Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(2):176-185. doi:10.7150/ijbs.10235 This issue Cite
Research Paper
Enhancement of Larval RNAi Efficiency by Over-expressing Argonaute2 in Bombyx mori
1. Key Laboratory of Insect Developmental and Evolutionary Biology, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China.
2. University of Chinese Academy of Sciences, Beijing 100049, China.
Received 2014-7-31; Accepted 2014-11-16; Published 2015-1-5
Abstract
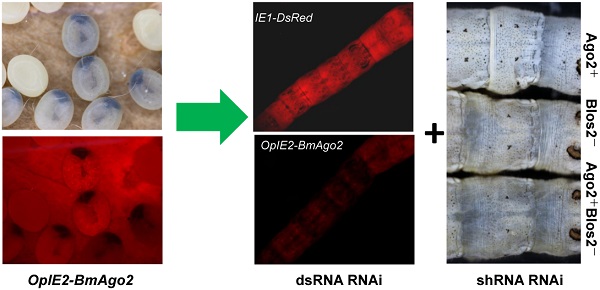
RNA interference has been described as a powerful genetic tool for gene functional analysis and a promising approach for pest management. However, RNAi efficiency varies significantly among insect species due to distinct RNAi machineries. Lepidopteran insects include a large number of pests as well as model insects, such as the silkworm, Bombyx mori. However, only limited success of in vivo RNAi has been reported in lepidoptera, particularly during the larval stages when the worms feed the most and do the most harm to the host plant. Enhancing the efficiency of larval RNAi in lepidoptera is urgently needed to develop RNAi-based pest management strategies. In the present study, we investigate the function of the conserved RNAi core factor, Argonaute2 (Ago2), in mediating B. mori RNAi efficiency. We demonstrate that introducing BmAgo2 dsRNA inhibits the RNAi response in both BmN cells and embryos. Furthermore, we establish several transgenic silkworm lines to assess the roles of BmAgo2 in larval RNAi. Over-expressing BmAgo2 significantly facilitated both dsRNA-mediated larval RNAi when targeting DsRed using dsRNA injection and shRNA-mediated larval RNAi when targeting BmBlos2 using transgenic shRNA expression. Our results show that BmAgo2 is involved in RNAi in B. mori and provides a promising approach for improving larval RNAi efficiency in B. mori and in lepidopteran insects in general.
Keywords: RNAi, Bombyx mori, Argonaute2, dsRNA, shRNA
Introduction
RNA interference (RNAi) is a functional conserved process that regulating gene expression at the post-transcriptional level [1]. It is triggered by 21-23 nucleotides small interfering RNAs (siRNAs) which is phosphorylated at the 5' terminal and hydroxylated at the 3' terminal [2, 3]. The process of RNAi can be divided into two major steps: the initiation step and the effector step [4]. In the initial step, introduced long dsRNA or short-hairpin RNA (shRNA) are digested into siRNA by Dicer, an RNA-III family exonuclease [5]. In the subsequent effector step, the siRNA unwound and incorporated into the functional protein complex: RNA-induced silencing complex (RISC), guiding Argonaute (Ago) to degrade the target mRNA and disrupting target gene transcription [6]. In addition to Dicer and Ago, numerous affiliated factors, including R2D2 [7-9], transactivation region RNA binding protein (TRBP) [10], heat-shock protein 90 (HSP90) [11] and systemic RNA interference-deficient (SID) [12], play important roles in the RNAi machinery of many organisms.
Sequence-specific RNAi has been extensively applied in the functional analysis of insect genes and in pest management [13]. However, RNAi efficiency is highly variable between different insect species and even between different tissues and developmental stages within the same insect species. In the coleopteran insect Tribolium castaneum and the orthopteran insect Locusta migratoria, a robust systemic RNAi response can be induced by the direct injection of dsRNA into the hemocoel [14-18]. There has been little success with the direct injection of dsRNA into the body cavity of the model insect Drosophila melanogaster [19]; however, transgenic RNAi approaches have been well established to drive dsRNA or shRNA to silence target genes in vivo [20, 21]. Different tissues in the same species also show diverse RNAi responses. For example, introduced dsRNA is effective for most tissues in Anopheles gambiae and Caenorhabditis elegans, but the central nervous system displays a refractory RNAi response [22, 23]. In the lepidopteran insect Manduca sexta, genes expressed in immune cells are sensitive to RNA silencing, while those expressed in the epidermal tissue are rather refractory [24]. Thus, exploring the critical RNAi factors in insects is urgently needed to develop RNAi as a comprehensive tool for functional gene analysis, especially in non-drosophilid species.
The silkworm, Bombyx mori, is a lepidopteran model insect with economic importance. dsRNA- or shRNA-mediated RNAi has been effective in several B. mori cell lines and embryos [25-29]. However, little success has been reported for RNAi in B. mori larvae, although wandering stage larvae showed sensitivity to large amounts of dsRNA injection [30, 31]. Screening for genes in the RNAi pathway revealed that most of the conserved RNAi factors are present in B. mori, except two dsRNA-binding proteins, BmR2D2 and BmTranslin, show minimal expression levels [32]. shRNA-mediated RNAi was enhanced by introducing Escherichia coli RNase III into a silkworm cell line [33]. Furthermore, introduction of the C. elegans SID-1 gene, which is responsible for dsRNA transportation, into BmN cells enhanced the uptake of dsRNA [26, 34]. However, no significant silencing enhancement was observed when CeSID-1 was ectopically over-expressed in silkworms [26, 34]. Although numerous studies have been carried out to explore the factors that limit RNAi in B. mori, no endogenous RNAi enhancers have been identified [35].
Four Argonaute proteins, BmAgo1, BmAgo2, BmAgo3 and BmPIWI, have been identified in B. mori [36]. BmAgo1 and BmAgo2 are included in the Ago subfamily, which are responsible for siRNA- and miRNA-mediated post-transcriptional silencing, respectively [37]. However, BmAgo3 and BmPIWI are described as the PIWI subfamily members and express in the silkworm gonads significantly [38]. Our results demonstrate that BmAgo2, rather than the other silkworm Ago proteins, binds to siRNA, indicating that BmAgo2 functions as the core RNAi machinery in B. mori. Disruption of BmAgo2 by introducing BmAgo2-specific dsRNA inhibited the RNAi response in both BmN cultured cells and embryos. Furthermore, transgenic over-expression of BmAgo2 significantly facilitated both dsRNA- and shRNA-mediated larval RNAi in B. mori. In conclusion, our results demonstrate that over-expression of BmAgo2 improves RNAi efficiency in B. mori, particularly during the larval feeding stages.
Materials and Methods
Insect strains and cell lines
The silkworm strain, Nistari, was used for the germline transformations and other experiments. A bivoltine strain, Dazao, was used for embryonic RNAi experiments. Silkworm larvae were fed with mulberry leaves at 25°C [39].
The silkworm ovary-derived BmN cell line was cultured with Tc100 insect medium containing 10% fetal bovine serum (Gibco) at 27°C.
Electrophoretic mobility shift assays (EMSA) and MALDI-TOF/TOF analysis
Crude embryonic protein extracts were prepared as previously described [40]. Double-stranded oligonucleotides targeting the 5'-GCAGCACGACUUCUUCAAG-3' sequence of enhanced green fluorescence Protein (EGFP) were synthesized with FAM labeling at the 3' ends via an LC-LC linkage (RiboBio). The reaction mixture containing 5 μg extracted crude protein and 2 μl (10 mg/ml) labeled EGFP siRNA probe were incubated for 30 min at 37°C using the Thermo Scientific LightShift Chemiluminescent RNA EMSA Kit (Thermo). Subsequently, the reaction mixture was subjected to electrophoresis using a 6% native polyacrylamide gel at 100 V for 2 h, and the gel was photographed with a FUJIFILM FLA-9000 image reader (Fujifilm).
After photographing, the EMSA gel was stained with R-250 Coomassie Brilliant Blue (CBB), and the protein band whose position was shifted by the siRNA probe was excised. The excised gel bands were washed with ultrapure water three times, destained with 20 mM NH4HCO3 dissolved in 50% acetonitrile. Subsequently, the sample was dried with 100% acetonitrile and digested by trypsin (Promega) overnight at 37°C. The pooled and lyophilized peptides dissolved in 5 mg/mL CHCA with 0.1% TFA and 50% acetonitrile were sent for MALDI-TOF/TOF analysis subsequently.
Plasmid preparation
The promoter sequences in the phRL-TK and pGL3 plasmids (Promega) were replaced by the silkworm baculovirus immediate-early gene promoter IE1 (pIE1-RL and pIE1-GL) and used for the dual luciferase reporter assay in the BmN cell line [41]. The 5'-GTGCTCGTAGGAGTAGTGAAA-3' sequence of Renilla luciferase (RL) was used to design shRNA and was amplified with the primers (RL shRNA) listed in Supplementary Material: Table S1, as previously reported by Tanaka et al. [42].
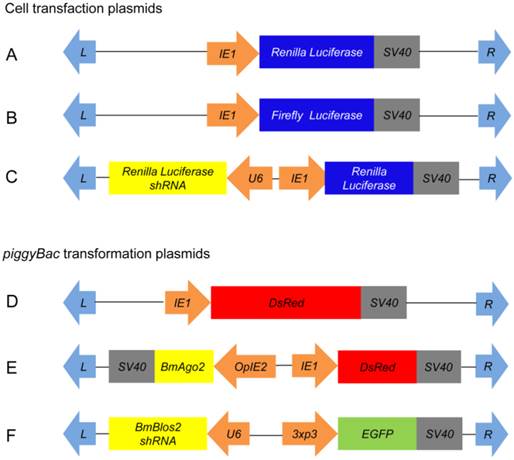
Three transformation plasmids were constructed based on the initial piggyBac vectors pBac[3xp3-DsRed] and pBac[3xp3-EGFP]: pBac[IE1-DsRed] (IE1-DsRed), pBac[IE1-DsRed-OpIE2-BmAgo2] (OpIE2-BmAgo2) and pBac[3xp3-EGFP-U6-Blos2 shRNA] (U6-Blos2 shRNA) [43].
The 3xp3 promoter in the pBac[3xp3-DsRed] plasmid was replaced with the IE1 promoter to generate the IE1-DsRed plasmid using ClonExpressTM II One Step Cloning Kit (Vazyme Biotech Co., Ltd.) [43]. The open reading frame (ORF) of BmAgo2 was amplified from silkworm embryonic cDNA according to the sequence deposited in GenBank (NM_001043530.2). To strongly induce the expression of BmAgo2, the 3116-bp BmAgo2 ORF was inserted into the pIZT/V5-His A plasmid (Invitrogen) downstream of the optimized silkworm baculovirus immediate-early gene promoter IE2 (OpIE2). Subsequently, the OpIE2-Ago2-SV40 cassette was sub-cloned into the IE1-DsRed plasmid via AscI and FseI double digestion to generate the plasmid OpIE2-BmAgo2. For the U6-Blos2 shRNA plasmid, the 467-bp silkworm small nuclear RNA promoter U6 was amplified from silkworm genomic DNA and used as a template to amplify the U6-Blos2 shRNA cassette using a primer pair BmBlos2 shRNA that contained the shRNA target in the reverse primer (Table S1). BmBlos2 shRNA targeting the sequence 5'-TCAAGCAGCATGTAGTGTTCC-3' was designed using the Invitrogen BLOCK-iTTM RNAi Designer (http://rnaidesigner.lifetechnologies.com/rnaiexpress/). The sequenced U6-Blos2 shRNA cassette was sub-cloned into the pBac[3xp3-EGFP] plasmid, which contains the selection marker EGFP gene driven by eye-specific promoter 3xp3, to generate the U6-Blos2 shRNA plasmid.
Double-stranded RNA (dsRNA) preparation and treatment
The following gene-specific sequences were used as the dsRNA templates: a 449-bp fragment targeting Renilla Luciferase (RL), a 504-bp fragment targeting BmAgo2, a 367-bp fragment targeting EGFP and a 416-bp fragment targeting DsRed. dsRNA was synthesized using the T7-MEGAScript kit (Ambion) with the primers listed in Table S1. We incubate the reaction mixture at 37°C for 3 h, heat shocked it at 75°C for 5 min and annealed the double strands at room temperature for 1 h. Subsequently, dsRNA were treated with DNase I to remove the templates and purified with phenol/chloroform.
Purified dsRNA was dissolved in distilled water and used to transfect the BmN cells using the Effectene Transfection Reagent (Qiagen) according to the manufacturer's instructions. Either 0.5 μg/μl or the indicated concentration of dsRNA targeting the silkworm genes white egg 2 (Bmw-2) and BmAgo2 was injected into preblastoderm embryos. For larval RNAi, 10 μg/larvae (5 μg/μl) of DsRed or EGFP dsRNA was injected into transgenic silkworms on day one of the third larval instar. All the subjected animals expressed comparable fluorescence levels and were staged at the third instar ecdysis. After injection, the animals were reared separately, and the fluorescence was observed via fluorescence microscopy (Nikon AZ100).
Quantitative real-time PCR (qRT-PCR)
TRIzol Reagent purchased from Invitrogen was used to extract the total RNA. Subsequently, the RNA was treated with DNase I (Takara) to remove the genome DNA. One microgram of extracted RNA was used as the templates to synthesize cDNA using the RevertAid First-Strand cDNA synthesis kit (Fermentas). Quantitative mRNA measurements were performed with TOYOBO SYBR Green Realtime PCR Master Mix. Three independent biological replicates were examined by Eppendorf Mastercycler ep realplex. The PCR program was the following: incubation at 94°C for 3 min, followed by 40 cycles of 94°C for 10 s and 55°C for 20 s. The standard curves were performed with 10-fold serial dilution cDNA. The data were normalized to B. mori ribosomal protein 49 (Bmrp49) and analyzed by GraphPad Prism version 5.01. All of the qRT-PCR primers are included in Table S1.
Dual luciferase assays
The pIE1-RL plasmid, which expresses RL, was used as a reporter to indicate RNAi efficiency, and the pIE1-GL plasmid containing the Firefly Luciferase (FL) reporter gene was used as a control to indicate the transfection efficiency. The relative luciferase activity was calculated by normalizing RL to FL (RL/FL).
BmN cells were placed in 96-well plates (Nunc), and the medium was replaced after 24 hours (150 μl). The cells were subsequently transfected with BmAgo2 dsRNA (20 ng/well, 24 h later) and later co-transfected with the luciferase plasmids (25 ng/well for pIE1-GL and 100 ng/well for pIE1-RL, 72 h later). RL dsRNA was co-transfected along with the pIE1-RL and pIE1-GL plasmids at 5 ng/well during the second transfection to determine dsRNA-mediated RNAi efficiency. The shRNA-mediated RNAi efficiency was determined by co-transfecting the U6-RL shRNA plasmid with two luciferase expression plasmids.
Germline transformation and inverse PCR
Plasmid transformations were performed by microinjection into the preblastoderm G0 embryos followed by incubation at 25°C in a humidified chamber before hatching [43]. The resulting G0 adults were inbred, and the G1 embryos were screened for the expression of the fluorescence protein using a Nikon AZ100 fluorescence microscope.
Silkworm genomic DNA was extracted with the UniversalGen DNA Kit (CWBio) from the G1 transgenic moths. The genomic DNA was then digested with DpnII and ligated at 4°C overnight. Primers listed in Table S1 were used for amplification, and PCR was performed using the following conditions: 94°C for 2 min, followed by 40 cycles of 94°C for 15 s, 55°C for 15 s, and 72°C for 1 min, followed by a final extension period of 72°C for 10 min. The purified PCR products were cloned into the pMD18-T vector and sequenced directly.
Western blots
The entire body of G1 fifth instar larva was homogenized and dissolved in PBS (phosphate-buffered saline). The protein concentration was quantified with PierceTM BCA Protein Assay Kit (Thermo). Samples were separated in 8% SDS-PAGE and then transferred to nitrocellulose membranes (GE Healthcare). The monoclonal mouse anti-BmAgo2 (1:500 dilution; Abmart) and anti-α-tubulin (1:1000 dilution; Vazyme Biotech Co., Ltd.) primary antibodies were used to detect the BmAgo2 and α-tubulin proteins [44], respectively. The secondary antibody used here was goat anti-mouse IgG (1:5000 dilution; Beyotime, Shanghai, China) with horseradish peroxidase-conjugated.
Statistics analysis of data
All the data was analyzed using GraphPad Prism (version 5.01) with One-way ANOVA and the Dunnett post-hoc test analysis. All error bars are the means ± S.E.M. p<0.05 was used to determine significance in all cases.
Results and Discussion
BmAgo2 is one of the core RNAi factors in B. mori
RNA EMSA and MALDI-TOF/TOF were used to screen siRNA-interacting proteins in B. mori embryos. Seven proteins, including the heat shock proteins HSP70 and HSP70B, Cellular Retinoic Acid Binding Acid (CRABA) protein and Enolase, were identified with significant abundance (Table 1). The number of matched peptides for these four detected proteins was 8, 2, 2 and 4, respectively (Table 1). Furthermore, the conserved RNAi components Ago2, Dicer2 and Tudor-sn were also detected in the protein complex with 5, 11 and 8 matched peptides, respectively (Table 1). For the sequence coverage, we calculated the number of matched amino acids and divided it by the length of each protein. The sequence coverage of the former four proteins was 14.33%, 4.5%, 16.54% and 8.08%, respectively, and the sequence coverage of BmAgo2, BmDicer2 and BmTudor-sn was 4.82%, 7.45% and 9.68%, respectively (Table 1). Detailed information on the matched peptides was presented in Supplementary Material: Table S2, and the peptide spectra were included in Supplementary Material: Fig. S1-S7. As Ago2 is one of the conserved components of the RNAi machinery in insects [25], we exploited the function of BmAgo2 in B. mori RNAi in the current study.
SiRNA interacting proteins in B. mori embryos identified by using MALDI-TOF/TOF analysis.
| Pro Name | Accession NO. | Score | Sequence coverage (%) | Peptides matched | pI/MW(Exp.) |
|---|---|---|---|---|---|
| BmHSP70 | gi|320526705 | 180 | 14.33% | 8 | 5.33/71359.3 |
| BmHSP70B | gi|336454474 | 145 | 4.5% | 2 | 5.47/70712.9 |
| Cellular Retinoic Acid Binding Acid | gi|108793850 | 85 | 16.54% | 2 | 5.66/14963.4 |
| BmEnolase | gi|119381542 | 66 | 8.08% | 4 | 5.62/47164.3 |
| BmAgo2 | gi|166706854 | 8 | 4.82% | 5 | 9.44/117243.8 |
| BmDicer2 | gi|302318907 | 17 | 7.45% | 11 | 6.15/192070.2 |
| BmTudor-sn | gi|302190081 | 20 | 9.68% | 8 | 8.56/99418 |
BmAgo2 affected the RNAi response in the BmN cell line
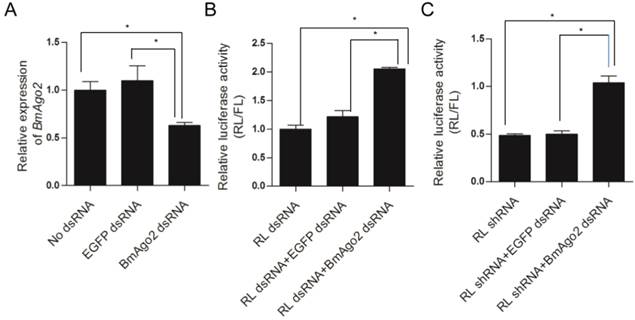
To explore the role of BmAgo2 in the B. mori RNAi machinery, we constructed several transfection plasmids (Fig. 1A, B and C) and performed double RNAi assays (Supplementary Material: Fig. S8A) in the BmN cell line to investigate whether depletion of BmAgo2 affecting the RNAi response. The transcription level of endogenous BmAgo2 was down-regulated to 60% of the control (no dsRNA or EGFP dsRNA) after transfection with dsRNA targeting BmAgo2 (Fig. 2A), suggesting that BmAgo2 dsRNA-mediated RNAi effect was significant in BmN cells. Subsequently, we investigated effect of BmAgo2 RNAi on the silencing potency of Renilla Luciferase (RL) dsRNA or shRNA using dual luciferase reporter assays. Transfection with RL dsRNA down-regulated the relative RL/FL level to 11% of the level observed in control cells that were not transfected with RL dsRNA (Supplementary Material: Fig. S9). In comparison, the relative luciferase activity was increased 2.2-fold (22% of the control) by co-transfection of BmAgo2 dsRNA with RL dsRNA (Fig. 2B), suggesting that the depletion of BmAgo2 inhibited the dsRNA-mediated RNAi response in BmN cells. In addition, RL shRNA expression directed by the U6 promoter (U6-RL shRNA, Fig. 1C) down-regulated the RL/FL level to 50% compared to the control (Fig. S9). When BmAgo2 dsRNA was co-transfected with the U6-RL shRNA, the relative RL/FL level recovered to that of the control, indicating that the RNAi effect was completely inhibited (Fig. 2C). In conclusion, our results demonstrate that BmAgo2 is involved in regulating both dsRNA- and shRNA-mediated RNA silencing in BmN cells.
Schematics of plasmids used in this study. A-C: Plasmids for cell transfection; D-F: Plasmids for germ line transformation. A. pIE1-RL; B. pIE1-GL; C. pIE1-RL-U6-RL shRNA; D. pBac[IE1-DsRed]; E. pBac[IE1-DsRed-OpIE2-BmAgo2]; and F. pBac[3xp3-EGFP-U6-Blos2 shRNA]. Orange arrows indicate promoters; light-blue arrows indicate piggyBac recombination sites; grey boxes indicate polyadenylation transcription terminal sequences; colored boxes indicate coding regions.
Assessment of BmAgo2 function in RNAi via dual luciferase reporter assay in BmN cells. A. Quantification of the endogenous BmAgo2 transcription level by quantitative real-time PCR (qRT-PCR); B. Inhibition of dsRNA-mediated RL silencing by disruption of BmAgo2 transcription; C. Complete inhibition of shRNA-mediated RL silencing by the disruption of BmAgo2 transcription. Three independent biological replicates were used for quantification. The relative luciferase activity in B and C were calculated by dividing the RL activity by the FL activity. The asterisks represent significant difference (p<0.05) between groups. Error bars are the means ± S.E.M.
BmAgo2 affected the RNAi response in embryos
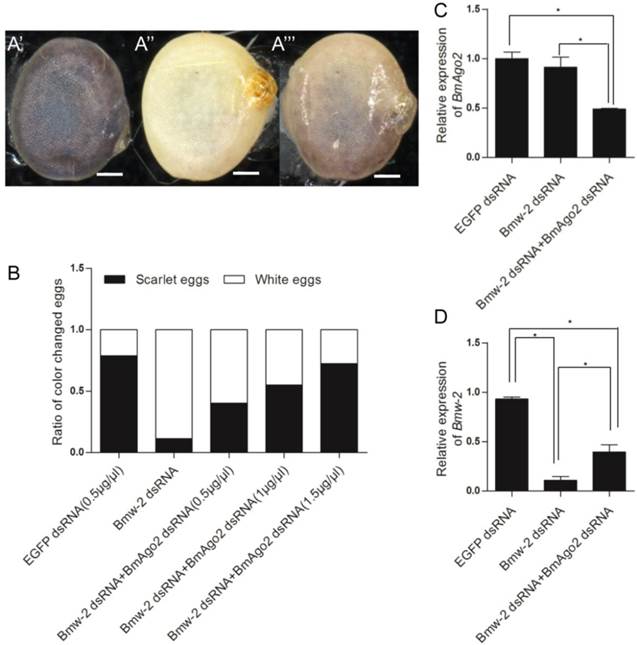
To investigate the role of BmAgo2 in the RNAi response in vivo, we performed embryonic double RNAi (Fig. S8B) targeting the silkworm white egg 2 (Bmw-2) gene, which is responsible for the accumulation of ommochrome pigments in silkworm eggs [45, 46]. We injected dsRNA targeting Bmw-2 into preblastoderm embryos of the Dazao strain, as the eggs of this strain become scarlet due to serosa pigmentation 24 h after oviposition. Pigments failed to accumulate in 89% (n=80) of the eggs after Bmw-2 dsRNA injection (Fig. 3A'' and B), showing that the RNAi response was effective in embryos, which was consistent with previous results [28]. When BmAgo2 dsRNA (0.5 μg/μl) was co-injected with Bmw-2 dsRNA, the ratio of white eggs decreased to 60% (n=80), and the remaining eggs were a light scarlet color (Fig. 3A''' and B). Furthermore, the ratio of white eggs decreased in a dose-dependent manner with increasing concentrations of BmAgo2 dsRNA (45% when treated with 1 μg/μl BmAgo2 dsRNA, and 28% when treated with 1.5 μg/μl BmAgo2 dsRNA, Fig. 3B). No egg pigmentation defects were observed when dsRNA targeting EGFP was co-injected with Bmw-2 dsRNA as the control (Fig. 3A'). When BmAgo2 dsRNA was introduced, the BmAgo2 transcription level was down-regulated to 50% of that in the control (injected with EGFP dsRNA) as determined by qRT-PCR (Fig. 3C). Bmw-2 transcription was down-regulated to 10% of the control (injected with EGFP dsRNA) when only the Bmw-2 dsRNA was injected and to 40% of the control after co-injection with the BmAgo2 and Bmw-2 dsRNAs (Fig. 3D), indicating that introducing BmAgo2 dsRNA inhibited the Bmw-2 RNAi efficiency in B. mori embryos.
Disruption of BmAgo2 transcription affected the RNAi response in embryos. A'- A'''. Eggs photographed under bright field on day 5 after dsRNA injection. Scale bars represent 0.25 mm. The eggs were injected with EGFP dsRNA (A'), with Bmw-2 dsRNA (A'') or with both BmAgo2 and Bmw-2 dsRNA (A'''); B. Statistic of the egg color. 80 eggs were injected for each group; C and D. qRT-PCR analysis of the level of BmAgo2 and Bmw-2 transcription. Three independent biological replicates were analyzed. The asterisks indicate statistical significance (p<0.05), and error bars are means ± S.E.M.
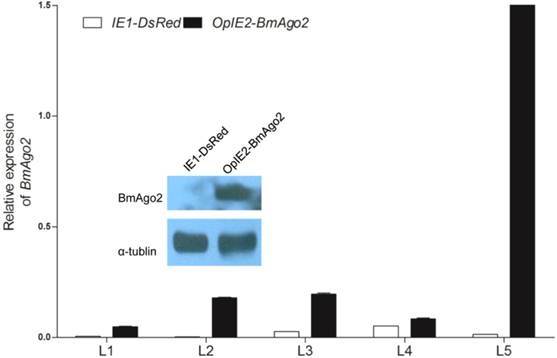
Over-expression of BmAgo2 in pBac[IE1-DsRed-OpIE2-BmAgo2] (OpIE2-BmAgo2) transgenic silkworms. Whole bodies of day one of each instar larvae were homogenized, and the total RNA was extracted. The plasmid that only expressed the selection marker (IE1-DsRed) was used as a control. For each stage, three transgenic animals were used to extract the total RNA. Error bars are means ± S.E.M, and the inset shows the increased translation of BmAgo2 in the final instar larvae.
Generation of transgenic silkworm lines
To investigate whether BmAgo2 functions in the B. mori RNAi response during the post-embryonic stages, especially the larval stages, we established transgenic silkworm lines that ectopically express BmAgo2 in a ubiquitous manner driven by the IE2 promoter (OpIE2-BmAgo2, Fig. 1E). A total of 960 preblastoderm eggs were injected and 333 survived to the adult stage. By inbreeding or crossing with the wild type moths, 56 individual G1 batches were screened based on red fluorescence and 7 batches had one or more positive transgenic silkworms. The transformation efficiency was accordingly 12.5%. Two independent lines were selected and genomic insertion sites were determined (Supplementary Material: Fig. S10A). Significant over-expression of BmAgo2 was detected and reached a maximum of 124-fold compared to the control animals (IE1-DsRed, Fig. 1D) in the fifth larval instar (Fig. 4). Additionally, a significant increase in the BmAgo2 protein level in the fifth larval instar was confirmed using western blotting analysis (Fig. 4).
Two more transgenic silkworm lines, IE1-DsRed (Fig. 1D) and U6-Blos2 shRNA (Fig. 1F), were also constructed with transformation efficiency of 7.6% and 8.5% separately. Three independent lines were created for each transgene, and all of the insertion sites were in non-functional sequences of the silkworm genome (Fig. S10). No deleterious phenotypes were observed during the growth and development of these transgenic silkworm lines and we chose one line for each transgene to use in the subsequent experiments.
Over-expression of BmAgo2 enhanced dsRNA-mediated RNAi
The IE1-directed DsRed gene was also introduced as a reporter gene and a selection marker. dsRNA was injected into the hemocoel of BmAgo2 transgenic animals to examine the larval RNAi efficiency. 72% (n=25, Table 2) of the OpIE2-BmAgo2 transgenic animals showed significantly decreased red fluorescence (Fig. 5B) 24 h after DsRed dsRNA injection. A further slight decrease in DsRed fluorescence was observed 48 h after injection (Fig. 5B, Table 2). qRT-PCR analysis revealed that dsRNA injection down-regulated the DsRed transcription level to 40% of the control (non-injected) after 24 h. Consistent with the fluorescence microscopy analysis, the RNAi efficiency was time-dependent, and the transcription level of DsRed was restored to 60% and 77% of the control at 48 h and 72 h after injection, respectively (Fig. 5D). No significant decrease in the red fluorescence signal was observed in OpIE2-BmAgo2 animals without injection or with EGFP dsRNA injection (Fig. 5B). In the control IE1-DsRed animals, no obvious fluorescence decrease was observed either with or without injection with EGFP and DsRed dsRNA (Fig. 5A and Table 2). The qRT-PCR results revealed that the transcription level of DsRed did not change significantly in IE1-DsRed animals (Fig. 5C). Both fluorescence investigation and qRT-PCR analysis suggest over-expression of BmAgo2 facilitates dsRNA-mediated RNAi in the larval silkworms.
DsRNA-mediated larval RNAi was enhanced in OpIE2-BmAgo2 transgenic silkworms. A and B. Day one of third instar larvae injected with dsRNA were photographed under UV light equipped with an RFP filter at 0 h, 24 h and 48 h after injection. Non-injected and EGFP dsRNA-injected larvae were used as controls, and DsRed dsRNA-injected larvae were used as reporters to indicate silencing efficiency. A. IE1-DsRed transgenic silkworms; B. OpIE2-BmAgo2 transgenic silkworms; C and D. qRT-PCR quantification of the DsRed gene transcription level of IE1-DsRed and OpIE2-BmAgo2 transgenic silkworms after dsRNA treatment. Three independent biological replicates were used for the analysis. Error bars are means ± S.E.M.
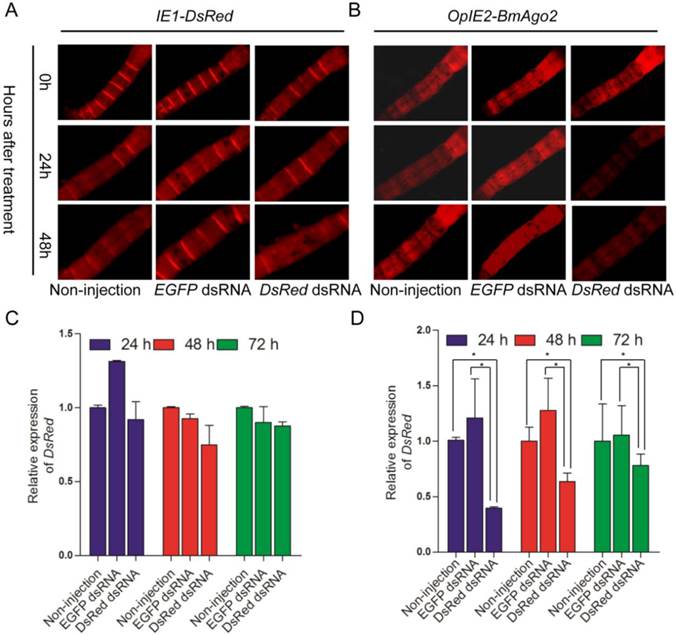
Fluorescence investigation after dsRNA injection at day one of third larval instar. For each group, 25 larvae were subjected to injection. Non-injection and EGFP dsRNA injected animals were used as the control. Red fluorescence was investigated at 24h and 48 h after treatment.
| Transgenic lines | DsRNA for target genes | No. of larvae injected | No. of larvae showed fluorescence decrease | ||
|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | |||
| IE1-DsRed | Non-injection | 25 | 0 | 0 | 0 |
| EGFP | 25 | 0 | 0 | 0 | |
| DsRed | 25 | 0 | 1 | 0 | |
| OpIE2-BmAgo2 | Non-injection | 25 | 0 | 1 | 0 |
| EGFP | 25 | 0 | 3 | 1 | |
| DsRed | 25 | 0 | 18 | 4 | |
BmAgo2 over-expression enhanced shRNA-mediated RNAi
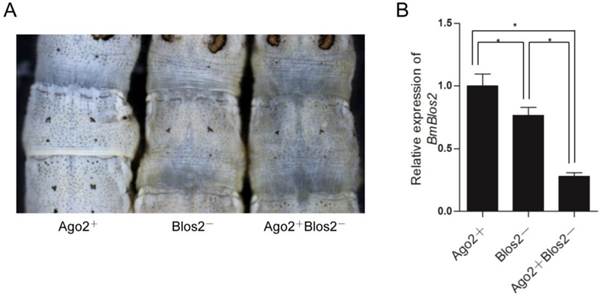
As shRNA is ultimately processed into mature siRNA to function in mRNA degradation similar to dsRNA, we speculate that BmAgo2 also plays an important role in the shRNA-mediated RNAi pathway. A transgenic silkworm line that expresses the BmBlos2 shRNA under the control of the U6 promoter (U6-Blos2 shRNA) was used to determine the shRNA-mediated silencing efficiency (Fig. 1F) [47, 48]. BmBlos2 plays important role in silkworm urate granule synthesis in the silkworm larval epidermis. The mutation of BmBlos2 results in translucent larval skin, which was designated as the oily skin phenotype (OSP) [49]. Expressing BmBlos2 shRNA in U6-Blos2 shRNA transgenic larvae resulted in OSP (Fig. 6A). We then crossed the OpIE2-BmAgo2 and the U6-Blos2 shRNA transgenic lines and investigated the frequency of OSP in the offsprings. Of the animals that only expressed the BmBlos2 shRNA (Blos2-), 28.9% (n=97) displayed OSP during the larval stages (Fig. 6A and Table 3). Of the animals that expressed both the BmAgo2 and BmBlos2 shRNA (Ago2+ Blos2-), 64.3% (n=56) showed OSP (Fig. 6A and Table 3). Furthermore, the OSP was more severe in the Ago2+Blos2- animals than in the Blos2- animals (Fig. 6A). We performed qRT-PCR analysis to quantify the RNAi efficiency as enhanced by BmAgo2 over-expression and showed that the BmBlos2 transcription level was down-regulated to 75% in Blos2- animals compared with their Ago2+ siblings (Fig. 6B). However, in Ago2+Blos2- animals, the BmBlos2 transcription level was down-regulated to 30% compared to their Ago2+ siblings (Fig. 6B). The results presented here indicate that over-expression of BmAgo2 significantly facilitates shRNA-mediated RNAi when targeting BmBlos2.
Although numerous factors are involved in the insect RNAi machinery, we have identified BmAgo2 as one of the important RNAi factors in B. mori and provided a promising approach for enhancing RNAi efficiency in B. mori larvae. The disruption of BmAgo2 in BmN cells and embryos reduced the efficiency of RNAi, while the over-expression of BmAgo2 facilitated the RNAi effect during the larval stages in transgenic animals. Furthermore, BmAgo2 was shown to be involved in both the dsRNA- and shRNA-mediated RNAi effect both in vitro and in vivo. Enhancing larval RNAi efficiency by over-expressing BmAgo2 will greatly contribute to both RNAi-based analysis of gene function and pest management in lepidopteran insects.
Numbers of the animals which presented OSP in different transgenic lines.
| Genotype | No. of total animals | No. of OSP animals | Ratio of OSP animals |
|---|---|---|---|
| Ago2+ | 50 | 0 | 0 |
| Blos2- | 97 | 28 | 28.9% |
| Ago2+Blos2- | 56 | 36 | 64.3% |
Enhancement of shRNA-mediated larval RNAi in OpIE2-BmAgo2 transgenic silkworms. A. Offspring resulting from crossing the OpIE2-BmAgo2 and the U6-Blos2 shRNA transgenic silkworms. Siblings that expressed only the BmBlos2 shRNA (Blos2-) and that co-expressed the BmAgo2 and BmBlos2 shRNA (Ago2+Blos2-) presented an oily skin phenotype (OSP). B. Quantification of the relative expression level of BmBlos2 by qRT-PCR. The asterisks indicate statistical significance (p<0.05), and error bars are means ± S.E.M.
Abbreviations
Ago2: Argonaute 2; RNAi: RNA interference; RISC: RNA-induced silencing complex; dsRNA: double-stranded RNA; shRNA: short-hairpin RNA; siRNA: small interfering RNA; TRBP: transactivation region RNA binding protein; HSP: heat shock protein; SID: RNA interference deficient; EMSA: Electrophoretic mobility shift assay; CRABA: Cellular Retinoic Acid Binding Acid; RL: Renilla Luciferase; FL: Firefly Luciferase; ORF: open reading frame; rp49: ribosome protein 49; PBS: phosphate-buffered saline; OSP: oily skin phenotype; CBB: Coomassie Brilliant Blue; PCR: polymerase chain reaction; qRT-PCR: quantitative real-time PCR; ABC: ATP-binding cassette; OpIE2: optimized silkworm baculovirus immediate-early gene promoter IE2; BmN: Bombyx mori ovary-derived cell line; EGFP: enhanced green fluorescent protein; w-2: white egg 2.
Supplementary Material
Tables S1-S2, Figures S1-S10.
Acknowledgements
This work was supported by grants from the National Science Foundation of China (31030060, 31272037 and 31372257), the External Cooperation Program of BIC, Chinese Academy of Sciences (Grant No. GJHZ201305) and the Shanghai Committee of Science and Technology, China (11JC1413900).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hannon GJ. RNA interference. Nature. 2002;418:244-51
2. Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25-33
3. Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschi T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877-88
4. Dueck A, Meister G. Assembly and function of small RNA-Argonaute protein complexes. Biol Chem. 2014;395:611-29
5. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363-6
6. Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146-50
7. Liu Q, Rand TA, Kalidas S, Du F, Kin HE, Smith DP. et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921-5
8. Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514-20
9. Okamura K, Robine N, Liu Y, Liu Q, Lai EC. R2D2 organizes small regulatory RNA pathways in Drosophila. Mol Cell Biol. 2011;31:884-96
10. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K. et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740-4
11. Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17:1024-6
12. Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456-9
13. Katoch R, Sethi A, Thakur N, Murdock LL. RNAi for insect control: current perspective and future challenges. Appl Biochem Biotechnol. 2013;171:847-73
14. Tomoyasu Y. Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008;9:R10
15. Posnien N, Schinko J, Grossmann D, Shippy TD, Konopova B, Bucher G. RNAi in the red flour beetle (Tribolium). Cold Spring Harb Protoc. 2009
16. Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera). Curr Biol. 2002;12:R5-6
17. Luo Y, Wang X, Yu D, Kang L. The SID-1 double-stranded RNA transporter is not required for systemic RNAi in the migratory locust. RNA Biol. 2012;9:663-71
18. Ren D, Cai Z, Song J, Wu Z, Zhou S. dsRNA uptake and persistence account for tissue-dependent susceptibility to RNA interference in the migratory locust, Locusta migratoria. Insect Mol Biol. 2014;23:175-84
19. Dzitoyeva S, Manev H. Reduction of Cellular Lipid Content by a Knockdown of Drosophila PDP1γ and Mammalian Hepatic Leukemia Factor. J Lipids. 2013;2013:297932
20. Yamamoto-Hino M, Goto S. In vivo RNAi-based screens: studies in model organisms. Genes (Basel). 2013;4:645-65
21. Miller SC, Brown SJ, Tomoyasu Y. Larval RNAi in Drosophila? Dev Genes Evol. 2008;218:505-10
22. Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E. et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One. 2010;5:e9471
23. Fimhaber C, Hammarlund M. Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLoS Genet. 2013;9:e1003921
24. Eleftherianos I, Xu M, Yadi H, Ffrench-Constant RH, Reynolds SE. Plasmatocyte-spreading peptide (PSP) plays a central role in insect cellular immune defenses against bacterial infection. J Exp Biol. 2009;212:1840-8
25. Kolliopoulou A, Swevers L. Functional analysis of the RNAi response in ovary-derived silkmoth Bm5 cells. Insect Biochem Mol Biol. 2013;43:654-63
26. Xu J, Mon H, Kusakabe T, Li Z, Zhu L, Iiyama K. et al. Establishment of a soaking RNA interference and Bombyx mori nucleopolyhedrovirus (BmNPV)-hypersensitive cell line using Bme21 cell. Appl Microbiol Biotechnol. 2013;97:10435-44
27. Wei L, Li Z, Cheng D, Kusakabe T, Pan M, Duan J. et al. RNAi silencing of the SoxE gene suppresses cell proliferation in silkworm BmN4 cells. Mol Biol Rep. 2014;41:4769-81
28. Quan G, Kanda T, Tamura T. Induction of the white egg 3 mutant phenotype by injection of the double-stranded RNA of the silkworm white gene. Insect Mol Biol. 2002;11:217-22
29. Yamaguchi J, Mizoguchi T, Fujiwara H. siRNAs induce efficient RNAi response in Bombyx mori embryos. PLoS One. 2011;6:e25469
30. Tian L, Ma L, Guo E, Deng X, Ma S, Xia Q. et al. 20-hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy. 2013;9:1172-87
31. Huang J, Zhang Y, Li M, Wang S, Liu W, Couble P. et al. RNA interference-mediated silencing of the bursicon gene induces defects in wing expansion of silkworm. FEBS Lett. 2007;581:697-701
32. Swevers L, Liu J, Huvenne H, Smagghe G. Search for limiting factors in the RNAi pathway in silkmoth tissues and the Bm5 cell line: The RNA-binding proteins R2D2 and Translin. PLoS One. 2011;6:e20250
33. Lee JM, Kojin Y, Tatsuke T, Mon H, Miyagawa Y, Kusakabe T. Coexpression of Escherichia coli RNase III in silkworm cells improves the efficiency of RNA interference induced by long hairpin dsRNAs. Insect Sci. 2013;20:69-77
34. Mon H, Kobayashi I, Ohkubo S, Tomita S, Lee J, Sezutsu H. et al. Effective RNA interference in cultured silkworm cells mediated by over-expression of Caenorhabditis elegans SID-1. RNA Biol. 2012;9:40-6
35. Terenius O, Papanicolaou A, Garbutt JS. et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231-45
36. Wang G, Jiang L, Zhu L, Cheng T, Niu W, Yan Y. et al. Characterization of Argonaute family members in the silkworm, Bombyx mori. Insect Sci. 2013;20:78-91
37. Zhu L, Masaki Y, Tatsuke T, Li Z, Mon H, Xu J. et al. A MC motif in silkworm Argonaute 1 is indispensible for translation repression. Insect Mol Biol. 2013;22:320-30
38. Kawaoka S, Minami K, Katsuma S, Mita K, Shimada T. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem Biophys Res Commun. 2008;367:755-60
39. Tan A, Tanaka H, Tamura T, Shiotsuki T. Precocious metamorphosis in transgenic silkworms overexpressing juvenile hormone esterase. Proc Natl Acad Sci U S A. 2005;102:11751-6
40. Zhong B, Li J, Lin J, Liang J, Su S, Xu H. et al. Possible effect of 30K proteins in embryonic development of silkworm Bombyx mori. Acta Biochimica Biophys Sin. 2005;37:355-61
41. Masumoto M, Ohde T, Shiomi K, Yaqinuma T, Niimi T. A Baculovirus immediate-early gene, ie1, promoter drives efficient expression of a transgene in both Drosophila melanogaster and Bombyx mori. PLoS One. 2012;7:e49323
42. Tanaka H, Fujita K, Sagisaka A, Tomimoto K, Imanishi S, Yamakawa M. shRNA expression plasmids generated by a novel method efficiently induce gene-specific knockdown in a silkworm cell line. Mol Biotechnol. 2009;41:173-9
43. Tan A, Fu G, Jin L, Guo Q, Li Z, Niu B. et al. Transgene-based, female-specific lethality system for genetic sexing of the silkworm, Bombyx mori. Proc Natl Acad Sci U S A. 2013;110:6766-70
44. Njaine B, Martins RA, Santiago MF, Linden R, Silveira MS. Pituitary adenylyl cyclase-activating polypeptide controls the proliferation of retinal progenitor cells through downregulation of cyclin D1. Eur J Neurosci. 2010;32:311-21
45. Abraham EG, Sezutsu H, Kanda T, Suqasaki T, Shimada T, Tamura T. Identification and characterisation of a silkworm ABC transporter gene homologous to Drosophila white. Mol Gen Genet. 2000;264:11-9
46. Tatematsu K, Yamamoto K, Uchino K, Narukawa J, Iizuka T, Banno Y. et al. Positional cloning of silkworm white egg 2 (w-2) locus shows functional conservation and diversification of ABC transporters for pigmentation in insects. Genes Cells. 2011;16:331-42
47. Tanaka H. Construction of shRNA expression plasmids for silkworm cell lines using single-stranded DNA and Bst DNA polymerase. Methods Mol Biol. 2013;942:347-55
48. Ando T, Fujiwara H. Electroporation-mediated somatic transgenesis for rapid functional analysis in insects. Development. 2013;140:454-8
49. Wang Y, Li Z, Xu J, Zeng B, Ling L, You L. et al. The CRISPR/Cas System mediates efficient genome engineering in Bombyx mori. Cell Res. 2013;23:1414-6
Author contact
![]() Corresponding authors: Anjiang Tan, Tel: 86-2154924046; Fax: 86-2154924015; Email: ajtan01ac.cn. Yongping Huang, Tel: 86-2154924047; Fax: 86- 2154924015; Email: yphuangac.cn.
Corresponding authors: Anjiang Tan, Tel: 86-2154924046; Fax: 86-2154924015; Email: ajtan01ac.cn. Yongping Huang, Tel: 86-2154924047; Fax: 86- 2154924015; Email: yphuangac.cn.

 Global reach, higher impact
Global reach, higher impact