (A). Alignment of AcrAP1 and AcrAP2 with the most similar AMP found in the NCBI database, AamAP1 (Androctonus amoreuxi) (B). Sites of amino acid differences are underlined in each case.
Discussion
Molecular products from natural sources have long been the mainstay of drug lead discovery and, while originally these were largely constituted by secondary metabolites from plants and microbes, currently there is a greater emphasis on more complex natural molecules, most notably proteins and peptides [3-5, 15, 16].
Animal venoms are a unique and largely untapped resource for this procedure and contain an abundance of proteins and peptides whose often highly-targeted functions have been subjected to natural selective processes for aeons [3-5, 15, 16]. While usage of the standard twenty or so proteogenic amino acids can provide the basis for a large number of primary structural permutations in oligopeptides and proteins, the addition of one or more of the many known post-translational chemical modifications to peptide chains, can elegantly provide several orders of higher molecular complexity and diversity [8, 9, 17, 18]. Thus while the proteins/peptides of animal venoms can in some instances provide templates for drug design, in a broader sense, they can be used by pharmacologists to reveal the complexity of regulation of known disease-associated targets or to identify novel targets [1-5, 15, 16]. Differential functionality and receptor targeting can often be provided through just single site-specific amino acid substitutions or post-translational modifications and most of the known modifications have been found within the proteomes/peptidomes of animal venoms [8, 9]. Additionally, natural templates can be rationally modified to design analogues with enhanced properties for discrete biological end-points and that was the strategy adopted in the present study.
Synthetic replicates of both AcrAP1 and AcrAP2 were found to possess relatively potent antimicrobial activity against a reference Gram-positive bacterium (MIC 8 µM) and a yeast (MIC 16 µm) but exhibited no activity against a reference Gram-negative bacterium at concentrations up to and including, 250 µM. These values would be similar to those reported for other members of this AMP family from the venoms of other scorpions [10, 12].
Effects of the cationicity-enhanced analogues, AcrAP1a and AcrAP2a, on the proliferation of four human cancer cell lines, H460 (A), MB435s (B), MCF-7 (C) and PC3 (D). *p<0.05, **p<0.01, ***p<0.001. Each panel represents the mean and standard error of 24 replicates.
As many AMPs exhibit anti-proliferative actions on cultured cancer cell lines in vitro [19], the synthetic replicates of both natural peptides were incubated at a range of concentrations with four different human cancer cell lines but with little if any observed effects. Thus the natural AMPs described here possessed a relatively narrow spectrum of action. However, while the natural templates may have relatively low potencies and spectra of actions with respect to both prokaryotic and eukaryotic cell proliferation, they nevertheless provide templates or lead structures for the rational design of analogues with potential enhanced properties for these end-points [20]. Helical wheel modelling of AcrAP1 and AcrAP2 revealed their amphipathic nature with one hydrophobic face and an opposite hydrophilic face (Figure 6A and B). Analogues of both were synthesised, in which the residues whose side-chains constituted the neutral hydrophilic face, were substituted with lysyl (K) residues, thus producing classical cationic amphipathic peptide constructs (Figure 6C and D). This manipulation produced peptides with enhanced antimicrobial potencies (MICs of 4 µM) against both the reference Gram-positive bacterium and the yeast, and a broadening of spectrum of action to the reference Gram-negative bacterium (MIC 8 µM). The MBC against the yeast was particularly potent between 4 and 8 µM. Perhaps the most startling biological effect observed was the manifestation of growth modulating effects on each of the four human cancer cell lines employed that were not observed for the synthetic replicates of the natural peptides. The cationicity-enhanced analogues were isobaric and differed by just a single amino acid residue - I/L at position 14. Despite this small structural difference, both peptide analogues were different in their proliferative effects on the four cancer cell lines. Both peptides exhibited highly-significant (p<0.001) anti-proliferative effects on all four cancer cell lines tested at concentrations of 10-4 and 10-5 M. These anti-proliferative effects are probably due to lysis of cells as has been reported for several other amphibian skin AMPs and also AMPs from other sources [19-22]. However, an unexpected observation with the analogue, AcrAP1a, was a selective and highly-significant increase in proliferation of both H460 and PC3 cells at concentrations a low as 10-9 M (p<0.001). While the cause of this unusual effect is unknown and undoubtedly warrants further in-depth investigation, there could be several explanations, albeit speculative at present. Perhaps the most compelling relates to the observations that some AMPs from amphibian skin secretions, at concentrations several orders of magnitude below those that produce cytolysis, are capable of inducing insulin release from insulinoma-derived cell lines [23] and cytokine release from macrophages [24]. It is proposed that binding of these AMPs at low, non-cytolytic concentrations to secretory cell membranes, can cause localised perturbation effects, such as depolarisation, that can lead to stimulation of release of endogenous secretory vesicles. Some cancer cells are known to produce autocrine/paracrine growth factors [25] and it is possible that the cancer cell lines, H460 and PC3, are among these. If so, then increased secretion of such growth factors could explain the highly-significant increases in proliferation observed here. Recent reports have suggested that some established and widely-used anticancer drugs, such as doxorubicin, while manifesting obvious anti-cancer effects, can promote the growth of cells in the cancer which exhibit cancer stem cell phenotypes and these are often drug-resistant and metastatic [26]. This effect can in part be explained by activation of the TGFβ signalling pathway. This cytokine acts in a paracrine fashion and plays a fundamental role in cell proliferation and differentiation [27]. The study of this cytokine in H460 and PC3 cells is thus warranted in light of these facts.
In summary, the isolation of two novel AMPs from the venom of the scorpion, Androctonus crassicauda, is described. Following cloning of biosynthetic precursor-encoding cDNAs and identification/structural characterisation of both mature peptides in venom, each was chemically-synthesised along with respective cationicity-enhanced analogues. Biological testing of all four peptides, revealed that the synthetic natural peptides had a moderately potent but narrow spectrum of antimicrobial activity. In contrast, the cationicity-enhanced analogue of each exhibited a broader spectrum of activity with an associated increase in potency. Modulation of proliferative activity of several human cancer cell lines was not observed for the synthetic natural peptides but was for their respective analogues. While both analogues exhibited highly-significant anti-proliferative cytotoxic effects on all cell lines tested at micromolar (10-6 M) concentrations, the analogue AcrAP1a caused a highly-significant proliferative effect on two cell lines, H460 and PC3, at nanomolar (10-9 M) concentrations. This contradictory concentration-dependent effect would thus constitute a serious problem in their further development as anti-cancer agents. Nevertheless, these data demonstrate that the natural peptide templates found in animal venoms, while possessing intrinsic biologically-active properties, can be chemically-manipulated through analogue design, to enhance such or indeed to produce novel actions. Both facilitations readily illustrate the considerable potential of these natural peptide templates in drug design and development.
Data deposition footnote
The nucleotide sequences of AcrAP1 and AcrAP2, from the venom of the scorpion, Androctonus crassicauda, have been deposited in the EMBL Nucleotide Sequence Database under the accession codes, HG939518 and HG939519.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Migdalof BH, Antonaccio MJ, McKinstry DN, Singhvi SM, Lan SJ, Egli P, Kripalani KJ. Captopril: pharmacology, metabolism and disposition. Drug Metab Rev. 1984;15:841-869
2. Iltz JL, Baker DE, Setter SM, Keith Campbell R. Exenatide: an incretin mimetic for the treatment of type 2 diabetes mellitus. Clin Ther. 2006;28:652-665
3. Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discov. 2003;2:790-802
4. Kapoor VK. Natural toxins and their therapeutic potential. Indian J Exp Biol. 2010;48:228-237
5. Fox JW, Serrano SM. Approaching the golden age of natural product pharmaceuticals from venom libraries: an overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr Pharm Des. 2007;13:2927-2934
6. Zhou M, Liu Y, Chen T, Fang X, Walker B, Shaw C. Components of the peptidome and transcriptome persist in lin wa pi: the dried skin of the Heilongjiang brown frog (Rana amurensis) as used in traditional Chinese medicine. Peptides. 2006;27:2688-2694
7. Liang S. Proteome and peptidome profiling of spider venoms. Expert Rev Proteomics. 2008;5:731-746
8. Zhijian C, Feng L, Yingliang W, Xin M, Wenxin L. Genetic mechanisms of scorpion venom peptide diversification. Toxicon. 2006;47:348-355
9. Erspamer V. Bioactive Secretions of the Integument. In: (ed.) Heatwole H, Barthalmus GT. Amphibian Biology, vol 1. Chipping Norton: The Integument. Surrey Beatty & Sons. 1994:179-350
10. Almaaytah A, Albalas Q. Scorpion venom peptides with no disulfide bridges: a review. Peptides. 2014;51:35-45
11. Zeng XC, Zhou L, Shi W, Luo X, Zhang L, Nie Y, Wang J, Wu S, Cao B, Cao H. Three new antimicrobial peptides from the scorpion Pandinus imperator. Peptides. 2013;45:28-34
12. Almaaytah A, Zhou M, Wang L, Chen T, Walker B, Shaw C. Antimicrobial/cytolytic peptides from the venom of the North African scorpion, Androctonus amoreuxi: biochemical and functional characterization of natural peptides and a single site-substituted analog. Peptides. 2012;35:291-299
13. Gautier R, Douquet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific. α-helical properties. Bioinformatics. 2008;24:2010-2012
14. Wu Y, Wang L, Lin C, Lin Y, Zhou M, Chen L, Connolly B, Zhang Y, Chen T, Shaw C. Vasorelaxin: a novel arterial smooth muscle-relaxing eicosapeptide from the skin secretion of the Chinese piebald odorous frog (Odorrana schmackeri). PLoS One. 2013;8:e55739
15. Omar HEDM. The biological and medical significance of poisonous animals. J Biol Earth Sci. 2013;3:M25-M40
16. Mebs D. Venomous and Poisonous Animals: A Handbook for Biologists, Toxicologists and Toxinologists, Physicians and Pharmacists. Medpharm Scientific Publisher. 2000
17. Escoubas P, Diochot S, Corzo G. Structure and pharmacology of spider venom neurotoxins. Biochimie. 2000;82:893-907
18. Doley R, Kini RM. Protein complexes in snake venom. Cell Mol Life Sci. 2009;66:2851-2871
19. Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim Biophys Acta. 2008;1778:357-375
20. Gaspar D, Veiga AS, Castanho MA. From antimicrobial to anticancer peptides. A review. Front Microbiol. 2013;4:294
21. Leuschner C, Hansel W. Membrane disrupting lytic peptides for cancer treatments. Curr Pharm Des. 2004;10:2299-2310
22. van Zoggel H, Hamma-Kourbali Y, Galanth C, Ladram A, Nicolas P, Courty J, Amiche M, Delbé J. Antitumor and angiostatic peptides from frog skin secretions. Amino Acids. 2012;42:385-395
23. Abdel-Wahab YH, Power GJ, Ng MT, Flatt PR, Conlon JM. Insulin-releasing properties of the frog skin peptide pseudin-2 and its [Lys18]-substituted analogue. Biol Chem. 2008;389:143-148
24. Roelants K, Fry BG, Ye L, Stijlemans B, Brys L, Kok P, Clynen E, Schoofs L, Cornelis P, Bossuyt F. Origin and functional diversification of an amphibian defense peptide arsenal. PLoS Genet. 2013;9:e1003662
25. Heasley LE. Autocrine and paracrine signaling through neuropeptide receptors in human cancer. Oncogene. 2001;20:1563-1569
26. Bandyopadhyay A, Wang L, Agyin J, Tang Y, Lin S, Yeh IT, De K, Sun LZ. Doxorubicin in combination with a small TGFbeta inhibitor: a potential novel therapy for metastatic breast cancer in mouse models. PLoS One. 2010;5:e10365
27. Dinarello CA. Historical Review of Cytokines. Eur J Immunol. 2007;37:S34-S45
Author contact
![]() Corresponding authors: 1073075810com or l.wangac.uk.
Corresponding authors: 1073075810com or l.wangac.uk.

 Global reach, higher impact
Global reach, higher impact