10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(10):1171-1180. doi:10.7150/ijbs.9598 This issue Cite
Research Paper
The Insect Ecdysone Receptor is a Good Potential Target for RNAi-based Pest Control
State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China
* These authors contributed equally to this work.
Received 2014-5-7; Accepted 2014-9-9; Published 2014-11-7
Abstract
RNA interference (RNAi) has great potential for use in insect pest control. However, some significant challenges must be overcome before RNAi-based pest control can become a reality. One challenge is the proper selection of a good target gene for RNAi. Here, we report that the insect ecdysone receptor (EcR) is a good potential target for RNAi-based pest control in the brown planthopper Nilaparvata lugens, a serious insect pest of rice plants. We demonstrated that the use of a 360 bp fragment (NlEcR-c) that is common between NlEcR-A and NlEcR-B for feeding RNAi experiments significantly decreased the relative mRNA expression levels of NlEcR compared with those in the dsGFP control. Feeding RNAi also resulted in a significant reduction in the number of offspring per pair of N. lugens. Consequently, a transgenic rice line expressing NlEcR dsRNA was constructed by Agrobacterium- mediated transformation. The results of qRT-PCR showed that the total copy number of the target gene in all transgenic rice lines was 2. Northern blot analysis showed that the small RNA of the hairpin dsNlEcR-c was successfully expressed in the transgenic rice lines. After newly hatched nymphs of N. lugens fed on the transgenic rice lines, effective RNAi was observed. The NlEcR expression levels in all lines examined were decreased significantly compared with the control. In all lines, the survival rate of the nymphs was nearly 90%, and the average number of offspring per pair in the treated groups was significantly less than that observed in the control, with a decrease of 44.18-66.27%. These findings support an RNAi-based pest control strategy and are also important for the management of rice insect pests.
Keywords: RNA interference, pest control, Nilaparvata lugens, ecdysone receptor (EcR), transgenic rice
Introduction
The utilization of transgenic technology has led to amazing progress in modern agriculture through the manipulation of traits such as resistance to herbicides, environmental stress, insects and viruses. Since 1996, when transgenic crops were first commercialized, the global industrialization of transgenic crops has continued to progress rapidly. A more than 100-fold increase in hectarage has occurred, from 1.7 million hectares in 1996 to over 175 million hectares in 2013, along with a sharp increase in the number of farmers planting transgenic crops, from 1.3 million farmers in 6 countries in 1996 to 18 million farmers in 27 countries in 2013 [1]. Bacillus thuringiensis (Bt) genes are the most widely used exogenous genes that confer insect resistance to transgenic crops [2-7]. It has been universally accepted that Bt toxins function as insecticides by destroying the permeability of the cell membrane in the midgut cells of the target insect; however, they have little or no effect on vertebrates and most other organisms [8-9]. In October 2009, Bt Huahui No.1 and Shanyou 63 rice lines were issued biosafety certificates for commercial production in Hubei Province by the Ministry of Agriculture of China [10]. There is no doubt that Bt transgenic crops have made a great contribution to the control of agricultural insect pests such as the cotton bollworm Helicoverpa armigera [11]. Nevertheless, as with any technology, potential risks may be associated with these Bt transgenic crops. Increasing numbers of insect populations have been reported to have developed resistance to Bt toxin proteins and Bt transgenic crops, such as Heliothis zea, Spodoptera frugiperda and Busseola fusca [12-14]. In addition, secondary pests, such as mirids and spider mites, may pose a new challenge in transgenic crop fields [15-23]. Furthermore, the negative impacts of Bt toxins derived from Bt transgenic crops on soil organisms should not be neglected [24-27]. Therefore, it is essential to identify new types of exogenous genes and even alternative pest control strategies for transgenic plants.
RNA interference (RNAi) has become a powerful tool for studying gene function in recent years and has been exploited in plants for pest control [28]. The pioneer RNAi studies were first published in 2007. Transgenic corn expressing dsRNA targeted to a subunit of the midgut enzyme vacuolar ATPase (V-ATPase) decreased the damage caused by the western corn rootworm (WCR) Diabrotica virgifera virgifera [29]. Meanwhile, the P450 monooxygenase gene was used as a target to develop transgenic Arabidopsis thaliana, tobacco and cotton that reduced the tolerance of cotton bollworm to gossypol [30-31]. More recently, transgenic tobacco plants expressing dsRNA of an insect gene, ecdysone receptor (EcR), were ingested at significantly lower levels by H. armigera larvae, thus causing molting defects and larval lethality [32]. DvSnf7 RNAi caused the accumulation of ubiquitinated proteins and the impairment of autophagic processes in the midgut and fat body tissues of dsRNA-fed WCR larvae [33]. Because the spectrum of activity for DvSnf7 is narrow and this activity is only evident in a subset of beetles within the Galerucinae subfamily of Chrysomelidae, DvSnf7 may be a good target gene for transgenic maize [34].
The brown planthopper (BPH) Nilaparvata lugens is one of the most devastating insect pests of rice in the temperate and tropical regions of Asia and Australia, with a special piercing-sucking mouthpart that causes a large yield reduction and major economic losses [35]. Although transgenic rice lines expressing the dsRNA of 3 BPH midgut genes were developed, no phenotype was observed [36]. Here, we tested whether transgenic rice lines expressing dsRNA from the EcR gene in BPH possess any pest control properties. Our results demonstrated that the number of BPH offspring was significantly reduced after BPH nymphs fed on the transgenic rice lines expressing the EcR dsRNA. These findings suggest that NlEcR is a good candidate target gene for the development of RNAi-based transgenic rice lines with resistance against N. lugens.
Materials and Methods
Insects
The N. lugens (BPH) colony was obtained from the Guangdong Academy of Agricultural Sciences (GAAS). The insects were reared in wooden cages (60 cm x 60 cm x 80 cm) on BPH-susceptible rice plants (Fenghua Zhan, purchased from GAAS, Guangzhou, China) in a greenhouse at 27±2°C with 80±10% humidity and a light/dark cycle of L14:D10 h.
Bioassays for feeding-based RNAi
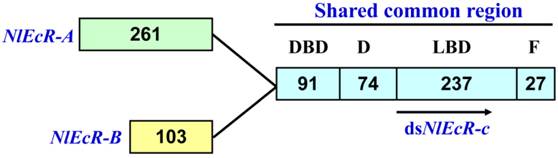
A 360 bp fragment common to both NlEcR-A (2393 bp, GenBank accession number FJ263048.1) and NlEcR-B (1963 bp, GenBank accession number FJ263049.1), referred to as NlEcR-c, was used for the synthesis of dsRNA (Fig. 1). The synthesis was conducted according to the manufacturer's instructions provided with the T7 RiboMAXTM Express RNAi System Kit (Promega, Madison, Wisconsin, USA) following previously described methods [37-38]. The forward and reverse primers are listed in Table 1. All sequences were verified by sequencing (Invitrogen Company, Shanghai, China).
The common fragment between the two EcR isoforms in Nilaparvata lugens used for RNAi. The number of amino acids in each domain is indicated.
The primers used in this study.
| Primer name | Forward primer | Reverse primer |
|---|---|---|
| Vector construction | ||
| Hairpin-sense | CACTGGATCCACTTCCAGAACGAATTCGAGC | GACGAGCTCCCTCCACCACATAACCCATG |
| Hairpin-antisense | GACGTCGACCCTCCACCACATAACCCATG | CACAAGCTTACTTCCAGAACGAATTCGAGC |
| qRT-PCR | ||
| QNlEcR-c | CCGAAGAAGCATAACCAAGAACGCG | CTCTGGTACAACACACTCAGGCCTCATTC |
| QNlβ-ACTIN | TGCGTGACATCAAGGAGAAGC | AGCCTTCCTTCTTGGGTATGG |
| QT-DNA | GCGAAGAACTCCAGCATGAGATCC | CGCCTTCTTGACGAGTTCTTCTGAG |
| QSPS | GCCACGGACTCCTCTAATTCTCTCTC | GATCGAGAGAGAGAATCGGCGAG |
| dsRNA synthesis | ||
| dsNlEcR-c | ACTTCCAGAACGAATTCGAGC | CCTCCACCACATAACCCATG |
| dsNlEcR-cT7 | TAATACGACTCACTATAGGGACTTCCAGAACGAATTCGAGC | TAATACGACTCACTATAGGGCCTCCACCACATAACCCATG |
| dsGFP | GCACCATCTTCTTCAAGGACG | ACTCCAGCAGGACCATGTGAT |
| dsGFP-T7 | TAATACGACTCACTATAGGGGCACCATCTTCTTCAAGGACG | TAATACGACTCACTATAGGGACTCCAGCAGGACCATGTGAT |
| RT-PCR | ||
| Tr-NlEcR-c | ACTTCCAGAACGAATTCGAGC | CCTCCACCACATAACCCATG |
The feeding RNAi experiment was conducted according to a previously described method [37]. Third instar BPH nymphs were fed an artificial diet [39] containing the above dsRNA (0.1 μg/μl or 0.5 μg/μl) for 10 days. Each replicate consisted of 30 individuals in 2 glass tubes (15 individuals in each tube), and three replicates were analyzed. The survival and phenotype of the insects were recorded daily.
Three nymphs were selected randomly on days 2, 4, 6 and 8 post-ingestion at 0.1 μg/μl and on days 1, 2, 6 and 8 post-ingestion at 0.5 μg/μl to detect NlEcR expression levels by qRT-PCR using a LightCycler480 system (Roche Diagnostics, Indianapolis, IN, USA) and SYBR Premix Ex Taq (TaKaRa, Kyoto, Japan). The procedure was performed as previously described [40]. The mRNA expression level in the dsGFP group was used as a control. Total RNA was extracted from BPH using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Next, the reverse transcriptase XL (AMV) (TaKaRa, Kyoto, Japan) protocol with an oligo dT18 primer was used to synthesize first-strand cDNA. qRT-PCR amplification was performed with the primers QNlEcR-c and QNlβ-ACTIN (Table 1) as follows: denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 40 s, with a final extension at 72°C for 10 min.
To measure the offspring of the surviving brachypterous adults, the fifth instar nymphs were removed from the glass tubes and placed on rice plants (Fenghua Zhan) in a plastic cage (diameter 14 cm, height 20 cm) in a basin (diameter 22 cm) so that they were able to emerge in the absence of the dsRNA. One male and one female brachypterous adult emerged within 24 hours and were paired in a plastic cage for egg laying. On day 15, the two insects, if living, were removed from the cage. The number of nymphs was counted 10 days later.
Construction of transgenic rice
Using two sets of primers that are listed in Table 1, an inverted repeat derived from NlEcR-c with the restriction enzyme sites BamHI/SacI at the ends of the sense arm and HindIII/SalI at the ends of the antisense arm was amplified by RT-PCR. The PCR products were inserted as inverted repeats on either side of the intron of the pSK-int vector to obtain a hairpin RNAi construct, which was then cloned into the final expression vector pCanG-HA using the SacI/SalI restriction enzyme sites such that it was under the control of the CaMV35S promoter and the nopaline synthase (nos) terminator cassette (Fig. 6A). After introducing the final vector into the Agrobacterium tumefaciens strain LBA4404 by the freeze-thaw method, the japonica rice variety Nipponbare was subjected to Agrobacterium-mediated transformation as previously described [41], and transformed calli were selected based on G418 resistance. Finally, transgenic plants were regenerated from the transformed calli and grown in a greenhouse at 27±2°C with 80±10% humidity and a light/dark cycle of L14:D10 h.
Identification of transgenic rice
Genomic DNA was isolated from the fresh leaves of transgenic plants using the CTAB method [42]. DNA from non-transgenic plants was used as a negative control. Plants containing the transgene were selected by RT-PCR of the target fragment with the set of primers listed in Table 1, and the T-DNA copy number was estimated by quantitative real-time PCR (qRT-PCR) using a LightCycler480 system (Roche Diagnostics Indianapolis, IN, USA) and SYBR Premix Ex Taq (TaKaRa, Kyoto, Japan) [43]. The reaction system was the same as that described above. The T-DNA fragment was amplified with QTDNA-F and QTDNA-R primers (Table 1) to detect the target gene. The sucrose phosphate synthase (SPS) gene fragment was amplified with QSPS-F and QSPS-R primers (Table 1) and was used as an endogenous reference gene. Standard curves were obtained using a 10-fold serial dilution of pooled DNA.
Northern blot analysis was used to detect the expression level of small RNA fragments of the target gene in transgenic rice lines. The hairpin dsRNA in the transgenic rice lines was used as the probe and was prepared using the pGEM®-T and pGEM®-T Easy Vector Systems (Promega, Madison, Wisconsin, USA) with the restriction enzymes SacI and ApaI (Supplementary Material: Fig. S1). The probe was then labeled with the DIG RNA Labeling Kit (SP6/T7) (Roche Diagnostics Indianapolis, IN, USA). Small RNA was analyzed by electrophoresis on a 2% agarose gel containing formaldehyde after being isolated from the fresh leaves of transgenic rice using the NucleoSpin miRNA kit (Macherey-Nagel, Germany). In accordance with the operation manual provided with the DIG Northern Starter Kit (Roche Diagnostics Indianapolis, IN, USA), Hybond N+ (Amersham, PRN303B) was prehybridized with siRNAs at 50°C for 2 h, and hybridizations were performed at 50°C for more than 16 h. Next, the blots were washed with 2×SSC/0.1% SDS for 2×5 min at 37°C, followed by 0.1×SSC/0.1% SDS for 2×15 min at 65-68°C. U6 was used as a hybridization reference gene, while non-transgenic plants were used as a control.
Bioassays for N. lugens fed on transgenic rice
The same T1 generations of transgenic rice lines were used for all bioassays. In each replicate, 25 newly hatched N. lugens nymphs were reared on transgenic rice plants (approximately 40 cm in height) or control rice plants (japonica variety Nipponbare) in a plastic cage (diameter 14 cm, height 80 cm) in a basin (diameter 22 cm) in a greenhouse until emergence. In each replicate, three 3rd instar, three 4th instar and three 5th instar nymphs were selected randomly for subsequent RNA extraction and detection of NlEcR mRNA levels using the method described above. Three replicates were analyzed.
As described above, another patch of newly hatched N. lugens nymphs were reared to observe survival rates. The number of rice plants of the same line was considered to be the number of replicates.
To measure the number of offspring of the brachypterous adults that survived on the transgenic rice lines, one male and one female brachypterous adult that emerged within 24 hours were paired in a plastic cage with rice plants for egg laying. On day 15, the two insects, if living, were removed from the cage. The number of nymphs was counted 10 days later. The number of rice plants of the same line was considered to be the number of replicates. Insects fed on non-transgenic rice plants were used as a control.
Statistical analysis
The data are expressed as the means ± SE. Significant differences between the means were determined by t-test at p < 0.05 or 0.01.
Results
Ingestion of EcR dsRNA led to RNAi effects
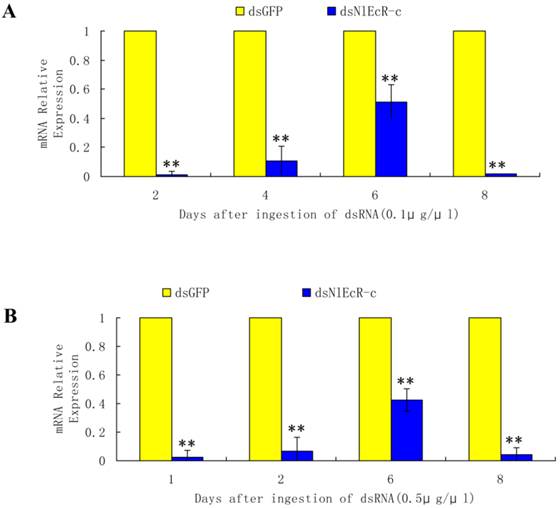
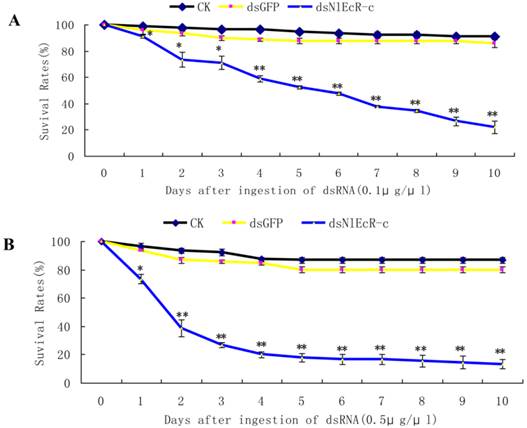
Based on the sequence of two N. lugens EcR genes (NlEcR-A and NlEcR-B) that were previously cloned in the lab, a 360 bp fragment common to both genes (NlEcR-c) was chosen and used for the synthesis of double-stranded RNA (dsRNA) (Fig. 1). On days 2, 4, 6 and 8 after ingestion of dsNlEcR-c at 0.1 μg/μl and on days 1, 2, 6 and 8 after ingestion of dsNlEcR-c at 0.5 μg/μl, the relative mRNA expression levels of NlEcR were significantly decreased compared with those in the dsGFP control (t-test, p < 0.01) (Fig. 2 A&B). In addition, some nymphs exhibited difficulty in molting (data not shown). The survival rate of BPH nymphs was significantly decreased 2 days after ingestion of 0.1 μg/μl dsRNA, while a significant reduction in the survival rate occurred only 1 day after ingestion of 0.5 μg/μl dsRNA (Fig. 3). Likewise, mortality increased with an increase in dsRNA concentration. These results indicated that the RNAi was effective.
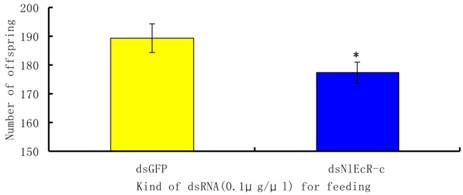
To evaluate the effects of RNAi on the fecundity of the pest, we chose to analyze brachypterous adults following ingestion of 0.1 μg/μl of dsNlEcR-c. The surviving fifth instar nymphs were removed from glass tubes and placed on rice plants in a cage for emergence in the absence of dsRNA. The number of offspring from each pair of brachypterous adults was recorded. The results showed that the average number of offspring per pair in the treated group was 177.45, which was significantly less than that in the dsGFP group (t-test, p < 0.05) (Fig. 4).
Construction and identification of transgenic rice lines expressing EcR dsRNA
Based the results of the feeding RNAi experiment; we chose the NlEcR-c fragment for the construction of the hairpin dsRNA. The inverted repeat in the target fragment was inserted on either side of the intron of the pSK-int vector using the BamHI/SacI and HindIII/SalI restriction enzyme sites, and the hairpin dsRNA structure was cloned into the expression vector pCanG-HA using the SacI/SalI restriction enzyme sites (Fig. 5A) before being introduced into LBA4404 by the freeze-thaw method. Calli from the japonica rice variety Nipponbare were subjected to Agrobacterium-mediated transformation and selected based on G418 resistance. Transgenic plants were identified by RT-PCR through the amplification of NlEcR-c (Fig. 5B). The confirmed transgenic rice lines were grown in a greenhouse for further experiments.
The EcR mRNA expression level in Nilaparvata lugens after ingestion of dsNlEcR-c. (A) 0.1 μg/μl. (B) 0.5 μg/μl. * indicates a significant difference in the expression level between the treated and control groups as determined by t-test at p < 0.05; ** indicates differences at the p < 0.01 level; all error bars represent the standard error of the mean as determined from three independent replicates.
The survival rates of Nilaparvata lugens after ingestion of dsNlEcR-c. (A) 0.1 μg/μl. (B) 0.5 μg/μl. Each data point is the mean ± SE of three independent experiments with thirty normal individuals. * indicates significant differences in the expression level between the treated and control groups as determined by t-test at p < 0.05; ** indicates differences at the p < 0.01 level.
The effects of feeding RNAi on the number of offspring of Nilaparvata lugens. An asterisk indicates a significant difference in the number of offspring between the treated and control groups (t-test, p < 0.05). All error bars represent the standard error of the mean as determined by more than three independent replicates.
Generation of transgenic rice lines expressing dsNlEcR-c. (A) Construction of the expression construct pCanG-dsNlEcR-c from the intermediate vector psk-dsNlEcR-c. (B) Transgenic rice lines were identified by amplifying the 360 bp fragment of NlEcR-c. 1-28 represent the transgenic rice lines, M represents DL2000.
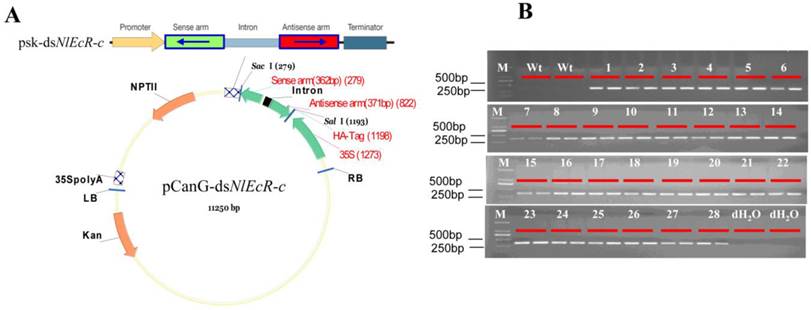
Northern blot analysis of dsNlEcR-c expression in transgenic rice lines. For each line, 20 μg of small RNA in the same volume was loaded. Non-transgenic plants were used as a control, and U6 was used as a reference gene.
Standard curves (Supplementary Material: Fig. S2) were constructed by amplifying the reference gene SPS and the target gene fragment T-DNA using the appropriate primers (Table 1). The qRT-PCR results showed that the total copy number of the target gene in all transgenic rice lines was 2 (Table 2). Northern blot analysis was used to detect the target siRNAs (Fig. 6), and the results showed that small RNA of the hairpin dsNlEcR-c was expressed successfully in each transgenic rice line.
Control of N. lugens using the transgenic rice lines
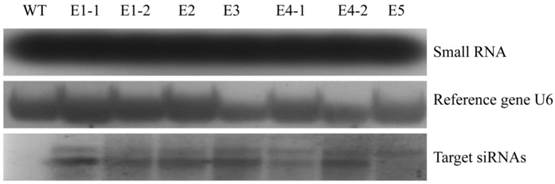
The ability of the transgenic rice lines (E1-E5) confirmed by northern blot to control N. lugens was studied. After the newly hatched N. lugens nymphs fed on the transgenic rice lines, third, fourth and fifth instar nymphs were randomly selected from three plants of each transgenic rice line, and their NlEcR mRNA expression levels were detected. The results revealed that the expression level of NlEcR in all samples analyzed was decreased significantly when compared with that in the control (t-test, p < 0.05). Specifically, the relative reductions in mRNA expression levels were 24.48-40.41%, 23.24-37.72% and 24.02-39.96% for the third, fourth and fifth instars, respectively, when they fed on the transgenic rice line E1 (Fig. 7). These results showed that the transgenic rice lines expressing NlEcR dsRNA could cause RNAi in insects.
After newly hatched N. lugens nymphs fed on the transgenic rice lines, the survival rate of the nymphs fed on all lines was nearly 90%. A significant reduction in the survival rates was only observed in the nymphs fed on lines E1-E3 on day 10 and in the nymphs fed on lines E1-E4 on day 11 (t-test, p < 0.05) (Fig. 8).
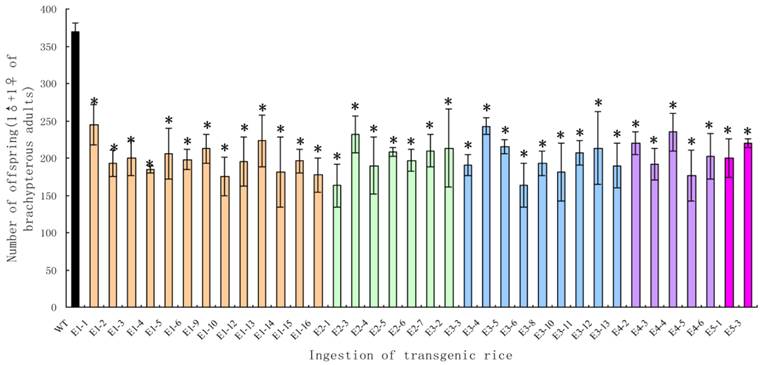
Although the survival rates were very high, the brachypterous adults fed on the transgenic rice lines produced far fewer offspring. The average numbers of offspring per pair in the treated groups were significantly lower than those in the control groups (t-test, p < 0.05), with a decrease of 44.18-66.27% (Fig. 9).
An estimation of the copy number of T-DNA fragments in the transgenic rice lines. T-DNA0 and SPS0 indicate the initial copy numbers of T-DNA and the SPS gene in the PCR reactions, respectively, and 2×T-DNA0/SPS0 is shown as the mean ± 95% confidence limit.
| Line | T-DNA0 | SPS0 | 2xT-DNA0/SPS0 | Copy number |
|---|---|---|---|---|
| E1 | 52531.891 | 635653.4 | 1.628±0.019 | 2 |
| E2 | 50592.823 | 1007072 | 1.67±0.008 | 2 |
| E3 | 79307.675 | 564049.3 | 1.703±0.009 | 2 |
| E4 | 26840.951 | 595118.4 | 1.534±0.013 | 2 |
| E5 | 62137.849 | 646039.9 | 1.65±0.014 | 2 |
| WT | 2.3277739 | 398051.1 | 0.075±0.19 | 0 |
NlEcR expression levels in Nilaparvata lugens after feeding on transgenic rice lines. An asterisk indicates a significant difference in the expression level between treated and control groups (t-test, p < 0.05). Error bars represent the standard error of the mean as determined from three independent replicates.
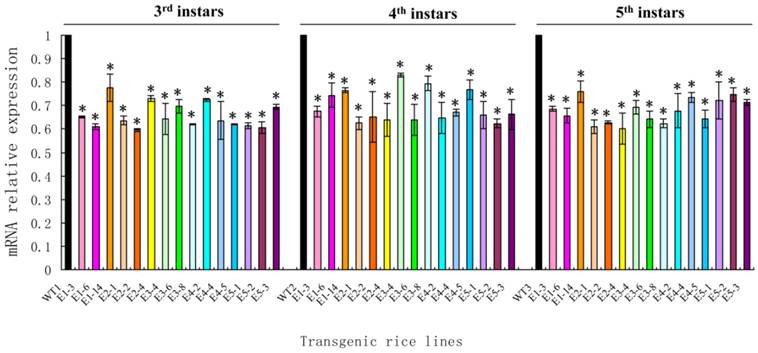
The survival rates of Nilaparvata lugens nymphs after feeding on transgenic rice lines. Each data point is the mean ± SE from more than three replicates with the same line. An asterisk indicates a significant difference in the survival rate between treated and control groups (t-test, p < 0.05).
The number of offspring from a pair of Nilaparvata lugens brachypterous adults after feeding on transgenic rice lines. Each data point is the mean ± SE from more than three replicates with the same line. An asterisk indicates a significant difference in the number of offspring between treated and control groups (t-test, p < 0.05).
Discussion
EcR as a target for biorational insecticide development
Insect molting and metamorphosis are governed by ecdysteroids and the juvenile hormone. In most insects, 20-hydroxyecdysone (20E) is the natural molting hormone; it binds and activates a heterodimer composed of two nuclear receptor proteins: the ecdysone receptor (EcR) and ultraspiracle (USP) [44]. Compounds acting on EcR will only affect insects and other arthropods, making EcR an interesting target for safer and more specific insecticides [45]. This finding has led to the development of several successful commercial ecdysteroid agonistic insecticides with a dibenzoylhydrazine (DBH) structure [46]. Currently, there are four DBH ecdysone agonists available on the market. The success of these compounds in insect control programs, in turn, has validated EcR as a valuable target for the development of environmentally friendly biorational insecticides [47].
To successfully search for new compounds that disrupt the insect endocrine system, high-throughput screening systems have been developed [46, 48-49]. These approaches will aid the development of ecdysone agonists that are specific for EcRs in insect orders, such as Lepidoptera, Diptera or Coleoptera [48].
Screening of target genes for RNAi-based insect pest control
With the rapid development of transgenic and RNAi technology, plant-mediated RNAi shows great potential for insect pest control. Today, researchers are engineering a variety of crops to produce small RNAs that will silence essential genes in insects, nematodes and pathogens, an approach called host-delivered RNAi [50]. However, some significant challenges must be overcome before RNAi-based pest control can become a reality. One of these challenges is the complexity of selecting a good target for RNAi. The criteria for such a decision should involve at least 2 aspects: safety and efficacy. For insect species with artificial diets, target genes can be screened through feeding dsRNA [37]. Different screening approaches have been used. Baum et al. (2007) identified a series of potential targets for RNAi in the western corn rootworm based on effectiveness in terms of LC50, including vacuolar ATPase [29]. In this study, we selected an EcR gene that has already been approved as a good target for insect pest management, as discussed above.
A high pest mortality rate is often used as an indicator for the assessment of pest control measures. However, from an ecological point of view, secondary insect pests are likely to become primary insect pests after the previous primary insect pests have been efficiently controlled in the field. For example, secondary pests, such as mirids, became new challenges in transgenic Bt cotton fields after the cotton bollworm H. armigera was successfully controlled [20, 23]. In this study, the mortality rates of N. lugens were quite low after these pests fed on the transgenic rice lines (Fig. 8); however, they had a 22.78%-77.83% decrease in the number of offspring (Fig. 9, Supplementary Material: Fig. S3). These results suggest that the abundance of the major insect pest N. lugens will gradually decrease in paddy fields, and secondary insect pests may have less of a chance to become primary insect pests, which is beneficial for maintaining an ecological balance. Additionally, it has been suggested that the low fecundity of an insect may be an alternative indicator that can be used for screening promising target genes for RNAi-based insect pest control.
Non-target analysis of transgenic rice expressing EcR dsRNA
Ecological risk assessments are important for both genetically modified (GE) crops and RNAi crops [51-52]. Six questions were formulated for special attention, including non-target effects, off-target effects, and the environmental persistence of small RNA molecules [51]. Unlike protein-based GM crops, RNAi-induced traits are generated precisely based on nucleic acid sequence identity; hence, the use of bioinformatics analysis is imperative in identifying the risk impacts on potential non-target organisms [52]. Induction of insect resistance in maize by downregulation of the V-ATPase gene of western corn rootworm had potentially adverse effects on non-target organisms sharing sequence homology with the target gene [29]. A similar result was observed in endeavors to achieve reduced production of allergen P34 in soybean [53]. The specificity of the DvSnf7 dsRNA was characterized through bioassays, and the results of these analyses showed that the likelihood of adverse effects on non-target arthropods due to realistic environmental exposure to DvSnf7 dsRNA is predicted to be extremely low [34]. In the bioinformatics analysis in this study, all available genomic databases of rice and insects (including N. lugens) were used for sequence specificity alignment, and 19 bp consecutive identical sequences were not found between the target fragment of the EcR gene and all other genes. The sequence specificity of dsRNAs theoretically ensures that the non-target risk of transgenic rice expressing EcR dsRNA is low. However, the potential risks of these plants must be assessed through bioassays in the near future.
Supplementary Material
Fig.S1 - S3.
Acknowledgements
We are grateful to Dr. Sheng Li (Institute of Plant Physiology and Ecology, Shanghai, China), Dr. Xiaoqiang Yu (University of Missouri-Kansas City, USA) for their suggestions. This work was supported by the National Natural Science Foundation of China (30930061) and the National Key Program on Transgenic Organisms of China (2009ZX08001-018B).
Author Contributions
Conceived and designed the experiments: RY, XX, WZ. Performed the experiments: RY, XX, YL, ZP, HJ, NW. Analyzed the data: RY, HT. Contributed reagents/materials/analysis tools: RY, XX. Wrote the paper: RY, WZ.
Competing Interests
The authors have declared that no competing interest exists.
References
1. James C. Global Status of Commercialized Biotech/GM Crops: 2013. ISAAA Brief No. 46. ISAAA: Ithaca, NY. 2013
2. Wang DP. Current status and future strategies for development of transgenic plants in China. J Integr Plant Biol. 2007;49(9):1281-1283
3. Brookes G, Barfoot P. GM Crops: Global Socio-Economic and Environmental Impacts 1996-2008. PG Economics Ltd. UK. 2010
4. Choudhary B, Gaur K. Bt Cotton in India: A Country Profile. ISAAA Series of Biotech Crop Profiles. ISAAA, Ithaca, NY. 2010
5. Huang J, Mi J, Lin H. et al. A decade of Bt cotton in Chinese fields: assessing the direct effects and indirect externalities of Bt cotton adoption in China. Sci China C Life Sci. 2010;53(8):981-991
6. Hutchison WD, Burkness EC, Mitchell PD. et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Sci. 2010;330(6001):222-225
7. Tabashnik BE. Communal benefits of transgenic corn. Sci. 2010;330:189-190
8. Knowles BH, Ellar DJ. Colloidos moticlysis is a general feature of the mechanism of action of Bacillus thuringiensis δ-endotoxins with different insect specificity. BBA. 1987;924:507-518
9. Schnepf E, Crickmore N, Van Rie J. et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62(3):775-806
10. Chen M, Shelton A, Ye GY. Insect-resistant genetically modified rice in China: from research to commercialization. Annu Rev Entomol. 2011;56:81-101
11. Wu KM, Lu YH, Feng HQ. et al. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Sci. 2008;321(5896):1676-1678
12. Tabashnik BE, Liu YB, Finson N. et al. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc Natl Acad Sci U S A. 1997;94(5):1640-1644
13. Tabashnik BE. Delaying insect resistance to transgenic crops. Proc Natl Acad Sci U S A. 2008;105(49):19029-19030
14. Andow DA, Zwahlen C. Assessing environmental risks of transgenic plants. Ecol Lett. 2006;9(2):196-214
15. Gouse M, Pray C, Schimmelpfennig D. et al. The distribution of benefits from Bt cotton adoption in South Africa. AgBioForum. 2004;7:187-194
16. Sharma OP, Bambawale OM, Dhandapani A. et al. Assessing of severity of important diseases of rainfed Bt transgenic cotton in southern Maharashtra. Indian Phytopathol. 2005;58:483-485
17. Williams MR. Cotton insect losses 2005. Proceedings of the Beltwide Cotton Conferences. Memphis, TN: National Cotton Council, USA. 2006:1151-1204
18. Wilson L, Hickman M. et al. Research update on IPM and secondary pests. Proceedings of the 13th Australian Cotton Research Conference, Broadbeach, Queensland, Australia. 2006 249-258
19. Xue K, Deng S, Wang R. et al. Leaf surface factors of transgenic Bt cotton associated with the feeding behaviors of cotton aphids: a case study on non-target effects. Sci China C Life Sci. 2008;51(2):145-156
20. Lu Y, Qiu F, Feng H. et al. Species composition and seasonal abundance of pestiferous plant bugs (Hemiptera: Miridae) on Bt Cotton in China. Crop Prot. 2008;27:465-472
21. Li YH, Romeis J. Bt maize expressing Cry3Bb1 does not harm the spider mite, Tetranychus urticae, or its ladybird beetle predator, Stethorus punctillum. Biol Contr. 2010;53:337-344
22. Zhao JH, Ho P, Azadi H. Benefits of Bt cotton counterbalanced by secondary pests? Perceptions of ecological change in China. Environ Monit Assess. 2011;173(1-4):985-994
23. Yu H, Li Y, Wu K. Risk assessment and ecological effects of transgenic Bacillus thuringiensis crops on non-target organisms. J Integr Plant Biol. 2011;53(7):520-538
24. Zwahlen C, Hilbeck A, Howald R. et al. Effects of transgenic Bt corn litter on the earthworm Lumbricus terrestris. Mol Ecol. 2003;12(4):1077-1086
25. Zwahlen C, Hilbeck A, Nentwig W. et al. Field decomposition of transgenic Bt maize residue and the impact on non-target soil invertebrates. Plant Soil. 2007;300:245-257
26. Clark BW, Phillips TA, Coats JR. et al. Environmental fate and effects of Bacillus thuringiensis (Bt) proteins from transgenic crops: a review. J. Agric. Food Chem. 2005;53(12):4643-4653
27. Saxena D, Pushalkar S, Stotzky G. Fate and effects in soil of Cry proteins from Bacillus thuringiensis: Influence of physicochemical and biological characteristics of soil. Open Toxinol J. 2010;3:133-153
28. Gordon KH, Waterhouse PM. RNAi for insect-proof plants. Nat Biotechnol. 2007;25(11):1231-1232
29. Baum JA, Bogaert T, Clinton W. et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25(11):1322-1326
30. Mao Y, Cai W, Wang J. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25(11):1307-1313
31. Mao Y, Tao X, Xue X. et al. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res. 2011;20(3):665-673
32. Zhu JQ, Liu S, Ma Y. et al. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS ONE. 2012;7(6):e38572
33. Ramaseshadri P, Segers G, Flannagan R. et al. Physiological and cellular responses caused by RNAi- mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLoS One. 2013;8:e54270
34. Bachman PM, Bolognesi R, Moar WJ. et al. Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res. 2013;22(6):1207-1222
35. Cheng JA, Zhu ZR. Analysis on the key factors causing the outbreak of brown planthopper in Yangtze Area, China in 2005. Plant Prot. 2006;32(4):1-4
36. Zha W, Peng X, Chen R. et al. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One. 2011;6(5):e20504
37. Chen J, Zhang D, Yao Q. et al. Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol Biol. 2010;19(6):777-786
38. Zhai YF, Zhang JQ, Sun ZX. et al. Proteomic and transcriptomic analyses of fecundity in the brown planthopper Nilaparvata lugens (Stål). J Proteome Res. 2013;12(11):5199-5212
39. Fu Q, Zhang Z, Hu C. et al. A chemically defined diet enables continuous rearing of the brown planthopper,Nilaparvata lugens (Stål) (Homoptera:Delphacidae). Appl Entomol Zool. 2001;36(1):111-116
40. Yao Q, Zhang DW, Tang B. et al. Identification of 20-hydroxyecdysone late-response genes in the chitin biosynthesis pathway. PLoS ONE. 2010;5(11):e14058
41. Hiei Y, Ohta S, Komari T. et al. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6(2):271-282
42. Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual, 3rd ed. New York: Cold Spring Harbor Laboratory Press. 2001:633-662
43. Omar AA, Dekkers MG, Graham JH. et al. Estimation of transgene copy number in transformed citrus plants by quantitative multiplex real-time PCR. Biotechnol Prog. 2008;24(6):1241-1248
44. Yao TP, Forman BM, Jiang Z. et al. Functional ecdysone receptor is the product of EcR and ultraspiracle genes. Nat. 1993;366(6454):476-479
45. Dhadialla TS, Carlson GR, Le DP. New insecticides with ecdysteroidal and juvenile hormone activity. Annu Rev Entomol. 1998;43:545-569
46. Zotti MJ, Geyter ED, Swevers L. et al. A cell-based reporter assay for screening for EcR agonist/antagonist activity of natural ecdysteroids in Lepidoptera (Bm5) and Diptera (S2) cell cultures, followed by modeling of ecdysteroid-EcR interactions and normal mode analysis. Pestic Biochem Physiol. 2013;107(3):309-320
47. Nakagawa Y. Nonsteroidal ecdysone agonists. Vitam. Horm. 2005;73:131-173
48. Thomas S, Masatoshi L, Luc S. et al. Towards Coleoptera-specific high-throughput screening systems for compounds with ecdysone activity: development of EcR reporter assays using weevil (Anthonomus grandis)-derived cell lines and in silico analysis of ligand binding to A. grandis EcR ligand-binding pocket. Insect Biochem Mol Boil. 2009;39(8):523-534
49. Thomas S, Swevers L, Georgia K. et al. Comparison of the activity of non-steroidal ecdysone agonists between dipteran and lepidopteran insects, using cell-based EcR reporter assays. Pest Manag Sci. 2010;66(11):1215-1229
50. Price DRG, Gatehouse JA. RNAi-mediated crop protection against insects. Trends in Biotechnol. 2008;26:393-400
51. Auer C, Frederick R. Crop improvement using small RNAs: Applications and predictive ecological risk assessments. Trends in Biotechnol. 2009;27:644-651
52. Ramesh SV. Non-coding RNAs in crop genetic modification: Considerations and predictable environmental risk sssessments (ERA). Mol Biotechnol. 2013;55:87-100
53. Herman E. Soybean allergenicity and suppression of the immunodominant allergen. Crop Sci. 2005;45:462-467
Author contact
![]() Corresponding author: Wenqing Zhang, Tel: +86-20-39332963, Fax: +86-20-39943515, Email: lsszwqsysu.edu.cn
Corresponding author: Wenqing Zhang, Tel: +86-20-39332963, Fax: +86-20-39943515, Email: lsszwqsysu.edu.cn

 Global reach, higher impact
Global reach, higher impact